Abstract
A number of recent studies have begun to explore a new and exciting area: the interaction between the gut microbiome and renal physiology. In particular, multiple studies have focused on the role of microbially produced short chain fatty acids, which are generally thought to promote health. This review will focus on what is known to date regarding the influence of the microbiome on renal function, with emphasis on the cell biology, physiology, and clinical implications of short chain fatty acids and short chain fatty acid receptors. It is clear that microbe-host interactions are an exciting and ever-expanding field, which has implications for how we view diseases such as hypertension, acute kidney injury, and chronic kidney disease. However, it is important to recognize that although the potential promise of this area is extremely enticing, we are only the very edge of this new field.
Keywords: acute kidney injury, antibiotics, cell signaling, chronic kidney disease
In recent years it has become increasingly clear that the physiology of a number of organisms—from humans1,2 and mice3,4 to Drosophila5 and Caenorhabditis elegans6—can be influenced by the gut microbiota. In particular, metabolites of the gut microbiota have been shown to play a role in influencing “host” physiology. These metabolites include those thought to have both positive and negative effects on host health. For example, in patients with chronic kidney disease, the gut microbiome produces uremic toxins, including indoles,7 ammonia,8,9 and trimethylamine N-oxide.10 On the other hand, other metabolites—notably, short chain fatty acids (SCFAs)—are generally thought to promote health.11–14 This review will emphasize the role of SCFA receptors with regards to the cell biology, physiology, and clinical relevance of renal SCFA signaling.
SCFAs: where do they come from, and what is the evidence?
The gut microbiota, sometimes referred to as the “forgotten organ,” are a significant component of the physiology of the host. The total wet weight of gut microbes in humans is estimated to be 175 g to 1.5 kg,15,16 and the gut microbial population is a dynamic one whose composition and activity change over time. A significant body of research points to the fact that the gut microbial composition changes in disease (dysbiosis).3,9,16–34 In addition, it is clear that the gut microbiota naturally undergoes dynamic changes during different stages of life35,36 and in response to changes in habits of the host.37,38 For example, dietary changes in the host lead to significant alterations in gut microbial composition37,38 and subsequently to changes in microbial metabolite production.39 One type of microbial metabolite, which we will focus on in this review, is SCFAs. SCFAs most commonly refers to the straight-chain 2-4 carbon variety (acetate, propionate, and butyrate). SCFAs are produced from dietary fiber: because most dietary fiber cannot be degraded by host enzymes in the upper gastrointestinal tract, this fiber instead travels intact to the colon and cecum, where it is broken down by the gut microbiota. As a byproduct of this fermentation process, gut microbes produce a number of metabolites, with SCFAs being a major product. Gut microbial production is so robust that the concentration of SCFAs in the colon lumen itself is ~100 mmol/l.15
SCFAs produced by the gut microbiota enter the circulation of the host via monocarboxylate transporters40 as well as by diffusion. Although it is commonly stated that SCFAs found in the bloodstream of the host are produced by the gut microbiota, it is worth carefully considering the potential contribution of the gut microbiota toward plasma SCFA levels. Analysis of SCFAs in conventionally raised (with gut microbiota) versus germ-free animals (no gut microbiota) revealed that SCFA concentrations in the cecum are enhanced by over 100-fold by gut microbiota.41 Furthermore, a recent study reported that the most abundant microbial SCFA, acetate, is virtually undetectable in the plasma of germ-free mice.42 Additionally, it has been shown that serum SCFA levels in the host (conventional mice) correlate with the amount of fiber in the diet, again implicating gut microbiota as playing an important role in determining serum SCFA levels.43 It has been demonstrated that suppressing microbial SCFA production using a low-fiber diet, or increasing microbial SCFA production using a high-fiber diet, caused drastic changes in not only cecal SCFAs, but also in serum SCFAs. For example, on a low-fiber diet serum SCFAs decreased by ~75% (from ~1.3 mmol/l to approximately 300 mmol/l), whereas on a high-fiber diet serum SCFAs levels are roughly doubled. Therefore, gut microbial production is responsible for setting circulating SCFA levels. In plasma, SCFA concentrations are between 0.1 mmol/l and 10 mmol/l.22,24,43,44 Although acetate is generally reported to be the most abundant SCFA produced, the precise ratios of acetate:butyrate:propionate reported vary. For example, acetate:butyrate:propionate ratios have been reported to be 55:35:1043 or 55:23:22,45 but it is clear from the literature that these ratios change quite dramatically with dietary manipulation (i.e., 40:26:34, 43:20:37, 73:5:22, 78:17:5, etc.).39,46
Cell biology of SCFA signaling: how does it work?
Once absorbed into the bloodstream of the host, there are multiple ways in which SCFAs can have cellular effects. Here, we will discuss these in 2 major categories: G-protein coupled receptor (GPCR)–mediated (Figure 1) and non-GPCR–mediated effects. For the purpose of this review, we will pay particular attention to what is known of these mechanisms in the kidney.
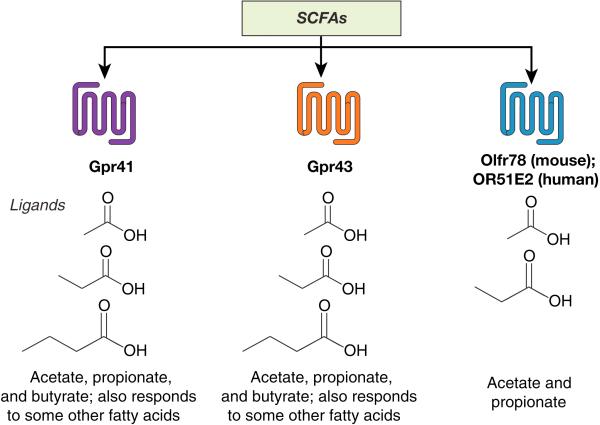
Figure 1. One way in which the gut microbiota and the host communicate is via host receptors that recognize microbial metabolites.
Three receptors for short chain fatty acids have been reported in the kidney, all of which are 7-transmembrane G-protein–coupled receptors. Gpr41 and Gpr43 respond to formate, acetate, and butyrate (as well as a few other fatty acids), whereas Olfr78 rather strictly responds to acetate and propionate.
GPCR-mediated mechanisms
To date, 4 SCFA receptors have been described: Gpr41, Gpr43, Gpr109a, and Olfr78. Here, we will first introduce what is known about the ligands and activities of each receptor, and then will discuss what (if anything) has been reported about their expression in the kidney.
Gpr41/Gpr43
Gpr41 (free fatty acid receptor 3, Ffar3) and Gpr43 (free fatty acid receptor 2, Ffar2) are the most well studied of the SCFA GPCRs and were initially described as SCFA receptors in 2003 by 2 separate groups.24,47 Both studies found that Gpr41 and Gpr43 are most responsive to propionate, although both receptors are activated by a number of other SCFA ligands. Among these, the strongest are acetate, butyrate, and isobutyrate, although they both also respond more weakly to other compounds with similar chemical structures.24,47 Although the exact EC50 values vary with both the assay and the species tested, Gpr41 and Gpr43 generally are reported to respond to acetate, propionate, and butyrate in the micromolar range, with propionate being the strongest ligand (Tables 1 and 2). In addition, beta-hydroxybutyrate has been reported, by different groups, to be either an agonist48 or an antagonist49 for Gpr41. Gpr41 couples to Gi, whereas Gpr43 can couple to both Gi and Gq.24,47 It is noteworthy that the human and rat orthologs of Gpr41 respond similarly to SCFAs,47 indicating that Gpr41 signaling is evolutionarily conserved.
Table 1.
Reported Gpr41 EC50 values
| Receptor | hGPR41 | rGPR41 | rGPR41 | hGPR41 | hGPR41 |
|---|---|---|---|---|---|
| Method | GTPƴS binding | GTPƴS binding | Yeast | cAMP | Ca2+ |
| Formate | 7760 mmol/l | 1000 mmol/l | > 10,000 mmol/l | No response | No response |
| Acetate | 1020 mmol/l | 393 mmol/l | 422 mmol/l | 1023 mmol/la | 1072 mmol/la |
| Propionate | 127 mmol/l | 41 mmol/l | 8 mmol/l | 6 mmol/la | 20 mmol/la |
| Butyrate | 158 mmol/l | 33 mmol/l | 64 mmol/l | 42 mmol/la | 58 mmol/la |
| Citation (yr) | Brown et al.47 (2003) | Brown et al.47 (2003) | Brown et al.47 (2003) | Le Poul et al.24 (2003) | Le Poul et al.24 (2003) |
Standard errors omitted for simplicity; additional (weaker) ligands omitted for simplicity.
Le Poul et al.24 reported pEC50 values; values here were converted to EC50.
Table 2.
Reported Gpr43 EC50 values
| Receptor | hGPR43 | hGPR43 | hGPR43 | hGPR43 |
|---|---|---|---|---|
| Method | FLIPR | GTPƴS binding | cAMP | Ca2+ |
| Formate | > 1300 mmol/l | 5640 mmol/l | 2455 mmol/la | 10233 mmol/la |
| Acetate | 52 mmol/l | 431 mmol/l | 35 mmol/la | 102 mmol/la |
| Propionate | 31 mmol/l | 290 mmol/l | 14 mmol/la | 79 mmol/la |
| Butyrate | 100 mmol/l | 371 mmol/l | 28 mmol/la | 339 mmol/la |
| Citation (yr) | Brown et al.47 (2003) | Brown et al.47 (2003) | Le Poul et al.24 (2003) | Le Poul et al.24 (2003) |
Standard errors omitted for simplicity; additional (weaker) ligands omitted for simplicity.
Le Poul et al.24 reported pEC50 values; values here were converted to EC50.
Subsequent work has outlined a number of roles for Gpr41 and Gpr43 in physiology—perhaps most prominently, Gpr41 has a role in metabolism,50 whereas Gpr43 plays a role in immune responses.22,24 Within the kidney, Gpr41 and Gpr43 have both been reported to be expressed in whole kidney and in the renal artery by reverse transcriptase-polymerase chain reaction.51
OR51E2/Olfr78
OR51E2 was initially deorphanized in 2009 by 2 groups: one reported that OR51E2 responded to propionate,52 whereas the other reported that OR51E2 was a receptor for β-ionone as well as several androgens.53 A subsequent study by a separate group in 201351 confirmed that OR51E2 (and its murine ortholog, Olfr78) responded to propionate as well as acetate, but did not detect any response of Olfr78 or OR51E2 to β-ionone. In 2015, it was reported54 that Olfr78 responded to acetate, propionate, and lactate (OR51E2 was not examined in this study).
Although OR51E2/Olfr78 have been reported to respond to other compounds (as described above), for the purpose of this review we will focus on the role of OR51E2/Olfr78 as an SCFA receptor. It is intriguing to note that there are very few examples of “functional” murine-human orthologs for ORs, that is, orthologs that respond to the same ligand. Unfortunately, the large number of similar OR genes in both mice (1000)55 and humans (300)56 makes it difficult to bioinformatically assign ortholog pairs with certainty. However, Olfr78 rather uniquely has a clear human ortholog (OR51E2) based on sequence similarity, and we know that these 2 receptors both recognize SCFAs. In fact, this OR is extremely well conserved among mammalian species and has a clear ortholog in mice, rats, rabbits, elephants, horses, and 5 species of primates,57 further implying that the role of this receptor is evolutionarily important and conserved. It should be noted that in contrast to Gpr41/43, OR51E2/Olfr78 responds to acetate and propionate but not to butyrate and has a much higher EC50 than does Gpr41/43. The reported ligands of Olfr78 and OR51E2 are summarized in Table 3.
Table 3.
Reported Olfr78/hOR51E2 EC50 values
| Receptor | hOR51E2 | hOR51E2 | Olfr78 (mouse) | hOR51E2 | Olfr78 (mouse) |
|---|---|---|---|---|---|
| Method | Ca2+ | cAMP/luciferase | cAMP/luciferase | cAMP/luciferase | cAMP/luciferase |
| Formate | No response | ||||
| Acetate | 2.35 mmol/l | 2.93 mmol/l | 2.01 mmol/l | ||
| Propionate | ~0.1 mmol/lb | 0.92 mmol/l | 2.16 mmol/l | 0.63 mmol/l | |
| Butyrate | No response | ||||
| β-iononea | ~2–3 mmol/la,b | No response | No response | ||
| Lactate | 4.04 | ||||
| Citation (yr) | Neuhaus et al.53 (2009) | Saito et al.52 (2009) | Pluznick et al.51 (2013) | Pluznick et al.51 (2013) | Chang et al.54 (2015) |
Standard errors omitted for simplicity; boxes left blank indicate that the ligand was not tested.
Neuhaus et al.53 also reported that OR51E2 responded to several androgens.
EC50 not reported (value estimated based on published data).
Within the kidney, Olfr78 has been localized by use of an lacZ reporter construct, which found that Olfr78 is expressed in the renal afferent arteriole as well as in a subset of large renal vessels. OR51E2 has not been localized in kidney on the protein level; however, RNA for OR51E2 is found in tissues that are consistent with the LacZ localization of Olfr78 (kidney as well as heart, skeletal muscle, etc.).58
Gpr109a
Gpr109a has also been reported as an SCFA receptor that responds to butyrate but not to acetate or propionate.59 The reported EC50 for butyrate is ~1 mmol/l. Gpr109a is arguably better known as a receptor for niacin and additionally has been reported to be a receptor for beta-D-hydroxybutyrate.59–61 Gpr109a was reported to be expressed (at a relatively low level) in the kidney by an reverse transcriptase-polymerase chain reaction Taqman array screen.62
SCFA GPCRs in the kidney
As for expression of these 4 GPCRs in the kidney, to date Gpr41, Gpr43, and Olfr78 have all been reported to be expressed in the kidney by reverse transcriptase-polymerase chain reaction.51 Gpr41 and Gpr43 were localized to major blood vessels (including the renal artery), but otherwise the specific localization of these receptors within the kidney has not been reported. Olfr78 was localized in the kidney with an lacZ reporter mouse (in which the native promoter of Olfr78 is used to drive expression of an lacZ reporter gene) and was found to localize to the renal afferent arteriole as well as to large renal arteries. Outside the kidney, Olfr78 has been found in vascular smooth muscle cells in a variety of vascular resistance beds.
Non-GPCR mechanisms
Although this review is focused on GPCR-mediated SCFA signaling, it would be remiss to not briefly discuss other aspects of cell biology related to SCFAs. For example, SCFAs can be transported across membranes by transporters: in the colon, SCFAs are transported across the apical membrane by SLC5A8, a sodium-coupled monocarboxylate transporter40,63–65; however, there is evidence that other transporters may be involved as well.66 Potential candidates include SCFA-permeant ion channels,67 organic anion transporters (i.e., mOat2, which has been found in the kidney68), or monocarboxylate transporter 1, which transports lactate and butyrate.69 In future work it will be important to understand how SCFA transporters contribute to the renal effects of SCFAs.
In addition to interactions with receptors and transporters, SCFAs can affect cell biology through additional pathways, including effects on cell proliferation, apoptosis, and histone acetylation. Propionate and butyrate both are known to inhibit cell proliferation14,70,71 and to induce apoptosis.72–74 Butyrate, in particular,75 is a strong inhibitor of histone deacetylases, and thus by altering histone acetylation, SCFAs can alter gene expression.76–80 SCFA-mediated inhibition of HDACs is thought to be independent of GPCRs,81 although it has been suggested that Gpr41 may play a role.82
Physiology of SCFA signaling: what does it do?
Here, we will discuss reported links between the microbiota and microbial SCFAs, and 3 different aspects of renal pathophysiology: blood pressure regulation, acute kidney injury, and chronic kidney disease (CKD). This section will attempt to summarize the chief findings to date for each of these areas.
Blood pressure regulation
Work on the microbiota and blood pressure regulation was encouraged by the realization that Olfr78 is expressed in cell types associated with blood pressure regulation.51 Olfr78 was localized to 2 specific subsets of renal blood vessels: (i) very large renal blood vessels (i.e., the renal artery and major branches of the renal artery) as well as (ii) the renal afferent arteriole. Intriguingly, Olfr78 was not found in intermediately sized vessels. Outside the kidney, Olfr78 was found in peripheral vascular beds in a variety of tissues (including muscle, skin, etc.). By polymerase chain reaction, Gpr41 and Gpr43 were shown to be expressed in the renal artery and in other large vessels (aorta, iliac artery). Subsequent experiments revealed that SCFAs act through both Olfr78 and Gpr41 to modulate blood pressure regulation, using 2 different pathways. First, SCFAs act via Olfr78 in the renal afferent arteriole to increase renin release, thereby contributing to basal renin levels (Olfr78 null mice have lowered plasma renin) and baseline blood pressure (Olfr78 null mice are hypotensive). However, SCFAs also act in the peripheral vasculature to acutely alter blood pressure. SCFAs act via Gpr41 to lower blood pressure (presumably via vasodilation, as SCFAs had been previously shown to dilate vessels ex vivo83–86), whereas Olfr78 counters this response. Although it seems counterintuitive to have 2 different GPCRs responding to the same ligand but modulating blood pressure in opposing ways (that is, to have Gpr41 promoting hypotension and Olfr78 promoting hypertension), it has been suggested51,87 that the “logic” behind this system is related to the very different EC50s of these 2 receptors (Table 1 vs. Table 3). Gpr41 is much more likely to be active at lower, even basal levels of SCFAs, and thus would primarily drive the vasodilatory response seen with SCFAs. However, as SCFAs rise appreciably, Olfr78 (which has a much higher EC50) would eventually be activated, thus serving as a brake on this pathway to prevent an inappropriate level of hypotension. In subsequent basic science studies, hypertension has been associated with changes in the microbiota88–90 as well as in SCFAs.88–90 However, it is clear that we still have much to learn in this area. Although one study90 reported that rats that had altered microbiota and increased blood pressure also had an increase in plasma acetate, other studies reported that butyrate-producing bacteria88 or both acetate and butyrate-producing bacteria89 tended to decrease in a hypertension model. In the future, it will be important to show that changes in SCFA-producing bacteria truly translate to changes in plasma levels of SCFAs, and to what degree.
It is also intriguing to note that there are hints in the clinical literature that there may indeed be a connection between blood pressure regulation and microbial SCFAs. For example, it has been reported that high-fiber diets91,92 and probiotic use93 (both of which should increase microbial SCFA production) are associated with lowered blood pressure. In fact, dietary fiber not only increases SCFA production, but can dramatically alter the acetate:butyrate:propionate ratio.39 In one study, which fed rats a diet with a daily fiber intake of 0, 1, 2, or 4 g/d, the acetate:butyrate:propionate ratio changed from 65:12:23 (0 g) to 40:26:34 (4 g)46 when dietary fiber was increased. Given the fact that Gpr41 (which lowers blood pressure) and Olfr78 (which increases blood pressure) have different affinities for the different SCFAs (i.e., Gpr41 but not Olfr78 responds to butyrate), one can imagine scenarios in which the particular choice of dietary fiber may help to further promote the hypotensive benefits of SCFAs. In addition, a study of > 4000 individuals reported that lower urinary formate (a microbial SCFA) correlates with increased blood pressure.94 Thus, although the majority of the clinical data to date are correlative, these data nevertheless underscore the need for further study in this new area. Indeed, there is an intriguing connection between the microbiota and blood pressure regulation, which likely involves SCFAs at least in part. However, it is clear that more studies must be done to truly understand this connection, let alone to investigate if and how we can manipulate it for potential benefit.
Acute kidney injury
Another area of renal (patho)physiology in which microbial SCFAs have been implicated is acute kidney injury.30 In a mouse model of renal ischemia-reperfusion injury, it was found that treatment with acetate, propionate, or butyrate was able to reduce kidney injury (measured by plasma creatinine) after ischemia-reperfusion injury.12 Intriguingly, a similar effect was also seen by treating mice with acetate-producing bacteria. The authors discuss potential roles for either SCFA GPCRs or for HDAC in ameliorating ischemia-reperfusion injury, although they state that this most likely occurs through “modulation of epigenetic processes.” Considering the difficulty of these studies, these are exciting and impressive findings. However, going forward it will be critical to reproduce these findings while (i) using, as a control, a closely related microbiota that does not produce high amounts of SCFAs, (ii) measuring the increase in plasma SCFAs as well as fecal SCFAs, and (iii) examining the underlying mechanism. In addition, it will be important to optimize conditions for colonization of the “new” microbiota. Therefore, although there is still much work to be done, this is an exciting new area with obvious clinical implications. It is also interesting to note that in ischemia-reperfusion models of other tissues, SCFAs11 or SCFA-producing bacteria95 have also been shown to be protective, implying that the underlying protective mechanism may be common across tissues.
Chronic kidney disease
Finally, microbiota have also been implicated in studies of chronic kidney disease.2,7,10,30,96–104 Multiple studies (reviewed in Ramezani et al.7 and Ramezani and Raj105) have reported a correlation between dysbiosis and CKD; that is, patients with CKD have altered microbiota as compared with control groups.103 Several known uremic toxins are produced by the microbiota (e.g., p-cresol sulfate, indoxyl sulfate, ammonia, trimethylamine N-oxide8,9,105–108), and most studies of the microbiota and CKD have focused on these toxins, with only a few references to SCFAs.107 Because this review is focused on SCFAs we will not cover the general literature on uremic toxins in the depth that it deserves; for a comprehensive review, please refer to the article by Ramezani and Raj.105 Regarding SCFAs, it has been shown that patients with CKD have an expansion of the microbiota that produce uremic toxins, and a contraction of microbiota that produce SCFAs.107 Of note, recent studies have highlighted the benefits of a high-fiber diet (which can act as a prebiotic for SCFAs) in CKD.98,109 In 2015, a metaanalysis of controlled feeding trials (143 participants in total) found that increased dietary fiber intake tended to reduce serum urea and creatinine.110 This is particularly noteworthy in light of the fact that CKD patients are often instructed not to consume a high-fiber diet (because of concerns regarding plasma potassium and phosphorus).
Clinical implications of SCFA signaling: what can we do?
Clearly, this entire field is still in its infancy, and many more studies need to be done to better understand these pathways and their potential implications. However, it is tempting to speculate that there may be opportunities in the future to take advantage of microbial signaling pathways for clinical benefit, on several possible fronts. First, understanding how and why dysbiosis correlates with disease may give us novel and unexpected insights into pathophysiology. Second, understanding whether dysbiosis may contribute to or perpetuate disease may give us new opportunities to intervene for clinical benefit. Clearly, the host genome is a major risk determinant for a number of different diseases (i.e., hypertension); however, unlike the host genome, the microbial genome present in a host can be modified. For example, by using prebiotics (foods that stimulate the growth or activity of certain microbes), probiotics (live bacteria), synbiotics (combinations of pre- and probiotics), or antibiotics, one can modify the content of the gut microbiota itself, and thus change the metabolites produced. But, there are a dizzying number of microbes to consider: which are the most crucial to “correct”? (Here, we must be careful to consider not only changes in genus, which are often what is reported because of sequencing limitations, but changes in species.) If we do correct them, would this necessarily result in a sustained improvement of an established phenotype? Clearly, much more needs to be done in this area, but there are encouraging hints in the literature; for example, an intriguing case report111 outlined a case in which antibiotics appeared to alleviate hypertension. Finally, regardless of whether we are able to develop clinical therapeutics, it is crucial that we better understand how these pathways already may be influencing human health: many patients (both with and without disease) are exposed to antibiotics, prebiotics (i.e., fiber), and probiotics (i.e., yogurt) on a regular basis. Are these exposures (re)shaping their microbiome, and/or altering their risk for disease? It is critical that more research be done, both in animal models as well as in human populations in order to understand the impact of these prevalent modifiers of microbiota on health, and whether and how they might be manipulated for health benefit.
In summary, the role of the gut microbiome in renal physiology and pathophysiology is an exciting new area that has exploded in recent years (53 of the citations for this review were published in 2013–2016). Although it is clear that changes in gut microbiota correlate with disease, we do not yet know whether we can purposefully manipulate microbiota in order to alter the course of disease (or whether they, or we, are simply along for the ride). In coming years, careful studies will be needed to explore and uncover the exciting potential in this new area.
FURTHER POINTS.
Any (grey) halftones (photographs, micrographs, etc.) are best viewed on screen, for which they are optimized, and your local printer may not be able to output the greys correctly.
If the PDF files contain colour images, and if you do have a local colour printer available, then it will be likely that you will not be able to correctly reproduce the colours on it, as local variations can occur.
If you print the PDF file attached, and notice some ‘non-standard’ output, please check if the problem is also present on screen. If the correct printer driver for your printer is not installed on your PC, the printed output will be distorted.
ACKNOWLEDGMENTS
The author would like to thank Niranjana Natarajan for helpful discussions related to this article. This work was supported by National Institutes of Health grant R01DK107726, National Institutes of Health grant R01HL128512, AHA 16IRG27260265, and Hopkins Conte Digestive Diseases Basic and Translational Research Core Center.
Footnotes
DISCLOSURES
All the author declared no competing interests.
REFERENCES
- 1.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2016;12:169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- 3.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell PD, Chaston JM, Wang Y, et al. In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Front Microbiol. 2014;5:1–15. doi: 10.3389/fmicb.2014.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabreiro F, Gems D. Worms need microbes too: microbiota, health and aging in. Caenorhabditis elegans. EMBO Mol Med. 2013;5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramezani A, Massy ZA, Meijers B, et al. Role of the gut microbiome in uremia: a potential therapeutic target. Am J Kidney Dis. 2016;67:483–498. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaziri ND, Zhao YY, Pahl MV. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant. 2016;31:737–746. doi: 10.1093/ndt/gfv095. [DOI] [PubMed] [Google Scholar]
- 9.Vaziri ND, Yuan J, Norris K. Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am J Nephrol. 2013;37:1–6. doi: 10.1159/000345969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moraes C, Fouque D, Amaral AC, et al. Trimethylamine N-oxide from gut microbiota in chronic kidney disease patients: focus on diet. J Ren Nutr. 2015;25:459–465. doi: 10.1053/j.jrn.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Aguilar-Nascimento JE, Salomao AB, Nochi RJ, Jr, et al. Intraluminal injection of short chain fatty acids diminishes intestinal mucosa injury in experimental ischemia-reperfusion. Acta Cir Bras. 2006;21:21–25. doi: 10.1590/s0102-86502006000100006. [DOI] [PubMed] [Google Scholar]
- 12.Andrade-Oliveira V, Amano MT, Correa-Costa M, et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol. 2015;26:1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellahcene M, O'Dowd JF, Wargent ET, et al. Male mice that lack the G-protein-coupled receptor GPR41 have low energy expenditure and increased body fat content. Br J Nutr. 2013;109:1755–1764. doi: 10.1017/S0007114512003923. [DOI] [PubMed] [Google Scholar]
- 14.Bindels LB, Porporato P, Dewulf EM, et al. Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br J Cancer. 2012;107:1337–1344. doi: 10.1038/bjc.2012.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugaut M. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol B. 1987;86:439–472. doi: 10.1016/0305-0491(87)90433-0. [DOI] [PubMed] [Google Scholar]
- 16.Hill MJ, Drasar BS. The normal colonic bacterial flora. Gut. 1975;16:318–323. doi: 10.1136/gut.16.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8:523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- 18.Dahlqvist G, Piessevaux H. Irritable bowel syndrome: the role of the intestinal microbiota, pathogenesis and therapeutic targets. Acta Gastroenterol Belg. 2011;74:375–380. [PubMed] [Google Scholar]
- 19.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang JS, Im CR, Im SH. Immune disorders and its correlation with gut microbiome. Immune Netw. 2012;12:129–138. doi: 10.4110/in.2012.12.4.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 22.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaziri ND. CKD impairs barrier function and alters microbial flora of the intestine: a major link to inflammation and uremic toxicity. Curr Opin Nephrol Hypertens. 2012;21:587–592. doi: 10.1097/MNH.0b013e328358c8d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le PE, Loison C, Struyf S, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinje S, Stroes E, Nieuwdorp M, et al. The gut microbiome as novel cardio-metabolic target: the time has come!. Eur Heart J. 2014;35:883–887. doi: 10.1093/eurheartj/eht467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puddu A, Sanguineti R, Montecucco F, et al. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediators Inflamm. 2014;2014:1–9. doi: 10.1155/2014/162021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noel S, Martina-Lingua MN, Bandapalle S, et al. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin Pract. 2014;127:139–143. doi: 10.1159/000363209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mafra D, Fouque D. Gut microbiota and inflammation in chronic kidney disease patients. Clin Kidney J. 2015;8:332–334. doi: 10.1093/ckj/sfv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto Y, Kurashima Y, Kiyono H. The gut microbiota and inflammatory bowel disease. Curr Opin Rheumatol. 2015;27:388–396. doi: 10.1097/BOR.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 33.Dowds CM, Blumberg RS, Zeissig S. Control of intestinal homeostasis through crosstalk between natural killer T cells and the intestinal microbiota. Clin Immunol. 2015;159:128–133. doi: 10.1016/j.clim.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 35.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 37.Dey N, Wagner VE, Blanton LV, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.den Besten G, van Eunen K, Groen AK, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coady MJ, Chang MH, Charron FM, et al. The human tumour suppressor gene SLC5A8 expresses a Na+-monocarboxylate cotransporter. J Physiol. 2004;557:719–731. doi: 10.1113/jphysiol.2004.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 42.Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 44.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cummings JH, Hill MJ, Bone ES, et al. The effect of meat protein and dietary fiber on colonic function and metabolism. II. Bacterial metabolites in feces and urine. Am J Clin Nutr. 1979;32:2094–2101. doi: 10.1093/ajcn/32.10.2094. [DOI] [PubMed] [Google Scholar]
- 46.Levrat MA, Remesy C, Demigne C. High propionic acid fermentations and mineral accumulation in the cecum of rats adapted to different levels of inulin. J Nutr. 1991;121:1730–1737. doi: 10.1093/jn/121.11.1730. [DOI] [PubMed] [Google Scholar]
- 47.Brown AJ, Goldsworthy SM, Barnes AA, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 48.Won YJ, Lu VB, Puhl HL, et al. Beta-hydroxybutyrate modulates N-type calcium channels in rat sympathetic neurons by acting as an agonist for the G-protein-coupled receptor FFA3. J Neurosci. 2013;33:19314–19325. doi: 10.1523/JNEUROSCI.3102-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura I, Inoue D, Maeda T, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeyasu K, Tamkun MM, Renaud KJ, et al. Ouabain-sensitive (Na+ + K+)-ATPase activity expressed in mouse L cells by transfection with DNA encoding the alpha-subunit of an avian sodium pump. J Biol Chem. 1988;263:4347–4354. [PubMed] [Google Scholar]
- 51.Pluznick JL, Protzko RJ, Gevorgyan H, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito H, Chi Q, Zhuang H, et al. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9, 1–14. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuhaus EM, Zhang W, Gelis L, et al. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang AJ, Ortega FE, Riegler J, et al. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature. 2015;527:240–244. doi: 10.1038/nature15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2584–2589. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–1496. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flegel C, Manteniotis S, Osthold S, et al. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taggart AK, Kero J, Gan X, et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005;280:26649–26652. doi: 10.1074/jbc.C500213200. [DOI] [PubMed] [Google Scholar]
- 60.Tunaru S, Kero J, Schaub A, et al. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- 61.Wise A, Foord SM, Fraser NJ, et al. Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem. 2003;278:9869–9874. doi: 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- 62.Rajkumar P, Aisenberg WH, Acres OW, et al. Identification and characterization of novel renal sensory receptors. PLoS One. 2014;9:e111053. doi: 10.1371/journal.pone.0111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyauchi S, Gopal E, Fei YJ, et al. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na(+)-coupled transporter for short-chain fatty acids. J Biol Chem. 2004;279:13293–13296. doi: 10.1074/jbc.C400059200. [DOI] [PubMed] [Google Scholar]
- 64.Gopal E, Fei YJ, Sugawara M, et al. Expression of slc5a8 in kidney and its role in Na(+)-coupled transport of lactate. J Biol Chem. 2004;279:44522–44532. doi: 10.1074/jbc.M405365200. [DOI] [PubMed] [Google Scholar]
- 65.Iwanaga T, Takebe K, Kato I, et al. Cellular expression of monocarboxylate transporters (MCT) in the digestive tract of the mouse, rat, and humans, with special reference to slc5a8. Biomed Res. 2006;27:243–254. doi: 10.2220/biomedres.27.243. [DOI] [PubMed] [Google Scholar]
- 66.Frank H, Groger N, Diener M, et al. Lactaturia and loss of sodium-dependent lactate uptake in the colon of SLC5A8-deficient mice. J Biol Chem. 2008;283:24729–24737. doi: 10.1074/jbc.M802681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Georgi MI, Rosendahl J, Ernst F, et al. Epithelia of the ovine and bovine forestomach express basolateral maxi-anion channels permeable to the anions of short-chain fatty acids. Pflugers Arch. 2014;466:1689–1712. doi: 10.1007/s00424-013-1386-x. [DOI] [PubMed] [Google Scholar]
- 68.Islam R, Anzai N, Ahmed N, et al. Mouse organic anion transporter 2 (mOat2) mediates the transport of short chain fatty acid propionate. J Pharmacol Sci. 2008;106:525–528. doi: 10.1254/jphs.sc0070291. [DOI] [PubMed] [Google Scholar]
- 69.Ritzhaupt A, Wood IS, Ellis A, et al. Identification and characterization of a monocarboxylate transporter (MCT1) in pig and human colon: its potential to transport L-lactate as well as butyrate. J Physiol. 1998;513(Pt 3):719–732. doi: 10.1111/j.1469-7793.1998.719ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan S, Jena GB. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: study in juvenile diabetic rat. Chem Biol Interact. 2014;213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 71.Siavoshian S, Segain JP, Kornprobst M, et al. Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut. 2000;46:507–514. doi: 10.1136/gut.46.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aoyama M, Kotani J, Usami M. Butyrate and propionate induced activated or non-activated neutrophil apoptosis via HDAC inhibitor activity but without activating GPR-41/GPR-43 pathways. Nutrition. 2010;26:653–661. doi: 10.1016/j.nut.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 73.Chen TH, Chen WM, Hsu KH, et al. Sodium butyrate activates ERK to regulate differentiation of mesenchymal stem cells. Biochem Biophys Res Commun. 2007;355:913–918. doi: 10.1016/j.bbrc.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 74.Fung KY, Brierley GV, Henderson S, et al. Butyrate-induced apoptosis in HCT116 colorectal cancer cells includes induction of a cell stress response. J Proteome Res. 2011;10:1860–1869. doi: 10.1021/pr1011125. [DOI] [PubMed] [Google Scholar]
- 75.Cousens LS, Gallwitz D, Alberts BM. Different accessibilities in chromatin to histone acetylase. J Biol Chem. 1979;254:1716–1723. [PubMed] [Google Scholar]
- 76.Hinnebusch BF, Meng S, Wu JT, et al. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 77.Medina V, Edmonds B, Young GP, et al. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 1997;57:3697–3707. [PubMed] [Google Scholar]
- 78.Tan J, McKenzie C, Potamitis M, et al. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 79.Ulven T. Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/ GPR41 as new potential therapeutic targets. Front Endocrinol (Lausanne) 2012;3:1–9. doi: 10.3389/fendo.2012.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De VF, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 81.Singh N, Thangaraju M, Prasad PD, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu J, Zhou Z, Hu Y, et al. Butyrate-induced GPR41 activation inhibits histone acetylation and cell growth. J Genet Genomics. 2012;39:375–384. doi: 10.1016/j.jgg.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Nutting CW, Islam S, Daugirdas JT. Vasorelaxant effects of short chain fatty acid salts in rat caudal artery. Am J Physiol. 1991;261:H561–H567. doi: 10.1152/ajpheart.1991.261.2.H561. [DOI] [PubMed] [Google Scholar]
- 84.Mortensen FV, Nielsen H, Mulvany MJ, et al. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut. 1990;31:1391–1394. doi: 10.1136/gut.31.12.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nutting CW, Islam S, Ye MH, et al. The vasorelaxant effects of acetate: role of adenosine, glycolysis, lyotropism, and pHi and Cai2+. Kidney Int. 1992;41:166–174. doi: 10.1038/ki.1992.23. [DOI] [PubMed] [Google Scholar]
- 86.Knock G, Psaroudakis D, Abbot S, et al. Propionate-induced relaxation in rat mesenteric arteries: a role for endothelium-derived hyperpolarising factor. J Physiol. 2002;538:879–890. doi: 10.1113/jphysiol.2001.013105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014;5:202–207. doi: 10.4161/gmic.27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Durgan DJ, Ganesh BP, Cope JL, et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension. 2016;67:469–474. doi: 10.1161/HYPERTENSIONAHA.115.06672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang T, Santisteban MM, Rodriguez V, et al. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mell B, Jala VR, Mathew AV, et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat model. Physiol Genomics. 2015;47:187–197. doi: 10.1152/physiolgenomics.00136.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson JW, Baird P, Davis RH, Jr, et al. Health benefits of dietary fiber. Nutr Rev. 2009;67:188–205. doi: 10.1111/j.1753-4887.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 92.Weisz OA, Gibson GA, Leung SM, et al. Overexpression of frequenin, a modulator of phosphatidylinositol 4-kinase, inhibits biosynthetic delivery of an apical protein in polarized madin-darby canine kidney cells. J Biol Chem. 2000;275:24341–24347. doi: 10.1074/jbc.M000671200. [DOI] [PubMed] [Google Scholar]
- 93.Khalesi S, Sun J, Buys N, et al. Effect of probiotics on blood pressure: a systematic review and meta-analysis of randomized, controlled trials. Hypertension. 2014;64:897–903. doi: 10.1161/HYPERTENSIONAHA.114.03469. [DOI] [PubMed] [Google Scholar]
- 94.Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H, Zhang W, Zuo L, et al. Bifidobacteria may be beneficial to intestinal microbiota and reduction of bacterial translocation in mice following ischaemia and reperfusion injury. Br J Nutr. 2013;109:1990–1998. doi: 10.1017/S0007114512004308. [DOI] [PubMed] [Google Scholar]
- 96.Poesen R, Windey K, Neven E, et al. The Influence of CKD on colonic microbial metabolism. J Am Soc Nephrol. 2016;27:1389–1399. doi: 10.1681/ASN.2015030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeng YQ, Dai Z, Lu F, et al. Emodin via colonic irrigation modulates gut microbiota and reduces uremic toxins in rats with chronic kidney disease. Oncotarget. 2016;7:17468–17478. doi: 10.18632/oncotarget.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kieffer DA, Piccolo BD, Vaziri ND, et al. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am J Physiol Renal Physiol. 2016;310:F857–F871. doi: 10.1152/ajprenal.00513.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoshifuji A, Wakino S, Irie J, et al. Gut Lactobacillus protects against the progression of renal damage by modulating the gut environment in rats. Nephrol Dial Transplant. 2016;31:401–412. doi: 10.1093/ndt/gfv353. [DOI] [PubMed] [Google Scholar]
- 100.Ranganathan N, Patel BG, Ranganathan P, et al. In vitro and in vivo assessment of intraintestinal bacteriotherapy in chronic kidney disease. ASAIO J. 2006;52:70–79. doi: 10.1097/01.mat.0000191345.45735.00. [DOI] [PubMed] [Google Scholar]
- 101.Moraes C, Borges NA, Mafra D. Resistant starch for modulation of gut microbiota: promising adjuvant therapy for chronic kidney disease patients? Eur J Nutr. 2016;55:1813–1821. doi: 10.1007/s00394-015-1138-0. [DOI] [PubMed] [Google Scholar]
- 102.Koppe L, Mafra D, Fouque D. Probiotics and chronic kidney disease. Kidney Int. 2015;88:958–966. doi: 10.1038/ki.2015.255. [DOI] [PubMed] [Google Scholar]
- 103.Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–315. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 104.Vaziri ND, Yuan J, Rahimi A, et al. Disintegration of colonic epithelial tight junction in uremia: a likely cause of CKD-associated inflammation. Nephrol Dial Transplant. 2012;27:2686–2693. doi: 10.1093/ndt/gfr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25:657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang WH, Wang Z, Kennedy DJ, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wong J, Piceno YM, Desantis TZ, et al. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol. 2014;39:230–237. doi: 10.1159/000360010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ellis RJ, Small DM, Vesey DA, et al. Indoxyl sulphate and kidney disease: causes, consequences and interventions. Nephrology (Carlton) 2016;21:170–177. doi: 10.1111/nep.12580. [DOI] [PubMed] [Google Scholar]
- 109.Vaziri ND, Liu SM, Lau WL, et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One. 2014;9:e114881. doi: 10.1371/journal.pone.0114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chiavaroli L, Mirrahimi A, Sievenpiper JL, et al. Dietary fiber effects in chronic kidney disease: a systematic review and meta-analysis of controlled feeding trials. Eur J Clin Nutr. 2015;69:761–768. doi: 10.1038/ejcn.2014.237. [DOI] [PubMed] [Google Scholar]
- 111.Qi Y, Aranda JM, Rodriguez V, et al. Impact of antibiotics on arterial blood pressure in a patient with resistant hypertension - A case report. Int J Cardiol. 2015;201:157–158. doi: 10.1016/j.ijcard.2015.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]