Abstract
Sialic acids (Sias) are abundantly displayed on the surfaces of vertebrate cells, and particularly on all mucosal surfaces. Sias interact with microbes of many types, and are the targets of specific recognition by many different viruses. They may mediate virus binding and infection of cells, or alternatively can act as decoy receptors that bind virions and block virus infection. These nine-carbon backbone monosaccharides naturally occur in many different modified forms, and are attached to underlying glycans through varied linkages, creating significant diversity in the pathogen receptor forms. Here we review the current knowledge regarding the distribution of modified Sias in different vertebrate hosts, tissues, and cells, their effects on viral pathogens where those have been examined, and outline unresolved questions.
Keywords: virus, sialic acids, modification, receptor, inhibitor.
Trends
Sialic acids (Sias) are components of cell-surface glycoproteins and glycolipids, as well as secreted glycoproteins and milk oligosaccharides. Sias play important roles in cell signaling, development, and host–pathogen interactions. Cellular enzymes can modify Sias, yet how modifications vary between tissues and hosts has not been fully elucidated.
Many viruses use Sias as receptors, with different modifications aiding or inhibiting virus infection. How modified Sias influence viral protein evolution and determine host/tissue tropism are poorly understood, and are important areas of research.
New advances in molecular glycobiology using pathogen proteins to detect varied forms allows for improved study of modified Sias that have otherwise proven difficult to isolate. This opens new avenues of inquiry for virology, as well as host interactions with bacterial and eukaryotic pathogens.
Interaction between Viruses and Sias
Sialic acids (see Glossary) generally occur as terminal monosaccharides on many different cell surfaces and secreted glycoconjugates and are therefore involved in key interactions between cells and many different viruses (as well as other pathogens) at various points in their infection and transmission cycles. Many viruses specifically bind to host Sias and use them as primary receptors for cell infection, or as components in a series of interactions that lead to infection. In vertebrates, Sias may be present as constituents of a variety of different complexes on the cell surface, including dense layers of glycoconjugates referred to as the glycocalyx. Mucosal surfaces are further protected by a viscous secretion, termed mucus, which varies in structure and composition depending on the location in the body [1]. However, mucus is formed in complex layers of cell-associated and secretory forms 2, 3. Sias in mucus and at cell surfaces can bind and trap viruses and prevent them from accessing their target tissues, and can also remove the bound virions in an active process mediated by mucocilliary transport [4]. Our understanding of virus–Sia interactions continues to be illuminated by new structural and biochemical analysis of viral proteins and bound glycans 5, 6. Currently we still do not clearly know which specific Sia structures are present on different cells or tissues of many viral hosts, including humans. It is clear that Sias exist in many diverse forms, and that their synthesis is tightly regulated yet highly variable on the cells and tissues of individual animals. The displayed glycan combinations vary under differing physiological conditions, stages of development, and also between different animal species. Many viruses also encode Sia-specific enzymes that alter the host Sia and therefore their specific interactions, including sialidases that remove the terminal Sia from glycans or esterases that remove ester-bonded acetyl modifications from the Sias where those are present.
Here we review examples of the various interactions between Sias and viruses in vertebrate animals, and examine how those can be modulated by variations in Sia structures, leading to differences in recognition and/or viral enzyme activity. We further summarize what is known about the diversity and distribution of modified Sias, the known and potential impacts of these specific glycan variants on different viruses, and define some questions that remain to be resolved.
Sias: Modifications and Variation
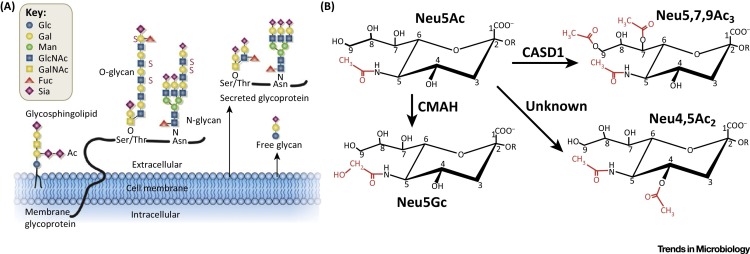
Sias are a highly diverse family of monosaccharides that serve as terminal residues of N- and O-linked glycoproteins and glycolipids, and they are also components of various polysaccharides. The core Sia structure contains nine carbons, and modifications are most commonly found at the 4, 5, 7, 8, and 9 positions. Modifications at the 5-carbon position determine the primary Sia forms: N-acetylneuraminic acid (Neu5Ac), N-glycolylneuraminic acid (Neu5Gc), and 2-keto-3-deoxynononic acid (KDN) [7]. These primary Sia forms serve as core structures, and additional modifications can be generated enzymatically to form more than 30 variant types of Sia [8]. Those include acetyl, sulfate, lactoyl, and methyl group additions to various positions. Display of the different modified forms clearly varies between different organisms, tissues, and cell types, and as the modifications may be present in myriad combinations, there may be a wide diversity of glycan forms (Figure 1 ). In addition, the linkage of the Sia to the underlying glycan structure may also vary, with α2,3 or α2,6 linkages being most common, as well as α2,8 linkages in the formation of poly-Sia structures which are primarily found in the brain and on certain cells of the immune system [9].
Figure 1.
Modified Sialic Acids. (A) Diverse glycan forms exist at the cell surface in both bound and secreted (or free) forms. Sias predominantly terminate carbohydrate chains, while encompassing their own chemical diversity. Reprinted from [33], with permission. (B) Sias are diverse structures, including modified forms induced by enzymatic functions that vary between species, tissues, and even individual cells. Representative hydroxylation and acetylations discussed in this review are shown on the base Sia (Neu5Ac) chemistry. We note the known (and unknown) encoded-enzymes that modify the CMP-Sia precursor molecule.
Our understanding of the expression and biological roles of this wide diversity of variant Sias is rather limited for various practical and technical reasons. However, the modifications are widespread and clearly allow the Sia to serve a variety of physiologically important roles in normal cell and tissue development and in controlling cell–cell interactions. For example, the constitutive expression of influenza C virus 9-O-acetylesterase in transgenic mice resulted in complete developmental arrest of embryos as early as the two-cell stage [10]. Polysialic acids play an important role in neural development and are involved in neuron growth, axon guidance, and synapse formation, and their synthesis is tightly regulated [11]. Transgenic mice lacking expression of synthases and transferases necessary for oligosialic acid expression in the central nervous system showed atypical neuronal development and a ‘sudden death’ phenotype [12]. In addition, Sias are involved in immune cell maturation and activation. Regulated 9-O-acetylation of ganglioside GD3 (also called CD60) on T cells correlates with cell differentiation and blocking of pro-apoptotic pathways in proliferating T cells [13]. Similarly, regulated O-acetylation at the C9-position of Sia on B cells is required for the correct development and activation of those cells, as 9-O-acetylation blocks recognition by the Sia-binding immunoglobulin-type lectin CD22 (SIGLEC-2), which acts as an inhibitor of B cell receptor signaling and activation [14]. Given the regulatory role of Sias in so many different cell processes and pathways, it is not surprising that abnormal expression of modified Sias has been identified as a hallmark of many cancers, including some types of leukemia 15, 16 and colorectal cancers [17].
Formation of specific glycans is regulated by the expression of a variety of enzymes, many of which are localized to the endoplasmic reticulum and Golgi apparatus, where they direct co- and post-translational modification of the glycans on glycoproteins or glycolipids. Enzymes specific for Sia addition and modification are expressed in the Golgi apparatus while others may be expressed in the cytoplasm and modify the Sia precursors. The varying expression of sialyltransferases, sialidases, esterases, and other Sia-modifying enzymes makes Sia synthesis and modification highly variable and dynamic, and as a result their display is influenced by both external and internal cellular stimuli. Several of the genes encoding Sia-modifying enzymes have been identified and analyzed in detail (Figure 1). Those include cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), the enzyme responsible for formation of Neu5Gc from the CMP-Neu5Ac precursor [18]. The CASD1 gene (capsule structure 1 domain containing 1) encodes a 9-O-acetyltransferase enzyme that mediates the formation of 9-O-acetyl Sia (9-O-Ac) from a CMP-Neu5Ac precursor 13, 19, 20. However, while enzymatic activity for a 4-O-acetyltransferase has been demonstrated, a candidate gene has not yet been identified [21]. It is also still not clear what activities control 7-O-acetyl Sia (7-O-Ac), as in vitro studies have shown acetyl groups added at the C7-position migrate to the C9-position by way of C8-position 22, 23. Whether CASD1 adds acetyl groups at the C7-position or directly to the C9-position is not clear, and it is possible that a migrase enzyme is involved in that modification. A variety of sialyltransferases (likely 20 in humans) regulate the specific addition and linkage of the Sia to the underlying N- or O-linked glycan on glycoproteins or glycolipids 24, 25.
Many sialylated glycans are synthesized in and presented on vertebrate cells, but detailed information about how those are produced and displayed is often lacking; however, it is clear that the relative display patterns of each modified Sia form can vary widely between even closely related animal species, as well as between and within different tissue types 26, 27. For example, 4-O-acetyl Sia (4-O-Ac) appears to be restricted to only a few tissue types in mice, including the colon, spleen, stomach, thymus, and uterus 27, 28. One factor determining the species-specific display of modified Sia is the presence and expression of the necessary enzymes – as some vertebrate lineages may lack the intact genes encoding certain Sia-modifying enzymes. The best documented case is a lack of Neu5Gc due to independent loss of the functional CMAH gene in humans and in some new world primates, as well as a lack of CMAH expression or activity in birds and reptiles (sauropsids), seals (pinnipeds), ferrets (mustelids), and possibly in western breeds of dogs 18, 29, 30, 31, 32. In many other mammalian species CMAH is actively expressed and Neu5Gc is generally abundant, but with observed variation between tissues and cell types. Further, there appears to be no enzyme that causes reversion of the glycolyl modification [33]. It is still unclear why such wide differences in Neu5Gc expression exist between different animals. It has been suggested that the loss of Neu5Gc in humans following divergence from our common ancestor with chimpanzees may have been beneficial by allowing resistance to Neu5Gc-specific pathogens, such as ancestral strains of Plasmodium malaria [34]. Another interesting case is the high variability of 4-O-Ac in many mammals, including apparent low levels or loss of that Sia form in cows, pigs, sheep, goats, and humans 26, 27. Again, the biological reasons for the highly variable synthesis and distribution of modified Sias are not known.
Viral Interactions: An Overview
Many different viruses bind to Sia-terminated glycans either as primary receptors for infection or by using them as accessory receptors in host cell binding and uptake (Table 1 ), and the evolution of the viruses has selected for specific interactions with particular Sia forms and linkages on different hosts and tissues. Where these have been defined, the evolved specificities for the host receptor variants and alternative linkages often play important roles in the tropism of the virus at the level of host, tissue, and specific differentiated cell types. In addition, the relative distribution of modified forms of Sia in given hosts and tissues may shape the evolution of Sia-interacting viral proteins, including hemagglutinins. Determining the exact nature of virus–Sia interactions, as well as the impact of varying Sia chemistry on those interactions, is therefore critical to understanding the complex impacts of host glycan variation on virus systems. Here we review some of the Sia modifications and their known interactions with viruses, while also identifying areas where more information is required. In addition to the Sia modifications discussed, other modifications may also be present on Sias that potentially interact with viruses (lactoyl, phosphate, sulfate, or methyl additions), yet little is known about their relative synthesis and distribution [7].
Table 1.
Representative Viruses Known to Bind Sialic Acidsa
| Family | Virus Genus | Viral Sialolectin | Sia Enzyme | Sia Specificity | Refs |
|---|---|---|---|---|---|
| Reoviridae dsRNA |
Reovirus | Sigma-1 | Neu5Acα2-3 Gal (Type 1) | [72] | |
| Rotavirus | VP4 | Neu5Ac (RRV, UK) Neu5Gc (NCDV, OSU, SA11) |
49, 50, 51, 73 | ||
| Orbivirus | VP2 | Not known | [74] | ||
| Coronaviridae +ssRNA |
Betacoronavirus | Spike, HE | Esterase (HE) | Neu5,7,9Ac3 (BCoV-Mebus) Neu4,5Ac2 (MHV-S) |
27, 38, 41 |
| Toroviruses | Spike, HE | Esterase (HE) | Neu5,9Ac2 (PToV-P4) | [27] | |
| Picornaviridae +ssRNA |
Enterovirus | VP1, VP3; VP1 only (CVA24v) | Neu5Acα2-3Gal Neu5Acα2-6 Gal (EV-D68); Neu5Acα2-3 Gal (EV-70, CVA24v) |
75, 76, 77 | |
| Carbovirus | VP1, VP3 | Neu5Acα2-3Gal | [78] | ||
| Orthomyxoviridae -ssRNA |
Influenza A | HA, NA | Sialidase (NA) | Neu5Acα2-3 Gal (Avian, Porcine) Neu5Acα2-6 Gal (Human, Porcine) |
[79] |
| Influenza B | HA, NA | Sialidase (NA) | Neu5Acα2-3Gal Neu5Acα2-6 Gal |
[80] | |
| Influenza C | HEF | Esterase (HE) | Neu5,9Ac2 | [81] | |
| Paramyxoviridae -ssRNA |
Rubulavirus | HN | Sialidase (HN) | Neu5Acα2-3 Gal Neu5Acα2-6Gal Neu5Acα2,8polySia (Mumps) |
[82] |
| Respirovirus | HN | Sialidase (HN) | Neu5Acα2-3 Gal (hPIV-1); Neu5Acα2-3 Gal, Neu5Acα2-6 Gal, Neu5Gcα2-3 Gal (hPIV-3) |
[83] | |
| Avulavirus | HN | Sialidase (HN) | Neu5Ac (NDV) | [84] | |
| Parvoviridae ssDNA |
Bocavirus | VP1 | 2,3-N- and 2,3-O-linked Sia (BPV) | [85] | |
| Protoparvovirus | VP1 | Neu5Acα2-3 Gal (MVM) | [86] | ||
| Polyomaviridae dsDNA |
Polyomavirus | VP1 | Neu5Acα2-3 Gal (BKV, SV-40), Neu5Acα2-6 Gal (JCV), Neu5Gcα2-3 Gal (SV-40) | 46, 47, 48 |
Abbreviations: dsRNA, double-stranded RNA; +ssRNA, positive-sense single-stranded RNA; −ssRNA, negative-sense single-stranded RNA; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA.
4-O-Acetylated Sia
The distribution and roles of 4-O-Ac Sias are not well understood as, until recently, there were few means to detect their presence in cells and tissues. Mass spectrometry to identify glycan structures may use methods (permethylation) that remove the O-acetyl modifications, and many approaches that can detect the 4-O-Ac modification do not characterize its distribution at the cellular or subcellular level. Recently the hemagglutinin-esterases (HEs) of the mouse hepatitis virus (MHV) S strain and of infectious salmon anemia virus (ISAV) were found to specifically recognize 4-O-Ac Sias, and esterase-inactive forms of the MHV-S protein can be used to detect the presence of that modified Sia in cells or tissue sections (Box 1 ) 26, 27. These probes showed that 4-O-Ac-modified Sias are displayed at high levels on the erythrocytes and other tissues of some mammals, including guinea pigs and horses, and in many ‘lower’ vertebrates, particularly fish.
Box 1. Detection of Modified Sialic Acid (Sia).
Traditional characterization of Sia composition of biological samples has involved chemical release, purification, and chemical derivatization (DMB labeling) for analysis by high-performance liquid chromatography (HPLC), mass spectrometry, thin-layer chromatography, or nuclear magnetic resonance. However, many of the methods for sample preparation result in the loss of labile modifications of Sia. Further, these methods do not show the relative distribution of the modified Sia forms in the context of an intact biological sample, such as tissue, its various cell populations, or mucosal surfaces. Histochemical analyses have been previously undertaken with mild periodate oxidation and saponification to generally visualize Sia and 9-O-acetylated forms [7].
The accurate identification of modified Sia in biological samples requires new and robust molecular methodologies. The last two decades have seen steady progress in the field of glycobiology, including the use of polyclonal and monoclonal antibodies, naturally occurring lectins, sialidases, esterases, as well as Sia lysases to interrogate and manipulate Sia in their native state on cells and tissues. Monoclonal antibody generation against specific carbohydrate forms is difficult and often results in low affinity antibody forms with specificities for particular linkages or other specific variants, and is hampered by the need for a naïve animal for immunization that does not present that particular Sia form (so that it is not a self-antigen). The use of plant-derived lectins (SNA, MAL, and MAH), as well as others from various sources, has greatly contributed to the analysis of linkage-variant Sia species, and has revealed the variable Sia linkages that control the specificities and tropisms of many viruses. More recently, the development of recombinant expressed sialolectins with various fusion and post-translational labels have expanded the molecular toolkit for probing Sia diversity. These expressed sialolectins include those of vertebrate (siglecs) and microbial origin, as well as a number of virus-derived virolectins. This approach allows for the harnessing of naturally coevolved specificities, under a recombinant molecular platform that would allow for rational or artificial-selection engineering. Recent publications have verified the use of nidovirus hemagglutinin-esterase (HE) probes for analysis of O-acetyl Sia forms in the tissues of humans and mice [27]. A robust survey and cataloging of virus-encoded sialolectins would contribute to our understanding of virus–Sia interactions while aiding in the development of key tools for interrogating the ‘sialome’ of cellular systems.
The physiological role of 4-O-Ac Sias and the reasons for the apparent loss or altered modifying enzyme expression in so many animals is unclear, but their ability to act as an inhibitor for many sialidases may be involved. 4-O-Ac Sias have been known since the 1950s to influence influenza tropisms due to their role as gamma inhibitors in horse and guinea pig sera, which interfere with infection and replication of some human influenza A H2 and H3 strains by inhibiting the activity of the viral neuraminidase (NA). Those inhibited viruses can escape by selecting variants in the viral hemagglutinin (HA) that do not bind the modified Sias 35, 36, 37. In contrast to their inhibitory effects on influenza viruses, some other viruses use 4-O-Ac Sias as specific receptors. The presence of 4-O-Ac Sias in the gastrointestinal tract, particularly the colon, and brain of mice likely contributes to the tropism seen in some MHV-S infections which target those organs [38]. As 4-O-Ac Sia is highly enriched in the mucus present on fish skin, some piscine pathogens (including paramyxoviruses and ISAV) may bind to or interact with this modified form 26, 39. Apart from ISAV, it is unclear how 4-O-Ac interacts with fish viruses or other pathogens, in part due to the difficulty of studying this Sia variant. Overall, very little is currently known about the synthesis and distribution of this modified Sia in most vertebrates, or how its presence shapes tropism of other Sia-binding viruses (or of other pathogens which bind or cleave Sia).
9-O-Acetylated (and 7-O-Acetylated) Sia
The addition of acetyl or other groups to the C9 position of Sia is widely seen in cells and tissues of a variety of animals, including humans, and 9-O-acetylated (9-O-Ac) Sias have previously been detected using the HE protein of human influenza C virus as a probe [40]. As discussed previously, acetyl modifications of the 7-position also occur, creating 7,9-O-Ac2 modified Sias in addition to 9-O-Ac modifications. 7-O-Ac modifications alone are rarely observed, likely due to the natural tendency for migration of the acetyl group to the C9-position. Besides influenza C virus, 9-O-Ac Sia can be used as a specific receptor for many coronaviruses (including in the 7,9-O-Ac2 form) and toroviruses 27, 41. It appears to be the receptor for the recently discovered influenza D virus (sometimes referred to as bovine influenza virus) which is 50% homologous with human influenza C virus and which also has a HE receptor-binding and esterase protein 42, 43. The presence of the 9-O-Ac Sia modification has been shown to affect the activities of the NA of influenza viruses, reducing its activity by 3- to 4-fold compared to the nonmodified forms [44], although the significance of that activity for natural viral fitness is not yet known.
Neu5Gc
Variations in modified Sia between related species may underlie major evolutionary host variability and differences in pathogen specificity. As mentioned above, humans differ from most nonhuman primates by the loss of the enzyme CMAH, which produces Neu5Gc from CMP-Neu5Ac precursors. Among viruses that infect both human and nonhuman primate species (as well as bacteria and macroparasites), divergence of strains and differential tropisms for Neu5Ac versus Neu5Gc have been observed 34, 45. The polyomaviruses are small double-stranded DNA (dsDNA) viruses that infect a wide range of hosts, and they bind Sia via the major capsid protein, VP1, which is arranged within the particle as 72 pentamers. The polyomaviruses from humans (JC virus, BK virus, and Merkel Cell polyomavirus) all have a strong preference for Neu5Ac Sia in cell attachment, while viruses from nonhuman primates and other mammalian hosts (simian virus 40) show increased flexibility in Sia recognition as they have a larger, more polar pocket allowing binding to Neu5Gc Sia 46, 47, 48. Similarly, rotaviruses, double-stranded RNA (dsRNA) viruses in the Reovirus family, also show a strain-dependent specificity for Neu5Ac or Neu5Gc binding – bovine strain NCDV and porcine strains OSU and CRW-8 have specificity for Neu5Gc while rhesus macaque strain RRV prefers Neu5Ac [49]. Whether human rotaviruses interact with Sias has been an area of debate as they are classically termed sialidase-insensitive, meaning removal of terminal Sia via sialidases does not inhibit infection. However, some Sia forms are resistant to sialidase (including complex branching, internally linked Sia, and further modifications) and it appears likely that at least some human rotaviruses interact with Sias during cell entry 50, 51. It is also possible that the sialidase insensitivity that distinguishes human rotaviruses from other animal strains is due to the loss of Neu5Gc in humans and subsequent adaptation of the rotavirus to a different modified Sia form [50]. However, even within a single host there may be different specificities, such as bovine strains that include the Neu5Ac-binding UK or Neu5Gc-binding NCDV [49]. It is unknown whether the loss of Neu5Gc in humans has impacted the relative emergence potential of Sia-binding viruses from Neu5Gc-positive zoonotic reservoirs.
Viral Proteins Altering Sia: Sialidases and Esterases
Viruses can encode one of two varieties of Sia-modifying enzymes: sialidases or esterases. Viral sialidases (often referred to as neuraminidases) complement the Sia-binding glycoproteins to coordinate receptor interaction activity by removing the Sia from glycans. Viral esterases remove O-acetyl groups from modified Sia and are often present as HEs where both activities are expressed as separate functions of the same protein. Both sialidase and esterase activities appear to provide similar benefits to the viruses, as they alter the host environment by removing receptors and improving viral penetration of mucus, favoring cell infection, and enhancing virion release during the final stages of the virus lifecycle 52, 53. While some viral sialidases may be blocked by modified Sia (as discussed previously), viral esterases have evolved to remove specific modifications. We discuss two examples of how modified Sias influence viral infection.
Sialidases
Influenza A Viruses. The HA and NA are encoded in distinct genomic segments, so that re-assortment of HA and NA variants can occur readily. However, the resulting viruses appear to need a balance between the HA Sia-binding and NA Sia-cleaving activities, which may be accomplished by coevolution of either or both genes [54]. It has been shown that sialidases from many pathogens (bacterial and influenza) have optimal enzymatic activities for α2,3-linked Neu5Ac Sia, and the shift to the α2,6 linkage, hydroxylation of Neu5Ac to Neu5Gc, or addition of O-acetyl groups to C4, 7, or 9 reduce the capacity of sialidases to cleave in vitro 44, 55, 56. The natural in vivo activity of human or avian influenza NA towards modified Sia appears to be consistently reduced, while ‘drift’ in linkage specificity has been observed over large influenza evolutionary timescales [57]. It appears that, for influenza viruses, HA compensatory mutations are commonly used to avoid inhibitory or ‘decoy’ Sias, rather than changes in the sialidase specificity of NA, and also that sialidases cannot readily gain the ability to avoid inhibition by at least the 4-O-Ac Sia [58]. Similarly, NA resistance alleles to sialidase inhibitors (e.g., Oseltamivir-resistant influenza) can arise, but may require favorable compensatory epistatic alleles in HA to allow fixation 59, 60. For example, mutations that allow human influenza viruses to replicate in the presence of high levels of 4-O-Ac Sia occurred in the Sia binding site of the HA and prevented HA binding to the 4-O-Ac Sia. Those mutations were seen in the H3N8 and H7N7 equine influenza viruses, as well as in human influenza viruses experimentally adapted to grow in the presence of horse serum proteins [58]. It is not known whether selection of mutations in HA to avoid these inhibitory modified Sias also alters HA receptor binding specificities for other Sias, or alters antigenic epitopes, and thereby shapes the adaptive potential of the viruses in different hosts. However, it is likely that the relative abundance of modified Sia forms in different hosts contributes to host-specific HA and NA selection, and may also constrain the relative transmission potential in novel emergent viruses.
Paramyxoviruses. Many paramyxoviruses encode a hemagglutinin-neuraminidase (HN) glycoprotein, which carries both receptor-binding and sialidase functions [61]. The HN of paramyxoviruses is therefore analogous to both the HA and NA in influenza A viruses. The NA-domain likely plays a part in the release of budding virions and in the clearing of Sia decoys, similar to influenza NA. However, unlike influenza HA, HN does not directly cause fusion of the virus with its target cell; instead, the HN binding of Sia activates fusion through an additional protein, the fusion (F) protein [62]. Structural models show that paramyxovirus HN sialidase domains share similarities with influenza NA despite having divergent polypeptide sequences; whether or not they share a common origin is unknown [63]. Recent studies have shown that the NA-active site also has functional receptor-binding activity. These secondary Sia binding sites aid in balancing the receptor-binding–receptor-destroying–F activation activities of HN 64, 65. Whether these secondary binding sites also avoid binding to modified forms requires further research, particularly as the presence and location of secondary sites varies between paramyxovirus species [65]. Overall, little is known about how modified Sias interact with paramyxoviruses or how their presence shapes viral evolution in this complex and highly diverse family [61].
Esterases
Nidovirales. This Order contains the evolutionarily related families of coronaviruses and toroviruses, which are both enveloped positive-strand RNA viruses that commonly infect the respiratory and/or gastrointestinal tracts of many vertebrates. Both toroviruses and group II coronaviruses often express an HE glycoprotein on the surface of their envelope in addition to the large binding and fusion active spike (S) protein [66]. Coronavirus and torovirus HEs appear to have originated through multiple horizontal gene transfer and recombination events between strains 67, 68. Interestingly, these HEs also appear homologous to the Orthymyxovirus family member influenza C HE glycoprotein, suggesting a common evolutionary ancestor for the protein resulting from gene exchange between these different viral taxa [69].
The modified Sia specificities of S and HE are determinants of tissue and cell tropism for both type II coronaviruses and for toroviruses. For example, the coronavirus MHV strains S and JHM preferentially bind to, as well as de-acetylate, 4-O-Ac modified Sia, while ancestral strains of MHV prefer 9-O-Ac Sia similar to other nidovirus members 23, 38. Both 9-O-Ac and 4-O-Ac are displayed in the colon of mice [27], and 4-O-Ac-specific MHV strains also exhibit a novel neurotropic phenotype likely due to the presence of this modified Sia in the brain [38]. The matching of S and HE binding specificities is further seen in bovine coronaviruses and toroviruses, porcine toroviruses, and many others, with Sia specificity matching that of their target tissue 27, 68. The HE is not an essential protein for all coronaviruses [70], but appears to increase the efficiency of cell infection, either by improving viral binding initially or by enhancing virion release [71]. However, the specific roles of HE in penetrating or altering host mucus, facilitating membrane fusion, or aiding in release of budding virions from infected cells are currently poorly understood. It also remains to be seen what role changes in HE specificity may play in viral adaptation during infection of new host species; this is particularly relevant considering the zoonotic potential of coronaviruses.
Concluding Remarks
Sias are one of the first points of contact between many different pathogens and the host organism due to their presence on the outer surfaces of cells and mucosal tissues. The diversity of Sia forms results from modifications that can be added singly or in combinations (Figure 1), leading to complex barriers that pathogens must navigate to reach their target cells or tissues. The influence of Sia variation on viral adaptation is seen in the diverse viral families that use Sia as a receptor or coreceptor, often showing specificity for some modified forms over others. This is also shown in the number of viruses that carry Sia-modifying enzymes such as sialidases or esterases, which allow them to control their binding and release with high specificity (Table 1). Sia interactions and modifications also extend to bacterial and eukaryotic pathogens, as well as commensal colonizers (Box 2 ). While Sias have been known as pathogen receptors for many decades, much remains to be learned about the many modifications and how they are synthesized and distributed in different organisms and tissues. The recent development of new tools (Box 1) that allow for in situ analysis or removal of modified Sias has expanded our ability to study more complex forms and to answer questions about the roles of these modifications in shaping viral evolution, tissue tropism, and host range. These tools will also allow us to address how Sias with different modifications, including 4-O-Ac, 9-O-Ac, and Neu5Gc (and combinations), shape organismal development and cell–cell communication (see Outstanding Questions). While Sias have a great variety of forms, it is important to remember that they are part of an existing mosaic of glycans that vary from organism to organism, tissue to tissue, and even within a single cell over time. The roles played by modified Sias in these complex environments and how they vary between organisms are key questions in glycobiology and in host–pathogen interaction studies.
Outstanding Questions.
What genes encode enzymes involved in Sia modification (e.g., 4-O-acetyltransferase)? Why have some enzymes been lost in some lineages? Are any modifying enzymes novel in given lineages? How does gene expression vary between species/evolutionary lineages and contribute to differential ‘sialomes’?
How is the expression of Sia-modifying enzymes regulated during development and within and between tissues? What are the mechanisms that result in enrichment of modified Sia forms in cancerous cells?
How do modified Sias alter host specificity and tropism of different virus families? How do modified Sias contribute to the evolution of Sia-interacting viral proteins? Does differential Sia presence and distribution between lineages impact viral emergence in new hosts?
In cases of antagonism, how do viruses avoid modified Sias during infection?
What Sia forms are present on the N- and O-linked glycans present on virus glycoproteins? Do modified Sias on viral glycoproteins interfere with folding or function of viral proteins, or sialidase release of viral particles? Might modified Sia forms on pathogen glycans increase or mask immune detection?
What is the role of diverse Sia forms in cases of transkingdom coinfection? What are their effects on bacterial or eukaryotic pathogen infections?
Box 2. Non-Viral Pathogens and Modified Sialic Acids (Sias).
Many bacteria and eukaryotic microbes that must bind to or pass through mucosal surfaces also interact with Sias. The gut microbiome contains many groups that utilize mucin glycans as carbohydrate sources, including Sia 87, 88. Since modified Sias may pose as difficult metabolic substrates, bacterial pathogens with Sia metabolic operons often express O-acetylesterases along with sialidases to enhance their function by removing modifications [89]. Sias can also be used as an adhesion point for bacterial colonization or as a method of decorating the bacterial surface and avoiding immune detection 7, 90, 91. For example, Neisseria gonorrhoeae displays synthesized or acquired Sias to mask their presence from innate immune responses [92]. For toxins produced by bacterial pathogens, such as Salmonella enterica serovar Typhi, the Neu5Ac Sia can act as a toxin receptor and determine susceptibility to certain bacterial infections [93]. Modifications of the target Sia could lead to resistance to that toxin. The activities of different pathogen sialidases and esterases can result in complex interactions during mixed infections of various pathogens, as seen in viral–bacterial superinfections 94, 95, 96. In the case of influenza, the activity of NA is known to contribute to secondary bacterial pneumatic infections – the removal of Sia molecules appears to enhance the binding of Streptococcus pneumoniae via alveolar galectins as well as promoting bacterial growth by enhancing free Sia carbon substrates 97, 98. In addition to bacterial species, many eukaryotic parasites also use Sia as a receptor or adhesion point. Both Plasmodium falciparum and Trypanosoma cruzi use Sias expressed on erythrocytes to facilitate infection and express multiple Sia-binding proteins 7, 99. The relative effects of modified Sias on macroparasites are unexplored.
Acknowledgments
We thank Ajit Varki (UC-San Diego), members of the Parrish laboratory, and two anonymous reviewers for feedback and critical analysis of the manuscript. Support for this work was received from the NIAID Centers of Excellence in Influenza Research and Surveillance (HHSN272201400008C), NIH grant R01 GM080533-05 to CRP, and NIH Common Grant (U01CA199792).
Glossary
- Esterase (acetylesterase)
in this context, an enzyme that removes O-acetyl groups from Sia sugars by cleaving the ester bond. Esterases have specificity for the position of the O-acetyl group in the Sia, typically 4-O-Ac or 9-O-Ac. Esterases are encoded by influenza C and nidovirus family viruses in the same glycoprotein (related-origin) encoding a hemagglutinin domain that has specificity for the same modified Sia.
- Hemagglutinin (HA)
a viral lectin that binds to Sia (sialolectin), historically characterized by the ability of a virus to agglutinate erythrocytes in vitro. Orthomyxo-, paramyxo-, and nidoviruses encode hemagglutinating glycoproteins either alone or fused to esterase or sialidase domains. Many surface capsid proteins of nonenveloped viruses also have hemagglutination sialolectin function.
- Sialic acid (Sia)
nine-carbon α-keto acidic sugar whose name is derived from its presence in saliva. It was discovered from human samples as primarily N-acetylneuraminic acid (Neu5Ac). It can be enzymatically modified at multiple positions. Sia is typically found at the termini of N- and O-glycans on glycoproteins, as well as components of glycolipids, where they perform important recognition reactions for other cellular factors and/or environmental agents such as pathogens.
- Sialidase
an enzyme that removes the Sia sugar from glycocarbohydrate structures. Endo-sialidases cleave poly-Sia linkages. Sialidases of viruses have been traditionally defined as neuraminidases (NA) for their cleavage of Neu5Ac. Historically also identified as a virus-encoded receptor-destroying enzyme (RDE).
- Tropism
the preferential viral infection of target hosts, tissues, or cells. Tropism is often related to permissiveness resulting from the presence of a specific receptor. Sia-binding virus tropisms may be greatly influenced by the relative distribution of Sia species on a given tissue. The degree of Sia diversity (including modified inhibitors or decoys) in alternative hosts or tissues may influence potential viral emergence.
References
- 1.Corfield A.P. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochem. Biophys. Acta. 2015;1850:236–252. doi: 10.1016/j.bbagen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Deplancke B., Gaskins H.R. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001;73 doi: 10.1093/ajcn/73.6.1131S. 1131S–1141A. [DOI] [PubMed] [Google Scholar]
- 3.Thornton D.J. Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 2008;70:459–486. doi: 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 4.Knowles M.R., Boucher R.C. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin Invest. 2002;109:571–577. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stehle T., Khan Z.M. Rules and exceptions: sialic acid variants and their role in determining viral tropism. J. Virol. 2014;88:7696–7699. doi: 10.1128/JVI.03683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stencel-Baerenwald J.E. The sweet spot: defining virus-sialic acid interactions. Nature Rev. Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varki A. Cold Spring Harbor Laboratory Press; 2009. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 8.Angata T., Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 9.Muhlenhoff M. Polysialic acid: three-dimensional structure, biosynthesis and function. Curr. Opin. Struct. Biol. 1998;8:558–564. doi: 10.1016/s0959-440x(98)80144-9. [DOI] [PubMed] [Google Scholar]
- 10.Varki A. Developmental abnormalities in transgenic mice expressing a sialic acid-specific 9-O-acetylesterase. Cell. 1991;65:65–74. doi: 10.1016/0092-8674(91)90408-q. [DOI] [PubMed] [Google Scholar]
- 11.Bruses J.L., Rutishauser U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie. 2001;83:635–643. doi: 10.1016/s0300-9084(01)01293-7. [DOI] [PubMed] [Google Scholar]
- 12.Kawai H. Mice expressing only monosialoganglioside GM3 exhibit lethal audiogenic seizures. J. Biol. Chem. 2001;276:6885–6888. doi: 10.1074/jbc.C000847200. [DOI] [PubMed] [Google Scholar]
- 13.Wipfler D. Differentially regulated expression of 9-O-acetyl GD3 (CD60b) and 7-O-acetyl-GD3 (CD60c) during differentiation and maturation of human T and B lymphocytes. Glycobiology. 2011;21:1161–1172. doi: 10.1093/glycob/cwr050. [DOI] [PubMed] [Google Scholar]
- 14.Cariappa A. B cell antigen receptor signal strength and peripheral B cell development are regulated by a 9-O-acetyl sialic acid esterase. J. Exp. Med. 2009;206:125–138. doi: 10.1084/jem.20081399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal C. Regulation of O-acetylation of sialic acids by sialate-O-acetyltransferase and sialate-O-acetylesterase activities in childhood acute lymphoblastic leukemia. Glycobiology. 2012;22:70–83. doi: 10.1093/glycob/cwr106. [DOI] [PubMed] [Google Scholar]
- 16.Shi W.X. Sialic acid 9-O-acetylation on murine erythroleukemia cells affects complement activation, binding to I-type lectins, and tissue homing. J. Biol. Chem. 1996;271:31526–31532. doi: 10.1074/jbc.271.49.31526. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y. O-acetylation and de-O-acetylation of sialic acids in human colorectal carcinoma. Eur. J. Biochem. 2004;271:281–290. doi: 10.1046/j.1432-1033.2003.03927.x. [DOI] [PubMed] [Google Scholar]
- 18.Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconjugate J. 2009;26:231–245. doi: 10.1007/s10719-008-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arming S. The human Cas1 protein: a sialic acid-specific O-acetyltransferase? Glycobiology. 2011;21:553–564. doi: 10.1093/glycob/cwq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann A.-M.T.M. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nature Commun. 2015;6:7673. doi: 10.1038/ncomms8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwersen M. Solubilisation and properties of the sialate-4-O-acetyltransferase from guinea pig liver. Biol. Chem. 2003;384:1035–1047. doi: 10.1515/BC.2003.116. [DOI] [PubMed] [Google Scholar]
- 22.Kamerling J.P. Migration of O-acetyl groups in N,O-acetylneuraminic acids. Eur. J. Biochem. 1987;162:601–607. doi: 10.1111/j.1432-1033.1987.tb10681.x. [DOI] [PubMed] [Google Scholar]
- 23.Langereis M.A. The murine coronavirus hemagglutinin-esterase receptor-binding site: a major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012;8:e1002492. doi: 10.1371/journal.ppat.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harduin-Lepers A. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl. Environ. Microbiol. Biotechnol. 2012;94:887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aamelfot M. The in situ distribution of glycoprotein-bound 4-O-Acetylated sialic acids in vertebrates. Glycoconjugate J. 2014;31:327–335. doi: 10.1007/s10719-014-9529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langereis M.A. Complexity and diversity of the mammalian sialome revealed by Nidovirus Virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinninger A. Localisation and distribution of O-acetylated N-acetylneuraminic acids, the endogenous substrates of the hemagglutinin-esterases of murine coronaviruses, in mouse tissue. Glycoconjugate J. 2006;23:73–84. doi: 10.1007/s10719-006-5439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto Y. Further studies on the red cell glycolipids of various breeds of dogs. A possible assumption about the origin of Japanese dogs. J. Biochem. 1984;96:1777–1782. doi: 10.1093/oxfordjournals.jbchem.a135010. [DOI] [PubMed] [Google Scholar]
- 30.Ng P.S. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nature Commun. 2014;5:5750. doi: 10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samraj A.N. A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2015;112:542–547. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springer S.A. Parallel evolution of a self-signal: humans and new world monkeys independently lost the cell surface sugar Neu5Gc. Immunogenetics. 2014;66:671–674. doi: 10.1007/s00251-014-0795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varki N.M., Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab. Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M.J. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biddle F. Properties of horse serum gamma inhibitor. Nature. 1965;207:381–383. doi: 10.1038/207381a0. [DOI] [PubMed] [Google Scholar]
- 36.Pepper D.S. The sialic acids of horse serum with special reference to their virus inhibitory properties. Biochem. Biophys. Acta. 1968;156:317–326. doi: 10.1016/0304-4165(68)90261-4. [DOI] [PubMed] [Google Scholar]
- 37.Rogers G.N. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131:394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 38.Kazi L. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 2005;79:15064–15073. doi: 10.1128/JVI.79.24.15064-15073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X. O-acetylation of sialic acids in N-glycans of Atlantic salmon (Salmo salar) serum is altered by handling stress. PROTEOMICS. 2008;8:2849–2857. doi: 10.1002/pmic.200701093. [DOI] [PubMed] [Google Scholar]
- 40.Martin L.T. Recombinant influenza C hemagglutinin-esterase as a probe for sialic acid 9-O-acetylation. Meth. Enzymol. 2003;363:489–498. doi: 10.1016/S0076-6879(03)01074-7. [DOI] [PubMed] [Google Scholar]
- 41.Vlasak R. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hause B.M. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5:14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song H. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified Influenza D Virus: basis for its broad cell tropism. PLoS Pathog. 2016;12:e1005411. doi: 10.1371/journal.ppat.1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz-Barroso I. Increased influenza A virus sialidase activity with N-acetyl-9-O-acetylneuraminic acid-containing substrates resulting from influenza C virus O-acetylesterase action. Virus Res. 1992;25:145–153. doi: 10.1016/0168-1702(92)90106-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byres E. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–652. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campanero-Rhodes M.A. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J. Virol. 2007;81:12846–12858. doi: 10.1128/JVI.01311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neu U. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Hara S.D. Glycan receptors of the Polyomaviridae: structure, function, and pathogenesis. Curr. Opin. Virol. 2014;7:73–78. doi: 10.1016/j.coviro.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Yu X. Structural basis of rotavirus strain preference toward N-Acetyl- or N-glycolylneuraminic acid-containing receptors. J. Virol. 2012;86:13456–13466. doi: 10.1128/JVI.06975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banda K. ‘Sialidase sensitivity’ of rotaviruses revisited. Nature Chem. Biol. 2009;5:71–72. doi: 10.1038/nchembio0209-71. [DOI] [PubMed] [Google Scholar]
- 51.Blanchard H. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*) J. Mol. Biol. 2007;367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 52.Palese P. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 53.Wagner R. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 54.Mitnaul L.J. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000;74:6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corfield A.P. The specificity of viral and bacterial sialidases for alpha(2-3)- and alpha(2-6)-linked sialic acids in glycoproteins. Biochem. Biophys. Acta. 1983;744:121–126. doi: 10.1016/0167-4838(83)90080-8. [DOI] [PubMed] [Google Scholar]
- 56.Corfield A.P. The action of sialidases on substrates containing O-acetylsialic acids. Biol. Chem. Hoppe-Seyler. 1986;367:433–439. doi: 10.1515/bchm3.1986.367.1.433. [DOI] [PubMed] [Google Scholar]
- 57.Xu G. Sialidase of swine influenza A viruses: variation of the recognition specificities for sialyl linkages and for the molecular species of sialic acid with the year of isolation. Glycoconjugate J. 1995;12:156–161. doi: 10.1007/BF00731360. [DOI] [PubMed] [Google Scholar]
- 58.Matrosovich M. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J. Virol. 1998;72:6373–6380. doi: 10.1128/jvi.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kryazhimskiy S. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet. 2011;7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neverov A.D. Coordinated evolution of influenza a surface proteins. PLoS Genet. 2015;11:e1005404. doi: 10.1371/journal.pgen.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poehlmann S., Simmons G. Springer-Verlag; 2013. Viral Entry Into Host Cells. [Google Scholar]
- 62.Porotto M. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J. Virol. 2012;86:5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iorio R.M. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 2001;75:1918–1927. doi: 10.1128/JVI.75.4.1918-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porotto M. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J. Virol. 2006;80:1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Streltsov V.A. Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN) Antiviral Res. 2015;123:216–223. doi: 10.1016/j.antiviral.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Gorbalenya A.E. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Groot R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconjugate J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smits S.L. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng Q. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popova R., Zhang X. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology. 2002;294:222–236. doi: 10.1006/viro.2001.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desforges M. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helander A. The viral sigma1 protein and glycoconjugates containing alpha2-3-linked sialic acid are involved in type 1 reovirus adherence to M cell apical surfaces. J. Virol. 2003;77:7964–7977. doi: 10.1128/JVI.77.14.7964-7977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haselhorst T. Sialic acid dependence in rotavirus host cell invasion. Nature Chem. Biol. 2009;5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X. Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6292–6297. doi: 10.1073/pnas.0913403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y. Sialic acid-dependent cell entry of human enterovirus D68. Nature Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nokhbeh M.R. Enterovirus 70 binds to different glycoconjugates containing alpha2,3-linked sialic acid on different cell lines. J. Virol. 2005;79:7087–7094. doi: 10.1128/JVI.79.11.7087-7094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zocher G. A sialic acid binding site in a human picornavirus. PLoS Pathog. 2014;10:e1004401. doi: 10.1371/journal.ppat.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lipton H.L. Differential usage of carbohydrate co-receptors influences cellular tropism of Theiler's murine encephalomyelitis virus infection of the central nervous system. Glycoconjugate J. 2006;23:39–49. doi: 10.1007/s10719-006-5436-x. [DOI] [PubMed] [Google Scholar]
- 79.Xiong X. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr. Top. Microbiol. Immunol. 2014;385:63–91. doi: 10.1007/82_2014_423. [DOI] [PubMed] [Google Scholar]
- 80.Velkov T. The specificity of the influenza B virus hemagglutinin receptor binding pocket: what does it bind to? J. Mol. Recognit. 2013;26:439–449. doi: 10.1002/jmr.2293. [DOI] [PubMed] [Google Scholar]
- 81.Rogers G.N. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 82.Santos-Lopez G. Structure-function analysis of two variants of mumps virus hemagglutinin-neuraminidase protein. Braz. J. Infect. Dis. 2009;13:24–34. doi: 10.1590/s1413-86702009000100007. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki T. Receptor specificities of human respiroviruses. J. Virol. 2001;75:4604–4613. doi: 10.1128/JVI.75.10.4604-4613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaitsev V. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson F.B. Attachment of bovine parvovirus to sialic acids on bovine cell membranes. J. Gen. Virol. 2004;85:2199–2207. doi: 10.1099/vir.0.79899-0. [DOI] [PubMed] [Google Scholar]
- 86.Nam H.-J.J. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 2006;281:25670–25677. doi: 10.1074/jbc.M604421200. [DOI] [PubMed] [Google Scholar]
- 87.Pudlo N.A. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. MBio. 2015;6 doi: 10.1128/mBio.01282-15. e01282-01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vimr E.R. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phansopa C. Characterization of a sialate-O-acetyl esterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH its cognate sialidase. Biochem. J. 2015;472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 90.Severi E. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 91.Vimr E., Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10:254–257. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 92.Gulati S. Utilizing CMP-sialic acid analogs to unravel Neisseria gonorrhoeae lipooligosaccharide-mediated complement resistance and design novel therapeutics. PLoS Pathog. 2015;11:e1005290. doi: 10.1371/journal.ppat.1005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song J. Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature. 2013;499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bosch A.A. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nature Rev. Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 96.Rynda-Apple A. Influenza and bacterial superinfection: illuminating the immunologic mechanisms of disease. Infect. Immun. 2015;83:3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nita-Lazar M. Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectin binding. Mol. Immunol. 2015;65:1–16. doi: 10.1016/j.molimm.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siegel S.J. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe. 2014;16:55–67. doi: 10.1016/j.chom.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lehmann F. Sialic acid-specific lectins: occurrence, specificity and function. Cell. Mol. Life Sci. 2006;63:1331–1354. doi: 10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]