Abstract
How differentiation between cell types evolved is a fundamental question in biology, but few studies have explored single-gene phenotypes that mediate first steps towards division of labour with selective advantage for groups of cells. Here, we show that differential expression of the FLO11 gene produces stable fractions of Flo11+ and Flo11− cells in clonal Saccharomyces cerevisiae biofilm colonies on medium with intermediate viscosity. Differentiated Flo11+/− colonies, consisting of adhesive and non-adhesive cells, obtain a fourfold growth advantage over undifferentiated colonies by overgrowing glucose resources before depleting them, rather than depleting them while they grow as undifferentiated Flo11− colonies do. Flo11+/− colonies maintain their structure and differentiated state by switching non-adhesive cells to adhesive cells with predictable probability. Mixtures of Flo11+ and Flo11− cells from mutant strains that are unable to use this epigenetic switch mechanism produced neither integrated colonies nor growth advantages, so the condition-dependent selective advantages of differentiated FLO11 expression can only be reaped by clone-mate cells. Our results show that selection for cell differentiation in clonal eukaryotes can evolve before the establishment of obligate undifferentiated multicellularity, and without necessarily leading to more advanced organizational complexity.
Keywords: differentiation, multicellularity, division of labour, cooperation
1. Introduction
Multicellularity has evolved many times, both in the prokaryotes and the eukaryotes, but most lineages have not progressed beyond the facultative expression of multicellular phenotypes with no or very limited differentiation of cell types [1,2]. Examples are unicellular Myxococcus bacteria and Dictyostelium slime moulds that form non-clonal aggregations upon starvation to cooperate for spore dispersal [3,4]. This involves differentiation between spore-forming cells and altruistic sacrifices by non-reproducing cells [5,6], but without affecting that unicellularity remains the standard life form. Reproductive altruism is favoured by high relatedness between donor and recipient cells, which is best secured in clonal aggregations, but cell-type differentiation remained very limited in cyanobacteria [7] and volvocine algae [8,9] in spite of clonality being secured. This suggests that synergistic, fitness-enhancing differentiation benefits normally evolve with clonal multicellularity or after it is established [8,10–12] rather than being already present as preadaptation in unicellular progenitors; in other words, cells must first stick to each other in undifferentiated form before they can reap any selective benefits of dividing labour between differentiated cell types [1,13].
Comparative data reconstructions of evolutionary transitions in multicellularity and division of labour among cell types usually rely on sister-lineage comparisons [2,7,8,14,15] between extant clades where entire life-history syndromes have been modified. This implies that the multicellular phenotypes that are compared often represent secondary elaborations rather than very first origins of multicellularity [13,16], precluding formal tests of whether multicellularity always became obligate before cell differentiation evolved. Studying the selective benefits of genetic mutations that initiate multicellularity is therefore most feasible in lineages where evolution does not progress to more advanced stages. Saccharomyces cerevisiae baker's yeast is a eukaryotic microorganism that has these characteristics as it can switch between different unicellular and multicellular growth forms via the expression and regulation of flocculin (FLO) genes [17–23]. Flocculation phenotypes normally involve adhesion of undifferentiated cells in response to resource limitation [22], when clumping may provide protection against toxic ethanol challenges [19], but FLO11 has a gene-expression polymorphism with the potential to induce cell differentiation benefits. This FLO gene is only distantly related to other cell adhesion genes, such as FLO1, FLO5, FLO9 and FLO10 [17,24,25], and has one of the most complex promotor regions in the genome of S. cerevisiae [22], encoding a Flo11p cell-wall glycoprotein that is essential for development of surface spreading biofilm phenotypes [26]. These biofilms can both be haploid or diploid [26], but their selective advantages have not been evaluated as possible examples of incipient multicellularity driven by cell differentiation benefits without prior adhesion as undifferentiated cells.
For an incipient facultative differentiation trait in a clonal unicellular microorganism to be maintained by natural selection, its expression would need to (i) depend on specific and thus predictable habitat (medium) conditions, (ii) offer unambiguous growth benefits under these conditions and (iii) be immune to invasion by unrelated cheater cells whose chimeric exploitation of enhanced colony growth rates would annihilate these fitness gains. FLO11 expression has several aspects suggesting that these conditions might be fulfiled. First, the FLO11 gene is periodically turned on and off by two oppositely acting transcription factors Slf1p and Flo8p [27–29], a stochastic expression switch that differentiates cells and creates mixtures of clonal Flo11+ and Flo11− cells in several growth forms of S. cerevisiae [18,21,22,29]. Second, the FLO11 gene becomes expressed only in specific environments such as low glucose medium, so that any multicellular phenotype initiated by this gene is likely to be predictably condition dependent [30]. Third, protein-level (Flo11p-Flo11p) adhesion interactions are strongly homophilic, providing a very direct ligand-to-ligand self-recognition system [25], which might better preserve clonal integrity in growing colonies than the heterophilic adhesion mechanisms via oligo/polysaccharides on the surface of neighbouring cells that other Flo proteins have [22].
2. Material and methods
(a). Strains
The S. cerevisiae Σ1278b YS-11 (MATa can1Δ::STE2p-SpHIS5 lyp1Δ::STE3p-LEU2 his3::HisG leu2Δ ura3Δ) was used as wild-type strain, and our flo8 strain was a flo8::KanMX deletion mutant in the Σ1278b wild-type background, whereas our sfl1 strain was a sfl1::KanMX deletion mutant in the Σ1278b wild-type background. All three strains were described in detail previously [20]. A green fluorescent protein (GFP)-labelled wild-type strain was constructed by inserting a PTEF1-GFP at chromosome IX into Σ1278b (provided by Rasmus K. Bojsen). The natural isolates YJM269, CLIB219, 114, M22, T73, UC8, 273614X, WE372, Y9 J, Y55, YJM978, YJM981, T7, NC_02, IL_01, UC1, YPS1009, Y3, Y9, Y10, Y12, CLIB413, CLIB294, YPS163, YPS1000, EM93, K12, DBVPG6861, DBVPG4651, DBVPG3591, DBVPG1794, DBVPG1788, CECT10109, CBS7960, YJM678, YJM653, YJM454, YJM440, YJM436, YJM434, YJM428, YJM421, YJM413, YJM326, YJM320, YJM280, YJM145, CLIB326 and CLIB192 were provided by Joseph Schacherer.
(b). Surface spreading biofilms and their biomass
Biofilm colonies on yeast extract peptone dextrose (YPD) medium were made with 0.3% agar as described previously [31] unless specified otherwise. Colonies were inoculated with 500 cells and grown at room temperature for 7 days (or as indicated). Mixtures of Flo11− (flo8) and Flo11+ (sfl1) mutants were made by placing 500 cells on the centre of semisolid complex medium plates in different inoculation proportions, and colony biomass (g dry weight; DW) was subsequently measured across YPD media of different viscosity (% w/v agar) (see electronic supplementary material, figure S2 for representative images). Details were as in [32] except that biofilm colonies were soaked in water, loosened with a Drigalski spatula and transferred to filters by pipette. All experiments were conducted in triplicate.
(c). RNA FISH
RNA FISH was conducted as described in [21] except that RNA FISH from 7-day-old biofilm colonies was made by soaking and washing plates with the same fixation medium and continuing fixation at 4°C as described in [21]. ACT1 served as a positive control and only ACT1 mRNA-positive cells were investigated for the amount of FLO11 mRNA using images similar to those in figure 2c that were blinded to avoid observer bias. Error bars are s.d. based on three replicates.
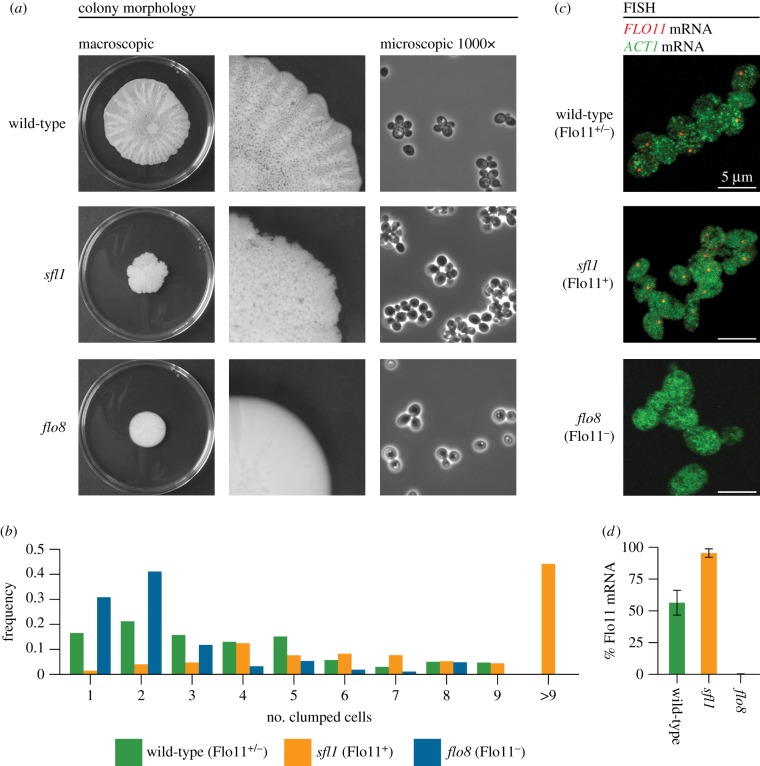
Figure 2.
Differentiation of S. cerevisiae biofilms in Flo11+ and Flo11− cells. (a) Colony morphology at different magnifications of the genetically tractable wild-type strain Σ1278b, and isogenic sfl1 and flo8 mutant strains. (b) The average cell cluster sizes of wild-type (n = 1350), sfl1 (n = 1844) and flo8 (n = 1490) colonies. (c) FLO11 transcripts visualized after RNA FISH for wild-type Flo11+/−, and sfl1 and flo8 strains, hybridized with FLO11 (red) and ACT1 (green) probes. (d) Percentage of cells expressing FLO11 mRNA in wild-type (n = 756), sfl1 (n = 574) and flo8 (n = 509) colonies.
(d). Glucose measurements
Glucose measurements were conducted on 5 µl medium collected right below the rim of biofilm colonies when these had reached a diameter of 3.5 cm on YPD, 0.3% agar. The medium was diluted in 45 µl H2O and heated for 2 min at 90°C to kill cells after which the glucose concentrations were measured with a Contour Blood glucose meter (Bayer). Error bars are s.d. based on three replicates.
(e). Switching rate from non-adhesive to adhesive cells
Thirty-four non-adhesive mother cells were isolated by collecting solitary cells from the rim of a wild-type colony and placing them on YPD plates with a dissection needle. Cells were allowed to divide, after which daughter cells were removed from the mother cell with a micromanipulator at every cell division. The cell divisions in which a mother cell could be separated from her daughter cell were counted as non-adhesive divisions, and when the daughter cell could no longer be removed from the mother cell, we inferred that a switch to an adhesive cell had taken place.
(f). Images
Pictures of colonies were taken with a Canon EOS 1100D camera and microscope images were obtained with a Nikon Eclipse E600 microscope mounted with an Optronics camera at 400× magnification.
3. Results
(a). Saccharomyces cerevisiae biofilm colonies are competitively superior and differentiated in two cell types
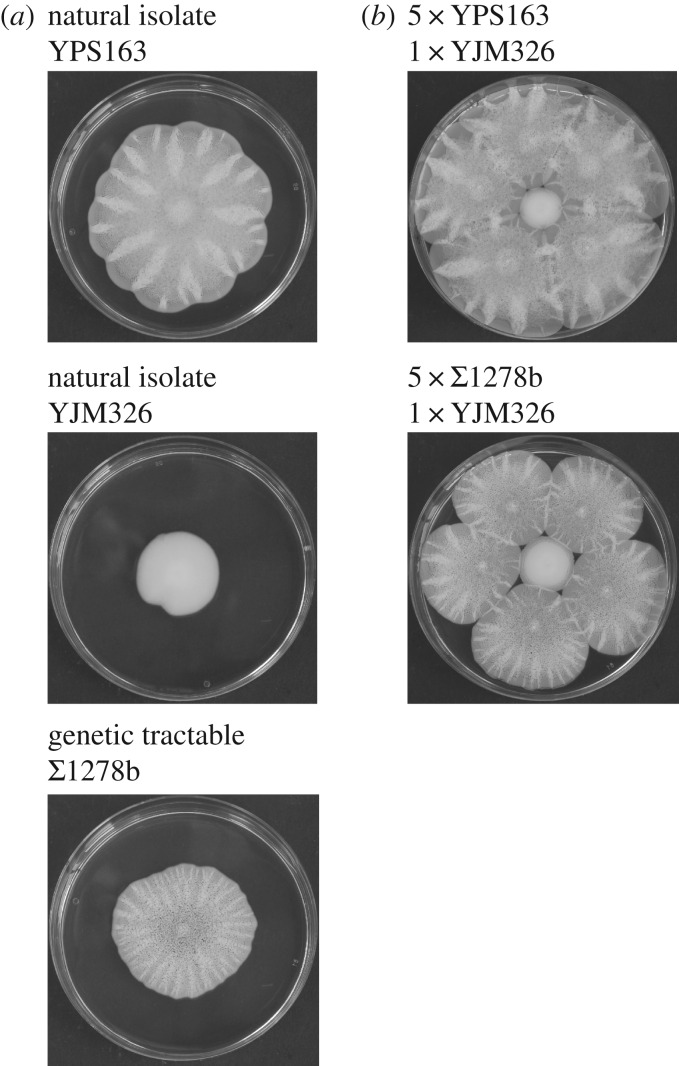
We first tested whether natural isolates of S. cerevisiae formed surface spreading colonies similar to the biofilms of the genetically tractable haploid isolate Σ1278b in which FLO11 is the only expressed FLO gene [17]. Six out of 49 natural isolates formed large morphologically structured biofilms (electronic supplementary material, figure S1) while 41 isolates formed smaller smooth colonies and two grew smaller rough colonies (electronic supplementary material, figure S1). Σ1278b biofilms thus appeared to be representative of natural biofilms (e.g. YPS163; figure 1a) and to grow larger than natural non-biofilm-forming colonies such as the clinical isolate YJM326. Large biofilm colonies were also competitively superior when we grew Σ1278b and YPS163 together with the non-biofilm-forming isolate YJM326 (figure 1b), as both biofilm-forming isolates inhibited the growth of smooth non-biofilm-forming YJM326 colonies.
Figure 1.
Growth phenotypes and competition between biofilm-forming and non-biofilm-forming isolates of Saccharomyces cerevisiae. (a) Morphology of the three isolates used in our study: the biofilm-forming YPS163 isolate from soil, the non-biofilm-forming clinical isolate YJM326, and the biofilm-forming genetically tractable isolate Σ1278b. (b) Competition experiments between non-biofilm-forming YJM326 (centre) and biofilm-forming YPS163 and Σ1278b, showing that biofilm-forming strains constrain the growth of a non-biofilm strain. All colonies were initiated on 0.3% agar from 500 cells, and growth was recorded after 7 days at room temperature (22°C–25°C). Pictures are representative for three independent experiments.
More than 60% of cells in the growing rim of biofilm colonies formed small clumps of three or more yeast cells while the remaining 38% were single yeast cells or cells in the process of dividing (figure 2a,b). To test whether differentiation between cells expressing FLO11 mRNA (Flo11+) or not (Flo11−) affected colony size and morphology, we compared these biofilm colonies with two recessive undifferentiated biofilm mutants, sfl1 and flo8, that are known to have very high proportions of Flo11+ and Flo11− cells, respectively, when grown in liquid medium [29]. We used RNA FISH to investigate whether differential FLO11 expression could be responsible for the mixture of adhesive and non-adhesive cells, which showed that FLO11 expression was restricted to a similar subset (59%) of the biofilm-forming cells (figure 2c,d). Differential expression of FLO11 thus appears to explain why some cells formed small clumps whereas others remain free living.
The sfl1 mutant, depleted of the FLO11 repressor, formed small wrinkled colonies (figure 2a) with FLO11 mRNA being expressed in 98% of the cells, confirming highly adhesive Flo11+ phenotypes (figure 2c,d). The flo8 mutant, depleted of the Flo8p transcriptional activator of FLO11, formed small smooth colonies (figure 2a) that did not produce FLO11 mRNA, similar to the smooth unstructured colony morphology of the flo11 mutant (electronic supplementary material, figure S2). Colonies of flo8 were thus 100% Flo11− (figure 2c,d) and had much higher proportions of free-living cells than wild-type colonies (figure 2b). Median cell cluster sizes were five (wild-type), seven (sfl1, Flo11+) and two (flo8, Flo11−), differences that were statistically significant (H = 341.14; d.f. = 2; Crit-H0.05 = 5.99) and positively correlated with FLO11 mRNA expression levels (figure 2c,d) (H = 7.26; d.f. = 2; Crit-H0.05 = 5.99).
(b). Cooperation in differentiated clonal biofilm colonies at intermediate medium viscosity
Biomass in differentiated Flo11+/− wild-type colonies was up to four times higher than the biomass of separate Flo11+ (sfl1) and Flo11− (flo8) colonies (figure 3a), and these growth advantages were restricted to intermediate medium viscosities (0.25–1.0% agar; H = 7.54; d.f. = 2; Crit-H(0.05) = 5.99 for 0.25% agar; figure 3a; electronic supplementary material, figure S3). Differentiated colonies with both Flo11+ and Flo11− cells thus obtain condition-dependent growth benefits by some form of synergistic cell-type division of labour compared with undifferentiated colonies that were purely Flo11+ or Flo11−. Validation of this result in liquid medium (0% agar) and on high-viscosity medium (2% agar) confirmed that Flo11+/− differentiation does not affect clonal biomass under these growth conditions, and also showed that the sfl1 and flo8 mutants are not generally compromised in their growth (H = 2.66 (0% agar) and 2.11 (2% agar); d.f. = 2; Crit-H(0.05)= 5.99; figure 3a; electronic supplementary material, figure S3). Flo11+/− induced biofilm formation thus appears to represent a lifestyle to rapidly colonize favourable habitat patches on semisolid substrate.
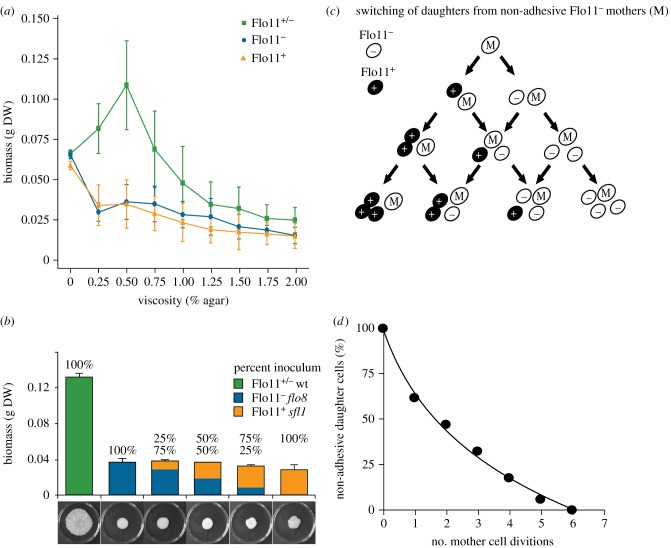
Figure 3.
Condition-dependent growth advantage of biofilm colonies relies on cell-type switching. (a) Dry weight biomass (g DW) of wild-type Flo11+/− (green), sfl1 (orange, Flo11+) and flo8 (blue, Flo11−) colonies ± s.d. on medium of different viscosity. (b) Biomass of differentiated wild-type (Flo11+/−) clones and mixtures of mutants that are either flo8 (Flo11−) or sfl1 (Flo11+). (c) The expected trajectory of Flo11− mother cells (M) in a wild-type colony, assuming that Flo11+ (black) and Flo11− (white) can either produce daughter cells of their own type or daughter cells of the opposite type so that a mixed population of Flo11+/− cells arises de novo. The observed trajectories of non-adhesive Flo11− mother cells producing non-adhering Flo11− daughter cells are plotted in (d), showing that Flo11− daughter cell production exponentially decays with a half-life of 1.8 cell divisions. Flo11− mother cells were recorded by microdissection until they produced an adhesive daughter cell.
Growth rate benefits at intermediate viscosity are consistent with social synergies being most likely to arise under intermediate spatial structure [33,34]. Yeast cells in low-viscosity liquid medium are less likely to interact with cells of their own clone so differentiation will be selected against when non-relatives benefit, whereas high-viscosity medium will maintain clonal population structure but physically preclude differentiated clones from growing faster. However, at intermediate medium viscosity (figure 3a) cheater clones that fail to invest in Flo11p proteins could invade fast-growing Flo11+/− biofilms because both adhesive and non-adhesive cells were found in the rim (figure 2a,b). We therefore tested whether non-clonal mixtures of Flo11+ (sfl1) and Flo11− (flo8) cells could produce the structured biofilm phenotype with the ensuing higher biomass and found that this was not the case (figure 3b). Such combined sfl1–flo8 colonies always formed a central structured hub that appeared to be composed of adhesive Flo11+ cells surrounded by a smooth zone of non-adhesive Flo11− cells (electronic supplementary material, figure S4), consistent with Flo11+ cells not offering resource acquisition advantages to unrelated Flo11− cells. This lack of cooperation between Flo11+ (sfl1) and Flo11− (flo8) cells underlines that clonality is essential for the expression of synergistic division of labour in differentiated Flo11+/− biofilms and appears to exclude any green beard explanations for the cooperative Flo11+/− biofilms [19].
(c). Cell-type switching ensures stable mixtures that monopolize local resources
Results so far indicated that synergistic growth benefits in Flo11+/− colonies need to be generated via differential gene expression within clones and that Flo11− cells in wild-type colonies generate Flo11+ cells de novo in the actively growing periphery to ensure a stable beneficial mixture of the two cell types. To simulate this process, we sampled non-adhesive peripheral cells of wild-type colonies, dissecting them from their daughter cells, and followed their trajectory (figure 3c,d). This showed that non-adhesive cells produced only 1.8 non-adhesive daughters on average before they generated an adhesive daughter cell (figure 3d), supporting previous data for epigenetic switching of the FLO11 promoter in conjunction with the reporter gene YFP [28]. Stochastic switching thus allows biofilm clones to maintain stable proportions of adhesive and non-adhesive cells in the growing rim to reap differentiation benefits without risking invasion by free-riding Flo11− cells from other clones that would require costly kin-discrimination mechanisms to eliminate.
To resolve how clonal Flo11+/− colonies obtain a threefold to fourfold increase in growth rate, we reared a series of them in competition with same-phenotype colonies. This confirmed the growth rate advantages of Flo11+/− colonies (figure 1): differentiated Flo11+/− biofilms covered plates almost entirely within a week, whereas undifferentiated Flo11+ and Flo11− colonies never reached each other and never overgrew entire plates (figure 4a), suggesting that they access nutrients in the medium in a fundamentally different way. Fast-growing Flo11+/− biofilms created contact zones where neighbouring colonies overgrew each other, but apparently without merging (figure 4b).
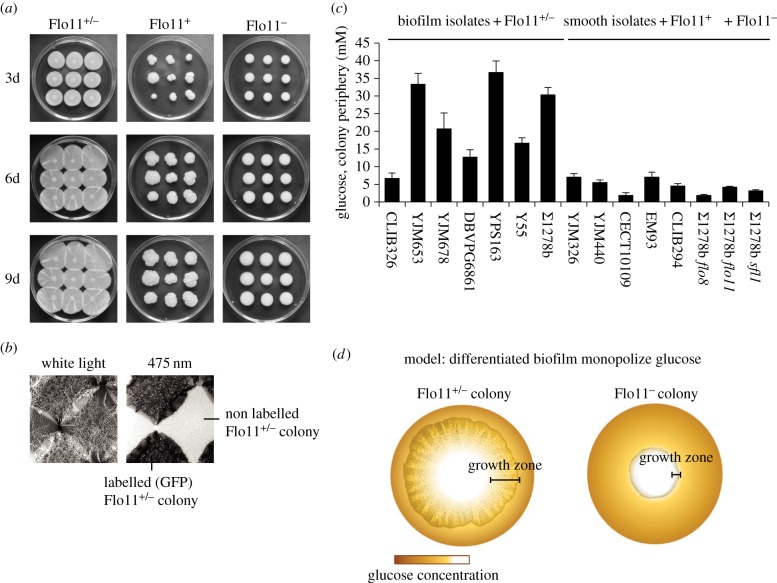
Figure 4.
Differentiated biofilm colonies monopolize local resources before exhausting them. (a) Growth of wild-type Σ1278b, sfl1 and flo8 colonies after 3, 6 and 9 days. (b) Contact zones between four Flo11+/− colonies of which two expressed green fluorescent proteins (GFP) (white light, left; 475 nm light, right). (c) Glucose concentrations (means ± s.d.) below the rim of natural biofilm-forming isolates (CLIB326, YJM653, YJM678, DBVPG6861, YPS163, Y55) and Σ1278b compared with smooth (Flo11−) colonies (YJM326, YJM440, CECT10109, EM93, CLIB294, flo8, flo11) and the sfl1 (Flo11+) mutant. (d) Schematic diagram reconstructing how differentiated biofilm colonies monopolize glucose resources. High glucose concentration is shown in dark brown and low glucose concentration in white. Scale bars indicated the growing edge of a differentiated biofilm colony (left) and an undifferentiated smooth colony (right).
Further measurements showed that glucose was almost depleted in the periphery of undifferentiated Flo11− colonies and natural smooth isolate colonies (mean 4.5 ± 2.2 mM glucose compared with the original 111 mM glucose; n = 7), but that glucose concentrations were up to 15 times higher (mean 33.2 ± 11.0 mM; n = 7) in the periphery of differentiated Flo11+/− colonies and similar natural biofilm isolates (figure 4c; ANOVA: F(14,35) = 68.6; p < 0.0001). Flo11+/− biofilms thus appear to monopolize territories on agar plates before depleting them, whereas smooth Flo11− colonies use glucose in direct proportion to immediate availability as yeast colonies normally do [35] (figure 4c,d). This unusual growth pattern offers consistent opportunities for enhanced cell division within Flo11+/− biofilms relative to what can be achieved by Flo11− colonies (figure 4d). The epigenetic FLO11 expression switch thus enables clonal biofilms to practise a form of pre-emptive contest competition by optimizing the frequency of adhesive and non-adhesive cell production. Under specific growth conditions, this must provide significant fitness advantages relative to the scramble competition strategies that are practised by unicellular or adhesive growth forms that are unable to produce Flo11+ and Flo11− cells simultaneously.
4. Discussion
Our results show that clonal but differentiated Flo11+/− biofilm phenotypes fulfil the three conditions that should apply for a latent synergistic division of labour trait to be evolutionary stable: condition-dependent expression at low glucose concentration, unambiguous fitness benefits but only on medium of intermediate viscosity, and robustness against exploitation because undifferentiated strains that do not invest in the simultaneous production of both cell types cannot invade. Intermediate medium viscosity as a condition for realizing cooperative growth benefits (figure 3a) may be related to solid medium precluding niche pre-emption via fast surface growth and liquid medium having too much clonal mixing to favour Flo11+/− phenotypes. The mechanics by which the complementary cell types realize synergistic growth are unknown, but it would seem likely that Flo11+ cells form a kind of scaffold for the subsequent spread of clone-mate Flo11− cells while generating the structured appearance of biofilm colonies. The stochastic gene-expression switch in clonal Flo11+/− biofilms [18,29] may thus function as a feedback system to maximize growth rate through somatic differentiation, but without realizing true multicellularity as biofilms remain fragmented mixtures of adhesive and non-adhesive cells.
A central question is whether the low glucose concentration and intermediate viscosity conditions that might select for differentiated biofilm colonies and allow for preemptive contest competition exist in natural habitats. We believe this is likely to be the case. Saccharomyces cerevisiae is the dominant yeast in many spontaneous fermentation processes of domesticated and natural stands of grape, cacao and other types of fruits [36,37], and is known to be dispersed through insect vectors [38]. Thus, it seems likely that the semisolid surface of natural fruit pulp provides the advantageous environment and that insect vectors will inoculate clonal biofilm colonies soon after such patches reach the appropriate stage of decay. However, quantitative field studies of S. cerevisiae biofilm are lacking, so it is at present impossible to evaluate the extent to which reproductive fitness of differentiated FLO11 gene expression is realized in natural environments.
The ability of Flo11+/− S. cerevisiae biofilms to maintain clonality in a highly efficient homophilic way (figure 1) may have been crucial for the evolution and maintenance of differentiation without proper multicellularity. This is consistent with a large comparative study of obligate and facultative multicellular organisms revealing that obligate multicellular lineages are all clonal and that the highest level of organizational complexity, measured as the number of different cell types, is also found in clonal lineages [2]. Theoretical studies also predict that clonality is a key condition for the evolution of obligate multicellularity, because separation between germline and sterile somatic cells required clonality and reduced the mutation load from selfishly over-replicating mutant cell lines (cancer) in complex multicellular organisms such as the bilaterian Metazoa [1,16]. The importance of clonality has not been directly tested in the present experiments, but its importance can be inferred. Given the cohesion of the FLO11 biofilms and our consistently negative results when attempting to create differentiated biofilms from non-clonal FLO11 genotypes (figure 3), it must be true that differentiated biofilms maintain clonality. Even biofilm cultures of the same clonal stock that were inoculated separately overgrew each other rather than becoming mixed (figure 4b), suggesting that recognition mechanisms are present that even exclude subtle somatic mutations to be secondarily mixed.
It is important to realize that multicellularity in Flo11+/− S. cerevisiae biofilms differs fundamentally from several other model systems that have been used to study facultative multicellularity. First, the biofilms of baker's yeast differ from the multicellular structures in Dictyostelium discoideum slime moulds [3,5] and Myxococcus xanthus bacteria [6] in being clonal rather than arising via cell aggregation, making the latter much more vulnerable to chimeric invasion by free-riding cells. Second, S. cerevisiae biofilms differ fundamentally from the undifferentiated S. cerevisiae aggregates known as flocs that can form between both related and non-related yeast cells [17,19]. Flocculation protects S. cerevisiae cells from environmental stress [19] but does not provide a novel and potentially synergistic growth form as the FLO11 biofilm phenotype does. Flocculation relies on the production of Flo proteins with a PA4 domain, such as Flo1p, Flo5p, Flo9p and Flo10p that form Ca2+-dependent heterophilic interactions with mannose residues on the cell walls to adhere to neighbouring cells [24], whereas the biofilm phenotype of our present study relies on homophilic protein (Flo11p–Flo11p) interactions [25].
Finally, the organization of S. cerevisiae biofilms is also clearly different from any form of filamentous multicellularity such as found in Streptomyces [39], where clonal cell differentiation has evolved to enhance spore dispersal rather than resource acquisition. Yet any form of more advanced multicellularity is lacking in Saccharomyces, confirming that clonal integrity is a necessary but not sufficient condition for establishing obligate multicellularity [2]. We suspect that the condition dependence (intermediate viscosity media only) of synergistic growth advantages has constrained S. cerevisiae to remain unicellular by default, because its mode of dispersal and the media available throughout its natural niche would not select for irreversibly multicellular phenotypes. Overall, the incipient cell-type division of labour characteristics that our present study discovered underline that Flo11+/− biofilm is a highly suitable model for evaluating evolutionary trade-offs that likely applied to most single-gene very first steps towards differentiated multicellularity. Other recent experimental evolution studies of S. cerevisiae have also considerably advanced our understanding of the selection pressures and proximate mechanisms that can make differentiated multicellularity evolve [40]. The addition of Flo11+/− biofilms to this spectrum appears to further enhance the status of S. cerevisiae as a genetic and phenotypic model system for incipient multicellularity that recent authors of major evolutionary transitions reviews have found wanting [12,13,16].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Sefa Alizadeh and Annika N. Jeppe for technical assistance, Roberta Fisher, Hans-Ulrich Mösch, David Nash and Stuart West for comments and discussion, and Rasmus K. Bojsen and Joseph Schacherer for kindly providing a number of strains.
Authors' contributions
K.S.A., K.E.H and B.R. performed experiments and analysed data; B.R. and J.J.B designed the study and wrote the paper. All authors discussed the results and commended on the manuscript.
Data accessibility
All supplementary data are presented in the electronic supplementary material for this article.
Competing interests
The authors declare no competing interest.
Funding
We were supported by grants from the Faculty of Science, University of Copenhagen (B.R.) and the Danish National Research Foundation (DNRF57) (J.J.B.).
References
- 1.Grosberg RK, Strathmann RR. 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654. ( 10.1146/annurev.ecolsys.36.102403.114735) [DOI] [Google Scholar]
- 2.Fisher RM, Cornwallis CK, West SA. 2013. Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. 23, 1120–1125. ( 10.1016/j.cub.2013.05.004) [DOI] [PubMed] [Google Scholar]
- 3.Strassmann JE, Queller DC. 2011. Evolution of cooperation and control of cheating in a social microbe. Proc. Natl Acad. Sci. USA 108(Suppl 2), 10 855–10 862. ( 10.1073/pnas.1102451108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaiser D. 2008. Myxococcus: from single-cell polarity to complex multicellular patterns. Annu. Rev. Genet. 42, 109–130. ( 10.1146/annurev.genet.42.110807.091615) [DOI] [PubMed] [Google Scholar]
- 5.Foster KR, Fortunato A, Strassmann JE, Queller DC. 2002. The costs and benefits of being a chimera. Proc. R. Soc. Lond. B 269, 2357–2362. ( 10.1098/rspb.2002.2163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velicer GJ, Kroos L, Lenski RE. 2000. Developmental cheating in the social bacterium Myxococcus xanthus. Nature 404, 598–601. ( 10.1038/35007066) [DOI] [PubMed] [Google Scholar]
- 7.Schirrmeister B, Antonelli A, Bagheri H. 2011. The origin of multicellularity in cyanobacteria. BMC Evol. Biol. 11, 45 ( 10.1186/1471-2148-11-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herron MD, Michod RE. 2008. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin's eye. Evolution 62, 436–451. ( 10.1111/j.1558-5646.2007.00304.x) [DOI] [PubMed] [Google Scholar]
- 9.Kirk DL. 2005. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 27, 299–310. ( 10.1002/bies.20197) [DOI] [PubMed] [Google Scholar]
- 10.Rossetti V, Schirrmeister BE, Bernasconi MV, Bagheri HC. 2010. The evolutionary path to terminal differentiation and division of labor in cyanobacteria. J. Theor. Biol. 262, 23–34. ( 10.1016/j.jtbi.2009.09.009) [DOI] [PubMed] [Google Scholar]
- 11.Koschwanez JH, Foster KR, Murray AW. 2013. Improved use of a public good selects for the evolution of undifferentiated multicellularity. Elife 2, e00367 ( 10.7554/eLife.00367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West SA, Fisher RM, Gardner A, Kiers ET. 2015. Major evolutionary transitions in individuality. Proc. Natl Acad. Sci. USA 112, 10 112–10 119. ( 10.1073/pnas.1421402112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke E. 2014. Origins of evolutionary transitions. J. Biosci. 39, 303–317. ( 10.1007/s12038-013-9375-y) [DOI] [PubMed] [Google Scholar]
- 14.Bell G, Mooers AO. 1997. Size and complexity among multicellular organisms. Biol. J. Linn. Soc. 60, 345–363. ( 10.1111/j.1095-8312.1997.tb01500.x) [DOI] [Google Scholar]
- 15.Bonner JT. 2003. On the origin of differentiation. J. Biosci. 28, 523–528. ( 10.1007/BF02705126) [DOI] [PubMed] [Google Scholar]
- 16.Bourke AFG. 2011. Principles of social evolution. New York, NY: Oxford University Press. [Google Scholar]
- 17.Guo B, Styles CA, Feng Q, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl Acad. Sci. USA 97, 12 158–12 163. ( 10.1073/pnas.220420397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halme A, Bumgarner S, Styles C, Fink GR. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116, 405–415. ( 10.1016/S0092-8674(04)00118-7) [DOI] [PubMed] [Google Scholar]
- 19.Smukalla S, et al. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 726–737. ( 10.1016/j.cell.2008.09.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan O, et al. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337, 1353–1356. ( 10.1126/science.1224339) [DOI] [PubMed] [Google Scholar]
- 21.Andersen KS, Bojsen R, Sørensen LGRR, Nielsen MW, Lisby M, Folkesson A, Regenberg B. 2014. Genetic basis for Saccharomyces cerevisiae biofilm in liquid medium. G3 4, 1671–1680. ( 10.1534/g3.114.010892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brückner S, Mösch HU. 2012. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 36, 25–58. ( 10.1111/j.1574-6976.2011.00275.x) [DOI] [PubMed] [Google Scholar]
- 23.Bojsen RK, Andersen KS, Regenberg B. 2012. Saccharomyces cerevisiae—a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 65, 169–182. ( 10.1111/j.1574-695X.2012.00943.x) [DOI] [PubMed] [Google Scholar]
- 24.Veelders M, Brückner S, Ott D, Unverzagt C, Mösch HU, Essen LO. 2010. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl Acad. Sci. USA 107, 22 511–22 516. ( 10.1073/pnas.1013210108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraushaar T, Brückner S, Veelders M, Rhinow D, Schreiner F, Birke R, Pagenstecher A, Mösch HU, Essen LO. 2015. Interactions by the fungal Flo11 adhesin depend on a fibronectin type III-like adhesin domain girdled by aromatic bands. Structure 23, 1005–1017. ( 10.1016/j.str.2015.03.021) [DOI] [PubMed] [Google Scholar]
- 26.Reynolds TB, Fink GR. 2001. Bakers’ yeast, a model for fungal biofilm formation. Science 291, 878–881. ( 10.1126/science.291.5505.878) [DOI] [PubMed] [Google Scholar]
- 27.Pan X, Heitman J. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22, 3981–3993. ( 10.1128/MCB.22.12.3981-3993.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Octavio LM, Gedeon K, Maheshri N. 2009. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genet. 5, e1000673 ( 10.1371/journal.pgen.1000673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. 2012. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell. 45, 470–482. ( 10.1016/j.molcel.2011.11.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchin S, Vyas VK, Carlson M. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22, 3994–4000. ( 10.1128/MCB.22.12.3994-4000.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman F. 1991. Guide to yeast genetics and molecular biology (eds Guthrie C, Fink GR). San Diego, CA: Academic Press. [Google Scholar]
- 32.Bro C, Regenberg B, Förster J, Nielsen J. 2006. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 8, 102–111. ( 10.1016/j.ymben.2005.09.007) [DOI] [PubMed] [Google Scholar]
- 33.Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. ( 10.1038/nature02744) [DOI] [PubMed] [Google Scholar]
- 34.Brockhurst MA, Buckling A, Gardner A. 2007. Cooperation Peaks at Intermediate Disturbance. Curr. Biol. 17, 761–765. ( 10.1016/j.cub.2007.02.057) [DOI] [PubMed] [Google Scholar]
- 35.Kamath RS, Bungay HR. 1988. Growth of yeast colonies on solid media. J. Gen. Microbiol. 134, 3061–3069. ( 10.1099/00221287-134-11-3061) [DOI] [PubMed] [Google Scholar]
- 36.Goddard MR. 2008. Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89, 2077–2082. ( 10.1890/07-2060.1) [DOI] [PubMed] [Google Scholar]
- 37.Meersman E, Steensels J, Mathawan M, Wittocx PJ, Saels V, Struyf N, Bernaert H, Vrancken G, Verstrepen KJ. 2013. Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS ONE 8, e81559 ( 10.1371/journal.pone.0081559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefanini I, et al. 2012. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl Acad. Sci. USA 109, 13 398–13 403. ( 10.1073/pnas.1208362109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. ( 10.1038/nrmicro1968) [DOI] [PubMed] [Google Scholar]
- 40.Ratcliff WC, Denison RF, Borello M, Travisano M. 2012. Experimental evolution of multicellularity. Proc. Natl Acad. Sci. USA 109, 1595–1600. ( 10.1073/pnas.1115323109) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supplementary data are presented in the electronic supplementary material for this article.