Abstract
Serine β-lactamases are bacterial enzymes that hydrolyze β-lactam antibiotics. They utilize an active-site serine residue as a nucleophile, forming an acyl-enzyme intermediate during hydrolysis. In this study, thermal denaturation experiments as well as X-ray crystallography were performed to test the effect of substitution of the catalytic serine by glycine on protein stability in serine β-lactamases. Six different enzymes comprising representatives from each of the three classes of serine β-lactamases were examined including TEM-1, CTX-M-14, and KPC-2 of class A, P99 of class C, and OXA-48 and OXA-163 of class D. For each enzyme, the wild type and a serine-to-glycine mutant were evaluated for stability. The glycine mutants all exhibited enhanced thermostability compared to the wild type. In contrast, alanine substitutions of the catalytic serine in TEM-1, OXA-48 and OXA-163 did not alter stability, suggesting removal of the Cβ atom is key to the stability increase associated with the glycine mutants. The X-ray crystal structures of P99 S64G, OXA-48 S70G and S70A, and OXA-163 S70G suggest that removal of the side chain of the catalytic serine releases steric strain to improve enzyme stability. Additionally, analysis of the torsion angles at the nucleophile position indicates that the glycine mutants exhibit improved distance and angular parameters of the intra-helical hydrogen bond network compared to the wild-type enzymes, which is also consistent with increased stability. The increased stability of the mutants indicates that the enzyme pays a price in stability for the presence of a side chain at the catalytic serine position but that the cost is necessary in that removal of the serine drastically impairs function. These findings support the stability-function hypothesis, which states that active-site residues are optimized for substrate binding and catalysis but that the requirements for catalysis are often not consistent with the requirements for optimal stability.
Keywords: Structure-function, activity-stability trade off, steric strain, active-site nucleophile, serine β-lactamases
INTRODUCTION
The active sites of enzymes are organized to effectively bind substrates, stabilize transition states, and release products. Achieving these tasks requires the precise placement of residues in configurations that may not be optimal for the overall stability of the enzyme. 1–6 The tension between the requirements for optimal function versus optimal stability has been articulated as the stability-function trade-off hypothesis.7 For example, there is often internal strain in the active site of enzymes created by the arrangement of the residues involved in catalysis. Functional residues in the active site, typically polar or charged, can be found buried in hydrophobic clefts and often adopt unfavorable, strained conformations.8–13 Additionally, hydrophobic patches that evolve to bind substrate are often exposed and destabilizing in the free enzyme. Residues of the same charge may also cluster together to improve substrate binding or transition state stabilization.14–16 The acquisition of amino acid substitutions that improve enzyme function by enhancing the specific interactions with bound substrates, transition states or products are often coupled with a decrease in stability.13, 17–20
β-Lactamases are bacterial enzymes that hydrolyze and inactivate β-lactam antibiotics. Based on mechanism, they are classified as either metallo (Class B) or serine β-lactamases (Classes A, C, and D).21 All serine β-lactamases have the same general two-domain (α and α/β) fold but they differ greatly in amino acid sequence identity across classes (as little as 15% sequence identity in some cases).22, 23 Despite the amino acid sequence variability, the active-site residues involved in hydrolysis are almost completely conserved. Serine β-lactamases are mechanistically similar to serine proteases.24 Both types of enzymes are hydrolases and both use acylation and deacylation of an active site serine in their mechanism of catalysis (Figure 1). Additionally, they have a residue strategically positioned to act as a general base to activate the catalytic serine for the nucleophilic attack on the substrate. Lys73 and Glu166 activate the active-site serine in class A enzymes while Lys67 and Tyr150 perform this task in class C β-lactamases.25, 26 Class D β-lactamases have an unusual N-carbamylated lysine (Lys73) that acts as the general base.27
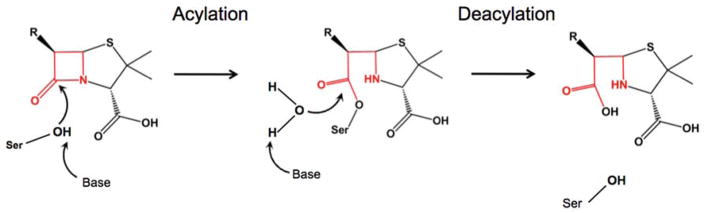
Figure 1.
General scheme of the catalytic mechanism of serine β-lactamases. The β-lactam ring is shown in red. The serine nucleophile is activated via a general base and attacks the carbonyl of the β-lactam ring in the acylation step. This results in formation of the covalent acyl-enzyme complex. In the deacylation step the complex is resolved by a catalytic water molecule, which is activated by the general base.
β-Lactamases are an excellent model system to assess questions on structure, function, and stability. The class C AmpC β-lactamase was previously used as a model to investigate the stability-function trade-off hypothesis.12 In that study, several key active site residues, including the catalytic Ser64 residue, were substituted and tested for activity and stability. It was found that the S64G substitution resulted in significantly increased thermal stability (ΔTm=6.5°C) and greatly reduced catalytic activity compared to the wild-type enzyme. This result provided evidence that the side chain of the nucleophilic Ser64 residue is associated with a cost in stability. The structure of the S64G enzyme suggested that relieving the steric strain from a close contact in the AmpC enzyme between Cβ of Ser64 and Oη of Tyr150 is responsible for the observed increase in stability. However, this has been the only study examining a mutant of the active-site serine for its effect on stability in serine β-lactamases. It is unknown whether the increased stability associated with removal of the active-site serine side chain in AmpC β-lactamase is a general property of all serine β-lactamases.
In this study, a series of thermal stability experiments were performed on six serine β-lactamases (from classes A, C, and D) and their respective active-site serine mutants (Ser to Gly or Ala). The serine-to-glycine mutant enzymes exhibited increased (2.1–13.3 °C) thermal stability compared to the wild-type enzymes. In addition, the crystal structures of P99 S64G, OXA-48 S70G and S70A, and OXA-163 S70G mutant enzymes were examined. The crystal structures suggest the increase in stability obtained by removal of the side chain is due to relief of a steric strain caused by the presence of the serine nucleophile side chain in the active site, specifically a steric clash involving the methylene group of the serine side chain. Consistent with this hypothesis, substitution of the catalytic serine by alanine in TEM-1, OXA-48 and OXA-163 does not increase stability. In addition, an improvement of the intra-helical hydrogen bond distance (C=O…H-N) and angular (O…H-N) parameters in the glycine mutants may further contribute to the increased thermostability. The increase in stability due to removal of the side chain at the nucleophile residue position occurs across a wide range of enzymes from multiple β-lactamase classes, suggesting that the local environment of the catalytic serine is similar across these enzymes and that the strained conformation of the catalytic nucleophile may be an evolutionarily conserved feature of serine β-lactamases.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis PCR
The following plasmids that have been described previously were used in this study: pET24a encoding blaTEM;20 pTP12328 encoding blaCTX-M-14 and blaKPC-2;29, 30 and pET29a encoding blaP99, blaOXA-48, and blaOXA-163.31, 32 The S70G (TEM-1, CTX-M-14, KPC-2, OXA-48, and OXA-163), S64G (P99), and S70A (TEM-1, OXA-48, and OXA-163) amino acid substitutions were introduced by Quikchange PCR with Phusion® DNA Polymerase (New England BioLabs, Ipswich, MA, USA) according to the manufacturer guidelines. DNA sequencing of the entire bla gene for each mutant was performed to ensure the absence of extraneous mutations.
Protein expression and purification
The TEM-1, P99, OXA-48, and OXA-163 β-lactamases and their respective mutants were expressed in E. coli BL21(DE3) cells as described previously.33 In brief, cells were grown in 500 mL LB broth containing 300 mM sorbitol, 2.5 mM betaine, and 30 μg/mL kanamycin to an OD600 of 0.6–0.8 before induction with 0.4 mM IPTG. The culture was then grown at 23°C for 18–20 hours with shaking. Cell pellets were obtained by centrifugation and the culture supernatant was concentrated 10-fold by ultrafiltration using Vivaflow50 10MWCO (Sartorius, Goettingen, Germany) followed by dialysis overnight at 4°C against buffer containing 20 mM Tris, 0.4 M NaCl pH 8.0 for TEM-1, 10 mM MES pH 6.0 for P99, and 20 mM Tris, 0.4 M NaCl pH 8.2 for OXA-48 and OXA-163. TEM-1 was purified using a Fast Flow Chelating Sepharose™ column (GE Healthcare, Pittsburgh, PA) loaded with zinc and eluted with a linear gradient of 200 mM of imidazole. P99 was purified using a HiTrap SPFF Sepharose™ column (GE Healthcare) eluted with linear gradient of 1.0 M NaCl. OXA-48 and OXA-163 were purified using a Fast Flow Chelating Sepharose™ column (GE Healthcare) loaded with zinc and eluted with a linear gradient of 150 mM imidazole.32 Purity was determined by SDS-PAGE and protein fractions were concentrated with Amicon centrifugal filters with a 10,000 MW cut-off (Merck KGaA, Darmstadt, Germany). TEM-1 protein concentrations were determined using the Bio-Rad Bradford protein assay reagent with a standard curve that was calibrated using quantitative amino acid analysis of the wild-type TEM-1 enzyme. The P99 protein concentration was determined by absorbance at 280 nm using an extinction coefficient of 84,340 M−1 cm−1.34 The protein concentration of OXA-48 and OXA-163 was determined by absorbance measurements at 280 nm using an extinction coefficient of 63,940 M−1 cm−1.34
The CTX-M-14 and KPC-2 β-lactamases and their respective serine to glycine mutants were expressed in E. coli RB791 cells as previously described.29, 30 In brief, 1.5 L of LB medium containing 12.5 μg/mL of chloramphenicol was inoculated with overnight cell culture at final dilution of 1:100 and grown to OD600 of 0.6–0.8 at 37°C with shaking before induction with addition of 1 M IPTG at 0.2 mM final concentration. Following induction, the cultures were incubated overnight at 23°C with shaking. Cells were harvested by centrifugation and the cell pellet was frozen at −80°C for at least one hour. The periplasmic contents were released by resuspending the cell pellet in 30 mL of 10 mM Tris pH 8.0, 20% sucrose, 1 mM EDTA before adding 50 mL of sterile water and shaking vigorously. The mixture was centrifuged and the supernatant was filtered before passing it through a HiLoad SP column (Amersham, GE Healthcare, Piscataway, NJ). CTX-M-14 enzymes were eluted with a linear gradient of NaCl. KPC-2 enzymes were bound to the column by adjusting the buffer to pH 5.5 using MES acid and eluted using a linear NaCl gradient. Fractions were pooled and concentrated with Vivaspin® Turbo centrifugal filters, 10,000 MW cut-off (Sartorius). Subsequent size exclusion was performed using a HiLoad Superdex 75 column (GE Healthcare). Protein purity was determined by SDS-PAGE, and fractions with greater than 90% purity were pooled and dialyzed overnight against 50 mM sodium phosphate buffer pH 7.2. CTX-M-14 and KPC-2 protein concentration was determined by absorbance measurements at 280 nm using an extinction coefficient of 25,565 M−1 cm−1 and 39,545 M−1 cm−1, respectively.34
Thermal Denaturation
Two-state van’t Hoff thermodynamic parameters were determined from circular dichroism (CD)-thermal melts. CD experiments were performed as previously described.35–37 In brief, 0.075 μg/mL of protein in 50 mM sodium phosphate pH 7.2 was denatured by raising the temperature in 0.1 °C increments at a ramp rate of 2 °C min−1 using a Jasco J-815 spectropolarimeter (Jasco, Essex, UK) with a Peltier effect temperature controller and an in-cell temperature monitor. Denaturation was marked by an obvious transition in the far-UV CD (223 nm) signal. The β-lactamases were shown to refold in that 95% of the signal was recovered when cooled back to the starting temperature at a protein concentration of 0.075 μg/mL. Experiments were performed in duplicates or more. The melting temperature (Tm) is the temperature mid-point of protein unfolding and was determined by fitting the data to a single Boltzmann model. All data were fitted using GraphPad Prism 6 (GraphPad Software, Inc. La Jolla, CA).
Enzyme Kinetic Studies
Assays were performed on a DU800 spectrophotometer at 30°C in 50 mM sodium phosphate buffer pH 7.2 as previously described.20, 32 Substrate hydrolysis was followed at 235 nm for ampicillin (Δε = −900 M−1 cm−1) and 262 nm for cephalothin (Δε = −7,660 M−1 cm−1). Enzyme kinetics data was analyzed with GraphPad Prism 6 (GraphPad Software, Inc. La Jolla, CA) and fitted to the Michaelis-Menton equation.
Crystallization and data collection
Crystal conditions were screened with 10 mg/mL protein using commercially available screens. Crystallization was performed by the vapor diffusion hanging-drop method. P99 S64G crystals formed in 0.1M SPG buffer (succinic acid, sodium dihydrogen phosphate, and glycine) and 25% PEG 1500. OXA-48 S70G and S70A crystals formed in 0.2M sodium formate, 0.1M cadmium chloride, and 25% w/v PEG 3, 350, and 0.1M Tris-HCl pH 8.5 and 25% w/v PEG 1000, respectively. OXA-163 S70G crystals formed in 0.2M potassium acetate and 20% w/v PEG 3350. Crystals were cryo-protected with the well solution containing 30% v/v MPD and flash-cooled in liquid nitrogen. Data sets were collected at beamlines 5.0.1 and 8.2.2 (OXA-48 S70A) of the Berkeley Center for Structural Biology in the context of the Collaborative Crystallography Program.
Structure determination and refinement
Diffraction data was processed using the CCP4 suite.38 iMOSFLM was used to process and integrate the images.39 The data was scaled using AIMLESS.40 The crystal structures were determined by molecular replacement with wild-type P99 (PDB ID: 1XX2), OXA-48 (PDB ID: 3HBR), and OXA-163 (PDB ID: 4S2L) as the phasing model using MOLREP.41 Structure refinement was done using several cycles of the REFMAC542 program and phenix.refine43 from the PHENIX suite.44 Concurrently, the model was inspected manually with COOT.45 TLS groups were determined using the TLSMD46, 47 server and final refinement was done using either REFMAC5 or phenix.refine. The final structure was validated using PDB_REDO48 and MolProbity.49.
The structure atomic coordinates were deposited in the Protein Data Bank50 with accession code 5HAI (P99 S64G), 5HAQ (OXA-48 S70G), 5HAP (OXA-48 S70A), 5HAR (OXA-163 S70G). Alignment and RMSD calculations were performed by SSM procedure.51 All structural figures were generated with the UCSF Chimera graphics program.52
Angle measurements and hydrogen bonding distances of serine β-lactamases
All of the angle measurements and bond distances were determined as implemented in VMD and UCSF Chimera programs.52, 53 The structures used for the measurements were the following: TEM-1 (PDB ID: 1ZG4); TEM-1 S70G (PDB ID: 1ZG6); CTX-M-14 (PDB ID: 1YLT) CTX-M-14 S70G (PDB ID: 4PM6); P99 (PDB ID: 1XX2); P99 S64G (this study); OXA-48 (PDB ID: 3HBR); OXA-48 S70G (this study); OXA-48 S70A (this study); OXA-163 (PDB ID: 4S2L); and OXA-163 S70G (this study).
RESULTS
Serine β-lactamase stability
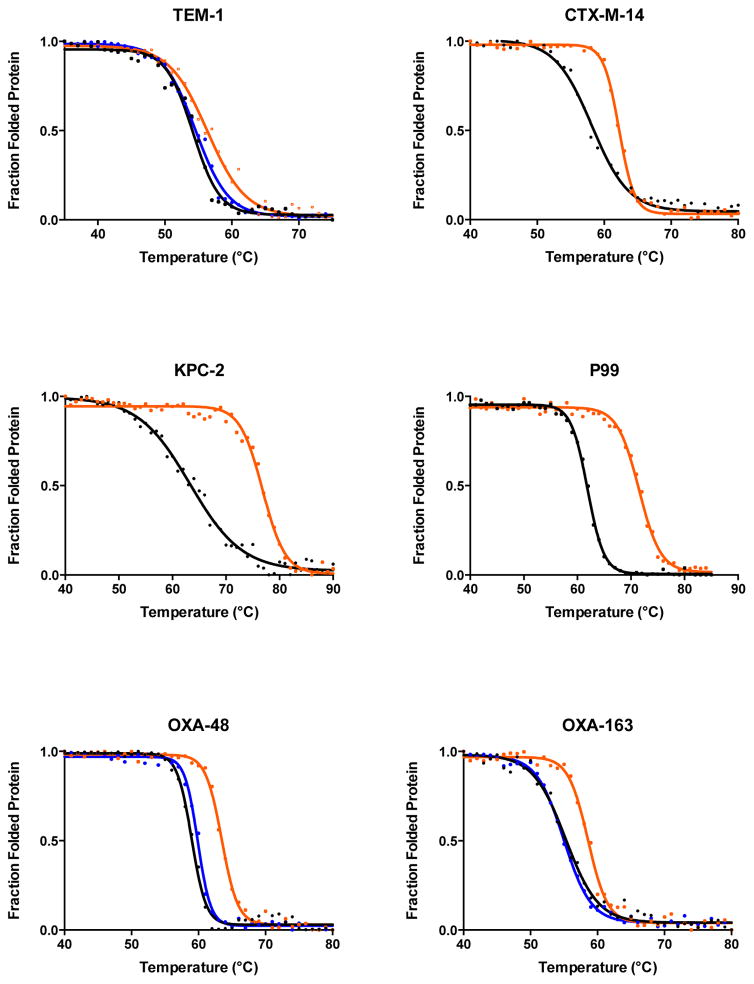
The effect of glycine substitutions of the active-site serine on the stability of serine β-lactamases was examined by CD-monitored thermal denaturation experiments. The thermal denaturation measurements were done with the following β-lactamases and their respective active-site serine-to-glycine mutants: TEM-1, CTX-M-14, and KPC-2 of Class A, P99 from Class C, and OXA-48 and OXA-163 from Class D. In all cases, the glycine mutants showed higher melting temperatures than their wild-type counterparts (Figure 2, Table 1).
Figure 2.
Thermal denaturation curves of six serine β-lactamases. The denaturation curves for the wild-type enzymes are shown in black and the curves for the active-site serine-to-glycine mutants are colored orange. The denaturation curves for the S70A mutants of TEM-1, OXA48, and OXA-163 are shown in blue.
Table 1.
Melting temperature values of serine β-lactamases and their respective serine nucleophile mutants.
| β-Lactamase | Tm (°C) | ΔTm (°C) |
|---|---|---|
| TEM-1 | 54.2 | - |
| TEM-1 S70G | 56.3 | 2.1 |
| TEM-1 S70A | 54.6 | 0.4 |
| CTX-M-14 | 58.2 | - |
| CTX-M-14 S70G | 62.2 | 4.0 |
| KPC-2 | 63.3 | - |
| KPC-2 S70G | 76.6 | 13.3 |
| P99 | 61.9 | - |
| P99 S64G | 71.5 | 9.6 |
| OXA-48 | 58.9 | - |
| OXA-48 S70G | 63.5 | 4.6 |
| OXA-48 S70A | 59.5 | 0.6 |
| OXA-163 | 55.2 | - |
| OXA-163 S70G | 58.6 | 3.4 |
| OXA-163 S70A | 55 | −0.2 |
The class A serine-to-glycine mutants all exhibited increased stability, however, the magnitude of the increase in ΔTm was different. The ΔTm of the TEM-1 S70G mutant increased 2.1°C while the CTX-M-14 S70G mutant increased 4.0°C and the KPC-2 S70G mutant increased a remarkable 13.3°C compared to their wild-type counterparts. The role of the Cβ at position 70 on stability was also assessed for TEM-1 with an S70A mutant. The mutant exhibited similar stability as wild-type TEM-1 suggesting that removal of Cβ is important for the observed increased stability of the glycine mutant (Figure 2, Table 1).
The S64G mutant of the class C P99 enzyme also showed a large, 9.6°C increase in Tm. This agrees with a previous study of the related class C AmpC S64G enzyme, which exhibited a 6°C increase compared to the wild-type enzyme.12 P99 β-lactamase has a very similar overall structure (overall r.m.s.d. 0.61 Å) and 71% sequence identity with the AmpC β-lactamase. These findings suggest that the stabilizing effect of the S64G mutation is due to a similar mechanism in both enzymes. It is also noteworthy that the AmpC study showed an S64A mutant exhibits a 0.7 °C decrease in Tm compared to wild type, consistent with removal of Cβ being important for the improved stability of the glycine mutant. 12
The class D enzymes have not been studied as extensively as the other two classes of serine β-lactamases. The class D family is the most diverse in terms of sequence identity and possesses an unusual spontaneous post-translational carbamylated lysine that serves as the general base in the catalytic mechanism.27, 54 The class D enzymes examined in this study, OXA-48 and OXA-163 are closely related and differ by only five residues.55, 56 One substitution (S212D) and a four-amino acid deletion (214-RIEP-217) converts OXA-48 into OXA-163 and results in a different substrate profile for OXA-163 compared to OXA-48.32 In terms of thermal stability, the S70G mutants of OXA-48 and OXA-163 show increases in Tm of 4.6 °C and 3.4°C, respectively. In addition, the role of the Cβ at position 70 in class D enzymes was examined by measuring the stability of the S70A mutants of OXA-48 and OXA-163, which had Tm values similar to the wild-type enzyme (Figure 2, Table 1). Thus, comparison of the effects of the glycine and alanine substitutions consistently shows the increase in stability observed after removing the serine side chain is associated with removal of the β-methylene group.
Steady-state kinetics
In order to determine the impact on catalytic activity upon substitution of the active-site serine to glycine, steady-state kinetics were performed with a penicillin (ampicillin) and a cephalosporin (cephalothin) substrate. The steady-state kinetic parameters are listed in Table 2. All of the mutant enzymes exhibited a greatly reduced catalytic efficiency (kcat/Km) for both substrates compared to the wild-type enzymes (up to 9000-fold), which is not surprising given that the serine residue is a key component of the hydrolytic mechanism. This result further confirms the stability-function trade-off for the S64G/S70G substitutions of serine β-lactamases in that the substitution increases stability but decreases catalytic function indicating the sequence requirements for optimal stability (Gly64/Gly70) and catalysis (Ser64/Ser70) do not correspond.
Table 2.
β-Lactamase steady-state kinetics parameters.
| Ampicillin | Cephalothin | |||
|---|---|---|---|---|
|
| ||||
| Wild type | S70G | Wild type | S70G | |
| TEM-1 | ||||
| kcat (sec−1) | 1653 ± 370 | 8.36 ± 0.1 | 142 ± 5 | 1.33 ± 0.1 |
| Km (μM) | 63 ± 14 | 2500 ± 430 | 218 ± 15 | 179 ± 28 |
| kcat/Km (sec−1μM−1) | 26.2 | 0.003 | 0.65 | 0.007 |
|
| ||||
| CTX-M-14 | ||||
| kcat (sec−1) | 97.6 ± 4 | ND | 572 ± 8 | 0.17 ± 0.01 |
| Km (μM) | 71 ± 16 | ND | 35 ± 6 | 50 ± 10 |
| kcat/Km (sec−1μM−1) | 1.37 | - | 16.3 | 0.004 |
|
| ||||
| KPC-2 | ||||
| kcat (sec−1) | 39.5 ± 0.95 | 0.08 ± 0.01 | 105 ± 6 | 0.06 ± 0.01 |
| Km (μM) | 98 ± 2 | 105 ± 16 | 95 ± 8 | 29 ± 6 |
| kcat/Km (sec−1μM−1) | 0.4 | 0.001 | 1.1 | 0.002 |
|
| ||||
| P99 | ||||
| kcat (sec−1) | 0.78 ± 0.01 | 0.39 ± 0.01 | 145 ± 5 | 0.05 ± 0.01 |
| Km (μM) | 10 ± 2 | 248 ± 13 | 20 ± 2 | 19 ± 2 |
| kcat/Km (sec−1μM−1) | 0.08 | 0.002 | 7.25 | 0.003 |
|
| ||||
| OXA-48 | ||||
| kcat (sec−1) | 208 ± 3.8 | 0.68 ± 0.05 | 2.8 ± 0.1 | ND |
| Km (μM) | 1630 ± 217 | 345 ± 22 | 140 ± 10 | ND |
| kcat/Km (sec−1μM−1) | 0.13 | 0.002 | 0.02 | - |
|
| ||||
| OXA-163 | ||||
| kcat (sec−1) | 86 ± 8.8 | 1.73 ± 0.11 | 1.7 ± 0.1 | 0.02 ± 0.01 |
| Km (μM) | 737 ± 23 | 522 ± 20 | 3.4 ± 0.4 | 4 ± 1 |
| kcat/Km (sec−1μM−1) | 0.12 | 0.003 | 0.5 | 0.005 |
ND - No detectible hydrolysis with up to 10 μM enzyme and 500 μM substrate.
An interesting additional finding to emerge from the kinetic analysis is that all of the mutant enzymes had very similar kcat/Km values (0.001–0.007 sec−1μM−1) for both ampicillin and cephalothin, regardless of the activity of the wild-type enzyme towards these substrates. For example, TEM-1 is a penicillinase with a kcat/Km value of 26.2 sec−1μM−1 for ampicillin hydrolysis that is reduced 8700-fold for the S70G enzyme (0.003 sec−1μM−1). In contrast, the P99 β-lactamase has a low catalytic efficiency (kcat/Km, 0.08 sec−1μM−1) for ampicillin hydrolysis while the P99 S64G mutant has a kcat/Km value of 0.002 sec−1μM−1, which is very similar to that of the TEM-1 S70G mutant but represents only a 40-fold decrease in activity compared to wild-type P99 (Table 2). Similarly, both CTX-M-14 and P99 are excellent cephalosporinases with kcat/Km values for cephalothin hydrolysis of 16.3 and 7.3 sec−1μM−1, respectively, while the TEM-1 and OXA-163 enzymes exhibit modest kcat/Km values of 0.7 and 0.5 sec−1mM−1. Nevertheless, the serine to glycine mutants of these four enzymes have very similar kcat/Km values for cephalothin hydrolysis (0.003–0.007 sec−1mM−1) (Table 2). Therefore, it appears that the change in catalytic mechanism due to the loss of the side chain of the serine nucleophile creates a baseline of catalytic activity that is similar across the serine β-lactamases. The similar, low-level, activity among the mutant enzymes is likely due to the loss of contributions of residues that are involved in the acylation and deacylation reactions. The mechanism of the Ser-to-Gly mutants presumably involves a direct attack on the β-lactam by a catalytic water molecule.57 This water molecule may be positioned and coordinated in the active site by structural elements common to all serine β-lactamases. Another interesting finding is that hydrolysis activity of the S70A mutants of OXA-48 and OXA-163 was not detectable using the same conditions as for the S70G mutants (data not shown). In addition, the S70A mutant of TEM-1 exhibited 30- and 70-fold decreases in catalytic efficiencies for ampicillin (0.0001 sec−1μM−1) and cephalothin (0.0001 sec−1μM−1) compared to TEM-1 S70G (0.003 sec−1μM−1and 0.007 sec−1μM−1) mainly due to decreased kcat (0.21 sec−1 for ampicillin and 0.02 sec−1 for cephalothin). The Km values of the TEM-1 S70A mutant were similar to S70G mutant with less than a 2-fold difference (1500 μM for ampicillin and 149 μM for cephalothin). These results suggest that the cavity formed by the removal of the serine side chain by substitution with glycine may accommodate a water molecule that is involved in the hydrolysis mechanism of the S70G mutants but is displaced when an alanine is present.
To evaluate the possibility that the steady-state parameters for the S70G mutants are due to wild-type enzyme contamination, the activity of the S70G mutants for ampicillin (OXA-48 and OXA-163) and cephalothin (TEM-1, CTX-M-14, KPC-2, and P99) were tested in the presence of avibactam (20 μM).58, 59 Avibactam is a potent non-β-lactam covalent inhibitor of serine β-lactamases and inhibits the wild-type enzymes with a range of Ki values between 2–20 nM.59 We reasoned that since the serine-to-glycine mutants no longer have the serine nucleophile they would not be affected by the presence of avibactam. The enzymes were tested under kcat/Km and Vmax conditions and it was found that the activities of all of the enzymes tested were not affected by avibactam, except KPC-2 S70G for which a decrease in activity was observed. Since the Km value of KPC-2 S70G is 3-fold lower in comparison to KPC-2 (Table 2), it is unlikely that there is a wild-type KPC-2 present. One plausible explanation for the reduction of activity of KPC-2 S70G is that avibactam can enter and bind non-covalently in the active site of KPC-2 and thereby inhibit the enzyme. However, further investigation of this observation is beyond the scope of this study.
Crystal structures of the P99 S64G, OXA-48 S70G and S70A, and OXA-163 S70G mutants
X-ray crystallography was performed to evaluate structural changes resulting from the serine-to-glycine substitutions. Crystal structures of the class A enzymes TEM-1 and CTX-M-14 and their S70G mutant equivalents are available.29, 57 However, crystal structures of the P99, OXA-48 and OXA-163 S70G mutants are not available and were therefore determined. Attempts to crystallize the S70G mutant of KPC-2 were not successful.
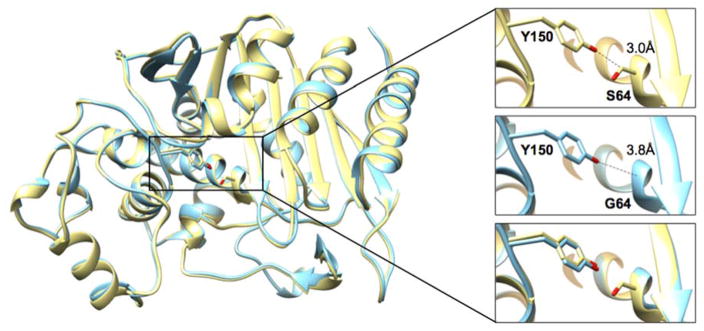
The P99 S64G crystal structure was determined to a resolution of 2.74 Å (Table 3). The P99 S64G mutant crystallized in the P22121 space group with one molecule in the asymmetric unit. Overall, the structure is almost identical with the P99 wild-type structure (PDB ID: 1XX2)60 with an r.m.s.d. of the Cα’s of 0.36 Å. Additionally, the active-site residues adopt very similar conformations in both structures. The absence of the serine side chain in the mutant eliminates a close contact of 3 Å present in wild-type P99 between Cβ of Ser64 and the Oη of Tyr150 (Figure 3). This observation is consistent with a similar observation in a previous study of the AmpC β-lactamase and its S64G mutant.12 Furthermore, the S64G substitution creates space for binding of a single phosphate ion that is present in the crystallization condition at 0.2 M concentration. The binding of the phosphate ion was also observed in the AmpC S64G crystal structure where it was shown not to have an effect on the stability of the enzyme.12
Table 3.
Crystallography data collection and refinement statistics.
| Protein | P99 S64G | OXA48 S70G | OXA48 S70A | OXA-163 S70G |
|---|---|---|---|---|
| Data collection | PDB ID: 5HAI | PDB ID: 5HAQ | PDB ID: 5HAP | PDB ID: 5HAR |
|
| ||||
| Wavelength (Å) | 0.997 | 0.997 | 0.919 | 0.997 |
| Resolution range (Å) | 61.9 - 2.74 (2.84 - 2.74) | 60 - 2.14 (2.22 - 2.14) | 57.1 - 1.89 (1.96 - 1.89) | 62.38 - 1.74 (1.80 - 1.74) |
| Space group | P 2 21 21 | P 21 21 21 | P 65 2 2 | C 2 2 21 |
| Unit cell | ||||
| a, b, c (Å) | 61.9, 69.4, 78.1 | 72.3, 73.6, 106.3 | 121.9, 121.9, 161.5 | 44.3, 88.0, 124.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 1200.0 | 90.0, 90.0, 90.0 |
| Unique reflections | 9252 (896) | 32093 (3160) | 57087 (5591) | 25438 (2507) |
| Multiplicity | 6.9 (6.9) | 6.5 (6.7) | 25.5 (21.7) | 7.1 (7.5) |
| Completeness | 99.5 (98.1) | 99.8 (99.7) | 99.9 (99.8) | 99.7 (100) |
| Mean I/sigma(I) | 5.9 (2.3) | 9.2 (2.3) | 10.9 (2.1) | 13.8 (1.7) |
| Wilson B-factor (Å2) | 30.2 | 28.6 | 23.2 | 19.5 |
| Rmerge (%) | 6.4 (25) | 5.0 (32) | 3.8 (35) | 3.7 (41) |
| Mean I half-set correlation CC1/2 | 0.99 (0.84) | 1.0 (0.84) | 1.0 (0.85) | 0.99 (0.58) |
|
| ||||
| Refinement | ||||
| Rwork (Rfree) (%) | 21.9 (25.2) | 19.1 (21.6) | 17.2 (19.5) | 18.9 (22.6) |
| Number of non-hydrogen atoms | 2819 | 4181 | 4374 | 2228 |
| Protein | 2751 | 3944 | 3952 | 1939 |
| Ligands | 2 | 12 | 129 | 7 |
| Waters | 68 | 225 | 293 | 282 |
| Protein residues | 359 | 482 | 482 | 237 |
| r.m.s.d. bond length (Å) | 0.011 | 0.009 | 0.014 | 0.003 |
| r.m.s.d. bond angle (°) | 1.36 | 1.18 | 1.59 | 0.75 |
| Ramachandran favored (%) | 98 | 97 | 98 | 97 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
| Average B-factor (Å2) | 26.6 | 36.1 | 30.8 | 25.2 |
| Protein | 26.7 | 35.9 | 29.8 | 23.5 |
| Ligands | 41.6 | 36.5 | 55.4 | 31.2 |
| Waters | 24.7 | 39.9 | 33.7 | 36.8 |
Figure 3.
Structure alignment of P99 wild type and S64G mutant β-lactamases. The wild-type P99 structure (PDB ID: 1XX2) is shown in yellow and the P99 S64G mutant structure determined in this study is shown in cyan. The inset contains a detailed view of residues 64 and 150. The absence of the side chain at position 64 in the S64G mutant (middle panel) increases the distance between Oη of the Y150 and position 64 to 3.8 Å compared to 3.0 Å in the wild type (top panel).
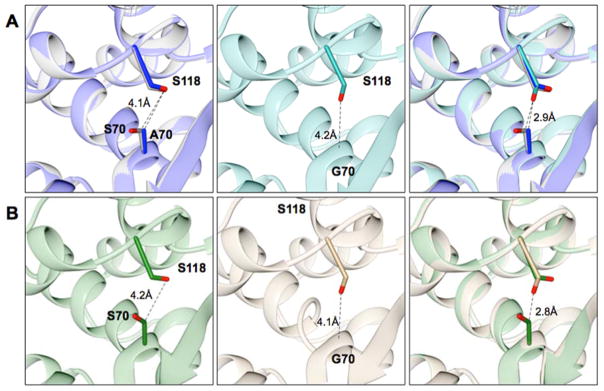
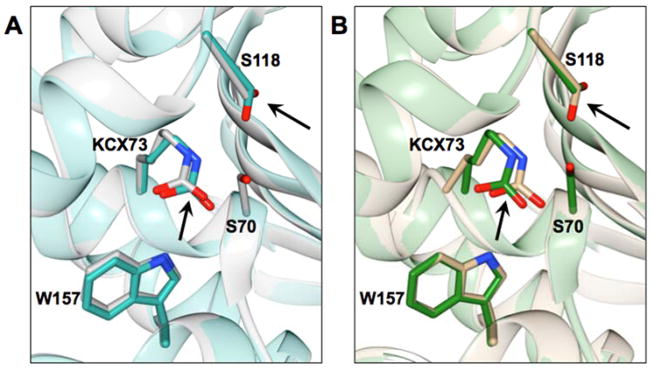
The crystal structures of the class D S70G mutant enzymes of OXA-48 and OXA-163 were determined at 2.14 Å and 1.74 Å resolution, respectively. Superimposition of the wild-type structures with the S70G mutant structures gave an r.m.s.d. of 0.32 Å for OXA-48 (PDB ID: 1HBR) and 0.23 Å for OXA-163 (PDB ID: 4S2L) for the matching Cα atoms. As in the structures of the wild-type enzymes, extra electron density was observed extended from Lys73 in the glycine mutants indicating that it is in carbamylated form. Despite these similarities there are some differences in the active site of the S70G mutants. The most prominent change is in the conformation of Ser118 (Figure 4). In the wild-type structures, Ser118 adopts a conformation pointing at Lys208 located on the β5-strand away from the Ser70. The absence of a side chain at position 70 in the S70G mutants allows Ser118 to adopt a different conformation in which it points down at the main chain towards Gly70 (Figure 4). In this conformation, the Oγ of Ser118 is within hydrogen bond distance (3.3 Å) to the Nζ of the carbamylated lysine. This conformation would not be favorable in the wild-type structures mainly because of a close contact (2.9 Å) with the Cβ of Ser70, which will introduce a steric strain (Figure 4). Also, in the active site of the S70G mutants, the carbamylated Lys73 is shifted approximately 0.5 Å towards the active-site pocket (Figure 5). Lastly, in both S70G structures (OXA-48 and OXA-163) there is an anion present in the active site of the enzyme. In the OXA-48 S70G structure a formate ion is positioned in close proximity to the main chain of Gly70 and interacts with the carboxyl of the carbamylated lysine and the hydroxyl of Ser118. In the OXA-163 S70G structure an acetate ion is present at the same position.
Figure 4.
Overlay of class D enzymes OXA-48 and OXA-163 and their Ser70 mutants. Positions 70 and Ser118 are shown in stick representation. Distances between selected atoms are labeled and represented as dashed lines. (A) Left panel, structure alignment of OXA-48 β-lactamase (PDB ID: 3HBR) in grey and the S70A mutant structure in blue. Middle panel, the S70G mutant structure shown in dark cyan. Right panel, the structure alignment of OXA-48 wild type with OXA-48 S70A, and S70G mutants. (B) Left panel, the OXA-163 (PDB ID: 4S2L) structure shown in green. Middle panel, the OXA-163 S70G structure shown in tan. Right panel, structural alignment of OXA-163 with the S70G mutant. In both the OXA-48 and OXA-163 enzymes, Ser118 adopts a different conformation when Ser70 is substituted to glycine (middle panels). This conformation is not possible in the wild-type enzymes because of close contact between Ser70 and Ser118 (right panels).
Figure 5.
The position of Lys73 is shown in the crystal structure of the S70G mutant of OXA-48 (dark cyan) aligned with wild-type OXA-48 (PDB ID: 3HBR) in grey (left panel). The right panel shows the S70G mutant of OXA-163 in tan aligned with OXA-163 (PDB ID: 4S2L) in green. Selected residues are represented in stick and labeled. Black arrows point at the Lys73 and Ser118 residues outlining the movement of these residues in the S70G mutants in comparison to the wild-type enzymes.
To evaluate if the Cβ at position 70 plays a role in inducing a different conformation of Ser118 in the S70G mutant structures of the OXA enzymes, the X-ray crystal structure of an OXA-48 S70A mutant was determined at 1.89 Å resolution. OXA-48 S70A crystallized in a P6522 (hexagonal) space group with two molecules (one dimer) in the asymmetric unit, which differs from the two other crystal structures obtained with this protein: monoclinic (wild type)61 and orthorhombic (S70G mutant in this study). The r.m.s.d of the Cα atoms for the S70A enzyme are 0.30 Å versus wild type and 0.22 Å versus S70G. In the crystal structure of OXA-48 S70A, Ser118 adopts the same conformation as in the structure of the wild-type enzyme where it is pointing towards Lys208 and away from the Cβ of Ala70 (Figure 4A). This observation indicates that Ser118 is constrained to this conformation because of the steric strain imposed by the close contact between Oγ of Ser118 and Cβ of position 70 (Ala and Ser)(Figure 4A). This observation, along with the finding described above that the S70A substitution does not significantly stabilize the enzyme relative to wild type OXA-48, suggests that the movement of Ser118 seen in the OXA-48 S70G structure is associated with the increased stability of OXA-48 S70G and this movement requires removal of both the hydroxyl and Cβ atom of Ser70.
Analysis of the crystal structures of TEM-1 S70G, CTX-M-14 S70G and KPC-2 β-lactamases
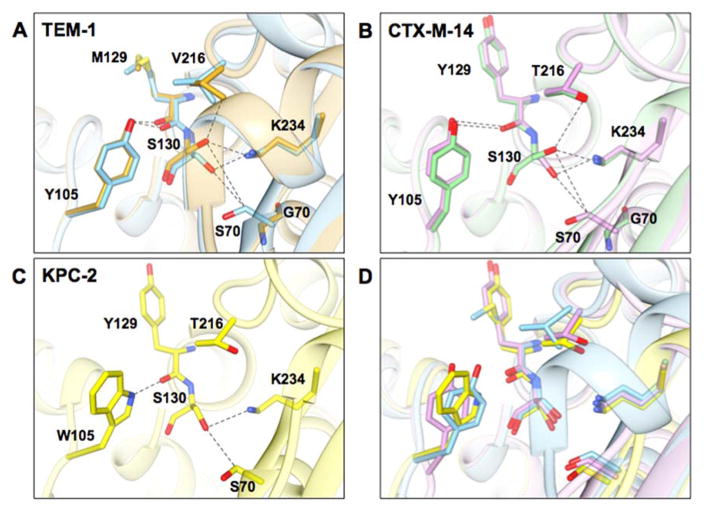
X-ray crystal structures of the class A enzymes TEM-1 and CTX-M-14 and their S70G mutant equivalents are available.29, 57 To better understand the structural basis of the increases in stability of the S70G mutants, we examined the structures of these enzymes. Both enzymes, when superimposed with their S70G counterparts are almost identical, aside from the absence of the side chain of Ser70 in the glycine mutants. However, there are differences in the conformation of Ser130, which is the structural and mechanistic equivalent of Ser118 in the class D enzymes.
In the case of TEM-1, a prominent change is the altered conformation of Ser130, which is pointing down towards Ser70 in the wild-type enzyme resulting in a mild steric clash between Oγ of Ser130 and Cβ of Ser70 (3.15 Å) (Figure 6). In the S70G mutant, Ser130 adopts a different conformation where it points away from Gly70 increasing the distance between these residues (6.47 Å Oγ to Cα). Similarly, the position of Ser130 in the CTX-M-14 wild-type structure also points down towards Ser70 and its Oγ is placed 3.36 Å away from the Cβ of Ser70 (Figure 6). In the structure of the S70G CTX-M-14 enzyme, Ser130 adopts a different conformation and is oriented away from the Gly70 increasing the distance between the residues to 5.61Å (6.47 Å Oγ to Cα). The change in conformation of Ser130 in CTX-M-14 S70G is identical with the change in conformation of TEM-1 S70G (Figure 6).
Figure 6.
Structural representations of the active sites of class A β-lactamases from this study. A) Wild-type TEM-1 (PDB ID: 1ZG4) is shown in blue and TEM-1 S70G in gold (PDB ID: 1ZG6). B) Wild-type CTX-M-14 (PDB ID: 1YLT) is shown in purple and the S70G mutant (PDB ID: 4PM6) in light green. C) Wild type KPC-2 (PDB ID: 2OV5) is shown in yellow. D) Structural alignment of TEM-1, CTX-M-14, and KPC-2 β-lactamases. Selected residues are represented in stick and labeled. Dashed lines represent distances between atoms from 2.8 to 3.8 angstroms. Note that KPC-2 has a tryptophan at position 105 while CTX-M-14 and TEM-1 have tyrosine at this position. In addition, CTX-M-14 and KPC-2 have tyrosine and threonine at positions 129 and 216 while TEM-1 has methionine and valine at these positions.
In the case of KPC-2 β-lactamase, only the crystal structure of the wild-type enzyme is available.62 When superimposed to the other two class A enzymes, the active site of KPC-2 has Ser70 and Ser130 closer to each other (Figure 6) with a distance of 3.12 Å between Cβ of Ser70 and Oγ of Ser130, which is the shortest distance observed between these residues among the three class A enzymes examined. By analogy with the TEM-1 and CTX-M-14 S70G structures, the elimination of the side chain and Cβ atom in particular, would be expected to relieve the steric strain and contribute to the observed increased stability of the KPC-2 S70G mutant.
Analysis of the nucleophile residue main-chain hydrogen bonds and angle measurements of wild-type enzymes and S70G mutants
To further examine the internal strain introduced by the serine nucleophile in the active site of serine β-lactamases, we analyzed the angles (omega and phi/psi) of the nucleophile position as well as the hydrogen bonds formed by the main chain (C=O(Nu)…H-N(Nu+3)) in the wild type and glycine mutants. The conformation of an ideal peptide unit is planar with an omega angle value of 180 degrees.63 Deviations from this value will introduce strain on the peptide unit that may result in compromised stability. In addition, for all of the enzymes in this study, the serine nucleophile is located at the N-terminus of an α-helix where the first five residues of the helix form a 310-helix characterized with i to i+3 main-chain hydrogen bonds.64 The phi/psi angles of the nucleophile are rather far away from the phi/psi angles of an ideal 310-helix (φ ~−49°, ψ ~−27°).65 As a result, the intrahelical hydrogen bond that is formed by its main-chain carbonyl atom has less optimal distance (C=O…H-N) and angular (O…H-N) parameters. Additionally, serine compared to glycine has a side chain, which further restrains the conformational space that can be adapted by this amino acid.66 Based on the aforementioned, we reasoned that in the wild-type enzymes the serine (–COOH and –NH) would be restrained to a conformation that is suboptimal for stability and by substituting the serine to glycine this restraint will be relaxed leading to an increase in stability. Subsequently, the relaxation in conformation by the glycine substitution will enhance the main chain hydrogen bonding of this position (2.8 Å distance and 180° angle) leading to improvement of the overall hydrogen bond network in the α-helix.
The values of the angles for the nucleophile position together with the respective hydrogen bonds of the main chain are shown in Table 4. In all of the analyzed enzymes, the omega values of the nucleophile position in the wild-type enzymes showed greater deviation from 180° compared to the glycine mutants suggesting increased strain of the peptide unit of the serine nucleophile in the wild-type enzymes. The largest improvement of the omega angles of the glycine mutants is observed in CTX-M-14 (4°) followed by OXA-48 and OXA-163 (3°), TEM-1 (2°) and lastly P99 had an improvement of the omega angle by 1°.
Table 4.
Phi/Psi values of the nucleophile position (Nu) and the position after the nucleophile (Nu+1) in wild type and glycine mutants extracted from their respective crystal structures. Additionally, distances of main chain hydrogen bonds formed by these positions and their angles are listed.
|
|
|||||
|---|---|---|---|---|---|
| Nucleophile (Nu) position
|
|||||
| Protein | omega (ω) [-Cα(Nu)-CO(Nu)- NH(Nu+1)-Cα(Nu+1)-] (°) | phi (φ) (°) | psi (ψ) (°) | C=O(Nu)…H–N(Nu+3) (Å) | H-bond Angle (°) |
| TEM-1 WT | 177 | −65.2 | −8.15 | 3.01 | 140 |
| TEM-1 S70G | 179 | −54.4 | −15.4 | 2.91 | 147 |
| CTX-M-14 WT | 172 | −71.3 | −4.76 | 1.96 | 138 |
| CTX-M-14 S70G | 176 | −67.2 | −4.19 | 2.00 | 134 |
| P99 WT | 176 | −61.2 | −14.9 | 1.97 | 156 |
| P99 S64G | 177 | −58.9 | −12.8 | 3.19 | 163 |
| OXA-48 WT | 174 | −68.1 | −10.8 | 2.94 | 136 |
| OXA-48 S70A | 175 | −67.1 | −10.3 | 2.97 | 135 |
| OXA-48 S70G | 177 | −53.5 | −21.5 | 3.04 | 163 |
| OXA-163 WT | 176 | −72.4 | −10.4 | 2.91 | 135 |
| OXA-163 S70G | 179 | −58.8 | −19.9 | 2.94 | 160 |
In addition to improved omega angles, there is a general trend of the glycine substitutions improving the phi/psi angles by shifting them to the more favorable regions in the Ramachandran plot.66, 67 The class D enzymes (OXA-48 and OXA-163) have the largest change in the phi/psi angles where the S70G mutants shift to a more favorable conformation by ~14° (phi) and ~10° (psi). It is worth noting that the S70A mutant of OXA-48 has almost identical phi/psi angles and hydrogen bond distances with OXA-48, which is consistent with the wild-type-like thermostability for this mutant (Table 1 and Figure 2). Additionally, in the glycine mutants the hydrogen bond distances and/or the angles associated with them are also improved. In general, there is a slight decrease in the hydrogen bond distance (0.03–0.21 Å) and increase in the respective angle (7–15°). However, in the case of CTX-M-14 there is not much difference between the wild-type enzymes and the glycine mutants. Furthermore, the P99 S64G mutant compared to P99 has very similar phi/psi angles and exhibits longer hydrogen bond distance (Table 4) indicating little effect on the stability. These observations are most likely because of the questionably close distances of the hydrogen bonds in P99 wild type (1.97–2.02 Å) and CTX-M-14 wild type (1.96 Å and 2.32 Å) and S70G mutant (2.00 Å and 2.33 Å) crystal structures and might be viewed as a relief of a close contact and steric strain in the S70G mutants. Furthermore, this also illustrates the complexity of protein stability and the role of each element when deciphering their contribution to the overall thermostability. Nevertheless, considering the flexibility introduced by the glycine mutants it is plausible to postulate that a relief of an internal strain, elimination of a close contact by the glycine substitution, improved omega and phi/psi angles and improved hydrogen bond parameters taken together result in increased thermostability.
DISCUSSION
Mechanism of hydrolysis of S70G mutants
Considering the critical role of the serine nucleophile for the function of these enzymes, it is not surprising that the substitution of serine to glycine results in enzymes with poor catalytic activity. The mutant enzymes must bypass the covalent acyl-enzyme intermediate and presumably function by direct attack of a water molecule on the carbonyl carbon of the amide bond in the ring (Figure 1). In this case, a strategically positioned catalytic water molecule is needed for the hydrolysis to occur.
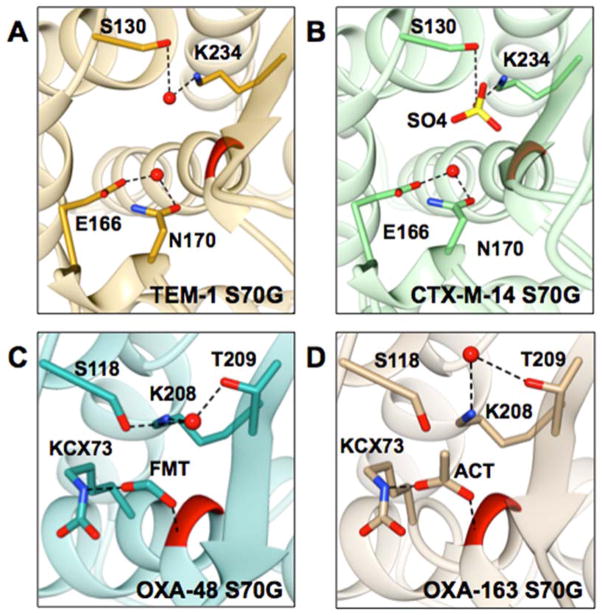
Based on the crystal structure of the TEM-1 S70G mutant, Stec et al. suggested that in class A enzymes this catalytic water is coordinated by the side chains of Ser130 and Lys234 (Figure 7A).56 In the structure of the class A enzyme CTX-M-14, the presence of a sulfate ion in the active site of the S70G would preclude the observation of a water molecule located approximately in the same position (Figure 7B).29 In the Class D structures, we did observe a water molecule coordinated by the side chains of Ser118 and Lys208, which are the equivalents of Ser130 and Lys234 in class A enzymes (Figure 7C and 7D). The equivalent residues for class C P99 enzyme are Tyr150 and Lys315, however, the 2.74 Å resolution of the S64G structure limits the ability to locate a water. Another possibility is that the water responsible for deacylation in the mechanism of the serine β-lactamases serves as the catalytic water in the glycine mutants and this water is present in the TEM-1 and CTX-M-14 S70G structures (Figure 7A and 7B).29, 57 The lower resolution of the P99 S64G structure prevented accurate identification of the deacylation water.25 In class D β-lactamases the deacylation water is coordinated by the N-carbamylated lysine.68 This water, however, was not observed in the crystal structures of OXA-48 S70G and OXA-163 S70G (Figure 7C and 7D). The detailed mechanism of hydrolysis by the serine to glycine mutants of the serine β-lactamases will require additional mechanistic studies.
Figure 7.
Representations of the active sites of class A and class D S70G mutants. Position 70 is colored in red, selected residues and respective anions are represented in stick and labeled; water molecules are represented as red balls, and dashed lines represent selected interactions. A) TEM-1 S70G (PDB ID: 1ZG6) is shown in gold, the deacylating water is coordinated by E166 and N170. Stec et al. proposed that the water molecule coordinated by S130 and K234 is involved in the hydrolysis mechanism of the S70G mutant.57 B) CTX-M-14 S70G (PDB ID: 4PM6) is shown in green, the deacylating water is coordinated by E166 and N170. S130 and K234 coordinate a sulfate ion that is shown in yellow. C) OXA-48 S70G (PDB ID: 5HAQ) is shown in dark cyan. S118, K208, and T209 coordinate a water molecule. Additionally, a formate ion is coordinated by the N-carbamylated K73 and the –NH of G70. D) OXA-163 S70G (PDB ID: 5HAR) is shown in tan. K208 and T209 coordinate a water molecule. Also, an acetate ion is coordinated by N-carbamylated K73 and the –NH of G70.
Relief of a close contact with the nucleophilic serine
A common theme in the structures of the glycine mutants is the elimination of a close contact between the catalytic serine and a hydroxyl group that occupies a similar position in all of the enzymes. In the class A enzymes, the hydroxyl is from Ser130 while in class D it is the mechanistically equivalent Ser118 and in class C it is Tyr150. In all cases, removal of the catalytic serine side chain and, more specifically, the Cβ atom, relieves the close contact. For the class A and D enzymes, the glycine substitution results in a movement of Ser130/Ser118 while Tyr150 in class C P99 does not change position (Figs. 3–6). Interestingly, the movement of Ser130 in the class A mutants is different from that in the class D mutants. In the class A TEM-1 and CTX-M-14 mutants the Ser130 side chain adopts a conformation where the hydroxyl group points away from Gly70 and towards Lys234. This brings the Oγ of Ser130 and Nε of Lys234 about 0.2 Å closer in the class A S70G mutants compared to the wild-type enzymes. This closer interaction results in strengthening of the hydrogen bond between these two residues that may also contribute to increased stability. In the glycine mutants of OXA-48 and 163 the Ser118 conformation orients the side chain towards Gly70, which is opposite to the change in the class A enzymes. The different orientation of the Ser118 side chain brings the Oγ into hydrogen bond distance (3.00 Å) with the Nε of the N-carboxylated Lys73 and creates a hydrogen bond network between Lys73-Ser118-Lys208 that is not present in the wild-type enzymes and could also contribute to increased stability.
Strained conformation of the serine nucleophile
In addition to the close contact introduced by the active-site serine, the position of the nucleophile is located at the N-terminus of a 310-helix and introduces strained interactions in the active site of serine β-lactamases. This strained conformation of the serine is not unique to β-lactamases and has been observed in many structures of proteins where an α-helix begins with a serine residue.67, 69, 70 In a detailed study of a large set of crystal structures (363 polypeptide chains) Pal et al observed that out of the twenty amino acids, serine has the highest propensity to be in a disallowed region of the Ramachandran plot and contributes to strained interactions when is located at the N-terminus of an α-helix.70 Although the nucleophilic serine is not in a disallowed region in the wild-type β-lactamase structures, its omega and phi-psi angles are not ideal and are improved in the glycine mutants. The analysis of the crystal structures of the β-lactamases in this study suggest that by replacing serine with a conformationally less restrained glycine to a large extent relieves the strained interactions in the mutant enzymes, consistent with increased stability. Substitution of serine with a glycine residue also allows the placement of its main chain carbonyl oxygen atom in position to form an intra-helical hydrogen bond with improved distance (C=O…H-N) and angular (O…-H-N) parameters thereby further contributing to increased protein stability.71
CONCLUSIONS
Serine β-lactamases from multiple classes (A, C, and D) are stabilized by removal of the side chain at the position of the active-site nucleophile. All of the S70G mutants tested were more stable than their wild-type counterparts. The increased thermostability ranged from 2.1–13.3 °C, most likely due to variations in the local environment near the catalytic serine and the spatial arrangement of the enzyme’s active site. Structural analysis suggests that in all of these enzymes the serine nucleophile introduces a steric strain in the active site, which compromises stability. Removing the serine side chain and particularly the Cβ atom relieves this steric strain. Additionally, improvement of the intra-helical hydrogen bond network and torsion angles of the glycine mutants relative to wild type may also contribute to the increase in the thermostability. Taken together, these findings suggest that the contacts of the active-site serine with other catalytic residues in the vicinity that result in internal strain are a common feature of all serine β-lactamases. The results also support the stability-function trade-off hypothesis that active-site residues are selected for optimal catalysis even if this is not consistent with optimal enzyme stability. 7, 19, 72
Acknowledgments
Funding Sources
This work was supported by NIH grant AI32956 to TP and Robert Welch Grant Q1279 to BVVP. The Berkley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, and Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH1123. VS is supported by training grant T32 AI55449 from the National Institute of Allergy and Infectious Diseases.
The authors thank Dr. Paul Leonard, Dr. Todd Link and the Center for Biomolecular Structure and Function, University of Texas MD Anderson Cancer Center for the use of their Circular Dichroism instrument. The authors also thank Dr. Hiram Gilbert for comments on the manuscript. The authors acknowledge the Collaborative Crystallography Program at the Berkley Center for Structural Biology, which is in part supported by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. VS acknowledges the CCP4/APS Crystallographic School, 2015 and all of the developers and educators involved in the process of successful determination and validation of protein crystal structures.
References
- 1.Herzberg O, Moult J. Analysis of the steric strain in the polypeptide backbone of protein molecules. Proteins. 1991;11:223–229. doi: 10.1002/prot.340110307. [DOI] [PubMed] [Google Scholar]
- 2.Ota M, Isogai Y, Nishikawa K. Structural requirement of highly-conserved residues in globins. FEBS Lett. 1997;415:129–133. doi: 10.1016/s0014-5793(97)01110-1. [DOI] [PubMed] [Google Scholar]
- 3.Garcia C, Nishimura C, Cavagnero S, Dyson HJ, Wright PE. Changes in the apomyoglobin folding pathway caused by mutation of the distal histidine residue. Biochemistry. 2000;39:11227–11237. doi: 10.1021/bi0010266. [DOI] [PubMed] [Google Scholar]
- 4.Liang S, Zhang J, Zhang S, Guo H. Prediction of the interaction site on the surface of an isolated protein structure by analysis of side chain energy scores. Proteins. 2004;57:548–557. doi: 10.1002/prot.20238. [DOI] [PubMed] [Google Scholar]
- 5.Mukaiyama A, Haruki M, Ota M, Koga Y, Takano K, Kanaya S. A hyperthermophilic protein acquires function at the cost of stability. Biochemistry. 2006;45:12673–12679. doi: 10.1021/bi060907v. [DOI] [PubMed] [Google Scholar]
- 6.Chen YC, Lim C. Common physical basis of macromolecule-binding sites in proteins. Nucleic Acids Res. 2008;36:7078–7087. doi: 10.1093/nar/gkn868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci U S A. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warshel A, Sussman F, Hwang JK. Evaluation of catalytic free energies in genetically modified proteins. J Mol Biol. 1988;201:139–159. doi: 10.1016/0022-2836(88)90445-7. [DOI] [PubMed] [Google Scholar]
- 10.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber G, Buckle AM, Fersht AR. Stability and function: two constraints in the evolution of barstar and other proteins. Structure. 1994;2:945–951. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- 12.Beadle BM, Shoichet BK. Structural bases of stability-function tradeoffs in enzymes. J Mol Biol. 2002;321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 13.Thomas VL, McReynolds AC, Shoichet BK. Structural bases for stability-function tradeoffs in antibiotic resistance. J Mol Biol. 2010;396:47–59. doi: 10.1016/j.jmb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhi W, Srere PA, Evans CT. Conformational stability of pig citrate synthase and some active-site mutants. Biochemistry. 1991;30:9281–9286. doi: 10.1021/bi00102a021. [DOI] [PubMed] [Google Scholar]
- 15.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 16.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci U S A. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. P Natl Acad Sci USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 20.Stojanoski V, Chow DC, Hu L, Sankaran B, Gilbert HF, Prasad BV, Palzkill T. A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis. J Biol Chem. 2015;290:10382–10394. doi: 10.1074/jbc.M114.633438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambler RP. The structure of beta-lactamases. Philos T Roy Soc B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 22.Galleni M, Lamotte-Brasseur J, Raquet X, Dubus A, Monnaie D, Knox JR, Frere JM. The enigmatic catalytic mechanism of active-site serine beta-lactamases. Biochem Pharmacol. 1995;49:1171–1178. doi: 10.1016/0006-2952(94)00502-d. [DOI] [PubMed] [Google Scholar]
- 23.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 25.Hata M, Fujii Y, Tanaka Y, Ishikawa H, Ishii M, Neya S, Tsuda M, Hoshino T. Substrate deacylation mechanisms of serine-beta-lactamases. Biol Pharm Bull. 2006;29:2151–2159. doi: 10.1248/bpb.29.2151. [DOI] [PubMed] [Google Scholar]
- 26.Fisher JF, Mobashery S. Three decades of the class A beta-lactamase acyl-enzyme. Curr Protein Pept Sci. 2009;10:401–407. doi: 10.2174/138920309789351967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard DA, Bonomo RA, Powers RA. Class D beta-lactamases: a reappraisal after five decades. Acc Chem Res. 2013;46:2407–2415. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrosino J, Rudgers G, Gilbert H, Palzkill T. Contributions of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of beta-lactamases. J Biol Chem. 1999;274:2394–2400. doi: 10.1074/jbc.274.4.2394. [DOI] [PubMed] [Google Scholar]
- 29.Adamski CJ, Cardenas AM, Brown NG, Horton LB, Sankaran B, Prasad BV, Gilbert HF, Palzkill T. Molecular basis for the catalytic specificity of the CTX-M extended-spectrum beta-lactamases. Biochemistry. 2015;54:447–457. doi: 10.1021/bi501195g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta SC, Rice K, Palzkill T. Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability. PLoS Pathog. 2015;11:e1004949. doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Yu Y, Musser JM, Palzkill T. Amino acid sequence determinants of extended spectrum cephalosporin hydrolysis by the class C P99 beta-lactamase. J Biol Chem. 2001;276:46568–46574. doi: 10.1074/jbc.M102757200. [DOI] [PubMed] [Google Scholar]
- 32.Stojanoski V, Chow DC, Fryszczyn B, Hu L, Nordmann P, Poirel L, Sankaran B, Prasad BV, Palzkill T. Structural basis for different substrate profiles of two closely related class D beta-lactamases and their inhibition by halogens. Biochemistry. 2015;54:3370–3380. doi: 10.1021/acs.biochem.5b00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosa-Peinado A, Mustafi D, Makinen MW. Overexpression and biosynthetic deuterium enrichment of TEM-1 beta-lactamase for structural characterization by magnetic resonance methods. Protein Expr Purif. 2000;19:235–245. doi: 10.1006/prep.2000.1243. [DOI] [PubMed] [Google Scholar]
- 34.Gasteiger EHC, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 35.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown NG, Pennington JM, Huang W, Ayvaz T, Palzkill T. Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM beta-lactamases. J Mol Biol. 2010;404:832–846. doi: 10.1016/j.jmb.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Z, Huang W, Bakkalbasi E, Brown NG, Adamski CJ, Rice K, Muzny D, Gibbs RA, Palzkill T. Deep sequencing of systematic combinatorial libraries reveals beta-lactamase sequence constraints at high resolution. J Mol Biol. 2012;424:150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 42.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 43.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 47.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 48.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EFGT, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;13:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.Evans BA, Amyes SG. OXA beta-lactamases. Clin Microbiol Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. OXA-163, an OXA-48-related class D beta-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stec B, Holtz KM, Wojciechowski CL, Kantrowitz ER. Structure of the wild-type TEM-1 beta-lactamase at 1.55 A and the mutant enzyme Ser70Ala at 2.1 A suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Crystallogr D Biol Crystallogr. 2005;61:1072–1079. doi: 10.1107/S0907444905014356. [DOI] [PubMed] [Google Scholar]
- 58.Stachyra T, Pechereau MC, Bruneau JM, Claudon M, Frere JM, Miossec C, Coleman K, Black MT. Mechanistic studies of the inactivation of TEM-1 and P99 by NXL104, a novel non-beta-lactam beta-lactamase inhibitor. Antimicrob Agents Chemother. 2010;54:5132–5138. doi: 10.1128/AAC.00568-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. Kinetics of avibactam inhibition against Class A, C, and D beta-lactamases. J Biol Chem. 2013;288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lobkovsky E, Moews PC, Liu H, Zhao H, Frere JM, Knox JR. Evolution of an enzyme activity: crystallographic structure at 2-A resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc Natl Acad Sci U S A. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. Crystal structure of KPC-2: insights into carbapenemase activity in class A beta-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chellapa GD, Rose GD. On interpretation of protein X-ray structures: Planarity of the peptide unit. Proteins. 2015;83:1687–1692. doi: 10.1002/prot.24854. [DOI] [PubMed] [Google Scholar]
- 64.Khrustalev VV, Barkovsky EV, Khrustaleva TA. The influence of flanking secondary structures on amino Acid content and typical lengths of 3/10 helices. Int J Proteomics. 2014;2014:360230. doi: 10.1155/2014/360230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Armen R, Alonso DO, Daggett V. The role of alpha-, 3(10)-, and pi-helix in helix-->coil transitions. Protein Sci. 2003;12:1145–1157. doi: 10.1110/ps.0240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho BK, Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Struct Biol. 2005;5:14. doi: 10.1186/1472-6807-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho BK, Thomas A, Brasseur R. Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the alpha-helix. Protein Sci. 2003;12:2508–2522. doi: 10.1110/ps.03235203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc Natl Acad Sci U S A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stites WE, Meeker AK, Shortle D. Evidence for strained interactions between side-chains and the polypeptide backbone. J Mol Biol. 1994;235:27–32. doi: 10.1016/s0022-2836(05)80008-7. [DOI] [PubMed] [Google Scholar]
- 70.Pal D, Chakrabarti P. On residues in the disallowed region of the Ramachandran map. Biopolymers. 2002;63:195–206. doi: 10.1002/bip.10051. [DOI] [PubMed] [Google Scholar]
- 71.Pace CN, Fu H, Lee Fryar K, Landua J, Trevino SR, Schell D, Thurlkill RL, Imura S, Scholtz JM, Gajiwala K, Sevcik J, Urbanikova L, Myers JK, Takano K, Hebert EJ, Shirley BA, Grimsley GR. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23:652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palzkill T, Le QQ, Venkatachalam KV, LaRocco M, Ocera H. Evolution of antibiotic resistance: several different amino-acid substitutions in an active-site loop alter the substrate profile of beta-lactamase. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]