Abstract
Background & Aims
Peptic ulcer disease and gastric cancer are most often caused by Helicobacter pylori strains that harbor the cag pathogenicity island (cagPAI), which encodes a type IV secretion system (T4SS) that injects the CagA oncoprotein into host cells. cagY is an essential gene in the T4SS and has an unusual DNA repeat structure that predicts in-frame insertions and deletions. These cagY recombination events typically lead to a reduction in T4SS function in mouse and primate models. We examined the role of the immune response in cagY-dependent modulation of T4SS function.
Methods
H pylori T4SS function was assessed by measuring CagA translocation and the capacity to induce interleukin-8 (IL8) in gastric epithelial cells. cagY recombination was determined by changes in PCR restriction fragment-length polymorphisms. T4SS function and cagY in H pylori from C57BL/6 mice were compared to strains recovered from Rag1−/− mice, T and B cell deficient mice, mice with deletion of IFNGR or IL10, and Rag1−/− mice that received adoptive transfer of control or Ifng−/− CD4+ T cells. To assess relevance to humans, T4SS function and cagY recombination were assessed in strains obtained sequentially from a patient after 7.4 years of infection.
Results
H pylori infection of T-cell deficient and Ifngr1−/− mice, and transfer of CD4+ T cells to Rag1−/− mice, demonstrated that cagY-mediated loss of T4SS function requires a T-helper 1-mediated immune response. Loss of T4SS function and cagY recombination were more pronounced in Il10−/− mice, and in control mice infected with H pylori that expressed a more inflammatory form of cagY. Complementation analysis of H pylori strains isolated from a patient over time demonstrated changes in T4SS function that were dependent on recombination in cagY.
Conclusions
Analysis of H pylori strains from mice and from a chronically infected patient showed that CagY functions as an immune-sensitive regulator of T4SS function. We propose that this is a bacterial adaptation to maximize persistent infection and transmission to a new host under conditions of a robust inflammatory response.
Keywords: IL8, bacteria, adaptation, stomach
INTRODUCTION
Approximately 10% of those infected with Helicobacter pylori will develop peptic ulcer disease and 1–3% will progress to gastric adenocarcinoma1, the third most common cause of cancer death worldwide. The bacterial genetic locus most closely associated with development of peptic ulcer and gastric cancer is the H pylori cag pathogenicity island (cagPAI), a 40kb DNA segment that encodes a type IV secretion system (T4SS) that is essential for translocation of the CagA oncoprotein into host gastric epithelial cells2. A series of complex, T4SS-dependent changes in host cell signaling lead to actin cytoskeletal rearrangements, disruption of tight junctions, alterations in cell polarity, and the induction of proinflammatory cytokines, including interleukin-8 (IL8)3.
A functional T4SS that translocates CagA and induces IL8 requires 18 genes on the cagPAI, including cagY4. CagY is an orthologue of VirB10, an essential component in the canonical T4SS of Agrobacterium tumefaciens and closely related systems in Escherichia coli and other Gram-negative bacteria. Protein-protein interaction studies5 and negative stain electron microscopy6 in H pylori suggest that CagY also forms part of a 41 nm core complex, which is substantially larger than in E. coli or A. tumefaciens5. CagY is also much larger than VirB10, ~220 kDa depending on the H pylori strain, and it is encoded by a gene that contains an extraordinary number of direct DNA repeats. In silico predictions suggest that these DNA repeats would generate in-frame insertions or deletions via homologous recombination, yielding numerous theoretical variants of the cagY allele7. Immunogold labeling of CagY demonstrates that this repeat region is localized to the bacterial surface8. Thus, CagY has several features that distinguish it from other VirB10 orthologs, which suggests that it may be functionally unique.
It has been known for many years that passage of H pylori in mice leads to loss of T4SS function9, though the mechanism was unknown. We recently demonstrated that recombination in the cagY repeat region during colonization of mice often yields cagY variants that form a non-functional T4SS pilus that does not translocate CagA or induce IL8, though the CagY protein is expressed8. Similar observations were made in the rhesus macaque model, where we could also demonstrate CagY-mediated gain of T4SS function. Loss of T4SS function and recombination of cagY did not occur in Rag1−/− mice, which do not have functional B or T cells, suggesting that CagY-mediated modulation of T4SS function occurs in response to selective pressure by the adaptive immune system8.
H pylori infection of the gastric mucosa triggers a predominantly CD4+ T cell response that differentiates towards a Th1 phenotype, with expression of interferon gamma (IFNγ) and other proinflammatory cytokines that are essential for development of H pylori induced gastritis and control of bacterial burden10, 11. Here we used the mouse model to test the hypothesis that this Th1-biased immune response is also required for selection of cagY variants that have lost T4SS function during persistent H pylori infection. Using knockout mice and adoptive transfer experiments, we demonstrate that IFNγ and CD4+ T cells are essential for selection of cagY-mediated loss of T4SS function. Moreover, we show that cagY recombination and loss of T4SS function rescues H pylori colonization in Il10−/− mice, which have an exaggerated inflammatory response to H pylori infection. Analysis of paired patient isolates collected over many years demonstrates that cagY recombination can modulate T4SS function during chronic H pylori infection in humans. These results suggest that CagY functions as an immune-sensitive molecular regulator that modulates T4SS function.
METHODS
H pylori strains and culture
H pylori strains (Table S1) were cultured on brucella agar (BBL/Becton Dickinson, Sparks, MD) supplemented with 5% heat-inactivated newborn calf serum (NCS, Invitrogen, Carlsbad, CA) and ABPNV antibiotics (amphotericin B, 20 μg/ml; bacitracin, 200 μg/ml; polymyxin B, 3.3 μg/ml; nalidixic acid, 10.7 μg/ml; vancomycin, 100 μg/ml), unless otherwise indicated. Cultures were incubated at 37°C under microaerophilic conditions generated by a 5% CO2 incubator or by a fixed 5% O2 concentration (Anoxomat, Advanced Instruments, Norwood, MA).
Animals and experimental challenge
Specific-pathogen free female mice (Table S3) from Jackson Laboratories were housed in microisolator cages and provided with irradiated food and autoclaved water ad libitum. At 10 to 12 weeks of age mice were challenged with 2.5 × 109 CFU of H pylori suspended in 0.25 ml of brucella broth administered by oral gavage. Mice were euthanized between 2 and 16 weeks PI with pentobarbital sodium injection (50 mg/ml IP). Stomachs were cut longitudinally, and half was homogenized and plated by serial dilution on brucella agar supplemented with 5% NCS and ABPNV. Multiple single colony isolates (3-6/mouse) were characterized by cagY PCR-RFLP and for their capacity to induce IL8 in AGS gastric epithelial cells.
Study Approval
Experiments were carried out at the University of California, Davis under protocols approved by U.C. Davis Institutional Animal Care and Use Committee, which has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animal experiments were performed in accordance with NIH guidelines, the Animal Welfare Act, and U.S. federal law.
IL8 ELISA
H pylori induction of IL8 was measured as described previously12. WT H pylori PMSS1, its isogenic cagY deletion, and brucella broth were included on every plate as positive and negative controls. IL8 values were normalized to WT H pylori, arbitrarily set to 1.0.
Adoptive Transfer
Rag1−/−mice were reconstituted with 1×106 CD4+ T cells isolated from WT or Ifnγ−/− C57BL/6 mice, which had been infected for 8 weeks with PMSS1. Mice were euthanized with pentobarbital sodium injection (50 mg/ml IP) and single cell suspensions were obtained by passing spleens through a 40μm cell mesh into PBS. Cells were pelleted by centrifugation and erythrocytes lysed for 2 min at room temperature with AKC buffer (0.15M NH4Cl, 10mM KHCO3, 0.1mM EDTA, pH7.35). Cells were washed with PBS, pooled, and CD4+ T cells were isolated by using anti-CD4+ magnetic beads (Miltenyi, San Diego, CA) over a magnetic column, resulting in > 90% purity of CD3+CD4+ cells demonstrated by flow cytometry. CD4+ T cells were resuspended in PBS at 5×106 cells/ml per 200μl and injected into uninfected Rag1−/− mice via tail vein.
Flow Cytometry
Following RBC lysis, splenocytes were resuspended and washed once in FACS buffer (1XPBS, 0.5% BSA). Cells were stained with anti-CD3 FITC (clone 17A2, BD Biosciences) and anti-CD4 PE (clone RM4-5, BD Biosciences) in FACS stain buffer (FACS buffer, 20% mouse serum) for 15 minutes at room temperature. Cells were washed two times in FACS buffer and fixed with 2% PFA. Data were collected on a BD FACSCalibur and analyzed using FlowJo software 8.8.7 (Treestar).
Immunoblots and CagA translocation
Immunoblots of CagA and CagA translocation were performed as described previously using an MOI of 100:1 and 22 hours of culture at 37°C in 5% CO28. CagY expression was detected by electrophoresis of sonicated bacterial proteins on a 7.5% polyacrylamide gel, incubating with rabbit antiserum (1:10,000) to CagY7 as primary antibody and HRP-conjugated anti-rabbit IgG (1:20,000) as secondary antibody.
cagY PCR-RFLP
cagY genotyping was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using primers in Table S4 as previously described8. Changes in PCR-RFLP patterns are referred to as recombination based on DNA sequence analysis from prior experiments8, although this was not formally demonstrated.
Contraselection for genetic exchange of cagY
Alleles of cagY were exchanged between H pylori strains using contraselectable streptomycin susceptibility as described previously8 using primers in Table S4 and plasmid constructs in Table S2.
Competition experiment
cagY was deleted from PMSS1 and replaced with either wild type PMSS1 cagY (PMSS1ΔcagY[PMSS1]) or SS1 cagY (PMSS1ΔcagY[SS1]) using contraselection. Strains expressing cagY from PMSS1 or SS1 were also marked by replacing bases 343–360 of the rdxA locus with an antibiotic resistance gene encoding either kanamycin or chloramphenicol resistance, respectively. Briefly, plasmid pJ318 (Table S2) was constructed in pBluescript SK- by amplifying fragments 1194 bp upstream and 904 bp downstream of the rdxA deletion site in PMSS1, and ligating to a kanamycin resistance cassette13. pJ318 was then used to naturally transform PMSS1ΔcagY[PMSS1] with selection on 25 μg/ml kanamycin. pJ319 (Table S2), which was created in the same fashion but with a chloramphenicol resistance gene, was similarly used to naturally transform PMSS1ΔcagY[SS1], with selection on 5 μg/ml chloramphenicol. Gastric tissue from mice challenged with a 1:1 mixture of the marked PMSS1ΔcagY[PMSS1] and PMSS1ΔcagY[SS1] strains was plated separately on brucella agar with ABPNV plus either kanamycin or chloramphenicol to enumerate the relative abundance of each strain. This was expressed as a log10 competition index calculated as CFU of PMSS1ΔcagY [SS1]/[PMSS1], normalized to the abundance of each strain in the input inoculum.
Histology Scoring
Sections of the glandular stomach were formalin fixed and stained with hematoxylin and eosin. Each microscopic field was scored separately for the presence or absence of neutrophilic infiltration (polymorphonuclear leukocytes), gastritis, and epithelial metaplasia using a system previously validated in mice14. The results were reported as the mean percentage of fields affected for each mouse averaged over the three histologic criteria (minimum10 fields/sample).
Statistics
H pylori colonization (CFU/g) was analyzed using a 2-tailed Student's t test (Prism 6.0) after log10 transformation. Normalized IL8 levels were compared between groups using Wilcoxon rank-sum tests. Analysis of gastritis and proportions of samples with changed cagY were compared between groups using chi-square tests. In experiments with more than two groups, logistic regression was used to evaluate pairwise differences in proportion of output strains with recombination in cagY. A P value ≤ 0.05 was considered statistically significant.
RESULTS
CD4+ T cells are required for in vivo selection of cagY recombination and loss of T4SS function
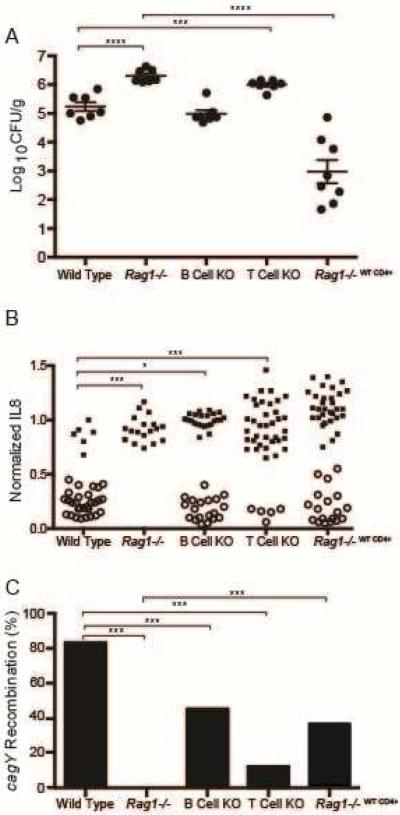
H pylori SS1 is a mouse-passaged strain that was proposed as the standard for H pylori studies in mice15, and was later found to have a defective T4SS that we showed was a result of cagY recombination8. PMSS1, which has a functional T4SS and readily colonizes mice, is the original H pylori human isolate that gave rise to the SS1 strain after serial passage mice16. Unlike in wild type (WT) mice, PMSS1 does not undergo cagY recombination or lose T4SS function when recovered from Rag1−/− mice8, which do not have functional B or T cells. To identify which arm of the adaptive immune response is responsible for loss of T4SS function, knockout (KO) mice lacking functional B cells (μMT) or T cells (TCR β/δ−/−) were challenged with PMSS1 and sacrificed 8 weeks post infection (PI). WT and Rag1−/− mice were challenged simultaneously as controls. The H pylori bacterial burden in Rag1−/− and T cell KO mice was approximately 10-fold higher than in WT mice and in B cell KO mice (Figure 1A). Loss of T4SS function (reduced IL8 induction) and recombination in cagY (defined as a change in PCR-RFLP) occurred commonly during infection of WT and B cell KO mice, but never in RAG−/− mice and only occasionally in T cell KO mice (Figure 1B, C).
Figure 1. CD4+ T cells are required to control H pylori colonization density and select strains with loss of T4SS function and recombination in cagY.
(A) H pylori colonization density was significantly greater in Rag1−/− and T cell KO mice than in wild type mice. Adoptive transfer of WT CD4+ T cells into Rag1−/− mice markedly reduced H pylori colonization compared to Rag1−/−. Each data point represents CFU/g for an individual mouse 8 weeks PI (N=7–8 mice/group). Horizontal lines indicate mean ± standard error of the mean (SEM). (B) Single colonies recovered from WT and B cell KO mice, and mice adoptively transferred with WT CD4+ T cells, showed marked loss in the capacity to induce IL8 that was accompanied by recombination in cagY (open circles). In contrast, all colonies from Rag1−/− mice and most from T cell KO mice induced IL8 and had the same cagY RFLP (closed circles) as WT H pylori PMSS1. Each data point represents the result from a single colony (N=3-6 colonies/mouse). (C) Percent of colonies that underwent cagY recombination (open circles divided by total colonies for each group in panel B). *P≤0.05, **P≤0.01, ***P≤0.001.
Since H pylori infection in mice primarily triggers a CD4+ Th1 immune response10, 11, we also asked if CD4+ T cells alone could select for H pylori with cagY alleles that encode a nonfunctional T4SS that is no longer capable of inducing IL8. To examine this possibility, we performed adoptive transfer experiments in which CD4+ T cells were isolated from WT mice that were infected with H pylori for 8 weeks, and then transferred into Rag1−/− mice (Rag1−/−WT CD4+) 24 hours before H pylori challenge. Flow cytometry on splenocytes from adoptively transferred mice demonstrated engraftment, with a mean of 8.9% (± 1.1% SEM) CD3+CD4+ cells in the lymphocyte gate. The bacterial burden 8 weeks PI was significantly lower in Rag1−/−WT CD4+ mice than in Rag1−/− mice (Figure 1A). It was also lower than in WT mice, which has been observed previously and likely reflects a relative failure of Treg engraftment17. Loss of T4SS function (Figure 1B) and recombination in cagY (Figure 1C) were also more common in Rag1−/−WT CD4+ mice than in RAG−/− mice, though the difference in IL8 induction did not reach statistical significance. Together, these data suggest that CD4+ T cells are essential for control of bacterial burden and for selection of H pylori with CagY variants that form a non-functional T4SS.
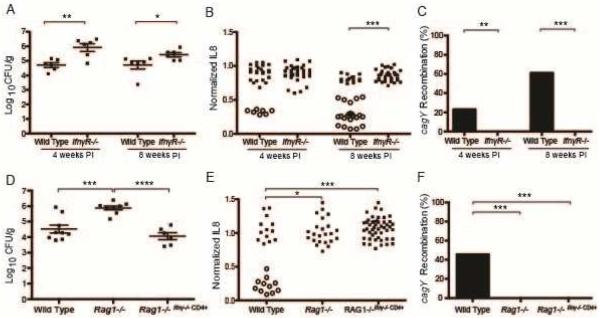
Selection of cagY variants and loss of T4SS function requires IFNγ signaling
The development of gastritis and control of H pylori bacterial burden are mediated by CD4+ T cells18, which also drive CagY-mediated loss of T4SS function (Figure 1). Since CD4+ T cells are a major source of IFNγ, we next asked if loss of T4SS function is mediated downstream of IFNγ. WT mice and mice lacking the IFNγ receptor (IfnγR−/−) were infected with H pylori PMSS1 and sacrificed 4 or 8 weeks PI. Similar to the bacterial burden in Rag1−/− mice, IfnγR−/− mice were colonized at approximately 10-fold higher levels than WT mice (Figure 2A). H pylori isolated from WT mice 4 and 8 weeks PI showed gradual loss of T4SS function associated with recombination in cagY. In contrast, H pylori from IfnγR−/− mice retained T4SS function (Figure 2B) and showed no cagY recombination (Figure 2C). To determine if IFNγ from CD4+ T cells alone is sufficient for selection of cagY variants and loss of T4SS function, we performed adoptive transfer. CD4+ T cells from Ifnγ−/− mice infected with PMSS1 for 8 weeks were adoptively transferred into Rag1−/−mice (Rag1−/−Ifnγ−/− CD4+), which were then infected with H pylori PMSS1 and sacrificed 8 weeks PI. Flow cytometry on splenocytes from adoptively transferred mice demonstrated engraftment, with a mean of 9.3% (± 0.7% SEM) CD3+CD4+ cells in the lymphocyte gate. Adoptive transfer of Ifnγ−/− CD4+ T cells was sufficient to control bacterial load (Figure 2D), but did not select H pylori variants with loss of T4SS function (Figure 2E) or recombination in cagY (Figure 2F). These results indicate that signaling downstream of IFNγ derived from CD4+ T cells is essential for CagY-mediated loss of T4SS function, but is not strictly required for control of H pylori bacterial load.
Figure 2. Selection of CagY variants is mediated downstream of IFN-γ signaling.
(A) H pylori colonization density was significantly higher in IfnγR−/− mice compared to WT at both 4 and 8 weeks PI. Each data point represents CFU/g from an individual mouse (N=6/group). (B) Single colonies (N=3-6/mouse) recovered from WT mice showed loss in the capacity to induce IL8 that was associated with recombination in cagY (open circles), but colonies from IfnγR−/− mice induced IL8 similarly to WT PMSS1 and had no changes in cagY (closed circles). (C) Percent of colonies that underwent cagY recombination (open circles divided by total colonies for each group in panel B). Adoptive transfer of Ifnγ−/− CD4+ T cells into Rag1−/− mice was sufficient to control bacterial load 8 weeks PI (D), but did not select H pylori variants with loss of IL8 induction or change in cagY PCR-RFLP (E,F). Horizontal lines indicate mean ± SEM. *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001
Variation in cagY functions as a molecular rheostat to alter the inflammatory capacity of H pylori
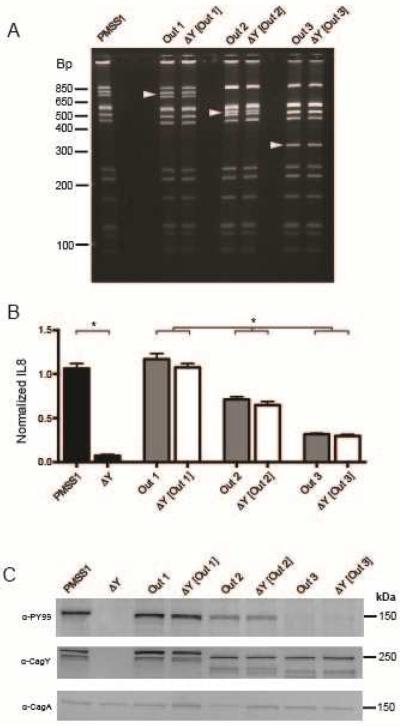
Most H pylori strains that recombined cagY showed a markedly reduced capacity to induce IL8, though we occasionally identified strains with changes in cagY but intermediate levels of IL8 induction (e.g., WT infected mice 8 weeks PI, Figure 2B). This suggests that recombination in cagY may function to modulate T4SS function rather than eliminate it. In other words, cagY may function more like a rheostat than a switch. To test this hypothesis, we first identified mouse output strains with unique cagY RFLP patterns (Figure 3A) that reproducibly showed high (Out1, cagY PCR-RFLP equivalent to WT PMSS1), intermediate (Out2), or low (Out3) induction of IL8 (Figure 3B, grey bars). We next used contraselection8 to replace the cagY gene in PMSS1 with that from each output, which was confirmed by PCR-RFLP (Figure 3A). Transformants complemented with cagY from output strains (ΔY[Out1], ΔY[Out2], ΔY[Out3]) restored the capacity to induce IL8 (Figure 3B, white bars) and translocate CagA (Figure 3C) to levels similar to that of the respective output strain. These results suggest that different cagY alleles vary in the extent to which they enable the bacterial cell to induce IL8 and translocate CagA, and that recombination in cagY functions as a molecular rheostat to modulate H pylori T4SS function.
Figure 3. CagY is a molecular rheostat that alters the inflammatory capacity of H pylori.
Three single colonies recovered from WT mice infected with PMSS1 (Out1, with cagY PCR-RFLP equivalent to wild type cagY from PMSS1; Out2; Out3) had unique cagY PCR-RFPL patterns (A), and induced high, intermediate, or low IL8, respectively (B) (gray bars) compared to PMSS1 and its cagY deletion mutant (black bars). Complementation of ΔcagY with Out1 (ΔY [Out1]), Out2 (ΔY [Out2]), or Out3 (ΔY [Out3]) phenocopied the IL8 induction of the respective output strain (white bars). Data represent mean ± SEM of four replicates. *P≤0.05. (C) Out1, Out2 and Out3 also demonstrated decreasing translocation of phosphorylated CagA (α-PY99), which was phenocopied when PMSS1ΔcagY was complemented with the respective cagY gene. Differences in CagY (α-CagY) were also apparent by immunoblot. Arrowheads in panel A indicate unique bands.
cagY recombination and loss of T4SS function are strain-dependent and associated with the capacity of H. pylori to induce inflammation
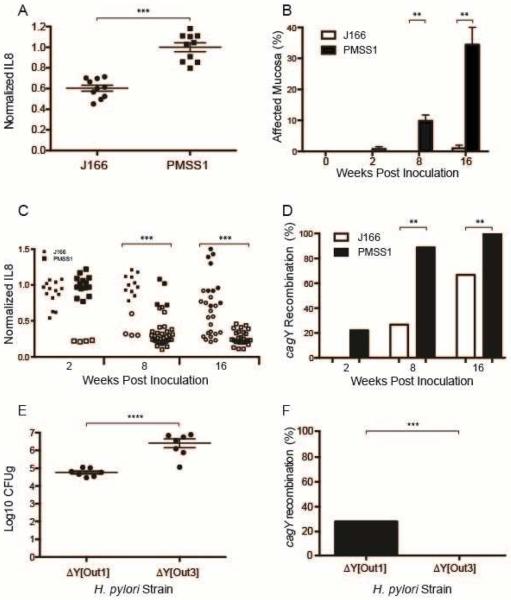
H pylori strains encoding a T4SS can differ markedly in their capacity to induce IL8, despite having an intact cagPAI19. Since loss of T4SS function and cagY recombination are immune driven, we hypothesized that the capacity of H pylori to induce IL8 would be inversely related to cagY recombination and loss of T4SS function during in vivo infection. To test this hypothesis, we first compared the in vitro response of AGS gastric epithelial cells to H pylori strains J166 and PMSS1, which show relatively low and high induction of IL8, respectively (Figure 4A). Mice infected with PMSS1 had significantly more inflammation in the gastric mucosa compared to J166 (Figure 4B, Figure S1), though bacterial loads were similar (data not shown). Consistent with the greater capacity of H pylori PMSS1 to induce IL8 in vitro, and induce inflammation in vivo, PMSS1 infected mice also showed more rapid and more complete loss of T4SS function that was associated with cagY recombination (Figure 4C,D).
Figure 4. Kinetics of cagY recombination and loss of T4SS function are associated with the capacity of H. pylori to induce inflammation.
(A) Replicate IL8 assays (N=10) for WT H pylori strains PMSS1 and J166. Data for both strains are normalized to PMSS1. (B) Mice infected with H pylori PMSS1 showed increased inflammation in gastric tissue compared to J166, which was statistically significant at 8 and 16 weeks PI (B). (C) Colonies recovered from PMSS1-infected WT mice lost the capacity to induce IL8 and changed cagY (open symbols) more rapidly and more completely than colonies recovered from J166-infected mice. Data for each strain are normalized to their respective WT. (D) Percent of colonies that underwent cagY recombination (open circles divided by total colonies for each group in panel C. H pylori recovered 8 weeks after challenge of WT mice with ΔY[Out1], which induces high IL8, were at a lower bacterial density (E) and underwent cagY recombination more frequently (F) than H pylori from mice colonized with ΔY[Out3]. Bars represent mean ± SEM. **P≤0.01, ***P≤0.001, ****P≤0.0001.
To examine this more systematically using isogenic strains, we next infected WT C57BL/6 mice with PMSS1 bearing the cagY from Out1 or Out3, which have high and low T4SS function (Figure 3), respectively, and sacrificed them 8 weeks PI. Colonization density was significantly greater in mice challenged with ΔY[Out3], which has poor T4SS function, compared to ΔY[Out1] (Figure 4E). Recombination in cagY occurred only in output colonies from mice infected with ΔY[Out1] (Figure 4F), though the frequency in this experiment was lower than observed previously. Similarly, complete elimination of T4SS function by deletion of cagE16, which encodes an ATPase that is essential for T4SS function, increased bacterial load and eliminated recombination in cagY 8 weeks PI (Figure S2). These data suggest that control of bacterial load and selection of H pylori with a nonfunctional T4SS are enhanced in H pylori strains that induce a more robust host immune response.
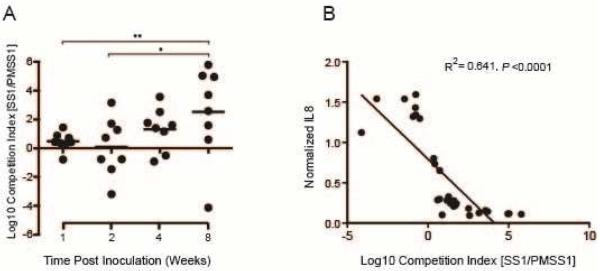
Competitive advantage of CagY-mediated loss of T4SS function increases progressively during H pylori infection
Recombination in cagY and loss of T4SS function increase over time during infection of WT mice, beginning around 4 weeks PI. Since loss of T4SS function is immune-mediated, this may simply reflect the time required for development of adaptive immunity. On the other hand, we previously reported that early during infection of rhesus macaques we could detect cagY-mediated gain of T4SS function8, suggesting the possibility that there may be selection for a functional T4SS very early during infection. To address this question, we used contraselection to construct isogenic strains of PMSS1 bearing either the WT cagY (PMSS1ΔcagY[PMSS1]) or the non-functional cagY from SS1 (PMSS1ΔcagY[SS1]), which were marked respectively in the neutral rdxA locus with antibiotic resistance to kanamycin or chloramphenicol. We then performed a competition experiment in which WT C57BL/6 mice were inoculated with a 1:1 mixture of both strains and sacrificed between 1 and 8 weeks PI. Gastric contents were plated on kanamycin and chloramphenicol to permit calculation of a competition index. The results demonstrated progressive selection for loss of T4SS function beginning 4 weeks PI, with > 300-fold competitive advantage by 8 weeks PI (Figure 5A). As expected, competition index showed a strong inverse correlation (R2=0.64, P≤0.0001) with IL8 induction performed on colony sweeps from each mouse (Figure 5B), which confirms that loss of T4SS function was due to selection for PMSS1ΔcagY[SS1], and not to a mutation in PMSS1ΔcagY[PMSS1]. These results are consistent with progressive loss of T4SS function that results from development of adaptive immunity, with no fitness advantage to a functional T4SS early during infection in mice.
Figure 5. Competitive advantage of CagY-mediated loss of T4SS function increases progressively over time.
(A) Output colonies from mice infected with an equal mixture of isogenic H pylori PMSS1 strains bearing either the functional (PMSS1) or non-functional (SS1) cagY allele were enumerated by selective plating, and used to calculate the log10 competition index. Each data point represents a single mouse; horizontal lines=geometric mean. At early time points there was no selective advantage, but by 8 weeks PI the PMSS1 strain bearing the SS1 cagY was present at > 300-fold greater abundance. *P≤0.05, **P≤0.01. (B) Normalized IL8 induction of a sweep culture from each mouse showed a strong inverse correlation with log10 competition index.
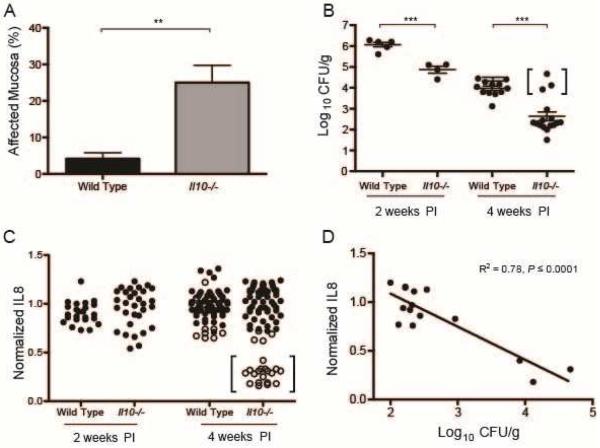
CagY-mediated loss of T4SS function promotes bacterial persistence in the setting of increased inflammation
CagY-mediated loss of T4SS function occurs less commonly in mice with impaired immunity (Figures 1, 2). This suggests that mice with an enhanced immune response might have a greater selection for H pylori strains with loss of T4SS function, which might be a bacterial strategy to persist in the face of inflammation. To test this hypothesis, we infected Il10−/− mice with H pylori, which triggers a robust inflammatory response with severe gastritis20, 21 and increased levels of IFNγ and other Th1 cytokines22 compared to WT mice. Because we anticipated that the aggressive inflammatory response would clear the infection at later time points, mice were sacrificed at 2 and 4 weeks PI, rather than 8 weeks PI as usual. Il10−/− mice infected with H pylori PMSS1 showed more inflammation (Figure 6A) and a lower bacterial burden (Figure 6B) compared to WT mice. Colonization in Il10−/− mice was significantly lower than in WT—often near the limit of detection (~100 CFU/g) 4 weeks PI (Figure 6B)—and was undetectable in 3 mice. At 4 weeks PI, recombination in cagY occurred in 22 of 82 colonies recovered from Il10−/− mice but only 10 of 75 colonies from WT mice (chi-square=4.23, P≤0.05, Figure 6C). All colonies recovered 4 weeks PI from the 3 mice that showed the highest colonization levels (similar to WT mice), also induced a low level of IL8 and showed recombination in cagY (Figure 6B,C; bracketed data points). Moreover, the average IL8 induction of H pylori isolates from Il10−/− mice showed a highly significant inverse correlation with the bacterial burden (Figure 6D). These data suggest that CagY-mediated loss of T4SS function allows for increased H pylori colonization in the face of a robust immune response.
Figure 6. CagY-mediated loss of T4SS function promotes bacterial persistence during an intense inflammatory response.
(A) Il10−/− mice inoculated with H pylori PMSS1 showed significantly increased gastritis compared to WT mice 4 weeks PI. (B) H pylori bacterial burden was significantly higher in WT compared to Il10−/− mice. By 4 weeks PI, bacterial burden in Il10−/− mice was frequently near the level of detection and 3 mice were uninfected (not shown). Mice whose CFU are shown in brackets yielded the colonies whose IL8 induction is shown in brackets in panel C. (C) Loss of the capacity to induce IL8 associated with changes in cagY PCR RFLP (open circles) was more apparent in H pylori colonies recovered from Il10−/− compared to WT mice, particularly in colonies from mice that showed colonization density that resembled that in WT mice. All colonies whose IL8 induction is shown in brackets in panel C were recovered from the mice whose CFU are bracketed in panel B. (D) Average normalized IL8 induction of all colonies from each mouse showed a strong inverse correlation with bacterial burden (R2=0.78, P≤0.0001). **P≤0.01,***P≤0.001.
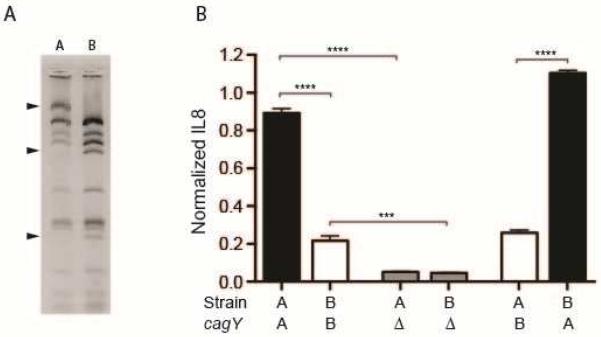
Recombination in cagY modulates T4SS function during chronic infection in humans
T4SS function (IL8 induction) is highly variable among H pylori clinical isolates, even when the cagPAI is fully intact19. The explanation for this is unknown, but it is intriguing that cagY is under strong diversifying selective pressure—second only to cagA among genes on the cagPAI19. To examine the possible role of cagY recombination in modulating T4SS function during chronic infection of humans, we examined paired isolates that were previously collected from 14 patients over intervals ranging from 3.0 to 10.2 yrs (mean=6.1 yrs). Multilocus sequencing typing analysis demonstrated that each pair was clonal, but showed microevolution during prolonged infection23. Of the 14 pairs, one showed changes in cagY PCR-RFLP (Figure 7A) together with a significant decrease in the capacity to induce IL8 between the A and B isolates, which were collected 7.4 years apart (Figure 7B). Deletion of cagY completely eliminated IL8 induction in both the A and B isolates (Figure 7B), which demonstrated that the T4SS was intact in both. To determine if recombination in cagY was responsible for the change in T4SS function, we used contraselection to exchange cagY genes between the A and B isolates, and confirmed it by PCR RFLP. Exchange of cagY genes demonstrated that change in cagY was sufficient to explain the differences in IL8 induction of the A and B isolates (Figure 7B). Control experiments in which cagY was deleted from the A and B strains, and then reinserted by contraselection, recovered the IL8 induction of the parent strain (data not shown). These results demonstrate that recombination in cagY during chronic human infection can modulate T4SS function.
Figure 7. Recombination in cagY modulates T4SS function during chronic infection in humans.
(A) cagY PCR-RFLP analysis of sequential A and B H pylori isolates. Arrowheads denote bands that changed in isolate A and B, which were collected from the same patient 7.4 years apart. (B) IL8 induction normalized to strain PMSS1 for sequential H pylori isolates A and B, their cagY knockouts (Δ), and strains in which their cagY genes have been exchanged. ***P≤0.001,****P≤0.0001.
DISCUSSION
The T4SS system encoded on the cagPAI is the key bacterial virulence factor associated with progression to peptic ulcer disease or gastric cancer, rather than asymptomatic gastritis. Analysis of the PAI in vivo has been hampered by the observation that T4SS function is lost during experimental infection of mice9, which was initially viewed as an artifact of infecting mice with a bacterium that is naturally found only in humans and some non-human primates. The mechanism was unknown. We recently demonstrated that loss of T4SS in mice is typically due to in-frame recombination in the middle repeat region of the cagY gene, which encodes an essential component of the H pylori T4SS4. Loss of T4SS does not occur in RAG−/− mice, which lack functional B or T cells8. While indels or SNPs in any of the essential genes can result in loss of T4SS function, cagY seems specifically designed for recombinatorial variation, suggesting that it is a bacterial contingency locus24 that modulates or “tunes” the host inflammatory response.
Here we have further investigated the immunologic basis and functional significance for loss of T4SS function during H pylori infection of mice and humans. Previous investigators speculated that recombination in cagY was a form of antigenic variation to avoid antibody responses directed against a surface-exposed component of the H pylori T4SS pilus7. However, this was difficult to reconcile with the general lack of human antibody response to CagY7 and the evidence that humoral immunity is generally not thought to play an important role in control of H pylori infection25. Our results suggest that loss of T4SS function and recombination in cagY are largely independent of B cells, but instead require CD4+ T cells expressing IFNγ (Figures 1, 2). The modest loss of T4SS function we observed in B cell KO mice (Figure 1B) may actually reflect a decrease in B cell-mediated immunoregulation26, rather than B cell control of infection. Together, these observations are consistent with seminal vaccine studies demonstrating that MHC class II-restricted, Th1-polarized T cells are essential to control H pylori infection27, 28.
Several lines of evidence suggest that the variation in cagY and T4SS function that we have observed in animal models is relevant to human H pylori infection. First, it is not simply an artifact of the mouse model, because cagY mediated loss of T4SS function also occurs in rhesus macaques8, which most closely mimic human infection. Second, cagY recombination can both up- and down-modulate T4SS function8, in a graded fashion (Figure 3), suggesting that this observation is a window into the biology of H pylori, which actually has the capacity to “tune” or optimize the host inflammatory response to achieve a homeostatic balance. Third, there is marked variability in the capacity of different PAI positive H pylori strains to induce IL8, varying by up to 20-fold19. Since cagY is second only to cagA in the percentage of codons under positive selection19, CagY diversity may be important for adaptation to chronic human infection. Most importantly, here we demonstrate that cagY recombination within an individual patient can modulate T4SS function (Figure 7). The cagY variants in the sequential isolates might represent the dominant population present at each time point, which underwent recombination and functional change under pressure from changes in, for example, host physiology. Alternatively, we cannot exclude the possibility that the cagY variants we observed represent diversity that was present at each time point, because only a single A and B isolate were examined. Regardless of which scenario is correct, these results provide proof-of-principle that cagY recombination can modulate T4SS function during H pylori infection in humans. H pylori adaptation during acute and chronic human infection has also been demonstrated at other virulence loci, such as the babA adhesion29, 30, and more broadly using whole genome sequencing31.
When might cagY recombination occur during human infection, and why? Clearly, a functional T4SS enhances bacterial fitness, probably by increasing its capacity to acquire iron and other nutrients32–34. But rearrangement in cagY that confers loss of T4SS function may be more advantageous under conditions that are unfavorable for H pylori growth, because loss of T4SS function decreases the host inflammatory response, increases bacterial load16, 35, and thus increases the likelihood of transmission to a new host. It may occur soon after acquisition, as in mice and monkeys, or perhaps during some environmental event, which from the bacterial perspective tips the balance for or against inflammation. If so, it may be difficult to “catch it in the act” because acute H pylori infection is rarely detected in humans and the hypothetical environmental events are unknown. One possibility is that cagY-mediated down regulation of T4SS function is a bacterial strategy to persist in the setting of an intercurrent infectious disease such as malaria, tuberculosis or typhoid—which, along with H pylori, have evolved with humans since antiquity and might cause sufficient systemic inflammation that would otherwise reduce H pylori colonization and perhaps even clear the infection. A similar phenomenon has been observed experimentally in the mouse model of herpes viruses, where viral infection can non-specifically protect against bacterial challenge with Listeria or Yersinia pestis by inducing IFNγ and systemic activation of macrophages36. This hypothesis is also supported by the results in Il10−/− mice, which have an exaggerated inflammatory response to H pylori and demonstrate a strong inverse correlation between bacterial load and cagY-mediated loss of T4SS function (Figure 6). Alternatively, perhaps cagY-mediated loss of T4SS enhances bacterial persistence in the setting of atrophic gastritis, in which elevated gastric pH reduces H pylori burden and sometimes leads to bacterial clearance37. Both are testable hypotheses that we are currently examining.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grants R01 AI081037 and R01 AI108713 to JS from the National Institutes of Health. RB was partially supported by NIH training grant T32AI060555 to JS. Statistical support was provided by the National Center for Advancing Translational Sciences, National Institutes of Health grant UL1 TR000002. We thank Angela Green for technical assistance with flow cytometry. The study sponsor had no role in the design, data collection, analysis, or interpretation of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no conflict of interest exists.
Author contributions: RB, LH, AL, ES, MM, LC, YL, AL, and DC performed experiments, and collected and analyzed data. RB, LH, ES, MM, YL, and SS edited the manuscript. RB and JS planned and designed the experiments, analyzed data, and wrote the manuscript. JS obtained funding.
REFERENCES
- 1.Wroblewski LE, Peek RM, Jr., Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–39. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odenbreit S, Püls J, Sedlmaier B, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 3.Segal ED, Lange C, Covacci A, et al. Induction of host signal transduction pathways by Helicobacter pylori. Proc Natl Acad Sci U S A. 1997;94:7595–9. doi: 10.1073/pnas.94.14.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer W, Püls J, Buhrdorf R, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 5.Kutter S, Buhrdorf R, Haas J, et al. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190:2161–71. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frick-Cheng AE, Pyburn TM, Voss BJ, et al. Molecular and Structural Analysis of the Helicobacter pylori cag Type IV Secretion System Core Complex. MBio. 2016;7 doi: 10.1128/mBio.02001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aras RA, Fischer W, Perez-Perez GI, et al. Plasticity of repetitive DNA sequences within a bacterial (Type IV) secretion system component. J Exp Med. 2003;198:1349–60. doi: 10.1084/jem.20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrozo RM, Cooke CL, Hansen LM, et al. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9:e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philpott DJ, Belaid D, Troubadour P, et al. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell Microbiol. 2002;4:285–96. doi: 10.1046/j.1462-5822.2002.00189.x. [DOI] [PubMed] [Google Scholar]
- 10.Bamford KB, Fan X, Crowe SE, et al. Lymphocytes in the human gastric mucosa during Helicobacter pylori have a T helper cell 1 phenotype. Gastroenterol. 1998;114:482–92. doi: 10.1016/s0016-5085(98)70531-1. [DOI] [PubMed] [Google Scholar]
- 11.Lundgren A, Trollmo C, Edebo A, et al. Helicobacter pylori-specific CD4+ T cells home to and accumulate in the human Helicobacter pylori-infected gastric mucosa. Infect Immun. 2005;73:5612–9. doi: 10.1128/IAI.73.9.5612-5619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Israel DA, Salama N, Arnold CN, et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 2001;107:611–20. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton KA, Danon SJ, Krakowka S, et al. A reproducible scoring system for quantification of histologic lesions of inflammatory disease in mouse gastric epithelium. Comp Med. 2007;57:57–65. [PubMed] [Google Scholar]
- 15.Lee A, O'Rourke J, De Ungria MC, et al. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterol. 1997;112:1386–97. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 16.Arnold IC, Lee JY, Amieva MR, et al. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterol. 2010;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray BM, Fontaine CA, Poe SA, et al. Complex T cell interactions contribute to Helicobacter pylori gastritis in mice. Infect Immun. 2013;81:740–52. doi: 10.1128/IAI.01269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayi A, Kohler E, Hitzler I, et al. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J Immunol. 2009;182:7085–101. doi: 10.4049/jimmunol.0803293. [DOI] [PubMed] [Google Scholar]
- 19.Olbermann P, Josenhans C, Moodley Y, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Shu D, Chadwick VS. Helicobacter pylori infection: mechanism of colonization and functional dyspepsia Reduced colonization of gastric mucosa by Helicobacter pylori in mice deficient in interleukin-10. J Gastroenterol Hepatol. 2001;16:377–83. doi: 10.1046/j.1440-1746.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- 21.Ismail HF, Fick P, Zhang J, et al. Depletion of neutrophils in IL10(−/−) mice delays clearance of gastric Helicobacter infection and decreases the Th1 immune response to Helicobacter. J Immunol. 2003;170:3782–9. doi: 10.4049/jimmunol.170.7.3782. [DOI] [PubMed] [Google Scholar]
- 22.Lee CW, Rao VP, Rogers AB, et al. Wild-type and interleukin-10-deficient regulatory T cells reduce effector T-cell-mediated gastroduodenitis in RAG2−/− mice, but only wild-type regulatory T cells suppress Helicobacter pylori gastritis. Infect Immun. 2007;75:2699–707. doi: 10.1128/IAI.01788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morelli G, Didelot X, Kusecek B, et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6:e1001036. doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–33. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 25.Ermak TH, Giannasca PJ, Nichols R, et al. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted responses. J Exp Med. 1998;188:2277–88. doi: 10.1084/jem.188.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayi A, Kohler E, Toller IM, et al. TLR-2-activated B cells suppress Helicobacter-induced preneoplastic gastric immunopathology by inducing T regulatory-1 cells. J Immunol. 2011;186:878–890. doi: 10.4049/jimmunol.1002269. [DOI] [PubMed] [Google Scholar]
- 27.Akhiani AA, Pappo J, Kabok Z, et al. Protection against Helicobacter pylori infection following immunization is IL-12-dependent and mediated by Th1 cells. J Immunol. 2002;169:6977–6984. doi: 10.4049/jimmunol.169.12.6977. [DOI] [PubMed] [Google Scholar]
- 28.Myers GA, Ermak TH, Georgakopoulos K, et al. Oral immunization with recombinant Helicobacter pylori urease confers long-lasting immunity against Helicobacter felis infection. Vaccine. 1999;17:1394–403. doi: 10.1016/s0264-410x(98)00387-9. [DOI] [PubMed] [Google Scholar]
- 29.Moonens K, Gideonsson P, Subedi S, et al. Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori. Cell Host Microbe. 2016;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nell S, Kennemann L, Schwarz S, et al. Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans. MBio. 2014;5 doi: 10.1128/mBio.02281-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linz B, Windsor HM, McGraw JJ, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat Commun. 2014;5:4165–4172. doi: 10.1038/ncomms5165. [DOI] [PubMed] [Google Scholar]
- 32.Noto JM, Lee JY, Gaddy JA, et al. Regulation of Helicobacter pylori virulence within the context of iron deficiency. J Infect Dis. 2014;211:1790–94. doi: 10.1093/infdis/jiu805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan S, Noto JM, Romero-Gallo J, et al. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7:e1002050. doi: 10.1371/journal.ppat.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rieder G, Merchant JL, Haas R. Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils. Gastroenterol. 2005;128:1229–42. doi: 10.1053/j.gastro.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 36.Barton ES, White DW, Cathelyn JS, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–9. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 37.Karnes WE, Jr., Samloff IM, Siurala M, et al. Positive serum antibody and negative tissue staining for Helicobacter pylori in subjects with atrophic body gastritis. Gastroenterol. 1991;101:167–74. doi: 10.1016/0016-5085(91)90474-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.