Abstract
The family Rhabdoviridae consists of mostly enveloped, bullet-shaped or bacilliform viruses with a negative-sense, single-stranded RNA genome that infect vertebrates, invertebrates or plants. This ecological diversity is reflected by the diversity and complexity of their genomes. Five canonical structural protein genes are conserved in all rhabdoviruses, but may be overprinted, overlapped or interspersed with several novel and diverse accessory genes. This review gives an overview of the characteristics and diversity of rhabdoviruses, their taxonomic classification, replication mechanism, properties of classical rhabdoviruses such as rabies virus and rhabdoviruses with complex genomes, rhabdoviruses infecting aquatic species, and plant rhabdoviruses with both mono- and bipartite genomes.
Keywords: Rhabdovirus, Negative-sense RNA virus, Genome organization, Diversity, Replication, Taxonomy
Graphical abstract
1. Introduction – Properties and diversity of rhabdoviruses
Rhabdoviruses are a large and ecologically diverse group of viruses, which infect terrestrial and aquatic vertebrates, invertebrates and plants. They include many pathogens of significance to public health, agriculture and fisheries (Dietzgen and Kuzmin, 2011). The majority of rhabdoviruses are transmitted by arthropods to vertebrate or plant hosts, but lyssaviruses (e.g. rabies virus) and novirhabdoviruses (e.g. infectious hematopoietic necrosis virus) have evolved to circulate among vertebrates without a biological vector, and sigmaviruses (e.g. Drosophila melanogaster sigma virus) are congenitally transmitted in fruit flies. High throughput host genome sequencing has revealed the integration of rhabdovirus-like elements into the genomes of some arthropods and plants (Ballinger et al., 2012; Fort et al., 2012; Chiba et al., 2011; Katzourakis and Gifford, 2010; Li et al., 2015), suggesting an ancient evolutionary origin and long-standing association of rhabdoviruses with their hosts.
Rhabdoviruses are taxonomically classified in thirteen genera in the family Rhabdoviridae, order Mononegavirales, which also includes families Bornaviridae, Filoviridae, Paramyxoviridae and recently established Nyamiviridae (Dietzgen et al., 2011; Walker et al., 2015). The name is based on their characteristic shape and derived from the Ancient Greek word rhabdos meaning “rod or wand”. Classic rhabdoviruses typified by vesicular stomatitis virus (VSV) and sonchus yellow net virus (SYNV) form characteristic bullet- or cone-shaped (animal host; Ge et al., 2010) or bacilliform (plant host; Jackson et al., 2005) enveloped virions that contain non-segmented, negative-sense, single-stranded (ss) RNA genomes of 11–16 kb in length (Figs. 1 and 2). Virions range in size from 100 to 430 × 45 to 100 nm. The basic genome organisation shared by all rhabdoviruses includes five canonical genes that encode (from 3′ to 5′) the nucleoprotein (or nucleocapsid protein, N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and large protein (L, RNA-dependent RNA polymerase) (Fig. 2). This group of structural protein genes is flanked by regulatory 3′ leader and 5′ trailer sequences that show terminal complementarity and contain promoter sequences to initiate replication. The individual genes are flanked by conserved transcription stop and start signals separated by short untranscribed intergenic sequences (Dietzgen and Kuzmin, 2012). The infectious nucleocapsid core [a ribonucleoprotein (RNP) complex], which is active in transcription and replication, consists of the genomic RNA that is always tightly associated with N protein, together with P and L proteins. M protein is responsible for condensation of RNP complex during virion assembly at the host plasma membrane, and the transmembrane spike protein G likely plays an important role in assembly, budding and host cell entry (Dietzgen and Kuzmin, 2012).
Fig. 1.
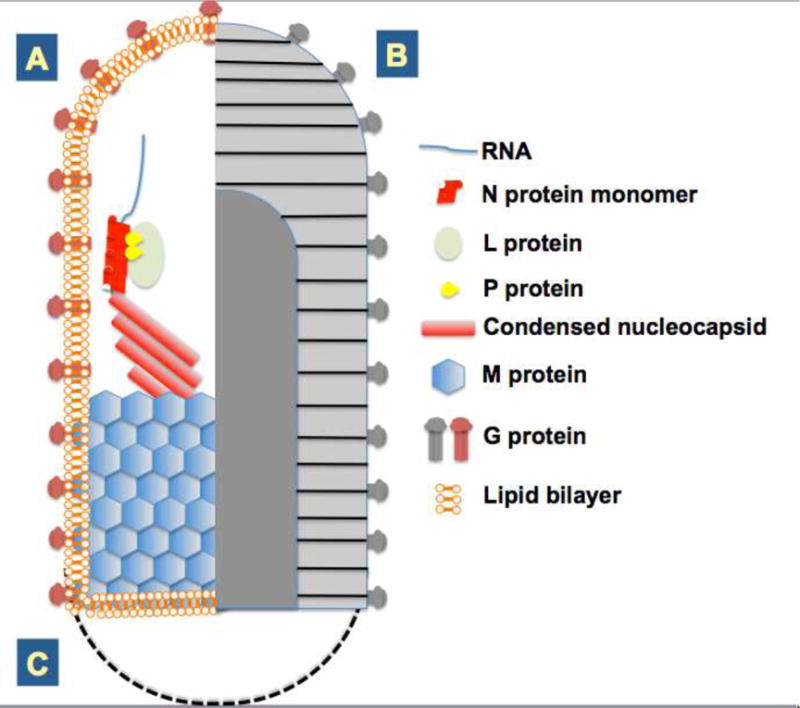
A. (left half of figure). Schematic representation of rhabdovirus particle internal structure. The single-stranded negative-sense genomic RNA is encapsidated along its entire length by the nucleoprotein N. Associated with the L polymerase and P phosphoprotein, transcriptionally-competent nucleocapsids represent the minimal infectious unit of rhabdoviruses. During virion maturation the nucleocapsid is condensed by the matrix protein, and this complex ultimately buds through host membranes to acquire the lipid envelope and transmembrane glycoprotein present in mature virus particles. B. (right half of figure). External virus particle appearance. Negatively-stained particles appear striated when examined by electron microscopy. Glycoprotein spikes decorate the surface of virions and the central cavity fills with stain which contrasts the space occupied by the matrix-protein condensed nucleocapsid. C. While A and B together show the “rhabdoid” or bullet shape typical of rhabdoviruses adapted to mammalian cells, the virions of plant-adapted rhabdoviruses are typically bacilliform in shape, represented here by a dashed line.
Fig. 2.
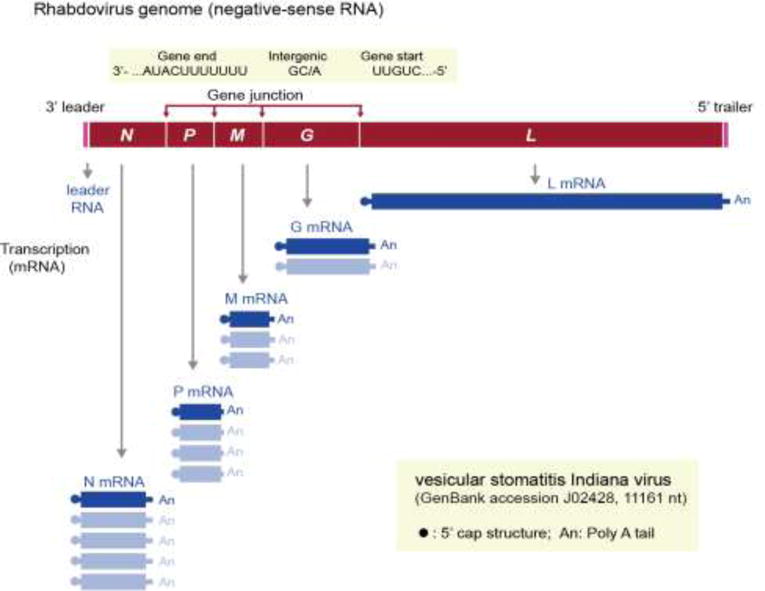
Schematic representation of the 3′ to 5′ genome organization (negative-sense) and gene expression of vesicular stomatitis Indiana virus (VSIV, genus Vesiculovirus). VSIV genome encodes the following five canonical rhabdovirus proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and large protein (L, polymerase). The genes are sequentially transcribed presumably through a “stop-start” mechanism, resulting in a 3′–5′ polar gradient of mRNA production (N>P>M >G>L) (see section 3).
Rhabdovirus genomes are diverse and often complex because the five canonical genes may be overprinted, overlapped or interspersed with a range of novel accessory genes, such that the number of genes range from 5–10 or more (Walker et al., 2011; 2015; Fig. 3). Among RNA viruses, rhabdovirus genomes have an unusual capacity for plasticity and appear to have frequently acquired and lost new genes during evolution (Walker et al., 2015). Furthermore, two groups of plant-infecting rhabdoviruses (genera Dichorhavirus and Varicosavirus) were recently found to have a divided genome with the L gene located on a separate RNA segment (Dietzgen et al., 2014; Kormelink et al., 2011). For other reviews on rhabdoviruses see Blondel et al. (2015), Walker et al. (2011), Dietzgen and Kuzmin (2011), Dietzgen et al. (2011), Ammar et al. (2009) and Jackson et al. (2005).
Fig. 3.
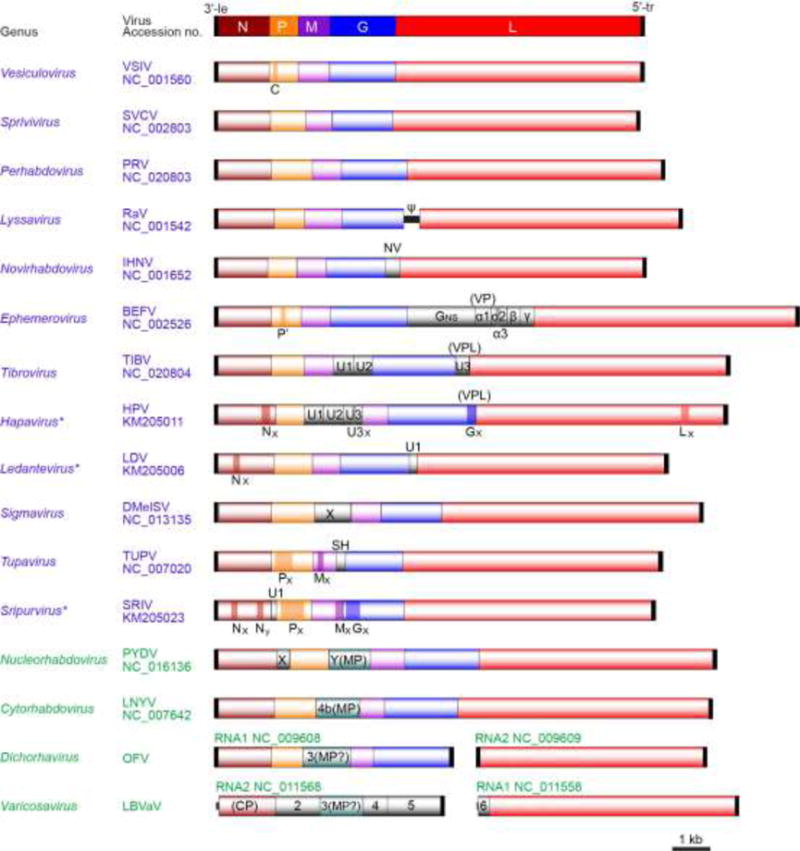
Comparative genome organization of representative members from sixteen genera of the family Rhabdoviridae (Dietzgen et al., 2011; 2014, Walker et al., 2015). The five canonical structural protein genes (N, P, M, G and L) are shaded in different colors. Other genes including movement protein (MP), viroporin (VP) or viroporin-like protein (VPL) and unknown function protein genes are shaded in grey. Several overlapping and consecutive ORFs (>150 nt) within each transcriptional unit are also shown with dark color (see Walker et al., 2015). The 3′-leader (le) and 5′-trailer (tr) regions are not scaled. Virus names (abbreviation of the member of type species of genera) and Refseq numbers are shown. Asterisks indicate proposed genera.
2. Taxonomic classification – past, present and future
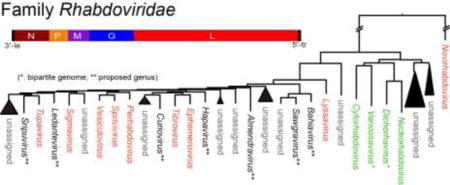
Rhabdovirus taxonomy is evolving rapidly in an effort to incorporate ever-increasing numbers of new viral sequences obtained through high throughput technologies and to harmonize classifications across genera and families in the order Mononegavirales. In the 9th report of the International Committee on Taxonomy of Viruses (ICTV) the family Rhabdoviridae comprised six genera (Dietzgen et al., 2011). In the 2015 Taxonomy Release [http://www.ictvonline.org/virusTaxonomy.asp], thirteen genera were recognised: Cytorhabdovirus, Dichorhavirus, Ephemerovirus, Lyssavirus, Novirhabdovirus, Nucleorhabdovirus, Perhabdovirus, Sigmavirus, Sprivivirus, Tibrovirus, Tupavirus, Varicosavirus and Vesiculovirus (Fig. 4). Among these, in the recently recognized Dichorhavirus and Varicosavirus genera (Afonso et al., 2016; Adams et al., 2016) for the first time, virus species containing plant-infecting negative-sense ssRNA viruses with bipartite genomes have been classified as rhabdoviruses based on significant genome sequence identities with nucleo- and cytorhabdoviruses, respectively. Furthermore, binomial species names that indicate which genus the species belongs to have been introduced for all rhabdovirus species, to facilitate easier differentiation between virus names and the names of taxonomic species these viruses are classified in (Afonso et al., 2016; Adams et al., 2016). For example, rabies virus is a virus in the species Rabies lyssavirus, and potato yellow dwarf virus is a virus in the species Potato yellow dwarf nucleorhabdovirus. Based on the recent large-scale discovery and analysis of vertebrate and arthropod rhabdoviruses, several new genera have been proposed (e.g. Almendravirus, Bahiavirus, Curiovirus, Hapavirus, Ledantevirus, Sawgravirus and Sripuvirus) (Walker et al., 2015; Longdon et al., 2015; Li et al, 2015) (Fig. 4).
Fig. 4.
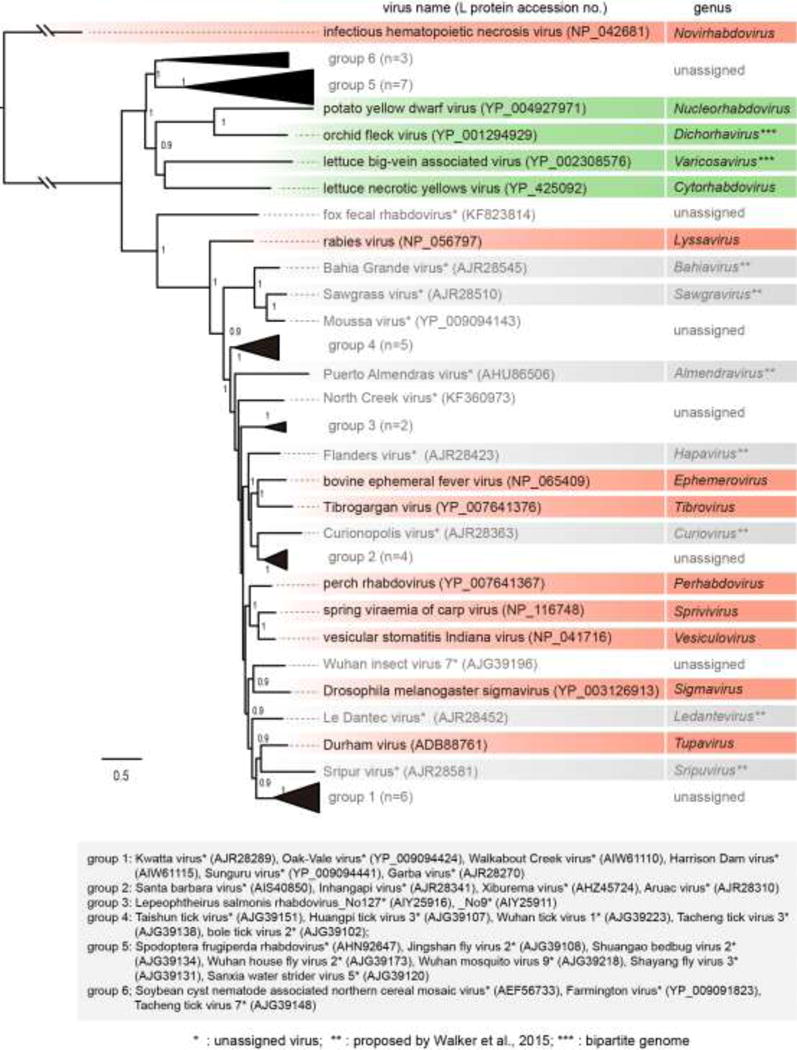
Phylogenetic relationships of members of the family Rhabdoviridae. The tree was constructed by the maximum likelihood (ML) method as described previously (Kondo et al., 2015). The entire L polymerase sequences were aligned using MAFFT 7.0 (Katoh and Toh, 2008) under default settings and ambiguously aligned regions were removed using Gblocks 0.91b (Talavera and Castresana, 2007) with all the options of less stringent selection. A model LG with + I + G + F was selected as the best fit model using PhyML 3.0 (Guindon et al., 2010) with automatic model selection by Smart Model Selection (SMS). The resulting tree was visualized with the FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/). Numbers at the nodes indicate aLRT values determined using an SH-like calculation (values less than 0.9 are not displayed). Virus names (the member of type species of genera and other selected unclassified rhabdoviruses) and GenBank/Refseq accession numbers (within parentheses) of L proteins are shown. Distantly related rhabdoviruses (group 1 to 6; n, number of sequences) that formed monophyletic clades, probably establishing additional genera in the family, were collapsed into a black triangle. The names and accession numbers of these viruses are shown below the tree.
3. Replication, transcription and translation
The rhabdovirus replication mechanism is almost universal across the family. Replication mechanisms of some plant-infecting rhabdoviruses differ slightly due to the plant cell environment and the establishment of replication factories in the nucleus (rather than the cytoplasm) for nucleorhabdoviruses and dichorhaviruses. Otherwise, the universal pathway of the cytoplasmic replication cycle follows (i) cell entry, facilitated by clathrin-mediated or receptor-binding endocytosis (or vector-mediated penetration of the plant cell wall); (ii) uncoating; (iii) transcription and translation; (iv) genome replication and encapsidation; and (v) assembly and release (budding). Fusion of endocytosed virus with endosomes and its subsequent lysis releases the RNP complex into the cytoplasm allowing for the initiation of early transcription and replication events. A critical step in this process is the dissociation of the M protein from the nucleocapsid (Mire et al., 2010), which is required for the initiation of viral transcription (Clinton et al., 1978; Pal et al., 1985) also called primary transcription. The term denotes the short nature of transcription from parental templates as opposed to subsequent prolonged transcription events from progeny templates (secondary transcription) following genome replication. Transcription of the negative-stranded genome is facilitated by a transcriptase complex and occurs progressively on a decreasing molar gradient based on gene distance from the genomic 3′ end (for example, N→P→M→G→L) (Fig. 2). The relative abundance of each viral mRNA and thus each protein is regulated by the disassociation of the transcriptase from the RNA template once it reaches the respective gene transcription termination polyadenylation (TTP) signal at the end of each viral gene. This stop-start (‘stuttering’) sequential gene transcription mechanism is governed by the interaction of cis-acting signals (e.g. TTP) located on the genome template and the transcriptase complex (Abraham and Banerjee 1976). The cis-acting signals are well conserved with some minor variations (Walker et al., 2015). In canonical genome architectures each gene junction consists of conserved sequence motifs, originally identified for vesicular stomatitis viruses as (i) a TTP (3′-AUACUUUUUUU-5′), whose function is to polyadenylate and terminate the upstream mRNA (Rose 1980; Barr et al., 1997a; Hwang et al., 1998); (ii) a non-transcribed intergenic dinucleotide (G/CA) (Rose 1980; Stillman and Whitt, 1997; Barr et al., 1997b); and (iii) a transcription initiation (TI) pentanucleotide sequence (3′-UUGUC-5′) (Rose 1980), whose function is to initiate, cap and methylate the down stream mRNA. Exception to this mechanism is observed in the initiation of transcription between the 3′ leader sequence and the N gene. The leader is encoded by the 3′-terminal nucleotides of the genome (47 nt for VSIV) that lack the U7 sequence, resulting in transcription of a short leader RNA that lacks a polyA tail and cap (Whelan and Gertz, 1999) (Fig. 2). The leader sequence primary function is to serve as promoter for the initiation of RNA synthesis, whereas its newly synthesised complementary sequence facilitates the encapsidation of nascent RNA, thus allowing transcriptional read-through to generate full-length complementary genomes (i.e. antigenomes). Thus there are two possible outcomes for the polymerase complex at the end of each gene junction: (i) either traverse the intergenic dinucleotide and initiate again at the TI motif of the downstream gene; or (ii) dissociate from the RNA template at the gene junction resulting in an apparent attenuation of the downstream gene and its protein expression, as a function of each gene’s distance from the 3′ end of the genome (Iverson and Rose, 1981; reviewed in: Lyles et al., 2013).
The production and accumulation of the virally encoded proteins signals a switch in the polymerase function, from viral mRNA transcription to genome replication, in which N plays a critical role. An essential step in the viral replication of the nascent positive-sense genome (antigenome) relies on its encapsidation, a process facilitated by cis-acting conserved sequences located on the 3′ ends of viral genome and antigenome (Whelan and Wertz, 1999; Li and Pattnaik, 1999). Additionally, N and P proteins are critical in promoting genome replication, as the N/P complex provides the structural and chaperone support for the nascent RNA to bind via sugar-phosphate interactions to the N protein (Albertini et al., 2006). The bound antigenome will then function as template for the synthesis of encapsidated negative-sense genomes, which will be assembled into progeny virions.
Virion assembly is a staggered process where the various components [nucleocapsid core (RNP), G and M proteins] are sequestered in different cellular compartments and converge in the final steps of the process. The nucleocapsid is assembled during RNA replication in the cytoplasm, as is observed for members of the genera Vesiculovirus, Lyssavirus, Ephemerovirus and Novirhabdovirus. Viral G protein is inserted into the endoplasmic reticulum where chaperones (BiP and calnexin) (Hammond and Helenius, 1994) facilitate its proper folding and assembly into trimers (Doms et al., 1988), prior to transport and fusion into the Golgi complex. As it traffics through the cell it undergoes further posttranslational modifications including glycosylations (Schmidt and Schlesinger, 1979), prior to its transport to cholesterol- and sphingolipid-rich lipid rafts in the baso-lateral plasma membrane. M protein is synthesized mostly as a soluble protein in the cytoplasm (McCreedy et al., 1990) and is also membrane bound, albeit at lower amounts (Ogden et al., 1986). However both forms of the M protein are recruited for assembly of nucleocapsid/M complexes at the host plasma membrane from where virions will bud (Odenwald et al., 1986). This budding process is facilitated by the interaction of M with host-encoded proteins responsible for the formation of multivesicular bodies (MVB), and their release from the plasma membrane (Harty et al., 2001).
4. ‘Classical’ vertebrate rhabdoviruses
For historical reasons any reference to classical vertebrate rhabdoviruses denotes members of the genera Vesiculovirus and Lyssavirus, represented by the prototype species vesicular stomatitis Indiana virus (VSIV) and rabies virus (RABV), respectively. Vesiculoviruses have a wide host range among mammals and are transmitted by hematophagous insects (sandflies and/or mosquitoes). Lyssaviruses utilize mostly bats as their principal reservoir hosts as well as various terrestrial carnivores as terminal hosts. Viruses of each genus form a monophyletic clade in a maximum likelihood (ML) tree inferred from complete L protein sequences (Dietzgen et al., 2011; Walker et al., 2015). Structurally both demonstrate the classic rhabdovirus enveloped bullet-shaped virions (Fig. 1) packaging a genome consisting of five genes (3′-N-P-M-G-L-5′), each separated by a short gene junction (intergenic region), and flanked by highly conserved 3′ leader (le) and 5′ trailer (tr) sequences (Fig. 3). In vesiculoviruses the P gene mRNA contains 2 additional alternate start codons that initiate translation at alternative open reading frames (ORFs) that encode two small basic proteins C and C′ (55-aa and 65-aa, respectively) of unknown function (Spiropoulou and Nichol, 1993; Peluso et al., 1996). Suppression of C/C′ expression has no apparent effects in virus replication or pathogenicity in vivo (Kretzschmar et al., 1996). Of note is that not all members of the genus express alternative ORFs in P [e.g. vesicular stomatitis Alagoas, Maraba, Malpais Spring, Morreton viruses] (Walker et al., 2015), and additional ORFs (≥ 150 nt) may be present in alternative reading frames in other genes than P (Walker et al., 2015).
For a long time lyssaviruses were considered antigenically unique represented by various RABV isolates. However the discovery of other RABV-like viruses in expanded geographic regions of the globe allowed initially the establishment of the genus Lyssavirus, followed by its demarcation into four distinct groups anchored by RABV, Lagos bat, Mokola and Duvenhage viruses, based solely on their serological relationships. The advent of Sanger sequencing permitted the determination of lyssavirus genetic signatures, thus allowing phylogenetic relationships to be further refined and their classification into the currently recognized 11 species was aided by a number of species demarcation criteria including genetic distances, phylogenies, antigenic relationships and biological properties (Lyles et al, 2013). Vesiculoviruses have been classified into 10 species using similar criteria (Dietzgen et al., 2011). The massive generation of sequencing data based on new technologies (Walker et al., 2015) and the need for consistency across taxa necessitates refinement of these criteria. This will undoubtedly recalibrate the richness of vesiculovirus diversity in the future (Peter Walker, personal communication).
While the level of diversity within the family Rhabdoviridae is being realized at an unprecedented rate due to new detection and sequencing methods (e.g. next generation sequencing, metagenomics), reverse genetics of the prototype vesiculoviruses (Schnell et al., 1994; Whelan et al., 1995) allowed exploration of rhabdovirus plasticity and evolution at the genetic level, as well as an understanding of the factors influencing gene expression and molecular and cellular basis of pathogenesis, and their use as vaccine delivery vehicles (reviewed in: Whitt et al., 2016). The error-prone nature of their RNA-dependent RNA polymerase results in the generation of a rich diversity of genetic variants following each replication cycle, which are subject to adaptation due to selective pressures under different conditions, such as alternative hosts (vector vs vertebrate) during the course of their natural transmission (Novella et al., 2010; Novella et al., 2011; Wasik et al., 2016). Similarly recombinant RABVs with targeted mutations in P gene demonstrated its critical role in suppression of interferon signaling through blocking the interaction of the transcription factor IRF-3 (interferon regulatory factor 3) with two cell-expressed protein kinases (Brzozka et al., 2005), ultimately altering pathogenesis in vivo (Rieder et al., 2011). On the other hand, recombinant VSVs were critical in demonstrating the role of M in inhibiting host gene expression without affecting virus assembly, a process facilitated through the formation of complexes with the mRNA export factor Rae1 (Faria et al., 2005), which interferes with the function of downstream factors (Connor et al., 2006) essential for host gene expression.
5. Vertebrate rhabdoviruses with complex genomes
Several of the currently recognized genera within the family Rhabdoviridae show associations with a dominant group of vertebrate hosts, such as the ephemeroviruses and tibroviruses that are hosted by cattle, with many viruses either isolated from cattle and/or from mosquitoes or biting midges that feed on cattle (Walker 2005; Gubala et al., 2011). Others like the recently designated ledanteviruses (Blasdell et al., 2015), not only exhibit a strong ecological association with bats, but also a broader natural host specificity (e.g. ungulates, rodents and humans) suggesting spill over from their natural reservoir. A recent landmark study extended the range of the known rhabdovirus genome complexity to include viruses in proposed new genera Hapavirus, Ledantevirus and Sripuvirus among others (Walker et al., 2015). While the canonical genome organization of the prototype rhabdoviruses features the five ORFs arranged in the order 3′-N-P-M-G-L-5′, rhabdovirus genomes may also be more complex and contain additional ORFs encoding putative proteins of unknown function. These may occur in alternative and/or overlapping ORFs within the major structural protein genes or as independent ORFs flanked by TI or TTP sequences in the regions between the structural protein, some of which may have arisen by gene duplication.
Ephemeroviruses have among the largest genomes of all rhabdoviruses. The prototype virus, bovine ephemeral fever virus (BEFV) has a genome approximately 14.8 kb in length that contains 10 genes separated by short intergenic regions (Walker 2005) (Fig. 3). Due to an apparent duplication of the viral G gene there is an additional gene for a non-structural glycoprotein (GNS) that has a significant amino acid sequence homology and is synthesized at same levels as the G protein, but it is not incorporated into the virion envelope and thus does not induce any antibody responses (Hertig et al., 1996). GNS is followed by a viroporin gene (α1), probably encoding a viral ion channel protein, and several other accessory genes (α2, β and γ) (Walker et al., 2015).
Hapaviruses infect birds, reptiles or mammals and many have been isolated from culicine mosquitoes. They exhibit complex genomes, form a monophyletic group based on well-supported ML trees generated from complete L protein sequences and are anchored by the prototype Hart Park virus (HPV) (Walker et al., 2015) (Fig. 3). In addition to the five canonical genes encoding the structural proteins, three long ORFs, U1, U2 and U3, with significant amino acid sequence homology with each other, are located in independent transcriptional units between the P and M genes (Walker et al., 2015). A fourth ORF lies within the G gene transcriptional unit and is thought to encode a class IA viroporin. Alternative long ORFs in different reading frames in the N gene, U3 gene and L gene are observed but may not be expressed as functional proteins due to poor Kozak context and distal location far from the start of their transcriptional units (Walker et al., 2015). Ngaingan virus (NGAV) has the longest genome of any rhabdovirus described to date (Gubala et al., 2010) with 15,764 nt containing 13 genes. Three of the eight additional ORFs are located in two transcriptional units between the P and M genes, and another lies in a transcriptional unit between the M and G genes, likely encoding a unique protein of unknown function. Additionally, there are four ORFs between the G and L genes, including GNS, which encodes a class I transmembrane protein that is related in sequence to the NGAV G protein and the G proteins of other rhabdoviruses, ORFs likely encoding two unique proteins of unknown function, and a viroporin-like protein. Lastly, alternative ORFs are present within the P and M genes, but likely are not expressed due to poor Kozak context and distal location far from the start of their transcriptional units (Walker et al., 2015).
Ledanteviruses infect humans, rodents, and ungulates, may be vectored by arthropods and have a strong association with bats, suggesting spill over from their natural reservoir (Blasdell et al., 2015; Walker et al., 2015). They are anchored by the prototype Le Dantec virus (LDV), whose genome comprises the five canonical structural protein genes, an additional transcriptional unit (U1), encoding a small protein between the G and L genes, and a small ORF that occurs in an alternative reading frame in the N gene, which likely is not expressed (Fig. 3).
Tibroviruses infect ungulates and are anchored by the prototype Tibrogargan virus (TIBV). Members of the genus Tibrovirus share the same unique genome organisation comprising five genes encoding the canonical rhabdovirus structural proteins and three additional genes encoding ORFs (U1, U2 and U3) (Fig. 3). ORF U1 and ORF U2 encode small proteins of unknown function, whereas ORF U3 encodes a small viroporin-like protein (Walker et al., 2015). Each ORF lies within an independent transcriptional unit bounded by consensus TI and TTP sequences.
While the genus Tupavirus consists of only three species, Tupaia tupavirus, Durham tupavirus and Klamath tupavirus (Allison et al., 2011; Johnson 1965; Kurz et al., 1986; Walker et al., 2015), tupaviruses are characterized by their extensive geographic distribution and host range. The genome of the prototype Tupaia virus (TUPV) includes in addition to the five genes encoding the canonical rhabdovirus structural proteins, an alternative ORF in the P gene and an ORF encoding a small hydrophobic protein located in an independent transcriptional unit between the M and G genes, as observed among vesiculoviruses (Fig. 3). Uniquely amongst tupaviruses, Klamath virus also contains an additional ORF encoding a small protein (U2) within an independent transcriptional unit between the G and L genes. Although the functions of these novel genes are currently unknown, it has been speculated that some may play a role in enhancement of transcriptional activity, host pathogenicity or insect transmission (Allison et al., 2011; Kretzschmar et al., 1996; Peluso et al., 1996).
The proposed genus Sripuvirus consists of five new species. Almpiwar, Chaco and Sena madueira viruses were isolated from reptiles, while Niakha and Sripur viruses were isolated from sandflies (Causey et al., 1966; McAllister et al., 2014; Monath et al., 1979; Vasilakis et al., 2013; Walker et al., 2015). Sripuvirus genomes are anchored by the prototype Sripur virus (SRIV), are similar in size and contain multiple ORFs encoding likely accessory proteins (Fig. 3). In addition to the five canonical rhabdovirus genes encoding the structural proteins, sripuvirus genomes also feature six other ORFs. Two reside within alternative reading frames in the N gene, encoding small proteins which likely are not expressed due to poor Kozak context and distal location far from the start of their transcriptional units (Walker et al., 2015). U1 resides as an independent transcriptional unit between the N and P genes, whereas Px resides within an alternative reading frame near the start of the P gene. Interestingly, the Mx resides within the M gene with its initiation codon overlapping the termination codon of the M gene. Gx resides within an alternative ORF near the start of the G gene, encoding a small double-membrane-spanning protein likely expressed by ‘leaky’ ribosomal scanning (Walker et al., 2015) (Fig. 3).
The genome complexity and plasticity observed in the genera described above reiterates the richness and diversity of the family Rhabdoviridae and suggests that different mechanisms may drive their genome expansion and evolution. These mechanisms may include: (i) function acquisition and rapid adaptation, (ii) gene duplication, (iii) loss of redundant ORFs through mutation, and (iv) deletion of redundant genome sequences followed by optimization of gene expression levels. A comprehensive overview of these mechanisms is presented in Walker et al. (2015).
6. Rhabdoviruses in aquatic systems
Rhabdoviruses of aquatic hosts (reviewed in Kurath and Winton, 2008) include important fish pathogens in three genera: Novirhabdovirus, Sprivivirus, and Perhabdovirus (Dietzgen et al., 2011). To date these genera contain only viruses of finfish hosts, and there are no fish viruses in other rhabdovirus genera. Due to the poikilothermic (cold-blooded) nature of their hosts, these viruses have temperature optima ranging from 15–25°C. In addition to these distinguishing ecological factors, genera of fish rhabdoviruses differ from other rhabdovirus genera, and from each other, by genetic divergence levels and placement of their constituent viruses in strongly supported monophyletic clades in phylogenetic analyses of both nucleotide and amino acid sequences. All viruses in these three genera have non-segmented negative-sense ssRNA genomes of approximately 11,000 nt. They are transmitted horizontally as waterborne viruses, and from parent to offspring as egg-associated viruses, without requiring a vector. They have been well studied due to the long history of severe disease impacts caused by some fish rhabdoviruses in freshwater and marine fish farms, netpen aquaculture, and conservation fish hatcheries. In the Aquatic Animal Health Code of the International Organization for Animal Health, (World Organization for Animal Health, 2014) fish rhabdoviruses cause three of the ten globally reportable aquatic diseases: infectious haematopoietic necrosis, viral haemorrhagic septicaemia, and spring viraemia of carp.
The Sprivivirus and Perhabdovirus genera currently have 2 and 3 viral species, respectively, and each species includes numerous viruses, mostly isolated from fish in Asia and Europe, including the United Kingdom (reviewed in Stone et al., 2013). Spriviviruses infect primarily freshwater fish species in the order Cypriniformes (e.g. common carp), and perhabdoviruses have broader host ranges, infecting many diverse species. The genomes of spriviviruses and perhabdoviruses are comprised of the conserved five canonical rhabdovirus genes and their genome organization, including transcription start and stop signals, is very similar to that of mammalian vesiculoviruses (Fig. 3). They are also most closely related to vesiculoviruses in phylogenetic analyses (Fig. 4). Indeed, the type species of the Sprivivirus genus, Spring viraemia of carp virus (Carp sprivivirus), was formerly classified in the Vesiculovirus genus until sequences of additional fish viruses revealed the two clearly distinct monophyletic groups comprising these fish rhabdovirus genera (Stone et al., 2013). Spring viraemia of carp viruses (SVCV) cause severe disease in farmed carp in Europe and Asia, and are reviewed in Ahne et al. (2002).
The Novirhabdovirus genus includes viruses in four species that differ in both geographic and host range (Leong and Kurath, 2012). Infectious haematopoietic necrosis viruses (IHNV) infect primarily salmon and trout species in the fish order Salmoniformes, and originated in western North America (Bootland and Leong, 2011). In contrast, viral haemorrhagic septicaemia viruses (VHSV) have an extremely broad host range, infecting over 60 host species from diverse taxonomic families, comprising a large marine fish reservoir in both the north Atlantic and north Pacific oceans (Skall et al., 2005; Smail and Snow, 2011). Viruses of the other two novirhabdovirus species occur in Asia, where hirame rhabdoviruses (HIRRV) infect cultured hirame (olive flounder), and snakehead virus has been less well characterized (Leong and Kurath, 2012). Novirhabdovirus genomes differ from those of the viruses in other fish rhabdovirus genera in having an additional gene encoding a non-virion (NV) protein between the G and L genes (Kurath and Leong, 1985; Kurath et al., 1997) (Fig. 3). The NV protein is expressed at low levels in infected cells (Schuetze et al., 1996), where it localizes to the nucleus and interferes with the host interferon response (Choi et al., 2011), as well as it triggers apoptosis (Ammayappan and Vakharia, 2011). Despite having a virion structure, genome organization, and sequence similarities clearly related to those of other rhabdoviruses, novirhabdoviruses fall phylogenetically far basal to the other genera, confirming their separation from the other fish rhabdovirus genera, and potentially indicating an ancestral role in the evolution of the rhabdovirus family (Fig. 4).
Due to the economic impact of IHNV, VHSV, and SVCV, routine surveillance for these viruses is conducted in cultured host populations, and molecular epidemiology is a valuable tool for fish health management (Breyta et al., 2016; Emmenegger et al., 2011; Jonstrup et al., 2009). Local, regional, and global phylogenies have revealed sources of disease outbreaks, virus transmission routes, virus emergence events and host jumps (e.g. Einer-Jensen et al., 2004; Enzmann et al., 2010; Kurath et al., 2003; Kurath, 2012; Nishizawa et al., 2006). Virus traffic between wild and cultured fish populations has also been documented, and risk factors for potential virulence evolution in aquaculture have been described (Kennedy et al., 2016; Kurath and Winton 2011). Genotyping of virus strains has confirmed global spread of fish rhabdoviruses due to aquaculture (reviewed in Kurath, 2012).
In laboratory studies fish viruses are investigated using cultured fish cell lines and in in vivo infection experiments in numerous fish host species. The host immune response to fish rhabdovirus infection consistently shows both rapid, strong innate immunity based on stimulation of interferon-related genes, and adaptive immunity based on antibody and cellular defences (Purcell et al., 2012; Verrier et al., 2011). An interesting aspect of host immunity is the dramatic effect of temperature, which impacts both virus replication and persistence, and the speed of the immune response in poikilothermic hosts. Numerous vaccines have been investigated for protection against IHNV, VHSV, SVCV, and HIRRV (reviewed in Winton, 2007). DNA vaccines containing the viral glycoprotein gene have been shown to be highly efficacious for IHNV, VHSV, and HIRRV, providing relative survival rates greater than 90% in numerous studies (Kurath, 2008; Lorenzen and LaPatra, 2005; Takano, 2004). Due to their consistent high efficacy the VHSV and IHNV DNA vaccines have also been studied extensively as models for DNA vaccines in vertebrate hosts (Kurath et al., 2007; Lorenzen et al., 2002).
In other laboratory work a research model for in vivo viral fitness and competition studies has been developed based on IHNV in rainbow trout (Troyer et al., 2008; Wargo et al., 2011). Reverse genetics systems have been developed for viruses in three of the four novirhabdovirus species, facilitating investigation of questions such as the role of the accessory NV gene, and determinants of virulence and host-specificity (Johnson et al., 2000; Biacchesi et al., 2000; Ammayappan et al., 2011). These studies have shown great flexibility of fish rhabdovirus genomes, such that gene exchanges between different viruses reliably generate viable chimeric viruses, and foreign genes can be inserted and expressed for various purposes (reviewed in Biacchesi, 2011). In a particularly interesting study a recombinant IHNV carrying a reporter luciferase gene was used to infect fish, and bioluminescence imaging on the living fish revealed that the fin bases were a major portal of viral entry (Harmache et al., 2006). Thus aquatic rhabdoviruses are both important veterinary pathogens and models for basic studies of rhabdovirus biology at the molecular and landscape levels.
7. Monopartite plant rhabdoviruses: Cyto- and nucleorhabdoviruses
Classic plant rhabdoviruses, like the majority of viruses in the family Rhabdoviridae have a non-segmented negative-sense ssRNA genome. They have been taxonomically separated into the genera Cytorhabdovirus and Nucleorhabdovirus based on their site of replication and morphogenesis in the cytoplasm or nucleus of infected plant cells, respectively (Fig. 5). This classification based on cytopathology has been confirmed by phylogenetic studies of available genome sequences. The biology of plant rhabdoviruses including ecology, cytopathology, vector associations, particle structure, genome organization and genetic variability have been described in a comprehensive review by Jackson et al. (2005).
Fig. 5.
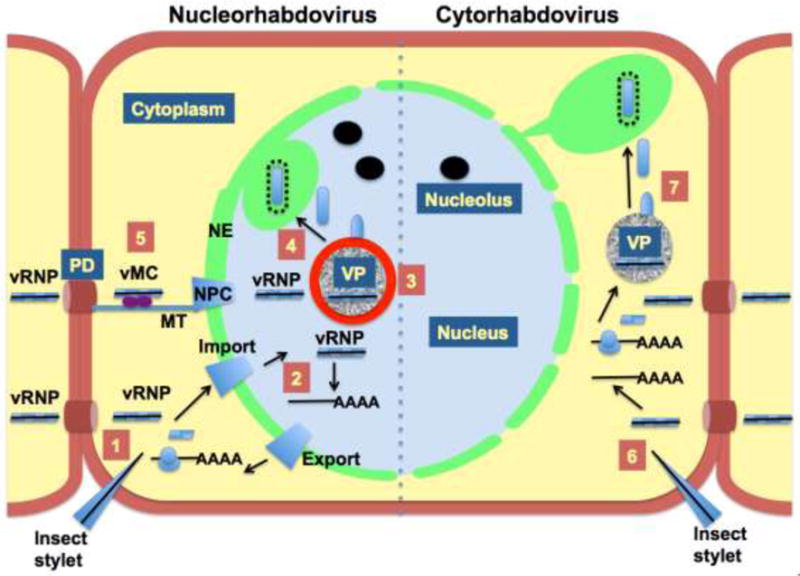
Comparison of the infection cycles of nucleo- and cytorhabdoviruses in plant cells. 1. Nucleorhabdoviruses are initially introduced into plant cells via a mechanical breach of the cell wall by insect vectors or experimentally by abrasion. The introduced viral ribonucleoprotein (vRNP) is imported into nucleus due to interaction with the nuclear import receptor importin-alpha. 2. Within nucleus the vRNP establishes transcription of poly(A)+ transcripts that are exported from the nucleus. Translation of the viral mRNAs in the cytoplasm produces proteins that are imported into nucleus where they aggregate to form the nuclear viroplasms where replication occurs (VP). 3. Nascent nucleocapsids are condensed on the perimeter of the VP, which is enriched in the phosphoprotein (sonchus yellow net virus, SYNV; Goodin, unpublished). 4. Matrix protein condensed nucleocapsids bud into the perinuclear space where mature virions accumulate. In the case of SYNV, the inner membrane of the nuclear envelope (NE) invaginates into the nuclear VP to form virus-containing spherules. 5. Mature virions do not move cell to cell, but may be transmitted to other plants via insect feeding. The vRNP exits the nucleus using the nuclear pore complex (NPC). In the cytoplasm the viral movement complex (vMC) is composed of the vRNP, the viral movement protein, and host factors, which are trafficked towards plasmodesmata (PD) on microtubules (MT), resulting in cell-to-cell spread of the virus. 6. Cytorhabdoviruses are introduced into cells in a manner similar to nucleorhabdoviruses, but carry out their replication cycles in the cytoplasm. 7. Transcription and translation of cytorhabdovirus vRNPs produce the protein pools required to form cytoplasmic VPs for replication. Condensed vRNPs mature in cytoplasmic proliferations on endoplasmic membranes. As above, mature virions do not move from cell to cell, and cytorhabdovirus vRNP are expected to traffic to the PD via a similar vMC mechanism.
Since then, the number of completely sequenced genomes has increased to 8 and 9 for cyto- and nucleorhabdoviruses, respectively. A series of plasmid vectors for transient agrobacterium-mediated expression of autofluorescent protein fusions and confocal microscopy have allowed comparative studies of intracellular localisation and interactions of rhabdoviral proteins in planta (e.g. Bandyopadhyay et al., 2010; Martin et al., 2012). These studies revealed essential protein-protein interactions like N:P that were observed for all studied rhabdoviruses, as well as interactions unique to a particular virus, and allowed the construction of specific interactome maps. Additionally, expression of tagged proteins in the context of virus-infected cells has provided insight into the differential localization of viral proteins within nuclei. SYNV (a nucleorhabdovirus) P protein localizes to ring structures that appear to define the viroplasm/nucleoplasm border. The N protein localizes onto both viroplasm and membranes, while the G and M proteins are confined solely to membranes, consistent with their roles in morphogenesis (Goodin et al., 2007). It is important to note that different nucleorhabdoviruses can induce markedly different cytological transformations. Infection by SYNV results in the formation of intranuclear spherules derived from the inner nuclear membrane, whereas potato yellow dwarf virus (PYDV) does so to a much lesser extent (Goodin et al., 2005). Similarly, the PYDV M protein induces intranuclear accumulation of the inner nuclear membrane when expressed alone (Bandyopadhyay, et al., 2010), an activity not observed with M proteins of other characterized plant-infecting rhabdoviruses.
Cell-to-cell movement function of the presumed viral movement proteins (MP) (accessory gene located between P and M genes) has been demonstrated for both selected cyto- and nucleorhabdoviruses using heterologous movement trans-complementation assays (Huang et al., 2005; Mann et al., 2016a). A recent study showed that P3 of two cytorhabdoviruses localised to plasmodesmata, the intercellular cytoplasmic bridges that the cross cell walls between plant cells, and acted as 30K-superfamily-like MP requiring a conserved LxD/N50-70G motif. Furthermore, involvement of a host microtubule-associated transcription factor has been suggested in cell-to-cell movement of both a cyto- and nucleorhabdovirus (Min et al., 2010; Mann et al., 2016a). In fact, several host factors were identified as potentially playing critical roles in SYNV (a well studied nucleorhabdovirus) nucleocapsid export from the nucleus and cell-to-cell transport by cytoplasm-tethered transcription activators (Min et al., 2010). Regarding other accessary genes, three transcriptional units are present between the P and M genes of a cereal cytorhabdovirus (barley yellow striate mosaic virus) and one of them has a second ORF that may encode a small hydrophobic (SH) protein, reminiscent of the SH proteins of several animal rhabdoviruses (Walker et al., 2015; Yan et al., 2015).
Plant viruses including rhabdoviruses have evolved RNA silencing suppressors (RSS) to interfere with host RNA silencing defences (Csorba et al., 2015). The P protein of a cytorhabdovirus, lettuce necrotic yellows virus, was recently identified as a local RSS that does not effect small-interfering RNA (siRNA) accumulation but that interacts with plant RNA silencing machinery proteins by inhibiting micro RNA-guided ARGONAUTE 1 cleavage and translational repression, as well as RNA-dependent RNA Polymerase 6/Suppressor of Gene Silencing 3 (RDR6/SGS3)-dependent silencing amplification (Mann et al., 2016b). The P6 accessary gene product of a rice nucleorhabdovirus, rice yellow stunt virus, was also shown to have RSS activity. P6 did not interfere with local RNA silencing, but with systemic RNA silencing by affecting the function of RDR6 during secondary siRNA synthesis (Guo et al., 2013).
The recent development of the first-ever reverse genetics system for a plant rhabdovirus provided a major technical breakthrough and a guide for the future study of other plant negative-sense ssRNA viruses (Ganesan et al., 2013; Wang et al., 2015; Jackson and Li, 2016). It allowed the recovery of infectious recombinant virus (rSYNV) from agroinfiltrated plants and the generation of rSYNV stably expressing a green fluorescent protein (GFP) reporter. Deletion analyses of rSYNV-GFP demonstrated the involvement of the sc4 (P3) protein in cell-to-cell movement and the importance of the G protein in virion morphogenesis (Wang et al., 2015).
8. Bipartite plant rhabdoviruses: Dichorha- and varicosaviruses
Within the Dichorhavirus genus, orchid fleck virus (OFV) and coffee ringspot virus (CoRSV) cause local chlorotic or necrotic spot symptoms (and/or systemic symptoms in some cases of OFV) in susceptible plant host species. They share several characteristics with nucleorhabdoviruses including nuclear cytopathological effects, structural protein composition, gene order, significant sequence identity and transcriptional mechanism, but they have a bipartite genome and their particles do not appear to be enveloped, although they may be found associated with host membranes (Dietzgen et al., 2014; Kondo et al., 2006; 2009; Ramalho et al., 2014). Dichorhaviruses have short bacilliform virions (40 × ~100–110 nm) and their genomes consist of two negative-sense ssRNA segments. RNA1 (~ 6.4 kb) encodes 3′-N-P-P3-M-G-5′ while RNA2 (~ 6.1 kb) encodes the L polymerase (Kondo et al., 2006; Ramalho et al., 2014). Both termini of each RNA segment are complementary, and all genes are separated by conserved intergenic regions, similar to other rhabdoviruses (Kondo et al., 2014). The proteins encoded by RNA1 appear to be nucleophilic based on in silico predictions, intracellular localisation of in planta expressed proteins and yeast nuclear import assays, suggesting viral replication in nuclear viroplasms, similar to nucleorhabdoviruses (Kondo et al., 2013; Ramalho et al., 2014). All known dichorhaviruses are transmitted by false spider mites (Brevipalpus spp.) in a persistent and probably propagative manner (reviewed in Dietzgen et al., 2014). OFV has been found worldwide due to the global exchange and trade of orchid plants. In contrast to OFV, CoRSV has been reported in only two countries, Brazil and Costa Rica (Bittancourt 1938; Chagas et al., 1981; Rodrigues et al., 2002). Whereas phylogenetic analysis of OFV showed little geographical relationship between isolates, genetic variation in CoRSV is largely dependent on the distance between collection sites (Ramalho et al 2016). This suggests that CoRSV-infected plants are not transported across great distances in coffee producing areas (the virus is not seed-transmitted), and that spread may be limited by short-range movement of the mite vectors. In support of the latter, evidence suggests that the population structure of CoRSV is likely defined by habitat expansion of thelytokous populations of its Brevipalpus vector (Ramalho et al., 2016).
Citrus leprosis virus nuclear type (CiLV-N) and citrus necrotic spot virus appear to be strains of OFV based on 90% or higher genome sequence identity (Cruz-Jaramillo et al., 2014; Dietzgen et al., 2014; Roy et al., 2015). By degradome sequencing, a distinct CiLV-N isolate was also found in a herbarium citrus specimen from Florida in 1948 (Hartung et al., 2015). These citrus strains and all other known dichorhaviruses have been restricted to the Americas.
In the Varicosavirus genus, lettuce big-vein associated virus (LBVaV) has non-enveloped, flexuous rod-shaped virions, 18 × 320–360 nm in size, which in electron micrographs appear similar to the nucleocapsid core of classic rhabdoviruses. LBVaV is transmitted by a soil-inhabiting chytrid fungus (Olpidium virulentus) and is distributed worldwide (Maccarone, 2013). This virus is frequently associated with lettuce big-vein disease that is caused by Mirafiori lettuce big-vein virus (a multi-segmented, negative-sense ssRNA virus, genus Ophiovirus). The LBVaV genome consists of two negative-sense ssRNA segments; RNA1 (6.7 kb) encodes a small ORF of unknown function and the L polymerase, whereas RNA 2 (6.1 kb) encodes the coat protein (CP, the N protein homolog) followed by 4 ORFs of unknown function thought to be equivalent to the plant rhabdoviral P, P3, M and G proteins (Kormelink et al., 2011; Walsh and Verbeek, 2011). The genome structure and transcription mechanism appears similar to the other rhabdoviruses with a moderate level of amino acid sequence identities in the CP/N and L proteins (Sasaya et al., 2004).
9. Rhabdovirus-like sequences in host genomes
Several rhabdovirus-like sequences integrated into plant and arthropod genomes have been identified (Ballinger et al., 2012; Fort et al., 2012; Chiba et al., 2011; Katzourakis and Gifford, 2010; Li et al., 2015), while no such sequences were detected in vertebrate genomes, despite the fact that several rhabdoviruses are able to infect vertebrates. In the case of plant genomes, at least four different types of rhabdovirus N protein-like sequences (RNLS1–4) have been identified (Chiba et al., 2011). Plant RNLSs are phylogenetically related to the CP (or N protein) genes of members of the genus Varicosavirus or Cytorhabdovirus (Fig. 6). These findings provide an interesting insight into the origin and deep evolution of plant rhabdoviruses with mono- and bipartite genomes.
Fig. 6.
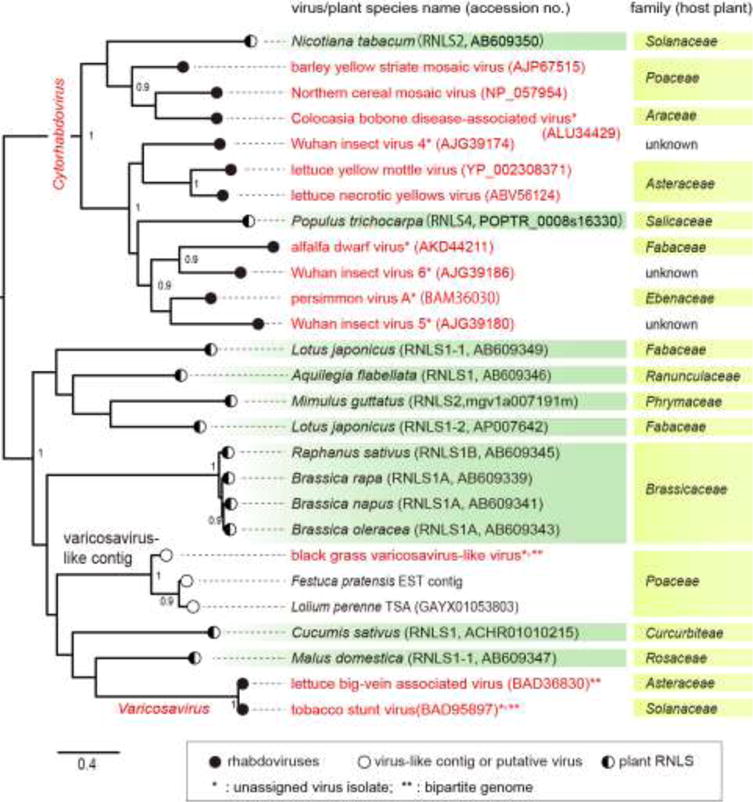
Phylogenetic relationships of the N (CP) proteins of varicosaviruses, cytorhabdoviruses and rhabdovirus N-like sequences (RLNSs) identified in plant host genomes. The ML tree was constructed using PhyML 3.0 with a best fit model “LG + G”. A part of this analysis was reported by Chiba et al. (2011). The accession numbers of rhabdoviruses and plant RLNSs (RLNS1, 2 and 4) are shown next to the virus or plant species name in the figure (within parentheses). Analyzed sequences were from 13 rhabdoviruses (varicosa- and varicosa-like viruses, cyto- and unclassified rhabdoviruses), 2 varicosavirus-like-contig sequences and 12 plant RNLSs (Chiba et al., 2011). Numbers at the nodes indicate aLRT values (values less than 0.9 are not displayed).
10. Conclusions and Perspectives
The rhabdoviruses comprise an extremely varied family of viruses that have successfully adapted and evolved to infect ecologically diverse hosts including mammals, birds, reptiles, fish and insects, and a wide array of both dicot and monocot plants. Although they share common morphological features and a canonical gene organization encoding five structural proteins, rhabdovirus genomes may also exhibit more complex organization and contain additional ORFs encoding putative accessory proteins. Genome organisation has been a useful taxonomic tool since similarities of genome architecture appear to be the result of significant evolutionary events that provide resolution between the genera and species levels. The inclusion of new rhabdovirus genera and of endogenous rhabdovirus elements will likely facilitate a deeper understanding of rhabdovirus evolution and allow further refinement of Rhabdoviridae taxonomy in the near future.
Within the family there are well known “classical” vertebrate rhabdoviruses such as VSV and rabies virus that have been studied in great detail for many decades and serve as model systems for elucidating the molecular mechanisms of rhabdovirus replication cycles in host cells. In contrast, there are also many novel rhabdoviruses discovered very recently using next generation sequencing, for which the genome sequences are known but most biological and ecological features remain to be elucidated. Those rhabdoviruses that are economically important pathogens in their respective plant and animal hosts have received the most attention, in many cases leading to extensive knowledge of their molecular biology, pathogenesis and host responses, as well as geographic and host ranges, ecology, phylogeography and evolutionary histories. Thus, the nature of current research efforts with different rhabdoviruses varies as much as the viruses themselves, providing promise of important discoveries on many levels in the future.
Highlights.
Family Rhabdoviridae is comprised of thirteen genera
Rhabdoviruses are mostly enveloped and infect ecologically diverse hosts
Rhabdovirus negative-sense RNA genome has five canonical structural protein genes
Rhabdovirus genomes may contain additional ORFs encoding putative accessory proteins
Diverse genome structure may guide taxonomic resolution at genus and species levels
Acknowledgments
This work was jointly supported by the Queensland Government Department of Agriculture and Fisheries and the University of Queensland. Author contributions were also supported in part by the International Collaboration Research Program on Joint Usage/Research Center Program at the IPSR, Okayama University and by a grant from the National Institutes of Health R24AI120942 to NV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham G, Banerjee AK. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MJ, et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2016 doi: 10.1007/s00705-016-2977-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, et al. Taxonomy of the order Mononegavirales – update 2016. Arch Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahne W, Bjorklund H, Essbauer S, Fijan N, Kurath G, Winton J. Spring viremia of carp – review. Dis Aquat Organ. 2002;52:261–272. doi: 10.3354/dao052261. [DOI] [PubMed] [Google Scholar]
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Allison AB, Palacios G, Travassos da Rosa A, Popov VL, Lu L, Xiao SY, DeToy K, Briese T, Lipkin WI, Keel MK, Stallknecht DE, Bishop GR, Tesh RB. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Res. 2011;155:112–122. doi: 10.1016/j.virusres.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar E-D, Tsai C-W, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu Rev Entomol. 2009;54:447–468. doi: 10.1146/annurev.ento.54.110807.090454. [DOI] [PubMed] [Google Scholar]
- Ammayappan A, Vakharia VN. Nonvirion protein of novirhabdovirus suppresses apoptosis at the early stage of virus infection. J Virol. 2011;85:8393–8402. doi: 10.1128/JVI.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammayappan A, Kurath G, Thompson TM, Vakharia VN. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Marine Biotechnol (NY) 2011;13:672–683. doi: 10.1007/s10126-010-9329-4. [DOI] [PubMed] [Google Scholar]
- Anderson G, Wang R, Bandyopadhyay A, Goodin M. The nucleocapsid protein of Potato yellow dwarf virus: protein interactions and nuclear import mediated by a non-canonical nuclear localization signal. Front Plant Sci. 2012;3:14. doi: 10.3389/fpls.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger MJ, Bruenn JA, Taylor DJ. Phylogeny, integration and expression of sigma virus-like genes in Drosophila. Mol Phylogenet Evol. 2012;65:251–258. doi: 10.1016/j.ympev.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Kopperud K, Anderson G, Martin K, Goodin M. An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology. 2010;402:61–71. doi: 10.1016/j.virol.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Whelan SP, Wertz GW. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997a;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Whelan SP, Wertz GW. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997b;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerman N, Giolitti F, de Breuil S, Trucco V, Nome C, Lenardon S, Dietzgen RG. Complete genome sequence and integrated protein localization and interaction map for alfalfa dwarf virus, which combines properties of both cytoplasmic and nuclear plant rhabdoviruses. Virology. 2015;483:275–283. doi: 10.1016/j.virol.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Biacchesi S. The reverse genetics applied to fish RNA viruses. Vet Res. 2011;42:12. doi: 10.1186/1297-9716-42-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, Thoulouze MI, Bearzotti M, Yu YX, Bremont M. Recovery of NV knockout infectious hematopoietic necrosis virus expressing foreign genes. J Virol. 2000;74:11247–11253. doi: 10.1128/jvi.74.23.11247-11253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittancourt AA. A mancha anular, uma nova ameaça do cafeeiro. O Biol. 1938;4:404–405. [Google Scholar]
- Blasdell KR, Guzman H, Widen SG, Firth C, Wood TG, Holmes EC, Tesh RB, Vasilakis N, Walker PJ. Ledantevirus: a proposed new genus in the Rhabdoviridae has a strong ecological association with bats. Am J Trop Med Hyg. 2015;92:405–410. doi: 10.4269/ajtmh.14-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D, Maarifi G, Nisole S, Chelbi-Alix MK. Resistance to Rhabdoviridae infection and subversion of antiviral responses. Viruses. 2015;7:3675–3702. doi: 10.3390/v7072794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootland LM, Leong JC. Infectious haematopoietic necrosis virus. In: Woo PTK, Bruno DW, editors. Fish diseases and disorders, Vol 3, Viral, bacterial, and fungal infections. 2nd. Wallingford, U.K.: CAB International; 2011. pp. 66–109. [Google Scholar]
- Breyta R, Samson C, Blair M, Black A, Kurath G. Successful mitigation of viral disease based on a delayed exposure rearing strategy at a large-scale steelhead trout conservation hatchery. Aquaculture. 2016;450:213–224. doi: 10.1016/j.aquaculture.2015.07.014. [DOI] [Google Scholar]
- Brzozka K, Finke S, Conzelmann KK. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol. 2005;79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causey OR, Shope RE, Bensabath G. Marco, Timbo, and Chaco, newly recognized arboviruses from lizards of Brazil. Am J Trop Med and Hyg. 1966;15:239–243. doi: 10.4269/ajtmh.1966.15.239. [DOI] [PubMed] [Google Scholar]
- Chagas CM, July JR, Alba APC. Mechanical transmission and structural features of Coffee ringspot virus (Crv) Phytopathol Z. 1981;102:100–106. [Google Scholar]
- Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MK, Moon CH, Ko MS, Lee UH, Cho WJ, Cha SJ, Do JW, Heo GJ, Jeong SG, Hahm YS, Harmache A, Bremont M, Kurath G, Park JW. A nuclear localization of the infectious haematopoietic necrosis virus NV protein is necessary for optimal viral growth. PLoS One. 2011;6:e22362. doi: 10.1371/journal.pone.0022362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton GM, Little SP, Hagen FS, Huang AS. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell. 1978;15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- Connor JH, McKenzie MO, Lyles DS. Role of residues 121 to 124 of vesicular stomatitis virus matrix protein in virus assembly and virus-host interaction. J Virol. 2006;80:3701–3711. doi: 10.1128/JVI.80.8.3701-3711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Jaramillo JL, Ruiz-Medrano R, Rojas-Morales L, López-Buenfil JA, Morales-Galván O, Chavarín-Palacio C, Ramírez-Pool JA, Xoconostle-Cázares B. Characterization of a proposed dichorhavirus associated with the citrus leprosis disease and analysis of the host response. Viruses. 2014;6:2602–2622. doi: 10.3390/v6072602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- Dietzgen RG, Kuzmin IV. Rhabdoviruses: Molecular taxonomy, evolution, genomics, ecology, host-vector interactions, cytopathology and control. Caister Academic Press; Norfolk, UK: 2012. p. 276. [Google Scholar]
- Dietzgen RG, Calisher CH, Kurath G, Kuzmin IV, Rodriguez LL, Stone DM, Tesh RB, Tordo N, Walker PJ, Wetzel T, Whitfield AE. Family Rhabdoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Elsevier Academic Press; Oxford: 2011. pp. 686–714. (Ninth Report of the International Committee on Taxonomy of Viruses). [Google Scholar]
- Dietzgen RG, Kuhn JH, Clawson AN, Freitas-Astua J, Goodin MM, Kitajima EW, Kondo H, Wetzel T, Whitfield AE. Dichorhavirus: a proposed new genus for Brevipalpus mite-transmitted, nuclear, bacilliform, bipartite, negative-strand RNA plant viruses. Arch Virol. 2014;159:607–619. doi: 10.1007/s00705-013-1834-0. [DOI] [PubMed] [Google Scholar]
- Doms RW, Ruusala A, Machamer C, Helenius J, Helenius A, Rose JK. Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J Cell Biol. 1988;107:89–99. doi: 10.1083/jcb.107.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einer-Jensen K, Ahrens P, Forsberg R, Lorenzen N. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J Gen Virol. 2004;85:1167–1179. doi: 10.1099/vir.0.79820-0. [DOI] [PubMed] [Google Scholar]
- Emmenegger EJ, Kentop E, Thompson TM, Pittam S, Ryan A, Keon D, Carlino JA, Ranson J, Life RB, Troyer RM, Garver KA, Kurath G. Development of an aquatic pathogen database (AquaPathogen X) and its utilization in tracking emerging fish virus pathogens in North America. J Fish Dis. 2011;34:579–587. doi: 10.1111/j.1365-2761.2011.01270.x. [DOI] [PubMed] [Google Scholar]
- Enzmann PJ, Castric J, Bovo G, Thiery R, Fichtner D, Schutze H, Wahli T. Evolution of infectious hematopoietic necrosis virus (IHNV), a fish rhabdovirus, in Europe over 20 years: implications for control. Dis Aquatic Org. 2010;89:9–15. doi: 10.3354/dao02182. [DOI] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Fort P, Albertini A, Van-Hua A, Berthomieu A, Roche S, Delsuc F, Pasteur N, Capy P, Gaudin Y, Weill M. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol Biol Evol. 2012;29:381–390. doi: 10.1093/molbev/msr226. [DOI] [PubMed] [Google Scholar]
- Ganesan U, Bragg JN, Deng M, Marr S, Lee MY, Qian S, Shi M, Kappel J, Peters C, Lee Y, Goodin MM, Dietzgen RG, Li Z, Jackson AO. Construction of a Sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. J Virol. 2013;87:10598–10611. doi: 10.1128/JVI.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Chakrabarty R, Yelton S, Martin K, Clark A, Brooks R. Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J Gen Virol. 2007;88:1810–1820. doi: 10.1099/vir.0.82698-0. [DOI] [PubMed] [Google Scholar]
- Gubala A, Davis S, Weir R, Melville L, Cowled C, Walker P, Boyle D. Ngaingan virus, a macropod-associated rhabdovirus, contains a second glycoprotein gene and seven novel open reading frames. Virology. 2010;399:98–108. doi: 10.1016/j.virol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Gubala A, Davis S, Weir R, Melville L, Cowled C, Boyle D. Tibrogargan and Coastal Plains rhabdoviruses: genomic characterization, evolution of novel genes and seroprevalence in Australian livestock. J Gen Virol. 2011;92:2160–2170. doi: 10.1099/vir.0.026120-0. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo H, Song X, Xie C, Huo Y, Zhang F, Chen XY, Geng Y, Fang R. Rice yellow stunt rhabdovirus protein 6 suppresses systemic RNA silencing by blocking RDR6-mediated secondary siRNA synthesis. Mol Plant-Microbe Interact. 2013;26:927–936. doi: 10.1094/MPMI-02-13-0040-R. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Harmache A, LeBerre M, Droineau S, Giovaninni M, Bremont M. Bioluminescence imaging if live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus. J Virol. 2006;80:3655–3659. doi: 10.1128/JVI.80.7.3655-3659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung JS, Roy A, Fu S, Shao J, Schneider W, Brlansky RH. History and diversity of Citrus leprosis virus recorded in herbarium specimens. Phytopathol. 2015;105:1277–1284. doi: 10.1094/PHYTO-03-15-0064-R. [DOI] [PubMed] [Google Scholar]
- Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig C, Pye AD, Hyatt AD, Davis SS, McWilliam SM, Heine HG, Walker PJ, Boyle DB. Vaccinia virus-expressed bovine ephemeral fever virus G but not G(NS) glycoprotein induces neutralizing antibodies and protects against experimental infection. J Gen Virol. 1996;77:631–640. doi: 10.1099/0022-1317-77-4-631. [DOI] [PubMed] [Google Scholar]
- Huang Y-W, Geng Y-F, Ying X-B, Chen X-Y, Fang R-X. Identification of a movement protein of rice yellow stunt rhabdovirus. J Virol. 2005;79:2108–2114. doi: 10.1128/JVI.79.4.2108-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LN, Englund N, Pattnaik AK. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LE, Rose JK. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- Jackson AO, Li Z. Developments in plant negative-strand RNA virus reverse genetics. Annu Rev Phytopathol. 2016;54 doi: 10.1146/annurev-phyto-080615-095909. in press. [DOI] [PubMed] [Google Scholar]
- Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. Biology of plant rhabdoviruses. Annu Rev Phytopathol. 2005;43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- Johnson HN. Disease derived from wildlife. California Health. 1965;23:35–39. [Google Scholar]
- Jonstrup SP, Gray T, Kahns S, Skall HF, Snow M, Olesen NJ. FishPathogens.eu/vhsv: a user-friendly viral haemorrhagic septicaemia virus isolate and sequence database. J Fish Dis. 2009;32:925–929. doi: 10.1111/j.1365-2761.2009.01073.x. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DA, Kurath G, Brito IL, Purcell MK, Read AF, Winton JR, Wargo AR. Potential drivers of virulence evolution in aquaculture. Evol Applic. 2016;9:344–354. doi: 10.1111/eva.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Maeda T, Shirako Y, Tamada T. Orchid fleck virus is a rhabdovirus with an unusual bipartite genome. J Gen Virol. 2006;87:2413–2421. doi: 10.1099/vir.0.81811-0. [DOI] [PubMed] [Google Scholar]
- Kondo H, Maeda T, Tamada T. Identification and characterization of structural proteins of orchid fleck virus. Arch Virol. 2009;154:37–45. doi: 10.1007/s00705-008-0268-6. [DOI] [PubMed] [Google Scholar]
- Kondo H, Chiba S, Andika IB, Maruyama K, Tamada T, Suzuki N. Orchid fleck virus structural proteins N and P form intracellular viroplasm-like structures in the absence of viral infection. J Virol. 2013;13:7423–7434. doi: 10.1128/JVI.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Maruyama K, Chiba S, Andika IB, Suzuki N. Transcriptional mapping of the messenger and leader RNAs of orchid fleck virus, a bisegmented negative-strand RNA virus. Virology. 2014;452–453:166–174. doi: 10.1016/j.virol.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Kondo H, Chiba S, Suzuki N. Detection and analysis of non-retroviral RNA virus-like elements in plant fungal and insect genomes. Methods Mol Biol. 2015;1236:73–88. doi: 10.1007/978-1-4939-1743-3_7. [DOI] [PubMed] [Google Scholar]
- Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni AL. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 2011;162:184–202. doi: 10.1016/j.virusres.2011.09.028. [DOI] [PubMed] [Google Scholar]
- Kretzschmar E, Peluso R, Schnell MJ, Whitt MA, Rose JK. Normal replication of vesicular stomatitis virus without C proteins. Virology. 1996;216:309–316. doi: 10.1006/viro.1996.0066. [DOI] [PubMed] [Google Scholar]
- Kurath G. Biotechnology and DNA vaccines for aquatic animals. Revue Scientifique et Technique (International Office of Epizootics) 2008;27:175–196. [PubMed] [Google Scholar]
- Kurath G. Fish Novirhabdoviruses. In: Dietzgen RG, Kuzman IV, editors. Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Host-Vector interactions, Cytopathology, and Control. Caister Academic Press; 2012. pp. 89–117. http://pubs.er.usgs.gov/publication/70045590. [Google Scholar]
- Kurath G, Leong JC. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. J Virol. 1985;53:462–468. doi: 10.1128/jvi.53.2.462-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath G, Winton JR. Fish rhabdoviruses. In: Mahy BWJ, VanRegenmortel MHV, editors. Encyclopedia of Virology. Oxford, UK: Elsevier; 2008. pp. 221–227. [Google Scholar]
- Kurath G, Winton JR. Complex dynamics at the interface between wild and domestic viruses of finfish. Curr Opin Virol. 2011;1:73–80. doi: 10.1016/j.coviro.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Kurath G, Higman KH, Bjorklund HV. Distribution and variation of NV genes in fish rhabdoviruses. J Gen Virol. 1997;78:113–117. doi: 10.1099/0022-1317-78-1-113. [DOI] [PubMed] [Google Scholar]
- Kurath G, Garver KA, Troyer RM, Emmenegger EJ, Einer-Jensen K, Anderson ED. Phylogeography of infectious hematopoietic necrosis virus in North America. J Gen Virol. 2003;84:803–814. doi: 10.1099/vir.0.18771-0. [DOI] [PubMed] [Google Scholar]
- Kurath G, Purcell M, Garver K. Fish rhabdovirus models for understanding host response to DNA vaccines. CAB Reviews: perspectives in agriculture, veterinary sciences, and natural resources. 2007;2:1–12. [Google Scholar]
- Kurz W, Gelderblom H, Flügel RM, Darai G. Isolation and characterization of a tupaia rhabdovirus. Intervirol. 1986;25:88–96. doi: 10.1159/000149661. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Novella IS, Dietzgen RG, Padhi A, Rupprecht CE. The rhabdoviruses: biodiversity, phylogenetics and evolution. Infect Genet Evol. 2009;9:541–553. doi: 10.1016/j.meegid.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Leong JC, Kurath G. The Springer Index of Viruses. 2nd. Springer-Verlag; Berlin Heidelberg New York: 2012. Novirhabdovirus, Rhabdoviridae; pp. 1731–1740. Part 79. [Google Scholar]
- Li T, Pattnaik AK. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J Virol. 1999;73:444–452. doi: 10.1128/jvi.73.1.444-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Murray GGR, Palmer WJ, Day JP, Parker DJ, Welch JJ, Obbard DJ, Jiggins FM. The evolution, diversity, and host associations of rhabdoviruses. Virus Evol. 2015;1 doi: 10.1093/ve/vev014. vev014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen N, LaPatra SE. DNA vaccines for aquacultured fish. Revue Scientifique et Technique (International Office of Epizootics) 2005;24:201–213. [PubMed] [Google Scholar]
- Lorenzen N, Lorenzen E, Einer-Jensen K, LaPatra SE. DNA vaccines as a tool for analysing the protective immune response against rhabdoviruses in rainbow trout. Fish Shellfish Immunol. 2002;12:439–453. doi: 10.1006/fsim.2002.0422. [DOI] [PubMed] [Google Scholar]
- Lyles DS, Kuzmin IV, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B, editors. Fields Virology. Sixth. Wolters Kluwer Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 885–922. [Google Scholar]
- Maccarone LD. Relationships between the pathogen Olpidium virulentus and viruses associated with lettuce big-vein disease. Plant Dis. 2013;97:700–707. doi: 10.1094/PDIS-10-12-0979-FE. [DOI] [PubMed] [Google Scholar]
- Mann KS, Bejerman N, Johnson KN, Dietzgen RG. Cytorhabdovirus P3 genes encode 30K-like cell-to-cell movement proteins. Virology. 2016a;489:20–33. doi: 10.1016/j.virol.2015.11.028. [DOI] [PubMed] [Google Scholar]
- Mann KS, Johnson KN, Carroll BJ, Dietzgen RG. Cytorhabdovirus P protein suppresses RISC-mediated cleavage and RNA silencing amplification in planta. Virology. 2016b;490:27–40. doi: 10.1016/j.virol.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Martin KM, Dietzgen RG, Wang RY, Goodin MM. Lettuce necrotic yellows cytorhabdovirus protein localization and interaction map and comparison with nucleorhabdoviruses. J Gen Virol. 2012;93:906–914. doi: 10.1099/vir.0.038034-0. [DOI] [PubMed] [Google Scholar]
- McAllister J, Gauci PJ, Mitchell IR, Boyle DB, Bulach DM, Weir RP, Melville LF, Davis SS, Gubala AJ. Genomic characterisation of Almpiwar virus, Harrison Dam virus and Walkabout Creek virus; three novel rhabdoviruses from northern Australia. Virol Rep. 2014;3:1–17. doi: 10.1016/j.virep.2014.09.001. [DOI] [Google Scholar]
- McCreedy BJ, Jr, McKinnon KP, Lyles DS. Solubility of vesicular stomatitis virus M protein in the cytosol of infected cells or isolated from virions. J Virol. 1990;64:902–906. doi: 10.1128/jvi.64.2.902-906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B-E, Martin K, Wang R, Tafelmeyer P, Bridges M, Goodin M. A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol Plant-Microbe Interact. 2010;23:1420–1432. doi: 10.1094/MPMI-04-10-0097. [DOI] [PubMed] [Google Scholar]
- Mire CE, White JM, Whitt MA. A spatio-temporal analysis of matrix protein and nucleocapsid trafficking during vesicular stomatitis virus uncoating. PLoS Pathog. 2010;6:e1000994. doi: 10.1371/journal.ppat.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP, Cropp CB, Frazier CL, Murphy FA, Whitfield SG. Viruses isolated from reptiles: identification of three new members of the family Rhabdoviridae. Arch Virol. 1979;60:1–12. doi: 10.1007/BF01318092. [DOI] [PubMed] [Google Scholar]
- Nishizawa T, Kinoshita S, Kim W-S, Higashi S, Yoshimizu M. Nucleotide diversity of Japanese isolates of infectious hematopoietic necrosis virus (IHNV) based on the glycoprotein gene. Dis Aquat Organ. 2006;71:267–272. doi: 10.3354/dao071267. [DOI] [PubMed] [Google Scholar]
- Novella IS, Presloid JB, Zhou T, Smith-Tsurkan SD, Ebendick-Corpus BE, Dutta RN, Lust KL, Wilke CO. Genomic evolution of vesicular stomatitis virus strains with differences in adaptability. J Virol. 2010;84:4960–4968. doi: 10.1128/JVI.00710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Presloid JB, Smith SD, Wilke CO. Specific and nonspecific host adaptation during arboviral experimental evolution. J Mol Microbiol Biotechnol. 2011;21:71–81. doi: 10.1159/000332752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald WF, Arnheiter H, Dubois-Dalcq M, Lazzarini RA. Stereo images of vesicular stomatitis virus assembly. J Virol. 1986;57:922–932. doi: 10.1128/jvi.57.3.922-932.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden JR, Pal R, Wagner RR. Mapping regions of the matrix protein of vesicular stomatitis virus which bind to ribonucleocapsids, liposomes, and monoclonal antibodies. J Virol. 1986;58:860–868. doi: 10.1128/jvi.58.3.860-868.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Grinnell BW, Snyder RM, Wagner RR. Regulation of viral transcription by the matrix protein of vesicular stomatitis virus probed by monoclonal antibodies and temperature-sensitive mutants. J Virol. 1985;56:386–394. doi: 10.1128/jvi.56.2.386-394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso RW, Richardson JC, Talon J, Lock M. Identification of a set of proteins (C′ and C) encoded by the bicistronic P gene of the Indiana serotype of vesicular stomatitis virus and analysis of their effect on transcription by the viral RNA polymerase. Virology. 1996;218:335–342. doi: 10.1006/viro.1996.0202. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Laing K, Winton JR. Immunity to fish Rhabdoviruses. Viruses. 2012;4:140–166. doi: 10.3390/v4010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho TO, Fugueira AR, Sotero AJ, Wang R, Geraldino Duarte PS, Farman M, Goodin MM. Characterization of Coffee ringspot virus-Lavras: A model for an emerging threat to coffee production and quality. Virology. 2014;464–465:385–396. doi: 10.1016/j.virol.2014.07.031. [DOI] [PubMed] [Google Scholar]
- Ramalho TO, Figueira AR, Wang R, Jones O, Harris LE, Goodin MM. Detection and survey of coffee ringspot virus in Brazil. Arch Virol. 2016;161:335–343. doi: 10.1007/s00705-015-2663-0. [DOI] [PubMed] [Google Scholar]
- Rieder M, Brzozka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. Genetic dissection of interferon- antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J Virol. 2011;85:842–852. doi: 10.1128/JVI.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JCV, Rodriguez CM, Moreira L, Villalobos W, Rivera C, Childers CC. Occurrence of Coffee ringspot virus, a Brevipalpus miteborne virus in coffee in Costa Rica. Plant Dis. 2002;86:564–564. doi: 10.1094/PDIS.2002.86.5.564B. [DOI] [PubMed] [Google Scholar]
- Rose JK. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- Roy A, Stone AL, Shao J, Otero-Colina G, Wei G, Choudhary N, Achor D, Levy L, Nakhla MK, Hartung JS, Schneider WL, Brlansky RH. Identification and molecular characterization of nuclear Citrus leprosis virus, a member of the proposed Dichorhavirus genus infecting multiple Citrus species in Mexico. Phytopathol. 2015;105:564–575. doi: 10.1094/PHYTO-09-14-0245-R. [DOI] [PubMed] [Google Scholar]
- Sasaya T, Kusaba S, Ishikawa K, Koganezawa H. Nucleotide sequence of RNA2 of Lettuce big-vein virus and evidence for a possible transcription termination/initiation strategy similar to that of rhabdoviruses. J Gen Virol. 2004;85:2709–2717. doi: 10.1099/vir.0.80061-0. [DOI] [PubMed] [Google Scholar]
- Schmidt MF, Schlesinger MJ. Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetze H, Enzmann PJ, Mundt E, Mettenleiter TC. Identification of the non-virion (NV) protein of fish rhabdoviruses viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus. J Gen Virol. 1996;77:1259–1263. doi: 10.1099/0022-1317-77-6-1259. [DOI] [PubMed] [Google Scholar]
- Skall HF, Olesen NJ, Mellergaard S. Viral haemorrhagic septicaemia virus in marine fish and its implications for fish farming – a review. J Fish Dis. 2005;28:509–529. doi: 10.1111/j.1365-2761.2005.00654.x. [DOI] [PubMed] [Google Scholar]
- Smail DA, Snow M. Viral haemorrhagic septicaemia. In: Woo PTK, Bruno DW, editors. Fish diseases and disorders, Vol 3, Viral, bacterial, and fungal infections. 2nd. Wallingford, U.K.: CAB International; 2011. pp. 110–142. [Google Scholar]
- Spiropoulou CF, Nichol ST. A small highly basic protein is encoded in overlapping frame within the P gene of vesicular stomatitis virus. J Virol. 1993;67:3103–3110. doi: 10.1128/jvi.67.6.3103-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DM, Kerr RC, Hughes M, Radford AD, Darby AC. Characterization of the genomes of four putative vesiculoviruses: tench rhabdovirus, grass carp rhabdovirus, perch rhabdovirus and eel rhabdovirus European X. Arch Virol. 2013;158:2371–2377. doi: 10.1007/s00705-013-1711-x. [DOI] [PubMed] [Google Scholar]
- Takano T, Iwahori A, Hirono I, Aoki T. Development of a DNA vaccine against hirame rhabdovirus and analysis of the expression of immune-related genes after vaccination. Fish Shellfish Immunol. 2004;17:367–374. doi: 10.1016/j.fsi.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Troyer RM, Garver KA, Ranson JC, Wargo AR, Kurath G. In vivo virus growth competition assays demonstrate equal fitness of fish rhabdovirus strains that co-circulate in aquaculture. Virus Res. 2008;137:179–188. doi: 10.1016/j.virusres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Vasilakis N, Widen S, Mayer SV, Seymour R, Wood TG, Popov V, Guzman H, Travassos da Rosa AP, Ghedin E, Holmes EC, Walker PJ, Tesh RB. Niakha virus: a novel member of the family Rhabdoviridae isolated from phlebotomine sandflies in Senegal. Virology. 2013;444:80–89. doi: 10.1016/j.virol.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier ER, Langevin C, Benmansour A, Boudinot P. Early antiviral response and virus-induced genes in fish. Dev Comp Immunol. 2011;35:1204–1214. doi: 10.1016/j.dci.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Walker PJ. Bovine ephemeral fever in Australia and the world. Curr Top Microbiol Immunol. 2005;292:57–80. doi: 10.1007/3-540-27485-5_4. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. Rhabdovirus accessory genes. Virus Res. 2011;162:110–125. doi: 10.1016/j.virusres.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, Paradkar PN, Holmes EC, Tesh RB, Vasilakis N. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog. 2015;11:e1004664. doi: 10.1371/journal.ppat.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JA, Verbeek M. Genus Varicosavirus. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Elsevier Academic Press; Oxford: 2011. pp. 777–781. (Ninth Report of the International Committee on Taxonomy of Viruses). [Google Scholar]
- Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, Zhou X, Jackson AO, Li Z. Rescue of a plant negative-strand RNA virus from cloned cDNA: Insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 2015;11:e1005223. doi: 10.1371/journal.ppat.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo AR, Kurath G. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J Virol. 2011;85:3959–3967. doi: 10.1128/JVI.01891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Muñoz-Rojas AR, Okamoto KW, Miller-Jensen K, Turner PE. Generalized selection to overcome innate immunity selects for host breadth in an RNA virus. Evolution. 2016;70:270–281. doi: 10.1111/evo.12845. [DOI] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt MA, Geisbert TW, Mire CE. Single-vector, single-injection recombinant vesicular stomatitis virus vaccines against high-containment viruses. Meth Mol Biol. 2016;1403:295–311. doi: 10.1007/978-1-4939-3387-7_16. [DOI] [PubMed] [Google Scholar]
- Winton JR. Molecular approaches to fish vaccines. J Applied Ichthyology. 2007;14:153–158. [Google Scholar]
- World Organization for Animal Health. Aquatic animal health code. 2014 http://www.oie.int/international-standard-setting/aquatic-code/
- Yan T, Zhu JR, Di D, Gao Q, Zhang Y, Zhang A, Tan C, Miao H, Wang XB. Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology. 2015;478:112–122. doi: 10.1016/j.virol.2014.12.042. [DOI] [PubMed] [Google Scholar]


