Abstract
Microsporidia are highly successful parasites that infect virtually all known animal lineages, including the model Danio rerio (zebrafish). The widespread use of this aquatic model for biomedical research has resulted in an unexpected increase in infections from the microsporidium Pseudoloma neurophilia, which can lead to significant physical, behavioral and immunological modifications resulting in non-protocol variation during experimental procedures. Here, we seek to obtain insights into the biology of P. neurophilia by investigating its genome content, which was obtained from only 29 nanograms of DNA using the Miseq technology and paired-end Illumina sequencing. We found that the genome of P. neurophilia is phylogenetically and genetically related to other fish-microsporidians, but features unique to this intracellular parasite are also found. The small 5.25-Mb genome assembly includes 1,139 unique open reading frames and an unusually high number of transposable elements for such a small genome. Investigations of intragenomic diversity also provided strong indications that the mononucleate nucleus of this species is diploid. Overall, our study provides insights into the dynamics of microsporidian genomes, and represents a solid sequence reference to be used in future studies of host-parasite interactions using the zebrafish Danio rerio and P. neurophilia as a model.
Keywords: Cryptomycota, Microsporidia, Transposable elements, Zebrafish, host-parasite interactions, Parasite, Effectors, Ploidy, RNAi
INTRODUCTION
Microsporidia are highly successful obligate intracellular parasites that infect many ecologically and economically important terrestrial animal lineages, such as honey bees (Higes et al. 2013), humans (Lobo et al. 2012), and aquatic research models like the zebrafish (Matthews et al. 2001; Sanders, Watral, and Kent 2012; Sanders and Kent 2014; Spagnoli, Xue, and Kent 2015). These parasites have been proposed to propagate via both vertical and horizontal modes of transmission (Ardila-Garcia and Fast 2012; Izquierdo et al. 2011; Sanders et al. 2013), and have been recently included in the phylum Cryptomycota (James et al. 2013). Microsporidia are best known for their reductive and derived features, which include a genome-less mitochondrion (the mitosome), prokaryote-sized ribosomal RNA (rRNA) genes, and an unconventional Golgi apparatus (Williams et al. 2002; Vossbrinck et al. 1987; Beznoussenko et al. 2007).
Genome sequencing projects of these successful parasites recently included in the new superphylum Opisthosporidia (Karpov et al. 2014) have provided essential insights into their basic biology and infection mechanisms, and have also unraveled the evolutionary dynamics of their genomes (Corradi 2015). As an adaptation to their obligate parasitic life cycle, these have undergone massive reductions in their genome content, particularly for genes involved in nucleic and amino acids metabolism (de novo biosynthesis of amino acids and nucleotides and the tricarboxylic acids cycle pathways (Higes et al. 2013; Corradi and Slamovits 2011)). In some species, intergenic regions and even proteins have been shortened in size, resulting at the extreme in genomes that are not only gene-poor but also very compact. Evidently, the miniaturization of many cellular activities has rendered all microsporidia very dependent on their hosts, possibly even for energy production (Keeling et al. 2010). Although the biochemical repertoire of known microsporidian species is always reduced, their genome sizes can vary substantially within the group, ranging from only 2.3 (Encephalitozoon intestinalis) to a known maximum of 51.3 Mb (Edhazardia aedis)(Corradi et al. 2010; Desjardins et al. 2015). These drastic variations in genome size have been correlated with gene density and the amount of transposable elements (TE) - i.e. larger genomes tend to harbor more TEs (with the exception of E. aedis) and much larger intergenic regions. To date, the genomes of 20 known species have been sequenced. This wealth of data has enabled a broad understanding of the microsporidian’s biology, but as more species get sequenced it becomes clear that the definition of a typical microsporidian genome often differs from its original description - e.g. not always small and compact, nor always with reduced protein length, biochemical repertoires and low counts of transposable elements).
The present paper aims to obtain first-hand knowledge of a notorious parasite of the zebrafish (Danio rerio); namely Pseudoloma neurophilia (Matthews et al. 2001). This parasite mostly targets the central nervous system and peripheral nerves of its host (Kent, Harper, and Wolf 2012), and significant increases in infection cases caused by this particular microsporidian species have been described in recent years (Murray et al. 2011; Sanders, Watral, and Kent 2012). Such infections have been detected in approximately 75% of the Zebrafish International Resource Centre facilities, and their presence has been demonstrated to result in significant physical, behavioral and neurological (Sisson et al. 2006) modifications; all of which are likely to affect reproducibility in experimental procedures. Here, we show that the haploid genome size of P. neurophilia is on the lower end compared to relative species in the group, and we describe several of its atypical features. These include an unusual expansion of TEs, and the presence of large numbers of inserts of different sequence and size located in protein-coding genes that are otherwise highly conserved in sequence among eukaryotes (including microsporidian species). Besides improving our knowledge of a microsporidian that is difficult to cultivate and study, this sequence data will also be helpful for future analyses of host-parasite interactions in the zebrafish, particularly for gene expression studies based on next generation sequencing, such as RNA-seq.
MATERIAL AND METHODS
Sample collection and DNA extraction
Spores of Pseudoloma neurophilia strain MK1were extracted from 50 infected adult Danio rerio at Oregon State University (Fig 1). Fish were euthanized using an overdose of MS222, brains and spinal cords were removed and placed in 5 ml of deionized water with antibiotics (pen-strep). This solution was passed through progressively smaller-gauge needles using a 5-ml syringe. An additional 5 ml of deionized water and antibiotics was added and the mixture was vortexed and allowed to sit overnight at room temperature. The following day, the mixture was again vortexed and passed through a 40-um cell strainer into a 50-ml conical tube. Water and antibiotics were added to bring the total volume to 20 ml then 20 ml of filter-sterilized Percoll was added. The mixture was vortexed and centrifuged at 1200 g for 1 hr. The pellet was recovered and placed in a 1.5 ml tube where it was washed two times with 1% SDS followed by two washes in PBS. The spores were resuspended in 200 ul of TE buffer and quantified using a hemocytometer. DNA was extracted from approximately 1.5 x 106 spores using the MasterPure DNA purification kit (Epicentre kit; Illumina, San Diego, California). Spores were first pelleted by centrifugation; resuspended in 300 Kl of Tissue & Cell lysis buffer (Epicentre kit; Illumina, San Diego, California) with 1Kl of Proteinase K and vortexed thoroughly. Glass beads (150–212 microns) were added to the solution and incubated at 65 °C for 15 minutes and shaken at 2500 rpm for 30 seconds every 5 minutes. Following the incubation, the sample was pelleted by centrifugation; cooled to 37°C and incubated for 30 min at the same temperature after the addition of 1Kl of 5Kg/Kl of RNAse A (Epicentre kit; Illumina, San Diego,California). The sample was then put on ice for 5 minute and 150 Kl of MPC Protein Precipitation Reagent (Epicentre) were added. The new solution was vortexed vigorously for 10 seconds and the debris were pelleted by centrifugation for 15 minutes at ≥10,000 × g at 4°C. Following the centrifugation, the supernatant was transferred to a clean microcentrifuge tube and 500 Kl of Isopropanol were added to the sample and incubated at −20 °C for 30 minutes. The incubated sample was then centrifuged at 4°C for 20 min at ≥10,000 × g. The pelleted DNA was rinsed twice with cold 70% ethanol and resuspended in Tris-EDTA buffer. DNA extraction resulted in a total of 29 ng of DNA.
Figure 1.
Adult zebrafish, Danio rerio, infected with Pseudoloma neurophilia. This fish shows grossly visible signs of infection with associated spinal curvature. Most P. neurophilia infections are chronic and fish show little to no signs of infection, confounding the detection of this parasite. Bar = 1 mm.
DNA sequencing and de novo assembly
Extracted DNA of P. neurophilia spores was sent to Illumina sequencing using the MiSeq technology by Fasteris S.A. (Geneva, Switzerland); which resulted in a library of 7,230,699 paired-end reads for a total of 14,461,398 reads of 250 bp length. Paired-end reads were trimmed using the PERL script trim-fastq.pl from the PoPoolation toolkit (Kofler et al. 2011) to remove adaptors and used to assemble the genome draft using Spades v3.0.0 (Bankevich et al. 2012) with k-mers ranging from 23 to 113 (23,33,43,53,63,73,83,93,103,113); giving 31,043 scaffolds totalling 30 Mb. Resulting contigs were screened for contaminant based on their GC content and average read coverage (Supp. Fig 2). Highly supported contigs (average coverage above 44) were retained and used to perform a BLAST analysis of all ORFs (e-value cut-off of 1xE−5) against the nr database in order to identify contigs demonstrating obvious microsporidian origin. Here, contigs with at least one ORF demonstrating homology with sequenced microsprodian species were retained and manually analyzed to confirm its potential microsporidian origin. The genome sequences have been deposited at DDBJ/EMBL/GenBank under the accession LGUB00000000.
Genome annotation
Potential open reading frames (ORFs) were predicted using an in-house script that combines Glimmer’s ab initio gene prediction algorithm and with the detection of CCC and GGG motifs found in close proximity of microsporidian transcription initiation sites (Peyretaillade et al. 2012). The gene function of each ORF was predicted using homology, which we inferred by performing BLAST searches against the NCBI nr database with an e-value cut-off of 1xE−10. ORFs were annotated using Geneious R8 (Kearse et al. 2012). To further describe the function of P. neurophilia predicted genes, a search for secretion signals and subcellular location prediction was performed using SignalP (Petersen et al. 2011) and TargetP (Emanuelsson et al. 2000), respectively. Eukaryotic Orthologous Groups (KOG) gene enrichments analyses were further conducted to categorize and identify orthologues and paralogs among microsporidian genomes.
Homology search of spliceosomal genes from closely related species and Saccharomyces cerevisiae listed listed in Watson et al. (Watson et al. 2015) revealed the presence of 13 splicesomal genes in P. neurophilia, which is significantly smaller than the repertoire of T. hominis, V. culicis and S. cerevisiae (22, 25 and 89 genes, respectively). Spliceosomal introns were searched in P. neurophilia based on homology with introns identified by others in closely related microsporidia (Watson et al. 2015; Campbell et al. 2013).
Genome data used in this study
The following microsporidian genomes were retrieved: Anncaliia algerae PRA109 (AOMV00000000.2), Edhazardia aedis USNM 41457 (AFBI00000000.3), Encephalitozoon cuniculi GB-M1 (AL391737.2), Encephalitozoon hellem ATCC 50504 (CP002713.1), Encephalitozoon intestinalis ATCC 50506 (CP001942.1), Encephalitozoon romaleae SJ-2008 (CP003518.1), Enterocytozoon bieneusi H348 (NZ_ABGB00000000.1), Mitosporidium daphniae (JMKJ00000000.1), Nematocida parisii ERTm1 (AEFF00000000.2), Nematocida sp. 1 ERTm2 (AERB00000000.1), Nosema antheraeae YY (http://silkpathdb.swu.edu.cn/silkpathdb/ftpserver, last accessed July 1, 2014), Nosema apis BRL 01 (ANPH00000000.1), Nosema bombycis CQ1 (ACJZ00000000.1), Nosema ceranae BRL01 (NZ_ACOL00000000.1), Ordospora colligata OC4 (JOKQ00000000.1), Spraguea lophii 42_110 (ATCN00000000.1), Trachipleistophora hominis (ANCC00000000.1), Vavraia culicis subspecies floridensis (AEUG00000000.1) and Vittaforma corneae ATCC 50505 (AEYK00000000.1). Additionally, the genome of the cryptomycotan species Rozella allomycis (ATJD00000000.1) was also retrieved.
Gene clustering and phylogenetic tree reconstruction
OrthoMCL v.2 (Li, Stoeckert, and Roos 2003) was used to conduct comparative genomic analyses of P. neurophilia predicted proteome with 20 other published microsporidian proteomes by identifying orthologous proteins within microsporidian genomes. Families were reconstructed using OrthoMCL with an e-value cut-off of 1e-10. Only families with more than one member were kept for further analyses. In house script was then used to convert the OrthoMCL standard output into a numeric table (Supp Table 4). 62 families were represented by a single member from each genome and were retained for phylogenetic analysis. To this end, members of each family were aligned using Muscle V3.8.31 (Edgar 2004) and the resulting 62 alignments were concatenated using the bioinformatics software Geneious R9 (Kearse et al. 2012). Concatenated alignments were trimmed using TrimAl V1.2 (Capella-Gutierrez, Silla-Martinez, and Gabaldon 2009) and the best-fit model for phylogenetic analysis was estimated using ProtTest V3.4 (Darriba et al. 2011) based on the Aikaike Information Criterion. To reconstruct the microsporidian phylogenetic tree, the best phylogenetic model was implemented in PhyloBayes-MPI V1.5 (Lartillot, Lepage, and Blanquart 2009) and PhyML 3.0 (Guindon et al. 2010) using posterior probabilities and 100 bootstraps branch support for the Bayesian and Maximum Likelihood analyses respectively.
Detection of putative transposable elements
Transposable elements from P. neurophilia were obtained following BLASTx search against the NCBI nr database to identify candidates with similarities with published TEs (evalue cut-off of 1e–10). Only candidates demonstrating the presence of previously described microsporidian TEs conserved domains (Parisot et al. 2014) within putative ORF were retained for further analyses. Based on the presence of specific conserved domains, P. neurophilia transposable elements were classified among 4 major families; LTR retrotransposon, Piggyback, Mariner and Merlin family. For comparison purpose, TEs in other microsporidians were identified using the same methodology. Bayesian phylogenetic analyses were performed using previously described transposable elements (Parisot et al. 2014) and reconstructed using the previously described phylogenetic tree reconstruction methodology (Bayesian approach).
Heterozygosity in P. neurophilia
Polymorphic sites were identified by two independent variant callers FreeBayes v0.9.18–3 and PoPoolation V1.2.2 following the alignment of the reads to the reference genome assembly of P. neurophilia using the Burrows-Wheeler Alignment tool BWA V0.7.12 with the BWA-ALN algorithm. The resulting sequence alignment map (SAM) file was then converted to the FreeBayes required input sorted BAM file using SAMtools. Polymorphic sites were then identified using the variant caller Freebayes. Variants were filtered using vcffilter from the C++ library vcflib to retain SNPs (TYPE = snp) found within a 25% interval of the average genome coverage (0.75 X > DP < 1.25 X genome coverage) that possessed only one alternative allele (NUMALT = 1). Resulting SNPs were retrieved and plotted to evaluate their frequency as well as their distribution within the genome using environment software for statistical computing and graphics R V3.2.1. K-mer coverage distribution was also performed to infer P. neurophilia’s ploidy using K-mergenie V1.6982
Gene order conservation
Region with potential gene order conservation were identify using SynMap tool from the CoGe platform (Lyons and Freeling 2008). To this end, previously published annotated microsporidian genomes were transferred into the platform and analyzed using the Quota Align Merge Algorithm with the DAGChainer Option “Relative Gene Order”. Flanking regions of identified regions were then manually inspected for potential expansion of the predicted region with gene synteny.
PCR and Sanger sequencing
Selected regions with variable single nucleic polymorphic allele frequencies were amplified by PCR (Table S10) using standard parameters and sequenced using Sanger Sequencing (McGill University and Génome Québec Innovation, Montréal, Canada).
Meiosis Related Genes (MRGs) distribution in microsporidia
Meiosis related genes, as described in Lee et al. (Lee, Heitman, and Ironside 2014), were retrieved from the NCBI database and used as queries against the Pseudoloma neurophilia’s genome (tBlastn; E-value cut-off of 1x e−20). Search of MRGs in other microsporidian genomes were conducted using the same methodology. True homology was confirmed using reciprocal blast procedures and by reconstructing the phylogenies of each MRG found in microsporidian genomes and distantly related species. To this end, MRGs homologs were aligned using Muscle V3.8.31 (Edgar 2004) and the resulting alignments were trimmed using TrimAl V1.2 (Capella-Gutierrez, Silla-Martinez, and Gabaldon 2009). The best-fit model for phylogenetic analysis was estimated for each alignments using ProtTest V3.4 (Darriba et al. 2011) based on the Aikaike Information Criterion and respective phylogenetic trees were reconstructed using PhyloBayes-MPI V1.5 (Lartillot, Lepage, and Blanquart 2009) with posterior probabilities.
RESULTS
A typical, small and likely diploid microsporidian genome
P. neurophila cannot be presently cultured outside of its original host, and its spores are located in the central nervous system of infected zebrafish, hampering spore isolation. In this study, a mere 29 ng of DNA was extracted from spores isolated from the neural system of 50 infected Danio rerio individuals (Fig. 1). This small amount of DNA was subjected to paired-end Illumina sequencing using the Miseq platform, resulting in 7,230,699 paired-end reads with an average length of 250 bp. Reads were assembled using Spades v3.0.0, resulting in an assembly of 31,043 contigs. Because of the obligate intracellular lifestyle of P. neurophilia, a large number of contigs were of obvious contaminant origin. Plotting coverage against GC content revealed a large number of highly covered contigs, as well as many others with low coverage and high GC content (Fig S1–2). Obvious contaminants contigs harbor lower coverage, and a GC content higher than what is typically observed in microsporidia (average GC: 38.16, (Selman et al. 2013)). Interestingly, low covered contaminants of microsporidian origin were also found (e.g. Encephalitozoon cuniculi. E. romaleae, Nosema ceranae,). Although it is possible that these represent bona-fide microsporidian infection that are less frequent (much lower coverage), they most probably originate from low-amounts of contaminant DNA from sequencing projects in parallel performed in our lab. Unsurprisingly, non-microsporidian contaminations with high coverage included animal mitochondrial genomes (e.g. the D. rerio mtDNA), as well as nuclear sequences affiliated with aquatic animals (the water snail Lottia gigantea, Hydra vulgaris, Branchiostoma sp).
All highly covered contigs with expected GC content were manually inspected for their microsporidian origin and were chosen as representative of the P. neurophilia genome. The total genome assembly of P. neurophilia is 5.25 Mb (Table 1), and is composed of 1603 contigs. These numbers compare well with similar studies performed on other microsporidia with similar genome sizes (Pan et al. 2013; Akiyoshi et al. 2009; Campbell et al. 2013).
Table 1.
Genome statistics of Pseudoloma neurophilia
| GENOME STATISTICS OF P. neurophilia | |
|---|---|
| Haploid genome size (Mb) | 5.3 |
| Total length >= 1000 bp (Mb) | 4.9 |
| Contigs | 1603 |
| N50 | 6148 |
| Largest contig | 172544 |
| Average genome coverage | 138X |
| GC content (%) | 29.6 |
| GC content coding (%) | 31.4 |
| Coding regions (Mb) | 2.9 |
| Genes | 3633 |
| SNPs frequency (SNPs/kb) | 1.98 |
| Candidate effectors* | 111 |
| *Defined as predicted secreted proteins of unknown function | |
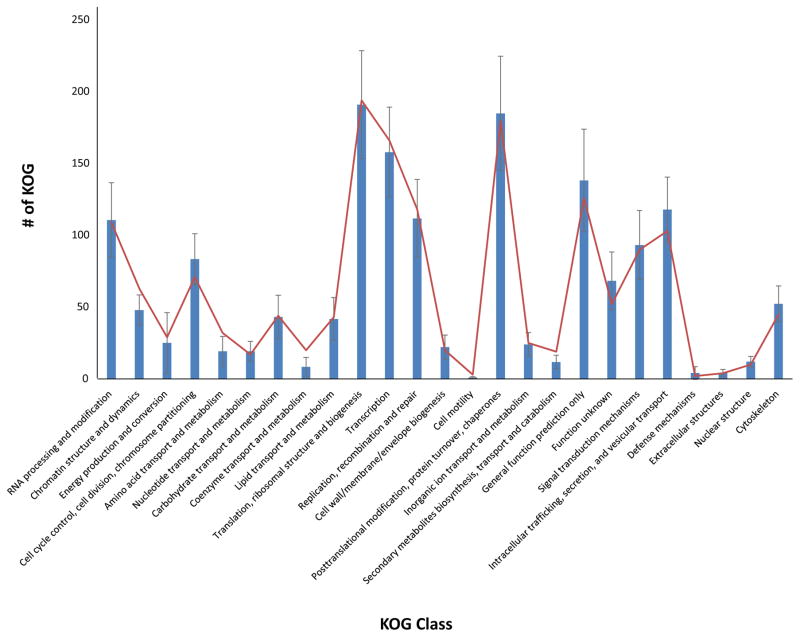
The genome is gene-dense, with 3645 predicted genes and coding regions encompassing 51% of the genome. Comparing Open Reading Frames (ORFs) counts and biological categories with publicly available microsporidian genome reveals that the P. neurophilia contigs harbor a typical microsporidian protein repertoire, and indicates that most of the genome space has likely been sampled (Fig. 2, Table S1). No evidence for the existence of spliceosomal introns was found. Interestingly, a third of the predicted ORFs (1,139) has no homologues in publicly available database, and can therefore be considered as Pseudoloma-specific. Protein domain analyses identified 196 proteins with signal peptide cleavage sites, indicating that these are probably secreted (Table S2) (Petersen et al. 2011), and more than half of these (111) are Pseudoloma-specific genes. These unique P. neurophilia homologues may represent candidate effectors (Campbell et al. 2013), whose function could be tested in the future using this host-parasite system (P. neurophilia- Danio rerio).
Figure 2.
Mean KOG Class distribution among microsporidian genomes (blue; with standard deviation) with observed KOG domain in Pseudoloma neurophilia (red).
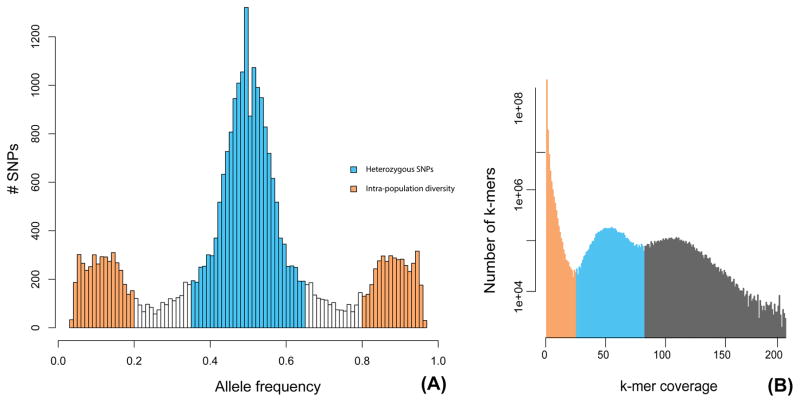
To date, genome analyses of microsporidia have suggested that the genomes of these intracellular parasites are mainly diploid, although evidence of polyploidy has also been observed in species with diplokaryons (Cuomo et al. 2012; Adrian Pelin et al. 2015).. Here, we tested the ploidy levels of P. neurophilia by plotting the allele frequency of single nucleotide polymorphisms (SNPs) along all contigs. These analyses identified 7,246 SNPs with a 0.5 frequency which is strongly indicative of diploidy (Fig. 3–A), and this pattern was independently confirmed using a K-mer coverage distribution (Fig. 3–B). The remaining 3,162 SNPs were found to have frequencies between 0.1–0.4. Sanger sequencing of selected regions confirmed the heterozygous nature of SNPs with a 0.5 frequency, but did not confirm the existence of SNPs with lower-frequencies. It has been recently shown that the number of low-frequency variants correlate with Illumina sequence quality - i.e. lower quality result in more sequencing errors, which are often of low frequency (A. Pelin et al. 2016). Therefore, these low-frequency most likely represent a similar mixture of sequencing errors and bona-fide polymorphisms present among spores isolated from the 50 infected zebrafish individuals (Fig S3). The overall SNP frequency in P. neurophilia is 1.98 SNP/Kb, in line to what has been reported from other microsporidian species (Table S3; (Cuomo et al. 2012; Desjardins et al. 2015; James et al. 2013; Adrian Pelin et al. 2015; Selman et al. 2013)). With 58.85% of SNPs occurring in coding regions of this gene-dense genome, the SNPs distribution also reflects what is observed in other microsporidian species (Cuomo et al. 2012).
Figure 3.
A. SNP allele frequency distribution within the genome of Pseudoloma neurophilia. B. K-mer calculated for k= 71. Proposed heterozygous variants (blue) and homozygous variants (orange) are highlighted.
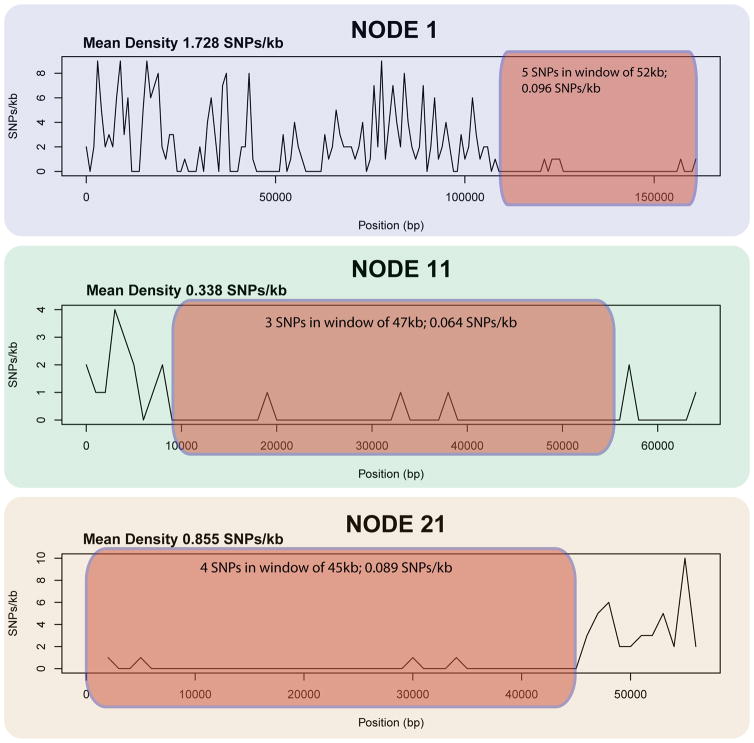
Finally, our SNP analyses also revealed regions with loss of heterozygosity (LOH, Fig. 4). None of the genomic regions affected by LOH showed signs of aneuploidy– i.e no drops in coverage was identified along these contigs - therefore, there is no evidence that LOH is linked with hemizygosity. LOH has been hypothesized to play a role in pathogenicity of distant pathogens, such as the yeast Candida albicans (Diogo et al. 2009) or the stramenopile Phytophtora capsici (Lamour et al. 2012). In these species, LOH has been proposed to facilitate adaptation to external negative stimuli (new host or chemical modification) through allele fixation, but its effect on microsporidia is unknown. In P. neurophilia, these homozygous regions can attain a maximum of 52 kb and harbor allele frequencies 30 times lower than the genome average. Genes affected by LOH have a variety of functions, and include 7 hypothetical highly similar ORFs with secretion signals that may have originated from successive duplication events as indicated by their phylogenetic clustering and high similarity (Fig S4, Table S4–S5).
Figure 4.
Large contigs with LOH (red). SNP count within region is shown.
Unique and lineage specific genes in P. neurophilia
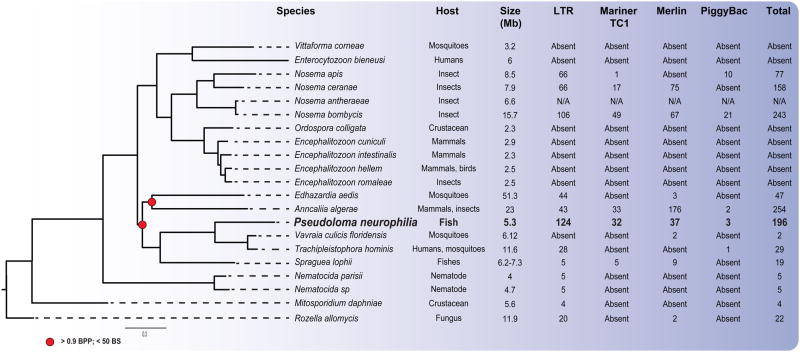
Comparing gene content between P. neurophilia and 20 publicly available microsporidian genomes (Table S6) allowed us to identify 62 single copy orthologues shared by all sequenced species. These were used to reconstruct a reliable phylogeny of the group, and showed that P. neurophilia groups with fish/mosquitoes microsporidians (Fig. 5). The tree topology is consistent with published phylogenies, and demonstrates that P. neurophilia is closely related to Trachipleistophora hominis, Vavraia culicis floridensis, Spraguea lophii ( Vossbrinck and Debrunner-Vossbrinck 2005). Consistent with this, our analysis revealed that the genome of P. neurophilia shares a substantially higher number of gene families with its most closely related species (T. hominis and V. culicis) than with any other member of the group (n=1,256 out of 1,558 orthologous groups present in P. neurophilia). Families shared by these species but absent in distant lineages include some with similarities to metabolically relevant proteins include the N-arginine dibasic convertase NRD1 and the glutaredoxin protein, as well as several ORFs with unknown function (Table S7). All species in this subclade also share the machinery required for the RNA interference pathway, as well as several putative effector proteins (Paldi et al. 2010; Heinz et al. 2012; Campbell et al. 2013). One example of effector proteins includes the ricin b lectin proteins, which were previously proposed to play a role in the pathogenicity of the microsporidia by facilitating its adhesion on the host cell during infection (Campbell et al. 2013). Consistent with what has been reported in the microsporidium S. lophii, signal peptide cleavage sites are also present in P. neurophilia ricin b lectin genes, suggesting that these proteins are probably also secreted into the host cell and play a role in the host-parasite interaction.
Figure 5.
Consensus phylogenetic tree of microsporidia reconstructed using maximum likelihood and Bayesian approaches with Rozella allomycis serving as an outgroup (Cryptomycota). The total numbers of transposable elements in each microsporidian with a sequenced genome are shown. Support values for nodes over 85 bootstrap values and 1 Bayesian posterior probability for Maximum Likelihood and Bayesian algorithms, respectively, are omitted.
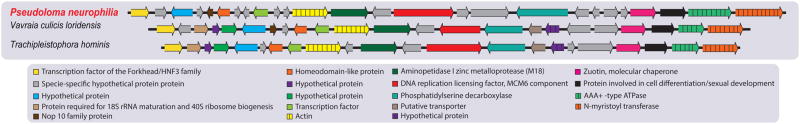
The level of synteny among P. neurophilia and closely related species appears extensive, confirming that the maintenance of gene order is common in microsporidia, and is in sharp contrast with their notoriously elevated sequence divergence (Fig. 6, Fig. S5) (Nakjang et al. 2013). At the extreme, up to 18 genes encompassing 44 kb have been found in highly conserved order among P. neurophilia, T. hominis and V. culicis. As in other microsporidia, several meiosis-related genes appear to be absent in P. neurophilia, another indication that this universal sexual process may have been heavily streamlined in these parasites. When compared to its closely related species, P. neurophilia is the microsporidian species where the fewest meiosis-related genes (MRG) have been found (Table S8). Targeted searches for meiosis-specific motifs within sequence reads and small contigs has been unsuccessful, so this slightly reduced MRG numbers is unlikely to result from genomic regions missing from data our genome assembly. To date, only three microsporidian assemblies contain less MRGs than P. neurophilia (Nosema apis, Mitosporidium daphniae and Enterocytozoon bieneusi). Phylogenies of P. neurophila MRGs highlight the common descent and likely orthology of all microsporidian MRG homologues, and underpin their extensive sequence divergence (Fig. S6).
Figure 6.
Examples of synteny identified between the genome of P. neurophilia and closely related microsporidians. A. Schematic representation of gene order conservation of thirteen proteins shared between P. neurophilia and closely related microsporidians found in contig 22.
RNA interference and in-frame inserts in conserved protein encoding genes
Interestingly, P. neurophilia harbors homologues of three key proteins involved in RNA-induced silencing complex. These include the argonaute, the dicer protein homologues and the RNA-dependent RNA polymerase. These homologues are also found in closely related species and other fungi (Heinz et al. 2012; Campbell et al. 2013; Dang et al. 2011) and have been hypothesised to be involved in transposon silencing through RNA-interference (Heinz et al. 2012; Obbard et al. 2009). The presence of these orthologues in almost all microsporidian genera (Table 2) suggests that such defense mechanism may be common to most microsporidians. While components of the RNA-induced silencing complex have not been demonstrated to be directly involved in gene regulation or affect transposons in microsporidia, their role in RNA interference pathway has been recently proposed in the bee pathogen Nosema ceranae (Paldi et al. 2010).
Table 2.
Distribution of the RNA interference key components within the microsporidian and cryptomycotan clade.
| Micropsoridian | Argonaute | Dicer | RNA-dependent RNA polymerase |
|---|---|---|---|
| Pseudoloma neurophilia | ✓ | ✓ | ✓ |
| Vavraia culics floridensis | ✓ | ✓ | ✓ |
| Trachipleistophora homins | ✓ | ✓ | ✓ |
| Spraguea lophii | ✓ | ✓ | ✓ |
| Anncaliia algerae | ✓ | ✓ | ✓ |
| Edhazardia aedis | ✓ | ✓ | ✓ |
| Nosema apis | ✓ | ✓ | ✓ |
| Nosema ceranae | ✓ | ✓ | ✓ |
| Vittaforma corneae | ✓ | ✓ | ✓ |
| Rozella allomycis | ✓ | - | ✓ |
| Mitosporidium daphniae | ✓ | ✓ | - |
| Nosema bombycis | ✓ | ✓ | - |
| Nosema antheraeae | - | ✓ | - |
| Nematocida parisii | - | - | - |
| Nematocida sp. | - | - | - |
| Encephalitozoon culiculi | - | - | - |
| Encephalitozoon hellem | - | - | - |
| Encephalitozoon intestinalis | - | - | - |
| Encephalitozoon romaleae | - | - | - |
| Enterocytozoon bieneusi | - | - | - |
| Ordospora colligata | - | - | - |
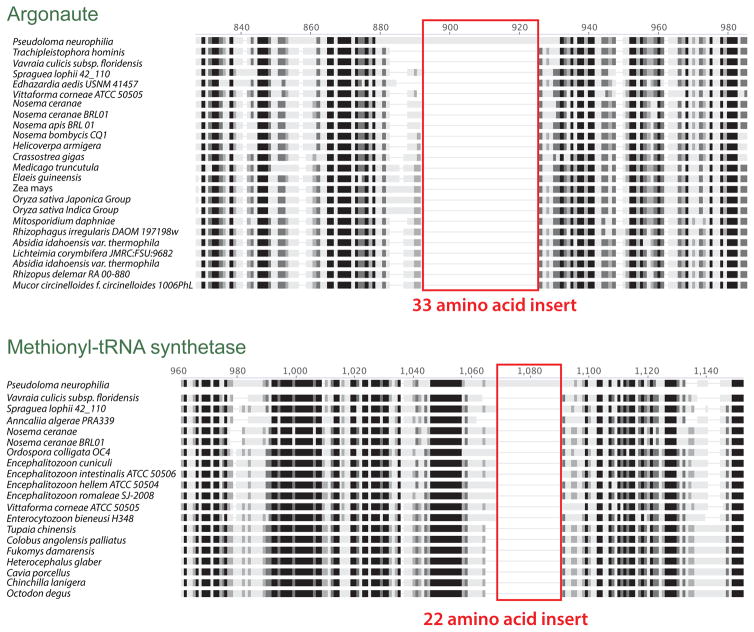
Surprisingly, we found that the argonaute protein harbors a 99-bp sequence insert within a region that is essential for its function in model organisms (the PIWI domain –Höck and Meister 2008) and that is conserved among microsporidian orthologues (Fig. 7). Genome explorations exposed many similar inserts elsewhere in the genome. In all cases, these inserts are found in-frame (i.e. none lead to stop codons), and do not demonstrate similarities with known introns (i.e. no obvious splicing sites, always in frame). These are probably analogous to inserts first reported in several genes of Hamiltosporidium tvaerminnensis (Corradi et al. 2009); a distant microsporidium that infects the crustacean Daphnia magna. In P. neurophilia, 30 conserved protein-encoding genes have been observed to harbor such features (Table S9). These insertions are surrounded by amino-acid sequences that are otherwise highly conserved among microsporidia and other eukaryotes, so there is a possibility that these affect the function of these proteins. Searching for similar inserts in other microsporidian lineages reveals that these intriguing genomic signatures are ubiquitous in this group of parasites, although their sequence and location always varies among species (Table S9). Indeed, protein encoding genes affected by these insertions are always different, with no clear functional pattern emerging from our analyses.
Figure 7.
Protein alignments highlighting the presence of large in-frame inserts identified in this study. The location of inserts is shown in red. The alignments highlight the general sequence conservation of these protein-encoding genes among microsporidia and members of other phyla.
Transposable element family expansion in a reduced genome
Sequence homology searches showed that the putative biochemical repertoire of P. neurophilia is very similar to that of other sequenced microsporidian species (Fig. 2, Table S1). However, multiple gene family expansions within P. neurophilia of notable interest are also found, the most compelling including transposable families. Indeed, the genome of P. neurophilia harbors a total of 142 ORFs whose sequences encode for proteins with conserved microsporidian transposable element domains, including DNA and retrotransposons (Parisot et al. 2014). The number of TEs in P. neurophilia is much more elevated than in closely related species with which it shares high sequence and gene content similarities (Fig. 5).
While TEs are numerous in a few select microsporidia with very large genomes (Parisot et al. 2014), the presence of TEs are otherwise rare in many members of the group; including the basal Nematocida sp. and the more derived Encephalitozoon sp. (Fig. 5). In this context, P. neurophilia is notable, as approximately 3% of its genome sequences (or 196 ORFs) are occupied by TEs. Most of these transposons have homologs in distant members of the group, but none of them originated from horizontal gene transfer (Parisot et al. 2014; Watson et al. 2015). In fact, there is evidence that some TE families may have been fuelled by numerous, and probably recent, duplication events, as highlighted by the high sequence similarity of several members of this family (Fig S7–S10).
The very high numbers of TEs in P. neurophilia (compared to most species in the group) almost certainly affect the biology of this parasite. Indeed, TEs possess the ability to relocate and/or duplicate along genome and can have potential negative repercussion on the organism by creating pseudogenes (Biémont 2010), so ensuring that their number do not inflate beyond control is an essential defense mechanism for any organism. One gene involved in gene silencing of TEs is the Argonaute gene (McCue et al. 2015; Thomson and Lin 2009). This gene is present in P. neurophilia and contains one of the abovementioned insertions. However, the presence of the insertion in a key element of the Argonaute gene does not seem to be associated with the expansion of TE in P. neurophilia, because TEs are also abundant in species with more conventional Argonaute genes (i.e. Nosema ceranae, N. bombycis, Figure 5,7). TE proliferation is also linked with the existence of sexual reproduction (Arkhipova and Meselson 2000; Lee, Heitman, and Ironside 2014; Wright and Finnegan 2001), a notion that is supported by the presence of diploidy and MRGs in P. neurophilia.
DISCUSSION
Acquiring a genome reference for P. neurophilia is a first step in understanding its biology. These sequences also pave the road for future analyses of host-parasite molecular interactions, using RNA-seq or other NGS tools. We demonstrated that this relevant parasite of zebrafish harbors a genome of small haploid size relative to most members of the group, and that it is closely related in both content and structure with those of T. hominis and V. culicis. Despite these similarities, a third of the P. neurophilia genome is specific to this species, with many of these unique genes harboring signal peptide cleavage sites that indicate their putative secretion and involvement in host manipulation (e.g. effectors).
Our analyses also support P. neurophilia as a diploid organism that harbors genomic signatures that are linked with both sexual (ploidy, TE abundance) and clonal modes of evolution (LOH, possibly the reduced number of MRG). Comparative analysis between P. neurophilia and other sequenced microsporidia demonstrated the presence of an unusual high amount of TEs inside this small genome. This is surprising as all sequenced microsporidian genomes with similar sizes harbor no, or low amounts of TE. TEs abundance does not seem to be affected by the presence of key components of the RNAi pathways (RNA-dependent RNA polymerase, Dicer and Argonaute proteins) that silence the expression of TEs and subsequently and slow-down their expansion (Obbard et al. 2009). This machinery has been demonstrated to be functioning in the bee pathogen N. ceranae, and future studies on P. neurophilia will hopefully provide more insight into the regulatory role of RNA interference in other microsporidians. Lastly, the discovery of in-frame inserts in coding regions usually highly conserved, and their ubiquitous presence in this group may also be of primary relevance for understanding these parasites. In particular, their universal presence in microsporidia, coupled with their localization in many homologues with known cellular function, suggest that these may have important effects on the biology of these parasites. Hopefully, recent advances in the development of genetic techniques in intracellular parasites may soon be applicable to study microsporidia (Vinayak et al. 2015). Such techniques will be essential to determine the function of any microsporidia genomic region, including those atypical insertions.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their comments on a previous version of this manuscript. Nicolas Corradi is a Fellow of the Canadian Institute for Advanced Research. This work was supported by the Discovery program from the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery).
Footnotes
Data deposition: Pseudoloma neurophilia MK1 (LGUB00000000.1)
LITERATURE CITED
- Akiyoshi Donna E, Morrison Hilary G, Lei Shi, Feng Xiaochuan, Zhang Quanshun, Corradi Nicolas, Mayanja Harriet, et al. Genomic Survey of the Non-Cultivatable Opportunistic Human Pathogen, Enterocytozoon bieneusi. PLOS Pathog. 2009;5(1):e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila-Garcia AM, Fast NM. Microsporidian Infection in a Free-Living Marine Nematode. Eukaryotic Cell. 2012;11(12):1544–51. doi: 10.1128/EC.00228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova Irina, Meselson Matthew. Transposable Elements in Sexual and Ancient Asexual Taxa. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14473–77. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich Anton, Nurk Sergey, Antipov Dmitry, Gurevich Alexey A, Dvorkin Mikhail, Kulikov Alexander S, Lesin Valery M, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznoussenko Galina V, Dolgikh Viacheslav V, Seliverstova Elena V, Semenov Petr B, Tokarev Yuri S, Trucco Alvar, Micaroni Massimo, et al. Analogs of the Golgi Complex in Microsporidia: Structure and Avesicular Mechanisms of Function. Journal of Cell Science. 2007;120(7):1288–98. doi: 10.1242/jcs.03402. [DOI] [PubMed] [Google Scholar]
- Biémont Christian. A Brief History of the Status of Transposable Elements: From Junk DNA to Major Players in Evolution. Genetics. 2010;186(4):1085–93. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell Scott E, Williams Tom A, Yousuf Asim, Soanes Darren M, Paszkiewicz Konrad H, Williams Bryony AP. The Genome of Spraguea lophii and the Basis of Host-Microsporidian Interactions. PLoS Genet. 2013;9(8):e1003676. doi: 10.1371/journal.pgen.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez Salvador, Silla-Martinez Jose M, Gabaldon Toni. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics. 2009;25(15):1972–73. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi Nicolas. Microsporidia: Eukaryotic Intracellular Parasites Shaped by Gene Loss and Horizontal Gene Transfers. Annual Review of Microbiology. 2015;69(1):167–83. doi: 10.1146/annurev-micro-091014-104136. [DOI] [PubMed] [Google Scholar]
- Corradi Nicolas, Haag Karen L, Pombert Jean-Francois, Ebert Dieter, Keeling Patrick J. Draft Genome Sequence of the Daphnia Pathogen Octosporea bayeri: Insights into the Gene Content of a Large Microsporidian Genome and a Model for Host-Parasite Interactions. Genome Biology. 2009;10(10):R106–R106. doi: 10.1186/gb-2009-10-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi Nicolas, Pombert Jean-Francois, Farinelli Laurent, Didier Elizabeth S, Keeling Patrick J. The Complete Sequence of the Smallest Known Nuclear Genome from the Microsporidian Encephalitozoon intestinalis. Nature Communications. 2010;1:77–77. doi: 10.1038/ncomms1082. Journal Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi Nicolas, Slamovits Claudio H. The Intriguing Nature of Microsporidian Genomes. Briefings in Functional Genomics. 2011;10(3):115–24. doi: 10.1093/bfgp/elq032. [DOI] [PubMed] [Google Scholar]
- Cuomo Christina A, Desjardins Christopher A, Bakowski Malina A, Goldberg Jonathan, Ma Amy T, Becnel James J, Didier Elizabeth S, et al. Microsporidian Genome Analysis Reveals Evolutionary Strategies for Obligate Intracellular Growth. Genome Research. 2012;22(12):2478–88. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Yunkun, Yang Qiuying, Xue Zhihong, Liu Yi. RNA Interference in Fungi: Pathways, Functions, and Applications. Eukaryotic Cell. 2011;10(9):1148–55. doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba Diego, Taboada Guillermo L, Doallo Ramon, Posada David. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics. 2011;27(8):1164–65. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins Christopher A, Sanscrainte Neil D, Goldberg Jonathan M, Heiman David, Young Sarah, Zeng Qiandong, Madhani Hiten D, Becnel James J, Cuomo Christina A. Contrasting Host–pathogen Interactions and Genome Evolution in Two Generalist and Specialist Microsporidian Pathogens of Mosquitoes. Nature Communications. 2015;6(May) doi: 10.1038/ncomms8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo Dorothée, Bouchier Christiane, d’Enfert Christophe, Bougnoux Marie-Elisabeth. Loss of Heterozygosity in Commensal Isolates of the Asexual Diploid Yeast Candida albicans. Fungal Genetics and Biology. 2009;46(2):159–68. doi: 10.1016/j.fgb.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Research. 2004;32(5):1792–97. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson Olof, Nielsen Henrik, Brunak Søren, von Heijne Gunnar. Predicting Subcellular Localization of Proteins Based on Their N-Terminal Amino Acid Sequence. Journal of Molecular Biology. 2000;300(4):1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Guindon Stephane, Dufayard Jean-Francois, Lefort Vincent, Anisimova Maria, Hordijk Wim, Gascuel Olivier. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology. 2010;59(3):307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Heinz Eva, Williams Tom A, Nakjang Sirintra, Noël Christophe J, Swan Daniel C, Goldberg Alina V, Harris Simon R, et al. The Genome of the Obligate Intracellular Parasite Trachipleistophora hominis: New Insights into Microsporidian Genome Dynamics and Reductive Evolution. PLoS Pathog. 2012;8(10) doi: 10.1371/journal.ppat.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higes Mariano, Meana Aranzazu, Bartolome Carolina, Botias Cristina, Martin-Hernandez Raquel. Nosema ceranae (Microsporidia), a Controversial 21st Century Honey Bee Pathogen. Environmental Microbiology Reports. 2013;5(1):17–29. doi: 10.1111/1758-2229.12024. [DOI] [PubMed] [Google Scholar]
- Höck Julia, Meister Gunter. The Argonaute Protein Family. Genome Biology. 2008;9(2):210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo Fernando, Antonio Jose, Hermida Castro, Fenoy Soledad, Mezo Mercedes, Gonzalez-Warleta Marta, del Aguila Carmen. Detection of Microsporidia in Drinking Water, Wastewater and Recreational Rivers. Water Research. 2011;45(16):4837–43. doi: 10.1016/j.watres.2011.06.033. [DOI] [PubMed] [Google Scholar]
- James Timothy Y, Pelin Adrian, Bonen Linda, Ahrendt Steven, Sain Divya, Corradi Nicolas, Stajich Jason E. Shared Signatures of Parasitism and Phylogenomics Unite Cryptomycota and Microsporidia. Current Biology. 2013;23(16):1548–53. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- Karpov Sergey A, Mamkaeva Maria A, Aleoshin Vladimir V, Nassonova Elena, Lilje Osu, Gleason Frank H. Morphology, Phylogeny, and Ecology of the Aphelids (Aphelidea, Opisthokonta) and Proposal for the New Superphylum Opisthosporidia. Frontiers in Microbiology. 2014;5(March) doi: 10.3389/fmicb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse Matthew, Moir Richard, Wilson Amy, Stones-Havas Steven, Cheung Matthew, Sturrock Shane, Buxton Simon, et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics. 2012;28(12):1647–49. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling Patrick J, Corradi Nicolas, Morrison Hilary G, Haag Karen L, Ebert Dieter, Weiss Louis M, Akiyoshi Donna E, Tzipori Saul. The Reduced Genome of the Parasitic Microsporidian Enterocytozoon bieneusi Lacks Genes for Core Carbon Metabolism. Genome Biology and Evolution. 2010;2(January):304–9. doi: 10.1093/gbe/evq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent Michael L, Harper Claudia, Wolf Jeffrey C. Documented and Potential Research Impacts of Subclinical Diseases in Zebrafish. Ilar Journal. 2012;53(2):126–34. doi: 10.1093/ilar.53.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler Robert, Orozco-terWengel Pablo, De Maio Nicola, Pandey Ram Vinay, Nolte Viola, Futschik Andreas, Kosiol Carolin, Schlötterer Christian. PoPoolation: A Toolbox for Population Genetic Analysis of Next Generation Sequencing Data from Pooled Individuals. PLoS ONE. 2011;6(1):e15925. doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour Kurt H, Mudge Joann, Gobena Daniel, Hurtado-Gonzales Oscar P, Schmutz Jeremy, Kuo Alan, Miller Neil A, et al. Genome Sequencing and Mapping Reveal Loss of Heterozygosity as a Mechanism for Rapid Adaptation in the Vegetable Pathogen Phytophthora capsici. Molecular Plant-Microbe Interactions3: MPMI. 2012;25(10):1350–60. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot Nicolas, Lepage Thomas, Blanquart Samuel. PhyloBayes 3: A Bayesian Software Package for Phylogenetic Reconstruction and Molecular Dating. Bioinformatics. 2009;25(17):2286–88. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lee Soo Chan, Heitman Joseph, Ironside Joseph E. Sex and the Microsporidia. In: Weiss Louis M, Becnel James J., editors. Microsporidia. John Wiley & Sons, Inc; 2014. pp. 231–43. http://onlinelibrary.wiley.com/doi/10.1002/9781118395264.ch8/summary. [Google Scholar]
- Li Li, Stoeckert Christian J, Roos David S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Research. 2003;13(9):2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo Maria Luísa, Xiao Lihua, Antunes Francisco, Matos Olga. Microsporidia as Emerging Pathogens and the Implication for Public Health: A 10-Year Study on HIV-Positive and -Negative Patients. International Journal for Parasitology. 2012;42(2):197–205. doi: 10.1016/j.ijpara.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Lyons Eric, Freeling Michael. How to Usefully Compare Homologous Plant Genes and Chromosomes as DNA Sequences. The Plant Journal. 2008;53(4):661–73. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Matthews Jennifer L, Brown Amanda MV, Larison Karen, Bishop-Stewart Janell K, Rogers Paul, Kent Michael L. Pseudoloma neurophilia N. G., N. Sp., a New Microsporidium from the Central Nervous System of the Zebrafish (Danio Rerio) The Journal of Eukaryotic Microbiology. 2001;48(2):227–33. doi: 10.1111/j.1550-7408.2001.tb00307.x. [DOI] [PubMed] [Google Scholar]
- McCue Andrea D, Panda Kaushik, Nuthikattu Saivageethi, Choudury Sarah G, Thomas Erica N, Slotkin R Keith. ARGONAUTE 6 Bridges Transposable Element mRNA-Derived siRNAs to the Establishment of DNA Methylation. The EMBO Journal. 2015;34(1):20–35. doi: 10.15252/embj.201489499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray Katrina N, Dreska Mathew, Nasiadka Andrzej, Rinne Miranda, Matthews Jennifer L, Carmichael Carrie, Bauer Justin, Varga Zoltan M, Westerfield Monte. Transmission, Diagnosis, and Recommendations for Control of Pseudoloma Neurophilia Infections in Laboratory Zebrafish (Danio Rerio) Facilities. Comparative Medicine. 2011;61(4):322–29. [PMC free article] [PubMed] [Google Scholar]
- Nakjang Sirintra, Williams Tom A, Heinz Eva, Watson Andrew K, Foster Peter G, Sendra Kacper M, Heaps Sarah E, Hirt Robert P, Martin Embley T. Reduction and Expansion in Microsporidian Genome Evolution: New Insights from Comparative Genomics. Genome Biology and Evolution. 2013;5(12):2285–2303. doi: 10.1093/gbe/evt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard Darren J, Gordon Karl HJ, Buck Amy H, Jiggins Francis M. The Evolution of RNAi as a Defence against Viruses and Transposable Elements. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364(1513):99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paldi Nitzan, Glick Eitan, Oliva Maayan, Zilberberg Yaron, Aubin Lucie, Pettis Jeffery, Chen Yanping, Evans Jay D. Effective Gene Silencing in a Microsporidian Parasite Associated with Honeybee (Apis mellifera) Colony Declines. Applied and Environmental Microbiology. 2010;76(17):5960–64. doi: 10.1128/AEM.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Guoqing, Xu Jinshan, Li Tian, Xia Qingyou, Liu Shao-Lun, Zhang Guojie, Li Songgang, et al. Comparative Genomics of Parasitic Silkworm Microsporidia Reveal an Association between Genome Expansion and Host Adaptation. BMC Genomics. 2013;14(March) doi: 10.1186/1471-2164-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisot Nicolas, Pelin Adrian, Gasc Cyrielle, Polonais Valerie, Belkorchia Abdel, Panek Johan, El Alaoui Hicham, et al. Microsporidian Genomes Harbor a Diverse Array of Transposable Elements That Demonstrate an Ancestry of Horizontal Exchange with Metazoans. Genome Biology and Evolution. 2014;6(9):2289–2300. doi: 10.1093/gbe/evu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelin Adrian, Selman Mohammed, Aris-Brosou Stéphane, Farinelli Laurent, Corradi Nicolas. Genome Analyses Suggest the Presence of Polyploidy and Recent Human-Driven Expansions in Eight Global Populations of the Honeybee Pathogen Nosema ceranae. Environmental Microbiology. 2015 Jul; doi: 10.1111/1462-2920.12883. n/a – n/a. [DOI] [PubMed] [Google Scholar]
- Pelin A, Moteshareie H, Sak B, Selman M, Naor A, Eyahpaise M-È, Farinelli L, Golshani A, Kvac M, Corradi N. The Genome of an Encephalitozoon cuniculi Type III Strain Reveals Insights into the Genetic Diversity and Mode of Reproduction of a Ubiquitous Vertebrate Pathogen. Heredity. 2016 Feb; doi: 10.1038/hdy.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen Thomas Nordahl, Brunak Søren, von Heijne Gunnar, Nielsen Henrik. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nature Methods. 2011;8(10):785–86. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Peyretaillade Eric, Parisot Nicolas, Polonais Valerie, Terrat Sebastien, Denonfoux Jeremie, Dugat-Bony Eric, Wawrzyniak Ivan, et al. Annotation of Microsporidian Genomes Using Transcriptional Signals. Nature Communications. 2012;3(October) doi: 10.1038/ncomms2156. [DOI] [PubMed] [Google Scholar]
- Sanders Justin L, Kent Michael L. The Zebrafish as a Model for Microsporidiosis. In: Weiss Louis M, Becnel James J., editors. Microsporidia. John Wiley & Sons, Inc; 2014. pp. 357–70. http://onlinelibrary.wiley.com/doi/10.1002/9781118395264.ch14/summary. [Google Scholar]
- Sanders Justin L, Watral Virginia, Clarkson Keri, Kent Michael L. Verification of Intraovum Transmission of a Microsporidium of Vertebrates: Pseudoloma neurophilia Infecting the Zebrafish, Danio Rerio. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0076064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders Justin L, Watral Virginia, Kent Michael L. Microsporidiosis in Zebrafish Research Facilities. Ilar Journal. 2012;53(2):106–13. doi: 10.1093/ilar.53.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman Mohammed, Sak Bohumil, Kváč Martin, Farinelli Laurent, Weiss Louis M, Corradi Nicolas. Extremely Reduced Levels of Heterozygosity in the Vertebrate Pathogen Encephalitozoon cuniculi. Eukaryotic Cell. 2013;12(4):496–502. doi: 10.1128/EC.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson M, Cawker J, Buske C, Gerlai R. Fishing for Genes of Vertebrate Behavior: Zebrafish as an Upcoming Model System.” In. Lab Anim. 2006;35:33–39. doi: 10.1038/laban0506-33. [DOI] [PubMed] [Google Scholar]
- Spagnoli Sean, Xue Lan, Kent Michael L. The Common Neural Parasite Pseudoloma neurophilia Is Associated with Altered Startle Response Habituation in Adult Zebrafish (Danio Rerio): Implications for the Zebrafish as a Model Organism. Behavioural Brain Research. 2015;291(September):351–60. doi: 10.1016/j.bbr.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson Travis, Lin Haifan. The Biogenesis and Function PIWI Proteins and piRNAs: Progress and Prospect. Annual Review of Cell and Developmental Biology. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak Sumiti, Pawlowic Mattie C, Sateriale Adam, Brooks Carrie F, Studstill Caleb J, Bar-Peled Yael, Cipriano Michael J, Striepen Boris. Genetic Modification of the Diarrhoeal Pathogen Cryptosporidium parvum. Nature. 2015;523(7561):477–80. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossbrinck Charles R, Debrunner-Vossbrinck Bettina A. Molecular Phylogeny of the Microsporidia: Ecological, Ultrastructural and Taxonomic Considerations. Folia Parasitologica. 2005;52(1–2):131–42. doi: 10.14411/fp.2005.017. discussion 130. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal RNA Sequence Suggests Microsporidia Are Extremely Ancient Eukaryotes. Nature. 1987;326(6111):411–14. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- Watson Andrew K, Williams Tom A, Williams Bryony AP, Moore Karen A, Hirt Robert P, Martin Embley T. Transcriptomic Profiling of Host-Parasite Interactions in the Microsporidian Trachipleistophora hominis. BMC Genomics. 2015;16(1) doi: 10.1186/s12864-015-1989-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Bryony AP, Hirt Robert P, Lucocq John M, Martin Embley T. A Mitochondrial Remnant in the Microsporidian Trachipleistophora hominis. Nature. 2002;418(6900):865–69. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Wright Stephen, Finnegan David. Genome Evolution: Sex and the Transposable Element. Current Biology. 2001;11(8):R296–99. doi: 10.1016/S0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.