Abstract
The genus Aeromonas is ubiquitous in aquatic environments encompassing a broad range of fish and human pathogens. Aeromonas strains are known for their enhanced capacity to acquire and exchange antibiotic resistance genes and therefore, are frequently targeted as indicator bacteria for monitoring antimicrobial resistance in aquatic environments. This study evaluated temporal trends in Aeromonas diversity and antibiotic resistance in two adjacent semi-intensive aquaculture facilities to ascertain the effects of antibiotic treatment on antimicrobial resistance. In the first facility, sulfadiazine-trimethoprim was added prophylactically to fingerling stocks and water column-associated Aeromonas were monitored periodically over an 11-month fish fattening cycle to assess temporal dynamics in taxonomy and antibiotic resistance. In the second facility, Aeromonas were isolated from fish skin ulcers sampled over a 3-year period and from pond water samples to assess associations between pathogenic strains to those in the water column. A total of 1200 Aeromonas isolates were initially screened for sulfadiazine resistance and further screened against five additional antimicrobials. In both facilities, strong correlations were observed between sulfadiazine resistance and trimethoprim and tetracycline resistances, whereas correlations between sulfadiazine resistance and ceftriaxone, gentamicin, and chloramphenicol resistances were low. Multidrug resistant strains as well as sul1, tetA, and intI1 gene-harboring strains were significantly higher in profiles sampled during the fish cycle than those isolated prior to stocking and these genes were extremely abundant in the pathogenic strains. Five phylogenetically distinct Aeromonas clusters were identified using partial rpoD gene sequence analysis. Interestingly, prior to fingerling stocking the diversity of water column strains was high, and representatives from all five clusters were identified, including an A. salmonicida cluster that harbored all characterized fish skin ulcer samples. Subsequent to stocking, diversity was much lower and most water column isolates in both facilities segregated into an A. veronii-associated cluster. This study demonstrated a strong correlation between aquaculture, Aeromonas diversity and antibiotic resistance. It provides strong evidence for linkage between prophylactic and systemic use of antibiotics in aquaculture and the propagation of antibiotic resistance.
Keywords: Aeromonas, aquaculture, antibiotic resistance, antibiotic resistance gene, integron
Introduction
Aeromonas spp. are Gram negative, facultative anaerobic, non-spore forming rods that often inhabit water and soil habitats (Martin-Carnahan and Joseph, 2005). They are also commonly isolated from food, invertebrates, plants, and animals (Araujo et al., 2002; Janda and Abbott, 2010; Joseph et al., 2013). Although Aeromonas are not directly linked to human pathogenicity, they have been encountered in numerous cases such as gastroenteritis, chronic diarrhea, wound infections, respiratory tract infections, peritonitis, urinary tract infections, septicemia, extra intestinal infections, food and water borne disease, and traveler’s diarrhea (Janda and Abbott, 1996; Altwegg, 1999; Vasaikar et al., 2002; Igbinosa et al., 2012). Comprehensive phylogenetic classification of Aeromonas strains is critical in view of their clinical and ecological importance (Colston et al., 2014), but the genus Aeromonas has been found to be taxonomically challenging due to the occurrence of microheterogeneities in the 16S rRNA gene (Alperi et al., 2008). This obstacle can be circumvented by targeting the rpoD gene, which is substantially more accurate for phylogenetic classification of Aeromonas strains (Alperi et al., 2008). Aeromonas outbreaks commonly occur in aquaculture facilities, as these bacteria are highly ubiquitous in freshwater bodies. Most often Aeromonas salmonicida and A. hydrophila are known to cause ulcerative and hemorrhagic skin ulcers (Igbinosa et al., 2012) in fish under stress, which is often associated with poor sanitation and nutritional deficiencies (Noga, 2011).
Antibiotic resistance genes (ARG) in Aeromonas are often harbored on mobile genetic elements including class 1 integrons, plasmids, IS elements, transposons, genomic islands carrying single or multidrug resistance (MDR) genes and gene cassettes (Hossain et al., 2013; Huddleston et al., 2013; Piotrowska and Popowska, 2015). Horizontal gene transfer of these elements can significantly expand their epidemiological capacity leading to the potential transfer of mobile genetic elements across microbiomes of common ecological niches. Many of these mobile elements harbor multiple antimicrobial resistance determinants resulting in the propagation of antibiotic resistance. For example, previous studies have indicated that Aeromonas strains frequently harbor extended spectrum beta lactamase (ESBL) genes (Lu et al., 2010; Girlich et al., 2011; Wu et al., 2011; Maravić et al., 2013), while others have demonstrated mobilization of genetic elements such as plasmids from fish to human pathogens (Kruse and Sørum, 1994; Sorum, 1998).
Antibiotics that are most commonly used in aquaculture around the world include sulfonamides potentiated with trimethoprim, ormethoprim, oxytetracycline, florfenicol, and erythromycin (Serrano, 2005). In addition to systemic application of these compounds to treat diseased fish, prophylactic use of antibiotics in aquaculture is widespread due to the belief that this practice enhances growth and reduces mortality thereby increasing production and revenues (Pridgeon and Klesius, 2013). These compounds are commonly administered through fish feed for the prevention and/or treatment of Aeromonas-related diseases such as motile Aeromonas septicemia (MAS), hemorrhagic septicemia, ulcer disease, and red-sore disease (Swann and White, 1991). Although antibiotics may ultimately contribute to aquaculture productivity, there is rising concern that the extensive application of antibiotics in aquaculture selects for antibiotic resistant bacteria (ARB) as well as ARG, especially when inadequate- or over-doses of antibiotics are administered (Kolar et al., 2001). These “hotspots” of antibiotic resistance can ultimately contribute to antibiotic resistance on a global level through water discharge and fish consumption. Nonetheless, data on bacterial community composition and antibiotic resistance dynamics in aquaculture environments subjected to antibiotic use are still limited.
The goal of this study was to determine if and to what extent semi-intensive aquaculture facilities contribute to freshwater resistomes. In order to elucidate this goal, we evaluated the phylogeny and antibiotic resistance dynamics of Aeromonas strains isolated from two adjacent semi-intensive aquaculture facilities that were prophylactically and systemically treated with antibiotics. Aeromonas were isolated from the water column of one fish pond for the duration of an 11-month fish fattening cycle prior and subsequent to fingerling stocking and prophylactic application of trimethoprim potentiated sulfonamides. In tandem, we evaluated the phylogeny and antibiotic resistance phenotypes of Aeromonas strains isolated from fish skin ulcers over a 3 year period and compared these to water column Aeromonas strains that were isolated from the same facility.
Materials and Methods
Site Description and Sampling Procedure
Aeromonas isolates were obtained from two adjacent aquaculture facilities (5.4 km from each other) located in the northern coastal region of Israel (Figure 1) as detailed below.
FIGURE 1.
Map of sampling sites in Israel. Top of the distance measuring scale indicate Dor aquaculture pond and the lower end indicates Madan fish pond. Sources: https://www.google.co.il/maps/@32.5828127,34.9243102,5661m/data=!3m1!1e3.
Dor aquaculture research station (32°36′25.4″N 34°55′54.7″E). The Dor Aquaculture research facility is a semi-commercial system consisting of 87 earthen fish ponds as well as several isolated concrete and plastic pools used for special treatments and quarantine. All of the ponds in the system are connected to a reservoir (∼80,000 m3), which is used to regulate the water levels in the fish ponds. Water is supplied to the reservoir from a local spring and from floodwater in winter months.
In this study, we focused on a complete fish fattening cycle in Dor fish pond 03 due to the standard prophylactic antibiotic protocol applied, which was as follows: On May 11th 2015 the pond was stocked with approximately 90,000 common carp (Cyprinus carpio carpio) fingerlings (average weight 1.35 g). Ninety days later on July 22nd 2015, 20,470 fingerlings were removed from the pond and temporarily transferred to vinyl-lined pools enclosed in a greenhouse and immersion vaccinated against cyprinid herpesvirus-3 (CyHV-3). A second group of 31,800 fish was transferred to the greenhouse pools without vaccination (due to financial considerations not all fish were vaccinated). To prevent vaccination stress-related secondary infections, fingerlings were treated with feed amended with sulfamethoprim (25% sulfadiazine; 5% trimethoprim) for 5 days in the greenhouse pool. A total of 19,000 of these fingerlings (ave. wt 19 g) were returned to pond 03 where sulfamethoprim was administered for an additional 5 days. At the end of the fish fattening cycle between March 29th and April 3rd 2016, around 9370 fish (Avg. wt 270 g) were removed from pond 03 for further growth and marketing.
Overall seven profiles were sampled from the upper 10 cm of the water column of Dor earthen pond 03 (∼20,000 m3) from April 2015 to March 2016. One profile (April 2015) was sampled prior to fingerling stocking, one (July 2015) after fish stocking but before prophylactic antibiotic treatment (see below), and five (August 2015, October 2015, December 2015, February 2016, and March 2016) at different stages of the fish farming cycle. Water samples were collected in three replicates from the North-East (NE), North-West (NW), and South-West (SW) corners of the fish pond for all profiles except for the April 15 profile, where only one replicate was collected. During the duration of this cycle, diseased fish from the Dor system were quarantined in concrete pools and treated through feed with oxytetracycline (50%). The quarantined water was discharged into the Dor system reservoir, which supplies water to pond 03. While we cannot calculate exact amounts of oxytetracycline, it is likely that residual concentrations of this antibiotic reached the pond.
Madan fish breeding center, Kibbutz Maagan Michael (32°33′24.83″N 34°55′1.2″E). An additional set of water column samples were taken from earthen ponds 35–37 (∼52,000 m3, ∼94,500 m3, and ∼112,500 m3, respectively) at the Madan fish breading center (Figure 1). A single profile (February 2016) was sampled during the late stage of a carp fattening cycle. During this cycle diseased fish were quarantined in concrete pools and systemically treated through feed with a total of 18.5 Kg of sulfadiazine-trimethoprim (30%); 77 Kg of oxytetracycline (50%), and 19 Kg of Aquaflor (50% Florfenicol). While it is difficult to calculate exact amounts of antibiotic that re-entered the fish ponds, it can be inferred that a fraction of these antibiotic used for treatment were released into the fish ponds though transferred water and fish excretions.
Isolation of Aeromonas Strains
Aeromonas strains were quantified and isolated from the Dor and Madan water column samples described above by filtering 100 ml through 0.45 μm nitrocellulose membranes (Whatman, UK), re-suspending in 10 ml of sterile distilled water, serially diluting and plating on commercially available Aeromonas medium base or Ryan’s medium (OXOID Ltd., England) with and without sulfadiazine (initial screening of the isolates on plates amended with this antibiotic was performed due to the fact that the fish fingerlings were treated with sulfadiazine as a prophylactic antibiotic). The plates were incubated at 30°C for 18–24 h; and dark green, opaque with darker center colonies characteristic of Aeromonas were selected as per the manufacturer’s instructions. All isolates were stored in tubes containing Luria-Bertani (LB) broth with 25% v/v glycerol at -80°C for further analysis. Preliminarily, isolates were validated as members of the genus Aeromonas using PCR with genus-specific 16S rRNA gene markers (Table 1).
Table 1.
Primers and PCR conditions used in this study.
| Gene target | Primer sequence | Annealing temp/time | Amplicon (bp) | Reference |
|---|---|---|---|---|
| sul1 | CGCACCGGAAACATCGCTGCAC | 55.9°C for 30 s | 163 | Pei et al., 2006 |
| TGAAGTTCCGCCGCAAGGCTCG | ||||
| sul2 | TCCGATGGAGGCCGGTATCTGG | 60.8°C for 30 s | 191 | Pei et al., 2006 |
| CGGGAATGCCATCTGCCTTGAG | ||||
| tetA | GCTACATCCTGCTTGCCTTC | 55°C for 45 s | 210 | Wu et al., 2010 |
| CATAGATCGCCGTGAAGAGG | ||||
| intI1 | CCTCCCGCACGATGATC | 58°C for 30 s | 293 | Levesque and Roy, 1993 |
| TCCACGCATCGTCAGGC | ||||
| Aero 16S | CTACTTTTGCCGGCGAGCGG | 68°C for 30 s | 953 | Lee et al., 2002 |
| TGATTCCCGAAGGCACTCCC | ||||
| ERIC | CAGCCATGAACAACTGGTGGCG | 52°C for 60 s | Size range: 0.1–3 kb | Versalovic et al., 1991 |
| TGCTTTGCGCAGGGAAGATTCC | ||||
| rpoD | ACGACTGACCCGGTACGCATGTA | 55°C for 30 s | 840 | Yamamoto et al., 2000 |
| ATAGAAATAACCAGACGTAAGTT |
A total of 1183 Aeromonas strains were isolated from the Dor and Madan water columns using the procedure described above. In addition to these, we acquired 17 additional Aeromonas strains that were isolated from common carp skin ulcers from the Madan ponds over a 3 year period (2013–2015) at the National Fish Research Center (Nir David, Israel). Water column isolates and pathogens were further screened to determine antibiotic resistance phenotypes and taxonomic affiliations as described below.
Antibiotic Resistance Profiling of Aeromonas Isolates
Since Dor and Madan water column samples were exposed to sulfonamides and trimethoprim, and the Dor pond 03 was exposed to prophylactic treatment with sulfamethoprim (25% sulfadiazine; 5% trimethoprim), the overall resistance of the above isolates to sulfonamides was initially determined by concomitantly streaking on LB agar with and without sulfadiazine (SDZ; 120 μg/ml) and trimethoprim (TMP; 200 μg/ml). Agar plates were incubated at 30°C for 18–24 h and resistance of the isolates was judged by comparing growth on SDZ- and TMP-amended vs. non-amended plates.
The scope of MDR amongst the SDZ-resistant isolates described above was determined using the disk diffusion method with the following disks (Oxoid, UK): chloramphenicol (30 mcg), ceftriaxone (30 mcg), gentamicin (10 mcg), tetracycline (30 mcg), sulfamethoxazole (25 mcg), and trimethoprim (5 mcg); which were selected on the basis of antibiotics used in aquaculture as well as for the treatment of Aeromonas-associated infections in humans. Briefly, five milliliters of LB broth was inoculated with 6–8 colonies from overnight growth of isolates from LB agar plates and incubated at 30°C with shaking at 180 rpm for 18–24 h. Cells were then pelleted, resuspended in saline and adjusted to a turbidity equivalent of 0.5 McFarland standards (Igbinosa, 2014). The culture suspension was spread onto Muller Hinton agar (HiMedia, India) in three directions using sterile swabs (∼3 rotations of 60° angle). Plates were allowed to dry for few minutes and impregnated with the above mentioned antibiotic disks using a disk dispenser (Oxoid, UK). Plates were then incubated at 30°C for 18–24 h (Igbinosa, 2014), and the diameter of inhibition zones were measured and compared with Escherichia coli sp. DH5α strain as a non-resistant control.
E Test assays were performed on a total of 18 random isolates representing all of the sampling points to determine minimum inhibition concentrations (MIC) of the following antibiotics: ceftriaxone (0.016–256 μg/ml), chloramphenicol (0.016–256 μg/ml), tetracycline (0.016–256 μg/ml), and sulfamethoxazole + trimethoprim (0.002–32 μg/ml). The culture suspension equivalent of 0.5 McFarland standards was used to inoculate Mueller-Hinton agar plates (Oxoid, Basingstoke, UK) as described for the disk diffusion assays above. After drying, E test strips were applied to the plates and incubated at 30°C for 18–24 h. The MICs on both ends were read on the intersection of the inhibition ellipse and the E Test strip edge in concurrence of the manufacturer’s instructions. E. coli sp. DH5α was used as control. These tests were performed according to CLSI (United States) and EUCAST (European) reference methods and standardized disk diffusion methods (Jorgensen and Turnidge, 2015).
Antibiotic resistance phenotypes were interpreted using MIC breakpoints (mg/L) for members of genus Aeromonas (Goni-Urriza et al., 2000; Aravena-Román et al., 2012) and isolates were classified as resistant, intermediate, and sensitive.
Selection and Phylogenetic Characterization of Multidrug Resistant Aeromonas Strains
Isolates co-resistant to sulfadiazine and trimethoprim were initially screened by Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR on boiled agar plate colonies (Table 1) to determine the clonal variation among the isolates and select unique strains. Subsequently, phylogenetic characterization of selected unique strains was determined by PCR amplification and sequencing (Macrogen Genomic Division, Korea) a fragment of the rpoD gene using the primers described in Table 1. A total of 55 water-associated isolates and 11 fish skin ulcer-associated isolates were characterized. Phylogenetic analysis of the Aeromonas isolates and closely related reference strains was conducted using MEGA version 6 (Tamura et al., 2013). The length and nucleotide positions of the sequences used in the phylogenetic analysis correspond to rpoD (gi| 117617447:895117-896973) positions 525–903 in the Aeromonas hydrophila subsp. hydrophila ATCC-7966 type strain. Initially, the rpoD sequences were aligned using Muscle (Tamura et al., 2013), and subsequently phylogenetic trees were generated using the neighbor-joining algorithm with 1000 bootstrap values (Tamura et al., 2013). The partial rpoD gene sequences were deposited to GenBank under accession numbers KX607409 – KX607474.
PCR Based Screening of Selected Antibiotic Resistance Genes
Aeromonas isolates were screened for the presence of mobilome- and antibiotic resistance-associated genes by colony PCR. Two genes associated with sulfonamide resistance (sul1 and sul2), one gene encoding tetracycline resistance (tetA) and the class 1 integron integrase (intI1) gene were targeted, applying the primers and conditions outlined in Table 1.
Temporal Concentrations of Sulfadiazine and Trimethoprim in Water Column Profiles
The concentrations of sulfadiazine and trimethoprim (S/T) in water column samples were analyzed using a liquid chromatography system consisting of a Prominence HPLC instrument (Shimadtzu Corporation, Kyoto, Japan) equipped with a degasser, a quaternary pump, a cooled autosampler, and a column oven connected to a 3200 QTRAP mass spectrometer (Applied Biosystem/MDS SCIEX, Darmstadt/Germany) equipped with an ESI interface. The analysis was performed according to Lopez de Alda et al. (2003) and Bouyou et al. (2014) with few modifications as described below. The injection volume was 20 μL. Separation was performed on a C18 column Phenomenex (150 mm × 4.60 mm, 5 μm) along with a guard C18 column applying an isocratic flow rate of 1 mL/min at 30°C. The elution was performed with 0.1% acetic acid in water and methanol (v/v) with composition of 20:80 and analysis time of 15 min. Electrospray ionization was performed in positive mode (ESI+). The desolation temperature was set to 400°C with a nebulizer gas pressure of 50 psi, while the curtain and collision gas pressures were 10 psi and medium, respectively. The ion spray voltage was 4000 v. The multiple reaction monitoring (MRM) function was applied in all analysis (SDZ- Q1: 251.07, Q3: 156.1, and TMP- Q1: 291.04, Q3: 123.1).
Statistical Analysis
Statistical significance of correlations between antibiotic concentrations, antibiotic resistance phenotypes, and ARG was performed using Pearson’s coefficient correlation analysis. Correlations were considered significant for p-values < 0.05.
Ethics Statement
The fish associated bacteria analyzed in this study were isolated in accordance with the recommendations of the Central Fish Health Laboratory using protocols approved by the Israeli Ministry of Agriculture and Rural Development Fisheries Department.
Results
Isolation of Aeromonas Strains
A total of 1103 Aeromonas strains were isolated from fish pond 03 at the Dor site (Figure 1) in seven profiles between April 2015 and March 2016. This pond was specifically targeted due to the prophylactic administration of sulfadiazine-trimethoprim at the beginning of the cycle and the relatively controlled conditions in the pond. An additional 80 Aeromonas strains were isolated from the water columns of three fish ponds from the Madan site (Figure 1) during a single sampling time in February 2016 as a reference. In addition, we analyzed 17 fish skin ulcer isolates acquired from 15 independent samples from the Madan aquaculture facility sampled from 2013 to 2015 as a taxonomic marker of prominent Aeromonas fish pathogens in the region.
Antibiogram Profiling
Preliminary antibiotic susceptibility screening indicated that 67 and 27% of the Aeromonas isolates were resistant to sulfadiazine and trimethoprim, respectively. The prophylactic administration of sulfadiazine-trimethoprim in Dor pond 03 led us to initially focus on isolates that were resistant to these two antibiotics (Table 2). The occurrence of sulfadiazine-trimethoprim resistant Aeromonas strains was substantially higher in the water column profiles sampled during the fish fattening cycle than in those taken prior to fish stocking (Tables 2 and 3), although significant variation in resistance was observed between post-stocking profiles. In the Madan site, sulfadiazine-trimethoprim resistance in the water column samples was similar to that observed in the Dor water column (26.3% vs. 20.8%), whereas co-resistance in the skin ulcer isolates was considerably higher (64.7%; Table 3).
Table 2.
Total and sulfadiazine-trimethoprim resistant Aeromonas strains isolated from different profiles of the Dor fish pond 03 water column.
| Site and sampling details | Total Aeromonas isolates | S/T co-resistant Aeromonas isolates (%) |
|---|---|---|
| Dor-Apr-15 | 53 | 7.5 |
| Dor-Jul-15 | 136 | 30.1 |
| Dor-Aug-15 | 151 | 10.6 |
| Dor-Oct-15 | 338 | 10.9 |
| Dor-Dec-15 | 106 | 8.5 |
| Dor-Feb-16 | 172 | 47.1 |
| Dor-Mar-16 | 147 | 23.1 |
Sulfadiazine, S; Trimethoprim, T.
Table 3.
Summary of total and sulfadiazine-trimethoprim resistant Aeromonas strains isolated from the Dor and Madan aquaculture facilities.
| Number of Aeromonas isolates | Dor |
Madan |
||
|---|---|---|---|---|
| Water associated pre-stocking | Water associated during fish farming | Water associated | Fish skin ulcers | |
| Total | 53 | 1050 | 80 | 17 |
| S/T co-resistant isolates (%) | 7.5 | 20.8 | 26.3 | 64.7 |
Sulfadiazine, S; Trimethoprim, T.
Disk diffusion assays were applied to further explore multi drug resistance in sulfadiazine-resistant Aeromonas isolates. In the Dor water column, sulfadiazine-resistant isolates were frequently resistant to trimethoprim and tetracycline as well, and substantially higher levels of resistance to these antibiotics were detected in isolates from the fish fattening cycle profiles relative to those sampled prior to fish stocking (Figure 2A). One exception was the August 2015 profile, which was characterized by high temperatures and cyanobacterial blooms, where co-resistance to tetracycline was found to be considerably lower than in other fish fattening cycle profiles (Figure 2A). Pearson’s coefficient analyses showed a significant positive correlation between sulfadiazine and trimethoprim resistance (p < 0.01; R: 0.3363; n = 1121) as well as between sulfadiazine/trimethoprim co-resistance and tetracycline resistance (p < 0.01; R: 0.3234; n = 324). Similarly, a positive correlation was observed between ceftriaxone and chloramphenicol resistance levels (p < 0.01; R: 0.5591; n = 324). Interestingly, co-resistance to ceftriaxone and chloramphenicol was actually higher in the pre-stocking isolates than during the fish fattening cycle; and gentamicin resistance was not detected at all in the samples (Figure 2A).
FIGURE 2.
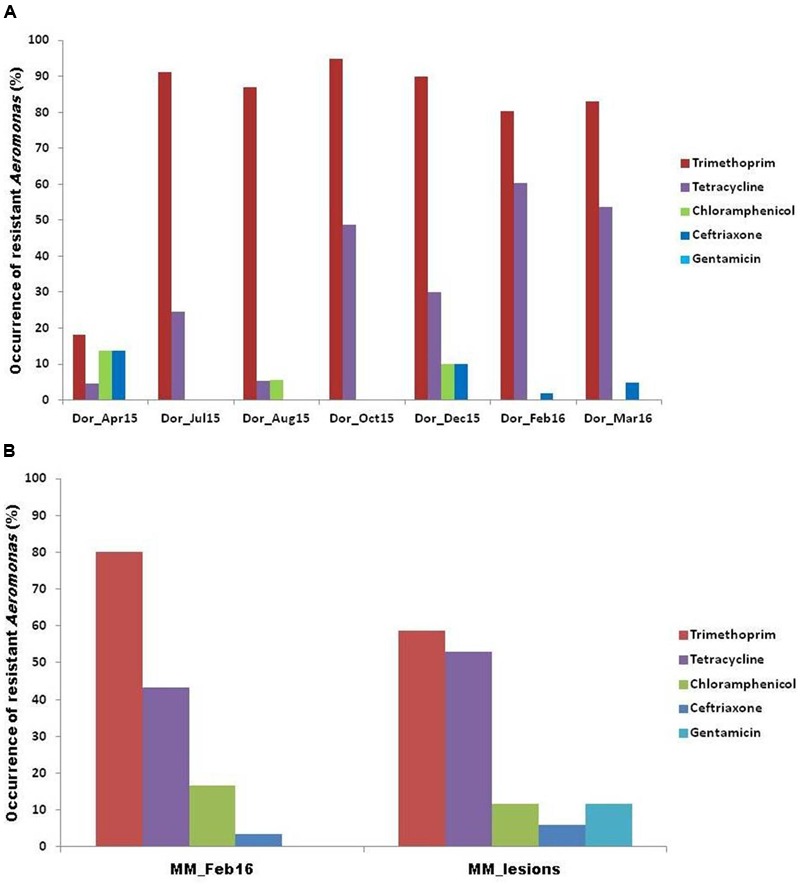
Antibiogram profiling of the Aeromonas isolates from (A) Dor fish pond, and (B) Madan fish pond.
Disk diffusion assays were also applied to determine co-resistance in sulfadiazine-resistant Aeromonas isolates from the Madan aquaculture facility water column and skin ulcer isolates (Supplementary Table S2). Similar to the Dor isolates, the strongest correlations were observed for trimethoprim and tetracycline resistance (Figure 2B). Overall, water column isolates exhibited higher levels of resistance for trimethoprim and chloramphenicol than isolates from fish skin ulcers; while the opposite was observed for tetracycline and ceftriaxone (Figure 2B). Gentamicin resistance was recorded in several fish skin ulcer isolates, contrary to all of the analyzed water column isolates.
E test assays were applied to determine MIC’s of four screened antibiotics against eighteen of the sulfadiazine-resistant Aeromonas isolates (Table 4; Supplementary Table S1). Similar to the disk diffusion assays, co-resistance to trimethoprim was highest (83%), followed by co-resistance to tetracycline (66%), chloramphenicol (28%), and ceftriaxone (11%).
Table 4.
Minimal inhibitory concentration breakpoints (μg/ml) of Aeromonas isolates based on E test (n = 18).
| Antibiotic (mcg) | ≤1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ≥512 | MIC (mg/L) | R (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chloramphenicol | ||||||||||||
| # resistant CFU | 11 | 2 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 16 | 28 |
| Ceftriaxone | ||||||||||||
| # resistant CFU | 16 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 11 |
| Tetracycline | ||||||||||||
| # resistant CFU | 2 | 3 | 3 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 66 |
| Trimethoprim-sulfamethoxazole | ||||||||||||
| # resistant CFU | 3 | 0 | 0 | 1 | 1 | 13 | 0 | 0 | 0 | 0 | 2/38 | 83 |
Resistance, R; Minimal inhibitory concentration, MIC.
Phylogenetic Characterization of Aeromonas Strains
Partial rpoD gene sequence analysis of 66 representative isolates revealed their close similarity to nine different Aeromonas species: A. veronii (46.3%), A. salmonicida (17.9%), A. media (16.4%), A. culicicola (6.0%), A. bestiarum (4.5%), A. sobria (3.0%), A. allosaccharophila (3.0%), Aeromonas sp. SHGW9 (1.5%), and A. fluvialis (1.5%). These sequences were aligned and used to generate a phylogenetic tree that contained five primary clades (Supplementary Figure S1). All of the post-stocking Dor isolates clustered within two (A. veronii and A. culicicola) clades (Figure 3A). In contrast, isolates from the pre-stocking profile were considerably more diverse and were also associated with A. salmonicida, A. jandai, A. enteropelogens, A. bestiarum, and A. media clades (Figure 3A).
FIGURE 3.
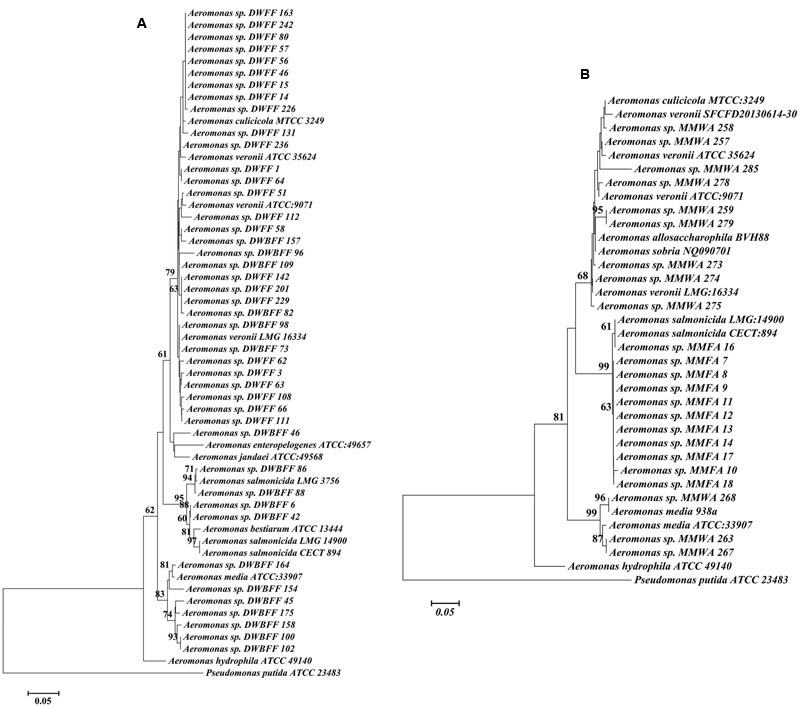
Consensus tree based on alignment of rpoD gene sequences of Aeromonas isolates from this study, along with reference sequences from NCBI. (A) Dor fish pond, and (B) Madan fish pond [DWBFF: Dor water before fish fattening; DWFF: Dor water fish fattening; MMWA: Madan water associated; and MMFA: Madan fish associated].
In the Madan aquaculture facility (Figure 3B) all fish skin ulcer samples grouped with pathogenic A. salmonicida strains, whereas the water column samples clustered with A. allosaccharophila, A. culicicola, A. media, A. sobria, and A. veronii and were similar to several of the Dor water column samples (Supplementary Figure S1).
PCR Based Genotyping
The prevalence and distribution of sul1, sul2, tetA, and the class 1 integron gene intI1 were analyzed in 77 isolates from Dor water column (Figure 4A; Supplementary Table S3) and 37 isolates from Madan fish pond (Figure 4B). In the Dor water column isolates fish fattening cycle profiles were characterized by substantially higher levels of sul1, tetA, and intI1 than in the pre-stocking water column samples (Figure 4A). Also, statistical analysis revealed positive correlations between sul1 and intI1 (p < 0.01; R: 0.977).
FIGURE 4.
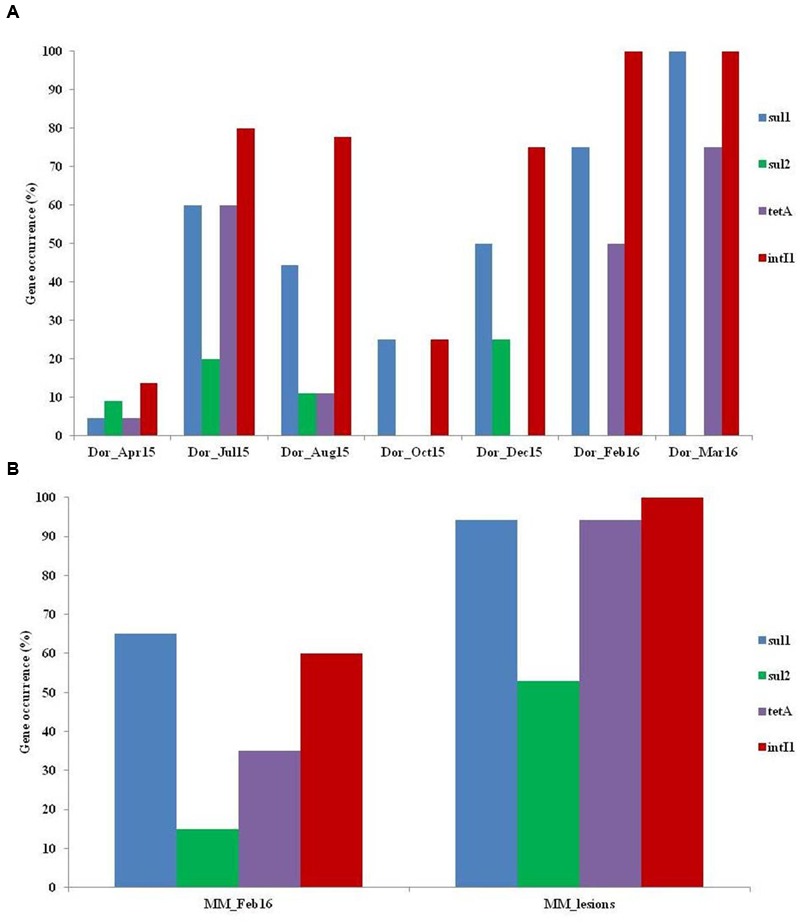
Antibiotic resistance gene (ARG) profiling of the Aeromonas isolates from (A) Dor fish pond, and (B) Madan fish pond.
In Madan isolates, all of the analyzed genes were substantially more abundant in the fish skin ulcers compared to those from water column (Figure 4B). Interestingly, 7.9% of the screened isolates from both sites carried all of the screened ARGs as well as intI1 gene.
Temporal Concentrations of Sulfadiazine and Trimethoprim in the Dor Water Column Profiles
Sulfadiazine and trimethoprim concentrations were measured in the seven Dor water column profiles in order to assess the persistence of these antibiotics over time and elucidate correlations between prophylactic antibiotic treatment and Aeromonas resistance (Table 5). Statistical analysis revealed positive but insignificant correlation between aquatic concentrations of trimethoprim and respective resistance levels. In contrast, sulfadiazine resistance levels were high regardless of the aquatic sulfadiazine.
Table 5.
Aquatic concentration of antibiotics and resistance level in Aeromonas isolates (n = 66).
| Sampling interval | n | Average aquatic concentration (ng/L) |
% resistant Aeromonas |
||
|---|---|---|---|---|---|
| S | T | S | T | ||
| Dor-Apr-15 | 1 | 200.0 | 4200.0 | 82.9 | 18.2 |
| Dor-Jul-15 | 3 | 0.0 | 9633.3 | 70.8 | 91.1 |
| Dor-Aug-15 | 3 | 4933.3 | 6966.7 | 54.1 | 87.0 |
| Dor-Oct-15 | 3 | 0.0 | 4533.3 | 34.3 | 94.9 |
| Dor-Dec-15 | 3 | 0.0 | 3566.7 | 39.0 | 90.0 |
| Dor-Feb-16 | 3 | 100.0 | 1933.3 | 65.3 | 80.2 |
| Dor-Mar-16 | 3 | 0.0 | 2833.3 | 53.9 | 82.9 |
Sulfadiazine, S; Trimethoprim, T.
Discussion
Two specific objectives were pursued in this study. The first aimed to elucidate associations between systemic and prophylactic antibiotic exposure and antibiotic resistance in Aeromonas strains from aquaculture systems; while the second attempted to determine taxonomic and antibiotic resistance dynamics of Aeromonas isolates from aquaculture pond water as a function of conventional commercial fish farming practices.
In the Dor system, clear phenotypic, genotypic and taxonomic distinctions were observed between pre-stocked and fish-stocked water column Aeromonas strains. Fish fattening cycle-associated strains were characterized by significantly higher resistance to sulfonamide, trimethoprim, and tetracycline antibiotics (approved for use in aquaculture in Israel and applied in fish ponds targeted in this study) and by higher levels of sulfonamide and tetracycline resistance genes and class 1 integron-encoding genes. In contrast, these isolates were less resistant to chloramphenicol, ceftriaxone, and gentamicin than Aeromonas strains isolated from the water column prior to fish stocking. Interestingly, chloramphenicol resistance was significantly higher in the Madan water column isolates than in those from Dor, potentially due to the fact that florfenicol (a derivative of chloramphenicol) was only used in the Madan facility. Similar results were obtained in antibiogram analyses of the Madan fish pathogens, which displayed high levels of resistance (Supplementary Figure S2) to tetracycline, sulfonamide, trimethoprim, and chloramphenicol antibiotics; versus very low or negligible resistance to aminoglycoside, quinolone, and macrolides that are not approved for use in aquaculture in Israel. Although β-lactam antibiotics are not used in aquaculture, Aeromonas strains are generally considered to be intrinsically resistant to this antibiotic family, explaining the high levels of amoxicillin and ampicillin resistance in the skin ulcer-associated strains (Bakken et al., 1988).
Furthermore, we observed a positive correlation between class 1 integron (intI1) levels and aquatic sulfadiazine concentrations, as well as a positive correlation between intI1 and the occurrence of sul1. Class 1 integrons are known to harbor sul1, and the close association between sul1 and class 1 integrons has been reported earlier (Antunes et al., 2005).
These results demonstrate a strong link between antibiotic use and resistance in both environmental and pathogenic Aeromonas strains in aquaculture ecosystems. Linkage between antibiotic use and resistance has been demonstrated for other aquaculture-associated bacteria (Verner-Jeffreys et al., 2009; Tamminen et al., 2010; Laganà et al., 2011; Scarano et al., 2014), and similar data has been reported in animal husbandry facilities. These findings have resulted in calls to ban non-medical use of antibiotics (Meek et al., 2015). Alternatively, it may be possible to develop accurate dosing guidelines for antibiotic treatment of fish if clear pathogen pharmacokinetics/pharmacodynamics is established within the framework of complex aquaculture ecosystems (Xu et al., 2013).
Interestingly, high levels of antibiotic resistance and ARG were detected in particular fish fattening cycle water column profiles characterized by low sulfonamide and trimethoprim concentrations suggesting persistence of antibiotic resistance phenotypes long after subsidence of selective pressure. This phenomenon was similarly demonstrated by Tamminen et al. (2010) who detected high levels of tetracycline resistance genes in aquaculture sediments despite the discontinued application of tetracycline in these systems (tetracycline gene occurrence was considerably lower in sediments not impacted by aquaculture). It may be suggested that additional factors beyond antibiotic concentrations such as fish density, temperature, and cyanobacterial blooms (prevalent in the July and August profiles of this study) also impact antibiotic resistance in the sampled Aeromonas strains. This is supported by a recent study that showed high levels of antibiotic resistance in bacterial isolates from aquaculture facilities that did not use antibiotics (Huang et al., 2015).
The rpoD gene based phylogenetic analysis demonstrated high diversity among Aeromonas strains in the Dor and Madan water columns The diversity of the Aeromonas isolates significantly declined in the Dor water column after stocking of fish pond 03 resulting in predominance of A. veronii-associated strains in all of the fish fattening cycle profiles. A. veronii has been previously detected in aquaculture water (Hu et al., 2012) and lakes (Skwor et al., 2014). The antibiotic resistance phenotypes of the A. veronii isolates from this study varied both within and between profiles and therefore the prevalence of these strains during the fish cycle is most likely not associated with selective pressure, and based on current literature it is difficult to infer what factors are associated with the observed proliferation of A. veronii.
Aeromonas salmonicida and Aeromonas veronii are widely known for their pathogenic attributes toward fish and humans, respectively (Robertson et al., 2014; Tang et al., 2014). A. salmonicida are widespread in non-salmonid fish (Wiklund and Dalsgaard, 1998); and are highly prevalent in skin ulcers in goldfish and pond-bred carp in Israel (Sinyakov et al., 2001). Interestingly, all of the Madan A. salmonicida strains isolated from fish ulcers over a 3 year period were highly similar to each other suggesting that pathogenicity in these systems is strongly associated with specific A. salmonicida genotypes. Nonetheless, the fact that the antibiotic resistance phenotypes and genotypes of these A. salmonicida strains varied considerably indicates that they readily acquire mobile genetic elements that harbor ARG. Strains with the same ERIC profile and further characterized as A. salmonicida by rpoD gene similarity were identified in Dor water column isolates from the pre-stocking profile. However, no such strains were identified in the Dor or Madan water columns during the fish fattening cycle suggesting that during fish growth either these strains cannot compete with the other Aeromonas strains, or they are strongly associated with the fish and therefore cannot be detected in the water column.
Conclusion
This study comprehensively assessed diversity, antibiotic resistance, and presence of selected ARG in a large collection of Aeromonas strains isolated from commercial earthen pond aquaculture facilities. The generated data divulges a strong link between antibiotic use in aquaculture and antibiotic resistance in both environmental and pathogenic Aeromonas strains. These findings suggest that current antibiotic application practices in aquaculture may have detrimental environmental and public health ramifications, implying that the efficacy of these practices may be significantly compromised in the future. Future research should focus on specifically linking selective pressure to antibiotic resistance in more controlled microcosm experiments and on screening plasmids and other mobile genetic elements from Aeromonas isolates in order assess the scope of genetic linkage between ARG associated with antibiotics that are readily applied in aquaculture. Ultimately, sustainable dosing practices that efficiently combat fish disease while concomitantly reducing spread of antibiotic resistance need to be developed.
Author Contributions
HP participated in sampling, conducted all of the culture-based and molecular experiments and participated in writing the manuscript; AlN and AB-P participated in sampling and aquaculture system maintenance; MS and TO isolated Aeromonas from diseased fish; AhN assisted in chemical analyses; DM co-supervised HP and assisted in experimental design; EC co-supervised HP and participated in the research design, sampling, and writing of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by Israeli Ministry of Science and Technology (MOST) Infrastructure grant 3-10342. HP was funded by the Planning and Budgeting Committee (PBC) postdoctoral fellowship fund.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01875/full#supplementary-material
References
- Alperi A., Figueras M. J., Inza I., Murcia J. M. (2008). Analysis of 16S rRNA gene mutations in a subset of” Aeromonas” strains and their impact in species delineation. Int. Microbiol. 11 185–194. [PubMed] [Google Scholar]
- Altwegg M. (1999). “Aeromonas and Plesiomonas,” in Manual of Clinical Microbiology eds Murray P., Baron E., Pfaller M., Tenover F., Yolken R. (Washington, DC: ASM Press; ) 507–516. [Google Scholar]
- Antunes P., Machado J., Sousa J. C., Peixe L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49 836–839. 10.1128/AAC.49.2.836-839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo V. S., Pagliares V. A., Queiroz M. L. P., Freitas-Almeida A. C. (2002). Occurrence of Staphylococcus and enteropathogens in soft cheese commercialized in the city of Rio de Janeiro, Brazil. J. Appl. Microbiol. 92 1172–1177. 10.1046/j.1365-2672.2002.01656.x [DOI] [PubMed] [Google Scholar]
- Aravena-Román M., Inglis T. J., Henderson B., Riley T. V., Chang B. J. (2012). Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob. Agents Chemother. 56 1110–1112. 10.1128/AAC.05387-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken J. S., Sanders C. C., Clark R. B., Hori M. (1988). Beta-lactam resistance in Aeromonas spp. caused by inducible beta-lactamases active against penicillins, cephalosporins, and carbapenems. Antimicrob. Agents Chemother. 32 1314–1319. 10.1128/AAC.32.9.1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyou P. L., Weisser J. J., Strobel B. W. (2014). Determination of sulfadiazine in phosphate-and DOC-rich agricultural drainage water using solid-phase extraction followed by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 406 5019–5030. 10.1007/s00216-014-7921-6 [DOI] [PubMed] [Google Scholar]
- Colston S. M., Fullmer M. S., Beka L., Lamy B., Gogarten J. P., Graf J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5:e2136-14. 10.1128/mBio.02136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2011). Diversity of clavulanic acid-inhibited extended-spectrum β-lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Agents Chemother. 55 1256–1261. 10.1128/AAC.00921-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni-Urriza M., Pineau L., Capdepuy M., Roques C., Caumette P., Quentin C. (2000). Antimicrobial resistance of mesophilic Aeromonas spp. isolated from two European rivers. J. Antimicrob. Chemother. 46 297–301. 10.1093/jac/46.2.297 [DOI] [PubMed] [Google Scholar]
- Hossain M. J., Waldbieser G. C., Sun D., Capps N. K., Hemstreet W. B., Carlisle K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 8:e80943 10.1371/journal.pone.0080943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Wang N., Pan Z. H., Lu C. P., Liu Y. J. (2012). Identity and virulence properties of Aeromonas isolates from diseased fish, healthy controls and water environment in China. Lett. Appl. Microbiol. 55 224–233. 10.1111/j.1472-765X.2012.03281.x [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhang L., Tiu L., Wang H. H. (2015). Characterization of antibiotic resistance in commensal bacteria from an aquaculture ecosystem. Front. Microbiol. 6:914 10.3389/fmicb.2015.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J. R., Brokaw J. M., Zak J. C., Jeter R. M. (2013). Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 36 224–234. 10.1016/j.syapm.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Igbinosa I. H. (2014). Antibiogram profiling and pathogenic status of Aeromonas species recovered from Chicken. Saudi J. Biol. Sci. 21 481–485. 10.1016/j.sjbs.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa I. H., Igumbor E. U., Aghdasi F., Tom M., Okoh A. I. (2012). Emerging Aeromonas species infections and their significance in public health. ScientificWorldJournal 2012:625023 10.1100/2012/625023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (1996). “Human pathogens,” in The genus Aeromonas eds Austin B., Altwegg M., Gosling P. J., Joseph S. W. (Chichester: John Wiley and Sons; ) 151–170. [Google Scholar]
- Janda J. M., Abbott S. L. (2010). The genus Aeromonas, taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen J. H., Turnidge J. D. (2015). “Susceptibility test methods: dilution and disk diffusion methods,” in Manual of Clinical Microbiology 11th Edn eds Jorgensen J., Pfaller M., Carroll K., Funke G., Landry M., Richter S., et al. (Washington, DC: American Society of Microbiology; ) 1253–1273. [Google Scholar]
- Joseph A. V., Sasidharan R. S., Nair H. P., Bhat S. G., Joseph A. V., Sasidharan R. S., et al. (2013). Occurrence of potential pathogenic Aeromonas species in tropical seafood, aquafarms and mangroves off Cochin coast in South India. Vet. World 6 300–306. 10.5455/vetworld.2013.300-306 [DOI] [Google Scholar]
- Kolar M., Urbánek K., Látal T. (2001). Antibiotic selective pressure and development of bacterial resistance. Int. J. Antimicrob. Agents 17 357–363. 10.1016/S0924-8579(01)00317-X [DOI] [PubMed] [Google Scholar]
- Kruse H., Sørum H. (1994). Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60 4015–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganà P., Caruso G., Minutoli E., Zaccone R., Delia S. (2011). Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol. 34 53–63. [PubMed] [Google Scholar]
- Lee C., Cho J. C., Lee S. H., Lee D. G., Kim S. J. (2002). Distribution of Aeromonas spp. as identified by 16S rDNA restriction fragment length polymorphism analysis in a trout farm. J. Appl. Microbiol. 93 976–985. 10.1046/j.1365-2672.2002.01775.x [DOI] [PubMed] [Google Scholar]
- Levesque C., Roy P. H. (1993). “PCR analysis of integrons,” in Diagnostic Molecular Microbiology: Principles and Applications eds Persing D. H., Smith T. F., Tenover F. C., White T. J. (Washington, DC: American Society for Microbiology; ) 590–594. [Google Scholar]
- Lopez de Alda M. J., Diaz-Cruz S., Petrovic M., Barcelo D. (2003). “Analysis of selected emerging pollutants (steroid sex hormones, drugs and alkylphenolic surfactants) in the aquatic environment by LC-MS and LC-MS-MS,” in New Horizons and Challenges in Environmental Analysis and Monitoring eds Namieśnik J., Chrzanowski W., Zmijewska P. (Gdañsk: Centre of Excellence in Environmental. Analysis and Monitoring (CEEAM)) 181–201. [Google Scholar]
- Lu S. Y., Zhang Y. L., Geng S. N., Li T. Y., Ye Z. M., Zhang D. S., et al. (2010). High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appl. Environ. Microbiol. 76 5972–5976. 10.1128/AEM.00711-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravić A., Skočibušić M., Šamanić I., Fredotović Ž, Cvjetan S., Jutronić M., et al. (2013). Aeromonas spp. simultaneously harbouring bla CTX-M-15, bla SHV-12, bla PER-1 and bla FOX-2, in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 166 301–308. 10.1016/j.ijfoodmicro.2013.07.010 [DOI] [PubMed] [Google Scholar]
- Martin-Carnahan A., Joseph S. W. (2005). “Genus I. Aeromonas. Stanier 1943, 213AL,” in Bergey’s Manual of Systematic Bacteriology 2nd Edn Vol. 2 Part B eds Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (New York, NY: Springer; ) 557–578. [Google Scholar]
- Meek R. W., Vyas H., Piddock L. J. V. (2015). Nonmedical uses of antibiotics: time to restrict their use? PLoS Biol. 13:e1002266 10.1371/journal.pbio.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noga E. J. (2011). Fish Disease: Diagnosis and Treatment. Hoboken, NJ: John Wiley and Sons. [Google Scholar]
- Pei R., Kim S.-C., Carlson K. H., Pruden A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40 2427–2435. 10.1016/j.watres.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Piotrowska M., Popowska M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494 10.3389/fmicb.2015.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon J. W., Klesius P. H. (2013). Major bacterial diseases in aquaculture and their vaccine development. Anim. Sci. Rev. 2012:141. [Google Scholar]
- Robertson B. K., Harden C., Selvaraju S. B., Pradhan S., Yadav J. S. (2014). Molecular detection, quantification, and toxigenicity profiling of Aeromonas spp. in source-and drinking-water. Open Microbiol. J. 8:32 10.2174/1874285801408010032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarano C., Spanu C., Ziino G., Pedonese F., Dalmasso A., Spanu V., et al. (2014). Antibiotic resistance of Vibrio species isolated from Sparus aurata reared in Italian mariculture. New Microbiol. 37 329–337. [PubMed] [Google Scholar]
- Serrano P. H. (2005). Responsible Use of Antibiotics in Aquaculture Vol. 469 Rome: Food and Agriculture Organization. [Google Scholar]
- Sinyakov M., Dror M., Margel S., Avtalion R. R. (2001). Immunogenicity of Aeromonas salmonicida A-protein in Goldfish (Carassius auratus L.). Isr. J. Aquacul. 53 110–114. [Google Scholar]
- Skwor T., Shinko J., Augustyniak A., Gee C., Andraso G. (2014). Aeromonas hydrophila and Aeromonas veronii predominate among potentially pathogenic ciprofloxacin-and tetracycline-resistant Aeromonas isolates from Lake Erie. Appl. Environ. Microbiol. 80 841–848. 10.1128/AEM.03645-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorum H. (1998). Mobile drug resistance genes among fish bacteria. APMIS 106 74–76. 10.1111/j.1600-0463.1998.tb05652.x [DOI] [PubMed] [Google Scholar]
- Swann L., White M. R. (1991). Diagnosis and treatment of “Aeromonas hydrophila” infection of fish. Aquaculture Extension, Illinois-Indiana Sea Grant Program (Fact Sheet AS-461). [Google Scholar]
- Tamminen M., Karkman A., Lõhmus A., Muziasari W. I., Takasu H., Wada S., et al. (2010). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45 386–391. 10.1021/es102725n [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Tao P., Tan J., Mu H., Peng L., Yang D., et al. (2014). Identification of bacterial community composition in freshwater aquaculture system farming of Litopenaeus vannamei reveals distinct temperature-driven patterns. Int. J. Mol. Sci. 15 13663–13680. 10.3390/ijms150813663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasaikar S., Saraswathi K., De A., Varaiya A., Gogate A. (2002). Aeromonas species isolated from cases of acute gastroenteritis. Indian J. Med. Microbiol. 20:107–109. [PubMed] [Google Scholar]
- Verner-Jeffreys D. W., Welch T. J., Schwarz T., Pond M. J., Woodward M. J., Haig S. J., et al. (2009). High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS ONE 4:e8388 10.1371/journal.pone.0008388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 19 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund T., Dalsgaard I. (1998). Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis. Aquat. Organ. 32 49–69. 10.3354/dao032049 [DOI] [PubMed] [Google Scholar]
- Wu C. J., Chuang Y. C., Lee M. F., Lee C. C., Lee H. C., Lee N. Y., et al. (2011). Bacteremia due to extended-spectrum-β-lactamase-producing Aeromonas spp. at a medical center in Southern Taiwan. Antimicrob. Agents Chemother. 55 5813–5818. 10.1128/AAC.00634-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Qiao M., Zhang B., Cheng W. D., Zhu Y. G. (2010). Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 44 6933–6939. 10.1021/es1007802 [DOI] [PubMed] [Google Scholar]
- Xu L., Wang H., Yang X., Lu L. (2013). Integrated pharmacokinetics/ pharmacodynamics parameters-based dosing guidelines of enrofloxacin in grass carp Ctenopharyngodon idella to minimize selection of drug resistance. BMC Vet. Res. 9:126 10.1186/1746-6148-9-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Kasai H., Arnold D. L., Jackson R. W., Vivian A., Harayama S. (2000). Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146 2385–2394. 10.1099/00221287-146-10-2385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



