Abstract
Mutations in Pantothenate kinase 2 and Coenzyme A (CoA) synthase (COASY), genes involved in CoA biosynthesis, are associated with rare neurodegenerative disorders with brain iron accumulation. We showed that zebrafish pank2 gene plays an essential role in brain and vasculature development. Now we extended our study to coasy. The gene has high level of sequence identity with the human ortholog and is ubiquitously expressed from the earliest stages of development. The abrogation of its expression led to strong reduction of CoA content, high lethality and a phenotype resembling to that of dorsalized mutants. Lower doses of morpholino resulted in a milder phenotype, with evident perturbation in neurogenesis and formation of vascular arborization; the dorso-ventral patterning was severely affected, the expression of bone morphogenetic protein (Bmp) receptors and activity were decreased, while cell death increased. These features specifically correlated with the block in CoA biosynthesis and were rescued by the addition of CoA to fish water and the overexpression of the human wild-type, but not mutant gene. These results confirm the absolute requirement for adequate levels of CoA for proper neural and vascular development in zebrafish and point to the Bmp pathway as a possible molecular connection underlining the observed phenotype.
Coenzyme A (CoA) is an essential cofactor in all living organisms being involved in about 4% of cellular biochemical processes. Its level is regulated by different extracellular stimuli and affected by various pathological conditions1,2. The biochemical steps of cellular CoA biosynthesis include five reactions. Pantothenic acid is firstly phosphorylated by pantothenate kinase (PANK) to generate 4′-phosphopantothenic acid (4′-PPA). Then, 4′-phosphopantothenoylcysteine synthase condenses cysteine with 4′-PPA forming 4′-phosphopantothenoylcysteine, which is subsequently decarboxylated to 4′-phosphopantetheine (4′-PP) by phosphopantothenoylcysteine decarboxylase. Finally, a bifunctional enzyme, CoA synthase (COASY), catalyzes the last two steps leading to the conversion of 4′-PP to dephospho-CoA and subsequently CoA3. The reactions controlled by PANK and CoA synthase are the limiting steps in the regulation of the whole process4. Quite unexpectedly, this fundamental biochemical pathway has been recently linked to cases of early onset neurodegeneration belonging to the Neurodegeneration with Brain Iron Accumulation (NBIA) category. This is a heterogeneous group of genetic neurologic disorders, characterized by dystonia, parkinsonism and spasticity, often with early onset and accumulation of iron in the brain, more typically in the basal ganglia5. In 2001, Zhou and colleagues6 identified nucleotide variations in the gene coding for PANK2 protein, one of the human isoforms of such enzyme. Pantothenate Kinase Associated Neurodegeneration (PKAN, MIM 234200) is the most common form of NBIA, representing almost two thirds of the cases included in this heterogeneous category7. More recently, mutations in COASY were found in patients with clinical manifestation and magnetic resonance imaging (MRI) signs typical of NBIA (MIM 615643)8,9. One subject carried a homozygous nucleotide substitution, c.1495 C > T, that affected a conserved arginine (p.Arg499Cys) in the nucleotide binding site of the dephospho-CoA kinase (DPCK) domain. The other case was a compound heterozygous carrying the same c.1495 C > T transition and the c.175 C > T variation, leading to a premature p.Gln59* stop codon in the N-terminal regulatory region of the protein. In both cases the mutations were associated with significant reduction of protein levels and enzymatic activity. Quite surprisingly, acetyl-CoA but not total CoA levels were significantly different in fibroblasts from patients versus controls and de-novo CoA biosynthesis was clearly reduced, but not absent.
The association between defects in PANK2 and COASY and specific types of NBIA obviously suggests a central role for CoA metabolism in neural cell development and maintenance, although the pathogenic mechanisms underlining this connection are not defined yet. The simplest interpretation of existing evidences links defects in PANK2 and COASY with shortage of cellular CoA that, in turn, acts as initial trigger of a cascade of events culminating with neural death in specific brain areas. This hypothesis is supported by the rescue capacity of pantethine and CoA documented both in cellular and animal models10,11,12,13. On the other hand, measurement of CoA concentrations in fibroblasts or blood samples from patients did not evidence reduced levels of the metabolite8,14. While this result is hard to explain for COASY mutants, compensating mechanisms can exist for PANK2 defects, since mammalian cells express other isoforms of the enzyme (PANK1a and b and PANK3). It may well be that distinctive features of PANK2, and particularly its localization in the mitochondrial intermembrane space, play a relevant role in determining the starting pathogenic mechanism and the specificity of PKAN neuropathology. Interestingly, morphological and functional perturbations of mitochondria have been found in fibroblasts from patients as well as in Drosophila and mouse knock-out models10,15,16. These features are often associated with signs of lipid dyshomeostasis, implying possible perturbation in mitochondrial membrane remodeling, and disruption of iron balance15,17. To gain new information about the functional connection between enzyme defects and pathology, we recently performed a thorough analysis of pank2 role during zebrafish embryonic development13. When we down-regulated larval pank2 expression by microinjection of a specific splice-inhibiting morpholino oligomer we observed a drastic reduction in neurogenin1 neurog1 and neurod1 positive neurons, particularly in telencephalon and midbrain, impaired development of main arteria, vein and intersegmental vessels and edema both in brain and at the tail plexus. Since defects of PANK2 and COASY affect the same biochemical pathway, we reasoned that a comparison of their involvement during zebrafish development could provide functional details about their specific role in the disease pathogenesis. To investigate the role of coasy during zebrafish development we microinjected a specific splice-inhibiting morpholino at 2-cell stage and followed the morphological consequences up to 72 hpf. At high morpholino doses the expression of coasy appeared completely abrogated and this resulted in a drastic decrease of CoA content and perturbation of embryos morphology, with severe reduction of the antero-posterior axis. At lower morpholino doses, the expression of coasy was reduced to about 60% and a milder phenotype was evident, partially resembling the one observed in pank2 morphants. The use of a transgenic bone morphogenetic protein (Bmp)-reporter line evidenced a relevant reduction of bmp activation, associated with lower levels of different Bmp-receptor isoforms, a severe perturbation in antero-posterior patterning, somitogenesis, and neurogenesis, as evidenced by in situ hybridization with a variety of markers. This complex phenotype was rescued by treatment of embryos with CoA in fish water or by injection of the human COASY mRNA. The study demonstrates the essential role of coasy and CoA biosynthesis in normal zebrafish development but also indicates that neurogenesis and Bmp signaling are particularly sensitive to partial CoA depletion. This piece of information could be of relevance for the understanding of the specific phenotype and pathogenic process associated with mutations of the human COASY gene.
Results
Identification and in silico analysis of the coasy gene in zebrafish
The Gene and HomoloGene databases at NCBI18 indicate the presence of one putative ortholog of the human COASY gene in zebrafish, namely coasy, on the reverse strand of chromosome 24. This finding was confirmed by a homology-based search performed with the human COASY protein sequence (NP_001035997.2, 593 aa, c isoform) versus the latest assembly of zebrafish genomic sequences (Genome Reference Consortium Zebrafish Build 10, Sept. 2014).
The Danio rerio coasy gene (ZFIN Id: ZDB-GENE-040912-137, Ensembl Id: ENSDARG00000042747), similarly to the human one, is organized in ten coding exons. It expresses a main transcript isoform encoding for a 554 aa protein (NP_001004577.1) with 51% identity and 69% sequence similarity to the human counterpart (Supplementary Fig. S1). The analysis of the chromosomal regions surrounding the human and zebrafish genes, carried out using the Synteny Database19, showed a conserved synteny between the human chromosome 17 region harboring the COASY gene and Danio rerio chromosome 24 region where the ortholog is located (Supplementary Fig. S1). A single coasy gene is present in other teleosts (data not shown). Analysis of the UniGene database allowed the retrieval of 25 Expressed Sequence Tag (EST) derived from coasy zebrafish transcripts, indicating that the gene is expressed in different tissues and developmental stages. RNA-Seq data from the Wellcome Trust Sanger Institute Zebrafish Transcriptome Sequencing Project20 evidence the presence of RNA-Seq reads at all developmental stages analyzed, including 2-cell stage embryos, indicating that the gene is maternally expressed (data not shown).
Temporal and spatial expression pattern of coasy during zebrafish development
We investigated the spatial and tissue-specific localization of coasy by whole mount mRNA in situ hybridization (WISH) in embryos from 0.2 to 36 hpf (Supplementary Fig. S2). A probe signal was detected since the earliest stages of development. At the sphere stage, coasy was expressed broadly in the entire blastoderm and, at the end of gastrulation, in ventral marginal and animal regions (presumptive ventral mesoderm and non-neural ectoderm, respectively) and, finally, in the animal pole and dorsal region. At the 20-somite stage, coasy displayed strong expression throughout the whole body but particularly in developing central nervous system (CNS) and somites. At later stages (24 and 36 hpf) the probe signal was detectable in specific brain regions, such as hindbrain, midbrain-hindbrain boundary, hindbrain ventricle tegmentum and persisted in newly formed somites. A sense probe was used in parallel control experiments at all stages without detecting any staining (Supplementary Fig. S2).
The expression of coasy gene was also investigated by real time RT-PCR. Coasy mRNA was detectable as early as 2-cell stage. Subsequently, its level remained rather constant throughout development with a slight decrease from 24 to 72 hpf (Supplementary Fig. S3). We then evaluated coasy expression in organs dissected from adult zebrafish, detecting ubiquitous expression, with higher levels in liver, muscles, eyes and brain (Supplementary Fig. S3). These results are in good agreement with available RNA Seq data. Overall, our study reveals that zebrafish coasy is a maternal gene being detected since the earliest stages of development. Consistent with the central role in cellular metabolism, its expression is rather ubiquitous throughout the stages of development and in adult organs, and more abundant in liver, muscles and head.
Knock-down of coasy expression in zebrafish
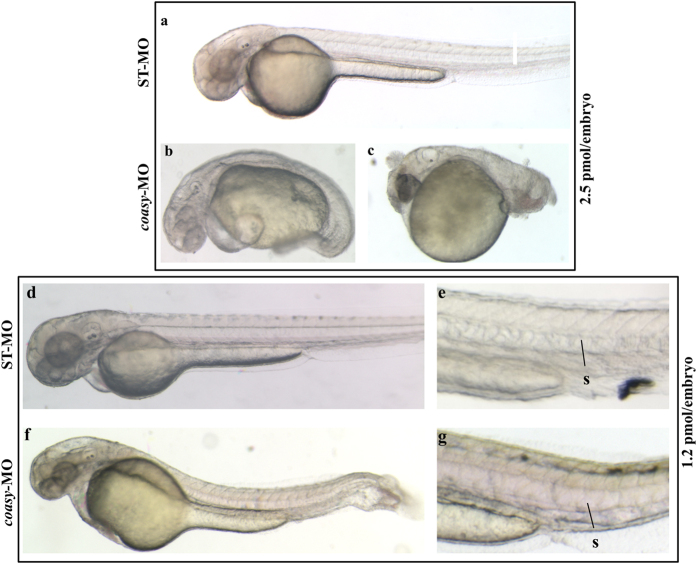
We designed an antisense morpholino (coasy-MO) complementary to intron 1-exon 2 junction in the coasy gene to specifically inhibit the correct mRNA splicing and hence reduce Coasy protein level since the zygotic stage, without affecting the very early maternal expression. In parallel, we used a standard morpholino (ST-MO, Gene Tools, USA) with no target sequence in the zebrafish genome as negative control. In a dose-curve experiment (0.5 to 2.5 pmol/embryo), we observed a survival rate of about 70% in embryos injected with coasy-MO at 1.0, 1.2, 1.5 pmol/embryo. Higher doses (2.0–2.5 pmol/embryo) resulted in a significant mortality (more than 50%) (Supplementary Fig. S4). The effects of morpholino injection upon in vivo mRNA processing were verified by analyzing the specific mature mRNA by RT-PCR at 24 hpf. Primers were designed to amplify an 852 bp cDNA fragment from normally-spliced coasy mRNA and a shorter one (281 bp) when exon 2 was skipped. In embryos injected with higher doses of coasy-MO the longer band was absent and completely substituted by the shorter one, thus indicating a complete abrogation of coasy expression. At lower doses (from 0.5 to 1.5 pmol/embryo) both bands were visible with different intensities, indicating an incomplete suppression of the normal splicing (Supplementary Fig. S4). The doses of 2.5 and 1.2 pmol/embryo were selected to investigate the morphological changes induced by total or partial knock-down of coasy expression at 24 and 48 hpf. At 2.5 pmol/embryo only about 40% of embryos survived till 48 hpf; the vast majority of surviving embryos (79%, n = 211/263 from at least three independent experiments) exhibited a severely aberrant phenotype with marked alteration of the antero-posterior axis and features recalling either a class 3 (64%, n = 134/211) (Fig. 1b) or a class 4 (36%, n = 77/211) (Fig. 1c) dorsalized mutant21,22. In general, structures of ventral origin in the blastoderm were altered or absent whereas the anteriormost head regions were better conserved albeit not normal. The trunk was twisted or reduced with abnormal somites, the tail and posterior somites were absent and the yolk significantly enlarged in most embryos; at the same time also the head development was perturbed, with poor definition of most brain structures. We developed a specific Liquid Chromatography/Mass Spectrometry (LC-MS/MS) method to measure CoA content in embryos (Supplementary Fig. S5). As expected, the absence of coasy led to a significant reduction of CoA levels (0.087 vs 0.36 pmol/embryo, P < 0.001, Fig. 2g) and appeared to be not compatible with normal embryonic development and survival into adulthood. Interestingly, the observed phenotype let infer that specific developmental pathways and tissues exhibit a stronger sensibility to the shortage of such essential metabolite. To further investigate this possibility, we focused our analysis on the effects of a lower dose of coasy-MO (1.2 pmol/embryo), associated with a milder reduction of the level of coasy mRNA and of CoA content (0.17 vs 0.36 pmol/embryo, P < 0.01, Fig. 2g). At this dose, most of the embryos showed an almost normal morphology at 24 hpf (237/298 out of three experiments). Actually, CNS structures were mildly altered, initial head and heart edema were evident, and somites and tail were quite normal or only partially curved (not shown). At 48 hpf the phenotype appeared more severe: morphants often showed a reduction in size (Supplementary Table S1), but no or modest delay in development as confirmed by the careful observation of the head-trunk angle (HTA) size, that was very similar to that of controls at 48 and 60 hpf23. In addition, 81% of coasy-MO injected embryos (n = 242/298 out of three independent experiments) had poorly defined brain structures (particularly hindbrain and midbrain) and head edema. The trunk was thinner with loss or perturbation of the chevron-like structure of somites, the tail was often curved or twisted, the ventral fin was reduced or absent and finally, edema was often present in the tail region (Fig. 1f,g). Altogether, our findings seem to indicate that defects in CoA availability severely interfere with neurogenesis, differentiation and with proper establishment of the antero-posterior axis in zebrafish embryos.
Figure 1. Phenotypic changes due to coasy-MO microinjection.
Representative images of embryos injected with 2.5 or 1.2 pmol/embryo of ST-MO (a,d,e) or coasy-MO (b,c,f,g) at 48 hpf. Embryos injected with high doses of coasy-MO showed a severely aberrant phenotype [(B, 64%) and (C, 36%)]. Embryos injected with 1.2 pmol/embryo of coasy-MO (f,g) showed a milder phenotype.
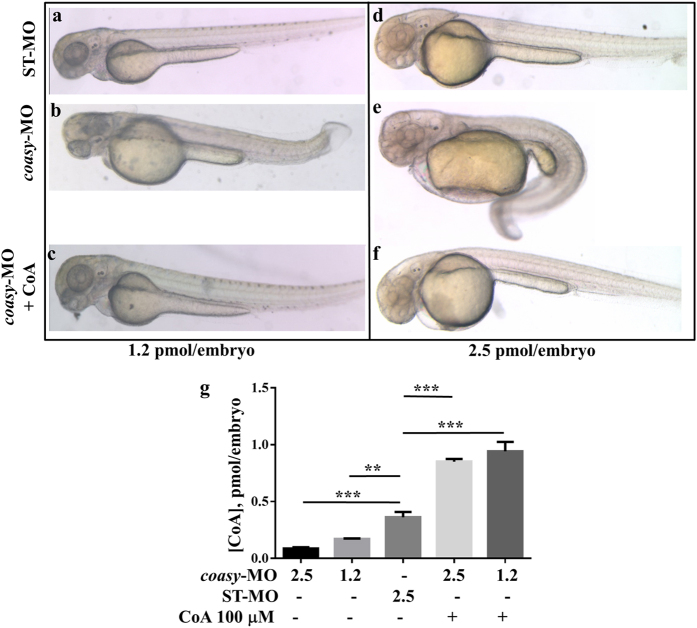
Figure 2. Morphological rescue by treatment with CoA.
Morphological comparison of ST-MO- (a,d), coasy-MO-injected embryos (b,e) and morphants treated with 100 μM CoA at 48 hpf (c,f). Embryos were injected with 1.2 (a–c) or 2.5 pmol/embryo (d–f). Embryos exposed to CoA showed a significant correction of the aberrant phenotype, in particular the antero-posterior development, brain and somites formation. All images are lateral views. The graph in panel G shows the mean CoA content (pmol/embryo) as measured by a specific LC-MS/MS method on extracts from embryos injected with ST-MO or coasy-MO, eventually treated with 100 μM CoA.
To demonstrate the specificity of the observed defects, we tested whether exogenous supply of CoA could prevent the development of the phenotype in embryos exposed to coasy-MO. In a dose-curve experiment, CoA added at 5 hpf showed some toxicity (50% of death) at doses higher than 200 μM (not shown), while at 100 μM it was not toxic and induced a relevant increase of CoA concentration in larvae (Fig. 2g). When embryos were injected with 1.2 pmol of coasy-MO and supplemented with exogenous 100 μM CoA, a very large percentage of embryos (83%, n = 189/227, at least three independent experiments) showed normal morphology at 24 and 48 hpf (Fig. 2c). Similar results (72%) were obtained in embryos injected with 2.5 pmol of coasy-MO (Fig. 2f), thus suggesting that prevention of CoA depletion results in a significant correction of the aberrant phenotype, in particular the antero-posterior axis development and the brain formation.
Over-expression of human COASY mRNA re-establish normal development
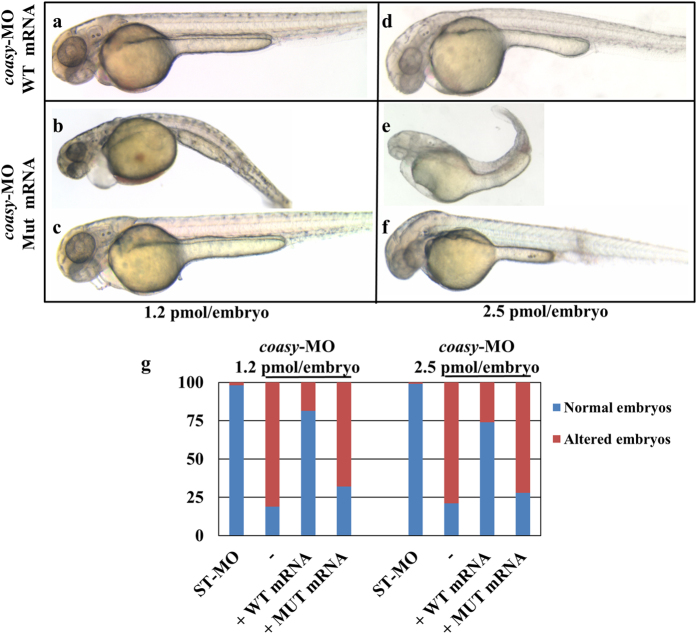
To further confirm the specificity of the observed morphological changes we injected coasy-MO (1.2 and 2.5 pmol/embryo) together with the human COASY mRNA (200 ng), either wild-type or carrying the c.1495 C > T missense mutation identified in two COASY protein-associated neurodegeneration (CoPAN) patients8,9. The co-injection of the wild-type mRNA fully re-established the normal development of most embryos in both experimental conditions (81.5% n = 72/89 for 1.2 pmol/embryo, and 74% n = 64/86 for 2.5 pmol/embryo) (Fig. 3a). Most of the embryos co-injected with the mutant COASY mRNA showed an aberrant phenotype, only partially overlapping with that induced by coasy-MO by itself (68% n = 101/148 and 72% n = 89/124 for 1.2 and 2.5 pmol/embryo respectively, Fig. 3b). Interestingly, 32% and 28% of the embryos showed a normal phenotype as opposed to 19% and 21% observed in embryos injected with coasy-MO alone. The difference observed at 1.2 pmol/embryo was statistically significant (P < 0.01), suggesting a residual biological function of the over-expressed mutant COASY protein. Altogether these results confirmed the specific relationship between the absence of Coasy enzymatic activity and the aberrant development observed in coasy-morphants.
Figure 3. Rescue of coasy knock-down by injection of human mRNA.
Morphological analysis of morphants co-injected with human COASY mRNA, either wild type (a,d) or mutated (b,c,e,f) at 48 hpf. Morpholinos were used at either 1.2 (a–c) or 2.5 pmol/embryo (d–f). Images are from a representative experiment out of four independently performed. The graph in G shows the total distribution of the different phenotypes observed in injected embryos (4 experiments).
Knock-down of coasy and brain development
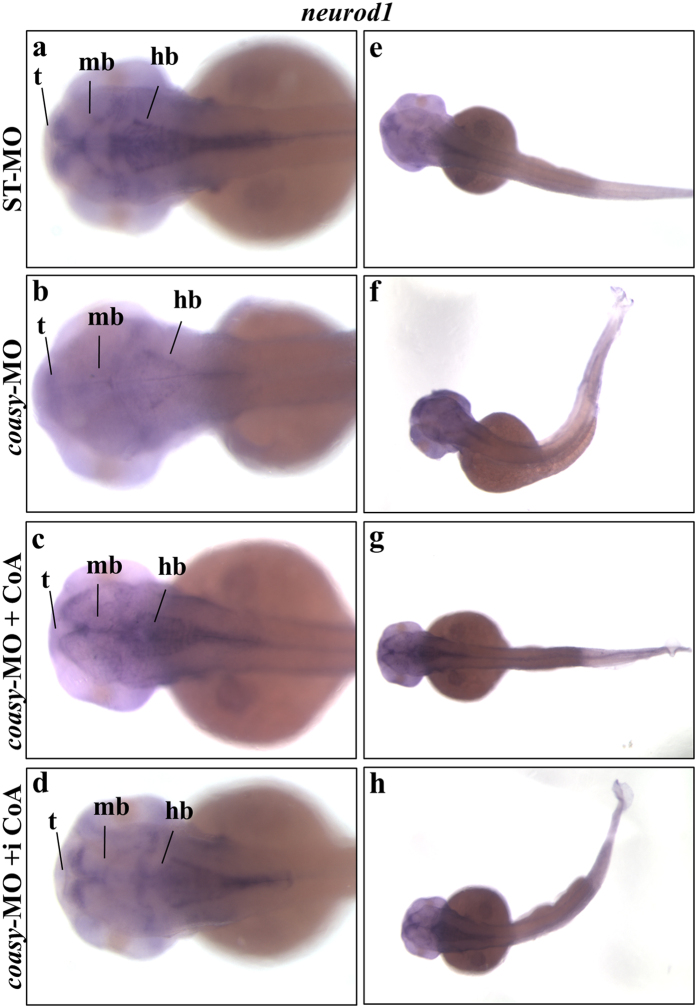
Down-regulation of pank2 in zebrafish embryos resulted in significant perturbation of CNS and vasculature structures13. We were interested in verifying the possible overlap between these two types of morphants that affect the same biochemical pathway. As shown in Figs 1 and 2, the injection of coasy-MO at 1.2 and 2.5 pmol/embryo caused evident alteration of brain morphology. We further investigated this aspect by performing WISH with different neuro-specific riboprobes. Firstly, we analyzed neurod1 expression. Neurod1 gene is specifically expressed during central nervous system development in zebrafish and is a key differentiation factor during neurogenesis; it is also involved in the determination of neural subtypes in the ganglia24. At 48 hpf the expression of neurod1 is more specifically localized at tegmentum, diencephalon, hindbrain and lateral line gaglia25,26. The vast majority of coasy morphants (85%, n = 45/53) showed altered neurod1 expression. The main anterior structures such as telencephalon and midbrain-hindbrain boundary as well as the lateral line ganglia were reduced in size and less intensively decorated by the specific probe as compared to controls (Fig. 4a,b), suggesting an overall reduction in the amount of neurod1 transcript in these neural regions. As expected, treatment with 100 μM CoA prevented the appearance of the altered phenotype (Fig. 4c). Interestingly, also the direct injection of CoA (0.5 pmol/embryo) in the brain ventricle at 24 hpf lead to a normal expression of neurod1 (Fig. 4d). This was not associated with a full recovery of the trunk morphology (Fig. 4e–h), suggesting that the depletion of CoA alters larval development by a cell-autonomous mechanism.
Figure 4. Analysis of neurod1 neural marker in coasy-morphants.
Representative images of WISH analyses of neurod1 expression in coasy morphants at 48 hpf. Dorsal views of ST-MO- (a,e) and coasy-MO-injected embryos (b–h), eventually exposed to CoA in fish water (c,g) or injected with CoA in the brain ventricle at 24 hpf (d,h). For each panel, results of one representative experiment with at least 40 embryos out of two independent replicates are shown. Abbreviations: d, diencephalon; tg, tegmentum; t, telencephalon; hb, hindbrain; hbn, hindbrain neurons; llg, lateral line ganglia; mb, midbrain; snc, spinal cord neurons.
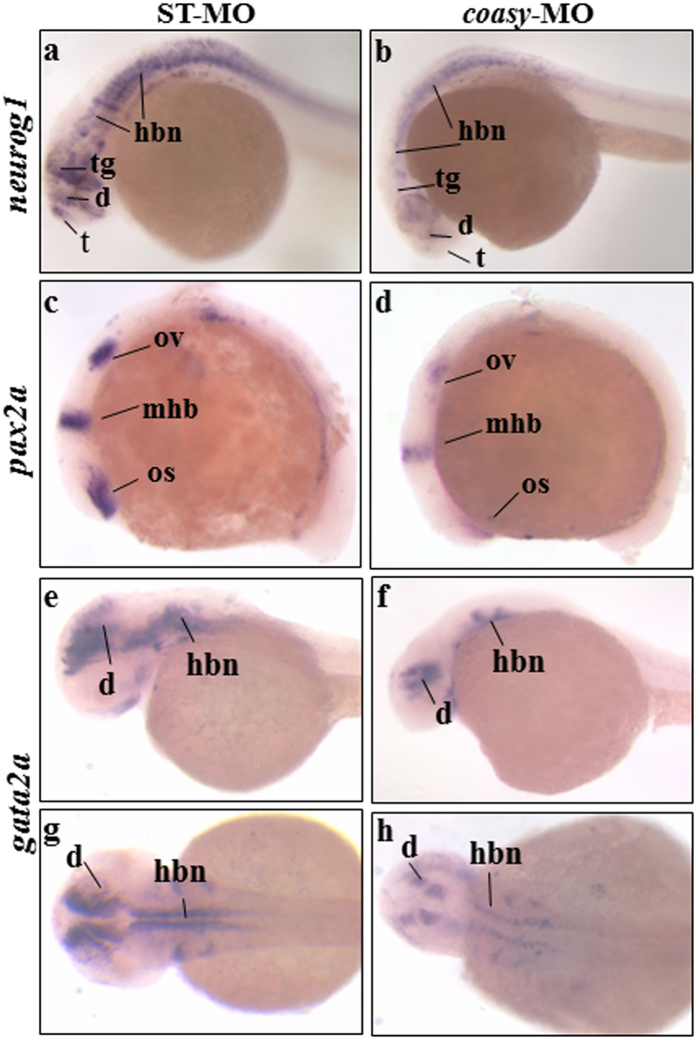
To corroborate the results obtained with neurod1 WISH, we investigated the expression of other neural markers. The onset of neurogenesis in the zebrafish neural plate becomes apparent during late gastrulation by the expression of specific proneural genes27. Neurog1 is among the first of these proneural transcription factors expressed in restricted proneural cell clusters28,29. In addition, ngn1 is an upstream regulator of neurod124, initially expressed in the neural plate and detectable at later stages in tegmentum, dorsal diencephalon, posterior midbrain, midbrain-hindbrain boundary (mhb), hindbrain area, optic stalk and spinal cord. At 24 hpf we observed a strong down-regulation of neurog1 staining in anterior brain areas (tegmentum, diencephalon, telencephalon) of most coasy-MO-injected embryos (n = 51/62, two microinjections) (Fig. 5a,b). Down-regulation of this transcription factor was also evident in hindbrain and spinal cord neurons.
Figure 5. Analysis of neurog1, pax2a and gata2a neural markers in coasy morphants.
Representative images of WISH analyses with neurog1, pax2a and gata2a riboprobes. neurog1 in ST-MO- (a), and coasy-MO-injected embryos (b) at 24 hpf, lateral views. Pax2a expression in ST-MO- (c), and coasy-MO-injected embryos (d) at 15 hpf, lateral views. Gata2a expression in ST-MO- (e,f), and coasy-MO-injected embryos (g,h) at 17 hpf (e,f lateral views and g,h dorsal views). Results are from one representative experiment with at least 40 embryos out of two independent replicates. Abbreviations: d, diencephalon; hb, hindbrain; mhb, midbrain-hindbrain boundary; os optic stalk; ov, otic vesiscle; pd, pronephritic ducts.
Given the brain structural alteration observed in morphants, it was of interest to explore mechanisms that maintain and refine regional identity in brain during development, that is, the formation of local boundaries between regions such as hindbrain and midbrain. These boundaries often co-localize with local organizing centers within the neural tube mediating specific positional information along the antero-posterior (AP) axis30. Pax2a gene encodes a paired-box transcription factor involved in this mechanism31,32. We analyzed the expression of pax2a in coasy-morphants at early stages of development (12 somites) and found it to be significantly reduced at the mhb (n = 37/47) (Fig. 5c d). Interestingly, the signal was also less intense in the pronephric duct formation (not shown), suggesting an interference also with lateral and ventral mesoderm development. Finally, we analyzed the expression of gata2a, a transcription factor required for vascular development and circulation in zebrafish33. Three independent enhancers drive the specific expression of gata2a in blood precursors, EVL, and CNS of zebrafish embryos34. At 48 hpf, the coasy-morphants (n = 34/40) showed marked reduction of gata2a specific signal in diencephalon and hindbrain when compared to controls (Fig. 5e,h).
Altogether, our WISH analysis by neural-specific riboprobes showed that even a mild reduction of coasy mRNA levels perturbs the expression of different proneural transcription factors thereby limiting normal development of many brain structures. Even though some brain regions appeared to be similarly affected both by pank2 and coasy down-regulation, the extent of perturbation induced by reduced levels of coasy was significantly larger.
Knock-down of coasy causes defects in vascular development
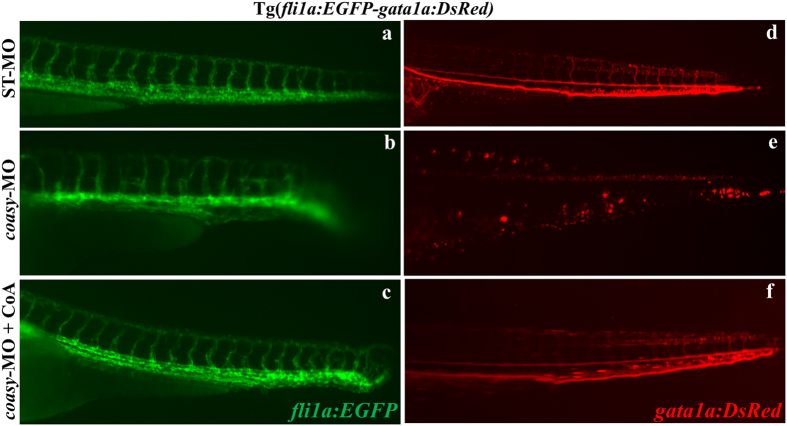
Since our previous findings revealed that pank2 down-regulation affected the formation of the normal vasculature, we extended our analysis by investigating the requirement for coasy during vascular system development. For this purpose, we used the transgenic line Tg(fli1a:EGFP-gata1a:DsRed). Transgenic embryos were injected with 1.2 pmol/embryo of ST-MO or coasy-MO and analyzed at 30 hpf (not shown) and 40 hpf at the fluorescence microscope. The vascular system was severely compromised and the intersegmental vessels were not well formed in coasy-MO injected embryos (n = 65/78). The addition of CoA to fish water efficiently rescued the aberrant phenotype (Fig. 6a and c). This phenotype was confirmed also by in situ hybridization with the ve-cadherin35 riboprobe (not shown) and is clearly reminiscent of the one observed in pank2 morphants. Interestingly, the same transgenic line allowed us to document that coasy morphants had a lower number of red cells when compared to controls and this was associated with signals of reduced blood flow (Fig. 6 d-f). These results suggest that coasy function is required for normal development of ventral mesoderm and/or its derivatives.
Figure 6. Effects of coasy-MO microinjection in the transgenic line Tg(fli1a:EGFP- gata1a:DsRed).
Lateral views of the trunk/tail regions of Tg(fli1a:EGFP-gata1a:DsRed) embryos injected with either ST-MO (a,d) or coasy-MO (b,c,e,f) at 1.2 pmol/embryo, eventually exposed to CoA in fish water (c,f) and analyzed at 40 hpf. fli1a (EGFP) marks vascular development, and gata-1 (DsRed) marks red blood cells. In coasy morphants compromission of the vascular system and particularly of the intersegmental vessels is evident (c,d) and associated with reduction in the number of blood cell (e,f). Results are from one representative experiment with at least 80 embryos out of three independent replicates.
Knock-down of coasy alters no tail and myod1, but not vmhc expression
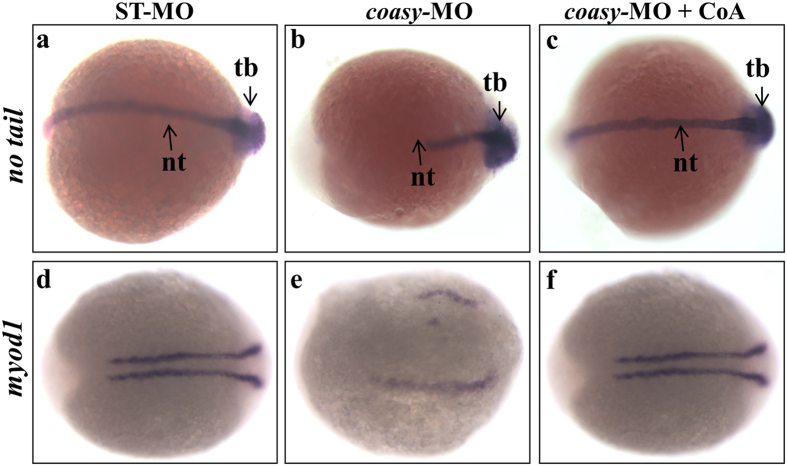
To study whether coasy down-regulation interferes with cellular fate in the dorso-ventral patterning, we evaluated the expression of different markers either involved in or related to this process. T-box genes are transcription factors that pattern the vertebrate mesoderm36,37. No tail is one of such genes. In no tail mutant embryos, the segmentation of paraxial mesoderm into somites occurs normally, but somite patterning at later stages is abnormal and somites are block-shaped rather than chevron-shaped. The notochord is aberrant and not formed and results in a dramatic shortening of the body axis. We investigated the expression of no tail in embryos injected with 1.2 pmol/embryo of coasy-MO at 16 somites (17 hpf). As shown in Fig. 7 at 17 hpf structures such as notochord and tail bud are strongly reduced in a large percentage (84% n = 67/80) of coasy-morphants compared to ST-MO-injected embryos. The morphological analysis documented that coasy-MO injection induced alteration of somite development (Fig. 1). To analyze in deeper detail this feature, we hybridized morphants with a probe for myod1, a gene required for musculature differentiation, morphogenesis and somite formation. Myod1 expression was reduced and indicative of structurally perturbed somites in morphants (76% n = 59/78) as compared to controls at 16 hpf (Fig. 7d,e). Both no tail and myod1 expression were normal when morphants were treated with exogenous CoA (Fig. 7c,f). To analyze whether any structures of mesodermal origin was affected by coasy knock-down, we investigated also the expression of ventricular myosin heavy chain (vmhc), a cardiac marker, in ST-MO and coasy-MO injected embryos at 48 hpf. No significant difference in the expression level of vmhc was evident when morphants were compared to control embryos (Supplementary Fig. S6).
Figure 7. Analysis of no tail and myod1 expression in coasy-morphants.
Representative dorsal view images of WISH analyses of no tail (at 17 hpf) and myod1 (at 16 hpf) expression in ST-MO- (a,d) and coasy-MO-injected embryos (b,c,e,f), eventually exposed to CoA in fish water (c,f). Results are from one representative experiment with about 80 embryos out of two independent replicates.
Knock-down of coasy reduces bmp activity
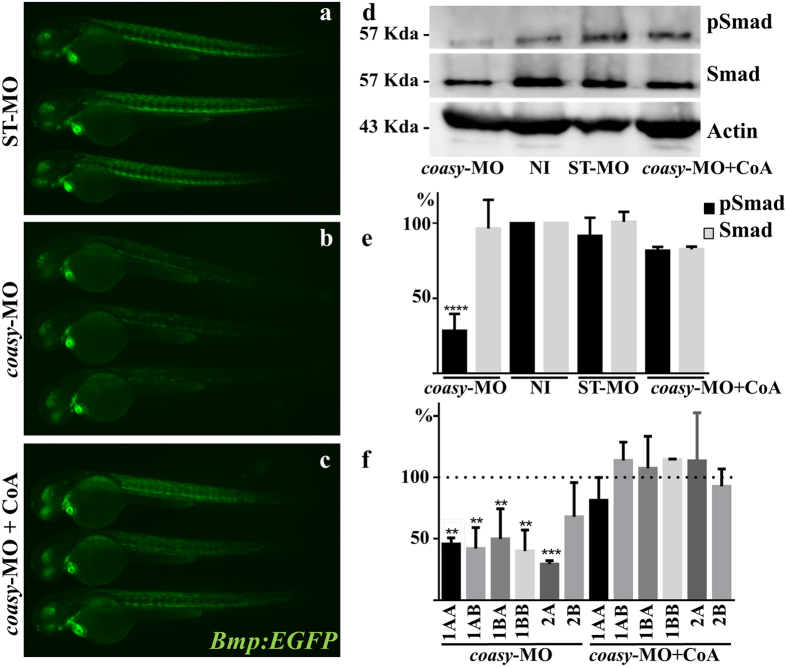
The Bmp signaling cascade plays a central role in dorso-ventral patterning of zebrafish embryo21,38. Different maternal and zygotic factors concur to establish a signaling gradient so that high levels of Bmp induce the development of most ventral mesodermal derivatives such as posterior tail, somites and blood39, whereas low levels are required for dorsal fates. Given the dorsalized phenotype observed in morphants both at morphological and functional level, we investigated the activity of this signaling pathway by microinjection of coasy-MO (1.2 pmol/embryo) in the transgenic line Tg(Bmp:EGFP), which reports Smad-mediated Bmp signaling in embryos and adults. This line expresses enhanced green fluorescent protein (EGFP), under the Bmp Response Element40,41. Down-regulation of coasy resulted in a significant decrease of the fluorescent signal at 60 hpf (Fig. 8 a and b), but the decrease was already evident at 30 hpf (Supplementary Fig. S7). The reduction was particularly evident in somites and eye, whereas less pronounced or absent in the heart. Treatment of embryos with CoA prevented the decrease of Bmp signaling (Fig. 8 c) either when added to fish water (100 μM) or directly injected (0.5 pmol/embryo) in the craniofacial region of morphants at 24 hpf (Supplementary Fig. S7). To confirm the result, we analyzed the phosphorylation of Smad5 protein (pSmad5), a downstream effector of Bmp-receptor activation in the same embryos. The immunoblot analysis showed a robust reduction in the intensity of pSmad5 band (about 70%, P < 0.0001) in coasy-MO-injected embryos as compared to not-injected embryos (Fig. 8d and e). No decrease in pSmad phosphorylation was evident in morphants exposed to 100 μM CoA in the water. In order to dissect the mechanism underlining this phenomenon, we quantified mRNA levels of ligands and receptors potentially involved in activation of the signaling cascade. While no difference was evident for Bmp-2A, Bmp-2B and Bmp-4 expression levels (Supplementary Fig. S9), a robust reduction was present for all the Bmp receptor isoforms we investigated, with the exception of BmpR-2B (Fig. 8f). On the basis of the fundamental involvement of Bmp signaling during vertebrate development as well as in adult tissues homeostasis, the results may be of extreme relevance for the interpretation of the phenotype of coasy-morphants and eventually indicates a new perspective for the analysis of CoPAN pathogenesis.
Figure 8. Analysis of Bmp activity in coasy-morphants.
Representative images of Tg(Bmp:EGFP) embryos injected with ST-MO (a) or coasy-MO (1.2 pmol/embryo, b and c), eventually exposed to CoA in fish water (c), at 60 hpf. A net reduction of the fluorescence intensity is evident in coasy-morphants. Results are from one representative experiment with at least 25 embryos out of two independent replicates. (d) Western blotting analysis of the amount of phosporylated Smad-5, total Smad, and Actin in embryos injected with coasy-MO (±CoA) or ST-MO (1.2 pmol/embryo), and not-injected (NI). Full size images of the gels are shown in Supplementary Fig. S7) The graph shows the quantitative analysis of the immunoblotting. (f ) Real-time RT-PCR quantification of different Bmp receptor mRNAs in morphants at 48 hpf, eventually treated with CoA in fish water. Mean values are form three independent experiments and expressed as percentage of the value in ST-MO injected embryos.
Knock-down of coasy induces cell death
Given the central role of CoA in energetic metabolism and the clear lethality associated with the strong reduction in coasy expression, we evaluated cell death events induced by the lower dose of coasy-MO using acridine orange staining42 (Supplementary Fig. S10). Florescence imaging showed a clear increase of the staining intensity in morphants, particularly evident in the head and tail regions. The quantitative analysis performed on lysates from pooled embryos documented a 2.5-fold increase in coasy-MO versus control embryos.
Discussion
NBIA is a set of clinically and genetically heterogeneous disorders, characterized by progressive neurodegeneration and accumulation of iron in the brain, most commonly in the basal ganglia. In the recent years an intense effort to identify the genetic basis of this type of diseases led to the discovery of mutations in at least 10 different genes5 and opened the way to the study of the molecular mechanisms underlining the neurodegenerative processes. Among this set of genes, two codes for enzymes catalyzing limiting steps in CoA biosynthesis: PANK2, linked to PKAN6 controls the phosphorylation of pantothenate to 4′-PPA while COASY, linked to CoPAN8,9, mediates the two final steps of the pathway, from 4′-PP to dephospho-CoA and finally to CoA. This drove the attention to the role played by this metabolite in neural development and functioning3. CoA and its derivatives are essential for many biochemical processes, such as energy production, lipid metabolism and heme synthesis, involved both in neurodegeneration and iron metabolism43. Quite unexpectedly, defects in CoA production results in disorders with prominent, if not exclusive, involvement of the CNS and particularly of the nuclei involved in movement control. This is surely one of the central aspects of the pathogenesis of these disorders that remains poorly understood, together with the connection with brain iron homeostasis and the very early onset of symptoms.
We tried to get more insights into the pathogenesis of these disorders by performing morpho-functional studies of pank2 and coasy genes during the early phases of zebrafish development. Pank2 mRNA was detected at highest levels in the CNS of fish embryos and larvae; the injection of a specific morpholino leads to a significant reduction of pank2 expression associated with evident perturbation of brain and vasculature development. Particularly affected was the telencephalon, presumably containing nuclei corresponding to the human globus pallidum44, where the expression of the neuronal differentiation marker neurod1 was dramatically reduced. This indicated an essential role for pank2 in the development and differentiation of specific sets of neural network, not compensated by other pank isoforms, but strongly linked to the production of CoA, as documented by the rescue capability of CoA and pantethine supplemented to the fish water. Interestingly, also the development of the vasculature arborization was perturbed, particularly in the head and the tail, thus suggesting a putative involvement of defective angio- and/or vasculogenesis in the pathogenesis of the human disease. On the basis of these results, we extended our analysis to coasy gene. As in most vertebrates, also in Danio rerio there is a single coasy gene, with high level of identity to the human homolog. It is a maternal gene with stable expression throughout the different stages of zebrafish embryonal development and ubiquitously transcribed in the adult, with higher levels in liver, brain and eye. The apparently complete down-regulation of coasy expression obtained by high doses of morpholino (2.5 pmol/embryo) leads to a severe alteration of development with death occurring within 72 hpf. Interestingly a large percentage of embryos showed a phenotype recalling C3 and C4 dorsalized mutants of ventral specifying genes21. Together with the alteration of tissues of ventral origin, also the brain was poorly defined. The partial down-regulation of coasy expression resulted in a milder phenotype that confirmed the important sensibility of neurogenesis and ventralizing pathways. These phenotypic features were specifically associated with a shortage of CoA availability, as measured by a specific LC-MS/MS method. Indeed, CoA supplementation to fish water or overexpression of the human wild-type COASY mRNA could prevent the abnormal phenotype. The injection of CoA in the brain ventricle of morphants allowed the local rescue of neurod1 expression, but did not seem to be able to efficiently prevent the alteration of mesodermal structures. This allows to infer that decrease of CoA level in each cell is the main trigger of the altered development, but the experiments so far performed cannot exclude possible cross-talks among different tissues.
Interestingly, the co-injection of human mutant mRNA with 1.2 pmol/embryo of coasy-MO ushered in a modest, yet statistically significant prevention of the altered morphology, thus indicating a partial preservation of the biological function of the mutated allele in vivo. This is in line with data from Dusi and coauthors8 showing loss of activity for the recombinant mutant protein in vitro, yet the persistence of CoA levels and de novo production in fibroblasts from patients carrying the same Arg499Cys substitution. While showing a modest rescuing capacity, the co-injection of mutant COASY mRNA lead to an aberrant phenotype that often differed from that of coasy morphants. Since no changes in morphology were observed when either wild-type or mutant mRNA were injected, the interpretation of this results is rather puzzling and will require further studies.
The in situ hybridization with neural specific probes confirmed the results of the morphological analysis on coasy morphants. Reduction in size and poor definition of brain structures was clearly associated with marked decrease of neurogenin1, neurod1, pax2a and gata2a signal. Contrary to what observed with pank2 suppression, all investigated brain regions and structures were affected, suggesting a more generalized perturbation of neural differentiation. When we evaluated the development of the vasculature by a specific probe (ve-cadherin) and in a fli1a trangenic line, we detected an altered vasculature structure, particularly in brain and trunk, with reduction in ISV formation associated to decreased red cells and blood flow. These findings overlap with the phenotype observed in pank2 morphants and highlights the pivotal role of adequate levels of CoA for normal vasculogenesis and angiogenesis. There is no proof yet that this holds true in the human pathology, but it could be of interest to investigate this aspect in mammalian experimental models of PKAN and CoPAN.
The shortage of CoA dramatically affected the development of zebrafish, with appearance of phenotypes with graded levels of dorsalization. This could be in part explained with the significant down-regulation of the Bmp signalling pathway, as clearly evidenced by the reduced fluorescent signal and Smad 5 phosphorilation observed in the Bmp-reporter transgenic line injected with coasy-MO (Fig. 8). BMPs comprise a large family of secreted signaling factors belonging to the transforming growth factor-beta (TGF-β) superfamily and are expressed in both developing and adult tissues. In zebrafish they are necessary for specification of posterior tail and ventral fates such as somites and blood38,45. The reduction in Bmp transcriptional activity we documented was obtained with injection of 1.2 pmol/embryo of coasy-MO and can explain the alteration observed in morphants, but could be more severe at higher morpholino doses, when the morphology more clearly resembled that of dorsalized mutants. While we did not observe significant changes in the mRNA level of the main Bmp isoforms, we found lower levels of Bmp-receptors expression, which could explain the initial result. We are not aware of any mechanism directly linking CoA metabolism and Bmp-receptors expression, and further work is necessary to better understand this result. Other mechanisms could contribute to the suppression of Bmp signaling. Interestingly, Miyares and coauthors46 documented the requirement for long-chain acyl-CoA synthetase 4a (Acsl4a) for proper Bmp signaling and patterning of the zebrafish dorso-ventral axis. G.
Indeed, gene expression patterns (gata2a, pax2a, myod1) and morphological features (dorsalization) of ascl4a morphants overlap with those of coasy ones. Ascl4a converts long chain-polyunsaturated fatty acids (LC-PUFA) to acyl-CoA that are fundamental for lipid metabolism and cell signaling. Mutations in the human ACSL4 ortholog are associated with X-linked mental retardation47 and the Drosophila hypomorphs show defects in segmentation48 and development of CNS, with reduced number of glial and neural cells in the brain49, as well as diminished synaptic growth and function50,51. These features could be explained, at least in part, by the reduction in Bmp activation observed both in zebrafish and in Drosophila. The enzyme can actually control the levels of Smad transcription factors by blocking the inhibiting activity of glycogen synthase kinase 3 and p38 mitogen-activated protein kinase, as shown in zebrafish46, but can also affect vesicles trafficking and membrane recycling, and hence the availability of Bmp receptor, as observed in Drosophila51. The reduction of CoA levels induced by down-regulation of Coasy can surely have an impact on LC-PUFA activation process and therefore affect Bmp signaling.
Perturbation of the ventral-to-dorsal Bmp gradient can be directly linked with many of the morphological and functional defects observed in coasy morphants. The decreased expression of makers such as gata2, myod1 and pax2a in tissues of mesodermal origin as well as the reduction in blood cells and ISVs formation can be ascribed to a defective Bmp signaling.
Restriction of Bmp expression to the ventral part of the embryo by the onset of gastrulation is instrumental for proper ectoderm neutralization39,52. Yet Bmp activity persists in the CNS at later stages and exerts a fundamental role in neurogenesis and neural differentiation, as documented in the retina53 or in the inner ear54. This is indirectly confirmed by the spatial analysis of EGFP expression in the Bmp-reporter line41 that documented a clear signal in eye and other brain regions (mhb) from 24 to 72 hpf. Hence, the reduction of Bmp expression induced by coasy down-regulation may concur to the insurgence of the brain structural and functional defects observed in morphants.
We did not explore iron homeostasis in pank2- and coasy-morphants, and zebrafish embryos are not the most suitable system to investigate iron accumulation, but it is interesting to observe that Bmp-signaling is the main regulator of hepcidin expression, the master regulator of systemic and cerebral iron balance55. Our findings may indicate a new perspective to investigate the mechanisms underlining brain iron accumulation in these disorders.
Given the central role of CoA in cellular metabolism and energetic processes, it is possible that other mechanisms and signaling pathways contribute to the observed phenotype, but the analysis performed at lower doses of morpholino and with milder reduction in coasy expression suggests a high sensibility of the Bmp signaling cascade to the shortage of CoA.
It is possible that perturbed lipid metabolism, including LC-PUFA, and membrane homeostasis is one of the immediate consequences of limited CoA availability. This seems to be a unifying theme for many types of NBIA disorders, and particularly for CoPAN, PKAN, PLAN, FAHN and MPAN, which are characterized by alteration of phospholipids and sphingolipids metabolism, membrane remodeling and mitochondrial morphology and functioning56. Further investigation of the possible connection between lipid dyshomeostasis and Bmp signaling cascade may provide relevant insight into the pathogenesis of these disorders and the specific neuronal involvement.
Materials and Methods
Zebrafish maintenance and stocks
Zebrafish were maintained and used following protocols approved by the Local Committee (Organismo Per il Benessere Animale–Committee for Animal Health) (No. 211B5-10) and in accordance with the Italian and European regulations on animal use. Breeding, and staging were performed as previously described23. Beginning from 24 hpf, embryos were cultured in fish water containing 0.003% 1-phenyl-2-thiouera (PTU) to prevent pigmentation. The following strains were used: the wild type AB, and the transgenics Tg(fli1a:EGFP;gata1a:DsRed)57 and Tg(BRE-AAVmlp:EGFP)mw29 [hereafter Tg(Bmp:EGFP)]41.
Bioinformatic analysis
Zebrafish genomic sequences were analyzed using the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/) on the Zv10 (Sept 2014) Danio rerio assembly and the Ensembl zebrafish genome database (http://www.ensembl.org/Danio_rerio/Info/Index).
Synteny analysis was achieved using the Synteny Database19. Nucleotide and amino acid sequences were compared to the non-redundant sequences present at the NCBI (National Center for Biotechnology Information) using the Basic Local Alignment Search Tool (BLAST)58. Multiple sequences alignment was performed using the MUSCLE algorithm59.
RNA extraction and Real Time RT-PCR
Total RNA was extracted from 30 embryos for each different developmental stage analyzed, frozen in liquid nitrogen, using TRI-Reagent (Sigma) according to manufacturer’s protocol. For tissues dissection, the adult fishes were euthanized by an excess of ethyl 3-aminobenzoate methanesulfonate salt solution (Sigma). RNA was quantified using the My Spect spectrophotometer (Biomed) and controlled by electrophoretic separation on a 1% TAE-agarose gel. 1.0 μg of total RNA was retro-transcribed to cDNA using Im-Prom Reverse Transcriptase (Promega) and oligo (dT) primers following the manufacturer’s protocol. Primers were designed by the PrimerQuest and Real Time PCR Tool from IDT (Supplementary Table S2). Real-Time PCR was performed using the Eco-Illumina system. Reactions were performed in a 10 μl volume, with 0.5 μM of each primer (sequences in Supplementary Table S2), 5 μl of Syber Green Master Mix (Biorad) and 20 ng of cDNA. The amplification profile consisted of a denaturation program (95 °C for 1 min), 40 cycles of two steps amplification (95 °C for 15s and 60 °C for 30s) followed by a melting cycle. Each reaction was performed in triplicate and when required actin beta 1 was used as a reference gene. Relative levels of expression were calculated by the ΔΔCT method.
Riboprobes synthesis and whole-mount in situ hybridization
Total RNA was extracted from a pool of 2-cell stage embryos using TRI-Reagent (Sigma) according to manufacturer’s protocol. RNA was quantified using the My Spect spectrophotometer (Biomed) and controlled by electrophoretic separation on a 1% TAE-agarose gel. 1.0 μg of total RNA was retro-transcribed to cDNA using Im-Prom reverse transcriptase (Promega) and oligo(dT) primers following the manufacturer’s protocol. To synthesize the riboprobes for the detection of zebrafish coasy transcripts, we amplified a specific region by PCR using as template the cDNA from the 2-cell stage and oligonucleotide primers (P3 and P4, Table S2). The amplification conditions were the following: 2 min at 95 °C, 30 cycles at 94 °C for 15 sec, 60 °C for 30 sec, 72 °C for 1 min, followed by final extension at 72 °C for 5 min. The PCR product was subcloned with the pGEM-T-Easy system (Promega), and verified for sequence and orientation of the inserts. Antisense and sense RNA probes were obtained by in vitro transcription of the cloned cDNAs with T7 or SP6 RNA polymerase, using a digoxigenin labeling mixture according to manufacturer’s instructions (Roche). Whole-mount in situ hybridization (WISH) was performed according to a standard method60. Briefly, embryos and larvae were collected, dechorionated and incubated at 28 °C at different stages. Embryos were fixed overnight in 4% paraformaldehyde (PFA) at 4 °C, dehydrated through an ascending methanol series and stored at −20 °C. After treatment with proteinase K (10 μg/ml, Roche), the embryos were hybridized overnight at 68 °C with DIG-labeled antisense or sense RNA probes (400 ng). Embryos were washed with ascending scale of Hybe Wash/PBS and SSC/PBS, then incubated with anti-DIG antibody conjugated with alkaline phospatase over night at 4 °C. The staining was performed with NBT/BCIP (blue staining solution, Roche) alkaline phosphatase substrates. When different type of probes (sense vs antiense) or fish (injected vs not-injected) had to be compared, all incubations were carried out at the same time, at the same probe concentration and, when required, with the same reagents and solutions. WISH images were taken with a Leica MZ16F stereomicroscope equipped with DFC 480 digital camera and LAS Leica Imaging software (Leica). Magnification: 50X, 63X, and 80X.
Morpholino injections
Injections were carried out on one/two-cell-stage embryos (with Eppendorf FemtoJet Micromanipulator 5171); the dye tracer phenol-red was co-injected as a control. To repress coasy mRNA translation we designed a splicing-inhibiting morpholino oligo (coasy-MO, Table S2); as a negative control we injected a standard control morpholino oligo (ST-MO) (Gene Tools) which targets a human beta-globin intron (Supplementary Table S2). Morpholinos were injected in Danieau buffer (pH 7.6) as previously described61. After microinjection, embryos were incubated in egg water supplemented with 0.003% PTU at 28 °C to prevent pigmentation processes. Embryo development was evaluated at 24 and 48 hpf. RT-PCR experiments were performed on RNA extracted from coasy-MO- or ST-MO-injected and wild type embryos at 24 hpf and 48 hpf, with specific primers (P5 and P6, Supplementary Table S2). Control RT-PCR amplification on the same RNAs was carried out with actin beta 1 primers (A1 and A2, Supplementary Table S2). When indicated, the comparison between morphants and controls was performed by selecting morphants with the mildest phenotype (embryos injected with 1.2 pmol of CoA-MO) and no evident delay in development (head-trunk angle).
Rescue of phenotype by COASY human wild type and mutant mRNA
The pGEM plasmids containing the coding region for human wild type and c.1495 C > T mutant COASY allele were kind gifts from Dr. Valeria Tiranti, Fondazione I.R.C.C.S. Istituto Neurologico Carlo Besta, Milan. The plasmids were digested with NcoI restriction enzyme and then transcribed using the mMESSAGE mMACHINE in vitro Transcription Kit (Life Technologies) according to the manufacturer’s instructions. Dose-response curve experiments were performed in wild type embryos to identify the maximum amount of coasy mRNA that did not induce phenotypic alterations. The rescue experiments were carried out by co-injecting coasy-MO together with 200 pg/embryo of human wild-type or mutant COASY mRNA.
CoA treatment and LC-MS/MS analysis of CoA
Control embryos and embryos injected with coasy-MO at 1.2 and 2.5 pmol were treated with 100 μM CoA in fish water at 5 hpf. At 48 hpf survived embryos were decoryonated, rinsed thoroughly with water and prepared for morphological analysis at the microscope or pooled in groups of 50 and frozen for CoA content analysis. Frozen samples were thawed and 150 μl of 5% perchloric acid (PCA) was immediately added. Samples were homogenized using a pipette, vortexed, sonicated and centrifuged at 18,000 × g for 5 minutes at 4 °C. The injection mixture was filtered through a 0.2 μm PVDF filter. The filtrates were brought to a volume of 0.5 ml with water and 10 μl were injected into the system LC-MS/MS. Calibration curves (100, 10, 1, and 0.1 pmol/μl) were prepared by dilution of the CoA standard (Supplementary Fig. S5). Linearity was determined by least-squares regression. An HPLC 1260 System equipped with an infinity system model 1260 injector and paired with an Agilent Technologies (Waldbronn, Germany) 6460 Triple Quad LC/MS triple quadruple mass spectrometer were used for the quantitative dosage of CoA. Chromatographic separation of CoA was achieved using INERTSIL ODS-3 C18 column (100 × 3 mm × 3μm) coupled with a security guard column. The flow rate was 0.3 mL/min with mobile phase of 0.01% Formic Acid in water (A) and Methanol (B). The column was equilibrated with 2% B for 3 min, a linear gradient to 95% B was applied over 12 min. The total run time was 15 min per sample. The optimal source parameters were: source temperature: 300 °C, drying gas flow: 12 L/min, drying gas temp: 350 °C, nebulizer pressure: 40 psi, capillary voltage: 3,500 V. Mass spectrometry was performed in the positive ion mode, monitoring the MS/MS transitions m/z 768.1 → 428.2 and 768.1 → 261.
Staining with Acridine Orange
Acridine orange (AO) staining is a nucleic acid selective metachromatic stain technique that can identify cell death. For AO staining, embryos at 48 hpf were incubated for 30 min in fish water containing acridine orange at 10 mg/l. Embryos were then rinsed three times in fish water, anesthetized with tricaine, and quickly imaged by epifluorescent microscopy. Pools of 10 embryos were than lysed in 100% ethanol and fluorescence was measured at λex 490 nm/λem 525 nm with a microplate reader (Ensight Perkin Elmer).
Immunoblotting
20 embryos for each condition were manually dechorionated. Proteins were extracted by homogenizing embryos in buffer (200 mM Tris/HCl, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% Glycerol, 1 mM NaFl and sodium orthovanadate) on ice and quantified by a standard bichinonic acid method. 80 μg of total protein extract were then separated on two 10% polyacrylamide gels, one for pSmad and the other for total SMAD detection, transferred on to membranes (GE AmershamTM HybondTM P 0.45 PVDF). Membranes were cut at the level of the molecular weight of interest, blocked with 2% skim milk in TBST, incubated with the primary antibody diluted in 2% skim milk in TBST (phospho-Smad5, 1:1000, Abcam #ab92698, Smad5, 1.1000, Cell signaling Technology #9517) and then with the anti-rabbit IgG secondary antibody also prepared in 2% skim milk (1:2000). ECL Western blotting detection reagent (Pierce) was used for detection. Images were acquired with Li-Cor Odissey image station, and band intensity quantified by ImageJ software without any modification of the original data. Original images were partially cropped to fit to the final figure.
Statistical analysis
The data are reported as the sum or as representative of at least three independent experiments with similar results. Statistically significant differences between different types of embryos were calculated by one-way ANOVA analyses; *, ** and *** indicate P < 0.05, <0.01 and <0.001), respectively. The Chi square test was applied to compare frequencies of morphants and normal embryos in different types of samples.
Additional Information
How to cite this article: Khatri, D. et al. Down-regulation of coasy, the gene associated with NBIA-VI, reduces Bmp signaling, perturbs dorso-ventral patterning and alters neuronal development in zebrafish. Sci. Rep. 6, 37660; doi: 10.1038/srep37660 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are grateful to Dr. Valeria Tiranti and Dr. Sabrina Dusi, (Fondazione I.R.C.C.S. Istituto Neurologico Carlo Besta, Milan) for providing plasmids and primers for the human COASY gene, either wild type or mutant. This work was partially supported by Fondazione Telethon (Grant No. GGP11088 to DF), by University of Brescia (ex 60%-2015 funds) to DZ, EM. NT was supported by the Italian Ministry of Health project Trambigen (RF-2010-2309484), the AFM-Telethon project Polygon (18572), and the EU ZF-HEALTH grant CT-2010-242048. DK is a student of the PhD program in Genetics, Biotechnology and Experimental Medicine at the University of Brescia and was also funded by Consorzio Interuniversitario Biotecnologie (CIB); NF is a Junior Post-doc fellow of the University of Padova (grant CPDR154779).
Footnotes
Author Contributions D.Z., N.T. and D.F. conceived and designed the experiments and wrote the manuscript. D.K., D.Z. and N.F. performed the experiments and analyzed the data; S.V. and A.G. performed CoA measurements in embryos; G.B. performed the in silico analysis; D.K., D.Z., N.T., N.F., S.V., A.G., M.M., E.M., G.B. and D.F. analyzed the data and corrected the manuscript.
References
- Tahiliani A. G. & Beinlich C. J. Pantothenic acid in health and disease. Vitam Horm 46, 165–228 (1991). [DOI] [PubMed] [Google Scholar]
- Smith C. M. & Savage C. R. Regulation of coenzyme A biosynthesis by glucagon and glucocorticoid in adult rat liver parenchymal cells. Biochem J 188, 175–184 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi R., Zhang Y. M., Rock C. O. & Jackowski S. Coenzyme A: back in action. Prog Lipid Res 44, 125–153, doi: 10.1016/j.plipres.2005.04.001 (2005). [DOI] [PubMed] [Google Scholar]
- Zhyvoloup A. et al. Molecular cloning of CoA Synthase. The missing link in CoA biosynthesis. J Biol Chem 277, 22107–22110, doi: 10.1074/jbc.C200195200 (2002). [DOI] [PubMed] [Google Scholar]
- Levi S. & Finazzi D. Neurodegeneration with brain iron accumulation: update on pathogenic mechanisms. Front Pharmacol 5, 99, doi: 10.3389/fphar.2014.00099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. et al. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat Genet 28, 345–349, doi: ng572 [pii] 10.1038/ng572 (2001). [DOI] [PubMed] [Google Scholar]
- Meyer E., Kurian M. A. & Hayflick S. J. Neurodegeneration with Brain Iron Accumulation: Genetic Diversity and Pathophysiological Mechanisms. Annu Rev Genomics Hum Genet 16, 257–279, doi: 10.1146/annurev-genom-090314-025011 (2015). [DOI] [PubMed] [Google Scholar]
- Dusi S. et al. Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am J Hum Genet 94, 11–22, doi: 10.1016/j.ajhg.2013.11.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annesi G., Gagliardi M., Iannello G. & Quattrone A. Mutational analysis of COASY in an Italian patient with NBIA. Parkinsonism Relat Disord, doi: 10.1016/j.parkreldis.2016.03.011 (2016). [DOI] [PubMed] [Google Scholar]
- Rana A. et al. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc Natl Acad Sci USA 107, 6988–6993, doi: 10.1073/pnas.0912105107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti D. et al. Pantethine treatment is effective in recovering the disease phenotype induced by ketogenic diet in a pantothenate kinase-associated neurodegeneration mouse model. Brain 137, 57–68, doi: 10.1093/brain/awt325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B. et al. Extracellular 4′-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat Chem Biol 11, 784–792, doi: 10.1038/nchembio.1906 (2015). [DOI] [PubMed] [Google Scholar]
- Zizioli D. et al. Knock-down of pantothenate kinase 2 severely affects the development of the nervous and vascular system in zebrafish, providing new insights into PKAN disease. Neurobiol Dis 85, 35–48, doi: 10.1016/j.nbd.2015.10.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni V. et al. Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations. Mol Genet Metab 105, 463–471, doi: 10.1016/j.ymgme.2011.12.005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio P. et al. Mitochondrial iron and energetic dysfunction distinguish fibroblasts and induced neurons from pantothenate kinase-associated neurodegeneration patients. Neurobiol Dis, doi: 10.1016/j.nbd.2015.02.030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti D. et al. Pantothenate kinase-associated neurodegeneration: altered mitochondria membrane potential and defective respiration in Pank2 knock-out mouse model. Hum Mol Genet 21, 5294–5305, doi: 10.1093/hmg/dds380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli M. et al. Pantothenate kinase-2 (Pank2) silencing causes cell growth reduction, cell-specific ferroportin upregulation and iron deregulation. Neurobiol Dis 39, 204–210, doi: 10.1016/j.nbd.2010.04.009 (2010). [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 43, D6–17, doi: 10.1093/nar/gku1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J. M., Conery J. S. & Postlethwait J. H. Automated identification of conserved synteny after whole-genome duplication. Genome Res 19, 1497–1505, doi: 10.1101/gr.090480.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. E., White S., Searle S. M. & Stemple D. L. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res 22, 2067–2078, doi: 10.1101/gr.137901.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins M. C. et al. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123, 81–93 (1996). [DOI] [PubMed] [Google Scholar]
- Kondo M. Bone morphogenetic proteins in the early development of zebrafish. FEBS J 274, 2960–2967, doi: 10.1111/j.1742-4658.2007.05838.x (2007). [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310, doi: 10.1002/aja.1002030302 (1995). [DOI] [PubMed] [Google Scholar]
- Mueller T. & Wullimann M. F. Expression domains of neuroD (nrd) in the early postembryonic zebrafish brain. Brain Res Bull 57, 377–379 (2002). [DOI] [PubMed] [Google Scholar]
- Korzh V., Sleptsova I., Liao J., He J. & Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn 213, 92–104, doi: (1998). [DOI] [PubMed] [Google Scholar]
- Sato A. & Takeda H. Neuronal subtypes are specified by the level of neurod expression in the zebrafish lateral line. J Neurosci 33, 556–562, doi: 10.1523/JNEUROSCI.4568-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B. & Chitnis A. Neurogenesis and specification of neuronal identity. Results Probl Cell Differ 40, 237–251 (2002). [DOI] [PubMed] [Google Scholar]
- Blader P., Fischer N., Gradwohl G., Guillemot F. & Strähle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development 124, 4557–4569 (1997). [DOI] [PubMed] [Google Scholar]
- Korzh V. & Strähle U. Proneural, prosensory, antiglial: the many faces of neurogenins. Trends Neurosci 25, 603–605 (2002). [DOI] [PubMed] [Google Scholar]
- Rhinn M. & Brand M. The midbrain–hindbrain boundary organizer. Curr Opin Neurobiol 11, 34–42 (2001). [DOI] [PubMed] [Google Scholar]
- Shima T., Znosko W. & Tsang M. The characterization of a zebrafish mid-hindbrain mutant, mid-hindbrain gone (mgo). Dev Dyn 238, 899–907, doi: 10.1002/dvdy.21916 (2009). [DOI] [PubMed] [Google Scholar]
- Lun K. & Brand M. A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 125, 3049–3062 (1998). [DOI] [PubMed] [Google Scholar]
- Zhu C. et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development 138, 4555–4564, doi: 10.1242/dev.066779 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng A., Tang H., Ong B. A., Farrell M. J. & Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci USA 94, 6267–6272 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Balaguer M. et al. Stable vascular connections and remodeling require full expression of VE-cadherin in zebrafish embryos. PLoS One 4, e5772, doi: 10.1371/journal.pone.0005772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering L. M. et al. An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc Natl Acad Sci USA 100, 9410–9415, doi: 10.1073/pnas.1633548100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amacher S. L., Draper B. W., Summers B. R. & Kimmel C. B. The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development 129, 3311–3323 (2002). [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M. & Mullins M. C. Dorsoventral patterning in the zebrafish: bone morphogenetic proteins and beyond. Results Probl Cell Differ 40, 72–95 (2002). [DOI] [PubMed] [Google Scholar]
- Schier A. F. & Talbot W. S. Molecular genetics of axis formation in zebrafish. Annu Rev Genet 39, 561–613, doi: 10.1146/annurev.genet.37.110801.143752 (2005). [DOI] [PubMed] [Google Scholar]
- Moro E. et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol Genet Genomics 288, 231–242, doi: 10.1007/s00438-013-0750-z (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collery R. F. & Link B. A. Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev Dyn 240, 712–722, doi: 10.1002/dvdy.22567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderström K.-O., Parvinen L.-M. & Parvinen M. Early detection of cell damage by supravital acridine orange staining. Experientia. 15; 33(2):265-6. (1977). [DOI] [PubMed]
- Gatta L. B., Vitali M., Verardi R., Arosio P. & Finazzi D. Inhibition of heme synthesis alters Amyloid Precursor Protein processing. J Neural Transm 116, 79–88, doi: 10.1007/s00702-008-0147-z (2009). [DOI] [PubMed] [Google Scholar]
- Ganz J. et al. Subdivisions of the adult zebrafish subpallium by molecular marker analysis. J Comp Neurol 520, 633–655, doi: 10.1002/cne.22757 (2012). [DOI] [PubMed] [Google Scholar]
- Tucker J. A., Mintzer K. A. & Mullins M. C. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell 14, 108–119, doi: 10.1016/j.devcel.2007.11.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyares R. L. et al. Long-chain Acyl-CoA synthetase 4A regulates Smad activity and dorsoventral patterning in the zebrafish embryo. Dev Cell 27, 635–647, doi: 10.1016/j.devcel.2013.11.011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renieri A. et al. Non-syndromic X-linked mental retardation: from a molecular to a clinical point of view. J Cell Physiol 204, 8–20, doi: 10.1002/jcp.20296 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao Y., Zhao X. & Wang Z. Drosophila long-chain acyl-CoA synthetase acts like a gap gene in embryonic segmentation. Dev Biol 353, 259–265, doi: 10.1016/j.ydbio.2011.02.030 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen D. & Wang Z. Analyses of mental dysfunction-related ACSl4 in Drosophila reveal its requirement for Dpp/BMP production and visual wiring in the brain. Hum Mol Genet 18, 3894–3905, doi: 10.1093/hmg/ddp332 (2009). [DOI] [PubMed] [Google Scholar]
- Liu Z., Huang Y., Zhang Y., Chen D. & Zhang Y. Q. Drosophila Acyl-CoA synthetase long-chain family member 4 regulates axonal transport of synaptic vesicles and is required for synaptic development and transmission. J Neurosci 31, 2052–2063, doi: 10.1523/JNEUROSCI.3278-10.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. dAcsl, the Drosophila ortholog of acyl-CoA synthetase long-chain family member 3 and 4, inhibits synapse growth by attenuating bone morphogenetic protein signaling via endocytic recycling. J Neurosci 34, 2785–2796, doi: 10.1523/JNEUROSCI.3547-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainter J. J., Boos A. & Kroll K. L. Neural induction takes a transcriptional twist. Dev Dyn 222, 315–327, doi: 10.1002/dvdy.1210 (2001). [DOI] [PubMed] [Google Scholar]
- Valdivia L. E. et al. Antagonism between Gdf6a and retinoic acid pathways controls timing of retinal neurogenesis and growth of the eye in zebrafish. Development 143, 1087–1098, doi: 10.1242/dev.130922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci H., Edlund R. K., Groves A. K. & Riley B. B. Tfap2a promotes specification and maturation of neurons in the inner ear through modulation of Bmp, Fgf and notch signaling. PLoS Genet 11, e1005037, doi: 10.1371/journal.pgen.1005037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrow N. L. & Fleming R. E. Bone morphogenetic proteins as regulators of iron metabolism. Annu Rev Nutr 34, 77–94, doi: 10.1146/annurev-nutr-071813-105646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombelli C., Aoun M. & Tiranti V. Defective lipid metabolism in neurodegeneration with brain iron accumulation (NBIA) syndromes: not only a matter of iron. J Inherit Metab Dis 38, 123–136, doi: 10.1007/s10545-014-9770-z (2015). [DOI] [PubMed] [Google Scholar]
- Lawson N. D. & Weinstein B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248, 307–318 (2002). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. & Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3, 59–69, doi: 10.1038/nprot.2007.514 (2008). [DOI] [PubMed] [Google Scholar]
- Nasevicius A. & Ekker S. C. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 26, 216–220, doi: 10.1038/79951 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.