Abstract
In addition to their antioxidant function, the eukaryotic peroxiredoxins (Prxs) facilitate peroxide-mediated signaling by undergoing controlled inactivation by peroxide-driven over-oxidation. In general, the bacterial enzyme lacks this controlled inactivation mechanism, making it more resistant to high H2O2 concentrations. During peroxide reduction, the active site alternates between reduced, fully folded (FF), and oxidized, locally unfolded (LU) conformations. Here we present novel insights into the divergence of bacterial and human Prxs in robustness and sensitivity to inactivation, respectively. Structural details provide new insights into sub-steps during the catalysis of peroxide reduction, enabling the transition from an FF to a LU conformation. Complementary to mutational and enzymatic results, these data unravel the essential role of the C-terminal tail of bacterial Prxs to act as a molecular switch, mediating the transition from an FF to a LU state. In addition, we propose that the C-terminal tail has influence on the propensity of the disulphide bond formation, indicating that as a consequence on the robustness and sensitivity to over-oxidation. Finally, a physical linkage between the catalytic site, the C-terminal tail and the oligomer interface is described.
Reactive oxygen species (ROS) are inevitable byproducts of normal aerobic metabolism, which at high levels can inflict damages on DNA, lipids and proteins. The intracellular concentration of peroxides is tightly maintained at very low levels by the oxidant-specific sensors that regulates the expression of oxidant-scavenger enzymes1,2. Peroxiredoxins (Prxs), a class of thiol-based peroxidases, present in all biological kingdoms from bacteria to human and consisting of six evolutionary subfamilies (Prx1, Prx5, Prx6, Tpx, PrxQ, and AhpE), are the dominant enzymes responsible for the reduction of over 90% of mitochondrial and cytoplasmic H2O23,4. These enzymes have recently grabbed attention not only due to their antioxidant function, but also for their additional significance in a broad range of cellular events including modulation of local intracellular H2O2 level required for signaling events5, and peroxide sensing for activation of transcription factors that control the expression of antioxidant enzymes6,7. Besides the importance of Prxs in keeping thioredoxins in a reduced state for maintaining cell viability under oxidative stress8, they act as peroxinitrite reductases9, chaperone holdases10, protein foldases11, and regulators of circadian rhythm12. Because of their fundamental cellular functions, Prxs are implicated in aging, cancer, cardiovascular diseases, diabetes and neurodegeneration, and are therefore appealing as therapeutic targets13,14.
The Prx subfamily enzyme Prx1, also typically called 2-Cys Prx, is among the most highly expressed, soluble proteins in the cell15, and is characterized by two conserved cysteine residues. One is represented by the peroxidatic cysteine (CP), which becomes selectively oxidized by H2O2 to the CP–SOH intermediate. This CP further reacts with the so-called resolving cysteine (CR). The CR is located at the C-terminus of the other subunit of the basic functional dimer, forming an intermolecular disulphide with the CP. Regeneration of the intermolecular disulphide bridge occurs via the Peroxiredoxin reductases (PrxR), enabling a continuous catalytic cycle16,17.
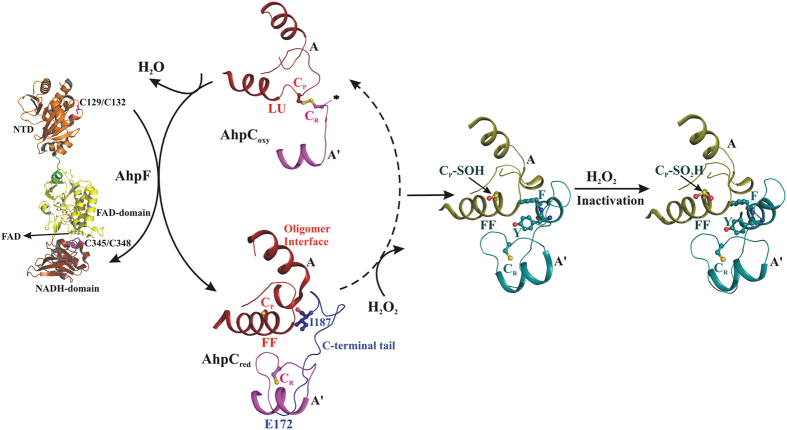
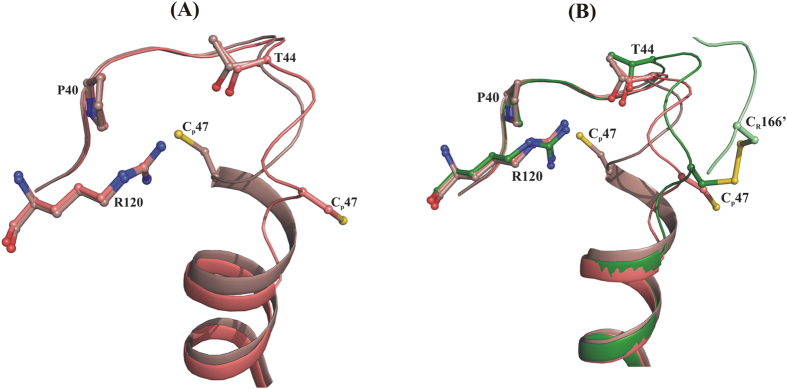
As shown in Fig. 1, the detailed structural analysis of bacterial Prx1, called Alkyl hydroperoxide reductase subunit C (AhpC), reveals two distinct active site conformations linked with their catalytic cycle. In the reduced state, the active site is competent for productive substrate binding and is in a so-called fully folded (FF) conformation, where the peroxidatic cysteine, CP47 (according to the Escherichia coli subunit AhpC numbering), is part of the α2-helix and is located at the bottom of the catalytic cavity, formed by the conserved residues, P40, T44 and R120. In parallel, the resolving cysteine (CR166), placed at the C-terminal tail of an adjacent subunit, is folded across the active site, placing CP and CR 14 Å away and in opposite orientation18 (Fig. 1). In the oxidized state, the CP and CR are disulphide bonded in the so-called locally unfolded (LU) conformation, where (i) CP of helix α2 is partially unwound, orienting CP towards CR, and (ii) the C-terminal tail unfolds from the active site region and becomes disordered19,20 (Fig. 1). Additionally, the redox-state dependent active site conformations are proposed to modulate the quaternary structure of Prxs, whereas the reduced FF active site favors the decamer formation and the oxidized LU active site weakens the decamer into a dimer19. Our recent studies on the Escherichia coli AhpC (EcAhpC) explored the different functional role of its C-terminal tail, as influenced by the redox-state of the enzyme. In the oxidized state, the C-terminal tail of EcAhpC, which includes amino acids 172 to 187, is essential for regeneration by its specific Alkyl hydroperoxide reductase subunit F (AhpF) (Fig. 1). At the same time, the C-terminal tail is essential for the formation of a stable doughnut-shaped decamer under reduced conditions21,22,23.
Figure 1. The catalytic cycle of 2-Cys Prxs.
The basic functional dimeric unit of 2-Cys Prxs and their active site region is shown. During peroxide reduction, the reduced CP assumes the FF conformation and reacts with H2O2 to form a CP-SOH intermediate. This local unfolding of the active site enables the formation of a disulphide bond with the CR located in the C-terminal tail of adjacent subunit. AhpC is then reduced by AhpF to its FF conformation for future catalytic cycles, with AhpF being oxidized in this process. AhpF is regenerated with the utilization of NADH molecules for further catalytic cycles. In comparison, human Prx is stabilized in the FF active site conformation even after the formation of the intermediate CP-SOH form, which eventually promotes over-oxidation.
Based on the elucidation of distinct redox-state-linked active-site conformations, it has been proposed that during the formation of the CP-SOH intermediate, the active site should move out of the FF conformation to facilitate the formation of the disulphide bond with CR16. Nevertheless, a detailed picture of the structural transition(s) between the FF and LU state during the catalysis is missing. In contrast, the structural studies on eukaryotic Prxs revealed that the FF active site conformation persists during and after oxidation of CP to CP-SOH (Fig. 1), which eventually promotes the over-oxidation of CP-SOH into CP-SO2H/CP-SO3H, and finally inactivates the enzyme24,25. The presence of two conserved sequence features in human and other eukaryotic Prxs, the so-called GGLG motif and the extended C-terminal helix containing the YF motif, are thought to be essential for regulating H2O2 mediated signal transduction5,18. However, the greater importance of the C-terminal helix for sensitivity to over-oxidation has been confirmed25,26. The C-terminal helix with its YF motif folds across the active site, thereby delaying the conformational change from FF to LU, and favoring over-oxidation15 (Fig. 1). It has been implicated that the disulphide formation or over-oxidation depends on the rate of FF to LU transitions. According to the proposed floodgate model, over-oxidation of Prxs facilitates the local accumulation of intracellular H2O2 required for signaling events18. Finally, the enzyme sulfiredoxin (Srx) catalyzes the repair of over-oxidized Prxs, to restore their peroxidase activity27.
In order to (i) gain deep insight into the delicate balance between the FF and LU active site during the catalytic cycle, (ii) to explore the underpinning role of the C-terminal tail in active site conformation, and (iii) to understand the features making the bacterial and human Prxs more robust and sensitive to inactivation, respectively, a combination of genetic engineering, enzyme kinetics and crystallography were used to establish the values of kinetic parameters and sensitivity for inactivation using the well-established E. coli AhpC and its mutants. By generating and determining the structure of a chimeric EcAhpC1-186-YFSKHN, which includes the extended C-terminal helix with the YF motif of human Prx2, we deduced for the first time the intermediate active-site conformations, lying between the FF and LU conformations. Furthermore, the studies reveal that the C-terminal tail acts as a molecular switch to mediate the structural transition between the FF and LU state. Finally, we propose the detailed conformational transition states that accompany the peroxide reduction and over-oxidation cycle, providing novel insight into the evolutionary divergence of the resistant and sensitive Prxs catalysts of bacteria and human, respectively.
Results
Sensitivity to oxidative inactivation of a novel chimeric EcAhpC1-186-YFSKHN protein
The bacterial Prx is less sensitive to inactivation by hyper-oxidation, whereas the human Prx is sensitive towards inactivation with its C-terminal helix (YFSKHN-helix), including the conserved residues YF, playing a vital role18. The question arises, whether the apparent sensitivity to over-oxidation exerted by the C-terminal helix of human Prx can be transformed into Prxs of prokaryotes. We used the mechanistically well understood E. coli as a prototype to construct a chimeric Prx, EcAhpC1-186-YFSKHN, composed of EcAhpC (residues 1–186) and the C-terminal YFSKHN-segment of human Prx 2, to be compared with wild-type (WT) EcAhpC (Fig. 2A). The E. coli TrxR-Trx system, the common reductase system to characterize Prxs, was used in the peroxidase assay, in which NADPH-oxidation by TrxR provides the electrons via Trx to EcAhpC for H2O2 reduction.
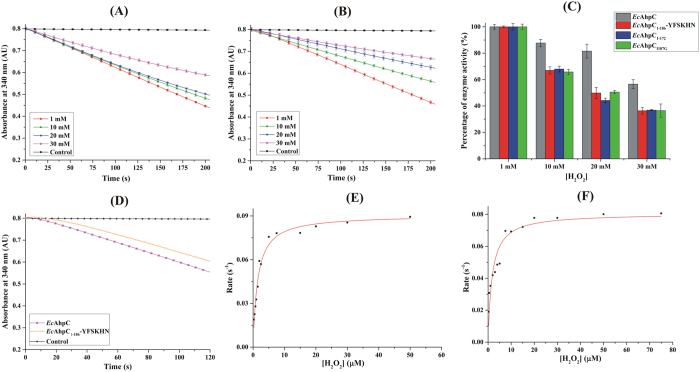
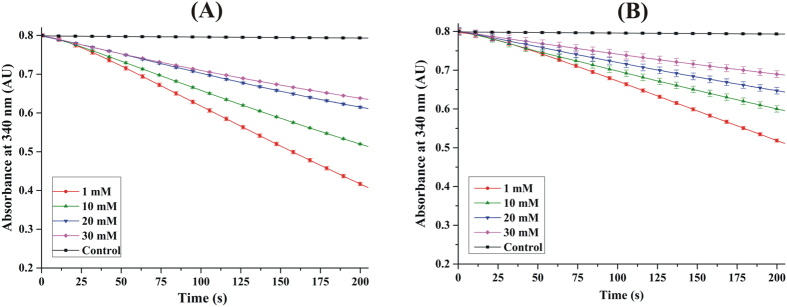
Figure 2. NADPH-dependent peroxidase activity of EcAhpC and EcAhpC1-186-YFSKHN.
Sensitivity of (A) EcAhpC and (B) EcAhpC1-186-YFSKHN to over-oxidation was determined by the measurement of NADPH oxidation at 1 mM (red), 10 mM (green), 20 mM (blue), and 30 mM H2O2 (magenta). The background oxidation without Prxs is shown as a control in black. The decrease in NADPH-oxidation with increasing concentrations of H2O2 is visible for both EcAhpC and EcAhpC1-186-YFSKHN. (C) Percentage of enzyme activity at each H2O2 concentration was calculated for WT EcAhpC and its mutants (Mean ± 1 SD) using the rate of NADPH oxidation at each concentration of H2O2. The rate of activity at the lowest concentration of H2O2 was taken as 100%. With increasing H2O2 concentrations, the percentage of enzyme activity decreased. (D) NAPDH oxidation of EcAhpC and EcAhpC1-186-YFSKHN measured at 30 μM H2O2 is shown as a representative for all other measurements done at various H2O2 concentrations to establish the enzyme kinetic parameters. Michaelis-Menten plot of (E) EcAhpC and (F) EcAhpC1-186-YFSKHN was done by fitting data of at least ten concentrations of H2O2.
As shown in Fig. 2A, WT EcAhpC reacts with 1 mM H2O2 and actively consumes NAPDH. Increasing the H2O2 concentration to 10 and 20 mM caused an activity drop of ~12% and ~18%, respectively. The inhibitory effect of H2O2 to EcAhpC increased significantly at a concentration of 30 mM, resulting in an enzyme activity reduction of ~44%. In comparison, an increase of 10 mM H2O2 decreased the chimeric EcAhpC1-186-YFSKHN enzyme activity by already ~33% (Fig. 2B,C), followed by a further drastic decrease of ~50% in the presence of 20 mM H2O2, and finally to a ~64% drop with 30 mM H2O2 (Fig. 2C). The data indicate that WT EcAhpC is resistant to inactivation up to 20 mM H2O2, as it shows little variation for peroxidase activity. In contrast, the peroxidase activity of chimeric EcAhpC1-186-YFSKHN is affected at H2O2 concentrations above 1 mM, revealing that the transformation of the human C-terminal YFSKHN-helix inside the bacterial EcAhpC increased its sensitivity to over-oxidation.
Effect of the human YFSKHN-segment on catalysis
In order to address the question, whether the YFSKHN-helix affects the peroxidase activity, the catalytic turnover number (kcat), and the Michaelis-constant (Km) for substrate-binding were determined. The peroxide-dependent NAPDH-oxidation of EcAhpC and EcAhpC1-186-YFSKHN were measured with varying concentrations of H2O2 and Fig. 2D shows the NAPDH-oxidation at 30 μM H2O2 as a representative of all other measurements done. WT EcAhpC and EcAhpC1-186-YFSKHN exhibited typical substrate saturation kinetics of the Michaelis-Menten type (Fig. 2E,F). The determined kinetic constants and catalytic efficiency are reported in Table 1. The Km- and kcat-value of EcAhpC were determined to be 1.44 μM and 0.09 s−1, respectively. In case of EcAhpC1-186-YFSKHN a higher Km of 1.77 μM and slightly lower kcat value of 0.082 s−1 was calculated, resulting in a lower catalytic efficiency of EcAhpC1-186-YFSKHN [kcat/Km (H2O2)], 4.8 × 104 M−1 s−1 when compared to the wild-type enzyme (6.2 × 104 M−1 s−1).
Table 1. Kinetic parameters of WT EcAhpC and its mutants.
| EcAhpC | kcat ± SD (s−1) | Km ± SD (μM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| WT EcAhpC | 0.090 ± 0.001 | 1.44 ± 0.1 | 6.2 × 104 |
| EcAhpC1–172 | 0.082 ± 0.004 | 9.45 ± 0.5 | 8.6 × 103 |
| EcAhpCI187G | 0.089 ± 0.003 | 8.26 ± 0.4 | 1.0 × 104 |
| EcAhpCS86A,T88A | 0.089 ± 0.004 | 9.74 ± 0.5 | 9.0 × 103 |
| EcAhpC1-186-YFSKHN | 0.082 ± 0.001 | 1.77 ± 0.1 | 4.6 × 104 |
Crystallographic structure of oxidized and decameric-shaped EcAhpC1-186-YFSKHN
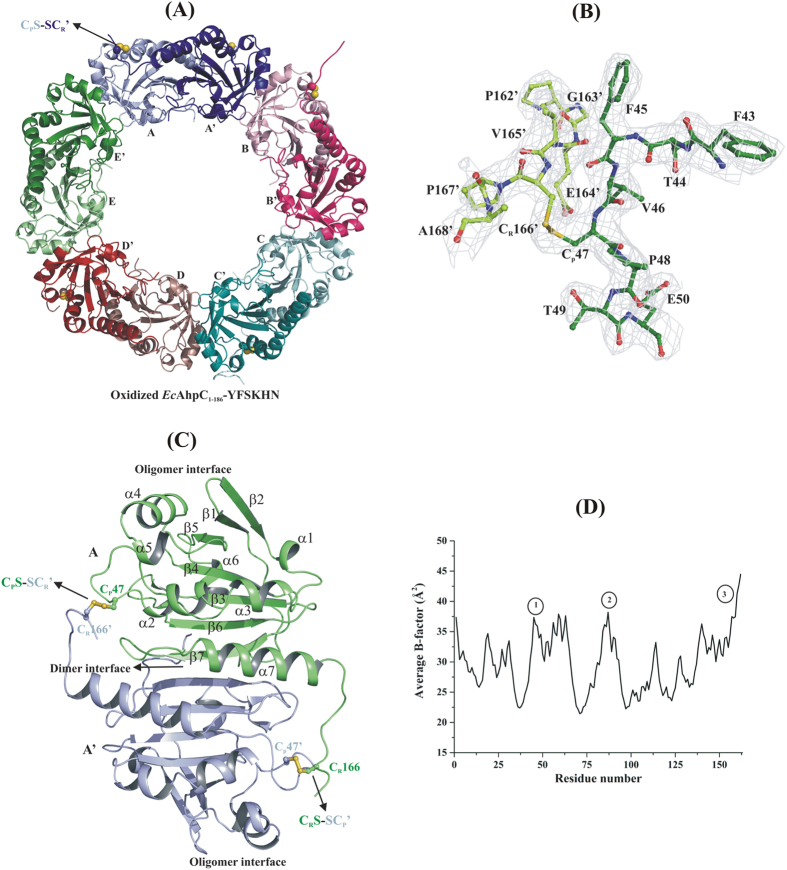
To understand the structural effect of the human YFSKHN-helix in general, in particular inside the chimeric EcAhpC1-186-YFSKHN, as well as to identify the individual amino acids essential for sensitivity to oxidative inactivation of Prxs, the crystal structure of oxidized EcAhpC1-186-YFSKHN has been solved at 2.7 Å resolution. The asymmetric unit contains one decamer composed of five catalytic dimers (α2)5 with each subunit containing one catalytic active peroxidatic and one resolving cysteine (Fig. 3A,B). The subunit consists of a central seven-stranded β-sheet, flanked at one side by four and at the other side by two α-helices (Fig. 3C). Each subunit has a dimer and oligomer interface. The dimer interface is mainly stabilized by salt bridge and hydrogen bond interactions, while the oligomer interface is mainly stabilized by hydrophobic interactions. Superposition of the recently resolved oxidized EcAhpC structure20 with the corresponding dimer of the oxidized EcAhpC1-186-YFSKHN one resulted in an r.m.s.d. value of 0.4 Å. The intermolecular disulphide bond observed between CP47 and CR166′ in the dimer interface (Fig. 3B,C) confirms the oxidized state of the EcAhpC1-186-YFSKHN structure. The peroxidatic cysteine (CP47), located in the first turn of helix α2, adopts an LU conformation in the oxidized structure (Fig. 3B,C and Supplementary Fig. S1A). In most of the disulphide bonded active sites of the decameric and oxidized EcAhpC1-186-YFSKHN, the C-terminal arm beyond the resolving cysteine (CR166) is disordered, indicating the high flexibility of the C-terminus.
Figure 3. Structural features of oxidized EcAhpC1-186-YFSKHN.
(A) Crystal structure of oxidized EcAhpC1-186-YFSKHN in decameric form (α2)5. Each subunit in the basic functional dimeric unit is shown in light and bright colors denoted as A and A’. The intermolecular disulphide bond between the peroxidatic (CP) and resolving (CR’) cysteine is shown in ball representation. (B) The 2FO-FC map contour at 1 σ level around the CP and CR’ in disulphide bond conformation. (C) Each subunit is composed of two interface regions, namely the dimer and oligomer interface. The secondary structural features are highlighted according to their position in the structure. (D) The average main chain B-factor of ten chains showed three highly dynamic regions in the structure.
However, the structure in chain (H) could be resolved until residue 177, showing amino acids 167 to 177 oriented away from the active site and stacked against the neighboring symmetry molecule. Similarly, the C-termini of chains A, G and J are also stabilized by the packing interactions with the symmetry related molecules (Supplementary Fig. S1B,C). The average main chain B-factors analysis revealed three dynamic regions of oxidized EcAhpC1-186-YFSKHN: (i) the CP47 containing α2-helix, (ii) helix α5 that contains residues S86 and T88, and (iii) the C-terminal tail region (Fig. 3D). The sulfate ions observed in the interface region of the structure, emphasis their role in stabilizing the oxidized AhpC in a decameric form in solution23.
The active site conformation in reduced EcAhpC1-186-YFSKHN
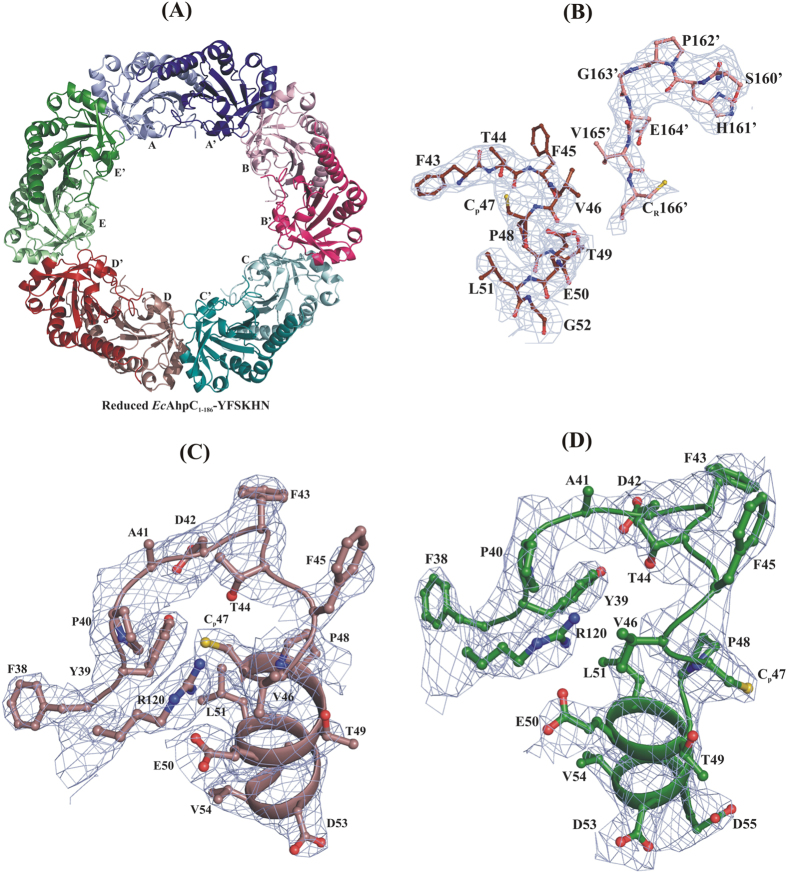
To shine a light on the redox modulated structural alterations in the active site, the crystallographic structure of reduced EcAhpC1-186-YFSKHN was determined to 3.1 Å resolution (Fig. 4A). Except for the C-terminus, clear electron density was observed for all the residues and the resolving cysteine (chains C, E, F, H and J), which is located about 10 Å away from the peroxidatic one (Fig. 4B). No disulphide bond between CP47 and CR166′ was observed in any of the active site interfaces of the decameric EcAhpC1-186-YFSKHN, confirming that the structure fairly represents the reduced form of EcAhpC1-186-YFSKHN. In the reduced state, the typical FF active site is arranged in a way that the CP47 positions in the first turn of helix α2 and CR166′, containing the C-terminal arm, is folded across the dimer interface to come in proximity to the first turn of helix α2 of another subunit. This conformational alteration brings the CP and CR’ to face opposite directions and to become separated by about 10 Å.
Figure 4. Active site conformation in reduced EcAhpC1-186-YFSKHN.
(A) Decameric structure of the reduced form of EcAhpC1-186-YFSKHN in cartoon representation. (B) A representative 2FO-FC electron density map around the CP and the CR region confirms the reduced state. (C) Ribbon representation of the active site, where CP47 adopts helical conformation in the reduced EcAhpC1-186-YFSKHN structure. The 2FO-FC electron density map is shown around the region. (D) Ribbon representation with 2FO-FC map is shown for the active site region observed once in the EcAhpC1-186-YFSKHN reduced structure, wherein CP adopts a loop conformation and is exposed outwards.
Interestingly, two different active site conformations were observed in the reduced EcAhpC1-186-YFSKHN structure (Supplementary Fig. S2A), where the residues in the active site region are clearly defined in the electron density map except for the side chains of F45 and V46 (Fig. 4C,D). The first conformation (chains B-J) reveals that the peroxidatic CP47 adopts helical conformation, bringing the CP47 in a narrow solvent accessible pocket, which is surrounded by the highly conserved active site residues P40, T44 and R120 (Fig. 5A). In the second active site conformation (chain A), the first turn of α2-helix is partially unfolded to place CP47 in the loop conformation (Fig. 5A). However, no significant influence of crystal packing on the conformations of CP47 was observed in the reduced EcAhpC1-186-YFSKHN structure (Supplementary Fig. S2B,C). A comparison of these two reduced active sites with the oxidized LU state of EcAhpC1-186-YFSKHN reflects a significant difference in the loop region (T44-P48) (Fig. 5B), where the FF active site residues of the first active site of reduced EcAhpC1-186-YFSKHN are rotated, to expose CP47 towards the CR166′, and finally forming a disulphide bond in the oxidized structure. While in the second active site of reduced EcAhpC1-186-YFSKHN, the unfolded CP47 adopts a lower magnitude of rotation, thereby preventing its full exposure towards CR166′ to form a disulphide (Fig. 5B).
Figure 5. Comparison of FF and LU states from EcAhpC1-186-YFSKHN crystal structures.
(A) Superimposition of two different active sites, the FFlike- (brown) and LUlike state (salmon) which were observed in the reduced EcAhpC1-186-YFSKHN structure. (B) Comparison of FFlike- (brown) and LUlike state (salmon) in the reduced EcAhpC1-186-YFSKHN structure and the LU state of the oxidized EcAhpC1-186-YFSKHN (green) structure.
FF to LU transition state like active site conformation
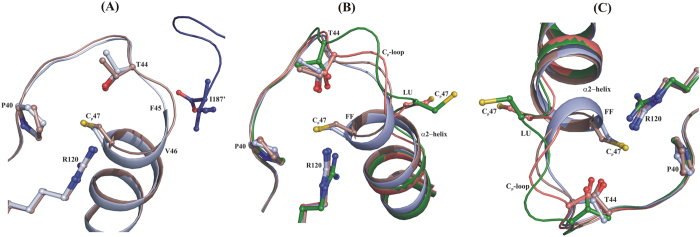
To gain insight into (i) the structural transition pathway between the FF and LU state, and (ii) the role of the C-terminal tail on the active site conformation, the two presented distinct active sites of reduced EcAhpC1-186-YFSKHN were compared with a typical reduced FF and oxidized LU conformation, represented by the crystallographic structure of the Salmonella typhimurium AhpC28 (PDB ID: 4MA9) and EcAhpC1-186-YFSKHN (see above), respectively. For clarity, we termed the first and second active sites of the reduced EcAhpC1-186-YFSKHN as FFlike and LUlike, respectively, to denote the folded and unfolded CP47 in the active site environments (Fig. 6A).
Figure 6. FF to LU transition state like conformation.
(A) Comparison of the FFlike active site of EcAhpC1-186-YFSKHN (brown) with the typical FF conformation in the reduced StAhpC (light blue), where the C-terminal tail is folded across the active site (blue), which is disordered in reduced EcAhpC1-186-YFSKHN. (B) The oxidized- (green) and reduced (salmon) active sites observed in the EcAhpC1-186-YFSKHN structures are compared with the reduced StAhpC (light blue) FF active site. The intermediate FFlike (brown) and LUlike (salmon) conformations, represent the transition state from the FF to the LU state. (C) Another view to close look at the comparison of FF, LU, FFlike and LUlike active sites.
Firstly, the reduced EcAhpC1-186-YFSKHN and the S. typhimurium AhpC structure (PDB ID: 4MA9) share an overall similarity indicated by an r.m.s.d. of 0.5 Å for the superposition of the catalytic dimers. When zoomed into the catalytic relevant α2-helix, the FF and FFlike active sites differ significantly (Fig. 6A). While the residues F45 and V46 are placed in the starting position of helix α2 inside the FF state, the α2-helix becomes unwound in the FFlike conformation and the main chain Cα atoms F45 and V46 shifted by about 0.9 Å. Similarly, in the FFlike conformation the main chain Cα atoms of CP47 and P48 moved about 0.6 Å and 0.5 Å, respectively, compared to their corresponding positions in the FF state (Fig. 6A). Furthermore, no obvious difference in the main chain conformations of the loop amino acids P40, T44 and R120 were observed between the two conformations.
The comparison of the FF (PDB ID: 4MA9) and LUlike active site (see above) shows, significant structural variation in the first turn of helix α2 and the loop region (Fig. 6B,C). Due to the partial unfolding of the α2-helix, the main chain Cα atoms of V44, F45, V46, C47 and P48 of the FF active site moved by 0.4, 1.8, 1.9, 5.1 and 1.1 Å, respectively, to achieve the LUlike state positions. As shown in Fig. 6B,C, the same residues moved during the transition from the FF to the LU state but with a higher magnitude of 1.4, 3.8, 3.8, 5.9 and 1.7 Å, respectively. Because of the magnitude of displacement, the data indicate that the FFlike and LUlike state, adopt intermediate conformations, lying between the catalytic FF and LU conformation. In addition, significant differences in the backbone torsion angles were determined between the FF, LUlike and LU active sites (Supplementary Table S1). Taken together, it can be proposed that the FFlike and LUlike active sites mimic the initial and the intermediate phase of the structural deformation that occurs during the redox-modulated FF to LU conformational switch.
Oligomer interface region
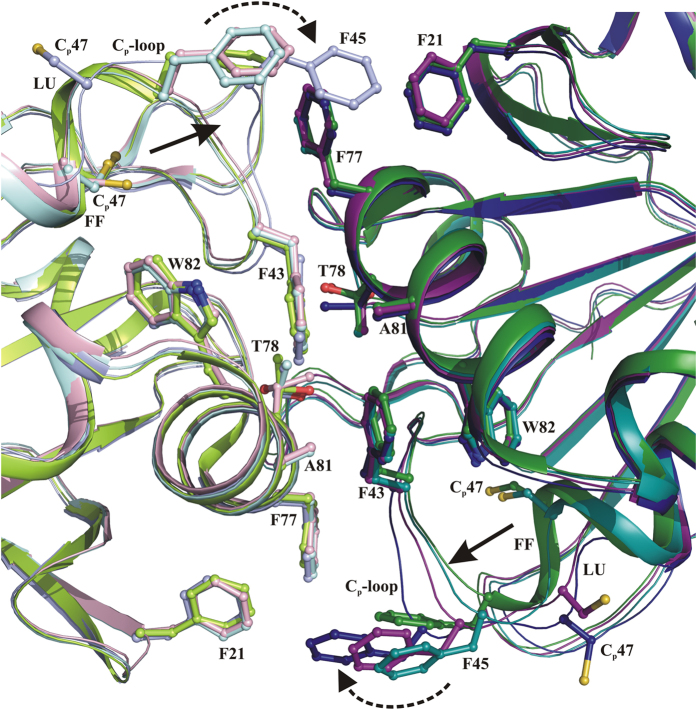
The oligomeric behavior of Prxs is proposed to be redox modulated29. Under reduced conditions, the FF active site buttresses the oligomer interface, and favors decameric ring formation. Whereas under oxidized conditions, the LU active site mediates the restructuring of the oligomer interface region, resulting into lower order oligomers19. A structural view to the oligomeric interface in Fig. 7 highlights differences of the distinct active site loop conformations observed between FF, FFlike, LUlike and LU, where the CP-containing loop residues of the LU conformation are pulled away from the oligomer interface. This facilitates the destabilization of the oligomer interface through structural rearrangements of F43 and F45, which participate in the interface region19,29. The rest of the interface region residues remain unchanged in the crystal structures.
Figure 7. The oligomer interface region.
The decamer building interface region between two subunits are shown. The significant structural difference in the interface region is observed only in the residues that form the CP-loop. Compared with “FF” (green) CP-loop structure, the “FFlike” (blue), “LUlike” (magenta) and “LU” (cyan) is shifted away (solid arrow) from the interface region and this facilitates the destabilization of the oligomeric interface through F43 and F45 (dotted arrow). The complementary interface region formed by the neighboring subunit is shown with light colors shade.
Effect of the C-terminal tail on substrate-binding and catalytic efficiency
By genetically engineering and enzymatically characterizing the EcAhpC mutants AhpC1–172, AhpCI187G and AhpCS86A,T88A, the questions were addressed, whether the folding of the C-terminal tail across the active site region as well as the dynamic region of helix α5 with residues S86 and T88 (Fig. 8A), influences the enzyme kinetics.
Figure 8. NADPH-dependent peroxidase activity of EcAhpC and C-terminal variants.
(A) Active site of a typical FF active site of reduced StAhpC, where the C-terminal tail folds into the active site and forms weak interactions with the active site residues. The residues S86 and T88 hold the C-terminal tail through hydrogen bond interactions with I187′. (B) NADPH oxidation of wild type EcAhpC and its C-terminal variants at 30 μM of H2O2 is shown as a representative of other measurements done with different concentrations of H2O2. The background oxidation of EcTrxR and EcTrx without EcAhpC is taken as a control. Michaelis-Menten plots for (C) EcAhpCI187G, (D) EcAhpCS86A,T88A, (E) EcAhpC1–172 were done with various concentrations of H2O2.
Like WT EcAhpC, all EcAhpC mutants exhibited Michaelis-Menten substrate saturation kinetics with a kcat of 0.089 s−1 for EcAhpCI187G and EcAhpCS86A,T88A, a value similar to EcAhpC (Table 1, Fig. 8B–E). In comparison, the kcat of 0.082 s−1 of the C-terminal truncated EcAhpC1–172 was slightly lower but similar to the one determined for the chimeric EcAhpC1-186-YFSKHN. In contrast to EcAhpC1-186-YFSKHN and WT EcAhpC, all three EcAhpC mutants showed a significant decrease in H2O2-binding, with Km values of 8.26 μM, 9.74 μM, and 9.45 μM for EcAhpCI187G, EcAhpCS86A,T88A and the C-terminal truncated EcAhpC1–172, respectively. The increased Km values suggest that the active site binding pocket is impaired in all EcAhpC mutants. The respective values of catalytic efficiency of the enzyme [kcat/Km (H2O2)] determined for these mutants are 1.0 × 104, 9.0 × 103, and 8.6 × 103 for EcAhpCI187G, EcAhpCS86A,T88A and EcAhpC1–172, respectively (Table 1).
The role of the C-terminus of bacterial AhpC in H2O2 robustness
Revealing that the transformation of the human C-terminal YFSKHN-helix inside the bacterial EcAhpC increased its sensitivity to over-oxidation, leads now to the question of whether the C-terminus of bacterial 2-Cys Prxs keeps the enzyme more robust and enabling the prokaryotes to survive under higher peroxide concentrations. To test this, the ability of the two C-terminal variants, EcAhpC1–172 and EcAhpCI187G, to inactivation was studied. As shown in Fig. 9A,B, EcAhpC1–172 and EcAhpCI187G showed NADPH consumption at 1 mM H2O2 concentration. Increase of H2O2 (10 mM) dropped the peroxidase activity of EcAhpC1–172 and EcAhpCI187G by ~32% and ~34%, respectively (Figs 2C and 9A,B). The inhibitory effect of H2O2 in EcAhpC1–172 and EcAhpCI187G increased significantly at a concentration of 30 mM, resulting in an enzyme reduction of ~64% and ~62%, respectively. These values are comparable to the once of the chimeric EcAhpC1-186-YFSKHN, which were determined to be ~33% at a concentration of 10 mM H2O2 followed by a further drastic decrease of 50% in the presence of 20 mM H2O2, and finally to a ~64% drop with 30 mM H2O2 (Fig. 2B). The results demonstrate that both EcAhpC1–172 and EcAhpCI187G are sensitized to inactivation in a magnitude similar to that of EcAhpC1-186- YFSKHN, and finally, that the C-terminus, in particular residue I187, is essential for the robustness at high H2O2 concentrations (Supplementary Fig. S3). The sensitivity of EcAhpC1–172 and EcAhpCI187G is surprising, since the destabilized active site, indicated by the increased Km value, is supposed to expose the CP for disulphide formation with CR’30. We propose, that the EcAhpC1–172, and EcAhpCI187G mutants might affect the propensity of the disulphide formation (Supplementary Fig. S3) which would indicate, that the C-terminus plays a role in aiding the disulphide bond formation.
Figure 9. Sensitivity of EcAhpCI187G and EcAhpC1–172 to inactivation by H2O2.
Sensitivity of (A) EcAhpC1–172 and (B) EcAhpCI187G to over-oxidation was determined by the measurement of NADPH oxidation at 1 mM (red), 10 mM (green), 20 mM (blue), and 30 mM H2O2 (magenta). The background oxidation without Prxs is shown as a control in black. The decrease in NADPH-oxidation with increasing concentrations of H2O2 is visible for both EcAhpC1–172 and EcAhpCI187G.
Discussion
Evolutionary adaptation to differing environmental conditions was fundamental to the formation of the different kingdoms of life. Eukaryotic 2-Cys Prxs show a high sensitivity to peroxide inactivation by over-oxidation of the peroxidatic cysteine, Cp, to sulphinic acid (Cp-SO2H). Due to this adaptation, the enzyme keeps resting levels of H2O2 low, while permitting higher levels during signal transduction5,18. The eukaryotic GGLG motif and its extended C-terminal helix, including the YF motif (residues YFSKHN), are responsible for regulating H2O2 mediated signal transduction18. The chimeric EcAhpC1-186-YFSKHN described here demonstrates, that transformation of the human C-terminal YFSKHN-helix into the related bacterial AhpC confers sensitivity to over-oxidation on the chimeric enzyme. As for human Prx2 (Fig. 1), the engineered YFSKHN-helix inside the chimeric EcAhpC1-186-YFSKHN falls across the active site of the chimeric enzyme, thereby delaying the conformational change from fully folded (FF) to locally unfolded (LU), thereby favoring over-oxidation.
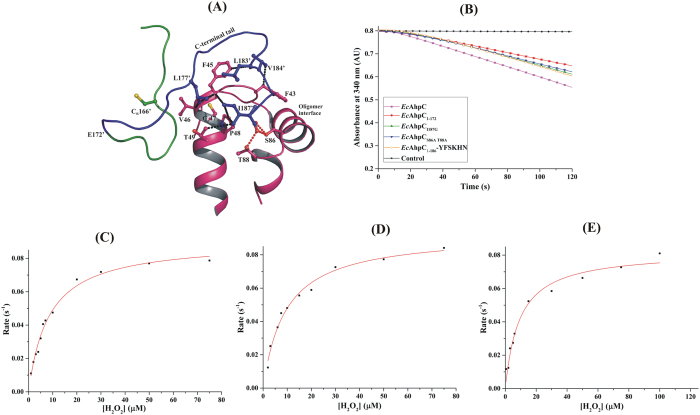
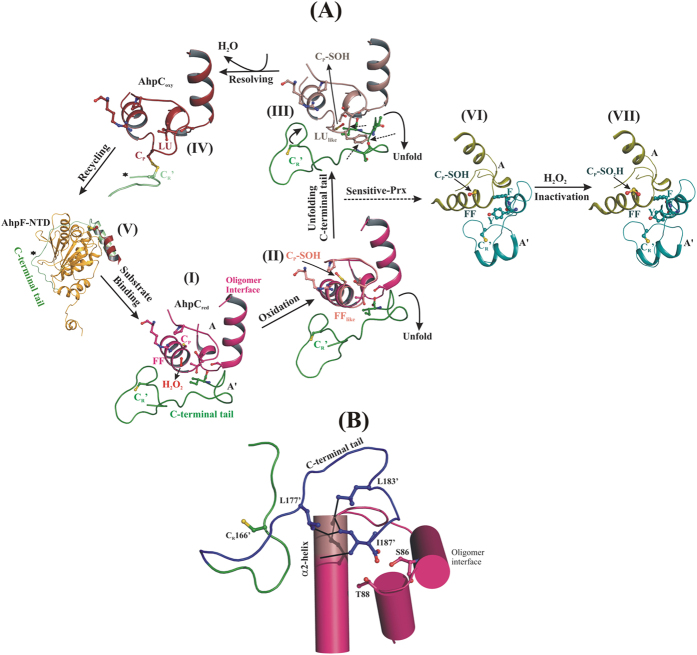
The crystallographic structures of the oxidized and reduced chimeric EcAhpC1-186-YFSKHN also provide new insights into the sub-steps during catalysis of peroxide reduction, characterized by the initial formation of a CP-SOH intermediate that is generally unstable and prone to further oxidation. This would indicate that the protective mechanism is likely to oppose over-oxidation. The local unfolding (LU) of the active site is such a protection mechanism, but the precise nature of the structural alterations that accompany LU was not known, due to the lack of structural details of this important intermediate CP-SOH state in the bacterial 2-Cys Prx family member AhpC. Although the existence of a CP-SO2H state in the FF active site was demonstrated in human Prx24, but did not provide the needed insights for FF to LU transition. As demonstrated by the reduced EcAhpC1-186-YFSKHN structure (Fig. 10A-I, -II), a shift of the peroxidatic cysteine, CP47, was observed combined with a first turn of the α2-helix, which is now called FFlike active site. This novel stage is proposed to mimic the early stage of conformational changes around the active site environment due to the oxidation of CP (Fig. 6A and Supplementary Fig. S4).
Figure 10. The proposed transition state(s) during the catalytic cycle of Prxs.
(A) (I) The reduced CP adopts FF conformation and is essential to activate and stabilize the CP-thiolate for peroxidation. (II) The oxidative modification of CP to CP-SOH leads to localized structural alterations around the conserved active site pocket as seen from the “FFlike” conformation of the reduced EcAhpC1-186-YFSKHN. (III) The “LUlike” conformation in the reduce EcAhpC1-186-YFSKHN structure, reveals a severe clash between the active site residues with the C-terminal tail. The entire C-terminal arm (aa 166–187) is needed to undergo alterations and to enable the disulphide formation between CR’ and CP (solid arrow). (IV) The disulphide bonded active site is stabilized in the LU conformation, wherein the C-terminal tail is disordered (denoted by asterisk). (V) The C-terminal tail of AhpC binds to its reducing partner, the N-terminal domain (NTD) of AhpF, to regenerate for another catalytic cycle. (VI) In human Prxs, the extra C-terminal YFSKHN-helix is folded across the active site, thereby delaying the FF to LU transition during the intermediate CP-SOH state. In such case, the CP-SOH form can further react with peroxide and leads to enzyme inactivation (VII). (B) The cartoon representation shows that the CP and the first turn of the α2-helix is stabilized by the weak interactions with the I187 and C-terminal tail, which is hold by residues S86 and T88. Disruption of weak interactions of the C-terminal tail, like between I187 and the α2-helix or between I187 with S86A and T88A, leads to unfolding of the C-terminal tail from the active site region.
The critical step, which decides about inactivation or continues resolving and recycling of AhpC, is combined with the newly described LUlike conformation in the reduced EcAhpC1-186-YFSKHN structure (Figs 6B and 10A-II). This structurally trapped conformation reveals a severe clash between the active site residues CP47 and F45 with the C-terminal tail, causing the C-terminal tail residues L183′, V184′ and I187′, to move away in order to accommodate the preceding structural alteration in the FF active site due to oxidative modification of the peroxidatic cysteine, CP. This dynamic behavior of C-terminal tail residues is supported by the FFlike structure (Fig. 6A), demonstrating that alterations in the CP and the first turn of the α2-helix caused by oxidation can disrupt interactions of the C-terminal tail, like between L183′, I187′ and the CP-loop residues or between I187 with S86A and T88A, the C-terminal tail unfolds partially and moves away from the active site (Fig. 10B). Therefore, alterations of the C-terminal tail confer instability on the FF active site, which in turn increases the rapid shift from the FF to the LU state. These structural rearrangements explain the significant increase of the binding constants observed for the EcAhpCI187G and EcAhpCS86A,T88A mutants (Table 1). Therefore, we propose that the side chain interactions of I187 with the CP-loop as well as with amino acids S86A and T88A affect binding of peroxide in the catalytic site.
The comparison of the presented LUlike and oxidized structure of EcAhpC1-186-YFSKHN also provides novel insights into the resolution of the CP-SOH form. As shown in Fig. 10A-III, the transfer from a LUlike to a LU state requires the entire C-terminal arm, including residues 166 to 187, to undergo a structural rearrangement that brings the resolving cysteine, CR166′, into proximity to finally resolve the CP-SOH form. Unfolding of the C-terminal tail might aid disulphide formation by favorably orienting both CP and CR’. During the LU conformation, the CP and CR’ disulphide bonded active site becomes stabilized (Fig. 10A-IV), which weakens the oligomer building interface19. In this oxidized state, the C-terminal tail is highly disordered and not visible in the EcAhpC1-186-YFSKHN structure (Fig. 3C), but essential for binding with its reducing partner, the N-terminal domain (NTD) of AhpF (Fig. 10A-V). In this essential regenerative step of the catalytic cycle, the C-terminus of EcAhpC wraps around the NTD and slows the dissociation rate for an efficient electron transfer process22. Once the disulphide bond is reduced by the NTD of AhpF, the active site is present in equilibrium between the FF and LU state. In this way, the folding of the C-terminal tail of AhpC back into the active site region and the formation of a stable decamer interface maintain the conformational integrity of the FF active site (Fig. 10A-I). Such a FF active-site pocket is essential to activate and stabilize the CP-thiolate for peroxidation16.
Establishing the delicate balance between FF and LU conformation and identifying the critical C-terminal tail (amino acids 172 to 187 according to the EcAhpC) and residues (S86, T88) are important as they provide insights not only into the catalytic cycle of Prxs, but also the divergence of the bacterial and human 2-Cys Prxs in robustness and sensitivity to inactivation, respectively. In the sensitive human and more generally eukaryotic Prxs, the two important segments identified here, namely the C-terminal tail and residues S86 and T88, of the less sensitive bacterial Prxs are structurally replaced by the conserved eukaryotic GGLG motif and the YF motif inside the extra C-terminal helix (YFSKHN -helix). Although both the eukaryotic as well as the bacterial enzyme reveal similar catalytic efficiency, the structurally distinct motifs/segments make the eukaryotic enzyme susceptible to over-oxidation31. As revealed in Fig. 10A-VI, the eukaryotic C-terminal YFSKHN -helix is complemented by the GGLG motif, forming a stable structural segment across the active site to securely pack the peroxidatic cysteine, CP, in the FF conformation, even after the oxidative modification of the CP to the sulfenic acid form24,25. Based on the proposed model of bacterial AhpC, the C-terminal tail is resistant to unfolding from the active site due to the oxidation induced structural perturbation, thereby delaying the unfolding step needed to switch from the FF active site to the LU conformation. Moreover, the resolution of CP-SOH depends on the availability of the resolving cysteine, CR’, which is also mediated by the unfolding of the C-terminal tail. This perspective is reflected in the rate of disulphide formation during catalysis, which in case of the bacterial AhpC is significantly higher (75 s−1)32, when compared to the low rate of 1.7 s−1 determined for human Prx233. Taken together, natural selection for the stable C-terminal tail over a stable FF active site reveals the essential role of the C-terminal tail to act as a molecular switch that mediates the structural transition between the FF and LU state during the catalytic cycle.
The oxidized EcAhpC1-186-YFSKHN structure in Fig. 3C,D reveals that the active site loop-helix-motif and the C-terminal tail region adopt structural alteration, bringing CP and CR’ to the disulphide bonded LU state. This structural feature is important, since the redox-linked active site conformations are proposed to regulate oligomerization. This regulative mechanism proposes that the disulphide bonded LU state destabilizes the decamer while the CP and CR’ reduced FF conformation favors decamer formation30. With the studies presented here, we delineate the linkage between the active site conformation and the C-terminal tail residues. Our earlier studies demonstrated that the EcAhpC1–172, EcAhpC1–182, and EcAhpCI187G mutants prevented decamer formation in solution21, leading to the conclusion that the destabilized active site, caused by these mutants, might confer instability to the oligomer building interface (Fig. 7). Taken together, we propose a physical linkage can be established between the three major regions, namely the CP containing active site, the C-terminal tail and the oligomer interface. In the reduced state, this physical linkage enables the folded C-terminal tail and stable oligomer interface to facilitate formation of the FF active site and vice versa. However, in the oxidized state, these three structural regions are destabilized19,30.
In summary, a combination of complementary approaches of genetic engineering, protein chemistry, enzymatic assays, and structural biology has provided new insights into the unique aspect of divergence between the bacterial and human 2-Cys Prxs in robustness and sensitivity to inactivation, respectively. Structural details of the oxidized and reduced chimeric EcAhpC1-186-YFSKHN provide novel insights into sub-steps during the catalysis of peroxide reduction, enabling the transition from a fully folded to a locally unfolded conformation. Together with mutational and enzymatic studies these data unravel the fundamental role of the C-terminal tail as a molecular switch that mediates the structural transition between the FF and LU state during the catalytic cycle. Finally, a physical linkage between the CP-containing active site, the C-terminal tail and the oligomer interface was established.
Materials and Methods
Cloning and overexpression
The chimeric protein EcAhpC1-186-YFSKHN, composed of the N-terminal 186 amino acids of E. coli AhpC (187 residues) and the C-terminal residues 193YFSKHN197 of human Prx2, was designed based on a protein sequence alignment. EcAhpC1-186-YFSKHN was amplified by polymerase chain reaction (PCR) using forward primer 5′-CAT GCC ATG GCA ATG TCC TTG ATT AAC ACC AAA ATT AAA CCT-3′ with an NcoI restriction site (bold) and reverse primer 5′-GCG AGC TCT TAG TTG TGC TTA CTA AAG TAT TTA CCA ACC AGG TCC AG-3′ with a SacI restriction site (bold), respectively, and using the pET9-d1-His6 plasmid34, which contains the gene encoding EcAhpC20. The double mutant, S86 and T88 to alanine (EcAhpCS86A,T88A) was amplified using site-directed mutagenesis with Hi-Fi KAPA DNA polymerase (KAPA Biosystems, USA) and the forward primer 5′-AGC AGC GCT GAA GCT ATC GCT AAA ATC AAA TAT GCG-3′ as well as the reverse primer 5′-GAT AGC TTC AGC GCT GCT GTG CCA TGC TTT-3′. The coding sequences of EcAhpC1-186-YFSKHN and EcAhpCS86A,T88A were verified by DNA sequencing.
Wild type E. coli thioredoxin (Trx) and thioredoxin reductase (TrxR) was amplified by PCR using forward primers 5′-CGT GCC ATG GCA ATG AGC GAT AAA ATT ATT CAC CTG-3′ and 5′-TAT GCC ATG GGC ACG ACC AAA CAC AGT-3′ with an NcoI restriction site (bold) and reverse primers 5′-TAG AGC TCG TTA CGC CAG GTT AGC GTC GAG GAA-3′ and 5′-GCG AGC TCG TTA TTT TGC GTC AGC TAA AC-3′ with a SacI restriction site (bold), respectively. E. coli genomic DNA was used as a template.
Liquid cultures were shaken in Luria Broth medium (LB-medium) containing kanamycin (30 μg/ml) at 310 K until an optical density OD600 of 0.6–0.7 was reached and isopropyl β-D-1-thiogalactopyranoside (IPTG; final concentration of 1 mM), was added for production of recombinant proteins. The incubation took 3 h at 310 K, except for EcAhpCS86A,T88A, which was grown overnight at 291 K after induction. Recombinant E. coli AhpC, the C-terminal truncated EcAhpC1–172 and mutant EcAhpCI187G were produced by the method described earlier21.
Purification of recombinant proteins
BL21 (DE3) E. coli cells containing recombinant EcAhpC, EcAhpC1–172, EcAhpCI187G, EcAhpCS86A,T88A and EcAhpC1-186-YFSKHN were purified according to Dip et al.21. Recombinant E. coli Trx (EcTrx) and -TrxR were purified with a slightly modified protocol, in which the purified Ni2+-NTA fractions were directly loaded on the Superdex 75 HR 10/30- (GE Healthcare) and Superdex 200 column (GE Healthcare), respectively. The purity and homogeneity of the protein samples were analyzed by a 17% SDS-gel35. The gels were stained with Coomassie Brilliant Blue G250. The gels of the generated and purified EcAhpC1-186-YFSKHN, EcAhpCS86A,T88A, EcTrx, and EcTrxR are shown in Supplementary Fig. S5. Protein concentrations were determined using a BioSpec-nano Spectrophotometer (Shimadzu, USA).
Enzymatic characterization using a peroxidase assay
Peroxide-dependent activity of the various forms of purified recombinant EcAhpC proteins was measured by coupling its activity with NADPH-oxidation (ɛ280 = 6220 M−1 s−1) catalyzed by EcTrxR and EcTrx. The peroxidase activity was carried out at 25 °C by monitoring the decrease in NADPH-absorbance at 340 nm for 120 sec using a stopped-flow spectrophotometer SX20 (Applied Photophysics, UK). The reaction mixture containing 100 μM NADPH, 50 mM HEPES buffer pH 7.0, 100 mM of ammonium sulfate, 0.5 mM EDTA, 0.25 μM of EcTrxR, 4 μM of EcTrx, and 4 μM of EcAhpC were mixed with varying concentrations of hydrogen peroxide (250 nM–100 μM) to initiate NADPH-oxidation. The rate of NADPH-oxidation was calculated by a least square fit to the linear portion of the curve. NADPH consumption measured in the absence of peroxiredoxin was taken as a control. The background rate was subtracted from the experimental rate to determine the activity due to EcAhpC. All rates reported here are the average of three independent experiments.
Inactivation assay
Peroxide-dependent overoxidation of EcAhpC, -AhpC1–172, -AhpC1187G and the chimeric EcAhpC1-186-YFSKHN were measured similar to the above mentioned condition, using hydrogen peroxide concentrations of 1–30 mM. NADPH-oxidation was monitored at 340 nm at 25 °C for 210 sec. Background NADPH-oxidation observed for EcTrx and EcTrxR in the absence of Prx, which is significant at higher H2O2 concentrations, was subtracted to estimate the activity due to Prx. The rate of reaction for each hydrogen peroxide concentration was calculated by fitting the linear portion of the curve. The rate observed for the lowest concentration of hydrogen peroxide is taken as 100% activity, to calculate the remaining enzyme activity in percentage at each concentration of hydrogen peroxide.
Peroxide reduction assay using SDS-PAGE
This assay is based on the observations that the reduced and oxidized 2-Cys Prxs run as a monomer and dimer, respectively, in a non-reducing SDS-PAGE33. Prior to each experiment, WT EcAhpC, EcAhpC1–172 and EcAhpC1187G were reduced with 20 mM dithiothreitol (DTT) in 50 mM phosphate buffer, pH 7.4 containing 1 mM diethylenetriaminepentaacetic acid for 1 h. Reduced proteins were separated from excess of DTT using a PD-10 desalting column (GE Healthcare). The different forms of EcAhpC (30 μM) were incubated with varying concentrations of H2O2 for 5 min. The reaction was stopped by adding 50 mMN‐ethyl maleimide in a sample buffer (4% SDS, 10% glycerol and 62.5 mM Tris–HCl, pH 6.8) and analyzed by a non-reducing 17% SDS-gel.
Crystallization of EcAhpC1-186-YFSKHN
Chimeric EcAhpC1-186-YFSKHN was concentrated to 8 mg/ml in buffer containing 50 mM Tris-HCl pH 7.5, 200 mM NaCl, using a Millipore spin concentrator with a molecular-mass cutoff of 10 kDa. An initial crystallization attempt was carried out using the recent protocol of EcAhpC (1.8 M ammonium sulfate, 100 mM MES (2- (N-morpholino) ethanesulfonic acid), pH 6.5 and 5% Dioxane)20, and the hanging-drop vapour diffusion method in 24-well VDX plates with sealant at 291 K. Diffraction quality crystals were obtained in the optimized condition of 1.6 M ammonium sulfate, 100 mM MES pH 6.5, 5% Dioxane and a protein concentration of 4.5 mg/ml, yielding rod shaped crystals of 0.3 mm × 0.2 mm × 0.1 mm. Reduced EcAhpC1-186-YFSKHN crystals were grown by soaking the oxidized crystals with 1 mM Tris(2-carboxyethyl) phosphine (TCEP) for 1–3 min. There was no cracking or disintegration of crystals observed during the soaking.
Data collection and structure determination
Crystals of EcAhpC1-186-YFSKHN were quick-soaked in a cryoprotectant solution containing 25% glycerol in the mother liquid and flash-cooled in liquid nitrogen at 100 K. A single wavelength dataset for both the oxidized and reduced EcAhpC1-186-YFSKHN were collected at 140 K, beamline 13B1 of the National Synchrotron Radiation Research Center (NSRRC, Hsinchu, Taiwan) using a ADSC Quantum 315 CCD detector. The diffraction data were indexed, integrated and scaled using Mosflm36 and HKL2000 suite37. Data collection and processing statistics for oxidized and reduced EcAhpC1-186-YFSKHN are summarized in Table 2. Oxidized EcAhpC1-186-YFSKHN crystals belong to the monoclinic space group P21 with the unit cell parameters a = 99.47 Å, b = 134.73 Å, c = 107.53 Å and β = 111.06°. The unit cell parameters of reduced EcAhpC1-186-YFSKHN are similar to that of the oxidized one (Table 2). The asymmetric unit contains 10 molecules with the solvent content of about 60%. The structure of oxidized EcAhpC1-186-YFSKHN was solved using the crystallographic structure of oxidized EcAhpC (PDB ID: 4O5R)20 as model for molecular replacement by the program PHASER38. The reduced form of EcAhpC1-186-YFSKHN was solved using the reduced structure of Salmonella typhimurium AhpC (PDB ID: 1N8J)18. Refinement39 was done until convergence and the geometry of the final model was validated with MolProbity40. The figures were generated using PyMOL41 and structural comparison analysis was carried out by SUPERPOSE42 as included in CCP4 suite43.
Table 2. Data collection, processing and refinement statistics of EcAhpC1-186-YFSKHN.
| Ox. EcAhpC1-186-YFSKHN | Red. EcAhpC1-186-YFSKHN | |
|---|---|---|
| Wavelength (Å) | 1.000 | 1.000 |
| Crystal-to-detector distance (mm) | 360 | 420 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 140 | 140 |
| Exposure time per image (s) | 5 | 5 |
| Space group | P21 | P21 |
| Unit cell parameters (Å,°) | ||
| a= | 99.47 | 100.49 |
| b= | 134.73 | 135.65 |
| c= | 107.53 | 106.49 |
| α= | 90 | 90 |
| β= | 111.06 | 111.22 |
| γ= | 90 | 90 |
| Molecules in asymmetric unit | 10 | 10 |
| Solvent content (%) | 60.72 | 60.96 |
| Resolution limits (Å) | 50.0–2.70 (2.85–2.70)a | 50.0–3.10 (3.21–3.10) |
| No. of reflections | 202994 | 138462 |
| Unique reflections | 69616 | 44808 |
| Multiplicity | 2.9 (2.9) | 3.1 (2.9) |
| Completeness (%) | 96.1 (91.3) | 92.9 (92.0) |
| Rmergeb(%) | 12.8 (58.4) | 11.7 (58.6) |
| <I//σ(I)> | 7.2 (1.8) | 8.9 (1.5) |
| CC1/2 | 98.4 (66.7) | 91.5 (66.3) |
| Refinement statistics | ||
| R-factorc (%) | 24.63 | 25.03 |
| R-freed (%) | 28.92 | 28.36 |
| Number of waters | 320 | 19 |
| Number of sulphates | 23 | 14 |
| Number of glycerol | 22 | − |
| MolProbity statistics | ||
| Ramachandran favoured (%) | 97.3 | 98.2 |
| Ramachandran outliers (%) | 0 | 0 |
| Clashscore | 0.61 | 0.39 |
| R.M.S. deviations | ||
| Bond lengths (Å) | 0.006 | 0.007 |
| Bond angles (˚) | 0.994 | 0.946 |
| Overall B values | ||
| From Wilson plot (Å2) | 48.20 | 89.60 |
| Mean B value (Å2) | 30.31 | 81.83 |
aValues in parentheses refer to the corresponding values of the highest resolution shell.
bRmerge = ΣΣi|Ih − Ihi|/ΣΣi Ih, where Ih is the mean intensity for reflection h.
cR-factor = Σ||FO| − |FC||/Σ|FO|, where FO and FC are measured and calculated structure factors, respectively.
dR-free = Σ||FO| − |FC|/Σ|FO|, calculated from 5% of the reflections selected randomly and omitted during refinement.
Additional Information
Accession codes: The atomic coordinates of the models and their corresponding structure factors of oxidized and reduced EcAhpC1-186-YFSKHN have been deposited in the Protein Data Bank (www.pdb.org) with the entry codes 5B8A and 5B8B, respectively.
How to cite this article: Kamariah, N. et al. Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins. Sci. Rep. 6, 37610; doi: 10.1038/srep37610 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the staff of beamline 13B1 at the National Synchrotron Radiation Research Centre (NSRRC), a national user facility supported by the National Science Council of Taiwan, ROC for expert help with data collection. The Synchrotron Radiation Protein Crystallography Facility at NSRRC is supported by the National Research Program for Genomic Medicine. This study was supported by an NTU-fund (Nanyang Technological University) to G.G. (M4080811.080). B.E. and F.E. kindly note the support from the Genome Informatics IAF311010 grant. Mun F. Sek thanks the Nanyang Technological University, Singapore, for awarding her a Nanyang Scholarship during her studies.
Footnotes
Author Contributions N.K., M.F.S., F.E., B.E. and G.G. designed the experiments. N.K. and M.F.S. performed the experiments. N.K., and M.F.S. and G.G. analyzed the data. N.K., M.F.S., F.E., B.E. and G.G. wrote the paper.
References
- Imlay J. A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autréaux B. & Toledano M. B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 (2007). [DOI] [PubMed] [Google Scholar]
- Winterbourn C. C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 (2008). [DOI] [PubMed] [Google Scholar]
- Cox A. G., Winterbourn C. C. & Hampton M. B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 425, 313–325 (2009). [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Woo H. A., Kil I. S. & Bae S. H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M. B., Vinh J. & Toledano M. B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 111, 471–481 (2002). [DOI] [PubMed] [Google Scholar]
- Sobotta M. C. et al. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 11, 64–70 (2015). [DOI] [PubMed] [Google Scholar]
- Day A. M. et al. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell. 45, 398–408 (2012). [DOI] [PubMed] [Google Scholar]
- Bryk R., Griffin P. & Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 407, 211–215 (2000). [DOI] [PubMed] [Google Scholar]
- Jang H. H. et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 117, 625–635 (2004). [DOI] [PubMed] [Google Scholar]
- Zito E. et al. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell. 40, 787–797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangherlin A. & Reddy A. B. Regulation of circadian clocks by redox homeostasis. J. Biol. Chem. 288, 26505–26511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. W., Rhee S. G., Chang T. S., Jeong W. & Choi M. H. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 11, 571–578 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T., Yang J. & Molin M. Peroxiredoxins, gerontogenes linking aging to genome instability and cancer. Genes Dev. 26, 2001–2008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Karplus P. A. & Poole L. B. Typical 2-Cys peroxiredoxins-structures, mechanisms and functions. FEBS J. 276, 2469–2477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Nelson K., Poole L. B. & Karplus P. A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 15, 795–815 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamariah N. et al. Crystallographic and solution studies of NAD+- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps. Biochim. Biophys. Acta. 1847, 1139–1152 (2015). [DOI] [PubMed] [Google Scholar]
- Wood Z. A., Poole L. B. & Karplus P. A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 300, 650–653 (2003). [DOI] [PubMed] [Google Scholar]
- Wood Z. A., Poole L. B., Hantgan R. R. & Karplus P. A. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 41, 5493–5504 (2002). [DOI] [PubMed] [Google Scholar]
- Dip P. V. et al. Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 70, 2848–2862 (2014a). [DOI] [PubMed] [Google Scholar]
- Dip P. V. et al. Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim. Biophys. Acta. 1837, 1932–1943 (2014b). [DOI] [PubMed] [Google Scholar]
- Nartey W. et al. NMR studies reveal a novel grab and release mechanism for efficient catalysis of the bacterial 2-Cys peroxiredoxin machinery. FEBS J. 282, 4620–4638 (2015). [DOI] [PubMed] [Google Scholar]
- Kamariah N., Nartey W., Eisenhaber B., Eisenhaber F. & Grüber G. Low resolution solution structure of an enzymatic active AhpC10:AhpF2 ensemble of the Escherichia coli Alkyl hydroperoxide Reductase. J. Struct. Biol. 193, 13–22 (2016). [DOI] [PubMed] [Google Scholar]
- Schröder E. et al. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 Å resolution. Structure. 8, 605–615 (2000). [DOI] [PubMed] [Google Scholar]
- Wang X., Wang L., Wang X., Sun F. & Wang C. C. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem. J. 441, 113–118 (2012). [DOI] [PubMed] [Google Scholar]
- Sayed A. A. & Williams D. L. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J. Biol. Chem. 279, 26159–26166 (2011). [DOI] [PubMed] [Google Scholar]
- Jönsson T. J., Johnson L. C. & Lowther W. T. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 451, 98–101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A. et al. The sensitive balance between the fully folded and locally unfolded conformations of a model peroxiredoxin. Biochemistry. 52, 8708–8721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S., Lázaro J. J. & Dietz K. J. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 583, 1809–1816 (2009). [DOI] [PubMed] [Google Scholar]
- Parsonage D. et al. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 44, 10583–10592 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A., Nelson K. J., Parsonage D., Poole L. B. & Karplus P. A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 40, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D. et al. Dissecting Peroxiredoxin Catalysis: Separating Binding, Peroxidation, and Resolution for a Bacterial AhpC. Biochemistry. 54, 1567–1575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin A. V. et al. Hyperoxidation of peroxiredoxins 2 and 3: rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. J. Biol. Chem. 288, 14170–14177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüber G. et al. Expression, purification, and characterization of subunit E, an essential subunit of the vacuolar ATPase. Biochem. Biophys. Res. Commun. 298, 383–391 (2002). [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- Battye T. G., Kontogiannis L., Johnson O., Powell H. R. & Leslie A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA (2002). [Google Scholar]
- Krissinel E. & Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.