Abstract
Interleukin-6 (IL-6) is a key pathogenic cytokine in multiple autoimmune diseases including rheumatoid arthritis and multiple sclerosis, suggesting that dysregulation of the IL-6 pathway may be a common feature of autoimmunity. The role of IL-6 in type 1 diabetes (T1D) is not well understood. We show that signal transducer and activator of transcription 3 (STAT3) and STAT1 responses to IL-6 are significantly enhanced in CD4 and CD8 T cells from individuals with T1D compared to healthy controls. The effect is IL-6-specific because it is not seen with IL-10 or IL-27 stimulation, two cytokines that signal via STAT3. An important determinant of enhanced IL-6 responsiveness in T1D is IL-6 receptor surface expression, which correlated with phospho-STAT3 levels. Further, reduced expression of the IL-6R sheddase ADAM17 in T cells from patients indicated a mechanistic link to enhanced IL-6 responses in T1D. IL-6-induced STAT3 phosphorylation was inversely correlated with time from diagnosis, suggesting that dysregulation of IL-6 signaling may be a marker of early disease. Finally, whole-transcriptome analysis of IL-6-stimulated CD4+ T cells from patients revealed previously unreported IL-6 targets involved in T cell migration and inflammation, including lymph node homing markers CCR7 and L-selectin. In summary, our study demonstrates enhanced T cell responses to IL-6 in T1D due, in part to, an increase in IL-6R surface expression. Dysregulated IL-6 responsiveness may contribute to diabetes through multiple mechanisms including altered T cell trafficking and indicates that individuals with T1D may benefit from IL-6-targeted therapeutic intervention such as the one that is being currently tested (NCT02293837).
Introduction
Type 1 diabetes (T1D) is a chronic, multifactorial autoimmune disease in which the pancreatic islet cells are destroyed, leading to lifelong dependence on exogenous insulin therapy. To date, there is no cure. However, understanding and treating factors of autoimmune inflammation may effectively treat or even prevent disease progression.
One of the factors involved in autoimmune inflammation is interleukin-6 (IL-6), a multifunctional cytokine with a role in chronic inflammatory and autoimmune diseases. IL-6 can be produced by many cell types including stromal cells and cells of the immune system, with monocytes and neutrophils being major sources of IL-6 after bacterial or viral infection (1). Elevated IL-6 serum/tissue concentrations are a hallmark of rheumatoid arthritis, systemic lupus erythematosus and multiple sclerosis and often correlate with disease activity (2–4). Mice deficient for IL-6 are protected from experimental autoimmune encephalomyelitis (5) and blockade of the IL-6 receptor (IL-6R) suppresses collagen-induced arthritis (6), indicating that IL-6 can drive autoimmunity. Additionally, the successful treatment of rheumatoid arthritis, systemic juvenile idiopathic arthritis, or Castleman’s disease with the anti-IL-6R antibody tocilizumab demonstrates the benefit of targeting the IL-6/IL-6R axis in humans (7).
IL-6 signals predominantly via the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (8). Binding of IL-6 to the cell surface-expressed IL-6R leads to recruitment and dimerization of gp130, the common signal-transducing subunit for the IL-6 family of cytokines. gp130 dimerization activates JAK family kinases, which phosphorylate tyrosine residues on gp130. Subsequent docking of the phosphatase SHP2 (Src homology 2 domain-containing tyrosine phosphatase 2) and STAT transcription factors (STAT3 and STAT1) stimulates the MAPK (mitogen-activated protein kinase) and STAT pathways, respectively. The STAT proteins are phosphorylated, dimerize, and translocate to the nucleus, where they induce transcription of target genes (8). A unique feature of IL-6 biology is a signaling mechanism termed “trans-signaling”, which facilitates activation of cells lacking membrane-bound IL-6R (mbIL-6R) (9). This mode of IL-6 signaling occurs through engagement of gp130 with a complex of IL-6 and a soluble form of the IL-6R (sIL-6R). Whereas small quantities of sIL-6R protein are generated by translation of an IL-6R splice variant, most of the soluble receptor arises from proteolytic cleavage of the IL-6R ectodomain from the cell surface, a process referred to as “shedding”(9, 10). ADAM17, also known as TACE, has been identified as the major protease that mediates IL-6R shedding (11, 12).
The pathological effects of IL-6 in autoimmunity are associated with the IL-6R-gp130-STAT3 axis; signaling via this pathway is essential for T helper 17 (TH17) cell differentiation and inhibition of regulatory T (Treg) cell development by suppression of FOXP3 expression (13, 14). Furthermore, IL-6 induced phosphorylation of STAT3 (pSTAT3) can mediate resistance of T effector (Teff) cells to suppression by Treg cells (15, 16). In T1D, the role of IL-6 is unclear. One early report demonstrated a significantly reduced incidence of diabetes with blockade of IL-6 in the nonobese diabetic (NOD)/Wehi mouse model of T1D (17). Data on serum IL-6 levels in T1D are inconsistent (18–20); however, other findings support the disease relevance of the IL-6 pathway, including increased IL-6 production by monocytes from type 1 diabetic subjects (21), increased numbers of TH17 cells in new-onset T1D (22), Teff resistance in T1D (23, 24), and association with a genetic variant in the IL6R gene (25).
Here, we find that IL-6-induced pSTAT3 and pSTAT1 are significantly increased in peripheral blood CD4 and CD8 T cells from patients with T1D compared with healthy controls, due, in part to, increased surface expression of the IL-6R. We show that enhanced IL-6 signaling associates with early clinical disease and that reduced expression of ADAM17 is a likely cause for elevated surface IL-6R. We also provide evidence for IL-6-induced upregulation of lymph node and peripheral tissue homing receptors, indicating increased T cell migratory capacity. Together, our results suggest a role for the IL-6 pathway in the immune dysregulation of T1D and also potential benefit from IL-6 targeted therapies in this disease, one of which is currently in clinical trial (http://www.extend-study.org/).
Results
IL-6 signaling is enhanced in T cells from type 1 diabetic patients
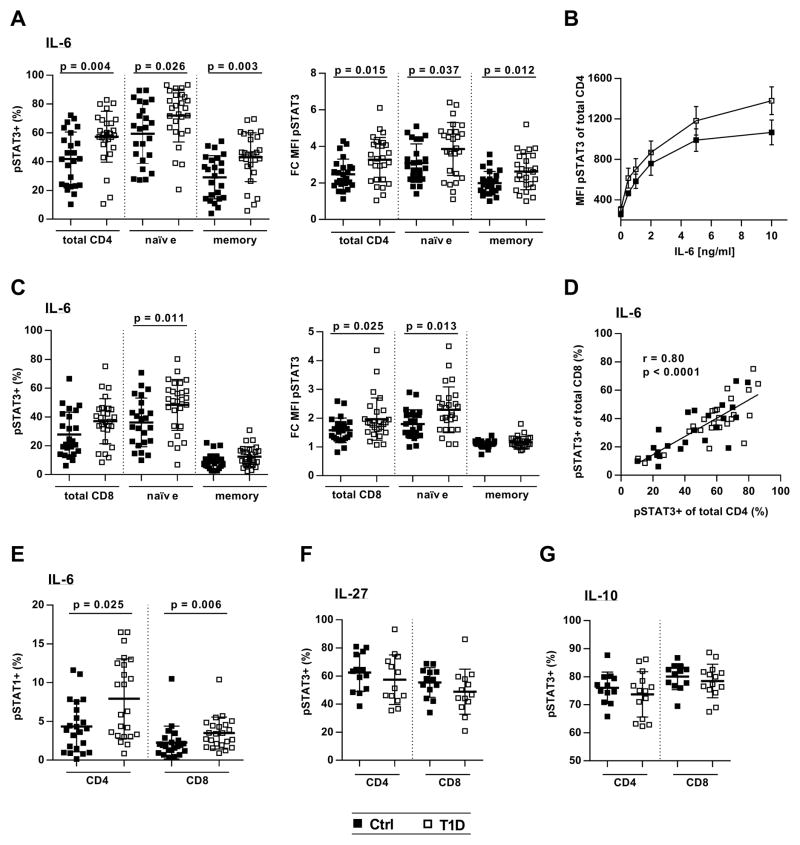
IL-6-mediated STAT3 phosphorylation was evaluated in peripheral blood mononuclear cells (PBMCs) stimulated with 2 ng of IL-6 for 10 min, a dose that consistently resulted in submaximal pSTAT3 signals. IL-6-responsive cells were detected by flow cytometry as a pSTAT3-positive population distinct from untreated or non-responsive cells (fig. S1). Although there was no difference in baseline pSTAT3 between controls and individuals with diabetes (fig. S2), we measured significantly increased IL-6-induced pSTAT3 signals in CD4 T cells from patients compared to controls (Fig. 1). This increase was observed in both the naïve (CD45RA+) and memory (CD45RA−) compartment and was evident by an increased frequency of pSTAT3-positive cells and a higher fold change in mean fluorescence intensity (MFI) (Fig. 1A). To exclude a dose-specific effect, we treated cells with varying IL-6 concentrations and found that pSTAT3 was consistently increased in CD4 T cells from T1D subjects (Fig. 1B). IL-6/pSTAT3 was also enhanced in CD8 T cells from patients, although the increase was restricted to the naïve CD8 subset (Fig. 1C). A strong positive correlation between IL-6 and pSTAT3 in CD4 and CD8 T cells from the same individuals demonstrated that enhanced IL-6 responsiveness was subject-specific and consistent across cell types (Fig. 1D). To validate the reproducibility of our phospho-flow assay, we measured IL-6/pSTAT3 in PBMCs from the same blood draw on two different occasions, which demonstrated excellent reproducibility of the assay. In addition, the IL-6/pSTAT3 signaling phenotype was stable over time (fig. S3). IL-6/pSTAT1 responses not only were relatively weak but also increased in T cells from T1D subjects and were highly correlated with IL-6/pSTAT3 (Fig. 1E and fig. S4). Furthermore, enhanced pSTAT3 appeared to be IL-6- specific because stimulation with IL-27 or IL-10, two cytokines that also signal via STAT3 and gp130 in the case of IL-27, showed similar pSTAT3 levels in controls and patients (Fig. 1, F and G). Together, these data suggest that T cells from patients with T1D are hyperresponsive to IL-6 stimulation.
Figure 1. IL-6-induced pSTAT3 and pSTAT1 are increased in T cells from patients with T1D.
Thawed and rested PBMCs from healthy controls and subjects with T1D were treated with recombinant cytokine (IL-6, IL-10, or IL-27) followed by staining for CD4, CD8, CD45RA, pSTAT3 (pY705), and pSTAT1 (pY701). (A) CD4 T cell response to IL-6 as determined by frequency of IL-6-induced pSTAT3-positive cells [pSTAT3+ (%)] and fold change in pSTAT3 MFI (FC MFI pSTAT3) compared to unstimulated cells (left and right panels, respectively); n = 24 (Ctrl) and n = 27 (T1D). (B) Dose-response curve showing IL-6-induced pSTAT3 (MFI) in total CD4 T cells from controls and patients; n = 5 (Ctrl) and n = 5 (T1D). (C) CD8 T cell response to IL-6 as determined by frequency of IL-6-induced pSTAT3-positive cells and fold change in pSTAT3 MFI (left and right panels, respectively); n = 24 (Ctrl) and n = 27 (T1D). (D) Linear regression showing positive correlation between IL-6-induced STAT3 activation in CD4 and CD8 T cells; n = 24 (Ctrl) and n = 27 (T1D). (E) pSTAT1 in response to IL-6 in total CD4 and CD8 T cells; n = 22 (Ctrl) and n = 23 (T1D). (F and G) pSTAT3 in total CD4 and CD8 T cells following stimulation with IL-27 (F) [n = 13 (Ctrl) and n = 13 (T1D)] or IL-10 (G) [n = 12 (Ctrl) and n = 14 (T1D)]. Statistical tests: Mann-Whitney U; r = Pearson correlation coefficient.
Enhanced T cell responses to IL-6 are associated with early disease
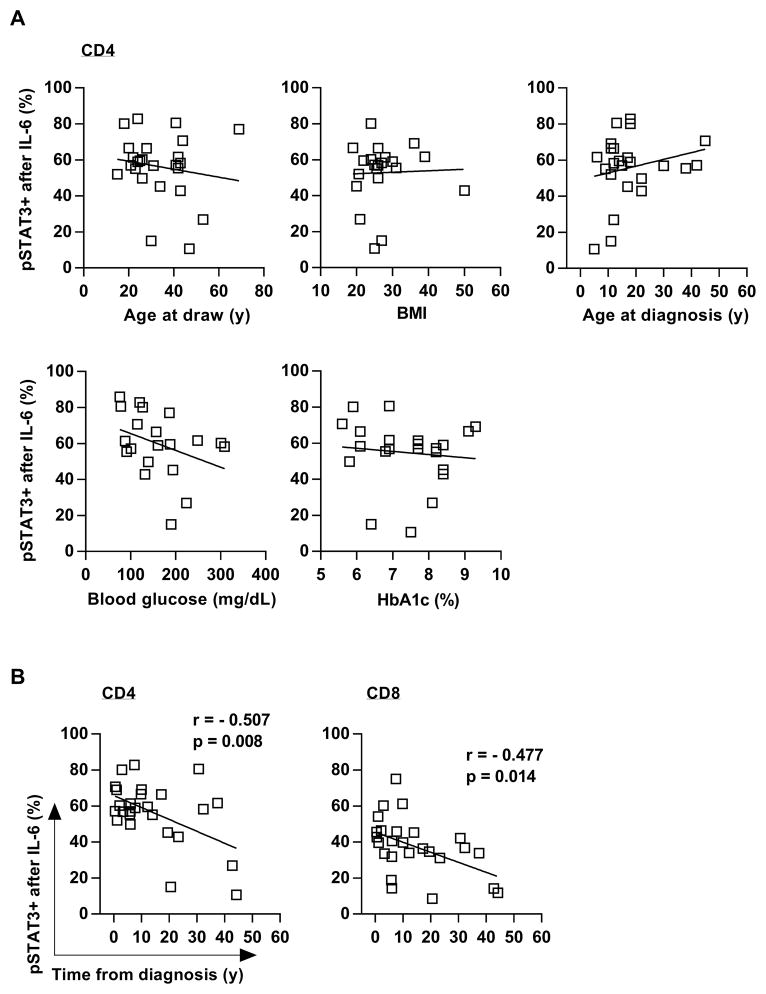
We next sought to examine clinical features of the individuals with T1D. We found no correlations between the IL-6-induced pSTAT3 signaling level to age, body mass index (BMI), age at diagnosis, glycated hemoglobin (HbA1c), or blood glucose levels (Fig. 2A). In contrast, we found that time from diagnosis negatively correlated with the frequency of pSTAT3+ CD4 and CD8 cells (Fig. 2B).
Figure 2. T cell responses to IL-6 decrease with time from diagnosis.
(A) Linear regression showing that IL-6-induced pSTAT3 is independent from age at draw, BMI, age at diagnosis, blood glucose levels, and glycated hemoglobin (HbA1c). (B) Linear regression showing the inverse correlation between time from diagnosis (disease duration) and IL-6/pSTAT3 in CD4 and CD8 T cells from subjects with T1D (left and right panels, respectively); 21≤ n ≤29 (A) and n = 26 (B) ; r = Pearson correlation coefficient.
IL-6R surface expression is increased in T cells from patients and correlates with IL-6-induced pSTAT3
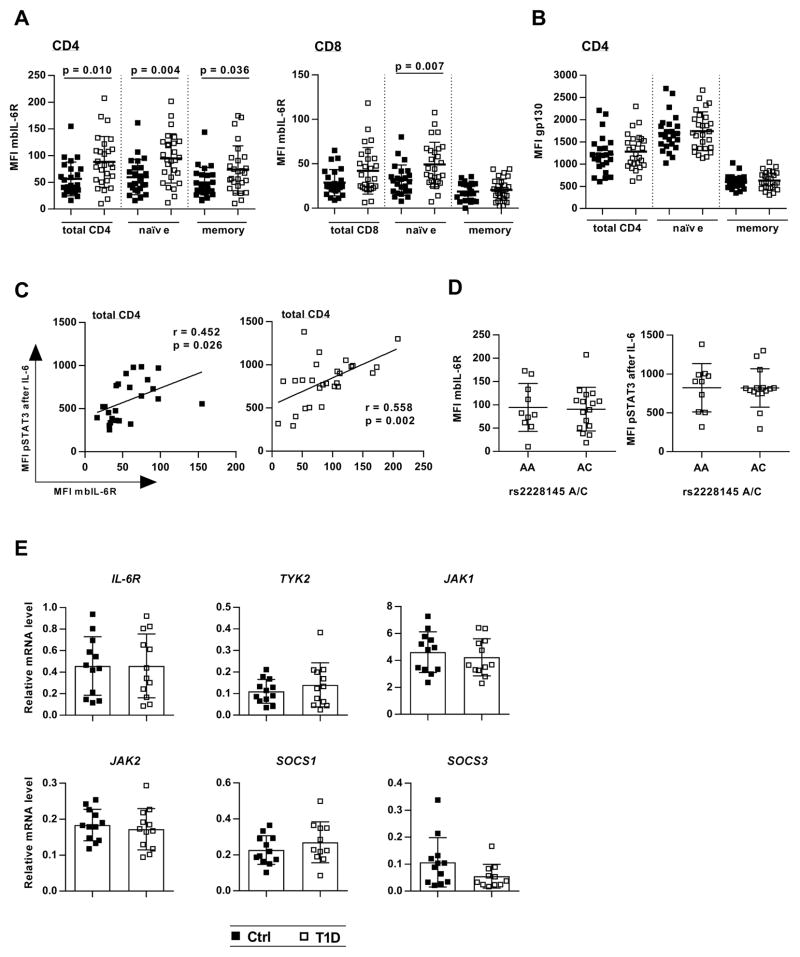
To determine the mechanisms that lead to enhanced T cell responses to IL-6 in T1D, we next assessed the expression of IL-6 signaling components by flow cytometry and real-time polymerase chain reaction (PCR). Analyzing samples from the same blood draws that had previously been assayed for pSTAT3 in response to IL-6, we found that baseline expression of mbIL-6R was significantly increased in individuals with diabetes (Fig. 3). This increase was seen in the naïve (CD45RA+) CD4 and CD8 T cell subsets, as well as in CD4 memory (CD45RA−) cells. CD8 memory cells displayed the lowest mbIL-6R expression, consistent with the low pSTAT3 response in this subset, and no difference was detected between controls and patients (Fig. 3A). Similar gp130 levels on T cells from control and diabetes subjects (Fig. 3B) implicated IL-6R expression in the enhanced IL-6 response. We found a significant positive correlation between mbIL-6R expression and IL-6/pSTAT3 in all T cell subsets, with the exception of the CD8 memory cells (Fig. 2C, and fig. S6).
Figure 3. Surface IL-6R levels are increased in T1D T cells and correlate with IL-6-induced pSTAT3.
Unstimulated PBMC from controls and patients obtained from the same blood draw as shown in Fig. 1 were stained for CD4, CD8, CD45RA, gp130 and IL-6R. IL-6R surface expression (mbIL-6R) was calculated as the antigen-specific MFI minus the MFI of a matched isotype control (see also fig. S5). (A) mbIL-6R expression in CD4 and CD8 T cells (left and right panels, respectively); n = 24 (Ctrl) and n = 27 (T1D). (B) gp130 expression (MFI) in CD4 T cells; n = 24 (Ctrl) and n = 27 (T1D). (C) Linear regression showing the positive relationship between mbIL-6R expression and IL-6-induced pSTAT3 in total CD4 T cells from subjects with T1D and controls (left and right panel, respectively); n = 24 (Ctrl) and n = 27 (T1D). (D) Effect of IL-6R rs2228145 A/C polymorphism on IL-6R surface levels and IL-6-induced pSTAT3 in CD4 T cells from subjects with T1D; n = 24. (E) Real-time PCR analysis for baseline expression of IL-6 signaling components in CD4+CD25− T cells; n = 12 (Ctrl) and n = 12 (T1D). Statistical tests: Mann-Whitney U; r = Pearson correlation coefficient.
To gain insight into potential cell intrinsic factors that contribute to altered IL-6 response in T1D, we examined the IL-6R variant 358Ala (rs2228145 A>C) that has been associated with reduced IL-6R surface levels and protection in T1D (25). To determine whether this variant contributed to the altered response to IL-6 or IL-6R surface expression in T cells, we stratified our data based on rs2228145 genotype. However, in our cohort of established T1D, we did not observe an effect of the rs228145 C allele on mbIL-6R expression or IL-6 induced pSTAT3 (Fig. 3D). We also performed quantitative real-time PCR for expression of signaling components in unstimulated CD4+CD25− T cells. Transcript levels of the IL-6R itself, the kinases TYK2, JAK1, and JAK2, as well as the negative regulators of the IL-6 pathway, SOCS1 and SOCS3, did not differ between groups (Fig. 3E).
IL-6, which induces transcription of IL-6R (26), was detected at similar levels in sera from controls and patients, and IL-1β and tumor necrosis factor (TNF), described to enhance IL-6R shedding (27) were not decreased in T1D (fig. S7). Together, the data show that enhanced T cell responses to IL-6 in T1D are largely determined by increased IL-6R surface levels, which appear to be caused by altered posttranslational regulation of the receptor, as opposed to soluble factors that induce gene expression.
ADAM17 is down-regulated in T cells from individuals with T1D
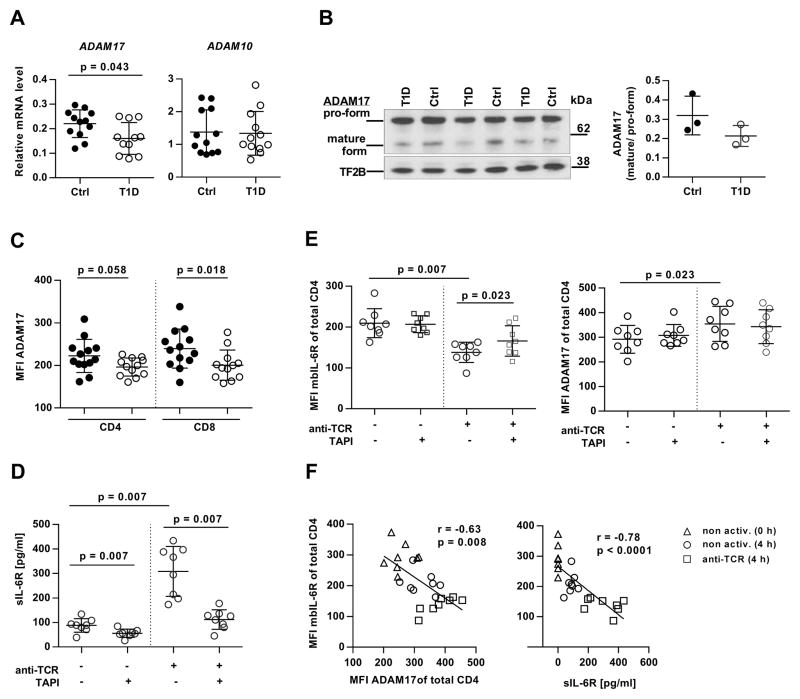
IL-6R surface expression is tightly regulated by proteolytic cleavage (shedding) through two proteases of the ADAM family, ADAM17 and ADAM10 (11). In T cells from healthy individuals, ADAM17 has been identified as the major protease that mediates IL-6R shedding after T cell receptor (TCR) activation (28). We performed quantitative real-time PCR and found that baseline messenger RNA (mRNA) expression of ADAM17, but not ADAM10, was significantly reduced in CD4+CD25− T cells from T1D patients (Fig. 4A). The T cells of T1D subjects also express less mature (active) ADAM17 by Western blot analysis of CD3 cell lysates (Fig. 4B and fig. S8), and protein expression was significantly reduced on the surface of resting CD8 T cells with a similar trend on CD4 T cells of T1D subjects (Fig. 4C). On the basis of these observations, we asked whether IL-6R processing by ADAM17 would be altered in T1D. To do this we designed an IL-6R shedding assay in which resting or anti-CD3/CD28-activated CD3 T cells from T1D subjects were cultured in the presence or absence of the ADAM17 inhibitor TAPI. After 4 hours, sIL-6R concentrations in the culture supernatant and surface expression of mbIL-6R and ADAM17 were measured by enzyme-linked immunosorbent assay (ELISA) and flow cytometry, respectively. The results of these experiments are shown in Fig. 4. Whereas sIL-6R could be detected in both the supernatant of resting and activated cells, TCR stimulation resulted in a 3.5-fold increase in the release of the soluble form of the receptor (88 pg/ml (resting) versus 308 pg/ml (activated), P = 0.007). However, in the presence of TAPI, anti-TCR-induced IL-6R shedding was efficiently blocked, with sIL-6R levels returning close to baseline concentrations (P = 0.007). Constitutive IL-6R shedding was also inhibited by TAPI, although to a lesser extent (Fig. 4D). Flow cytometric analysis showed that the accumulation of sIL-6R after TCR stimulation was matched by a significant decrease in mbIL-6R and up-regulated levels of surface ADAM17 (Fig. 4E and fig. S5). These data were comparable to results obtained from healthy controls (fig. S9, A and B), confirming a similar role of ADAM17 in IL-6R shedding from T cells of controls and patients. The relationship between sIL-6R, mbIL-6R and ADAM17 was further analyzed by linear regression demonstrating a significant inverse correlation between mbIL-6R and ADAM17, and between mbIL-6R and sIL-6R (Fig. 4F). A time course experiment suggested that the increased IL-6R surface expression in T cells from patients relative to controls is maintained in activated T cells over time (fig. S9C). Together, our data suggest that decreased ADAM17 expression, but not protease activity, in T cells from individuals with T1D contributes to higher IL-6R surface levels on T1D T cells.
Figure 4. Reduced expression of IL-6R sheddase ADAM17 in T cells from patients with T1D.
(A) Real-time PCR for ADAM17 and ADAM10 transcript in unstimulated CD4+CD25−_ cells form controls and patients with T1D; n = 12 (Ctrl) and n = 12 (T1D). (B) (Left) Western blot for ADAM17 in CD3 T cell lysates from controls and patients; TFIIB protein levels were determined as loading control. (Right) Densitometric analysis of the ratio between mature and pro-form of ADAM17; n = 3 (Ctrl) and n = 3 (T1D). (C) Flow cytometry showing ADAM17 surface expression on resting T cells from controls and patients; n = 13 (Ctrl) and n = 12 (T1D)] (D to F) IL-6R shedding assay demonstrating the role of ADAM17 in constitutive and TCR-activation induced shedding of mbIL-6R. CD3 T cells from patients with T1D were incubated for 4 hours in the presence or absence of anti-CD3/CD28 beads (anti-TCR) and the ADAM17 inhibitor TAPI. sIL-6R levels in the supernatant were determined by (D) ELISA and (E) surface expression of mbIL-6R and ADAM17 by flow cytometry. (F) Linear regression showing the inverse correlation between mbIL-6R and ADAM17 expression (left panel) and mbIL-6R and sIL-6R concentrations (right panel). Triangles, nonactivated cells at 0 hours; circles, nonactivated cells at 4 hours; squares, anti-CD3/CD28 bead-activated cells at 4 hours; n = 8 (Ctrl) and n = 8 (T1D). Statistical tests: Mann-Whitney U (A to C); Wilcoxon matched pairs (E and F); r = Pearson correlation coefficient.
Transcriptome analysis of IL-6-treated CD4 cells reveals previously unreported target genes of IL-6 associated with T cell migration
Having addressed mechanistic questions of enhanced IL-6 signaling in T1D, we sought to gain more insight into the functional consequences of our findings. Given the role of IL-6 in balancing Teff and Treg cells (13, 14), we measured frequency and cytokine production (IFN-γ, IL-17A) of both T cell subsets in PBMCs from controls and patients. For these studies we selected T1D individuals with enhanced IL-6 signaling and compared them to T1D and control subjects who do not have them this phenotype to specifically assess the impact of the IL-6/pSTAT3 response on T cell lineage. However, despite some variation and a trend toward reduced percentage of Treg cells in patients with higher IL-6/pSTAT3 signaling, no significant differences were detected (fig. S10).
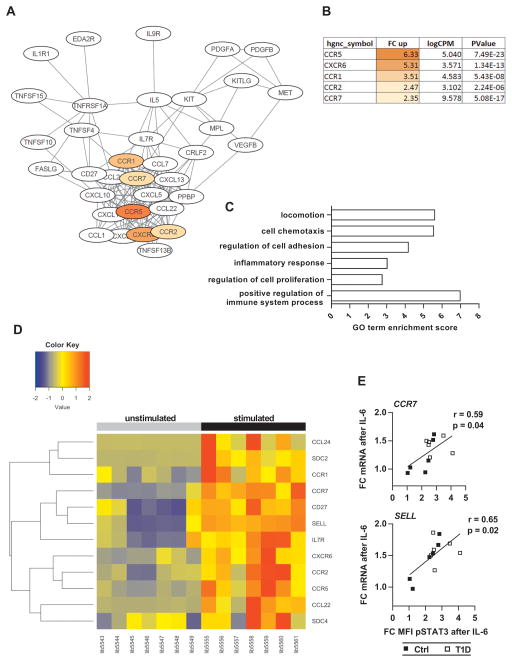
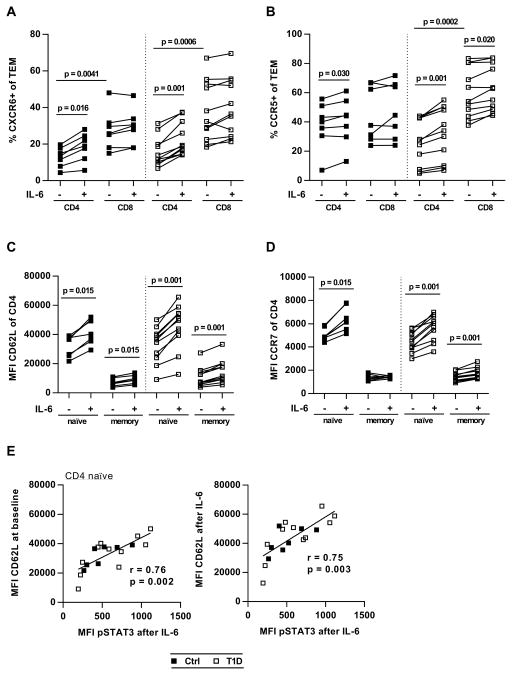
Because relatively little is known about IL-6-specific gene expression in human T cells, we next performed whole-transcriptome analysis of untreated and IL-6-stimulated CD4+CD25− T cells from a cohort of T1D patients studied above with available samples (n = 7). A total of 5836 Ensembl genes were significantly regulated by IL-6, with a fold change expression greater than 2 (in either direction). Of these, 3743 genes could be mapped by the Database for Annotation, Visualization and Integrated Discovery (DAVID) (29, 30) and were further analyzed: 56% of the 3743 genes were up-regulated, and 80% of all differentially expressed genes exhibited fold changes between 2 and 5 (Fig. S11). Performing Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping (31), we identified multiple enriched pathways in both the up-regulated and down-regulated gene sets (table S1). Whereas most of the up-regulated pathways were related to metabolic processes such as purine or fatty acid metabolism, the pathway “cytokine-cytokine receptor interaction” stood out because of its specific role in immune cell biology: 40 genes formed a network with a cluster of chemokines and chemokine receptors involved in T cell migration and inflammatory responses. The highest up-regulated receptors were CCR5 and CXCR6 (fold changes of 6.3 and 5.3, respectively), whereas CCR1, CCR2 and CCR7 were up-regulated between 2.3 and 3.5 fold (Fig. 5, A and B). IL-6 also triggered expression of several chemokines including CCL24, CCL22, and CXCL10, albeit at very low copy numbers (Fig. 5A and tables S2 and S3). In addition to pathway mapping we performed Gene Ontology (GO) enrichment analysis (32, 33) and found that 36 biological process GO terms were enriched in the up-regulated gene set. Of those, six were directly related to cell motility or regulation of the immune system, with gene counts between 13 and 28, and enrichment scores between 2.7 and 7.0 (Fig 5C and table S4). Besides the previously identified chemokine receptors, other IL-6 induced genes implicated in T cell migration and activity included SELL (encoding L-selectin), MMP-7, MMP-9, syndecan-2 and syndecan-4 (table S5). We next performed hierarchical clustering for a set of IL-6 regulated genes, which we considered most relevant for immune cell function. We found that the effect of IL-6 was largely consistent across subjects and that functionally related genes, for example CCR7, SELL and CD27, clustered together (Fig 5D). RNA sequencing (RNA-seq) data validation by quantitative reverse transcription PCR (qRT-PCR) confirmed the role of IL-6 in gene expression of most of the tested genes, including CCR7, SELL, CCR5 and CXCR6 (fig. S12). Last, by combining phospho-flow cytometry with qRT-PCR data from the same subjects, we demonstrated that IL-6-induced mRNA levels of CCR7 and SELL in T cells correlate with IL-6/pSTAT3 signaling strength (Fig 5E). In summary, our data show that IL-6 significantly enhances expression of cell migration- and inflammation-associated genes in CD4 Teff cells from patients with T1D. Many of the IL-6-regulated genes that were identified through the RNA-seq experiment encode surface expressed proteins, which interact with extracellular matrix (CD62L) or respond to chemokine gradients (CCR7, CCR5, CCR2, and CXCR6), thereby facilitating cell adhesion and tissue-specific homing. To assess the effect of IL-6 on surface expression of these markers, we cultured purified T cells in the presence or absence of IL-6 for 48 hours and evaluated cell surface expression by flow cytometric analysis. Samples for these assays were selected from T1D and control subjects who we had previously characterized for their T cell responses to IL-6, and which reflected the range of responses seen. The results of this experiment are shown in Fig. 6 and fig. S13. Using this approach, we found that the chemokine receptors CXCR6 and CCR5 were significantly up-regulated by IL-6 on effector memory (TEM) but not central memory (TCM) cells, whereas we detected virtually no expression of CCR2 on any T cell subset (Fig. 6A and B, and fig. S14). In contrast, we found that IL-6 significantly up-regulated surface levels of CD62L and CCR7 as measured by MFI in naïve and memory CD4 and CD8 T cells, and in patients as well as in controls, with the exception of CCR7 levels on CD4 memory T cells from healthy subjects, which remained largely unchanged in response to IL-6 (Fig. 6C and D, and fig. S13, A and B). Notably, there was a strong positive correlation between IL-6/pSTAT3 and surface expression of CD62L, in CD4 naïve T cells (r = 0.7, P = 0.003) (Fig. 6E). This relationship was also seen in total CD8 T cells, although in subjects with T1D only (fig. S13C). To assess the functional consequences of IL-6 exposure on migration we performed Transwell migration assays with CD4 T cells treated with IL-6 to establish their response to chemokines CCL19 (ligand for CCR7), CCL5 (ligand for CCR5) and CXCL16 (ligand for CXCR6). As shown in fig. S15, T cells that had been treated with IL-6 for 48 hours exhibited significantly increased migration toward CCL5 (P = 0.0002) and CCL19 (P = 0.039) compared to unstimulated cells. Consistent with this, we found a positive correlation between the MFI of CCR7 in naïve CD4 cells and cell migration toward CCL19. This data provide a mechanistic link between IL-6 and T cell migration that strengthens the possibility that enhanced T cell responses to IL-6 in T1D may contribute to disease pathogenesis by altering homing of T cells to the sites of islet inflammation.
Figure 5. Transcriptome analysis of IL-6 treated CD4+CD25− T cells from patients with T1D.
(A) Network of enriched KEGG pathway cytokine- cytokine receptor interaction illustrates cluster of IL-6-induced genes involved in T cell trafficking. Chemokine receptors are shown in color. (B) Fold change up-regulation, expression level (logCPM) and P value of the chemokine receptors identified in (A). (C) GO analysis of IL-6-induced genes shows enrichment of terms associated with cell migration and regulation of inflammatory responses. (D) Heat map of immune-relevant genes demonstrating consistent up-regulation by IL-6 across subjects. (Left) Unstimulated cells. (Right) IL-6-stimulated cells from the same subjects. (E) Linear regression showing positive correlation between IL-6/pSTAT3 and expression level of CCR7 and SELL in CD3+ T cells; n = 6 (Ctrl) and n = 6 (T1D). r = Pearson correlation coefficient.
Figure 6. IL-6 upregulates inflammatory homing makers CXCR6 and CCR5 on TEM cells.
Purified CD3 T cells were cultured in the absence or presence of IL-6 (10 ng/ml) for 48 hours, followed by surface staining and flow cytometric analysis. (A and B) Effect of IL-6 on frequency of CXCR6+ (A) and CCR5+ (B) CD4 and CD8 TEM cells. (C and D) Effect of IL-6 on CD62L (C) and CCR7 (D) expression in naïve (CD45RA+) and memory (CD45RA−) CD4 T cells. (E) Linear regression analysis demonstrating the relationship between IL-6/pSTAT3 and baseline CD62L (left panel) and IL-6-induced CD62L (right panel) in CD4 naïve T cells; n=7 (Ctrl) and n=11 (T1D) in all panels. Statistical tests: Wilcoxon matched pairs, Mann Whitney U; r = Pearson correlation coefficient.
Discussion
Here, we demonstrate enhanced IL-6 responsiveness in both CD4 and CD8 T cells of individuals with established T1D. Examining this in the context of time from diagnosis, we found that increased IL-6/pSTAT3 responses were seen in patients with a disease duration of less than 10 years, but that IL-6 signaling declined in subjects with long-standing disease (>20 years). This suggests that dysregulated IL-6 signaling in T1D is not due to the acute metabolic changes that occur at the time of diagnosis nor is it a trait acquired over time, for example a consequence of persistent hyperglycemia, but instead a hallmark for early events during clinical disease manifestation. Mechanistic studies showed that IL-6R surface expression is the major determinant for IL-6 signaling strength and that reduced expression of the IL-6R sheddase ADAM17 contributes to elevated IL-6R levels on T cells from patients with T1D. We provide insight into potential functional consequences of our findings by identifying IL-6-mediated upregulation of genes involved in T cell trafficking such as CCR7, L-selectin, CCR5 and CXCR6, and by demonstrating that T cell migration toward chemokine ligands of these receptors is enhanced by IL-6 in vitro.
IL-6 has been implicated in the development and progression of autoimmune diseases through both its role in the promotion of the innate immune response and its influence on T cell lineage, favoring the development of TH17 T cells and inhibiting Treg development (13, 14). Further, IL-6 and enhanced IL-6 signaling have been linked to Teff resistance to suppression by Treg (15, 16). In line with other reports (20, 34), we did not find convincing evidence of increased IL-6 production in T1D. Nonetheless, enhanced IL-6 responsiveness has the potential to promote inflammation and autoimmunity, even in the presence of homeostatic levels of IL-6. In relapsing-remitting multiple sclerosis patients, enhanced response to IL-6 as measured by pSTAT3 was found in CD4 T cells and also linked to increased IL-6R expression and a decrease in cleavage of mbIL-6R. In these subjects, Teff resistance to Treg-mediated suppression was observed and shown to be correlated to the enhanced IL-6R expression and IL-6-mediated STAT3 phosphorylation (16).
Multiple factors can influence the response to IL-6. Here, we demonstrated that increased expression of mbIL-6R correlated with enhanced IL-6-induced pSTAT3 in diabetic subjects independent of the clinical characteristics of patients. The lack of difference in IL-6R mRNA levels between controls and patients suggests that post-translational regulation of IL-6R is altered in T1D through shedding of the receptor. The T1D-associated IL-6R single-nucleotide polymorphism rs2228145 has been shown to influence plasma IL-6R and mbIL-6R levels (35) presumably through the process of altered shedding (36) but did not affect surface IL-6R expression in our cohort. Our finding of reduced transcript and protein levels of ADAM17 in peripheral blood T cells from subjects with T1D indicates a potential mechanism for enhanced IL-6 signaling and raises the possibility that activated T cells in vivo may retain their capacity to respond to mbIL-6R because of diminished ADAM17 expression or activity.
The increase in mbIL-6R is likely not the sole factor that contributes to the enhanced response to IL-6 in our T1D subjects. Cell-intrinsic factors may contribute to the altered response to IL-6 in some individuals, particularly those who do not demonstrate increased mbIL-6R but do show enhanced pSTAT3 in response to IL-6. To address this, we examined the expression of the major IL-6 signaling components in our subjects. We found similar expression of the JAKs and the negative regulators SOCS3 and SOCS1, but we cannot rule out that these may be important in individual cases, nor can we rule out the influence of genetic variants of signaling proteins including TYK2 (37) and STAT3 (38) or as-yet-unknown regulators of the IL-6 pathway.
The pathogenic effects of IL-6 on T cells in autoimmunity have been ascribed to the cytokine’s role in promoting TH17 cell differentiation and suppressing Treg cell function. In T1D, several lines of evidence support the involvement of TH17 immunity, including reports on increased frequencies of IL-17-producing CD4 and CD8 T cells in the circulation of new-onset patients (22) or increased numbers of TH17 cells in the pancreatic lymph nodes of long-term patients (40). Yet, overall, these effects are subtle and may be limited to specific subsets of TH17 and Treg cells. In our cohort, the frequency and cytokine profiles (IL-17A, IFN-γ) of CD4+Foxp3−Helios− Teff and CD4+Foxp3+Helios+ natural Treg cells in the blood were similar between IL-6/pSTAT3 “high” and ”low” T1D subjects, and no significant difference to healthy controls was observed. The limited number of subjects available for this analysis and the subtlety of these changes in the periphery may have made it difficult to demonstrate the impact of T cell responsiveness to IL-6 on the balance of Teff and Treg cells in the periphery. It is likely that enhanced IL-6 responses promote pathogenic T cell function locally in the inflamed islet or pancreatic lymph node in concert with other proinflammatory cytokines such as TNF, IL-1β, IL-21 and IL-23. In this setting, enhanced IL-6 responses may alter the fate and function of islet specific T cells, resulting in increased pathogenicity through impaired Treg function, the resistance of Teff to suppression by Treg (15, 16), and enhanced cytotoxicity via the induction of granzyme B linked to IL-6 trans-signaling (41, 42). In addition, IL-6 has been linked to reduced apoptosis of antigen-specific CD4 T cells in mice (43); this may be a mechanism by which autoantigen-specific T cells from individuals with diabetes prolong their survival.
In our study, STAT1 responses to IL-6 were also increased in T cells from individuals with diabetes. STAT1 and STAT3 are ascribed distinctive functions (44, 45), which may lead to multiple, subtle alterations in T cell differentiation and function, together significantly contributing to disease.
Whole-genome transcriptome analysis of unstimulated and IL-6-stimulated CD4+CD25− T cells from peripheral blood of patients with T1D yielded additional insight into the functional consequence of enhanced T cell responses to IL-6 in T1D subjects. Notably, a cluster of 40 genes involved in T cell trafficking and inflammatory responses was increased by exposure to IL-6. Using flow cytometry in combination with Transwell migration assays we were able to assess the impact of IL-6 on surface expression of these proteins in T cell subsets and on CD4 T cell migration in response to their ligands. We found that CXCR6, CCR5 and CCR7 were significantly up-regulated by IL-6 in CD4 and CD8 T cells, with CXCR6 and CCR5 expression largely restricted to the effector memory compartment. In agreement with this finding, we detected increased CD4 T cell migration toward their ligands after IL-6 treatment of the cells. CXCR6 and CCR5 are both chemokine receptors associated with TH1 or cytotoxic T cell function (46, 47) and, together with their ligands, have also been implicated in T1D pathogenesis. For example, circulating levels of the CCR5 ligand CCL5 were inversely correlated with β-cell function in children with T1D (48) and CXCL16, the ligand for CXCR6, was identified as one of the candidate genes in the Idd4 susceptibility locus of the NOD mouse (49). CCR7 directs recruitment of T cells into inflamed pancreatic islets (50). Together, this suggests that enhanced T cell responses to IL-6 may contribute to islet inflammation and destruction by enhancing the ability of islet specific T cells to access the islet. In keeping with this idea, IL-6 deficient mice exhibited reduced T cell recruitment to the site of acute inflammation, caused in part by dysregulation of T cell chemokine receptor expression (51).
We acknowledge that our study has limitations, one of which is that we are sampling peripheral blood, whereas the impact of the disease is at the islets and pancreatic lymph nodes. Further, this study focuses on the global T cell response and not on the response of islet-specific T cells. However, our observation likely applies to these cells in a manner similar to that seen globally, including having an impact on the fate, function, survival and localization of pathogenic islet-specific T cells in T1D subjects. In addition we have limited our studies to T cells but alterations in IL-6 signaling may extend beyond T cells. For example ADAM17 is expressed by myeloid cells; if ADAM17 expression is diminished on myeloid cells in T1D, this would have the potential to further enhance the response to IL-6 in T1D.
Enhanced IL-6 signaling in some subjects may be promoted by the immunologic and/or metabolic milieu before or at the time of diagnosis, which may wane over time. In other individuals enhanced IL-6 responses may be mediated through genetic mechanisms. Further exploration of these questions will require longitudinal studies to assess whether enhanced IL-6 signaling precedes clinical disease onset or changes during the course of disease. Additional work that explores whether there is altered IL-6R expression in other cell types will be important in evaluating therapeutic interventions that target this pathway. Collectively, these questions are pertinent to understand what factors lead to disease progression, and whether IL-6 responsiveness can predict disease progression or response to therapy. Understanding the mechanism and role of altered IL-6 signaling in T1D may assist in targeting this pathway for therapeutic intervention either through blockade of IL-6, and the IL-6R or through the use of small molecule inhibitors of the signaling pathway.
Material and Methods
Study design
Here, we posed the hypothesis that T cell responses to IL-6 are increased in T1D, thereby contributing to disease pathogenesis. The effects of IL-6 on CD4 and CD8 T cells were evaluated in PBMCs from healthy control and T1D subjects using a flow cytometry-based STAT phosphorylation assay. Our approach of using PBMCs as opposed to isolated T cells was validated by coculture experiments that show that the presence of antigen presenting cells did not modulate IL-6/pSTAT signals in the 10-min assays we performed. Mechanisms of dysregulated IL-6 signaling in T1D were interrogated by serum cytokine analyses and expression studies of IL-6 pathway components at the transcript and protein level. Functional consequences were assessed by immunophenotyping of peripheral blood lymphocytes and by transcriptome sequencing of IL-6-treated CD4 Teff cells from patients.
Control and T1D samples were selected randomly but matched for age and gender. No selection was made on human leukocyte antigen genotype or time from diagnosis. Samples were blinded for analysis but were provided in a manner that guaranteed that samples from both groups would be tested on each assay day. This was done to avoid batch effects between controls and T1D subjects. Power calculations were performed to determine the sample size required to have 80% power to detect a significant difference. These calculations indicated that a sample size of 25 per group would provide 80% power for differences ≥20 % in IL-6-induced pSTAT3.
Human samples
Frozen PBMCs and matched serum samples were obtained from participants in the Benaroya Research Institute (BRI) Diabetes and Immune Mediated Disease Registry and Repository. Control subjects (n = 58) were selected on the basis of the absence of autoimmune disease or any family history of autoimmunity. Patients with T1D (n = 60) had a disease duration between 0.2 and 51 years. Subjects were matched for age (mean age: controls, 35.2 ±13.2 years; T1D patients, 32.7 ±14.8 years), and all experiments were performed in a blinded manner. High baseline pSTAT3 levels in T cells from some controls and patients obscured the detection of IL-6 responses; those subjects were excluded from the study (fig. S2). Characteristics of study participants are listed in table S6. The research protocols were approved by the Institutional Review Board at BRI (#07109-136).
Flow cytometry
For analysis of STAT phosphorylation, thawed PBMCs were rested in serum-free X-VIVO-15 medium for 1 hour, washed with phosphate buffered saline (PBS), and stimulated at 106 cells per100 μl X-VIVO-15 with recombinant human IL-6 (rhIL-6) (2 ng/ml) (BD, catalog no. 550071) for 10 min, rhIL-10 (5 ng/ml) (BD, catalog no. 554611)for 20 min) or rhIL-27 (10 ng/ml) (eBioscience, catalog no. 14-8279) for 20 min. Cells were fixed and permeabilized using Fix Buffer I and Perm Buffer III (BD), respectively. Subsequently, cells were stained simultaneously for CD4 (BD, clone SK3), CD45RA (BD, clone HI100), CD8 (Beckman Coulter, clone SFCl121Thy2D3), pSTAT3 pY705 (BD, clone 4/P-STAT3), and pSTAT1 pY701 (BD, clone 4a) and incubated at room temperature for 45 min. Cell surface staining for IL-6R (BD, clone M5), gp130 (BD, clone AM64), CCR7 (BioLegend, clone G043H7), CD62L (BioLegend, clone DREG-56), CXCR6 (BioLegend, clone K041E5), CCR5 (BD, clone 2D7/CCR5), CCR2 (BD, clone 48607), ADAM17 (R&D Systems, clone #111633) and ADAM10 (BioLegend, clone SHM14) was carried out without previous cell fixation. Cells were acquired on a BD FACSCanto II and data were analyzed using FlowJo version vX.06 (Tree Star).
ELISA and serum cytokine analysis
sIL-6R serum concentrations were determined using the Human IL-6R Platinum ELISA Kit (eBioscience). IL-6, IL-1β, IFN-γ, and TNF serum levels were determined using the High Sensitivity Human Cytokine Magnetic Bead Kit (Milliplex Multi-Analyte Profiling, Millipore) in combination with the Luminex platform.
CD3 T cell isolation and culture
Total CD3 T cells were purified by negative selection from thawed PBMCs of healthy control and diabetes patients using magnetic-activated cell sorting (MACS) technology (Miltenyi). To assess the effect of IL-6 on expression of surface molecules, cells were cultured in 96-well round-bottom plates (Nunc) at 2 × 106 cells per well/ 200 μl of RPMI medium [10% fetal calf serum (FCS)] in the absence or presence of IL-6 (10 ng/ml) for 48 hours. Cells were washed with PBS and stained for cell surface markers for 30 min at room temperature.
IL-6R shedding assay
Purified CD3 T cells were distributed to 96-well round bottom plates at 106 cells/ 200μl of medium. Cells were activated with anti-CD3/CD28 beads (Dynabeads, Life Technologies) at a bead-to-cell ratio of 1:1 for 4 hours. In some conditions, cells were pretreated with the ADAM17 inhibitor TAPI-1 (20μM; Selleck Chemicals) for 30 min. After 4 hours, supernatants were collected and sIL-6R concentrations were determined by ELISA. For flow cytometric analysis, cells were magnetically separated from beads, washed in PBS and stained for CD4, CD8, CD45RA, IL-6R, ADAM17 and ADAM10.
Western blotting
Per subject, 2 × 106 CD3 T cells were lysed in 100 μl EBC buffer (50 mM tris (pH 8.0), 120 mM NaCl, 0.5 % NP-40, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1 mM 2-mercaptoethanol, and protease inhibitor (Roche complete Mini tablets)), and 10 μg of total protein was subjected to reducing SDS-polyacrylamide gel electrophoresis using NuPAGE Novex 4-12% Bis-Tris gels (Life Technologies). After protein transfer to polyvinylidene difluoride membrane, the membrane was blocked with 5% bovine serum albumin (BSA) in tris-buffered saline/0.05% Tween 20 (TBST) for 1 hour at room temperature and then probed with rabbit anti-ADAM17 polyclonal antibody (1 μg/ml) (Chemicon International) at 4°C overnight. For ADAM17 protein detection, the membrane was stained with a peroxidase-labeled anti-rabbit secondary antibody (Vector) at a 1:50,000 dilution in TBST for 1 hour at room temperature, followed by thorough washing with TBST and incubation with enhanced chemiluminescence substrate (ECL, Pierce). Signals were recorded by exposure to x-ray film (Research Products International). As endogenous control, protein levels of transcription factor IIB (TFIIB) were determined using rabbit anti-TFIIB polyclonal antibody C-18 (Santa Cruz Biotechnology).
Transwell migration assay
MACS purified CD4 T cells from healthy controls and subjects with T1D were seeded in a 96-well round-bottom plate at a density of 2 × 106 cells in RPMI medium supplemented with 10 % human serum. Cells were left untreated or were stimulated with IL-6 (10 ng/ml) for 48 hours. Cells were labeled with carboxyfluoroscein diacetate succinimidyl ester (CFSE), washed with PBS and added at 2 × 105 cells per 50 μl of serum-free RPMI (without phenol red, 1% BSA) to the inserts of a 96-well Fluoroblok Transwell plate (BD Falcon) fitted with a light-blocking 3 μm polyethylene terephthalate membrane. The bottom chamber of the Transwells was filled with medium containing 1 CCL5, CCL19 or CXCL16 (100 ng/ml). As standard, 1:2 serial dilutions of CFSE-labeled cells were added to some wells. Cell migration was quantified every half hour over a period of 3 hours by measuring fluorescence intensity from the bottom of the plate using a plate reader (EnSpire, Perkin Elmer).
Real-time qRT-PCR
CD4+CD25− T cells were isolated by negative selection from thawed PBMCs from healthy control and T1D subjects using MACS (Miltenyi). Cells were either left untreated or stimulated with IL-6 (10 ng/ml) for 24 hours in complete RPMI medium supplemented with 10% FCS. RNA was extracted from 1.5 × 106 to 2.0 × 106 cells using the RNeasy Mini Kit with on column DNA digestion (Qiagen). Superscript III (Life Technologies) was used to generate complementary DNA, and gene expression was measured by multiplex real-time PCR performed on an ABI 7500 Fast Real-Time PCR System. The following TaqMan expression assays were used: IL-6R (Hs01075666_m1), TYK2 (Hs00177464_m1), JAK1 (Hs01026983_m1), JAK2 (Hs00234567_m1), SOCS1 (Hs00705164_s1), SOCS3 (Hs02330328_s1), ADAM17 (Hs01041915_m1), ADAM10 (Hs00153853_m1), SELL (Hs00174151_m1), CCR7 (Hs1013469_m1), CCR5 (Hs00152917_m1), and CXCR6 (Hs00174843_m1). Expression of GTF2B (Hs00976258_m1) was measured for normalization. After log2 transformation of the ΔCt value, the resulting value was multiplied by an arbitrary number to obtain units of relative expression (52).
RNA processing for sequencing
CD4+ CD25− T cells from seven T1D patients were isolated and IL-6 stimulated as described above. RNA was extracted from 0.5 × 106 cells using the RNeasy Kit (Qiagen), and quality was assessed using the Bioanalyzer 2100 (Agilent). Sequencing libraries were constructed from total RNA using TruSeq RNA Library Prep Kit v2 (Illumina) and clustered onto a flow cell, using a cBOT amplification system with a HiSeq SR Cluster Kit v4 (Illumina). Single-read sequencing was carried out on a HiSeq 2500 sequencer (Illumina), using a HiSeq SBS Kit v4 to generate 58-base pair reads, with a target of about 10 million reads per sample.
RNA seq data analysis
FASTQ files were aligned to a human reference genome to generate gene counts. Analysis of the 64,253 Ensembl ID count data was performed using the edgeR package (53) in the R software environment (54). For each gene, a negative binomial general linear model (55) that is appropriate for count data was used for the two-group comparison (stimulated versus non-stimulated) while controlling for batch effects. The Ensembl IDs were filtered to those that had a trimmed mean of M values (TMM)- normalized count (53) of at least one in at least one library, which resulted in 21,129 Ensembl IDs used in the general linear model. A multi-dimensional scaling (56) plot was created to look at the major sources of variation in the data after filtering. The two-group comparison had 25.7% biological coefficient of variation and 5,836 Ensembl IDs had a false discovery rate (57) less than 0.05 and fold change greater than 2 (in either direction). Significantly regulated genes were subjected to enrichment analysis of KEGG pathways and GO terms using the bioinformatics resources DAVID (29, 30) and GOrilla (32), respectively. Redundancy of GO terms was removed with REViGO (33). Protein interaction networks were created using STRING (58) in combination with Cytoscape (59).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6. To assess statistical significance the Wilcoxon matched pairs test and the Mann-Whitney U test were used. Results were expressed as means +/− SD, and differences were considered statistically significant at P ≤ 0.05. Linear regression was performed by computing the Pearson correlation coefficient (r). Outliers were removed using the ROUT method (coefficient Q = 0.1%) (60).
Supplementary Material
Fig. S1. Gating strategy for analysis of IL-6/pSTAT3 by flow cytometry
Fig. S2. Baseline pSTAT3
Fig. S3. Reproducibility of phospho-flow assay and stability of IL-6/pSTAT3 phenotype
Fig. S4. Correlation of IL-6/pSTAT3 and IL-6/pSTAT1
Fig. S5. IL-6R and ADAM17 cell surface staining
Fig. S6. Correlation of mbIL-6R expression and IL-6/pSTAT3 in naïve and memory T cell subsets
Fig. S7. Serum cytokine concentrations
Fig. S8. Functional role of ADAM17 in basal and activation-induced IL-6R shedding
Fig. S9. Western blot for ADAM17 protein in CD3 T cells
Fig. S10. Frequency and cytokine profile of peripheral blood Teff and Treg cells
Fig. S11. RNA-seq: Overview of IL-6 regulated genes
Fig. S12. qRT-PCR validation of RNA-seq data
Fig. S13. Effect of IL-6 on surface expression of CCR7 and CD62L in CD8 T cells
Fig. S14. Gating strategy for the analysis of proinflammatory chemokine receptors on TCM versus TEM cells.
Fig. S15. IL-6 promotes CD4 T cell migration
Table S1. KEGG pathway mapping
Table S2. KEGG pathway cytokine-cytokine receptor interaction: IL-6 induced genes
Table S3. Normalized counts of IL-6 regulated genes
Table S4. GO enrichment analysis
Table S5. GO terms associated with T cell motility: IL-6 induced genes
Table S6. Study participant characteristics
Table S7. Source data (Excel)
Acknowledgments
We thank the BRI Clinical Core Laboratory, in particular T.-S. Nguyen, for coordinating the sample supply. We also thank the BRI Genomics and Bioinformatics Cores, in particular V. Gersuk and S. Presnell, for performing RNA sequencing and data analysis. We appreciate the work of Diabetes Clinical Research Program clinical coordinators and thank R. McMurry, I. Frank, and Anne Hocking for technical assistance. In addition, we thank the participants in the BRI Diabetes and Immune Mediated Disease Registries. Funding: We acknowledge support by U01 AI101990, DP3 DK104466 and Juvenile Diabetes Research Foundation Collaborative Center for Cell Therapy grant 2-5RA-2014-150-Q-R to JHB, U19 AI050864-11 (to KC) and the Benjamin and Margaret Hall Foundation. Author contributions: C.H. designed and performed the experiments, analyzed the data and wrote manuscript; A.R. performed experiments and analyzed data; E.W. analyzed RNA sequencing data and was the consulting statistician; J.C. performed experiments; A.S. performed preliminary experiments; A.L. assisted with flow cytometry experiments and reviewed the manuscript; S.W. performed genotyping; R.R. and M.K. performed experiments; S.P.E. and T.N.W. assisted with Transwell migration assays; CG recruited diabetes patients and reviewed the data with respect to clinical data, and reviewed the manuscript; K.C. designed and coordinated experiments, and wrote manuscript; J.H.B conceived and oversaw project. J.H.B. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Competing interests: The authors declare that they have no competing interests. Data and materials availability: RNAseq data have been deposited to the Gene Expression Omnibus database under accession number GSE78922.
References and notes
- 1.Calabrese LH, Rose-John S. IL-6 biology: Implications for clinical targeting in rheumatic disease. Nat Rev Rheumatol. 2014;10:720–727. doi: 10.1038/nrrheum.2014.127. [DOI] [PubMed] [Google Scholar]
- 2.Hirano T, Matsuda T, Turner M, Miyasaka N, Buchan G, Tang B, Sato K, Shimizu M, Maini R, Feldmann M. Excessive production of interleukin 6/B cell stimulatory factor-2 in rheumatoid arthritis. Eur J Immunol. 1988;18:1797–1801. doi: 10.1002/eji.1830181122. [DOI] [PubMed] [Google Scholar]
- 3.Linker-Israeli M, Deans RJ, Wallace DJ, Prehn J, Ozeri-Chen T, Klinenberg JR. Elevated levels of endogenous IL-6 in systemic lupus erythematosus. A putative role in pathogenesis. J Immunol. 1991;147:117–123. [PubMed] [Google Scholar]
- 4.Maimone D, Guazzi GC, Annunziata P. IL-6 detection in multiple sclerosis brain. J Neurol Sci. 1997;146:59–65. doi: 10.1016/s0022-510x(96)00283-3. [DOI] [PubMed] [Google Scholar]
- 5.Samoilova EB, Horton JL, Hilliard B, Liu TS, Chen Y. IL-6-deficient mice are resistant to experimental autoimmune encephalomyelitis: roles of IL-6 in the activation and differentiation of autoreactive T cells. J Immunol. 1998;161:6480–6486. [PubMed] [Google Scholar]
- 6.Fujimoto M, Serada S, Mihara M, Uchiyama Y, Yoshida H, Koike N, Ohsugi Y, Nishikawa T, Ripley B, Kimura A, Kishimoto T, Naka T. Interleukin-6 blockade suppresses autoimmune arthritis in mice by the inhibition of inflammatory Th17 responses. Arthritis Rheum. 2008;58:3710–3719. doi: 10.1002/art.24126. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka T, Narazaki M, Kishimoto T. Therapeutic targeting of the interleukin-6 receptor. Annu Rev Pharmacol Toxicol. 2012;52:199–219. doi: 10.1146/annurev-pharmtox-010611-134715. [DOI] [PubMed] [Google Scholar]
- 8.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J. 1994;300(Pt 2):281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Matthews V, Schuster B, Schutze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen KJ, Rose-John S. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J Biol Chem. 2003;278:38829–38839. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 12.Althoff K, Reddy P, Voltz N, Rose-John S, Mullberg J. Shedding of interleukin-6 receptor and tumor necrosis factor alpha. Contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur J Biochem. 2000;267:2624–2631. doi: 10.1046/j.1432-1327.2000.01278.x. [DOI] [PubMed] [Google Scholar]
- 13.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 14.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Goodman WA, Levine AD, Massari JV, Sugiyama H, McCormick TS, Cooper KD. IL-6 signaling in psoriasis prevents immune suppression by regulatory T cells. J Immunol. 2009;183:3170–3176. doi: 10.4049/jimmunol.0803721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider A, Long SA, Cerosaletti K, Ni CT, Samuels P, Kita M, Buckner JH. In active relapsing-remitting multiple sclerosis, effector T cell resistance to adaptive T(regs) involves IL-6-mediated signaling. Sci Transl Med. 2013;5:170ra115. doi: 10.1126/scitranslmed.3004970. [DOI] [PubMed] [Google Scholar]
- 17.Campbell IL, Kay TW, Oxbrow L, Harrison LC. Essential role for interferon-gamma and interleukin-6 in autoimmune insulin-dependent diabetes in NOD/Wehi mice. J Clin Invest. 1991;87:739–742. doi: 10.1172/JCI115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosa JS, Flores RL, Oliver SR, Pontello AM, Zaldivar FP, Galassetti PR. Sustained IL-1alpha, IL-4, and IL-6 elevations following correction of hyperglycemia in children with type 1 diabetes mellitus. Pediatr Diabetes. 2008;9:9–16. doi: 10.1111/j.1399-5448.2007.00243.x. [DOI] [PubMed] [Google Scholar]
- 19.Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK. Serum IL-1beta, IL-2, and IL-6 in insulin-dependent diabetic children. Mediators Inflamm. 2006;2006:59206. doi: 10.1155/MI/2006/59206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wedrychowicz A, Dziatkowiak H, Sztefko K, Wedrychowicz A. Interleukin-6 (IL-6) and IGF-IGFBP system in children and adolescents with type 1 diabetes mellitus. Exp Clin Endocrinol Diabetes. 2004;112:435–439. doi: 10.1055/s-2004-821189. [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw EM, Raddassi K, Elyaman W, Orban T, Gottlieb PA, Kent SC, Hafler DA. Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol. 2009;183:4432–4439. doi: 10.4049/jimmunol.0900576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q, Xu L, Priatel JJ, Levings MK, Tan R. Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunol. 2010;185:3814–3818. doi: 10.4049/jimmunol.1001860. [DOI] [PubMed] [Google Scholar]
- 23.Schneider A, Rieck M, Sanda S, Pihoker C, Greenbaum C, Buckner JH. The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J Immunol. 2008;181:7350–7355. doi: 10.4049/jimmunol.181.10.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson JM, Tremble J, Dayan C, Beyan H, Leslie RD, Peakman M, Tree TI. Increased resistance to CD4+CD25hi regulatory T cell-mediated suppression in patients with type 1 diabetes. Clin Exp Immunol. 2008;154:353–359. doi: 10.1111/j.1365-2249.2008.03810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira RC, Freitag DF, Cutler AJ, Howson JM, Rainbow DB, Smyth DJ, Kaptoge S, Clarke P, Boreham C, Coulson RM, Pekalski ML, Chen WM, Onengut-Gumuscu S, Rich SS, Butterworth AS, Malarstig A, Danesh J, Todd JA. Functional IL6R 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases. PLoS Genet. 2013;9:e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takizawa H, Ohtoshi T, Yamashita N, Oka T, Ito K. Interleukin 6-receptor expression on human bronchial epithelial cells: regulation by IL-1 and IL-6. Am J Physiol. 1996;270:L346–L352. doi: 10.1152/ajplung.1996.270.3.L346. [DOI] [PubMed] [Google Scholar]
- 27.Franchimont N, Lambert C, Huynen P, Ribbens C, Relic B, Chariot A, Bours V, Piette J, Merville MP, Malaise M. Interleukin-6 receptor shedding is enhanced by interleukin-1beta and tumor necrosis factor alpha and is partially mediated by tumor necrosis factor alpha-converting enzyme in osteoblast-like cells. Arthritis Rheum. 2005;52:84–93. doi: 10.1002/art.20727. [DOI] [PubMed] [Google Scholar]
- 28.Briso EM, Dienz O, Rincon M. Cutting edge: soluble IL-6R is produced by IL-6R ectodomain shedding in activated CD4 T cells. J Immunol. 2008;180:7102–7106. doi: 10.4049/jimmunol.180.11.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulseng B, Vatten L, Espevik T. Soluble tumor necrosis factor receptors in sera from patients with insulin-dependent diabetes mellitus: relations to duration and complications fo disease. Acta Diabetol. 1999;36:99–105. doi: 10.1007/s005920050152. [DOI] [PubMed] [Google Scholar]
- 35.van Dongen J, Jansen R, Smit D, Hottenga JJ, Mbarek H, Willemsen G, Kluft C, Penninx BW, Ferreira MA, Boomsma DI, de Geus EJ AAGC Collaborators. The contribution of the functional IL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. Behav Genet. 2014;44:368–382. doi: 10.1007/s10519-014-9656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbers C, Monhasery N, Aparicio-Siegmund S, Lokau J, Baran P, Nowell MA, Jones SA, Rose-John S, Scheller J. There interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim Biophys Acta. 2014;1842:1485–1494. doi: 10.1016/j.bbadis.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, Purcell S, Koivisto K, Tienari P, Sumelahti ML, Elovaara I, Pirttila T, Reunanen M, Aromaa A, Oturai AB, Sondergaard HB, Harbo HF, Mero IL, Gabriel SB, Mirel DB, Hauser SL, Kappos L, Polman C, De Jager PL, Hafler DA, Daly MJ, Palotie A, Saarela J, Peltonen L. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin WW, Yi Z, Stunz LL, Maine CJ, Sherman LA, Bishop GA. The adaptor protein TRAF3 inhibits interleukin-6 receptor signaling in B cells to limit plasma cell development. Sci Signal. 2015;8:ra88. doi: 10.1126/scisignal.aaa5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferraro A, Socci C, Stabilini A, Valle A, Monti P, Piemonti L, Nano R, Olek S, Maffi P, Scavini M, Secchi A, Staudacher C, Bonifacio E, Battaglia M. Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes. 2011;60:2903–2913. doi: 10.2337/db11-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottcher JP, Schanz O, Garbers C, Zaremba A, Hegenbarth S, Kurts C, Beyer M, Schultze JL, Kastenmuller W, Rose-John S, Knolle PA. IL-6 trans-signaling-dependent rapid development of cytotoxic CD8+ T cell function. Cell Rep. 2014;8:1318–1327. doi: 10.1016/j.celrep.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Bhela S, Kempsell C, Manohar M, Dominguez-Villar M, Griffin R, Bhatt P, Kivisakk-Webb P, Fuhlbrigge R, Kupper T, Weiner H, Baecher-Allan C. Nonapoptotic and extracellular activity of granzyme B mediates resistance to regulatory T cell (Treg) suppression by HLA-DR-CD25hiCD127lo Tregs in multiple sclerosis and in response to IL-6. J Immunol. 2015;194:2180–2189. doi: 10.4049/jimmunol.1303257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rochman I, Paul WE, Ben-Sasson SZ. IL-6 increases primed cell expansion and survival. J Immunol. 2005;174:4761–4767. doi: 10.4049/jimmunol.174.8.4761. [DOI] [PubMed] [Google Scholar]
- 44.Hirahara K, Onodera A, Villarino AV, Bonelli M, Sciume G, Laurence A, Sun HW, Brooks SR, Vahedi G, Shih HY, Gutierrez-Cruz G, Iwata S, Suzuki R, Mikami Y, Okamoto Y, Nakayama T, Holland SM, Hunter CA, Kanno Y, O’Shea JJ. Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity. Immunity. 2015;42:877–889. doi: 10.1016/j.immuni.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, Greenberg HB, Butcher EC. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer JM. CCR5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344–345. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 48.Pfleger C, Kaas A, Hansen L, Alizadeh B, Hougaard P, Holl R, Kolb H, Roep BO, Mortensen HB, Schloot NC D. Hvidore Study Group On Childhood. Relation of circulating concentrations of chemokine receptor CCR5 ligands to C-peptide, proinsulin and HbA1c and disease progression in type 1 diabetes. Clin Immunol. 2008;128:57–65. doi: 10.1016/j.clim.2008.03.458. [DOI] [PubMed] [Google Scholar]
- 49.Ivakine EA, Gulban OM, Mortin-Toth SM, Wankiewicz E, Scott C, Spurrell D, Canty A, Danska JS. Molecular genetic analysis of the Idd4 locus implicates the IFN response in type 1 diabetes susceptibility in nonobese diabetic mice. J Immunol. 2006;176:2976–2990. doi: 10.4049/jimmunol.176.5.2976. [DOI] [PubMed] [Google Scholar]
- 50.Shan Z, Xu B, Mikulowska-Mennis A, Michie SA. CCR7 directs the recruitment of T cells into inflamed pancreatic islets of nonobese diabetic (NOD) mice. Immuno Res. 2006;176:2976–2990. doi: 10.1007/s12026-014-8500-9. [DOI] [PubMed] [Google Scholar]
- 51.McLoughlin RM, Jenkins BJ, Grail D, Williams AS, Fielding CA, Parker CR, Ernst M, Topley N, Jones SA. IL-6 trans-signaling via STAT3 directs T cell infiltration in acute inflammation. Proc Natl Acad Sci U S A. 2005;102:9589–9594. doi: 10.1073/pnas.0501794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 53.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R. Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 55.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 58.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gating strategy for analysis of IL-6/pSTAT3 by flow cytometry
Fig. S2. Baseline pSTAT3
Fig. S3. Reproducibility of phospho-flow assay and stability of IL-6/pSTAT3 phenotype
Fig. S4. Correlation of IL-6/pSTAT3 and IL-6/pSTAT1
Fig. S5. IL-6R and ADAM17 cell surface staining
Fig. S6. Correlation of mbIL-6R expression and IL-6/pSTAT3 in naïve and memory T cell subsets
Fig. S7. Serum cytokine concentrations
Fig. S8. Functional role of ADAM17 in basal and activation-induced IL-6R shedding
Fig. S9. Western blot for ADAM17 protein in CD3 T cells
Fig. S10. Frequency and cytokine profile of peripheral blood Teff and Treg cells
Fig. S11. RNA-seq: Overview of IL-6 regulated genes
Fig. S12. qRT-PCR validation of RNA-seq data
Fig. S13. Effect of IL-6 on surface expression of CCR7 and CD62L in CD8 T cells
Fig. S14. Gating strategy for the analysis of proinflammatory chemokine receptors on TCM versus TEM cells.
Fig. S15. IL-6 promotes CD4 T cell migration
Table S1. KEGG pathway mapping
Table S2. KEGG pathway cytokine-cytokine receptor interaction: IL-6 induced genes
Table S3. Normalized counts of IL-6 regulated genes
Table S4. GO enrichment analysis
Table S5. GO terms associated with T cell motility: IL-6 induced genes
Table S6. Study participant characteristics
Table S7. Source data (Excel)