Abstract
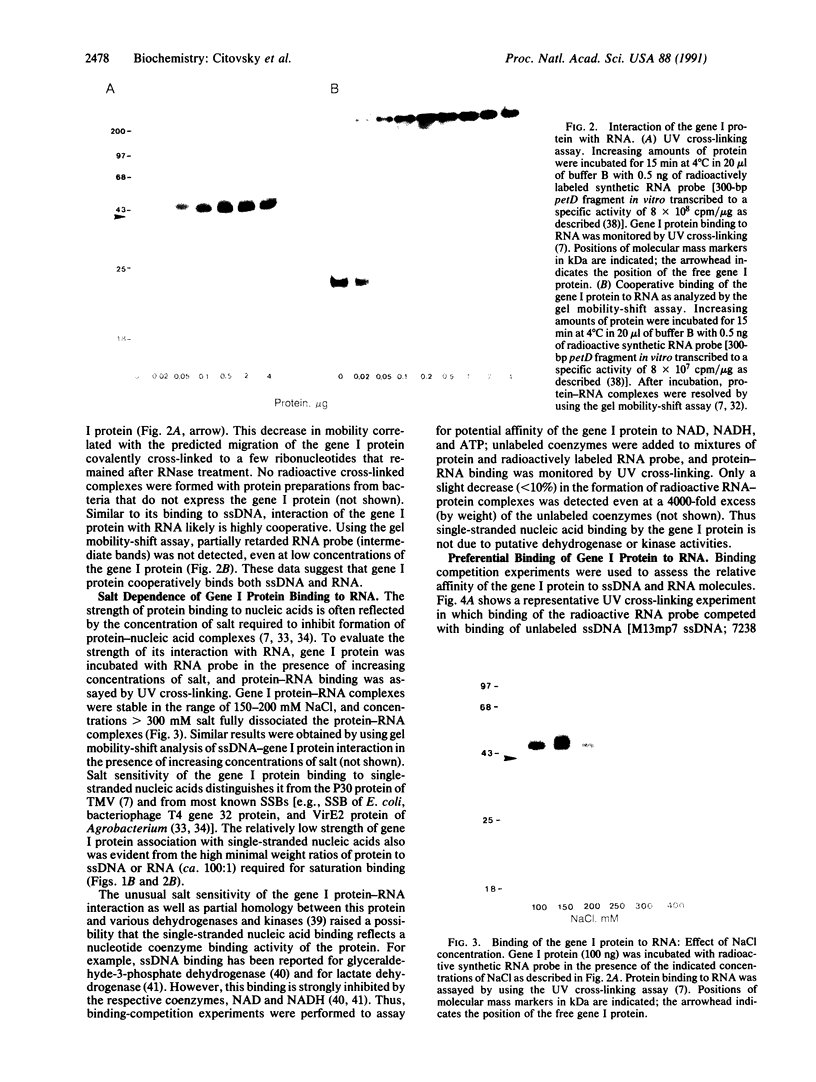
Cauliflower mosaic virus (CaMV) is a double-stranded DNA (dsDNA) pararetrovirus capable of cell-to-cell movement presumably through intercellular connections, the plasmodesmata, of the infected plant. This movement is likely mediated by a specific viral protein encoded by the gene I locus. Here we report that the purified gene I protein binds RNA and single-stranded DNA (ssDNA) but not dsDNA regardless of nucleotide sequence specificity. The binding is highly cooperative, and the affinity of the gene I protein for RNA is 10-fold higher than for ssDNA. CaMV replicates by reverse transcription of a 358 RNA that is homologous to the entire genome. We propose that the 35S RNA may be involved in cell-to-cell movement of CaMV as an intermediate that is transported through plasmodesmata as an RNA-gene I protein complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albrecht H., Geldreich A., de Murcia J. M., Kirchherr D., Mesnard J. M., Lebeurier G. Cauliflower mosaic virus gene I product detected in a cell-wall-enriched fraction. Virology. 1988 Apr;163(2):503–508. doi: 10.1016/0042-6822(88)90291-7. [DOI] [PubMed] [Google Scholar]
- Atabekov J. G., Dorokhov YuL Plant virus-specific transport function and resistance of plants to viruses. Adv Virus Res. 1984;29:313–364. doi: 10.1016/s0065-3527(08)60412-1. [DOI] [PubMed] [Google Scholar]
- Atabekov J. G., Taliansky M. E. Expression of a plant virus-coded transport function by different viral genomes. Adv Virus Res. 1990;38:201–248. doi: 10.1016/s0065-3527(08)60863-5. [DOI] [PubMed] [Google Scholar]
- Bonneville J. M., Sanfaçon H., Fütterer J., Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989 Dec 22;59(6):1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Williams K. R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Citovsky V., DE Vos G., Zambryski P. Single-Stranded DNA Binding Protein Encoded by the virE Locus of Agrobacterium tumefaciens. Science. 1988 Apr 22;240(4851):501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Knorr D., Schuster G., Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell. 1990 Feb 23;60(4):637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Wong M. L., Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. K., Hohn T. Initiation of translation of the cauliflower mosaic virus genome from a polycistronic mRNA: evidence from deletion mutagenesis. EMBO J. 1984 Dec 1;3(12):2731–2736. doi: 10.1002/j.1460-2075.1984.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Hahn P., Brown-Luedi M., Shepherd R. J., Messing J. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 1981 Jun 25;9(12):2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff G. P., Butler P. J., Akam M. E., Gait M. J., Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Wu F. C., Scholthof H. B., Shepherd R. J. Gene VI of figwort mosaic virus (caulimovirus group) functions in posttranscriptional expression of genes on the full-length RNA transcript. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9203–9207. doi: 10.1073/pnas.86.23.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Dudley R. K., Jonard G., Balàzs E., Richards K. E. Transcription of Cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell. 1982 Oct;30(3):763–773. doi: 10.1016/0092-8674(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Hull R., Sadler J., Longstaff M. The sequence of carnation etched ring virus DNA: comparison with cauliflower mosaic virus and retroviruses. EMBO J. 1986 Dec 1;5(12):3083–3090. doi: 10.1002/j.1460-2075.1986.tb04614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima E. W., Lauritis J. A. Plant virions in plasmodesmata. Virology. 1969 Apr;37(4):681–685. doi: 10.1016/0042-6822(69)90288-8. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lohman T. M., Overman L. B., Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986 Feb 20;187(4):603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- Mason W. S., Taylor J. M., Hull R. Retroid virus genome replication. Adv Virus Res. 1987;32:35–96. doi: 10.1016/s0065-3527(08)60474-1. [DOI] [PubMed] [Google Scholar]
- Maule A. J., Harker C. L., Wilson I. G. The pattern of accumulation of cauliflower mosaic virus-specific products in infected turnips. Virology. 1989 Apr;169(2):436–446. doi: 10.1016/0042-6822(89)90169-4. [DOI] [PubMed] [Google Scholar]
- Medberry S. L., Lockhart B. E., Olszewski N. E. Properties of Commelina yellow mottle virus's complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 1990 Sep 25;18(18):5505–5513. doi: 10.1093/nar/18.18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher U. Similarities between putative transport proteins of plant viruses. J Gen Virol. 1990 May;71(Pt 5):1009–1018. doi: 10.1099/0022-1317-71-5-1009. [DOI] [PubMed] [Google Scholar]
- Mesnard J. M., Kirchherr D., Wurch T., Lebeurier G. The cauliflower mosaic virus gene III product is a non-sequence-specific DNA binding protein. Virology. 1990 Feb;174(2):622–624. doi: 10.1016/0042-6822(90)90118-b. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Olszewski N. E., Guilfoyle T. J. Nuclei purified from cauliflower mosaic virus-infected turnip leaves contain subgenomic, covalently closed circular cauliflower mosaic virus DNAs. Nucleic Acids Res. 1983 Dec 20;11(24):8901–8914. doi: 10.1093/nar/11.24.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Hagen G., Guilfoyle T. J. A transcriptionally active, covalently closed minichromosome of cauliflower mosaic virus DNA isolated from infected turnip leaves. Cell. 1982 Jun;29(2):395–402. doi: 10.1016/0092-8674(82)90156-8. [DOI] [PubMed] [Google Scholar]
- Perucho M., Salas J., Salas M. L. Identification of the mammalian DNA-binding protein P8 as glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1977 Dec;81(3):557–562. doi: 10.1111/j.1432-1033.1977.tb11982.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stratford R., Covey S. N. Segregation of cauliflower mosaic virus symptom genetic determinants. Virology. 1989 Oct;172(2):451–459. doi: 10.1016/0042-6822(89)90187-6. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol. 1985 Nov;2(6):455–468. doi: 10.1093/oxfordjournals.molbev.a040365. [DOI] [PubMed] [Google Scholar]
- Volovitch M., Drugeon C., Yot P. Studies on the single-stranded discontinuities of the cauliflower mosaic virus genome. Nucleic Acids Res. 1978 Aug;5(8):2913–2925. doi: 10.1093/nar/5.8.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub M., Ragetli H. W., Leung E. Elongated virus particles in plasmodesmata. J Ultrastruct Res. 1976 Sep;56(3):351–364. doi: 10.1016/s0022-5320(76)90010-1. [DOI] [PubMed] [Google Scholar]
- Williams K. R., Reddigari S., Patel G. L. Identification of a nucleic acid helix-destabilizing protein from rat liver as lactate dehydrogenase-5. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5260–5264. doi: 10.1073/pnas.82.16.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. M. Plant viruses: a tool-box for genetic engineering and crop protection. Bioessays. 1989 Jun;10(6):179–186. doi: 10.1002/bies.950100602. [DOI] [PubMed] [Google Scholar]
- Wurch T., Kirchherr D., Mesnard J. M., Lebeurier G. The cauliflower mosaic virus open reading frame VII product can be expressed in Saccharomyces cerevisiae but is not detected in infected plants. J Virol. 1990 Jun;64(6):2594–2598. doi: 10.1128/jvi.64.6.2594-2598.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young M. J., Daubert S. D., Shepherd R. J. Gene I products of cauliflower mosaic virus detected in extracts of infected tissue. Virology. 1987 Jun;158(2):444–446. doi: 10.1016/0042-6822(87)90218-2. [DOI] [PubMed] [Google Scholar]