Abstract
A powerful new approach has become much more widespread and offers insights into aspects of DNA repair unattainable with billions of molecules. Single molecule techniques can be used to image, manipulate or characterize the action of a single repair protein on a single strand of DNA. This allows search mechanisms to be probed, and the effects of force to be understood. These physical aspects can dominate a biochemical reaction, where at the ensemble level their nuances are obscured. In this paper we discuss some of the many technical advances that permit study at the single molecule level. We focus on DNA repair to which these techniques are actively being applied. DNA repair is also a process that encompasses so much of what single molecule studies benefit – searching for targets, complex formation, sequential biochemical reactions and substrate hand-off to name just a few. We discuss how single molecule biophysics is poised to transform our understanding of biological systems, in particular DNA repair.
Keywords: DNA repair; Single molecule; FRET, Imaging; Laser tweezers; Magnetic tweezers; AFM
1. Introduction
1.1. The development of single molecule techniques
Studying systems at the single molecule level is becoming more widespread, however the field is still relatively new, finding its origins in the Nobel prize winning work on single ion channels [1]. These channel studies provided not only the concept that single molecules could be investigated, but also a huge number of tools; theoretical, practical and analytical. A key concept from this work is that ensemble level studies provide an overview of the average behavior of a molecular species. This seemingly obvious statement belies its importance, because during a biological process one or more proteins explore their energetic landscapes potentially through a multiplicity of pathways. The ensemble is the sum of all of these processes, however, a single molecule recording deconstructs this average; providing the discrete probability of passing through each constituent pathway. The importance of understanding molecular systems, at this level, is evident as one attempts to scale up toward more complex systems [2]. In the context of DNA repair, understanding systems one molecule at a time permits a view of the heterogeneity of the molecular states involved in a biochemical reaction. On the nanoscale both protein structures DNA structures fluctuate, it is only when these structural fluctuations coincide in the correct manner that a chemical reaction occurs. A structural snapshot of a process provides one final or intermediate state, but not the multiplicity of states, their significance, or their dynamics. Ensemble approaches to obtain dynamics require aligning the biochemistry of all of the molecules, for example during a ‘single turnover’ experiment [3]. This provides information on the behavior of proteins for a limited time, since they become asynchronous very quickly. In the steady state, each interaction at the single molecule level is a single turnover and therefore does not require synchronization. Furthermore, many repair reactions require multiple protein partners that assemble at the damaged site; therefore through single molecule analysis it is possible to follow the fate of a particular reaction from initial DNA damage location to completed repair. The ultimate goal of single molecule studies is the analysis of a complete reaction mechanism with dynamical information on the comings and goings of protein components. In other words, watching a full biochemical reaction unfold in real time.
1.2. Limitations of single molecule experiments
Although single molecule approaches are extremely useful there are crucial limitations. The main barrier to studying any system at the single molecule level is signal. Even in ensemble experiments with 1 mL of 1 mM solution, signal can be weak, despite the amplification offered by 1017 molecules. For single molecule experiments, no such inherent amplification is present. Instead amplification must come from external sources such as detectors. Therefore, dealing with such low signals above noise has required the development of new technologies. These utilize electron-multiplying CCD (charge coupled device) imaging cameras with unprecedented sensitivity and more recently large-sensor CMOS (complementary metal oxide semiconductor) cameras that possess incredibly fast frame rates [4]. From the aspect of fluorescence imaging, however, it is irrelevant how fast a camera records if the photons are not sufficient to generate a signal. Therefore new fluorophores have been derived that are extremely bright [5]; these however, in turn need to be balanced against photostability. This latter problem occurs when a fluorophore undergoes a covalent modification during the population of an excited state, resulting in loss of the ability to emit photons [6]. To offset this problem a number of groups have derived fluorophores with various emission wavelengths possessing enhanced quantum yields and photostability [7]. Also a broader understanding of fluorescence photophysics has generated solution additives that delay photobleaching [8]. One huge advance has been the development of quantum dots (Qdots) for biological applications [9–12]. These semi-conductor based fluorophores possess several unique properties: they are brighter and much more photostable than their organic counterparts and they emit over a more narrow wavelength range. In addition, their unique broad excitation spectra allow multiple Qdots to be excited by a single wavelength source; thus permitting true simultaneous multi-color imaging.
This review discusses how single molecule biophysics has been used to study the process of DNA repair. As a prelude to the articles within this special issue of DNA Repair, we aim to highlight the ingenious methods used to examine systems at this level focusing on imaging, manipulation and then how these may be used to study systems within cells.
2. Imaging based methods – a bright future
Real time imaging of protein#x02013;DNA interactions can provide information both on the compositional and dynamic aspects of repair. There exist a vast range of single molecule studies performed with DNA binding proteins, which have provided information on the search mechanism, as well as, the diffusion constants of the protein/complexes [13]. A few examples of the imaging techniques used to study different DNA repair systems and the data acquired using them are outlined here.
2.1. Tethered particle motion
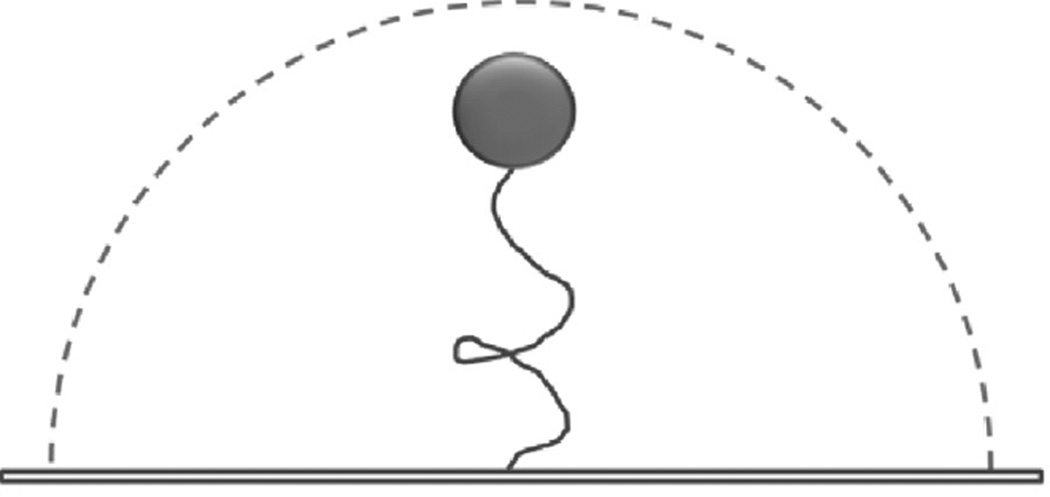
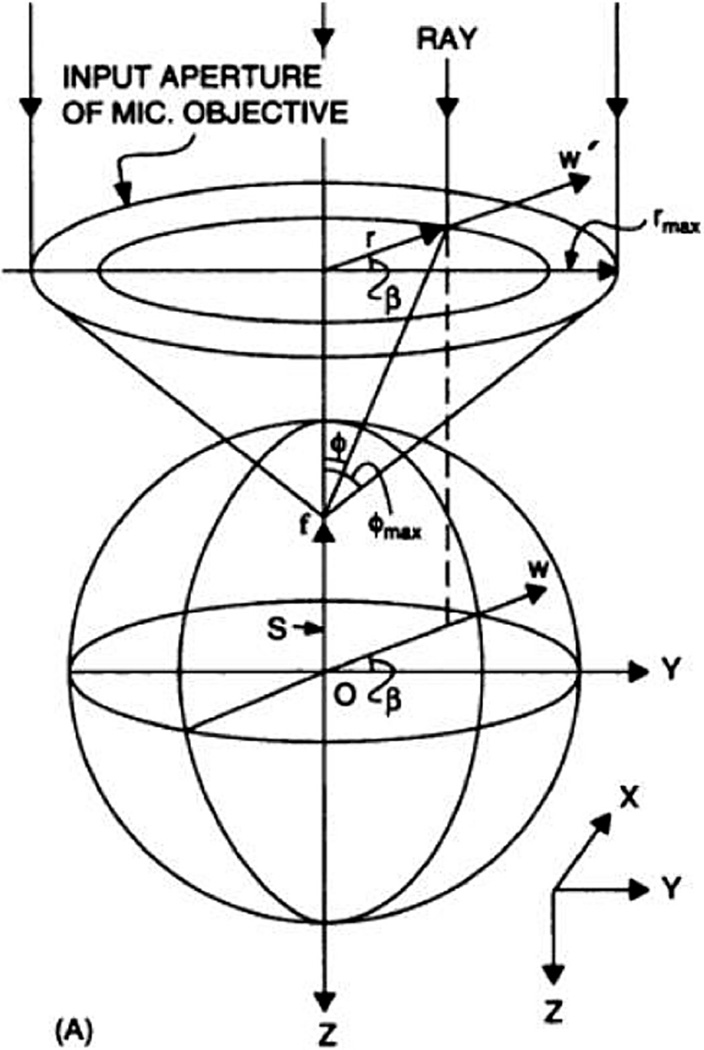
First used over two decades ago [14,15] tethered particle motion (TPM) detects the position of a bead attached to DNA at one end when the other end of the DNA is attached to a surface. The surface tethering of the DNA restricts the inherent Brownian motion of the bead in aqueous solution (Fig. 1). This restriction is proportional to the length of the tether [14], therefore the amount of Brownian motion provides a signal for DNA shortening events. This method is well suited to studying proteins that bend, loop or translocate along and hence shorten the DNA [16]. TPM has many advantages such as being simple and inexpensive with temporal resolution high enough to detect the rapid kinetic behavior of individual molecules. No external force such as flow is required during this type of investigation and TPM can be combined with other methods, such as optical trapping (see below). TPM has been used to monitor the processivity of RecBCD helicase, which is responsible for initiating double-strand break DNA repair through homologous recombination [17]. This investigation demonstrated that RecBCD can translocate along DNA, and its interaction with a regulatory 8nt Chi sequence (5′-GCTGGTGG-3′) did not alter its translocation velocity. However, subsequent alterations to TPM using stretching forces, induced by the introduction of a continuous buffer flow into the sample chamber, increased positional accuracy, such that reduced translocation velocity could be detected in the Chi regions [18].
Fig. 1.
The tethered particle movement assay. A single DNA molecule is tethered between the slide surface and a bead particle. The dash line represents the range of Brownian diffusion of the microsphere, which depends on the DNA contour length.
TPM has also been used to study RuvA, a protein involved in mediating branch migration and Holliday junctions created during homologous recombination in bacteria. Again a slight alteration to TPM was used where, instead of averaging an image over time, the individual trajectories of the beads were measured [19]. This allowed bead positioning with nanometer-scale precision. The study was able to view in real time RuvA-mediated unfolding of the Holliday junctions, detected as an increase in the length of the DNA tether.
2.2. Combing
DNA normally forms a collapsed bundle in solution with a diameter that can be approximated by random flight theory (for λ-DNA this is ~6 µm [20]). This makes imaging protein attachments to DNA particularly challenging, therefore investigators typically elongate or extend the DNA into a linear chain. One way this is performed is through the DNA combing technique. DNA is laid directly onto the surface of a cover slide in a combing assay. Firstly, the surface of the slide is activated with a hydrophobic moiety such as polystyrene or polymethylmethacrylate [21]. Then, either using continuous flow [22] or retracting a slide through an air–water interface [23]. DNA can be stretched between two or more points in low pH conditions. Imaging is performed using Total internal reflectance fluorescence (TIRF) microscopy, where the evanescent field provides high signal to noise [21,22]. Combing has the advantage of being technically simple, however, there are a few disadvantages; the DNA and the proteins are both imaged on the surface of the slide, meaning it can be difficult to distinguish fluorophores on the surface from those on the DNA. In addition, the surface itself may have profound effects on the protein–DNA interaction and/or the native states of both.
For DNA molecules that are surface bound from only one end, continuous flow can be applied to extend the DNA [24,25], over-coming some of limitations of pure combing. This approach has been used to study the base excision repair protein human oxoguanine DNA glycosylase1 (hOGG1) in a landmark study for single molecule imaging of DNA repair [24]. This study attempted to settle the facilitated diffusion paradox. By directly imaging the motion of hOGG1 on DNA it was possible to derive the diffusion constant of a molecule on DNA. From this value rotation-coupled diffusion was inferred and the energetics of the protein–DNA interaction could be derived. The observed movement occurred over hundreds of base pairs, and based on the earlier work of Von Hippel and colleagues [26], by using a broad range of salt concentrations, it was found that sliding dominated the 1D diffusional movement along DNA.
DNA combing has also been used to study Rad51, a key protein in homologous recombination [27]. It was shown that Rad51 can bind stably to dsDNA and diffuse one-dimensionally, consistent with a ring-like structure topologically linked to the DNA. This means the protein does not undergo rotation coupled diffusion and therefore possesses a significantly greater diffusion constant. However, the protein was found to stop at sites resembling double strand breaks. This work helped to suggest that the lateral diffusion of the protein may enable the recombinase to scan DNA for regions in need of repair. This particular form of DNA combing involved the alignment of multiple DNA strands in an array, this was later termed a DNA curtain.
2.3. DNA curtains
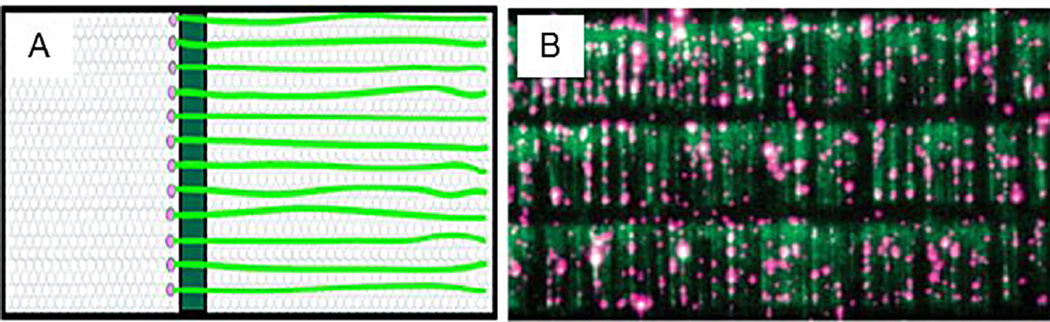
Described in more detail within this Special Issue [133,134,135] the work originating from the Greene laboratory has been based on a significant technical advancement to combing that allows DNA molecules to be spatially aligned on the coverslip, so that when flow is applied they elongate [28]. This has allowed for a number of useful investigations [29]. In this approach, a diffusional barrier is etched onto a slide surface perpendicular to the direction of buffer flow. The slide is then coated with a lipid bilayer attached to DNA molecules, as the bilayer slides due to buffer flow, it reaches the barrier where the molecules are impeded from further motion and the DNA aligns to this barrier, creating a DNA ‘curtain’ (Fig. 2). This technique offers enormous advantages over combing since multiple DNA molecules can be viewed simultaneously within a single field-of-view using TIRFM [29].
Fig. 2.
DNA curtains. (A) Schematic of a DNA curtain assay. Linear arrays of DNA attached to a lipid bilayer align to a linear barrier (B) YOYO stained DNA (green) with a Qdot-labeled (pink) protein complex.
Adapted with permission from [29]. Copyright 2013 American Chemical Society.
DNA curtains have been used to study many systems including the damage sensing protein complex in mismatch repair: Msh2-Msh6. This study showed Msh2-Msh6 uses thermally powered 1D diffusion to locate lesions rather than an ATP requiring mechanism [30]. It was also found that Msh2–Msh6 complexes were incapable of bypassing each other on the DNA, as expected for sliding molecules. However the Mlh1–Pms1 mismatch repair complex was capable of hopping, thus allowing protein complexes to move past each other. This has helped to demonstrate how proteins may bypass obstacles encountered within a crowded genome. Key fundamental questions pertaining once again to the mode by which targets on DNA are found are being currently addressed [31].
2.4. DNA tightropes
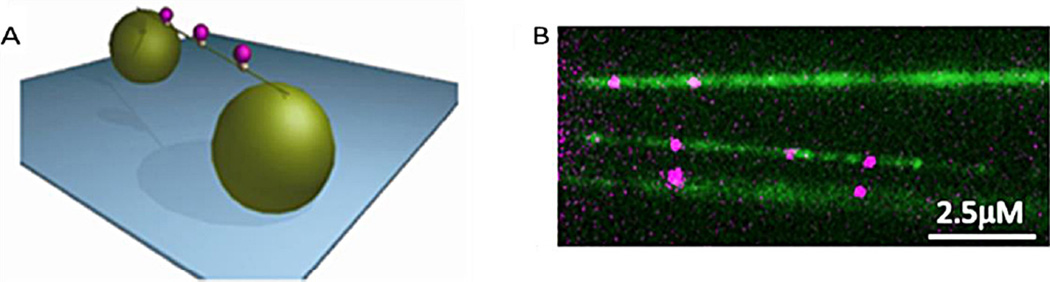
Another advance on DNA combing involves using DNA tightropes (elevated DNA platforms), in which lambda DNA is suspended between the tops of micron sized beads using a microfluidic flow based approach. Since the DNA is elevated from the surface, anything that appears within the focal plane at this height must be bound to the DNA, while all non-attached molecules contribute to the background noise. The method of flowing the components into the chamber is relatively trivial, requiring less than one hour to construct a usable tightrope setup [20,32,33]. Poly-l-lysine coated beads are used to strongly attach DNA, therefore no specific attachment strategy is required, and no flow is required once the tightropes have been formed. By tuning the density of the tightropes it is possible to have a single tightrope or many, offering multiple DNA–protein interactions within the field of view. Additionally as the proteins are elevated far from any surface-bound fluorophores, these do not interfere with fluorescent spot discrimination (Fig. 3). A near TIRF imaging technique is used to image the interaction of the protein to the DNA by steering the excitation laser to a sub-critical angle. This oblique angle fluorescence configuration provides an improved single to noise ratio relative to epi-fluorescence [34,35]. DNA tightropes have been used extensively to study the nucleotide excision repair pathway in bacteria [136]. UvrA was shown to utilize a three-dimensional search in NER for damage recognition [20]. However with the addition of UvrB, the resulting UvrAB slides when in contact with the DNA. Complex motion patterns were seen for UvrAB on DNA including unbiased diffusion, paused motion and directed motion. This indicates that more than one mechanism of movement can be utilized by proteins when moving on DNA. It was later shown that UvrC can independently form a stable DNA bound complex and that UvrC facilitates UvrB binding to DNA [32]. This was previously thought to only occur through UvrB’s interaction with UvrA in a forward reaction at the beginning of NER. It was found that this complex is abundant; suggesting that given the vast excess of UvrB over UvrC in the cell, all UvrC is present in an UvrBC complex in vivo [32]. Such complexes have not been characterized previously by ensemble techniques, highlighting the importance of single molecule methods.
Fig. 3.
DNA tightrope assay. (A) Schematic image of a DNA tightrope assay. Single DNA molecules are suspended between surface attached micron+ sized silica beads. (B) DNA tightropes labeled with YOYO-1 (green) bound with Qdot-labeled UvrC (pink). This assay is well suited to measuring the association, dissociation and movement of bound molecules.
Reprinted (adapted) with permission from [32] Copyright (2013) American Chemical Society.
Tightropes have also been used to investigate two glycosylase families involved in base excision DNA repair (see [33,137]. Members of these two structurally distinct oxidative damage recognizing families, helix–hairpin–helix (HhH) and Fpg/Nei, were labeled with single Qdots and imaged interacting with DNA tightropes. It was found that efficient target location was accomplished by rotational diffusion and that during the scanning process glycosylases examine DNA by interrogation with the wedge residue Phe111. This study has marked the beginning of a much deeper understanding of how DNA glycosylases find damage.
More recently, DNA tightropes have found application in the study of the telomere binding proteins TRF1 and TRF2 [36], reviewed by Lee et al. [134], where new insights into the mechanism by which proteins complexes are formed on DNA have been made. Together these investigations indicate that both DNA curtains and DNA tightropes offer extremely versatile platforms for the study of DNA–protein interactions.
2.5. Single molecule Förster resonance energy transfer (smFRET) and fluorescence quenching
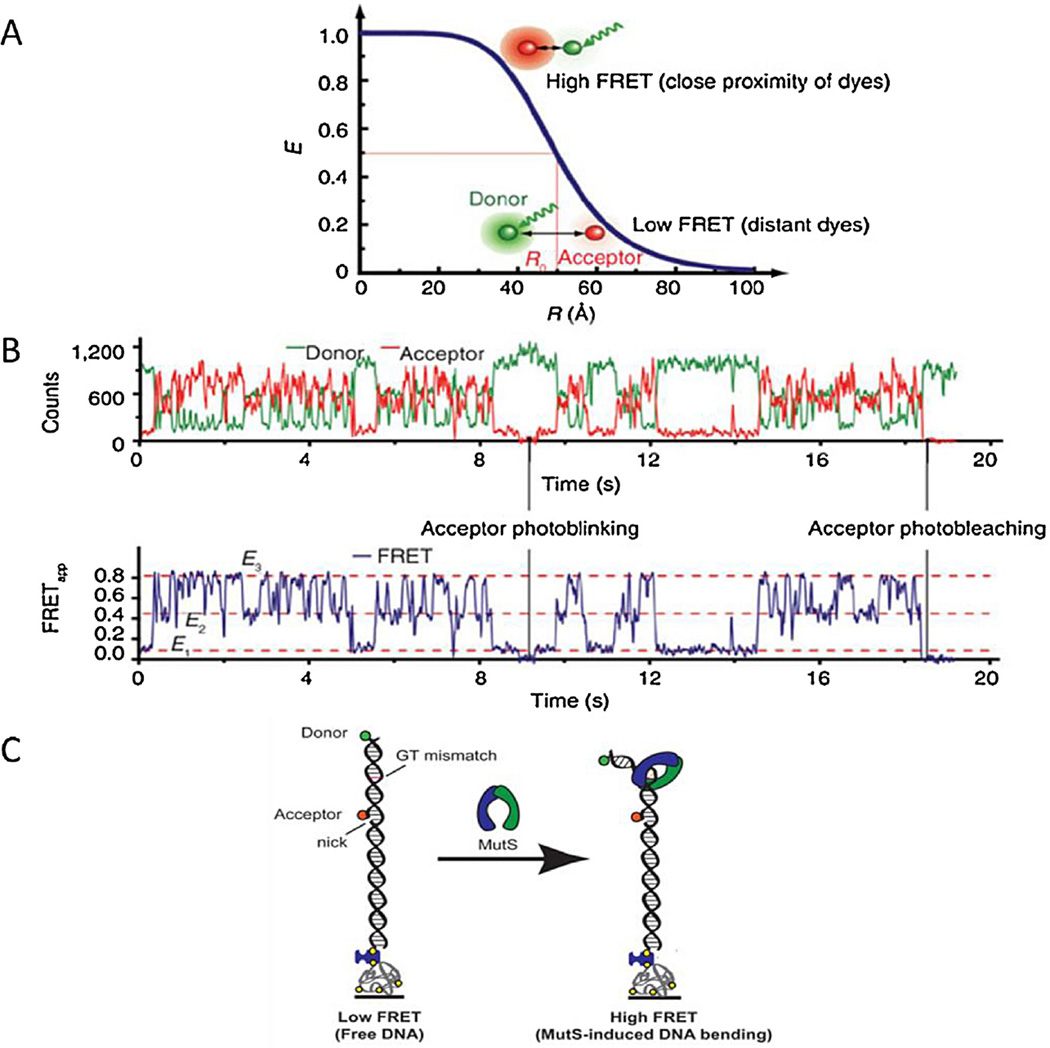
As evidenced by the multiple contributions across a range of topics within this Special Issue [133,134,138,139] smFRET has enjoyed a huge growth in its deployment for studying protein-DNA complexes at the single molecule level [37]. FRET is not only a direct imaging technique but can also be used to gauge distances between fluorophores by measuring the efficiency of energy transfer between two fluorophores. FRET involves the transfer of non-radiative energy to an acceptor molecule from a donor molecule via an induced dipole–dipole interaction (Fig. 4). The efficiency (E) of the transfer depends on the sixth power distance (r) between the donor and acceptor, summed up by the equation:
where r0 is the 50% efficiency distance. r0 is found to be strongly dependent on the properties of the dye molecules [37]. In practice the fluorophores need to be within 6–10 nm to get a strong FRET signal. For example the r0 value for the Cy3/Cy5 pair is ~6 nm.
Fig. 4.
Förster resonance energy transfer. (A) During FRET a donor dye is excited and in a non-radiative process this excitation energy is transferred to an acceptor dye. The FRET efficiency (E) of this process depends upon distance between dyes (R), accurate measurement of distance is made around the mid-point of FRET efficiency – the Förster distance. (B) An example of two color single molecule FRET data. The intensities of the individual donor and acceptor dyes (upper trace) permit calculation of the FRET efficiency (lower trace) to be calculated. Reprinted (adapted) with permission from [37]. Copyright (2013) American Chemical Society. (C) smFRET system with MutS-induced DNA bending. A 50 base pair strand of dsDNA with a FRET donor (green) and a FRET acceptor (red) is anchored to a surface through a biotin streptavidin link. A GT-mismatch is located approximately halfway between the dyes. As the MutS binds to the DNA and bends the DNA the distance between the dyes decreases which increases the FRET signal.
Reprinted (adapted) with permission from [39]. Copyright (2013) American Chemical Society.
Static heterogeneity (differences between molecules within the sample) or dynamic heterogeneity (time-dependent changes of individual molecules) can be present during an experiment. One of the advantages of using smFRET over ensemble FRET is that dynamic temporal resolution of an individual molecule is possible enabling these different populations to be detected [38]. In ensemble studies small amounts of a single species (either the donor or acceptor) can affect the interpretations of the results. This problem is not present in smFRET as only one complex is measured at a time [37].
smFRET imaging was used in the context of DNA repair to study the binding dynamics and diffusion of MutS on DNA containing a defined mismatch [39]. MutS was labeled with a single donor fluorophore (Cy3) and the DNA with an acceptor fluorophore (Cy5). The interaction between fluorophore-labeled MutS and the mis-match was studied (Fig. 4), as well as the rotational dynamics of MutS. This investigation provided insight into the MutS DNA binding conformation that had yet to be shown through structural analysis [39]. A smFRET approach called FRET TACKLE, where the dynamics of MutS-induced DNA bending of a GT-mismatch showed that the MutS-GT mismatch complex undergoes a con-formational transition between six different states, each with its own degree of DNA bending and lifetimes (see [133]). The results suggested that the efficiency of repair is directed by the dynamics of MutS-mismatch complex [39]. Additionally it was shown that ATP increased MutS diffusion, helping the protein to rotate freely around the DNA [40]. As the ATP depletes, the short-lived MutS lesion scanning becomes a highly stable MMR signaling clamp. These signaling clamps are capable of competing with chromatin and recruiting the MMR machinery [41]. An ingenious twist on the concept of FRET was employed to follow the translocation of the eukaryotic repair helicase XPD, by taking advantage of the fluorescence quenching effects of XPD’s intrinsic Fe–S cluster on Cy3 [42]. The Cy3 fluorophore was bound to DNA and as XPD translocated toward the Cy3, its fluorescence was quenched. The extent of quenching was proportional to the distance from the Fe–S cluster. Therefore the fluorescence intensity provided a marker for the translocation of the protein. This method also demonstrated that the translocation of XPD on ssDNA was differentially affected by the specific protein partners, suggesting that in some cases XPD utilizes hopping to bypass obstacles on the DNA [42], see also Spies [139].
smFRET has been used to study many protein interactions providing insights into the dynamics of biological systems. The method is constantly being modified and developed to improve data quality. Developments include using alternating laser excitation or ALEX to reject non-FRET background signals [43], offering exciting methods to study single protein-DNA complexes.
2.6. Atomic force microscopy
For many years atomic force microscopy (AFM) has been used extensively to image DNA-bound molecules on surfaces at high resolution to derive many properties such as DNA binding (see [136,140,141]), protein/DNA complex stoichiometries [133,140], mechanism of action [142] and specific binding locations (for a more extensive review see [44]).
AFM has proven invaluable in the study of DNA repair [45–49]. For example, UvrA and UvrAB, involved in NER, have been shown using AFM to preferentially bind to DNA ends on undamaged DNA substrates [45,46]. However, on substrates with nicked or damage sites, UvrA showed a preference for binding to the lesion site [45,46]. AFM also yields information about volumes which can be directly correlated to protein mass. For example UvrA has also been shown to form dimers, a process which is enhanced in the presence of ATP, probably due to conformational changes following hydrolysis [46]. We have recently shown that the human damage recognition protein which initiates NER, UV-DDB binds to DNA as a dimer, and about 17% of the cases bound as a dimer to two independent DNA molecules [50]. Furthermore, AFM was used to confirm observations from DNA tightrope experiments that UvrC binds to DNA alone and also in complex with UvrB; forming a novel UvrBC complex [32].
AFM can be combined with fluorescence imaging methods to provide both high resolution information of complex structure, and the spatial organization and stoichiometries of different proteins within a complex (see [141]). One such study combined AFM and TIRF imaging to observe UvrAB recognition of UV induced thymidine dimers on DNA using Qdots as fiducial markers for FIONA (fluorescence imaging with one nanometer accuracy) [47]. Similarly, the interaction of Rad54 with Rad51 nucleoprotein filaments involved in recombinational repair has been studied using a combined TIRF and SFM (scanning force microscopy) instrument [48]. Three types of Rad54 interaction with Rad51 filaments were observed; Rad54 at the end of a filament, Rad54 seemingly bridging multiple filaments together, and most commonly Rad54 interspersed along the filaments. All three interaction types were seen on filaments assembled on ss and dsDNA, but more (6 vs 1–4) Rad54 molecules bound along ssDNA-Rad51 filaments [48]. These observations have interesting implications for the role of Rad54 in Rad51 filament stabilization, disassembly and strand invasion.
2.7. Scanning near-field optical microscopy (SNOM): a future technique?
Due to diffraction, the wavelength of light provides a lower limit on the ability of a microscope to resolve objects. Unlike a visible light microscope SNOM is a surface probing technique that permits non-destructive imaging of surfaces with a resolution comparable to scanning electron microscopy [51,52]. This is made possible by the very close placement of the detector and illuminator to the surface being investigated; allowing for high spatial, spectral and temporal resolution. The technique works by funneling visible light through a small aperture at the end of an opaque probe, to illuminate an aperture sized region of the sample [53]. The resolution of the image is limited by the size of the detector aperture and not by the wavelength of the illuminating light [54,55]. The detector is also capable of simultaneously topographically mapping the surface much like AFM, thus providing even greater resolution. The resolution of SNOM lies between ~250nm (light microscopy) and 8 nm (FRET) [56]. This method therefore could be very useful in studying DNA repair proteins the future.
3. Manipulation - taking hold of the problem
Classically, to understand the kinetics and thermodynamics of a system, investigators alter various substrate, temperature and pressure dimensions. Single molecule biology permits these investigations, but also possess a crucial new dimension, force. From the very early ensemble level studies of muscle contraction, a connection between biochemistry and load was suggested [57]. However with the advent of single molecule manipulation, we were able to begin to understand the molecular origin of this phenomenon. By loading single molecules using optical traps, the effects of load could be explicitly located to specific biochemical steps during the catalytic cycle of muscle contraction [58,59]. Such load effects are also significant for DNA based systems, where working against a load is crucial to the role of specific proteins involved in various processes, such as transcription [14,60], and viral packaging where the most powerful molecular motors to date have been characterized [61,62]. The force dimension enables new insights, providing measurement of RNA polymerase motion with near base pair resolution [63], the effects of torque on formation of nucleoprotein structures [64,65], and the conformational changes that occur upon binding DNA [66]. Here we discuss some of the mechanisms of manipulating molecules.
3.1. Optical trapping - background
Optical trap mediated manipulation of DNA offers huge possibilities for single molecule investigation. Laser tweezers, also known as optical traps, are formed when a laser beam (usually infra-red) is brought to diffraction limited focus using a high numerical-aperture lens in the presence of micron sized dielectric objects such as silica beads. These beads interact with the beam, and due to the gradient force within the focused beam are then trapped near the beam focus [67] (Fig. 5; Movie 1). These provide physical handles that can be manipulated by moving the beam (Movie 2). In this simplest form coupling optical trapping with tethered particle motion (Fig. 6a and c) provides a simple approach to altering DNA tension by simply moving the trapped bead [68,69]. Multiple traps can be created using optical deflectors, movable mirrors or spatial light modulators [70–73]. Using just two traps, it is possible to create ‘dumbells’, where a single strand of DNA is stretched between two beads (Fig. 6b). This approach offers the unique ability to manipulate a single DNA molecule in three-dimensional space, demonstrated by the ability to tie a knot in DNA [47,74,75]. Furthermore, the isolation of the measurement system from the surface reduces environmental noise [76], making high resolution measurements down to the level of single base pair steps of RNA polymerase possible [63]. Optically trapped DNA can also be used in conjunction with microfluidics to change buffer composition, which is used to assemble protein complexes and alter substrate conditions rapidly [77–80]. The use of more than two traps in a large variety of positions allows more complex structures to be trapped, such as nanoprobes (see below).
Fig. 5.
The forces of gradient laser trap on a dielectric sphere in the ray optics regime. A focused laser beam attracts a dielectric sphere toward its waist. Repositioning of the laser beam results in repositioning of the trapped sphere.
Reprinted (adapted) with permission from [67]; see article for a full explanation. Copyright (2013) American Chemical Society.
Fig. 6.
Optical tweezer strategies in the study of DNA-protein interactions. Focused lasers (red cones) are used to trap beads (green) to which DNA can be attached. In (A) the other end of the bead is tethered to a surface permitting various amounts of tension to be applied to the DNA. (B) DNA is caught between two beads in a dumbbell configuration. This strategy allows for full three-dimensional position control of the bead, in addition to variable tension. (C) The DNA end shown in (A) can be brought toward the surface of a coverslip permitting TIRF illumination of the DNA for direct visualization of protein–DNA interactions.
3.1.1. Optical trapping in DNA repair
Laser tweezers have been used to study a range of DNA-based systems, including DNA translocases [62,81,82], DNA polymerases [66,83], RNA polymerases [63,68,84] and helicases [80,85,86]. These have revealed a number of important mechanistic facets and have led to a much deeper understanding of how these molecules interact with DNA. For example, measurements of RecBCD unwinding performed using optical tweezers revealed the rate of unwinding and the processivity, which agreed with previous data from ensemble studies [80]. However, further investigation of this process revealed pausing and backsliding activities that were found to be dependent on force applied to the DNA substrate [85]. Together these studies have led to an understanding of how these proteins find pause sites on the DNA and therefore produce the ssDNA tail required for homologous recombination. An important initial step is the formation of a RecA nucleoprotein filament on the ssDNA tail, created by the action of RecBCD, and unique insights into the dynamics of the nucleoprotein filament formation have been provided using laser tweezers in combination with surface-tethered dsDNA [87]. Further details revealed that RecA filament formation required 4–5 RecA molecules for nucleation, followed by the addition of 2 molecules at a time for bi-directional growth [75] Similarly, the eukaryotic homologue of RecA (Rad51), was found to form multiple stable nuclei consisting of 2–3 monomers [88]. In contrast to RecA, Rad51 did not continue to grow and form a filament from one nucleation event, rather multiple nuclei joined together to coat the DNA in Rad51 [65,75,88]. More recently using an imaging based approach, RecA filament formation has been studied on a more biologically relevant single stranded binding (SSB) protein coated ssDNA substrate [89]. On this substrate only 2 monomers were required for nucleation in contrast to the 5 required on dsDNA, presumably due to an increased affinity for ssDNA. However, consistent with previous data, growth was found to occur by the addition of monomers and could occur bi-directionally (although 5’–3’ growth was preferred) [75,89]. Addition of the mediator proteins RecOR was found to increase both nucleation and growth of filaments and it was suggested that this is achieved through weakening the interaction of SSB with the ssDNA [89]. Following filament formation, the nucleoprotein complex must find the complementary DNA sequence on the genome in order to undergo strand invasion, and ultimately repair the DNA. By labeling the nucleoprotein filament and changing the extension of a target DNA using optical tweezers, it has been elegantly shown that the RecA filament undergoes a 3D search for its homologous sequence. The filament was also found to bind more weakly to non-target sites suggesting that it interrogates multiple sites during its search [79].
Although assembly of nucleoprotein filaments is important so is their disassembly. Formation of Rad51 filaments is dynamic and aided by the binding of ATP, whereas ATP hydrolysis favors dissociation. This process of Rad51 filament disassembly has been observed using single molecule methods including laser tweezers. Increasing the tension applied to the DNA substrate or preventing ATP hydrolysis inhibited disassembly [65,88,90]. Van Memeren et al. observed bursts of Rad51 releasing from the DNA and have therefore suggested a model for filament disassembly akin to microtubule disassembly, whereby the terminal monomer in a filament hydrolyses ATP and releases; prompting adjacent monomers that have already undergone hydrolysis to release [90].
Recently, optical tweezers combined with TIRF imaging, “fleezers” have been used to study the translocase and helicase activities of the NER protein UvrD on ssDNA [91]. The optical tweezers were utilized to apply force to the DNA substrate in order to unzip secondary structures, which normally hinder measurements on ssDNA. Using particle tracking to monitor the position of a Cy3 labeled UvrD, it was found that the a UvrD monomer stopped at ssDNA/dsDNA junctions, and was unable to proceed to unwind the DNA unless joined by another monomer [91]. These results confirm an earlier study in which the fluorescence intensity was measured and monitoring the helicase activity versus the stepping of photobleaching, the authors propose that two monomers of UvrD are required for helicase activity.
Similarly, the eukaryotic NER protein, XPD, was found to undergo short, repetitive, non-processive burst of helicase activity when studied using optical tweezers [92]. This unwinding occurred in 1 bp steps, consisted of pauses and small backsteps, and could be enhanced by increasing the concentration of XPD, suggesting 2 or more monomers could act together to improve processivity [92]. Interestingly, the authors also observed large backsliding events, which they suggest correspond to the release of 5 bp of DNA normally held by the secondary binding site of the enzyme, allowing the substrate to reanneal. A model to encompass all data from this study suggests that XPD unwinds DNA in a ‘partially active’ manner, unwinding only the short sections of DNA required for its role in NER.
Laser tweezers can also be used to pull apart duplex DNA, permitting mismatch detection and can be used to detect whether proteins are bound to the DNA This method has been used to show that the MMR proteins Msh2–Msh6 bind preferentially to DNA mismatches in an orientation specific manner, without the need for other MMR factors [93]. Adding ATP to the Msh2–Msh6–DNA complex results in release of the proteins from the DNA, and by blocking the DNA ends, a fraction of these proteins could be trapped on the substrate. This suggests they are capable of sliding along the DNA as well as simply releasing [93].
3.1.2. Optically trapped nanoprobes
The use of optical traps is not restricted to simple spherical objects like the ones used in many of the studies highlighted above. Recently it has been shown that cylindrical objects can be trapped, and if these objects are ordered crystals they respond to the polarity of the trapping beam, enabling trap rotation [94–96]. This offers the possibility of studying twist and force simultaneously, in a similar manner to magnetic tweezers (see below). By time-sharing the laser beam to generate multiple traps simultaneously, more complex structures can be controlled, ranging from diatoms [97] to nanofabricated structures [98–100]. So far these structures have only been used for proof of principle experiments involving the characterization of the trapping, simple manipulations and interrogation of test substrates. We have recently begun using such structures in conjunction with DNA tightropes to understand the physical nature of the protein-DNA interaction. These structures offer the potential to be transformative in the way we probe protein–DNA interactions, particularly as they are combined with other technologies.
3.2. Magnetic tweezers
A technologically simpler alternative to directly manipulate single molecules, involves the use of magnetic beads attached to DNA. Using magnetic tweezers, a single molecule of DNA tethered between a surface and a paramagnetic bead can be manipulated by external magnets (Fig. 7). This enables the DNA to be stretched or relaxed, and also twisted, to create different DNA topologies [101]. Indeed this approach has been used to study properties, topology and the formation of DNA plectonemes (DNA supercoils) [102,103]. Magnetic tweezers certainly have the advantage over optical tweezers, in that they can easily be multiplexed, i.e. multiple DNA bound beads can be manipulated and visualized at one time. However, magnetic tweezers are limited to being able to manipulate all the beads isotropically, unlike laser tweezers which can move each trap independently.
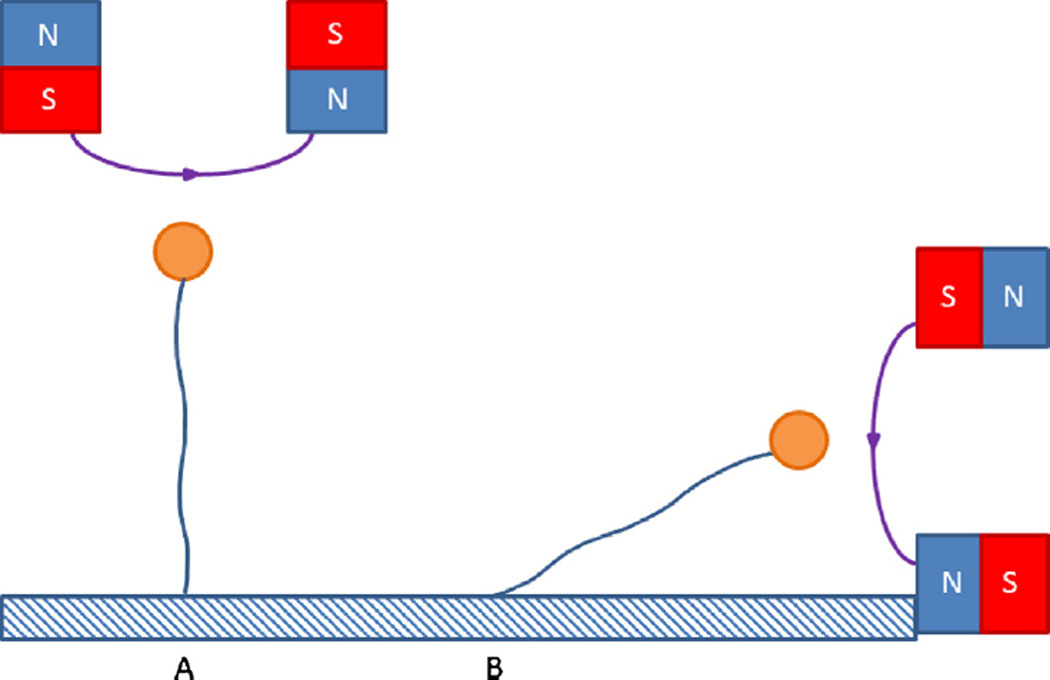
Fig. 7.
Magnetic tweezer strategies. (A) DNA (blue) is tethered between a surface and paramagnetic beads (orange) attracted toward the magnets. By rotating the magnets twist can be applied to the DNA forming plectonemes which in turn allow for very high resolution measurements of changes in the DNA structure. (B) In this strategy the magnet can be manipulated to bring DNA close to the surface and can therefore be used in conjunction with TIRF imaging. This has an advantage over laser tweezers in that all beads in the visual field will behave similarly, permitting multiplexing.
Magnetic tweezers have been used to study topoisomerases [104], polymerases [105], restriction enzymes [64,106] and helicases involved in DNA repair [107–109] amongst others. One such study investigated UvrD unwinding rate, lifetime, and base pairs unwound. From this data, the protein’s step size was estimated as ~6bp [107]. Following unwinding, the authors were also able to observe rehybridization of the DNA strands that was also dependent on enzyme translocation, which was attributed to strand switching of the enzyme.
Another helicase, AddAB helicase-nuclease is involved in DNA end processing in recombinational repair. As such, it degrades DNA ends until it recognizes a Chi sequence, when it stops degrading the 3′ strand but continues unwinding and degrading the 5′ strand, thereby creating a ssDNA overhang, which can be further processed to repair the DNA. This has recently been demonstrated at the single molecule level using magnetic tweezers [109]. In this study the authors observed pausing at Chi and Chi-like sequences and therefore propose a model for Chi recognition, whereby the enzyme pauses and undergoes a multistep conformational change involving the extrusion of a ssDNA loop, before resuming translocation after the Chi site [109]. This mechanism of target site search may be a general mechanism employed by many DNA binding proteins, whereby multiple weak binding events, to sites similar to the target, occur before a more stable protein-target complex is formed.
In another recent study described in more detail within this Special Issue [143], the first stage of transcription coupled DNA repair where a stalled RNA polymerase elongation complex is displaced by Mfd was investigated [108]. Using magnetic tweezers it was possible to observe transcription initiation, formation of the elongation complex and termination, as well as stalled elongation complexes formed by the introduction of damage or by nucleotide starvation. This allowed for the observation of a long lived stalled RNAP-Mfd complex that required ATP hydrolysis to remove the polymerase.
As with optical tweezers [91], magnetic tweezers can also be combined with fluorescence imaging to gain insight into forces and torque of the substrate, as well as location and movement of proteins bound to the DNA [110]. This is likely to represent a future development of the technique.
4. Studying DNA repair in vivo–cells sell
As the available technology develops, a deeper understanding of reconstituted complex systems in vitro can be achieved. A logical step therefore would be to begin studying these systems in vivo. The process of imaging single molecules in vivo has been made significantly easier by the discovery and development of fluorescent proteins (FPs) [111]. For imaging in live bacteria, FPs offer the simplest approach, since microinjection and endocytosis are not options given the size and behavior of these cells. Most FPs share a similar architecture; they are a β-barrel protein with a triad of residues that form the fluorescent core of the molecule (see Pymol session GFP.pse PDB: 2AWK). They are large compared to organic fluorophores, such as fluorescein, at ~25 kDa versus 1 kDa in size [112]. However, unlike organic dyes, FPs can be genetically encoded to a protein of interest thus providing 100% labeling in a wide possible spectrum of colors [113]. This not only permits imaging of single proteins within cells, but also enables multicolor labeling of different single molecules, aiding tracking and analysis of protein movements and interactions [114].
The key challenges in using FPs in cells are their brightness and solubility, as they are subject to photobleaching and often aggregate. Mutant FP’s are continually being developed to alter the extinction coefficient and the quantum yield, which in turn will affect the brightness and photostability, creating a more desirable fluorophore [115,116]. FPs are still widely used in cell biology to follow protein localization and gene expression using fluorescence microscopy [117]. However, cellular autofluorescence means the FP needs to be very bright, but tandem FP repeats can help to overcome this [118,119]. Conversely, if one is investigating single molecules having multiple FPs attached to the protein of interest can be undesirable. Therefore, several alternative strategies have been developed to distinguish single molecules within the dense population of a cell. For example an YFP variant, Venus, has been used to study gene expression. The low photostability of the FP allows for quick reduction in background fluorescence, so that upon expression and maturation of a new Venus molecule, a bright spot is detected [120]. When studying specific binding of transcription factors to DNA, a more photostable FP is advised. Different illumination strategies offer mechanisms to determine the residence time and position of a transcription factor on DNA. Since DNA is relatively immobile, any discrete fluorescent spots over a 1 s exposure would indicate DNA binding [118,121]. However, to detect faster motion, stroboscopic illumination can be used i.e. flashing the illumination at high intensity for a short period with the camera shutter open; this allows a snapshot of the protein’s movement within the cell [121].
In addition to standard FPs, further fluorescent protein developments include photoconvertible FPs. These fluorophores can be modulated to become fluorescently active or switch emission wavelength by exposure to a wavelength distinct from their excitation maximum. Such behavior is useful for single molecule cellular imaging, since single molecules can be isolated within a dense population of molecules [122]. This can be achieved using a technique known as photoactivated localization microscopy (PALM), which uses widefield fluorescence microscopy to visualize single molecules sequentially within a pool. Molecules are activated and then their position determined with high precision, by fitting a 2D Gaussian approximation of the point spread function, to the fluorescent spot profile before they photobleach. This technique has been extensively used to generate super-resolution images [123]. The main challenge in achieving super-resolution is overcoming the diffraction of light, which limits resolving capabilities to ~250nm in the x-y direction, PALM resolves at ~25 nm. 3D PALM can also be achieved by extending the resolution to the z-axis, using a custom 3 way beam splitter in a multiphase interferometric microscope [124]. PALM benefits from the use of FP probes, which are superior in identifying single molecules at high densities, aiding the study of their localization and kinetics [122,125].To address the issue of flu-orophore blinking a “clustering algorithm” is used group together spots identified to generate a super-resolution image [123].
In a recent study (discussed in more detail in this Special Issue [144]), PAFP mCherry was used to explore DNA polymerase I and DNA ligase activity in vivo during base excision repair [126]. Their use of PALM overcomes the limitations described above for single molecule detection within cells, by providing a constant source of newly activated molecules. mCherry is excited at 587 nm and emits at 610 nm and photoactivation was achieved using a continual low dose exposure to a 405 nm laser. This study represents a significant step forwards in following DNA repair in live bacteria in real time.
As mentioned previously the use of Qdots offers unprecedented levels of fluorescence emission and photostability. However, while Qdots are strongly resistant to photobleaching, they are liable to blink, which can be useful for determining the presence of a single molecule, but this also creates populations of Qdots with non-linear scaling of fluorescence with quantum dot number, complicating analysis [127]. Furthermore, introducing Qdots into cells is problematic often leading to aggregate formation as the Qdots are cycled to endosomes [128]. However in some cases this aggregation can be useful, enabling endosome transportation along microtubules to be tracked, unaffected by blinking due to the Qdot fluorescent integral overlap. Such studies have provided useful insights into molecular motor stepping. The displacement traces of endocytosed Qdots showed step sizes of 8 nm expected for kinesin, but also of 16 and 24 nm steps. This could be explained by unresolved multiple motors acting on a single cargo, highlighting an interesting facet of the complexity seen in vivo. This reinforces the importance and complexity of studying single molecules in cells. There are many more methods to introduce Qdots into cells to which we refer the reader to another review [118].
Overall, there are advantages for the use of either intrinsic or extrinsic labels. Due to their ease of use, FPs have been more popular, but, their size, photolability and brightness are distinct setbacks. Extrinsic labels such as Qdots are highly attractive due to their brightness, photostability and distinct emission profiles, which in turn allow for longer, faster and more accurate visualization. However, their size and toxicity may be problematic; although the biggest hurdle for Qdots is to get them into cells and specifically target a protein. Once this is achieved, the scope for investigation in cellulo will become a significantly more attainable goal.
5. A bright future for single molecule biology
In the context of DNA repair, what is to be learned using single molecule approaches? We have described the advantages of using single molecule detection approaches, from the perspective of obtaining an insight into mechanism and dynamics. It is also possible to use the multiplexing methods outlined above to measure the true stoichiometry and dynamics of transiently forming complexes during catalysis [126,129]. It is clear that many DNA repair processes are mediated by nano-machines which form at sites of repair through interacting protein partners. Thus these single-molecule techniques will allow us to directly watch the comings and goings of specific repair proteins. Furthermore, one can learn how proteins function together to hand-off specific DNA repair intermediates to the next proteins involved in the repair process. The classic observation of Riggs [130] that lac repressor could find its target faster than diffusion led to the concept of facilitated diffusion [26]. This process could occur by multiple mechanisms, but the over-riding concept is that a protein binds to a non-specific site on DNA and then uses the DNA to help guide it to its specific site. Despite many attempts at understanding this using ensemble approaches [131] it is still unclear how proteins find their targets on DNA. Do they slide to their target sites? Or perhaps jump? Or instead use a combination of both of these, or even switch between DNA regions using intersegmental transfer? Any of these mechanisms and their combinations could potentially underlie facilitated diffusion and single molecule approaches are poised to answer this long held question. Complicating this is that different proteins may utilize different strategies as highlighted by some of the studies in this special issue [20,24,32]. Another important aspect to consider as we transition into studying more complex systems is that of crowding, an Escherichia coli cell is an enormously crowded environment (Fig. 8). This crowding occurs both in solution and on DNA itself.
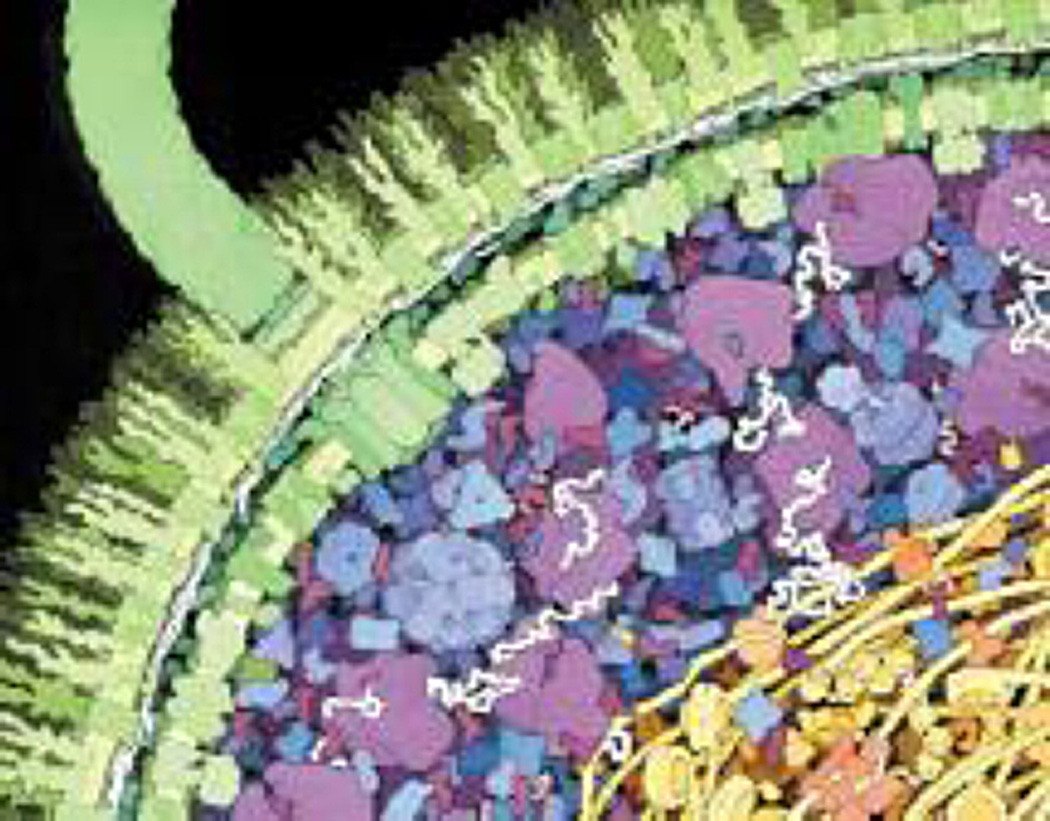
Fig. 8.
An artistic representation of the huge amount of crowding within an E. coli cell. Adapted from [132]. When studying the behavior of molecular systems, one cannot forget that cells are very crowded, how well do normal buffer solutions mimic this situation?
The future of single molecule investigation for complex processes such as that found during DNA repair is certainly very positive. As more groups begin to use these techniques their ubiquity will drive the development of fresh ideas and approaches. However, the importance of understanding the fundamental principles of Nature’s design using in vitro methods for the simpler prokaryotic systems should not be overlooked as we strive toward understanding DNA repair at the single molecule level in vivo. We are hopeful that this Special Issue will inform and inspire such work.
Supplementary Material
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2014.02.003.
References
- 1.Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260:799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends in Microbiology. 2007;15:45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman and Co; 1998. [Google Scholar]
- 4.Saurabh S, Maji S, Bruchez MP. Evaluation of sCMOS cameras for detection and localization of single Cy5 molecules. Optics Express. 2012;20:7338–7349. doi: 10.1364/OE.20.007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulrich G, Ziessel R, Harriman A. The chemistry of fluorescent bodipy dyes: versatility unsurpassed. Angewandte Chemie International Edition. 2008;47:1184–1201. doi: 10.1002/anie.200702070. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling C, Widengren J, Rigler R, Seidel CAM. Photobleaching of fluorescent dyes under conditions used for single-molecule detection: evidence of two-step photolysis. Analytical Chemistry. 1998;70:2651–2659. doi: 10.1021/ac980027p. [DOI] [PubMed] [Google Scholar]
- 7.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 8.Ha T, Tinnefeld P. Photophysics of fluorescent probes forsingle-molecule biophysics and super-resolution imaging. Annual Review of Physical Chemistry. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan WCW, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 10.Alivisatos AP. Naturally aligned nanocrystals. Science. 2000;289:736–737. doi: 10.1126/science.289.5480.736. [DOI] [PubMed] [Google Scholar]
- 11.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 12.Bruchez MP. Quantum dots find their stride in single molecule tracking. Current Opinion in Chemical Biology. 2011;15:775–780. doi: 10.1016/j.cbpa.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kad NM, Van Houten B. Dynamics of lesion processing by bacterial nucleotide excision repair proteins. In: Paul WD, editor. Progress in Molecular Biology and Translational Science. Academic Press; 2012. pp. 1–24. Chapter 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, Landick R, Gelles J. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophysical Journal. 1994;67:2468–2478. doi: 10.1016/S0006-3495(94)80735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schafer DA, Gelles J, sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:4. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 16.Monico C, Capitanio M, Belcastro G, Vanzi F, Pavone F. Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level. International Journal of Molecular Sciences. 2013;14:3961–3992. doi: 10.3390/ijms14023961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohoney KM, Gelles J. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 18.Fan H-F, Li H-W. Studying RecBCD helicase translocation along X-DNA using tethered particle motion with a stretching force. Biophysical Journal. 2009;96:1875–1883. doi: 10.1016/j.bpj.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pouget N, Dennis C, Turlan C, Grigoriev M, Chandler M, Salomé L. Single-particle tracking for DNA tether length monitoring. Nucleic Acids Research. 2004;32:e73. doi: 10.1093/nar/gnh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Molecular Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allemand JF, Bensimon D, Jullien L, Bensimon A, Croquette V. pH-dependent specific binding and combing of DNA. Biophysical Journal. 1997;73:2064–2070. doi: 10.1016/S0006-3495(97)78236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon WA, Fang MM, Haskins WE, Nie S. A dual-beam optical microscope for observation and cleavage of single DNA molecules. Analytical Chemistry. 1998;70:1743–1748. doi: 10.1021/ac980040+. [DOI] [PubMed] [Google Scholar]
- 23.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of {DNA} by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 24.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Physical Review Letters. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 26.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 27.Granéli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granéli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2005;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 29.Fazio T, Visnapuu M-L, Wind S, Greene EC. DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorman J, Plys A, Visnapuu JM, Alani E, Greene CE. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nature Structure Molecular Biology. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Redding S, Finkelstein IJ, Gorman J, Reichman DR, Greene EC. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nature Structure Molecular Biology. 2013;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes CD, Wang H, Ghodke H, Simons M, Towheed A, Peng Y, Van Houten B, Kad NM. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Research. 2013;41:4901–4912. doi: 10.1093/nar/gkt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Research. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant Journal. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, Countryman P, Buncher N, Kaur P, Zhang LEY, Gibson G, You C, Watkins SC, Piehler J, L Opresko P, Kad NM, Wang H. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Research. 2014;42(4):2493–2504. doi: 10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ, Kapani-dis AN. Defining the limits of single-molecule FRET resolution inTIRF microscopy. Biophysical Journal. 2010;99:3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sass LE, Lanyi C, Weninger K, Erie DA. Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry. 2010;49:3174–3190. doi: 10.1021/bi901871u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho W-K, Jeong C, Kim D, Chang M, Song K-M, Hanne J, Ban C, Fishel R, Lee J-B. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20:1264–1274. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong C, Cho W-K, Song K-M, Cook C, Yoon T-Y, Ban C, Fishel R, Lee J-B. MutS switches between two fundamentally distinct clamps during mismatch repair. Nature Structure Molecular Biology. 2011;18:6. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Molecular Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH, Weiss S. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophysical Journal. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buechner CN, Tessmer I. DNA substrate preparation for atomic force microscopy studies of protein-DNA interactions. Journal of Molecular Recognition. 2013;26:605–617. doi: 10.1002/jmr.2311. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Tessmer I, L Croteau D, Erie DA, Van Houten B. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Letters. 2008;8:1631–1637. doi: 10.1021/nl080316l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner K, Moolenaar G, van Noort J, Goosen N. Single-molecule analysis reveals two separate DNA-binding domains in the Escherichia coli UvrA dimer. Nucleic Acids Research. 2009;37:1962–1972. doi: 10.1093/nar/gkp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fronczek DN, Quammen C, Wang H, Kisker C, Superfine R, Taylor R, Erie DA, Tessmer I. High accuracy FIONA-AFM hybrid imaging. Ultramicroscopy. 2011;111:350–355. doi: 10.1016/j.ultramic.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez H, Kertokalio A, van Rossum-Fikkert S, Kanaar R, Wyman C. Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proceedings of the National Academy of Sciences USA. 2013;110:11385–11390. doi: 10.1073/pnas.1306467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buechner CN, Heil K, Michels G, Carell T, Kisker C, Tessmer I. Strand specific recognition of DNA damages by XPD provides insights into Nucleotide Excision Repair substrate versatility. Journal of Biological Chemistry. 2014;289(6):3613–3624. doi: 10.1074/jbc.M113.523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh JI, Levine AS, Du S, Chinte U, Ghodke H, Wang H, Shi H, Hsieh CL, Conway JF, Van Houten B, Rapic-Otrin V. Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair. Proceedings of the National Academy of Sciences USA. 2012;109:E2737–E2746. doi: 10.1073/pnas.1110067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tisler J, Oeckinghaus T, Stöhr RJ, Kolesov R, Reuter R, Reinhard F, Wrachtrup J. Single defect center scanning near-field optical microscopy on graphene. Nano Letters. 2013;13:3152–3156. doi: 10.1021/nl401129m. [DOI] [PubMed] [Google Scholar]
- 52.Wabuyele MB, Culha M, Griffin GD, Viallet PM, Vo-Dinh T. Near-field scanning optical microscopy for bioanalysis at nanometer resolution. Methods in Molecular Biology. 2005;300:437–452. doi: 10.1385/1-59259-858-7:437. [DOI] [PubMed] [Google Scholar]
- 53.Betzig E, Chichester RJ. Single molecules observed by near-field scanning optical microscopy. Science. 1993;262:1422–1425. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- 54.Durig U, Pohl DW, Rohner F. Near-field optical-scanning microscopy. Journal of Applied Physics. 1986;59:3318–3327. [Google Scholar]
- 55.Oshikane Y, Kataoka T, Okuda M, Hara S, Inoue H, Nakano M. Observation of nanostructure by scanning near-field optical microscope with small sphere probe. Science and Technology of Advanced Materials. 2007;8:181. [Google Scholar]
- 56.Betzig E, Lewis A, Harootunian A, Isaacson M, Kratschmer E. Near field scanning optical microscopy (NSOM): development and biophysical applications. Biophysical Journal. 1986;49:269–279. doi: 10.1016/S0006-3495(86)83640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fenn WO. A quantitative comparison between the energy liberated and the work performed by the isolated sartorius muscle of the frog. Journal of Physiology. 1923;58:175–203. doi: 10.1113/jphysiol.1923.sp002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nature Cell Biology. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 59.Kad NM, Patlak JB, Fagnant PM, Trybus KM, Warshaw DM. Mutation of a conserved glycine in the SH1-SH2 helix affects the load-dependent kinetics of myosin. BiophysicalJournal. 2007;92:1623–1631. doi: 10.1529/biophysj.106.097618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gelles J, Yin H, Wong O, Landick KR. Single-molecule kinetic studies on DNA transcription and transcriptional regulation. Biophysical Journal. 1995;68:1. [PMC free article] [PubMed] [Google Scholar]
- 61.Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage φ29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 62.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 63.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seidel R, Bloom JGP, van Noort J, Dutta CF, Dekker NH, Firman K, Szczelkun MD, Dekker C, Dynamics of initiation. termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO Journal. 2005;24:4188–4197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Research. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 67.Ashkin A. Forces of a single-beam gradient laser trap on a dielectric sphere in the ray optics regime. Biophysical Journal. 1992;61:569–582. doi: 10.1016/S0006-3495(92)81860-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophysical Journal. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Günther K, Mertig M, Seidel R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Research. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visscher K, Gross SP, Block SM. Construction of multiple-beam optical traps with nanometer-resolution position sensing. IEEE Journal of Selected Topics in Quantum Electronics. 1996;2:1066–1076. [Google Scholar]
- 71.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO Journal. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fällman E, Axner O. Design for fully steerable dual-trap optical tweezers. Applied Optics. 1997;36:2107–2113. doi: 10.1364/ao.36.002107. [DOI] [PubMed] [Google Scholar]
- 73.Grier DG. A revolution in optical manipulation. Nature. 2003;424:6. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 74.Arai Y, Yasuda R, Akashi K-I, Harada Y, Miyata H, Kinosita K, Itoh H. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 75.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 76.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proceedings of the National Academy of Sciences USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noom MC, van den Broek B, van Mameren J, Wuite GJL. Visualizing single DNA-bound proteins using DNA as a scanning probe. Nature Methods. 2007;4:1031–1036. doi: 10.1038/nmeth1126. [DOI] [PubMed] [Google Scholar]
- 78.Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation ofthe same DNA molecule. Science. 1999;286:120–123. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- 79.L Forget A, Kowalczykowski SC. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012;482:423–427. doi: 10.1038/nature10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 81.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage [phis]29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 82.Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification ofthe FtsK sequence-recognition domain. Nature Structure Molecular Biology. 2006;13:1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- 83.Ibarra B, Chemla YR, Plyasunov S, Smith SB, Lazaro JM, Salas M, Bustamante C. Proofreading dynamics of a processive DNA polymerase. EMBO Journal. 2009;28:2794–2802. doi: 10.1038/emboj.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proceedings of the National Academy of Sciences USA. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perkins TT, Li H-W, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Biophysical Journal. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proceedings of the National Academy of Sciences USA. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proceedings of the National Academy of Sciences USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee KS, Balci H, Jia H, Lohman TM, Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nature Communications. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qi Z, Pugh RA, Spies M, Chemla YR. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2:e00334. doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by Single-molecule unzipping force analysis. Molecular Cell. 2005;20:771–781. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 94.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nature Methods. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 95.Inman J, Forth S, Wang MD. Passive torque wrench and angular position detection using a single-beam optical trap. Optics Letters. 2010;35:2949–2951. doi: 10.1364/OL.35.002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olof SN, Grieve JA, Phillips DB, Rosenkranz H, Yallop ML, Miles MJ, Patil AJ, Mann S, Carberry DM. Measuring nanoscale forces with living probes. Nano Letters. 2012;12:6018–6023. doi: 10.1021/nl303585w. [DOI] [PubMed] [Google Scholar]
- 98.Pollard MR, Botchway SW, Chichkov B, Freeman E, Halsall RNJ, Jenkins DWK, Loader I, Ovsianikov A, Parker AW, Stevens R, Turchetta R, Ward AD, Towrie M. Optically trapped probes with nanometer-scale tips for femto-Newton force measurement. New Journal of Physics. 2010;12:113056. [Google Scholar]
- 99.Palima D, Bañas AR, Vizsnyiczai G, Kelemen L, Ormos P, Glückstad J. Wave-guided optical waveguides. Optics Express. 2012;20:2004–2014. doi: 10.1364/OE.20.002004. [DOI] [PubMed] [Google Scholar]
- 100.Phillips DB, Gibson GM, Bowman R, Padgett MJ, Hanna S, Carberry DM, Miles MJ, Simpson SH. An optically actuated surface scanning probe. Optics Express. 2012;20:29679–29693. doi: 10.1364/OE.20.029679. [DOI] [PubMed] [Google Scholar]
- 101.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 102.Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 103.Crut A, Koster DA, Seidel R, Wiggins CH, Dekker NH. Fast dynamics of supercoiled DNA revealed by single-molecule experiments. Proceedings of the National Academy of Sciences USA. 2007;104:11957–11962. doi: 10.1073/pnas.0700333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 105.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proceedings of the National Academy of Sciences USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwarz FW, Tóth J, van Aelst K, Cui G, Clausing S, Szczelkun MD, Seidel R. The helicase-like domains of type III restriction enzymes trigger long-range diffusion along DNA. Science. 2013;340:353–356. doi: 10.1126/science.1231122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dessinges M-N, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proceedings of the National Academy of Sciences USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, Darst SA, Savery NJ, Strick TR. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carrasco C, Gilhooly NS, Dillingham MS, Moreno-Herrero F. On the mechanism of recombination hotspot scanning during double-stranded DNA break resection. Proceedings of the National Academy of Sciences USA. 2013;110:E2562–E2571. doi: 10.1073/pnas.1303035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Graham JS, Johnson RC, Marko JF. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Research. 2011;39:2249–2259. doi: 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 112.Kremers G-J, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. Journal of Cell Science. 2011;124:157–160. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Valledor M, Hu Q, Schiller P, Myers RS. Fluorescent protein engineering by in vivo site-directed mutagenesis. IUBMB Life. 2012;64:684–689. doi: 10.1002/iub.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiological Reviews. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 115.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophysical Journal. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chemical Society Reviews. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: fluorescent protein structure and color variants. Cold Spring Harbor Protocols. 2009 doi: 10.1101/pdb.top63. pdb.top63. [DOI] [PubMed] [Google Scholar]
- 118.Xie XS, Choi PJ, Li G-W, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annual Review of Biophysics. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 119.Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 121.Elf J, Li G-W, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolu-tion imaging using single-molecule localization. Annual Review of Physical Chemistry. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coltharp C, Kessler RP, Xiao J. Accurate construction of photoactivated localization microscopy (PALM) images for quantitative measurements. PLoS ONE. 2012;7:e51725. doi: 10.1371/journal.pone.0051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proceedings of the National Academy of Sciences USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Galbraith CG, Galbraith JA. Super-resolution microscopy at a glance. Journal of Cell Science. 2011;124:1607–1611. doi: 10.1242/jcs.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uphoff S, Reyes-Lamothe R, Garza de Leon F, Sherratt DJ, Kapani-dis AN. Single-molecule DNA repair in live bacteria. Proceedings of the National Academy of Sciences USA. 2013;110:8063–8068. doi: 10.1073/pnas.1301804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walling M, Novak J, Shepard JRE. Quantum dots for live cell and in vivo imaging. International Journal of Molecular Sciences. 2009;10:441–491. doi: 10.3390/ijms10020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nan X, Sims PA, Xie XS. Organelle tracking in a living cell with microsecond time resolution and nanometer spatial precision. ChemPhysChem. 2008;9:707–712. doi: 10.1002/cphc.200700839. [DOI] [PubMed] [Google Scholar]
- 129.Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Riggs AD, Suzuki H, Bourgeois S. The lac repressor-operator interaction: I. Equilibrium studies. Journal of Molecular Biology. 1970;48:67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- 131.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Research. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends in Biochemical Sciences. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 133.Erie DA, Weninger KR. Single molecule Studies of DNA Mismatch Repair. DNA Repair. 2014;20:71–81. doi: 10.1016/j.dnarep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee J-B, Cho W-K, Park J-H, Jeon Y-M, Kim D-Y, Lee H, Fishel R. Single-molecule Views of MutS on Mismatched DNA. DNA Repair. 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Silverstein TD, Gibb B, Greene EC. Visualizing Protein Movement on DNA at the Single-molecule Level using DNA Curtains. DNA Repair. 2014;20:94–109. doi: 10.1016/j.dnarep.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Houten B, Kad N. Investigation of bacterial nucleotide excision repair using single-molecule techniques. DNA Repair. 2014;20:41–48. doi: 10.1016/j.dnarep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee AJ, Wallace DM. Insights into the glycosylase search for damage from single-molecule fluorescence microscopy. DNA Repair. 2014;20:23–31. doi: 10.1016/j.dnarep.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gubaev A, Klostermeier D. The mechanism of negative DNA supercoiling: A cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair. 2014;20:130–141. doi: 10.1016/j.dnarep.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 139.Spies M. Two steps forward, one step back: determining XPD helicase mechanism by single-molecule fluorescence and high-resolution optical tweezers. DNA Repair. 2014;20:8–70. doi: 10.1016/j.dnarep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin J, Kaur P, Countryman P, Opresko PL, Wang H. Unraveling secrets of telomeres: one molecule at a time. DNA Repair. 2014;20:142–153. doi: 10.1016/j.dnarep.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanchez H, Reuter M, Yokokawa M, Takeyasu K, Wyman C. Taking it one step at a time in homologous recombination repair. DNA Repair. 2014;20:110–118. doi: 10.1016/j.dnarep.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 142.Tessmer I, Fried MG. Insight into the cooperative DNA binding of the O6-alkylguanine DNA alkyltransferase. DNA Repair. 2014;20:14–22. doi: 10.1016/j.dnarep.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Howan K, Monnet J, Fan J, Strick TR. Stopped in its tracks: the RNA polymerase molecular motor as a robust sensor of DNA damage. DNA Repair. 2014;20:49–57. doi: 10.1016/j.dnarep.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 144.Uphoff S, Kapinidis AN. Studying the organization of DNA repair by single-cell and single-molecule imaging. DNA Repair. 2014;20:32–40. doi: 10.1016/j.dnarep.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.