Abstract
De novo experimental drug discovery is an expensive and time-consuming task. It requires the identification of drug-target interactions (DTIs) towards targets of biological interest, either to inhibit or enhance a specific molecular function. Dedicated computational models for protein simulation and DTI prediction are crucial for speed and to reduce the costs associated with DTI identification. In this paper we present a computational pipeline that enables the discovery of putative leads for drug repositioning that can be applied to any microbial proteome, as long as the interactome of interest is at least partially known. Network metrics calculated for the interactome of the bacterial organism of interest were used to identify putative drug-targets. Then, a random forest classification model for DTI prediction was constructed using known DTI data from publicly available databases, resulting in an area under the ROC curve of 0.91 for classification of out-of-sampling data. A drug-target network was created by combining 3,081 unique ligands and the expected ten best drug targets. This network was used to predict new DTIs and to calculate the probability of the positive class, allowing the scoring of the predicted instances. Molecular docking experiments were performed on the best scoring DTI pairs and the results were compared with those of the same ligands with their original targets. The results obtained suggest that the proposed pipeline can be used in the identification of new leads for drug repositioning. The proposed classification model is available at http://bioinformatics.ua.pt/software/dtipred/.
Author Summary
The emergence of multi-resistant bacterial strains and the existing void in the discovery and development of new classes of antibiotics is a growing concern. Indeed, some bacterial strains are now resistant to last-line antibiotics and considered untreatable. Drug repositioning has been suggested as a strategy to minimize time and cost expenses until the drug reaches the market, compared to traditional drug design. Drug-target interactions (DTIs) are the basis of rational drug design and thus, we proposed a computational approach to predict DTIs solely based on the primary sequence of the protein and the simplified molecular-input line-entry system of the ligand. In addition, network metrics are used to identify vital putative drug-targets in bacteria. Molecular docking experiments were performed to compare the binding affinities between a given ligand and a putative drug-target, as well as with their original targets. According to the docking results, the predicted DTIs have better or similar binding activities than the ligand and their real target, indicating the validity of the proposed model.
Introduction
Antibacterial resistance is becoming more frequent and is a growing concern, as bacterial resistance to last-line antibiotics has been steadily increasing and is already high globally [1,2]. Development of antibacterial resistance is the result of a cascade of events triggered by continued selective pressure of routinely used antibiotics, constituting a major medical and pharmaceutical challenge. In response to continued selective pressure the bacterial genome undergoes rapid evolution, which in turn is accelerated by the heavy focus on the same microbial pathways (protein synthesis, nucleic acid synthesis, cell wall synthesis and folate synthesis) [3,4]. Today, more than ever, new antibiotics or prodrugs able to neutralize antimicrobial resistant pathogens are necessary.
A growing strategy in drug screening for the past decade is drug repositioning, or repurposing. By focusing on one of the undesired effects of an already commercialized drug in an attempt to make it the main effect, it is possible to reposition that drug for new uses [5]. This strategy can greatly reduce the cost of lead screening and the time required for a drug to reach the market [6,7]. Some examples of successfully repositioned drugs for uses different from their original indications include bupropion, fluoxetine, thalidomide and sildenafil [8,9]. Sildenafil is probably the most popular example, which was initially used to treat hypertension, then angina, and currently for erectile dysfunction [10]. However, the repurposing of thalidomide should not be taken lightly, as it is an example of a withdrawn drug that could be reintroduced in the market [11]. From another perspective, neglected and rare diseases are also becoming increasingly attractive for pharmaceutical companies, which can be partially attributed to the smaller initial investment necessary to repurpose drugs for such diseases [9].
This was on the basis of the proposed works by Cheng et al. [12] and Yang et al. [13]. Yang et al. [13] developed the conditional random field (CRF) method, which integrates genomic, chemical, functional and pharmacological data, in addition to the topology of DTI networks. The CRF is a probabilistic graph model able to encode the drug-target network for DTI prediction. They apply a stochastic gradient ascent approach and the contrastive divergence algorithm to train their model and to identify the hidden associations between drugs and targets [13]. While this methodology may have the potential to be applied to reposition certain drugs, the use of functional similarity dismisses its use for the drug repositioning in infectious diseases. The most likely result of using functional similarity for this purpose would be the prediction of an antibacterial drug that would continue the selective pressure in the target microorganism, perpetuating antibacterial resistance.
The work by Cheng et al. [12] consisted in the construction of a bipartite graph of drugs approved by the United States Food and Drug Administration linked by binary associations to their respective protein targets to infer drug-target interactions (DTI). Their proposed method, network-based inference (NBI), uses known drug-target bipartite network topology similarity to predict unknown DTI. NBI considers transition processes over the bipartite graph, and thus, if a drug and a protein target interact, it is possible to compute the predictive score. The predictive score is calculated based on the number of drugs associated with each target, and on the degree of these drugs. Some of their predictions were validated by in vitro assays, confirming that five drugs had pharmacological effects on alternative targets [12]. While these results are very promising for drug repurposing, we believe their proposed methodology could not be used to solve the problem present here. Considering that Cheng et al. [12] used a DTI network composed of human protein targets and their respective drugs to construct their inference model, if they used a DTI network of a bacterial species of interest and the drugs targeting it (i.e., antibiotic drugs), the selective pressure problem would persist and result in antibacterial resistance.
Drug repositioning is especially challenging in infectious diseases for a number of reasons. First, most antibiotics were originally isolated by screening soil-derived Actinomycetes between 1940 and 1960 [14]. Shortly after, the productivity of this antibiotic discovery strategy started decaying rapidly, becoming obsolete. Second, the antibacterial “spectrum expansion” methodology, which consists of testing a drug able to suppress one bacterium species against other species [15], is too expensive and time-consuming. In addition, despite high-throughput screening against defined targets and rational drug design yielding several compounds, the compounds identified were not effective at penetrating bacterial cells [14]. Nonetheless, efforts to reposition drugs for infectious diseases are becoming increasingly attractive, especially those using computational methodologies [16,17]. Computational methodologies allow rapid and inexpensive screening of a broad spectrum of drugs and targets, either by screening ligands for a certain drug-target, or screening potential drug-targets for a specific ligand.
Nzila et al. [15] reviewed several strategies used to reposition drugs for the treatment of multi- and drug-resistant malaria and tuberculosis. Five strategies for drug repositioning were presented: 1) assess similarity in cell biology and biological processes, using compounds that target pathways that also exist in the microorganisms responsible for malaria and tuberculosis; 2) explore the microorganism genome information, aiming to identify putative drug targets already validated in another organism; 3) revisit data from failed drug reposition attempts, as inherent variables (e.g., animal model chosen, toxicity) could be poorly chosen or dealt with; 4) observe co-infection drug treatment efficacy thoroughly, as many diseases occur as co-infections with malaria and tuberculosis, and; 5) screen old and existing drugs. Indeed, in a recent approach Iwata et al. [16] proposed a statistical model to infer new drug-disease associations based on known drug-disease interaction knowledge. In their approach, each drug-disease pair was defined a descriptor based on the phenotypic effects of drugs (e.g., main effect and side effects) and with various molecular features of diseases (e.g., disease-causing genes, diagnostic markers, disease-related pathways, and environmental factors). Berenstein et al. [17] took advantage of extensively studied organisms to develop an integrative network model for the identification of bioactive drug-like molecules and candidate drug targets in neglected pathogen proteomes. More recently, Savoia [18] reviewed several promising experimental studies on drug repurposing of existing drugs for infectious diseases, most of them identified serendipitously or by exploring the side-effects of the drugs.
Computational drug repurposing approaches invariably make use of previously known drug-target associations. Finding alternative targets for known drugs has the added benefit of advancing into clinical trials sooner, as their pharmacokinetics and safety profiles are known by the regulatory authorities [19].
In this paper, we propose a methodology for screening putative DTIs for drug repositioning. The proposed pipeline allows the identification of potential drug-targets in any bacterial species of interest, and the prediction of putative DTIs between the identified drug-targets and already commercialized drugs. The newly identified DTIs can provide key leads for drug repurposing towards problematic pathogens, being a time and cost-effective strategy to support the development of new antibacterials.
Results and Discussion
Classification model performance assessment and comparison
We constructed our random forest classification model based on the training set, and using the values found by grid search for n_estimators (number of trees in the forest) and max_features (number of features to consider when looking for the best split). We performed five-fold cross-validation (internal validation) and tested the classification model against data sets independent of the training data (external validation) to evaluate classification performance. The AUCs for five-fold cross-validation and external data set validation were 0.99 and 0.91, respectively. After classifying the external validation data set we computed the confusion matrix for the predicted instances (Table 1). These results indicate that the presented model is valid and able to classify unseen data.
Table 1. Confusion matrix of external validation data set classification.
| Predicted positive | Predicted negative | |
|---|---|---|
| Condition positive | 2,792 (TP) | 542 (FN) |
| Condition negative | 69 (FP) | 5,126 (TN) |
TN–true negative; FP–false positive; FN–false negative; TP–true positive
Analysis of the impact of network metrics
The network metrics calculated for the proteins in the methicillin-resistant S. aureus (strain COL–MRSA COL) interactome were sorted by their betweenness centrality (BC) values in descending order and filtered for a subgraph centrality (SC) value greater than 1023 as the best putative drug-targets. Table 2 lists the ten best scoring proteins according to the calculated network metrics. The prokaryotic DNA-directed RNA polymerase is an enzyme with multiple subunits responsible for transcription in bacteria. It is an appealing drug target due to its essentiality for bacterial growth and survival and its different features from mammalian counterparts [20,21]. According to DrugBank, Rifabutin targets both the alpha and beta subunits in Escherichia coli strain K12, while the beta subunit is targeted by Rifapentine (in Mycobacterium tuberculosis), Rifampicin, Rifaximin, and Rifalazil (in E. coli strain K12).
Table 2. Top ten best putative drug-targets.
| STRING ID | UniProt ID | Protein name | SC | BC |
|---|---|---|---|---|
| SACOL2213 | Q5HDY4 | DNA-directed RNA polymerase subunit alpha | 1.85E+23 | 0.0329 |
| SACOL0591 | Q5HID0 | 30S ribosomal protein S12 | 2.76E+23 | 0.0198 |
| SACOL0588 | Q5HID3 | DNA-directed RNA polymerase subunit beta | 1.17E+23 | 0.0178 |
| SACOL2675 | Q5HCP4 | Accessory Sec system protein translocase subunit SecY2 | 1.01E+23 | 0.0128 |
| SACOL1292 | Q5HGF8 | 30S ribosomal protein S15 | 2.65E+23 | 0.0112 |
| SACOL0593 | Q5HIC8 | Elongation factor G | 2.82E+23 | 0.0093 |
| SACOL2234 | Q5HDW3 | 50S ribosomal protein L22 | 3.29E+23 | 0.0049 |
| SACOL2233 | Q5HDW4 | 30S ribosomal protein S3 | 3.11E+23 | 0.0047 |
| SACOL2207 | Q5HDZ0 | 50S ribosomal protein L13 | 2.94E+23 | 0.0046 |
| SACOL0545 | Q5HIH4 | 50S ribosomal protein L25 | 1.06E+23 | 0.0045 |
SC–Subgraph centrality; BC–Betweenness centrality
The ribosome is responsible for protein synthesis in the cell and is composed of two subunits, the 50S (larger) and 30S (smaller). Drugs that target ribosomal proteins to inhibit bacterial protein synthesis are either 50S inhibitors (chloramphenicol, clindamycin, macrolides, and pleuromutilins) or 30S inhibitors (tetracycline and aminoglycosides) [22–24].
The movement of tRNA and mRNA through the ribosome at the end of each round of polypeptide elongation is catalyzed by the prokaryotic elongation factor G (EF-G) [25]. Fusidic acid inhibits ribosomal peptide elongation (and ribosome recycling) by targeting EF-G, forming a strong complex when EF-G is ribosome-bound [26].
Protein secretion is crucial to export virulence factors and thus, to improve pathogenic survivability. The accessory Sec system is a specialized export system found in mycobacteria and some Gram-positive bacteria, where the common element is the accessory SecA protein SecA2. It was reported that in the specific case of S. aureus the SecA2/SecY2 system is required for the export of the serine-rich surface protein adhesion (SraP), an important virulence determinant in endovascular infection [27,28]. Inhibition of SecY2 was found to prevent SraP surface expression almost completely [28].
These literature findings suggest this heuristic is a good predictor to identify putative drug-targets in bacterial species of interest. To generate our test data set we combined the best ten proteins with the 3,081 unique ligands in our training and test data sets in an all-against-all fashion, resulting in 30,810 DTIs in the test set.
Analysis of predicted putative drug-target interactions
In our model we opted to predict the probability of each DTI pair to interact, i.e., the probability of a given DTI pair to be classified as a positive interaction. On a random forest classifier the predicted class probability is computed as the mean probability of the predicted class from the trees in the forest, where the single tree class probability is the fraction of samples of the same class in a leaf. This allowed us to sort DTI pairs by their class probabilities for easier identification of the most probable putative DTIs. We selected the five most probable DTIs according to our classification model for further analysis (Table 3). According to UniProt, the proteins involved in these DTIs did not have solved tertiary structures at the time of writing. Thus, we performed ab initio homology modeling following a well-established strategy [29]. First, we used the I-TASSER [30–32] online server to predict the tertiary structure of the proteins with UniProt IDs Q5HIC8, Q5HID3, and Q5HCP4, using the default parameters. The I-TASSER server uses three metrics to measure the confidence of each generated model: 1) C-score, which is calculated based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations; 2) RMSD, which is an average distance of all residue pairs in two structures, and; 3) TM-score, which weighs the small distance between all residue pairs stronger than the big distance, making the score insensitive to the local modeling error (disregarded in RMSD). However, RMSD and TM-score are used when the native structure is known, meaning their values in I-TASSER are predicted based on the C-score. The C-score, RMSD and TM-score values for Q5HIC8 are 1.53, 4.8±3.1Å, and 0.93±0.06, respectively. For Q5HID3 these values are 0.19, 8.8±4.6Å, and 0.74±0.11. Lastly, for Q5HCP4, the C-score is 1.20, RMSD is 4.3±2.9Å, and TM-score is 0.88±0.07. In general, C-score values are comprised between -5 and 2, with 2 being a good indicator of model confidence. Since the C-score of Q5HID3 fell short of those of Q5HIC8 and Q5HCP4 we decided to discard the modeled tertiary structure.
Table 3. Five best scoring putative drug-target interactions.
| UniProt ID | Protein Name | ZINC ID | Ligand name | Class probability |
|---|---|---|---|---|
| Q5HIC8 | Elongation factor G | ZINC85537089 | Proglumetacin maleate | 0.93 |
| Q5HID3 | DNA-directed RNA polymerase subunit beta | ZINC01550477 | Lapatinib | 0.93 |
| Q5HID3 | DNA-directed RNA polymerase subunit beta | ZINC85537027 | Tacrolimus | 0.92 |
| Q5HCP4 | Accessory Sec system protein translocase subunit SecY2 | ZINC19418959 | Trifluoperazine dihydrochloride | 0.92 |
| Q5HIC8 | Elongation factor G | ZINC01535101 | Rosuvastatin calcium | 0.91 |
The accuracy of the generated models was estimated using ProSA-web [33], an established tool used for the refinement and validation of experimental protein structures and in structure prediction and modeling.
This tool parses the coordinates of the structure and evaluates its energy using a distance-based pair potential and a potential capable of detecting solvent exposed residues. The Z-score, an indicator of overall model quality, is calculated using these energies. Specifically, it measures the deviation of the total energy of the model’s structure, considering an energy distribution derived from random conformations [33]. All possible conformations of a given protein have associated energy values. The number of conformations per energy interval, that is, the energy density N(E), characterizes the energy distribution of said protein. By the law of large numbers one can assume that the energy density follows a Gaussian distribution, defined by the average energy Ē, and standard deviation σ. Since every distribution has an average and a standard deviation, it is possible to normalize energy values, even without knowing the shape of this distribution. Thus,
| (1) |
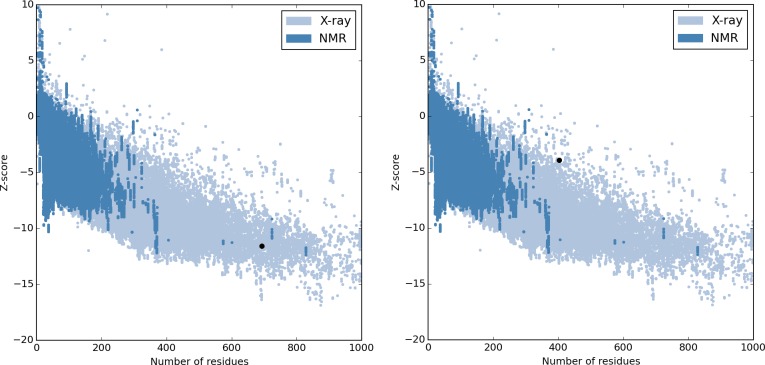
These normalized values are called z-scores [34]. Lower z-score values are correlated with typical native structures of similar size, while z-scores outside the characteristic range of the native proteins indicate erroneous structures [33]. Z-score values for Q5HIC8 and Q5HCP4 were -10.39 and -3.95, respectively, suggesting the quality of the modeled tertiary structures (Fig 1).
Fig 1. ProSA-web overall model quality output for Q5HIC8 (left) and Q5HCP4 (right), respectively.
Panels show these proteins are within the range of scores typically found for proteins of similar size.
Following ab initio structural modeling and validation, we performed docking experiments between our predicted DTIs to test the theoretical viability of the ligands to actually bind the modeled proteins. In addition, we also performed docking experiments between these ligands and their real targets to create a benchmark. All docking experiments were carried out using the SwissDock web-server [35] and AutoDock4 [36]. Table 4 summarizes the docking experiments and benchmarks performed, as well as their results. The results of SwissDock docking suggest that the ligands ZINC85537089 and ZINC01535101 have a greater binding affinity to Q5HIC8 than to the targets they were originally synthesized for. Although not possessing the lowest full fitness, the Q5HPC4-ZINC19418959 DTI pair still has a higher binding affinity than the P26447-ZINC19418959 pair. Trifluoperazine dihydrochloride was shown to have antiplasmid effects on a range of bacterial species [37,38]. Furthermore, it was reported that Prochlorperazine (ZINC19796018), an antipsychotic drug with MCS Tanimoto similarity of 0.8276 with Trifluoperazine dihydrochloride (ZINC19418959), also possesses antibacterial activity against several species [39]. Similar studies show evidence that statins, including Rosuvastatin calcium (ZINC01535101), also present activity against a range of bacterial species [40,41]. Proglumetacin maleate (ZINC85537089) belongs to the acetic acid derivatives and related substances class. While acetic acid has known antibacterial properties [42,43], Proglumetacin maleate does not have any antibacterial effects and had been actually labeled as non-antibacterial [44,45].
Table 4. Results of the molecular docking experiments performed for predicted and real (benchmark) DTIs.
| ZINC ID | UniProt ID | Target type | PDB ID | SD | AD4 |
|---|---|---|---|---|---|
| 85537089 | Q5HIC8 | Predicted | N/A | -2.69 | -2.03 |
| P23219 | Real | 1CQE | -2.06 | -3.93 | |
| P35354 | Real | 5F19 | -2.29 | -3.98 | |
| 19418959 | Q5HCP4 | Predicted | N/A | -0.89 | -5.69 |
| P63316 | Real | 1J1D | -1.50 | -5.28 | |
| P62158 | Real | 1CLL | -1.29 | -7.16 | |
| P26447 | Real | 2Q91 | -0.59 | -6.83 | |
| P14416 | Real | 5AER | -1.04 | -5.56 | |
| 01535101 | Q5HIC8 | Predicted | N/A | -2.90 | -1.59 |
| P04035 | Real | 1DQ8 | -2.20 | -2.63 |
Values are presented in cal/mol. SD–SwissDock; AD4 –AutoDock4.
The results of AutoDock4 docking were not so optimistic, with only ZINC19418959 showing greater binding affinity to the predicted protein Q5HCP4 than to two of its real targets (P63316 and P14416). Nonetheless, the evidence shown here is highly suggestive that the identified compounds are able to bind to the predicted drug-targets, attesting the performance of the proposed methodology. Indeed, we looked further into the antimicrobial activity of acetic acid and found reports of its ability to directly eradicate mature biofilms [46] and inhibit oral microorganisms [47].
Experimental testing will be decisive in validating the presented findings. Namely, if these DTIs actually occur, the identified ligands may not be able to cross the cell wall and cell membrane, which would most likely require lead optimization to improve selectivity to the target and efficiency of the ligand. Still, the robustness and reliability of the proposed pipeline can be attested, as it performed well in both internal validation and external validation data sets.
Overall, we show that the combined use of network metrics, namely subgraph centrality and betweenness centrality, are extremely useful for finding potential drug-targets in MRSA. Most of the ten best putative drug targets were part of the already heavy focused microbial pathways (protein synthesis, nucleic acid synthesis and cell wall synthesis), which demonstrates that this heuristic is able to identify essential proteins. Considering this, identification of the accessory Sec system protein translocase subunit SecY2 as a putative drug-target seems especially relevant, as it is part of a pathway that has not received much attention for antibacterial development. Future studies will focus on this and other less explored pathways for antimicrobial development.
Even though we used the MRSA interactome as a case-study, this pipeline was developed to be applied to any pathogen species of interest, as long as their interactome is at least partially known. By reducing the number of possible drug-targets it is possible to save time and funds to be directed to investigating the shorter drug-target pool. Moreover, DTI prediction further narrows the lead screening window, allowing the possibility of drug-repurposing. Finally, since these drugs are already commercialized there should be no inherent risks in using them as antibacterials.
Methods
Pipeline overview
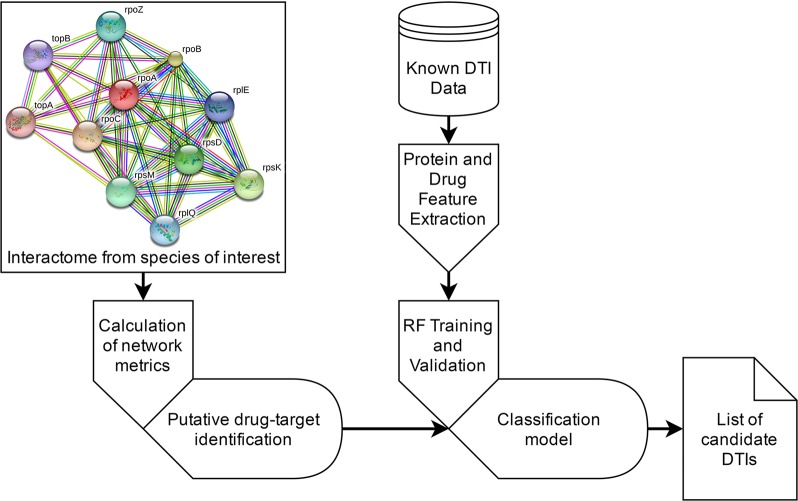
The proposed approach is schematized in Fig 2. Known DTI data were collected from publicly available databases. From the ligand’s simplified molecular-input line-entry system (SMILE) representation of a ligand the chemical structure data and physicochemical descriptors are retrieved and encoded. Similarly, from the primary sequence of a protein a variety of physicochemical descriptors are retrieved. These descriptors are used to generate the feature vectors that represent DTI pairs.
Fig 2. Diagram of the proposed pipeline.
The proposed classification model uses random forest (RF) [48], as these run efficiently on large data sets, provide accurate estimates, are able to estimate the most important features in the classification task, and are less prone to overfitting. Classifier validation includes internal (five-fold cross-validation) and external validation on an independent data set. The classification model is then used to predict putative DTIs between the estimated crucial drug-targets in the methicillin-resistant Staphylococcus aureus (strain COL–UniProt taxonomy ID: 93062) and all the drugs in our training and test data sets. The most essential drug-targets were estimated by a combination of subgraph centrality (SC) and the betweenness centrality (BC) of each protein in the bacterial interactome, as SC is highly correlated with the lethality of individual proteins removed from the proteome, and BC is likely to be associated with protein essentiality [49,50]. The methicillin-resistant S. aureus COL (MRSA) interactome was used to test and validate the proposed pipeline. Finally, we use the SwissDock server and AutoDock4 to perform docking simulations for the best scoring predicted DTIs. The docking process for SwissDock was set to “Accurate” and the region of interest was set to default, as this docking is flexible. For AutoDock4, the search parameters were set to long (25,000,000 evaluations) and carried out by a Genetic Algorithm. The docking process was performed using a Lamarckian Genetic Algorithm. Hydrogen was added to each protein undergoing docking testing and Gasteiger charges were assigned. The spacing between grid points used was the default value (0.375 Å). Additional docking simulations between the same ligands and their original drug-targets are performed to establish benchmarks for comparison.
SwissDock is based in the EADock DSS engine [35,51]. In this engine, binding models are generated and scored using a simple fitness function, minimized, and then clustered and evaluated according to their full fitness [51]. The most stable DTI complexes are those with the lowest docking score values. Since the SwissDock server [35] allows the direct upload of PDB codes (for target selection) and ZINC database accession identifiers (for ligand selection), pre-processing of the structures for the docking experiments by us was not required. Instead, this process is automatically performed in the SwissDock server, where the input molecules are converted to the CHARMM [52] format. This is the selected format since docking assays are performed in the CHARMM22/27 all-hydrogen force field. Protein target and ligand setup are thoroughly described in [35]. The EADock DSS engine generates between 5,000 and 15,000 binding models near the target cavities of the entire protein surface and simultaneously estimates their CHARMM energies on a grid. Binding models with the most favorable energies are then ranked, considering the solvent effect using the FACTS implicit solvation model [53]. The most favorable binding model clusters are then presented in the results file.
Positive data set construction
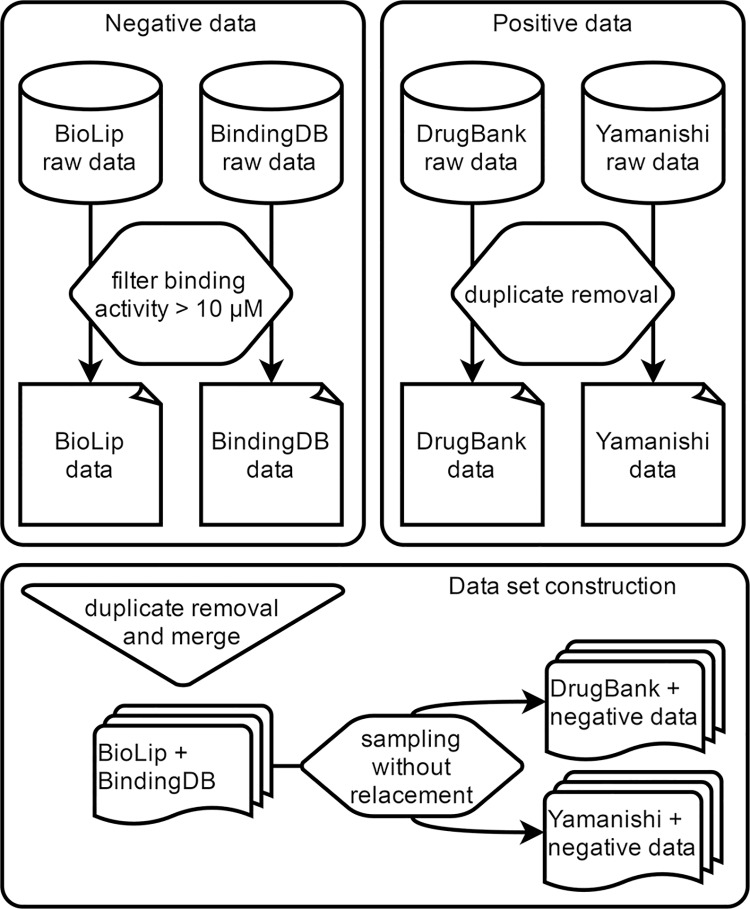
In this work we collected positive drug-target interaction (DTI) data from two different sources: (1) DrugBank [54] and (2) from a previous DTI prediction study by Yamanishi et al. [55]. The DrugBank database freely provides high-quality curated data regarding drugs and drug-targets for conducting in silico bio and chemoinformatics studies. DrugBank DTI data was downloaded on October 4 2015 (version 4.3). All DTIs were conveniently represented as a list of pairs, along with protein sequence information for each target, and SMILE format for each drug. Any drug or drug target without a valid SMILE or protein sequence, respectively, was removed from our data set. The latter comprises DTI data from KEGG BRITE [56,57], BRENDA [58], SuperTarget [59] and DrugBank [54] from November 2007 and has been used as a gold standard in several DTI prediction studies [12,60–63]. Since Yamanishi’s data [55] contains positive instances from DrugBank, all duplicated entries between the two data sets were removed (Fig 3). In this study we disregarded the specific classes of protein targets (i.e., enzymes, G-protein coupled receptors, ion channels, and nuclear receptors) and excluded proteins with unreviewed status in the UniProt Knowledgebase [64] (i.e., proteins automatically annotated in TrEMBL). The number of unique drugs in our positive data sets is 2,118, comprising 1,328 from DrugBank and 790 from Yamanishi’s data [55]. In the same data, the number of unique drug-targets is 2,077 (706 from DrugBank and 1,371 from Yamanishi [55]). Finally, the number of known DTIs between the drugs and targets in the positive data is 10,736 (3,530 from DrugBank and 7,206 from Yamanishi [55]).
Fig 3. Data set construction.
Negative data set construction
Ideally, the negative data set should also exclusively comprise experimentally determined non-DTIs. However, very few authors publish non-interacting protein data, as these are generally associated with failed hypothesis. To collect experimental negative data we screened the BindingDB [65] and BioLip [66] databases for DTI pairs with experimental bioactivity values greater than 10 μM (Fig 3). The same strategy was previously applied to compile negative examples in another DTI prediction methodology [67], since DTIs with bioactivity values above this threshold are considered to possess weak binding activity. BindingDB data was downloaded in December 2015, and BioLip data was downloaded in April 2016. The number of unique DTIs with experimental bioactivity values greater than 10 μM in BindingDB and BioLip is 14,985 and 1,223, respectively. In the former, the number of unique drugs and drug-targets is 12,454 and 404, respectively. The latter comprises 894 unique drugs and 636 unique drug-targets.
Machine learning data set construction
To ensure the discriminative power of the proposed approach, we used OpenBabel [68] to extract the molecular fingerprints of the drugs in our data sets and to compare their chemical similarity. We have found that within each data set (DrugBank, Yamanishi [55], BindingDB and BioLip) and across all data sets, less than 1% of all possible drug pairs had a sequence similarity score greater than 0.85. Then, we combined the described positive and negative data to construct the data sets for classification model training and external validation (Fig 3). The negative data collected from BindingDB and BioLip was merged and duplicated instances were removed, resulting in 16,209 unique negative DTIs. Each instance of these data was randomly selected and appended to one of the positive data sets (first to the Yamanishi [55] data set and then to the DrugBank data set) until all instances were exhausted, while maintaining a similar negative to positive ratio (approximately 1.5). Whenever a negative instance was randomly selected from the negative data to be appended in either positive data set, that instance was removed from the negative data set to ensure the absence of duplicated entries. This resulted in 18,118 instances in the training data set, consisting of 7,206 positive instances from the Yamanishi [55] data and 10,912 randomly selected negative instances. The external validation data set comprised 3,530 positive examples from DrugBank data and 5,297 randomly selected negative examples, totaling 8,827 DTIs.
Calculation of the bacterial interactome network metrics
The interactome of the methicillin-resistant S. aureus (strain COL–MRSA COL) was downloaded from the STRING database [69] in January 2016. The 300,477 protein-protein interactions (PPIs) downloaded included 36,230 unique proteins, comprised of 9,875 active proteins and 26,355 obsolete in UniProt. From the active proteins, only 1,074 were reviewed and manually annotated in Swiss-Prot [64]. To avoid the presence of false-positive proteins in our experiments, we only considered reviewed proteins in this study. As a result, the MRSA COL interactome filtered for reviewed proteins comprised 93,952 PPIs. To calculate the subgraph centrality (SC) and betweenness centrality (BC) of each protein in the MRSA COL interactome we used NetworkX (https://networkx.github.io/), a Python software package for the creation and study of complex networks.
The subgraph centrality (SC) metric can be calculated from the spectra of the adjacency matrix of the network and was found to be better at discriminating the nodes of a network than alternative measures (e.g., degree, closeness). In addition, it was shown that SC is more highly correlated with the lethality of individual proteins removed from the proteome, compared with the number of links per node [49,70]. For a given node u the SC is given by,
| (2) |
where vj is an eigenvector of the adjacency matrix A, corresponding to the eigenvalue λj obtained from the graph.
Bottlenecks in protein networks can be predicted by calculating the betweenness centrality, BC, with greater values suggesting a higher “bottleneck-ness”. These are networks nodes that have many shortest paths passing through them, making them key connector proteins. In comparison with degree centrality (i.e., “hub-ness”), bottlenecks are significantly better associated with essentiality [50]. For a node v, the BC is given by,
| (3) |
where V is the set of nodes, the denominator is the number of shortest paths in the network, and the numerator the number of those that pass through v.
Computation of drug and protein descriptors
Drug and protein descriptors were computed using PyDPI, a python package for chemogenomics studies [58]. PyDPI calculates the most frequently used structural and physicochemical properties of a protein given its amino acid sequence, molecular descriptors of a drug from its smile representation, protein-protein interaction (PPI) descriptors, and DTI descriptors.
Using PyDPI we calculated 755 descriptors for each DTI– 432 protein descriptors and 323 drug descriptors. The 432 protein descriptors are divided as follows: 20 amino acid composition descriptors; 240 Moran autocorrelation descriptors, and; 147 CTD (21 Composition, 21 Transition, and 105 Distribution) physicochemical descriptors. The amino acid composition group of descriptors represents the fraction of each amino acid type in the sequence. Autocorrelation descriptors express the level of correlation between two proteins regarding specific structural or physicochemical properties. The CTD descriptors group represent the amino acid distribution pattern of specific structural or physicochemical properties along the primary structure of a protein, including hydrophobicity, polarity, charge, polarizability, normalized van der Waals volume, secondary structures and solvent accessibility. Drug features comprise 30 molecular constitutional descriptors, 23 molecular connectivity indices, six molecular property descriptors, seven kappa shape descriptors, 12 charge descriptors, 166 Molecular Access System (MACCS) keys, and 79 E-state fingerprints. Constitutional descriptors characterize the chemical element and chemical bond type, path length, hydrogen bond and hydrogen acceptor, while molecular and valence connectivity are described with the connectivity indices. For instance, Kappa indices reflect shape attributes of the molecule, and charge descriptors express electronic features of the whole molecule and of particular regions (atoms, bonds, and molecular fragments). Molecular fingerprints encode chemical structures which consist of bins, with each bin being a substructure descriptor associated with a specific molecular feature. A detailed explanation of these and other descriptor groups is given in the original publication of the PyDPI package [58].
Drug-target interaction classification
The predictive model used in this study was implemented using scikit-learn, a Python package to perform data mining, data analysis and machine learning tasks [71]. To predict DTIs we implemented a classification model based on random forests of decision trees (RF) [48], which has been shown [72–74] as the best approach to solve complex classification problems in large data sets with a significant number of features. A random forest is an ensemble of many classifiers of the same base type (e.g., decision trees) which returns the class that is the mode of the classes across the output of the individual trees in the forest [48]. Each tree is fully constructed from a bootstrap sample drawn from the training set, by recursively splitting an upstream node. When splitting a node in the tree, the chosen split is only the best split among a random subset of features to prevent correlation between trees. This results in a split that is not the best split among all features, adding some randomness to the model and slightly increasing the forest bias. However, due to averaging between trees, the variance of the forest will also decrease, more than compensating the increase in bias and resulting in an overall better model. The trees are grown until a node cannot be split further. Conversely to the original model [48] where each tree votes for a single class, prediction of the class of input samples in the scikit-learn implementation is performed by averaging their probabilistic predictions. The number of trees (n_estimators) and the number of features to consider when looking for the best split (max_features) are important parameters when building an RF model. To define these parameters we used the grid search method and then adopted the parameters of the model with best mean accuracy after five-fold cross-validation. Thus, the parameters used were 150 n_estimators and 100 max_features. Since we only consider 100 features at each split, we believe over-parameterization does not occur.
The pipeline for the construction of our classification model is very straightforward: (1) train the RF; (2) assess internal classifier performance by five-fold cross-validation; (3) classify the external validation data set to evaluate classifier performance on out-of-sampling data, and; (4) classify the test data.
Predictive model validation
While a performance comparison with the method proposed by Cheng et al. [12] would be ideal to ascertain how our approach compares with the state-of-the-art, the links to their data sets are unavailable by the time of writing. Thus, to estimate the classification accuracy of the implemented predictive models we used internal and external validation. Internal validation was performed using five-fold cross validation, which consists of splitting the training set in five subsets, using four subsets to train the model, and testing on the remaining subset. This is done consecutively until every subset is used as the test set. External validation was performed by using a data set independent from the training data as test set for the classification model. This strategy is fundamental to better estimate the performance and generalizability of the classifier, as cross-validation estimates are usually biased towards over-performance [75,76].
Data Availability
The proposed classification model and related data files are available at http://bioinformatics.ua.pt/software/dtipred/.
Funding Statement
Fundação para a Ciência e Tecnologia (http://www.fct.pt/) funded EDC under grant SFRH/BD/86343/2012. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.ECDC (2015) Annual Epidemiological Report 2014—Antimicrobial Resistance and Healthcare-Associated Infections Stockholm: European Centre for Disease Prevention and Control. [Google Scholar]
- 2.Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, et al. (2015) The global threat of antimicrobial resistance: science for intervention. New Microbes and New Infections 6: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolář M, Urbánek K, Látal T (2001) Antibiotic selective pressure and development of bacterial resistance. International Journal of Antimicrobial Agents 17: 357–363. [DOI] [PubMed] [Google Scholar]
- 4.Haag NL, Velk KK, Wu C (2012) Potential Antibacterial Targets in Bacterial Central Metabolism. Int J Adv Life Sci 4: 21–32. [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley JT, Deshpande T, Butte AJ (2011) Exploiting drug-disease relationships for computational drug repositioning. Brief Bioinform 12: 303–311. 10.1093/bib/bbr013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburn TT, Thor KB (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3: 673–683. 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- 7.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, et al. (2011) Computational Repositioning of the Anticonvulsant Topiramate for Inflammatory Bowel Disease. Science Translational Medicine 3: 96ra76 10.1126/scitranslmed.3002648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley JT, Deshpande T, Butte AJ (2011) Exploiting drug–disease relationships for computational drug repositioning. Briefings in Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekins S, Williams AJ, Krasowski MD, Freundlich JS (2011) In silico repositioning of approved drugs for rare and neglected diseases. Drug Discovery Today 16: 298–310. 10.1016/j.drudis.2011.02.016 [DOI] [PubMed] [Google Scholar]
- 10.Cheng F, Zhou Y, Li J, Li W, Liu G, et al. (2012) Prediction of chemical-protein interactions: multitarget-QSAR versus computational chemogenomic methods. Mol Biosyst 8: 2373–2384. 10.1039/c2mb25110h [DOI] [PubMed] [Google Scholar]
- 11.Raje N, Anderson K (1999) Thalidomide—A Revival Story. New England Journal of Medicine 341: 1606–1609. 10.1056/NEJM199911183412110 [DOI] [PubMed] [Google Scholar]
- 12.Cheng F, Liu C, Jiang J, Lu W, Li W, et al. (2012) Prediction of Drug-Target Interactions and Drug Repositioning via Network-Based Inference. PLoS Comput Biol 8: e1002503 10.1371/journal.pcbi.1002503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Xu J, Zeng J (2014) Drug-target interaction prediction by integrating chemical, genomic, functional and pharmacological data. Pac Symp Biocomput: 148–159. [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12: 371–387. 10.1038/nrd3975 [DOI] [PubMed] [Google Scholar]
- 15.Nzila A, Ma Z, Chibale K (2011) Drug repositioning in the treatment of malaria and TB. Future Med Chem 3: 1413–1426. 10.4155/fmc.11.95 [DOI] [PubMed] [Google Scholar]
- 16.Iwata H, Sawada R, Mizutani S, Yamanishi Y (2015) Systematic Drug Repositioning for a Wide Range of Diseases with Integrative Analyses of Phenotypic and Molecular Data. Journal of Chemical Information and Modeling 55: 446–459. 10.1021/ci500670q [DOI] [PubMed] [Google Scholar]
- 17.Berenstein AJ, Magariños MP, Chernomoretz A, Agüero F (2016) A Multilayer Network Approach for Guiding Drug Repositioning in Neglected Diseases. PLoS Negl Trop Dis 10: e0004300 10.1371/journal.pntd.0004300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savoia D (2016) New Antimicrobial Approaches: Reuse of Old Drugs. Curr Drug Targets 17: 731–738. [DOI] [PubMed] [Google Scholar]
- 19.Chong CR, Sullivan DJ Jr. (2007) New uses for old drugs. Nature 448: 645–646. 10.1038/448645a [DOI] [PubMed] [Google Scholar]
- 20.Chopra I (2007) Bacterial RNA polymerase: a promising target for the discovery of new antimicrobial agents. Curr Opin Investig Drugs 8: 600–607. [PubMed] [Google Scholar]
- 21.Bai H, Zhou Y, Hou Z, Xue X, Meng J, et al. (2011) Targeting bacterial RNA polymerase: promises for future antisense antibiotics development. Infect Disord Drug Targets 11: 175–187. [DOI] [PubMed] [Google Scholar]
- 22.Poehlsgaard J, Douthwaite S (2005) The bacterial ribosome as a target for antibiotics. Nat Rev Micro 3: 870–881. [DOI] [PubMed] [Google Scholar]
- 23.Kohanski MA, Dwyer DJ, Collins JJ (2010) How antibiotics kill bacteria: from targets to networks. Nat Rev Micro 8: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson DN (2014) Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Micro 12: 35–48. [DOI] [PubMed] [Google Scholar]
- 25.Shoji S, Walker SE, Fredrick K (2009) Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chemical Biology 4: 93–107. 10.1021/cb8002946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borg A, Holm M, Shiroyama I, Hauryliuk V, Pavlov M, et al. (2015) Fusidic Acid Targets Elongation Factor G in Several Stages of Translocation on the Bacterial Ribosome. Journal of Biological Chemistry 290: 3440–3454. 10.1074/jbc.M114.611608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siboo IR, Chambers HF, Sullam PM (2005) Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in Binding to Human Platelets. Infection and Immunity 73: 2273–2280. 10.1128/IAI.73.4.2273-2280.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siboo IR, Chaffin DO, Rubens CE, Sullam PM (2008) Characterization of the Accessory Sec System of Staphylococcus aureus. Journal of Bacteriology 190: 6188–6196. 10.1128/JB.00300-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duarte-Pereira S, Silva SS, Azevedo L, Castro L, Amorim A, et al. (2014) NAMPT and NAPRT1: novel polymorphisms and distribution of variants between normal tissues and tumor samples. Scientific Reports 4: 6311 10.1038/srep06311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Yan R, Roy A, Xu D, Poisson J, et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Meth 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–410. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sippl MJ (1995) Knowledge-based potentials for proteins. Current Opinion in Structural Biology 5: 229–235. [DOI] [PubMed] [Google Scholar]
- 35.Grosdidier A, Zoete V, Michielin O (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Research 39: W270–W277. 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. (2009) AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. Journal of computational chemistry 30: 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster FL, Mandel N (1984) Phenothiazine compounds inhibit in vitro growth of pathogenic free-living amoebae. Antimicrobial Agents and Chemotherapy 25: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spengler G, Miczák A, Hajdú E, Kawase M, Amaral L, et al. (2003) Enhancement of plasmid curing by 9-aminoacridine and two phenothiazines in the presence of proton pump inhibitor 1-(2-benzoxazolyl)-3,3,3-trifluoro-2-propanone. International Journal of Antimicrobial Agents 22: 223–227. [DOI] [PubMed] [Google Scholar]
- 39.Rani Basu L, Mazumdar K, Kumar Dutta N, Karak P, Dastidar SG (2005) Antibacterial property of the antipsychotic agent prochlorperazine, and its synergism with methdilazine. Microbiological Research 160: 95–100. [DOI] [PubMed] [Google Scholar]
- 40.Bergman P, Linde C, Pütsep K, Pohanka A, Normark S, et al. (2011) Studies on the Antibacterial Effects of Statins—In Vitro and In Vivo. PLoS ONE 6: e24394 10.1371/journal.pone.0024394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masadeh M, Mhaidat N, Alzoubi K, Al-azzam S, Alnasser Z (2012) Antibacterial activity of statins: a comparative study of Atorvastatin, Simvastatin, and Rosuvastatin. Annals of Clinical Microbiology and Antimicrobials 11: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryssel H, Kloeters O, Germann G, Schäfer T, Wiedemann G, et al. The antimicrobial effect of acetic acid—An alternative to common local antiseptics? Burns 35: 695–700. [DOI] [PubMed] [Google Scholar]
- 43.Halstead FD, Rauf M, Moiemen NS, Bamford A, Wearn CM, et al. (2015) The Antibacterial Activity of Acetic Acid against Biofilm-Producing Pathogens of Relevance to Burns Patients. PLoS ONE 10: e0136190 10.1371/journal.pone.0136190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomás-Vert F, Pérez-Giménez F, Salabert-Salvador MT, Garcı, amp, et al. (2000) Artificial neural network applied to the discrimination of antibacterial activity by topological methods. Journal of Molecular Structure: THEOCHEM 504: 249–259. [Google Scholar]
- 45.Murcia-Soler M, Pérez-Giménez F, García-March FJ, Salabert-Salvador MT, Díaz-Villanueva W, et al. (2003) Discrimination and selection of new potential antibacterial compounds using simple topological descriptors. Journal of Molecular Graphics and Modelling 21: 375–390. [DOI] [PubMed] [Google Scholar]
- 46.Bjarnsholt T, Alhede M, Jensen PO, Nielsen AK, Johansen HK, et al. (2015) Antibiofilm Properties of Acetic Acid. Adv Wound Care (New Rochelle) 4: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CB, Alimova Y, Myers TM, Ebersole JL (2011) Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Archives of Oral Biology 56: 650–654. 10.1016/j.archoralbio.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breiman L (2001) Random Forests. Machine Learning 45: 5–32. [Google Scholar]
- 49.Estrada E, Rodríguez-Velázquez JA (2005) Subgraph centrality in complex networks. Physical Review E 71: 056103. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M (2007) The Importance of Bottlenecks in Protein Networks: Correlation with Gene Essentiality and Expression Dynamics. PLoS Comput Biol 3: e59 10.1371/journal.pcbi.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosdidier A, Zoete V, Michielin O (2011) Fast docking using the CHARMM force field with EADock DSS. Journal of Computational Chemistry 32: 2149–2159. 10.1002/jcc.21797 [DOI] [PubMed] [Google Scholar]
- 52.Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, et al. (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30: 1545–1614. 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haberthur U, Caflisch A (2008) FACTS: Fast analytical continuum treatment of solvation. J Comput Chem 29: 701–715. 10.1002/jcc.20832 [DOI] [PubMed] [Google Scholar]
- 54.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, et al. (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34: D668–672. 10.1093/nar/gkj067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M (2008) Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24: i232–i240. 10.1093/bioinformatics/btn162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, et al. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schomburg I, Hofmann O, Baensch C, Chang A, Schomburg D (2000) Enzyme data and metabolic information: BRENDA, a resource for research in biology, biochemistry, and medicine. Gene Function & Disease 1: 109–118. [Google Scholar]
- 59.Hecker N, Ahmed J, von Eichborn J, Dunkel M, Macha K, et al. (2012) SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res 40: D1113–1117. 10.1093/nar/gkr912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bleakley K, Yamanishi Y (2009) Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 25: 2397–2403. 10.1093/bioinformatics/btp433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Z, Zhang J, Shi X-H, Hu L-L, Kong X, et al. (2010) Predicting Drug-Target Interaction Networks Based on Functional Groups and Biological Features. PLoS ONE 5: e9603 10.1371/journal.pone.0009603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamanishi Y, Kotera M, Kanehisa M, Goto S (2010) Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics 26: i246–i254. 10.1093/bioinformatics/btq176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Liu M-X, Yan G-Y (2012) Drug-target interaction prediction by random walk on the heterogeneous network. Molecular BioSystems 8: 1970–1978. 10.1039/c2mb00002d [DOI] [PubMed] [Google Scholar]
- 64.Consortium TU (2015) UniProt: a hub for protein information. Nucleic Acids Research 43: D204–D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Liu M, Gilson MK (2001) BindingDB: a web-accessible molecular recognition database. Comb Chem High Throughput Screen 4: 719–725. [DOI] [PubMed] [Google Scholar]
- 66.Yang J, Roy A, Zhang Y (2013) BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Research 41: D1096–D1103. 10.1093/nar/gks966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu G, Ding Y, Seal A, Chen B, Sun Y, et al. (2016) Predicting drug target interactions using meta-path-based semantic network analysis. BMC Bioinformatics 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, et al. (2011) Open Babel: An open chemical toolbox. Journal of cheminformatics 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–416. 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Estrada E, Hatano N (2008) Communicability in complex networks. Physical Review E 77: 036111. [DOI] [PubMed] [Google Scholar]
- 71.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, et al. (2011) Scikit-learn: Machine learning in Python. The Journal of Machine Learning Research 12: 2825–2830. [Google Scholar]
- 72.Larrañaga P, Calvo B, Santana R, Bielza C, Galdiano J, et al. (2006) Machine learning in bioinformatics. Briefings in Bioinformatics 7: 86–112. [DOI] [PubMed] [Google Scholar]
- 73.Touw WG, Bayjanov JR, Overmars L, Backus L, Boekhorst J, et al. (2013) Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Briefings in Bioinformatics 14: 315–326. 10.1093/bib/bbs034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kotsiantis SB, Zaharakis I, Pintelas P (2007) Supervised machine learning: A review of classification techniques.
- 75.Bleeker SE, Moll HA, Steyerberg EW, Donders ART, Derksen-Lubsen G, et al. (2003) External validation is necessary in prediction research:: A clinical example. Journal of Clinical Epidemiology 56: 826–832. [DOI] [PubMed] [Google Scholar]
- 76.Simon R (2005) Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol 23: 7332–7341. 10.1200/JCO.2005.02.8712 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The proposed classification model and related data files are available at http://bioinformatics.ua.pt/software/dtipred/.