Abstract
Camptothecin (CPT) is a natural product discovered to be active against various cancers through its ability to inhibit Topoisomerase I (TOP1). CPT analogs also have anti-HIV-1 (HIV) activity that was previously shown to be independent of TOP1 inhibition. We show that a cancer inactive CPT analog (O2-16) inhibits HIV infection by disrupting multimerization of the HIV protein Vif. Antiviral activity depended on the expression of the cellular viral restriction factor APOBEC3G (A3G) that, in the absence of functional Vif, has the ability to hypermutate HIV proviral DNA during reverse transcription. Our studies demonstrate that O2-16 has low cytotoxicity and inhibits Vif-dependent A3G degradation, enabling A3G packaging into HIV viral particles that results in A3G signature hypermutations in viral genomes. This antiviral activity was A3G-dependent and broadly neutralizing against sixteen HIV clinical isolates from groups M (subtypes A-G), N, and O as well as seven single and multi-drug resistant strains of HIV. Molecular modeling predicted binding near the PPLP motif crucial for Vif multimerization and activity. O2-16 also was active in blocking Vif degradation of APOBEC3F (A3F). We propose that CPT analogs not active against TOP1 have novel therapeutic potential as Vif antagonists that enable A3G-dependent hypermutation of HIV.
Graphical Abstract
1. Introduction
Human APOBEC3G (A3G) is a member of the APOBEC family of cytidine/deoxycytidine deaminases that catalyze C to U deamination on single-stranded DNA (ssDNA) or RNA (Munk et al., 2012). The HIV antiviral mechanism for A3G includes: (i) packaging into virions by binding to Gag (Alce and Popik, 2004; Cen et al., 2004) and viral and host RNAs (Bach et al., 2008; Khan et al., 2007; Svarovskaia et al., 2004), (ii) impairing progression of Reverse Transcriptase (RT) by interfering with the viral replication machinery (Guo et al., 2006; Li et al., 2007) and (iii) deaminating dC to dU on proviral minus-strand DNA resulting in degradation through DNA repair pathways and dG to dA hypermutations in the plus-strand viral protein coding regions (Harris et al., 2003; Mangeat et al., 2003). Although A3G is the most potent and abundant anti-HIV APOBEC3 (A3), other A3 family members A3F, A3H (haplotype specific), and A3D are capable of blocking HIV (Chaipan et al., 2013; Li et al., 2010a). However, HIV has a defense factor against A3 proteins known as viral infectivity factor (Vif) (Sheehy et al., 2002). Vif targets A3 proteins for polyubiquitination and proteasomal degradation (Stopak et al., 2003; Yu et al., 2003). Vif has also been shown to affect A3G translation through mRNA binding (Mercenne et al., 2010) and A3G transcription through its interaction with transcription factor core-binding factor-β (CBFβ) (Anderson and Harris, 2015).
Multiple protein-protein interactions (PPI) are crucial for Vif-dependent A3G degradation and are potential therapeutic targets (Letko et al., 2015; Salter et al., 2014). Vif promotes A3G polyubiquitination by acting as a SOCS-box type substrate receptor for an E3 ubiquitin ligase complex comprising Cullin5 (Cul5), Rbx2, ElonginB/C (EloBC), and CBFβ (Hultquist et al., 2012; Jager et al., 2012; Mehle et al., 2006; Yu et al., 2003; Zhang et al., 2012). Another crucial PPI is Vif self-association involving the solvent-exposed 161-PPLP-164 motif (Salter et al., 2014). The PPLP motif is required for A3G binding and degradation (Bergeron et al., 2010; Donahue et al., 2008; Salter et al., 2014; Wolfe et al., 2010; Yang et al., 2001). Intracellular A3G abundance and its incorporation into viral particles were enhanced in the presence of peptides that were antagonists to the PPLP motif, leading to marked suppression of HIV infectivity (Miller et al., 2007). Mutating the PPLP motif produced a Vif multimerization dominant negative mutant that had significantly reduced interaction with A3G (Walker et al., 2010). Moreover, the PPLP motif highlights an interplay within the cell between Vif multimerization, A3G binding and RNA binding, as the size of Vif oligomers in the cellular context were reduced in the presence of A3G (Batisse et al., 2013) and PPLP has been shown to influence Vif’s RNA binding properties (Bernacchi et al., 2011). Overall, Vif’s multiple interactions make it difficult to fully dissect the process by which Vif oligomerizes and finds A3G in the cell and shuttles it to the E3 ligase complex away from viral particles, but the highly conserved PPLP motif seems to be a linchpin in this process.
CPT is a natural product that was isolated from the Asian tree, Camptotheca acuminate, and discovered by Wall and colleagues (Wall et al., 1966) to be active against various cancers through its ability to form a complex with Topoisomerase I (TOP1). Inhibition of TOP1 leads to double-stranded DNA breaks in actively replicating cells resulting in a G2 block and cell death (Burke and Adams, 2005). Many analogs of CPT have been tested clinically since its discovery, with two FDA-approved chemotherapeutic agents, Irinotecan and Topotecan, currently in use for a variety of cancers (Burke and Adams, 2005). Many analogs of CPT also were active against HIV, including 9-nitro CPT (Moulton et al., 1998), 7-hydroxy methyl CPT (Li et al., 2010b) and Topotecan (Zhang et al., 1997). Topotecan had strong anti-HIV activity in the CPT-K5 cell line. These cells express TOP1 with mutations at residues 533 and 583 that prevent Topotecan from binding and acting as a TOP1 inhibitor (Zhang et al., 1997). In this report, we demonstrate that the target for the antiviral mechanism of CPT derivatives in the absence of TOP1 inhibitory activity is Vif, and molecular modeling suggests the CPT analog interacts with Vif near the PPLP motif.
2. Materials and Methods
2.1. FqRET assay for Vif multimerization inhibition
The quenched fluorescence resonance energy transfer (FqRET) cell-based assay utilized expression of pNL4-3 Vif (accession no. M19921) tagged separately with either a EGFP fluorophore (EGFP-V5-Vif) or a REACh2 quencher (Vif-HA-REACh2) in the pIRES-P vector (Hobbs et al., 1998). The Vif multimerization assay was established through robotic co-transfection of HEK293T (293T) cells with Turbofect (Thermo) and optimized for input plasmid ratio (1:4, fluorophore to quencher) to achieve maximum quench of fluorescent signal. EGFP fluorescence was quantified 24 hours after transfection with a BioTek Synergy4 plate reader (ex = 485 nm, em = 528 nm) followed by the addition of 0–30 µM compound. The plates were read a second time 24 hours after compound addition. The differential between the first and second read in each well was quantified relative to the DMSO control to determine the change in relative fluorescence units (ΔRFU) (Fig. S2).
2.2. A3G and A3F degradation assay
A3G-V5-mCherry or A3F-V5-mCherry was stably expressed from the pIRES-P vector in 293T cells under puromycin selection. Fifty ng of plasmid expressing Vif-HA (accession no. AB573087) was transfected into the cells in 384-well format with Turbofect on a liquid handling robot. Four hours after transfection the compounds were added to cells in a range from 0–30 µM. Twenty-four hours after compound addition mCherry fluorescence was read on a BioTek Synergy4 plate reader (ex = 587 nm, em = 610 nm). Relative fluorescence units (RFU) from compound treated wells were quantified using thresholds set by the −Vif (positive control) and +Vif (negative control) (Fig. S3).
2.3. Pseudotyped virus infectivity
pDHIV3-GFP (+Vif virus) codes for all HIV genes except nef (replaced with EGFP) and env, and pΔVif-DHIV3-GFP (−Vif virus) contains a stop codon early within the vif gene. Producer cells of either 293T cells stably expressing A3G-V5-mCherry, A3F-V5-mCherry or transiently transfected with (+A3G) or without (−A3G) V5-A3G in pIRES-P were transfected with the viral vectors and VSV-G coat protein vector pVSV-G using Fugene HD (Promega). In the +A3G condition proviral DNA:VSV-G:A3G were added to cells with a ratio of 1:0.5:0.08, which establishes levels of A3G that are comparable to endogenous A3G (Soros et al., 2007). Virus producer cells were dosed with compound 4 hours after transfection and viral particles were harvested 24 hours later by filtering the media through a 0.45-micron syringe filter.
2.4. Viral particle analysis
Viral particles (30–50 ng p24) isolated from producer cells (section 2.3) were normalized with a p24 ELISA (Perkin Elmer) and sedimented by ultracentrifugation through a 20% sucrose cushion at 100,000 x g. Proteins were resolved by SDS-PAGE and western blotted for V5 (Life Technologies) and p24 with antibody obtained through the NIH AIDS Reagent Program, NIAID: HIV-1 p24 Monoclonal (183-H12-5C) (Wehrly and Chesebro, 1997).
2.5. Nested PCR and next generation sequencing (NGS) analysis
TZM-bl cells (Platt et al., 1998) were infected with equal loads of virus (section 2.3) and genomic DNA was isolated from the cells 48 hours post-infection using a Wizard gDNA Isolation Kit (Promega) according to manufacturer’s protocol. HIV proviral vector (− Control) and isolated gDNA from infected TZM-bl cells (infected with: (1) + Control, −Vif/+A3G virus; or (2) 33.3 µM O2-16 treated +Vif/+A3G and −Vif/−A3G viruses) were subjected to nested PCR and NGS, which is detailed in the Supplemental Materials and Methods.
2.6. CEM-SS and A3.01 Acute Infection
CEM-SS and A3.01 cells were infected in 96-well microtiter plates at ImQuest Biosciences (Frederick, MD). Fifty µL of solvent control and compound at final concentrations of 100, 200 and 400 nM were added in triplicate to 50 µL of cells at final volume plated density of 2.5 × 103 cells per well. HIV-1IIIB was added to the wells in a volume of 100 µL at predetermined titers to provide virus replication over the 14-day infection. Virus replication in the infected cells was monitored daily by RT assay as described previously (Buckheit and Swanstrom, 1991). Cultures were passaged on day 8 by performing a 1:5 split of the culture. Compound was replenished every other day. Uninfected cells at the same compound concentrations along with DMSO controls were evaluated in parallel with tetrazolium dye XTT to quantify the cell viability on days 5 and 14.
2.7. PBMC acute infection
PBMC assays were performed as previously described (Ptak et al., 2008). Briefly, PHA-stimulated cells from at least two normal donors were pooled, diluted in fresh media to a final concentration of 1 × 106 cells/mL, and plated in a 96-well round bottom microplate at 50 µL/well (5 × 104 cells/well). Each plate contained virus control wells and experimental wells using different viral isolates as described in Supplemental Materials and Methods. Test drug dilutions were prepared at a 2X concentration and 100 µL of each concentration was placed in appropriate wells. Fifty µL of a predetermined dilution of virus stock was placed in each test well (final MOI ≅ 0.1). On day seven, cell-free supernatant samples were collected for analysis of RT activity as described previously (Ptak et al., 2008). Toxicity assays were run in parallel and cell viability was measured as described previously (Ptak et al., 2008).
2.8. Molecular modeling on the Vif crystal structure
The published Vif crystal structure (PDB: 4N9F) (Guo et al., 2014) was used to create a computational model in PyMol for in silico docking to Vif. AutoDock Vina (Trott and Olson, 2010) was used to model Vif and O2-16 interactions and assess binding affinities (kcal/mol). Polar hydrogen atoms were added to the structure, and a rectangular grid was created covering the exposed surface of Vif to scan it with O2-16 without bias to interrogate the surface for potential O2-16 binding sites.
3. Results
3.1. O2-16 is a CPT analog that lacks TOP1 inhibition activity
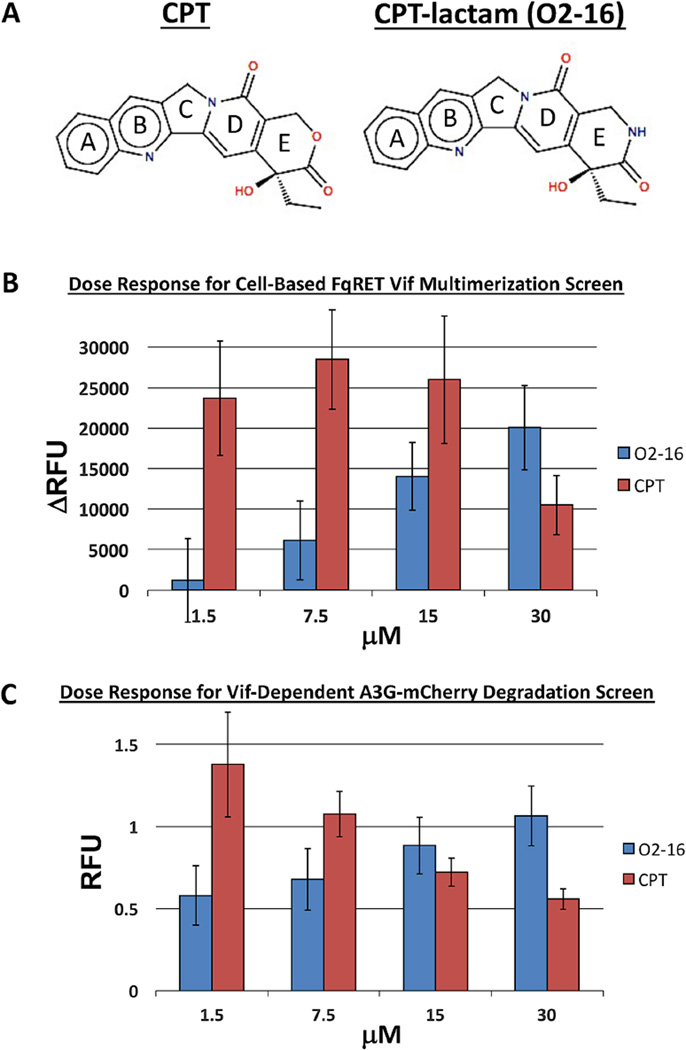
The crystal structure of Topotecan bound to TOP1 showed that the E-ring oxygen (Fig. 1A) is critical as a hydrogen bond acceptor in the catalytic pocket of the TOP1 enzyme (Staker et al., 2002). In order to isolate HIV antiviral activity from TOP1 inhibiting activity we contracted AsisChem to synthesize an analog of CPT (O2-16) in which the E-ring lactone of CPT was replaced with a lactam (Fig. 1A). This analog is ineffective as a TOP1 inhibitor and anti-cancer compound compared to native CPT and lactone-based analogs (e.g., Topotecan) (Jaxel et al., 1989). The critical hydrogen bond acceptor (O) is converted to a hydrogen bond donor (NH) in O2-16 (Fig. 1A) (Staker et al., 2002). The absence of TOP1 inhibition by O2-16 was confirmed by an in vitro TOP1 inhibition assay (Topogen) in which CPT trapped plasmid DNA in open nicked circles but O2-16 did not (Fig. S1A). This result was further confirmed through analysis of cell cycle progression by flow cytometry (Fig. S1B), which showed that whereas CPT induced a G2 block and apoptosis, O2-16 was no longer effective in arresting cells at the G2 phase of the cell cycle (Fig. S1B).
Fig. 1. A lactam analog of CPT (O2-16) inhibits Vif multimerization and A3G degradation.
(A) A depiction of CPT with the 5 rings labeled A-E showing the E-ring lactone (O, red) compared to CPT with an E-ring lactam (NH, blue). (B) A dose-response comparison of CPT and O2-16 (1.5-30 µM) displaying the change in relative fluorescence units (ΔRFU) compared to DMSO control in the cell-based FqRET Vif multimerization screen (described in Fig. S2), n=4 with error bars showing SD. (C) A dose-response comparison of CPT and O2-16 (1.5–30 µM) displaying RFU in a Vif-dependent A3G-mCherry degradation assay (described in Fig. S3), n=4 for CPT and n=6 for O2-16 with error bars showing SD.
3.2. O2-16 has a dose-dependent effect on Vif multimerization
The identification of O2-16 activity as a Vif multimerization antagonist relied on a cell-based FqRET assay that was developed as a high throughput screen for compounds that antagonize Vif multimerization (Fig. S2). The interaction of Vif molecules enables quenching of EGFP signal by REACh2, a mutant of the yellow fluorescent protein that is capable of accepting the EGFP donor fluorescence and quenching its signal without an acceptor signal (Ganesan et al., 2006). Compounds that inhibit Vif multimerization and relieve the quench manifest as a positive EGFP signal (Fig. S2).
We used the FqRET assay to determine whether O2-16 and CPT inhibition of Vif multimerization was dose-dependent. The fluorescence was measured before and after a 24hour incubation with increasing amounts of O2-16 and CPT and the differential between the two reads was calculated as ΔRFU. Above a concentration of 1.5 µM, O2-16 reversed quench in a dose-dependent manner compared to the negative control DMSO-treated cells and increased ΔRFU indicative of inhibition of Vif multimerization (Figs. 1B & S2D). Alternatively, CPT showed a strong reversal of quench at 1.5 µM but its toxicity reduced the effect at 30 µM (Fig. 1B).
3.3 O2-16 protects A3G and A3F from Vif-dependent degradation
A Vif-dependent mechanism for O2-16 antiviral activity would require that O2-16 is able to protect A3G from degradation. We determined the effect of O2-16 on A3G degradation using a cell-based, Vif-dependent A3G degradation assay similar to assays reported previously employing A3G-YFP (Cen et al., 2010; Nathans et al., 2008). The assay was optimized in a 293T cell line stably expressing A3G-V5-mCherry; degradation of A3G was induced by transfection of Vif-HA (Fig. S3). O2-16 displayed potent dose-dependent inhibition of Vif-dependent A3G degradation down to 1.5 µM (Fig. 1C). However, similar to the response in the Vif multimerization screen, CPT had a reduced effect on Vif-dependent A3G degradation with increasing concentrations (Fig. 1C) suggesting that, unlike O2-16, the cytotoxic effect due to CPT activity against TOP1 obfuscated the Vif inhibitory activity.
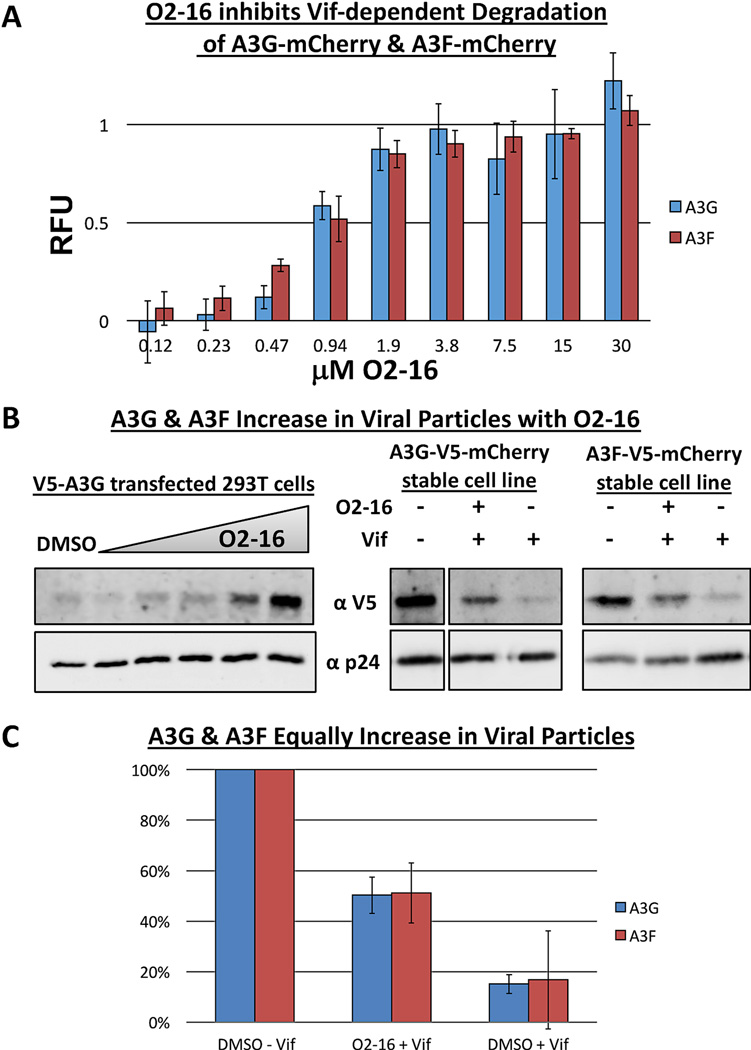
An A3F-V5-mCherry stable cell line also was created to test if O2-16 could block Vif-dependent A3F degradation (Fig. S3). In a side-by-side screen run in triplicate O2-16 was evaluated over an eight-point dose range (0.12–30 µM) and had nearly identical ability to protect A3F and A3G from Vif-dependent degradation (Fig. 2A).
Fig. 2. O2-16 inhibits Vif-dependent degradation and viral packaging of A3G and A3F.
(A) A dose-response for O2-16 (0.12–30 µM) displaying RFU in a Vif-dependent A3G-mCherry and A3F-mCherry degradation assay (described in Fig. S3), n=3 with error bars showing SD. (B) On the left shows the O2-16 dose-dependent increase in V5-A3G in pseudotyped HIV viral particles with p24 acting as the viral load control, with the DMSO control in lane 1 and lanes 2–6 showing increasing concentrations of O2-16 (2.1, 4.2, 8.3, 16.7, 33.3 µM); on the right shows viral particles isolated from A3G-V5-mCherry and A3F-V5-mCherry cells infected with −Vif virus, or +Vif virus with or without O2-16 (33 µM) treatment (NOTE: A3G-V5-mCherry −Vif lane is in a separate panel from the +Vif conditions, but all conditions are from the same gel and exposure with lanes in middle removed). (C) Quantification of densitometry measurements of the conditions from B (right) from three separate viral isolation experiments. The ratio of A3G or A3F to p24 in the −Vif control was set to 100%, + Vif conditions with and without O2-16 are shown with error bars representing SD (n=3).
3.4. O2-16 increases the amount of A3G and A3F recovered with HIV viral particles
Another hallmark of Vif’s activity is a reduction of A3G and A3F accumulation in viral particles (Stopak et al., 2003; Yu et al., 2003). We evaluated the ability of O2-16 to enhance the recovery of A3G in viral particles, by dosing O2-16 (2.1–33.3 µM) in virus producer 293T cells co-transfected with V5-A3G and plasmids encoding HIV proviral DNA to produce pseudotyped HIV. Viral particles were isolated and analyzed by western blotting for the HIV capsid p24 and V5-A3G. A3G recovery within virions increased with O2-16 in a dose-dependent manner (Fig. 2B, left). We also evaluated the effectiveness of 33.3 µM O2-16 to increase the amount of A3G and A3F in viral particles in the presence of Vif from virus produced in the A3G-V5-mCherry and A3F-V5-mCherry 293T cell lines used in Figures 1C and 2A. Representative western blots from virions isolated are shown in Figure 2B (right). The average viral recovery from three separate viral particle isolations revealed that whereas only 15% of A3G and 17% of A3F were incorporated into virions expressing Vif without O2-16 (DMSO control), both A3G and A3F viral levels increased to ~50% with O2-16 added when compared to the minus Vif maximum control for A3G packaging (Fig. 2C).
3.5. O2-16 enables A3G signature hypermutations in HIV proviral DNA
A3G mutations occur processively 3’ to 5’ on ssDNA (Chelico et al., 2006) with the highest proportion of mutations occurring in stretches of HIV DNA that remain single-stranded for the longest period of time during reverse transcription (Yu et al., 2004). Therefore, A3G-dependent hypermutation of HIV genomes is a critical proof of the mechanism of action (MOA) expected for Vif antagonists that enable A3G antiviral activity. Table 1 summarizes the results from NGS of proviral HIV DNA isolated from target cells in pseudotyped HIV infections.
Table 1.
A3G Signature Hypermutations Identified through Next Generation Sequencing of HIV
| − Controla | O2-16 −Vif /−A3G |
O2-16 +Vif /+A3G |
+ Control −Vif /+A3G |
|
|---|---|---|---|---|
| Hypermutation Rateb | 1.0% | 3.1% | 81.8% | 97.1% |
| GG to AGc | 14.4% | 17.2% | 79.6% | 86.6% |
| GA to AAc | 36.1% | 35.4% | 16.8% | 11.7% |
| GT to ATc | 37.8% | 35.1% | 3.0% | 1.6% |
| GC to ACc | 11.7% | 12.3% | 0.7% | 0.1% |
| G mutations increased | 1 | 1.1 | 20.1 | 303 |
| STOP | --- | --- | 46.2% | 35.9% |
| MISSENSE | --- | --- | 40.3% | 49.7% |
| NO CHANGE | --- | --- | 13.5% | 14.4% |
HIV proviral DNA to control for mutations that arise during PCR and NGS
Among reads with at least 1 mutation, the % of reads containing 3 or more G mutations within an individual sequence read (150–200 bp read length).
% of total G mutations with nearest neighbor preference in an 885 bp region of the pol gene of 5.0 ×105 to 1.0 ×106 reads generated by NGS.
The fold-increase in G mutations compared to Negative (−) Control.
An 885 base pair (bp) region of the pol gene was selected as a region highly mutated by A3G (Yu et al., 2004). The Ion Torrent sequencing methodology reads 150–200 bp fragment libraries, therefore the hypermutation rate is the calculation of the percentage of G mutations among all fragments with at least 1 mutation to have 3 or more mutations in the 150–200 bp read. Nested PCR of HIV proviral vector (– Control) and gDNA isolated from cells infected with O2-16 treated virus −Vif/−A3G both had a low percentage of sequences with 3 or more mutations (1.0% and 3.1%, respectively, Table 1). The slight increase in the O2-16 treated, -Vif/−A3G condition was likely due to contributions from low fidelity in HIV RT (Tripathi et al., 2012). As anticipated, the -Vif/+A3G (+ Control) infection condition had A3G hypermutation activity that was very high (97.1%, Table 1). Consistent with the MOA of O2-16, A3G hypermutation activity was enabled by O2-16 treatment of the +Vif/+A3G condition to levels that were comparable with those observed without drug when Vif was absent (81.8%, Table 1).
The nearest neighbor nucleotide preferences for dC that A3G mutates in HIV minus strand DNA are a 5’-dC and to a lesser extent 5’-dT with infrequent A3G mutations next to 5’-dA or -dG (Liddament et al., 2004). Table 1 compares the nearest neighbor preference of these four conditions in NGS to identify A3G signature mutations, which are manifested as dG to dA changes in the plus strand read. In the + Control this nearest neighbor preference was high for GG to AG (86.6%) and less so for GA to AA (11.7 %), with very low preference for AT or AC changes (1.7% and 0.1%, respectively, Table 1). The mutations resulting from O2-16 treatment of the +Vif/+A3G condition closely resembled the nearest neighbor preferences of the + Control (Table 1). As anticipated, O2-16 treatment −Vif/−A3G condition closely resembled the - Control pattern, with no apparent preference (Table 1).
A3G expression in the absence of Vif (+ Control) led to a 303-fold increase in G mutation rate compared to NGS of the HIV proviral vector (− Control) representing maximum mutation rate that could be expected in the pseudotyped HIV system (Table 1). In comparison, the O2-16 treatment +Vif/+A3G condition resulted in a 20-fold increase in G mutations over the - Control (Table 1), consistent with how much A3G was incorporated into these virions compared to the −Vif control (Fig. 2C). The mutation rates we have observed with O2-16 are likely to be catastrophic for HIV as hypothetically, only a 2- to 6-fold increase in HIV mutation rate would be required to exceed the threshold error rate necessary to induce nonviable HIV genomes (Tripathi et al., 2012). Supporting this prediction, the mutated sequences of the Gag-Pol protein suggested that O2-16 enabled A3G to produce mostly nonsense (stop) and missense mutations in similar percentages as the + Control (Table 1).
3.6. O2-16 has antiviral activity only in A3G expressing T-cells in a 14-day spreading HIV infection
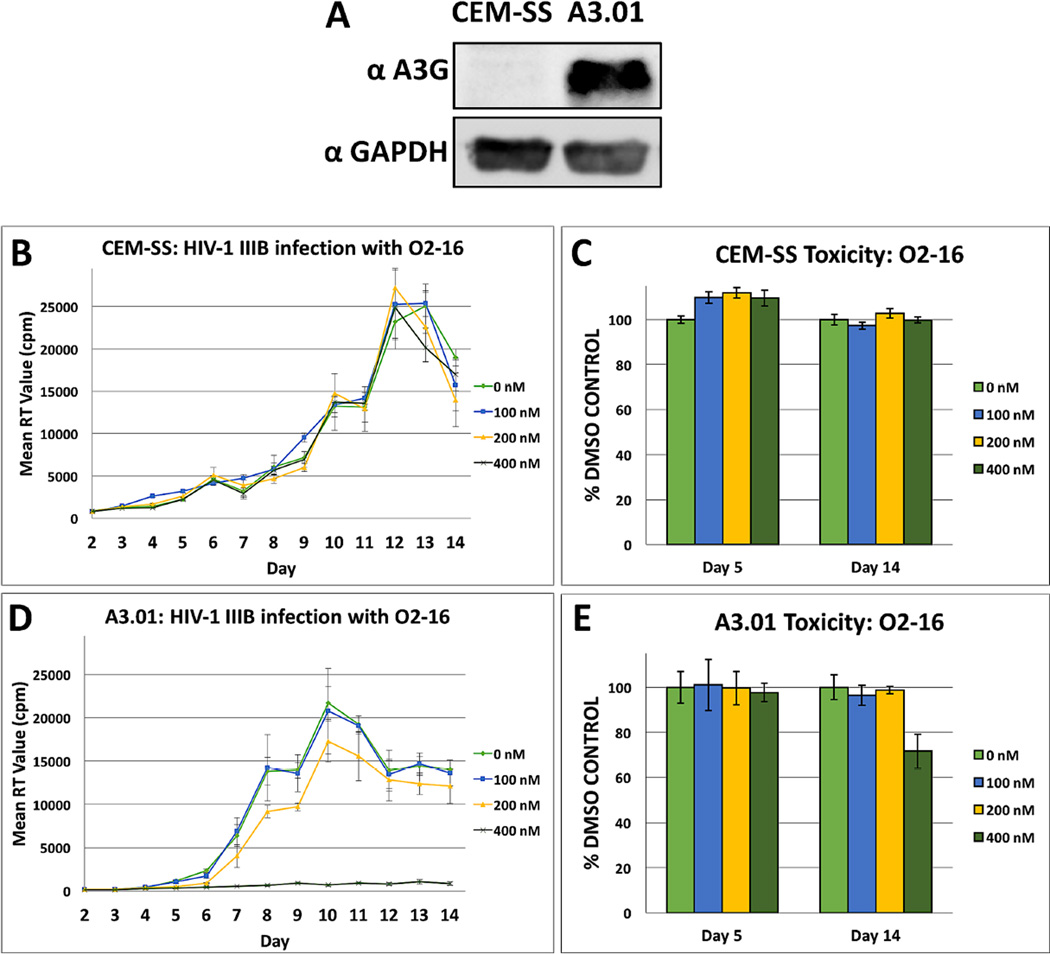
Two CEM-derived clonal cell lines, CEM-SS with low A3G expression and A3.01 with high A3G expression (Fig. 3A) were infected with the HIVIIIB isolate at low multiplicity of infection (MOI) to evaluate the A3G-dependency of the O2-16 antiviral effect during a 14-day spreading infection. Infected cells were dosed with 0 to 400 nM of O2-16, every other day. O2-16 had no effect on HIV production in the CEM-SS cell line (Fig. 3B) and no cytotoxicity in uninfected cultures treated with drug in parallel and monitored for changes in cell viability (Fig. 3C). Supporting the predicted MOA, 400 nM O2-16 strongly neutralized HIV infection in A3.01 cells throughout the 14-day study (Fig. 3D). In contrast, the HIV protease inhibitor (PI) Ritonavir showed no A3G-dependence and neutralized infectivity at 100 nM in both cell lines (Fig. S4). Following 5 days of treatment with 400 nM of O2-16 there was no cytotoxic effect on the cells and by Day 14 with treatment every other day, the viability of A3.01 cells treated with O2-16 was ~75% of the DMSO control (Fig. 3E). By contrast CPT was very toxic above 0.5 nM (data not shown), but was tested at 0.05, 0.1 and 0.5 nM (Fig. S5), and there was an antiviral effect that was not as robust as O2-16 and, consistent with the toxicity of CPT, was evident in both cell types (Fig. S5A & C). Also, opposed to 400 nM O2-16 (Fig. 3D) by day five 0.5 nM CPT treatment yielded cell viability that was down to ~75% (Fig. S5D). This data confirmed the data from Fig. 1 that TOP1 activity and Vif inhibitory activity cannot be separated with CPT, but the modifications in O2-16 eliminated TOP1-related toxicity, thus enabling A3G-dependent antiviral activity.
Fig. 3. O2-16 antiviral activity is dependent on A3G expression.
(A) A western blot with an A3G antibody obtained through the NIH AIDS Reagent Program, Anti-Human APOBEC3G C-terminal Polyclonal from Dr. Jaisri Lingappa and a GAPDH antibody as a loading control showing the relative A3G expression in two CEM-derived cell lines, with low to no expression in CEM-SS vs. strong expression in A3.01. (B) CEM-SS and (D) A3.01 14-day HIVIIIB infections at low MOI were treated every other day with 0, 100, 200, and 400 nM of O2-16. RT activity was measured with radioactive nucleotides and measured by radioactive counts per minute (cpm) to determine relative HIV abundance in the media. 400 nM O2-16 neutralized HIV infection only in A3G expressing A3.01, n=3 with error bars showing SD. (C) CEM-SS and (E) A3.01 cytotoxicity relative to DMSO control at days 5 and 14 measured by tetrazolium dye, n=3 with error bars showing SD.
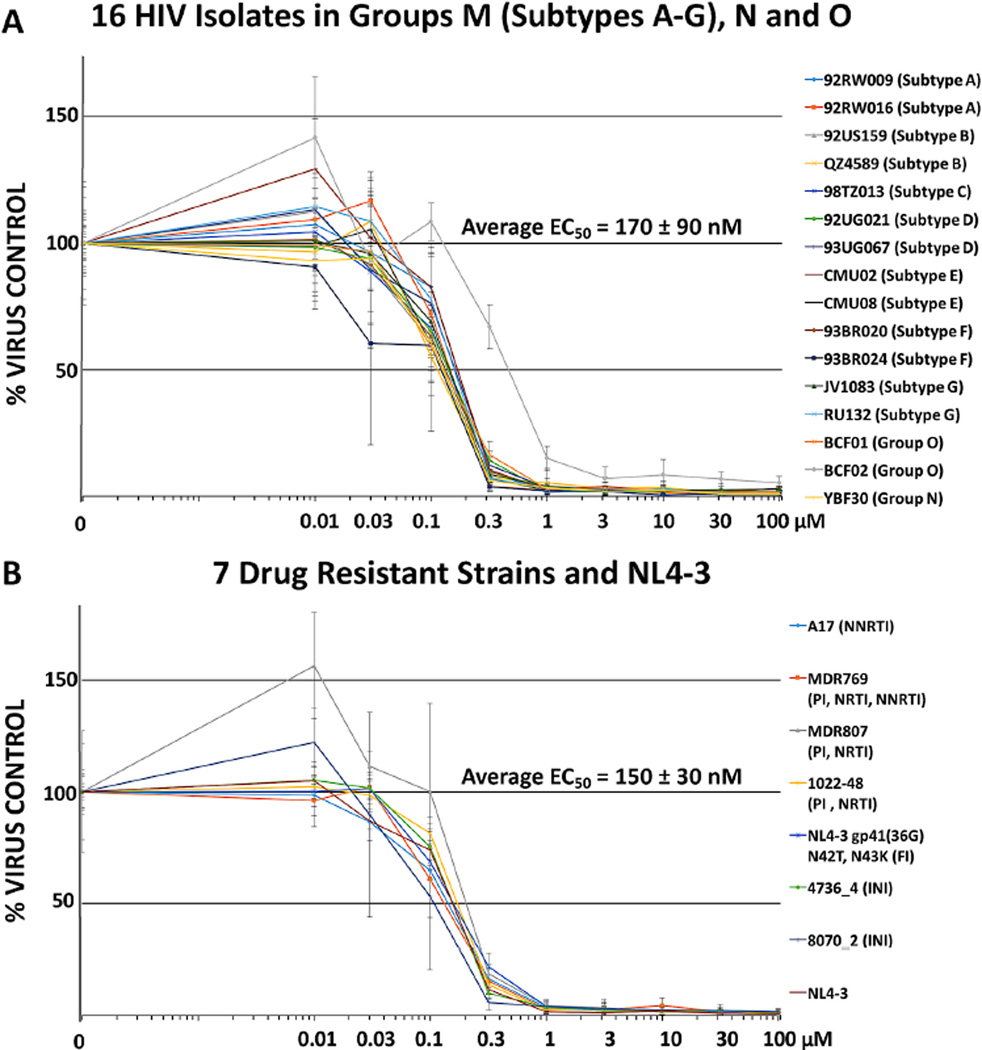
3.7. O2-16 was broadly neutralizing across HIV subtypes infecting PBMCs
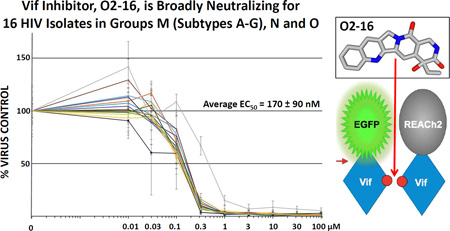
We evaluated the antiviral efficacy of O2-16 in PHA/IL-2 activated pooled patient peripheral mononuclear cells (PBMCs) in a 7-day infection using 16 HIV clinical isolates from 7 group M HIV subtypes as well as groups O and N. O2-16 neutralized all of the HIV isolates evaluated with an average 50% efficacy concentration (EC50) of 170 ± 90 nM (Fig. 4A). In PBMCs we observed a modest effect on cell viability with increasing concentration consistent with a cytostatic effect as opposed to overt cell toxicity (Fig. S6). In fact, multiple in vitro tests of cytostasis, changes in cell morphology, stress and apoptosis, and viability indicated that O2-16 treatment was not associated with cell death or necrosis, although in the micromolar range O2-16 had a cytostatic effect (Table S1).
Fig. 4. O2-16 antiviral activity is broadly neutralizing in PBMC.
Relative HIV abundance based on RT activity measured on day 7 compared to virus control of cells treated with a single dose of O2-16 on day 1. Doses ranged from 0.01 to 100 µM and all isolates were overlaid on a single graph for (A) 16 HIV isolates from 7 HIV-1 Group M envelope subtypes A, B, C, D, E, F, and G, as well as isolates from HIV-1 Groups O and N (subtypes/groups in parentheses listed on the right, detailed description of viral isolates are in Supplemental Materials and Methods), n=3 with error bars showing SD (average EC50 displayed with SD, n=16). (B) 7 drug resistant strains and NL4-3 (i.e., wild-type control) with abbreviations to drug types for which the isolates are resistant in parentheses listed on the right, n=3 with error bars showing SD (average EC50 displayed with SD, n=8).
3.8. O2-16 was broadly neutralizing of drug resistant HIV isolates and did not target other HIV proteins
Next we tested seven drug-resistant strains of HIV and the common laboratory strain pNL4-3 (wild-type control) to further establish that the antiviral activity of O2-16 was in fact Vif-dependent and not dependent on protein targets of other known HIV therapeutics, such as RT inhibitors (both nucleoside, NRTI and non-nucleoside-based RT inhibitors, NNRTI), PI, viral fusion inhibitors (FI), and Integrase inhibitors (INI). The antiviral EC50 (150 ± 30 nM) of O2-16 against multiple or single drug-resistant strains (Fig. 4B) were on par with the values seen for wild type HIV isolates (Fig. 4A). These findings corroborated those from A3.01 cells showing that the MOA for O2-16 was Vif- and A3G-dependent (Fig. 3) and confirmed that the antiviral mechanism of O2-16 was unique. To further establish Vif as the target, O2-16 was tested using in vitro assays that measure HIV Protease, Integrase and RT enzymatic activities. Whereas assays demonstrated activity with their cognate inhibitors, no inhibition was detected up to 100 µM with O2-16 (Table S2).
To establish the time in the viral life cycle O2-16 activity is manifest we employed a time of addition assay. In this assay an inhibitor’s efficacy depends on its addition to cells prior to and during the expression and function of the antiviral target. The inhibitor would be ineffective if added after this point. The control inhibitors included in the experiment began to lose antiviral efficacy at the following time points post-infection: FI Maraviroc at 1hr, NRTI AZT at 4hrs, NNRTI Nevirapine at 6hrs, INI Raltegravir after 8hrs (between 8hr and 24hr time points), and HIV RNA transcription inhibitor Temacrazine was effective through 24 hours of addition (Fig. S7). RLU from TZM-bl cells revealed that both the PI Darunavir and O2-16 had no activity when compound was added at 0, 1, 2, 4, 6, 8 and 24 hours after being infected with HIV Ba-L and RLU read at 48 hours (Fig. S7). The TZM-bl readout was dependent on HIV LTR reporter driven expression of luciferase (Fig. S7), therefore any inhibitor that has an effect downstream of viral translation would not be evident in the assay. These results showed that O2-16 had no effect at any stage of the viral life cycle prior to and including Tat transactivation of the HIV LTR, consistent with the lack of activity of the PI in this assay. Therefore, O2-16 is indeed acting late in the viral life cycle when Vif is blocking A3G packaging during viral particle assembly.
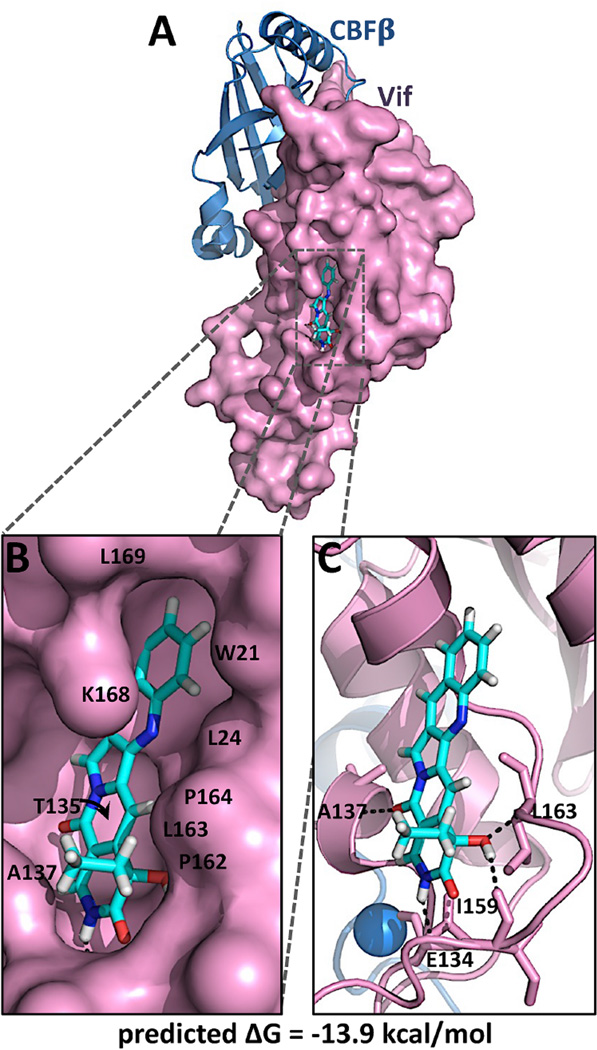
3.9. Modeling predicts that O2-16 occupies a pocket next to the PPLP motif
An unbiased docking of O2-16 to the exposed surface of the Vif crystal structure (Guo et al., 2014) was performed to assess whether O2-16 had the potential for binding to the surface of Vif. A low energy binding interaction was predicted with a ΔG of −13.9 kcal/mol (Fig. 5). O2-16 was predicted to bind a surface pocket on Vif, forming hydrophobic interactions with residues W21, L24, T135, A137, P162, L163, P164, K168 and L169 (Fig. 5B), and specific H-bonds with residues D134, A137, I159, and L163 that form the pocket (Fig. 5C). Interestingly, the unbiased docking predicted direct contact with 3 of the 4 residues in the 161-PPLP-164 motif described in section 1 as being crucial for Vif self-association, A3G degradation and RNA binding. Moreover, the PPLP motif enables crucial functional interactions in the cell for Vif to degrade A3F as well, thus the modeling correlates well the ability of O2-16 to inhibit Vif-dependent degradation of A3F along with A3G (Fig. 2).
Fig. 5. O2-16 docked to the crystal structure of Vif.
O2-16 depicted as a stick model is shown docked to a pocket on the solvent-exposed surface of the crystal structure of Vif. In (A) a surface rendering of Vif is depicted to illustrate the fit of O2-16 in the hydrophobic pocket of Vif, with (B) showing a close up of the region with interacting residues W21, L24, T135, A137, P162, L163, P164, K168 and L169 shown. In (C) Vif (purple) is depicted as a cartoon in the same orientation as A & B to illustrate the predicted H-bonds (black dashed lines) between O2-16 and Vif residues E134, A137, I159 and L163.
4. Discussion
We have established herein that a CPT analog that lacks TOP1 inhibitory activity was broadly neutralizing across HIV clinical groups and subtypes. Taking advantage of multiple assay methods, we demonstrated the MOA of the antiviral activity as an inhibitor of Vif multimerization and Vif-dependent A3G and A3F degradation. In the presence of Vif, O2-16 enabled an enhanced recovery of A3G and A3F with viral particles and A3G signature hypermutations of HIV proviral DNA. The observation that antiviral activity required A3G expression and that drug resistant strains were neutralized by O2-16 further suggested that the antiviral activity was A3G-dependent and was associated with a unique target. This conclusion was further strengthened by the lack of O2-16 activity against HIV Protease, Integrase and RT along with a time of addition assay showing that the target for O2-16 was late in the viral life cycle. Predictions from unbiased molecular docking further supported that the O2-16 binding site is adjacent to the PPLP motif in Vif.
A notable difference in effective concentrations of O2-16 for the different assays was observed. However, it must be noted that in the 293T based assays (e.g., Vif multimerization, Vif-dependent degradation and pseudotyped HIV viral production) cells were transfected with higher amounts of Vif than levels naturally expressed during an infection; requiring more compound to compensate for the increased number of Vif molecules. In contrast, A3.01 and PBMC live virus infections involved multiple rounds of infection over 7 to 14 days starting with a low MOI that was amplified through a spreading infection. Consistent with this explanation, nanomolar levels of O2-16 had effective antiviral activity in spreading infection assays compared with the micromolar levels required for 293T pseudotyped virus and high throughput analyses. Moreover, Refsland et al. showed that relative mRNA expression of A3G is 4-fold higher in PBMCs compared to CEM cells and 40-fold higher than CEM-SS. This observation correlates well with the apparent increased efficacy in PBMCs vs. A3.01 and CEM-SS (Refsland et al., 2010). A3F expression was higher in PBMCs as well, but to a lesser extent, and A3F was overall expressed much lower than A3G in both cell types (Refsland et al., 2010). Ultimately, the cell-based assays and CEM cell line studies established the compound’s target, while the benchmark for the potency of the antiviral efficacy may be more appropriately gauged from the more biologically relevant PBMC assay.
Removing TOP1 inhibitory activity not only aided in identifying the antiviral MOA of CPT analogs but also significantly decreased the compound’s cytotoxicity compared to CPT analogs currently in clinical use for treating cancer. The breadth of pre-clinical and clinical data on CPT analogs for cancer treatment will help guide the lactam analogs through formulation and clinical development of a therapeutic lead that exploits an untapped antiviral pathway.
Exploiting the antiviral activity of A3G is a long sought after but novel HIV therapeutic approach. Vif antagonists also present an opportunity to prevent the establishment of new reservoirs as HIV latent reservoirs are activated as part of ‘cure’ strategies to reveal and eliminate viral reservoirs (Kent et al., 2013; Ruelas and Greene, 2013; Xing and Siliciano, 2012). Vif inhibitors (alone or in combination with other antiretroviral drugs) would potentially ensure that viral particles of HIV budding from activated reservoirs will be defective because with Vif disabled they would contain A3G and A3F. A Vif inhibitor would be a significant weapon in the arsenal to combat HIV and identify the most effective combination of antiviral targets for therapeutic, preventative and eradication strategies.
Supplementary Material
Highlights.
This study identifies Vif as the TOP1 independent anti-HIV target of CPT analogs.
Vif multimerization and Vif-dependent A3G and A3F degradation are inhibited by the CPT analog, O2-16.
This Vif inhibition leads to A3G packaging and HIV hypermutations and is broadly neutralizing to all HIV clades.
A highly active Vif antagonist enables a potential new combinatorial strategy for therapy, prevention and eradication.
Acknowledgments
This work was supported in part by Public Health Service Grants R01 GM110119, R01 GM110038 and R21 NS067671 awarded to H.C.S. through OyaGen, Inc. This project was also conducted in part by Southern Research Institute using federal funds from the Division of AIDS, NIAID/NIH under contract HHSN272200700041C entitled “Confirmatory In Vitro Evaluations of HIV Therapeutics” and HHSN272201400010I entitled “In Vitro Testing Resources for HIV Therapeutics and Topical Microbicides”. We thank Junzhong Peng and Larry Ross of Southern Research Institute for their assistance in performing HIV RT (J.P.), Integrase (J.P.) and Protease (L.R.) enzyme inhibition assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
H.C.S. is the founder, CEO and president of OyaGen, Inc. and R.P.B. is the company’s CSO and lab director. Therefore, these individuals have a financial interest in the development of Vif antagonists as a treatment for HIV/AIDS.
References
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Anderson BD, Harris RS. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-beta/RUNX axis. Sci Adv. 2015;1:e1500296. doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batisse J, Guerrero SX, Bernacchi S, Richert L, Godet J, Goldschmidt V, Mely Y, Marquet R, de Rocquigny H, Paillart JC. APOBEC3G impairs the multimerization of the HIV-1 Vif protein in living cells. J Virol. 2013;87:6492–6506. doi: 10.1128/JVI.03494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JR, Huthoff H, Veselkov DA, Beavil RL, Simpson PJ, Matthews SJ, Malim MH, Sanderson MR. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 2010;6:e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S, Mercenne G, Tournaire C, Marquet R, Paillart JC. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2011;39:2404–2415. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckheit RW, Jr, Swanstrom R. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res Hum Retroviruses. 1991;7:295–302. doi: 10.1089/aid.1991.7.295. [DOI] [PubMed] [Google Scholar]
- Burke TG, Adams VR. Camptothecins in cancer therapy. Totowa, N.J: Humana Press; 2005. [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Cen S, Peng ZG, Li XY, Li ZR, Ma J, Wang YM, Fan B, You XF, Wang YP, Liu F, Shao RG, Zhao LX, Yu L, Jiang JD. Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem. 2010;285:16546–16552. doi: 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C, Smith JL, Hu WS, Pathak VK. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3’ --> 5’ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Donahue JP, Vetter ML, Mukhtar NA, D’Aquila RT. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Ameer-Beg SM, Ng TT, Vojnovic B, Wouters FS. A dark yellow fluorescent protein (YFP)-based Resonance Energy-Accepting Chromoprotein (REACh) for Forster resonance energy transfer with GFP. Proc Natl Acad Sci U S A. 2006;103:4089–4094. doi: 10.1073/pnas.0509922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hobbs S, Jitrapakdee S, Wallace JC. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1alpha promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem Biophys Res Commun. 1998;252:368–372. doi: 10.1006/bbrc.1998.9646. [DOI] [PubMed] [Google Scholar]
- Hultquist JF, Binka M, Larue RS, Simon V, Harris RS. Vif Proteins of Human and Simian Immunodeficiency Viruses Require Cellular CBFbeta To Degrade APOBEC3 Restriction Factors. J Virol. 2012;86:2874–2877. doi: 10.1128/JVI.06950-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaxel C, Kohn KW, Wani MC, Wall ME, Pommier Y. Structure-activity study of the actions of camptothecin derivatives on mammalian topoisomerase I: evidence for a specific receptor site and a relation to antitumor activity. Cancer Res. 1989;49:1465–1469. [PubMed] [Google Scholar]
- Kent SJ, Reece J, Petravic J, Martyushev A, Kramski M, De Rose R, Cooper DA, Kelleher AD, Emery S, Cameron PU, Lewin SR, Davenport MP. The search for an HIV cure: tackling latent infection. Lancet Infectious Diseases. 2013;13:614–621. doi: 10.1016/S1473-3099(13)70043-4. [DOI] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M, Booiman T, Kootstra N, Simon V, Ooms M. Identification of the HIV-1 Vif and Human APOBEC3G Protein Interface. Cell Rep. 2015;13:1789–1799. doi: 10.1016/j.celrep.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wu LI, Emerman M. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol. 2010a;84:88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem. 2007;44:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Li YY, Chen SW, Yang LM, Wang RR, Pang W, Zheng YT. The anti-HIV actions of 7- and 10-substituted camptothecins. Molecules. 2010b;15:138–148. doi: 10.3390/molecules15010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Current biology : CB. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- Mercenne G, Bernacchi S, Richer D, Bec G, Henriet S, Paillart JC, Marquet R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–646. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton S, Pantazis P, Epstein JS, Sadaie MR. 9-Nitrocamptothecin inhibits tumor recrosis factor-mediated activation of human immunodeficiency virus type 1 and enhances apoptosis in a latently infected T cell clone. AIDS Res Hum Retroviruses. 1998;14:39–49. doi: 10.1089/aid.1998.14.39. [DOI] [PubMed] [Google Scholar]
- Munk C, Willemsen A, Bravo IG. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Cao H, Sharova N, Ali A, Sharkey M, Stranska R, Stevenson M, Rana TM. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak RG, Gallay PA, Jochmans D, Halestrap AP, Ruegg UT, Pallansch LA, Bobardt MD, de Bethune MP, Neyts J, De Clercq E, Dumont JM, Scalfaro P, Besseghir K, Wenger RM, Rosenwirth B. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob Agents Chemother. 2008;52:1302–1317. doi: 10.1128/AAC.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter JD, Morales GA, Smith HC. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends in biochemical sciences. 2014 doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K, De Noronha C, Yonemoto W, Greene WC. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing both Its Translation and Intracellular Stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tripathi K, Balagam R, Vishnoi NK, Dixit NM. Stochastic simulations suggest that HIV-1 survives close to its error threshold. PLoS Comput Biol. 2012;8:e1002684. doi: 10.1371/journal.pcbi.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RC, Jr, Khan MA, Kao S, Goila-Gaur R, Miyagi E, Strebel K. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. Journal of virology. 2010;84:5201–5211. doi: 10.1128/JVI.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Wani MC, Cook CE, Palmer KH, McPhail AT, Sim GA. Plant Antitumor Agents. I. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata1,2. Journal of the American Chemical Society. 1966;88:3888–3890. [Google Scholar]
- Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- Wolfe LS, Stanley BJ, Liu C, Eliason WK, Xiong Y. Dissection of the HIV Vif interaction with human E3 ubiquitin ligase. J Virol. 2010;84:7135–7139. doi: 10.1128/JVI.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Siliciano RF. Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discovery Today. 2012;18:541–551. doi: 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang JL, Sharma PL, Li CJ, Dezube BJ, Pardee AB, Crumpacker CS. Topotecan inhibits human immunodeficiency virus type 1 infection through a topoisomerase-independent mechanism in a cell line with altered topoisomerase I. Antimicrob Agents Chemother. 1997;41:977–981. doi: 10.1128/aac.41.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.