Abstract
Protein arginine methyltransferase 3 (PRMT3) forms a stable complex with 40S ribosomal protein S2 (RPS2) and contributes to ribosome biogenesis. However, the molecular mechanism by which PRMT3 influences ribosome biogenesis and/or function still remains unclear. Using quantitative proteomics, we identified human programmed cell death 2-like (PDCD2L) as a novel PRMT3-associated protein. Our data suggest that RPS2 promotes the formation of a conserved extraribosomal complex with PRMT3 and PDCD2L. We also show that PDCD2L associates with 40S subunit precursors that contain a 3′-extended form of the 18S rRNA (18S-E pre-rRNA) and several pre-40S maturation factors. PDCD2L shuttles between the nucleus and the cytoplasm in a CRM1-dependent manner using a leucine-rich nuclear export signal that is sufficient to direct the export of a reporter protein. Although PDCD2L is not required for the biogenesis and export of 40S ribosomal subunits, we found that PDCD2L-null cells accumulate free 60S ribosomal subunits, which is indicative of a deficiency in 40S subunit availability. Our data also indicate that PDCD2L and its paralog, PDCD2, function redundantly in 40S ribosomal subunit production. Our findings uncover the existence of an extraribosomal complex consisting of PDCD2L, RPS2, and PRMT3 and support a role for PDCD2L in the late maturation of 40S ribosomal subunits.
INTRODUCTION
Proliferating cells greatly rely on protein synthesis to provide a continuous source of structural and catalytic proteins to grow, duplicate, and expand. The ribosome is an evolutionarily conserved macromolecular machine responsible for protein synthesis and consists of a ribonucleoprotein complex composed of four noncoding rRNAs and ∼80 ribosomal proteins (RPs) in eukaryotes. Ribosome biogenesis necessitates a major commitment in terms of cellular energy consumption, involving transcription from all three eukaryotic RNA polymerases, the spatial and temporal action of more than 300 ribosome assembly factors, and several quality control checkpoints to ensure proper ribosome assembly (1). In fact, given the substantial amount of resources invested in making ribosomes, the past few years have seen a growing body of evidence linking ribosome biogenesis to nutrient, growth factor, and stress responses (2), thereby implicating the regulation of ribosome biogenesis in stem cell homeostasis (3, 4) and cancer biology (5).
In eukaryotes, ribosome biogenesis begins in the nucleolus with the synthesis of a precursor rRNA (pre-rRNA) that is cotranscriptionally assembled into a large ribonucleoprotein particle (RNP) via the recruitment of specific RPs and early maturation factors (6). This large ∼90S preribosome complex is then rapidly converted into precursors of the 40S and 60S ribosomal subunits via a complex series of endonucleolytic and exonucleolytic RNA cleavage events (6). Pre-40S particles are rapidly exported from the nucleus and further processed in the cytoplasm, whereas maturation of pre-60S particles continues in the nucleus before export to the cytoplasm. The nuclear export of both pre-40S and pre-60S particles depends on the RanGTP-binding exportin CRM1, which recognizes a leucine-rich nuclear export signal (NES) in adaptor proteins to facilitate export from the nucleus (7). Interestingly, whereas the NES-containing adaptor Nmd3 has been shown to be essential for pre-60S export in yeast and vertebrates (8–11), the identification of a similar NES adaptor essential for pre-40S subunit export has remained elusive.
Historically, the ribosome is viewed as a generic machine that translates all mRNAs equally. However, findings in the past decade challenge this simplistic view and suggest that heterogeneity in ribosome composition may give rise to specialized ribosomes (12). One of the mechanisms proposed to promote ribosome heterogeneity is the addition of posttranslational modifications to RPs. Interestingly, whereas RPs are subject to a variety of posttranslational modifications (13–15), few RP-modifying enzymes have been identified. Methylation of RPs at arginines in both the eukaryotic 40S and 60S ribosomal subunits is evolutionarily conserved (16–18) and fluctuates depending on growth conditions (19, 20). Work in the fission yeast Schizosaccharomyces pombe identified the first eukaryotic RP methyltransferase, protein arginine methyltransferase 3 (PRMT3), which methylates 40S ribosomal protein S2 (RPS2) (21). PRMT3 is an evolutionarily conserved cytosolic arginine methyltransferase that contains a single C2H2-type zinc finger (22), which is required for interactions with RPS2 (23). Arginine methylation of RPS2 was also demonstrated in human cells (24) and in Saccharomyces cerevisiae (25), indicating the existence of a conserved RP modification. Consistent with a role in ribosome function, disruption of PRMT3 results in aberrant ribosome profiles in S. pombe and Arabidopsis (21, 23, 26). Furthermore, hypomorphic PRMT3 mice and PRMT3-null plants show developmental defects reminiscent of ribosome biogenesis mutants, including reduced embryo size and growth retardation (26, 27). As yet, however, the exact role of PRMT3 in ribosome biogenesis and/or ribosome function remains poorly understood.
Here, we used quantitative proteomics in human cells to identify novel PRMT3-associated proteins. High-resolution mass spectrometry (MS) analysis of PRMT3 purifications uncovered a stable complex consisting of PRMT3, RPS2, and the programmed cell death 2-like (PDCD2L) protein. PDCD2L belongs to a protein family that contains a TYPP (TSR4, YwqG, PDCD2L, and PDCD2) domain, which has been proposed to promote protein-protein interactions based on structure prediction (28). Although the S. cerevisiae ortholog of PDCD2L, Trs4p, is required for processing of the 20S pre-rRNA into mature 18S rRNA (29), the functional role of human PDCD2L had remained unknown. In this study, we show that a fraction of PDCD2L associates with late-stage 40S ribosomal subunit precursors that contain a 3′-extended form of 18S rRNA (18S-E pre-rRNA). PDCD2L contains a leucine-rich NES that is both necessary and sufficient for interactions with CRM1 and nucleocytoplasmic shuttling. Disruption of PDCD2L expression in human cells resulted in the accumulation of free 60S ribosomal subunits, a phenotype which is suggestive of defects in 40S ribosomal subunit availability. Our data also reveal some level of redundancy between PDCD2L and its paralog, PDCD2, in 40S ribosomal subunit biogenesis. Our findings uncover the existence of an extraribosomal complex consisting of PDCD2L, RPS2, and PRMT3 and support a role for PDCD2L in the late maturation of 40S ribosomal subunits.
MATERIALS AND METHODS
Cell culture.
HEK 293, U-2 OS, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% tetracycline-free fetal bovine serum (FBS). Inducible expression of green fluorescent protein (GFP), GFP-PRMT3, GFP-PDCD2L, GFP-PDCD2LNESmut, GFP-PABPN1, Flag-PDCD2L, and Flag-PABPN1 was achieved by site-directed recombination using the Flp-flippase recognition target system in HEK 293-FT and U-2 OS-FT cells, as previously described (30). Induction of GFP- and Flag-tagged proteins was achieved with 500 ng/ml of doxycycline for 20 h to 72 h. Small interfering RNAs (siRNAs) were transfected with Lipofectamine 2000 at a final concentration of 25 nM (control siRNA [siControl] and siRNA against PDCD2L [siPDCD2L]) or 32 nM (siBystin and siRPS2) for 72 h.
Generation of PDCD2L-null cells using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system.
For the deletion of PDCD2L in HeLa cells, 2 guide RNAs (gRNAs), the Cas9 nickase, and a template DNA were used. gRNA-A (5′-CGTGCACCGGCGCATCTCGAAGG-3′) and gRNA-B (5′-TGCCTGGACTGCTAGCAAGCTGG-3′) were designed via the CRISPR Design Web tool (available at http://crispr.mit.edu/). These sequences were inserted into the pSpCas9n(BB)-2A-GFP vector (Addgene) as previously described (31). For the construction of the template DNA construct containing the puromycin resistance gene (puromycin N-acetyltransferase [PAC]) under the control of the cytomegalovirus (CMV) promoter flanked by two PDCD2L homology regions, pEGFP-C1 (Clontech) was used as the backbone vector. The PAC sequence was amplified from pTRIPZ (GE Dharmacon), and the CMV promoter and immediate early enhancer sequences were amplified from pEGFP-C1 (Clontech). PDCD2L homology sequences were amplified from HeLa genomic DNA. For the 5′ PDCD2L homology arm, a 791-bp sequence ending at the nucleotide before gRNA-A was amplified. For the 3′ PDCD2L homology arm, a 784-bp sequence starting at the nucleotide after gRNA-B was amplified. Gibson assembly was used to insert the homology arms into the backbone vector. The PAC and CMV promoter sequences were joined by PCR fusion and inserted between the homology arms using BglII and NotI digestions.
HeLa cells were seeded into a 15-cm plate. The next day, cells were transfected with 10 μg of pSpCas9n(BB)-2A-GFP-gRNA-A, 10 μg of pSpCas9n(BB)-2A-GFP-gRNA-B, and 20 μg of the linearized DNA template using 80 μl of Lipofectamine 2000 (Life Technologies). At 48 h posttransfection, positive cells were selected by the addition of 2 μg/ml of puromycin (Wisent) to the cell culture medium. Following the visual detection of puromycin-resistant colonies, cells were detached, counted, and diluted sufficiently to obtain 1 cell/well in a 96-well plate. Following cellular expansion of individual clones in puromycin-supplemented medium, the cells were divided into two 24-well plates: one plate was used for screening by Western blotting, and the other was used for cell maintenance. In total, five independent PDCD2L-null clones were frozen and kept in liquid nitrogen. The inactivation of PDCD2L alleles was confirmed by DNA sequencing and Western blotting.
SILAC and label-free purifications.
For stable isotope labeling in cell culture (SILAC) experiments, proteins were labeled with stable isotopes of arginine and lysine in cell culture, as previously described (32). Briefly, HEK 293-FT cells expressing GFP- or Flag-tagged versions of proteins were grown in medium containing labeled amino acids (Cambridge Isotope Laboratories). Twenty-four hours to 48 h after induction with doxycycline, cells were collected in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 2 mM MgCl2, 1 mM dithiothreitol [DTT], and 1× Complete protease inhibitor cocktail [Roche]) and incubated at 4°C for 20 min. Lysates were centrifuged for 10 min at 13,000 rpm at 4°C, and equal amounts of proteins were incubated with GFP-trap agarose beads from ChromaTek (Martinsried, Germany) or anti-Flag M2 affinity gel (Sigma-Aldrich) for 3 h at 4°C. Beads were then washed 5 times with lysis buffer, and proteins were subjected to two rounds of elution by the addition of 100 μl of denaturing buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1 mM DTT) for 10 min at 90°C. Eluates were concentrated by using a SpeedVac and resuspended in reducing buffer (125 mM Tris-HCl [pH 8.0], 1.25% SDS, 37.5% glycerol, 60 mM DTT, 0.025% bromophenol blue). Gel electrophoresis, in-gel digestions, liquid chromatography-tandem mass spectrometry (LC-MS/MS), and analysis of SILAC ratios were performed as described previously (32).
Protein analysis and antibodies.
Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by immunoblotting using the following primary antibodies: anti-PABPN1 (Epitomics); antitubulin, antiactin, and anti-Flag (catalog numbers T5168, A5441, and F1804, respectively; Sigma-Aldrich); anti-PRMT3 (catalog number A302-526A; Bethyl); anti-PDCD2L (catalog number A303-783A; Bethyl); anti-hRRP12, anti-PNO1, and antibystin (catalog numbers sc-139043, sc-133263, and sc-271722, respectively; Santa Cruz Biotechnology); anti-hLTV1 (catalog number ab122100; Abcam); anti-RPL17 (catalog number GTX111934; GeneTex); and anti-RPS2 (a generous gift from Mark Bedford). Membranes were then probed with either a donkey anti-rabbit antibody conjugated to IRDye 800CW (catalog number 926-32213; Li-Cor) or a goat anti-mouse antibody conjugated to Alexa Fluor 680 (catalog number A-21057; Life Technologies). Protein detection was performed by using an Odyssey infrared imaging system (Li-Cor).
Yeast two-hybrid assays.
PRMT3, RPS2, and PDCD2L cDNAs were cloned into plasmids pVP16 and pLexNA and transformed in Saccharomyces cerevisiae strain L40. Levels of β-galactosidase activity were measured by liquid assays within the linear response range, as previously described (33).
Sucrose gradient analysis.
To analyze the cosedimentation of PDCD2L with ribosomal particles, sucrose gradient centrifugation was performed as previously described (21). Briefly, cells were washed with phosphate-buffered saline (PBS), centrifuged, resuspended, and incubated in lysis buffer (10 mM Tris-HCl [pH 7.4], 100 mM KCl, 10 mM MgCl2,1 mM DTT, 1% Triton X-100, Complete protease inhibitor cocktail [Roche], 40 U/ml RNase Out [Life Technologies], and 50 μg/ml cycloheximide [Sigma-Aldrich]) for 20 min at 4°C. Following clarification of the lysate by centrifugation at 13,000 rpm for 10 min at 4°C, 7 to 10 mg of total protein was loaded onto a 10 to 45% sucrose gradient and centrifuged for 6 h at 40,000 rpm at 4°C in an SW41 rotor (Beckman Coulter). The gradient was then fractionated by upward displacement with 55% (wt/vol) sucrose using a gradient fractionator (Brandel Inc.) connected to a UA-6 UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm. Fractions (0.6 ml) were collected, and proteins were precipitated with trichloroacetic acid (TCA) (15% final concentration) and analyzed by Western blotting.
For ribosome distribution profiles, cycloheximide-treated cells were washed twice with PBS, and lysis buffer was added directly to 15-cm dishes. Cells were scraped, incubated for 20 min at 4°C, and centrifuged. Five percent of the supernatant was kept for Western analysis, and the remainder (3.5 mg of total protein) was loaded onto a 5 to 50% sucrose gradient and centrifuged for 3 h at 40,000 rpm. 40S, 60S, and 80S subunit curves were reproduced in Excel, and the area under the curve was calculated by using the rectangle method.
RNA analysis.
Total RNA was prepared by using TRIzol (Life Technologies), and pre-rRNA species were analyzed by Northern blotting as previously described (34), with the following modifications: 4 μg/well of total RNA, mixed with 5 volumes of loading dye (10 mM EDTA, 60% glycerol, 0.04% bromophenol blue), was loaded onto a 0.8% denaturing agarose gel (0.8% agarose, 20 mM morpholinepropanesulfonic acid [MOPS], 5 mM sodium acetate, 1 mM EDTA, 6.3% formaldehyde) and run in MOPS buffer (20 mM; 5 mM sodium acetate, 1 mM EDTA) at 110 V. Northern blots were hybridized by using 32P-labeled probes (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′ for 5′ internal transcribed spacer 1 [ITS1], 5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′ for 18S, and 5′-CCCGTTCCCTTGGCTGTGGTTTCGCT-3′ for 28S). Signals were detected and quantified by using a Typhoon Trio instrument.
RNA coimmunoprecipitation (RIP) assay.
A 15-cm dish of HEK 293-FT cells conditionally expressing Flag-PDCD2L or Flag-PABPN1 was induced by using doxycycline for 72 h. Cells were washed twice with PBS, and 5% of cells were kept for total RNA extraction (input fraction). Cells were then resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 2 mM MgCl2, 1 mM DTT, Complete protease inhibitor cocktail [Roche], and 40 U/ml RNase Out [Life Technologies]) and incubated for 20 min at 4°C. The lysate was centrifuged at 13,000 rpm for 10 min at 4°C, and 5% of the lysate was kept for protein analysis. The remainder of the lysate was incubated with anti-Flag M2 affinity gel (Sigma-Aldrich) for 3 h at 4°C. The beads were washed 5 times with lysis buffer, and 10% of the beads were kept for protein analysis. RNA was extracted from the remainder of the beads by using TRIzol reagent (Life Technologies) and analyzed by Northern blotting.
rRNA pulse-chase assays.
Pulse-chase analyses of pre-rRNA processing were performed based on a previously described method (35) but with some modifications. Briefly, wild-type and PDCD2L-null cells were incubated in serum-free and methionine-free medium for 30 min, after which the cells were pulsed for 30 min by using the same medium but supplemented with 25 μCi of l-[methyl-3H]methionine (1 μCi/ml). The cells were then rinsed with regular medium and chased in complete DMEM for the indicated times. Cells were rinsed with ice-cold PBS and resuspended in 500 μl of TRIzol reagent for RNA extractions. A total of 15,000 to 20,000 cpm of radioactivity was resolved on 1.25% agarose–formaldehyde gels. RNAs were then transferred to Hybond-N+ membranes (GE Healthcare), UV cross-linked, and exposed to film for 3 to 4 weeks at −80°C.
Microscopy.
Leptomycin B (LMB) was purchased from Sigma-Aldrich and used at a final concentration of 10 nM in 70% methanol. Visual analysis of GFP-tagged proteins in human cells was performed as previously described (36). U-2 OS cells were washed twice with PBS, fixed with 4% paraformaldehyde for 10 min at room temperature, and washed twice again with PBS. Fixed cells were permeabilized by using a 0.2% Triton X-100–PBS solution for 10 min and washed 3 times with PBS. For the data shown in Fig. 5, cells were equilibrated for 20 min in PBS–1% bovine serum albumin (BSA) followed by 1 h of incubation with a primary antibody dilution in PBS–1% BSA (antifibrillarin at 1/400; Cell Signaling Technology). Cells were washed 3 times with PBS–1% BSA and incubated with a secondary antibody dilution (rabbit antibody-Alexa Fluor 568 [Invitrogen] at 1/1,000 in PBS–1% BSA) for 1 h. After 3 washes, coverslips were mounted onto a slide with SlowFade Gold antifade solution (Life Technologies). Images were captured by using a Zeiss Axio Observer microscope with either a 63× or a 100× oil objective.
FIG 5.
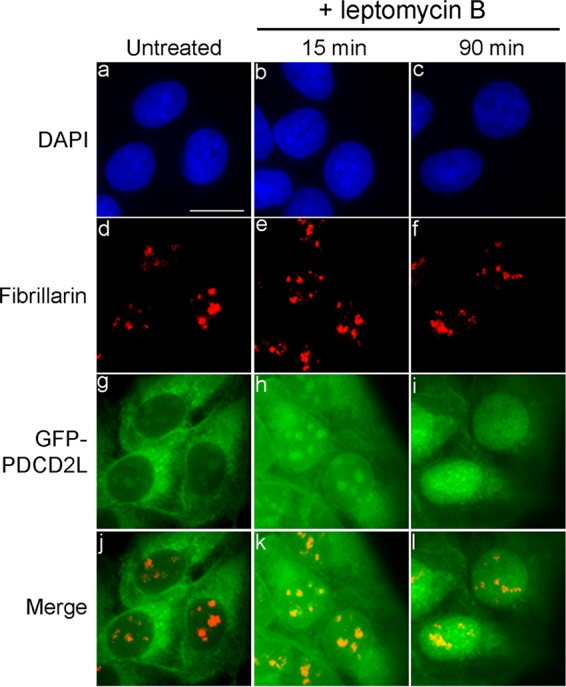
PDCD2L transits through the nucleolus. U-2 OS cells conditionally expressing GFP-PDCD2L were treated with LMB for 15 min (b, e, h, and k) and 90 min (c, f, i, and l) or left untreated (a, d, g, and j). Fixed cells were simultaneously analyzed by direct GFP fluorescence (g to i) and immunostaining for the nucleolar marker fibrillarin (d to f). DNA staining with DAPI shows the nucleus of each cell (a to c). Bar, 20 μm.
CRM1-binding assay.
Flow cytometry-based in vitro binding assays were performed as previously described (37). PDCD2L cDNA was transferred into plasmid pDEST15 by using LR Clonase II (Life Technologies). To obtain pDEST15-PDCD2LNESmut, we substituted Leu162, Leu165, and Leu167 for alanine residues by site-directed mutagenesis. Glutathione S-transferase (GST) alone, GST-PDCD2Lm and GST-PDCD2LNESmut were produced in Escherichia coli BL21 cells and purified by using glutathione-Sepharose beads. A total of 9 pmol Cy3-labeled CRM1 alone or in the presence of 180 pmol RanQ69L-GTP and 70 pmol Nup214 (amino acids [aa] 1916 to 2033) was used for binding assays.
RESULTS
Identification of PDCD2L as a new PRMT3-associated protein.
We and others previously reported the identification of a complex between PRMT3 and RPS2 (21, 24). However, with the exception of RPS2, the protein interaction network of PRMT3 has remained largely unknown. To investigate the network of PRMT3-associated proteins, we generated a Flp-In T-Rex HEK 293 cell line expressing a tetracycline-inducible and N-terminally GFP-tagged version of PRMT3. Specific PRMT3 interaction partners were determined by using the SILAC methodology (38), followed by GFP-PRMT3 purification and high-resolution MS. The presence of the GPF-PRMT3 bait protein in the eluate was confirmed by Western blotting before MS analysis.
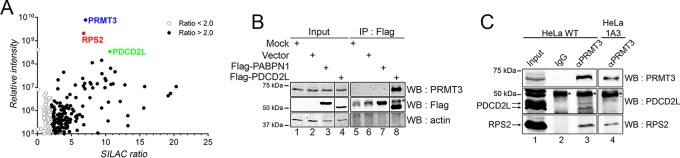
The SILAC-coupled MS approach classifies interactions by specificity (ratio of peptide intensities between the GFP-PRMT3 purification and a control purification) and protein abundance, as estimated from the sum of the peptide signal intensities of a given protein normalized to its molecular mass (Fig. 1A). As expected, RPS2 was the strongest PRMT3-associated protein identified by SILAC, detected at high levels in the PRMT3 pulldown. In total, 151 proteins showed at least a 2-fold enrichment of the SILAC ratio in the GFP-PRMT3 purification relative to the control (Fig. 1A; see also Table S1 in the supplemental material). Notably, besides RPS2, the PDCD2L protein was among the strongest PRMT3-associated proteins, showing a robust SILAC ratio and a high abundance (Fig. 1A; see also Table S1 in the supplemental material). A reciprocal immunoprecipitation assay validated the PRMT3-PDCD2L association, as PRMT3, but not actin, was detected in the eluate of an anti-Flag purification of Flag-PDCD2L (Fig. 1B, lane 8). In contrast, PRMT3 was not detected in eluates of control purifications (Fig. 1B, lanes 5 to 7). Importantly, we showed that immunoprecipitation of endogenous PRMT3 copurified endogenous PDCD2L and RPS2, whereas PDCD2L and RPS2 were not detected in a control immunoprecipitation (Fig. 1C, lanes 2 and 3). As an additional control, immunoprecipitation of endogenous PRMT3 using extracts prepared from PDCD2L-null cells (using CRISPR-Cas9-mediated genomic targeting [see Materials and Methods]) did not detect PDCD2L, yet RPS2 was still copurified with PRMT3 (Fig. 1C, lane 4). Consistent with this, the distribution of endogenous PRMT3 across a density gradient was not altered by a deficiency in PDCD2L (see Fig. S1 in the supplemental material). Together, these results uncover PDCD2L as a new PRMT3-associated protein and indicate that PDCD2L is not required for the association between PRMT3 and RPS2.
FIG 1.
PDCD2L is a PRMT3-associated protein. (A) GFP-PRMT3 SILAC copurification results plotted by SILAC ratios on the x axis (intensity of peptides originating from the GFP-PRMT3 purification versus the control purification), which reflects signal specificity, and signal intensities on the y axis (total peptide intensity for each protein normalized to molecular mass), which reflects the relative abundance of each protein in the purification. Proteins presenting SILAC ratios of >2.0 and <2.0 are indicated. GFP-PRMT3, RPS2, and PDCD2L are shown in color. (B) Western analysis of total extracts (Input) (lanes 1 to 4) and anti-Flag immunoprecipitates (IP) (lanes 5 to 8) prepared from HEK 293 cells transfected with the empty vector (lanes 2 and 6), Flag-PABPN1 (lanes 3 and 7), and Flag-PDCD2L (lanes 4 and 8) constructs. The asterisk indicates the IgG heavy chain. Areas from the same blot were spliced to remove irrelevant lanes (between lanes 3 and 4 and between lanes 7 and 8). (C) Western blot (WB) analysis of immunoprecipitates prepared by using a PRMT3-specific antibody (lanes 3 and 4) and a control IgG antibody (lane 2), using extracts prepared from wild-type (lanes 1 to 3) and PDCD2L-null (1A3) (lane 4) cells. The asterisk indicates the IgG heavy chain.
RPS2 is required for the association between PRMT3 and PDCD2L.
The predicted homolog of PDCD2L in Drosophila melanogaster, TRUS (CG5333), was previously identified as an RPS2-interacting protein by using a high-throughput two-hybrid screening approach (39). To test whether PDCD2L associates with RPS2 in human cells, we investigated the presence of RPS2 in immunoprecipitates of Flag-PDCD2L. As shown in Fig. 2A, both endogenous RPS2 and PRMT3 were detected after purification of Flag-tagged PDCD2L (lane 4) but not after purification of a control Flag-tagged protein (lane 3). We next analyzed the association between human RPS2, PRMT3, and PDCD2L by using the yeast two-hybrid system. We found that RPS2 strongly interacts with both PDCD2L (Fig. 2Bi) and PRMT3 (Fig. 2Bii). Interestingly, the interaction between PRMT3 and PDCD2L was roughly 5 times weaker than the interaction between PRMT3 and RPS2 (Fig. 2Bii), suggesting that RPS2 may promote and/or stabilize the association between PRMT3 and PDCD2L.
FIG 2.
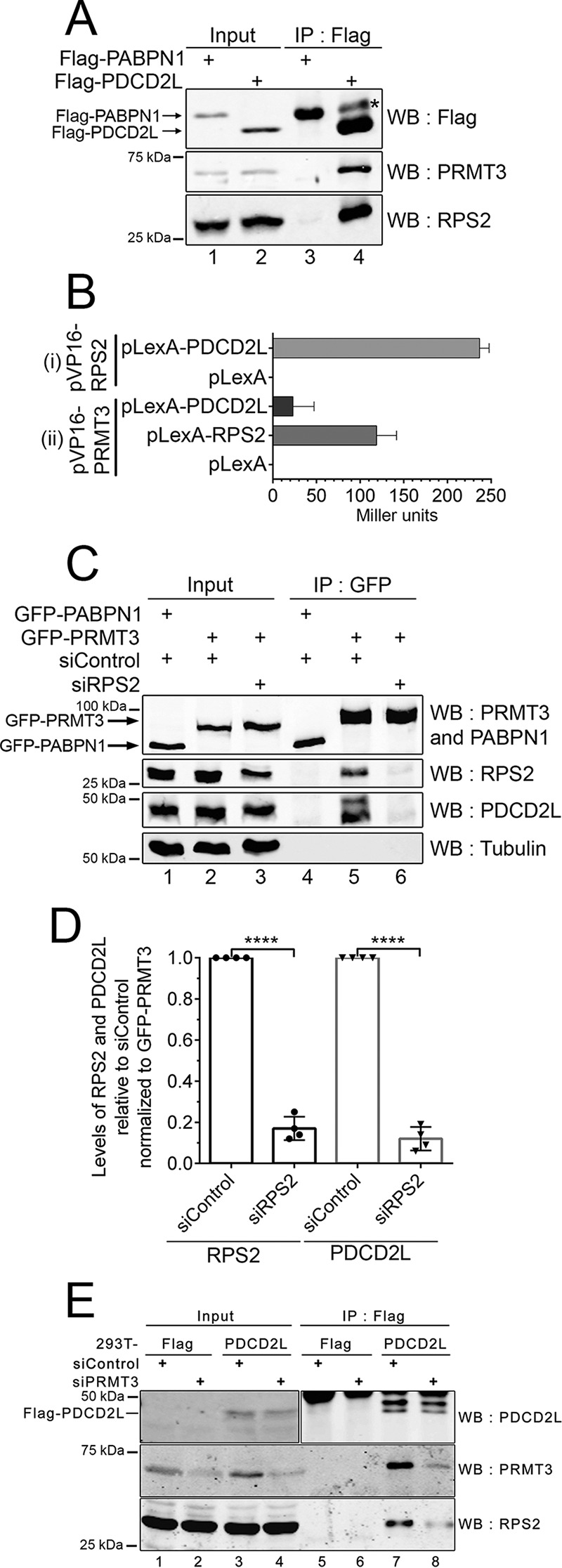
RPS2 is required for the association between PRMT3 and PDCD2L. (A) Western analysis of total extracts (lanes 1 and 2) and anti-Flag immunoprecipitates (lanes 3 and 4) prepared from HEK 293 cells that stably express Flag-PABPN1 (lanes 1 and 3) and Flag-PDCD2L (lanes 2 and 4). The asterisk indicates the IgG heavy chain. (B) Analysis of PDCD2L, RPS2, and PRMT3 interactions by a two-hybrid assay. (i) LexA and the LexA-PDCD2L fusion protein were coexpressed with the full-length VP16-RPS2 fusion protein. (ii) The LexA, LexA-RPS2, and LexA-PDCD2L proteins were coexpressed with full-length VP16-PRMT3. Protein interactions were measured by using liquid β-galactosidase assays in the S. cerevisiae L40 strain and are reported in Miller units. Data and error bars represent the means and standard deviations, respectively, from three independent experiments using two independent trans formants. (C) Western analysis of total extracts (lanes 1 to 3) and GFP immunoprecipitates (lanes 4 to 6) prepared from HEK 293 cells stably expressing GFP-PABPN1 (lanes 1 and 4) and GFP-PRMT3 (lanes 2, 3, 5, and 6) that were previously treated with RPS2-specific (lanes 3 and 6) and control nontarget (lanes 1, 2, 4, and 5) siRNAs. (D) Quantification of RPS2 and PDCD2L levels in GFP-PRMT3 immunoprecipitates. RPS2 and PDCD2L copurification levels (normalized to GFP-PRMT3 levels) were set to 1.0 for cells treated with control nontarget siRNAs. Data and error bars represent the means and standard deviations, respectively, from four independent experiments. ****, P < 0.0001, as determined by Student's t test. (E) Western analysis of total extracts (lanes 1 to 4) and Flag immunoprecipitates (lanes 5 to 8) prepared from HEK 293 cells stably expressing Flag-PDCD2L (lanes 3, 4, 7, and 8) and the empty vector control (lanes 1, 2, 5, and 6) that were previously treated with PRMT3-specific (lanes 2, 4, 6, and 8) and control nontarget (lanes 1, 3, 5, and 7) siRNAs.
To test the possibility that RPS2 bridges the association between PRMT3 and PDCD2L, we analyzed the effect of depleting RPS2 on the association between PRMT3 and PDCD2L. To this end, cells that stably expressed GFP-PRMT3 and a control GFP-tagged protein were treated with either RPS2-specific or control nontarget siRNAs. RNA interference (RNAi)-mediated depletion of endogenous RPS2 generally resulted in a 50% knockdown efficiency, yet total levels of PDCD2L and GFP-PRMT3 were not affected by the depletion of RPS2 (Fig. 2C, compare lane 3 to lanes 1 and 2). Notably, purification of GFP-PRMT3 from RPS2-depleted cells showed a significant decrease in the recovery of PDCD2L compared to purification of GFP-PRMT3 prepared from cells treated with control siRNAs (Fig. 2C, compare lanes 5 and 6, and D). We also examined whether the association between RPS2 and PDCD2L in human cells required the presence of PRMT3. As shown in Fig. 2E, depletion of PRMT3 reduced the levels of RPS2 copurified with PDCD2L (compare lanes 7 and 8). Together, these data suggest that both RPS2 and PRMT3 are required for the formation of a stable PRMT3-RPS2-PDCD2L complex.
PDCD2L associates with 40S ribosomal subunit precursors.
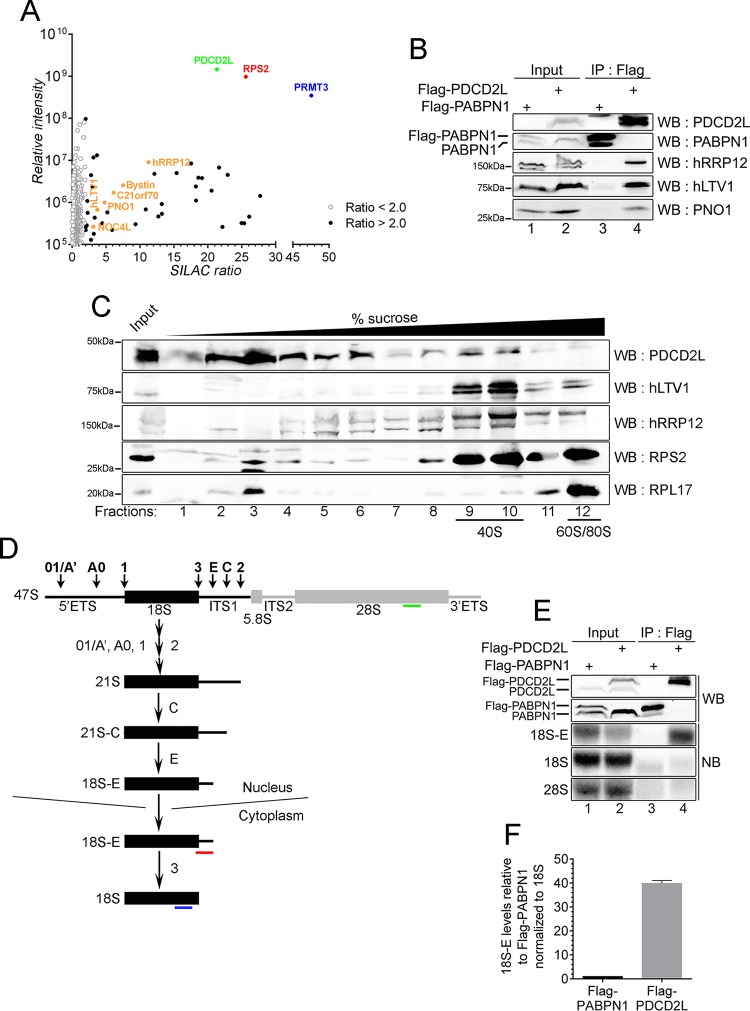
PDCD2L exhibits sequence homology to budding yeast Tsr4p, a protein identified in a genetic screen for factors required for the processing of the 20S rRNA precursor (29), which gives rise to the mature 18S rRNA of the 40S ribosomal subunit. As yet, however, a molecular function for human PDCD2L has remained unknown. To begin to explore the functional role of human PDCD2L, we used quantitative proteomics to define the protein-protein interaction network of PDCD2L via SILAC. As for PRMT3, we generated a HEK 293 cell line conditionally expressing a tetracycline-inducible and GFP-tagged version of PDCD2L. Data from the analysis of PDCD2L-associated proteins identified by SILAC-coupled MS are presented in Fig. 3A. In total, 25 proteins were copurified with PDCD2L with a SILAC enrichment ratio of >2.0 and a minimum of two detectable peptides (see Table S2 in the supplemental material). Notably, RPS2 and PRMT3 were the strongest interaction partners of PDCD2L, showing high specificity and peptide intensity (Fig. 3A), suggesting the existence of a stable PDCD2L-RPS2-PRMT3 complex. The purification of PDCD2L also recovered several factors that have been shown to be preribosomal factors composing the pre-40S particle or are yeast homologs of known pre-40S components (40–42): hRRP12, Bystin/ENP1, C21orf70, PNO1, hLTV1, and NOC4L (Fig. 3A; see also Table S2 in the supplemental material). Validation experiments confirmed that the enrichment of 40S preribosomal factors in the GFP-PDCD2L SILAC experiment was not dependent on the use of a GFP fusion protein, as a Flag-tagged version of PDCD2L specifically recovered hRRP12, hLTV1, and PNO1 (Fig. 3B). Furthermore, analysis of the SILAC/MS data revealed a significant enrichment of ribosomal proteins from the small subunit compared to the large ribosomal subunit (see Fig. S2 in the supplemental material). These results suggest that a fraction of PDCD2L is associated with pre-40S particles.
FIG 3.
PDCD2L associates with late 40S precursors. (A) GFP-PDCD2L SILAC copurification results plotted by SILAC ratios on the x axis (intensity of peptides originating from the GFP-PDCD2L purification versus the control purification), which reflects signal specificity, and signal intensity on the y axis (total peptide intensity for each protein normalized to molecular mass), which reflects the relative abundance of each protein in the purification. Proteins presenting SILAC ratios of >2.0 and <2.0 are shown. Factors known to be associated with late pre-40S particles are shown in orange. (B) Western analysis of total extracts (lanes 1 and 2) and Flag immunoprecipitates (lanes 3 and 4) prepared from HEK 293 cells that stably express Flag-tagged versions of PABPN1 (lanes 1 and 3) and PDCD2L (lanes 2 and 4). (C) Western analysis of endogenous proteins using fractions of centrifuged sucrose gradients that were prepared by using extracts of HEK 293 cells. The positions of the 40S, 60S, and 80S sedimentations are indicated at the bottom. (D) Schematic of 18S rRNA biogenesis in human cells. Following cleavage at sites 01/A′, A0, 1, and 2, the 21S pre-rRNA is matured into the 18S-E precursor in the nucleus. Final maturation of the 18S-E pre-rRNA into mature 18S rRNA occurs in the cytoplasm. Probes used to detect the 18S-E pre-rRNA (red) as well as mature 18S (blue) and 28S (green) rRNAs are indicated. ETS, external transcribed spacer; ITS, internal transcribed spacer. (E) Western blot (WB) (top two panels) and Northern blot (NB) (bottom three panels) analyses of protein and RNA, respectively, prepared from total cell extracts (lanes 1 and 2) and Flag immunoprecipitates (lanes 3 and 4) using HEK 293 cells that stably express Flag-tagged versions of PABPN1 (lanes 1 and 3) and PDCD2L (lanes 2 and 4). The 18S-E, 18S, and 28S RNAs were detected by using the specific probes shown in panel D. (F) Quantification of 18S-E pre-rRNA enrichment. 18S-E levels were normalized to levels of mature 18S rRNA. The values were then set to 1.0 for the control Flag-PABPN1 purification. Data and error bars represent the averages and standard deviations, respectively, from three independent experiments.
We next used sucrose gradients coupled to velocity sedimentation to examine the distribution of endogenous PDCD2L relative to known 40S maturation factors. As shown in Fig. 3C, the majority of PDCD2L was present in the low-density fractions, steadily decreasing from fractions 3 to 7. Notably, a fraction of PDCD2L clearly cosedimented with free 40S subunits/pre-40S particles, as demonstrated by the increased levels of PDCD2L detected in fractions 9 and 10, which are enriched in hLTV1, hRRP12, and RPS2. Fractions corresponding to free 60S, 80S monosomes, and polyribosomes showed barely detectable levels of PDCD2L (fractions 11 and 12) (Fig. 3C and data not shown).
The above-mentioned results suggest that a fraction of PDCD2L physically associates with immature pre-40S particles and/or mature 40S ribosomal subunits. To clarify the nature of the 40S particle associated with PDCD2L, we analyzed the type of rRNA associated with PDCD2L using RNA coimmunoprecipitation (RIP) assays. Two major forms of 3′-extended 18S rRNA precursors are associated with pre-40S particles in mammalian cells (Fig. 3D): the 21S pre-rRNA that is found in the nucleus and the 18S-E pre-rRNA that can be found in both the nucleus and the cytoplasm (43). Northern blot analysis of RNAs prepared from PDCD2L immunoprecipitates using a probe complementary to internal transcribed spacer 1 (ITS1) sequences (Fig. 3D, red probe) revealed a robust enrichment of the 18S-E rRNA precursor relative to the control purification (Fig. 3E, compare lanes 3 and 4, and F). A slight enrichment (1.5- to 2-fold) in 21S pre-rRNA was also detected after a long exposure (data not shown) but was substantially less than the marked enrichment of 18S-E pre-rRNA (Fig. 3F). In contrast, similar levels of mature 18S rRNA were detected in PDCD2L and control immunoprecipitates (Fig. 3E). Taken together, the data presented in Fig. 3 indicate that PDCD2L associates with pre-40S particles.
PDCD2L shuttles between the nucleus and the cytoplasm and interacts directly with CRM1.
Recent studies aimed at identifying shuttling substrates of CRM1, the major transport receptor for the export of proteins and RNPs from the nucleus, have identified peptides corresponding to endogenous PDCD2L (44, 45). To validate that PDCD2L is exported from the nucleus in a CRM1-dependent manner, we analyzed the localization of GFP-PDCD2L after incubation with the CRM1 inhibitor leptomycin B (LMB) (46). Whereas GFP-PDCD2L localized mainly to the cytoplasm in the absence of LMB (Fig. 4Aa to c), the majority of cells displayed PDCD2L localization throughout the nucleus after LMB treatment (Fig. 4Ad to f). Notably, most cells showed PDCD2L accumulation in the nucleus after only 40 min of LMB treatment (see Fig. S3 in the supplemental material). In contrast, PRMT3 did not show CRM1-dependent shuttling in U-2 OS cells, as the cytoplasmic localization of GFP-PRMT3 was not affected by the inhibition of CRM1 by LMB (Fig. 4Ag to l).
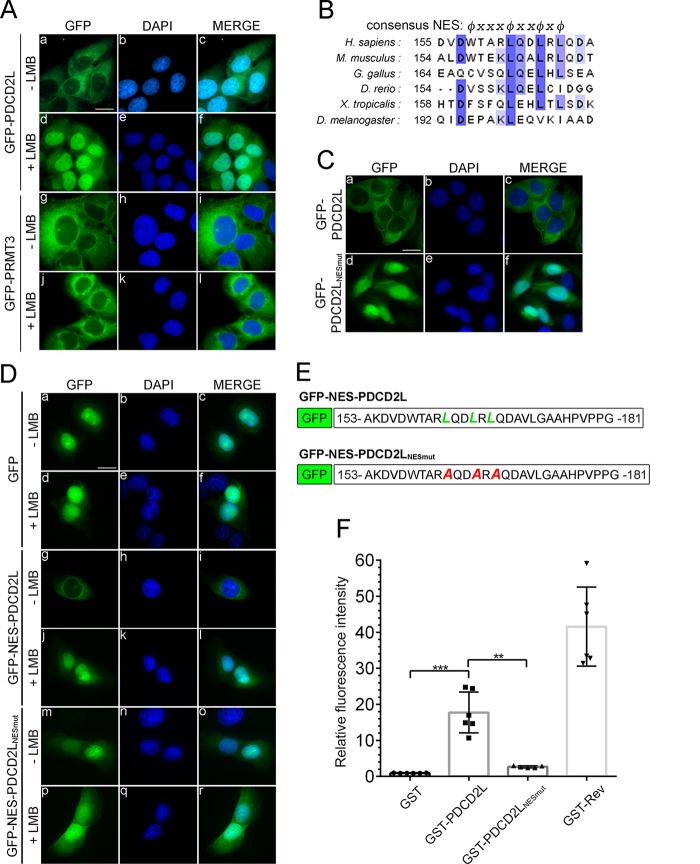
FIG 4.
PDCD2L is a nucleocytoplasmic shuttling protein that exports the nucleus by using a leucine-rich NES. (A) U-2 OS cells induced to express GFP-tagged versions of PDCD2L (a to f) and PRMT3 (g to l) were treated (d to f and j to l) or not treated (a to c and g to i) with LMB for 3 h. DNA staining with 4′,6-diamidino-2-phenylindole (DAPI) shows the nucleus of each cell (b, e, h, and k). Bar, 20 μm. (B) Alignment of putative NESs of PDCD2L proteins across multicellular organisms. The NES consensus sequence ϕx2-3ϕx2-3ϕxϕ is indicated at the top, where ϕ is a large, hydrophobic amino acid and x represents any amino acid. H. sapiens, Homo sapiens; M. musculus, Mus musculus; G. gallus, Gallus gallus; D. rerio, Danio rerio; X. tropicalis; Xenopus tropicalis. (C) Visual analysis of U-2 OS cells that were transiently transfected with DNA constructs expressing GFP-PDCD2L (a to c) and a version of GFP-PDCD2L with substitutions of leucines 162, 165, and 167 for alanines (NESmut) (d to f). (D) Visual analysis of U-2 OS cells transiently transfected with DNA constructs expressing GFP (a to f) as well as GFP fused to wild-type (g to l) and mutant (m to r) versions of the PDCD2L NES (amino acids 153 to 181) (see panel E). Cells were treated (+) or not treated (−) with LMB. (E) Schematic of wild-type and mutant versions of GFP-NES-PDCD2L fusions used for panel D. (F) Direct interaction between PDCD2L and CRM1 in vitro. Totals of 50 pmol GST, GST-PDCD2L, and GST-Rev fusion proteins were immobilized on beads and incubated with 9 pmol Cy3-labeled CRM1 alone or in the presence of 180 pmol RanGTPQ69L and 70 pmol Nup2141916–2033. Bound Cy3-CRM1 was analyzed by flow cytometry, and the fluorescence intensity relative to GST was plotted. Data and error bars represent the means and standard deviations, respectively, from six independent experiments. ***, P < 0.001; **, P < 0.01 (as determined by Student's t test).
The rapid retention of PDCD2L in the nucleus after LMB treatment suggested that PDCD2L might be a direct substrate of CRM1-mediated export. We therefore analyzed the amino acid sequence of PDCD2L using computer algorithms (47, 48) to predict the presence of a leucine-rich nuclear export signal (NES), which is recognized by CRM1 (49). Notably, both prediction tools identified a putative leucine-rich NES between amino acids 162 and 167 of human PDCD2L, a motif that appears to have been conserved in diverse PDCD2L homologs (Fig. 4B). To test the functional importance of this motif for PDCD2L nucleocytoplasmic shuttling, we substituted Leu162, Leu165, and Leu167 for alanine residues (PDCD2LNESmut). The triple-alanine substitution did not disrupt the folding of PDCD2L, as wild-type and NES mutant versions of PDCD2L recovered similar levels of RPS2 and PRMT3 after affinity purification (see Fig. S4 in the supplemental material). However, in contrast to wild-type PDCD2L, the NES mutant version was completely retained in the nucleus (compare Fig. 4Ca and d). We also addressed whether the NES of PDCD2L could promote CRM1-dependent nuclear export of a GFP reporter, which localizes predominantly to the nucleus at steady state (Fig. 4Da). We thus fused 28 amino acids (aa 153 to 181) of PDCD2L containing either the wild-type or mutant version of the NES to the C terminus of GFP (Fig. 4E). Amino acids 153 to 181 of PDCD2L were sufficient to stimulate the export of the GFP reporter to the cytoplasm (compare Fig. 4Da and g). The nuclear export of the GFP-NES-PDCD2L reporter was dependent on CRM1, as it accumulated in the nucleus after LMB treatment (compare Fig. 4Dg and j). Importantly, the GFP reporter fused to the mutant version of the PDCD2L NES remained primarily localized to the nucleus (Fig. 4Dm). We therefore conclude that the PDCD2L NES is necessary and sufficient for CRM1-dependent nuclear export.
Next, we examined whether the NES of PDCD2L can mediate a direct interaction with CRM1. To this end, we performed flow cytometry-based in vitro binding assays using fluorescently labeled recombinant CRM1 (37) together with recombinant versions of wild-type and mutant GST-PDCD2L. Binding assays were performed in the absence and presence of RanQ69L-GTP and a fragment of Nup214, which stabilize the interaction between CRM1 and its substrates (37). A GST-tagged version of HIV-1 Rev, which contains a leucine-rich NES (50), was used as a positive control and bound to CRM1 in a RanGTP-dependent manner, whereas GST alone did not (Fig. 4F). Notably, direct binding between CRM1 and PDCD2L was observed but was significantly reduced when the NES mutant version of PDCD2L was analyzed (Fig. 4F). These results indicate that PDCD2L is a nucleocytoplasmic shuttling protein that is an export substrate of CRM1.
PDCD2L transits through the nucleolus.
Although PDCD2L localizes predominantly to the cytoplasm at steady state, analysis of higher-magnification images showed punctate nuclear signals that were reminiscent of nucleolar staining (Fig. 5g), which are sites of ribosome biogenesis. To test the possibility that the punctate nuclear staining of PDCD2L corresponded to nucleoli, we combined GFP-PDCD2L localizations with an immunostaining procedure for endogenous fibrillarin, which is a nucleolar marker protein (Fig. 5d). Comparison of the different staining methods showed that GFP-PDCD2L was concentrated in nuclear regions that coincided with antifibrillarin staining (Fig. 5d, g, and j). The observation that GFP-PDCD2L localizes to nucleoli is consistent with data from proteomic studies that found that PDCD2L is part of the nucleolar proteome (51). Interestingly, the GFP-PDCD2L nucleolar signal increased shortly after LMB treatment (Fig. 5h) but was gradually lost over time (Fig. 5i). These results suggest that PDCD2L transits through the nucleolus before it is exported to the cytoplasm.
Deletion of PDCD2L results in the accumulation of free 60S ribosomal subunits.
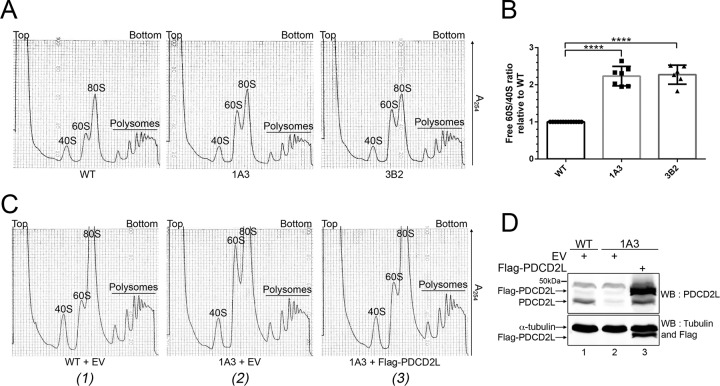
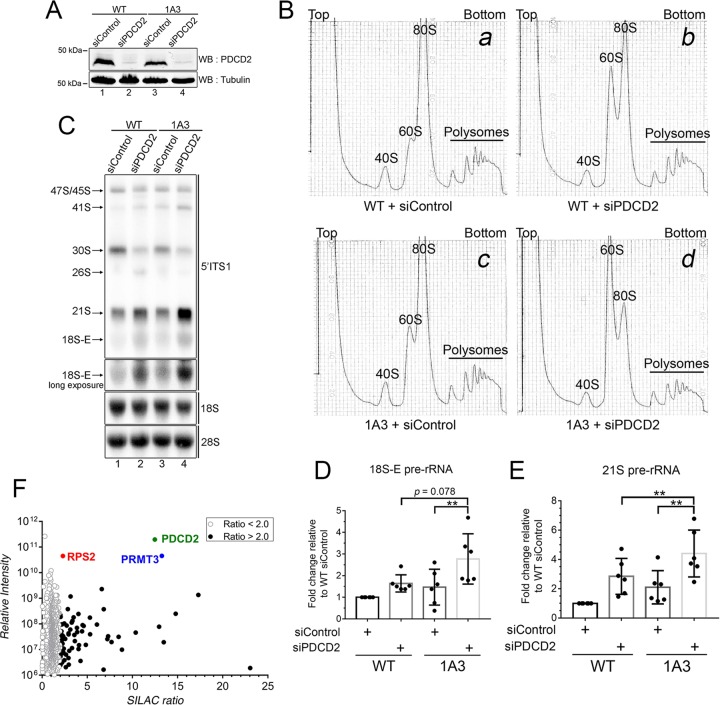
Our results showing that PDCD2L associates with factors involved in late stages of 40S ribosomal subunit maturation (Fig. 3A and B), copurifies with pre-40S particles (Fig. 3C to F), and shuttles between the nucleus and the cytoplasm using a leucine-rich NES (Fig. 4) suggested that PDCD2L could act as an adaptor protein for CRM1-mediated export of pre-40S particles. We therefore performed RNAi experiments to examine the effect of PDCD2L depletion on pre-40S export from the nucleus. Two independent assays were used to monitor pre-40S localization: (i) fluorescent in situ hybridization (FISH) using an ITS1-specific probe to detect the accumulation of 18S-E pre-rRNA in the nucleoplasm and/or the reduction of 18S-E pre-rRNA in the cytoplasm and (ii) immunostaining of the late pre-40S-associated factor hLTV1 for nuclear accumulation in PDCD2L-deficient cells. Whereas CRM1 inhibition using LMB showed the expected accumulation of 18S-E pre-rRNA and hLTV1 in the nucleoplasm, PDCD2L depletion did not result in a detectable change in the pre-40S subcellular localization (data not shown). We also analyzed the levels of 40S pre-rRNA species by Northern blotting using probes complementary to ITS1-specific sequences. Control experiments targeting the pre-40S maturation factor Bystin (Fig. 6A, lane 3) resulted in the accumulation of 21S pre-rRNA together with reduced levels of mature 18S rRNA (Fig. 6B, compare lane 3 to lane 1, to D), consistent with previously reported observations (34). In contrast, despite efficient PDCD2L depletion (Fig. 6A, lane 2), a deficiency in PDCD2L did not result in the accumulation of 21S and 18S-E rRNA precursors (Fig. 6B to D). To test the possibility that residual levels of PDCD2L after RNAi-mediated depletion are sufficient to promote pre-40S nuclear export, we used CRISPR-Cas9-mediated genomic targeting in HeLa cells to generate multiallelic mutations in PDCD2L, resulting in cells with undetectable levels of the PDCD2L protein (see Fig. S5 in the supplemental material). Consistent with siRNA-mediated depletions of PDCD2L, PDCD2L-null cells did not reveal major defects in the maturation of pre-40S particles as determined by Northern analysis (data not shown) and rRNA pulse-chase assays (see Fig. S6 in the supplemental material). However, analysis of the ribosome distribution by sucrose gradient centrifugation showed the accumulation of free 60S ribosomal subunits using two independent knockout clones of PDCD2L (Fig. 7A, compare clones 1A3 and 3B2 to the WT), yet the levels of free 40S ribosomal subunits did not appear to change between PDCD2L knockout and control parental cells (Fig. 7A). The accumulation of free 60S subunits in PDCD2L-null cells significantly altered the stoichiometry between free 40S and 60S ribosomal subunits (Fig. 7B) but not the overall content of 80S monosomes and polyribosomes (Fig. 7A).
FIG 6.
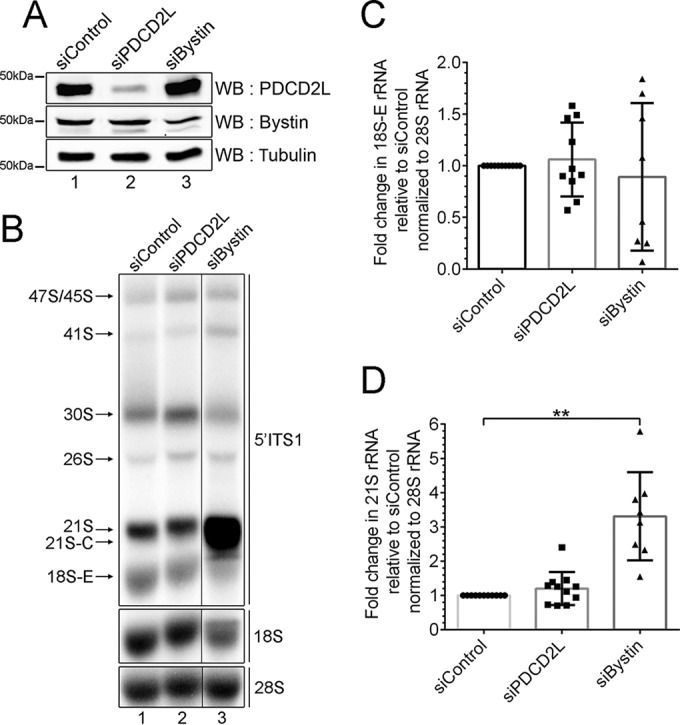
Analysis of pre-rRNA processing in PDCD2L-depleted cells. (A) Western blot analysis showing depletion of PDCD2L and bystin from HeLa cells. (B) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from PDCD2L- and bystin-depleted cells as well as control cells. Pre-rRNAs were detected by using a probe complementary to sequences in the 5′ ITS (red probe in Fig. 3D) region. Pre-rRNA species are indicated on the left. Areas from the same blot were spliced to remove irrelevant lanes (between lanes 2 and 3). (C and D) 18S-E (C) and 21S (D) pre-rRNA levels were normalized to the 28S rRNA level and are expressed relative to levels in cells treated with control siRNA. Data and error bars represent the means and standard deviations, respectively, from 8 independent (siBystin) and 10 independent (siPDCD2L) experiments. **, P value of <0.01, as determined by Student's t test.
FIG 7.
Deletion of PDCD2L causes the accumulation of free 60S ribosomal subunits. (A) Sucrose gradient analysis of total extracts prepared from WT and PDCD2L-null HeLa cells (clones 1A3 and 3B2). The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated. (B) Quantification of ratios of 60S/40S free ribosomal subunits expressed relative to values for wild-type HeLa cells. Data and error bars represent the means and standard deviations, respectively, from 7 independent (1A3) and 6 independent (3B2) experiments. ****, P value of <0.0001, as determined by Student's t test. (C) Sucrose gradient analysis of total extracts prepared from WT (1) and PDCD2L-deleted (clone 1A3) (2 and 3) HeLa cells that were previously transfected with an empty vector (EV) control (1 and 2) or a DNA construct expressing Flag-PDCD2L (3). (D) Western blot analysis showing the expression of Flag-PDCD2L in PDCD2L-null cells (lane 3). The band comigrating with endogenous PDCD2L in lane 3 is a degradation product generated from Flag-PDCD2L.
To ensure that the accumulation of free 60S ribosomal subunits in PDCD2L knockout cells was indeed caused by a deficiency in PDCD2L, we performed rescue experiments using a Flag-tagged version of PDCD2L. The expression of wild-type PDCD2L restored the increased levels of free 60S ribosomal subunits detected in PDCD2L-null cells (compare Fig. 7C2 and 3), resulting in levels of free 60S similar to those in parental control cells (Fig. 7C1). In contrast, PDCD2L-null cells transfected with the empty vector control showed the expected accumulation of free 60S ribosomal subunits (Fig. 7C2).
PDCD2L and its paralog, PDCD2, function redundantly in ribosome biogenesis.
In addition to PDCD2L, our analysis of PRMT3-associated proteins also identified PDCD2 (see Table S1 in the supplemental material), a paralog of PDCD2L that is 30% identical (52% similar) to human PDCD2L. Based on data from sequence analyses, PDCD2 is thought to have arose from a duplication of the PDCD2L gene prior to the divergence of animals, fungi, and plants from a common ancestor (28). Interestingly, it was recently found that the Drosophila homolog of PDCD2, Zfrp8, interacts with Rps2 (52). Considering the seemingly similar functions ascribed to human PDCD2L and Drosophila PDCD2 (Zfrp8), we examined whether human PDCD2 contributes to ribosome biogenesis and potentially acts redundantly to PDCD2L. As shown in Fig. 8A, PDCD2 was efficiently depleted from the parental HeLa cell line (lane 2) as well as from PDCD2L-null cells (lane 4). Analysis of ribosome production using extracts prepared from PDCD2-depleted cells showed reduced levels of free 40S ribosomal subunits together with increased levels of free 60S subunits (compare Fig. 8Ba and b), affecting the levels of 80S monosomes but not the levels of polysomes. We next examined the effect of depleting PDCD2 in PDCD2L-null cells to characterize the functional relationship between PDCD2 and PDCD2L. Notably, the combined deficiency of both PDCD2 and PDCD2L resulted in a synergistic increase in the ratio of 60S/40S free subunits relative to the single depletion of either PDCD2 or PDCD2L (compare Fig. 8Bd to b and c). Cells deficient for both PDCD2 and PDCD2L also showed reduced levels of 80S monosomes as well as a reduction in the amplitude of polysome peaks relative to single depletions of PDCD2 and PDCD2L (Fig. 8B). The functional relationship between PDCD2 and PDCD2L in 40S ribosomal subunit biogenesis demonstrated by sucrose gradient analyses was supported by data from Northern blotting assays of pre-rRNA processing using an ITS1-specific probe. Accordingly, cells deficient for PDCD2 showed an accumulation of 18S-E and 21S pre-rRNAs (Fig. 8C, lane 2), an accumulation which was further increased by the codepletion of both PDCD2 and PDCD2L (Fig. 8C, lane 4, to E). Cells deficient for both PDCD2 and PDCD2L also showed reduced levels of mature 18S rRNA (Fig. 8C, lane 4). From these results, we conclude that PDCD2 is important for optimal 40S ribosomal subunit production and that PDCD2 and PDCD2L have partially overlapping functions in 40S biogenesis.
FIG 8.
Human PDCD2 and PDCD2L proteins contribute to 40S ribosomal subunit production. (A) Western blot analysis showing depletion of PDCD2 from wild-type HeLa cells (lanes 1 and 2) as well as PDCD2L-null HeLa cells (1A3) (lanes 3 and 4). (B) Sucrose gradient analysis of total extracts prepared from wild-type (a and b) and PDCD2L-null (c and d) HeLa cells that were previously treated with PDCD2-specific (b and d) or control nontarget (a and c) siRNAs. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated. (C) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from WT (lanes 1 and 2) and PDCD2L-null (1A3) (lanes 3 and 4) cells that were previously treated with PDCD2-specific (lanes 2 and 4) or control nontarget (lanes 1 and 3) siRNAs. Pre-rRNAs were detected by using a probe complementary to sequences in the 5′ ITS region. Pre-rRNA species are indicated on the left. (D and E) 18S-E (D) and 21S (E) pre-rRNA levels were normalized to the 28S rRNA level and are expressed relative to values for cells treated with control siRNAs. Data and error bars represent the means and standard deviations, respectively, from six independent experiments. **, P value of <0.01, as determined by Student's t test. (F) GFP-PDCD2 SILAC copurification results plotted as SILAC ratios on the x axis and signal intensities on the y axis. Proteins presenting SILAC ratios of >2.0 and <2.0 are indicated. GFP-PDCD2, RPS2, and PRMT3 are shown in color.
To further investigate the functional relationship between PDCD2L and PDCD2, we used SILAC-based quantitative proteomics to determine the protein interaction network of human PDCD2. Using a stable cell line conditionally expressing a GFP-tagged version of PDCD2, we identified 80 proteins that copurified with PDCD2 with a SILAC enrichment ratio of >2.0 and a minimum of two detectable peptides (see Table S3 in the supplemental material). As for PDCD2L, RPS2 and PRMT3 were the strongest interaction partners of PDCD2, showing robust peptide intensities (Fig. 8F). Interestingly, despite the fact that both PDCD2 and PDCD2L associated with the RPS2-PRMT3 duo, PDCD2L peptides were not detected in the PDCD2 purification, and reciprocally, PDCD2 was not identified as a PDCD2L-associated protein. Furthermore, mass spectrometry analysis of PDCD2-associated proteins did not lead to the recovery of preribosomal maturation factors, which is in striking contrast to PDCD2L, for which several pre-40S components were identified and validated (Fig. 3). Taken together, our data suggest some level of redundancy between PDCD2L and PDCD2 paralogs in 40S ribosomal subunit biogenesis, contributing to parallel pathways that would converge to the RPS2-PRMT3 complex.
DISCUSSION
In this study, we report that human PDCD2L forms a stable complex with RPS2 and the protein arginine methyltransferase PRMT3. Our results also indicate that PDCD2L is a nucleocytoplasmic shuttling protein that associates with pre-40S particles and that a disruption of PDCD2L expression leads to the accumulation of free 60S ribosomal subunits, findings which suggest a role for PDCD2L in the supply of mature 40S ribosomal subunits.
RPS2 forms an extraribosomal complex with PRMT3 and PDCD2L.
Our analysis of PRMT3- and PDCD2L-associated proteins using SILAC-based proteomics revealed that both PRMT3 and PDCD2L strongly associate with RPS2. Notably, levels of RPS2 were 1,000 times and 200 times higher than the levels of the next most abundant ribosomal protein detected in the PRMT3 and PDCD2L purifications, respectively. Consistent with our results, data from a recent study using high-throughput affinity purification coupled to mass spectrometry detection also support the existence of a stable PRMT3-RPS2-PDCD2L complex in human cells (53). The robust levels of RPS2 detected after the purification of human PRMT3 (Fig. 1A) are also consistent with data for fission yeast, where RPS2 was the predominant ribosomal protein identified after purification of S. pombe Rmt3 (21). In addition, a two-hybrid-based interaction map of the Drosophila proteome (39) shows interactions between SOP and TRUS (RPS2 and PDCD2L) as well as between SOP and ART3 (RPS2 and PRMT3) but not between ART3 and TRUS (PRMT3 and PDCD2L). These observations are consistent with our data (Fig. 2C and D) indicating that RPS2 is required for the copurification of PDCD2L and PRMT3. Collectively, the above-mentioned interactions detected in S. pombe, Drosophila, and human support the existence of a conserved complex consisting of PRMT3, RPS2, and PDCD2L. An increasing number of studies suggest functional roles for ribosomal proteins outside the ribosome (54, 55). Although the extraribosomal function of RPS2 remains to be determined, its association with proteins that are functionally linked to ribosome biosynthesis, namely, PRMT3 and PDCD2L, suggests that the extraribosomal role of RPS2 will be related to the regulation of ribosome biogenesis, which appears to be the case for many of the extraribosomal functions of ribosomal proteins (56).
PDCD2L contributes to the biogenesis of small ribosomal subunits.
In addition to the identification of a complex between PDCD2L, RPS2, and PRMT3, our study suggests that a fraction of PDCD2L escorts late precursors of the 40S ribosomal subunit to the cytoplasm. The conclusion that PDCD2L physically associates with pre-40S ribosomal particles is supported by several observations: (i) SILAC-based proteomic analyses of PDCD2L-associated proteins identified several maturation factors found in late pre-40S ribosomal particles, including hRRP12, hLTV1, PNO1, bystin, NOC4L, and C21orf70 (Fig. 3A and B); (ii) immunoprecipitates of PDCD2L were preferentially enriched for ribosomal proteins of the 40S subunit relative to the 60S subunit (see Fig. S2 in the supplemental material); (iii) PDCD2L cosedimented with 40S/pre-40S particles on sucrose gradients (Fig. 3C); and (iv) PDCD2L-bound 40S particles were enriched for 18S-E pre-rRNA (Fig. 3E and F), the last precursor of mature 18S rRNA. Important to this study is also the demonstration that PDCD2L actively shuttles between the nucleus and the cytoplasm via binding to the major cellular exportin CRM1 using a leucine-rich NES (Fig. 4). Accordingly, the association of PDCD2L with pre-40S particles together with the ability of PDCD2L to rapidly export the nucleus via a direct interaction with CRM1 suggest a model in which PDCD2L serves as an adaptor protein for CRM1-mediated export of pre-40S particles. In this model, nucleolar PDCD2L (Fig. 5) would be recruited to 40S precursors via interactions with RPS2, a late-associating ribosomal protein that has been shown to be important for the nuclear export of pre-40S subunits (57–59). The leucine-rich NES of PDCD2L could then recruit CRM1 to nuclear pre-40S particles to stimulate transit through the nuclear pore complex to the cytoplasm. However, our data revealed that PDCD2L is not absolutely required for the export of 40S precursors in HeLa cells, as PDCD2L-null cells are viable and do not show a global retention of pre-40S particles in the nucleus.
In contrast to data from steady-state RNA analyses and FISH assays, which may not readily detect a modest reduction in the biogenesis and function of ribosomes, analysis of ribosome profiles by sucrose gradient sedimentation can provide a highly sensitive assay, as ribosomes are analyzed as free subunits, single monosomes, and polysomes. Since 5 to 10% of the ribosomal subunits are not assembled into ribosomes (60, 61), a slight deficiency in ribosomal subunit availability can result in a detectable change in the ratios between free ribosomal subunits, which may not be detected for the whole ribosome population. Accordingly, despite the absence of noticeable changes in monosome and polysome distributions, analysis of ribosome profiles using extracts from PDCD2L-null cells revealed the accumulation of free 60S ribosomal subunits. Such accumulations of free 60S subunits are generally the consequence of an alteration in 40S subunit production as a result of a deficiency in a factor that promotes small-subunit biogenesis (21, 35, 62–64). Consistent with this view, the depletion of the putative homolog of PDCD2L in S. cerevisiae, Tsr4p, results in a striking accumulation of free 60S subunits together with reduced levels of free 40S ribosomal subunits (29). Intriguingly, the accumulation of free 60S subunits detected in PDCD2L-null cells was not accompanied by an apparent decrease in the levels of free 40S ribosomal subunits. Although the core features of ribosome assembly are similar from yeast to humans and several proteins involved in the biogenesis of 40S subunits are evolutionarily conserved, data suggest that ribosome biogenesis has increased in complexity and flexibility during the course of evolution (65, 66). Accordingly, whereas the TSR4 gene is indispensable for the viability of budding yeast, our data indicate that PDCD2L is not essential in HeLa cells, suggesting the presence of redundant factors that can compensate for the absence of PDCD2L in mammalian cells or the acquisition of a specialized function.
One potential candidate that could compensate for the production of small ribosomal subunits in the absence of PDCD2L is its paralog, PDCD2. Indeed, the synergistic defect in 60S subunit accumulation in cells deficient for both PDCD2L and PDCD2 (Fig. 8) suggests functional redundancy between the two paralogs. Moreover, our data indicate that both PDCD2 and PDCD2L form a complex with RPS2 and PRMT3, yet they appear to form two independent complexes, as PDCD2L was not detected in the PDCD2 purification, and reciprocally, PDCD2 was not identified as a PDCD2L-associated protein. Members of a duplicate gene pair can follow different fates after a duplication event. For instance, one paralog might accumulate mutations and evolve a new function, whereas the other paralog retains its ancestral function; alternatively, mutations in both paralogous genes can lead to a partitioning of their ancestral function (67). PDCD2L and PDCD2 belong to a family of proteins containing a TYPP domain, which was interrupted by the insertion of an MYND zinc finger domain in PDCD2 (28). It is probable that the insertion of an MYND domain in PDCD2 allowed the acquisition of new functions, which may explain the lack of overlap between the protein interaction networks of PDCD2 and PDCD2L, with the exception of RPS2 and PRMT3. We therefore speculate that PDCD2L and PDCD2 contribute to 40S ribosomal subunit production via parallel pathways that converge to the RPS2-PRMT3 complex.
The exact mechanism underlying the PDCD2L-dependent accumulation of free 60S subunits still needs investigation but may include possible influences on the export of pre-40S particles. We see at least two plausible scenarios that could explain the accumulation of free 60S subunits without apparent changes in the levels of 40S subunits in PDCD2L-null cells: (i) PDCD2L functions as one of several redundant adaptors that promote the CRM1-mediated nuclear export of pre-40S particles, or (ii) PDCD2L contributes to the export and/or maturation of a subpopulation of 40S ribosomal particles that is involved in a specialized function. The first possibility is supported by evidence showing that Rio2, a conserved ribosome biogenesis factor that binds directly to CRM1 and associates with 40S precursors, promotes pre-40S export in yeast and humans (42, 68). Ltv1p and Dim2p, which are the homologs of mammalian hLTV1 and hDIM2/PNO1, respectively, have also been shown to promote pre-40S export in S. cerevisiae and were suggested to be possible adaptors for CRM1-mediated export of 40S precursors (69, 70). Evidence from yeast also indicates that the conserved Mex67-Mtr2 complex can facilitate the nuclear export of pre-40S particles (71) and that Rrp12p binds to pre-rRNA to promote the export of ribosomal subunits (72). It is therefore likely that redundant pre-40S export adaptors allow sufficient pre-40S export in the absence of PDCD2L to cause only minimal perturbations in 40S availability in the cytoplasm of PDCD2L-null cells. Consistent with the activation of compensatory pathways, we found that trans-acting factors involved in the late stages of 40S subunit maturation were upregulated in PDCD2L-null cells (see Fig. S7 in the supplemental material).
In summary, we provide evidence for the existence of an evolutionarily conserved extraribosomal complex that includes PDCD2L, RPS2, and PRMT3. Furthermore, our functional characterization of human PDCD2L suggests that it has a role in small-subunit biogenesis, possibly acting as an adaptor protein for CRM1-dependent nuclear export of pre-40S particles. Given that PDCD2L appears to contribute to cell cycle progression and cell proliferation in humans (73) and Drosophila (74, 75), elucidating the cellular functions of PDCD2L should provide significant insights into the mechanisms that coordinate ribosome biogenesis and cellular proliferation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Mark Bedford for generously sharing the RPS2 antibody and to Marlene Oeffinger for critical reading of the manuscript.
This work was supported by Canadian Institutes of Health Research grant MOP-273292 to F.B. and by a graduate scholarship from the Natural Sciences and Engineering Research Council of Canada to A.-M.L.-V. F.B. is a Canada Research Chair in the Quality Control of Gene Expression.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00303-16.
REFERENCES
- 1.Woolford JL Jr, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. 2015. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 556:27–34. doi: 10.1016/j.gene.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. 2016. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18:276–290. doi: 10.1016/j.stem.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, Shalaby NA, Buszczak M. 2014. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343:298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentilella A, Kozma SC, Thomas G. 2015. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 1849:812–820. doi: 10.1016/j.bbagrm.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, Panse VG. 2015. Eukaryotic ribosome assembly and nuclear export. Int Rev Cell Mol Biol 319:107–140. doi: 10.1016/bs.ircmb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho JH, Kallstrom G, Johnson AW. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas F, Kutay U. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 11.Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J 22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue S, Barna M. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. 2002. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A 99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louie DF, Resing KA, Lewis TS, Ahn NG. 1996. Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J Biol Chem 271:28189–28198. doi: 10.1074/jbc.271.45.28189. [DOI] [PubMed] [Google Scholar]
- 15.Odintsova TI, Muller EC, Ivanov AV, Egorov TA, Bienert R, Vladimirov SN, Kostka S, Otto A, Wittmann-Liebold B, Karpova GG. 2003. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem 22:249–258. doi: 10.1023/A:1025068419698. [DOI] [PubMed] [Google Scholar]
- 16.Kruiswijk T, Kunst A, Planta RJ, Mager WH. 1978. Modification of yeast ribosomal proteins. Methylation Biochem J 175:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lhoest J, Lobet Y, Costers E, Colson C. 1984. Methylated proteins and amino acids in the ribosomes of Saccharomyces cerevisiae. Eur J Biochem 141:585–590. doi: 10.1111/j.1432-1033.1984.tb08233.x. [DOI] [PubMed] [Google Scholar]
- 18.Ramagopal S. 1991. Covalent modifications of ribosomal proteins in growing and aggregation-competent Dictyostelium discoideum: phosphorylation and methylation. Biochem Cell Biol 69:263–268. doi: 10.1139/o91-040. [DOI] [PubMed] [Google Scholar]
- 19.Chang FN, Navickas IJ, Au C, Budzilowicz C. 1978. Identification of the methylated ribosomal proteins in HeLa cells and the fluctuation of methylation during the cell cycle. Biochim Biophys Acta 518:89–94. doi: 10.1016/0005-2787(78)90118-1. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Ji H, Doudna JA, Leary JA. 2005. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci 14:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachand F, Silver PA. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J 23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachand F. 2007. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot Cell 6:889–898. doi: 10.1128/EC.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perreault A, Gascon S, D'Amours A, Aletta JM, Bachand F. 2009. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J Biol Chem 284:15026–15037. doi: 10.1074/jbc.M109.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swiercz R, Person MD, Bedford MT. 2005. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem J 386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipson RS, Webb KJ, Clarke SG. 2010. Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae. Biochem Biophys Res Commun 391:1658–1662. doi: 10.1016/j.bbrc.2009.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hang R, Liu C, Ahmad A, Zhang Y, Lu F, Cao X. 2014. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc Natl Acad Sci U S A 111:16190–16195. doi: 10.1073/pnas.1412697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swiercz R, Cheng D, Kim D, Bedford MT. 2007. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem 282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 28.Burroughs AM, Aravind L. 2014. Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis. Front Genet 5:424. doi: 10.3389/fgene.2014.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergeron D, Pal G, Beaulieu YB, Chabot B, Bachand F. 2015. Regulated intron retention and nuclear pre-mRNA decay contribute to PABPN1 autoregulation. Mol Cell Biol 35:2503–2517. doi: 10.1128/MCB.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drissi R, Dubois ML, Douziech M, Boisvert FM. 2015. Quantitative proteomics reveals dynamic interactions of the minichromosome maintenance complex (MCM) in the cellular response to etoposide induced DNA damage. Mol Cell Proteomics 14:2002–2013. doi: 10.1074/mcp.M115.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachand F, Yao XJ, Hrimech M, Rougeau N, Cohen EA. 1999. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem 274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 34.Carron C, O'Donohue MF, Choesmel V, Faubladier M, Gleizes PE. 2011. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res 39:280–291. doi: 10.1093/nar/gkq734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. 2008. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet 17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 36.Mallet PL, Bachand F. 2013. A proline-tyrosine nuclear localization signal (PY-NLS) is required for the nuclear import of fission yeast PAB2, but not of human PABPN1. Traffic 14:282–294. doi: 10.1111/tra.12036. [DOI] [PubMed] [Google Scholar]
- 37.Port SA, Monecke T, Dickmanns A, Spillner C, Hofele R, Urlaub H, Ficner R, Kehlenbach RH. 2015. Structural and functional characterization of CRM1-Nup214 interactions reveals multiple FG-binding sites involved in nuclear export. Cell Rep 13:690–702. doi: 10.1016/j.celrep.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 38.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 40.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyler E, Zimmermann M, Widmann B, Gstaiger M, Pfannstiel J, Kutay U, Zemp I. 2011. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA 17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zemp I, Wild T, O'Donohue MF, Wandrey F, Widmann B, Gleizes PE, Kutay U. 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouquette J, Choesmel V, Gleizes PE. 2005. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirli K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Gorlich D. 2015. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 4:e11466. doi: 10.7554/eLife.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. 2013. Identification of CRM1-dependent nuclear export cargos using quantitative mass spectrometry. Mol Cell Proteomics 12:664–678. doi: 10.1074/mcp.M112.024877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A 96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu SC, Imai K, Horton P. 2011. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res 39:e111. doi: 10.1093/nar/gkr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 49.Kutay U, Guttinger S. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Meyer BE, Malim MH. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev 8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 51.Boisvert FM, Lam YW, Lamont D, Lamond AI. 2010. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minakhina S, Naryshkina T, Changela N, Tan W, Steward R. 2016. Zfrp8/PDCD2 interacts with RpS2 connecting ribosome maturation and gene-specific translation. PLoS One 11:e0147631. doi: 10.1371/journal.pone.0147631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. 2015. The BioPlex network: a systematic exploration of the human interactome. Cell 162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhavsar RB, Makley LN, Tsonis PA. 2010. The other lives of ribosomal proteins. Hum Genomics 4:327–344. doi: 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu H, Zhu YF, Xiong J, Wang R, Jia Z. 2015. Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol Res 177:28–33. doi: 10.1016/j.micres.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Warner JR, McIntosh KB. 2009. How common are extraribosomal functions of ribosomal proteins? Mol Cell 34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 58.O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perreault A, Bellemer C, Bachand F. 2008. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res 36:6132–6142. doi: 10.1093/nar/gkn625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moy TI, Silver PA. 2002. Requirements for the nuclear export of the small ribosomal subunit. J Cell Sci 115:2985–2995. [DOI] [PubMed] [Google Scholar]
- 61.Sivan G, Kedersha N, Elroy-Stein O. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol 27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fassio CA, Schofield BJ, Seiser RM, Johnson AW, Lycan DE. 2010. Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics 185:199–209. doi: 10.1534/genetics.110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kressler D, de la Cruz J, Rojo M, Linder P. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 17:7283–7294. doi: 10.1128/MCB.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ounap K, Kasper L, Kurg A, Kurg R. 2013. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS One 8:e75686. doi: 10.1371/journal.pone.0075686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lafontaine DL. 2015. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22:11–19. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 66.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell 51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Perez JM, Candela H, Micol JL. 2009. Understanding synergy in genetic interactions. Trends Genet 25:368–376. doi: 10.1016/j.tig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 68.Fischer U, Schauble N, Schutz S, Altvater M, Chang Y, Faza MB, Panse VG. 2015. A non-canonical mechanism for Crm1-export cargo complex assembly. eLife 4:e05745. doi: 10.7554/eLife.05745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seiser RM, Sundberg AE, Wollam BJ, Zobel-Thropp P, Baldwin K, Spector MD, Lycan DE. 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174:679–691. doi: 10.1534/genetics.106.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanrobays E, Leplus A, Osheim YN, Beyer AL, Wacheul L, Lafontaine DL. 2008. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA 14:2061–2073. doi: 10.1261/rna.1176708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. 2012. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet 8:e1002915. doi: 10.1371/journal.pgen.1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oeffinger M, Dlakic M, Tollervey D. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Q, Yan C, Yan Q, Feng L, Chen J, Qian K. 2008. The novel MGC13096 protein is correlated with proliferation. Cell Biochem Funct 26:141–145. doi: 10.1002/cbf.1410. [DOI] [PubMed] [Google Scholar]
- 74.Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 75.Liang L, Haug JS, Seidel CW, Gibson MC. 2014. Functional genomic analysis of the periodic transcriptome in the developing Drosophila wing. Dev Cell 29:112–127. doi: 10.1016/j.devcel.2014.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.