Abstract
Osteosarcoma is the most common primary malignant bone tumor in children and young adults. Lactate dehydrogenase (LDH) is considered as the key glycolytic enzyme and involved in tumor initiation and metabolism. Here, we firstly found that LDHB was highly expressed in osteosarcoma cell lines. Expression profiling indicated that LDHB mRNA was elevated in osteosarcoma tissues with metastasis versus without metastasis, and LDHB high expression predicted a poor prognosis in patients. After LDHB knockdown by siRNA transfection, cell growth and proliferation were inhibited and presented a dose-dependent cell death via MTT assay. Meanwhile, wound healing and matrigel invasion assay revealed that LDHB knockdown inhibited migration and invasion activities in osteosarcoma cells. We further constructed tissue microarray in 40 osteosarcoma tissues. Correlation between LDHB and clinicopathological features indicated that LDHB expressions were associated with tumor TNM stage, recurrence and survival. Kaplan-Meier survival curve revealed that overall survival was significantly decreased in patients with high expression of LDHB. Patients with recurrence or advanced stage showed an increased LDHB, suggesting that increased LDHB was closely associated with a poor prognosis in osteosarcoma patients. Thus, LDHB can be considered as a prognostic marker for tumor recurrence and poor overall survival in osteosarcoma.
Keywords: Osteosarcoma, Lactate dehydrogenase, LDHB, tissue microarray, prognosis
Introduction
Osteosarcoma, deriving from primitive bone-forming mesenchyme, is the most common primary malignant bone tumor in children and young adults [1], and accounts for 5% of all pediatric malignancies. It is a very aggressive type of cancer. Surgery alone cures only a small minority of patients who present with localized disease [2,3]. Recently, great progress in therapeutic strategies, including surgery, adjuvant chemotherapy, radiotherapy and biological therapy have resulted in an increased the disease-free survival rate to more than 60% [4], especially in patients with localized osteosarcoma approaching 75% [5,6]. But the prognosis of patients remains unsatisfactory due to bone destruction, neoplastic bone formation, and pulmonary metastasis [5-7]. Therefore, novel therapeutic strategies are needed to improve the survival rate of this population. As a kind of malignancy, the development and progression of osteosarcoma is a multistep process with accumulation of complex molecular changes, but the underlying molecular mechanism of this disease has not been fully elucidated [8]. Thus, a better understanding of the molecular biology of osteosarcoma will lead to the development of new therapeutic strategies [2,3].
Cancer cells prefer to produce energy by a high rate of glycolysis followed by lactic acid fermentation, resulting in increased lactate production instead of aerobic respiration in the mitochondria, even under oxygen-sufficient conditions, has been called the Warburg effect or aerobic glycolysis [9,10]. The Warburg effect is one of the most important hallmarks of cancer [11,12]. The up-regulation of a series of metabolic enzymes along the glycolytic pathway may contribute to the Warburg effect. Recent studies indicated that glycolysis was essential for tumor proliferation, invasion and metastasis [11,13,14]. Thus, the inhibition of these glycolytic enzymes along the glycolytic pathway can be severed as a promising therapeutic strategy for selectively killing cancers or preventing metastasis [11,13-15].
Lactate dehydrogenase (LDH) is a terminal enzyme for catalyzing the interconversion of pyruvate and lactate in anaerobic glycolytic pathway, and is considered as the key glycolytic enzyme [16,17]. Human LDH, including five isoenzymes, is tetramers that compose two types of subunits, LDHA and LDHB, and distribute in different tissues [18,19]. LDH is involved in tumor initiation and metabolism and has attracted attention as a potential target for cancer therapy and contraception. Recent studies have indicated that LDHA is overexpressed in many malignant tumors and plays an essential role in tumor metabolism [20-23], whereas the role of LDHB is not well-known. Some studies revealed that the expression of LDHB was suppressed in hepatocellular carcinoma, pancreatic cancer, prostate cancer, and gastric cancer [16,19,24,25]. Other studies demonstrated that LDHB could enhance the proliferation of tumor cells of lung adenocarcinoma and breast cancer and high LDHB expression was a significant predictor of poor prognosis in patients [26-28].
Currently, a recent study has reported that LDHB is overexpressed in osteosarcoma by integrated analysis of gene expression and genomic aberration data [29]. However, less definitive evidence has been reported in osteosarcoma, and the link between LDHB and osteosarcoma development is poorly understood. Further studies that focus on the roles of LDHB in the diagnosis and treatment for osteosarcoma are important and of great interest. In this study, we firstly examined the expression of LDHB in osteosarcoma cell lines. Subsequently, we evaluated the functional role of LDHB in growth/proliferation and migration/invasion of osteosarcoma cell lines. Furthermore, we determined the expression of LDHB in osteosarcoma tissues of patients and evaluated the relationship of LDHB expression with the prognostic significance of osteosarcoma patients. Our results suggest that LDHB can be considered as a prognostic marker for both tumor recurrence and poor overall survival in osteosarcoma.
Materials and methods
Cell lines and cell culture
The human osteosarcoma cell lines (MG63, U2OS, and SAOS) were purchased from the American Type Culture Collection (Rockville, MD). The human osteoblast cells HOBC was purchased from PromoCell GmbH (Heidelberg, Germany). The osteosarcoma cell lines were incubated in RPMI-1640 (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin (Life Technologies, Carlsbad, CA). HOBC were cultured in osteoblast growth medium (PomoCell) with 10% fetal bovine serum. All cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C. Light microscopic images were recorded by Zeiss microscope from Carl Zeiss, Inc. (Oberkochen, Germany) with an attached Nikon D40 digital camera. (New York, NY).
Western blotting assay
Total protein lysates from the osteosarcoma cells were harvested using RIPA Lysis Buffer (Upstate Biotechnology, Charlottesville, VA) supplemented with 10 mm NaF (Sigma, Germany). Protein Assay Reagents (Bio-Rad, CA) and a SPECTRAmax Microplate Spectrophotometer from Molecular Devices (Sunnyvale, CA) was applied to evaluate the protein concentrations. The primary antibodies for LDHB (1:1000 dilution) and actin (1:2000 dilution) were purchased from Abcam. Goat anti-mouse HRP antibodies were obtained from Zhongshan Jinqiao Company, Beijing. Western blot analysis was performed as previously reported [30]. Quantification analysis of western blot values was performed with Odyssey software 3.0 (Li-COR Bioscience, NE).
Expression data sets
A total of 52 patients from TCGA with LDHB mRNA expression data in osteosarcoma tissues were used to analyze LDHB mRNA levels. The set of microarray data (GSE21257) were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo).
A total of 52 patients with follow-up in the GSE21257 cohort were involved in the analyses of overall survival and clinical significance with LDHB mRNA level. Among them, a set of 214 patients with HK2 mRNA expression data in both their cancer and matched adjacent non-tumorous tissues was used to analyze HK2 mRNA expression level.
Lentiviral LDHB siRNA transduction; Synthetic LDHB siRNA transfection
The nonspecific siRNA (as a control) and siRNA against LDHB were purchased from RiboBio, and the transfection was carried out using lipofectamine™ 2000 (Invitrogen, Grand Island, NY) according to the manufacturer’s instructions. At 72 h after transfection, the effects of gene silencing were measured by western blot analysis.
MTT cell proliferation assay
U2OS and SOAS were transfected with LDHB siRNA with different doses (15, 30, 45 nmol/L). 5×103 cells in a volume of 100 μL were seeded into a 96-well plate. Each group included three repeated wells. After 72 hours of incubation, proliferation assays were performed with MTT solution (Promega, Madison, WI, USA) using the standard protocol. The absorbance at a wavelength of 490 nm (A490) was measured on a SPECTRAmax Microplate Spectrophotometer from Molecular Devices (Sunnyvale, CA). All results were analyzed by GraphPad Prism 5 software. (San Diego, CA).
Matrigel invasion assay
The matrigel invasion assay was conducted with the BD BioCoatTM MatrigelTM Invasion Chamber (Becton-Dickinson, MA) following the manufacturer’s instructions. After the LDHB siRNA or nonspecific siRNA administration, the transfected cells (5×104 cell/plate) were seeded into the upper chamber of each well in serum-free medium, and complete medium was put into the bottom chamber. The invasion chamber was incubated for 22 hours at 37°C under 5% concentration of CO2. Next, the non-invading cells were removed by scrubbing from the upper surface of the membrane with cotton-tipped swabs. Following the processes of fixation in 100% methanol and staining in hematoxylin, the invading cells were counted in three images of each membrane under a microscope using a 200× objective. The cells images were taken by fluorescence microscope (Nikon Eclipse Ti-U) and phase contrast microscope equipped with a SPOT RT digital camera.
Wound healing migration assay
Migration ability of cells was determined by multiple scratch wounds assay. 1×105 cells per well were plated into 12-well plates and transfected with LDHB siRNA. Three parallel lines were made in confluent cell cultures with a 200 μl tip. After suspended cells were washed away with serum-free medium, the cultures were fed with regular medium again. The scratch wounds were observed at 0, 8, and 24 hours after scratching separately, and photographed via microscope (Nikon Instruments, Inc.) each time. The distance between the two edges of the wound width was randomly quantified at 10 sites in each image. The cell migration distance was defined as the distance between the wound width at the 0 h time point and the wound width at each time point and then divided by two.
Construction of the osteosarcoma tissue microarray (TMA)
A retrospective study of 40 osteosarcoma patients was identified for TMA immunohistochemical staining. The study was approved by the human ethic committee of the Affiliated Cancer Hospital of Zhengzhou University. All patients signed the approval letters that their clinical information would be kept in the databases of Henan Province cancer hospital and utilized for research. Those patients with archival tissue blocks available through the Department of Pathology were selected. The data of age, gender, tumor sites, TNM stages, tumor size, metastasis and recurrence of patients were collected, as shown in Table 1. In the recipient master blocks, each case included two different areas of tumor parts to ensure accurate representation of the selected cores. The corresponding slides of the paraffin-embedded tumor specimens with hematoxylin-eosin (H&E) stained were reviewed in order to further confirm the original diagnosis and choose the most representative areas for the TMA construction. From one paraffin block of each tumor, a 5-μm diameter tissue section was cut and mounted consecutively on the recipient master blocks. The osteosarcoma TMA was constructed by the Tissue Microarray and Imaging Core at the Affiliated Cancer Hospital of Zhengzhou University.
Table 1.
Correlation of clinico-pathological features with LDHB expression levels in 40 osteosarcoma patients
| Clinicopathological features | No. of cases (%) | LDHB | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| Low (%) | High (%) | ||||
| Age (years) | ≤ Median | 20 (50.0) | 13 | 7 | 0.748 |
| > Median | 20 (50.0) | 11 | 9 | ||
| Gender | Male | 27 | 17 | 10 | 0.583 |
| Female | 13 | 7 | 6 | ||
| Tumor site | Femur | 19 | 12 | 7 | 1.000 |
| Tibia | 12 | 7 | 5 | ||
| Other | 9 | 5 | 4 | ||
| TNM stage | Stage I | 12 | 11 | 4 | 0.035 |
| Stage II | 24 | 13 | 8 | ||
| Stage III | 4 | 0 | 4 | ||
| Metastasis | Absent | 15 | 10 | 5 | 0.740 |
| Present | 25 | 14 | 11 | ||
| Recurrence | Absent | 25 | 19 | 6 | 0.018 |
| Present | 15 | 5 | 10 | ||
| Tumor size | ≥7.5 cm | 32 | 19 | 13 | 1.000 |
| ≤7.5 cm | 8 | 5 | 3 | ||
| Survival | Live | 20 | 16 | 4 | 0.022 |
| Dead | 20 | 8 | 12 | ||
Immunohistochemical (IHC) staining on TMA
The expression level of LDHB was determined based on the immunohistochemistry protocol (Paraffin) from Cell Signaling Technology (Beverly, MA) as previously described [31]. Briefly, 5-μm-thick array sections were baked at 60°C for 1 h, deparaffinized using xylene (three times for 5 minutes each), transferred through 100% ethanol (twice for 5 minutes each), rehydrated through graded alcohol, and then immersed in deionized water for 10 minutes. For antigen retrieval, the slides were immersed in boiling (95-100°C) citrate buffer (pH 6.0) for 20 min. Upon washing with phosphate-buffered saline (PBS), the slides were immersed into blocking solution (3% bovine serum albumin) at room temperature for 30 min. Next, the slides were incubated overnight at 4°C with primary LDHB antibodies (Abcam, Cambridge, MA, USA) diluted in blocking serum. Following rinsing with PBS, a horseradish peroxidase (HRP) polymer conjugated to the secondary antibody (SuperPicture™ Polymer Detection Kit, HRP, broad spectrum; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was added to the slides for 10 min, and 3,3’-iaminobenzidine chromogen was then added for 5 min. Following each incubation step, the slides were washed in PBS for 5 min. Mayer’s Hematoxylin Solution (Sigma-Aldrich, St. Louis, MO, USA) was utilized for counterstaining. Subsequently, the slides were dehydrated, air-dried and mounted with neutral resins.
Assessment of LDHB staining was conducted according to nuclear staining showing and calculated by the percentage of positive cells among the entire spot. Thereby, LDHB staining patterns were categorized into 6 groups: 0, no nuclear staining; A, 1+, <10% of cells stained positive; B, 2+, 10% to 25% positive cells; C, 3+, 26% to 50% positive cells; D, 4+, 51% to 75% positive cells; E, 5+, >75% positive cells. A total of 50% of the slides were examined by a second independent assessor who was blinded to the scores and clinicopathological criteria, and good concordance existed between the two scorers (single measure intraclass correlations, >0.8). The images of LDHB staining for IHC and H&E staining were obtained using the NanoZoomer 2.0-RS system (Hamamatsu Photonics Inc., Germany), and the digital slides were analyzed by the software of the NDP. view 2.5.14 version.
Statistical analysis
Continuous variables were expressed as mean ± standard deviation (SD). Student t test was used to assess parametric data between the two groups. The correlation between protein expression and clinicopathological characteristics was analyzed with Pearson’s χ2 test of association or Fisher’s exact test in a 2×2 table. The log-rank test was used to compare the differences in survival curves. Prognostic factors associated with overall survival were analyzed according to the Cox proportional hazards regression model, in a stepwise manner. Statistical analyses were performed using SPSS software version 17.0 (SPSS Inc., Chicago, IL) for Windows (Microsoft Corporation, Redmond, WA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
LDHB is highly expressed in osteosarcoma cell lines
We firstly analyzed the expression of LDHB protein in osteosarcoma cell lines by western blotting. In normal human osteoblast cell lines (HOBC), LDHB protein expressions were extremely low and almost undetectable, while the expression of LDHB protein was significantly increased in osteosarcoma cell lines (MG63, U2OS and SAOS) (Figure 1A). The grey value of protein bands were analyzed in each western blotting, and the data also indicated that LDHB protein presented a remarkably high expression in osteosarcoma cell lines including MG63, U2OS and SAOS, versus normal human osteoblast cell lines (HOBC) (all P<0.001) (Figure 1B).
Figure 1.
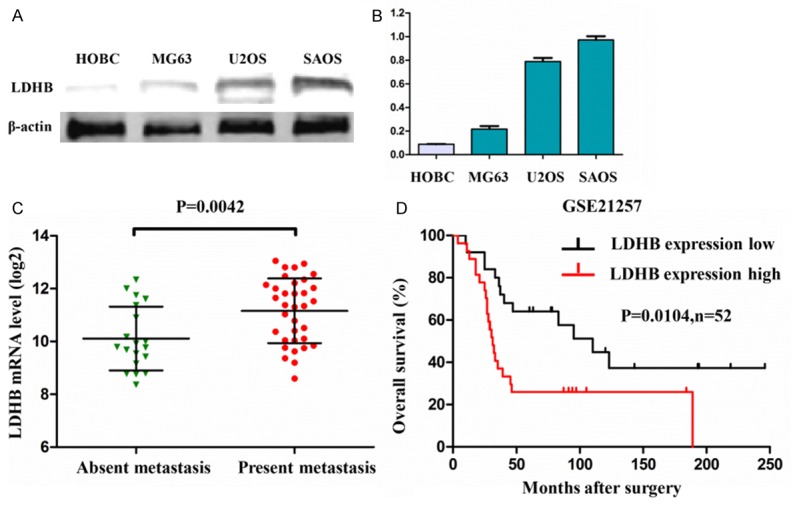
LDHB is highly expressed in osteosarcoma cell lines. A: The expressions of LDHB protein in osteosarcoma cell lines (MG63, U2OS and SAOS) and normal human osteoblast cell lines (HOBC). B: The grey values of protein bands were analyzed in each western blotting in osteosarcoma cell lines and normal human osteoblast cell lines. C: The expression profiling data (GSE21257) of LDHB mRNA in osteosarcoma from pubmed gene bank database indicated that the expression of LDHB mRNA was significantly elevated in human osteosarcoma tissues with metastasis versus without metastasis. D: The overall survival analysis indicated that the osteosarcoma patients with low expression of LDHB mRNA presented a significantly long-term survival, compared with the patients with high LDHB mRNA (n=52).
High expression of LDHB mRNA predicted a poor prognosis in osteosarcoma patients
To detect expression level of LDHB mRNA in human osteosarcoma tissues, we downloaded the expression profiling data (GSE21257) of LDHB mRNA in osteosarcoma. Our analysis results indicated that the expression level of LDHB mRNA was significantly elevated in human osteosarcoma tissues with metastasis versus without metastasis (P=0.0042) (Figure 1C). Meanwhile, according to the expression level of LDHB mRNA, we divided the osteosarcoma tissues into two groups: low expression group and high expression group. The overall survival analysis indicated that the osteosarcoma patients with low expression of LDHB mRNA presented a significantly long-term survival, compared with the patients with high LDHB mRNA (P=0.0104, n=52) (Figure 1D). These data suggested that the high expression of LDHB mRNA predicted a poor prognosis in clinical osteosarcoma patients.
LDHB is crucial for osteosarcoma cell growth and proliferation
To illustrate the function of LDHB in osteosarcoma cell growth and proliferation, lentiviral human LDHB shRNA was transfected in the U2OS and SAOS cell lines, respectively. The non-specific siRNA (N.S siRNA) was served as the control. Western blot analysis confirmed low expression of LDHB in lentiviral shRNA particles cell lines, and indicated that LDHB expressions were gradually decreased under siRNA administration in both U2OS and SAOS cell lines along with the gradual increase of LDHB siRNA concentration (from 15 to 45 nmol/L) (Figure 2A).
Figure 2.
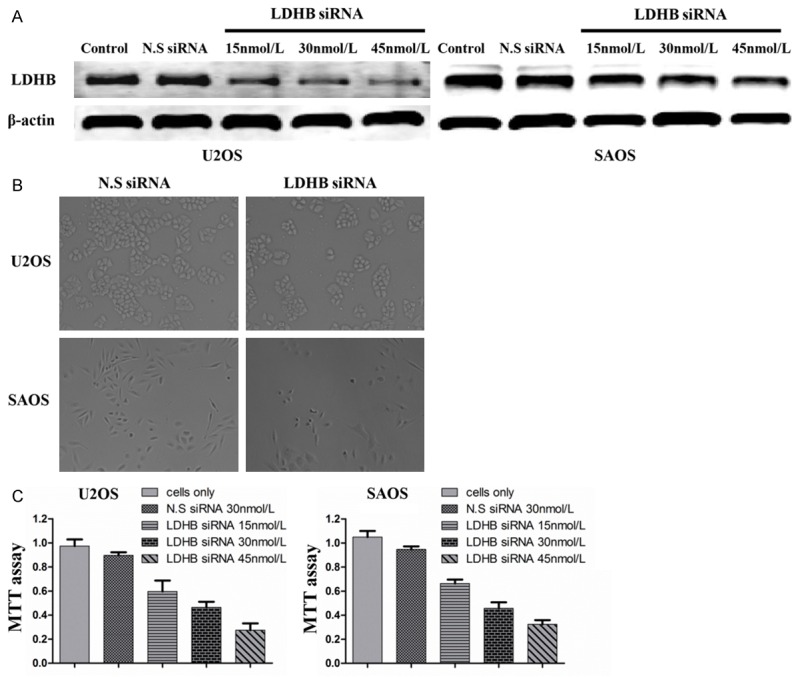
The knockdown of LDHB expression by lentiviral siRNA transfection inhibited cell growth and proliferation in U2OS and SAOS cell lines. A: The confirmation of LDHB knockdown by lentiviral siRNA transfection in the both U2OS and SAOS cell lines through western blot analysis. LDHB expressions were gradually decreased under siRNA administration in the both U2OS and SAOS cell lines along with the gradual increase of LDHB siRNA concentration (from 15 to 45 nmol/L). B: Cells growth were significantly inhibited in the both U2OS and SAOS cell lines after LDHB siRNA administration compared with the N.S siRNA administration. C: Both U2OS and SAOS cells were transfected with LDHB siRNA or non-specific siRNA, and cell proliferations after transfection were determined under the different LDHB siRNA concentrations (15, 30 to 45 nmol/L) by MTT assay.
Next, we observed cells growth in the U2OS and SAOS cell lines after LDHB siRNA administration. The results revealed that cells growth were significantly inhibited in both the U2OS and SAOS cell lines after LDHB siRNA administration compared with the N.S siRNA administration (Figure 2B).
Moreover, we used the MTT assay to evaluate cells proliferation after LDHB siRNA administration. The results indicated that cells proliferation in the U2OS cell line was reduced after the knockdown of LDHB, and cells proliferation were further inhibited in osteosarcoma cells along with the gradual increase of LDHB siRNA concentration (from 15 to 45 nmol/L) (Figure 2C). To further confirm the role of LDHB in osteosarcoma, we also examined the effects of LDHB siRNA on another osteosarcoma cell lines (SAOS). Depletion of LDHB by siRNA resulted in a dose-dependent cell death in SAOS cell line, which were not observed in the nonspecific siRNA transfection (Figure 2C). These data further suggested that LDHB expressions promoted cells growth and proliferation in osteosarcoma cells.
LDHB is important for osteosarcoma cell migration and invasion
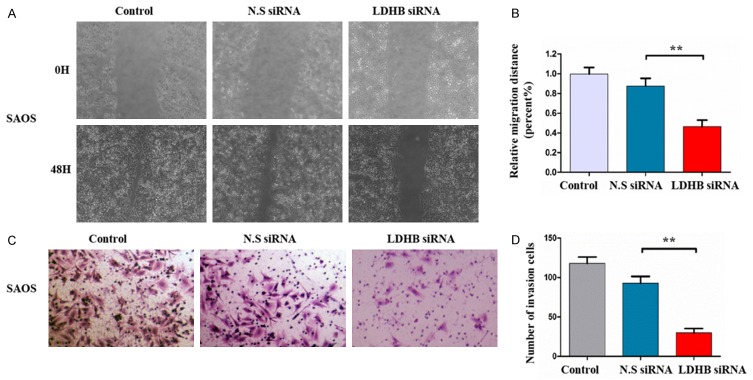
Migration and invasion ability are a crucial hallmark for metastatic osteosarcoma. After the confirmation of significant knockdown of LDHB expression by siRNA in SAOS cells, we examined whether LDHB knockdown by siRNA transfection could influence the migration and invasion ability of osteosarcoma cells by the wound healing assay and matrigel invasion assay. As shown in the wound healing assay, the wounds were almost recovered after 24 hours of migration in blank control and non-specific siRNA treated cells. In contrast, the wound healing and the cells migration were remarkably inhibited after LDHB siRNA administration (Figure 3A). We further recorded and calculated the migration distance of osteosarcoma cells after LDHB knockdown by siRNA transfection, and found that the relative migration distance were obviously decreased in the SAOS cells with LDHB siRNA, compared to the blank control and non-specific siRNA treated cells (P<0.01) (Figure 3B).
Figure 3.
The knockdown of LDHB expression by lentiviral siRNA transfection decreased cell migration and invasion ability in SAOS cell lines. A: Micrographs of osteosarcoma SAOS cells at 0, 8, and 24 hours after wounding. B: Migration distance of SAOS cells for each time point and condition. C: Micrographs of osteosarcoma SAOS cells transfected with LDHB siRNA or nonspecific siRNA. The invading cells were stained with hematoxylin. D: The average numbers of invasive osteosarcoma cells among those transfected LDHB siRNA or nonspecific siRNA. **P<0.01 (comparison transfected cells with control cells using Student’s t-test).
Moreover, in matrigel invasion assays, the invasion ability of SAOS cells after LDHB knockdown by siRNA transfection was remarkably reduced, compared with the blank control and the non-specific siRNA cells (Figure 3C). We also calculated the numbers of osteosarcoma cells that could invade through the matrigel after LDHB siRNA administration, and found that the average number of invasion cells was significantly lower in the LDHB siRNA group than that in the blank control and the non-specific siRNA group (P<0.01) (Figure 3D). Thus, both wound healing assay and matrigel invasion assay revealed that LDHB knockdown by siRNA transfection significantly inhibited the migration and invasion activities of osteosarcoma cells.
Correlation between LDHB expression and clinicopathological parameters in osteosarcoma patients
To further verify the functional role of LDHB in human osteosarcoma, we constructed the osteosarcoma TMA and analyzed the protein expression of LDHB in human osteosarcoma tissues by IHC. A total of 40 osteosarcoma samples were enrolled, and 80 tumor points were collected and stained by TMA, as shown in Figure 4A. LDHB staining was assessed according to nuclear staining intensity and the percentage of positive cells among the entire spot. The LDHB staining score ranged from score 1+ to score 5+, and staining intensity distributed from poor to strong. The details were shown in Figure 4B.
Figure 4.
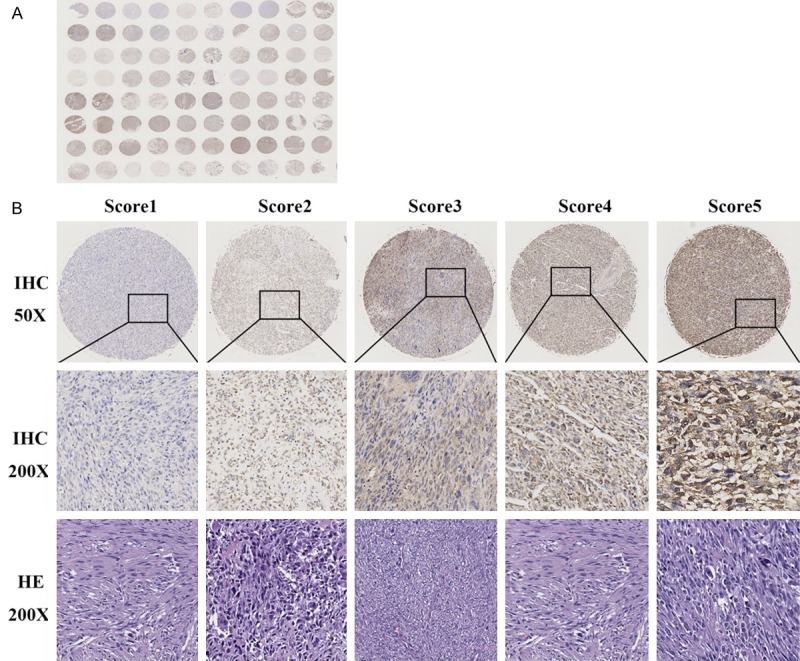
Construction of tissue microarray (TMA) in osteosarcoma and analysis of LDHB staining by immunohistochemistry (IHC). A: The construction of TMA in 40 osteosarcoma tissues samples. 80 tumor points were collected and stained by IHC. B: Representative images of different IHC staining intensities of LDHB or HE staining are shown in osteosarcoma tissues. Assessment of LDHB staining was conducted according to nuclear staining showing and calculated by the percentage of positive cells among the entire spot. Thereby, LDHB staining patterns were categorized into 6 groups: 0, no nuclear staining; A, 1+, <10% of cells stained positive; B, 2+, 10% to 25% positive cells; C, 3+, 26% to 50% positive cells; D, 4+, 51% to 75% positive cells; E, 5+, >75% positive cells.
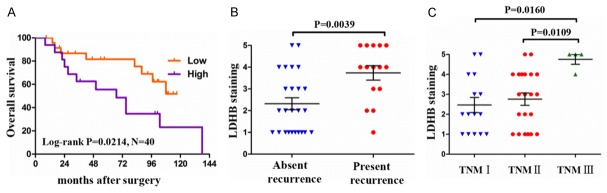
In 40 clinical patients with osteosarcoma, the median age for the patients in the TMA was 35 years (range from 15 to 65), and predominantly male (27/40, 67.5% of patients). To analyze the correlation between LDHB expression levels and clinicopathological features in 40 osteosarcoma patients, we further divided LDHB staining intensity into two grades: low expression (score 1+, 2+ and 3+) and high expression (score 4+ and 5+) among the evaluable specimens. Pearson’s χ2 test or Fisher’s exact test analysis indicated that the expression levels of LDHB had no significant correlation with patients age, gender distribution and tumor site, tumor metastasis and tumor size (all P>0.05). In contrast, the expression levels of LDHB were closely associated with tumor TNM stage (P=0.035), tumor recurrence (P=0.018) and patients survival (P=0.022) (Table 1).
To confirm the relationship between LDHB expression and osteosarcoma prognosis, Kaplan-Meier survival curve was conducted, and the result revealed that the overall survival was significantly decreased in the patients with high expression of LDHB, compared with the patients with low expression of LDHB (Figure 5A). Moreover, 15 out of 40 patients presented tumor recurrence, while 25 patients had no tumor recurrence. The patients with tumor recurrence showed an increased score of LDHB staining versus the patients without recurrence (P=0.0039, Figure 5B). Also, the analysis between tumor TNM stage and LDHB staining score revealed that the patients with TNM III presented an elevated score of LDHB staining versus the patients with TNM II (P=0.0109) and TNM I (P=0.016, Figure 5C). These results suggested that the high expression of LDHB was closely associated with a poor prognosis in patients with osteosarcoma.
Figure 5.
Correlation analysis between LDHB expression and clinicopathological features in osteosarcoma patients. A: Kaplan-Meier survival curve for the overall survival was conducted between the patients with high expression and low expression of LDHB. B: The score of LDHB staining in patients with tumor recurrence and the patients without recurrence. C: The score of LDHB staining in osteosarcoma patients with TNM I, TNM II and TNM III stages, respectively. The log-rank test was used to compare the differences in survival curves. Student t test was used to assess parametric data between the two groups. P<0.05 was considered to indicate a statistically significant difference.
Discussion
Osteosarcoma is a very aggressive type of cancer. Recently, great progress in therapeutic strategies, including surgery, adjuvant chemotherapy, radiotherapy, and biological therapy have resulted in an increased the disease-free survival rate [4-6]. But the prognosis of patients remains unsatisfactory due to bone destruction, neoplastic bone formation, and pulmonary metastasis [5-7]. Therefore, novel therapeutic strategies are needed to improve the survival rate of this population. A better understanding of the molecular biology of osteosarcoma will lead to the development of new therapeutic strategies [2,3]. In this study, we firstly found that LDHB is highly expressed in osteosarcoma cell lines. The expression profiling data of LDHB mRNA indicated that LDHB mRNA expression was significantly elevated in human osteosarcoma tissues with metastasis versus without metastasis, and the high expression of LDHB mRNA predicted a poor prognosis in clinical osteosarcoma patients. After LDHB knockdown by siRNA transfection, osteosarcoma cells growth and proliferation were significantly inhibited and presented a dose-dependent cell death via the MTT assay. Meanwhile, the wound healing assay and matrigel invasion assay revealed that LDHB knockdown significantly inhibited migration and invasion activities in osteosarcoma cells. To further verify the role of LDHB in osteosarcoma, we constructed tissue microarray in 40 osteosarcoma tissues. The correlation between LDHB expression and clinicopathological features in patients indicated that the expressions of LDHB were closely associated with tumor TNM stage, recurrence and patient survival. Kaplan-Meier survival curve revealed that the overall survival was significantly decreased in patients with high expression of LDHB. The patients with tumor recurrence or advanced stage showed an increased LDHB expression, suggesting that the high expression of LDHB was closely associated with a poor prognosis in patients with osteosarcoma. Our study suggests that LDHB can be considered as a prognostic marker for tumor recurrence and poor overall survival in osteosarcoma.
The Warburg effect is one of the most important hallmarks of cancer [11,12]. The inhibition of these glycolytic enzymes can be regarded as a novel strategy for anti-cancer treatment. LDH is a terminal enzyme for catalyzing the interconversion of pyruvate and lactate in anaerobic glycolytic pathway, and is considered as the key glycolytic enzyme [16,17]. It is a tetrameric enzyme which composes two types of subunits, LDHA (muscle-type, M subunit) and LDHB (heart-type, H subunit), and results in five isozymes: M4 (LDH5), M3H1 (LDH4), M2H2 (LDH3), M1H3 (LDH2), and H4 (LDH1) [18,19]. Concerning the expressions of LDHA in different types of malignant tumors, A similar result was found in tumors tissue versus normal tissue: it is overexpressed in colon cancer, bladder cancer, prostate cancer, lung adenocarcinomas, HCC and renal cell carcinoma, and plays critical roles in cancer development and progression [20-23,32-35]. Moreover, the knockdown of LDHA could inhibit cell proliferation, tumor growth and metastasis, by decreasing cell cycle and promoting apoptosis, and reducing the expression of matrix metalloproteinase (MMP)-2 and MMP-9 [34,36-39]. However, with regard to LDHB expression, there was a controversial in different types of malignant tumors. Some studies revealed a similar result with LDHA that the high expression of LDHB presented in lung adenocarcinoma and breast cancer which enhanced the proliferation of tumor cells, and that LDHB was a significant predictor of poor prognosis in patients [26-28]. Other studies demonstrated that the expression of LDHB was suppressed in HCC, prostate cancer, pancreatic cancer and gastric cancer, and that LDHB might function as a suppressor of glycolysis and suppressed pancreatic cancer and HCC progression [16,19,24,25]. Further studies investigated this phenomenon and found that the low expression of LDHB in pancreatic cancer, HCC, and prostate cancer, which was due to promoter hypermethylation and decreased expression of LDHB, might lead to glycolytic transition by converting lactate to pyruvate and the shift from LDH1 to LDH5 (more LDHA in a LDH tetrameric enzyme), thereby promoting cancer progression [19,24,25,40]. Thus, the roles and mechanism of LDHB in the development and progression of different types of tumors are obviously different. As a therapy target, LDHB should be regarded in different ways in different types of tumors.
In osteosarcoma, a recent study has reported that LDHB is overexpressed in osteosarcoma by the integrated analysis of gene expression and genomic aberration data [29]. However, further evidences are needed to understand the link between LDHB and osteosarcoma development. In our study, we first examined the expression of LDHB in osteosarcoma cell lines and found that the high expression of LDHB in osteosarcoma cell which consist with previous studies and analysis results (GSE21257) [29]. Meanwhile, the inhibition of LDHB could decrease cells growth, proliferation, migration and invasion of osteosarcoma cells in vitro. Moreover, we further analyzed the expression level of LDHB mRNA in human osteosarcoma tissues with the clinical feature from the download expression profiling data (GSE21257). We found that the expression level of LDHB mRNA was significantly elevated in human osteosarcoma tissues with metastasis versus without metastasis, and the osteosarcoma patients with low expression of LDHB mRNA presented a significantly long-term survival, compared with the patients with high LDHB mRNA. Furthermore, we recruited 40 osteosarcoma patients for further independent verification, and found the high expression of LDHB in osteosarcoma tissues of patients had a poor prognosis with low overall survival. In addition, we found that patients with recurrence possessed a high level of LDHB in osteosarcoma tissues, as well as patients in advanced tumor stage. Our results suggest that LDHB can be considered as a prognostic marker for both tumor recurrence and poor overall survival in osteosarcoma. Of course, further studies focusing on the roles and mechanism of LDHB in the development and progression of osteosarcoma should be performed.
In conclusion, the present study demonstrates that LDHB is overexpressed in osteosarcoma cell lines and tissues. The high expression of LDHB could be served as a biomarker of poor prognosis for osteosarcoma patients. Moreover, the knockdown of LDHB by siRNA transfection decreased the growth, proliferation, migration and invasion of osteosarcoma cells in vitro. Correlation analysis indicated that the high expressions of LDHB were closely associated with advanced TNM stage, recurrence and poor survival. Thus, LDHB may be considered as a prognostic marker for tumor recurrence and poor overall survival in patients with osteosarcoma, and may be a potential promising therapy target in the treatment of osteosarcoma in future.
Acknowledgements
This study was sponsored by grants from the Education Department S&T Project of Henan Province.
Disclosure of conflict of interest
None.
Authors’ contribution
Q.C. and C.L. designed the experiments; C.L., Y.C., P.B., J.W., Z.L. and Q.C. performed the experiments; C.L., Y.C., P.B. and T.W. analyzed the data; C.L. and Y.C. wrote the manuscript; all authors reviewed the manuscript.
References
- 1.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 2.Anninga JK, Gelderblom H, Fiocco M, Kroep JR, Taminiau AH, Hogendoorn PC, Egeler RM. Chemotherapeutic adjuvant treatment for osteosarcoma: where do we stand? Eur J Cancer. 2011;47:2431–2445. doi: 10.1016/j.ejca.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Khanna C, Fan TM, Gorlick R, Helman LJ, Kleinerman ES, Adamson PC, Houghton PJ, Tap WD, Welch DR, Steeg PS, Merlino G, Sorensen PH, Meltzer P, Kirsch DG, Janeway KA, Weigel B, Randall L, Withrow SJ, Paoloni M, Kaplan R, Teicher BA, Seibel NL, Smith M, Uren A, Patel SR, Trent J, Savage SA, Mirabello L, Reinke D, Barkaukas DA, Krailo M, Bernstein M. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin Cancer Res. 2014;20:4200–4209. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Adamson PC, Blaney SM. New approaches to drug development in pediatric oncology. Cancer J. 2005;11:324–330. doi: 10.1097/00130404-200507000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015;39:593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–307. [PubMed] [Google Scholar]
- 8.Zhao J, Chen F, Zhou Q, Pan W, Wang X, Xu J, Ni L, Yang H. Aberrant expression of microRNA-99a and its target gene mTOR associated with malignant progression and poor prognosis in patients with osteosarcoma. Onco Targets Ther. 2016;9:1589–1597. doi: 10.2147/OTT.S102421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Sheng H, Tang W. Glycolysis Inhibitors for Anticancer Therapy: A Review of Recent Patents. Recent Pat Anticancer Drug Discov. 2016;11:297–308. doi: 10.2174/1574892811666160415160104. [DOI] [PubMed] [Google Scholar]
- 13.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 14.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamed LA, Tachikawa H, Gao XD, Nakanishi H. Yeast cell-based analysis of human lactate dehydrogenase isoforms. J Biochem. 2015;158:467–476. doi: 10.1093/jb/mvv061. [DOI] [PubMed] [Google Scholar]
- 16.Leiblich A, Cross SS, Catto JW, Phillips JT, Leung HY, Hamdy FC, Rehman I. Lactate dehydrogenase-B is silenced by promoter hypermethylation in human prostate cancer. Oncogene. 2006;25:2953–2960. doi: 10.1038/sj.onc.1209262. [DOI] [PubMed] [Google Scholar]
- 17.Song T, Gan W, Chen J, Huang L, Yin H, He T, Huang H, Hu X. Antibodies against Clonorchis sinensis LDH could cross-react with LDHB localizing on the plasma membrane of human hepatocarcinoma cell SMMC-7721 and induce apoptosis. Parasitol Res. 2016;115:1595–1603. doi: 10.1007/s00436-015-4895-z. [DOI] [PubMed] [Google Scholar]
- 18.Drent M, Cobben NA, Henderson RF, Wouters EF, van Dieijen-Visser M. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur Respir J. 1996;9:1736–1742. doi: 10.1183/09031936.96.09081736. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Zhou X, Yu Z, Liu J, Huang G. Low Expression of LDHB Correlates With Unfavorable Survival in Hepatocellular Carcinoma: Strobe-Compliant Article. Medicine (Baltimore) 2015;94:e1583. doi: 10.1097/MD.0000000000001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Chen R, Xie W, Ni Y, Liu J, Huang G. Relationship between 18F-FDG accumulation and lactate dehydrogenase A expression in lung adenocarcinomas. J Nucl Med. 2014;55:1766–1771. doi: 10.2967/jnumed.114.145490. [DOI] [PubMed] [Google Scholar]
- 21.Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP, Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 2012;279:3898–3910. doi: 10.1111/j.1742-4658.2012.08748.x. [DOI] [PubMed] [Google Scholar]
- 22.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui J, Quan M, Jiang W, Hu H, Jiao F, Li N, Jin Z, Wang L, Wang Y, Wang L. Suppressed expression of LDHB promotes pancreatic cancer progression via inducing glycolytic phenotype. Med Oncol. 2015;32:143. doi: 10.1007/s12032-015-0589-8. [DOI] [PubMed] [Google Scholar]
- 25.Maekawa M, Taniguchi T, Ishikawa J, Sugimura H, Sugano K, Kanno T. Promoter hypermethylation in cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes 1-4. Clin Chem. 2003;49:1518–1520. doi: 10.1373/49.9.1518. [DOI] [PubMed] [Google Scholar]
- 26.McCleland ML, Adler AS, Deming L, Cosino E, Lee L, Blackwood EM, Solon M, Tao J, Li L, Shames D, Jackson E, Forrest WF, Firestein R. Lactate dehydrogenase B is required for the growth of KRAS-dependent lung adenocarcinomas. Clin Cancer Res. 2013;19:773–784. doi: 10.1158/1078-0432.CCR-12-2638. [DOI] [PubMed] [Google Scholar]
- 27.McCleland ML, Adler AS, Shang Y, Hunsaker T, Truong T, Peterson D, Torres E, Li L, Haley B, Stephan JP, Belvin M, Hatzivassiliou G, Blackwood EM, Corson L, Evangelista M, Zha J, Firestein R. An integrated genomic screen identifies LDHB as an essential gene for triple-negative breast cancer. Cancer Res. 2012;72:5812–5823. doi: 10.1158/0008-5472.CAN-12-1098. [DOI] [PubMed] [Google Scholar]
- 28.Dennison JB, Molina JR, Mitra S, Gonzalez-Angulo AM, Balko JM, Kuba MG, Sanders ME, Pinto JA, Gomez HL, Arteaga CL, Brown RE, Mills GB. Lactate dehydrogenase B: a metabolic marker of response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2013;19:3703–3713. doi: 10.1158/1078-0432.CCR-13-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong Y, Wu S, Du Q, Wang A, Wang Z. Integrated analysis of gene expression and genomic aberration data in osteosarcoma (OS) Cancer Gene Ther. 2015;22:524–529. doi: 10.1038/cgt.2015.48. [DOI] [PubMed] [Google Scholar]
- 30.Duan Z, Weinstein EJ, Ji D, Ames RY, Choy E, Mankin H, Hornicek FJ. Lentiviral short hairpin RNA screen of genes associated with multidrug resistance identifies PRP-4 as a new regulator of chemoresistance in human ovarian cancer. Mol Cancer Ther. 2008;7:2377–2385. doi: 10.1158/1535-7163.MCT-08-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Guo S, Schwab JH, Nielsen GP, Choy E, Ye S, Zhang Z, Mankin H, Hornicek FJ, Duan Z. Tissue microarray immunohistochemical detection of brachyury is not a prognostic indicator in chordoma. PLoS One. 2013;8:e75851. doi: 10.1371/journal.pone.0075851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutzky LP, Siciliano MJ. Various isozyme gene expression patterns among human colorectal adenocarcinoma cell lines and tissues. J Natl Cancer Inst. 1982;68:81–90. [PubMed] [Google Scholar]
- 33.Wang H, Zhao L, Zhu LT, Wang Y, Pan D, Yao J, You QD, Guo QL. Wogonin reverses hypoxia resistance of human colon cancer HCT116 cells via downregulation of HIF-1alpha and glycolysis, by inhibiting PI3K/Akt signaling pathway. Mol Carcinog. 2014;53(Suppl 1):E107–118. doi: 10.1002/mc.22052. [DOI] [PubMed] [Google Scholar]
- 34.Lea MA, Guzman Y, Desbordes C. Inhibition of Growth by Combined Treatment with Inhibitors of Lactate Dehydrogenase and either Phenformin or Inhibitors of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase 3. Anticancer Res. 2016;36:1479–1488. [PubMed] [Google Scholar]
- 35.Girgis H, Masui O, White NM, Scorilas A, Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH, Bjarnason GA, Jewett MA, Evans A, Al-Haddad S, Siu KM, Yousef GM. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Mol Cancer. 2014;13:101. doi: 10.1186/1476-4598-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Xu L, Wu Q, Liu M, Tang F, Cai Y, Fan W, Huang H, Gu X. Inhibition of LDHA Deliver Potential Anticancer Performance in Renal Cell Carcinoma. Urol Int. 2016 doi: 10.1159/000445125. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 37.He TL, Zhang YJ, Jiang H, Li XH, Zhu H, Zheng KL. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med Oncol. 2015;32:187. doi: 10.1007/s12032-015-0633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xian ZY, Liu JM, Chen QK, Chen HZ, Ye CJ, Xue J, Yang HQ, Li JL, Liu XF, Kuang SJ. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumour Biol. 2015;36:8093–8100. doi: 10.1007/s13277-015-3540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 40.Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424–1435. e1421–1425. doi: 10.1053/j.gastro.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]