ABSTRACT
Although HIV-2 does not encode a vpu gene, the ability to antagonize bone marrow stromal antigen 2 (BST-2) is conserved in some HIV-2 isolates, where it is controlled by the Env glycoprotein. We previously reported that a single-amino-acid difference between the laboratory-adapted ROD10 and ROD14 Envs controlled the enhancement of virus release (referred to here as Vpu-like) activity. Here, we investigated how conserved the Vpu-like activity is in primary HIV-2 isolates. We found that half of the 34 tested primary HIV-2 Env isolates obtained from 7 different patients enhanced virus release. Interestingly, most HIV-2 patients harbored a mixed population of viruses containing or lacking Vpu-like activity. Vpu-like activity and Envelope functionality varied significantly among Env isolates; however, there was no direct correlation between these two functions, suggesting they evolved independently. In comparing the Env sequences from one HIV-2 patient, we found that similar to the ROD10/ROD14 Envs, a single-amino-acid change (T568I) in the ectodomain of the TM subunit was sufficient to confer Vpu-like activity to an inactive Env variant. Surprisingly, however, absence of Vpu-like activity was not correlated with absence of BST-2 interaction. Taken together, our data suggest that maintaining the ability to antagonize BST-2 is of functional relevance not only to HIV-1 but also to HIV-2 as well. Our data show that as with Vpu, binding of HIV-2 Env to BST-2 is important but not sufficient for antagonism. Finally, as observed previously, the Vpu-like activity in HIV-2 Env can be controlled by single-residue changes in the TM subunit.
IMPORTANCE Lentiviruses such as HIV-1 and HIV-2 encode accessory proteins whose function is to overcome host restriction mechanisms. Vpu is a well-studied HIV-1 accessory protein that enhances virus release by antagonizing the host restriction factor BST-2. HIV-2 does not encode a vpu gene. Instead, the HIV-2 Env glycoprotein was found to antagonize BST-2 in some isolates. Here, we cloned multiple Env sequences from 7 HIV-2-infected patients and found that about half were able to antagonize BST-2. Importantly, most HIV-2 patients harbored a mixed population of viruses containing or lacking the ability to antagonize BST-2. In fact, in comparing Env sequences from one patient combined with site-directed mutagenesis, we were able to restore BST-2 antagonism to an inactive Env protein by a single-amino-acid change. Our data suggest that targeting BST-2 by HIV-2 Env is a dynamic process that can be regulated by simple changes in the Env sequence.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) and type 2 (HIV-2) infections are well defined as viral zoonoses. Phylogenetic analysis shows that HIV-1 is closely related to simian immunodeficiency virus (SIV) from chimpanzees (SIVcpz), and HIV-2 is closely related to SIV from sooty mangabeys (SIVsm) (1). At least nine lineages of HIV-2 have been identified, referred to as HIV-2 groups A through I. However, only groups A and B are known to cause human epidemics. In fact, group A viruses account for the vast majority of HIV-2 infections worldwide, which are concentrated mainly in West Africa, Europe, and some Asian countries (1–3). Like all primate retroviruses, HIV-2 encodes three structural proteins (Gag, Pol, and Env) and a set of accessory proteins (Vif, Vpx, Vpr, and Nef). Most, if not all, of the accessory proteins serve to antagonize host restriction factors, which are part of the host's innate immune system and are considered a first line of defense against viruses. Overall, the genomes of HIV-1 and HIV-2 are very similar. Two notable differences are (i) the presence of a vpu gene in HIV-1 which is absent from HIV-2 and (ii) the absence of a vpx gene in HIV-1 which is present in HIV-2. Vpu targets bone marrow stromal antigen 2 (BST-2) and induces degradation of CD4, while Vpx induces degradation of sterile alpha motif and HD domain-containing protein 1 (SAMHD1) (for a review, see reference 4). There is no known functional homolog to Vpx in HIV-1 to target SAMHD1, and while Nef is well known to downregulate CD4 from the cell surface (5), the ability to induce proteasomal degradation of CD4 is limited to viruses expressing Vpu (6, 7). Thus, the Vpu and Vpx proteins are not functional homologs. On the other hand, the ability to enhance virus release by antagonizing BST-2 is not limited to Vpu-encoding viruses. In fact, in HIV-2, antagonizing BST-2 is a functional property of the Env glycoprotein (8, 9), while in SIV this function is executed by the Nef protein (10–13). For the remainder of this work, we refer to the ability of HIV-2 to enhance virus release as Vpu-like activity.
BST-2, also known as tetherin or CD317, is a 30- to 36-kDa type II transmembrane protein that inhibits the release of retrovirus particles by physically tethering virions to the cell surface (14, 15). The exact mechanism of how Vpu antagonizes BST-2 is still unclear. However, it is thought to involve a process that interferes with the resupply of newly synthesized BST-2 from the endoplasmic reticulum (ER) to the cell surface (reviewed in reference 4). Similar to HIV-1 Vpu, the ability of HIV-2 Env to overcome the restrictive phenotype of Vpu-deficient HIV-1 was known long before the cellular target was identified (8, 9, 16, 17). Direct evidence that HIV-2 Env, like Vpu, antagonizes human BST-2 was provided for two HIV-2 laboratory isolates (ROD10 and RODA [16, 18, 19]) and for one SIVtan isolate, which was adapted for replication in a human CD4+ T cell line (20). It is also interesting that serial passaging of a nef-deleted SIV in rhesus macaques resulted in the acquisition of mutations in the cytoplasmic domain of gp41 that conferred resistance to rhesus BST-2 (21). In contrast, the Env proteins of HIV-2 and SIVtan were found to target BST-2 through ectodomain interactions (20, 22), leading to the recruitment of a clathrin adaptor AP2 complex via a membrane-proximal GYXXϕ motif in the cytoplasmic domain of gp41 and resulting in the sequestration of BST-2 in the trans-Golgi network (TGN) (23).
We had previously found that a single-amino-acid change in the ectodomain of the HIV-2 Env TM subunit can regulate the ability of HIV-2 Env to enhance virus release (24). However, these studies were done with highly laboratory-adapted virus isolates, and it was not clear how relevant the Vpu-like activity was in vivo. To address this question, we cloned primary HIV-2 env sequences from viruses that had been isolated by coculture of patient peripheral blood mononuclear cells (PBMCs) with PBMCs from uninfected individuals (25). In total, we isolated 35 full-length HIV-2 Env sequences from 8 patients. All 35 Env isolates were analyzed for Vpu-like activity in a virus release assay, and their envelope function was tested by pseudotyping Env-defective HIV-2. We found that all Env proteins were functional in the pseudotyping assay, although there was significant variability in the relative pseudotyping efficiency. Interestingly, almost half of the primary HIV-2 isolates also exhibited Vpu-like activity, and viruses with Env proteins capable or incapable of antagonizing BST-2 were found to coexist in the same patient. Finally, mutational analysis of an Env isolate lacking Vpu-like activity revealed that a single-amino-acid change could lead to gain of Vpu-like function. Interestingly, gain of Vpu-like activity was not caused by a gain of interaction with BST-2, since both inactive and active Envs interacted with BST-2 with similar efficiency. Taken together, our data reveal that the ability to target BST-2 is conserved not only in HIV-1 but also in HIV-2. Our data also show that the ability of HIV-2 to target BST-2 is a dynamic process that can be regulated by very subtle changes in the Env amino acid sequence. These changes can occur in the same patient in vivo without correlating to the functionality of the Env proteins with respect to producing infectious virus. Finally, consistent with our observations on Vpu, the ability of HIV-2 Env to interact with BST-2 is presumably necessary but not sufficient for antagonism.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa, HeLa-TZM-bl, and 293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). For transfection, cells were grown in 25-cm2 flasks to about 80% confluence. Cells were transfected using Lipofectamine Plus (Invitrogen Corp., Carlsbad, CA) by following the manufacturer's recommendations. A total of 6 μg of plasmid DNA per 25-cm2 flask was used. Total amounts of transfected DNA were kept constant in all samples of any given experiment by adding empty vector DNA as appropriate. Cells were harvested 24 h posttransfection.
Viral RNA extraction, HIV-2 envelope cloning, and sequence analysis.
Virus culture samples from 8 patients infected with HIV-2 were obtained from the Research Institute for Medicines (iMed.ULisboa), University of Lisbon, Lisbon, Portugal (25, 26). Patient data are summarized in Table 1. For each sample, 140 μl of culture supernatant was used to extract viral RNA using a QIAamp viral RNA minikit (Qiagen). RNA was eluted in 60 μl of elution buffer and immediately subjected to first-strand cDNA synthesis using the SuperScript III reverse transcriptase kit according to the manufacturer's instructions (Invitrogen Life Technologies). The resulting cDNA was subjected to first-round PCR using primers to conserved regions upstream or downstream of env (5′ primer F3 or A1m2F and 3′ primer R1 or NT5mR) (Table 2). PCR products were cloned into the pCR4-TOPO vector (Invitrogen) and sequenced. Specific primers were designed for subcloning of individual env isolates into a mammalian expression vector (Table 2). Note that the 3′ primers were designed to add a hemagglutinin (HA) tag to the C terminus of Env. Also, the 5′ primer (HIV-2 Rev-Xba-F) was designed to include the first exon of Rev. Using these primers, a 2,600- to ∼2,700-bp fragment encompassing the entire env gene and the rev gene was amplified from individual TOPO clones by 2nd-round PCR using Platinum Taq DNA polymerase high fidelity (Invitrogen) and cloned into the Env expression vector pCM10 (24). This vector allows for the expression of Env proteins in a Tat- and Rev-independent manner. As a control, we also created C-terminally HA-tagged variants of the HIV-2 ROD10 and ROD14 Env using primers listed in Table 2. For consistency, these vectors also included the upstream first exon of Rev. All PCR fragments were cloned via the primer-encoded XbaI and XhoI restriction sites into the corresponding sites in pCM10 (24).
TABLE 1.
Summary of clinical data of patients involved in this study
| Patient | Sample IDa | RNA copies/mlb | CD4 T cell count/μlb | Coreceptor usage | Genderc | Date (yr) of: |
Therapyd | ||
|---|---|---|---|---|---|---|---|---|---|
| Sample collection | Diagnosis | Starting therapt | |||||||
| P1 | HCC1.03 | <200 | 308 | CCR5 | F | 2003 | 2001 | 2001 | DDI, D4T, IDV |
| P2 | HCC6.03 | <200 | 615 | CCR5 | F | 2003 | 1992 | 1996 | AZT, 3TC, IDV |
| P3 | HCC10.03 | 160559 | 48 | CXCR4 | M | 2003 | 1996 | 1996 | DDI, AZT, SQV |
| P4 | HCC19.03 | <200 | 175 | CCR5 | F | 2003 | 2003 | 2005 | D4T, 3TC, LPVr |
| P5 | HCC20.03 | NAe | 78 | CXCR4 | F | 2003 | 1998 | 2005 | TDF, ABC, LPVr |
| P6 | HSM10.04 | 4792 | 265 | CXCR4 | F | 2004 | 2001 | 2002 | AZT, 3TC, NVF |
| P7 | HSMAK.10 | 1793 | 40 | Dual/mixed population | F | 2010 | 2009 | No ARTf | No ART |
| P8 | HSMNC.10 | <200 | 231 | CCR5 | F | 2010 | 2008 | NA | SQV, ABC, 3TC |
At the time of sample collection.
F, female; M, male.
3TC, lamivudine; ABC, abacavir; AZT, zidovudine; D4T, stavudine; DDI, didanosine; IDV, indinavir; LPVr, lopinavir; SQV, saquinavir.
NA, not available.
ART, antiretroviral therapy.
TABLE 2.
Primers used for construction and site-directed mutagenesis of HIV-2 envelope
| Primer ID | Gene | Sequencea (5′-3′) |
|---|---|---|
| F3 | vpr | 5′-TAGACATGGAGACACCCTTGAARGMGC-3′ |
| A1m2F | rev | 5′-GCGCTCTAGAGCCACCATGAACGAAAGGGCAGACGAAGAAGGACTCC-3′ |
| R1 | nef | 5′-TGTAAWACAKCCCTTCCAGTCCYCC-3′ |
| NT5mR | env | 5′-CYTCACAGGAGGGCRAKTTCTGC-3′ |
| ROD10/14-XbaI-F | env | 5′-GCGCTCTAGAATGAACGAAAGGGC-3′ |
| ROD10/14-XhoI-HAtag-R | env | 5′-GCGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGCT-3′ |
| HIV-2 Rev-XbaI-F | env | 5′-GCGCTCTAGAGCCACCATGAACGAAAGGGCAGACGAAGAAGGACTCC-3′ |
| HIV-2 Rev-NheI-F | env | 5′-GCGCGCTAGCGCCACCATGAACGAAAGGGCAGACGAAGAAGGACTCC-3′ |
| 1-SalI-HAtag-R | env | 5′-CGCGTCGACTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGAGTTCTGCTCC-3′ |
| 2-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATACACTATCCCGGCCAGTAAAG-3′ |
| 3-XhoI-HAtag-R | env | 5′-GCGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGAGTTCTGCCC-3′ |
| 3-s10-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATATGTCATATTGTCCCATTTAG-3′ |
| 3-s11-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATATTCTATCTGCCAAGGCCAGG-3′ |
| 4-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGAGTTCTGCTTC-3′ |
| 5/6-SalI-HAtag-R | env | 5′-CGCGTCGACTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGATTTCTGCTCC-3′ |
| 7-XhoI-HAtag-R | env | 5′-GCGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATACAGGAGGGCGAGTTCTGCCC-3′ |
| 8-s3-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATATTCCTTCTCTGTCTGGCTGT-3′ |
| 8-s4-XhoI-HAtag-R | env | 5′-CGCCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATATCGGCCAAGGCCAGGAGCTG-3′ |
| m1-F | env | 5′-GTGAATCACCTAAAGAAGGCACAAACACAACTAGCACACCTAGCACAGCTGTAAATGACA-3′ |
| m1-R | env | 5′-TGTCATTTACAGCTGTGCTAGGTGTGCTAGTTGTGTTTGTGCCTTCTTTAGGTGATTCAC-3′ |
| m2-F | env | 5′-GTGAATCACCTAAAGAAGGCAACACAACTAGCACACCTGTAAATGACAGT-3′ |
| m2-R | env | 5′-ACTGTCATTTACAGGTGTGCTAGTTGTGTTGCCTTCTTTAGGTGATTCAC-3′ |
| m3-F | env | 5′-GCAAAAACTAAATAGCTGGGATATTTTTGGCAACTGGTTTGACTTGACCT-3′ |
| m3-R | env | 5′-AGGTCAAGTCAAACCAGTTGCCAAAAATATCCCAGCTATTTAGTTTTTGC-3′ |
| m4-F | env | 5′-ACAGAACAGGACAAATCAGACAAAACGCAATTATGTGTC-3′ |
| m4-R | env | 5′-TTGCGTTTTGTCTGATTTGTCCTGTTCTGTACCCAATTG-3′ |
| m5-F | env | 5′-TTTACTGGCTGGGATAGTGCAGCAACAGCAACAGCTGTTG-3′ |
| m5-R | env | 5′-TGCTGTTGCTGCACTATCCCAGCCAGTAAAGTCCGGGAC-3′ |
| m6-F | env | 5′-AATTGTTAAGTAGACTTAGAAAGGGCTATAGGCCTGTTTTCTC-3′ |
| m6-R | env | 5′-TATAGCCCTTTCTAAGTCTACTTAACAATTGTACTATGTATATTAC-3′ |
| m7-F | env | 5′-AGAGAAGAAACAGAAGAAGACGTTGGAAACAGCGTTGGAGACAG-3′ |
| m7-R | env | 5′-TGTTTCCAACGTCTTCTTCTGTTTCTTCTCTGGCTGGCTG-3′ |
R = A or G, M = A or C, W = A or T, K = G or T, and Y = C or T.
Phylogenetic analysis.
Clonal envelope sequences from each patient were codon aligned with a set of reference sequences representative of HIV-2 groups A and B, obtained from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/) using MUSCLE (27), and the alignment was manually edited with GeneDoc (http://iubio.bio.indiana.edu/soft/molbio/ibmpc/genedoc-readme.html). Maximum likelihood (ML) phylogenetic analysis was performed using the best-fit model of molecular evolution estimated by Modeltest v3.7 using the Bayesian information criterion. The ML tree was inferred with program MEGA6 (28). To find the ML tree, the nearest neighbor interchange (NNI) iterative heuristic method was used. The reliability of the obtained topology was estimated by bootstrap (1,000 replicates).
Site-directed mutagenesis.
HIV-2 envelope point mutants were created using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) and primer pairs m1 to m7 (Table 2). Mutations were verified by sequencing.
Antibodies.
HIV-1 Gag proteins were identified using human HIV-1 Ig (catalog number 3957; NIH Research and Reference Reagent Program). HIV-2 Gag proteins were identified using HIV-2 patient serum (catalog number 1495 [discontinued]; NIH Research and Reference Reagent Program). Mouse anti-tubulin and mouse anti-HA monoclonal antibodies (MAbs) were from Sigma-Aldrich (catalog numbers T-9026 and H9658, respectively; St. Louis MO).
Western blotting.
Cells were washed with ice-cold phosphate-buffered saline (PBS) twice and lysed with 1× SDS protein loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 5% glycerol, 5% β-mercaptoethanol, and 0.01% bromophenol blue). Samples were then heated at 95°C for 10 min with occasional vortexing of the samples. The lysates were resolved by SDS-PAGE and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica MA). The membrane was blocked with dry milk (5% solution in 1× TNT buffer [10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.3% Tween 20]) and probed with the primary antibodies in TNT buffer, followed by incubation with alkaline phosphatase-conjugated secondary antibodies (Sigma-Aldrich, St. Louis, MO). Finally, signals were detected using chemiluminescence by following the manufacturer's recommendations (Applied Biosystems, Foster City, CA). α-Tubulin was used as a loading control.
Assessment of viral particle release.
Pulse-chase analysis was performed as described previously, with some modifications (9). Briefly, HeLa cells were cotransfected with 4 μg of Vpu-defective pNL4-3/Udel-1 (29) and 2 μg of one of the HA-tagged Env expression vectors using Lipofectamine Plus. Cells were pulse labeled 24 h later with [35S]-EXPRE35S35S-label (2 mCi/ml; PerkinElmer, Waltham, MA) for 30 min at 37°C and chased in 1 ml of prewarmed complete DMEM-FBS for 0, 2.5, or 5 h. At each time point, cells were collected and lysed in 400 μl of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100). Cell lysates were precleared by incubation at 4°C for 1 h with protein A-Sepharose beads (Sigma). The cell-free culture supernatants were mixed with 200 μl of lysis buffer. Cell lysates and detergent-treated supernatants were immunoprecipitated with HIV-IgG (catalog number 3957; NIH Research and Reference Reagent Program). Immunoprecipitates were solubilized by heating in sample buffer and separated by SDS-PAGE using 12% polyacrylamide gels. Gels were treated for 20 min with 1 M Na-salicylic acid and dried. Radioactive bands were visualized by fluorography using Bio-Max MR film (Eastman Kodak, Rochester NY). Quantitation of the relevant bands was performed with a Fujix BAS 2000 Bio-Image analyzer. The efficiency of particle release at each time point was calculated by dividing the amount of Gag proteins present in the virus fraction by the total of cell- and virus-associated Gag proteins. The ratio of virion-associated to total Gag protein then was plotted as a function of time.
Virus preparation.
Virus stocks were prepared by transfection of 293T cells with appropriate plasmid DNAs. Virus-containing supernatants were harvested 24 h after transfection. Cellular debris was removed by centrifugation (5 min; 1,500 rpm), and the clarified supernatants were filtered (0.45 μm) to remove residual cellular contaminants. Supernatants were quantified by reverse transcriptase (RT) assay (30) and used for infection of TZM-bl indicator cells.
Viral infectivity assay.
A 200-μl aliquot of viral stock was used to infect TZM-bl cells (CD4+, CCR5+, and CXCR4+) in a 24-well plate (5 × 104 cells were seeded 1 day prior to infection) in a total volume of 1 ml. Typically, infections were performed in duplicate. Infection was allowed to proceed for 48 h at 37°C. Medium was removed, and cells were lysed in 200 μl of Promega 1× reporter lysis buffer (Promega Corp., Madison, WI) and frozen at −80°C for a minimum of 30 min. To determine the luciferase activity in the lysates, 10 μl of each lysate was combined with 50 μl of luciferase substrate (Steady-Glo; Promega Corp., Madison, WI), and light emission was measured using a Modulus II microplate reader (Turner Biosystems Inc., Sunnyvale, CA). Values were corrected for differences in input virus (based on reverse RT assay).
Coimmunoprecipitation analyses.
293T cells were transfected with expression vectors for HIV-2 Env and BST-2 as indicated. Cells were harvested 24 h posttransfection, washed twice with cold PBS, lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40; supplemented with Complete protease inhibitor cocktail [Roche Life Science, Indianapolis, IN]) at 4°C for 20 min, and then clarified by centrifugation at 15,000 × g for 10 min. Ten percent of the lysate was used as the input control, and the remaining lysate was used for immunoprecipitation of HA-tagged antigens. Precleared cell lysates were mixed with anti-HA antibody-conjugated agarose beads (Sigma-Aldrich, Inc., St. Louis MO) and incubated at 4°C for 4 h. Samples were then washed three times with RIPA buffer. Proteins were eluted by boiling beads in sample buffer and subjected to immunoblot analysis with antibodies to HA and BST-2.
Accession number(s).
The nucleotide sequence data determined during the course of this work were deposited in GenBank under the following accession numbers: KX791206 to KX791239.
RESULTS
Phylogenetic analysis of primary HIV-2 isolates.
We obtained virus culture samples from eight HIV-2-infected individuals (P1 to P8; Table 1). env sequences were amplified by RT-PCR. Since we expected significant sequence variation in the env gene, we first amplified env sequences using PCR primers mapping to more conserved regions in the upstream vpr and downstream nef genes (Table 2). Resulting cDNAs were cloned into pCR4-TOPO, and individual clones from each sample were sequenced. env sequences isolated from a given patient were labeled according to the patient code followed by the clone number. For instance, sample P3-11 represents clone 11 from patient 3. Of the clones analyzed, 35 expressed detectable protein levels. Clones that did not express detectable protein because of deletions or truncations were excluded from further analysis. Also, despite several attempts we were unable to obtain more than a single clone from patient 5. This clone was severely truncated and nonfunctional; therefore, we decided to exclude it from our study as well.
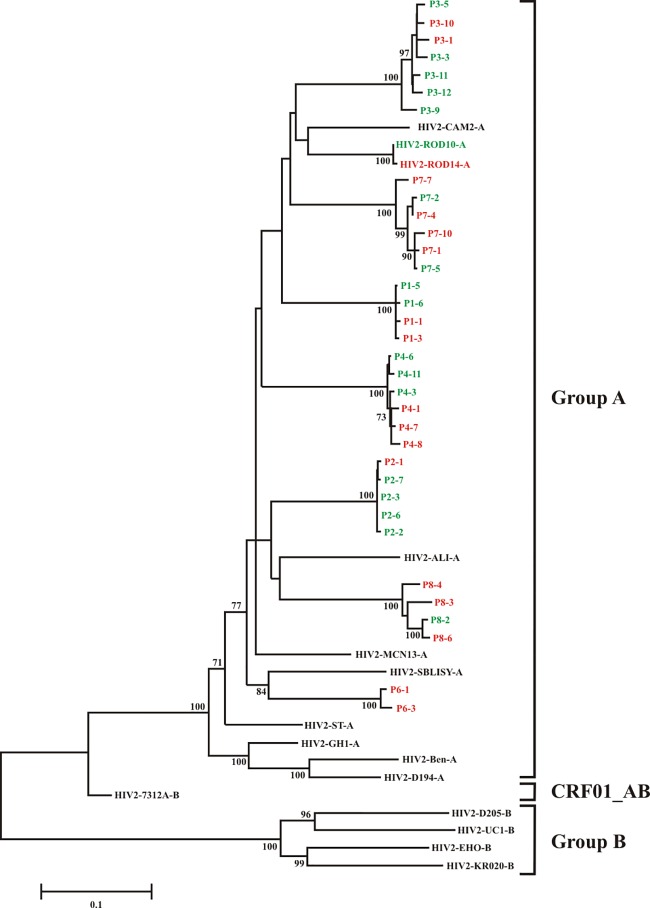
Phylogenetic analysis was performed based on 9 group A, 4 group B, and 1 AB reference sequence published in the NCBI database (http://www.ncbi.nlm.nih.gov), together with the 34 full-length HIV-2 env sequences identified in the present study (Fig. 1). We found that the env sequences from all seven HIV-2 patients clustered significantly with HIV-2 group A reference sequences. Sequences of Env variants isolated from the same patient were fairly conserved (96 to 99% at the amino acid level [data not shown]). Variation across the entire env gene sequence when samples from all patients were analyzed was as high as 20% at the nucleotide level and up to 25% at the amino acid level (data not shown).
FIG 1.
Molecular phylogenetic analysis of envelope gene sequences. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model (GTR+G+I) (28). The tree with the highest log likelihood (−25,625.8806) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Only values of ≥70% are displayed. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories; +G parameter, 0.6138). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 25.7204% of sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Each reference HIV-2 strain is represented by its genetic group and name at the right. HIV-2 isolates in green exhibit Vpu-like virus release activity (Fig. 3 and 4A); HIV-2 isolates in red do not exhibit Vpu-like activity.
Antagonism of BST-2 by HIV-2 envelope glycoproteins.
HIV-2 does not encode a vpu gene. Nevertheless, we and others previously reported that certain HIV-2 isolates, such as HIV-2 ROD10, encode a Vpu-like activity that results in enhanced virus release and maps to the HIV-2 Env protein (8, 16). Interestingly, the closely related ROD14 Env lacks a Vpu-like activity due to a single-amino-acid change in Env (24). Indeed, after the identification of BST-2 as the cellular target of Vpu (15, 31), it was confirmed that HIV-2 Env, like Vpu, antagonizes BST-2 to counteract BST-2-mediated tethering of virus particles to the host cell membrane (18, 19).
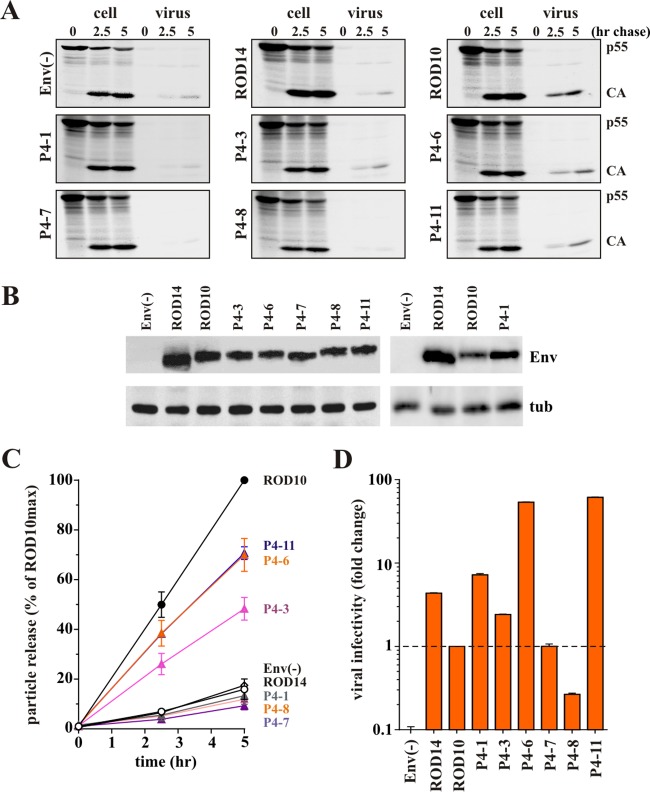
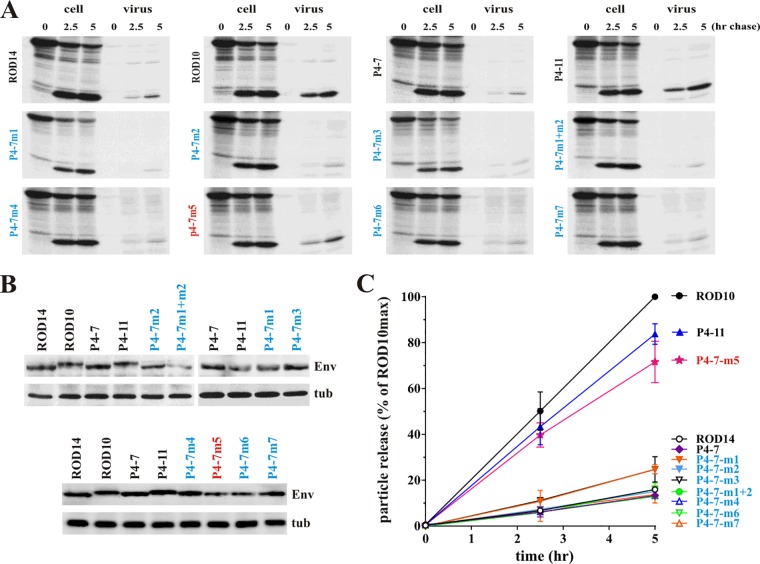
To assess the ability of our Env isolates to antagonize BST-2, we subcloned the full-length env genes into the Env expression vector pCM10 (24). To be able to track expression and virus incorporation of the Env products, all constructs, including ROD10 and ROD14, were modified to add a C-terminal HA tag. Rev independence was achieved by including the first exon of Rev upstream of the Env coding sequence. Vpu-like activity was determined by comparing the effects of ROD10 Env (positive control) and ROD14 Env (negative control) to the various primary HIV-2 envelope isolates on the release of Vpu-defective HIV-1 NL4-3 using a pulse-chase metabolic labeling assay described previously (8, 9). Vpu-deficient HIV-1 was chosen as a model system, since we had previously demonstrated the Vpu-like activity of HIV-2 Env in this system (9). Also, antibodies for immunoprecipitation of HIV-1 Gag proteins are more readily available than antibodies to HIV-2 Gag. Experiments were performed in transiently transfected HeLa cells, which express high levels of endogenous BST-2 (32). Representative experimental data are shown for 6 Env variants isolated from patient 4 (Fig. 2A and B). In all experiments cells were pulse labeled for 30 min and chased for up to 5 h as described in Materials and Methods. At each time point, equal aliquots of cells were harvested and virions released into the supernatant were collected. Each fraction was lysed in lysis buffer, and viral proteins were subjected to immunoprecipitation with an HIV-1 patient serum. Immunoprecipitated proteins were separated by SDS-PAGE and visualized by fluorography (Fig. 2A). Expression of comparable levels of HIV-2 Env was confirmed by immunoblotting (Fig. 2B). Quantitation of results from two independent experiments is presented in Fig. 2C. All other Env isolates were analyzed in a similar fashion, and quantitation of the data is summarized in Fig. 3 and 4A.
FIG 2.
HIV-2 envelope glycoprotein enhances HIV-1 particle release. (A) Kinetic analysis of viral particle release by Vpu-deficient HIV-1 in the presence of the different HIV-2 Env isolates. HeLa cells were transfected with pNL4-3/Udel-1 together with HA-tagged HIV-2 Env vectors pROD14-Env, pROD10-Env, and pHA vector [Env(−)] as controls, as well as vectors for the expression of HA-tagged Envs from HIV-2 patient 4 isolates P4-1, P4-3, P4-6, P4-7, P4-8, and P4-11. Samples were subjected to pulse-chase analysis, and viral proteins recovered by immunoprecipitation were separated by 12% SDS-PAGE. The major HIV-1 Gag proteins p55gag and p24CA are identified on the right. A representative experiment is shown. (B) Relative expression of Env in the transfected cells was verified by Western blot analysis using an HA-specific MAb. Expression of cellular α-tubulin served as a loading control (tub). (C) Efficiency of virus release was determined by quantifying bands in panel A corresponding to the precursor and mature Gag proteins at each time point. Results were plotted as a function of time. Maximal virus release by ROD10 at the 5-h time point was defined as 100%, and the remaining data points were normalized accordingly. Data are presented as means ± standard errors of the means (SEM) from two independent experiments. (D) To assess the ability of HIV-2 Env variants to produce infectious virus, 293T cells were transfected with 4 μg of envelope-deficient pROD10.env1 DNA in the presence of empty pHA vector [Env(−)] or HA-tagged pROD14-Env, pROD10-Env, or HIV-2 patient 4 isolates P4-1, P4-3, P4-6, P4-7, P4-8, and P4-11 as indicated. Virus-containing supernatants were harvested 24 h later, and a portion of the filtered culture supernatant was used for the infection of TZM-bl cells. Luciferase activity was measured 48 h after infection and normalized for input virus. The result shown is representative of two independent experiments. Infectivity of viruses pseudotyped with the ROD10 Env was defined as 1. Differences in viral infectivity of the other samples are expressed as fold change relative to ROD10 Env. Graphs represent the means ± SEM from duplicate infections.
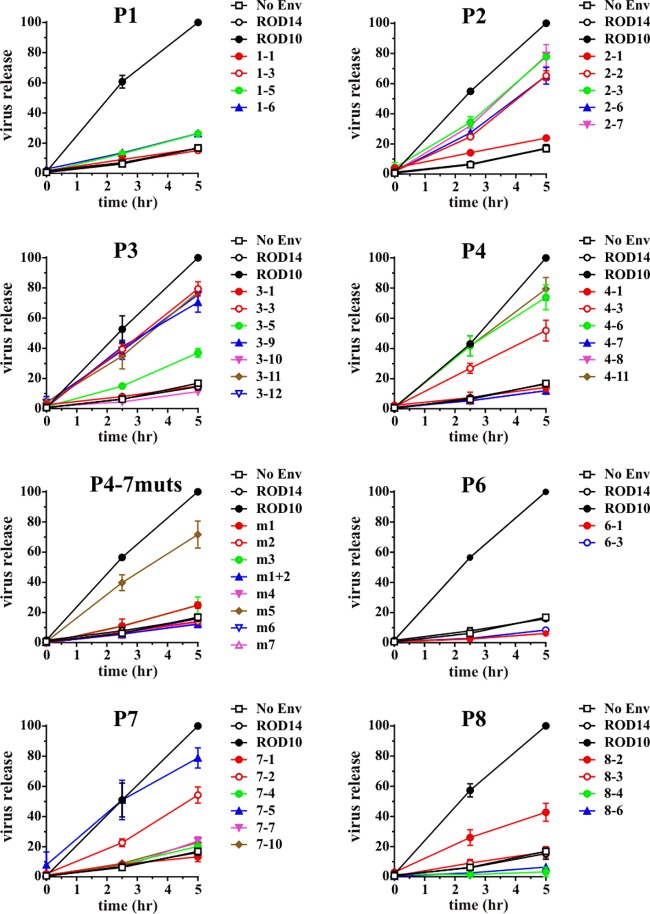
FIG 3.
Antagonism of BST-2 by HIV-2 Env variants. Pulse-chase analyses were performed for all Env variants as described for Fig. 2A. Quantitation was done as described for Fig. 2C. Data were grouped by patient and are presented as means ± SEM from two independent experiments.
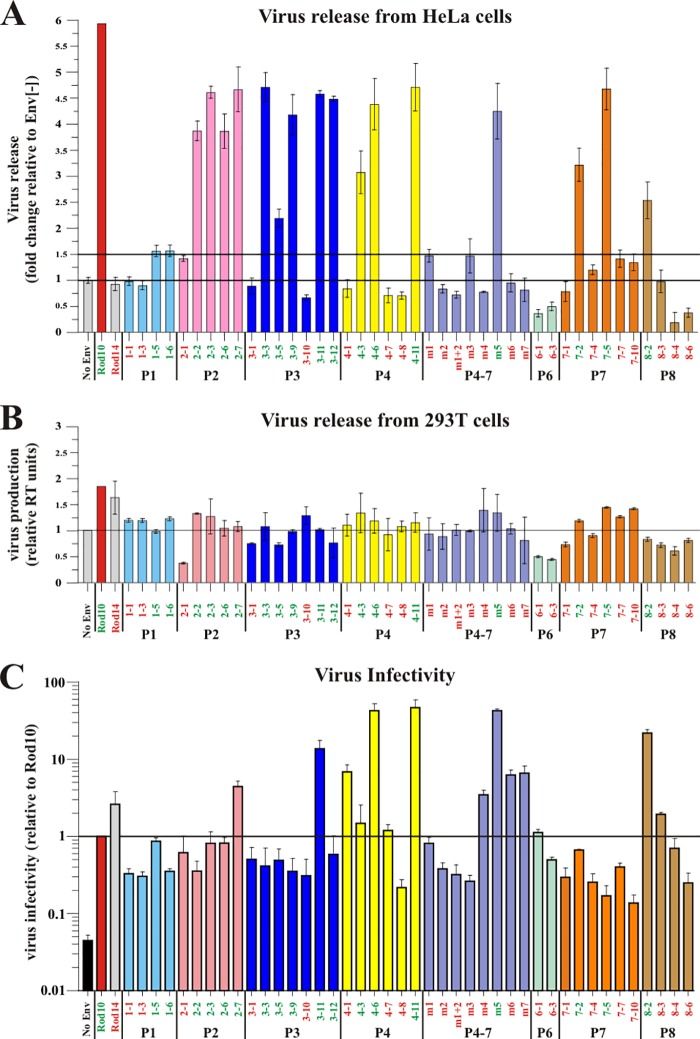
FIG 4.
Summary of the functional data for all HIV-2 Env variants. (A) Effect of HIV-2 Env on the release of HIV-1 from BST-2-expressing HeLa cells. Release of Vpu-deficient HIV-1 in the presence of the different HIV-2 Env variants was determined by pulse-chase analysis as described for Fig. 2A. Virus release observed after 5 h of chase was quantified as described for Fig. 2C. Virus release in the absence of Env was defined as 1 and is marked by a horizontal line. Virus release in the presence of individual Env variants was calculated as fold change relative to the Env-negative sample. Data are presented as means ± SEM from at least two independent experiments. A 1.5-fold increase is marked by a second horizontal line and represents an empirical cutoff to define Vpu-like activity. (B) Effect of HIV-2 Env on the release of HIV-2 in the absence of BST-2. BST-2-negative 293T cells were transfected with 4 μg of envelope-deficient pROD10.env1 DNA in the presence of empty pHA vector [Env(−)] or HA-tagged pROD14-Env, pROD10-Env, or HIV-2 patient isolates. Virus-containing supernatants were harvested 24 h later, and virus production was quantified by measuring the virus-associated reverse transcriptase activity. Virus production in the absence of Env was defined as 1 (marked by a horizontal line). Effects of individual Env proteins on virus release were calculated as fold difference relative to the Env-negative sample. Graphs represent the means ± SEM from two independent experiments. Colors indicate individual patients. (C) Effect of HIV-2 Env on viral infectivity. Virus samples from panel B were used for the infection of TZM-bl cells. Infections were done in duplicates. Luciferase activity was measured 48 h after infection and normalized for input virus. Infectivity of viruses pseudotyped with the ROD10 Env was defined as 1 and is marked by a horizontal line. Differences in viral infectivity of the other samples are expressed as fold change relative to ROD10 Env. Graphs represent the means ± SEM from at least two independent experiments performed in duplicate infections.
As expected, virus release in the presence of the ROD14 Env was poor and similar to that observed in the absence of Env [Fig. 2C, compare ROD14 to Env(−)]. In contrast, coexpression of the ROD10 Env significantly enhanced the release of viral Gag proteins. Of the 6 tested Env isolates from patient P4, three (P4-1, P4-7, and P4-8) behaved like the ROD14 Env and exhibited a Vpu(−) phenotype. Two of the Env isolates (P4-6 and P4-11) significantly enhanced virus release compared to ROD14 Env, although they were not quite as effective as the laboratory-adapted ROD10 Env (Fig. 2C). Finally, the Env protein from isolate P4-3 exhibited an intermediate phenotype. Thus, three of the six Env isolates derived from patient 4 exhibited some degree of Vpu-like activity. Overall, half (17/34, or 50%) of the Env isolates tested in this study were able to enhance the release of virus particles to various degrees and thus revealed Vpu-like activity (Fig. 3 and 4A). To ascertain that the observed effects of HIV-2 Env on virus release are dependent on BST-2, we assessed virus release from BST-2-negative 293T cells. 293T cells were transfected with the env-defective pROD10.env1 (4 μg) (8) either in the absence of Env (no Env) or together with 2 μg of individual Env variants. Virus release was quantified 24 h later by determining the virus-associated reverse transcriptase activity in the culture supernatants (Fig. 4B). As expected, the effects of HIV-2 Env proteins on virus release in the absence of BST-2 were small compared to their effects on virus release from BST-2-expressing cells (compare Fig. 4A and B). Some Env variants had a slight enhancing effect (e.g., ROD10 and ROD14), while other Env proteins had a modest inhibitory effect (e.g., P2-1, P6-1, or P6-3). We conclude that the ability to antagonize BST-2 is conserved in about half of the HIV-2 Env variants. The ability to antagonize BST-2 was not specific to Env variants from specific patients. Indeed, most patients harbored viruses with Env proteins that contained or lacked BST-2 antagonizing activity.
HIV-2 Envs differ in their ability to produce infectious viruses.
We next tested the ability of our Env isolates to support the production of infectious viruses by coexpression with the env-deficient pROD10.env1 and tested the infectivity of the resulting virus preparations in a single-round infectivity assay. To avoid interference of virus production by BST-2, we used BST-2-negative 293T cells for this experiment. Cells were transfected with pROD10.env1 (4 μg) either in the absence of Env (2 μg empty vector [Ctrl]) or together with 2 μg of individual Env variants. Virus-containing supernatants were used for the infection of TZM-bl cells, and virus-induced luciferase activity was determined 48 h later. We found that four of the six P4 Env variants (Fig. 2D, P4-1, P4-3, P4-6, and P4-11) produced particles with significantly higher infectivity than viruses containing the laboratory-adapted ROD10 Env. Interestingly, the two Env isolates from patient P4 with the highest Vpu-like activity (P4-6 and P4-11) also scored highest in Env function. Analysis of all Env isolates for their ability to produce infectious virus is summarized in Fig. 4C. We observed significant variation among different Env variants. Overall, however, there was no direct correlation between envelope function and the ability to antagonize BST-2 (compare Fig. 4A and C), suggesting that these functions of the HIV-2 Env protein evolved independently.
A naturally occurring substitution in HIV-2 Env regulates its Vpu-like virus release activity.
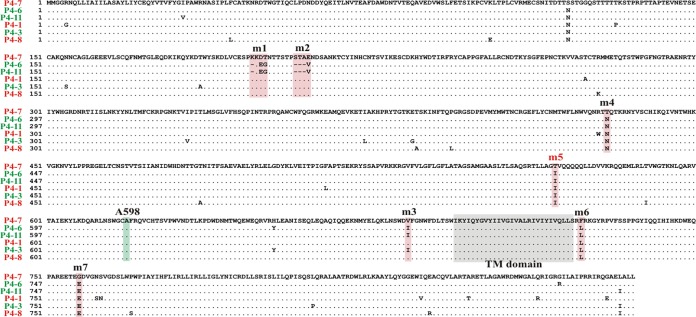
In a previous study, we observed that a single-amino-acid change in ROD14 Env to the corresponding residue in ROD10 (T598A) was sufficient to restore Vpu-like activity (24). Sequence analysis of patient 4 isolates using P4-7 Env, which exhibits a Vpu(−) phenotype, as a reference sequence revealed a number of amino acid differences among the individual isolates that were spread out across the entire Env sequence (Fig. 5). However, there were no common amino acid differences between variants with and without Vpu-like activity. Of note, the two Env isolates with the strongest Vpu-like phenotype (P4-6 and P4-11; Fig. 2C) differed from the P4-7 reference sequence in 2 identical small deletions and only 9 amino acid positions, 8 of which were common to P4-6 and P4-11 (Fig. 5, pink background). Most of the sequence differences indicated by the pink background, together with additional changes, were also found in the other patient 4 Env sequences.
FIG 5.
Sequence comparison of Env variants from patient P4. Amino acid sequences from all six patient 4-derived Env variants were aligned. Identical sequences appear as dots. The transmembrane domain (TM domain) is marked by a gray background. Regions tested in Fig. 6 for their ability to convey Vpu-like activity are marked by a pink background. Alanine 598 (A598), which is critical for the ability of ROD10 Env to antagonize BST-2 (24), is highlighted by a green background.
To test which of these sequence differences or deletions accounted for the Vpu-like phenotype of the P4-6/P4-11 Envs, we introduced amino acid changes/deletions into the P4-7 backbone (Fig. 5, m1-m7) either individually or in combination and assessed the resulting constructs in a gain-of-function analysis for their ability to enhance virus release using pulse-chase metabolic labeling as described for Fig. 2A (Fig. 6A). Analysis of Env expression by immunoblotting showed only minor variations in Env protein levels (Fig. 6B). Quantitation of the pulse-chase analysis data revealed that most of the Env mutants, including the deletions, retained the Vpu(−) phenotype associated with the parental P4-7 isolate (Fig. 6C). Interestingly, however, mutation of T568 in P4-7 Env to isoleucine (Fig. 5, m5) conferred Vpu-like activity to the P4-7 Env variant (Fig. 6C, P4-7m5). Of note, residue 568 is isoleucine in all P4 Env isolates except P4-7, even those without Vpu-like activity (Fig. 5), indicating that isoleucine at this position is important but not sufficient to confer Vpu-like activity to all Env variants.
FIG 6.
Ectodomain of the TM subunit of HIV-2 Env is critical for enhancing virus release. (A) Amino acid differences in P4-11 Env highlighted in Fig. 4 were transferred individually or in combination as indicated into the backbone of P4-7 Env. The ability of the resulting mutants to antagonize BST-2 was tested in HeLa cells by pulse-chase analysis as described for Fig. 2A. (B) Expression of Env mutants was verified by Western blot analysis using cellular α-tubulin as a loading control (tub). (C) Kinetic data from panel A were quantified as described for Fig. 2C. Maximal virus release by ROD10 at the 5-h time point was defined as 100%, and the remaining data points were adjusted accordingly. Data are presented as means ± SEM from two independent analyses.
Coimmunoprecipitation of HIV-2 Env with BST-2.
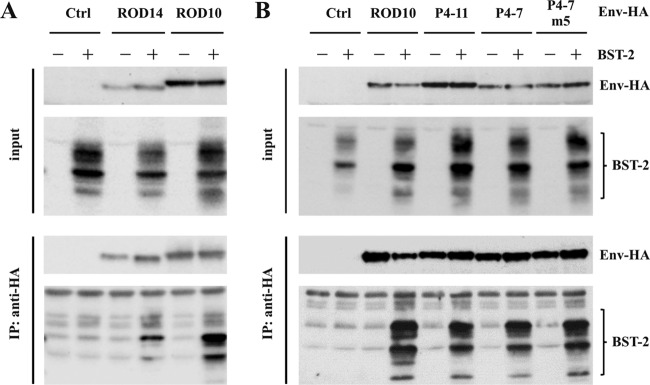
The inability of ROD14 Env to antagonize BST-2 was recently associated with a lack of physical interaction of the two proteins (33). To confirm this observation, we performed coimmunoprecipitation studies in 293T cells by coexpressing HA-tagged ROD10 or ROD14 Env with BST-2 (Fig. 7A). As a control, BST-2 was expressed in the absence of Env protein (Fig. 7A, Ctrl). Transfected cells were harvested 24 h posttransfection and lysed, and envelope proteins were immunoprecipitated with an anti-HA monoclonal antibody. Total input samples and immunoprecipitates were separated by SDS-PAGE and subjected to immunoblot analysis with antibodies to HA or BST-2 (Fig. 7A). We found that BST-2 efficiently interacted with the ROD10 Env protein. Consistent with the earlier report (33), interaction of BST-2 with ROD14 Env was significantly reduced but not entirely eliminated.
FIG 7.
Coimmunoprecipitation of BST-2 with HIV-2 Env. (A) 293T cells were transfected with 0.25 μg of pcDNA-BST-2 together with 4 μg of empty vector (Ctrl) or with HA-tagged pROD14-Env or pROD10-Env. Cell extracts were prepared 24 h later, and a fraction of total lysate was used as the input control (top). The remaining lysate was used for immunoprecipitation with anti-HA-coated beads (bottom). Samples were separated by SDS-PAGE and probed with antibodies to HA (Env-HA) or BST-2. (B) 293T cells were transfected with 0.25 μg of pcDNA-BST-2 together with 4 μg of empty vector (Ctrl) or the indicated Env expression vectors. Samples were processed as described for panel A. The experiment was performed independently three times. Shown is a representative result.
The interaction of P4-11, P4-7, and the P4-7m5 Env variants with BST-2 was determined in a similar manner (Fig. 7B). Empty vector (Ctrl) and ROD10 Env-expressing vector (ROD10) were included as controls. Interestingly, BST-2 interacted efficiently with the HIV-2 Env variants P4-7 and P4-11, as well as with the gain-of-function mutant P4-7m5, irrespective of their Vpu phenotype (Fig. 7B). Taken together, our data suggest that binding of Env to BST-2 is not sufficient to antagonize BST-2 activity.
DISCUSSION
The functional significance of BST-2/tetherin downmodulation by primate lentiviruses is still unclear. It has been suggested that BST-2 downmodulation serves to protect infected cells from antibody-dependent cellular cytotoxicity (ADCC) by minimizing cell surface exposure of viral antigen (34–36). It is also possible that downmodulation of BST-2 benefits the virus by increasing virus spread through cell-free transmission (reviewed in reference 37). There is, however, no doubt that controlling BST-2 is critical for primate lentiviruses, since HIV-1, HIV-2, SIV, feline immunodeficiency virus (FIV), and equine infectious anemia virus (EIAV) all have evolved mechanisms to antagonize BST-2. What is particularly striking is the fact that these viruses use distinct strategies to target and neutralize BST-2. In the case of HIV-1, Vpu has evolved as the BST-2 antagonist (14, 15). For most SIVs, Nef has acquired the ability to target BST-2 (10–13). The latter include SIVcpz, the presumed ancestor of HIV-1, which has a vpu gene yet uses Nef to control BST-2 (12, 38), suggesting that the original function of Vpu was not the targeting of BST-2. Like most SIV strains, HIV-2 lacks a vpu gene. While HIV-2 does have a nef gene, it does not use Nef to antagonize BST-2 but has found yet another way by using its Env protein (8, 16, 18). Finally, FIV and EIAV acquired Env-dependent strategies similar to those of HIV-2 (39, 40). Thus, there are at least three lentiviral proteins with the demonstrated capacity to target and antagonize BST-2.
The reasons why BST-2 is not targeted by a common lentiviral protein are unclear. However, it could be that in evolutionary terms, BST-2 represents a more recent challenge that lentiviruses have had to cope with in different ways. Since BST-2 does not impose an absolute restriction on virus replication, viruses may have had the luxury of gradually developing BST-2 resistance by expanding the functional breadth of available viral proteins. An interesting example is the acquisition of a Vpu-like activity by the Env protein of a nef-deleted SIV following serial passaging in rhesus macaques (21). Nevertheless, antagonism of BST-2 by any of the three viral factors follows more or less the same pathway and is initiated by the physical interaction with BST-2. For Vpu, this interaction clearly involves the TM domain (41–47), although the involvement of the Vpu cytoplasmic domain has also been reported (48–52). For Nef, the interaction with BST-2 is limited to the BST-2 cytoplasmic domain for the simple reason that Nef does not have a TM or ectodomain but is attached to membrane through a myristic acid moiety (10–12, 38). For HIV-2 Env, interactions with BST-2 have been reported to involve the membrane-proximal ectodomain (17, 18, 22). However, as with Vpu, the cytoplasmic domain may have a role in the antagonism of BST-2 as well (33). Exactly where in the cell the interaction of BST-2 with Vpu, Nef, or Env is initiated is currently unclear. The coexpression of BST-2 with Vpu, Env, or Nef can result in the surface downmodulation of BST-2 (reviewed in reference 53). However, whether surface downmodulation of BST-2 is an actual prerequisite or a downstream consequence of BST-2 antagonism is still unclear. We previously found that in the context of an acute spreading infection of T cells, Vpu-dependent enhancement of virus release does not coincide with BST-2 surface downmodulation (32). We also reported that antibody-based interference with BST-2 must occur prior to BST-2 reaching the cell surface (54), suggesting that the interaction of BST-2 with virus assembly complexes that ultimately results in membrane tethering is initiated inside the cells. This is true for HIV-1 as well as HIV-2 (54).
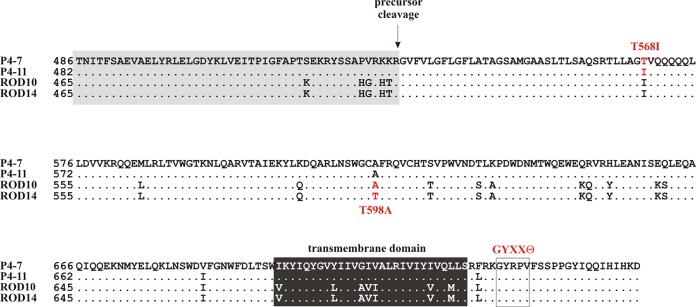
Our hypothesis that the ability to antagonize BST-2 is a more recent functional acquisition of HIV-2 is supported by the fact that only about half of the functional Env isolates characterized in our study have Vpu-like activity. Furthermore, the fact that there is significant variation in the extent to which individual Env proteins can antagonize BST-2 supports the model that antagonizing BST-2 is an ongoing evolutionary process. This is supported by the observation that we were able to isolate Env variants that contained or lacked Vpu-like activity from most patient samples (Fig. 1 and 4). More importantly, the ability or inability to antagonize BST-2 is not a stable functional property but was sensitive to single-amino-acid changes. Examples are the previously reported naturally occurring T598A mutation (24) as well as the naturally occurring T568I mutation described in the current study (Fig. 8). It is interesting that in both cases the presence of a threonine residue with its polar side chain was replaced by an amino acid with a hydrophobic side chain, suggesting structural changes are involved in the acquisition of Vpu-like activity. It was previously reported that mutations resulting in a loss of Vpu-like activity in HIV-2 Env were associated with a loss or at least a reduction in BST-2-Env binding (33). Our own results are in partial agreement with those data, in the sense that the T598A mutation in ROD10/14 appeared to reduce, although not completely abolish, the binding affinity to BST-2 (Fig. 7A). Interestingly, however, we did not observe a difference in the interaction of BST-2 with Env variants P4-7 [Vpu(−) phenotype] and P4-11 [Vpu(+) phenotype] or with the P4-7m5 back mutation (T568I) that restored the Vpu-like activity in P4-7 Env (Fig. 7B). These results are in line with results from experiments involving a glycosylphosphatidylinositol-anchored version of HIV-2 Env, which was able to interact with BST-2 but did not antagonize BST-2 function (33), and strongly suggest that binding of Env to BST-2 in itself is not sufficient to antagonize BST-2 function.
FIG 8.
Multiple changes in Env affect its Vpu-like activity. Shown is a partial amino acid alignment of four HIV-2 Env isolates. ROD10, ROD14, P4-7, and P4-11 sequences differ by deletions/insertions in the SU domain. Therefore, sequences were aligned using the transmembrane (TM) domains as references (black box with white lettering). Amino acid positions refer to the initiation codon of each Env protein as position 1. The presumed precursor cleavage site (55) is indicated, and the SU portion of the sequence is underlaid by a gray box. The boxed area downstream of the transmembrane domain delineates a tyrosine-based internalization motif (GYXXϴ) that includes a tyrosine (Y707) required for BST-2 antagonism (17, 18).
ACKNOWLEDGMENTS
We thank Sandra Kao, Sayaka Sukegawa, Eri Miyagi, and Angela Yoo for helpful discussions and critical comments on the manuscript. Many thanks to Alicia Buckler-White and Ron Plishka for their support with extensive sequence analyses. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. (catalog number 8129), HIV-1 immunoglobulin (catalog number 3957), and HIV-2 patient serum to detect p26 (CA).
This work was supported by the Intramural Research Program of the NIH, NIAID (1 Z01 AI000669).
REFERENCES
- 1.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Silva TI, Cotten M, Rowland-Jones SL. 2008. HIV-2: the forgotten AIDS virus. Trends Microbiol 16:588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, Li Y, Hahn BH, Delaporte E, Leendertz FH, Peeters M. 2013. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural Cote d'Ivoire. AIDS 27:2488–2491. doi: 10.1097/01.aids.0000432443.22684.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strebel K. 2013. HIV accessory proteins versus host restriction factors. Curr Opin Virol 3:692–699. doi: 10.1016/j.coviro.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 6.Willey RL, Maldarelli F, Martin MA, Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol 66:7193–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimitrov DS, Willey RL, Martin MA, Blumenthal R. 1992. Kinetics of HIV-1 interactions with sCD4 and CD4+ cells: implications for inhibition of virus infection and initial steps of virus entry into cells. Virology 187:398–406. doi: 10.1016/0042-6822(92)90441-Q. [DOI] [PubMed] [Google Scholar]
- 8.Bour S, Schubert U, Peden K, Strebel K. 1996. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J Virol 70:820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bour S, Strebel K. 1996. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J Virol 70:8285–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serra-Moreno R, Zimmermann K, Stern LJ, Evans DT. 2013. Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLoS Pathog 9:e1003487. doi: 10.1371/journal.ppat.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 15.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritter GD, Yamshchikov G, Cohen SJ, Mulligan MJ. 1996. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J Virol 70:2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abada P, Noble B, Cannon PM. 2005. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol 79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, Cannon PM. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. doi: 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci U S A 106:20889–20894. doi: 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serra-Moreno R, Jia B, Breed M, Alvarez X, Evans DT. 2011. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9:46–57. doi: 10.1016/j.chom.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble B, Abada P, Nunez-Iglesias J, Cannon PM. 2006. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J Virol 80:2924–2932. doi: 10.1128/JVI.80.6.2924-2932.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bour S, Akari H, Miyagi E, Strebel K. 2003. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology 309:85–98. doi: 10.1016/S0042-6822(02)00128-9. [DOI] [PubMed] [Google Scholar]
- 25.Marcelino JM, Borrego P, Rocha C, Barroso H, Quintas A, Novo C, Taveira N. 2010. Potent and broadly reactive HIV-2 neutralizing antibodies elicited by a vaccinia virus vector prime-C2V3C3 polypeptide boost immunization strategy. J Virol 84:12429–12436. doi: 10.1128/JVI.01102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borrego P, Calado R, Marcelino JM, Bartolo I, Rocha C, Cavaco-Silva P, Doroana M, Antunes F, Maltez F, Caixas U, Barroso H, Taveira N. 2012. Baseline susceptibility of primary HIV-2 to entry inhibitors. Antivir Ther 17:565–570. doi: 10.3851/IMP1996. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol 64:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol 62:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagi E, Andrew AJ, Kao S, Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci U S A 106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Exline CM, Yang SJ, Haworth KG, Rengarajan S, Lopez LA, Droniou ME, Seclen E, Cannon PM. 2015. Determinants in HIV-2 Env and tetherin required for functional interaction. Retrovirology 12:67. doi: 10.1186/s12977-015-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, Evans DT. 2014. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A 111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pham TN, Lukhele S, Hajjar F, Routy JP, Cohen EA. 2014. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 11:15. doi: 10.1186/1742-4690-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrew A, Strebel K. 2011. The interferon-inducible host factor bone marrow stromal antigen 2/tetherin restricts virion release, but is it actually a viral restriction factor? J Interferon Cytokine Res 31:137–144. doi: 10.1089/jir.2010.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang SJ, Lopez LA, Hauser H, Exline CM, Haworth KG, Cannon PM. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. doi: 10.1186/1742-4690-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison JH, Guevara RB, Marcano AC, Saenz DT, Fadel HJ, Rogstad DK, Poeschla EM. 2014. Feline immunodeficiency virus envelope glycoproteins antagonize tetherin through a distinctive mechanism that requires virion incorporation. J Virol 88:3255–3272. doi: 10.1128/JVI.03814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin X, Hu Z, Gu Q, Wu X, Zheng Y-H, Wei P, Wang X. 2014. Equine tetherin blocks retrovirus release and its activity is antagonized by equine infectious anemia virus envelope protein. J Virol 88:1259–1270. doi: 10.1128/JVI.03148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shingai M, Yoshida T, Martin MA, Strebel K. 2011. Some human immunodeficiency virus type 1 Vpu proteins are able to antagonize macaque BST-2 in vitro and in vivo: Vpu-negative simian-human immunodeficiency viruses are attenuated in vivo. J Virol 85:9708–9715. doi: 10.1128/JVI.00626-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida T, Kao S, Strebel K. 2011. Identification of residues in the BST-2 TM domain important for antagonism by HIV-1 Vpu using a gain-of-function approach. Front Microbiol 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi T, Ode H, Yoshida T, Sato K, Gee P, Yamamoto SP, Ebina H, Strebel K, Sato H, Koyanagi Y. 2011. Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J Virol 85:932–945. doi: 10.1128/JVI.01668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta RK, Hue S, Schaller T, Verschoor E, Pillay D, Towers GJ. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog 5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem 284:35060–35072. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, Ruiz A, Stephens EB, Opella SJ, Guatelli J. 2012. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem 287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, Xiong Y. 2014. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. eLife 3:e02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kueck T, Neil SJD. 2012. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog 8:e1002609. doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau D, Kwan W, Guatelli J. 2011. Role of the endocytic pathway in the counteraction of BST-2 by human lentiviral pathogens. J Virol 85:9834–9846. doi: 10.1128/JVI.02633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog 5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz A, Hill MS, Schmitt K, Guatelli J, Stephens EB. 2008. Requirements of the membrane proximal tyrosine and dileucine-based sorting signals for efficient transport of the subtype C Vpu protein to the plasma membrane and in virus release. Virology 378:58–68. doi: 10.1016/j.virol.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyagi E, Andrew A, Kao S, Yoshida T, Strebel K. 2011. Antibody-mediated enhancement of HIV-1 and HIV-2 production from BST-2/tetherin+ cells. J Virol 85:11981–11994. doi: 10.1128/JVI.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freed EO, Myers DJ. 1992. Identification and characterization of fusion and processing domains of the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 66:5472–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]