Abstract
The NAD+-dependent deacetylase SIRT1 plays key roles in numerous cellular processes including DNA repair, gene transcription, cell differentiation, and metabolism. Over-expression of SIRT1 protects against a number of age-related diseases including diabetes, cancer, and Alzheimer's disease. Moreover, overexpression of SIRT1 in the murine brain extends lifespan. A number of small-molecule sirtuin-activating compounds (STACs) that increase SIRT1 activity in vitro and in cells have been developed. While the mechanism for how these compounds act on SIRT1 was once controversial, it is becoming increasingly clear that they directly interact with SIRT1 and enhance its activity through an allosteric mechanism. Here, we present detailed chemical syntheses for four STACs, each from a distinct structural class. Also, we provide a general protocol for purifying active SIRT1 enzyme and outline two complementary enzymatic assays for characterizing the effects of STACs and similar compounds on SIRT1 activity.
1. INTRODUCTION
Overexpression of the NAD+-dependent histone deacetylase Sir2 extends the lifespan of diverse model organisms including yeast, worms, and flies (Hubbard & Sinclair, 2013, 2014; Morris, 2013). In mammals, seven Sir2 homologs have been identified (SIRT1-7), with SIRT1 bearing the closest phylogenetic relationship to yeast Sir2 (Hubbard & Sinclair, 2014). SIRT1 is involved in mediating numerous critical cellular processes such as DNA repair and apoptosis, muscle and fat differentiation, neuro-genesis, mitochondrial biogenesis, and various aspects of cell metabolism (Morris, 2013). Interestingly, deregulation of SIRT1 activity has been implicated in a number of age-related diseases including heart disease, diabetes, Alzheimer’s disease, and cancer (Hubbard & Sinclair, 2014). Moreover, overexpression of SIRT1 has been shown to be protective in models of colon cancer (Firestein et al., 2008), neurodegeneration (Kim et al., 2007), and age-related metabolic decline (Feige et al., 2008; Gomes et al., 2013; Price et al., 2012), and to extend the lifespan of mice when over-expressed in neurons (Satoh et al., 2013). In light of the wide array of health benefits it confers, SIRT1 has emerged as an attractive drug target for treating a variety of diseases in humans (Hubbard & Sinclair, 2014).
While a number of different strategies to increase SIRT1 enzymatic activity in cells have been proposed (Hubbard & Sinclair, 2014), including pansirtuin activators that block binding of the sirtuin inhibitor nicotinamide (Sauve, Moir, Schramm, & Willis, 2005), and molecules that displace the protein inhibitor of SIRT1, DBC1 (Hubbard, Loh, et al., 2013), the majority of research has focused on the development of allosteric SIRT1 activators (STACs) (Hubbard & Sinclair, 2014). The first STACs were discovered in 2003 using a high-throughput screen employing a fluorophoreconjugated peptide (Howitz et al., 2003). Several classes of polyphenols, including flavones, stilbenes, and anthocyanidins, were shown to increase SIRT1 activity through a mechanism involving a lowering of peptide substrate Km (Bhullar & Hubbard, 2015; Howitz et al., 2003). Resveratrol, the most efficacious SIRT1 activator identified in this screen, activated SIRT1 by up to 10-fold (Howitz et al., 2003). Later, chemically distinct compounds based on an imidazothiazole scaffold (eg, SRT1460, SRT1720) that are more potent were developed by Sirtris Pharmaceuticals, and these were also shown to elicit changes in cells consistent with SIRT1 activation (Milne et al., 2007). However, the validity of these early compounds was challengedby a series of reports showing that while STACs activated SIRT1 deacetylation on the fluorophore-tagged peptides used in the initial screen, no activity enhancement was observed when the corresponding untagged peptides were used (Borra, Smith, & Denu, 2005; Kaeberlein et al., 2005; Pacholec et al., 2010). Furthermore, these studies led to speculation that the effects of STACs in vivo might be due to off-target effects (Chung, 2012).
Recent work has supported the original assertion that STACs do indeed act as direct allosteric SIRT1 activators (Dai et al., 2015, 2010; Gertz et al., 2012; Hubbard, Gomes, et al., 2013). For example, STACs can activate SIRT1 deacetylation of natural amino acid peptides (Dai et al., 2010; Lakshminarasimhan, Rauh, Schutkowski, & Steegborn, 2013) and can enhance SIRT1 deacetylation of native peptides bearing hydrophobic amino acids adjacent to the acetyl-lysine (Hubbard, Gomes, et al., 2013). A model of “assisted allosteric activation” has been proposed to account for how STACs activate SIRT1 (Hubbard, Gomes, et al., 2013). In this model, binding of specific peptide substrate to SIRT1 induces the formation of an exosite that enhances drug binding. Once bound to SIRT1, the activator is thought to stabilize the substrate binding, resulting in a lowering of apparent peptide Km (Hubbard, Gomes, et al., 2013). This model has now been supported by additional crystallographic data (Cao et al., 2015; Dai et al., 2015). Here, we present detailed protocols for the synthesis and assay of STACs. We outline full syntheses for four structurally distinct classes of STACs, describe a general method for SIRT1 enzyme purification, and provide instructions on how to assay the effects of STACs on SIRT1 using two complementary activity assays (PNC1-OPT and RapidFire mass spectrometry).
2. MATERIALS
2.1 Synthesis of SIRT1-Activating Compounds
General organic chemistry glassware.
Reagent-grade chemicals outlined in the syntheses.
Equipment for Flash column chromatography (eg, ISCO CombiFlash Rf or similar system) with appropriate columns.
Analytical HPLC (eg, Agilent 1100 series), for compound verification.
Nuclear magnetic resonance (NMR) instrument (eg, Bruker Advance III), for compound verification.
High-resolution mass spectrometry (HRMS) (eg, Waters qTOF Premier Mass Spectrometer), for compound verification.
2.2 Expression and Purification of Recombinant His-Tagged SIRT1
pET-based His-tagged SIRT1 expression vector (or similar).
BL21 pLysS(DE3) or similar chemically competent bacteria.
Antibiotics (ampicillin or kanamycin, chloramphenicol).
LB media and LB-agar plates with appropriate antibiotics.
Large temperature-controlled bacterial incubator.
Centrifuge for spinning down large bacterial cultures (0.5–1 L volumes).
Protease inhibitor pellets (eg, Roche EDTA-free) (see Note 1).
Pipetteman with 10 or 25 mL serological pipettes.
Isopropyl β-D-1-thiogalactopyranoside (IPTG).
Ice buckets filled with ice.
Sonicator with wide-fitted head.
Ni-NTA agarose beads (eg, Qiagen).
Lysis buffer: 1% Triton X-100, 50 mM Tris pH 8.0, 150 mM NaCl, 20 mM imidazole, 3 mM β-mercaptoethanol.
Wash buffer: 1% Triton X-100, 50 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole, 3 mM β-mercaptoethanol.
Elution buffer: 50 mM Tris pH 8.0, 250 mM imidazole, 3 mM β-mercaptoethanol.
Biorad Polyprep columns.
Spectrophotometer capable of performing absorbance measurements.
2.3 Assay of SIRT1 Activators Using the PNC1-OPT Assay
Purified recombinant SIRT1 enzyme (produced as described earlier).
Purified recombinant PNC1 enzyme (yeast PNC1 (yPNC1) may be purified using the same procedure described for SIRT1) (Howitz et al., 2003; Hubbard, Gomes, et al., 2013).
Assay buffer: Gibco phosphate-buffered saline (PBS) pH 7.4 (1 mM KH2PO4, 155 mM NaCl, 3 mM Na2HPO4.7H2O) supplemented with 1 mM dithiothreitol (DTT) (add fresh from a frozen stock).
Peptide substrate—typically between 5 and 15 amino acids (eg, Ac-RHKK(ac)W-NH2), either synthesized in-house or purchased commercially (see Note 2).
β-Nicotinamide adenine dinucleotide (β-NAD)—prepare as 100 mM aliquots and store at −20°C.
OPT developer reagent: a 30% EtOH/70% PBS (pH 7.4) solution supplemented with 10 mM ortho-phthalaldehyde (OPT) and 10 mM DTT. This solution should be stored at −20°C until use and kept away from light (cover with aluminum foil).
Nicotinamide (NAM)—prepare 100 mM aliquots and store at −20°C.
37°C incubator.
Orbital shaker.
96-Well black opaque bottom plates for fluorometry.
Spectrophotometer with fluorescence capabilities and appropriate filters (excitation ~413 nm and emission ~476 nm).
2.4 Assay of SIRT1 Activators Using the RapidFire Mass Spectrometry Assay
Purified recombinant SIRT1 enzyme (produced as described earlier).
Bovine serum albumin (BSA).
Reaction buffer: 50 mM HEPES–NaOH, pH 7.5, 150 mM NaCl, 1 mM DTT, and 1 % DMSO.
β-NAD prepared as 100 mM aliquots and stored at −20°C.
Peptide substrate—typically between 5 and 15 amino acids (eg, Ac-RHKK(ac)W-NH2) either synthesized in-house or purchased commercially.
Agilent RapidFire System.
SPE cartridges.
Mass spectrometer fitter with an electrospray ionization source (eg, ABSciex API 4000).
Stop Reagent: 10% formic acid and 50 mM nicotinamide.
1:1 mixture of acetonitrile:methanol.
Buffer A: 90:10 acetonitrile:water with 0.1 mM ammonium acetate and 0.2% formic acid.
Buffer B: 60:40 acetonitrile:water supplemented with 1.6 mM ammonium acetate.
3. METHODS
3.1 Synthesis of SIRT1-Activating Compounds
A wide variety of natural molecules with the ability to increase SIRT1 activity have been reported (Hubbard & Sinclair, 2014). While thesecompounds comprise diverse structural scaffolds, including flavones, stilbenes, chalcones (Howitz et al., 2003), coumarins (Dao et al., 2012), and triterpenes (Yang et al., 2014), they all appear to activate SIRT1 through a common peptide Km-lowering mechanism (Hubbard & Sinclair, 2014). In addition to natural product SIRT1 activators, Sirtris Pharmaceutics (GSK) has described the development of an expanding collection of synthetic SIRT1 activators with EC50 values in the nM range (hundreds of times more potent natural compounds) over the past 10 years (Dai et al., 2015, 2010; Hubbard, Gomes, et al., 2013; Milne et al., 2007). These activators constitute structurally distinct compounds including imidazothiazoles, thiazolopyridines, imidazopyridines, and bridged ureas (Dai et al., 2015, 2010; Hubbard, Gomes, et al., 2013; Milne et al., 2007). Below we present detailed synthetic routes for four STACs, consisting of one example from each structural class.
General points
Flash column chromatography should be performed using silica gel with a particle size of 60Å, mesh of 230–400, using standard techniques. Unless otherwise indicated, chromatography refers to medium pressure chromatography performed on an ISCO CombiFlash Rf or similar system.
Following completion of each synthesis, final compound purities should be determined by analytical HPLC using the area percentage method on the UV trace recorded at a wavelength of 254 nm (see Note 3 for HPLC parameters). Expected purity is 95% purity unless otherwise specified.
1H NMR spectra should be obtained to characterize each product. Example of NMR parameters may be found in Note 4. NMR spectral data reported in this protocol follow these conventions: chemical shift (δ) in ppm (multiplicity, coupling constants in Hertz, number of protons, assignment), s—singlet, d—doublet, t—triplet, q—quartet, m—multiplet, br—broad.
To further verify successful product synthesis, routine mass spectral (MS) analysis of each compound can be performed (eg, using an Agilent 1100 series spectrometer integrated into an Agilent 1200 series HPLC system). In addition, HRMS can be performed (see Note 5 for parameters). The HRMS acceptable error is 3 mDa or 5 ppm, although most analyses are observed within 0.5 mDa with isotope fits in good agreement with the proposed structures.
3.1.1 Preparation of a Imidazo[1,2-b]thiazole STAC (Synthesis Adapted from Milne et al., 2007)
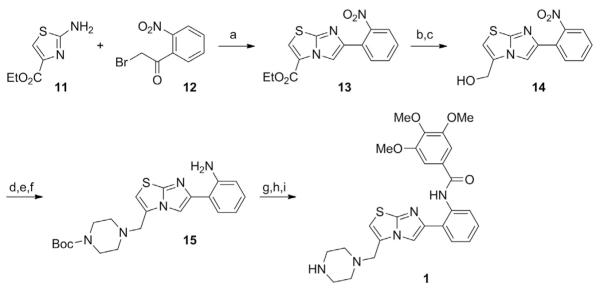
See Fig. 1.
Fig. 1.
Schematic outlining the synthesis of an imidazo[1,2–b]thiazole STAC. Compounds 11–15 represent intermediate products leading toward the production of the STAC 3,4,5–trimethoxy–N–(2–(3–(piperazin–1–ylmethyl)imidazo[2,1–b]thiazol–6–yl)phenyl) benzamide (1). Special reagents and conditions noted: (a) MEK, reflux (79%); (b) NaOH, 1:1 THF/H2O, RT, then HCl; (c) iBuOCOCl, NMM, THF, 0°C, then NaBH4, H2O, 0°C (74% for 2 steps); (d) MsCl, Et3N, CH2Cl2, 0°C; (e) Boc–piperazine, Et3N, CH3CN, RT; (f ) NaSH, 6:1 MeOH/H2O, reflux (60% for 3 steps); (g) 3,4,5–trimethoxybenzoyl chloride, pyridine; (h) TFA, CH2Cl2 (65%); (i) NaHCO3, H2O, then 3 N HCl, 1:1 CH3CN/H2O. Adapted from Milne, J. C., Lambert, P. D., Schenk, S., Carney, D. P., Smith, J. J., Gagne, D. J.,…., Westphal, C. H. (2007). Small–molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature, 450(7170), 712–716.
6-(2-Nitrophenyl)-imidazo[2,1-b]thiazole-3-carboxylic acid ethyl ester (13)
Stir a solution of ethyl 2-aminothiazole-4-carboxylate (11) (2.1 g, 12 mmol) and 2-bromo-2–nitroacetophenone (12) (3.0 g, 12 mmol) in methyl ethylketone (25 mL) under reflux for 18 h.
Cool the solution to room temperature and filter.
Concentrate the filtrate in vacuo to afford 3.10 g (79% yield) of 6–(2–nitrophenyl)–imidazo[2,1–b]thiazole–3–carboxylic acid ethyl ester (13). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 8.39 (br s, 1 H), 8.31 (br s,1 H), 7.92 (d, J=8 Hz, 1 H), 7.82 (d, J 7 Hz, 1 H), 7.3–7.7 (m, 2 H), 4.4 (q, J=7 Hz, 2 H), 1.37 (t, J=7 Hz, 3 H); MS m/z=318.0 (M+H)+.
[6–(2–Nitrophenyl)imidazo[2,1–b]thiazol–3–yl]methanol (14)
-
4.
Stir a solution of 6–(2–nitrophenyl)imidazo[2,1–b]thiazole–3–carboxylic acid ethyl ester (13) (14.50 g, 46 mmol) in THF (100 mL) and water (100 mL) containing NaOH (7.3 g, 4 equiv.) at room temperature for 18 h and then concentrate in vacuo.
-
5.
Wash the aqueous layer once with CH2Cl2 and then acidify with 6 N HCl. Collect the solids by filtration and dry to afford 7.4 g of the acid intermediate.
-
6.
Dissolve this material (7.4 g, 26 mmol) in anhydrous THF (200 mL) containing N–methylmorpholine (2.8 mL, 26 mmol) and cool to 0°C.
-
7.
Add isobutyl chloroformate (3.35 mL, 26 mmol) and stir the reaction mixture in an ice bath for 3 h.
-
8.
Add NaBH4 (0.97 g, 25.6 mmol) in water (30 mL) and stir the mixture at 0°C for 45 min. Warm to room temperature and concentrate.
-
9.
Extract the aqueous layer with CH2Cl2. Dry the combined organic layers (Na2SO4) and concentrate to afford the crude product.
-
10.
Purify by chromatography to afford 5.20 g (74% yield) of [6–(2–nitrophenyl)imidazo[2,1–b]thiazol–3–yl]methanol (14). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 8.14 (br s, 1 H), 7.2–7.9 (m, 4 H), 7.16 (br s, 1 H), 5.65 (t, J=7 Hz, 1 H), 4.6 (m, 2 H); MS m/z=276.0 (M+H)+.
4–[6–(2–Aminophenyl)imidazo[2,1–b]thiazol–3–ylmethyl] piperazine–1–carboxylic acid tert–butyl ester (15)
-
11.
Treat a chilled(0°C)solutionof[6–(2–nitrophenyl)imidazo[2,1–b]thiazol–3–yl]methanol (14) (1.0 g, 3.6 mmol) and Et3N (0.51 mL, 3.64 mmol) in CH2Cl2 (100 mL) with methanesulfonyl chloride (0.28 mL, 3.7 mmol) and allow the resulting reaction to warm to room temperature.
-
12.
Stir for 15 min.
-
13.
Quench the reaction with brine and extract with CH2Cl2.
-
14.
Dry (Na2SO4) the combined organic layers and concentrate in vacuo to afford the mesylate intermediate.
-
15.
Dissolve this material in CH3CN (4 mL) containing Et3N (0.51 mL, 3.6 mmol) and Boc–piperazine (680 mg, 3.6 mmol) and stir the resulting solution at room temperature for 1 day.
-
16.
Concentrate the reaction mixture and partition the resulting residue between CH2Cl2 and water.
-
17.
Dry (Na2SO4) the organic layer and concentrate to afford the crude product. Dissolve this material in a solution of MeOH (6 mL) and water (1 mL) containing sodium hydrosulfide hydrate (200 mg).
-
18.
Stir the resulting reaction mixture under reflux for 24 h, cool to room temperature, and concentrate. Dilute the residue with water(2 mL) and extract with CH2Cl2. Dry (Na2SO4) the combined organic layers and concentrate to afford 0.90 g (60% yield) of 4–[6–(2–aminophenyl)–imidazo[2,1–b]thiazol–3–ylmethyl]–piperazine–1–carboxylic acid tert–butyl ester (15). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 9.2 (br s, 1 H), 8.7 (br s, 1 H), 8.15 (s, 1 H), 8.10 (s, 1 H), 6.8–7.8 (m, 4 H), 6.16 (br s, 2 H), 3.72 (br s, 2 H), 1.39 (br s, 9 H); MS m/z=414.1 (M+H)+.
3,4,5–Trimethoxy–N–(2–(3–(piperazin–1–ylmethyl)imidazo[2,1–b] thiazol–6–yl)phenyl)benzamide (1)
-
19.
Dissolve 4–[6–(2–aminophenyl)imidazo[2,1–b]thiazol–3–ylmethyl] piperazine–1–carboxylic acid tert–butyl ester (15) (300 mg, 0.73 mmol) in pyridine (5 mL) and treat with 3,4,5–trimethoxybenzoyl chloride (167 mg, 0.73 mmol).
-
20.
Heat the reaction mixture in a microwave reactor (160°C × 10 min), cool to room temperature, and concentrate in vacuo.
-
21.
Purify the resulting crude product by chromatography (gradient elution, CH2Cl2 to 95% CH2Cl2, 4% MeOH, and 1% Et3N) and treat the purified product with a solution containing 25% TFA in CH2Cl2 (2 mL) for 2 h. Subsequently, concentrate and titrate the resulting residue with Et2O to afford 335 mg (65% yield) of 1 as the TFA salt.
-
22.
To prepare the corresponding HCl salt of 1, dissolve the TFA salt in water and neutralize with NaHCO3. Extract the resulting aqueous layer with CH2Cl2. Wash the combined organic layers with brine, dry (Na2SO4), and concentrate under reduced pressure.
-
23.
Take up the resulting residue in 50% aqueous CH3CN and add 1 mL of 3 N HCl. Lyophilize the mixture to obtain 1 as the HCl salt. Characterization properties: m.p.: 193.5°C (HCl salt). 1H NMR (300 MHz, DMSO–d6): δ 9.9 (br s, 1 H), 9.0 (br s, 1 H), 8.7–7.10 (m, 7 H), 8.5 (s, 1 H), 4.0 (br s, 9 H), 3.8 (m, 2 H), 3.2–2.8 (m, 8 H); 13C NMR (100 MHz, DMSO–d6): δ 47.56 50.01, 56.15,60.16, 104.68, 111.52, 120.70, 120.93, 123.63, 126.89, 128.13, 130.13, 136.01, 140.54, 144.43, 147.75, 152.86, 164.09; HRMS calculated for C26H29N5O4S: 508.2018; found: 508.2039.
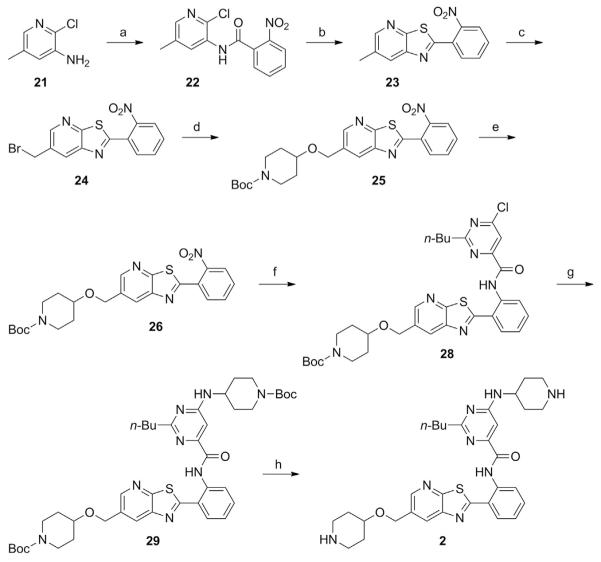
3.1.2 Preparation of a Thiazolopyridine STAC (Synthesis Adapted from Dai et al., 2010)
See Fig. 2.
Fig. 2.
Schematic outlining the synthesis of a thiazolopyridine STAC. Compounds 21–29 represent intermediate products leading toward the production of the STAC 2–butyl–6–(piperidin–4–ylamino)–N–(2–(6–((piperidin–4–yloxy)methyl)thiazolo[5,4–b]pyridin–2–yl)phenyl)pyrimidine–4–carboxamide (2). Special reagents and conditions noted: (a) 2–nitrobenzoyl chloride, pyridine 0°C to RT (91%); (b) P2S5, pyridine, p–xylene, 140°C (75%); (c) NBS, benzoyl peroxide, CCl4; (d) tert–butyl 4–hydroxypiperidine–1–carboxylate, (n–Bu)4N+HSO–4, NaOH, toluene, H2O (65%); (e) Fe, AcOH, THF, H2O, 50°C (88%); (f ) 2–butyl–6–chloropyrimidine–4–carbonyl chloride (27), Et3N, CH2Cl2 (91%); (g) tert–butyl 4–aminopiperidine–1–carboxylate, (i–Pr)2NEt, DMSO, 100°C (76%); (h) TFA, then NaHCO3, H2O (81%). Adapted from Dai, H., Kustigian, L., Carney, D., Case, A., Considine, T., Hubbard, B. P., …, Stein, R. L. (2010). SIRT1 activation by small molecules: Kinetic and biophysical evidence for direct interaction of enzyme and activator. Journal of Biological Chemistry, 285(43), 32695–32703.
N–(2–Chloro–5–methylpyridin–3–yl)–2–nitrobenzamide (22)
Add 2–nitrobenzoyl chloride (13.65 g, 73.6 mmol) dropwise to a solution of 5–amino–6–chloro–3–picoline (9.54 g, 66.9 mmol) (21) in pyridine (200 mL) at 0°C.
Stir the resulting mixture at room temperature for 18 h.
Dilute the dark mixture with water (1500 mL).
Add sat. NaHCO3 solution until pH 8.
Collect the precipitate by filtration, rinse with water (3 × 30 mL), and dry in an oven to afford N–(2–chloro–5–methylpyridin–3–yl)–2– nitrobenzamide (17.70 g, 91%) as a pale solid.
6–Methyl–2–(2–nitrophenyl)thiazolo[5,4–b]pyridine (23)
-
6.
Stir a mixture of N–(2–chloro–5–methylpyridin–3–yl)–2–nitrobenzamide (22; 5.0 g, 17.1 mmol) and P2S5 (7.6 g, 34.2 mmol) in pyridine (50 mL) and p–xylene (200 mL) at 140°C for 20 h.
-
7.
Transfer the hot solution to another flask and remove the solvent in vacuo.
-
8.
Purify the residue by recrystallization from EtOH to afford 6–methyl–2–(2–nitrophenyl)thiazolo[5,4–b]pyridine (3.5 g, 75%) as a yellow solid.
6–(Bromomethyl)–2–(2–nitrophenyl)thiazolo[5,4–b]pyridine (24)
-
9.
Add 6–methyl–2–(2–nitrophenyl)thiazolo[5,4–b]pyridine (23; 2.9 g, 10.7 mmol), N–bromosuccinimide (NBS; 1.91 g, 10.7 mmol), CCl4 (200 mL), and benzoyl peroxide (0.021 g) into a three–neck flask (500 mL) under argon.
-
10.
Reflux the resulting yellow mixture for 2 h.
-
11.
Add additional NBS (1.91 g) and benzoyl peroxide (0.021 g).
-
12.
Add more NBS (0.95 g) and benzoyl peroxide (0.021 g) 2 h later and continually reflux the mixture for 3 h.
-
13.
Cool the mixture to room temperature.
-
14.
Transfer the solution into another flask and concentrate in vacuo to afford crude 6–(bromomethyl)–2–(2–nitrophenyl)thiazolo[5,4–b]pyri–dine (4.0 g).
tert–Butyl 4–((2–(2–nitrophenyl)thiazolo[5,4–b]pyridin–6–yl) methoxy)piperidine–1–carboxylate (25)
-
15.
Add 15 mL of toluene and 5.4 mL of water to a mixture of 2.73 g (13.56 mmol) of tert–butyl 4–hydroxypiperidine–1–carboxylate, 4.75 g (13.56 mmol) of 6–bromomethyl–2–(2–nitrophenyl)thiazolo [5,4–b]pyridine (24),1 230 mg (0.678 mmol) of tetrabutylammonium bisulfate, and 5.42 g (135.6 mmol) of sodium hydroxide. Stir the reaction at ambient temperature at a rate sufficient to mix the two layers well.
-
16.
After 15 h, dilute the reaction with 100 mL of 1 M HCl and then extract with ethyl acetate (3 × 25 mL).
-
17.
Back–extract the combined ethyl acetate layers with water (1 × 25 mL), and brine (1 × 25 mL), dry over MgSO4, filter, and concentrate to an orange oil. Purify this via medium pressure silica gel chromatography (240 g prepacked column), eluting with an isocratic mixture of 50% ethyl acetate: heptanes. It is expected that tert–butyl 4–hydroxypiperidine–1–carboxylate and the product coelute.
-
18.
Pool and concentrate the product–containing fractions and take up the crude product in 20 mL of hot ethyl acetate and dilute with an equal volume of heptanes. The product should crystallize to give a yellow solid; the expected total yield is ~4.15 g (65%). Characterization prop erties: 1H NMR (300 MHz, DMSO–d6): δ 8.71 (d, J=1.9 Hz, 1 H), 8.41 (d, J=1.9 Hz, 1 H), 8.14 (dd, J=7.5, 2.3 Hz,1 H), 8.03 (dd, J=7.5, 2.3 Hz, 1 H), 7.90 (m, 2 H), =4.75 (s, 2 H), 3.65 (m, 3 H), 3.06 (m, 2 H), 1.87 (m, 2 H), 1.46 (m, 2 H), 1.40 (s, 9 H); MS m/z=471 (M+H)+.
tert–Butyl 4–((2–(2–aminophenyl)thiazolo[5,4–b]pyridin–6–yl) methoxy)piperidine–1–carboxylate (26)
-
19.
To a solution of 5.00 g (10.6 mmol) of tert–butyl 4–((2–(2–nitrophenyl) thiazolo[5,4–b]pyridin–6–yl) methoxy)piperidine–1–carboxylate (25) in 50 mL of THF add 12.5 mL of water, then 3.00 g (53.5 mmol) of iron powder, and 3.2 g (53.3 mmol) of glacial acetic acid. Heat the mixture at 50°C for 2–4 h, monitoring it by HPLC to determine when the reaction is complete.
-
20.
Next, filter the mixture through diatomaceous earth and wash the filter cake with CH2Cl2 (4 × 50 mL).
-
21.
Separate the phases and then extract the organic phase with saturated NaHCO3 (aq.) (1 × 50 mL) and brine (1 × 50 mL). Dry over Na2SO4, filter, and concentrate to 4.1 g (88%) of a yellow solid. Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 8.53 (d, J=1.9 Hz, 1 H), 8.30 (d, J=1.9 Hz, 1 H), 7.64 (dd, J=8.0, 1.3 Hz, 1 H), 7.41 (s, 2 H), 7.25 (m, 1 H), 6.90 (dd, J=8.3, 0.8 Hz, 1 H), 6.66 (m, 1 H), 4.70 (s, 2 H), 3.65 (m, 3H), 3.07 (m, 2 H), 1.86 (m, 2 H), 1.46 (m, 2 H), 1.40 (s, 9 H); MS m/z=441 (M+H)+.
2–Butyl–6–chloropyrimidine–4–carbonyl chloride (27)
-
22.
To 21.0 g (100 mmol) of diethyl oxaloacetate sodium salt add 80 mL of water.
-
23.
Add 16 mL (100 mmol) of 6.25 M NaOH over 1 min at ambient temperature to the stirred suspension, and stir the mixture for 10 min, until only traces of undissolved oxaloacetate ester remain, giving an orange solution.
-
24.
Add a solution of 13.9 g (100 mmol) of n–pentanamidine hydrochlo–ride in 20 mL of water to this mixture.
-
25.
Monitor the reaction with a pH meter and add additional 6.25 M NaOH as necessary to keep the pH between 10 and 11.
-
26.
After stirring at ambient temperature for 22 h, cool the mixture using an ice bath and then add 12 M HCl until pH 2.0, yielding a white precipitate. Filter this precipitate, wash with 50 mL of water, and then dry on the filter for 1 h.
-
27.
Suspend the white solid in 50 mL of heptanes and distill at 1 bar with a Dean–Stark trap until no more water is collected in the trap.
-
28.
Cool the suspension, filter, and dry on the filter to give 8.13 g (41%) of 2–butyl–6–hydroxypyrimidine–4–carboxylic acid as a white solid. Characterization properties: 1H NMR (DMSO–d6): δ 13.37 (br s, 1 H), 12.82 (br s, 1 H), 6.69 (s, 1 H), 2.56 (t, J=7.7 Hz, 2 H), 1.64 (m, 2 H), 1.31 (m, 2 H), 0.89 (t, J=7.3 Hz, 3 H); MS m/z=197 (M+H)+.
-
29.
Add 35 mL of phosphorus oxychloride to 5.00 g (25.48 mmol) of 2–butyl–6–hydroxypyrimidine–4–carboxylic acid. Stir the reaction at 105°C for 1 h and then concentrate in vacuo.
-
30.
Suspend the dark residue in 50 mL of heptanes and then concentrate in vacuo to remove most of the remaining phosphorus oxychloride.
-
31.
Next, suspend the residue in 100 mL of heptanes and extract with water (3 × 25 mL) and then brine (1 × 25 mL). Dry the organic layer over MgSO4, filter, and concentrate in vacuo to give 5.1 g (86%) of acid chloride 27 as an amber oil.
tert–Butyl 4–((2–(2–(2–butyl–6–chloropyrimidine–4–carboxamido) phenyl)thiazolo[5,4–b]pyridin–6–yl)methoxy)piperidine–1–carboxylate (28)
-
32.
To a solution of 795 mg (1.80 mmol) of tert–butyl 4–((2–(2–aminophenyl)thiazolo[5,4–b]pyridin–6–yl)methoxy)piperidine–1–carboxylate (26) in 8 mL of CH2Cl2 add 0.45 mL (3.2 mmol) of triethylamine, followed by a solution of 505 mg (2.17 mmol) of 2–butyl–6–chloropyrimidine–4–carbonyl chloride (27) in 2 mL of CH2Cl2.
-
33.
After 1.25 h, dilute the reaction with 30 mL of methanol to give a crystalline precipitate.
-
34.
Filter the precipitate, wash with 15 mL of methanol, and dry on the filter to give 1.04 g of 28 (91%) as colorless needles. Characterization properties: 1H NMR (300 MHz, CDCl3): δ 13.42 (s, 1 H), 8.98 (dd, J=8.4, 1.0 Hz, 1 H), 8.60 (d, J=1.9 Hz, 1 H), 8.35 (d, J=1.9 Hz, 1 H), 8.10 (s, 1 H), 7.93 (dd, J=7.9,=1.4 Hz, 1 H), 7.59 (m, 1=H), 7.30 (td, J=1.0, 7.6 Hz, 1 H), 4.75 (s, 2 H), 3.87 (m, 2 H), 3.66 (m, 1 H), 3.18 (t, J=7.8 Hz, 2 H), 3.08 (m, 2 H), 1.90 (m, 4 H), 1.60 (m, 2 H), 1.47 (s, 9 H), 1.41 (m, 2 H), 0.90 (t, J=7.3 Hz, 1 H).
tert–Butyl 4–((2–(2–(6–(1–(tert–butoxycarbonyl)piperidin–4–ylamino)–2–butylpyrimidine–4–carboxamido)phenyl)thiazolo [5,4–b]pyridin–6–yl)methoxy)piperidine–1–carboxylate (29)
-
35.
To a mixture of 709 mg (1.11 mmol) of tert-butyl 4-((2-(2-(2-butyl-6-chloropyrimidine-4-carboxamido) phenyl)thiazolo[5,4-b]pyridin-6-yl)methoxy)piperidine-1-carboxylate and 267 mg (1.33 mmol) of tert-butyl 4-aminopiperidine-1-carboxylate add 7 mL of DMSO and 0.40 mL (2.24 mmol) of N,N-diisopropyl-N-ethylamine. Heat the mixture at 100°C under N2 for 4 h.
-
36.
Monitor the reaction by 1H NMR, taking an aliquot of the reaction and dissolving it in CDCl3, then observing the resonances in the region downfield of 6 ppm. After the starting material has been consumed, dilute the reaction with 35 mL of water to give a granular precipitate.
-
37.
Filter this precipitate and wash with 25 mL of water to give a light tan solid. Recrystallize the crude solid from 30 mL of isopropanol, and wash with 60 mL of cold isopropanol to give 674 mg (76%) of a light yellow solid. Characterization properties: 1H NMR (300 MHz, CDCl3): δ 13.3 (br s, 1 H), 8.98 (d, J=8.4 Hz, 1 H), 8.59 (d, J=1.7 Hz, 1 H), 8.38 (d, J=1.6 Hz, 1 H), 7.90 (dd, J=7.8, 1.2 Hz, 1 H), 7.56 (td, J=7.3, 1.2 Hz, 1 H), 7.26 (m, 1 H), 7.11 (s, 1 H), 5.10 (br s, 1 H), 4.74 (s, 2 H), 4.09 (m, 2 H), 3.83 (m, 2 H), 3.64 (m, 1 H), 3.12 (m, 2 H), 2.94 (m, 4 H), 2.06 (m, 2 H), 1.91 (m, 2 H), 1.82 (quintet, J=7.6 Hz, 2 H), 1.62 (m, 4 H), 1.47 (s, 18 H), 1.41 (m, 3 H), 0.90 (t, J=7.3 Hz, 3 H); MS m/z=802 (M+H)+.
2–Butyl–6–(piperidin–4–ylamino)–N–(2–(6–((piperidin–4–yloxy) methyl)thiazolo[5,4–b]pyridin–2–yl)phenyl)pyrimidine–4–carboxamide (2)
-
38.
To 600 mg (0.750 mmol) of tert–butyl 4–((2–(2–(6–(1–(tertbutoxycarbonyl)piperidin–4–ylamino)–2–butylpyrimidine–4–carboxamido) phenyl)thiazolo[5,4–b]pyridin–6–yl)methoxy)piperidine–1–carboxylate (29) slowly (to control the vigorous evolution of gas) add 5 mL of trifluoroacetic acid.
-
39.
Stir the reaction for 10 min at ambient temperature and then remove the solvent in vacuo at 50°C.
-
40.
Dilute this residue with 12 mL of saturated NaHCO3, to give an oily suspension with pH 8, and then stir and heat at 80°C for 60 min, prior to cooling to ambient temperature, affording a pale yellow precipitate.
-
41.
Filter the precipitate, then suspend in water, and filter again (3 × 20 mL). Dry the wet solid by suction on the filter for 18 h to give 364 mg (81%) of a pale yellow solid (2). Characterization properties: 1H NMR(300 MHz, CDCl3): δ 13.28 (s, 1 H), 8.98 (d, J=8.3 Hz, 1 H), 8.60 (d, J=1.6 Hz, 1 H), 8.40 (d, J=1.6 Hz, 1 H), 7.90 (dd, Hz, 1 J=7.8, 1.1 H), 7.56 (m, 1 H), 7.26 (m, 1 H [CHCl3 overlap]), 7.11 (s, 1 H), 5.23 (br s, 1 H), 4.74 (s, 2 H), 3.81 (br s, 1 H), 3.56 (m, 1 H), 3.14 (m, 4 H), 2.93 (t, J=7.7 Hz, 2 H), 2.77 (m, 2 H), 2.65 (m, 2 H), 2.04 (m, 4 H), 1.82 (m, 4 H [H2O overlap]), 1.50 (m, 6 H), 0.90 (t, J=7.3 Hz, 3 H); MS m/z=301 (M+2H)2+, 601 (M+H)+.
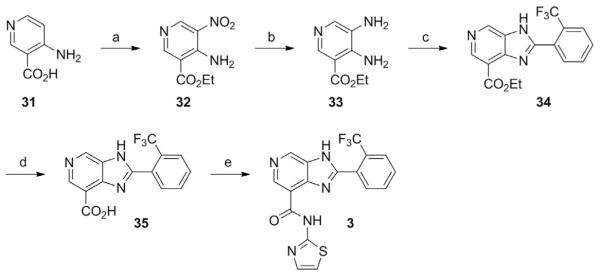
3.1.3 Preparation of an Imidazo[4,5–c]pyridine STAC (Synthesis Adapted from Hubbard, Gomes, et al., 2013)
See Fig. 3.
Fig. 3.
Schematic outlining the synthesis of a imidazo[4,5–c]pyridine STAC. Compounds 31–35 represent intermediate products leading toward the production of the STAC N–(thiazol–2–yl)–2–(2–trifluoromethyl–phenyl)–3H–imidazo[4,5–c]pyridine–7–carboxamide (3). Special reagents and conditions noted: (a) KNO3, H2SO4, 0–75°C, then add EtOH, RT to 60°C (35%); (b) H2, 10% Pd/C, MeOH (80%); (c) 3–trifluoromethylbenzaldehyde, Na2S2O5, DMF, 120°C (70%); (d) 10% aq. NaOH, EtOH, reflux, then 5 N HCl (85%); (e) thiazol–2–amine, HATU, (i–Pr)2NEt, DMF, RT (12%). Adapted from Hubbard, B. P., Gomes, A. P., Dai, H., Li, J., Case, A. W., Considine, T., …, Sinclair, D. A. (2013). Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science, 339(6124), 1216–1219.
4–Amino–5–nitro–nicotinic acid ethyl ester (32)
Add potassium nitrate (20.5 g, 200 mmol) to a stirred solution of 31 (27.6 g, 200 mmol) in concentrated H2SO4 (200 mL) at 0°C.
Stir the resulting mixture at 0°C for 30 min and then at 75°C for 3 h. Cool the reaction to ambient temperature and then add EtOH (540 mL).
Stir the resulting mixture at 60°C for 18 h and then slowly add it to an ice–cold solution of potassium acetate (800 g, in 1.5 L of water).
Collect the resulting precipitate by filtration, wash with water, and dry over Na2SO4 to afford 32 (14.6 g, 35%), for direct use in step 5. Characterization properties: 1H NMR (CDCl3): δ 9.31(s, 1 H), 9.07 (s, 1 H), 9.05 (br, 1 H), 8.28 (br, 1 H), 4.42 (q, 2 H), 1.43 (t, 3 H); MS m/z=211.92 (M+H)+.
4,5–Diamino–nicotinic acid ethyl ester (33)
-
5.
Stir a mixture of 32 (15 g, 71 mmol) and 10% Pd/C (500 mg) in MeOH (500 mL under 1 atm. of hydrogen at ambient temperature for 18 h).
-
6.
Filter the resulting mixture through celite® and concentrate to afford 33 (10 g, 80%) to be used in the following step without additional purification. Characterization properties: 1H NMR (CDCl3): δ 8.62 (s, 1 H), 7.96 (s, 1 H), 6.14 (br, 2 H), 4.36 (q, 2 H), 3.15 (br, 2 H), 1.40 (t, 3 H).
2–(2–Trifluoromethyl–phenyl)–3H–imidazo[4,5–c]pyridine–7–carboxylic acid ethyl ester (34)
-
7.
Stir a mixture of 33 (1.81 g, 10 mmol), trifluoromethylbenzaldehyde (1.9 g, 11 mmol), and Na2S2O5 (9.5 g, 50 mmol) in DMF (50 mL) at 120°C for 18 h. Cool the resulting mixture to ambient temperature and pour into cold water (100 mL).
-
8.
Filter the resulting solid, wash with water, and dry in vacuo to provide 34 (2.35 g, 70%).
2–(2–Trifluoromethyl–phenyl)–3H–imidazo[4,5–c]pyridine–7–carboxylic acid (35)
-
9.
Heat at reflux for 30 min a solution of 4 (2.35 g, 7 mmol) in 10% aqueous NaOH (40 mL) and ethanol (20 mL). Allow the mixture to cool to ambient temperature and acidify with 5 N aqueous HCl.
-
10.
Filter, wash with water, and dry the resulting yellow solid in vacuo to afford 35 (1.77 g, 85%). Characterization parameters: 1H NMR (DMSO–d6): δ 13.4 (br, 1 H), 9.18 (s, 1 H), 8.87 (s, 1 H), 7.95 (m, 1 H), 7.82 (m, 3 H); MS m/z=308.0 (M+H)+.
N–(Thiazol–2–yl)–2–(2–trifluoromethyl–phenyl)–3H–imidazo [4,5–c]pyridine–7–carboxamide (3)
-
11.
Stir a mixture of 35 (64.0 mg, 0.21 mmol), HATU (160 mg, 0.42 mmol), DIPEA (70 μL, 0.42 mmol), and thiazol–2–amine (21.0 mg, 0.21 mmol) in DMF (2 mL) at room temperature for 18 h.
-
12.
Remove the solvent in vacuo and purify the residue by chromatography (CH2Cl2/MeOH 50:1 to 5:1 gradient) to give 3 (10 mg, 12%) as a pale yellow solid. Characterization parameters: 1H NMR (CH3OD): δ 9.1(s, 2 H), 8.00 (d, 1 H), 7.94 (d, 1 H), 7.90–7.85 (m, 2 H), 7.49 (d, 1 H), 7.22 (d, 1 H); HRMS calculated for C17H11N5OSF3: 390.0634; found: 390.0635.
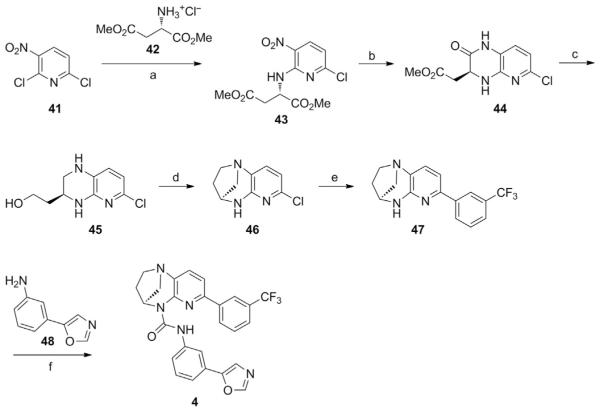
3.1.4 Preparation of a Bridged–Urea STAC (Synthesis Adapted from Dai et al., 2015)
See Fig. 4.
Fig. 4.
Schematic outlining the synthesis of a bridged–urea STAC. Compounds 41–48 represent intermediate products leading toward the production of the STAC (4S)–N–(3–(oxazol–5–yl)phenyl)–7–(3–(trifluoromethyl)phenyl)–3,4–dihydro–1,4–methanopyrido [2,3–b][1,4]diazepine–5(2H)–carboxamide (4). Special reagents and conditions noted: (a) NaHCO3, THF, 40°C (quantitative); (b) Fe, AcOH, i–PrOH, H2O, 40–70°C (68%); (c) LiAlH4, THF, 0°C to reflux (81%); (d) POCl3, Et3N, CH2Cl2, 0°C to RT (66%); (e) 3–trifluoromethylphenylboronic acid, Pd(OAc)2, X–Phos, Cs2CO3, dioxane, H2O, reflux (81%); (f ) triphosgene, Et3N, CH2Cl2, reflux, then add 3–(oxazol–5–yl)aniline (48), reflux (63%). Adapted from Dai, H., Case, A. W., Riera, T. V., Considine, T., Lee, J. E., Hamuro, Y., …, Ellis, J. L. (2015). Crystallographic structure of a small–molecule SIRT1 activator–enzyme complex. Nature Communications, 6, 7645.
(S)–Dimethyl 2–((6–chloro–3–nitropyridin–2–yl)amino)succinate (43):
To a 2–L flask equipped with a thermometer, a reflux condenser, and a mechanical stirrer add 2,6–dichloro–3–nitropyridine (41; 100 g, 0.52 mol), (S)–aspartic acid dimethyl ester hydrochloride (42; 205 g, 1.04 mol), NaHCO3 (174 g, 2.07 mol), and tetrahydrofuran (1 L).
Stir the reaction at 40°C for 16 h and monitor for the disappearance of 2,6–dichloropyridine by HPLC.
Following completion of the reaction, filter away solids and wash with ethyl acetate (3 × 300 mL). Concentrate the combined filtrate and wash ings to dryness, and take up the residue in 1 L of ethyl acetate.
Stir the solution with charcoal (200 g) at ambient temperature for 2 h, filter away the charcoal, and wash with additional ethyl acetate (3 × 200 mL).
Concentrate the combined filtrate and washings in vacuo to obtain crude (S)–dimethyl 2–((6–chloro–3–nitropyridin–2–yl)amino)succinate (43; 180 g, >100%) as a yellow oil. Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 9.00 (d, J=7.9 Hz, 1 H), 8.50 (d, J=8.6 Hz, 1 H), 6.92 (d, J=8.6 Hz, 1 H), 5.23 =(m, J=5.7, 7.9 Hz, 1 H),3.67 (s, 3 H), 3.63 (s, 3 H), 3.06 (m, J=5.8 Hz, 2 H); 13C NMR (APT) (75 MHz, DMSO–d6): δ 170.93 (C), 170.65 (C), 154.65 (C), 150.59 (C), 138.82 (CH), 127.28 (C), 112.81 (CH), 52.23 (CH3), 51.74 (CH3), 50.20 (CH), 35.31 (CH2); MS m/z=318.0 (M+H)+; HRMS calculated for C11H13N3O6Cl: 318.0493; found: 318.0492.
(S)–Methyl 2–(6–chloro–2–oxo–1,2,3,4–tetrahydropyrido[2,3–b] pyrazin–3–yl)acetate (44)
-
6.
Charge a 5–L three–necked flask equipped with a thermometer, a reflux condenser, and a mechanical stirrer with crude (S)–dimethyl 2–((6–chloro–3–nitropyridin–2–yl)amino)succinate (43; 180 g, 0.52 mol), iron powder (146 g, 2.59 mol), 2–propanol (2 L), and water (700 mL).
-
7.
Stir the mixture at 40°C and then add acetic acid (15.5 g, 0.259 mmol) at a rate sufficient to keep the internal temperature below 70°C. Stir at 70°C for 30 min, or until HPLC indicates that the reaction is complete.
-
8.
Cool the mixture to 40°C, then add Na2CO3 (165 g, 1.55 mol), and stir the mixture for 1 h. Filter the solids and wash with tetrahydrofuran (3 × 500 mL).
-
9.
Concentrate the combined filtrate, wash in vacuo, and then stir the residue in ethanol (1 L) for 12 h.
-
10.
Filter the solid, wash with cold ethanol, and dry in vacuo to obtain (S)–methyl 2–(6–chloro–2–oxo–1,2,3,4–tetrahydropyrido[2,3–b]pyrazin–3–yl)acetate as an off–white solid (44; 91 g, 68%). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 10.55 (br s, 1 H), 7.35 (br s, 1 H), 6.92 (d, J=7.9 Hz, 1 H), 6.57 (d, J=7.8 Hz, 1 H), 4.43 (m, J=1.4, 5.1 Hz, 1 H), 3.57 (s, 3 H), 2.79 (m, J=5.1, 16.4 Hz, 2 H); 13C NMR (APT) (75 MHz, DMSO–d6): δ 170.32 (C), 164.96 (C), 146.13 (C), 140.32 (C), 122.41 (CH), 119.47 (C), 111.31 (CH), 51.81 (CH), 51.39 (CH3), 37.01 (CH2); MS m/z=256.0 (M+H)+; HRMS calculated for C10H11N3O3Cl: 256.0489; found: 256.0487.
(S)–2–(6–Chloro–1,2,3,4–tetrahydropyrido[2,3–b]pyrazin–3–yl) ethanol (45)
-
11.
Charge a 5–L three–necked flask equipped with a mechanical stirrer, a reflux condenser, and a nitrogen inlet with LiAlH4 (60 g, 1.58 mol). Cool the flask with an ice bath and add tetrahydrofuran (500 mL).
-
12.
Cool the stirred mixture to 0°C and then add a solution of (S)–methyl 2–(6–chloro–2–oxo–1,2,3,4–tetrahydropyrido[2,3–b]pyrazin–3–yl)acetate (44; 81 g, 0.32 mol) in tetrahydrofuran (2 L), while keeping the internal temperature below 5°C.
-
13.
Heat the reaction at reflux for 16 h, while monitoring for the appearance of product by HPLC. The ester reduction should occur rapidly, while the lactam reduction may require longer for complete reduction.
-
14.
Cool the reaction to 5°C and then add water (60 mL) while keeping the internal temperature below 10°C.
-
15.
Stir the reaction for 15 min. Add 15% (w/w) aqueous NaOH (60 mL) while keeping the internal temperature below 5°C.
-
16.
Stir the reaction for 15 min, then add water (180 mL), and stir at ambient temperature for 1 h.
-
17.
Filter off and wash the solids with tetrahydrofuran (3 × 150 mL). Concentrate the filtrate, wash, in vacuo, and dry the solid residue in vacuo to obtain (S)–2–(6–chloro–1,2,3,4–tetrahydropyrido[2,3–b]pyrazin–3–yl)ethanol as a brown solid (45; 55 g, 81%). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 6.60 (br s, 1 H), 6.58 (d, J=7.8 Hz, 1 H), 6.32 (d, J=7.8 Hz, 1 H), 5.69 (m, 1 H), 4.57 (t, J=5.0 Hz, 1 H), 3.56 (m, J=5.8 Hz, 2 H), 3.47 (m, 1 H), 3.22 (m, J=2.7, 11.1 Hz, 1 H), 2.84 (m, J=1.6, 6.7, 11.1 Hz, 1 H), 1.65 (m, J=6.7 Hz, 1 H), 1.54 (m, J=6.3 = Hz, 1 H); 13C NMR (APT) (75 MHz, DMSO–d6): δ 146.75 (C), 134.44 (C), 128.20 (C), 118.97 (CH), 110.59 (CH), 57.97 (CH2), 47.47 (CH), 43.99 (CH2), 36.60 (CH2); MS m/z=214.1 (M+H)+; HRMS calculated for C9H13N3OCl: 214.0747: found: 214.0743.
(4S)–7–Chloro–2,3,4,5–tetrahydro–1,4–methanopyrido[2,3–b][1,4] diazepine (46)
-
18.
Add triethylamine (95 g, 0.936 mol) to a solution of (S)–2–(6–chloro–1,2,3,4–tetrahydropyrido[2,3–b]pyrazin–3–yl)ethanol (45; 50 g, 0.234 mol) in CH2Cl2 (500 mL). Stir the mixture at ambient temperature until it is homogeneous, then cool to 0°C.
-
19.
Add POCl3 (54 g, 0.351 mol) dropwise to the reaction mixture while maintaining the temperature between 0 and 5°C. Remove cooling and stir the reaction at ambient temperature for 2 h while monitoring for the disappearance of the starting alcohol by HPLC.
-
20.
After the reaction is complete, add 1.2 M aqueous NaHCO3 (200 mL). Separate the layers and extract the aqueous layer with CH2Cl2.
-
21.
Extract the combined CH2Cl2 layers with 1 M HCl (4 × 300 mL), adjust the pH of the combined HCl layers to 8 with solid NaHCO3. Extract the resulting mixture with CH2Cl2 (4 × 300 mL) and dry (Na2SO4) this set of CH2Cl2 layers, filter, and treat with charcoal (50 g).
-
22.
Stir the mixture at ambient temperature for 3 h, filter, and wash the charcoal with CH2Cl2 (200 mL). Concentrate the combined filtrate and wash solution to dryness, and dry the solid residue in vacuo to obtain (4S)–7–chloro–2,3,4,5–tetrahydro–1,4–methanopyrido[2,3–b] [1,4]diazepine as an off–white crystalline solid (46; 30 g, 66%). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 7.47 (br d, J=4.5 Hz, 1 H), 7.09 (d, J=7.7 Hz, 1 = 1 H), 6.39 (d, J=7.7 Hz, 1 H), 3.89 (m, J=5.0 Hz, H), 2.95–3.13 (m, 2 H), 2.77 (m, 2 H), 1.98 (m, J=5.0 Hz, 1 H), 1.86 (m, J=6.9 Hz, 1 H); 13C NMR (APT) (75 MHz, DMSO–d6): δ 153.45 (C), 144.50 (C), 134.32 (CH), 133.19 (C), 109.73 (CH), 59.88 (CH2), 53.07 (CH2), 50.08 (CH), 38.38 (CH2); MS m/z=196.1 (M+H)+; HRMS calculated for C9H11N3Cl: 196.0642; found: 196.0637.
(4S)–7–(3–(Trifluoromethyl)phenyl)–2,3,4,5–tetrahydro–1,4–methanopyrido[2,3–b][1,4]di–azepine (47)
-
23.
Heat at reflux a solution of (4S)–7–chloro–2,3,4,5–tetrahydro–1, 4–methanopyrido[2,3–b][1,4]diazepine (46; 5.0 g, 25.6 mmol), 3–trifluoromethylphenylboronic acid (7.3 g, 38 mmol), Pd(OAc)2 (0.14 g, 0.63 mmol), 2–dicyclohexylphosphino–2′,4′,6′–triisopropylbiphenyl (0.61 g, 1.3 mmol), and Cs2CO3 (24.9 g, 76.4 mmol) in a mixture of dioxane (100 mL) and water (10 mL) for 2.5 h and cool to room temperature.
-
24.
Filter the reaction mixture through celite® and concentrate.
-
25.
Dilute the residue with ethyl acetate, wash the organic layer with aqueous sat. NaHCO3, water, and brine, then dry (Na2SO4), and concentrate to dryness.
-
26.
Purify by silica gel chromatography (50–100% ethyl acetate gradient in pentane) to afford (4S)–7–(3–(trifluoromethyl)phenyl)–2,3,4,5–tetrahydro–1,4–methanopyrido[2,3–b][1,4]diazepine as a white solid(47; 6.31 g, 81%). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 8.27 (s, 1 H), 8.19 (d, J=7.44 Hz, 1 H), 7.69 (d, J=7.9 Hz, 1 H), 7.64 (t, J=7.6 Hz, 1 H), =7.27 (d, J=4.5 Hz, 1 H), 7.20 (d, J=7.7 Hz, 1 H), 7.09 (d, J=7.7 Hz, 1 H), 3.93 =(m, J=2.3 Hz, 1 H), 3.00–3.17 (m, 2 H), 2.85 (d, J=11.2 Hz, 1 H), 2.80 (dd, J=2.0, 11.2 Hz, 1 H), 1.96–2.07 (m, 1 H),1.85–1.96 (m, 1 H); 13C NMR = (APT) (75 MHz, DMSO–d6): δ 153.31 (C), 149.41 (C), 140.10 (C), 134.64 (C), 132.42 (CH), 129.74 (CH), 129.54 (CH), 129.29 (q, JCF=31.5 Hz, C), 124.42 (q, JCF=3.7 Hz, CH), 124.33 (q, JCF=271.9 Hz, C), 122.41 (q, JCF=3.9 Hz, CH), 108.35 (CH), 59.85 (CH2), 53.52 (CH2), 50.31 (CH), 38.30 (CH2); MS m/z=305.1 (M+H)+; HRMS calculated for C16H15N3F3: 306.1218; found: 306.1219.
(4S)–N–(3–(Oxazol–5–yl)phenyl)–7–(3–(trifluoromethyl)phenyl)–3,4–dihydro–1,4–methanopyrido[2,3–b][1,4]diazepine–5(2H)–carboxamide (4)
-
27.
To a solution of (4S)–7–(3–(trifluoromethyl)phenyl)–2,3, 4,5–tetrahydro–1,4–methanopyrido[2,3–b][1,4]diazepine (47; 3.05 g, 10 mmol) and triphosgene (2.37 g, 8.0 mmol) in CH2Cl2 (30 mL) add triethylamine (4.17 mL, 30 mmol).
-
28.
Heat the solution at reflux for 1.5 h and then add 3–(oxazol–5–yl)aniline (48; 2.40 g, 15 mmol) as a solid. Then heat the reaction mixture at reflux for 1 h, cool to room temperature, and dilute with CH2Cl2.
-
29.
Wash the resulting organic layer with sat. aqueous NaHCO3, water, and brine, then dry (Na2SO4), and concentrate to dryness. During the aqueous workup the by–product (1,3–bis(3–(oxazol–5–yl)phenyl) urea) will likely form a rag layer, which can be removed by filtration.
-
30.
Purify the crude product by silica gel chromatography (0–6% MeOH gradient in CH2Cl2) to obtain 4 as a foam. Sonicate the foam in pentane, concentrate, and dry under high vacuum to obtain (4S)–N–(3–(oxazol–5–yl)phenyl)–7–(3–(trifluoromethyl)phenyl)–3,4–dihydro–1,4–methanopyrido[2,3–b][1,4]diazepine–5(2H)–carboxamide as a free–flowing white solid (4; 3.08 g, 63%). Characterization properties: 1H NMR (300 MHz, DMSO–d6): δ 12.96 (s, 1 H), 8.46 (s, 1 H), 8.26 (d, J=7.7 Hz, 1 H), 8.20 (m, 1 H), 7.93 (m, 1 H), 7.90 (d, J=7.9 Hz, 1 H), 7.82 (t, J=7.7 Hz, 1 H), 7.72 (d, J=7.9 Hz, 1 H), 7.65 (d, J=7.9 Hz, 1 H), 7.61 (s, 1 H), 7.38–7.46 (m, 3 H), 5.51 (dd, J=2.9, 5.7 Hz, 1 H), 3.05–3.25 (m, 3 H), 2.98 (dd, J=3.2, 12.0 Hz, 1 H), 2.19–2.34 (m, 1 H), 1.91–2.00 (m, 1 H); 13C=NMR (APT) (75 MHz, DMSO–d6): δ 151.77 (C), 151.26 (CH assigned based on HSQC), 150.23 (C), 148.81 (C), 148.62 (C), 139.36 (C), 139.19 (C), 137.24 (C), 135.89 (CH), 130.65 (CH), 130.26 (CH), 129.91 (q, JCF=31.7 Hz, C), 129.59 (CH), 128.05 (C), 125.69 (m, JCF=4.2 Hz, CH), 123.97 (q, JCF=272.5 Hz, C), 122.91 (m, JCF=3.9 Hz, CH), 122.03 (CH), 119.28 (CH), 118.82 (CH), 115.59 (CH), 114.67 (CH), 58.73 (CH2), 53.59 (CH2), 51.66 (CH), 34.98 (CH2); MS m/z=491.9 (M+H)+; HRMS calculated for C26H21N5O2F3: 492.1647; found: 492.1646.
3.2 Expression and Purification of Recombinant His–Tagged SIRT1
The SIRT1 expression and purification protocol below has been adapted from the previous works (Howitz et al., 2003; Hubbard, Gomes, et al., 2013; Schneider et al., 1998; Wood et al., 2004). While this procedure produces relatively crude SIRT1 protein (>75% purity), it does not require any specialized equipment (eg, fast–protein liquid chromatography (FPLC) machine), and it is both quick and cost–effective. Moreover, it can be readily adapted to incorporate FPLC or other chromatography–based approaches (starting at step #9), as previously described (Feldman, Baeza, & Denu, 2013).
Transformation of BL21 bacteria
Transform the pET His–SIRT1 plasmid into BL21 pLysS(DE3) bacteria according to the manufacturer’s instructions and grow the transformed cells on LB–agar plates supplemented with the appropriate antibiotics overnight at 37°C.
Once colonies have formed (usually 14–16 h later), inoculate 10 mL of antibiotic–supplemented LB with a colony in a Falcon tube, and grow the culture overnight at 37°C.
The next day, inoculate a larger culture (eg, 2 L) with 1 mL of this culture and grow up in the presence of antibiotics. Take OD600 culture readings periodically using a spectrophotometer.
Once an OD of 0.6–0.8 has been reached, induce expression of His–SIRT1 by adding IPTG to a final concentration of 1 mM.
Immediately following addition of IPTG, transfer the flask into a shaker and grow the culture overnight ( ~16 h) at 16°C.
Purification of recombinant His–SIRT1
Supplement the lysis and wash buffers with protease inhibitor pellets (eg, Roche EDTA–free cocktail tablets—1 tablet/10 mL lysis buffer). Approximately 15 mL of lysis buffer is needed for each liter of bacterial culture that was spun down. Allow the pellets to dissolve completely (eg, rotate for ~5 min at 4°C to dissolve) while keeping solutions chil led on ice.
Spin down cultures in a centrifuge capable of handling large volumes (eg, 2500 ×g for 20 min) at 4°C.
Discard the supernatant and keep the bacterial pellet on ice.
Add lysis buffer containing the protease inhibitors to the cell pellet and pipette the mixture up and down thoroughly to resuspend cells.
Allow the mixture to incubate on ice for ~30 min, until it becomes viscous. Viscosity can be monitored by drawing up the mixture in a Pasteur pipette.
Sonicate the mixture on ice for 30 s using a sonicator with a wide–fitted head (perform sonication in 50 mL Falcon tubes) set to 60%. Allow the sample to cool for 1 min and then repeat this process 4 (total of 5).
Transfer the sonicated lysate into centrifuge tubes and spin down at 27,000 × g(16,000 rpm using a Sorvall SS34 rotor) for 30 min at 4° C to pellet cell debris.
While the sample is centrifuging, prepare the Ni–NTA resin. Aliquot approximately 1.5 mL of slurry (0.75 mL packed beads) per L of culture (see Note 6) into a 15–mL Falcon tube. Spin down the slurry (eg, 100 × g) and aspirate off the supernatant. Next, add 10 mL of cold lysis buffer (supplemented with protease inhibitor pellet) to the resin, invert to mix, spin down, and aspirate off the supernatant. Perform one additional wash.
Once centrifugation is complete, collect the sample supernatant into 50 mL Falcon tubes, add the appropriate amount of washed Ni–NTA resin, and rotate for 1 h at 4°C (eg, in a cold room).
Spin down the sample (eg, 100–200 × g) sufficiently to pellet the resin. Pipette or aspirate off the supernatant and discard.
Add twice the bead volume of ice–cold wash buffer (with protease inhib. pellet) to the resin, invert to resuspend, and centrifuge the resin (100–200 ×g). Pipette or aspirate off the supernatant and discard. Repeat this process at least four times.
Elute the bound protein by adding 1.5 times the bead volume of chilled elution buffer to the resin and allowing the mixture to rotate for 1 h at 4°C.
Spin down the beads (100 × g at 4°C) and transfer the supernatant to a Biorad Polyprep column. Allow the supernatant to filter through the column by gravity into a collector tube (eg, Falcon tube).
To reduce the concentration of imidazole for certain downstream applications, dialysis using Millipore Microcon columns may be performed according to the manufacturer’s instructions.
Dilute the eluate 1:1 with glycerol (mix well), quantitate, and aliquot and store at 20°C.
To obtain a higher purity protein prep. (>90%), His–SIRT1 may be further purified by size exclusion chromatography in SEC buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.1 mM TCEP) using a Hi–load Superdex 200 16/60 column (GE LifeSciences, United States) via FPLC.
3.3 Assay of SIRT1 Activators Using the PNC1–OPT Assay
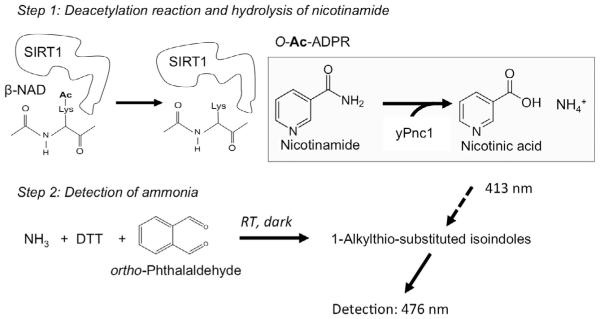
The PNC1–OPT assay affords a quick and reliable method to measure the deacetylase activity of SIRT1 without the need for any specialized equipment (Hubbard & Sinclair, 2013). Moreover, in contrast to previously developed SIRT1 assays which require the use of a fluorophore–conjugated peptide substrate, any custom peptide may be used in this assay (Hubbard & Sinclair, 2013). As outlined in Fig. 5, two distinct steps are involved in the measurement of SIRT1 activity using this assay. First, SIRT1 is incubated with acetylated peptide in the presence of β–NAD, reaction buffer, and saturating amounts of PNC1. As nicotinamide is produced from the deacetylation reaction, it is converted into free ammonia (NH3) by PNC1. Second, the reaction is stopped and the amount of ammonia present is quantified via a chemical reaction with OPT and DTT. This assay can be used to reliably study the effects of any STAC that does not interfere with the fluorescence signal or inhibit PNC1 (Hubbard & Sinclair, 2013).
Fig. 5.
Outline of the PNC1–OPT assay (Hubbard & Sinclair, 2013). In the first step, deacetylation of a custom peptide substrate by SIRT1 results in the production of nicotinamide (NAM), which is subsequently converted into nicotinic acid and ammonia by the nicotinamidase PNC1. In the second step, the reaction is quenched and ammonia is reacted with o–phthalaldehyde and dithiothreitol (DTT) (in the dark) to produce fluorescent adducts that are quantified using a spectrophotometer.
Preparation of a nicotinamide standard curve
Thaw an aliquot of nicotinamide and perform a serial dilution to yield solutions that are 10 × of the final reaction concentrations. A series of final concentrations that cover the dynamic range of the assay are 0,5,10, 20, 30, 40, and 50 μM. Pipette 10 μL of each of these solutions into appropriately labeled Eppendorf tubes.
Prepare a reaction mastermix corresponding to the total number of reactions to be performed (it is advisable to perform all reactions in triplicate). For each desired reaction, add 100 μL of assay buffer and 1 μg of PNC1 and mix the solution by gentle vortex.
Pipette 90 μL of the reaction mastermix into each tube containing the nicotinamide standards, mix by pipetting, and close the lid on each tube.
Place all of the sample tubes in a holder rack and shake using an orbital shaker at 37°C in an incubator for 1 h.
During the incubation period, thaw the OPT developer reagent by heating the stock at 42°C. Ensure that the solution is well mixed by vortexing, and that no DTT precipitate is present.
Remove the samples from the incubator, and under dim light, add 100 μL of the OPT developer reagent to each reaction as quickly as possible. Vortex all samples to mix on the highest setting for 5 s. Put the tubes back into a holder, cover all samples with aluminum foil, and incubate at room temperature on an orbital shaker for 1 h (see Note 7).
Transfer samples to a 96–well plate, under dim light, and read the fluorescence using a spectrophotometer with excitation and emission wavelengths set to 413 and 476 nm (Sugawara & Oyama, 1981), respectively (in practice an λex of 420 ± 10 nm and λem 460 ± 10 nm work fine a 0.1– 1–s read with or time).
Subtract the background fluorescence (0 μM NAM) from all samples and plot a graph of normalized fluorescence vs concentration of NAM (standard curve).
Assay of SIRT1 activators
-
1.
Thaw aliquots of β–NAD, peptide substrate, and PNC1 and SIRT1 enzymes on ice.
-
2.
Arrange and label a series of Eppendorf tubes in a holder corresponding to twice the number of samples to be assayed: Label one set “+NAD” and the second set “−NAD.” The latter set of samples will be used as background fluorescence control reactions (see Note 8). In addition, as stated earlier, it is advised that all samples be measured in triplicate (including the −NAD controls).
-
3.
Pipette the various test compounds into each corresponding tube and include a vehicle control (eg, DMSO).
-
4.
Prepare a reaction master mix corresponding to the total number of samples to be assayed (both +NAD and −NAD samples). For each reaction add the following (prepare on ice in a 15–mL Falcon tube): Reaction Buffer (100 μL minus the volume of other components), peptide substrate (typically 10–30 μM), PNC1 (1 μg), and SIRT1 purified as described in Section 3.1 (1 μg) (see Note 9). Mix by pipetting followed by gentle vortex (50% efficiency for 5 s).
-
5.
Divide the mastermix into two Falcon tubes. To one tube add β–NAD to the appropriate final concentration (typically 100 μM) (+β–NAD mastermix), and to the second tube add an identical volume of water (for the no NAD control). Mix briefly.
-
6.
Aliquot 100 μL of the +NAD mastermix to each experimental reaction tube, and 100 μL of the −NAD mastermix to each corresponding neg ative control reaction. Close the lids on each tube, and very gently vortex to mix (20% amplitude).
-
7.
Incubate reactions at 37°C for 1 h.
-
9.
During the incubation period, thaw the OPT developer reagent and incubate at 42°C for ~15 min (keep covered in aluminum foil). Ensure that the solution is well mixed by vortexing, and that no DTT precipitate is observed. If particulates are visible, vortex and continue to heat.
-
8.
Once the incubation period is complete, under dim light, quickly remove each sample tube from the holder and add 100 μL of OPT developer reagent. If available, a multichannel pipette may be used. It is imperative that the developer be added to each sample as quickly as possible to ensure consistency. Vortex all samples on the highest setting for 5 s.
-
9.
Place the tubes back into the holder, cover all samples with aluminum foil (to prevent exposure to light), and incubate at room temperature on an orbital shaker for 1 h (see Note 7).
-
10.
Following the development phase, remove the foil under dim light and transfer 150–200 μL from each tube into 1 well of a 96–well dark bottom plate.
-
11.
Read the fluorescence using a spectrophotometer with excitation and emission wavelengths set to 413 and 476 nm (Sugawara & Oyama, 1981), respectively (as noted earlier, λex of 420 10 nm and λem 460 ± 10 nm work fine with a 0.1– or 1–s read time). Calculate the background fluorescence for each condition by taking the mean of the arbitrary fluorescence (AF) readings for the NAD samples. Next, calculate the net fluorescence for each reaction condition by subtracting the mean background fluorescence from each reading, Fcorrected Fcorrected=F+NAD–F−NADcontrol (mean value). The resulting value is proportional to the amount of NAM produced during the deacetylation reaction.
-
12.
AFU can be converted into amounts of NAM production using the linear equation obtained from the standard curve above.
3.4 Assay of SIRT1 Activators Using the RapidFire O–Ac–ADPR Detection Assay
RapidFire (Agilent) is a solid–phase extraction–based system that enables high sensitivity detection using mass spectrometry (MS) (Lim, Ozbal, & Kassel, 2010). SIRT1 activity can be assayed using RapidFire/MS technology by monitoring the production of either nicotinamide or 3′–OAc–ADPR following a deacetylation reaction. Since this assay does not rely on any type of fluorescence or require any additional coupling enzymes, it produces highly reliable results. Like the PNC1–OPT assay, the RapidFire detection assay can be performed using any custom native substrate, and it is amenable to medium to high–throughput compound screening. Below we outline a procedure for assaying SIRT1 activity using the RapidFire/MS system to detect production of O–Ac–ADPR.
Detection parameters and O–Ac–ADPR standard curve
Optimize MS parameters for the detection of O–Ac–ADPR (see Note 10).
Thaw an aliquot of O–Ac–ADPR and perform dilutions in Reaction buffer supplemented with 0.05% BSA and 4 mM β–NAD to yield 100 μL solutions that cover the dynamic range of the assay. All standards should be run in triplicate.
Mix 10 μL of each sample with 40 μL of a 1:1 acetonitrile:methanol solution.
Run the samples on the Agilent RapidFire system coupled to a mass spectrometer (eg, ABSciex API 4000) fitted with an electrospray ionization source: aspirate samples for 250 ms using the RapidFire system andabsorb 10 μL of each sample onto an SPE cartridge with Buffer A (3 s). Elute in Buffer B for 5 s.
Integrate peak data using the RapidFire Integrator software.
Subtract the background signal from all samples, plot a graph of peak area vs O–Ac–ADPR concentration, and fit the standard curve using linear regression.
Assay of SIRT1 activators
Thaw aliquots of β–NAD and peptide substrate, and SIRT1.
Dispense 1 μL of each test compound dissolved in DMSO into one well of a 96– or 384–well microtiter plate. In addition, prepare a vehicle–only control reaction (eg, DMSO) (in triplicate).
Add 50 μL of Reaction buffer supplemented with 0.05% BSA and SIRT1 (at a final concentration of 5 nM) to each well.
Incubate the enzyme–compound mixtures for 20 min at 25°C before starting the reaction with substrates.
Prepare a mastermix containing 2 × the final concentration of β–NAD and peptide substrate (see Note 11) in Reaction buffer supplemented with 0.05% BSA, and pipette 50 μL of this master mix into each reaction tube.
Allow the reaction to proceed for 30 min at 25°C (room temperature).
Add Stop reagent to each reaction (final concentrations should be 1% formic acid and 5 mM NAM).
Dilute the quenched reactions fivefold with a solution of 1:1 acetonitrile:methanol.
Spin down samples at 5000 × g for 10 min to precipitate protein.
Run the samples on the Agilent RapidFire as described earlier: Aspirate samples for 250 ms and absorb 10 μL of each sample onto an SPE cartridge with Buffer A (3 s), then elute in Buffer B for 5 s.
Reequilibrate the system using Buffer A for 500 ms.
Use the standard curve produced above to calculate the amount of O–acetyl–adenosine diphosphate ribose (O–Ac–ADPR) produced.
ACKNOWLEDGMENTS
The authors thank Dr. W.H. Miller for critically reading the manuscript and providing suggestions for improvement. D.S. is supported by an NIA/NIH MERIT award 5R37AG028730–09 and generous support from the Glenn Foundation for Medical Research and Edward Schulak.
4. NOTES
The protease inhibitor cocktail must be EDTA–free since EDTA is incompatible with Ni–NTA resin.
In order to minimize background due to peptide fluorescence, peptides that are shorter are preferred (aromatic groups also increase background fluorescence). Also, only certain peptides support STAC–mediatedSIRT1 activation, such as those with hydrophobic groups adjacent to the acetyl–lysine (Hubbard, Gomes, et al., 2013), so these are recommended for studying SIRT1 activation.
Analytical HPLC can be performed on an Agilent 1100 Series HPLC machine equipped with a 3.5 μm Eclipse XDB–C18 (4.6 mm × 100 mm) column using the following parameters: CH3CN/H2O, modified with 0.1% formic acid mobile phase, Gradient elution: 5% CH3CN hold (2 min), 5–95% CH3CN gradient (11 min), 95–5% CH3CN gradient (0.3 min), 5% CH3CN hold (2.7 min), 15 min total run time with a flow rate of 0.8 mL/min.
Proton NMR spectra may be obtained using any standard spectrometer (eg, Bruker Advance III 300 MHz spectrometer) and should be referenced to internal TMS (0.00 ppm), CHCl3 (7.26 ppm), or DMSO (2.49 ppm).
HRMS can be completed on a Waters qTOF Premiere Mass Spectrometer operating in W mode positive ionization with a resolving power of approximately 15,000. A Waters nano–acquity LC may be used for flow injection.
To facilitate pipetting of the Ni–NTA resin, cut several mm off the pipette tip using scissors or a razor blade.
The length of the incubation period may need to be adjusted depending on signal strength. However, it is important to use a consistent time for all samples to minimize variance.
A background control reaction is performed to subtract out inherent substrate or small–molecule fluorescence and also NAD+–independent deacetylase activity. Subtracting the AFU reading of the −NAD samples from the +NAD samples yields fluorescence values that are proportional to the amount of NAM produced (Fcorrected=F+NAD–F–NADcontrol). Importantly, β–NAD also exhibits fluorescence in this assay at high concentrations (~200 μM). In these instances, control reactions lacking enzyme rather than NAD may be more appropriate. An alternative approach is to perform a parallel set of reactions using the corresponding nonacetylated peptide and use these as background controls.
The quantities of PNC1 and SIRT1 that are added to each reaction may vary depending on the specific activity of each preparation.
To optimize MS conditions use a direct infusion of O–Ac–ADPR in a 1:1 ethanol:water mixture containing 0.1% formic acid at a rate of 10 μL/min. MS parameters: curtain gas 20, probe temperature 550 °C, ion source gas 150, ion source gas 250, interface heater on, collision gas 10, ion source voltage 3800 V, declustering potential 85 entrance potential −10 V collision energy −37 V, and collision exit potential −20 V. Negative MRM mode was used monitoring the tran sition 600.1/345.9 for the parent/daughter ion under low–resolution conditions.
Peptide substrate concentrations of approximately 1/10th of their Km value should be used when assaying Km–modulating activators.
REFERENCES
- Bhullar KS, Hubbard BP. Lifespan and healthspan extension by resveratrol. Biochimica et Biophysica Acta. 2015;1852(6):1209–1218. doi: 10.1016/j.bbadis.2015.01.012. http://dx.doi.org/10.1016/j.bbadis.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. Journal of Biological Chemistry. 2005;280(17):17187–17195. doi: 10.1074/jbc.M501250200. http://dx.doi.org/10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang N, Xu RM. Structural basis for allosteric, substrate–dependent stimulation of SIRT1 activity by resveratrol. Genes & Development. 2015;29(12):1316–1325. doi: 10.1101/gad.265462.115. http://dx.doi.org/10.1101/gad.265462.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH. Using PDE inhibitors to harness the benefits of calorie restriction: Lessons from resveratrol. Aging (Albany, NY) 2012;4(3):144–145. doi: 10.18632/aging.100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Case AW, Riera TV, Considine T, Lee JE, Hamuro Y, Ellis JL. Crystallographic structure of a small molecule SIRT1 activator–enzyme complex. Nature Communications. 2015;6:7645. doi: 10.1038/ncomms8645. http://dx.doi.org/10.1038/ncomms8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Stein RL. SIRT1 activation by small molecules: Kinetic and biophysical evidence for direct interaction of enzyme and activator. Journal of Biological Chemistry. 2010;285(43):32695–32703. doi: 10.1074/jbc.M110.133892. http://dx.doi.org/10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TT, Tran TL, Kim J, Nguyen PH, Lee EH, Park J, Oh WK. Terpenylated coumarins as SIRT1 activators isolated from Ailanthus altissima. Journal of Natural Products. 2012;75(7):1332–1338. doi: 10.1021/np300258u. http://dx.doi.org/10.1021/np300258u. [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet–induced metabolic disorders by enhancing fat oxidation. Cell Metabolism. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. http://dx.doi.org/10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long–chain fatty acids and widespread deacylation by mammalian sirtuins. Journal of Biological Chemistry. 2013;288(43):31350–31356. doi: 10.1074/jbc.C113.511261. http://dx.doi.org/10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesisand colon cancer growth. PLoS One. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. http://dx.doi.org/10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz M, Nguyen GT, Fischer F, Suenkel B, Schlicker C, Franzel B, Steegborn C. A molecular mechanism for direct sirtuin activation by resveratrol. PLoS One. 2012;7(11):e49761. doi: 10.1371/journal.pone.0049761. http://dx.doi.org/10.1371/journal.pone.0049761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear–mitochondrial communication during aging. Cell. 2013;155(7):1624–1638. doi: 10.1016/j.cell.2013.11.037. http://dx.doi.org/10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. http://dx.doi.org/10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Sinclair DA. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339(6124):1216–1219. doi: 10.1126/science.1231097. http://dx.doi.org/10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Loh C, Gomes AP, Li J, Lu Q, Doyle TL, Sinclair DA. Carboxamide SIRT1 inhibitors block DBC1 binding via an acetylation–independent mechanism. Cell Cycle. 2013;12(14):2233–2240. doi: 10.4161/cc.25268. http://dx.doi.org/10.4161/cc.25268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Sinclair DA. Measurement of sirtuin enzyme activity using a substrate–agnostic fluorometric nicotinamide assay. Methods in Molecular Biology. 2013;1077:167–177. doi: 10.1007/978-1-62703-637-5_11. http://dx.doi.org/10.1007/978–1–62703–637–5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Sinclair DA. Small molecule SIRT1 activators for the treatment of aging and age–related diseases. Trends in Pharmacological Sciences. 2014;35(3):146–154. doi: 10.1016/j.tips.2013.12.004. http://dx.doi.org/10.1016/j.tips.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Kennedy BK. Substrate–specific activation of sirtuins by resveratrol. Journal of Biological Chemistry. 2005;280(17):17038–17045. doi: 10.1074/jbc.M500655200. http://dx.doi.org/10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO Journal. 2007;26(13):3169–3179. doi: 10.1038/sj.emboj.7601758. http://dx.doi.org/10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Rauh D, Schutkowski M, Steegborn C. Sirt1 activation by resveratrol is substrate sequence–selective. Aging (Albany NY) 2013;5(3):151–154. doi: 10.18632/aging.100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KB, Ozbal CC, Kassel DB. Development of a high–throughput online solid–phase extraction/tandem mass spectrometry method for cytochrome P450 inhibition screening. Journal of Biomolecular Screening. 2010;15(4):447–452. doi: 10.1177/1087057110362581. http://dx.doi.org/10.1177/1087057110362581. [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450(7170):712–716. doi: 10.1038/nature06261. http://dx.doi.org/10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free Radical Biology & Medicine. 2013;56:133–171. doi: 10.1016/j.freeradbiomed.2012.10.525. http://dx.doi.org/10.1016/j.freeradbiomed.2012.10.525. [DOI] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. Journal of Biological Chemistry. 2010;285(11):8340–8351. doi: 10.1074/jbc.M109.088682. http://dx.doi.org/10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin–Montalvo A, North BJ, Sinclair DA. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metabolism. 2012;15(5):675–690. doi: 10.1016/j.cmet.2012.04.003. http://dx.doi.org/10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metabolism. 2013;18(3):416–430. doi: 10.1016/j.cmet.2013.07.013. http://dx.doi.org/10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2–dependent silencing by relief of nicotinamide inhibition. Molecular Cell. 2005;17(4):595–601. doi: 10.1016/j.molcel.2004.12.032. http://dx.doi.org/10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Schneider A, Smith RW, Kautz AR, Weisshart K, Grosse F, Nasheuer HP. Primase activity of human DNA polymerase alpha–primase. Divalent cations stabilize the enzyme activity of the p48 subunit. Journal of Biological Chemistry. 1998;273(34):21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- Sugawara K, Oyama F. Fluorogenic reaction and specific microdetermination of ammonia. Journal of Biochemistry. 1981;89(3):771–774. doi: 10.1093/oxfordjournals.jbchem.a133257. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. http://dx.doi.org/10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang JL, Ha TK, Dhodary B, Kim KH, Park J, Lee CH, Oh WK. Dammarane triterpenes as potential SIRT1 activators from the leaves of Panax ginseng. Journal of Natural Products. 2014;77(7):1615–1623. doi: 10.1021/np5002303. http://dx.doi.org/10.1021/np5002303. [DOI] [PubMed] [Google Scholar]