Summary
The nucleolus and other ribonucleoprotein (RNP) bodies are membrane-less organelles that appear to assemble through phase separation of their molecular components. However, many such RNP bodies contain internal sub-compartments, and the mechanism of their formation remains unclear. Here, we combine in vivo and in vitro studies, together with computational modeling, to show that sub-compartments within the nucleolus represent distinct, coexisting liquid phases. Consistent with their in vivo immiscibility, purified nucleolar proteins phase separate into droplets containing distinct non-coalescing phases that are remarkably similar to nucleoli in vivo. This layered droplet organization is caused by differences in the biophysical properties of the phases – particularly droplet surface tension – which arises from sequence-encoded features of their macromolecular components. These results suggest that phase separation can give rise to multilayered liquids that may facilitate sequential RNA processing reactions in a variety of RNP bodies.
Introduction
The cellular interior is organized into organelles whose structures have evolved to facilitate their functions. The most well known examples are the canonical membrane-bound organelles such as secretory vesicles, the Golgi apparatus, and the endoplasmic reticulum. However, many intracellular compartments are membrane-less bodies comprised of RNA and protein, often referred to as RNP bodies; these include stress granules and processing bodies in the cytoplasm, and Cajal bodies and nucleoli in the nucleus, among many others. Despite their lack of a delimiting membrane, these organelles nevertheless maintain a coherent size and shape, with a well-defined boundary that compartmentalizes different types of proteins and RNA. By concentrating molecules within a micro-compartment, while allowing dynamic molecular interactions, these RNP bodies may function to control reaction efficiencies much like conventional membrane bound cytoplasmic organelles (Balagopal and Parker, 2009, Spector, 2001).
Many of these RNP bodies exhibit liquid-like biophysical properties, and growing evidence suggests they assemble via liquid-liquid phase separation (Brangwynne et al., 2009, Nott et al., 2015, Li et al., 2012, Weber and Brangwynne, 2015). Intracellular phase transitions can result in switch-like changes in molecular organization and the spontaneous formation of micron-scale membrane-less organelles. Such behavior is reminiscent of well-known in vitro observations in protein crystallization, where soluble proteins are observed to condense into concentrated liquid phases or crystalline solid phases. A number of recent papers suggest that intrinsically disordered proteins or low complexity sequences (IDP/LCS) drive phase transitions underlying assembly of the nucleolus (Berry et al., 2015), stress granules (Wippich et al., 2013, Molliex et al., 2015, Patel et al., 2015), P granules and nuage bodies (Elbaum-Garfinkle et al., 2015, Wang et al., 2015), and nuclear speckles (Hennig et al., 2015).
The liquid-like nature of the nucleolus may facilitate its function in ribosome biogenesis. The nucleolus forms around regions of chromosomes containing stretches of tandem ribosomal DNA (rDNA) gene repeats, known as nucleolar organizer regions (NORs). In most eukaryotes (including human, X. laevis, and C. elegans) a precursor ribosomal RNA (rRNA) transcript is generated from the rDNA gene, and contains each of the co-transcribed 18S, 5.8S, and 28S rRNA subunits, separated by two intervening transcribed sequences (Fig. 1A). The nucleolus may facilitate increased reaction rates by locally concentrating rRNA processing factors involved in transforming the precursor rRNA transcript into individual rRNA subunits. Due to its role in producing this protein translational machinery, the structure and function of the nucleolus are intimately connected with cell growth and size homeostasis (Derenzini et al., 2000, Frank and Roth, 1998).
Figure 1. Liquid-like behavior of biophysically distinct nucleolar sub-compartments.
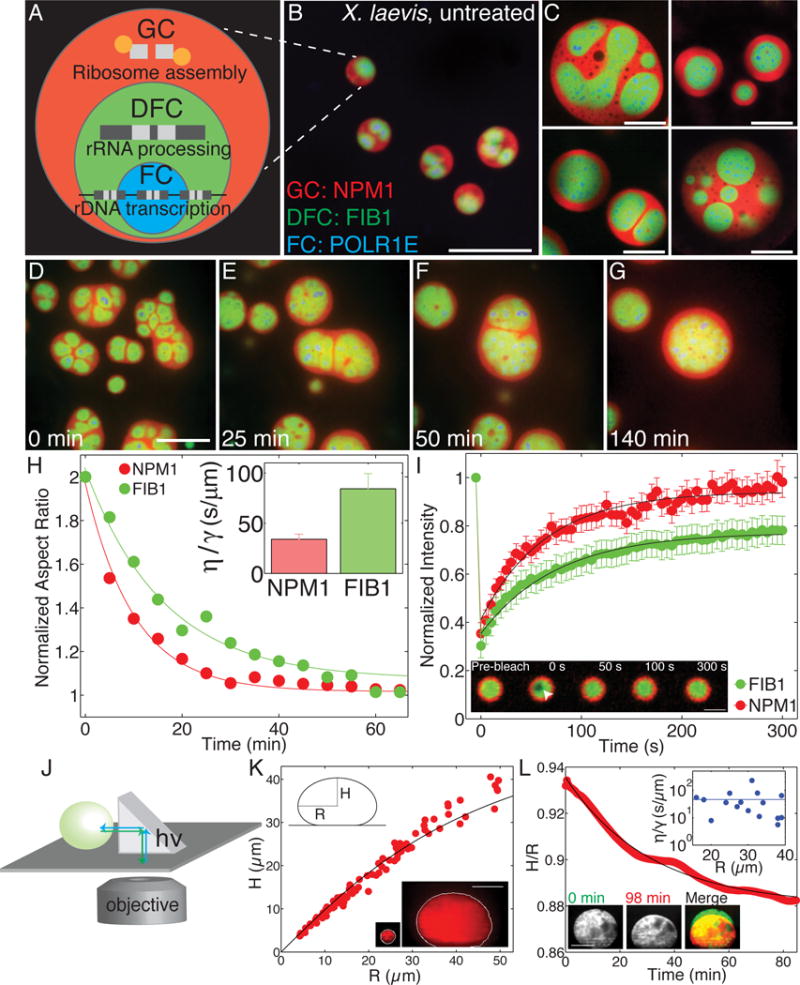
(A) Schematic diagram of ribosome biogenesis in nucleolus. (B) Nucleoli in an untreated X. laevis nucleus. Scale bar = 20 μm. For all images, granular component (GC) is visualized with NPM1 (red), dense fibrillar component (DFC) with FIB1 (green), and fibrillar center (FC) with POLR1E (blue). (C) Examples of nucleoli after coarsening in X. laevis nuclei treated with Lat-A. Scale bar = 20 μm. (D–G) Time-course of nucleolar component fusion after actin disruption by Lat-A. Scale bar = 20 μm. (H) Normalized aspect ratio vs. time for nucleolar components fusing after actin disruption. Inset shows η/γ for 59 nucleoli analyzed from 20 nuclei. (I) FRAP recovery curves for NPM1 (red) and FIB1 (green) in X. laevis oocytes. Inset: FRAP of FIB1-labeled DFC (green). Scale bar = 5 μm. (J) Schematic illustrating XZ imaging with a right angle prism. (K) Height, H, vs. radius, R, of different sized nucleoli at steady-state (91 nucleoli, from 61 nuclei). Black line is the fit from the average surface tension for all nucleoli. Bottom inset: example of the shape of a small vs. large nucleolus. Scale bar = 40 μm. (L) Example of nucleolar height to radius ratio, H/R, vs. time for one nucleolus deforming under gravity. Black line is an exponential fit. Top inset: η/γ for 16 nucleoli in 16 different nuclei. Blue line indicates average. Bottom inset shows example deforming nucleolus: Scale bar = 40 μm. See also Figure S1 and Movies S1, S2, S3, and S4.
Despite the biological importance of the nucleolus, a mechanistic biophysical understanding of its assembly and internal organization is lacking. The simplest picture of the nucleolus as a unitary liquid phase body becomes difficult to reconcile with its well-known complex and multi-component nature. Indeed, in addition to the various types of RNA in the nucleolus, the nucleolar proteome consists of hundreds of different proteins that are segregated into at least three distinct compartments (Boisvert et al., 2007). This layered tripartite organization consists of the fibrillar center (FC), where the RNA polymerase I (POL1) machinery is active; the dense fibrillar component (DFC) that is enriched in the protein fibrillarin (FIB1); and the granular component (GC) that is enriched in the protein nucleophosmin (NPM1/B23) (Fig. 1A,B). This multi-layered structure is not unique to the nucleolus, as stress granules and other liquid-like RNP bodies exhibit similar “core-shell” structuring (Jain et al., 2016, Hubstenberger et al., 2013).
The multi-layered structure of the nucleolus is thought to facilitate assembly-line processing of rRNA. Nascent rRNA transcripts undergo sequential processing steps by enzymes that localize to the distinct compartments, ultimately exiting the nucleolus, and being exported for final ribosome assembly in the cytoplasm (Fig. 1A). Although recent work has shown that the entire nucleolus can exhibit active liquid-like properties (Brangwynne et al., 2011) and its assembly may represent a type of liquid-liquid phase transition (Weber and Brangwynne, 2015), reconciliation of these findings with the multi-layered structure of the nucleolus has proven elusive. Indeed, if the nucleolus is a type of liquid, what mechanism prevents the three components from mixing and fusing to form a single liquid phase?
Here, we uncover a physical mechanism for intranucleolar organization: differences in miscibility between proteins from different nucleolar compartments keep the compartments phase separated, giving rise to the layered, multiphase droplet nature of nucleoli. By isolating protein domains from key nucleolar proteins, we provide evidence for a molecular mechanism whereby intrinsically disordered regions (IDRs) help drive protein condensation into droplets, while associated RNA binding domains confer sub-compartment specificity by making the two droplet phases immiscible with one another.
Results
Nucleolar sub-compartments behave as liquid-like phases in vivo
To gain insight into the biophysical assembly principles underlying nucleolar structure, we took advantage of the numerous large nucleoli, ranging in size from 1–10 microns, found in the nucleus (germinal vesicle, GV) of X. laevis oocytes (Fig. 1B). This system is also convenient because the nucleoli are extra-chromosomal, forming around amplified stretches of rDNA, allowing us to disentangle the confounding effects of somatic chromosome architecture on nucleolar structure. Nucleoli in X. laevis oocytes will fuse with one another when brought into contact (Brangwynne et al., 2011), but the frequency of such coalescence events is slowed significantly by the presence of a nuclear actin network (Fig. S1A–D) (Feric and Brangwynne, 2013, Feric et al., 2015).
We visualized intranucleolar organization by labeling individual components of nucleoli with fluorescent fusion proteins as follows: granular component with nucleophosmin (GC, NPM1::Cerulean), dense fibrillar component with fibrillarin (DFC, FIB1::RFP), and the fibrillar center with RNA polymerase 1E (FC, GFP::POLR1E). To test the role of nuclear actin in organizing these nucleolar substructures, we utilized the actin-disrupting compound Latrunculin-A (Lat-A). As shown previously, the entire nucleolus undergoes liquid-like coalescence events, which can be seen by fusion of the NPM1 (GC) region of one nucleolus with the NPM1 (GC) region from another nucleolus (Fig. 1D–G, Supplemental Movies S1–S3). The FC regions (POLR1E) rarely came into close contact with each other, and consequently, we did not observe fusion between multiple FCs, but we did see rearrangements into more spherical FC domains. Strikingly, however, we typically observe that the FIB1 (DFC) cores from one nucleolus will fuse when in close proximity with FIB1 (DFC) cores from a different nucleolus (Fig. 1D–G, S1E–L). These DFC regions, which were initially irregular in shape, would round up and coalesce, exhibiting classic liquid-like behavior. After long times (~1 hour), these coalescence events ultimately resulted in DFCs located in the center of the nucleolus, surrounded by one continuous phase of GC, representing an outer-most enveloping compartment (Fig. 1C).
In vivo sub-compartments have different biophysical properties
To gain insight into the biophysical properties of different nucleolar sub-compartments, we quantitatively analyzed fusion events. We found that homotypic fusion between the GC (NPM1) or DFC (FIB1) occurs by exponential relaxation to a single larger spherical shape; this is characteristic of coalescing liquid droplets and can be used to determine the ratio of droplet viscosity, η, to surface tension, γ, known as the inverse capillary velocity: η/γ (Brangwynne et al., 2011) (Fig. 1H). We find that FIB1-labeled DFC tends to exhibit slower fusion dynamics, with a larger value η/γ = 80±15 s/μm (mean ± s.e.m) compared to NPM1-labeled GC with η/γ = 30±5 s/μm (mean ± s.e.m.) (Fig. 1H inset, S1M,N). This behavior suggests that these nucleolar components behave as distinct liquid-like phases within the nucleolus, with different properties that could underlie nucleolar structural organization.
Interfaces represent sharp concentration gradients, and surface tension, with units of free energy per unit area, is the energetic cost of increasing the interfacial area. Surface tension is a key parameter that governs how two different droplets interact with one another. We therefore hypothesized that different surface tension values of the nucleolar sub-phases could explain their immiscibility and multi-layered organization. To measure the surface tension of the outermost GC compartment (i.e. interfacial energy of GC/nucleoplasm interface), we disrupted actin and allowed nucleoli to fuse for several hours. This resulted in a single large coalesced nucleolar droplet, which becomes measurably flattened at the bottom of the nucleus due to gravity. Since this flattening is resisted by surface tension, we could use a right-angle prism to measure the (XZ) shape of the droplet and determine the surface tension: 4±1×10−7 N/m (mean ± s.e.m) (Fig. 1J–K) (Supplemental Experimental Procedures); this value is very low, roughly five orders of magnitude lower than water-oil surface tension values (Than et al., 1988), but is comparable to values reported for colloidal liquids (Aarts et al., 2004). Complementary measurements of the droplet flattening timescale combined with droplet fusion measurements are consistent with this low value for the surface tension of the NPM1-rich GC compartment (Fig. 1L, Supplemental Movie S4) (Supplemental Experimental Procedures).
To confirm the liquid-like dynamics of the nucleolar sub-compartments, we performed fluorescence recovery after photobleaching (FRAP) experiments of X. laevis nucleoli in vivo (Fig. 1I). NPM1 exhibits fast dynamics with a nearly complete recovery on a timescale of τ=64±8 s for a bleach spot of 1 μm, consistent with the expected response for diffusion within a liquid. However, FIB1 recovery was slightly slower (τ=75±7 s). Moreover, the recovery of FIB1 was not complete, but only reached ~80%; this suggests that the DFC sub-compartment may not be a simple liquid, but instead may exhibit partially solid-like properties (i.e., viscoelasticity).
Purified FIB1 and NPM1 can phase separate into droplets similar to nucleoli in vivo
To gain further insight into how nucleolar proteins could give rise to distinct liquid-like nucleolar sub-phases, we purified recombinant FIB1 and NPM1, and studied their behavior in vitro. Consistent with our previous work, we find that FIB1::GFP (hereafter referred to simply as FIB1) can phase separate in vitro under near physiological protein and salt concentrations (Berry et al., 2015), giving rise to condensed liquid-phase droplets that are roughly 50-fold more concentrated with protein than the surrounding dilute phase (Fig. 2A). Indeed, in the presence of 5 μg/mL rRNA and 150 mM NaCl, FIB1 condenses into droplets at a protein concentration of roughly 600 nM. FIB1 can phase separate even in the presence of non-specific poly-U50 and heparin, suggesting that electrostatic interactions contribute to droplet assembly (Fig. S2A). NPM1 has also recently been demonstrated to undergo phase separation into liquid-like droplets (Fig. 2B) (Mitrea et al., 2016). However, at 150 mM NaCl, NPM1 requires significantly higher concentrations of protein (2 μM) and rRNA (100 μg/mL). Moreover, phase separation of NPM1 required rRNA and cannot be induced by the addition of heparin or poly-U50.
Figure 2. Purified nucleolar proteins can phase separate into droplets with different biophysical properties.
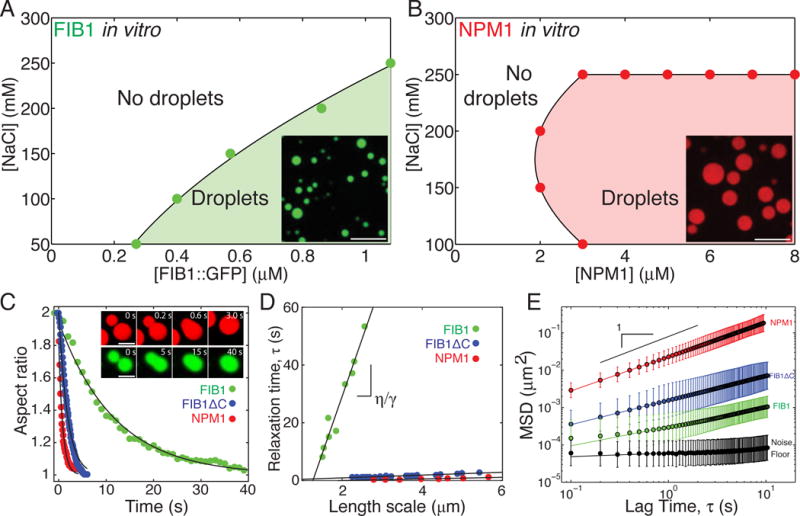
(A) Phase diagram of purified FIB1 in the presence of 5 μg/ml of rRNA. Inset: FIB1 droplets. Scale bar = 10 μm. (B) Phase diagram of purified NPM1 in the presence of 100 μg/ml of rRNA. Inset: NPM1 droplets. Scale bar = 10 μm. (C) Aspect ratio vs. time for fusing droplets of FIB1 (green), NPM1 (red), and FIB1ΔC (blue). Inset: FIB1 fusing (scale = 2 μm) and NPM1 fusing (scale = 5 μm). (D) Relaxation time versus length scale for fusion data from multiple FIB1 (green), NPM1 (red), and FIB1ΔC droplets (blue). (E) MSD vs. lag time of microrheological probe particles (R=50 nm) embedded in droplets of FIB1 (green), NPM1 (red), or FIB1ΔC (blue); black data points represent the noise floor (black). See also Figure S2.
Given the distinct biophysical properties of the nucleolar sub-compartments, we hypothesized that the two different types of in vitro droplets would also exhibit different material properties. As with the in vivo sub-compartments, in vitro droplets undergo homotypic fusion events when brought into close contact, but do so with markedly different time scales: FIB1 droplets typically take roughly a hundred times longer than NPM1 droplets of comparable size to coalesce and relax into a single larger sphere (Fig. 2C). Using an analysis similar to that performed in vivo, we find that FIB1 has an inverse capillary velocity of η/γ =40±10 s/μm (95% confidence interval), comparable to that measured in vivo (Fig. S2C). Also mirroring the in vivo data, purified NPM1 droplets have a lower value of 0.30±0.07 s/μm (Fig. 2D, S2C). A series of experiments confirmed that the presence of the GFP tag does effect droplet properties, but does not qualitatively impact our findings (Fig. S2B,C; Supplemental Experimental Procedures).
NPM1 readily formed large droplets in vitro, allowing for direct measurement of the surface tension, using a method similar to our in vivo set up with the prism (Fig. 1J). We estimated a surface tension of γNPM1=8±2×10−7 N/m (mean ± s.e.m) (Fig. S2D). This value is again surprisingly low and on the same order of magnitude as the value obtained for X. laevis nucleoli (Fig. S2C).
Viscoelasticity and time-dependence of in vitro droplets
To further shed light on the different properties of the two droplet subtypes, we performed microrheology experiments using the fluctuating motion of probe particles (R = 50 nm) (Elbaum-Garfinkle et al., 2015, Zhang et al., 2015). These data reveal that NPM1 droplets exhibit a diffusive exponent of α=0.92±0.06, (red symbols, Fig. 2E), consistent with that of a simple viscous liquid for which α= 1. We can thus calculate a viscosity of NPM1 droplets, η=0.74±0.06 Pa-s, which is several hundred times more viscous than water (Fig. S2C). By contrast, probe particle motion in FIB1 droplets is significantly reduced (green symbols, Fig. 2E), in agreement with the slowed coalescence dynamics observed with FIB1 droplets. Interestingly, FIB1 droplet microrheology reveals a sub-diffusive exponent (α=0.5±0.1), which implies that these are not simple viscous liquid droplets, but are instead viscoelastic.
To determine how FIB1 droplet viscoelasticity may arise, we performed FRAP experiments on phase-separated in vitro droplets (Fig. 3). After 30 minutes of initiating phase separation, we find that NPM1 has near complete recovery (84±3%) on very short time scales, with a recovery constant of τ=23 ±1 s (Fig. 3A, S3C); this is consistent with the purely viscous microrheology results, as well as the nearly complete NPM1 FRAP recovery in vivo. By contrast FIB1 has low recovery (37±2%) with a time scale of τ=56 ±5 s (Fig. 3B, S3C). Such incomplete FRAP recovery is expected for a viscoelastic material, since not all molecules exhibit dynamic, fluid-like exchange. Moreover, this in vitro FIB1 behavior agrees well with the incomplete FIB1 FRAP recovery in vivo, in both cultured mammalian cells expressing FIB::GFP and amphibian nucleoli (Fig. 3F).
Figure 3. Nucleolar protein droplets exhibit liquid-like dynamics, but FIB1 shows evidence for aging.
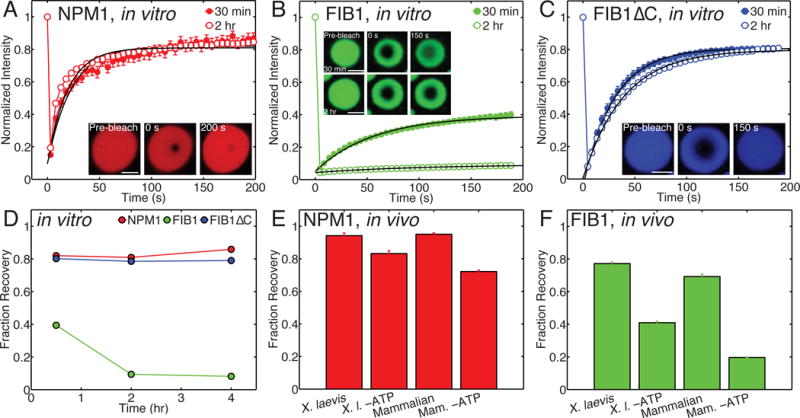
(A–C) FRAP recovery curves for NPM1 (red), FIB1 (green), and FIB1ΔC (blue) droplets, 30 minutes (closed circles) and 2 hours (open squares) after phase separation was initiated. (A) Inset: example FRAP timecourse. Scale bar = 5 μm. (B) Insets: example FRAP timecourses after 30 minutes (top) and 2 hours (bottom). Scale bar = 2 μm. (C) Inset: example FRAP timecourse. Scale bar = 2 μm. (D) Fraction recovery after FRAP experiment as a function of time after phase separation for NPM1 (red), FIB1 (green), and FIB1ΔC (blue) droplets. (E,F) Fraction recovery for NPM1 (E) and FIB1 (F) in X. laevis nucleoli and mammalian cell culture nucleoli in vivo, for native and ATP depletion conditions. See also Figure S3.
To test whether droplet material properties change with time, we performed FRAP experiments on in vitro droplets as a function of time. We find that NPM1 always exhibits a near complete recovery, even for droplets that have been sitting for several hours (Fig. 3A, 3D). However, FIB1 FRAP dynamics are strongly impacted by the droplet age. By 2 hours, the percent recovery has dropped by a factor of ~4, to 8±0.5% (Fig. 3B, 3D); this suggests that these droplets become increasingly solid-like with time, potentially due to the formation of fibers. Consistent with this, we see liquid-like FIB1 droplets evolve into sticky gel-like structures over a 2 hour time period (Fig. S3A). Also, we find that replacing the GFP tag with the monomeric GFP (A206K) did not alter the FIB1 aging behavior (Fig. S3B). As we describe further below, the N-terminal R/G-rich domain of FIB1, designated as FIB1ΔC, drives phase separation as an autonomous unit. However, it exhibits nearly complete FRAP recovery (Fig. 3C, 3D). Moreover, unlike full length FIB1, the percent recovery is stable over four hours (Fig. 3D). Therefore, we conclude that the C-terminal methyltransferase domain of FIB1 plays a key role in promoting viscoelastic maturation of FIB1 droplets in vitro.
ATP-dependent active processes have been hypothesized to play an important role in promoting dynamics within cells, in a process known as “active diffusion” (Brangwynne et al., 2011, Parry et al., 2014, Kroschwald et al., 2015). Depleting ATP from X. laevis ooctyes or mammalian cells (Fig. 3E) had relatively little effect on FRAP recovery of NPM1, with nearly full FRAP recovery, comparable to that seen with in vitro NPM1 droplets (Fig. 3A, 3D). By contrast, ATP-depletion resulted in significantly slowed FIB1 dynamics, with longer recovery times in X. laevis (Fig. S3C), and a 2-3-fold decrease in the percent recovery in both systems (Fig. 3F). Moreover, the low percent recovery of FIB1 in ATP-depleted cells (20–40%) is similar to that measured for in vitro FIB1 droplets (Fig. 3D, 3F). This suggests that ATP-dependent enzymatic activity is essential for actively maintaining the fluidity of the aging-prone, FIB1-rich DFC.
In vitro FIB1 and NPM1 coexist as multiphase droplets
Given that FIB1 and NPM1 individually phase separate into liquid droplets in the presence of rRNA, we next tested how these proteins behave when mixed together. At relatively low concentrations, both proteins colocalize in the same condensed droplets; depending on the relative amount of FIB1 to NPM1, these droplets are either enriched in FIB1 (FIB1-rich/NPM1-lean phase) (Fig. 4B, S4B) or they are enriched in NPM1 (FIB1-lean/NPM1-rich phase) (Fig. 4C, S4C). Thus, considering the soluble “buffer” phase, in these cases the system still resides within a two-phase region of the phase diagram (Fig. 4D). However, when these proteins are both mixed at relatively high concentrations, we observe a three-phase system with both FIB1-rich/NPM1-lean droplets coexisting with FIB1-lean/NPM1-rich droplets, surrounded by the buffer phase (Fig. 4A). Interestingly, the NPM1 rich phase tends to partially envelope the FIB1 rich phase (Fig. S4A); in the absence of the GFP tag, this envelopment becomes even more pronounced, with FIB1 droplets fully embedded within NPM1 (Fig. S4D). This droplet organization is very similar to what is observed in X. laevis and mammalian nucleoli, where the FIB1 DFC is always internalized within the NPM1 GC. A phase diagram can be constructed by determining the threshold concentrations of FIB1 and NPM1 required to phase separate into a three-phase system (Fig. 4D). These results suggest that the “layered” structural organization of nucleolar proteins could be self-organized by liquid-liquid phase separation alone.
Figure 4. FIB1 and NPM1 form immiscible droplets in vitro and in vivo.
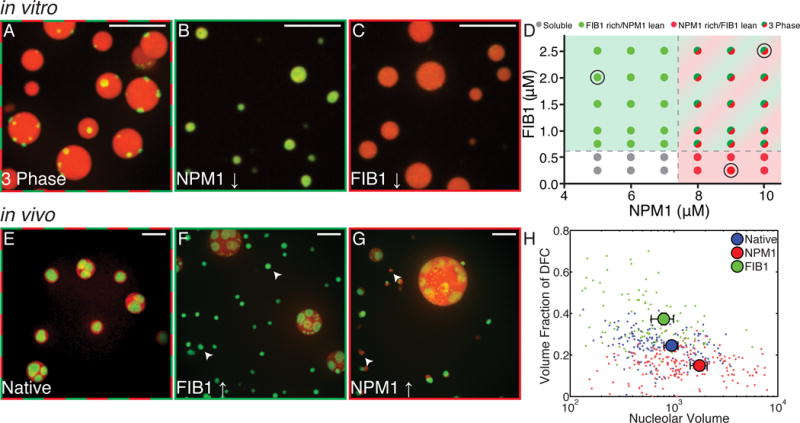
(A–C) In vitro images of mixtures of purified NPM1 and FIB1. Scale bar = 10 μm. (A) High concentrations of both proteins (FIB1: 2.5 μM, NPM1: 10 μM) give rise to FIB1-rich droplets (green) which are immiscible with and partially enveloped by NPM1-rich droplets (red). (B) For much lower concentrations of NPM1 (NPM1: 5 μM, FIB1: 2 μM) only FIB1-rich/NPM1-lean droplets are observed. (C) For much lower concentrations of FIB1 (FIB1: 0.25 μM, NPM: 9 μM) only NPM1-rich/FIB1-lean droplets are observed. (D) Phase diagram for varying concentrations of NPM1 and FIB1 in vitro. Colors represent observed phase (gray = soluble phase, green = FIB1 rich/NPM1 lean phase, red = NPM1 rich/FIB1 lean phase, and red/green = three phase). Black circles indicate concentrations shown in A, B, and C. (E–G) Images of nucleoli in X. laevis; red=NPM1, green=FIB1. Scale bar = 10 μm. (E) Untreated nuclei. (F) Nuclei after microinjection of FIB1 (G) Nuclei after microinjection of NPM1. (H) Volume fraction of the DFC (identified by FIB1) in each nucleolus for native nuclei (blue), after NPM1 injection (red) and after FIB1 injection (green). Large symbols represent mean ± s.e.m. See also Figure S4.
To test whether qualitatively similar phase behavior may be observed upon changing the relative protein concentrations in living cells, we injected nucleolar proteins into living X. laevis nuclei (Fig. 4E). Consistent with the expectation from equilibrium phase coexistence, we observed that the volume fraction of the corresponding component increased after microinjection (Fig. 4H). Typically, the DFC visualized by FIB1 is 25±2% (mean ± s.e.m.) of the volume in the nucleolus. When more FIB1 was injected, the protein localized preferentially to the DFC causing the fibrillar cores to increase in size, occupying about 37±3% (mean ± s.e.m.) of the volume. Conversely, when NPM1 was injected, the protein localized preferentially to the GC and caused the nucleoli to swell to large sizes, causing the fibrillar cores to occupy a lower volume fraction of only 15±1% (mean ± s.e.m.). Moreover, small extranucleolar droplets of the respective protein appeared to form de novo (Fig. 4F, 4G). This is possible if the saturation concentration in the nucleoplasm has been reached, causing spontaneous condensation of nucleolar proteins, without requiring NORs for nucleation (Berry et al., 2015).
Protein domains required for phase separation and immiscibility
To gain insight into the molecular-scale driving forces underlying phase separation and droplet immiscibility, we created deletion mutants of both FIB1 and NPM1 that contained individual domains. Full-length FIB1 consists of two domains: a disordered N-terminal arginine (R)/glycine (G)-rich domain with low-sequence complexity (R/G domain) and an RNA methyltransferase domain (MD) that together with small nucleolar RNA (snoRNA) can methylate substrate rRNA (Fig. 5A). We find that the R/G domain (FIB1ΔC) is sufficient to form liquid-like droplets in vitro, while the MD alone (FIB1ΔN) is unable to form droplets in vitro (Fig. 5A). Interestingly, FIB1ΔC can phase separate into liquid-like droplets in vitro, even in the absence of RNA (Fig. S2E). By contrast, full length FIB1 requires rRNA; however this may be a non-specific consequence of the polyanionic nature of rRNA since heparin can also drive phase separation of full length FIB1 (Fig. S2A, S2F). Furthermore, we find that FIB1ΔC droplets undergo homotypic fusion with an inverse capillary velocity of η/γ=0.5 ±0.04 s/μm (95% confidence interval); these dynamics are significantly faster than for full length FIB1 (η/γ≈40±10 s/μm), and comparable to NPM1 (η/γ≈0.3±0.07 s/μm) (Fig. 2C, 2D, S2C). Consistent with this, in microrheology experiments, FIB1ΔC droplets also exhibit faster dynamics than full length FIB1 (blue symbols, Fig. 2E, S2C).
Figure 5. Molecular mechanism of phase partitioning in X. laevis oocytes.
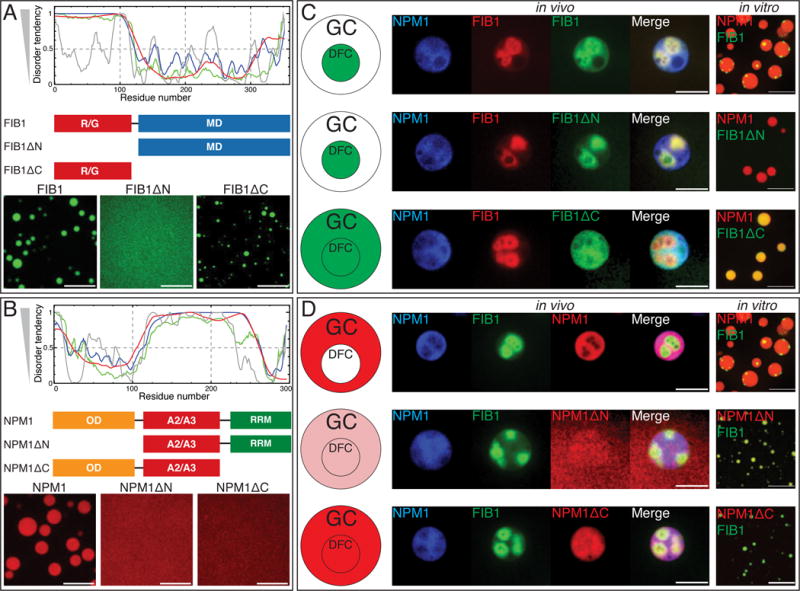
(A) Domain analysis of FIB1. Plot shows predicted disorder across full length FIB1 using various algorithms, P-FIT (green line), VSL2B (blue line), VL3 (red line), and VLXT (grey line). Schematic diagrams show three constructs: FIB1 full length, R/G deletion (FIB1ΔN), and deletion of MD (FIB1ΔC), with images below testing for constructs’ ability to form droplets. Scale bar = 10 μm. (B) Domain analysis of NPM1. Predicted disorder across full length NPM1 for the four algorithms. Schematic diagrams show three constructs: NPM1 full length, oligomerization deletion (NPM1ΔN), and RNA binding deletion (NPM1ΔC) with images below testing for constructs’ ability to form droplets. Scale bar = 10 μm. (C,D) The left most panel shows schematic summary of center panels. Center panels contain images from X. laevis nucleoli in vivo. Left channel contains expression of mRNA for NPM1::Cerulean, followed by expression of mRNA for FIB1::RFP or GFP, followed by injection of various constructs (FIB1, FIB1ΔN, FIB1ΔC, NPM1, NPM1ΔN, NPM1ΔC), and final image is the overlay of all three channels. Scale bar = 10 μm. The right most panel shows in vitro images of FIB1 or mutants (green) mixed with NPM1 droplets (red) or NPM1 or mutants (red) mixed with FIB1 droplets (green). Scale bar = 10 μm. See also Figure S5.
When these mutant proteins were injected into X. laevis nuclei, FIB1ΔN strongly partitions to the DFC, similar to the full-length FIB1 protein (Fig. 5C). Similarly, we observed that FIB1ΔN does not colocalize with NPM1 droplets in vitro (Fig. 5C, S5A); DFC enrichment of FIB1ΔN in vivo may not reflect immiscibility, but may instead reflect co-recruitment due to the presence of full length FIB1 in the native system. Moreover, we observed that injected FIB1ΔC localizes to the entire nucleolus, and has nonspecific interactions with the GC and DFC. Consistent with this, we find that the FIB1ΔC colocalizes with in vitro NPM1 droplets, rather than forming a third immiscible droplet phase (Fig. S5A). Taken together, these data suggest that the N-terminal R/G domain of FIB1 is sufficient for droplet formation, but does not encode for a separate liquid-like DFC sub-compartment; instead, the C-terminal MD of FIB1, which alone is not sufficient for droplet formation, confers immiscibility with proteins in the GC.
We next probed the importance of the three domains of NPM1: an N-terminal oligomerization domain (OD) which has been shown to be necessary to form an ordered pentameric structure (Mitrea et al., 2014), a central disordered domain containing acidic tracts (A2/A3), and a C-terminal RNA binding domain (RRM) (Fig. 5B). Furthermore, the OD of NPM1 can form a pentamer to generate multivalency and could potentially increase the affinity of its RRM domain for rRNA. We deleted the N-terminal oligomerization domain to create NPM1ΔN, and we deleted the C-terminal RNA binding domain to create NPM1ΔC. We find that neither mutant is able to form droplets in vitro, consistent with phase separation of NPM1 requiring the oligomerization of NPM1 into multivalent pentamers that can bind to rRNA (Mitrea et al., 2016).
When NPM1ΔN is injected into X. laevis nuclei, we find that it localizes only very weakly to the nucleolus (Fig. 5D). This is consistent with the finding that pentameric state of NPM1 is necessary to retain this protein in the nucleolus (Mitrea et al., 2016). When NPM1ΔN is mixed with FIB1 droplets in vitro, we see strong co-localization of NPM1ΔN within FIB1 droplets (Fig. 5D, S5A). To determine whether this strong co-localization is associated with the presence of rRNA in FIB1 droplets, we used poly-U50 to drive the phase-separation of FIB1. Interestingly, we find that the localization of NPM1ΔN within FIB1 droplets is reduced significantly with poly-U50 (Fig. S5B); this suggests that the NPM1ΔN can localize within FIB1 droplets through its RRM interacting with rRNA. When NPM1ΔC is injected into X. laevis oocytes, we see that the protein strongly localizes to both the GC and DFC. However, NPM1ΔC localizes very weakly to FIB1 droplets in vitro (Fig. 5D, S5A); this in vitro co-localization does not appear to be affected by the types of RNA used, suggesting that the interaction between NPM1ΔC and FIB1 in vivo is not driven by RNA, since NPM1ΔC lacks an RRM (Fig. S5C), but rather by interactions between the R/G domain of FIB1 and the OD/A2/A3 domains of NPM1 (Mitrea et al., 2014).
In summary, our domain analysis supports three key conclusions: 1) the disordered R/G domain of FIB1 can drive phase separation, but the time-dependent viscoelastic properties of full-length FIB1 require the RNA-binding MD; 2) the disordered domains of both FIB1 and NPM1 appear capable of localizing equally to both components of nucleoli; and 3) the RNA binding domains, multivalent in the case of NPM1, play a key role in driving each protein to their respective sub-compartment.
A minimalist computational model for three-phase behavior
Our data lead to the hypothesis that spatial organization within the nucleolus derives from the sequence-encoded interaction preferences of the different domains of nucleolar proteins. To test this hypothesis, we asked if the observed spatial organization could be reproduced in coarse-grained computer simulations. The simulation is comprised of 900 of each of the three polymers, performed on three-dimensional lattices to reduce the computational complexity. FIB1 and rRNA were modeled as linear polymers of interaction modules, while the pentameric nature of NPM1 was captured using a branched polymer with five arms, each comprising the appropriate number of interaction modules (Fig. 6A).
Figure 6. Preferential interaction model captures the formation of spatially organized droplets for the ternary system comprising of FIB1, NPM1, and rRNA.
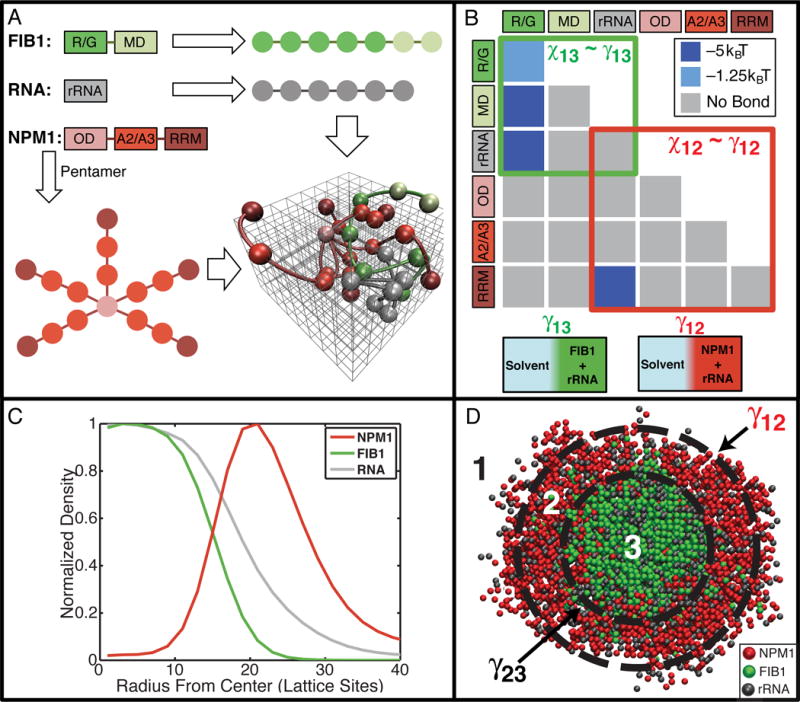
(A) Mapping of the sequences of FIB1, NPM1, and rRNA to linear/branched polymers of modules on three-dimensional lattices. FIB1 is modeled as a linear polymer comprising of seven modules, five corresponding to the R/G domain and two corresponding to the MD. Similarly, the rRNA sequence is modeled as a linear polymer comprising six modules. NPM1 is modeled as a branched polymer with five arms. Here, the ODs of five NPM1 molecules occupy the base for each branch; two modules correspond to the intrinsically disordered acid-rich regions (A2/A3) and a single module captures the RNA recognition module (RRM). A representative snapshot is shown of polymers on the cubic lattice. (B) The matrix of module interaction strength. (C) The normalized mean radial density of FIB1 (green), NPM1 (red), and RNA (grey) for representative largest cluster observed throughout a simulation. (D) Visual depiction of a slice through representative phase separated droplet; FIB1 (green) and NPM1 (red). See also Figure S6 and Movie S5.
Interactions between modules of each of the protein- and RNA-like polymers are governed by parameters of an interaction matrix. These parameters represent effective pairwise affinities in the presence of the competing effects of module-solvent and module-module interactions (Fig. 6B). The interaction matrix in Fig. 6B is sufficient to reproduce the totality of experimental observations. Fig. 6C shows the normalized density profiles of FIB1, NPM1, and rRNA within droplets that form in the simulations, revealing a FIB1-rich core and NPM1-rich outer shell, with rRNA distributed across the two regions; Fig. 6D shows a representative cutaway snapshot from the simulations (Supplemental Movie S5). Numerous distinct matrix parameterizations fail to reproduce one or more aspects of the in vitro data, although there are other specific choices of matrix parameters that do reproduce all of the experimental data (Fig. S6).
An exploration in the space of interaction matrix parameters suggests that the computational model must include three necessary features in order to reproduce all of the in vitro data. First, the R/G modules must have favorable homotypic interactions. Second, the OD of NPM1 should generate the requisite multivalency of RRM modules that drives the phase separation of NPM1 through interactions with rRNA. Third, the A2/A3 modules of NPM1 must be preferentially solvated, thus ensuring that they form weak or no bonds (Fig. 6B and S6L).
The minimalist model supports the presence of three distinct phases: Phase 1 is the solvent and includes water plus the solution ions; Phase 2 is NPM1 + rRNA; and Phase 3 is FIB1 + rRNA. The balance of interactions can be quantified in terms of pairwise interaction coefficients designated as χ12 (Solvent-NPM1), χ13 (Solvent-FIB1) and χ23 (NPM1-FIB1) that are derived from the Flory-Huggins theory for polymer solutions and blends (Rubinstein and Colby, 2003). The individual χ values quantify the free energy gained or lost when modules exchange homotypic interactions for heterotypic ones. A direct consequence of the structure of the interaction matrix (Fig. 6B) is that the values of each of χ12, χ13, and χ23 are positive. The three-phase behavior observed in experiments and reproduced in simulations implies that χ values must obey the relation: χ13 > χ12 > χ23 > 0. Since surface tension is directly proportional to the Flory parameter, γ~χ, it follows that γ13 > γ12 > γ23 i.e., the surface tension of FIB1 droplets is larger than that of NPM1 droplets. It is thus energetically more favorable to envelope the FIB1 droplet within the NPM1 droplet, as opposed to NPM1 being enveloped within a FIB1 droplet (Fig. S7D).
Nucleolar organization arises from differential surface tension of sub-compartments
The physical picture that emerges from our computational model is consistent with the very low values we obtained for the surface tension of the in vitro NPM1 droplets, as well as for the in vivo NPM1-rich GC. Unfortunately, the small size of in vitro FIB1 droplets, as well as their viscoelasticity, makes it difficult to undertake direct surface tension measurements. As an alternate route to evaluate the relative droplet surface tensions, we measured droplet wetting behavior on hydrophobic and hydrophilic coverslips. Wetting refers to the contact between liquids and surfaces and is a consequence of surface tension; for example, water droplets will spread over a Pluronic-treated hydrophilic surface (low contact angle), while water droplets will round up and avoid contact with a Sigmacote-treated hydrophobic surface (high contact angle), as shown in Fig. 7A,B.
Figure 7. Surface tension drives organization of multiphase droplets.
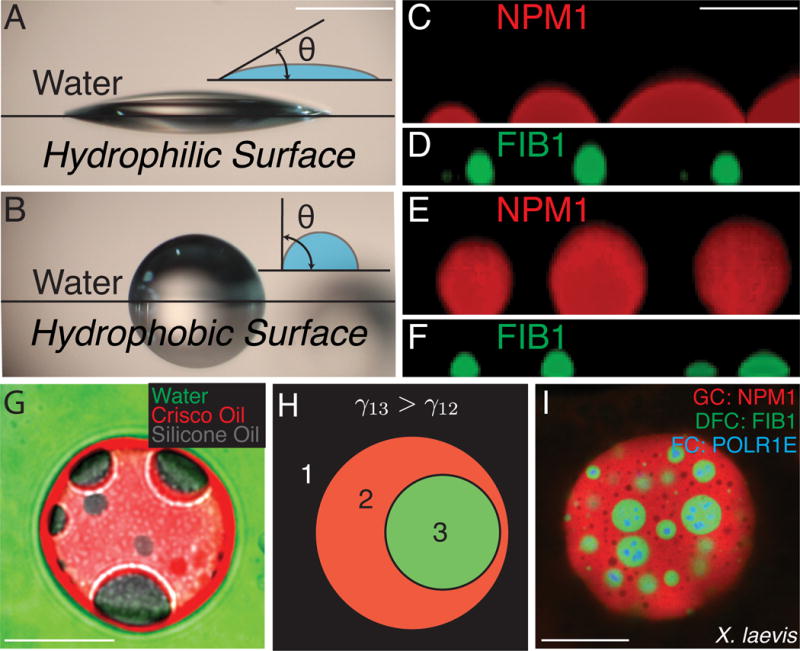
(A–F). Images of droplets on hydrophilic surfaces (Pluronic-treated, A) or hydrophobic surfaces (Sigmacote-treated, B). (A) Water droplet on hydrophilic surface. (B) Water droplets on hydrophobic surface. Scale bar for A, B = 1 mm. (C) NPM1 droplets on hydrophilic surface. (D) FIB1 droplets on hydrophilic surface. (E) NPM1 droplets on hydrophobic surface. (F) FIB1 droplets on hydrophobic surface. Scale bar for C–F= 5 μm. (G) Image of non-biological multiphase droplets: green=water, red=Crisco oil, and gray=silicone oil. Scale bar = 20 μm. (H) Schematic organization of immiscible multiphase droplets. The more hydrophobic phase (green), has a higher surface tension with water than the more hydrophilic phase (red), which has a lower surface tension with water. (I) Image of multiphase nucleoli after actin disruption in X. laevis. Scale bar = 20 μm. See also Figure S7.
On hydrophobic surfaces, we find that NPM1 droplets behave as water droplets and exhibit minimal wetting, with a contact angle of 130±10° (mean±s.d) (Fig. 7E, S7A,C). On hydrophilic surfaces they exhibit increased wetting, with a contact angle of 60±10° (Fig. 7C, S7A,C). In contrast, FIB1 droplets tended to better wet the hydrophobic coverslips, with a contact angle of 70±10° (mean±s.d) (Fig. 7F, S7A,C), as compared to the hydrophilic coverslips, on which they exhibited a contact angle of 130±10° (Fig. 7D, S7A,C). The differential hydrophobicity of NPM1 and FIB1 droplets explains our key observation, which we describe in vivo (Fig. 1 A–G), in vitro (Fig. 4A), and also in silico (Fig. 6C–D): FIB1 and NPM1 form multiphase droplets where FIB1 is at least partially encapsulated by NPM1 (Fig. 7I). This organization is quite similar to how immiscible liquids are organized in non-biological multiphase systems (Neeson et al., 2012). To demonstrate this with a simple example, we prepared a system of water, Crisco vegetable oil, and silicone oil, which are immiscible liquids with known surface tensions (Than et al., 1988) (Fig. 7G). Silicone oil is more hydrophobic than Crisco oil, i.e. γsilicone/water > γCrisco/water, and as a result, the silicone oil droplets are always enveloped by the Crisco oil droplet. Similarly, since FIB1 is more hydrophobic than NPM1, γFIB1/water > γNPM1/water, FIB1 droplets will tend to be encapsulated within NPM1 droplets (Fig. 7H, S7D). We note that in both cases there is also the requirement that a third surface tension, γsilicone/Crisco or γFIB1/NPM1, must not be too high, or the two droplets would never interact (Fig S7D).
Discussion
The nucleolus is the most prominent of numerous membrane-less RNP bodies and was recognized over 150 years ago by early microscopists (Pederson, 2011). However, a mechanistic biophysical understanding of the principles governing the well-known sub-compartmental organization has been elusive (Thiry and Lafontaine, 2005). Our findings suggest that these distinct compartments arise as a consequence of liquid phase immiscibility, supported by: 1) The different layers can undergo coalescence upon contact, relaxing toward round droplet-like structures; 2) Protein components of the different layers are highly dynamic as determined by FRAP; 3) When expressed and purified, key enriched nucleolar proteins undergo phase separation into droplets with properties comparable to those found in their corresponding nucleolar sub-compartment; 4) When mixed, purified proteins exhibit phase immiscibility, resulting in sub-compartmentalized in vitro droplets that are strikingly similar to in vivo nucleoli; 5) A minimalist coarse-grained model is sufficient for reproducing phase immiscibility and the observed colocalization of different truncation constructs, and further shows how a network of molecular interactions determines surface tensions, which dictate the core-shell droplet architecture; and 6) Biophysical characterization and droplet wetting behavior indicate that the FIB1/DFC phase exhibits a higher surface tension than the NPM1/GC phase, which results in the former being embedded within the latter.
What are the functional implications of liquid phase immiscibility and surface-tension driven sub-compartmentalization of the nucleolus? The most important nucleolar function is ribosome biogenesis (Boisvert et al., 2007), which involves the coordinated modification and assembly of rRNA through multiple processing steps (Tschochner and Hurt, 2003, Henras et al., 2008). Analogous to an assembly line or the staged processes in a modern chemical plant, the spatial separation and distinct physical and compositional features of the FIB1-rich DFC and NPM1-rich GC may tune the vectorial transport and associated processing of rRNA into mature pre-ribosomal particles. Indeed, continuous transcription within the FC causes radial flux of rRNA through the DFC into the GC and finally into the nucleoplasm. The DFC is effectively an enzymatic bath, which facilitates correct base-pairing with small nucleolar guide RNAs (snoRNAs), for example those associated with methylation by FIB1 as part of a box C/D snoRNP and pseudouridinylation by H/ACA snoRNPs, as well as cleavage reactions and other rRNA modifications (Henras et al., 2008). These modifications are critical for correct rRNA folding and stability, subsequent assembly with ribosomal proteins, and ultimately ribosome function (i.e., translational fidelity) (Gigova et al., 2014).
We propose that the viscoelastic properties of the DFC serve to lower the flux of incompletely or incorrectly processed/folded pre-ribosomal particles, ensuring that DFC-associated enzymatic processes are completed, before passage of pre-ribosomal particles into the outer GC layer, where they encounter NPM1 and early binding ribosomal proteins. Indeed, NPM1 phase separates with both rRNA and ribosomal proteins (Mitrea et al., 2016), consistent with fluorescence imaging studies suggesting that ribosomal proteins localize to the GC, but not the DFC (Kruger et al., 2007). Our findings indicate that pentameric NPM1 is integral to the fluid features of the GC, whose relatively low viscosity may allow ribosomal proteins to remain dynamically accessible to pre-ribosomal particles emerging from the DFC.
Our data show that FIB1 droplets, but not NPM1 droplets, are metastable and can age with time both in vitro and in vivo. These data are consistent with the hypothesis that disordered regions can facilitate the transition from liquid-like to solid-like structures (Weber and Brangwynne, 2012), which is supported by recent studies on a variety of RNA binding proteins (Zhang et al., 2015, Patel et al., 2015, Lin et al., 2015, Molliex et al., 2015, Xiang et al., 2015). We note that the name “Fibrillarin” was given due to its localization to fibrillar structures within the DFC (Ochs et al., 1985); these structures could reflect droplet aging/fibril formation. Consistent with this, we observe apparent aging of nucleoli in non-dividing C. elegans intestinal cells: FIB1 recovers less completely in older adult worms compared with younger larvae (Fig. S7E). However, given that RNA can impact the fluidity of related phase separated droplets (Elbaum-Garfinkle et al., 2015, Zhang et al., 2015), the rate of FIB1-rich DFC aging could be impacted by the flux of newly synthesized rRNA transiting through the DFC. Future studies will be required to elucidate the biophysical origin of nucleolar droplet maturation and the role of RNA transcription and other ATP-dependent processes.
Our findings underscore the importance of surface tension, whose role is well established in physical systems, as is readily visualized with immiscible oils in water (Fig. 7G). In living systems, effective surface tension may be important for the organized demixing of cell populations: cell types that exhibit a relatively high apparent surface tension will tend to be enveloped by cell types with a relatively low apparent surface tension (Foty et al., 1996). Our data show that this same basic principle is important for structuring the nucleolus, with possible implications for other RNP bodies. For example, histone locus bodies (HLBs) in the frog nucleus are commonly found to have B-snurposomes attached to their surface (Gall, 2000); incomplete internalization of B-snurposomes suggests that their surface tension may be similar to HLBs. Interestingly, this partial internalization is reminiscent of the altered nucleolar structure observed in actinomycin-D treated nucleoli, wherein rRNA transcriptional inhibition results in a more lobulated nucleolus (Shav-Tal et al., 2005, Wachtler and Stahl, 1993). A similar organization is also seen with processing bodies (Kedersha et al., 2005, Buchan and Parker, 2009, Hubstenberger et al., 2013). Recently, stress granules have been shown to contain less dynamic cores, which exhibit a qualitative similarity to the FIB1/DFC cores of the nucleolus (Jain et al., 2016). Building on the biophysical groundwork we have laid here, it may be possible to alter or even invert the organization of such RNP bodies, by using surfactants to modulate droplet surface tensions; this could significantly impact sequential RNA processing steps and the overall flow of genetic information.
Organelle sub-compartmentalization is well-known in membrane-bound organelles, such as mitochondria. Our data show that membrane-less liquid phase organelles can also generate significant substructure. Phase separation and the coexistence of multiple distinct liquid RNA/protein phases thus provide a simple but elegant mechanism for the cell to control the spatial localization and processing of molecules, without relying on membrane boundaries.
Experimental Procedures
Detailed methods are available in the Supplemental Experimental Procedures
Preparation of X. laevis, mammalian, and C. elegans nucleoli
Frogs were anesthetized with 0.1% MS-222 solution, and oocytes were surgically removed from female X. laevis frogs following an IACUC approved protocol. mRNA of endogenous proteins (FIB1, NPM1, and POLR1E) with fluorescent tags and recombinant proteins were microinjected into oocytes. Nuclei were manually dissected in mineral oil and subsequently imaged. Actin was disrupted using Lat-A, and ATP was depleted using Apyrase. Mammalian cells expressing fluorescent fusion proteins (FIB1 and NPM1) were maintained at 37°C using standard conditions, and ATP was depleted using sodium azide and deoxyglucose. C. elegans expressing intestinal FIB1::GFP were maintained at 20°C under standard conditions and anesthetized with levamisole in M9 prior to imaging.
Purification and phase separation of in vitro droplets
FIB1 and NPM1 variants were expressed using a standard E. coli expression system, purified using either a 6×-His or GST tag, and stored in a high salt buffer. Phase separation was initiated by lowering the salt concentration of stock protein in the presence or absence of rRNA. Non-biological multiphase droplets were obtained by vortexing water, Crisco oil, and silicone oil.
Biophysical characterization of in vivo and in vitro droplets
For fusion relaxation experiments, the aspect ratio was measured as a function of time for droplets of different size to obtain the inverse capillary velocity. Surface tension of non-wetting droplets with measured densities was estimated from non-spherical XZ shape profiles obtained using a right-angle prism. For microrheology experiments, time-lapse images of fluctuating R=50 nm particles inside protein droplets were acquired and analyzed using particle-tracking Matlab code to obtain the mean squared displacement as a function of lag time; from the Stokes-Einstein relation, the viscosity was determined. For FRAP experiments, 1 μm spots inside in vivo and in vitro droplets were photobleached, and percent fluorescent recovery and recovery times were determined using standard techniques. Wetting behavior of in vitro droplets was observed for surfaces treated with Sigmacote (hydrophobic) or Pluronic (hydrophilic), and the contact angle was measured at the interface between the glass and line tangent to the droplet.
Supplementary Material
Acknowledgments
We thank members of the Brangwynne laboratory for discussions and Adrienne Fung for preliminary work on surface tension measurements. We acknowledge funding from the Princeton Center for Complex Materials, a MRSEC supported by NSF Grant DMR 1420541. This work was also supported by an NIH Director’s New Innovator Award (1DP2GM105437-01) (C.P.B.), an NSF CAREER award (1253035) (C.P.B.), a Helen Hay Whitney Fellowship (N.V.), an NSF grant (MCB 1121867) (R.V.P.), an NIH grant (5RO1NS056114) (R.V.P.), an NIH grant (5R01GM115634) (R.W.K.), an NCI Cancer Center Support grant (P30CA21765 at St. Jude Children’s Research Hospital) (R.W.K.), and ALSAC (R.W.K.).
Footnotes
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five movies.
Author Contributions
M.F. performed and analyzed data in X. laevis, in vitro surface tension and microrheology experiments, a non-biological multiphase system, and contact angle experiments. N.V. performed and analyzed in vitro experiments with FIB1 and NPM1 and FRAP experiments in C. elegans. T.S.H. performed computational simulations and analysis. D.M.M. created in vitro NPM1 constructs and created NPM1 phase diagram. L.Z. analyzed experiments in mammalian tissue culture cells. T.M.R. performed and analyzed FRAP experiments in X. laevis with help from M.F. and ATP depletion in mammalian tissue culture cells with help from L.Z. C.P.B, M.F., and N.V. designed the study and wrote the paper with help from all authors.
References
- AARTS DG, SCHMIDT M, LEKKERKERKER HN. Direct visual observation of thermal capillary waves. Science. 2004;304:847–850. doi: 10.1126/science.1097116. [DOI] [PubMed] [Google Scholar]
- BALAGOPAL V, PARKER R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Current opinion in cell biology. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERRY J, WEBER SC, VAIDYA N, HAATAJA M, BRANGWYNNE CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proceedings of the National Academy of Sciences. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOISVERT FM, VAN KONINGSBRUGGEN S, NAVASCUES J, LAMOND AI. The multifunctional nucleolus. Nat Rev Mol Cell Bio. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- BRANGWYNNE CP, ECKMANN CR, COURSON DS, RYBARSKA A, HOEGE C, GHARAKHANI J, JULICHER F, HYMAN AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- BRANGWYNNE CP, MITCHISON TJ, HYMAN AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCHAN JR, PARKER R. Eukaryotic stress granules: the ins and outs of translation. Molecular cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DERENZINI M, TRERE D, PESSION A, GOVONI M, SIRRI V, CHIECO P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. The Journal of pathology. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- ELBAUM-GARFINKLE S, KIM Y, SZCZEPANIAK K, CHEN CC-H, ECKMANN CR, MYONG S, BRANGWYNNE CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences. 2015:201504822. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERIC M, BRANGWYNNE CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nature Cell Biology. 2013;15:1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERIC M, BROEDERSZ CP, BRANGWYNNE CP. Soft viscoelastic properties of nuclear actin age oocytes due to gravitational creep. Scientific reports. 2015;5 doi: 10.1038/srep16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOTY RA, PFLEGER CM, FORGACS G, STEINBERG MS. Surface tensions of embryonic tissues predict their mutual envelopment behavior. Development. 1996;122:1611–1620. doi: 10.1242/dev.122.5.1611. [DOI] [PubMed] [Google Scholar]
- FRANK DJ, ROTH MB. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. The Journal of cell biology. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALL JG. Cajal bodies: the first 100 years. Annual review of cell and developmental biology. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- GIGOVA A, DUGGIMPUDI S, POLLEX T, SCHAEFER M, KOS M. A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA. 2014;20:1632–1644. doi: 10.1261/rna.043398.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENNIG S, KONG G, MANNEN T, SADOWSKA A, KOBELKE S, BLYTHE A, KNOTT GJ, IYER KS, HO D, NEWCOMBE EA. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. The Journal of cell biology. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENRAS A, SOUDET J, GERUS M, LEBARON S, CAIZERGUES-FERRER M, MOUGIN A, HENRY Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and Molecular Life Sciences. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUBSTENBERGER A, NOBLE SL, CAMERON C, EVANS TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Developmental cell. 2013;27:161–173. doi: 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAIN S, WHEELER JR, WALTERS RW, AGRAWAL A, BARSIC A, PARKER R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016 doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEDERSHA N, STOECKLIN G, AYODELE M, YACONO P, LYKKE-ANDERSEN J, FRITZLER MJ, SCHEUNER D, KAUFMAN RJ, GOLAN DE, ANDERSON P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. The Journal of cell biology. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROSCHWALD S, MAHARANA S, MATEJU D, MALINOVSKA L, NUSKE E, POSER I, RICHTER D, ALBERTI S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRUGER T, ZENTGRAF H, SCHEER U. Intranucleolar sites of ribosome biogenesis defined by the localization of early binding ribosomal proteins. The Journal of cell biology. 2007;177:573–578. doi: 10.1083/jcb.200612048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI P, BANJADE S, CHENG HC, KIM S, CHEN B, GUO L, LLAGUNO M, HOLLINGSWORTH JV, KING DS, BANANI SF. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN Y, PROTTER DS, ROSEN MK, PARKER R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Molecular cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITREA DM, CIKA J, GUY C, BAN D, BANERJEE P, STANLEY C, NOURSE A, DENIZ A, KRIWACKI R. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R rich linear motifs and rRNA. Elife. 2016:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITREA DM, GRACE CR, BULJAN M, YUN MK, PYTEL NJ, SATUMBA J, NOURSE A, PARK CG, BABU MM, WHITE SW. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proceedings of the National Academy of Sciences. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOLLIEX A, TEMIROV J, LEE J, COUGHLIN M, KANAGARAJ AP, KIM HJ, MITTAG T, TAYLOR JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEESON MJ, TABOR RF, GRIESER F, DAGASTINE RR, CHAN DY. Compound sessile drops. Soft Matter. 2012;8:11042–11050. [Google Scholar]
- NOTT TJ, PETSALAKI E, FARBER P, JERVIS D, FUSSNER E, PLOCHOWIETZ A, CRAGGS TD, BAZETT-JONES DP, PAWSON T, FORMAN-KAY JD. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OCHS R, LISCHWE M, SPOHN W, BUSCH H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biology of the Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- PARRY BR, SUROVTSEV IV, CABEEN MT, O’HERN CS, DUFRESNE ER, JACOBS-WAGNER C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATEL A, LEE HO, JAWERTH L, MAHARANA S, JAHNEL M, HEIN MY, STOYNOV S, MAHAMID J, SAHA S, FRANZMANN TM. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- PEDERSON T. The nucleolus. Cold Spring Harbor perspectives in biology. 2011;3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUBINSTEIN M, COLBY RH. Polymer physics 2003 [Google Scholar]
- SHAV-TAL Y, BLECHMAN J, DARZACQ X, MONTAGNA C, DYE BT, PATTON JG, SINGER RH, ZIPORI D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Molecular biology of the cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPECTOR DL. Nuclear domains. Journal of cell science. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- THAN P, PREZIOSI L, JOSEPHL D, ARNEY M. Measurement of interfacial tension between immiscible liquids with the spinning road tensiometer. Journal of colloid and interface science. 1988;124:552–559. [Google Scholar]
- THIRY M, LAFONTAINE DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends in cell biology. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- TSCHOCHNER H, HURT E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends in cell biology. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- WACHTLER F, STAHL A. The nucleolus: a structural and functional interpretation. Micron. 1993;24:473–505. [Google Scholar]
- WANG JT, SMITH J, CHEN BC, SCHMIDT H, RASOLOSON D, PAIX A, LAMBRUS BG, CALIDAS D, BETZIG E, SEYDOUX G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2015;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBER SC, BRANGWYNNE CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- WEBER SC, BRANGWYNNE CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Current Biology. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIPPICH F, BODENMILLER B, TRAJKOVSKA MG, WANKA S, AEBERSOLD R, PELKMANS L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- XIANG S, KATO M, WU LC, LIN Y, DING M, ZHANG Y, YU Y, MCKNIGHT SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG H, ELBAUM-GARFINKLE S, LANGDON EM, TAYLOR N, OCCHIPINTI P, BRIDGES AA, BRANGWYNNE CP, GLADFELTER AS. RNA controls PolyQ protein phase transitions. Molecular cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


