Abstract
Multiple myeloma is incurable by standard approaches because of inevitable relapse and development of treatment resistance in all patients. In our prior work, we identified a panel of macropinocytosing human monoclonal antibodies against CD46, a negative regulator of the innate immune system, and constructed antibody-drug conjugates (ADCs). In this report, we show that an anti-CD46 ADC (CD46-ADC) potently inhibited proliferation in myeloma cell lines with little effect on normal cells. CD46-ADC also potently eliminated myeloma growth in orthometastatic xenograft models. In primary myeloma cells derived from bone marrow aspirates, CD46-ADC induced apoptosis and cell death, but did not affect the viability of nontumor mononuclear cells. It is of clinical interest that the CD46 gene resides on chromosome 1q, which undergoes genomic amplification in the majority of relapsed myeloma patients. We found that the cell surface expression level of CD46 was markedly higher in patient myeloma cells with 1q gain than in those with normal 1q copy number. Thus, genomic amplification of CD46 may serve as a surrogate for target amplification that could allow patient stratification for tailored CD46-targeted therapy. Overall, these findings indicate that CD46 is a promising target for antibody-based treatment of multiple myeloma, especially in patients with gain of chromosome 1q.
Introduction
The treatment of multiple myeloma (MM) has greatly improved in recent years with FDA approval of agents in the immunomodulatory imide drug (IMiD) and proteasome inhibitor drug classes. Nevertheless, myeloma remains incurable, and patients inevitably develop treatment-refractory disease. Furthermore, high-risk cytogenetic subgroups, including those with deletion of chromosome 17p or gain of chromosome 1q21, progress more rapidly through approved agents and have shortened overall survival (1, 2). Therefore, patients with relapsed/refractory (R/R) disease or with poor cytogenetic profiles are in dire need of novel therapies.
Antibody-based therapies have potential to fill this clinical need. Naked antibodies have recently shown increased promise with demonstration of single-agent activities of the anti-CD38 antibodies daratumumab and SAR650984 (3, 4). In addition, the anti–signaling lymphocyte activation molecule family member 7 (anti-SLAMF7) antibody elotuzumab was recently shown to improve outcome in combination with lenalidomide and dexamethasone in a randomized phase III trial (5). Antibody-drug conjugates (ADCs) have potential to further improve on the clinical efficacy of naked antibodies via targeted delivery of highly cytotoxic payloads directly to malignant plasma cells (6–8). ADCs have recently seen proof-of-concept clinical success in Hodgkin lymphoma (brentuximab vedotin) and human epidermal growth factor receptor 2–positive (HER2-positive) breast cancer (ado-trastuzumab emtansine) (9, 10). Because of the considerable potential for clinical benefit, novel ADCs should be evaluated in MM (11).
Our research objective is to identify a novel ADC for MM treatment, with an emphasis toward patients with R/R disease. We previously developed a novel antibody discovery method based on a phage antibody library selection on tissue using laser capture microdissection (12). By this method, antibodies were identified that bind to tumor cells residing in their natural microenvironment (12). The platform was pioneered on prostate cancer tissue. One novel antibody that showed excellent in vivo targeting properties (13) has been identified to target the CD46 antigen (also known as membrane cofactor protein, MCP; Y. Su and B. Liu, unpublished observations). CD46 is a multifunctional protein that has a role in complement inhibition, which may explain its overexpression on malignant cells (14), and cellular entry by pathogens including measles virus (15, 16). The latter quality has led to CD46 targeting in viral immunotherapy with the Edmonston strain of measles virus (17). In normal tissue, CD46 appears to have a low level of expression outside placenta and prostate (14).
The CD46 gene is located on the long arm of chromosome 1 (1q32.2), 50 Mbp from a clinically used FISH probe that may provide a surrogate marker for CD46. FISH is used clinically in MM for risk stratification, and amplification of 1q occurs commonly in MM (1, 18–20). Amplification of 1q21 (amp1q21) is considered a high-risk feature that becomes more prevalent at relapse (1, 2, 21). A recent report also suggests that amp1q21 remains a poor prognostic factor in the era of current protease inhibitor and IMiD therapies (22). Thus, antibody-based targeting of cell surface antigens such as CD46 that undergo genomic copy number gain on chromosome 1q is an attractive possibility in myeloma.
Our selection method preferentially identifies human monoclonal antibodies that are internalized via macropinocytosis, a tumor-selective pathway for cellular entry (23), and thus are well suited for ADC development (24). Therefore, we used these antibodies to develop a novel anti-CD46 ADC with the microtubule inhibitor monomethyl auristatin F (MMAF) (hereafter referred to as CD46-ADC). Herein we describe the preclinical characterization of CD46-ADC in MM using cell lines, orthometastatic xenograft models, and patient samples. We found high CD46 expression on MM cell lines and patient samples, and the expression level is further upregulated upon coculture with bone marrow (BM) stromal cells. CD46-ADC showed potent and specific myeloma cell killing activity in vitro and in vivo and on primary MM cells ex vivo. Thus, CD46-ADC has potential to be an efficacious treatment for MM. Finally, we evaluated the hypothesis that patients with disease demonstrating amp1q21 by FISH also coamplify the CD46 gene and upregulate antigen expression on the MM cell surface. The results support the use of 1q21 FISH as a biomarker for translation of CD46-targeting agents for use in MM.
Results
CD46 antigen is highly expressed in myeloma cell lines.
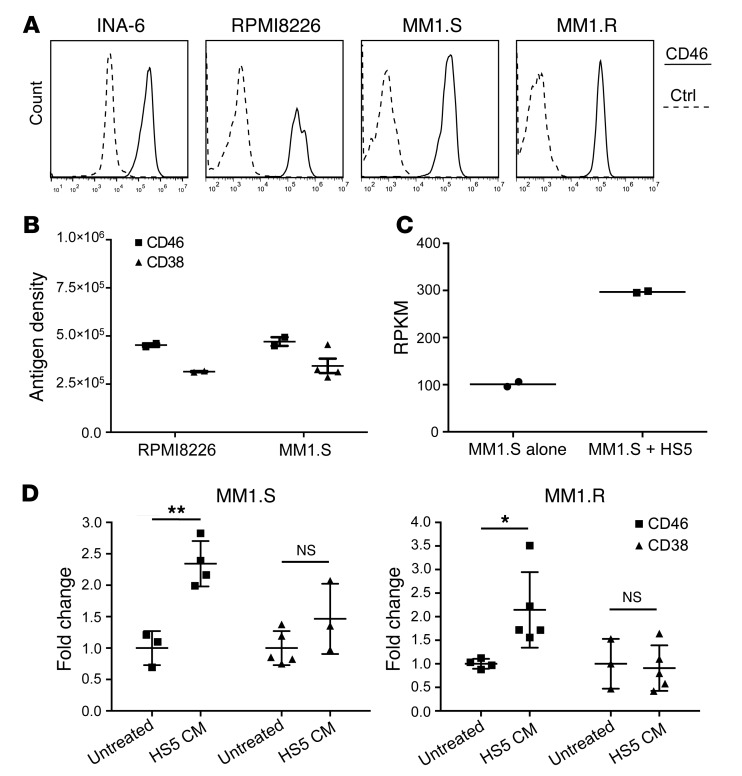
To evaluate whether CD46 was overexpressed in MM, we studied its cell surface expression by FACS on cell lines. CD46 was highly expressed on the cell surface of all MM cell lines tested (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI85856DS1). We next sought to quantify the CD46 antigen number per cell (referred to henceforth as antigen density), using methods described previously (25). The mean antigen density on MM cell lines RPMI8226 and MM1.S ranged from 454,668 to 470,991 for CD46, compared with 314,953 to 344,865 for CD38, a commonly used marker for MM (Figure 1B). It has previously been reported that extracellular CD46 antigen is shed from the cell surface of solid tumor cell lines (26). To assess whether MM cells shed CD46 antigen, we conducted Western blotting of cell lysates and supernatants from RPMI8226 cells. In either the presence or the absence of CD46 antibody stimulation, we found no appreciable shedding of CD46 antigen from MM cells into the culture supernatant (Supplemental Figure 2).
Figure 1. CD46 is highly expressed on MM cells and is further increased in the setting of BM microenvironment.
(A) CD46 expression in INA-6, RPMI8226, MM1.S, and MM1.R measured by FACS (solid lines), compared with nonbinding control (Ctrl = dashed lines, representative data, n = 3). (B) CD46 antigen density estimation compared with CD38 on RPMI8226 and MM1.S (data represent mean ± SEM, n = 3). (C) Coculture of MM1.S with HS5 BM stromal cells increases the expression of CD46 mRNA (n = 2). RPKM, reads per kilobase of transcript per million mapped reads. (D) CD46 and CD38 antigen densities in RPMI8226 and MM1.S, incubated with or without HS5-conditioned media (CM) for 3 days (data represent mean ± SEM, n = 3–5). Two-tailed Student’s t test, *P < 0.05, **P < 0.01.
CD46 is upregulated in the context of the BM microenvironment.
Myeloma is a disease in which the BM microenvironment promotes MM cell survival and chemotherapy resistance (27). The majority of MM patients have disease that is primarily localized to the BM. To assess whether the CD46 expression level in MM cells is impacted by this microenvironment, MM1.S cells were cocultured with the BM stromal cell line HS5. Analysis of mRNA expression by RNA sequencing (RNA-seq) showed the CD46 mRNA level increased in MM1.S when cocultured with HS5, compared with monocultures (Figure 1C). To assess whether this observation could be generalized to other MM cell lines and coculture conditions, MM1.S and MM1.R were incubated with HS5-conditioned media and analyzed by FACS for CD46. For comparison, CD38 was also studied in parallel. MM1.S and MM1.R cells showed upregulation of CD46 when cultured with HS5-conditioned media (P = 0.0031 and 0.02, respectively), suggesting that a factor from BM stromal cells may increase CD46 expression by MM cells (Figure 1D). In contrast, CD38 showed a variable response upon incubation of MM1.S and MM1.R cells with stromal cell–conditioned media (P = 0.15 and 0.8, respectively) (Figure 1D). The nature of the factor(s) and whether it is BM specific remains to be determined.
Generation of anti-CD46 antibody and internalization by myeloma cells.
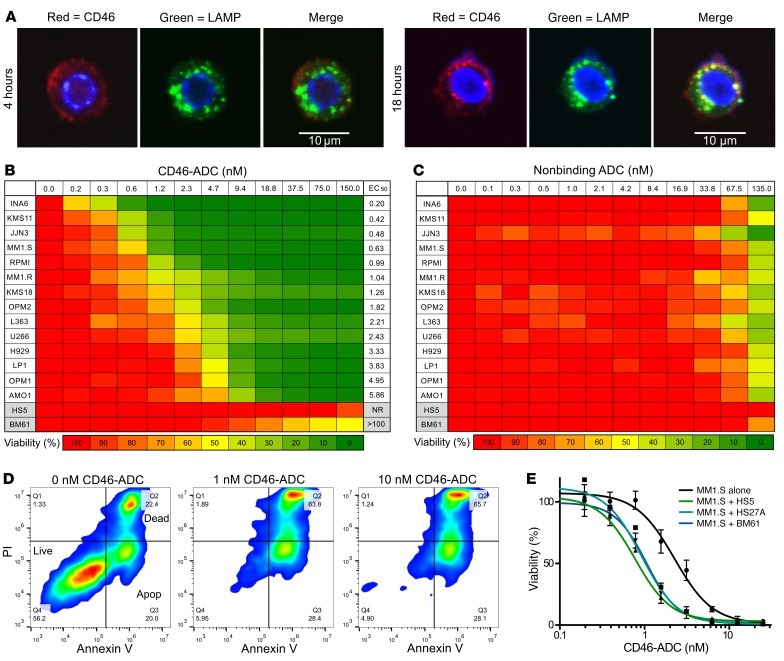
We generated a panel of human monoclonal antibodies binding to domains 1 and 2 of human CD46 by phage and yeast antibody display (28) and identified a lead antibody, 23AG2, that binds to the target antigen specifically with high affinity. The equilibrium dissociation constant (KD) of the antibody on a recombinant human CD46 protein fragment was 2.99 nM on the BLItz system (Supplemental Figure 3). Measured on living MM cells, 23AG2 KD values were 1.19 nM for RPMI8226 and 2.24 nM for MM1.S, respectively (Supplemental Figure 4). To determine whether the anti-CD46 antibody (23AG2) is internalized by MM cells, we incubated the antibody with MM1.R, analyzed internalization using confocal microscopy, and studied colocalization with lysosomal-associated membrane protein 1 (LAMP1). CD46 antibody was internalized and colocalized with LAMP1 (Figure 2A), indicating subcellular localization that is well suited for ADC delivery. This was also performed on RPMI8226 cells with similar results (data not shown).
Figure 2. Potent and selective activity of CD46-ADC on MM cells compared with BM stromal cells, with potentiation of ADC effect in the context of MM-stromal interactions.
(A) Confocal immunocytochemistry of MM1.R after 4 (left) and 18 (right) hours of incubation with anti-CD46 antibody (red). Late lysosomes shown with anti-LAMP1 antibody (green), and partial colocalization shown in merged panel (yellow). (B) Dose response for CD46-ADC inhibition of viability of MM cells compared with HS5 and BM61 BM stromal cells after 96-hour incubation (n = 3). NR: EC50 not reached due to lack of killing at the highest concentration tested. (C) Lack of effect of nonbinding control ADC until approximately 100 nM (n = 3). (D) Annexin V and PI staining of MM cell line INA-6 for 0–10 nM CD46-ADC, with apoptosis and death by 48 hours (representative data, n = 3). (E) Sensitivity of MM1.S cell line to CD46-ADC is increased in the presence of HS5, BM61, or HS27A BM stromal cells. EC50 was 2.25 nM on MM1.S alone and 0.77 nM, 0.92 nM, and 1.05 nM for MM1.S in the presence of HS5, HS27A, and BM61, respectively (data represent mean ± SEM, n = 3).
Anti–CD46-ADC has potent and selective cytotoxicity against myeloma cell lines.
To provide an initial assessment of CD46 as a suitable ADC target, we conjugated our antibody to a membrane-impermeant plant toxin (saporin) to form an immunotoxin and evaluated its effect on RPMI8226 cells. The immunotoxin showed strong inhibition of cell proliferation with EC50 in the picomolar range, with no effect observed from toxin alone (Supplemental Figure 5). We next covalently conjugated the MMAF to our anti-CD46 antibody via a lysosomal protease–sensitive valine-citrulline linker (8, 29). HPLC analysis with hydrophobic interaction chromatography of the final conjugate showed an average drug per antibody of 3.3 (Supplemental Figure 6). We tested the CD46-ADC on the RMPI8226 cell line and found that it also potently killed MM cells with EC50 in the picomolar range (Figure 2B).
On the panel of MM cell lines, CD46-ADC showed an EC50 range of 150 pM to 5 nM (Figure 2B). On BM stromal cells, CD46-ADC had an EC50 greater than 100 nM for patient-derived BM61 cells (generated via culture of CD138-negative BM mononuclear cells) and essentially no effect on HS5 cells across all concentrations tested (up to 150 nM, no EC50 estimated) (Figure 2B). Isotype control ADC constructed with a nonbinding antibody showed little to no effect on MM cell line proliferation at concentrations up to 67 nM (Figure 2C). The level of both CD46 transcript and cell surface expression measured by FACS correlated inversely with the EC50 of CD46-ADC (Supplemental Figure 7, A and B), suggesting that surface expression is a reasonable surrogate for potency. The induction of apoptosis and death was detectable in MM cells tested for annexin V and propidium iodide (PI) after 48 hours of ADC treatment (Figure 2D). Next, we examined the effect of BM microenvironment interactions on the efficacy of CD46-ADC. Cocultures of MM1.S cells with BM61, HS5, or HS27A BM stromal cells enhanced the potency of CD46-ADC (Figure 2E), consistent with our observations of increased CD46 expression in coculture and correlation of potency to CD46 levels described above.
We next sought to validate that CD46-ADC cytotoxicity is dependent on binding to CD46 for specific tumor cell killing. First, competitive cell binding of the anti-CD46 antibody was measured and detected in the presence of increasing amounts of the recombinant CD46-Fc fusion protein (Supplemental Figure 8A). In addition, the CD46-Fc blocked the cytotoxic effect of CD46-ADC (Supplemental Figure 8B). Next, the effect of CD46 knockdown on myeloma cell cytotoxicity of CD46-ADC was tested. The MM cell line H929 was infected with lentivirus coexpressing green fluorescent protein (GFP) and shRNA against CD46. By quantitative FACS analysis, CD46 antigen density was knocked down by 82% from 146,647 in uninfected (GFP-negative) cells to 25,847 in GFP-positive cells (Supplemental Figure 8C). The knockdown of CD46 reduced the induction of cell death by CD46-ADC compared with scrambled shRNA control (Supplemental Figure 8D), providing additional evidence that cytotoxicity of CD46-ADC is dependent on CD46 expression. In summary, CD46-ADC is potent and specifically cytotoxic to MM cells in vitro, and is further potentiated in models of the BM microenvironment.
Anti–CD46-ADC potently eliminates MM cell line xenografts in vivo.
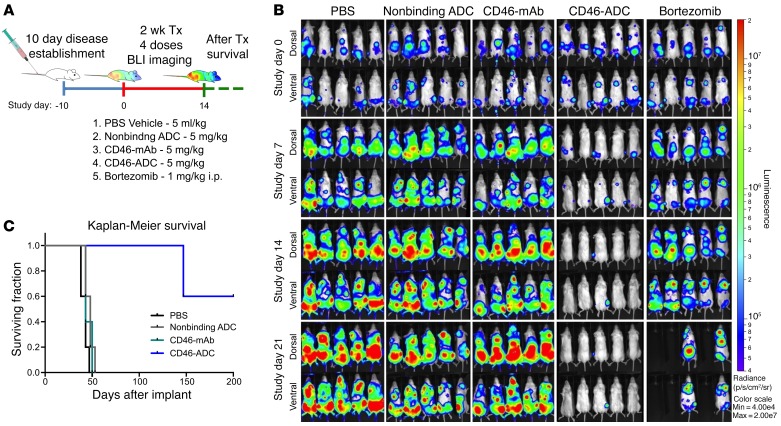
RPMI8226 cells expressing firefly luciferase (RPMI8226-Luc) were used to establish an orthometastatic xenograft model in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice, as previously described (30). A total of 4 injections of CD46-ADC at 5 mg/kg were given once every 3–4 days. CD46-ADC resulted in near-complete elimination of myeloma cell bioluminescent signal, whereas controls (vehicle, nonbinding ADC, naked antibody) did not (Figure 3, A and B). A comparison group was treated with bortezomib, which delayed increase in the bioluminescence activity but failed to reverse growth (Figure 3B). It should be noted that this bortezomib schedule was chosen for comparison with the ADC, not to simulate clinical use, which would be continuous. The survival of mice treated with CD46-ADC was significantly improved over that of control groups (hazard ratio [HR] = 0.151 between control-ADC and CD46-ADC; P = 0.004), with most of the treated animals living until study was discontinued at day 200 (Figure 3C).
Figure 3. In vivo CD46-ADC antimyeloma activity in the RPMI8226-Luc disseminated xenograft model.
(A) Study treatment scheme. RPMI8226-Luc cells were injected i.v. and established for 10 days. Starting on the tenth day (treatment day 1), a total of 4 injections of PBS, control nonbinding ADC (5 mg/kg), naked CD46 antibody (5 mg/kg), CD46-ADC (5 mg/kg), or bortezomib (1 mg/kg) were given twice per week (n = 5 mice per group). (B) Disease was monitored by bioluminescence imaging (BLI) (top views, dorsal; bottom views, ventral). BLI measurement in photons per second per cm2 per steradian (p/s/cm2/sr) was translated to color to indicate disease activity in the mice by the legend shown at far right. Tx, treatment; mAb, naked antibody. (C) Kaplan-Meier survival curves of NSG xenografts transplanted with RPMI-Luc and treated with CD46-ADC or controls.
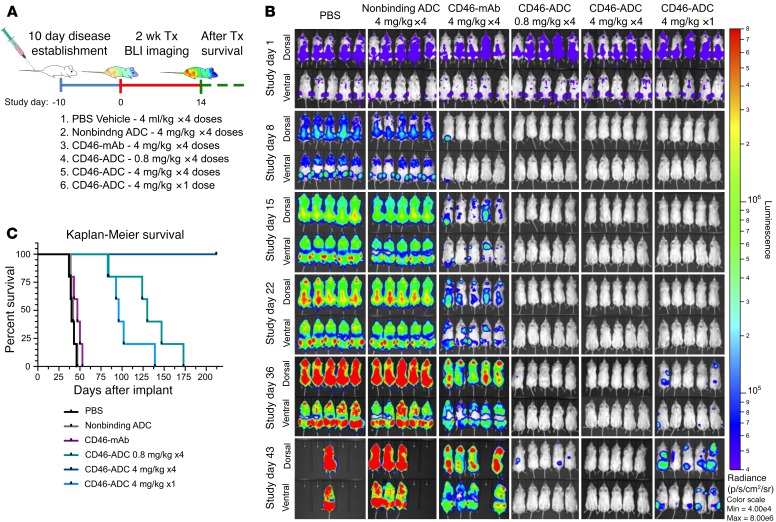
Next, a luciferase reporter–bearing MM1.S cell line was used in a second xenograft model of orthometastatic MM. Two different ADC doses and a single-dose regimen were investigated (Figure 4). Mice were treated once every 3–4 days at either 4 mg/kg or 0.8 mg/kg for a total of 4 injections. A third group was treated with a single dose of 4 mg/kg. Tumor burden continually increased in the control groups treated with vehicle and control ADC (Figure 4A). Naked CD46 antibody delayed increase in bioluminescence, but mice succumbed by day 53. The single dose showed elimination of bioluminescence by day 36, but all mice relapsed and succumbed by day 139 (Figure 4C). The 0.8-mg/kg (4-dose) regimen eliminated bioluminescence through day 43, but all succumbed by day 173. The 4-mg/kg (4-dose) regimen eliminated bioluminescent activity throughout the duration of the study, and all mice survived to study discontinuation at day 212. Together, these studies showed potent in vivo activity of CD46-ADC. This effect is dose dependent, with significant activity at a very low dose (0.8 mg/kg) (HR = 0.215 compared with control-ADC; P = 0.004) and apparent curative potential at a moderate dose of 4 mg/kg.
Figure 4. Dose- and schedule-dependent in vivo activity of CD46-ADC in a disseminated MM xenograft model with MM1.S cell line.
(A) Study treatment scheme. MM1.S-Luc cells were injected and established for 10 days. Starting on day 11 (treatment day 1), a total of 4 injections were given twice a week at the concentrations shown for all groups, except for the single-dose group. For each group, n= 5 mice per group. (B) BLI rapidly increased in negative control groups, but decreased to undetectable levels with all CD46-ADC treatment regimens (top views, dorsal; bottom views, ventral). Relapse of disease activity was observed progressively at single-dose 4-mg/kg and low-dose 0.8-mg/kg groups. No detectable BLI signal and no relapse after treatment was observed for the 4-mg/kg, 4-dose schedule, suggesting complete elimination of MM1.S xenografts in vivo. BLI in photons per second per cm2 per steradian (p/s/cm2/sr) was translated to color to indicate disease activity by the legend shown at far right. Tx, treatment; mAb, naked antibody. (C) Kaplan-Meier survival curves of NSG xenografts transplanted with MM1.S-Luc and treated with varying dose levels of CD46-ADC.
Establishing a clinical biomarker for CD46 expression in MM.
The CD46 gene is located on 1q32, in proximity to a clinically used FISH probe at 1q21. We thus postulated that patients with amp1q21 may also amplify 1q32 and therefore carry increased CD46 expression. To explore this we mined data sets from previous gene expression analyses in MM (31–33). By Affymetrix array, CD46 transcript increased 3-fold in MM cells compared with normal plasma cells (P = 6.375 × 10–5) and was also significantly increased in a sequential manner between monoclonal gammopathy of unclear significance (MGUS) and smoldering myeloma (asymptomatic proliferative plasma cell states) (Supplemental Figure 9, A–C). Furthermore, data annotated by 1q21 FISH status (33) demonstrated coamplification of CD46 by array comparative genomic hybridization (Supplemental Figure 9D) and a concomitant 2.8-fold increase in CD46 transcript expression (P = 0.002) compared with MM samples that were 1q21 normal by FISH (Supplemental Figure 9E).
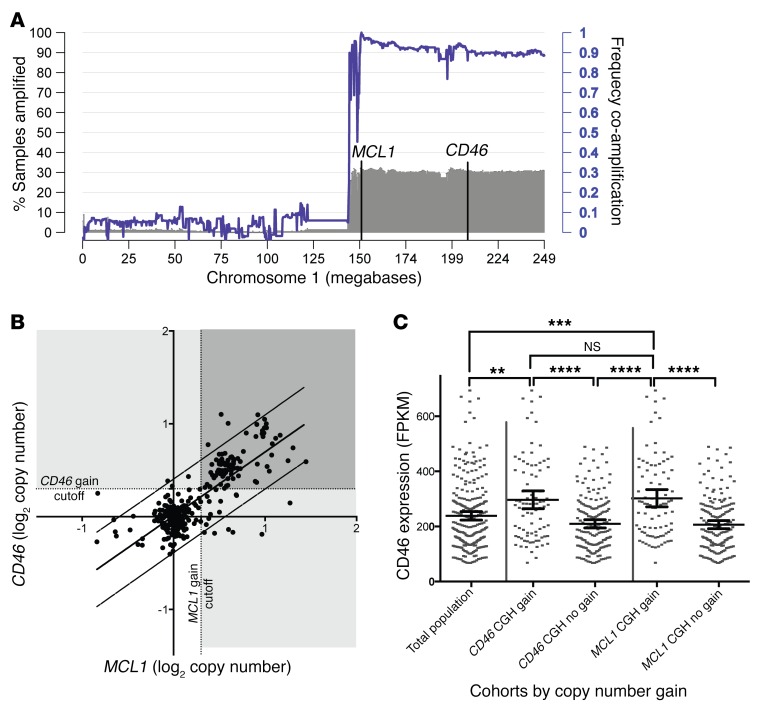
Analysis of additional data derived from the CoMMpass Study (Interim Analysis 6) also confirms that about 30% of newly diagnosed patients demonstrate focal amplification of the myeloid cell leukemia-1 (MCL1) gene located at 1q21 (Figure 5A). In addition to 1q21, the entirety of the 1q arm is amplified for a similar fraction of patients (Figure 5A, gray track). Furthermore, coamplification analysis for individual samples demonstrates that 85.4% of patients carrying MCL1 amplification also amplified CD46, and the majority of the 1q arm (purple line). Thus, there is a high level of coamplification between a negative prognostic factor (amp1q21) and the gene for the CD46-ADC target. Additionally, comparison of the correlations of MCL1 with CD46 copy number and MCL1 with CKS1B (another gene located at 1q21) demonstrates that the 1q21 and 1q32 loci correlate similarly (Figure 5B and Supplemental Figure 10, A and B). Lack of correlation between CCND1 (chromosome 11) copy number and CD46 or MCL1 indicates that coamplification is unique to genes on the 1q arm (Supplemental Figure 10, C and D). Samples with copy gain on either CD46 or MCL1 also demonstrate significantly increased CD46 transcript expression, compared with samples not demonstrating copy gain at these loci and with the overall population (Figure 5C). Thus, data from both published studies and the CoMMpass Study indicate that nearly all patients with amp1q21 also demonstrate CD46 gene amplification and increased CD46 mRNA expression.
Figure 5. CD46 locus is frequently coamplified with 1q21, and copy gain for either MCL1 or CD46 identifies patients with high CD46 expression.
(A) About 30% of newly diagnosed patients demonstrate 1q21 copy gain overlapping the MCL1 locus by array comparative genomic hybridization (CGH), defined as log2 copy number >0.3, and a similar proportion demonstrate amplification along the 1q arm, including CD46 (gray shaded histogram; left axis). High frequency of coamplification along the 1q arm with MCL1 amplification is shown by the purple line and quantified on the right axis (n = 322 patients). (B) Dot plot of log2 CGH values in patient samples indicates that copy numbers for CD46 and MCL1 loci are highly correlated and cluster in regular copy number intervals (n = 322 patients). (C) Mean CD46 transcript expression values (quantified as fragments per kilobase per million mapped reads [FPKM]) for CD46 or MCL1 copy-amplified cohorts, defined by ≥0.3 log2 CGH value, versus nonamplified cohorts indicate high CD46 expression in MCL1 or CD46 amplified patient samples, compared with total population or samples without copy gain (n = 260 patients). Data represent mean ± 95% CI. One-way ANOVA with Tukey’s multiple comparison correction, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cell surface CD46 is increased in myeloma samples with gain of 1q21 by FISH.
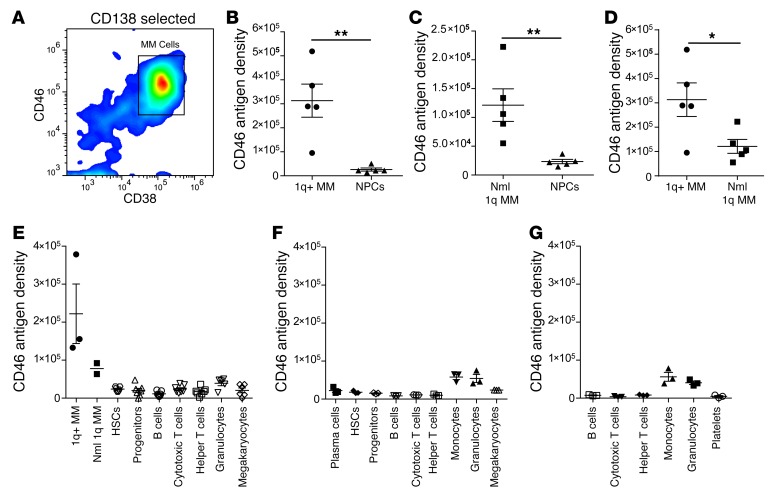
To validate 1q21 FISH as a clinical biomarker, we next measured CD46 cell surface expression in a cohort of patients at diagnosis or at relapse with a pure monoclonal population of MM cells (Supplemental Table 1). An initial cohort of 10 MM patient samples was evaluated, 7 of which had amp1q21 and 3 of which did not (Supplemental Table 1, samples 1–10). CD46 was coexpressed on the MM cell surface with well-known myeloma antigens CD38 and CD138 (Figure 6A), but expressed at low levels on nonplasma cells (NPCs) that have negative/low CD38 and CD138 expression and represent a heterogeneous mixture of normal mononuclear cells (MNCs) (Supplemental Figure 11, A and B). The average mean fluorescence intensity (MFI) (anti-CD46) of CD138-positive/CD38-positive cells by FACS was 152,049 (SEM 22,767) with amp1q21, significantly higher compared with 37,113 (SEM 9,926) in patients with normal 1q21 (P = 0.014, 2-tailed t test; Supplemental Figure 11C). Thus, CD46 was overexpressed on MM cells from all patients and further amplified in patients with amp1q21, with low expression on nonmalignant NPCs.
Figure 6. CD46 is overexpressed on cell surface of primary MM patient cells and further amplified in patients with amp1q21 compared with normal 1q.
(A) FACS plot showing that CD46 surface expression correlates with CD38 in CD138-selected cells to identify the MM population by FACS (representative data, n = 25). (B) Quantitative FACS results for CD46 antigen density from MM versus NPCs from patients with normal (Nml) 1q (n = 5). (C) Quantitative FACS results for CD46 antigen density from MM versus NPCs from patients with amp1q21 (1q+, n = 5). (D) CD46 antigen density is further increased in amp1q21 patients (n = 5) compared with patients with normal 1q (n = 5). (E) CD46 antigen density on various BM normal cell populations compared with MM cells from 7 additional patients. (F) CD46 antigen density on various BM cell populations from 3 normal donors. (G) CD46 antigen density on various peripheral blood cell populations from 3 normal donors. Data represent mean ± SEM. Two-tailed t test, *P < 0.05, **P < 0.01.
We next measured cell surface antigen density by FACS to quantify the expression difference between amp1q21 and normal 1q21 patients. Samples were analyzed for CD46 cell surface expression on MM cells and matching NPCs from patients with and without amp1q21. In a second cohort of 10 patients, unselected MNCs were analyzed (Supplemental Table 1, samples 11–20). In 5 patients with amp1q21 the mean CD46 antigen density on MM cells was 313,190 (SEM 68,849), and on NPCs was 26,214 (SEM 6,329) (Figure 6B; 2-tailed t test, P = 0.0032). In 5 patients with normal 1q21 the mean CD46 antigen density on MM cells was 121,316 (SEM 28,352), and on NPCs was 23,388 (SEM 3,729) (Figure 6C; 2-tailed t test, P = 0.009). CD46 antigen density was significantly higher on amp1q21 MM samples compared with MM with normal 1q (Figure 6D; 2-tailed t test, P = 0.032), supporting the hypothesis that CD46 cell surface expression variability in MM is due to selective genomic amplification in patients with amp1q21.
We next sought to better differentiate the levels of CD46 on various nontumorigenic or normal hematopoietic cell populations. CD46 antigen density was measured on hematopoietic stem cells, progenitors, B cells, T cells, granulocytes, monocytes, megakaryocytes, and platelets (Supplemental Figure 12). In BM from 7 subsequent patients with MM, granulocytes had the highest CD46 antigen density (mean 39,248, SEM 6,492), but other nontumor cell populations all had low CD46 expression (antigen density range 11,593–23,764) (Figure 6E and Supplemental Table 2). In BM from normal donors, monocytes (mean 58,320, SEM 6,874) and granulocytes (mean 54,439, SEM 10,688) had the highest CD46 antigen density, whereas others again were relatively low (range 8,443–23,772) (Figure 6F and Supplemental Table 3). Similar results were obtained from peripheral blood (PB) samples from normal donors, showing that CD46 antigen density is highest on monocytes (mean 56,237, SEM 11,649) and granulocytes (mean 40,523, SEM 8,165), but otherwise low (range 3,698–8,256) (Figure 6G and Supplemental Table 4).
In summary, CD46 expression was high on MM cells from 100% of patients (n = 25, cumulatively) and further amplified in patients with amp1q21. The overall CD46 expression on normal hematopoietic cells is low. Notably, monocytes and granulocytes expressed relatively higher levels of CD46 compared with other normal cell populations. Interestingly, benign plasma cells from normal donors also had low CD46 antigen density (mean 22,475, SEM 4,869; Figure 6F and Supplemental Table 3), suggesting that high CD46 on MM cells occurs with malignant transition.
Anti–CD46-ADC potently and selectively induces primary MM cell apoptosis and death.
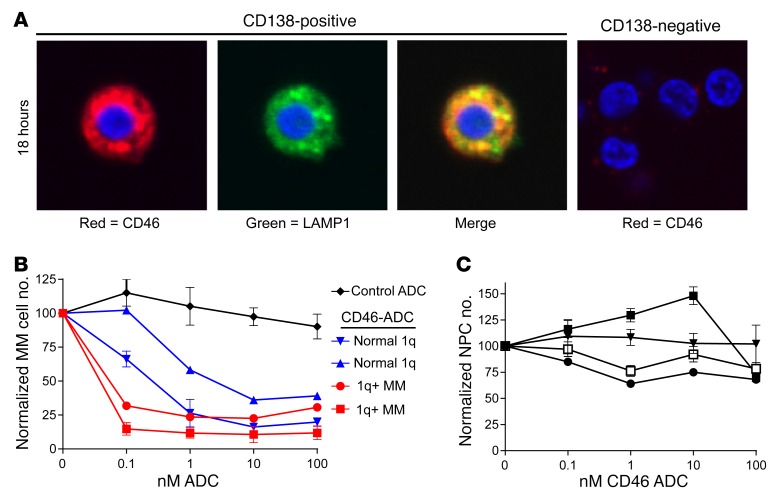
We first studied internalization of our anti-CD46 antibodies by primary MM cells. As shown in Figure 7A (left panel), the anti-CD46 antibody was internalized and partially colocalized with LAMP1. In contrast, no internalization into CD138-negative MNCs was observed (Figure 7A, right). We next evaluated whether CD46-ADC was specifically cytotoxic to primary MM cells ex vivo. BM MNCs were treated with 0–100 nM in triplicate for each condition and evaluated by FACS after 48 hours. CD46-ADC treatment consistently reduced the number of MM cells, with no effect from nonbinding ADC control (Figure 7B). CD46-ADC had no effect on the number of NPCs in all patients tested (Figure 7C). Furthermore, the MM cells with gain of chromosome 1q21 showed higher sensitivity to CD46-ADC (lower EC50) compared with patients with normal 1q21 (Figure 7B). Notably, only weak correlation with known 1q21 copy number and CD46-ADC potency was found in a limited number of myeloma cell lines for which information on both was available (Supplemental Figure 7C) (2). The analysis is limited because of the narrow range of CD46-ADC potency observed, and the fact that all but 1 of the MM cell lines have 1q21 gain. However, in patient samples, ADC sensitivity ex vivo also suggests that 1q21 FISH and antigen density may be potential biomarkers for response in MM patients.
Figure 7. Ex vivo evaluation of CD46-ADC in patient sample MM cells.
(A) Internalization of CD46 antibody (red) into MM patient cells ex vivo. CD138 positively selected cells from a patient with MM were incubated with CD46 antibody for 18 hours and costained with anti-LAMP1 antibody (green) and Hoechst dye (blue). Representative cell illustrates the intracellular localization of CD46 antibody, partially colocalizing with the late lysosomal marker LAMP1 (left). CD138-negative cells were treated in the same fashion and showed minimal binding of CD46 antibody without discernible internalization (right). Images were taken using a digital confocal microscope (FluoView, Olympus) at ×60 magnification. (B) CD46-ADC depletes the number of CD138-positive, CD38-positive MM cells more potently in patients with amp1q21. Mean values with SEM are shown for CD46-ADC treatment of 2 patient samples with amp1q21 (red lines) and 2 patient samples with normal 1q21 (blue lines), compared with cells treated with nonbinding control ADC (black). (C) CD46-ADC does not affect the number of NPCs up to a concentration of 100 nM (n = 4) (data represent mean ± SEM).
We also evaluated ADC induction of apoptosis and death of CD138-positive and CD138-negative fractions by FACS. Gating specifically on CD38-positive, CD138-selected cells, FACS for annexin V and PI was performed after 48-hour treatment of MM patient cells ex vivo with graded concentrations CD46-ADC. Gating for viable annexin V–negative/PI-negative cells, MM cell killing at 48 hours was consistent with cell line proliferation assays at 96 hours, with EC50 less than 10 nM (Supplemental Figure 13). No toxic effect on nonmalignant CD138-negative cells was observed up to 100 nM (Supplemental Figure 13). Overall, these data suggest that CD46-ADC will be selectively cytotoxic to MM cells and may have improved potency in patients with amp1q21.
Tolerability evaluation of CD46-ADC in human CD46–expressing transgenic mice.
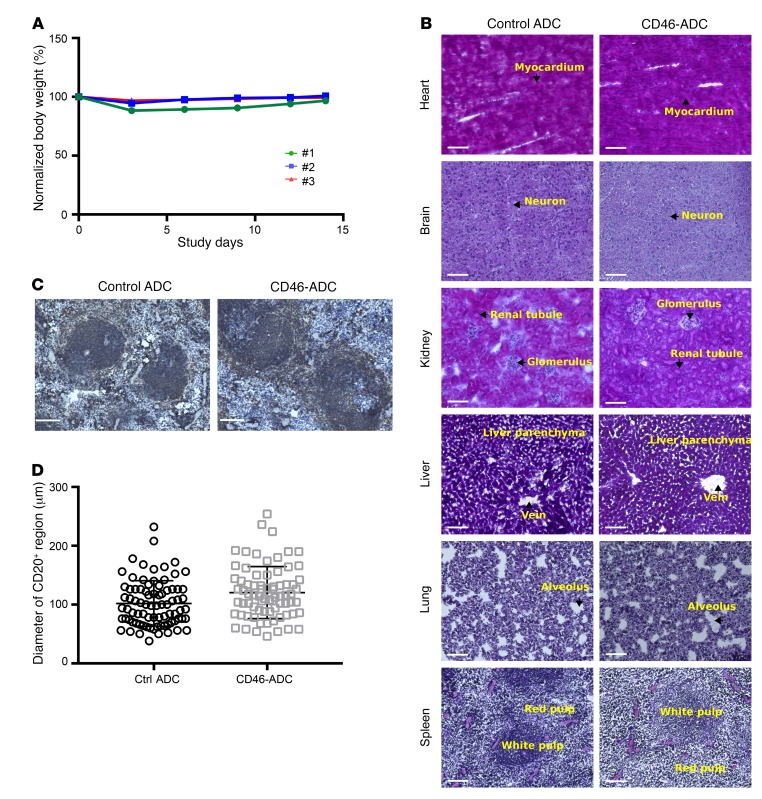
We sought to study tolerability of CD46-ADC in vivo using a relevant animal model. The expression of murine CD46 is restricted to the inner acrosomal membrane of testes and the retina (34, 35). In addition, human CD46 and murine CD46 share weak homology, and our anti-CD46 antibody does not bind the mouse CD46 protein. For these reasons, the mouse xenograft experiments shown above provide limited data on potential toxicity of CD46-ADC. To circumvent this limitation, we studied in vivo tolerability of CD46-ADC using human CD46 transgenic mice (36). Following a single i.v. bolus injection of 6 mg/kg CD46-ADC, animals were monitored for body weight loss and sign of overt toxicity for 14 days. No significant body weight loss or overt sign of toxicity was observed (Figure 8A). At study discontinuation on day 14, necropsy study was performed. All organs appeared to be morphologically normal except for a slight increase in spleen size in CD46-ADC–treated animals. Histologic analysis of major organs showed no notable tissue damage (Figure 8B at ×20 and Supplemental Figure 14 at ×40 magnification). To assess whether CD46-ADC caused any notable effect on B cells in the spleen, we stained spleen sections with anti–mouse CD20 (Figure 8C). We measured diameters of CD20-positive regions in CD46-ADC– and control ADC–treated groups, and found no statistically significant difference (Figure 8D). Thus, in this relevant small-animal model, CD46-ADC appears to be tolerated in vivo.
Figure 8. Evaluation of tolerability of CD46-ADC in transgenic mice expressing human CD46.
Three mice were treated with CD46-ADC or isotype (nonbinding) control ADC at 6 mg/kg i.v. and monitored for 14 days. (A) Body weight analysis over the monitoring period showed no weight loss greater than 12% following CD46-ADC injection. (B) Histological analysis at the time of experiment discontinuation. Images were taken by a Keyence digital microscope at ×20 magnification. Histological features (arrows) are indicated on the images. No notable difference was observed between CD46-ADC– and control ADC–treated samples. Scale bars: 100 μm. (C) Immunohistochemistry analysis of CD20-positive region in CD46-ADC– and control ADC–treated spleens. Scale bars: 100 μm. (D) Diameters of CD20-positive regions in CD46-ADC– and control ADC–treated spleens (n = 74 for CD46-ADC–treated and 81 for control ADC–treated, respectively). No significant difference was observed (2-tailed t test, P > 0.05).
Discussion
MM is incurable, and patients inevitably develop resistance to current therapies. New molecular targets and targeting agents have the potential to overcome limitations associated with current therapeutic strategies. Our study shows that CD46 is an attractive novel target for development of antibody therapy against MM. We have shown that CD46 is highly expressed on the surface of MM cell lines and primary MM cells (with relatively low expression on various normal BM and PB cell populations). In addition, the expression level increases in the presence of a model of the BM microenvironment. Previous studies have characterized the CD46 gene as a transcriptional target of signal transducer and activator of transcription 3 (STAT3) (37). This finding is consistent with the notion that exposure of MM cells to marrow-derived cytokines, commonly associated with the activation of STAT3 activity, results in subsequent induction of CD46 transcript expression.
CD46 is an attractive cell surface target for ADC development because of its original identification from an unbiased screening strategy that prioritized macropinocytosing antigens in their native conformations (12, 23). Our novel CD46-ADC potently and selectively eliminates MM cell lines in vitro and in vivo, and primary MM cells ex vivo. A unique distinction of CD46 is that the gene is located near a FISH probe already in routine clinical use (1). This 1q21 probe frequently undergoes genomic gain in patients with MM, especially in high-risk and R/R disease (2, 21). We have found that the genomic gain correlates well with CD46 antigen expression level and susceptibility to CD46-ADC. The discovery of a readily obtainable biomarker for target overexpression can prove extremely valuable for patient selection and stratification in clinical trials. Overall, our data suggest potential for CD46-ADC as a novel therapeutic for MM, with particular benefit predicted for patients with genomic amplification of the region where the CD46 gene resides (1q).
The finding of a putative drug target in high-risk or R/R MM with amp1q21 could be highly advantageous for the treatment of advanced disease. Putative oncogenes located on 1q include CKS1B and MCL1. In addition, CD46 overexpression itself is protective for cancer cell evasion of complement activation (38). MCL-1 is a prosurvival member of the B cell lymphoma 2 (BCL-2) family that is also overexpressed in high-risk MM patients and those with relapsed disease (39, 40). Importantly, MCL-1 upregulation has been described to evoke bortezomib resistance, which adds significant difficulty in treating advanced myeloma (41–43). These features of MM cells with 1q21 gain make CD46 an attractive target to combat drug resistance and oncogene accumulation in advanced disease.
If successful, there are several myeloma disease settings that may benefit from treatment with an efficacious ADC: (a) R/R MM with resistance to IMiDs and proteasome inhibitors, for which there is great clinical need for new therapies; (b) patients with high-risk disease features that tend to progress rapidly through current therapies — patients with amp1q21 at diagnosis fit into this category (2), making a particularly attractive group for treatment with CD46-ADC; and (c) addition of CD46-ADC to standard induction regimens for patients with newly diagnosed MM, with or without subsequent autologous stem cell transplant. Although standard approaches often achieve deep remission, they are not curative treatment for MM. The addition of a targeted cytotoxic therapy, such as an ADC that has a different mechanism of action, could potentially improve the odds of attaining cures that have thus far been elusive.
Current efforts on antibody targeting against MM are focused on a few lineage markers such as CD138 and CD38. Currently, 3 ADCs have been developed and entered clinical trials for MM. These include the anti-CD138 ADC indatuximab ravtansine (maytansinoid toxin DM4, disulfide cleavable linker), the anti-CD56 ADC lorvotuzumab mertansine (maytansinoid toxin DM1), and the anti-BCMA ADC GSK2857916 (MMAF toxin) (44–46). The linker/toxin combination used by CD46-ADC is most similar to the anti-BCMA ADC, except that the linker used is cleavable. The linker/toxin combination for CD46-ADC is not used by any ADCs currently in clinical trials, but has been described previously (29). A key differentiation between CD46 and the other targets of ADCs in clinical trials for myeloma is that CD46 is a functional antigen and its expression is increased as the disease progresses. CD46 has a known role in negative regulation of complement, rendering cells dependent on its expression (14). In addition, the genomic region where CD46 resides is amplified in R/R MM (2, 21). This FISH biomarker for CD46 amplification is thus far unique in MM. Finally, the internalization mechanism of other MM antigens has not been characterized to assess whether they are optimally suited for ADC development (11). On the other hand, we have shown that our anti-CD46 antibodies enter cells through macropinocytosis (Y. Su and B. Liu, unpublished observations), an inherently tumor-selective pathway (23, 24), and our CD46-ADC showed high potency against MM cells, but selectively spared normal BM cells.
Interestingly, CD46 has been independently targeted for MM and other malignancies using oncolytic measles virus (17). An early report of 2 patients treated in phase I study in myeloma described substantial decreases in disease activity (47). It would be of interest to evaluate whether patients who tend to respond have amp1q21, as would be suggested by our study finding of higher CD46 expression levels in these patients. While promising, the approach needs to address immunogenicity of measles virus that limits repeat dosing. Human antibody-based approaches such as our CD46-ADC have less potential for immunogenicity. Nonetheless, the oncolytic measles virus approach has shown that CD46-targeted therapy can lead to tumor-specific effects (47, 48). Other than infusion-related reactions, oncolytic measles virus was well tolerated, showing that CD46 can be engaged in humans without triggering unacceptable toxicity (47, 48). It remains to be tested whether our CD46-ADC is safe in phase I trials.
We have found thus far that CD46-ADC was tolerable in mice, including transgenic mice that express the human CD46 gene. Our ex vivo study of CD46-ADC on primary cells showed potent cytotoxicity on myeloma cells, but not NPCs. Further study will determine potential differential effects of the ADC on subpopulations of NPCs. Because CD46 is expressed at low levels on all nucleated cells, there is potential for a tissue sink that could also impact the pharmacokinetics of CD46-ADC. We acknowledge this potential problem but believe it will be a minor issue as our quantitative measurement has shown that the CD46 antigen density on NPC subpopulations is low (typically ≤25,000 per cell). Ultimately, the pharmacokinetics and toxicity of CD46-ADC need to be investigated in nonhuman primates.
Mutations in CD46 are known to be a predisposing factor for about 10% of cases of atypical hemolytic uremic syndrome, with incomplete penetrance (49). CD46 expression is low on most normal cells, including endothelial cells (14, 50). A potential toxicity concern is that an antibody-based therapy targeting CD46 may block complement inhibition and potentially induce atypical hemolytic uremic syndrome. However, our ADC binds a conformational epitope residing in Sushi domains 1 and 2, outside of the interface where the complement pathway components bind (51). Consistent with this, we have observed that our CD46 antibody does not compete with complement binding and does not enhance complement activity in in vitro assays (Y. Su and B. Liu, unpublished observations). Therefore the ADC is not expected to lead to excessive terminal complement cascade activation on normal cells.
In addition to MM, CD46 is overexpressed in prostate cancer, ovarian cancer, cervical cancer, breast cancer, endometrial cancer, lung cancer, and leukemia (38, 52, 53). Intriguingly, 1q gain is also common in cancers from multiple tissue types (54, 55). We speculate that 1q gain may be the mechanism for CD46 upregulation in other cancers. Thus, CD46-ADC may have efficacy in other malignancies, particularly those that commonly have copy number gain of 1q.
In conclusion, we have identified a novel functional antigen, CD46, for ADC targeting of MM, with unique potential for high-risk and R/R disease that has genomic amplification at the CD46 gene locus and is in dire need of therapy. We have developed a novel CD46-ADC and shown that it is highly potent and selective in eliminating MM cells (cell lines and primary tumor cells) in preclinical models. We have found that CD46 genomic gain on chromosome 1q correlates with antigen amplification, and further identified a biomarker based on a clinical FISH test that can be used for patient stratification. Thus, our study could lead directly to the application of a novel ADC therapeutic for treating MM.
Methods
MM patient and normal donor samples.
BM samples from MM patient samples 1–10 were separated into 2 fractions based on selection for CD138 expression with EasySep magnetic bead columns (Stem Cell Technologies). Primary cells were cultured in RPMI1640 media with 100 U/ml penicillin, 100 μg/ml streptomycin, 10% FBS, and 2 ng/ml IL-6. BM and PB controls from normal donors were purchased from AllCells.
Antibody generation.
We have previously identified a phage antibody binding to tumor cells in situ by laser capture microdissection–based selection using prostate cancer specimens (12). We later identified that CD46 is the target antigen and the original antibody binds to a conformational epitope residing in Sushi domains 1 and 2 (Y. Su and B. Liu, unpublished observations). A combined phage and yeast antibody display library selection approach (28) was used to identify additional anti-CD46 human antibodies. Briefly, the human CD46 gene fragment containing domains 1 and 2 was cloned into the pFUSE-Fc vector (Invivogen) to create a recombinant Fc fusion molecule that was produced in HEK293a cells, and purified by protein A affinity chromatography. A nonimmune phage antibody display library containing 1 billion members (Y. Su and B. Liu, unpublished observations) was incubated with CD46-Fc fusion coated on polystyrene beads in PBS/2% dry milk at room temperature for 1 hour, followed by washing 3 times with PBS/2% dry milk, elution with 100 mM triethylamine, and neutralization with 1 M Tris-HCl, pH 6.8. After 3 rounds of selection, binding phages were screened by FACS on Du145 cells and sequenced. Unique phage antibodies were retested on recombinant CD46-Fc fusion–coated microtiter plates to confirm binding specificity. In parallel, an alternative strategy was used selecting the phage antibody library on live tumor cells, followed by transfer of the output into a yeast surface display vector, and then FACS-based selection using low-concentration ligands to enrich high-affinity binders to the recombinant CD46-Fc fusion protein as previously described (28). A panel of novel anti-CD46 antibodies was identified and further studied for internalization and macropinocytosing activities (23), and the 23AG2 lead antibody was chosen for ADC development. Full-length human 23AG2 IgG1 was constructed from phage antibody sequence, produced in HEK293a, and purified by protein A chromatography as previously described (23). Affinity of 23AG2 IgG1 to recombinant CD46-Fc (domains 1 and 2) was measured by the label-free BLItz system (ForteBio) with the biosensor coated with the 23AG2 antibody. Affinity for living myeloma cells was measured by FACS on RPMI8226 and MM1.S cell lines with MFI values curve-fit using 1-site specific binding model (GraphPad). A control IgG1 (YSC10) was created from a phage antibody picked randomly from the nonselected library, which does not bind to any cell surface antigen and hence is designated as a nonbinding antibody in this article.
MM cell lines.
MM cell lines bearing the firefly luciferase reporter gene were provided by Constantine Mitsiades (Dana-Farber Cancer Institute, Boston, Massachusetts, USA) (56). RPMI8226, MM1.S, and MM1.R were purchased from American Type Culture Collection (ATCC). All MM cell lines were maintained in RPMI1640 with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. INA-6 was supplemented with IL-6 (2 ng/ml). Patient stromal culture, BM61, was derived from CD138-negative MNC fractions derived from BM aspirate.
FACS analysis of cell surface CD46 expression.
To determine CD46 expression on MM cells, FACS was performed using biotin-labeled human anti-CD46 IgG1 followed by detection with Alexa Fluor 647–conjugated streptavidin (Life Technologies). All samples were analyzed using an Accuri C6 (BD Biosciences) with a 96-well autosampler. Nonspecific Fc receptor binding was minimized by preincubation with Clear Back reagent (Medical and Biological Laboratories Co.). To determine CD46 expression on patient samples, multicolor analysis was performed using FITC-conjugated anti-CD38 antibody (clone AT1, Stem Cell Technologies) to identify MM cells in CD138-selected samples. For FACS of CD46 on various BM and PB cell populations, an extended antibody panel was used with anti-CD38 (clone HIT2) PerCP-Cy5.5, anti-CD138 (clone MI15) BV421, anti-CD45 (clone HI30) BV510, anti-CD19 (clone HIB19) BB515, anti-CD4 (clone RPA-T4) PerCP-Cy5.5, anti-CD8 (clone RPA-T8) Pacific Blue, anti-CD3 (clone HIT3a) FITC, anti-Lineage FITC, anti-CD34 (clone 581) PE, anti-CD61 (clone VI-PL2) PE, anti-CD33 (clone HIM3-4) FITC, anti-CD14 (clone Hop9) PerCP-Cy5.5 (BD Biosciences), and Live/Dead-Near IR (Life Technologies/Thermo Fisher Scientific) on a FACSCanto II (BD Biosciences) flow cytometer.
Cell surface antigen density determination.
Quantitative flow cytometry was performed to determine CD46 antigen density. Anti-CD46 and anti-CD38 antibodies were labeled with Alexa Fluor 647 (Molecular Probes/Life Technologies) according to the manufacturer’s recommendations. MFI conversion to molecules of equivalent soluble fluorophores (MESF) was done by generation of a standard curve with Quantum beads (Bangs Laboratories) (25). The fluorophore-to-antibody ratio of the labeled antibodies was determined using Simply Cellular anti-human (for CD46) or anti-mouse (for CD38) IgG beads (Bangs Laboratories). Finally, conversion of MESF to cell surface antigen density was done by division of the fluorophore-to-antibody ratio.
Antigen shedding.
To assay for CD46 antigen shed into cell culture media, 4 × 105 RPMI8226 cells were seeded in a 6-well plate (Falcon) and cultured in a CO2 incubator at 37°C overnight. Next, 5 ml serum-free culture medium containing 10 μg/ml anti-CD46 antibody or nonbinding control antibody was added for an additional 24-hour incubation. The cells were separated by centrifugation at 491 g for 10 minutes, and lysed with cell lysis buffer (20 mM Tris-HCl, pH 7.4, 0.3 M NaCl, 1% NP-40) supplemented with complete protease inhibitor cocktail (Roche). The supernatants were concentrated 50 times by centrifuging in a Centricon filter unit (Millipore) at 3,488 g for 30 minutes at 4°C. Samples were boiled in SDS sample buffer and analyzed by SDS-PAGE. After semi-dry transfer to Immobilon-P membrane (Millipore), Western blotting was performed using anti-CD46 antibody H-294 (sc-9098, Santa Cruz Biotechnology) followed by anti-rabbit HRP (Jackson ImmunoResearch Laboratories) and detected by chemiluminescence with Pierce ECL Western Blotting Substrate (Pierce/Thermo Fisher Scientific) according to the manufacturer’s instructions. Images were captured using a C-DiGit blot scanner (LI-COR Biosciences).
Antibody internalization by confocal microscopy.
Alexa Fluor 647–labeled anti-CD46 antibody was incubated with MM cell lines for 4 or 18 hours, washed with PBS, fixed with 4% PFA, permeabilized with PBS with 0.1% Triton X-100 and 1% BSA, and analyzed by confocal microscopy (Olympus FluoView). Nonbinding isotype antibody was studied in parallel as a control. For internalization by patient cells ex vivo, CD138-positive and CD138-negative (control) cells were incubated with Alexa Fluor 647–labeled anti-CD46 or nonbinding isotype control antibodies for 18 hours, processed, and analyzed as described above. Subcellular localization to lysosomes was assessed by costaining with LAMP1 antibody (clone D2D11, Cell Signaling Technology).
ADC generation and characterization.
MMAF was conjugated to anti-CD46 IgG1 via an mcvcpab linker (29). To create the mcvcpab linker, N-ε-maleimidocaproyloxysuccinimide ester (Pierce) was dissolved in anhydrous dimethylformamide (DMF) (final 0.14 mM). This solution was then added to valine-citrulline-p-aminobenzylalcohol (Concortis Biosystems) (final 0.14 mM). After brief agitation to dissolve all components, diisopropylethylamine (DIPEA) was added (final 0.41 mmol), incubated at room temperature for 1 hour, and precipitated with cold ethyl acetate (EtOAc) to form mcvcpab. Bis-nitrophenyl carbonate (0.41 mmol) was added along with DIPEA (0.41 mmol) to mcvcpab dissolved in DMF and incubated at room temperature for 4 hours followed by EtOAc precipitation as before to yield maleimidocaproyl-valine-citrulline-p-aminobenzylalcohol p-nitrophenol carbonate (mcvcpab-PNP). MMAF hydrochloride salt (Concortis Biosystems), N-hydroxysuccinimide, and DIPEA were added to DMF-dissolved mcvcpab-PNP and incubated at room temperature for 16 hours to form mcvcpabMMAF (29). CD46 IgG1 was reduced by tris(2-carboxyethyl)phosphine (TCEP) at 37°C for 2 hours, purified by Zeba spin column (Pierce/Thermo Fisher Scientific), buffer-exchanged into PBS with 5 mM EDTA, and incubated with linker-conjugated MMAF (mcvcpabMMAF) at room temperature for 1 hour. Conjugation products were purified by running twice though the spin column to remove free MMAF and analyzed by HPLC using hydrophobic interaction chromatography with Infinity 1220 LC System (Agilent). The drug-to-antibody ratio is estimated from area integration using OpenLab CDS software (Agilent).
Apoptosis assay.
Induction of apoptosis and cell death in myeloma cell lines was evaluated using an annexin V–FITC Early Apoptosis Detection Kit (Cell Signaling Technology). Cell lines were seeded into 96-well plates at 4 × 104 cells per well and incubated with varying concentrations of CD46-ADC or nonbinding control ADC at 37°C for 48 hours. Apoptosis and cell death were assessed in triplicate by FACS for annexin V–FITC and PI. For primary samples, induction of apoptosis and cell death was also studied using the above approach, with the following modifications. Magnetic bead column-separated CD138-positive and -negative cell fractions were each plated at 4 × 104 cells per well and incubated with CD46-ADC or nonbinding ADC at 37°C. After 48 hours, cells were analyzed by FACS using annexin V–FITC, PI, and CD38 (AT1, Santa Cruz Biotechnology). Annexin V staining and PI staining were gated separately for the CD138-positive, CD38-positive MM cells and the CD138-negative, CD38-negative NPCs.
Cell proliferation assays.
The firefly luciferase reporter–expressing lines were used to determine ADC potency in vitro. Cells were plated into 96-well plates at 2,000 per well and incubated for 96 hours with CD46-ADC. Following administration of luciferin, firefly luciferase activity was measured as an indicator of viability using a BioTek Synergy 2. The data were normalized against untreated control wells and EC50 estimated using GraphPad Prism version 6.0c.
For assessment of patient cell sensitivity to ADC, unselected MNC samples were plated at 4 × 104 cells per well in 96-well plates and treated with CD46-ADC or nonbinding ADC at 37°C for 48 hours. Cells were harvested after treatment, washed, and stained with phycoerythrin-conjugated anti-CD138 (BD Biosciences) and FITC-conjugated anti-CD38 antibodies. The numbers of CD138-positive, CD38-positive MM cells and CD138-negative, CD38-negative NPCs were gated and counted, with the curve constructed following normalization to the cell numbers in untreated wells. Treatments were performed in triplicate and plotted with SEM.
BM stromal cell coculture and conditioned media.
BM coculture experiments evaluating ADC activity in MM1.S cells were performed using compartment-specific bioluminescence as previously described (56). Following ammonium-chloride-potassium lysis, CD138-positive cells were isolated from MNCs using the EasySep Human Whole Blood and BM CD138 Positive Selection Kit (Stem Cell Technologies). HS5 and HS27A were purchased from ATCC. BM61 cells were generated from culture of CD138-negative BM MNCs from a myeloma patient sample. HS5, HS27A, or BM61 cells were grown in RPMI1640-supported cultures for later use in monoculture or coculture assays. Coculture assays were initiated using a 2:1 ratio of bone marrow stromal cells to MM cells.
CD46 gene expression of MM cells in coculture was evaluated as follows: MM1.S cells were seeded in monoculture at a density of 5 × 105 cells/ml, or, for coculture, seeded at the same density over 70% confluent stromal cells. After 24 hours, cells were collected and treated with trypsin-free dissociation solution (Accumax), and CD138+ magnetic bead separation (Miltenyi) was used to isolate MM1.S cells. FACS for mCherry expression verified <5% contamination by stromal cells (that do not carry the mCherry gene). mRNA-Seq was performed on MM1.S cells as previously described (57), and sequenced on an Illumina HiSeq 2500. Normalized sequencing reads, in fragments per kilobase per million mapped reads (FPKM), mapping to CD46 canonical transcript sequence were measured. A genome-wide sequencing data set from this experiment will be published separately (A. Wiita and B.T. Aftab, unpublished observations).
BM stromal cell–conditioned medium was collected from HS5 cells cultured under serum-free conditions for 48 hours, as previously described (58). MM1.S and MM1.R cell CD46 and CD38 antigen density was measured with or without the addition of conditioned media for 72 hours.
CD46 knockdown.
CD46 was targeted for knockdown with shRNA oligo with sequence 5′-ATTGGAGAGAGCACGATTTAT-3′. The shRNA sequence was cloned into pLKO.1-GFP vector, containing a U6 promoter to drive shRNA expression and an IRES-GFP as previously described (59). Lentivirus particles were produced in HEK293T cells by cotransfection of shRNA constructs with pPax2 and pMD2.G. H929 MM cell lines were incubated with lentivirus and assessed for infection by GFP expression and CD46 knockdown by FACS 7 days after infection.
In vivo animal study.
For in vivo efficacy assessment of CD46-ADC, 5 × 105 RPIM8226-Luc or MM1.S-Luc cells were injected i.v. into NSG mice (4–6 weeks of age, male and female) (Jackson Laboratory) to create orthometastatic MM xenograft models. Bioluminescence imaging (BLI) was used to monitor graft status (typically injected tumor cells established themselves in the BM and joint in 10 days). Four mouse groups were treated with CD46-ADC, control ADC (MMAF-conjugated to a nonbinding human IgG1), naked anti-CD46 antibody, or vehicle control (PBS). Tumor status was assessed by BLI and results analyzed by Living Image (PerkinElmer). After treatment, mice were continuously monitored for survival endpoints over a period of at least 200 days. HR was determined by log-rank method and significance by Wilcoxon test. For tolerability assessment, transgenic mice expressing human CD46 under its native promoter (MCP) (36) were used (created in strain C57BL/6, backcrossed into C57BL/6 background for over 8 generations). The following primers were used for genetic screening: hMCPTg1 (5′-ATTGTTGCGTCCCATATCT-3′) and hMCPTg2 (5′-CGGAGAAGGAGTACAGC-3′). Eight-week-old male mice were used in the study. Both the CD46-ADC and the YSC10 nonbinding control ADC were tested. Animals were injected with an i.v. bolus of the testing agent at 6 mg/kg (n = 3) and monitored for body weight loss and other overt signs of stress for 14 days. At the end of the experiment, major organs were harvested, formalin-fixed, frozen in liquid nitrogen, cryosectioned by Cryostat (Leica Biosystems), and stained with H&E (ScyTek Laboratories). To assess the status of CD20-positive region in the spleen, we stained the spleen sections with goat anti–mouse CD20 antibody (clone M-20, Santa Cruz Biotechnology), and measured diameters of the CD20-positive regions following treatment (n = 74 for CD46-ADC–treated and 81 for control ADC–treated regions, respectively). P value was calculated by 2-tailed Student’s t test.
Data mining.
CD46 mRNA expression and copy number data were mined from 3 published data sets (31–33). Data from the CoMMpass Study (Interim Analysis 6) were provided by the Multiple Myeloma Research Foundation (https://research.themmrf.org). A detailed explanation for these data mining procedures is provided in the Supplemental Methods.
Statistics.
All data are presented as mean and SEM unless noted. Significance was determined using GraphPad Prism version 6.0c. Two-tailed Student’s t test was used for comparing 2 means. For comparing multiple groups, ANOVA was used with multiple comparison correction. Levels of significance are categorized as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Mouse model sample sizes were determined by preliminary in vivo experience with CD46-ADC and other ADCs published in the literature, rather than power calculation (46), and statistically significant results were observed.
Study approval.
BM samples from MM patients were obtained from the UCSF and University of Colorado Anschutz Medical Campus hematologic malignancies tissue banks with approval from the UCSF and Western Institutional Review Boards, respectively. Informed consent was obtained from all who donated samples. Identifying patient information was replaced with sequentially assigned numbers, in accordance with Health Insurance Portability and Accountability Act guidelines. Mouse studies were approved by the UCSF Animal Care and Use Committee (AN092211-01) and Washington University in St. Louis Animal Studies Committee (20100272A1).
Author contributions
DWS, BTA, YS, CRB, and BL designed experiments. DWS, BTA, YS, CRB, AW, and BCH performed experiments. DAA, PW, and MAS aided in model system development. ACL, JLW, and TGM developed tissue banks and contributed patient samples. XW and JPA conducted the transgenic mice experiment. DWS, BTA, YS, CRB, and BL wrote the manuscript. BL conceived the overall project idea. All authors contributed to manuscript revision.
Supplementary Material
Acknowledgments
We thank David Quigley and Martin Kampmann for statistical and data analysis, Scott Bidlingmaier and Nam-Kyung Lee for help with experiments and discussion, and Emma McMahon, Mike Flanders, Megan Murnane, and Zachary Walker for technical assistance. We thank Constantine Mitsiades of the Dana-Farber Cancer Center for providing cell lines, and Joan Levy and Daniel Auclair at the Multiple Myeloma Research Foundation and all patients, clinical sites, and investigators for contributing to the CoMMpass Study. We also thank Jonathan Keats of the Translational Genomics Research Institute for mRNA-Seq data used from the MMRF MM HMCL69 Cell Line Characterization Project. DWS was supported by a Young Investigator award and BTA by a Grand Scholar award from the UCSF Stephen and Nancy Grand Multiple Myeloma Translational Initiative (MMTI). DAA received support from a Collaborative Innovation Award from the Howard Hughes Medical Institute. PW is an Investigator of the Howard Hughes Medical Institute. The work is supported by grants from the MMTI (to BL, DWS, and BTA), the Multiple Myeloma Research Foundation (to BL and BTA), the National Institute of General Medical Sciences (R01 GM099111 to JPA), and the National Cancer Institute (R01 CA118919, R01 CA171315, and R01 CA129491 to BL).
Footnotes
Conflict of interest: B. Liu, Y. Su, and C.R. Behrens hold patents (PCT/US2015/049492)on novel internalizing anti-CD46 human monoclonal antibodies and therapeutic targeting of cancers overexpressing CD46. B. Liu holds stocks in a biotech company that licensed the patented technology from the University of California. The company did not sponsor this study.
Reference information:J Clin Invest. 2016;126(12):4640–4653. doi:10.1172/JCI85856.
Contributor Information
Yang Su, Email: Yang.Su@ucsf.edu.
Arun Wiita, Email: Arun.Wiita@ucsf.edu.
Diego Acosta-Alvear, Email: diego.acosta.alvear@gmail.com.
Peter Walter, Email: pwalter@biochem.ucsf.edu.
Xiaobo Wu, Email: xwu@wustl.edu.
Bin Liu, Email: bin.liu@ucsf.edu.
References
- 1.Fonseca R, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanamura I, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108(5):1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin TG, et al. SAR650984, a CD38 monoclonal antibody in patients with selected CD38+ hematological malignancies — data from a dose-escalation phase I study. Blood. 2013;122:284 [Google Scholar]
- 4.Plesner T, et al. Daratumumab, a CD38 monoclonal antibody in patients with multiple myeloma-data from a dose-escalation phase I/II study. Blood. 2012;120:73 [Google Scholar]
- 5.Lonial S, et al. Elotuzumab therapy for relapsed or refractory multiple myeloma. N Engl J Med. 2015;373(7):621–631. doi: 10.1056/NEJMoa1505654. [DOI] [PubMed] [Google Scholar]
- 6.Behrens CR, Liu B. Methods for site-specific drug conjugation to antibodies. MAbs. 2014;6(1):46–53. doi: 10.4161/mabs.26632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber HP, Koehn FE, Abraham RT. The antibody-drug conjugate: an enabling modality for natural product-based cancer therapeutics. Nat Prod Rep. 2013;30(5):625–639. doi: 10.1039/c3np20113a. [DOI] [PubMed] [Google Scholar]
- 8.Senter PD, Sievers EL. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol. 2012;30(7):631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 9.Verma S, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367(19):1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Younes A, et al. Results of a pivotal phase II study of brentuximab vedotin for patients with relapsed or refractory Hodgkin’s lymphoma. J Clin Oncol. 2012;30(18):2183–2189. doi: 10.1200/JCO.2011.38.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherbenou DW, Behrens CR, Su Y, Wolf JL, Martin TG, Liu B. The development of potential antibody-based therapies for myeloma. Blood Rev. 2015;29(2):81–91. doi: 10.1016/j.blre.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan W, Sassoon A, An F, Simko JP, Liu B. Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol Cell Proteomics. 2006;5(12):2364–2373. doi: 10.1074/mcp.M600246-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.He J, et al. Targeting prostate cancer cells in vivo using a rapidly internalizing novel human single-chain antibody fragment. J Nucl Med. 2010;51(3):427–432. doi: 10.2967/jnumed.109.069492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 15.Dörig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75(2):295–305. doi: 10.1016/0092-8674(93)80071-L. [DOI] [PubMed] [Google Scholar]
- 16.Le Friec G, et al. The CD46-Jagged1 interaction is critical for human TH1 immunity. Nat Immunol. 2012;13(12):1213–1221. doi: 10.1038/ni.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong HT, et al. Oncolytic measles virus targets high CD46 expression on multiple myeloma cells. Exp Hematol. 2006;34(6):713–720. doi: 10.1016/j.exphem.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Avet-Loiseau H, et al. Molecular cytogenetic abnormalities in multiple myeloma and plasma cell leukemia measured using comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19(2):124–133. doi: 10.1002/(SICI)1098-2264(199706)19:2<124::AID-GCC8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Cigudosa JC, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91(8):3007–3010. [PubMed] [Google Scholar]
- 20.Gutiérrez NC, et al. Differences in genetic changes between multiple myeloma and plasma cell leukemia demonstrated by comparative genomic hybridization. Leukemia. 2001;15(5):840–845. doi: 10.1038/sj.leu.2402116. [DOI] [PubMed] [Google Scholar]
- 21.Balcárková J, et al. Gain of chromosome arm 1q in patients in relapse and progression of multiple myeloma. Cancer Genet Cytogenet. 2009;192(2):68–72. doi: 10.1016/j.cancergencyto.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Biran N, Malhotra J, Bagiella E, Cho HJ, Jagannath S, Chari A. Patients with newly diagnosed multiple myeloma and chromosome 1 amplification have poor outcomes despite the use of novel triplet regimens. Am J Hematol. 2014;89(6):616–620. doi: 10.1002/ajh.23705. [DOI] [PubMed] [Google Scholar]
- 23.Ha KD, Bidlingmaier SM, Zhang Y, Su Y, Liu B. High-content analysis of antibody phage-display library selection outputs identifies tumor selective macropinocytosis-dependent rapidly internalizing antibodies. Mol Cell Proteomics. 2014;13(12):3320–3331. doi: 10.1074/mcp.M114.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kay S, et al. Quantitative flow cytometry of ZAP-70 levels in chronic lymphocytic leukemia using molecules of equivalent soluble fluorochrome. Cytometry B Clin Cytom. 2006;70(4):218–226. doi: 10.1002/cyto.b.20078. [DOI] [PubMed] [Google Scholar]
- 26.Hakulinen J, Junnikkala S, Sorsa T, Meri S. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur J Immunol. 2004;34(9):2620–2629. doi: 10.1002/eji.200424969. [DOI] [PubMed] [Google Scholar]
- 27.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 28.Bidlingmaier S, Su Y, Liu B. Combining phage and yeast cell surface antibody display to identify novel cell type-selective internalizing human monoclonal antibodies. Methods Mol Biol. 2015;1319:51–63. doi: 10.1007/978-1-4939-2748-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doronina SO, et al. Enhanced activity of monomethylauristatin F through monoclonal antibody delivery: effects of linker technology on efficacy and toxicity. Bioconjug Chem. 2006;17(1):114–124. doi: 10.1021/bc0502917. [DOI] [PubMed] [Google Scholar]
- 30.Rozemuller H, et al. A bioluminescence imaging based in vivo model for preclinical testing of novel cellular immunotherapy strategies to improve the graft-versus-myeloma effect. Haematologica. 2008;93(7):1049–1057. doi: 10.3324/haematol.12349. [DOI] [PubMed] [Google Scholar]
- 31.Zhan F, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99(5):1745–1757. doi: 10.1182/blood.V99.5.1745. [DOI] [PubMed] [Google Scholar]
- 32.Zhan F, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109(4):1692–1700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agnelli L, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48(7):603–614. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- 34.Holers VM, Kinoshita T, Molina H. The evolution of mouse and human complement C3-binding proteins: divergence of form but conservation of function. Immunol Today. 1992;13(6):231–236. doi: 10.1016/0167-5699(92)90160-9. [DOI] [PubMed] [Google Scholar]
- 35.Lyzogubov V, et al. Complement regulatory protein CD46 protects against choroidal neovascularization in mice. Am J Pathol. 2014;184(9):2537–2548. doi: 10.1016/j.ajpath.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kemper C, et al. Membrane cofactor protein (MCP; CD46) expression in transgenic mice. Clin Exp Immunol. 2001;124(2):180–189. doi: 10.1046/j.1365-2249.2001.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buettner R, et al. Activated signal transducers and activators of transcription 3 signaling induces CD46 expression and protects human cancer cells from complement-dependent cytotoxicity. Mol Cancer Res. 2007;5(8):823–832. doi: 10.1158/1541-7786.MCR-06-0352. [DOI] [PubMed] [Google Scholar]
- 38.Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol Immunol. 2003;40(2–4):109–123. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 39.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90(8):3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuillème-Toumi S, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19(7):1248–1252. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- 41.Podar K, et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27(6):721–731. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Bougie P, et al. Noxa up-regulation and Mcl-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer Res. 2007;67(11):5418–5424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- 43.Hu J, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119(3):826–837. doi: 10.1182/blood-2011-07-366492. [DOI] [PubMed] [Google Scholar]
- 44.Ikeda H, et al. The monoclonal antibody nBT062 conjugated to cytotoxic Maytansinoids has selective cytotoxicity against CD138-positive multiple myeloma cells in vitro and in vivo. Clin Cancer Res. 2009;15(12):4028–4037. doi: 10.1158/1078-0432.CCR-08-2867. [DOI] [PubMed] [Google Scholar]
- 45.Tassone P, et al. In vitro and in vivo activity of the maytansinoid immunoconjugate huN901-N2’-deacetyl-N2’-(3-mercapto-1-oxopropyl)-maytansine against CD56+ multiple myeloma cells. Cancer Res. 2004;64(13):4629–4636. doi: 10.1158/0008-5472.CAN-04-0142. [DOI] [PubMed] [Google Scholar]
- 46.Tai YT, et al. Novel anti-B-cell maturation antigen antibody-drug conjugate (GSK2857916) selectively induces killing of multiple myeloma. Blood. 2014;123(20):3128–3138. doi: 10.1182/blood-2013-10-535088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell SJ, et al. Remission of disseminated cancer after systemic oncolytic virotherapy. Mayo Clin Proc. 2014;89(7):926–933. doi: 10.1016/j.mayocp.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galanis E, et al. Oncolytic measles virus expressing the sodium iodide symporter to treat drug-resistant ovarian cancer. Cancer Res. 2015;75(1):22–30. doi: 10.1158/0008-5472.CAN-14-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fremeaux-Bacchi V, et al. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2006;17(7):2017–2025. doi: 10.1681/ASN.2005101051. [DOI] [PubMed] [Google Scholar]
- 50.Noone DG, et al. Von Willebrand factor regulates complement on endothelial cells. Kidney Int. 2016;90(1):123–134. doi: 10.1016/j.kint.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forneris F, et al. Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 2016;35(10):1133–1149. doi: 10.15252/embj.201593673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng KW, TenEyck CJ, Galanis E, Kalli KR, Hartmann LC, Russell SJ. Intraperitoneal therapy of ovarian cancer using an engineered measles virus. Cancer Res. 2002;62(16):4656–4662. [PubMed] [Google Scholar]
- 53.Anderson BD, Nakamura T, Russell SJ, Peng KW. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004;64(14):4919–4926. doi: 10.1158/0008-5472.CAN-04-0884. [DOI] [PubMed] [Google Scholar]
- 54.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMillin DW, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16(4):483–489. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiita AP, et al. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C, et al. Paracrine factors produced by bone marrow stromal cells induce apoptosis and neuroendocrine differentiation in prostate cancer cells. Prostate. 2011;71(2):157–167. doi: 10.1002/pros.21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei S, et al. Targeting aberrant glutathione metabolism to eradicate human acute myelogenous leukemia cells. J Biol Chem. 2013;288(47):33542–33558. doi: 10.1074/jbc.M113.511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.