Abstract
Publication of the genome from the clade I organism, Trichinella spiralis, has provided us an avenue to address more holistic problems in parasitology; namely the processes of adaptation and the evolution of parasitism. Parasitism among nematodes has evolved in multiple, independent events. Deciphering processes that drive species diversity and adaptation are keys to understanding parasitism and advancing control strategies. Studies have been put forth on morphological and physiological aspects of parasitism and adaptation in nematodes; however, data is now coming available to investigate adaptation, host switching and parasitism at the genomic level. Herein we compare proteomic data from the clade I parasite, Trichinella spiralis with data from Brugia malayi (clade III), Meloidogyne hapla and Meloidogyne incognita (clade IV), and free-living nematodes belonging to the genera Caenorhabditis and Pristionchus (clade V). We explore changes in protein family birth/death and expansion/reduction over the course of metazoan evolution using Homo sapiens, Drosophila melanogaster and Saccharomyces cerevisiae as out-groups for the phylum Nematoda. We further examine relationships between these changes and the ability and/or result of nematodes adapting to their environments. Data are consistent with gene loss occurring in conjunction with nematode specialization resulting from parasitic worms acclimating to well-defined, environmental niches. We observed evidence for independent, lateral gene transfer events involving conserved genes that may have played a role in the evolution of nematode parasitism. In general, parasitic nematodes gained proteins through duplication and lateral gene transfer, and lost proteins through random mutation and deletions. Data suggest independent acquisition rather than ancestral inheritance among the Nematoda followed by selective gene loss over evolutionary time. Data also show that parasitism and adaptation affected a broad range of proteins, especially those involved in sensory perception, metabolism, and transcription/translation. New protein gains with functions related to regulating transcription and translation, and protein family expansions with functions related to morphology and body development have occurred in association with parasitism. Further gains occurred as a result of lateral gene transfer and in particular, with the cyanase protein family In contrast, reductions and/or losses have occurred in protein families with functions related to metabolic process and signal transduction. Taking advantage of the independent occurrences of parasitism in nematodes, which enabled us to distinguish changes associated with parasitism from species specific niche adaptation, our study provides valuable insights into nematode parasitism at a proteome level using T. spiralis as a benchmark for early adaptation to or acquisition of parasitism.
Keywords: Adaptation, Genomics, Parasitism, Proteomics, Brugia, Caenorhabditis, Meloidogyne, Trichinella
1. Introduction
Parasitic nematodes comprise the majority of eukaryotic parasites and have a broad host range which includes plants, insects, and vertebrates. In humans, nematode parasitism triggers a variety of diseases (Liu et al., 1999; de Silva et al., 2003; Bethony et al., 2006), which pose both physical and financial burdens on healthcare. Indeed, parasitic nematodes infect over 50% of the world's population (Stepek et al., 2006) and impose substantial strain on the global food supply where economic losses in Australia alone exceed 1 billion US dollars; a number that increases 10 fold when losses are examined worldwide (Roeber et al., 2013).
Today, issues have surfaced on control strategies that rely heavily on anthelmintics, such as rapid re-infection, drug resistance, and water and soil contamination. Further, as in the case of parasites in the genus Trichinella, sylvatic hosts can act as reservoirs to introduce or maintain the parasite in the local food supply. Because of the escalation of drug-resistance and the inability to generate immune strategies to curb animal parasitism, producers are once again seeking alternative approaches to controlling parasitic nematodes. In as much as new biochemical approaches are both target-and system-specific, they are intimately linked to basic nematode biology, and in particular, the genetic/genomic mechanisms of parasitism.
It is clear that adaptation, exaptation and host switching have had profound effects on parasitism at the genetic, physiological and ecological levels. As example, plant parasites developed stylets to pierce host cell walls for secretion and feeding, and Tylenchids and Aphelenchids have evolved enlarged secretory cells to produce proteins critical for plant parasitism (Hussey, 1989; Davis et al., 2004;). The parasite cuticle allows for the worm to escape the physiologic and immune defenses of the host (Jones et al., 1993). Parasites of the genus Trichinella use an oral stylet for mechanical rather than enzymatic host cell penetration. Further, Trichinella spp. have adapted to a simple life cycle that is confined to a single host and does not involve a free-living stage; both of which are quite unique among the parasitic nematodes.
Although it is well known that parasitism among the Nematoda evolved multiple times spanning hundreds of millions of years (Dorris et al., 1999), and tropism is not a delineating factor that links parasite clades, questions remain as to whether or not there are conserved, intrinsic genetic factors or external genetic events that link nematode parasites beyond physiological shape and host associations. As example, lateral or horizontal gene transfer appears to have played a role in the evolution of plant parasitism (Kikuchi et al., 2004; Jones et al., 2005; Mitreva et al., 2009). Wang et al. (2010) observed a number of parasitic specific insertions and deletions (indels) in a variety of proteins suggesting that this genetic character could be one mechanism related to the evolution of parasitism. Further, among the sequences currently available, parasite genomes tend to encode fewer genes than those of free-living nematodes suggesting gene deletion and the evolution of a genetic profile more focused on a well-defined and confined life-style are important in parasitism (Mitreva et al., 2011).
Parasitism is a process involving a complex set of genes. Exploring parasitism, therefore, requires data at a genome level that spans the phylum. A recent paper by Zarowiecki and Berriman (2015) showed that few similarities exist among independently evolving parasitic worms; however, they go on to suggest that the process of adaptation is not without some level of conservation or independent convergence on a common outcome, in particular the use of later gene transfer. Fortunately, the number of available draft genomes that cover several major lineages and span both parasitic and free-living nematodes is increasing. In addition, the deduced proteomes of these species have been well characterized based on primary sequence similarity. These data make available a systematic analysis of genetic linkages among parasitic nematodes for determining if specific proteins favor parasitism and for characterizing the parasitic proteins involved. This may also provide critical information for designing alternative treatments to control nematode infections. In this study, we investigated parasitism through protein family dynamics utilizing genomes of species spanning the phylum Nematoda. The genome from Trichinella spiralis appeared prominently in our analysis because it represents a unique organism and a member of the most diversified clade in the Nematoda. Our analyses provide new insights into parasitism and it identified potential targets for better parasite control. Clearly, findings and hypotheses presented in this paper will be tested with more exhaustive comparisons as the nematode genomic database expands.
2. Materials and methods
2.1. The proteomes
The sequenced and annotated genomes that comprised the analysis included 6 free-living nematodes and 4 parasitic nematodes. The 6 free-living nematode genomes were from Caenorhabditis elegans (Consortium, 1998), Caenorhabditis briggsae (Stein et al., 2003), Caenorhabditis remanei, Caenorhabditis brenneri, Caenorhabditis japonica, and Pristionchus pacificus (Dieterich et al., 2008). The 4 parasitic nematode genomes were T. spiralis (Mitreva et al., 2011), Brugia malayi (Ghedin et al., 2007), Meloidogyne incognita (Abad et al., 2008) and Meloidogyne hapla (Opperman et al., 2008). All nematode deduced proteomes were obtained from WormBase [version 204], or the above citations. In addition, the proteomes of Saccharomyces cerevisiae, Homo sapiens, and Drosophila melanogaster were downloaded from BioMart (Smedley et al., 2009) and used as outgroup genomes. Isoforms of the collected proteomes were screened out and only the longest sequences were utilized.
2.2. Family reconstruction
Protein families were defined using Tribe-MCL, a robust algorithm for protein family reconstruction at a genome level (Enright et al., 2002; Enright et al., 2003). Non-clustered proteins from each species as well as clusters composed of single protein members were treated as singleton families.
2.3. Phylogeny reconstruction
The phylogeny of the 10 nematodes together with the 3 outgroup species was inferred based on 94 universal orthologous protein families generated using ORTHOMCL (Li et al., 2003) each occurring exactly once in each of the 13 species. A phylogenetic tree was reconstructed for each family using PROTDIST and FITCH (PHYLIP; Felsenstein, 1989) after having been aligned using MUSCLE (Edgar, 2004). A single consensus tree derived from these family trees was used as the working phylogeny. Branch lengths of the consensus tree were computed using PROML (PHYLIP) based on concatenated sequences from the 94 protein families.
2.4. Inference of protein duplication and deletion
Family member duplication and deletion was inferred using URec (Gorecki and Tiuryn, 2007), which employs tree reconciliation. A species tree was generated using all protein families having one sequence per species and spanning all 13 species (hereafter, referred to as “single member universal families”); the gene trees in this study were those derived from each of the families. Specifically, family sequences were aligned using MUSCLE (Edgar, 2004), protein distances were computed using PROTDIST (Felsenstein, 1989) and family gene trees were reconstructed using FITCH (Felsenstein, 1989). For the species tree, the aligned sequences from all the single member universal families were merged. Subsequently, protein duplications and deletions within each family were inferred using URec by reconciling the species and gene family trees.
2.5. Protein family birth/death inference
Protein family birth and death events were inferred using DOLLOP (Felsenstein, 1989) by treating each family as a character where presence and absence were defined as individual states. This reconstruction of protein family birth/death events permitted exploration of adaptation. Because of the phylogenetic distribution of our species where all free-living organisms were from the “crown” clade i.e., clade V, and all parasitic nematodes were from “root” clades i.e., clades I, III, and IV (Blaxter et al., 1998), death events uniformly occurring among all parasites would be inferred mistakenly as birth events at the ancestor of the clade V free-living nematodes using DOLLOP. Protein sequence similarities were therefore used to correct this misconception. Specifically, for the families inferred as born at the ancestor of clade V free-living nematodes, pairwise protein distance (i.e. similarities) of all the family member sequences were examined. If there existed any pair of paralogs whose distance was more than twice the length (2×) of the longest distance of orthologous pairs from one of the paralogs, the family was treated as ancestral and assumed to have gone through uniform parasite death. A cutoff of 2× was chosen because the phylum first diverged in the Precambrian approximately 543 million years ago (Mya) whereas those of the crown clades are estimated to have begun diversification 300–380 Mya (Blaxter, 2009). Therefore ancestral paralogs diverged 543 Mya while orthologs among the clade V free-living nematodes diverged no more than 300–380 Mya.
2.6. Inferring parasite uniform protein deletions
Tree reconciliation can track duplication and deletion events; however, with respect to multiple copy families, the program is unable to assign gene deletions to a specific copy. Thus, we examined the family protein distance matrix. If a protein within the free-living nematodes exhibited distances with all other homologs that were larger than the average distances of all orthologs between free-living nematodes and T. spiralis (the most highly diverged among the group examined), we assumed these orthologs were uniformly deleted among all other parasites examined. Because the branch distances of orthologs follow speciation, all homologs with large distances was interpreted as the deletion of its orthologs.
Insertions and deletions were inferred from sequence alignments using MUSCLE (Edgar, 2004). Indels at the ends of sequences were excluded as were those that were less than 4 residues in length. Indels specific to parasites were further explored through functional analyses.
In order to construct the phylogenetic tree based upon the cyanase protein family, a BLAST search of NCBI sequences was performed using default parameters and based upon a truncated T. spiralis protein sequence. The longest sequence from each taxonomic order was then used to build the tree. Protein sequences were aligned with MUSCLE (Edgar, 2004), and phylogenetic tree was reconstructed using PROTML (Felsenstein, 1989).
2.7. Functional analysis and gene ontology (GO) enrichment
Putative evolutionary events were detected and assimilated to parasitism and adaptation through functional analyses of the associated protein families and/or proteins. The functional analyses utilized either the GO annotation of C. elegans proteins (from WormBase), or the GO annotation derived from InterProScan (Zdobnov and Apweiler, 2001). GO enrichment/depletion probabilities (multiple test corrected) were computed using FUNC (Prufer et al., 2007).
3. Results
3.1. Reconstruction of protein families and inference of species phylogeny
From the proteomes of the 13 species collected (total of 265,454 proteins) 139,901 protein families were built including 118,174 singleton protein families (Table 1). The 21,727 non-singleton families contained members from C. elegans, C. brenneri, C. japonica, C. remanei, C. briggsae, P. pacificus, M. hapla, M. incognita, B. malayi, T. spiralis, H. sapiens, D. melanogaster, and S. cerevisiae, (Supplemental Table 1). Among these 1589 families were universal to all nematodes, and 739 families were universal to all species. In addition, there were 683 families universal and specific to free-living nematodes i.e., present in all free-living nematodes in this study. In contrast, there were only 8 families universal and specific to all non-parasitic species (including outgroups), and only three families were universal and specific to parasites (Supplemental Table 2). One of these three families is a LEM domain containing protein that shares similarity with the human MAN1 gene which regulates the expression of several fundamental genes; the second family is an RNA recognition motif domain containing protein (T. spiralis annotation), that shares similarities with a splice factor in Schistosoma japonicum; the third family is cyanase, a gene family evolutionarily linked to plants, bacteria and fungi.
Table 1.
Reconstruction of protein families from the proteomes of 6 free-living nematodes, 4 parasitic nematodes and 3 outgroup species.
| Species | Total proteins | Proteins in OPFsa | Clustered singletons | OPFs with >1 member | Proteins in OPFs with >1 member | |
|---|---|---|---|---|---|---|
| Outgroups | Homo sapiens | 24,013 | 17,807 | 6206 | 5463 | 11,669 |
| Drosophila melanogaster | 14,141 | 9466 | 4675 | 4274 | 8949 | |
| Saccharomyces cerevisiae | 6698 | 3343 | 3355 | 1725 | 5080 | |
| Nematodes | Caenorhabditis elegans | 20,173 | 18,602 | 1571 | 9234 | 10,805 |
| C. brenneri | 30,702 | 26,949 | 3753 | 9650 | 13,403 | |
| C. japonica | 25,870 | 20,695 | 5175 | 8672 | 13,847 | |
| C. remanei | 31,518 | 28,675 | 2843 | 10,171 | 13,014 | |
| C. briggsae | 21,974 | 19,194 | 2780 | 9626 | 12,406 | |
| Pristionchus pacificus | 29,201 | 18,649 | 10552 | 6401 | 16,953 | |
| Meloidogyne hapla | 14,421 | 11,824 | 2597 | 6141 | 8738 | |
| M. incognita | 19,212 | 17,109 | 2103 | 5833 | 7936 | |
| Brugia malayi | 11,407 | 8376 | 3031 | 4686 | 7717 | |
| Trichinella spiralis | 16,124 | 10,804 | 5320 | 4064 | 9384 |
OPF, orthologous protein families.
To better explore parasitism and adaptation, a reliable phylogeny was needed. Current nematode phylogeny is mainly based on the 18 s rRNA study of Blaxter et al. (Blaxter et al., 1998; Holterman et al., 2006; Meldal et al., 2007). In addition, the relationship between the nematodes and the outgroups included is also not well resolved (Wolf et al., 2004; Philip et al., 2005;). For this purpose, we identified 94 single copy orthologous universal protein families using OrthoMCL (Li et al., 2003) to reconstruct a consensus tree. Branch lengths of the tree were calculated using PROML of PHYLIP (Felsenstein, 1989) based on the concatenated sequences from the 94 single copy universal orthologous families. This tree (Supplemental Fig. 1) differs from that based upon the 18 S rRNA (Blaxter et al., 1998; Meldal et al., 2007) which depicted B. malayi as an outgroup of Meloidogyne and other nematodes.
It is clear that parasitic nematodes do not have extended branch lengths, suggesting that positive selection on parasites did not lead to increased amino acid substitutions at least among conserved proteins. Indeed, the branch lengths of the free-living nematodes in particular those of the genus Caenorhabditis, are longer than those of parasites.
3.2. Protein family birth/death
Protein family birth/death events are defined as the emergence and disappearance of protein families, respectively. In the process of nematode evolution, protein encoding genes were very dynamic where thousands of protein births and deaths occurred in conjunction with each speciation event (Fig. 1). Generally and coincident with speciation, organisms tend to experience net gains in the number of protein families they acquire. However, parasitic nematodes exhibited more protein family deaths than those observed in the free-living organisms we examined (except for P. pacificus and C. brenneri). More than 1000 families disappeared after the splits of T. spiralis, B. malayi, and the common ancestor of M. hapla and M. incognita (Fig. 1). Further, parasitic nematodes showed fewer protein family births, though it is not clear if this is related to the higher sampling density of the free-living nematodes. Thus, an increase in deaths is one contributing factor to parasites having fewer protein families (Table 1).
Fig. 1.
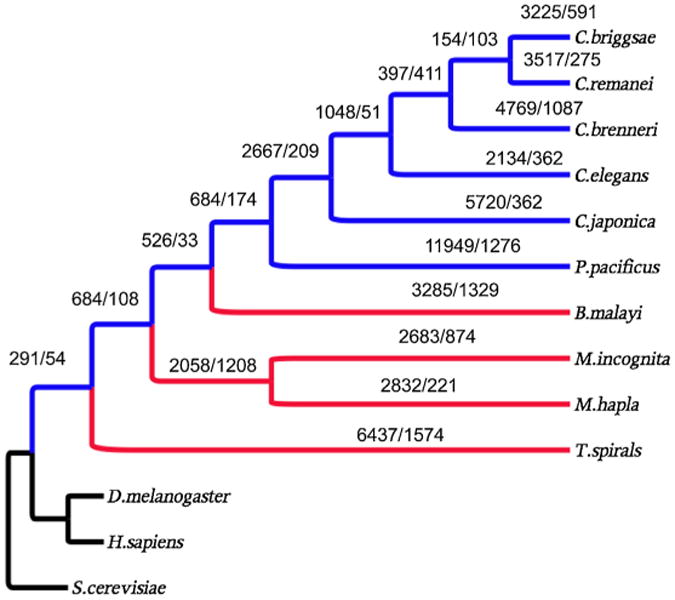
Protein family birth (emergence)/death (disappearance) over the course of evolution. Blue = free-living nematodes; Red = parasitic nematodes; Black = outgroups.
As the number of family deaths increases along each individual parasite lineage, there is also an increase in family deaths at the last common ancestor (LCA) of the free-living nematodes of clade V if considering the branch lengths. The branch length of the LCA of free-living nematodes is 0.13, and that of the LCA of free-living nematodes and B. malayi is 0.07; however, the family deaths at these two LCAs are 174 and 33, respectively (Fig. 1). These increased deaths are again related to parasitism and adaptation since 113 of these 174 families are parasite specific.
Because of their phylogenetic distribution, protein families born after the split of the nematodes and arthropods, and that subsequently died in all nematode parasites could be inferred as families born at the LCA of the clade V, free-living nematodes. Given that parasitism evolved numerous times within the phylum Nematoda, and that diversification only supports free-living nematodes as the ancestral state, we corrected these inferences by comparing the distances between orthologs and paralogs of these families. Among the 684 families inferred as arising at the ancestor of the free-living nematodes examined herein, 398 have paralogs in some of the free-living nematodes and 225 of them possess old paralogs (more than twice the age of their oldest orthologs, (see Materials and Methods)) indicating their births occurred before the split of the basal groups i.e., T. spiralis. We suggest that these families died uniformly in parasites.
3.3. Protein family birth/death and their parasitic association
The association between parasitism and the birth and death of protein families can be illustrated by the GO term enrichments of families related to parasites. The above 113 parasite specific families coincided with 28 GO terms in biological process and 44 GO terms in molecular function (Table 2). These biological process GO terms have functional enrichment in regulation of translational fidelity (GO:0006450), cyanate metabolic process (GO:0009439), and transcription initiation (GO:0006352). And the 225 families that died uniformly in parasites have enrichments in GO terms related to free-living, and depletions in GO terms related to basic cellular functions (Table 3). The most highly enriched biological process GO terms are G-protein coupled receptor protein signaling pathway (GO:0007186), neuropeptide signaling pathway (GO:007218), and activation of MAPKK activity (GO:0000186).
Table 2.
GO term enrichment of protein families died at the ancestor of free-living nematodes and not present in the outgroup species.
| GO Category | GO term | GO descriptor | Under-represent P value | Over-represent P value |
|---|---|---|---|---|
| Biological processes | ||||
| GO:0006139 | nucleobase, nucleoside, nucleotide and na metabolic process | 0.137 | 1.000 | |
| GO:0006260 | DNA replication | 0.977 | 0.215 | |
| GO:0006281 | DNA repair | 0.964 | 0.265 | |
| GO:0006351 | transcription, DNA-dependent | 0.409 | 0.872 | |
| GO:0006352 | transcription initiation | 1.000 | 0.006 | |
| GO:0006397 | mRNA processing | 0.998 | 0.065 | |
| GO:0006450 | regulation of translational fidelity | 1.000 | 0.002 | |
| GO:0006461 | protein complex assembly | 0.996 | 0.092 | |
| GO:0006508 | proteolysis | 0.933 | 0.249 | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 0.983 | 0.186 | |
| GO:0006810 | transport | 0.249 | 1.000 | |
| GO:0006811 | ion transport | 0.439 | 1.000 | |
| GO:0006886 | intracellular protein transport | 0.967 | 0.251 | |
| GO:0007049 | cell cycle | 0.988 | 0.154 | |
| GO:0007154 | cell communication | 0.292 | 1.000 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.900 | 0.424 | |
| GO:0008152 | metabolic process | 0.193 | 1.000 | |
| GO:0008154 | actin polymerization and/or depolymerization | 1.000 | 0.016 | |
| GO:0009058 | biosynthetic process | 0.395 | 1.000 | |
| GO:0009439 | cyanate metabolic process | 1.000 | 0.004 | |
| GO:0009987 | cellular process | 0.131 | 0.975 | |
| GO:0016192 | vesicle-mediated transport | 0.987 | 0.160 | |
| GO:0016998 | cell wall catabolic process | 0.999 | 0.041 | |
| GO:0032259 | methylation | 0.990 | 0.143 | |
| GO:0043283 | biopolymer metabolic process | 0.204 | 0.953 | |
| GO:0044238 | primary metabolic process | 0.326 | 0.874 | |
| GO:0044249 | cellular biosynthetic process | 0.179 | 1.000 | |
| GO:0050794 | regulation of cellular process | 0.096 | 0.983 | |
| Molecular function | ||||
| GO:0000166 | nucleotide binding | 0.924 | 0.217 | |
| GO:0003676 | nucleic acid binding | 0.847 | 0.262 | |
| GO:0003700 | transcription factor activity | 0.462 | 0.780 | |
| GO:0003735 | structural constituent of ribosome | 0.424 | 1.000 | |
| GO:0003824 | catalytic activity | 0.137 | 1.000 | |
| GO:0003896 | DNA primase activity | 1.000 | 0.016 | |
| GO:0004175 | endopeptidase activity | 0.452 | 1.000 | |
| GO:0004252 | serine-type endopeptidase activity | 0.941 | 0.328 | |
| GO:0004298 | threonine endopeptidase activity | 0.999 | 0.036 | |
| GO:0004871 | signal transducer activity | 0.151 | 1.000 | |
| GO:0005083 | small GTPase regulator activity | 0.941 | 0.328 | |
| GO:0005179 | hormone activity | 0.885 | 0.444 | |
| GO:0005198 | structural molecule activity | 0.962 | 0.169 | |
| GO:0005215 | transporter activity | 0.384 | 1.000 | |
| GO:0005509 | calcium ion binding | 0.407 | 1.000 | |
| GO:0005515 | protein binding | 0.996 | 0.010 | |
| GO:0005524 | ATP binding | 0.178 | 0.959 | |
| GO:0008073 | ornithine decarboxylase inhibitor activity | 1.000 | 0.020 | |
| GO:0008158 | hedgehog receptor activity | 0.999 | 0.048 | |
| GO:0008234 | cysteine-type peptidase activity | 0.899 | 0.420 | |
| GO:0008270 | zinc ion binding | 0.957 | 0.089 | |
| GO:0008324 | cation transmembrane transporter activity | 0.321 | 1.000 | |
| GO:0008824 | cyanate hydratase activity | 1.000 | 0.007 | |
| GO:0009055 | electron carrier activity | 0.450 | 1.000 | |
| GO:0016301 | kinase activity | 0.477 | 0.831 | |
| GO:0016491 | oxidoreductase activity | 0.061 | 1.000 | |
| GO:0016740 | transferase activity | 0.198 | 0.952 | |
| GO:0016787 | hydrolase activity | 0.428 | 0.857 | |
| GO:0016788 | hydrolase activity, acting on ester bonds | 0.262 | 1.000 | |
| GO:0016836 | hydro-lyase activity | 0.996 | 0.094 | |
| GO:0016874 | ligase activity | 0.483 | 1.000 | |
| GO:0016986 | transcription initiation factor activity | 1.000 | 0.003 | |
| GO:0017111 | nucleoside-triphosphatase activity | 0.437 | 1.000 | |
| GO:0022857 | transmembrane transporter activity | 0.498 | 1.000 | |
| GO:0022892 | substrate-specific transporter activity | 0.414 | 1.000 | |
| GO:0030234 | enzyme regulator activity | 0.386 | 1.000 | |
| GO:0030528 | transcription regulator activity | 0.312 | 1.000 | |
| GO:0042578 | phosphoric ester hydrolase activity | 0.489 | 1.000 | |
| GO:0043169 | cation binding | 0.459 | 1.000 | |
| GO:0043565 | sequence-specific DNA binding | 0.150 | 1.000 | |
| GO:0046872 | metal ion binding | 0.320 | 1.000 | |
| GO:0046983 | protein dimerization activity | 0.901 | 0.416 | |
| GO:0048037 | cofactor binding | 0.427 | 1.000 | |
| GO:0051537 | 2 iron, 2 sulfur cluster binding | 1.000 | 0.032 | |
Table 3.
GO enrichment of protein families disappeared uniformly in all nematodes (free-living nematode specific families having born time before the split of Trichinella spiralis).
| GO Category | GO terms | GO descriptor | Under-represent P value | Over-represent P value |
|---|---|---|---|---|
| Biological processes | ||||
| GO:0000186 | activation of MAPKK activity | 1.00E + 00 | 2.45E − 02 | |
| GO:0002119 | nematode larval development | 2.90E − 02 | 9.83E − 01 | |
| GO:0006303 | double-strand break repair via non-homologous end joining | 9.99E − 01 | 4.85E − 02 | |
| GO:0006412 | translation | 1.74E − 02 | 1.00E + 00 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 9.99E − 01 | 4.96E − 03 | |
| GO:0007218 | neuropeptide signaling pathway | 9.99E − 01 | 1.92E − 02 | |
| GO:0009792 | embryonic development ending in birth or egg hatching | 2.28E − 03 | 9.99E − 01 | |
| GO:0040017 | positive regulation of locomotion | 3.40E − 02 | 1.00E + 00 | |
| GO:0048268 | clathrin coat assembly | 9.99E − 01 | 4.85E − 02 | |
| Molecular function | ||||
| GO:0001584 | rhodopsin-like receptor activity | 9.96E − 01 | 1.69E − 02 | |
| GO:0003707 | steroid hormone receptor activity | 1.00E + 00 | 5.07E − 04 | |
| GO:0003735 | structural constituent of ribosome | 4.83E − 02 | 1.00E + 00 | |
| GO:0003993 | acid phosphatase activity | 9.99E − 01 | 1.64E − 02 | |
| GO:0004190 | aspartic-type endopeptidase activity | 1.00E + 00 | 3.39E − 03 | |
| GO:0004497 | monooxygenase activity | 9.98E − 01 | 1.79E − 02 | |
| GO:0004792 | thiosulfate sulfurtransferase activity | 9.99E − 01 | 4.47E − 02 | |
| GO:0004930 | G-protein coupled receptor activity | 9.98E − 01 | 2.88E − 02 | |
| GO:0005158 | insulin receptor binding | 9.99E − 01 | 4.47E − 02 | |
| GO:0005506 | iron ion binding | 9.96E − 01 | 1.79E − 02 | |
| GO:0005545 | phosphatidylinositol binding | 9.99E − 01 | 4.47E − 02 | |
| GO:0008191 | metalloendopeptidase inhibitor activity | 9.99E − 01 | 4.47E − 02 | |
| GO:0008825 | cyclopropane-fatty-acyl-phospholipid synthase activity | 1.00E + 00 | 2.26E − 02 | |
| GO:0009055 | electron carrier activity | 9.99E − 01 | 3.99E − 03 | |
| GO:0020037 | heme binding | 9.98E − 01 | 1.16E − 02 | |
| GO:0045296 | cadherin binding | 9.99E − 01 | 4.47E − 02 | |
| GO:0051015 | actin filament binding | 1.00E + 00 | 9.87E − 03 | |
The limited numbers of shared families suggests the independent occurrences or evolution of parasitism and adaptation. However, the functional enrichments of protein families that were born or that died at each parasitic lineage illustrated the commonality of functional adaptation. In spite of the fact that these families appeared or disappeared in distinct lineages, they shared functional enrichments. Protein families born at different parasitic lineages exhibited significant enrichment in molecular function terms relative to biological process terms. These families shared enrichments in zinc ion binding (GO:0008270), chromatin binding (GO:0003682), protein dimerization activity (GO:0046983), and cyclin-dependent protein kinase inhibitor activity (GO:0004861) (Fig. 2, and Supplemental Table 3). In contrast, the protein families that died at different parasitic lineages exhibited greater significant enrichment in biological process GO terms than in molecular function GO terms. These families did not show common functional enrichment over all four parasitic lineages, but have three enriched terms shared by B. malayi, T. spiralis, and Meloidogyne (Fig. 3, and Supplemental Table 4). As expected, the enrichment patterns of B. malayi and T. spiralis are close, and those of M. incognita and M. hapla are similar. In addition, those of the families that died at the LCA of Meloidogyne share high commonality with those of B. malayi and T. spiralis. This suggests that among this group, parasitism occurred at the LCA of M. hapla and M. incognita, and that M. hapla and M. incognita acquired host specificity after becoming species.
Fig. 2.
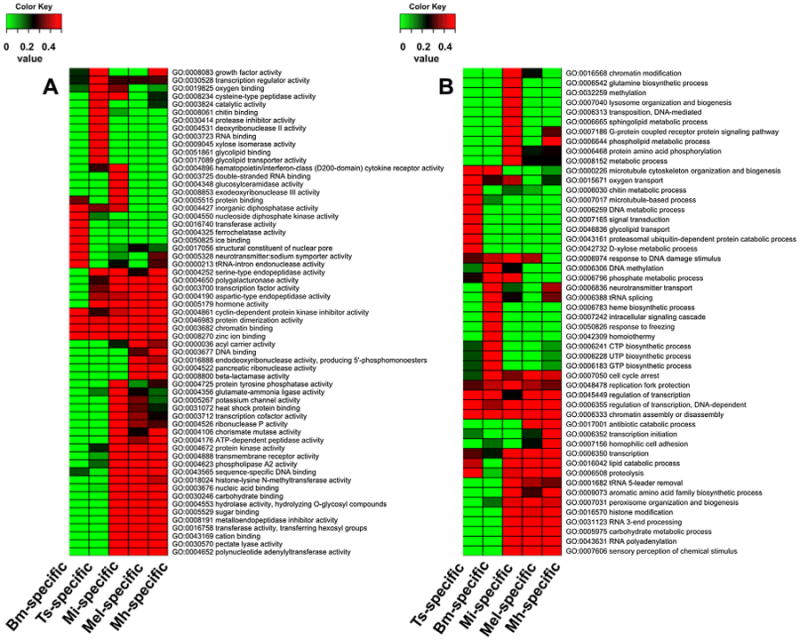
Heatmap showing the GO enrichment of protein families born at each parasite lineage A = molecular function, B = biological process. Bm = B. malayi; Ts = T. spiralis; Mi = M. incognita; Mel = all Meloidogyne; Mh = M. hapla.
Fig. 3.
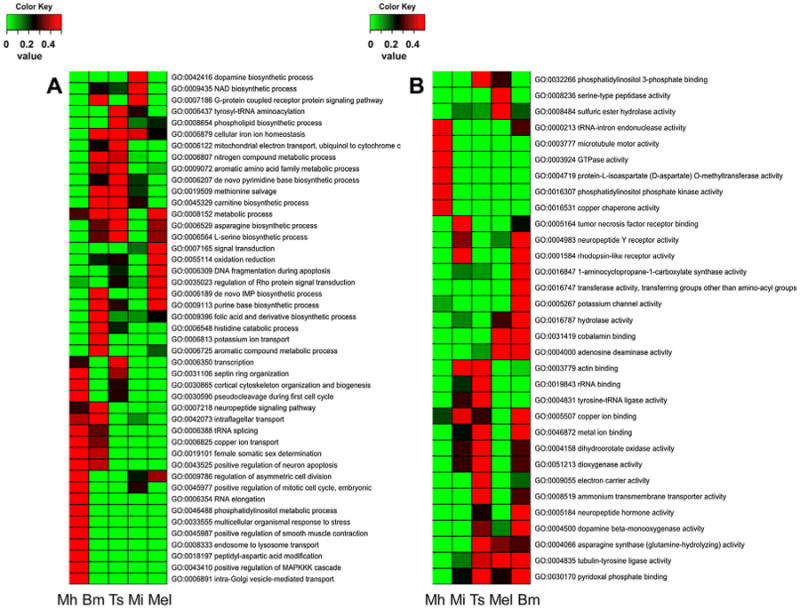
Heatmap showing GO enrichment of families that died at the different parasitic lineages. A = Biological process; B = molecular function.
3.4. Protein family expansion and reduction
The dynamic nature of whole protein families is mirrored in the plasticity of individual protein family members. Examining member duplication and deletion within families universal to nematodes revealed that each family underwent many duplication and deletion events (Fig. 4) over evolution. This in turn resulted in changes to the sizes of the families. Unlike changes in protein family births and deaths, greater numbers of deletions occurred relative to duplications in association with each speciation event suggesting a reduction in family sizes. Parasitic and free-living nematodes are somewhat similar in the numbers of deletion events that occurred; however, parasitic nematodes tend to have fewer duplications than free-living nematodes (with the exception of M. incognita) which leads to smaller protein families in parasites (P < 0.008, TTEST) (Table 1).
Fig. 4.

Duplications/deletions of all universal nematode protein families (1572 families). Blue = Free living nematodes; Red = parasitic nematodes; Black = outgroups.
More than 80% of the families were reduced in size over the course of nematode evolution and parasitism. In fact, more than 50% of the families were reduced in size in all the parasites examined. In contrast, less than 10% of the protein families expanded in size in B. malayi, T. spiralis and M. hapla. Expansion in M. incognita is different where 30% of the families expanded predominantly due to an increase in duplications (1336 duplications in 725 families), no reduced deletions. The top three expanded families are those homologous to C. elegans R09B3.4 (ubc-12, ubiquitin-conjugating enzyme; 24 duplications; 4 deletions), K03E6.3 (ncs-3, neuronal calcium sensor, calcium binding; 14 duplications; 1 deletion) and C39E9.6 (scl-8, SCP-like extracellular protein, defense related protein; 20 duplications; 5 deletions).
Consistent with the independence of parasitism and adaptation, only one family has shown gene expansion over all four parasites examined. Meanwhile these independent events also lead to an excessive number of reductions in common families, though parasitic nematodes do not show increased numbers of total protein deletions. Among the 1572 families exhibiting inferences of duplications and deletions, 790 families showed reductions over all four parasites examined. Also, this appeared to be non-random protein deletion wherein the orthologs of 524 C. elegans genes were uniformly deleted in all four parasites. These orthologs were distributed only over 93 families.
3.5. Protein family expansion and reduction and their parasitic association
We examined the GO term enrichment of families that expanded or reduced over individual parasite lineages and over all parasite lineages investigated. The GO enrichments of the above 524C. elegans orthologs were also examined. The families that expanded at the different parasitic lineages exhibited different functional enrichments (Fig. 5) while the families that were reduced at different parasitic lineages showed similar functional enrichments (Fig. 6). In fact, the functional enrichments of reduced families in parasites have a more than 60 percent overlap with those also reduced in C. elegans. Fig. 6 displays only the parasite specific enrichments. Many of these parasite specific enrichments are related to signal processing, such as two-component sensor activity (GO:0000155), protein prenyltransferase activity (for receptor localization, GO:0008318), serotonin receptor activity (GO:0004993), drug binding (GO:0008144) and extracellular-glutamate-gated chloride channel activity (GO:0008068). Despite the different enrichment patterns, the majority of the families found to contain enriched terms in parasites are related to growth and development possibly associated with the unique food sources. Glycolysis was not enriched in the protein families that expanded in T. spiralis though it is enriched in those of the other three parasites (Fig. 5A). The 524C. elegans genes where their orthologs were uniformly deleted in all parasites were predominantly kinases and involved channel activities such as ATP binding (GO:0005524), nicotinic acetylcholine-activated cation-selective channel activity (GO:0004889), protein serine/threonine kinase activity ()GO:0004674, protein tyrosine kinase activity (GO:0004713), etc. (Table 4).
Fig. 5.

GO term enrichment pattern of protein families expanded at different parasite lineages C. elegans lineage was added as reference; A = biological process; B = molecular function; Bm = B. malayi; Ts = T. spiralis; Mi = M. incognita; Mel = all Meloidogyne; Mh = M. hapla; Ce = C. elegans.
Fig. 6.

GO enrichment pattern (molecular function) of families that were reduced over different parasite lineages. Bm = B. malayi; Ts = T. spiralis; Mi = M. incognita; Mel = all Meloidogyne; Mh = M. hapla; All = all parasites.
Table 4.
Molecular function GO enrichment of the 524 Caenorhabditis elegans genes whose orthologs were uniformly deleted in all parasitic species.
| Molecular functions (12426) | GO Descriptor | Under-represent P value | Over-represent P value |
|---|---|---|---|
| GO:0005524 | ATP binding | 1.00E + 00 | 9.61E − 26 |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | 1.00E + 00 | 1.55E − 25 |
| GO:0004674 | protein serine/threonine kinase activity | 1.00E + 00 | 9.24E − 19 |
| GO:0004713 | protein tyrosine kinase activity | 1.00E + 00 | 2.68E − 18 |
| GO:0016787 | hydrolase activity | 1.00E + 00 | 9.48E − 17 |
| GO:0004386 | helicase activity | 1.00E + 00 | 9.84E − 16 |
| GO:0005230 | extracellular ligand-gated ion channel activity | 1.00E + 00 | 3.68E − 12 |
| GO:0005328 | neurotransmitter:sodium symporter activity | 1.00E + 00 | 1.21E − 10 |
| GO:0015464 | acetylcholine receptor activity | 1.00E + 00 | 5.93E − 09 |
| GO:0019001 | guanyl nucleotide binding | 1.00E + 00 | 8.36E − 08 |
| GO:0004222 | metalloendopeptidase activity | 1.00E + 00 | 2.10E − 04 |
| GO:0004725 | protein tyrosine phosphatase activity | 1.00E + 00 | 4.18E − 04 |
| GO:0004879 | ligand-dependent nuclear receptor activity | 1.00E + 00 | 5.80E − 04 |
| GO:0019787 | small conjugating protein ligase activity | 1.00E + 00 | 7.09E − 04 |
| GO:0022848 | acetylcholine-gated cation channel activity | 1.00E + 00 | 9.93E − 04 |
| GO:0005388 | calcium-transporting ATPase activity | 1.00E + 00 | 1.31E − 03 |
| GO:0003677 | DNA binding | 9.99E − 01 | 1.60E − 03 |
| GO:0004016 | adenylate cyclase activity | 1.00E + 00 | 5.71E − 03 |
| GO:0004402 | histone acetyltransferase activity | 9.99E − 01 | 8.75E − 03 |
| GO:0004871 | signal transducer activity | 9.97E − 01 | 9.42E − 03 |
| GO:0016706 | oxidoreductase activity, acting on paired donors | 9.99E − 01 | 9.63E − 03 |
| GO:0017111 | nucleoside-triphosphatase activity | 9.96E − 01 | 9.69E − 03 |
| GO:0005509 | calcium ion binding | 9.92E − 01 | 1.52E − 02 |
| GO:0005330 | dopamine:sodium symporter activity | 1.00E + 00 | 3.15E − 02 |
| GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity | 1.00E + 00 | 3.15E − 02 |
| GO:0035034 | histone acetyltransferase regulator activity | 1.00E + 00 | 3.15E − 02 |
| GO:0016849 | phosphorus-oxygen lyase activity | 9.91E − 01 | 3.95E − 02 |
| GO:0003712 | transcription cofactor activity | 9.92E − 01 | 4.74E − 02 |
| GO:0003723 | RNA binding | 1.76E − 02 | 9.96E − 01 |
| GO:0004888 | transmembrane receptor activity | 1.63E − 02 | 9.96E − 01 |
| GO:0003707 | steroid hormone receptor activity | 3.01E − 03 | 9.99E − 01 |
| GO:0003824 | catalytic activity | 8.97E − 04 | 1.00E + 00 |
| GO:0005215 | transporter activity | 3.18E − 04 | 1.00E + 00 |
| GO:0016491 | oxidoreductase activity | 1.26E − 04 | 1.00E + 00 |
| GO:0005488 | binding | 2.06E − 05 | 1.00E + 00 |
| GO:0003735 | structural constituent of ribosome | 3.38E − 05 | 1.00E + 00 |
| GO:0004497 | monooxygenase activity | 1.73E − 02 | 1.00E + 00 |
| GO:0008270 | zinc ion binding | 2.40E − 09 | 1.00E + 00 |
| GO:0009055 | electron carrier activity | 2.27E − 04 | 1.00E + 00 |
| GO:0016301 | kinase activity | 2.31E − 02 | 1.00E + 00 |
| GO:0016747 | transferase activity, transferring non amino-acyl groups | 2.03E − 02 | 1.00E + 00 |
| GO:0016758 | transferase activity, transferring hexosyl groups | 1.52E − 02 | 1.00E + 00 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 2.31E − 02 | 1.00E + 00 |
| GO:0020037 | heme binding | 1.96E − 03 | 1.00E + 00 |
| GO:0046872 | metal ion binding | 2.90E − 02 | 1.00E + 00 |
| GO:0050660 | FAD binding | 3.09E − 02 | 1.00E + 00 |
3.6. Origination of parasitic proteins
Birth of parasite specific protein families can occur through sub-functionalization, lateral gene transfer (LGT) or protein family expansion. As stated above, we found only 3 families that were common to all parasitic nematodes. The majority of the families (94/113) were common among B. malayi and M. incognita or M. hapla (77 were common to all three). Consistent with our inferred tree (Supplemental Fig. 1), very limited numbers of families are common among B. malayi and T. spiralis and even less so with Meloidogyne. The limited number of protein families that may be linked to parasitism, and their consistency within the phylogeny suggest that parasitism and adaptation occurred independently over different species, and that parasitism proteins were acquired mainly from speciation through duplication and through sub-functionalization or exaptation.
It should also be noted that LGT played a role in the birth of some of these families. The phylogenetic tree of cyanase clearly illustrates this. Despite the independent acquisition of parasitism, all the parasites in our studies appear to have acquired cyanase through LGT. Reconstructing the evolution of cyanase showed no less than 4 independent acquisitions of cyanase in nematodes, one for the ancestor of Trichinella, one for B malayi, one for the ancestor of Meloidogyne, and possibly one additional acquisition for T. spiralis (Fig. 7). Cyanase was also found in Loa loa, a filarial nematode that was highly similar to that of B. malayi suggesting a common origin for these acquisitions (data not shown)
Fig. 7.
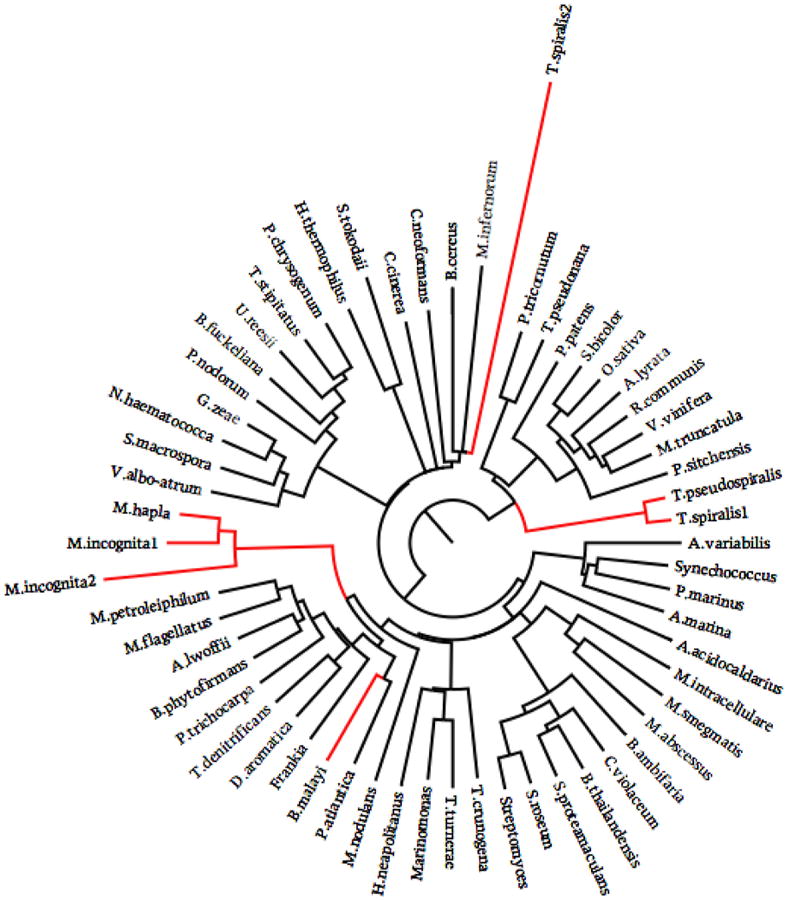
Evolutionary tree of cyanase showing 4 independent events of lateral gene transfer.
4. Discussion
Studies on nematode parasitism and adaptation usually focus on a specific species, and explore only a single protein or protein family. Consequently, there is a lack of understanding of these events at the holistic level. In this study, we explored at the genome level, similarities and distinctions among a subset of parasitic and free-living nematodes as they relate to parasitism and adaptation. We used a subset of genomes from organisms representing different clades and tropisms among the Nematoda. Our results revealed some genetic mechanisms underlying different modes of adaptation and validated the multiple independent adaptation events that occurred during the evolution of parasitism.
4.1. Independent adaptation among parasitic nematodes
Parasitism independently originated multiple times over the evolution of the nematodes (Dorris et al., 1999; Blaxter, 2003). These independent events are supported by the observation that only three protein families were exclusively shared by all parasitic nematodes, and that only one family expanded in this group of nematodes. One of the families exclusively shared by all parasites examined here is cyanase. The independent origins of these parasitic cyanases, reflected by their phylogenetic relationships with other cyanases, provides direct evidence for the acquisition of the gene via LGT, which was key to the evolution of the parasitic character of these organisms (Jones et al., 2005; Mitreva et al., 2009). The three plant parasitic cyanases appear to have similar origins, meaning that LGT occurred before the split of M. hapla and M. incognita. It is interesting to note that this gene was either not acquired by the free-living nematodes, or that after early acquisition, it was lost in the process of further diversification of the free-living state.
Meanwhile, the free-living nature of nematode ancestor is illustrated by examining the functions of the protein families that emerged before the split of the clade I nematode, T. spiralis and that were lost in the evolution of the nematode parasites studied herein. The top enriched biological process GO terms are G-protein coupled receptor protein signaling pathway (GO:0007186), neuropeptide signaling pathway (GO:007218), and activation of MAPKK activity (GO:0000186). One can envision where all three processes are crucial for free-living nematodes by permitting more rapid responses to environmental change e.g., olfactory perception and the localization of food resources (Thomas and Robertson, 2008). Further, MAPKK activity is key to osmolarity sensing and thus important for free-living organisms. These functional enrichments in free-living worms further support the ancestral character of the Nematoda was the free-living state and that adaptation associated with parasitism resulted in the independent death or loss of these families in the parasites examined here.
4.2. Parasitic adaptation through gene loss
The proteomes of nematodes have exhibited a high level of plasticity over the course of evolution. Parasite-based adaptations resulted in reductions in the sizes of the proteomes investigated to date (Mitreva et al., 2011), including fewer protein families and a reduction in the number of members of those protein families. More protein families disappeared in the evolution of parasitic nematodes than in the adaptation of free-living nematodes to their local environments. However, the protein families that disappeared at the lineages of T. spiralis, B. malayi and the common ancestor of Meloidogyne seemed to exhibit conservation in function. The molecular functions of the families that disappeared (relative to free-living nematodes) were significantly enriched in asparagine synthase activity, tubulin-tyrosine ligase activity, and pyridoxal phosphate binding. Among the biological processes examined, the most significant enriched GO term was metabolic processes.
One can envision where adaptation to parasitism can lead to reductions in genes involved in metabolic processes because of a simplification in food and energy resources for parasites as their life cycles become more predictable, confined and less dependent upon the outside environment (Blaxter, 2003). Examining shared molecular functional enrichments provides more enlightenment on this process of adaptation. Asparagine synthase involves the biosynthesis of l-asparagine from l-aspartate in an ATP-dependent reaction. It has been shown that asparagine synthase expression correlates with nutrient stress (Fafournoux et al., 2000) where increasing nutrients decreases gene expression. Thus the disappearance of these proteins in the parasitic nematodes may relate to the abundance of available asparagine in the host and a reduced need for biosynthesis by the parasite. Pryodoxal phosphate binding involves vitamin B6 activity, where vitamin B6 is a cofactor in many biological processes, including antioxidant activity (Denslow et al., 2005). Again, the disappearance of these proteins may reflect reduced oxidative stress of the parasite within the host environment compared to that of free-living nematodes that are exposed to a larger number of more complex metabolic substrates and other metabolizing organisms (Dieterich and Sommer, 2009; Zarowiecki and Berriman, 2015). While parasite-based, adaptation-associated gene loss can consist of functionally related proteins; such loss can also trigger ortholog deletion over all parasitic nematodes. We detected a loss of 524C. elegans orthologs within the 93 nematode universal members.
In addition to the individual families that disappeared at each parasitic lineage, there were 250 protein families that emerged prior to the split of T. spiralis then disappeared uniformly across all parasites. As stated previously, these families were enriched in functions that probably exhibit redundancy in parasitic nematodes. Besides the disappearances of whole protein families, a large number of the remaining protein families also lost members. Nearly 50% of the families that emerged before the split of T. spiralis were reduced in size in all the parasites examined. When checking individually, more than 80% percent of those families were reduced in size in at least one parasite group. This group of protein families possesses functional enrichment patterns biased towards signal processing which uniformly disappeared in all nematode parasites in this study. Interestingly, enrichments in drug binding and extracellular-glutamate-gated chloride channel activity among the reduced families suggest the build-up of drug resistant character. It has been shown that mutations in extracellular-glutamate-gated chloride channel decrease the sensitivity to the anthelmintics (Njue et al., 2004). Further, the enrichment of ATP binding and kinases suggests a deeper involvement of cellular signaling in the process of parasite adaptation. Similar GO term enrichments were found in the nematode intestinal genes (Yin et al., 2008). It is possible that genes expressed in nematode intestines experienced significant change related to adaptation in relation to the changes in food and energy resources during the evolution of parasitism.
4.3. Parasitism and adaptation through gene duplication and LGT
During the evolution of parasitism, gene gain appears to have occurred far less extensively than did gene loss. Proteins exhibited significantly lower numbers of duplications among the parasitic nematodes possibly reflecting less of a need for redundancy in a more predictable host environment when compared to free-living nematodes. Constant environment imposes less selective pressure on protein diversification. Nevertheless, gene gain has played a role in parasitism and adaptation. This is reflected by functional enrichment in the protein families specific to parasites and in the protein families that expanded in the parasite species. The protein families that expanded in parasites were predominantly related to growth and development. As example, many parasites have very well controlled free-living and parasitic stages where the free-living stages are characterized by developmental arrest prior to transitioning to the parasitic stages. Consistent with this, the parasite-associated protein families showed functional enrichment in transcription and translation related terms in addition to cyanate metabolism, much of which was acquired via LGT. It is not clear yet what role cyanase played in the origins or persistence of parasitism. Clearly this is an evolutionarily conserved protein with foundations in detoxification in early organisms during a period in the earth's development when hydrogen cyanide was a key component of our atmosphere. Cyanide undergoes rapid oxidation to cyanate which in turn is degraded to NH3 and CO2. Bacteria, early plants and fungi subsequently utilized the nitrogen for growth and development. It is possible that the detoxifying components of cyanase were important in early parasites and the protein has since been repurposed (Elleuche and Poggeler, 2008). Surprisingly, to date it has not been identified in any free-living or clade V nematodes.
The majority of gene gains are species specific; however, these have common functional enrichments in zinc ion binding, chromatin binding, protein dimerization activity, and cyclin-dependent protein kinase inhibitor activity. Chromatin binding and cyclin-dependent protein kinase inhibitors are associated with gene expression and thus to a great extent, growth and development. A key function of zinc ion binding involves control of gene transcription through selective DNA binding (Iuchi, 2001). Zinc ion binding is also a GO term that is enriched in the transcripts expressed in the L3 and adult A. caninum (Wang et al., 2010) and in excretory-secretory products of microfilariae of B. malayi (Moreno and Geary, 2008) suggesting a link to parasitism and/or adaptation.
5. Conclusions
It is well known that parasitism evolved independently at least 18 times over the course of evolution of the Nematoda. Given the uniqueness in lifecycles, origins and tropisms, it can be difficult to ask and answer questions related to genetic and proteomic conservation among parasitic nematodes associated with parasitism and adaptation. And yet, one must wonder if in fact certain well-rooted mechanisms or biological processes act prominently to advance host-pathogen associations, disparate thought they may be. Using T. spiralis to represent parasites that diversified early in the evolution of the Nematoda (clade I), along with a group of free-living clade V nematodes and a collection of parasites that represent members of clades III and IV, we attempted to look for similarities that might link the parasitic nematodes, and differentiate this group from the most recently evolved free-living nematodes. Although one must temper these data with the proviso that only a small subset of organisms was used in our analysis, we were intrigued to find a few similarities among the parasites. We identified three protein families that were shared by all parasitic nematodes:1) a LEM domain-containing protein which is an inner nuclear membrane protein that can function as a specific repressor of Tgf-β, activin, and BMP signaling through interaction with the R-SMAD proteins, clearly associated with growth and development; 2) an RNA recognition motif domain-containing protein that in S. japonicum has been linked to alternative splicing and would certainly advance the parasite directive on adaptation; and 3) a cyanase family which was putatively acquired via LGT. The most intriguing questions to ask and answer are why would such a peculiar gene such as cyanase, which functions in the detoxification of cyanate, be independently acquired by parasites of such unique tropisms and yet not appear in free-living nematodes or in other metazoa? This finding has spurned a new set of studies to: 1) identify the depth of conservation in this independent event among the Nematoda; 2) determine if the nematode cyanase family is a non-functional relic or a biologically relevant protein in the life of these parasites; 3) better discern why such a gene would remain functional long after the potential need for detoxification of cyanate in the life cycles of nematodes has ceased; and 4) determine if LGT played a larger role than currently believed in adaptation and the parasitic character among the Nematoda.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health NIAID (R01 AI081803) and NIGMS (R01 GM097435) to M.M.
Footnotes
Conflict of interest: All authors express no financial, personal or organizational conflicts of interest relative to the content of this manuscript.
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetpar.2016.07.003.
References
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, Hotez PJ. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367:1521–1532. doi: 10.1016/S0140-6736(06)68653-4. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas WK. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Blaxter ML. Nematoda: genes, genomes and the evolution of parasitism. Adv Parasitol. 2003;54:101–195. doi: 10.1016/s0065-308x(03)54003-9. [DOI] [PubMed] [Google Scholar]
- Blaxter ML. Nematodes (Nematoda) In: Hedges SB, Kumar S, editors. The TimeTree of Life. Oxford University Press; New York: 2009. pp. 247–250. [Google Scholar]
- Consortium, T.C.e.S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Baum TJ. Getting to the roots of parasitism by nematodes. Trends Parasitol. 2004;20:134–141. doi: 10.1016/j.pt.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Denslow SA, Walls AA, Daub ME. Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol Mol Plant Pathol. 2005;66:244–255. [Google Scholar]
- Dieterich C, Sommer RJ. How to become a parasite—lessons from the genomes of nematodes. Trends Genet. 2009;25:203–209. doi: 10.1016/j.tig.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, Mitreva M, Roeseler W, Tian H, Witte H, Yang SP, Wilson RK, Sommer RJ. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris M, De Ley P, Blaxter ML. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol Today. 1999;15:188–193. doi: 10.1016/s0169-4758(99)01439-8. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuche S, Poggeler S. A cyanase is transcriptionally regulated by arginine and involved in cyanate decomposition in Sordaria macrospora. Fungal Genet Biol. 2008;45:1458–1469. doi: 10.1016/j.fgb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucl Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Kunin V, Ouzounis CA. Protein families and TRIBES in genome sequence space. Nucl Acids Res. 2003;31:4632–4638. doi: 10.1093/nar/gkg495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafournoux P, Bruhat A, Jousse C. Amino acid regulation of gene expression. Biochem J. 2000;351:1–12. doi: 10.1042/0264-6021:3510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP-Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CKS, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DMA, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrin-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki P, Tiuryn J. URec: a system for unrooted reconciliation. Bioinformatics. 2007;23:511–512. doi: 10.1093/bioinformatics/btl634. [DOI] [PubMed] [Google Scholar]
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol Biol Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Hussey RS. Disease-inducing secretions of plant parasitic nematodes. Annu Rev Phytopathol. 1989;27:123–141. [Google Scholar]
- Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JT, Perry RN, Johnston MRL. Changes in the ultrastructure of the cuticle of the potato cyst-nematode, Globodera rostochiensis, during development and infection. Fundam Appl Nematol. 1993;16:433–445. [Google Scholar]
- Jones JT, Furlanetto C, Kikuchi T. Horizontal gene transfer from bacteria and fungi as a driving force in the evolution of plant parasitism in nematodes. Nematology. 2005;7:641–646. [Google Scholar]
- Kikuchi T, Jones JT, Aikawa T, Kosaka H, Ogura N. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004;572:201–205. doi: 10.1016/j.febslet.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang X, Qiu D, Xiao S, Hotez PJ, Zhen D, Zhen H, Li M, Ren H, Zhan B, Xue H, Hawdon J, Feng Z. Epidemiology of human hookworm infections among adult villagers in Hejiang and Santai Counties Sichuan Province. China Acta Trop. 1999;73:243–249. doi: 10.1016/s0001-706x(99)00035-2. [DOI] [PubMed] [Google Scholar]
- Meldal BH, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, Bert W, Borgonie G, Moens T, Tyler PA, Austen MC, Blaxter ML, Rogers AD, Lambshead PJ. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Smant G, Helder J. Role of horizontal gene transfer in the evolution of plant parasitism among nematodes. Methods Mol Biol. 2009;532:517–535. doi: 10.1007/978-1-60327-853-9_30. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, Yang SP, Warren WC, Fulton RS, Bhonagiri V, Zhang X, Hallsworth-Pepin K, Clifton SW, McCarter JP, Appleton J, Mardis ER, Wilson RK. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Y, Geary TG. Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl Trop Dis. 2008;2:e326. doi: 10.1371/journal.pntd.0000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njue AI, Hayashi J, Kinne L, Feng XP, Prichard RK. Mutations in the extracellular domains of glutamate-gated chloride channel α3 and β subunits from ivermectin-resistant Cooperia oncophora affect agonist sensitivity. J Neurochem. 2004;89:1137–1147. doi: 10.1111/j.1471-4159.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, Houfek TD, Liu Q, Mitros T, Schaff J, Schaffer R, Scholl E, Sosinski BR, Thomas VP, Windham E. Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc Nat Acad Sci USA 105. 2008;105:14802–14807. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip GK, Creevey CJ, McInerney JO. The Opisthokonta and the Ecdysozoa may not be clades: stronger support for the grouping of plant and animal than for animal and fungi and stronger support for the Coelomata than Ecdysozoa. Mol Biol Evol. 2005;22:1175–1184. doi: 10.1093/molbev/msi102. [DOI] [PubMed] [Google Scholar]
- Prufer K, Muetzel B, Do HH, Weiss G, Khaitovich P, Rahm E, Paabo S, Lachmann M, Enard W. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinf. 2007;8:e41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeber F, Jex AR, Gasser RB. Impact of gastrointestinal parasitic nematodes of sheep, and the role of advanced molecular tools for exploring epidemiology and drug resistance—an Australian perspective. Parasit Vectors. 2013;6:e153. doi: 10.1186/1756-3305-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—biological queries made easy. BMC Genomics. 2009;10:e22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, Coulson A, Eustachio P, Fitch DHA, Fulton LA, Fulton RE, Griffiths-Jones S, Harris TW, Hillier LW, Kamath R, Kuwabara PE, Mardis ER, Marra MA, Miner TL, Minx P, Mullikin JC, Plumb RW, Rogers J, Schein JE, Sohrmann M, Spieth J, Stajich JE, Wei C, Willey D, Wilson RK, Durbin R, Waterston RH. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepek G, Buttle DJ, Duce IR, Behnke JM. Human gastrointestinal nematode infections: are new control methods required? Int J Exp Pathol. 2006;87:325–341. doi: 10.1111/j.1365-2613.2006.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Robertson H. The Caenorhabditis chemoreceptor gene families. BMC Biol. 2008;6:e42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Abubucker S, Martin J, Wilson RK, Hawdon J, Mitreva M. Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation. BMC Genomics. 2010;11:e307. doi: 10.1186/1471-2164-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Koonin EV. Coelomata and not ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Martin J, Abubucker S, Scott AL, McCarter JP, Wilson RK, Jasmer DP, Mitreva M. Intestinal transcriptomes of nematodes: comparison of the parasites Ascaris suum and Haemonchus contortus with the free-living Caenorhabditis elegans. PLoS Negl Trop Dis. 2008;2:e269. doi: 10.1371/journal.pntd.0000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarowiecki M, Berriman M. What helminth genomes have taught us about parasite evolution. Parasitology. 2015;142:S85–S97. doi: 10.1017/S0031182014001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- de Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol. 2003;19:547–551. doi: 10.1016/j.pt.2003.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


