Crystal structure analysis reveals the mode of substrate radical binding and indicates a previously unrecognized catalytic function for dirigent proteins during enantioselective pinoresinol formation.
Abstract
Dirigent proteins impart stereoselectivity to phenoxy radical coupling reactions in plants and, thus, play an essential role in the biosynthesis of biologically active natural products. This includes the regioselective and enantioselective coupling and subsequent cyclization of two coniferyl alcohol radicals to pinoresinol as the committed step of lignan biosynthesis. The reaction is controlled by dirigent proteins, which, depending on the species and protein, direct the reaction to either (+)- or (−)-pinoresinol. We present the crystal structure of the (−)-pinoresinol forming DIRIGENT PROTEIN6 (AtDIR6) from Arabidopsis (Arabidopsis thaliana) with data to 1.4 Å resolution. The structure shows AtDIR6 as an eight-stranded antiparallel β-barrel that forms a trimer with spatially well-separated cavities for substrate binding. The binding cavities are two lobed, exhibiting two opposing pockets, each lined with a set of hydrophilic and potentially catalytic residues, including essential aspartic acids. These residues are conserved between (+) and (−)-pinoresinol-forming DIRs and required for activity. The structure supports a model in which two substrate radicals bind to each of the DIR monomers. With the aromatic rings fixed in the two pockets, the propionyl side chains face each other for radical-radical coupling, and stereoselectivity is determined by the exact positioning of the side chains. Extensive mutational analysis supports a previously unrecognized function for DIRs in catalyzing the cyclization of the bis-quinone methide reaction intermediate to yield (+)- or (−)-pinoresinol.
Dirigent proteins (DIRs) play important roles in plant natural product biosynthesis. By mechanisms largely unknown, they impart stereochemical control onto reactions that otherwise lack regioselectivity and stereoselectivity (Davin and Lewis, 2005a; Pickel and Schaller, 2013). The dirigent phenomenon was discovered by Davin et al. (1997), who studied the coupling of two coniferyl alcohol radicals (CA·) generated from coniferyl alcohol by single-electron transfer (e.g. by laccases or peroxidases) as the first step in the biosynthesis of lignans. Random coupling of the resonance-stabilized radicals yielded a variety of 8-8′-, 8-5′-, and 8-O-4′-linked racemic reaction products in vitro. In the presence of a protein preparation from the cell walls of Forsythia spp., however, coupling was regiospecific (8-8′) and resulted in optically active (+)-pinoresinol (Davin et al., 1997). The purified protein (FiDIR1) lacked oxidizing activity but was able to direct the coupling mode of coniferyl alcohol radicals, hence the name DIR (Davin et al., 1997).
The discovery of DIRs provided a rationale for the formation of optically pure 8-8′-linked lignans in plants and led to a new model for stereoselective phenoxy radical coupling in biological systems in general (Davin and Lewis, 2005b). According to this hypothesis, DIRs with different specificities are responsible for the regioselective and stereoselective formation of the many 8-8′, 8-5′, and 8-O-4′ lignans, norlignans, and ellagitannins in numerous plant species (Niemetz and Gross, 2005; Vassao et al., 2010). Consistent with a broader role of DIRs in plant secondary metabolism, large gene families for DIRs and DIR-like proteins have been identified in all plant species investigated so far (35 genes in spruce [Picea abies], 25 in Arabidopsis [Arabidopsis thaliana], 54 in rice [Oryza sativa], and 19 in Isatis indigotica; Ralph et al., 2007; Li et al., 2014).
However, a general role for DIRs as mediators of stereoselectivity is not undisputed. The pioneering work of Zenk as well as subsequent studies showed that intramolecular phenol coupling during alkaloid biosynthesis is catalyzed regioselectively and stereoselectively by cytochrome P450 enzymes (Gerardy and Zenk, 1992; Gesell et al., 2009). Furthermore, there is a growing number of examples also for bimolecular couplings that are mediated by cytochrome P450s, laccases, or peroxidases in a stereospecific manner (Schlauer et al., 1998; Niemetz and Gross, 2003; Fang et al., 2012; Aldemir et al., 2014; Mazzaferro et al., 2015). Stereochemical control by the oxidizing enzymes alleviates the need for any DIR activity. Therefore, to sustain the dirigent hypothesis, the identification of DIRs for other coupling modes or substrates was urgently needed.
First support for a more general contribution of DIRs to stereoselective natural product formation was provided by AtDIR6 from Arabidopsis (Pickel et al., 2010). AtDIR6 acts on the same substrate as FiDIR1 but directs the coupling reaction toward (−)- rather than (+)-pinoresinol (Pickel et al., 2010; Kazenwadel et al., 2013). In addition to the prototypical FiDIR1 and AtDIR6, further (+)- and (−)-pinoresinol-forming DIRs have been identified in several other plant species (Davin and Lewis, 2005a; Kim et al., 2012; Dalisay et al., 2015; Seneviratne et al., 2015). Moreover, we recently found a novel type of DIR in cotton (Gossypium spp.) conferring atropselectivity to the oxidative coupling of hemigossypol (Effenberger et al., 2015). This reaction constitutes the final step in the biosynthesis of the sesquiterpenoid dimer gossypol, an important natural product and defense compound in cotton plants (Wagner et al., 2012; Tian et al., 2016). These findings indicate that the role of DIRs is not restricted to lignan biosynthesis but extends to entirely different biosynthetic pathways in plant secondary metabolism.
Despite the well-documented ability of DIRs to mediate stereoselective phenol coupling in vitro, their mode of action is still poorly understood. Ligand-binding studies and kinetic modeling of FiDIR1 suggested that two phenoxy radicals are bound, one to each of the two subunits of an active DIR homodimer. The bound radicals are then oriented in a way that favors a specific coupling mode (e.g. 8-8′ for pinoresinol formation in the lignan pathway), and, by excluding other reaction channels, DIRs mediate enantioselective product formation (Halls and Lewis, 2002; Halls et al., 2004; Davin and Lewis, 2005a). The proposed mode of action explains the DIR-mediated formation of optically active products, in competition with racemic product formation by random coupling of unbound radicals in solution (Halls et al., 2004; Pickel and Schaller, 2013). Structural data are needed to further substantiate this hypothesis.
As a first approximation, we exploited the distant homology of DIRs with allene oxide cyclase (AOC; Hofmann et al., 2006), an enzyme in the biosynthetic pathway of jasmonates (Schaller and Stintzi, 2009), to model the AtDIR6 structure (Pickel et al., 2012). Consistent with the proposed DIR mode of action, the model (Protein Model Database accession no. PM0078038) shows AtDIR6 as an eight-stranded antiparallel β-barrel with a central binding pocket, in which many of the potential ligand-binding positions are conserved between AOCs and DIRs and largely occupied by hydrophobic residues (Pickel et al., 2012). The model suggests that DIRs evolved within the calycin superfamily from binding proteins for hydrophobic ligands, and it is consistent with the proposed DIR mode of action. Binding of CA· with its aromatic ring in the hydrophobic cavity, one in each subunit of the proposed DIR homodimer, would allow the propenyl side chains to protrude and to engage in the radical-radical coupling reaction (Pickel et al., 2012). However, this view was challenged by a first crystal structure recently reported for DRR206, a (+)-pinoresinol-forming DIR from pea (Pisum sativum; Kim et al., 2015). Generally consistent with the structure modeled for AtDIR6, DRR206 crystallized as an eight-stranded β-barrel with a large hydrophobic cavity (Kim et al., 2015). However, the crystal structure is inconsistent with the proposed oligomeric state and mechanism of DIR action. DRR206 is not dimeric but rather forms a tightly packed homotrimer, in which the three potential substrate-binding pockets are too far apart for intermolecular coupling between sites (Kim et al., 2015). Unfortunately, most of the surface loops at the barrel opening were unresolved; therefore, the DRR206 structure did not provide conclusive insight into alternative modes of substrate binding or radical coupling.
We report here the structure of the enantiocomplementary, (−)-pinoresinol-forming AtDIR6 in its glycosylated state. All loops were clearly resolved with low B-factors, allowing us to propose a model for CA· binding. The presence of highly conserved, invariant hydrophilic residues at the potential binding site suggests an acid base-catalyzed reaction mechanism.
RESULTS AND DISCUSSION
Overall Structure
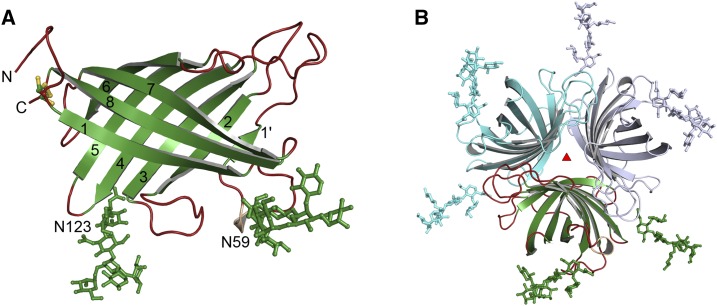
The structure of Arabidopsis DIR6 was solved by molecular replacement and refined to a resolution of 1.4 Å (Protein Data Bank [PDB] accession code 5LAL; Fig. 1; Table I). AtDIR6 forms an eight-stranded antiparallel β-barrel, similar to the previously reported (+)-pinoresinol-forming DIR DDR206 from pea (Kim et al., 2015) and to the distantly related AOCs from Arabidopsis and Physcomitrella patens (Hofmann et al., 2006; Neumann et al., 2012). N and C termini are located next to each other and are connected by an intramolecular disulfide bridge between Cys-40 and Cys-186, as predicted by structural modeling and confirmed by mass spectrometry (Pickel et al., 2012). The asymmetric unit contains two monomers, A and B (Supplemental Fig. S1A), including AtDIR6 residues 35 to 187 and 33 to 187, respectively (numbering refers to the full-length sequence including a 29-amino acid cleavable signal peptide). Each monomer forms a trimer with its respective crystallographic symmetry mates, enclosing an extensive interaction surface of almost 1,200 Å2 (Fig. 1B). In addition to many hydrophobic contacts at the barrel interface, the mutual orientation of subunits is maintained by eight hydrogen bonds (HBs) and the ionic interaction between Arg-159 and Glu-185. Arg in position 159 and an acidic residue in position 185 are conserved among other pinoresinol-forming DIRs (DIRs from now on; Supplemental Fig. S2), suggesting that this electrostatic interaction and, hence, the trimeric structure is a general property of these proteins. The interface of the dimer between monomers A and B is only 420 Å2 and, thus, considerably smaller than for the trimer. In contrast to DDR206 (Kim et al., 2015), the interchange of a β-strand between adjacent subunits is not observed in our AtDIR6 structure. Strand β1′ of AtDIR6, rather than inserting into the neighboring barrel, interacts directly with β2 within the same subunit.
Figure 1.
Structure of AtDIR6. A, Structure of monomer B of AtDIR6, showing strands 1 to 8 and the paucimannose modifications on Asn-59 and Asn-123. B, Cartoon representation of the crystallographic trimer that is formed by symmetry mates (cyan and light blue) of monomer B (green).
Table I. X-ray crystallographic statistics.
| Synchrotron | Swiss Light Source X10SA |
|---|---|
| Processing | |
| Wavelength (Å) | 1.0 |
| Resolution (Å) | 40.11–1.4 (1.44–1.4) |
| Space group | R3:H |
| Unit cell (Å) | a = b = 101.09, c = 90.24 |
| Total reflections | 675,500 (47,151) |
| Unique reflections | 64,669 (5,013) |
| Completeness (%) | 100 (100) |
| I/σ(I) | 9.13 (1.05) |
| CC1/2 | 99.6 (45.6) |
| Rmeas (%) | 14.3 (232.3) |
| Wilson B-factor | 20.6 |
| Refinement | |
| Rwork/Rfree (%) | 16.9/18.6 |
| Ramachandran favored (allowed; %) | 97 (3) |
| RMS, bonds (Å) | 0.014 |
| RMS, angles (°) | 1.82 |
| Number of TLS groups | 18 |
| Average B-factor | |
| Total | 29.1 |
| Macromolecules | 26.4 |
| Ligands | 50.6 |
| Solvent | 33.0 |
| No. of atoms | |
| Macromolecules | 2,629 |
| Ligands | 300 |
| Solvent | 189 |
AtDIR6 is a glycoprotein carrying N-linked paucimannose-type glycans at Asn-59 and Asn-123 that are required for protein solubility and function (Pickel et al., 2010; Kazenwadel et al., 2013). Glycans at Asn-59 are well ordered (Supplemental Fig. S1E), as they fold back onto the surface of the trimer. They are stabilized by extensive protein contacts to the same subunit as well as to symmetry-related molecules (Fig. 1A). The glycan chains at Asn-123, on the other hand, protrude straight into the solvent. In monomer A, these glycans are highly disordered, so that only the first two GlcNAc rings could be modeled. In monomer B, they are stabilized by contacts to another protein molecule and, thus, are better resolved (Supplemental Fig. S1, C and D).
The interaction of monomers A and B within the crystallographic asymmetric unit is stabilized by a HB network between the main chains of residues 33 to 36 and 184 to 186. Interestingly, residue Thr-33B, with its free α-amino group, is embedded inside a loop between the N and C termini of monomer A, indicating that Thr-33B represents the N terminus of the polypeptide chain (Supplemental Fig. S1B). Apparently, since the signal peptide was shown to be cleaved between residues 29 and 30 (Pickel et al., 2010), additional processing resulted in the loss of three more amino acids (residues 30–32) from the N terminus of the protein. In monomer A, the first two residues, Thr-33 and Ile-34, are disordered and, thus, not resolved in the structure. The extensive interaction between the protein termini within the asymmetric unit is regarded as a crystal artifact and most likely not seen in solution. Thus, we consider the trimer to be the functionally relevant form of the protein.
AtDIR6 Features Two Binding Pockets for CA·
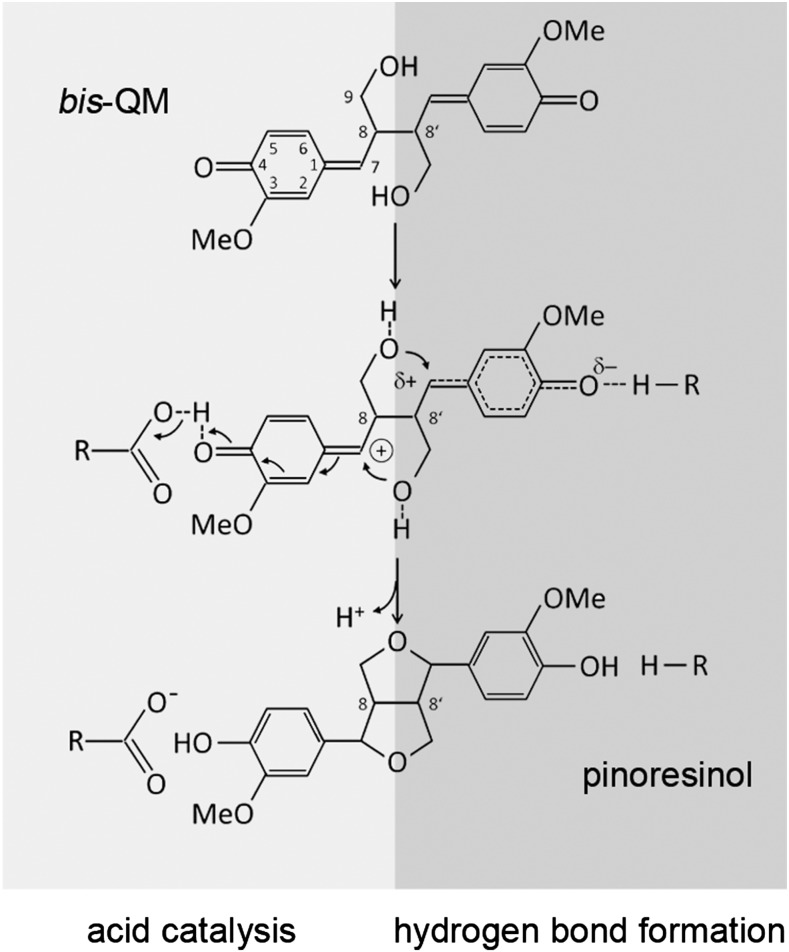
The regioselectivity of DIRs has been explained by a model in which the two subunits of the DIR homodimer sequentially bind two CA· and orient them in a way that favors 8-8′ coupling over other coupling modes (Halls et al., 2004; Davin and Lewis, 2005a; Pickel et al., 2012). Radical-radical coupling then results in a bis-quinone methide (bisQM) intermediate (Fig. 2) that is electron deficient at C7 and, thus, susceptible to nucleophilic attack (Freudenberg, 1959; Ralph et al., 2009). During pinoresinol formation, the terminal OH groups of the propionyl side chains act as nucleophiles. The 1,6-addition to the p-quinone methides leads to cyclization of the furan rings and rearomatization of the cyclohexadienones (Fig. 2). The latter results in a large increase in π stabilization energy, providing the driving force for the 1,6-addition and contributing to the instability of the reaction intermediate (Toteva et al., 2003; Ralph et al., 2009). As compared with the reactivity of the bisQM, the mode of coupling control and the role of DIRs in the cyclization reaction are unclear. The proposed quaternary structure and 1:1 stoichiometry of binding, with two CA· bound to the DIR homodimer (Halls and Lewis, 2002; Halls et al., 2004), was challenged recently by the crystal structure of DRR206, a (+)-DIR from pea (Kim et al., 2015). The three potential binding sites of the DRR206 trimer are so far apart that bimolecular coupling of CA· between sites is impossible (Kim et al., 2015). This is observed here also for the enantiocomplementary AtDIR6 trimer; therefore, we considered within-site coupling after the binding of two CA· to the same AtDIR6 subunit as an alternative possibility.
Figure 2.
Cyclization of the bisQM reaction intermediate. Nucleophilic addition of the terminal OH groups leads to ring closure of the furan rings and rearomatization of the cyclohexadienone rings. Nucleophilic attack is facilitated by the full or partial positive charge on C7 as a result of acid catalysis (left, light gray) or HB formation (right, dark gray).
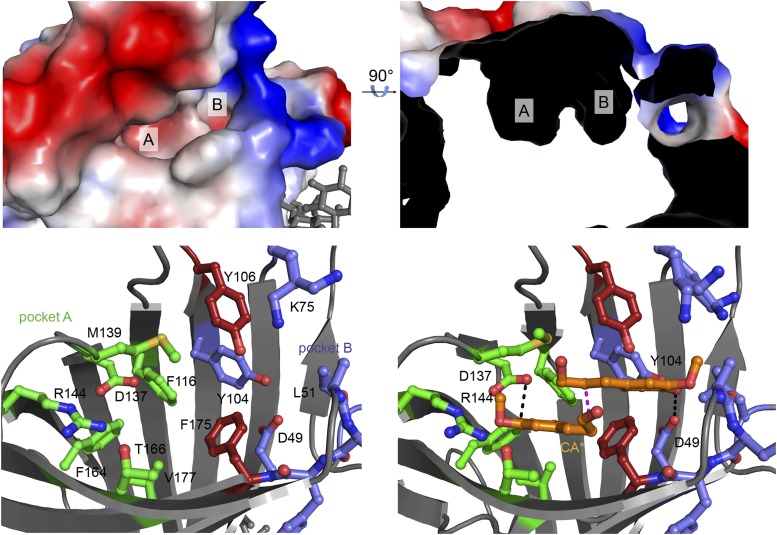
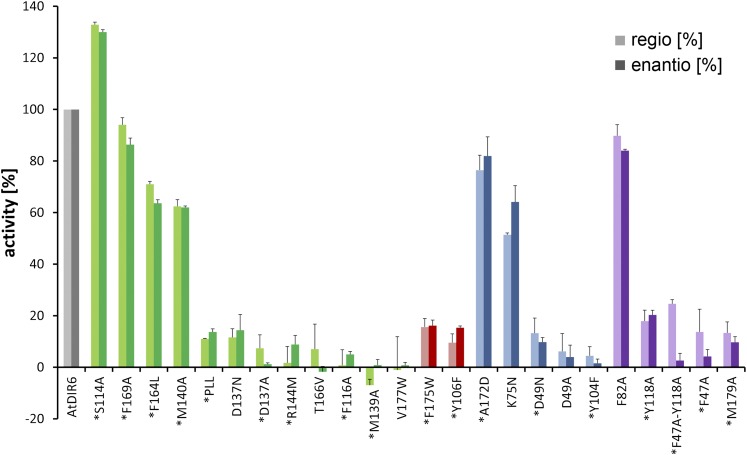
AtDIR6 exhibits a two-lobed cavity at the open end of the barrel (Fig. 3, A and B), which is consistent with the binding of two substrate molecules to each of the AtDIR6 monomers. The cavity is closed at the bottom by bulky aliphatic side chains and, therefore, does not protrude very deeply into the barrel. Two conserved residues (Phe-175 and Tyr-106) part the cavity into two differently sized pockets, A and B (Fig. 3, A–C). The relevance of the two pockets for DIR function was tested by site-directed mutagenesis. A centrally located hydrophobic amino acid of each pocket was substituted by Trp, thereby blocking access to either. Both mutations, V177W for pocket A and F175W for pocket B, resulted in a dramatic loss of activity without affecting protein expression or stability (Fig. 4), indicating that both pockets are functionally important.
Figure 3.
Active site of AtDIR6. A, Surface representation of monomer A showing the two pockets of the active site. Red, Negative surface charge; blue, positive surface charge. B, Slice through the surface of AtDIR6, rotated by 90° to show pockets A and B. C, View of the active site showing important residues of pocket A in green, pocket B in blue, and Tyr-106 and Phe-175 separating the two pockets in red (a stereo view is shown in Supplemental Fig. S3A). D, Potential binding mode of two CA· substrate radicals supported by energy minimization of the manually placed ligands (a stereo view is shown in Supplemental Fig. S3B). The color code is as in C.
Figure 4.
Mutational analysis of active site residues. The effect of point mutations on the regioselectivity and enantioselectivity of AtDIR6 variants is shown in percentage of wild-type AtDIR6 activity. Regioselectivity was analyzed by reverse-phase (RP)-HPLC as relative increase in pinoresinol formation upon the addition of DIRs (light colors). Enantioselectivity was determined by chiral HPLC as the enantiomeric excess of (−)- over (+)-pinoresinol. It is shown for the mutants in percentage of the enantiomeric excess generated by wild-type AtDIR6 (dark colors). Values represent means ± sd of activity assays performed in triplicate for each of the mutant proteins. The experiment was repeated twice with independent protein preparations for 18 of the 24 mutants (indicated by asterisks), yielding equivalent results. The color coding corresponds to Figure 3, showing residues of pocket A in green, pocket B in blue, dividing residues in red, and nonpocket residues in purple.
The two pockets differ slightly between the two monomers of the asymmetric unit. In monomer A, the side chain of Phe-169 (Phe-169A) is rotated toward the entrance, while Phe-169B points into the cavity, drastically reducing accessibility (Supplemental Fig. S1F). This also is the case for Met-140, which is rotated outward in monomer A and inward in monomer B. Thus, the pockets are more open in monomer A, potentially allowing substrate binding. Interestingly, the two pockets are lined with several hydrophilic residues, most of which are conserved among DIRs (Supplemental Fig. S2). Pocket A contains Asp-137, Arg-144, and Thr-166, whereas Asp-49, Lys-75, Lys-78, Tyr-104, and Tyr-106 are found in pocket B (Fig. 3C; Supplemental Fig. S3A). Site-directed mutagenesis showed that the invariant Asp-137, Arg-144, and Thr-166 in pocket A, in addition to Asp-49, Tyr-104, and Tyr-106 in pocket B, are critical for DIR function (Fig. 4). Hence, we find two sets of potentially catalytic groups in both pockets, which suggested to us that each may bind one of the substrate radicals.
Testing this possibility, we manually placed CA· into pockets A and B, so that the oxygens at the C4 ring positions would be coordinated by the two salient Asp residues in either one of the two pockets. We tested and optimized the position and geometry of the ligands by running a simulated annealing refinement, in which the carbonyl oxygens of the two CA· were each tied to one of the Asp oxygens at 2.9 ± 0.2 Å distance. During refinement and energy minimization, the two CA· align in a way that juxtaposes C8 and C8′. The protein side chains barely change upon refinement, with minor differences observed for Met-139, Tyr-104, Phe-175, and Lys-75. In this conformation, Arg-144 is located in hydrogen-bonding distance to the C4 carbonyl in pocket A, and the hydroxyl group of Tyr-106 is located in close proximity to the C9-OH of both CA· (Fig. 3D; Supplemental Fig. S3B). Hence, the two-lobed pocket is perfectly sized to accommodate two CA· in their extended, planar all-trans-conformation, in which they are precisely positioned to enable 8-8′ coupling at the rere face (Fig. 3D; Supplemental Fig. S3B). This would lead to the S,S-configured bisQM and, consistent with the enantioselectivity of AtDIR6, (−)-pinoresinol after cyclization (compare with Fig. 1 in Davin and Lewis, 2005a).
We further modeled and energy minimized the bisQM-bound intermediate and the final product-bound state with (−)-pinoresinol in the binding pocket. In a similar approach as for the CA·, we defined a bond between the carbonyl oxygens of the bisQM (or the hydroxyl oxygens of pinoresinol) and the carboxyl groups of Asp-137 and Asp-49. Again, the protein pocket required only minor movements for optimal bisQM or pinoresinol binding (Supplemental Fig. S3C). The distance between Asp-137 and Asp-49 is perfect for accommodation of the reaction intermediate (Fig. 5A). The flexible hydroxyls at C9 and C9′ of the bisQM find their minimum in an optimal position for the nucleophilic attack on C7 and C7′, respectively (Fig. 5A; Supplemental Fig. S3C). For (−)-pinoresinol, energy minimization resulted in a conformational change ensuing a rotation of the two tetrahydrofuran rings. In this conformation, the position of (−)-pinoresinol in the active site of AtDIR6 is highly similar to that of the bisQM reaction intermediate (Supplemental Fig. S3C). The results of these simulated annealings are fully consistent with the binding mode we propose, according to which two phenoxy radicals are captured and coupled within the cavity of each of the three AtDIR6 subunits. They also evoke a reaction mechanism in which AtDIR6 takes an active role in catalyzing the cyclization of the reaction intermediate.
Figure 5.
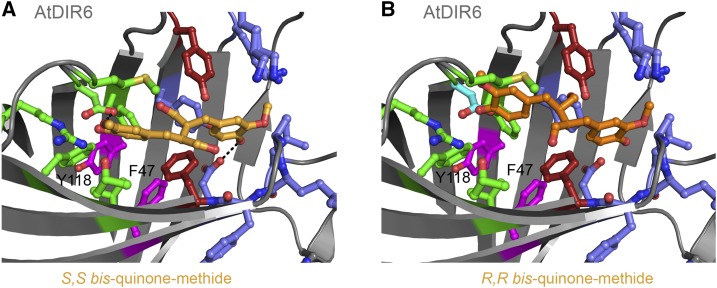
Modeling of reaction intermediates for (+)- and (−)-pinoresinol in the active site of AtDIR6. A, Model of the S,S-bisQM intermediate (light orange) in the active site of AtDIR6. Conformation and position were optimized by energy minimization after manual placement of the ligand. Pocket A, Green; pocket B, blue; Tyr-106 and Phe-175, red; Tyr-118 and Phe-47, purple. B, Model of manually placed R,R-bisQM (dark orange) in the active site of AtDIR6. The quinone ring in pocket B was overlaid onto that of S,S-bisQM from A.
AtDIR6 Catalyzes Cyclization of the bisQM Intermediate
The proposed mode of binding provides a rationale for the regioselectivity and enantioselectivity of AtDIR6 and, importantly, suggests a previously unrecognized catalytic function for DIRs in the cyclization reaction of the bisQM intermediate. Nucleophilic additions to p-quinone methides are catalyzed by reversible protonation of the carbonyl oxygen, resulting in a carbocation at C7 that is captured by the nucleophile (Chiang et al., 2002; Toteva et al., 2003; Ralph et al., 2009). Similarly, nucleophilic additions also are facilitated by the partial positive charge that develops on C7 when the carbonyl is engaged in an HB (Toteva et al., 2003; Fig. 2).
In pocket A, Asp-137 and Arg-144, both strictly conserved in (+)-DIRs as well as in (−)-DIRs (Supplemental Fig. S2), are available as potential HB donors. Consistent with such a role for Arg-144, the R144M mutant, in which the general space-filling, hydrophobic properties of the side chain are maintained but the ability to serve as an HB donor is lost, was completely inactive (Fig. 4). D137A also was inactive, highlighting the functional relevance of this residue. A pronounced loss of activity also was observed when its carboxyl group was replaced by a carboxamide (D137N), suggesting that the acid function rather than the HB donor is important in this position (Fig. 4).
However, a potential role in acid catalysis would require Asp-137 to be in its undissociated form; therefore, its pKa would need to be considerably higher than that of free Asp. Large pKa shifts have been reported for aspartyls in the vicinity of negatively charged groups or embedded in a hydrophobic microenvironment, which favors the uncharged state of the side chain (Urry et al., 1994; Yamazaki et al., 1994; Mehler et al., 2002). The latter appears to be the case in AtDIR6, where Asp-137 is flanked by Phe-116 and Phe-164 (Fig. 3C; Supplemental Fig. S3A). In (+)-DIRs, these aromatic residues are replaced by aliphatic amino acids with large hydrophobic side chains (Leu and Ile; Supplemental Fig. S2). Consistent with the relevance of hydrophobicity rather than the presence of aromatic rings, the F164L mutant was only marginally less active than wild-type AtDIR6, while F116A showed a complete loss of activity (Fig. 4). Further support for the importance of a hydrophobic microenvironment for Asp-137 is provided by the S114A mutant, the only one for which an increase in activity over the wild-type protein was observed (Fig. 4). Interestingly, most other DIRs possess the more hydrophobic Ala in this position (Supplemental Fig. S2).
The presence of bases next to the essential Asp-137 and Asp-49, Arg-144 in pocket A and Lys-75 in pocket B (Fig. 3D; Supplemental Fig. S3B), may support the catalytic function of the Asp residues by donating a proton that is then shared between the substrate radicals and the Asp residues. Alternatively, the proton may be donated by Tyr-104 in pocket B and the resulting phenoxide ion stabilized by Lys-75 or by Asn in (+)-DIRs and the K75N mutant (Fig. 4). Also functionally important are the hydroxyl groups of Thr-166 in pocket A and the centrally localized Tyr-106. Substitution with Val (T166V) or Phe (Y106F) abolished activity (Fig. 4). These hydroxyls may assist other residues in lining up the side chains for catalysis or, in the case of Tyr-106, facilitate nucleophilic attack and ring closure by accepting an HB from the bisQM hydroxyl (Figs. 3D and 5).
The fact that DIR function depends on residues of both pockets supports the proposal that two CA· bind to one DIR monomer. Acid catalysis and/or H-bond donation as the critical factors for ring closure of the bisQM intermediate are substantiated by our mutational analysis and are in general agreement with the acidic pH optimum of DIRs (no activity at pH > 6; Halls et al., 2004). The proposed role for DIRs as catalysts of this intramolecular Michael addition is in agreement with the remarkable stability of some bisQMs (estimated half-life of 60 min; Freudenberg, 1959; Ralph et al., 2009). It also is consistent with kinetic modeling demonstrating that neither substrate binding nor the coupling reaction, but rather subsequent events (ring closure or product release), are rate limiting during pinoresinol formation (Halls et al., 2004).
AtDIR6 Is in a Precatalytic Conformation
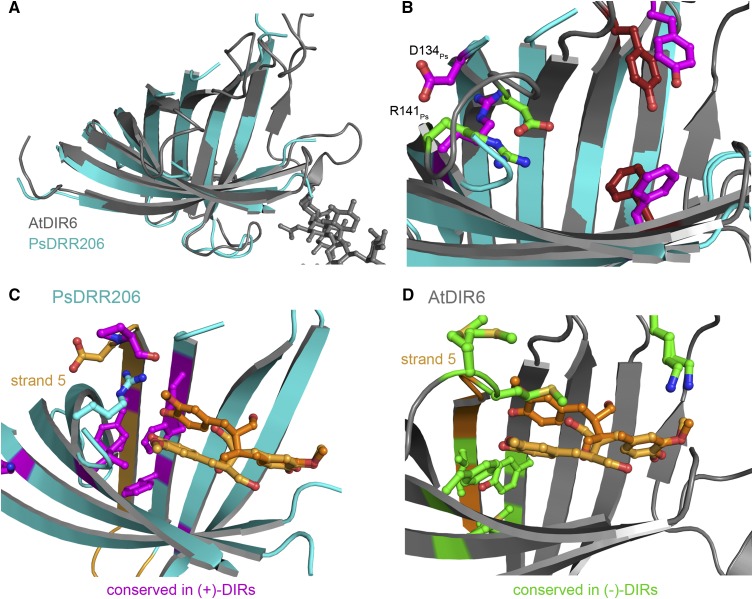
The previously published DRR206 structure (Kim et al., 2015) is highly similar to that of AtDIR6. With the exception of the swapped β-strand in DRR206, β-sheets are similarly arranged and, thus, the barrel diameter is the same in both proteins (Fig. 6A). However, there are striking differences with respect to the putative active site. In AtDIR6, all the loops at the open end of the barrel are well resolved, show good density, and could be refined with appropriate B-factors. In DRR206, on the other hand, the loop connecting β1′ and β2 (loop1′-2), loop3-4, and loop5-6 are flexible and, thus, not resolved in the structure (Kim et al., 2015). Unlike AtDIR6, where the strands leading to these loops are bent inward, they point more toward the solvent in DRR206 (Fig. 6B). This also is observed for the extended loop1-1′. As a result, the binding pocket of DRR206 appears more flat and open (Fig. 6B; Supplemental Fig. S4).
Figure 6.
Comparison of AtDIR6 with pea DRR206. A, Overlay of monomer A of AtDIR6 (gray) with monomer A of DRR206 (cyan; PDB accession code 4REV). Both proteins have a root-mean-square deviation of 1.71 Å. B, Overlay of AtDIR6 and DRR206 active sites. DRR206 residues are depicted in purple and those of AtDIR6 in green (pocket A) and red. C, Cartoon representation of DRR206 (cyan) with residues conserved only in (+)-DIRs shown in purple. D, Cartoon representation of AtDIR6 (gray) with residues conserved only in (−)-DIRs depicted in green.
There also are significant differences with respect to the localization of active site residues. In AtDIR6, critical pocket A residues, Asp-137At and Arg-144At, are ideally placed for catalysis. In DRR206, on the other hand, Asp-134Ps protrudes into the solvent and Arg-141Ps forms an HB with β5 (Fig. 6B). Similarly, Tyr-103Ps and Phe-172Ps are shifted outward in the pea enzyme, while the corresponding Tyr-106At and Phe-175At are in separate pockets A and B of AtDIR6 (Fig. 6B). Apo-DRR206, therefore, seems to be in a noncatalytic state and will have to rearrange crucial residues upon binding of the two CA·. Apo-AtDIR6, on the other hand, is in a precatalytic conformation, where active site residues are lined up for catalysis. Assuming that the two structures correspond to two different conformations of DIRs, they are suggestive of a possible reaction mechanism. Prior to substrate binding, DIRs appear to be in an open conformation, as represented by DRR206. Sequential binding of the two substrate radicals first locks the centrally located Tyr-106At/Tyr-103Ps into position. Binding of the second CA· in pocket A may then induce a rearrangement of loop5-6, thereby positioning Asp-137At/Asp-134Ps and Arg-144At/Arg-141Ps for catalysis.
Enantioselectivity Is a Feature of the Whole Enzyme
The enantioselectivity of DIRs is a direct consequence of regioselective 8-8′ coupling: if coupling occurs at the rere face, the S,S-configured bisQM is formed as the precursor of (−)-pinoresinol (Fig. 3D; Supplemental Figs. S3B and S4A). Si-si coupling, on the other hand, yields the R,R-bisQM and, consequently, (+)-pinoresinol (Davin and Lewis, 2005a). Therefore, not surprisingly, the regioselectivity and enantioselectivity of AtDIR6 are tightly linked (Fig. 4; Supplemental Fig. S5). With the aromatic rings fixed in the two pockets, 8-8′ is the only remaining coupling possibility, and the stacking of the propionyl side chains, rere versus si-si, is the decisive factor for enantioselectivity.
In AtDIR6, the room for the side chains is restricted by Met-139 and Met-140 forming the upper rim of pocket A. Single point mutations showed that only Met-139 is essential for activity (Fig. 4). Hoping to free up some space and allow opposite stacking of propionyls, we generated an L138P, M139L, and M140L triple mutant, in which the LeuMetMet motif of AtDIR6 was replaced by the corresponding ProLeuLeu of the (+)-DIR ScDIR1 from Schizandra chinensis (Kim et al., 2012). However, the triple mutant was almost inactive, lacking both regioselectivity and enantioselectivity (Fig. 4), indicating that other parts of the enzyme contribute to enantioselectivity. This likely includes residues that lie deeper within pocket A and require a certain flexibility to allow cyclization of the bisQM. Cyclization ensues a rotation and flipping of the cyclohexadienone rings, and the direction of flipping is opposite for the S,S and R,R enantiomers. Consistently, we could place and energy minimize both the position and conformation of S,S-bisQM in pockets A and B of AtDIR6 (Fig. 5A), while the R,R enantiomer had different constraints with respect to the binding pocket (Fig. 5B).
Our interpretation is consistent with data from Kim et al. (2012), who were able to alter the enantioselectivity of ScDIR1 by introducing certain regions of AtDIR6. Replacing residues 90 to 138 (region B) of ScDIR1 by the corresponding sequence of AtDIR6 resulted in a reversal of enantioselectivity, while substitution of residues 79 to 116 (region A) did not. It was concluded that the nonoverlapping part of region B is likely to be responsible for enantioselectivity (Kim et al., 2012). Interestingly, this includes strands β4 and β5 that form part of pocket A. Furthermore, β5 merges into the MM motif that restricts the room for the propionyl side chains in (−)-DIRs, and β4 includes three residues that are differentially conserved between (+)- and (−)-DIRs (Phe-116At/Leu-113Ps, Tyr-118At/Phe-115Ps, and Leu-120At/Phe-117Ps; Supplemental Fig. S2, red dots). The first two are found in a ring-like arrangement below pocket A, which also includes the differentially conserved Phe-164At/Ile-161Ps and Met-179At/Val-176Ps (Fig. 6, C and D). Site-directed mutagenesis confirmed a contribution to enantioselectivity for one of them, Tyr-118, and for the neighboring Phe-47. The F47A single mutant and, more so, the F47A/Y118A double mutant showed partial uncoupling of regioselectivity and enantioselectivity. The double mutant retained 25% of the wild type’s regioselectivity but did not show any preference for the (−)-enantiomer anymore. Apparently, a fraction of substrate radicals were still able to bind with their aromatic rings in pockets A and B; thus, regioselective 8-8′ coupling was maintained. The observed decrease in enantioselectivity suggests that the propionyl side chains had sufficient space and flexibility for either rere or si-si stacking. Hence, imperfect precoordination of the substrate radicals may be responsible for the loss of enantioselectivity. We conclude that enantioselectivity is provided by discontinuous regions of the enzyme that act together to determine the stacking of the propionyl substituents and provide the flexibility for cyclization to occur.
CONCLUSION
DIRs have hitherto not been considered as enzymes in sensu stricto. Lacking oxidizing activity, they rely on laccases or peroxidases for the generation of substrate radicals. Also, the cyclization of the primary coupling product did not seem to require any lowering of an activation energy barrier, since the bisQM cyclizes spontaneously in solution. The binding and proper orientation of substrate radicals seemed to be sufficient for DIR activity. The structure of AtDIR6 indicates that DIRs are more than passive binding proteins but, rather, actively involved in catalysis. We propose that two coniferyl alcohol-derived radicals bind to the two-lobed active site of the enzyme in a way that places the propionyl side chains in position for 8-8′ coupling via the rere face in AtDIR6 or the si-si face in (+)-DIRs. DIRs then catalyze pinoresinol formation by donating a proton to the hexadienone ring carbonyls of the bisQM reaction intermediate via HB formation or acid catalysis. Structures of DIRs complexed with substrate analogs, reaction intermediates, or products will be required to confirm and fully trace the reaction mechanism. Future studies will show whether the proposed mode of action extends to dirigents of other substrates or coupling modes.
MATERIALS AND METHODS
Site-Directed Mutagenesis of AtDIR6
Site-directed mutagenesis was performed on an Arabidopsis (Arabidopsis thaliana) AtDIR6 expression construct comprising the cauliflower mosaic virus 35S promoter, the open reading frame for AtDIR6 (At4g23690; GI:1063724484), and the OCS terminator in pART7 (Gleave, 1992) that was amplified in methylation-competent (dam+) Escherichia coli DH10B. Mutations were introduced by PCR using complementary primers that contained the altered codon in a central position, flanked on either side by at least 10 bp matching the AtDIR6 sequence (primer sequences are given in Supplemental Table S1). PCRs contained 50 ng of the template DNA, 0.18 µm of the two mutagenic primers, and 0.5 units of Pfu DNA polymerase (Fermentas/ThermoFisher Scientific) in a reaction volume of 10 µL. After 15 to 17 PCR cycles, 10 units of DpnI was added to digest residual template DNA overnight at 37°C. Aliquots of the reaction were transformed into E. coli DH10B to amplify the PCR-generated plasmid. The mutation was confirmed by sequence analysis. The expression cassette was then mobilized with NotI and cloned into the NotI site of the binary vector pART27 (Gleave, 1992).
Expression of AtDIR6 and of Site-Directed AtDIR6 Mutants
AtDIR6 was expressed in a transgenic plant cell culture system and purified from cell wall extracts by fractionated ammonium sulfate precipitation followed by size-exclusion and cation-exchange chromatography as described (Pickel et al., 2010) and dialyzed against 0.1 m MES, pH 5. AtDIR6 mutants and the wild-type control were transiently expressed in Nicotiana benthamiana by agroinfiltration as described previously for cotton (Gossypium spp.) DIRs (Effenberger et al., 2015). The leaves were harvested after 3 d, cut into pieces of 3 to 5 cm2, and rinsed in ice-cold deionized water. The leaf pieces were vacuum infiltrated with 0.1 m MES, pH 5, and 300 mm NaCl at 200 mbar. The infiltrate was recovered by centrifugation through glass wool at 1,500g for 7 min. The apoplastic wash fluid was cleared by centrifugation (14,000g, 10 min), and its volume was reduced to one-fifth by ultrafiltration (Vivaspin 20 [10-kD cutoff]; Sartorius). A total of 275 µL of the concentrate was used in the dirigent activity assay.
Assay of Regioselectivity and Enantioselectivity
The dirigent activity of transiently expressed AtDIR6 and AtDIR6 mutants was analyzed as described (Pickel et al., 2010). The assay contained 1.13 mm coniferyl alcohol, 30 milliunits of Trametes versicolor laccase, and 275 µL of aploplastic protein extract in a total volume of 300 µL. Reaction products were analyzed by HPLC. In a first chromatographic step, RP-HPLC was employed to separate the different coupling products. The pinoresinol peak was collected and subjected to chiral HPLC to separate (+)- and (−)-pinoresinol. Regioselectivity was derived from the RP-HPLC scan by comparing the peak areas for pinoresinol and dehydrodi-CA as one of the random coupling products. It is expressed as percentage increase in the pinoresinol-dehydrodi-CA ratio in the presence of DIRs as compared with the undirected control (apoplastic extracts from mock-infiltrated plants). Enantioselectivity was determined by chiral HPLC as an enantiomeric excess of (−)- over (+)-pinoresinol. To compare specific activities, the expression level of each AtDIR6 mutant was quantified by comparing its western-blot signal with a standard curve of AtDIR6 wild-type protein on the same membrane. A C-DiGit blot scanner (LI-COR) was used for quantification of the enhanced chemiluminescence signal. The experiment was repeated twice with independent protein preparations for 18 of the 24 mutants with equivalent results.
Crystallization and Data Collection
AtDIR6 at 6 mg mL−1 was incubated in 0.1 m succinic acid-phosphate-Gly buffer, pH 7, and 22.5% polyethylene glycol-1000 at 4°C employing the hanging drop method, and elongated plates grew within approximately 14 d. Using these crystals as the seeding source, final crystals (trigonal space group H3) of size 150 × 150 × 100 µm were grown in the same conditions within 3 to 4 d. The crystals were cryoprotected in mother liquor supplemented with 30% (v/v) ethylene glycol and flash frozen in liquid nitrogen. Data were collected at 100 K in house on a Bruker FR591 rotating anode with the MAR345dtb detector system and at the X10SA beamline at the Swiss Light Source using a Pilatus detector.
Structure Solution and Refinement
Atomic coordinates were deposited in the PDB (Berman et al., 2003) with accession code 5LAL. Data sets were processed using XDS and XSCALE (Kabsch, 2010) and converted into appropriate formats by XDSCONV. The AtDIR6 structure was solved by molecular replacement using the in-house data set and a monomer of the pea (Pisum sativum) dirigent protein DRR206 (4REV; Kim et al., 2015) as a search model.
A clear solution was found by MOLREP (Vagin and Teplyakov, 1997), and initial phases were obtained using REFMAC (Murshudov et al., 1997) giving initial Rwork and Rfree values of 0.4346 and 0.4452, respectively. After transfer of Rfree flags to the high-resolution data set with phenix.reflection_file_editor, further refinement was done with phenix.refine from the PHENIX suite (Adams et al., 2010). For manual model building, the molecular graphics program COOT (Emsley and Cowtan, 2004) was employed. All structural figures shown were generated with PyMOL (PyMOL Molecular Graphics System, version 1.8; Schrödinger). Analyses of intermolecular interactions were performed with PDBePISA (Krissinel and Henrick, 2007).
Energy Minimization of Manually Placed Ligands
Two coniferyl alcohol radicals, S,S-bisQM and (−)-pinoresinol, were manually placed into the active site using COOT. Geometry cif files for refinement of the ligands were created using JLigand (Lebedev et al., 2012). In order to test for the optimal placement and geometry of the ligands, phenix.refine from the PHENIX suite was used. Fake fobs for the protein structure without ligand were calculated using phenix.fmodel to 1.5 Å. Refinement was done using the following parameters: simulated annealing using default parameters, individual site refinement, and a refinement target weight of 0.1 in order to minimize the x-ray term. Bond restraints of 2.9 ± 0.2 Å were introduced between the carbonyl oxygens of the ligands (hydroxyl oxygens of pinoresinol) and the OD2 oxygens of both Asp-137 and Asp-49.
Accession Numbers
Structural coordinates were deposited in the PDB with accession code 5LAL.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Structural features of AtDIR6.
Supplemental Figure S2. Sequence of AtDIR6 aligned with those of other (−)- and (+)-DIRs.
Supplemental Figure S3. Active site in free and ligand-bound states (stereo views of Fig. 3, C and D).
Supplemental Figure S4. Surface representations of AtDIR6- and DRR206-binding cavities.
Supplemental Figure S5. Regioselectivity and enantioselectivity are tightly linked.
Supplemental Table S1. Primers for site-directed mutagenesis.
Supplementary Material
Acknowledgments
We thank the beamline staff of X10SA at the Swiss Light Source and at beamlines at the European Synchrotron Radiation Facility as well as our colleagues from the Max-Planck-Institute for Molecular Physiology for support during data collection; Martin Feigel (Ruhr University Bochum) for fruitful discussions on the reaction mechanism; and Bernhard Hauer (University of Stuttgart) for critical reading of the article.
Glossary
- PDB
Protein Data Bank
- bisQM
bis-quinone methide
- HB
hydrogen bond
- RP
reverse-phase
Footnotes
This work was supported by the German Research Foundation (grant no. SCHA591/10 to A.S. and grant no. HO2600/1–2 to E.H.).
Articles can be viewed without a subscription.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldemir H, Richarz R, Gulder TAM (2014) The biocatalytic repertoire of natural biaryl formation. Angew Chem Int Ed Engl 53: 8286–8293 [DOI] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10: 980. [DOI] [PubMed] [Google Scholar]
- Chiang Y, Kresge AJ, Zhu Y (2002) Flash photolytic generation and study of p-quinone methide in aqueous solution: an estimate of rate and equilibrium constants for heterolysis of the carbon-bromine bond in p-hydroxybenzyl bromide. J Am Chem Soc 124: 6349–6356 [DOI] [PubMed] [Google Scholar]
- Dalisay DS, Kim KW, Lee C, Yang H, Rübel O, Bowen BP, Davin LB, Lewis NG (2015) Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: integrated omics and MALDI mass spectrometry imaging. J Nat Prod 78: 1231–1242 [DOI] [PubMed] [Google Scholar]
- Davin LB, Lewis NG (2005a) Dirigent phenoxy radical coupling: advances and challenges. Curr Opin Biotechnol 16: 398–406 [DOI] [PubMed] [Google Scholar]
- Davin LB, Lewis NG (2005b) Lignin primary structures and dirigent sites. Curr Opin Biotechnol 16: 407–415 [DOI] [PubMed] [Google Scholar]
- Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275: 362–366 [DOI] [PubMed] [Google Scholar]
- Effenberger I, Zhang B, Li L, Wang Q, Liu Y, Klaiber I, Pfannstiel J, Wang Q, Schaller A (2015) Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol. Angew Chem Int Ed Engl 54: 14660–14663 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fang W, Ji S, Jiang N, Wang W, Zhao GY, Zhang S, Ge HM, Xu Q, Zhang AH, Zhang YL, et al. (2012) Naphthol radical couplings determine structural features and enantiomeric excess of dalesconols in Daldinia eschscholzii. Nat Commun 3: 1039. [DOI] [PubMed] [Google Scholar]
- Freudenberg K. (1959) Biosynthesis and constitution of lignin. Nature 183: 1152–1155 [DOI] [PubMed] [Google Scholar]
- Gerardy R, Zenk MH (1992) Formation of salutaridine from (R)-reticuline by a membrane-bound cytochrome P-450 enzyme from Papaver somniferum. Phytochemistry 32: 79–86 [Google Scholar]
- Gesell A, Rolf M, Ziegler J, Díaz Chávez ML, Huang FC, Kutchan TM (2009) CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J Biol Chem 284: 24432–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Halls SC, Davin LB, Kramer DM, Lewis NG (2004) Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 43: 2587–2595 [DOI] [PubMed] [Google Scholar]
- Halls SC, Lewis NG (2002) Secondary and quaternary structures of the (+)-pinoresinol-forming dirigent protein. Biochemistry 41: 9455–9461 [DOI] [PubMed] [Google Scholar]
- Hofmann E, Zerbe P, Schaller F (2006) The crystal structure of Arabidopsis thaliana allene oxide cyclase: insights into the oxylipin cyclization reaction. Plant Cell 18: 3201–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazenwadel C, Klebensberger J, Richter S, Pfannstiel J, Gerken U, Pickel B, Schaller A, Hauer B (2013) Optimized expression of the dirigent protein AtDIR6 in Pichia pastoris and impact of glycosylation on protein structure and function. Appl Microbiol Biotechnol 97: 7215–7227 [DOI] [PubMed] [Google Scholar]
- Kim KW, Moinuddin SGA, Atwell KM, Costa MA, Davin LB, Lewis NG (2012) Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J Biol Chem 287: 33957–33972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Smith CA, Daily MD, Cort JR, Davin LB, Lewis NG (2015) Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95 Å resolution with three isolated active sites. J Biol Chem 290: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN (2012) JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr 68: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chen J, Xiao Y, Di P, Zhang L, Chen W (2014) The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance. BMC Genomics 15: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzaferro LS, Hüttel W, Fries A, Müller M (2015) Cytochrome P450-catalyzed regio- and stereoselective phenol coupling of fungal natural products. J Am Chem Soc 137: 12289–12295 [DOI] [PubMed] [Google Scholar]
- Mehler EL, Fuxreiter M, Simon I, Garcia-Moreno EB (2002) The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins 48: 283–292 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neumann P, Brodhun F, Sauer K, Herrfurth C, Hamberg M, Brinkmann J, Scholz J, Dickmanns A, Feussner I, Ficner R (2012) Crystal structures of Physcomitrella patens AOC1 and AOC2: insights into the enzyme mechanism and differences in substrate specificity. Plant Physiol 160: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemetz R, Gross GG (2003) Ellagitannin biosynthesis: laccase-catalyzed dimerization of tellimagrandin II to cornusiin E in Tellima grandiflora. Phytochemistry 64: 1197–1201 [DOI] [PubMed] [Google Scholar]
- Niemetz R, Gross GG (2005) Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry 66: 2001–2011 [DOI] [PubMed] [Google Scholar]
- Pickel B, Constantin MA, Pfannstiel J, Conrad J, Beifuss U, Schaller A (2010) An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew Chem Int Ed Engl 49: 202–204 [DOI] [PubMed] [Google Scholar]
- Pickel B, Pfannstiel J, Steudle A, Lehmann A, Gerken U, Pleiss J, Schaller A (2012) A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J 279: 1980–1993 [DOI] [PubMed] [Google Scholar]
- Pickel B, Schaller A (2013) Dirigent proteins: molecular characteristics and potential biotechnological applications. Appl Microbiol Biotechnol 97: 8427–8438 [DOI] [PubMed] [Google Scholar]
- Ralph J, Schatz PF, Lu F, Kim H, Akiyama T, Nelsen SF (2009) Quinone methides in lignification. In Rokita SE, ed, Quinone Methides. John Wiley & Sons, Hoboken, NJ, pp 385–420 [Google Scholar]
- Ralph SG, Jancsik S, Bohlmann J (2007) Dirigent proteins in conifer defense. II. Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 68: 1975–1991 [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A (2009) Enzymes in jasmonate biosynthesis: structure, function, regulation. Phytochemistry 70: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Schlauer J, Rückert M, Wiesen B, Herderich M, Assi LA, Haller RD, Bär S, Fröhlich KU, Bringmann G (1998) Characterization of enzymes from Ancistrocladus (Ancistrocladaceae) and Triphyophyllum (Dioncophyllaceae) catalyzing oxidative coupling of naphthylisoquinoline alkaloids to michellamines. Arch Biochem Biophys 350: 87–94 [DOI] [PubMed] [Google Scholar]
- Seneviratne HK, Dalisay DS, Kim KW, Moinuddin SG, Yang H, Hartshorn CM, Davin LB, Lewis NG (2015) Non-host disease resistance response in pea (Pisum sativum) pods: biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 113: 140–148 [DOI] [PubMed] [Google Scholar]
- Tian X, Ruan J, Huang J, Fang X, Mao Y, Wang L, Chen X, Yang C (2016) Gossypol: phytoalexin of cotton. Sci China Life Sci 59: 122–129 [DOI] [PubMed] [Google Scholar]
- Toteva MM, Moran M, Amyes TL, Richard JP (2003) Substituent effects on carbocation stability: the pK(R) for p-quinone methide. J Am Chem Soc 125: 8814–8819 [DOI] [PubMed] [Google Scholar]
- Urry DW, Gowda DC, Peng S, Parker TM, Jing N, Harris RD (1994) Nanometric design of extraordinary hydrophobic-induced pKa shifts for aspartic acid: relevance to protein mechanisms. Biopolymers 34: 889–896 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vassao DG, Kim KW, Davin LB, Lewis NG (2010) Lignans (neolignans) and allyl/propenyl phenols: biogenesis, structural biology, and biological/human health considerations. In Townsend CA, ed, Comprehensive Natural Products. II. Chemistry and Biology. Elsevier, Oxford, UK, pp 815–928 [Google Scholar]
- Wagner TA, Liu J, Stipanovic RD, Puckhaber LS, Bell AA (2012) Hemigossypol, a constituent in developing glanded cottonseed (Gossypium hirsutum). J Agric Food Chem 60: 2594–2598 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Eyermann CJ, Hodge CN, Lam PYS, Ru Y, et al. (1994) NMR and X-ray evidence that the HIV protease catalytic aspartyl groups are protonated in the complex formed by the protease and a non-peptide cyclic urea-based inhibitor. J Am Chem Soc 116: 10791–10792 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.