During developmental leaf senescence in Arabidopsis, mitochondria simultaneously maintain primary energy processes and are the site of a number of catabolic processes, thus ensuring effective nutrient reallocation.
Abstract
The functions of mitochondria during leaf senescence, a type of programmed cell death aimed at the massive retrieval of nutrients from the senescing organ to the rest of the plant, remain elusive. Here, combining experimental and analytical approaches, we showed that mitochondrial integrity in Arabidopsis (Arabidopsis thaliana) is conserved until the latest stages of leaf senescence, while their number drops by 30%. Adenylate phosphorylation state assays and mitochondrial respiratory measurements indicated that the leaf energy status also is maintained during this time period. Furthermore, after establishing a curated list of genes coding for products targeted to mitochondria, we analyzed in isolation their transcript profiles, focusing on several key mitochondrial functions, such as the tricarboxylic acid cycle, mitochondrial electron transfer chain, iron-sulfur cluster biosynthesis, transporters, as well as catabolic pathways. In tandem with a metabolomic approach, our data indicated that mitochondrial metabolism was reorganized to support the selective catabolism of both amino acids and fatty acids. Such adjustments would ensure the replenishment of α-ketoglutarate and glutamate, which provide the carbon backbones for nitrogen remobilization. Glutamate, being the substrate of the strongly up-regulated cytosolic glutamine synthase, is likely to become a metabolically limiting factor in the latest stages of developmental leaf senescence. Finally, an evolutionary age analysis revealed that, while branched-chain amino acid and proline catabolism are very old mitochondrial functions particularly enriched at the latest stages of leaf senescence, auxin metabolism appears to be rather newly acquired. In summation, our work shows that, during developmental leaf senescence, mitochondria orchestrate catabolic processes by becoming increasingly central energy and metabolic hubs.
Senescence is the terminal phase of leaf development and is thought to culminate in a form of vacuolar programmed cell death (PCD; Smart, 1994; van Doorn et al., 2011). The entire process facilitates the tightly regulated degradation of intracellular structures and the subsequent reallocation of the resulting nutrients to the remainder of the plant. Evidence of senescence-like processes has been found in the whole green lineage, ranging from angiosperms and gymnosperms to bryophytes, green algae, and even photosynthetic prokaryotes (Thomas et al., 2009). Genes encoding proteins with functions relating to the turnover of the photosynthetic machinery make up some of the most ancient senescence processes conserved across multiple evolutionary clades of plants. In contrast, proteins associated with the regulation of senescence processes and their integration with developmental and stress-signaling networks constitute some of the most recent additions (Thomas et al., 2009). The carefully orchestrated dismantling of chloroplasts is one of the defining features of developmental leaf senescence (DLS) and necessitates a drastic reordering of cellular metabolism, as the leaves undergo a transition from autotrophic to heterotrophic functions. During this time, the remaining organelles producing energy (i.e. the peroxisomes and the mitochondria) are thought to acquire additional functions in order to compensate for this shift in metabolic demand and maintain cell viability, thus allowing for the optimum remobilization of nutrients (Keech et al., 2007). The mitochondrion often is characterized by its role in the production of cellular energy through the process of oxidative phosphorylation and in being the site of the metabolism of amino acids and the biosynthesis of many crucial vitamins and cofactors. However, it is important to consider that mitochondria also occupy a critical role in the signaling and regulatory landscape of the cell. Recent additions to the literature are redefining our view of the mitochondrion as a signaling hub, capable of perceiving and consolidating numerous signals relating to stress (Vanlerberghe, 2013; Ivanova et al., 2014) and developmental processes (Carlsson et al., 2008; Law et al., 2012).
Surprisingly, the role played by mitochondria during DLS appears poorly documented, even though a thorough understanding of the major cellular players involved is crucial in the optimization and management of this process. A number of studies, however, have put forward some interesting observations in Medicago truncatula cells (Zottini et al., 2006), Arabidopsis (Arabidopsis thaliana; Keech et al., 2007; Keech, 2011; Woo et al., 2016), grapevine (Vitis vinifera; Ruberti et al., 2014), and poplar (Populus spp.; Bhalerao et al., 2003; Keskitalo et al., 2005), revealing the persistent presence of mitochondria in the cell, long after the chloroplasts have commenced disassembly and whole-cell metabolism has been thoroughly modified. In Arabidopsis, an earlier study utilized microscopy and physiological assays to examine the differing fates of mitochondria and chloroplasts during senescence induced by placing either the whole plant or an individual leaf into extended darkness (Keech et al., 2007). In individually darkened leaves, an experimental setup that is analogous for many aspects of DLS, it was observed that, even though chloroplast and mitochondrial numbers diminished significantly and photosynthetic capacity deteriorated rapidly, mitochondrial respiration was maintained at high levels in individually darkened leaves even after 6 d (Keech et al., 2007). An increase in respiration also has been reported in senescing leaves of the American aspen (Populus tremuloides) and the northern red oak (Quercus rubra; Collier and Thibodeau, 1995). Together, these observations suggest the importance of mitochondria in supporting leaf viability while the remobilization of nutrients is carried out. Yet, the manner in which this is facilitated remains unknown.
In this study, we employed a number of experimental and analytical approaches to test the hypothesis that the mitochondrion has an integral role in the execution of DLS and to define the manner in which this takes place. First, confocal and electron microscopy were utilized to assemble a picture of the morphological changes occurring in mitochondrial populations as leaf senescence progresses. In addition, measurements of adenylate phosphorylation state and mitochondrial respiratory capacity were carried out to assess the energy status of leaf cells during this time period. Then, these morphological and physiological observations were complemented with transcriptomic and metabolomic analyses. A manually curated list of mitochondrial genes was established, and differential expression and hierarchical clustering analyses from a publicly available DLS transcriptomic data set (Breeze et al., 2011) were utilized to identify the timing and coordination of the major transcriptomic changes. This formed the foundation of a more focused analysis supported by metabolomic profiles of leaves during DLS, with particular attention paid to genes encoding proteins involved in catabolic processes.
RESULTS
Confocal and Electron Microscopy Analyses of Mitochondria during Leaf Senescence
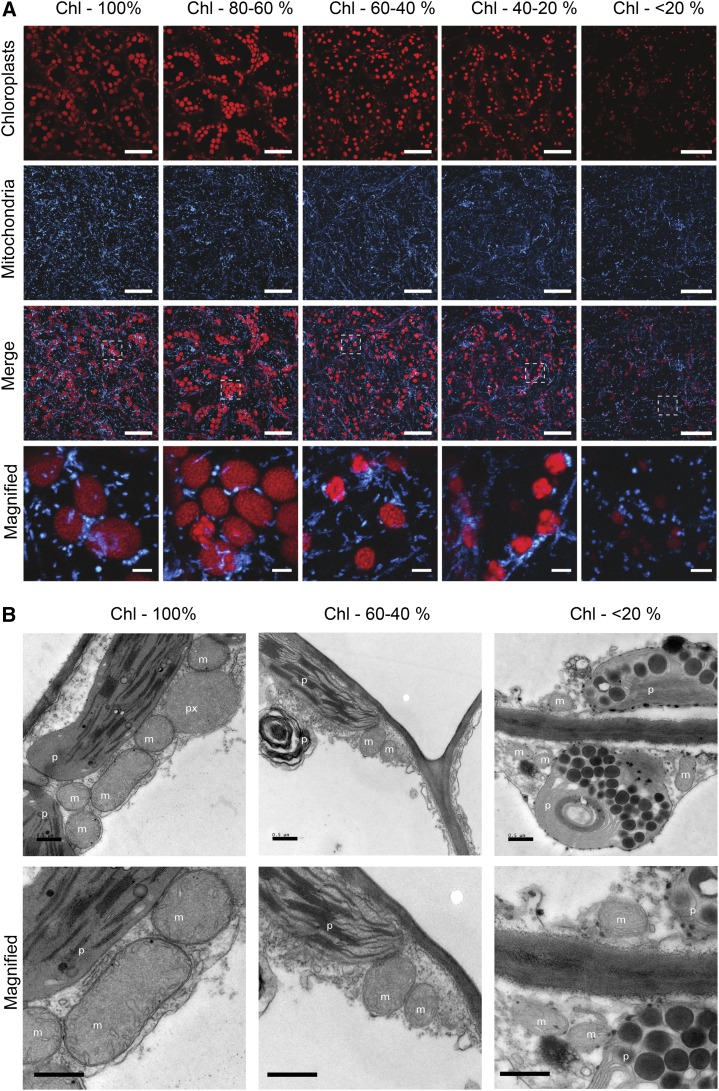
Microscopic analysis of mitochondria during DLS was performed using two complementary methods. First, confocal laser microscopy of chloroplasts and fluorescently labeled mitochondria (β-ATPase-GFP; Logan and Leaver, 2000) revealed that, throughout senescence, the populations of these energy organelles varied differentially (Fig. 1A; Supplemental Fig. S1). The number of chloroplasts did not decrease particularly during the process, while a two-dimensional estimation of the chloroplast size pointed toward an acute size reduction of these organelles as senescence progressed (Fig. 1; Supplemental Fig. S1, A and B). This was shown by plotting the distribution of plastids according to their projected two-dimensional area (Supplemental Fig. S1A). In contrast, the number of mitochondria underwent a significant reduction, which represented a decrease of about 30% compared with the population density of mitochondria in leaves with 100% chlorophyll (Fig. 1; Supplemental Fig. S1C). These data about the mitochondrial population corroborate earlier studies obtained with Arabidopsis undergoing dark-induced leaf senescence (Keech et al., 2007) and DLS (Keech 2011) as well as with grapevine leaves undergoing DLS (Ruberti et al., 2014). In a second step, electron microscopy was carried out to better visualize the ultrastructural changes occurring in organelles in mesophyll cells (Fig. 1B; for higher resolution images, see Supplemental Fig. S2). Early in the time course, some chloroplasts underwent a massive rearrangement, which included the disassembly of grana and an increase in the size and number of plastoglobuli, as a result of membrane degradation (Besagni and Kessler, 2013). As the time course progressed, more chloroplasts were affected, plastoglobuli became larger, and the overall number of functional chloroplasts decreased. Toward the end of senescence, small chloroplasts, also sometimes referred to as gerontoplasts, containing abnormally circularized thylakoids and large plastoglobuli, remained visible. In contrast, throughout the process of senescence, the ultrastructure of mitochondria appeared unchanged, with clearly demarcated cristae and membrane boundaries (Fig. 1B; Supplemental Fig. S2).
Figure 1.
In folio visualization of mitochondria by laser confocal microscopy (A) and electron microscopy (B) techniques during DLS in Arabidopsis. Stages of DLS are indicated by chlorophyll (Chl) content above each column of micrographs (for more details, see Fig. 2, A and B). For A, chlorophyll autofluorescence depicts chloroplasts, while mitochondria were labeled fluorescently with a β-ATPase-GFP construct (Logan and Leaver, 2000). White rectangles in the merge field indicate the magnified area. Bars = 50 μm and 5 μm for magnified. For B, m, mitochondrion; p, plastid; px, peroxisome in the respective senescing leaves. Bars = 0.5 μm.
Mitochondrial Energy Status and Respiratory Properties
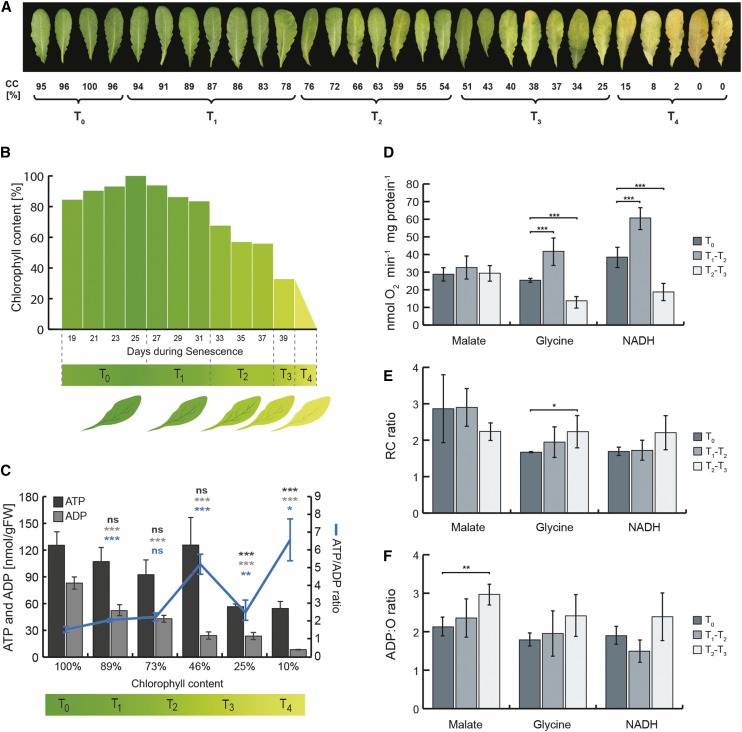
Within a single senescing leaf, cells can be at differing stages of senescence progression (Keech et al., 2010). A typical time course for the progress of leaf senescence, from rosettes 8 to 10 of Arabidopsis plants grown under a short-day photoperiod, is shown in Figure 2A. This demonstrates that, from leaves with 75% chlorophyll content and higher, a certain heterogeneity in the progress of senescence can be observed. In order to circumvent the heterogeneity of the DLS process and also to standardize our data throughout this study with that of the transcriptomic analysis outlined by Breeze et al. (2011), a referential time course was established based on the degradation of chlorophyll in senescing leaves (Fig. 2, A and B). Using the time course (in days) outlined by Breeze et al. (2011) for which leaf chlorophyll content was recorded (Fig. 2B), we identified five sequential periods delineating the progression of senescence in leaves: T0, leaf expansion to mature leaf; T1, mature leaf to early senescence; T2, early senescence to middle senescence; T3, middle senescence to late senescence; and T4, latest stage of senescence. For the Breeze et al. (2011) data set, the fifth period, T4, corresponding to leaves containing less than 20% chlorophyll, was extrapolated (Fig. 2B; see “Materials and Methods”) in order to corroborate our data sets for microscopy, adenylate pool measurements, and metabolomic analysis.
Figure 2.
Energy status and mitochondrial respiratory properties during DLS. A, Photographs of developmentally senescing Arabidopsis leaves under a short-day photoperiod. Below each leaf, the chlorophyll content (CC) is given as well as its classification into the following time periods: T0, T1, T2, T3, and T4. B, Graph displaying chlorophyll degradation during a time course of DLS (based on a time course defined by Breeze et al. [2011]). In designing these experiments, an additional time point (T4) was implemented, corresponding to the latest stage of leaf senescence. C, Graph of ATP and ADP abundance (nmol g−1 fresh weight [FW]) in total leaf extract, over a time course of DLS, and the corresponding ATP-ADP ratios. D to F, Respiratory capacities of mitochondria isolated from senescing leaves when incubated with malate, Gly, or NADH. Specifically, respiratory rate (D), respiratory control (RC) ratio (E), and ADP-O ratio (F) are shown. Each measurement displays the mean ± sd, and asterisks indicate significant differences after Student’s t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For more details about specific sample sizes, see “Materials and Methods.”
In light conditions, chloroplasts are the main energy producers of the cell, but the ATP produced by photophosphorylation is consumed mainly within the organelle for CO2 fixation. In contrast, mitochondria seem to provide most of the ATP for the cytosol, at least under conditions with high rates of photosynthesis (Gardeström and Igamberdiev, 2016). Thus, initial experiments were carried out to monitor the overall adenylate pool status in response to the senescence-associated dismantling of chloroplasts. By the end of the time course, the total content of ATP + ADP had dropped to about one-third of the starting amount. However, while the abundance of cellular ADP declined significantly in a progressive manner during the time course of senescence, ATP levels remained rather stable up to T2 and then decreased significantly in the latest stages of the process (i.e. T3 and T4; Fig. 2C). Consequently, the ATP-ADP ratio increased globally throughout the process, although with some fluctuations (Fig. 2C), suggesting that the cellular production of ATP by respiration is active and that a satisfactory energy status for the cell is being maintained. In addition, respiratory measurements were performed with isolated mitochondria (Fig. 2, D–F). The isolation and enrichment of intact mitochondria from senescing leaves was a technically demanding experiment, notably regarding the heterogeneity within the material sampled, as mentioned previously (Fig. 2A). These experiments required a minimum of 5 g fresh weight per extraction. Since leaf senescence is a very heterogenous process, only a few leaves per plant could be used at a time to ensure proper validation of the experimental data. As a consequence, about 20 to 40 plants were required per extraction. Here, different substrates were provided to the isolated organelles, and the rates of oxygen consumption, respiratory control ratios (RCRs), and ADP-O ratios were calculated. While respiratory rates did not change throughout the time course with malate provided as a substrate, we could observe a significant transient increase in mitochondrial respiratory capacity when Gly and NADH were added. This ability to oxidize Gly and NADH dropped significantly in the final T3 time points (Fig. 2D). In a second step, RCRs were calculated. RCR provides an indication of the coupling efficiency between the oxidative and phosphorylative mechanisms of the mitochondrial electron transport chain (mETC) and, therefore, represents a good proxy for mitochondrial integrity. In Figure 2E, we show that the RCR for isolated mitochondria, regardless of the substrate provided, remained high during leaf senescence. This indicated that isolated mitochondria conserved their integrity even at late stages of leaf senescence and, therefore, confirmed that the substrate-dependent modulation of respiratory capacities was not an artifact from the isolation procedure. Finally, the ADP-O ratio, which reflects the efficiency of ATP production by building up a proton gradient across the inner membrane and reducing oxygen with the electrons from the substrate, had values from about 2 up to 3 and remained stable for all tested substrates as leaf senescence progressed (Fig. 2F). Yet, a minor increase in this ratio, which even appeared significant at P < 0.05 with malate, was observed and could perhaps indicate an increased efficiency in ATP production toward the end of the senescence process. Taken together, our data clearly show that mitochondrial integrity, as well as its ability to provide energy to the cell, are maintained during leaf senescence, a function that arguably could happen in a substrate-dependent manner.
Overview of Transcriptomic Changes in Genes Encoding Mitochondrial Proteins during Developmental Leaf Senescence
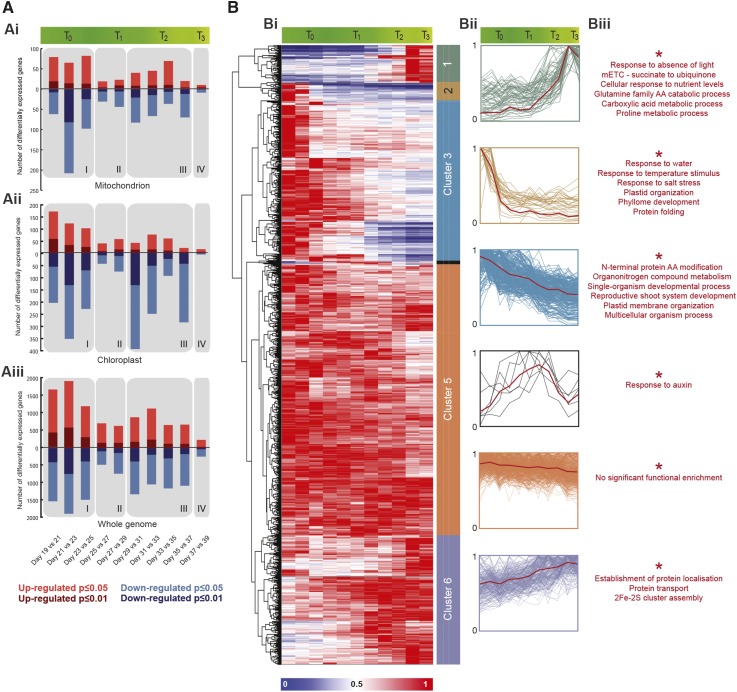
To gain a comprehensive insight into the role of the mitochondrion during leaf senescence, data from a detailed transcriptomic study of DLS (Breeze et al., 2011) were selected, as they represents the most experimentally robust and exhaustive sampling of DLS in Arabidopsis available. This data set was reanalyzed (see “Materials and Methods”; Supplemental Table S1) and further cross-referenced with a comprehensive list of genes encoding proteins targeted to the mitochondrion (mitochondrial set), which was prepared by combining highly stringent computational prediction tools and experimental data, prior to manual curation (Supplemental Table S2). This mitochondrial set, which is publicly available (http://www.upsc.se/olivier_keech/resources), will be updated regularly. Breeze et al. (2011) outlined a transcriptomic analysis that encompasses 11 time points during DLS, ranging from leaves collected 19 d after sowing (early into leaf expansion; 50% of final size) to leaves collected 39 d after sowing (visibly senescing leaf; 50% of leaf area yellowed). Note that in all cases, leaf 7 was collected, and for this study, only morning-period (7 h into the light period) samples were selected for further analysis. During this time course, 1,060 genes of the mitochondrial set (version 1.0) were observed to be expressed at one or more time points, with 49% of transcripts from these genes changing significantly in abundance (following false discovery rate correction; P < 0.05) from the beginning to the end of the time course. Interestingly, the overall pattern of differential transcript abundance could be divided into four distinct transcriptomic phases. The majority of the changes occur at the beginning of the time course, overlapping with T0 (named phase I), and toward the end of the time course, comprising the end of T1 and all of T2 (phase III), with a period of highly sustained expression in the middle of the time course, the end of T0 and beginning of T1 (phase II), leading to only a small number of significant changes (Fig. 3Ai). The final time point (phase IV) was characterized by the lowest number of differentially expressed genes of the time course and corresponded with T3. This same pattern of differential expression also was observed in curated lists of genes encoding proteins targeted to the chloroplast and the whole present set (Fig. 3A, ii and iii). The largest number of differentially expressed genes (P < 0.01) was observed in phase I, with the majority of these changes associated with decreases in transcript abundance (Fig. 3Ai). This peak in transcription abundance change could represent a genetic hallmark of the leaf transitioning from autotrophy to heterotrophy.
Figure 3.
Overview of transcriptomic changes in genes encoding proteins targeted to mitochondria during DLS. A, Differential expression analysis of transcripts encoding proteins targeted to mitochondria (Ai), chloroplast (Aii), and whole genome (total present set; Aiii) during DLS (false discovery rate by PPDE threshold was not applied). Transcript data from Breeze et al. (2011) were utilized as a foundation for the analysis of transcripts during DLS. Differential expression analysis was carried out on consecutive time points, with P value cutoffs of 0.05 and 0.01 as indicated at bottom. Four distinct phases of transcriptomic activity were identified and are marked with gray boxes. B, Hierarchical cluster of transcripts encoding proteins targeted to the mitochondrion during DLS. Bi, Transcript data from Breeze et al. (2011) were filtered with a manually curated list of mitochondrial transcripts. These values were normalized to the highest level of expression over the time course and hierarchically clustered using average linkage based on Euclidean distance. Bii, Expression profiles for each distinct cluster were plotted, with mean expression profiles represented by solid red lines. Biii, Each cluster was interrogated individually to assess the presence of significant (P < 0.05) overrepresentation of specific Gene Ontology (GO) categories in the gene lists.
In a second step, the expression values of the mitochondrial set were made relative to their maximum expression over the time course and hierarchically clustered (Fig. 3Bi). This analysis distributed the genes into six primary clusters. Clusters 2 and 3 represented approximately 30% of the mitochondrial set and were characterized by high transcript abundance in the early stages of DLS followed by decreasing levels of abundance as the time course progressed (Fig. 3Bi). GO analysis of these clusters revealed them to be significantly (P < 0.05) enriched in categories pertaining to temperature, salinity, and water stress responses and protein folding (cluster 2) and to single-organism developmental process, organic cyclic compound binding, and organo-nitrogen compound metabolic process (cluster 3; Fig. 3Biii). Here, it is worth noting that GO analyses of clusters 2 and 3 also were enriched significantly in genes associated with chloroplast and chloroplast part, respectively (Fig. 3Biii). The main reason for this is that several of these genes encode for products putatively dual targeted to chloroplasts and mitochondria. The fact that those genes are part of the two down-regulated clusters thus strengthens the idea that the main isoform of the encoded protein is mostly plastid associated, but it does not exclude an additional targeting to mitochondria. In contrast, cluster 1 was characterized by transcripts with low abundance in the early stages of the time course (T0–T1) before increasing to maximal levels in the final stages of DLS (end of T2 and T3). Transcripts belonging to this cluster were enriched significantly (P < 0.05) in functions associated with complex II of the mETC, in particular, components involved in the transfer of electrons from succinate to ubiquinone (coenzyme Q). In addition, this cluster was enriched with functions linked to responses to the absence of light and nutrient level depletion. Finally, this cluster also was enriched in functions associated with the metabolism of amino acids and derivatives, in particular Pro metabolic processes. Cluster 4 contained only a limited number of genes that were expressed transiently, with maximal expression observed at the transition of T1 to T2. This small cluster was enriched significantly in genes associated with response to auxin. Cluster 5 represented 44% of the total mitochondrial set. It is characterized by consistently high transcript abundance throughout the entire time course and is not enriched significantly in any functions, likely as it is populated by myriad housekeeping functions. This supports our earlier observations that mitochondria largely maintain their activity during senescence, even while chloroplasts are actively dismantled and whole cell metabolism is hugely readjusted. Cluster 6 was similar to cluster 5, except that it is characterized by an increasing trend in transcript abundance, culminating at the final two time points. This cluster was enriched in functions associated with protein localization, protein transport, and, interestingly, 2Fe-2S cluster assembly.
Mitochondrial Primary Metabolism Is Maintained through Dynamic Transcriptional Regulation during Developmental Leaf Senescence
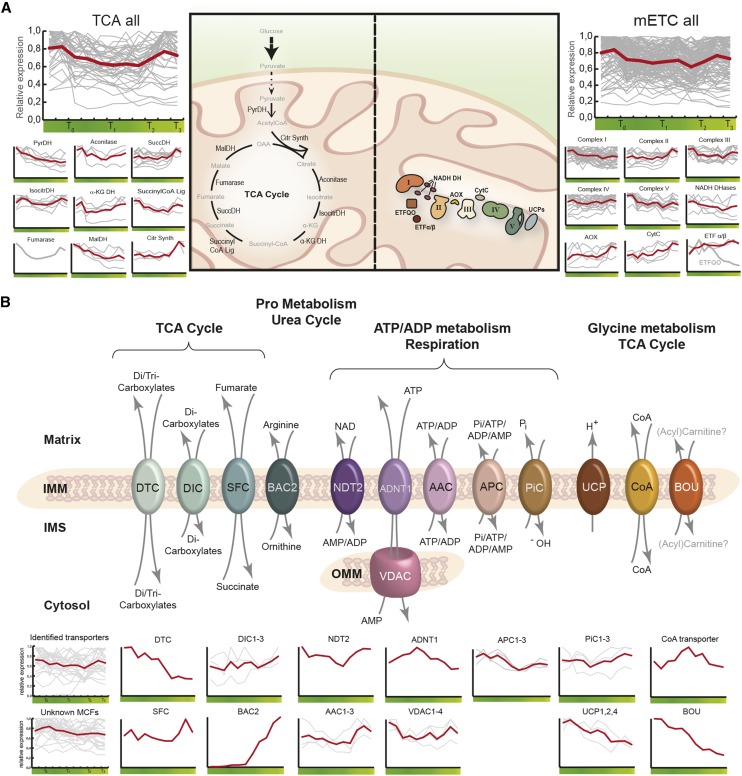
The biochemical processes carried out by the tricarboxylic acid cycle and mETC are arguably the most significant metabolic functions of mitochondria and collectively make up the most abundant proteins in the mitochondrion. Therefore, we analyzed in isolation a manually curated subset of genes encoding the proteins associated with these metabolic functions (Supplemental Table S3). The transcript profiles of these subsets are summarized throughout DLS in Figure 4A, and it was observed that the general trend for genes involved in mitochondrial primary metabolism is that of rather stable expression levels throughout the progression of DLS, as shown with the two summary graphs in Figure 4A (TCA all and mETC all). When studied in more detail, many transcripts encoding subunits of primary metabolic protein complexes displayed significant up or down variations in abundance during the time course. With a few exceptions, the transcript abundance of several genes (e.g. succinate dehydrogenase, citrate synthase, cytochrome c, or alternative oxidase [AOX]) was observed to increase slightly toward the end of the time course, particularly at the penultimate time point, day 37.
Figure 4.
Transcriptomic changes to mitochondrial primary metabolism and mitochondrial membrane transporters during DLS. A, Changes in the transcript abundance of the central enzymes and protein complexes of the tricarboxylic acid (TCA) cycle and the mETC during DLS. B, Visualization of the expression profiles of a defined subset of MCF members (DTC, DIC, SFC, BAC2, NDT2, ADNT1, AAC, PiC, UCP, and BOU) in addition to a CoA transporter and the VDAC. The mean expression profiles of the known MCF transporters and the unknown MCF members are visualized at bottom left. AAC, ADP/ATP carrier; ADNT1, ADENINE NUCLEOTIDE TRANSPORTER1; AOX, alternative oxidase; APC, ATP/inorganic phosphate carrier; BAC2, BASIC AMINO ACID CARRIER2; BOU, Bout de Souffle mitochondrial carnitine acyl carrier-like protein; CitrSynth, citrate synthase; CytC, cytochrome C; DIC, dicarboxylic acid carrier; DTC, ditricarboxylic acid carrier; IMS, intermembrane space; IsocitrDH, isocitrate dehydrogenase; α-KG DH, α-ketoglutarate dehydrogenase; MalDH, malate dehydrogenase; NDT2, NAD+ TRANSPORTER2; OAA, oxaloacetate; PiC, phosphate carrier; PyrDH, pyruvate dehydrogenase; SFC, succinate-fumarate carrier; SuccDH, succinate dehydrogenase; SuccinylCoA Lig, succinyl-CoA ligase; UCPs, uncoupling proteins.
Fumarase, the only single-copy gene encoding a component of the tricarboxylic acid cycle, had an expression profile characterized by down-regulation between T0 and T2 followed by an increase of transcript abundance during the final part of the time course. All the isoforms of citrate synthase were progressively up-regulated throughout the process of leaf senescence. In contrast, other enzymes encoded by multiple gene isoforms showed more contrasting expression patterns. This was the case for cytochrome c, AOX, and the α-ketoglutarate (α-KG) dehydrogenase, for which several isoforms were strongly up-regulated at the final stages of DLS while others displayed down-regulation rather early in the process. Both internal and external NADH-dependent dehydrogenases also could be included in this cluster, for which dramatically different expression profiles were observed: two genes (NDB1 and NDA1) were down-regulated throughout the DLS time course, two (NDB4 and NDA2) stayed rather stable, and one (NDB2) displayed maximal transcript abundance in the final time points. NDB2 is a core component of the alternative respiratory pathway and was identified previously as one of the most stress-responsive mitochondrial proteins in Arabidopsis (Van Aken et al., 2009). Together with AOX1a, NDB2 can form a complete alternative respiratory system (Clifton et al., 2006), bypassing ATP (and reactive oxygen species [ROS]) production while promoting heat generation. Interestingly, there was distinct heterogeneity in the expression profiles of the different AOX isoforms during the time course, with the expression profiles of AOX1b reducing and AOX1c remaining consistently high during the time course. AOX1a and AOX1d showed the opposite of this, with the transcript abundance of these two genes increasing consistently throughout the time course and reaching maximum levels in the final two time points, a finding that has been noted in previous studies documenting expression profiles of AOX family members during developmental and dark/starvation-induced senescence in Arabidopsis (Buchanan-Wollaston et al., 2005; Clifton et al., 2006). This observed variation in the gene expression for members of multigene families is suggestive of a genetic program that facilitates senescence-specialized roles. Succinate dehydrogenase (or complex II) has a dual role in the tricarboxylic acid cycle and the mETC. Similar to AOX1a and NDB2, all but one of the subunits of succinate dehydrogenase were observed to display positive expression profiles with the highest transcript abundance in the penultimate time point (as observed in the hierarchical clustering and subsequent GO analysis; Fig. 3Biii, cluster 1). Finally, the genes coding for the electron-transfer flavoprotein:ubiquinone oxidoreductase (ETFQO) and ETFα/β complexes, known to provide additional electrons to the mETC under carbon starvation (Ishizaki et al., 2005; Araujo et al., 2010), displayed opposite expression profiles. While the ETFα/β complex displayed a slight increase in transcript abundance during DLS, the ETFQO clearly showed a decrease in its transcript levels.
Intrinsically associated with both the tricarboxylic acid cycle and mETC functions, the maturation of the iron-sulfur (Fe-S) proteins represents another crucial function supported by mitochondria. Indeed, seven iron-sulfur clusters (ISCs) are found in complex I, three in complex II, and one in complex III of the mETC, but various types also are present in aconitase, lipoate and biotin synthases, or ferredoxins, to cite only a few examples. Moreover, whereas the chloroplastic sulfur mobilization (SUF) machinery operates independently (Van Hoewyk et al., 2007), the mitochondrial iron-sulfur cluster (ISC) machinery controls the maturation of both mitochondrial and cytosolic/nuclear Fe-S proteins, including crucial proteins involved in ribosome biogenesis and DNA metabolism. This is due to the fact that mitochondria provide the sulfur required to build ISCs onto cytosolic/nuclear proteins via the cytosolic iron-sulfur cluster assembly machinery. This is done in the form of an as yet unidentified compound conveyed by an ATP-binding cassette transporter present on the inner membrane of mitochondria called ATM3/STA1 (Couturier et al., 2013; Balk and Schaedler, 2014). Here, we established a list of genes coding for components of the ISC machinery as well as for known Fe-S proteins (Supplemental Table S3). The transcripts encoding the majority of the 22 putative ISC components were not much affected during leaf senescence. The trend for genes encoding known mitochondrial Fe-S proteins was somewhat more contrasted. With the exception of genes encoding subunits of complexes II and III, for some complex I subunits, for one aconitase, and for cnx2, most of the other genes had their expression levels decreased as the time course progressed (Supplemental Table S3).
Isoforms of Mitochondrial Transporters Are Differentially Expressed to Maintain Effective Transport during Leaf Senescence
In the above sections, it was highlighted that mitochondria are dynamic in their ability to modify their metabolism as the cellular environment changes. The involvement and connection of the energy-producing organelles to various other parts of the cell implies that a high metabolic flux exists across the mitochondrial membranes. The mitochondrial carrier family (MCF) is a gene family that consists of transport proteins with specific structural features that localize to the inner mitochondrial membrane. This family accounts for more than 50 transport proteins, of which a few also are localized to organelles other than mitochondria (Haferkamp and Schmitz-Esser, 2012). In Figure 4B, the identified and characterized MCF members are shown (Palmieri et al., 2011; Monne et al., 2015) in addition to a mitochondrial CoA transporter (Zallot et al., 2013) and the voltage-dependent anion channel (VDAC) complex (Robert et al., 2012). The 26 transport proteins depicted are implicitly involved in various metabolic pathways inside the mitochondrion (tricarboxylic acid cycle, aerobic respiration, nucleotide transport, and amino acid metabolism) and connect them to the metabolic networks of the cell. The degradation of nucleic acids, proteins, lipids, and fatty acids results in the accumulation of various products that can be utilized downstream for energy production and the reorganization of the metabolic status of the cell during DLS. Here, the transcript levels of most of the identified transporters as well as of the as yet uncategorized MCF members (i.e. unknown MCFs; Fig. 4B) were rather stable during senescence. Interestingly, stable or slight up-regulation of transcript abundance was observed for the transporters supporting the tricarboxylic acid cycle (i.e. SFC, DICs, and CoA transporter) and for those involved in ATP/ADP/AMP and inorganic phosphate shuffling across the inner mitochondrial membrane (i.e. ADNT1, NDT2, PICs, AACs, and APCs). In contrast, a few transporters had their transcript abundance clearly down-regulated during DLS. This was particularly the case for the DTC and the putative carnitine/(acyl)carnitine carrier (BOU) and to some extent to all UCPs. UCPs have been linked to roles in maintaining the redox balance of the mETC, and more specifically, UCP1 would contribute to the dissipation of the proton gradient, relieving both respiration and photorespiration on some electrochemical constraint while bypassing the production of ATP (Sweetlove et al., 2006). As such, the observed decline in abundance of transcripts encoding UCPs is in accordance with the decline in photosynthetic ability. Although the down-regulation of BOU was expected, as the function of this transporter was recently also associated with photorespiration and linked to Gly decarboxylase activity (Eisenhut et al., 2013), the down-regulation of UCPs was unexpected, since these proteins are often thought to play an important role in the mitochondrial redox balance and would protect against high ROS levels (Van Aken et al., 2009; Nogueira et al., 2011; Barreto et al., 2014), which were reported to increase during leaf senescence (Zimmermann and Zentgraf, 2005). One of the most interesting expression profiles relates to BAC2, with minimal transcript abundance at the beginning of the time course, followed by a sudden increase in abundance at the onset of leaf senescence, reaching a maximal peak in expression at the end of the time course. Finally, working in tandem with several adenylate transporters (see above), the VDACs also displayed rather stable expression profiles. VDACs are crucial for energy-related processes (Pan et al., 2014, and refs. therein), and it is important to note that a functional interplay between these transport proteins is essential for mitochondrial activity and energy supply for the cell. Interestingly, in mammals, VDACs have been associated with PCD (Pan et al., 2014), while in plants, their contribution to cell death mechanisms remains elusive (Robert et al., 2012; Godbole et al., 2013).
Isoforms of Mitochondrial Import Components Are Differentially Regulated during Leaf Senescence
The data presented above suggest that, in addition to mitochondrial populations being actively maintained, mitochondria also acquire novel functions in order to compensate for the shift in metabolic demand brought on by the senescence-mediated chloroplast degradation. This augmentation must be facilitated at multiple regulatory levels, but one of the most pivotal is the gatekeepers of the mitochondrial membranes: the protein import components. As such, a subset of nuclear genes encoding mitochondrial import components (in addition to chaperones, heat shock proteins [HSPs], and respiratory chain assembly proteins [RCAPs]) was isolated from the Breeze et al. (2011) data set and analyzed (Supplemental Fig. S3; Supplemental Table S4). From two earlier studies (Lister et al., 2004; Van Aken et al., 2009), a number of mitochondrial import components and assembly proteins were defined as highly responsive to stress (expression profiles of these genes are colored red in Supplemental Fig. S3). In accordance with the genes encoding components of mitochondrial primary metabolism, expression profiles for the genes encoding import components showed differential regulation over the time course, with many displaying their highest transcript abundances in the final time points of leaf senescence (as signified by the blue mean line on each graph in Supplemental Fig. S3). HSPs are an exception; they displayed almost exclusive down-regulation over the time course. It is important to note that the central pore of the translocase of the outer membrane (TOM), TOM40, displayed a declining expression profile, with the lowest transcript abundance for this gene observed in the final stage of leaf senescence. Whether this indicates that the corresponding protein levels of TOM40 also were down is unknown; however, as the steady-state abundance of TOMs usually exceeds that of the translocase of the inner membranes (TIMs), the rate of protein import is commonly viewed as being a product of TIM complex abundance in plants and yeast (Dekker et al., 1997; Wang et al., 2012).
It has been demonstrated previously that, although most mitochondrial import components are composed of small multigene families, in general, only one member is dominantly expressed at a given time (Lister et al., 2004). However, in response to certain stress conditions (Lister et al., 2004) or developmental reprogramming (Wang et al., 2014), these isoforms have been observed to switch. This suggests that the redundancy in isoforms lends the complement of mitochondrial import components a degree of plasticity, which can be modified under a given signal, to alter the import of mitochondrial proteins and thus modulate the protein content of mitochondria. Similar to TOM40, it was observed that TOM20-2, TOM20-3, and TOM5 displayed maximal transcript abundance at the beginning of the time course (T0) before declining consistently and reaching the lowest abundance in the final time point (T3). This is an interesting observation given that a number of genes encoding different TOMs (TOM20-4 and TOM22-1) showed the opposite of this, with low transcript abundance at the beginning of the time course before increasing to maximum levels in the final stages of leaf senescence (Supplemental Fig. S3B, TOMs and SAM). Similarly, members of the TIM also displayed discordant expression profiles, with some transcript species displaying declining expression profiles (TIM17-3 and AT3G25120) while others increased transiently at either the end of T0 or the beginning of T2 before declining in abundance (TIM17-1, TIM17-2, TIM23-1, TIM23-2, and TIM23-3; Supplemental Fig. S2B, TIM17:23).
For the successful import of precursor proteins into the mitochondrion, presequence cleavage and folding must take place for a functional protein to reach its destination. As such, mitochondrial chaperones, HSPs, and RCAPs also were analyzed alongside the protein import apparatus. For intermembrane space TIMs, chaperones, and RCAPs, it was observed that transcripts remained relatively abundant throughout the time course, with many genes reaching maximum transcript abundance in the final stages of leaf senescence (Supplemental Fig. S2B). In contrast, the subset of mitochondrially localized HSPs was observed to largely exhibit a declining expression profile, with the notable exceptions of HEAT SHOCK COGNATE70-5 and HSP70. HSP70 has an exhaustively well-documented role in a number of crucial processes taking place within the mitochondrion, such as protein folding and transcriptional regulation (Lin et al., 2001). As such, it is possible that it plays an increasingly important role in aiding protein metabolism as the cellular environment becomes progressively hostile (e.g. increase of ROS and various lytic vacuoles) during the final stages of DLS.
Taken together, these transcriptomic data support our earlier findings and strengthen the proposition that, in contrast to reported PCD events (for review, see Vianello et al., 2007), mitochondrial activity is maintained during DLS even though not all transcript levels are stable and several functions seem clearly down-regulated. Nevertheless, these data identify situations whereby members of multigene families are differentially regulated in response to DLS, suggesting that these genes may fulfill highly specialized roles during DLS that will require further investigation in the future.
The Metabolic Profile of Leaves during Developmental Leaf Senescence
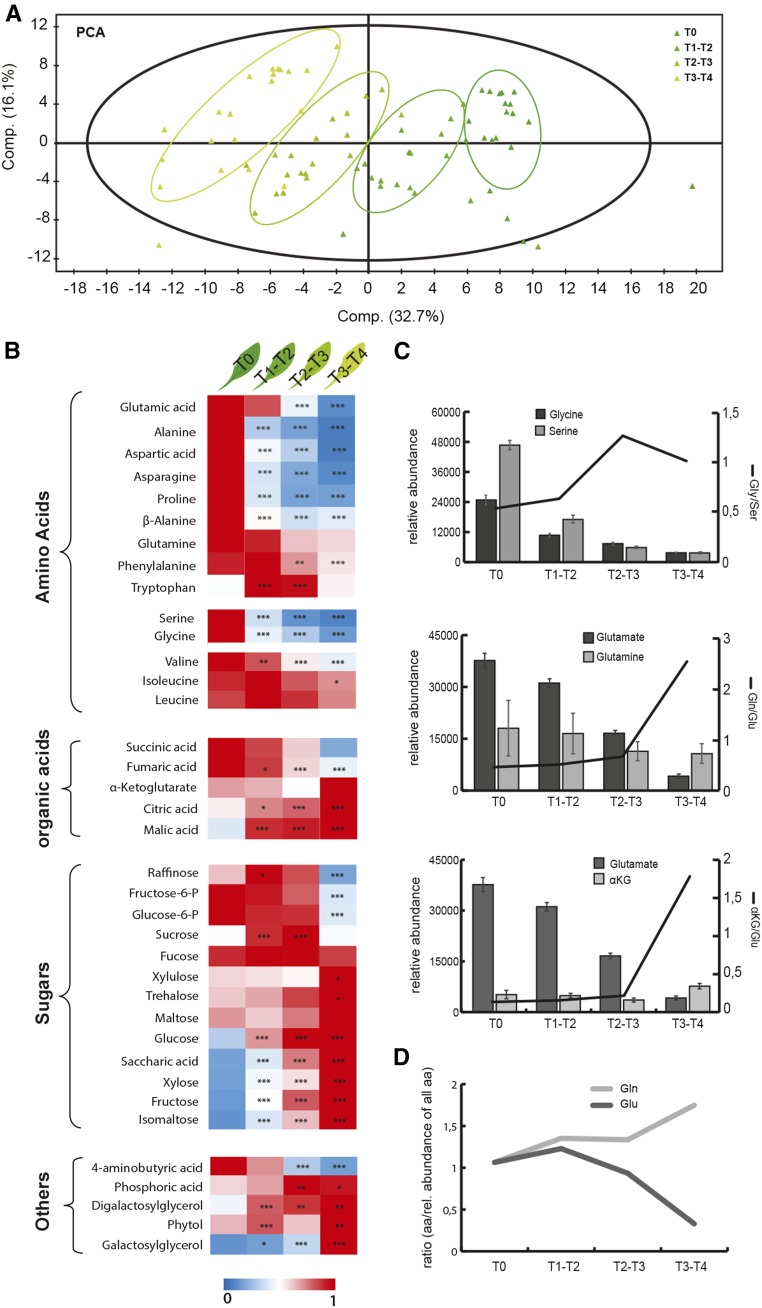
To support both our physiological data and targeted transcriptomic investigation, we undertook gas chromatography-mass spectrometry-based metabolic analysis using the same time course of DLS as outlined above. A principal component analysis (PCA) of all biological replicates showed a clear separation between the metabolic profiles at four time points of DLS (T0, T1-T2, T2-T3, and T3-T4; Fig. 5A). Furthermore, it was observed that, at the beginning of the time course (T0), the samples clustered together densely, whereas toward the end of this time course (T3-T4), the samples were more distantly separated, indicating an increasing variability among biological replicates as senescence progressed. As a matter of fact, this biological heterogeneity is quite difficult to circumvent and remains a major bottleneck when studying natural aging in leaves, which over the years has necessitated the use of alternative experimental designs, such as dark-induced senescence (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Keech et al., 2010).
Figure 5.
Metabolomic analysis of leaves undergoing DLS. A, PCA plot of samples over the time course of leaf senescence. B, Heat map of identified amino and organic acids, sugars, and other metabolites normalized to their maximal relative abundance. Statistically significant differences in abundance compared with time point T0 were tested using Student’s t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. C, Relative abundances and ratios of Gln-Glu, Gly-Ser, and α-KG-Glu throughout DLS. Error bars represent se. D, Ratio of Glu and Gln to the relative pool of all amino acids in senescing leaves.
In a second step, the observed metabolites were clustered based on functional groups, and their abundance was normalized to the maximum value and presented as a heat map (with significant differences identified when compared with T0; Fig. 5B). Most of the detectable amino acids decreased as senescence progressed. However, while the abundance of some of these amino acids decreased drastically (Glu, Ala, Pro, Asn, Ser, and Gly), several others, including branched-chain amino acids (BCAA) and aromatic amino acids, remained quite high in abundance. This also included the principal nitrogen-rich and senescence-associated amino acid Gln. Sugars, on the other hand, exhibited a more heterogenous pattern. While some sugars, such as Glc, Fru, Xyl, trehalose, or even maltose, showed an increased content toward the end of the DLS (T3-T4), others, including notably Suc and hexose phosphate, had a more transient abundance profile with a clear increase toward the middle of the process. Furthermore, a few organic acids also were detected, and while the abundance of fumarate and succinate decreased progressively, α-KG, citrate, and malate increased. Finally, several key metabolites were additionally quantified, namely, galactosylglycerol, digalactosylglycerol, and phytol, and the abundance of these was found to massively increase during leaf senescence, most likely as they represent downstream products of thylakoid and chlorophyll degradation.
With a focus on nitrogen metabolism, we also examined metabolites that often are substrate-product partners in a given reaction. In green leaves, Gly and Ser are seen mainly as two metabolic intermediates of the photorespiratory pathway and appear connected to each other by the tandem glycine decarboxylase complex (GDC) and serine hydroxymethyl transferase (SHMT). This reaction combines two Gly into one Ser and leads to the release of NH3, CO2, and NADH (Douce et al., 2001). Here, the abundance of both Gly and Ser decreased with time; however, the ratio of Gly to Ser increased as senescence progressed (Fig. 5C). Interestingly, the two pairs Gln/Glu and α-KG/Glu had their respective ratios increase during the latest stages of DLS. When compared with the relative pool of all amino acids over the DLS time course, the pool of Gln increased while the pool of Glu decreased (Fig. 5D). Together, this indicates that Glu becomes depleted faster than Gln and α-KG, which, in turn, suggests that Glu would become a limiting metabolic factor during DLS.
Mitochondria-Localized Catabolic Processes
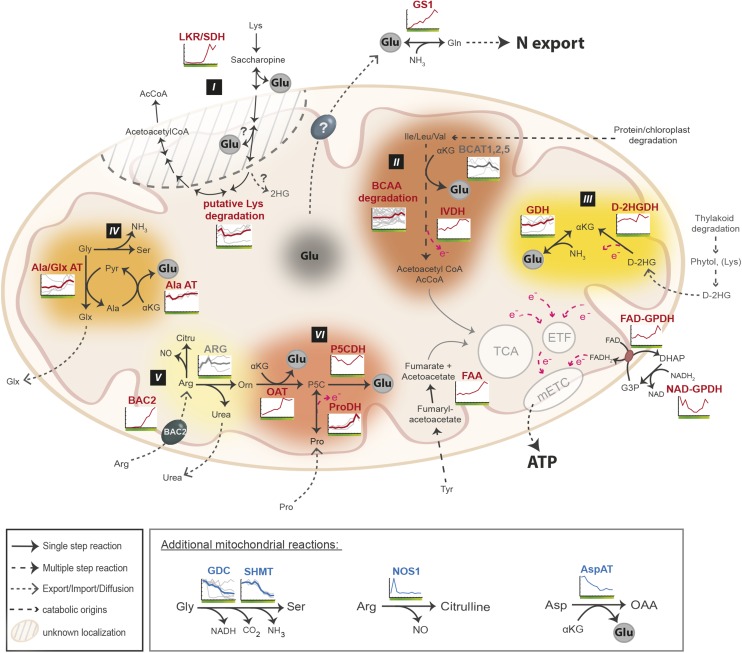
Perhaps the most important outcome of DLS is the procurement and subsequent redistribution of nutrients, and in particular nitrogen, from senescing tissues to the rest of the plant (Himelblau and Amasino, 2001; Guiboileau et al., 2010). The data presented above revealed that the primary metabolic contribution of mitochondria in the cellular landscape was maintained until very late into DLS. Furthermore, it is important to note that mitochondria also are the site of numerous important catabolic processes. Therefore, we questioned whether, during leaf senescence, mitochondria could become an essential player in the release and remobilization of nitrogen by hosting diverse catabolic processes. To challenge this hypothesis, we first identified several catabolic and conversion pathways for which mitochondrial steps were present. Second, a manually curated list of genes encoding mitochondrial proteins associated with these pathways was analyzed in detail, and six main metabolic routes involved in catabolic processes were mapped onto a summarizing figure. Figure 6 thus represents a distilled overview of the mitochondrial metabolic reactions likely contributing to nitrogen remobilization during leaf senescence.
Figure 6.
Production of Glu, reducing equivalents, and tricarboxylic acid (TCA) cycle intermediates from catabolic reactions occurring in the mitochondrion during DLS. Transcriptomic overviews are shown for the mitochondrially localized portion of the following metabolic pathways: Lys degradation (I), BCAA degradation (II), 2-hydroxyglutarate (d-2HG) metabolism (III), Gly and Ala metabolism (IV), urea cycle (V), and Pro metabolism (VI). Specific genes of these pathways and their transcript abundance during DLS are illustrated. The production of reducing equivalents is shown as an arrow with an electron (e−). AcCoA, Acetyl-CoA; AlaAT, Ala aminotransferase; Ala/GlxAT, Ala/glyoxylate aminotransferase; ARG, Arginase; AspAT, Asp aminotransferase; BCAT, branched-chain amino acid transaminase; Citru, citrulline; FAA, fumarylacetoacetase; FAD-GPDH, FAD-dependent glycerol-3-phosphate dehydrogenase; Fum, fumarate; Glx, glyoxylate; GS, Gln synthase; d-2HGDH, d-2-hydroxyglutarate dehydrogenase; IVDH, isovalerate dehydrogenase; LKR/SDH, Lys-ketoglutrarate reductase/saccharopine dehydrogenase; NAD-GPDH, NAD-dependent glycerol-3-phosphate dehydrogenase; NO, nitric oxide; NOS1, nitric oxide synthase; OAT, Orn-d-aminotransferase; ProDH, Pro dehydrogenase; Pyr, pyruvate; P5C, pyrroline-5-carboxylate; P5CDH, pyrroline-5-carboxylate dehydrogenase.
Lys is an essential amino acid that can be broken down to produce acetoacetyl-CoA and acetyl-CoA, a potential substrate of the tricarboxylic acid cycle (Fig. 6, pathway I). Concomitantly, two molecules of Glu are released. The first two steps of Lys degradation can be catalyzed either by two separate enzymes or a bifunctional enzyme called LKR/SaccDH. First, LKR catalyzes the conversion of Lys and α-KG to saccharopine, which is then converted to adipic semialdehyde and Glu via the SaccDH. It has been reported that the bifunctional LKR/SDH also encodes a functional equivalent of two monofunctional enzymes, LKR and SaccDH (Tang et al., 2002). Whereas these enzymes are known to localize to the cytosol, other enzymes in this pathway are proposed to localize to the mitochondrion. Regardless, the whole metabolic pathway of Lys degradation was up-regulated, including its mitochondrial component, leading to the release of Glu. Similar outcomes were observed in the degradation of BCAA (Fig. 6, pathway II; Supplemental Fig. S4). The transcript abundance of the majority of the genes associated with this catabolic pathway increased to maximum abundance in the penultimate time point, leading to the release of Glu and to the delivery of acetyl-CoA to the tricarboxylic acid cycle (Supplemental Fig. S4). Additionally, electrons released from this pathway can support the mETC via the ETF-ETFQO system (Ishizaki et al., 2006) and thereby contribute to further ATP generation in mitochondria, particularly under starvation conditions (Ishizaki et al., 2005; Binder, 2010). d-2HG appears as a product of phytol and potentially of Lys degradation (Engqvist et al., 2011). The conversion of d-2HG to α-KG by d-2HGDH leads to the release of electrons, which can be utilized by the mETC (Fig. 6, pathway III). α-KG can be provided directly to the tricarboxylic acid cycle or further converted to Glu in the presence of ammonium via NADH-dependent Glu dehydrogenase (GDH). The transcript abundance of both the d-2HGDH and GDH isoforms was maintained throughout DLS, while, interestingly, an isoform of GDH appeared strongly up-regulated in comparison with the other isoforms, suggesting a specific role for this isoform in DLS.
The expression profiles for the transcripts encoding the subunits of GDC and SHMT were clearly progressively down-regulated during DLS (Fig. 6, pathway IV and boxed section), even though at T2, Gly was still oxidized by isolated mitochondria (Fig. 2C). That said, a favorable alternate route for Gly conversion may take place during DLS, as shown by the increase in transcript abundance of two connected aminotransferases: Gly can be converted to glyoxylate via the activity of an Ala/glyoxylate aminotransferase; also coupled to this conversion, the Ala aminotransferase (AlaAT) converts Ala back to pyruvate and produces Glu.
Part of the urea cycle also takes place in the mitochondrion (Fig. 6, pathways V and VI). Urea is produced from the conversion of l-Arg to l-Orn by the enzyme arginase, which is encoded by two isoforms. Interestingly, the transcript abundance of these two isoforms display opposing expression profiles, with the transcript abundance of one isoform decreasing at the onset of DLS while the other isoform increases to maximal levels at day 37. The pool of l-Orn generated from l-Arg conversion can then yield two Glu molecules, via the conversion of l-Orn to P5C plus Glu by OAT, before being further converted to Glu by P5CDH. Similarly, Pro can be converted to P5C (in a reaction that also yields reducing equivalents that can be utilized by the mETC) by ProDH. Investigation of the transcript abundance of these two genes revealed that they were both up-regulated to maximum levels in the penultimate time point. It is worth noting that an additional reaction may occur, where Arg is converted to citrulline by a nitric oxide synthase-like activity with the concomitant release of nitric oxide. However, the enzyme responsible for this activity remains to be clearly identified (see “Discussion”). A few of these metabolic pathways are shown to be connected to primary metabolism, by providing electrons, acetyl-CoA, or other metabolites like fumarate. Indeed, the transcript abundance of the fumarylacetoacetase also was progressively up-regulated during DLS. This enzyme, localized in the mitochondrion, contributes to the final steps of the Tyr degradation pathway and ultimately leads to the production of fumarate, which, in turn, can be incorporated into the tricarboxylic acid cycle.
The Evolutionary Age of Mitochondrial Genes Expressed during Developmental Leaf Senescence
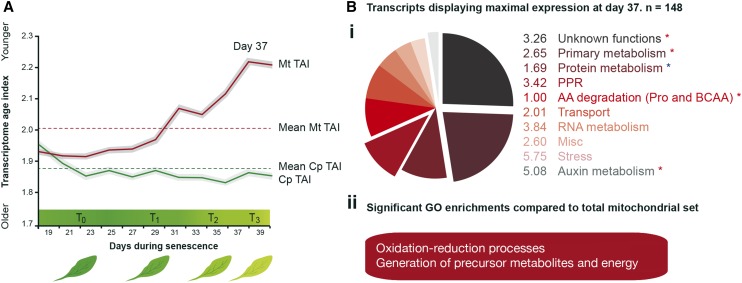
Finally, in order to investigate the relationship between the evolutionary age of a given gene and its expression during DLS, the phylostratigraphic ranking established by Quint et al. (2012) was matched with the mitochondrial set. In brief, each gene was assigned to one of 13 phylostrata (PS1–PS13), with members of PS1 making up the most evolutionarily ancient genes, with homologous sequences in prokaryotes, and with PS13 making up the most evolutionarily recent genes, with no orthologs outside of Arabidopsis. These data were then transformed to provide a weighted mean of the evolutionary age of each gene, based on its expression level, resulting in the Transcriptome Age Index (TAI) of each time point (see “Materials and Methods”; Fig. 7A). This analysis revealed that, as DLS progresses, the mean evolutionary age of mitochondrial genes being actively expressed becomes progressively younger, with the most evolutionarily ancient genes expressed primarily at the beginning of the time course (T0 and T1) and the youngest toward the final stages of senescence (T2 and T3). As a point of reference, this analysis also was carried out for a curated list of genes encoding proteins targeted to the chloroplast (Law et al., 2012), in which the opposite was observed, with the evolutionary age of expressed genes getting older as DLS progressed. Interestingly, the transcripts displaying the youngest TAI in the entire time course were observed in the penultimate time point, day 37. Functional analysis of transcripts expressed at maximal abundances at this time point (day 37) revealed a significant (P ≤ 0.05) enrichment in primary metabolism functions (components of the tricarboxylic acid cycle, TAI = 2.65; mETC, TAI = 4.15), in the degradation of amino acids (chiefly Pro and the BCAA Leu, Ile, and Val; TAI = 1), and in auxin metabolism (TAI = 5.08; Fig. 7Bi). In addition, GO enrichment analysis of these transcripts revealed a significant (Best per Parent, P < 0.005) overrepresentation of processes associated with oxidation reduction and the generation of precursor metabolites and energy (Fig. 7Bii).
Figure 7.
Transcriptome age profiling of genes encoding proteins targeted to mitochondria during DLS. A, Using data derived from Breeze et al. (2011), a subset of genes encoding proteins targeted to the mitochondrion was established and cross-referenced with the phylostratigraphic ranking established by Quint et al. (2012) to investigate the evolutionary age of mitochondrial genes expressed during DLS. These data were transformed to provide a weighted mean of the evolutionary age of each gene, based on its expression level, resulting in the TAI of each time point (see “Materials and Methods”). As a point of reference, a subset of genes encoding proteins targeted to the plastid also was analyzed. The mean evolutionary ages of mitochondrial (Mt) and chloroplastic (Cp) genes are plotted with dotted lines. The gray areas bordering the lines represent the se of the TAI, as estimated by bootstrap analysis. B, Transcripts displaying maximal expression at day 37 (n = 148) were further assessed for functional enrichments compared with the total mitochondrial list. Bi, Significantly enriched/depleted functional groups are identified with colored asterisks (red, enriched; blue, depleted). The mean TAI of each functional group is listed. Bii, In addition, GO enrichments were analyzed.
DISCUSSION
Mitochondria are highly adaptive organelles that fulfill a number of vital physiological, biosynthetic, and energetic roles. The study presented here provides, to our knowledge, the first in-depth analysis of the varied and complex roles of mitochondria during DLS. Under normal photosynthetic conditions, mitochondria work in concert with chloroplasts and peroxisomes to harvest and distribute energy to the cell (Gardeström and Igamberdiev, 2016). As chloroplasts are progressively dismantled during DLS, it has been postulated that mitochondria maintain their role as primary energy providers to fuel the energetically demanding reallocation of nutrients during DLS (Smart, 1994; Keech et al., 2007) and also during autumnal leaf senescence such as in poplar (Keskitalo et al., 2005). However, until now, the manner in which this was facilitated had remained unknown. The targeted transcriptomic analysis carried out here revealed that, for the most part, the transcript abundance of components of the tricarboxylic acid cycle, the ISC assembly machinery, and mETC remain stable throughout the time course, with many showing their maximal expression in the penultimate time point. This was further supported by our physiological assays presented in Figure 2, which clearly indicate that mitochondria maintain a complete and functioning complement of primary metabolism enzymes. A comparison of mitochondrial transcriptomic data sets from Breeze et al. (2011) and van der Graaff et al. (2006) also supports the robustness of our conclusions, particularly when regarding the catabolic reactions shown in Figure 6 (Supplemental Fig. S5A; Supplemental Table S5). Having said that, the Breeze et al. (2011) data set was utilized throughout our study, as it represents the most detailed publicly available transcriptomic analysis of DLS, as shown by the PCA plot in Supplemental Figure S5B. Furthermore, as our study dealt with a different photoperiod compared with the previous study (Breeze et al., 2011), we performed a set of complementary quantitative reverse transcription (qRT)-PCR assays for several key genes related to our study (Supplemental Fig. S6; Supplemental Table S6). The results of these assays were satisfactory and showed mostly the same trends as the microarray experiments, suggesting that the process of leaf senescence is conserved and largely independent from photoperiodic regimes. Interestingly, in the instances where a gene encoding a component of mitochondrial primary metabolism showed a declining expression profile, it was often accompanied by a corresponding increase in the transcript abundance of a complementing isoform. Isoform switching has been observed in other developmental processes, such as seed germination (Law et al., 2012), leaf development (Wang et al., 2015), and flowering (Wang et al., 2014); however, the observations made here offer a unique insight into the carefully coordinated stage transitions underlying mitochondrial activity in the final developmental stages of a plant.
Mitochondria Orchestrate Senescence-Associated Catabolism to Support Nitrogen Remobilization
The energetic contribution of mitochondria during DLS is undoubtedly a crucial process that compensates for the shift in metabolic demand and prolongs cell viability until the optimum reallocation of nutrients has been achieved. However, perhaps the most interesting findings to emerge from our analysis have been the numerous ways in which the mitochondrion is able to integrate catabolic reactions in order to optimize nitrogen reallocation. The assembly of different mitochondria-localized pathways is complex, and undoubtedly the scheme presented here remains fragmentary. That said, the uniform trend of increasing transcript abundances of the majority of the genes involved in these catabolic pathways coupled to the metabolic changes occurring at the leaf level strongly support the idea that, during DLS, mitochondria (1) provide carbon skeletons for the cell, (2) deliver energy in the form of reducing equivalents and ATP, and (3) have a central role in nitrogen metabolism. Specifically, we propose the amino acid Glu to play a crucial role in nitrogen remobilization during senescence, being released in many of the identified mitochondrial catabolic pathways (Fig. 6). The metabolomic analysis of senescing leaves showed that the Glu content in leaves decreases during senescence, while the ratio of Gln to Glu increases (Fig. 5, B–D). Furthermore, the α-KG-to-Glu ratio, which is involved in numerous transamination reactions, also increases during the process. These results are in agreement with the study from Watanabe et al. (2013), who reported metabolic changes in different parts of leaves undergoing senescence from plants grown under long-day conditions.
Besides Asn and Glu, Gln is the main amino acid present in the phloem sap and represents one of the main molecules for senescence-associated nitrogen export processes in Arabidopsis (Masclaux-Daubresse et al., 2010) or in poplar (Couturier et al., 2010). In addition, it has been shown in various species (Arabidopsis [Li et al., 2006; Keech et al., 2010], radish [Raphanus sativus] cotyledons [Kawakami and Watanabe, 1988], barley [Hordeum vulgare; Avila-Ospina et al., 2015], and tobacco [Nicotiana tabacum; Masclaux et al., 2000], to name a few) that Gln can be synthesized from Glu via the GS and that the protein content of the cytosolic GS1 increases strongly during DLS while the amount of chloroplastic GS2 drops quickly. Therefore, we propose that Glu and, to some extent, ammonium released from the catabolic reactions occurring in the mitochondrion during DLS are transported from the matrix to the cytosol and subsequently are converted into Gln via the GS1. The produced Gln can then leave the cell via the action of the amino acid exporters, GLUTAMINE DUMPER1 (GDU1) to GDU7, located at the plasma membrane (Pratelli et al., 2010). Compellingly, the transcript abundance of two of the seven GDU isoforms, AtGDU3 (At5g57685) and AtGDU4 (At2g24762), show a progressive up-regulation, with the highest transcript abundance at the penultimate time point, clearly pointing to a possible role during DLS (Supplemental Table S3). However, how Glu is exported out of the mitochondrion remains unclear. While mitochondrial Glu transporters have been characterized in humans (Fiermonte et al., 2002), no specific mitochondrial Glu transporter has been described yet in plants (Catoni et al., 2003). This is not surprising, as the transport specificities of about half of the MCF members have not yet been characterized (Fig. 4B).
In addition to its key role in providing the carbon skeleton for the fixation of ammonium and the subsequent export of nitrogen out of the cell, Glu plays an important role in supplying the primary energy metabolism via GDH, which can catalyze the oxidative deamination of Glu to α-KG. This reaction being reversible, some studies have discussed the physiological contexts under which the bidirectional function of this enzyme may play a role (Aubert et al., 2001; Dubois et al., 2003). Although at present no clear answer has emerged from this debate, we here propose that mitochondrial GDH can contribute to nitrogen metabolism during DLS either by producing Glu or by replenishing the pool of α-KG concomitantly to release reducing equivalents (NADH). This latter option would support the transaminations coupled to amino acid catabolism (Fig. 6).
The degradation of Pro (Fig. 6, pathway VI) also appears as a central pathway releasing Glu and reducing equivalents; therefore, it is advantageous in highly energy-demanding developmental processes such as leaf senescence (Faes et al., 2015; Zhang and Becker, 2015). Here, we observed that the increase of transcript abundance of the two Arabidopsis Columbia-0 Pro dehydrogenases during senescence is similar. Pro metabolism is coupled to the segment of the urea cycle (Fig. 6, pathway V) that localizes to the mitochondria and connects to the highly up-regulated mitochondrial transporter called BAC2. BAC2 transports Arg and Orn across the inner mitochondrial membrane, and it can be induced by stress or dark-induced senescence. Furthermore, ethyl methanesulfonate and T-DNA knockout mutants of BAC2 have been demonstrated to accumulate Pro under hyperosmotic stress conditions (Palmieri et al., 2006; Toka et al., 2010; Planchais et al., 2014), while overexpression of BAC2 led to higher Arg catabolism and accumulation of urea (Planchais et al., 2014). Several mitochondrial pathways lead to the degradation of imported Arg, which, in turn, can either be converted to citrulline or enter the Pro catabolic pathway. The conversion of Arg to citrulline potentially leads to the production of nitric oxide; a contentious issue, as it remains unclear whether a functional nitric oxide synthase (so far coined NOS1 [At3g47450]) exists in higher plants (Hancock, 2012, and refs. therein; Du et al., 2016; Fig. 6). The second pathway degrades Arg to Orn, which can then enter the Pro catabolic pathway. Here, since the genes involved in Pro degradation (OAT, ProDH, and P5CDH) are all up-regulated during DLS, while the putative NOS1 gene is clearly down-regulated, it is reasonable to propose that the imported Arg preferentially fuels Pro catabolism, which, in turn, produces Glu at various steps. This conversion from Arg to Pro via Orn also leads to the production of urea, a molecule that was recently reported to be exported from senescing leaves as a source of nitrogen (Bohner et al., 2015). These results, in line with the recently proposed fate of Pro during petal and leaf senescence (Zhang and Becker, 2015), are further supported by our metabolic data (Fig. 5) indicating a decrease in Pro levels during DLS.
We considered two additional catabolic pathways that can modify mitochondrial metabolism during DLS, the degradation of BCAA and Lys. It has been shown that, under starvation conditions, BCAA degradation can become an alternative source of energy (Araujo et al., 2010; Peng et al., 2015), notably due to the activity of IVDH. This enzyme oxidizes isovaleryl-CoA to methylcrotonyl-CoA and generates reducing equivalents (Daschner et al., 2001), which, in turn, can support the mETC through the ETF/ETFQO complex. Interestingly, through isotope tracer experiments, the role of IVDH also has been suggested to be coupled to Lys catabolism (Araujo et al., 2010), even though this still remains an unresolved matter (Engqvist et al., 2014). In general, Lys can be degraded via a number of different pathways. It originates from protein degradation, is converted to d-2HG (Engqvist et al., 2009; Araujo et al., 2010) and then into α-KG that is fed into the tricarboxylic acid cycle. It also can be degraded via the LKR/SaccDH pathway (Tang et al., 2002). While d-2HGDH is localized to the mitochondrion and releases electrons to the ETF/ETFQO complex, the localization of the Lys degradation pathway via LKR/SaccDH is not yet fully elucidated; some reactions clearly take place in mitochondria, while others might be localized in the cytosol. Regardless, the outcome of this pathway is the production of acetyl-CoA and two molecules of Glu. Even though the respective flux through these different pathways during DLS is particularly hard to estimate, it is obvious that they all participate in the release of Glu, the delivery of substrates to the tricarboxylic acid cycle, or feed the mETC with electrons, which once again supports our metabolic model (Fig. 6).
Surprisingly, we observed that, with Gly as a substrate, mitochondria isolated from leaves in the middle of the senescence process had a transient increase in oxygen consumption (Fig. 2D). In contrast, the transcript levels of both GDC and SHMT were progressively down-regulated (Fig. 6, boxed area), and both pools of Gly and Ser decreased concomitantly in senescing leaves (Fig. 5B). This seems to indicate that, despite the down-regulation of its transcripts, the GDC complex remains active in leaves and offers the possibility to decarboxylate Gly. Indeed, even though GDC is involved in photorespiration, which undoubtedly drops during leaf senescence, the complex also plays a role in C1 metabolism and was shown to be essential for plants (Engel et al., 2007). In addition, examination of the reactions coupled to Gly metabolism in mitochondria identified several aminotransferases suspected to be involved in Gly degradation. Among them, the Ala/glyoxylate aminotransferase and AlaAT were both up-regulated during leaf senescence, the latter leading to Glu production, while a mitochondrion-targeted Asp aminotransferase was clearly down-regulated (Fig. 6). Therefore, we propose a model where Gly conversion in senescing leaves would progressively switch from decarboxylation to transamination. Such transaminations would result, in addition to Glu, in the production of glyoxylate. Glyoxylate could be further exported toward the peroxisomes/glyoxysomes to integrate the glyoxylate cycle, known to be up-regulated during leaf senescence (Debellis and Nishimura, 1991; Pistelli et al., 1992; Pracharoenwattana and Smith, 2008). Ultimately, this would lead to the production of additional organic acids, which, in turn, could feed the tricarboxylic acid cycle. Consistent with this scheme, both malate and citrate were found to accumulate in senescing leaves (Fig. 5B). Another piece of evidence supporting that such a mechanism would contribute to nitrogen remobilization comes from the fact that introducing the barley ortholog AlaAT cDNA into rice (Oryza sativa) leads to a better nitrogen use efficiency in plants, with increased biomass and total nitrogen content (Shrawat et al., 2008). This suggests that the increase in AlaAT transcript levels could indeed be beneficial to senescing plants, to better remobilize nitrogen and efficiently reallocate it to developing organs.
Is the Maturation of Fe-S Proteins Still Active in Senescent Leaves?
Fe-S proteins ensure many vital functions, participating for instance in electron transfer reactions or being required for enzymatic catalysis. They are first synthesized as apoproteins, and the prosthetic groups are inserted into the polypeptide through the action of dedicated assembly machineries, namely SUF, ISC, and cytosolic ISC assembly, devoted to the maturation of plastidial, mitochondrial, and nuclear or cytosolic proteins, respectively (Couturier et al., 2013; Balk and Schaedler, 2014). Considering that mitochondria are central to the maturation of both mitochondrial and extramitochondrial Fe-S proteins, and that this is certainly one of the primary and ancestral functions of mitochondria, we hypothesized that ISC assembly should remain functional as long as possible. We provide below tentative explanations for the constant or variable expression of certain genes in light of known metabolic or physiological changes occurring during senescence.
Besides genes coding for accessory factors (Mge1, HscA, and HscB), a noticeable decrease in expression was observed only for the IndH/1 gene among the core ISC components. The IndH/1 gene is a late-acting factor that is specifically required for complex I assembly (Wydro et al., 2013). This decrease in expression might be consistent with the specific down-regulation observed for genes coding for Fe-S subunits present in complex I. Indeed, the expression of genes coding for Fe-S subunits of complexes II and III remains either unchanged or is increased, supported by the significant enrichment of 2Fe-2S cluster assembly factors in cluster 6 (increasing trend of transcript abundance over the time course) in Figure 3. Overall, this may point to a concerted regulation for favoring complex II and decreasing reliance of complex I, which may become bypassed during senescence in order to avoid ROS production (see above).
Cnx2 is a protein involved in the synthesis of the molybdenum cofactor, which occurs partly in mitochondria. In plants, these cofactors are found in extramitochondrial proteins, such as in cytosolic xanthine dehydrogenases, nitrate reductases, and aldehyde oxidases or in peroxisomal sulfite oxidases (Mendel and Leimkuhler, 2015). The fact that this gene is strongly up-regulated during the senescence process could point to the importance of one or several of these enzymes. In fact, this may fit with the requirement of xanthine dehydrogenase and nitrate reductase for nitrogen remobilization, which is a crucial metabolic adaptation occurring during senescence. In fact, the maintenance of a high expression of ISC genes confirms the view that mitochondrial and/or extramitochondrial Fe-S or molybdenum cofactor-containing proteins are likely required for crucial functions associated with senescence. By analogy to nonplant systems, where it was demonstrated that ISCs are present in enzymes involved in ribosome biogenesis or in DNA repair and replication (Paul and Lill, 2015), we can speculate that this maturation pathway has to be maintained as long as possible in all organs, including senescing ones. It is interesting that a similar analysis performed with SUF components or chloroplastic Fe-S proteins showed a rapid decline in transcript levels for most genes. This is in accordance with the fact that the chloroplast metabolism declines rapidly during DLS and that the SUF machinery is not required for nonchloroplastic Fe-S proteins (Van Hoewyk et al., 2007).
The Conserved and Newly Acquired Functions of Mitochondria during DLS
The evolutionary origin of plant senescence can be traced back to photosynthetic prokaryotes, and over time, it has been augmented with successive rounds of evolutionary innovation. Consequently, senescence can be viewed as an assembly of varied but complementary processes that have evolved gradually in response to diverse selective pressures (Thomas et al., 2009). To examine the influence of the evolutionary age of a gene on its expression during leaf senescence, phylostratigraphic analysis was carried out on the mitochondrial set during the time course established by Breeze et al. (2011). Previous studies have examined the evolutionary age of genes expressed during major developmental processes, such as seed embryogenesis (Quint et al., 2012) and seed germination (Dekkers et al., 2013). In both processes, a transcriptomic hourglass was identified, characterized by the expression of evolutionarily young genes at the beginning and end of these processes, with a transient period of repression of these young genes and expression of the most evolutionarily ancient genes at the midpoint of the respective time courses (known as a phylotypic stage). In both developmental programs, the transient dominance of evolutionarily ancient genes is thought to signify a pivotal period where cellular differentiation and spatiotemporal organization processes are coordinated (Quint et al., 2012; Dekkers et al., 2013). A recent review applied a similar methodology to examine the evolutionary age of mitochondrial genes during seed germination and revealed the opposite pattern of evolutionary age/expression, with the oldest mitochondrial genes expressed at the beginning and end of germination and the youngest expressed transiently during the middle of the time course (Law et al., 2014). This deviation from observations made for the total present set could be attributed to the presence of a large group of evolutionarily young PPR proteins that were expressed transiently during seed germination. In contrast to these observations, during DLS no such evolutionary hourglass was observed, with the mean TAI increasing steadily as the time course progressed. The observation that the most evolutionarily ancient genes are expressed predominantly at the onset of DLS while the expression of evolutionarily young genes is dominant at the final stages of DLS is interesting, although perhaps not surprising. A previous investigation has shown that the earliest senescence processes to be acquired were linked to catabolism of the light energy transduction machinery, while more recent evolutionary additions have been linked to the development of mechanisms for the perception and integration of signals regulating senescence initiation and execution. Closer examination of genes belonging to the mitochondrial set that were expressed maximally at day 37 (when transcripts displaying the youngest TAI were observed) showed a significant underrepresentation of protein metabolism, which had an average phylostratigraphic ranking of 1.69, which is on the evolutionarily ancient end of the spectrum (Fig. 7). In contrast, this set was significantly overrepresented with unknown functions (average TAI = 3.26), primary metabolism (average TAI = 2.65), amino acid degradation (average TAI = 1.00), and auxin metabolism (average TAI = 5.08). With the exception of amino acid degradation, which is relatively ancient in evolutionary terms, these results indicate that the execution of DLS has changed over evolutionary time as a consequence of mitochondria acquiring novel functions, which have fortified their increasingly central roles as hubs for signaling and energy production in the final stages of DLS.
To conclude, for several decades, the commercial impact of senescence to the agricultural industry has driven the exhaustive research into fruit, flower, and leaf senescence in plants (Schippers et al., 2015). However, little attention had been paid to the enigmatic role of mitochondria during this vital developmental process. In this study, we provide, to our knowledge, the first comprehensive analysis describing mitochondrial metabolic functions during DLS. Most interestingly, our work shows that, during DLS, in addition to becoming the primary site of ATP production, mitochondria also orchestrate a number of catabolic processes and, in doing so, assume an increasingly central energy and metabolic role in the cellular landscape. Particularly, through these reactions, mitochondria provide the carbon backbone essential for nitrogen remobilization, a process of utmost importance not only to ensure the quality of grain in crop species but also representing a valuable target for minimizing the negative ecological impact of fertilizer in current farming practices.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 plants were grown in a controlled environment growth chamber with a short-day photoperiod (8/16 h of light/dark, 22°C/17°C) at 75% relative humidity and 200 μmol m–2 s–1 white light at growth level. The short-day growth period served to increase both the number and size of the leaves, thus facilitating physiological analyses.
Chlorophyll Measurements
Leaves of rosettes 8 to 10 of Arabidopsis plants were followed throughout the process of senescence. The chlorophyll content of these leaves was measured with a CCM-200 Chlorophyll Content Meter from Opti-Sciences by clamping each leaf tip to the CCM-200 detector and measuring an area of 0.71 mm2. To avoid the main vain, the tips of the leaves were chosen for this measurement.
Breeze et al. (2011) published a time course of chlorophyll degradation during DLS under a long-day photoperiod. Based on our chlorophyll determination data for a short-day photoperiod, we could easily match four time points (T0, T1, T2, and T3) from our reference set (Fig. 2A) to the Breeze et al. (2011) reference set (Fig. 2B). Furthermore, in order to match our data for microscopy, adenylate pool determination, and the metabolomics of developmentally senescing leaves, we added a fifth time point (T4) to the Breeze et al. (2011) reference set, representing the latest stages of leaf senescence (i.e. less than 20% chlorophyll content in leaves).
Microscopy
Confocal Laser Microscopy
Images were acquired using a Zeiss LSM 780 confocal microscope with a C-Apochromat 40×/1.20 W Korr M27 objective. The pixel dwell was 0.64 µs, which corresponds to 671 ms per image. Only the 488-nm laser was used at 0.9%, and collection windows were as follows: ChS1, 499 to 542 for GFP; and Ch2, 650 to 700 for chlorophyll fluorescence.
Electron Microscopy
Leaves were placed in a petri dish filled with water, and leaf sections of approximately 1 mm2 were prepared. Squares of 1 mm2 were always cut approximately 2 to 3 mm to the right and left from the mid vain and at a distance from the leaf tip of 0.5 cm. Three biological replicates per leaf senescence stage were fixed overnight in 2.5% (w/v) glutaraldehyde/0.1 m cacodylate buffer, pH 7.2 (primary fixation). The samples were washed twice with cacodylate buffer and transferred to a 1% (w/v) osmium tetroxide solution for 2 h in darkness (secondary fixation). After washing twice for 15 min in 0.1 m cacodylate buffer, the samples were dehydrated in a graded ethanol series and sequentially transferred to a mix of anhydrous ethanol and Spurr embedding resin (TAAB Laboratories). The next day, the samples were transferred overnight to pure Spurr embedding resin, embedded in beem capsules, and incubated at 65°C for 24 h. After embedding, ultrathin sections (80 nm) were cut for transmission electron microscopy analysis. The sections were stained with 5% aqueous uranyl acetate for 60 min and then with lead citrate (Reynolds, 1963) for 6 min before being examined with the electron microscope (JEM 1230; JEOL). Images were taken using a Gatan MSC 600CW 2kX2k CCD camera.
Isolation of Mitochondria and Respiratory Measurements
Leaf mitochondria were isolated according to Keech et al. (2005), and respiratory measurements were performed as indicated therein. Several leaves per time point were sampled for the isolation of organelles, while a subset of this pool of leaves was frozen for chlorophyll determination. Three to five mitochondrial isolations per time point were performed. From each mitochondrial extraction, several respiratory measurements were conducted with malate, Gly, and NADH as substrates.
Quantification of Adenylate Pools
Total ATP and ADP pools of senescing leaves were determined with the ATP Kit SL purchased from BioThema Luminescent Assays. During DLS, the assessment of the chlorophyll content in leaves was performed with a CCM-200 Chlorophyll Content Meter from Opti-Sciences as described above. For all time points, at least four leaves per plant were harvested and at least three plants (i.e. biological replicates) were sampled. Samples were frozen in liquid nitrogen and further ground with a mortar and pestle. Finally, 25 mg of frozen ground plant material per sample was used to enzymatically determine the ATP and ADP content according to the manufacturer’s protocol. Two to three technical replicates were measured per biological replicate.
List of Genes Encoding for Mitochondria-Targeted Products
A comprehensive list of genes encoding proteins targeted to the mitochondrion was assembled, utilizing a combination of outputs from the subcellular localization database for Arabidopsis proteins (SUBA3; Tanz et al., 2013) and manual curation. The list was constructed as follows. First, SUBA3 was queried to isolate genes that were identified as encoding proteins targeted to the mitochondrion, as demonstrated using experimental and computational prediction, based on protein sequence. Next, to identify false negatives that did not meet these criteria, genes encoding proteins that were identified experimentally (GFP or mass spectrometry [MS]) as targeting to a nonmitochondrial location with only one example, but called mitochondrial by the Subcellular Arabidopsis consensus (SUBAcon) algorithm, were assessed manually and either included or dismissed. SUBAcon is a robust bioinformatic tool that integrates 22 computational prediction algorithms, experimental GFP and MS, coexpression data, and protein-protein interaction data (Hooper et al., 2014). To exclude false positives, genes encoding proteins that were identified as targeting to mitochondria experimentally (GFP or MS) with only one example, but having additional experimental targeting to other organelles and classed as nonmitochondrial by SUBAcon, were assessed manually and either included or dismissed. In this way, a list of 1,060 genes encoding proteins targeted to the mitochondrion was prepared.
Microarray Normalization, Differential Expression, and Hierarchical Clustering Analysis
The microarray experimental data detailing leaf senescence in Arabidopsis described by Breeze et al. (2011) was obtained from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). This experiment utilized a CATMA Arabidopsis 30K spotted cDNA array platform, and the data can be requisitioned under series GSE22982. These data were processed using quantile normalization in parallel with the array data set of van der Graaff et al. (2006; data obtained from the authors), which also detailed a leaf senescence time course, in the Bioconductor software project. This was carried out in parallel in order to facilitate downstream comparison and validation of these two leaf senescence data sets. For the Breeze et al. (2011) data set, intensity values of the four replicates for each time point were used as the input for differential expression analysis, which was carried out using the Cyber-T Web server (Kayala and Baldi, 2012). It utilizes a Bayesian method for the identification of significant changes in the transcript abundance of probe sets. All input criteria were set according to Cyber-T recommendations applicable for each experimental set. Step-wise differential expression analysis was carried out on consecutive time points, and a probe set was defined as significantly changing at P < 0.01 (false discovery rate by PPDE threshold was not applied). To visualize the expression profiles of our total set, the four replicates associated with each time point were averaged and the intensity value of each probe set was made relative to its maximum intensity. The data were then imported into The Institute for Genomic Research Multiexperiment Viewer and hierarchically clustered using average linkage based on Euclidean distance to generate the set of clusters in Figure 2.
GO
Assignment of GO subcategory annotations to the clusters was carried out using the Web-based tool g:Profiler (Reimand et al., 2007). Each cluster was analyzed in reference to the total mitochondrial gene list, filtering for significant calls only using the Best per Parent hierarchical filtering option.
Metabolomics
Sampling
Leaf blades from stages T0, T1-T2, T2-T3, and T3-T4, as determined using a CCM-200 Chlorophyll Content Meter, were cut from the plant, placed into a 2-mL Eppendorf tube, and flash frozen in liquid nitrogen. Samples were milled using two beads per tube (2.5 mm Ø) in two steps of 30 s each, at maximum speed, separated by a cooling step in liquid nitrogen. Samples were weighed (AT20; Mettler) into 1.5-mL tubes using precooled equipment.
Metabolite Extraction
Frozen and ground leaf material (19–21 mg) was extracted according to Gullberg et al. (2004). In brief, 11 stable isotope reference compounds (7 ng μL−1 l-Pro-13C5, succinic acid-d4, salicylic acid-d6, α-KG-13C4, Glu-13C5, putrescine-d4, myristic acid-13C3, d-Glc-13C6, hexadecanoic acid-13C4, Suc13C12, and cholesterol-d7) were added to a chloroform:methanol:water (20:60:20, v/v/v) extraction mixture. One milliliter of the spiked mixture was added to each sample in 1.5-mL tubes on ice. After adding a 3-mm tungsten carbide bead (Retsch) to each tube, they were shaken at 30 Hz for 3 min in an MM 301 Vibration Mill (Retsch). The beads were removed before centrifugation for 10 min at 14,000 rpm in a Mikro 220R centrifuge (Hettich). The supernatant from each tube (200 μL) was transferred to a 250-μL micro vial (Chromatol) and evaporated to dryness in a miVac quattro concentrator (Barnstead Genevac). Samples were derivatized by adding 30 μL of methoxyamine hydrochloride (15 mg mL−1) in pyridine and shaking for 10 min at 5°C in a VX-2500 Multitube Vortexer (VWR Scientific), followed by 16 h of incubation at room temperature, then adding 30 µL of MSTFA in 1% trimethyl chlorosilane for silylation, vortex mixing, then 1 h of incubation at room temperature. Heptane (30 μL, including 15 ng μL−1 methyl stearate) was added, and the samples were mixed by vortexing.
Gas Chromatography-Time of Flight-Mass Spectrometry Analysis
The gas chromatography-mass spectrometry analysis followed the gas chromatography-time of flight-mass spectrometry procedure published by Gullberg et al. (2004). Quality-control samples and an n-alkane series (C8 C40) were included in each analysis (Schauer et al., 2005). The derivatized samples (1 μL) were injected into a split/splitless injector in splitless mode by a CTC PAL systems autosampler (with a 10-μL syringe) into an Agilent Technologies 7890A gas chromatography system equipped with a 30-m- × 0.25-mm-diameter fused silica capillary column with a bonded 0.25-μm Durabond DB-5MSUI stationary phase (part no. 122-5222UI; Agilent J&W gas chromatography columns). The settings were as follows: injector temperature, 260°C; front inlet septum purge flow rate, 3 mL min−1; gas flow rate, 1 mL min−1; column temperature, 70°C for 2 min, then increased by 20°C min−1 to 320°C, and held for 8 min. The column effluent was introduced into the electron-impact ion source of a Pegasus HT gas chromatograph-high throughput time of flight-mass spectrometer (LECO), with the following settings: transfer line temperature, 270°C; ion source temperature, 200°C; detector voltage, 1,520 V; electron-impact electron beam, −70 V; ionization current, 2 mA. Twenty spectra were recorded per second with a 50 to 800 mass-to-charge ratio range and a 290-s solvent delay.
All nonprocessed MS files from the metabolic analysis were exported from the ChromaTOF software, in NetCDF format, to MATLAB software 7.0 (Mathworks), in which all data pretreatment procedures, such as baseline correction chromatogram alignment, data compression, and hierarchical multivariate curve resolution, were performed using custom scripts according to Jonsson et al. (2005). All manual integrations were performed using ChromaTOF 2.32 software (LECO) or custom scripts.
Analysis of the Evolutionary Age of Genes Expressed during Senescence
The phylostratigraphic ranking established by Quint et al. (2012) was matched with the mitochondrial set, assigning each gene an evolutionary age ranking from PS1 for the oldest genes, showing the strongest homology to prokaryotic sequences, to PS13 for the evolutionarily youngest genes, with no homologs in any other species. In order to examine the influence of gene expression on the evolutionary age of a gene, all genes i at a specific time point s were transformed to provide a weighted mean of the phylostratigraphic ranking psi, weighted by the expression level eis, leading to the TAI, where n is the total number of genes analyzed.
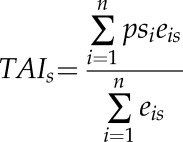 |
Similar to the phylostratigraphic ranking for individual genes, a low TAI indicates an evolutionarily ancient transcriptome and high TAI values correspond to evolutionarily young transcriptomes.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chloroplast and mitochondria occurrence during leaf senescence.
Supplemental Figure S2. High-resolution electron microscopy images.
Supplemental Figure S3. Mitochondrial import components and their transcriptomic regulations during DLS.
Supplemental Figure S4. Detailed transcriptomic regulation of BCAA catabolic pathways during DLS.
Supplemental Figure S5. Confirmation of observations utilizing an additional experimental survey of DLS (van der Graaff et al., 2006).
Supplemental Figure S6. qRT-PCR analysis of SAGs and a subset of relevant genes involved in mitochondrial metabolism during DLS.
Supplemental Table S1. Normalized Breeze et al. (2011) microarray data set.
Supplemental Table S2. Manually curated mitochondrial list (version 1.0).
Supplemental Table S3. List of genes employed for Figures 4 and 6 and the ISC machinery.
Supplemental Table S4. List of genes encoding mitochondrial import components.
Supplemental Table S5. Correlation analysis between the Breeze et al. (2011) and van der Graaff et al. (2006) microarray data sets.
Supplemental Table S6. List of primers used for qRT-PCR experiments.
Supplementary Material
Acknowledgments
We thank Dr. Nicolas Delhomme and the Umeå Plant Science Centre bioinformatics facility (https://bioinfomatics.upsc.se) for data analysis support and Vicky Buchanan-Wollaston for critical comments on the differential expression levels.
Glossary
- PCD
programmed cell death
- DLS
developmental leaf senescence
- RCR
respiratory control ratio
- mETC
mitochondrial electron transport chain
- GO
Gene Ontology
- ROS
reactive oxygen species
- SUF
sulfur mobilization
- ISC
iron-sulfur cluster
- MCF
mitochondrial carrier family
- VDAC
voltage-dependent anion channel
- PCA
principal component analysis
- BCAA
branched-chain amino acids
- TAI
Transcriptome Age Index
- qRT
quantitative reverse transcription
- MS
mass spectrometry
Footnotes
This work was supported by the Swedish Research Council (grant no. 621-2014-4688 to O.K.), the Kempe Foundations (Gunnar Öquist Fellowship to O.K.), and the Carl Tryggers Stiftelse; Unité Mixte de Recherche 1136 is supported by a grant overseen by the French National Research Agency as part of the Investissements d’Avenir program (grant no. ANR-11-LABX-0002-01).
References
- Araujo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Douce R, Gout E, Ratcliffe RG, Roberts JKM (2001) Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by C-13 and P-31 nuclear magnetic resonance. J Exp Bot 52: 37–45 [PubMed] [Google Scholar]
- Avila-Ospina L, Marmagne A, Talbotec J, Krupinska K, Masclaux-Daubresse C (2015) The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J Exp Bot 66: 2013–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Barreto P, Okura VK, Neshich IA, Maia Ide G, Arruda P (2014) Overexpression of UCP1 in tobacco induces mitochondrial biogenesis and amplifies a broad stress response. BMC Plant Biol 14: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besagni C, Kessler F (2013) A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 237: 463–470 [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Bjorkbacka H, Birve SJ, Karlsson J, Gardestrom P, Gustafsson P, Lundeberg J, et al. (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi/10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohner A, Kojima S, Hajirezaei M, Melzer M, von Wiren N (2015) Urea retranslocation from senescing Arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J 81: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Carlsson J, Leino M, Sohlberg J, Sundstrom JF, Glimelius K (2008) Mitochondrial regulation of flower development. Mitochondrion 8: 74–86 [DOI] [PubMed] [Google Scholar]
- Catoni E, Desimone M, Hilpert M, Wipf D, Kunze R, Schneider A, Flugge UI, Schumacher K, Frommer WB (2003) Expression pattern of a nuclear encoded mitochondrial arginine-ornithine translocator gene from Arabidopsis. BMC Plant Biol 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Collier DE, Thibodeau BA (1995) Changes in respiration and chemical content during autumnal senescence of Populus tremuloides and Quercus rubra leaves. Tree Physiol 15: 759–764 [DOI] [PubMed] [Google Scholar]
- Couturier J, Doidy J, Guinet F, Wipf D, Blaudez D, Chalot M (2010) Glutamine, arginine and the amino acid transporter Pt-CAT11 play important roles during senescence in poplar. Ann Bot (Lond) 105: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Touraine B, Briat JF, Gaymard F, Rouhier N (2013) The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front Plant Sci 4: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daschner K, Couee I, Binder S (2001) The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellis L, Nishimura M (1991) Development of enzymes of the glyoxylate cycle during senescence of pumpkin cotyledons. Plant Cell Physiol 32: 555–561 [Google Scholar]
- Dekker PJ, Martin F, Maarse AC, Bomer U, Muller H, Guiard B, Meijer M, Rassow J, Pfanner N (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J 16: 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Muller K, King JR, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Du S, Zhang R, Zhang P, Liu H, Yan M, Chen N, Xie H, Ke S (2016) Elevated CO2-induced production of nitric oxide (NO) by NO synthase differentially affects nitrate reductase activity in Arabidopsis plants under different nitrate supplies. J Exp Bot 67: 893–904 [DOI] [PubMed] [Google Scholar]
- Dubois F, Tercé-Laforgue T, Gonzalez-Moro MB, Estavillo MB, Sangwan R, Gallais A, Hirel B (2003) Glutamate dehydrogenase in plants: is there a new story for an old enzyme? Plant Physiol Biochem 41: 565–576 [Google Scholar]
- Eisenhut M, Planchais S, Cabassa C, Guivarc’h A, Justin AM, Taconnat L, Renou JP, Linka M, Gagneul D, Timm S, et al. (2013) Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J 73: 836–849 [DOI] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Parnik T, Keerberg O, Bauwe H (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M, Drincovich MF, Flugge UI, Maurino VG (2009) Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J Biol Chem 284: 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Esser C, Maier A, Lercher MJ, Maurino VG (2014) Mitochondrial 2-hydroxyglutarate metabolism. Mitochondrion 19 Pt B: 275–281 [DOI] [PubMed] [Google Scholar]
- Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EE, Jakobs C, Weber AP, Maurino VG (2011) Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J Biol Chem 286: 11382–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faes P, Deleu C, Ainouche A, Le Caherec F, Montes E, Clouet V, Gouraud AM, Albert B, Orsel M, Lassalle G, et al. (2015) Molecular evolution and transcriptional regulation of the oilseed rape proline dehydrogenase genes suggest distinct roles of proline catabolism during development. Planta 241: 403–419 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 277: 19289–19294 [DOI] [PubMed] [Google Scholar]
- Gardeström P, Igamberdiev AU (2016) The origin of cytosolic ATP in photosynthetic cells. Physiol Plant 157: 367–379 [DOI] [PubMed] [Google Scholar]
- Godbole A, Dubey AK, Reddy PS, Udayakumar M, Mathew MK (2013) Mitochondrial VDAC and hexokinase together modulate plant programmed cell death. Protoplasma 250: 875–884 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Sormani R, Meyer C, Masclaux-Daubresse C (2010) Senescence and death of plant organs: nutrient recycling and developmental regulation. C R Biol 333: 382–391 [DOI] [PubMed] [Google Scholar]
- Gullberg J, Jonsson P, Nordstrom A, Sjostrom M, Moritz T (2004) Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal Biochem 331: 283–295 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Schmitz-Esser S (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT. (2012) NO synthase? Generation of nitric oxide in plants. Period Biol 114: 19–24 [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH (2014) SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30: 3356–3364 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee JH, Zhang B, Van Aken O, Radomiljac JD, van der Merwe M, Yi K, et al. (2014) A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates ALTERNATIVE OXIDASE1a expression in Arabidopsis. Plant Physiol 165: 1233–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P, Johansson AI, Gullberg J, Trygg JAJ, Grung B, Marklund S, Sjostrom M, Antti H, Moritz T (2005) High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem 77: 5635–5642 [DOI] [PubMed] [Google Scholar]
- Kawakami N, Watanabe A (1988) Senescence-specific increase in cytosolic glutamine synthetase and its mRNA in radish cotyledons. Plant Physiol 88: 1430–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayala MA, Baldi P (2012) Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res 40: W553–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O. (2011) The conserved mobility of mitochondria during leaf senescence reflects differential regulation of the cytoskeletal components in Arabidopsis thaliana. Plant Signal Behav 6: 147–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardestrom P (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardestrom P (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Gutierrez L, Ahad A, Bellini C, Smith SM, Gardestrom P (2010) Leaf senescence is accompanied by an early disruption of the microtubule network in Arabidopsis. Plant Physiol 154: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskitalo J, Bergquist G, Gardestrom P, Jansson S (2005) A cellular timetable of autumn senescence. Plant Physiol 139: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Narsai R, Whelan J (2014) Mitochondrial biogenesis in plants during seed germination. Mitochondrion 19 Pt B: 214–221 [DOI] [PubMed] [Google Scholar]
- Li RJ, Hua W, Lu YT (2006) Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem Biophys Res Commun 342: 119–126 [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugiere N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211: 510–518 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel RR, Leimkuhler S (2015) The biosynthesis of the molybdenum cofactors. J Biol Inorg Chem 20: 337–347 [DOI] [PubMed] [Google Scholar]
- Monne M, Miniero DV, Obata T, Daddabbo L, Palmieri L, Vozza A, Nicolardi MC, Fernie AR, Palmieri F (2015) Functional characterization and organ distribution of three mitochondrial ATP-Mg/Pi carriers in Arabidopsis thaliana. Biochim Biophys Acta 1847: 1220–1230 [DOI] [PubMed] [Google Scholar]
- Nogueira FT, Sassaki FT, Maia IG (2011) Arabidopsis thaliana uncoupling proteins (AtUCPs): insights into gene expression during development and stress response and epigenetic regulation. J Bioenerg Biomembr 43: 71–79 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, Fernie AR (2011) Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J 66: 161–181 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Todd CD, Arrigoni R, Hoyos ME, Santoro A, Polacco JC, Palmieri F (2006) Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim Biophys Acta 1757: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Pan X, Chen Z, Yang X, Liu G (2014) Arabidopsis voltage-dependent anion channel 1 (AtVDAC1) is required for female development and maintenance of mitochondrial functions related to energy-transaction. PLoS ONE 9: e106941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul VD, Lill R (2015) Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim Biophys Acta 1853: 1528–1539 [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL (2015) The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiol 169: 1807–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistelli L, Perata P, Alpi A (1992) Effect of leaf senescence on glyoxylate cycle enzyme-activities. Aust J Plant Physiol 19: 723–729 [Google Scholar]
- Planchais S, Cabassa C, Toka I, Justin AM, Renou JP, Savoure A, Carol P (2014) BASIC AMINO ACID CARRIER 2 gene expression modulates arginine and urea content and stress recovery in Arabidopsis leaves. Front Plant Sci 5: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Smith SM (2008) When is a peroxisome not a peroxisome? Trends Plant Sci 13: 522–525 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G (2010) Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol 152: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Drost HG, Gabel A, Ullrich KK, Bonn M, Grosse I (2012) A transcriptomic hourglass in plant embryogenesis. Nature 490: 98–101 [DOI] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J (2007) g:Profiler: a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35: W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, d’Erfurth I, Marmagne A, Erhardt M, Allot M, Boivin K, Gissot L, Monachello D, Michaud M, Duchene AM, et al. (2012) Voltage-dependent-anion-channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Mol Biol 78: 431–446 [DOI] [PubMed] [Google Scholar]
- Ruberti C, Barizza E, Bodner M, La Rocca N, De Michele R, Carimi F, Lo Schiavo F, Zottini M (2014) Mitochondria change dynamics and morphology during grapevine leaf senescence. PLoS ONE 9: e102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schippers JH, Schmidt R, Wagstaff C, Jing HC (2015) Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol 169: 914–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG (2008) Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol J 6: 722–732 [DOI] [PubMed] [Google Scholar]
- Smart CM. (1994) Gene-expression during leaf senescence. New Phytol 126: 419–448 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR (2006) Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci USA 103: 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GL, Zhu XH, Gakiere B, Levanony H, Kahana A, Galili G (2002) The bifunctional LKR/SDH locus of plants also encodes a highly active monofunctional lysine-ketoglutarate reductase using a polyadenylation signal located within an intron. Plant Physiol 130: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the subcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Huang L, Young M, Ougham H (2009) Evolution of plant senescence. BMC Evol Biol 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toka I, Planchais S, Cabassa C, Justin AM, De Vos D, Richard L, Savoure A, Carol P (2010) Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol 152: 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Abdel-Ghany SE, Cohu CM, Herbert SK, Kugrens P, Pilon M, Pilon-Smits EA (2007) Chloroplast iron-sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc Natl Acad Sci USA 104: 5686–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianello A, Zancani M, Peresson C, Petrussa E, Casolo V, Krajnakova J, Patui S, Braidot E, Macri F (2007) Plant mitochondrial pathway leading to programmed cell death. Physiol Plant 129: 242–252 [Google Scholar]
- Wang T, Li R, Wen L, Fu D, Zhu B, Luo Y, Zhu H (2015) Functional analysis and RNA sequencing indicate the regulatory role of Argonaute1 in tomato compound leaf development. PLoS ONE 10: e0140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Law SR, Ivanova A, van Aken O, Kubiszewski-Jakubiak S, Uggalla V, van der Merwe M, Duncan O, Narsai R, Whelan J, et al. (2014) The mitochondrial protein import component, TRANSLOCASE OF THE INNER MEMBRANE17-1, plays a role in defining the timing of germination in Arabidopsis. Plant Physiol 166: 1420–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Koo HJ, Kim J, Jeong H, Yang JO, Lee IH, Jun JH, Choi SH, Park SJ, Kang B, et al. (2016) Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol 171: 452–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J (2013) The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis. Plant Cell 25: 4014–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R, Agrimi G, Lerma-Ortiz C, Teresinski HJ, Frelin O, Ellens KW, Castegna A, Russo A, de Crecy-Lagard V, Mullen RT, et al. (2013) Identification of mitochondrial coenzyme A transporters from maize and Arabidopsis. Plant Physiol 162: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Becker DF (2015) Connecting proline metabolism and signaling pathways in plant senescence. Front Plant Sci 6: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Zentgraf U (2005) The correlation between oxidative stress and leaf senescence during plant development. Cell Mol Biol Lett 10: 515–534 [PubMed] [Google Scholar]
- Zottini M, Barizza E, Bastianelli F, Carimi F, Lo Schiavo F (2006) Growth and senescence of Medicago truncatula cultured cells are associated with characteristic mitochondrial morphology. New Phytol 172: 239–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.