The barley MIR1 E3 ubiquitin ligase interacts with MLA immune receptors to promote their ubiquitination and proteasomal degradation, thus regulating receptor stability and immunity.
Abstract
Following the detection of pathogen cognate effectors, plant Nod-like receptors (NLRs) trigger isolate-specific immunity that is generally associated with cell death. The regulation of NLR stability is important to ensure effective immunity. In barley (Hordeum vulgare), the allelic Mildew locus A (MLA) receptors mediate isolate-specific disease resistance against powdery mildew fungus (Blumeria graminis f. sp. hordei). Currently, how MLA stability is controlled remains unknown. Here, we identified an MLA-interacting RING-type E3 ligase, MIR1, that interacts with several MLAs. We showed that the carboxyl-terminal TPR domain of MIR1 mediates the interaction with the coiled-coil domain-containing region of functional MLAs, such as MLA1, MLA6, and MLA10, but not with that of the nonfunctional MLA18-1. MIR1 can ubiquitinate the amino-terminal region of MLAs in vitro and promotes the proteasomal degradation of MLAs in vitro and in planta. Both proteasome inhibitor treatment and virus-induced gene silencing-mediated MIR1 silencing significantly increased MLA abundance in barley transgenic lines. Furthermore, overexpression of MIR1 specifically compromised MLA-mediated disease resistance in barley, while coexpression of MIR1 and MLA10 attenuated MLA10-induced cell death signaling in Nicotiana benthamiana. Together, our data reveal a mechanism for the control of the stability of MLA immune receptors and for the attenuation of MLA-triggered defense signaling by a RING-type E3 ligase via the ubiquitin proteasome system.
Plants rely on two major classes of immune receptors for pathogen recognition and defense activation. The pathogen-associated molecular pattern (PAMP) recognition receptors mediate PAMP-triggered immunity (PTI) following the detection of PAMP molecules (Zipfel, 2014), while plant Nod-like receptors (NLRs; also known as resistance proteins) initiate effector-triggered immunity (ETI) upon the detection of strain-specific pathogen effectors that are delivered into host cells to inhibit host immunity (Dodds and Rathjen, 2010; Dou and Zhou 2012; Cui et al., 2015). Although PTI and ETI are triggered by different types of immune receptors, both types of immunity can converge in various signaling pathways and culminate in similar cellular responses that can lead to cell death (Tsuda and Katagiri, 2010; Cook et al., 2015). Inappropriate cell death triggered by immune receptor overaccumulation or autoactivation is detrimental to plant growth and development (Cheng et al., 2011; Chae et al., 2014; Rodriguez et al., 2016). Therefore, the stability and activity of these immune receptors must be tightly controlled in plants (Shirasu, 2009; Vierstra, 2009; Cheng and Li, 2012; Duplan and Rivas 2014; Li et al., 2015b). Thus far, the mechanisms controlling NLR stability have not been fully elucidated.
The ubiquitin (Ub)-26S proteasome system (UPS) is mechanistically conserved in eukaryotes, and its major function is to control the abundance of key regulatory proteins and enzymes through proteolysis (Smalle and Vierstra, 2004). The UPS targets protein substrates for ubiquitination via an ATP-dependent reaction cascade that typically involves the sequential action of three enzymes: E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme), and E3 (Ub ligase). The polyubiquitinated proteins are finally degraded via the 26S proteasome (Vierstra, 2009). The UPS is involved in many biological processes in plants, including hormone signaling, chromatin structure and transcription, development, and responses to biotic and abiotic stresses (Vierstra, 2009). The UPS plays important roles in the regulation of immune receptors and immune signaling components in plant-pathogen interactions (Trujillo and Shirasu, 2010; Marino et al., 2012; Li et al., 2014). For example, in Arabidopsis (Arabidopsis thaliana), the detection of bacterial flagellin by FLS2 induces the recruitment of two U-box E3 ligases, PUB12 and PUB13, to the FLS2 receptor complex in a BAK1-dependent manner, and PUB12/PUB13 polyubiquitinate FLS2 and promote flagellin-induced FLS2 degradation (Lu et al., 2011). Interestingly, the rice (Oryza sativa) homolog of PUB12/PUB13, SPL11, also acts as a negative regulator of plant immunity by targeting a RhoGAP protein, SPIN6, for ubiquitination and degradation (Zeng et al., 2004; Liu et al., 2015). Although different E3 ligases and the UPS have been shown to play key roles in plant innate immunity, most targets of these E3 ligases have yet to be identified (Marino et al., 2012; Duplan and Rivas, 2014).
Plant NLRs are intracellular immune receptors that are divided into two major subclasses according to their N-terminal structure, which contains either a Toll/IL receptor or a coiled-coil (CC) domain (Maekawa et al., 2011b; Li et al., 2015b). NLR activity and stability are believed to be tightly regulated to ensure appropriate defense responses and avoid adverse effects on plant growth and development (Li et al., 2015b). Recent studies have suggested that multiple mechanisms control NLR stability, and UPS-mediated proteolysis plays a key role in this process (Bieri et al., 2004; Holt et al., 2005; Kim et al., 2010; Li et al., 2010; Cheng et al., 2011; Gou et al., 2012; Huang et al., 2014; Xu et al., 2015; Huang et al., 2016; Park et al., 2016). For example, two Arabidopsis NLRs, SNC1 and RPS2, are substrates of the F-box protein CPR1/CRP30 (SCFCPR1) and are targeted for proteasomal degradation (Cheng et al., 2011; Gou et al., 2012). Two plant TRAF proteins, MUSE13 and MUSE14, also were shown to cooperate with SCFCPR1 to regulate NLR turnover, presumably through the formation of a plant-type TRAFasome (Huang et al., 2016). Furthermore, the polyubiquitination of SNC1 and RPS2 is facilitated by an E4 ligase that may act downstream of the SCFCPR1 E3 ligase to promote NLR degradation (Huang et al., 2014). These findings indicate the importance of the UPS in the intricate regulation of NLR stability.
The barley (Hordeum vulgare) MLA locus encodes ∼30 allelic CC-NB LR-type NLRs that each confers isolate-specific disease resistance against the barley powdery mildew fungus, Blumeria graminis f. sp. hordei (Bgh; Seeholzer et al., 2010). Previous studies have shown that overexpression and/or autoactivation of MLA10 can trigger cell death signaling in Nicotiana benthamiana (Maekawa et al., 2011a; Bai et al., 2012). However, how MLA triggers cell death in the cytoplasm and the regulation of MLA protein level are unclear. Several components were reported to increase MLA abundance; for example, the RAR1 cochaperone was shown to cooperate with the cellular chaperone machinery and increase MLA abundance, likely by facilitating receptor folding (Bieri et al., 2004). In contrast, other factors are expected to negatively affect MLA level; for example, we reported that four members of the barley microRNA family, miR9863, target a subset of Mla alleles and negatively regulate Mla expression at the posttranscriptional level in barley (Liu et al., 2014). These findings indicate the importance of controlling MLA levels through different mechanisms. Nevertheless, whether the UPS is involved in regulating MLA stability remains unknown.
To determine how MLA stability is regulated, we searched for components that may control MLA turnover by yeast two-hybrid (Y2H) screening. Here, we report a barley RING-type E3 ligase, MIR1, that interacts with several functional MLAs through the N-terminal CC-containing region of the receptors. In vitro assays demonstrated that MIR1 directly ubiquitinates the N terminus of MLAs. In vitro and in planta biochemical assays showed that functional MIR1 promotes MLA degradation via the 26S proteasome. Virus-induced gene silencing (VIGS) of the MIR1 gene resulted in higher accumulation of MLA1 in a barley transgenic line expressing an MLA1-hemagglutinin (HA) fusion protein. Functional analyses indicated that overexpression of MIR1 compromises MLA1- and MLA10-mediated isolate-specific resistance to Bgh in barley but does not affect PTI or MLG-mediated race-specific or mlo-mediated broad-spectrum resistance. Additionally, MLA10-triggered cell death signaling is attenuated by the coexpression of MIR1 in N. benthamiana. These data suggest that the barley MIR1 E3 ligase targets MLAs and regulates the stability of these receptors through the UPS.
RESULTS
Identification and Characterization of MIR1, a Barley E3 Ub Ligase
To identify interactors or signaling components of barley MLA immune receptors, we performed Y2H screenings using baits containing fragments of MLA1 and a prey cDNA library derived from Bgh-challenged barley leaf epidermis (Chang et al., 2013). We identified a cDNA clone harboring the sequence of the C-terminal half of the barley gene MLOC_54598.2 that encodes a putative E3 Ub ligase of the RING type, hereafter designated MIR1 (for MLA interacting RING-type E3 ubiquitin ligase1). The MIR1 open reading frame encodes a protein of 376 amino acids that has a deduced molecular mass of 40.65 kD. We identified close homologs of MIR1 in different plant species and constructed a phylogenetic tree using sequences from represented plant species (Supplemental Fig. S1). The homologs from monocots and dicots are clearly separated into two groups (Supplemental Fig. S1), and barley MIR1 is closely related to homologs from wheat (Triticum aestivum; 95% identity) and wild einkorn (Triticum urartu; 94% identity). Analysis using the SMART program (http://smart.embl-heidelberg.de/) revealed that MIR1 contains a C3HC4-type RING (Really Interesting New Gene) finger at the N terminus and a triple tetratricopeptide repeat (TPR) domain at the C terminus that predominantly functions in protein-protein interactions (Supplemental Fig. S2; Blatch and Lässle, 1999). Notably, sequence alignment showed that both the RING and TPR domains are almost identical among barley and the wheat homologs but diverged in the closely related dicot homologs.
Characterization of the Interactions between MLAs and MIR1 E3 Ubiquitin Ligase
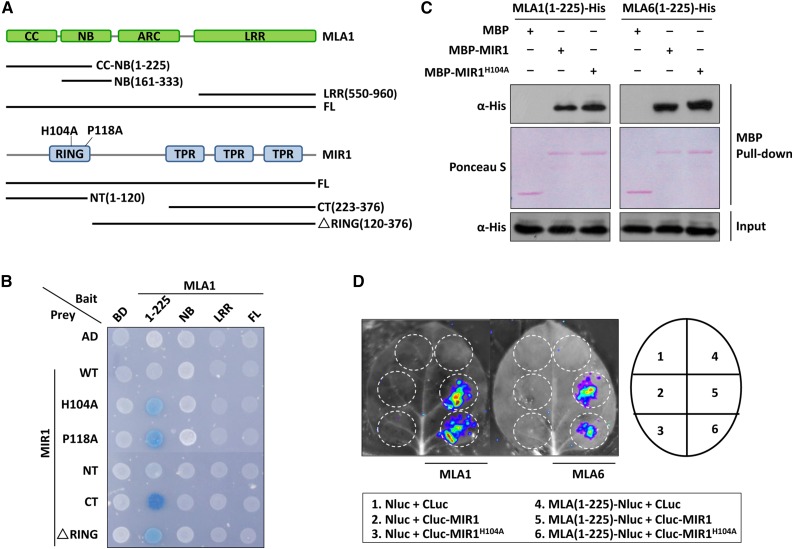
To further investigate the MIR1 interactions with MLA1 and other MLA members, we analyzed the interactions by targeted Y2H using baits and preys containing fragments of or full-length MLA1 and MIR1, respectively (Fig. 1A). Y2H analysis revealed that MLA1 relies on its N-terminal CC-NB(1-225) fragment to interact with MIR1 via the TPR domain (Fig. 1, A and B, blue yeast colony indicating positive interaction). MLA1 CC-NB(1-225) was further tested in yeast for interaction with full-length wild-type MIR1 and two MIR1 variants (H104A and P118A), each harboring a substitution of a conserved residue in the RING finger that should abolish the E3 ligase activity (Supplemental Fig. S2; Zheng et al., 2000). Unexpectedly, MLA1 CC-NB(1-225) was unable to interact with MIR1 wild type but did interact with the RING finger mutant variants, MIR1H104A and MIR1P118A (Fig. 1B). All baits and preys were confirmed to be properly expressed in yeast, and protein accumulation of MLA1 fragments or MIR1 wild type and mutants was detected using respective antibodies (Supplemental Fig. S3A). Since the barley Mla locus harbors both functional and nonfunctional alleles (Shen et al., 2003; Seeholzer et al., 2010; Jordan et al., 2011), we examined the MIR1-MLA interaction by Y2H screening using several MLAs that are confirmed to be functional, MLA1, MLA6, MLA10, and MLA25-1, as well as MLA18-1, which was shown to be nonfunctional (Jordan et al., 2011). Interestingly, MIR1H104A and MIR1P118A could interact with the CC-NB(1-225) fragment of functional MLAs but not with that of MLA18-1 (Supplemental Fig. S3B), suggesting that MIR1 associates only with functional MLAs. Similarly, MIR1 mutant variants did not interact with the equivalent region of Arabidopsis CC-type NLRs, RPM1 and RPS2, confirming the specificity of the MIR1 and MLA interaction (Supplemental Fig. S3B).
Figure 1.
MIR1 interacts with the N-terminal region of MLAs. A, Schematic diagram representing the domain structures of MLA1 and MIR1. Colored boxes represent individual domains, and fragments tested in Y2H analysis are shown as lines. The amino acid substitutions in the RING finger of MIR1 (H104A and P118A) are indicated. B, Y2H analysis of the MLA1 and MIR1 interaction. Baits of MLA1 were fused to the LexA DNA-binding domain (BD), and preys of MIR1 were fused to the B42 activation domain (AD). Colonies in blue represent positive interactions. CT, C terminus; FL, full length; LRR, Leu-rich repeat; NT, N terminus; WT, wild type. C, In vitro MBP pull-down assay for the MLA and MIR1 interaction. MBP, MBP-MIR1, and MLA-His were obtained from E. coli. MBP alone or MBP-MIR1 fusions were immobilized on amylose resin beads and incubated further with MLA fusions. Input represents equal amounts of purified MLA fusions before MBP pull down. D, LCI assay in N. benthamiana. The N-terminal half of luciferase (NLuc) was fused to MLA(1-225), and the C-terminal half of luciferase (CLuc) was fused to MIR1. Indicated fusion pairs were coexpressed in N. benthamiana by agroinfiltration. The luminescent signal was collected at 48 h post infiltration with a CCD imaging apparatus.
We also conducted in vitro MBP pull-down and in planta luciferase complementation imaging (LCI) assays to verify the MIR1-MLA interaction (Fig. 1; Supplemental Fig. S3). In vitro pull-down assays demonstrated that both MIR1 wild type and the H104A variant can interact with the CC-NB(1-225) fragment from MLA1, MLA6, and MLA10 (Fig. 1C; Supplemental Fig. S3C). Moreover, in planta LCI analysis also showed that coexpression of MIR1 wild type or the H104A variant with MLA1(1-225) or MLA6(1-225) generated strong luminescence signals that were not detected in the control pairs (Fig. 1D).
In summary, MIR1 can interact with the N-terminal region of multiple functional MLAs via the C-terminal TPR domain.
MIR1 Displays E3 Ub Ligase Activity and Can Ubiquitinate the MLA N Terminus in Vitro
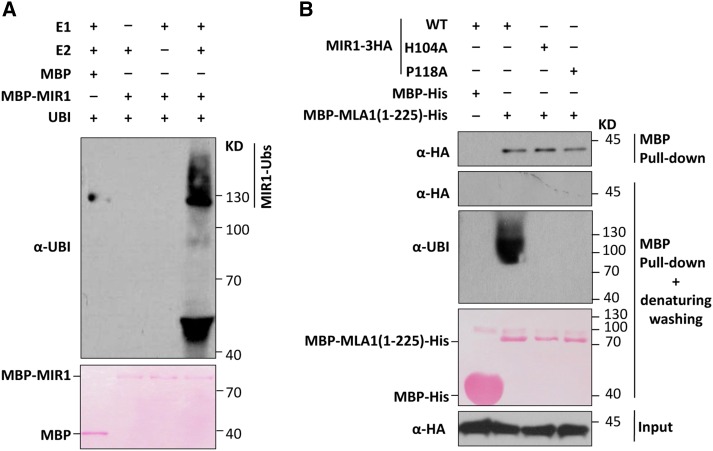
Since MIR1 contains a C3HC4-type RING finger, a feature of the RING-type E3 ligase (Deshaies and Joazeiro, 2009; Supplemental Fig. S2), we employed an in vitro ubiquitination assay to determine whether MIR1 possesses E3 activity (Zhao et al., 2013). For this assay, we used a wheat E1 (GI:136632), an Arabidopsis E2 (AtUBC10; At5g53300) and Ub (AtUBQ14; At4g02890), and the MIR1-MBP fusion and MBP alone, which were expressed and purified from Escherichia coli. In the presence of all ubiquitination components and MIR1-MBP, we detected a ladder of ubiquitination signals that are most likely autoubiquitinated MIR1 (Fig. 2A, lane 4). In the absence of E1 or E2, these ubiquitinated signals disappeared (Fig. 2A, lanes 2 and 3). These results confirmed that MIR1 possesses E3 Ub ligase activity.
Figure 2.
MIR1 possesses E3 Ub ligase activity and can ubiquitinate the MLA1 N terminus. A, MIR1 displays E3 Ub ligase activity in the ubiquitination assay in vitro. Polyubiquitinated MBP-MIR1 was analyzed by immunoblotting. E1 (wheat GI:136632), E2 (AtUBC10), Ub (AtUBQ14), MBP, and the MBP-MIR1 (E3) fusion were all obtained from E. coli. B, MIR1 ubiquitinates the MLA1 N terminus in vitro. MIR1-3HA fusions were expressed in N. benthamiana by agroinfiltration, and MBP-MLA1(1-225)-His was obtained from E. coli. Crude extract of N. benthamiana was incubated with MBP-His or MBP-MLA1(1-225)-His protein before being mixed with Ni-NTA beads for His pull down. These mixtures were subjected to pull down and then immunoblot analysis of MIR1-3HA fusions (top) or further washing with denaturing washing buffer and immunoblot analysis of MIR1-3HA (middle) or ubiquitinated MBP-MLA1(1-225)-His (bottom). WT, Wild type.
We next investigated whether MIR1 targets MLAs for ubiquitination (Wang et al., 2011; Choi and Harhaj, 2014). Since the full-length MLA proteins could not be successfully expressed and purified from E. coli (Maekawa et al., 2011a), for ubiquitination analyses, we expressed and purified the MLA N-terminal fusions MBP-MLA1(1-225)-His and MBP-MLA10(1-225)-His from E. coli and obtained crude extracts expressing MIR1-3HA, MIR1H104A-3HA, and MIR1P118A-3HA from agroinfiltrated N. benthamiana (Fig. 2B). Crude extracts of N. benthamiana expressing individual MIR1 fusions were incubated with MBP-MLA1(1-225)-His or MBP-MLA10(1-225)-His before His pull-down and immunoblot analyses (Fig. 2B; Supplemental Fig. S4). Indeed, the MLA1(1-225) fusion was able to pull down all MIR1 fusions (Fig. 2B, first gel), confirming their interactions (Fig. 1). To avoid nonspecific ubiquitination signals from MLA interactors, we further washed pull-down precipitates using denaturing washing buffer to eliminate any MLA-associated proteins prior to immunoblotting (Fig. 2B, second and third gels). As expected, the association of the MIR1 fusion with the MLA1(1-225) fusion protein was disrupted; thus, no signals were observed in the immunoblots with an anti-HA antibody (Fig. 2B, second gel). Nevertheless, polyubiquitination signals of MLA1(1-225) were clearly detected in the presence of wild-type MIR1 but not with the MIR1 mutant variants (Fig. 2B, third gel). The same experiment was repeated for the MLA10(1-225) fusion with similar results (Supplemental Fig. S4). These data demonstrated that MIR1 can ubiquitinate the N-terminal fragments of MLA1 and MLA10, and E3 activity was eliminated by the H104A or P118A substitution in the RING finger of MIR1.
MIR1 Promotes the Degradation of MLA in Vitro and in Planta
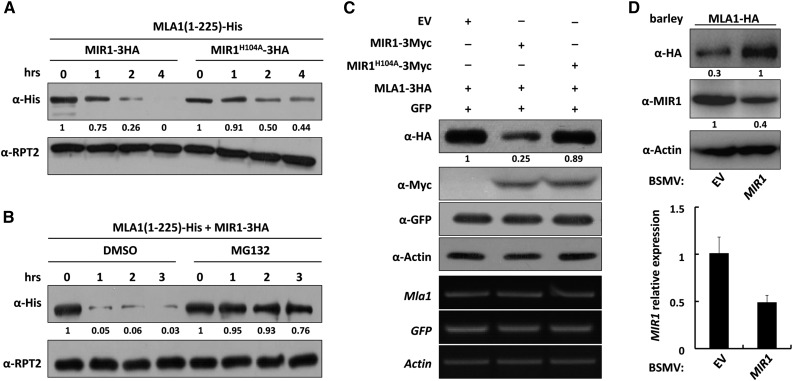
Because polyubiquitinated proteins generally are degraded by the 26S proteasome (Hershko and Ciechanover, 1998; Pickart and Eddins, 2004), we expected that MLA polyubiquitination by MIR1 would lead to proteasomal degradation. We tested this hypothesis by probing the stability of the E. coli-derived His fusion of MLA1(1-225) and MLA10(1-225) over time after mixing the proteins with the crude extracts from N. benthamiana expressing wild-type MIR1 or the MIR1H104A mutant (Fig. 3; Supplemental Fig. S5). Indeed, in the presence of MIR1 wild type, the MLA1(1-225) and MLA10(1-225) fusions were degraded to almost undetectable levels 4 h after incubation (Fig. 3A; Supplemental Fig. S5A, left), while the degradation of MLA1(1-225) or MLA10(1-225) was attenuated significantly in the presence of MIR1H104A (Fig. 3A; Supplemental Fig. S5A, right). Furthermore, addition of the proteasome inhibitor MG132 to the incubation mixture almost fully prevented the degradation of MLA1(1-225) or MLA10(1-225) at various time points compared with DMSO solution (Fig. 3B; Supplemental Fig. S5B). In this assay, the 26S proteasome regulatory subunit RPT2 was used as a loading control and detected on a separate gel (Fig. 3; Supplemental Fig. S5, A and B). Thus, these results indicate that MIR1 can promote the proteasomal degradation of the N-terminal fragments of MLA1 and MLA10 in vitro.
Figure 3.
MIR1 promotes the proteasomal degradation of MLA1 in vitro and in planta. A and B, MLA1(1-225)-His derived from E. coli was incubated with N. benthamiana crude extracts expressing MIR1-HA or MIR1H104A-HA fusion (A) or additionally treated with DMSO; left half) or MG132 (right half) for the indicated times (hrs; B). Immunoblot analysis shows the protein levels of MLA1(1-225) and RPT2, a 26S proteasome regulatory subunit that was detected on a separate gel and used as a loading control. C, MIR1 expression reduces the MLA1 protein level in N. benthamiana. The indicated MIR1-3Myc fusion was coexpressed with MLA1-3HA or GFP in N. benthamiana by agroinfiltration. Crude extracts obtained from leaf tissues at 60 h post infiltration were subjected to immunoblot analysis of MLA1 protein level (top). RT-PCR analysis shows equal expression of Mla1, GFP, and ACTIN (bottom) in this assay. GFP was used as an infiltration efficiency indicator and ACTIN as a loading control. D, Silencing of MIR1 leads to higher MLA1 protein levels in barley. Immunoblot analysis shows MLA1 and MIR1 protein levels in the barley transgenic line expressing MLA1-HA (top). Silencing of MIR1 (right lane) was achieved by BSMV-VIGS, and BSMV-empty vector (EV) treatment was used as a negative control (left lane). The silencing efficiency of MIR1 was quantified by qRT-PCR (bottom). Note that, in the experiments, wherever protein abundance needed to be quantified and compared, we determined the protein amount by digital imaging and analysis with ImageJ software.
Next, we asked whether the full-length MLA protein behaves similarly to the N-terminal fragment and also is a substrate of MIR1 in planta. To this end, we carried out degradation assays in N. benthamiana (Liu et al., 2010) by employing full-length MLA1 and MLA6. Myc-tagged MIR1 or MIR1H104A was coexpressed with 3× HA-tagged MLA1 or MLA6, respectively, in N. benthamiana (Fig. 3C; Supplemental Fig. S5C). In the presence of wild-type MIR1, the protein level of MLA1 or MLA6 was reduced significantly to ∼20% to 25% compared with the empty vector control (Fig. 3C; Supplemental Fig. S5C, lanes 1 and 2); in contrast, in the presence of MIRH104A, the level of MLA1 or MLA6 was reduced only marginally (Fig. 3C; Supplemental Fig. S5C, lane 3). Both GFP and actin, as an infiltration control and a loading control, respectively, remained unchanged (Fig. 3C; Supplemental Fig. S5C). These results indicate that MLA full-length proteins are genuine substrates of the MIR1 E3 ligase in planta.
To further verify the E3 ligase-substrate relationship between MIR1 and MLA1 in barley, we used a transgenic barley line expressing MLA1-HA (Bieri et al., 2004) and employed Barley stripe mosaic virus (BSMV) VIGS (Yuan et al., 2011) to silence MIR1 in barley (Fig. 3D). Silencing of MIR1 resulted in a significant reduction in the mRNA and protein levels to ∼50% of the control (Fig. 3D); in contrast, the MLA1 protein level was increased by at least 3-fold compared with the empty vector control (Fig. 3D, first gel). This reverse correlated abundance of MIR1 and MLA1 indicates that MLA1 is a bona fide substrate of MIR1.
In summary, our data obtained in vitro, in planta, and in vivo demonstrated that MIR1 promotes the degradation of MLA allelic receptors and that functional MLAs are bona fide substrates of MIR1.
MLA1 Turnover Is Regulated by the 26S Proteasome in Barley during Bgh Infection
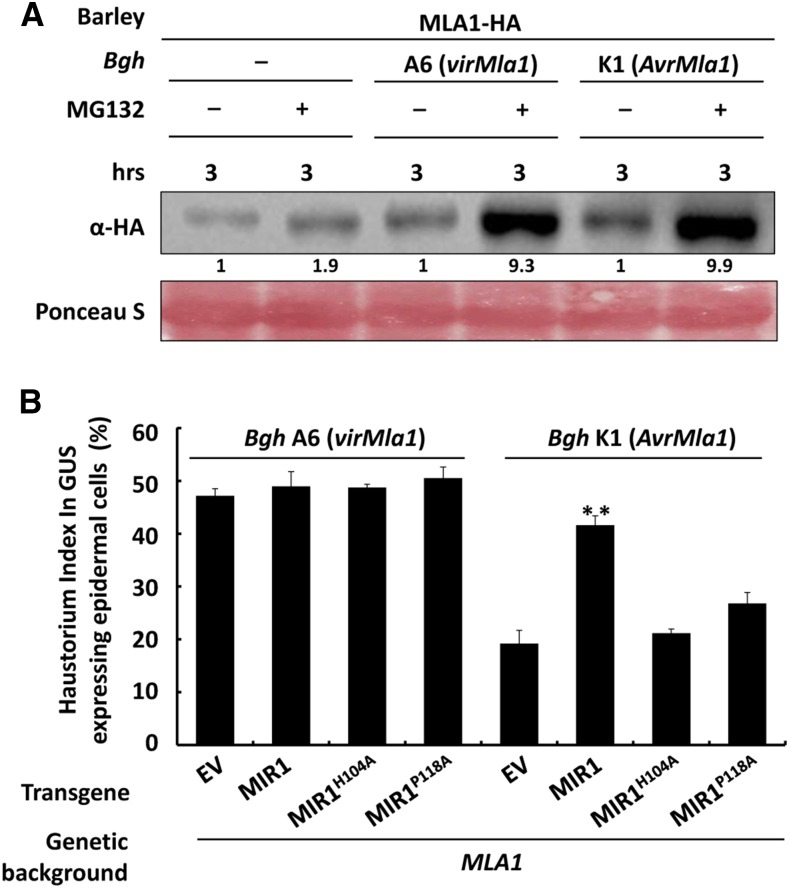
Similar to several other plant NLRs, barley MLAs are constitutively expressed at low levels and markedly induced during infection with B. graminis fungus (Caldo et al., 2006). SGT1 was implicated in participating in the process of MLA turnover, but other possible mechanisms for MLA turnover are largely unknown (Shirasu, 2009). Our data here suggested that MLA turnover may be regulated by the 26S proteasome. To elucidate this further, we again utilized the MLA1-HA transgenic barley line to examine MLA1 protein levels in healthy and Bgh-infected plants with or without MG132 treatment (Fig. 4A). Notably, MLA1 showed greater accumulation in Bgh-infected leaves than in unchallenged leaves without MG132 treatment (Fig. 4A, lane 1 compared with lanes 3 and 5), consistent with previous findings that Mla expression is induced by Bgh fungus (Caldo et al., 2006). Moreover, MG132 treatment significantly enhanced MLA1 accumulation in barley upon Bgh infection (Fig. 4A, lane 2 compared with lanes 4 and 6). This increased MLA1 accumulation after MG132 treatment in healthy and Bgh-infected barley leaves indicated that MLA1 is degraded via the 26S proteasome and that the proteasome constitutively regulates MLA turnover.
Figure 4.
MLA1 is degraded through the 26S proteasome, and MIR1 overexpression compromises MLA1-triggered immunity. A, Immunoblot analysis of the MLA1 protein level in the barley transgenic line expressing MLA1-HA. Barley leaves, noninoculated or inoculated with BghA6 (compatible) or BghK1 (incompatible) for 60 h, were infiltrated with DMSO or MG132 solution for 3 h prior to immunoblotting of MLA1 protein. B, MIR1 and it mutant variants were transiently overexpressed in leaf epidermal cells of the barley isogenic line containing Mla1 by particle bombardment. After particle delivery, the indicated Bgh isolates were inoculated for 48 h before the Bgh haustorium index (HI) was microscopically scored from more than 100 transformed cells. Every experiment was repeated at least three times. Asterisks indicate a significant difference (P < 0.001) between the empty vector (EV) control and this test.
MLA1-Mediated Disease Resistance Is Compromised by MIR1 Overexpression
Our results showed that MIR1 promotes proteasomal degradation of MLA1, MA6, and MLA10 (Fig. 3; Supplemental Fig. S5) and that the proteasome constitutively regulates the turnover of MLA1 in barley during Bgh infection (Fig. 4A). We next examined whether MIR1 regulates MLA-mediated immunity against Bgh. Therefore, we used a transient gene expression assay to overexpress the gene of interest together with a GUS reporter in barley epidermal cells through particle bombardment (Shen et al., 2003; Chang et al., 2013). We expressed MIR1 or its mutant, MIR1H104A or MIR1P118A, in the barley near-isogenic lines harboring MLA1, infected them with the virulent isolate BghA6 (Fig. 4B, left) or the avirulent isolate BghK1 (Fig. 4B, right) for 48 h, and determined the Bgh HI. Overexpression of MIR1 or the mutant variants had no effect on the HI in compatible interactions, similar to the empty vector control (Fig. 4B, bars 1–4), suggesting that MIR1 does not affect barley basal immunity. However, in incompatible interactions, overexpression of MIR1 significantly increased the HI from ∼19% in the empty vector control to 42% (Fig. 4B, bars 5 and 6), while overexpression of MIR1H104A or MIR1P118A only marginally increased the HI to ∼21% or 25% (Fig. 4B, bars 7 and 8). These results indicate that MLA1-mediated disease resistance, but not basal immunity, is specifically compromised by the overexpression of MIR1 but not by the RING mutants of MIR1.
MIR1 Expression Attenuates MLA10-Triggered Disease Resistance and Cell Death
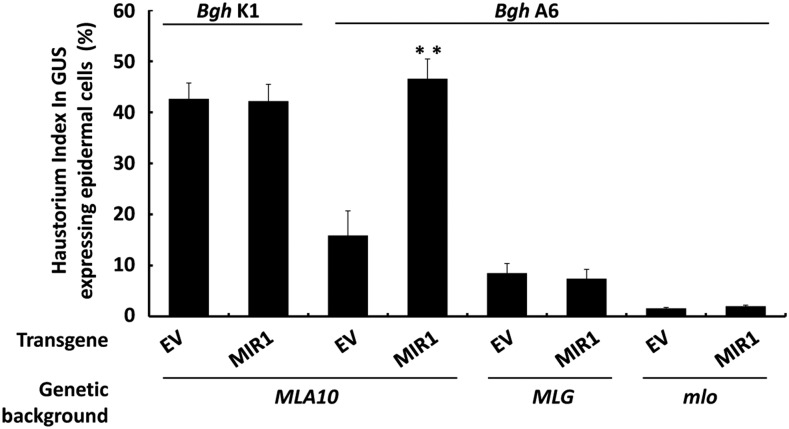
Barley MLA10 confers race-specific immunity against Bgh and triggers substantial cell death in N. benthamiana (Bai et al., 2012). To further establish the functional link between MIR1 and MLA-triggered immune responses, we extended the functional analysis of MIR1 in the MLA10-mediated immune response in barley and N. benthamiana. First, we utilized barley near-isogenic lines harboring MLA10 or an MLG race-specific resistance gene against Bgh (Caffier et al., 1996) and a barley mlo mutant line conferring broad-spectrum resistance (Büschges et al., 1997; Kim et al., 2002). As expected, overexpression of MIR1 in the MLA10 background did not affect basal immunity against BghK1 (Fig. 5, bars 1 and 2) but did significantly compromise MLA10-triggered immunity against BghA6 (Fig. 5, bars 3 and 4). However, overexpression of MIR1 had no impact on MLG-mediated race-specific and mlo-mediated broad-spectrum resistance against Bgh (Fig. 5, bars 5–8). Again, these data confirmed that MIR1 specifically compromises MLA-mediated resistance against Bgh.
Figure 5.
MIR1 overexpression compromises MLA10-triggered but not MLG- or mlo-mediated immunity. MIR1 was transiently overexpressed in leaf epidermal cells of the barley isogenic line harboring MLA10, or the MLG race-specific resistance gene, or the mlo broad-spectrum resistance gene by particle bombardment. BghK1 (virulent on MLA10) or BghA6 (avirulent on MLA10 or MLG) was used for inoculation. The Bgh HI was microscopically scored at 48 h postinoculation as described (Fig. 4B). Asterisks indicate a significant difference (P < 0.001). EV, Empty vector.
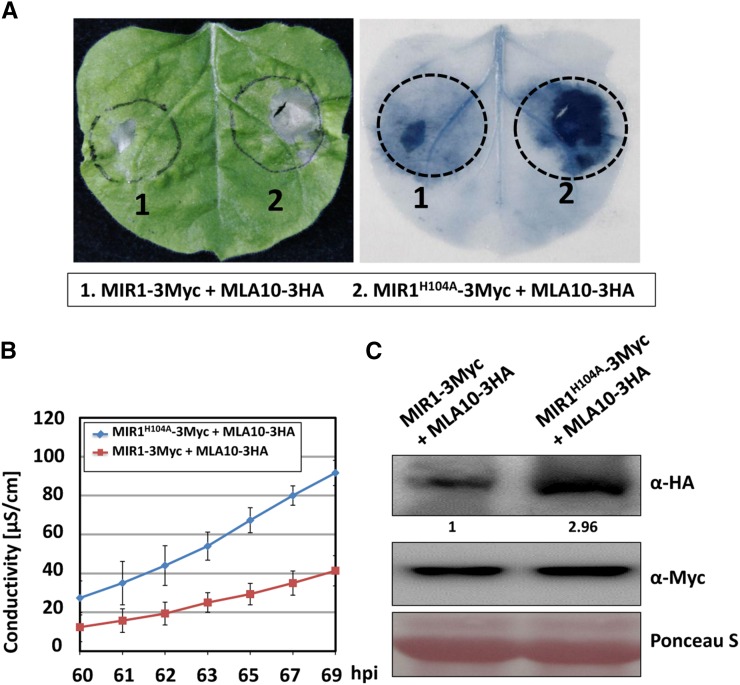
Second, we used a heterologous expression system in N. benthamiana in which MLA10-triggered cell death signaling is retained (Maekawa et al., 2011a; Bai et al., 2012). MLA10-3HA was coexpressed with MIR1-3Myc or MIR1H104A-3Myc in N. benthamiana by agroinfiltration. Indeed, MLA10-triggered cell death was attenuated by the expression of MIR1 but not by MIR1H104A (Fig. 6A). This was further confirmed by ion-leakage conductivity analyses (Fig. 6B). Immunoblot analysis of the MLA10 protein level revealed that MIR1 coexpression significantly reduced the MLA10 abundance to approximately one-third of that with the MIR1H104A mutant (Fig. 6C).
Figure 6.
MIR1 expression attenuates MLA10-triggered cell death in N. benthamiana. A, MIR1-3Myc or MIR1H104A-3Myc was coexpressed with MLA10-3HA in N. benthamiana. The photograph was taken at 70 h post infiltration (hpi; left), and then the leaf was stained with Trypan Blue solution (right). B, The electrolyte was measured using infiltrated leaf discs at the indicated time points. Error bars were calculated from three replicates per time point and construct. C, Immunoblot analysis of MLA10 protein level in N. benthamiana upon MIR1 and MLA10 coexpression. Total protein extract was obtained from N. benthamiana at 60 h post infiltration, and the experiment was done as described by Bai et al. (2012).
Together, these results show that MIR1 specifically compromises MLA10-triggered immunity and cell death signaling.
DISCUSSION
The Ub proteasome system has been shown to play an important role in both PTI and ETI as well as in systemic acquired resistance (Spoel et al., 2009). The UPS is involved predominantly in plant immunity by controlling the stability of immune receptors and regulators that act positively or negatively in immune signaling pathways. However, most of the findings were discovered from the interactions of the model plant Arabidopsis and bacterial pathogens. In recent years, studies have begun to reveal the importance of UPS and E3 ligases in crop resistance against fungal pathogens and in pathogen virulence (Li et al., 2011; Park et al., 2012, 2016; Liu et al., 2015; Zhu et al., 2015). In this study, we extensively characterized the relationship between barley MLA NLRs and a novel E3 Ub ligase, MIR1, in the context of the barley interaction with the obligate biotrophic powdery mildew fungus. We found that MIR1 via its TPR domain interacts with the N-terminal CC-containing region of functional MLAs. MIR1 possesses E3 Ub ligase activity and promotes MLA proteasomal degradation in vitro and in planta. We discovered that MLA turnover is a default process but is accelerated during Bgh infections. Single-cell transient expression of MIR1 in barley did not affect basal immunity but specifically compromised MLA-mediated disease resistance. Coexpression of MIR1 and MLA10 in N. benthamiana decreased the receptor protein level and attenuated MLA10-triggered cell death signaling. These data revealed that the MIR1 E3 ligase acts specifically on certain MLA immune receptors to control their stability and turnover and to attenuate their immune signaling.
Barley has ∼30 MLA allelic NLRs that are closely related. How does MIR1 target specific MLAs? The specificity may be determined by the physical interaction between MIR1 and individual MLAs. A previous sequence analysis of 29 full-length MLA proteins resulted in two distinct subfamilies, 1 and 2; subfamily 2 contains only three full-length proteins (i.e. MLA16-1, MLA18-1, and MLA25-1). Each of these three MLAs contains a CC domain whose sequence is diverged from that of subfamily 1 members and, thus, is believed to be nonfunctional (Seeholzer et al., 2010). However, domain swap using the CC-NB domain of MLA18-1 and MLA25-1 and the Leu-rich repeat domain of MLA1 showed that the MLA18-1 CC-NB is nonfunctional whereas the MLA25-1 CC-NB is functional in defense signaling (Jordan et al., 2011). Therefore, in our Y2H analysis to assess the MIR1 and MLA interaction, we included the N terminus of three functional MLAs from subfamily 1 (MLA1, MLA6, and MLA10) and two MLAs from subfamily 2 (MLA18-1 and MLA25-1; Supplemental Fig. S3B). Interestingly, MIR1 could interact with the N terminus of MLA1, MLA6, MLA10, and MLA25-1 but not with that of MLA18-1 (Supplemental Fig. S3B). These data indicate that MIR1 may interact only with functional N termini that adopt a signaling-competent conformation. In this regard, a potential feature shared by these active MLA N termini may be a CC homodimer that was identified in the MLA10 CC and required for downstream cell death signaling (Maekawa et al., 2011a; Bai et al., 2012). Future experiments should assess whether the homodimerization of functional CC domains is linked to its interaction with MIR1 and what other factors contribute to the specificity of MIR1 for different MLAs.
Using a barley transgenic line expressing the MLA1-HA fusion protein, we showed that MLA1 degradation via the proteasome appears to be a default process, as MG132 treatment significantly increased MLA1 protein levels in healthy, as well as in Bgh-challenged, barley leaves (Fig. 4A). Interestingly, although there is already a discernible increase in MLA1 level in barley leaves upon Bgh infection (Fig. 4A, compare lanes 1, 3, and 5), MG132 treatment resulted in a much higher increase of MLA1 in leaves infected with either Bgh isolate compared with that in healthy leaves (Fig. 4A, compare lanes 2, 4, and 6). These marked increases suggest that much more MLA1 is degraded, thus increasing the turnover in Bgh-challenged leaves over that in healthy ones. We currently do not know whether MIR1 determines the turnover rate of MLA, although our data demonstrated that MIR1 plays a key role in MLA degradation in barley. Similarly, we examined the relative expression level of MIR1 by qRT-PCR under challenge conditions. Unexpectedly, MIR1 expression was relatively unchanged from 16 to 60 h postinoculation with a virulent or avirulent Bgh isolate or the wheat powdery mildew isolate (Bgt) (Supplemental Fig. S6). These results indicate that MIR1 may be constitutively expressed at a specific level that is independent of pathogen inoculations, and this level of MIR1 may be sufficient for substrate targeting. The activation status of MLA1 did not appear to be related to its degradation, because the degradation level of MLA1 was similar in barley leaves challenged with either the virulent or avirulent isolate (Fig. 4A). What other factors may be involved in and probably control the rate of MLA degradation? Future studies should search for other factors and complexes that are involved in the degradation of MLA as well as where the degradation occurs at the subcellular level.
MLA1 or MLA10 also was degraded to some extent in N. benthamiana or in its crude extracts (Fig. 3; Supplemental Fig. S5); however, we are not sure whether this is partly because the N. benthamiana homolog of MIR1 also can target MLA and promote its degradation or because of the activity of some unspecific proteases. Our phylogenetic analysis and sequence alignment for the RING and TPR domains of selected MIR1 homologs from other species suggest that the Arabidopsis and N. benthamiana homologs are diverged from monocot homologs and likely are functionally different (Supplemental Figs. S1 and S2). Consistent with this prediction, the Arabidopsis homolog known as NO CATALASE ACTIVITY1 (NCA1) was identified previously in mutant screens for extremely low catalase activity, compromised cell death, and abnormal abiotic stress responses (Hackenberg et al., 2013; Li et al., 2015a). NCA1 was shown to regulate catalase activity through direct interaction with CATALASE2 (CAT2) via its C-terminal TPR domain, while zinc ion binding of its N-terminal RING finger domain is required for the increase in CAT2 activity (Li et al., 2015a). It is possible that several MIR1 homologs still possess E3 Ub ligase activity while others may have evolved to adopt a different function; for example, NCA1 acts as a molecular chaperone to assist in substrate folding (Li et al., 2015a), or the E3 ligase activity of MIR1 homologs may have evolved more recently.
MATERIALS AND METHODS
Plant and Fungal Materials
Barley (Hordeum vulgare) cultivars used in this study include ‘P01’ (nearly isogenic line from cv Pallas containing Mla1), ‘P09’ (near isogenic line from cv Pallas containing Mla10), ‘I10’ (near isogenic line from cv Ingrid containing Mla12), and the MLA1-HA transgenic line. All barley seedlings were grown in a growth chamber at 20°C with 16 h of light and 18°C with 8 h of darkness. Nicotiana benthamiana plants were grown in a greenhouse at 24°C ± 1°C with a 16-h light period.
The barley powdery mildew (Blumeria graminis f. sp. hordei) isolates A6 (AvrMla6, AvrMla10, AvrMla12, virMla1) and K1 (AvrMla1, vir6, vir10, vir12) used in this assay were maintained on barley ‘P01’ and ‘I10’ plants, respectively, and kept at 70% relative humidity in a 20°C light/18°C dark cycle.
Yeast Strains and Y2H Assay
The yeast strains EGY48 [MATa, his3, trp1, ura3, LexAop(x6)-LEU1 Plus p8op-lacZ] and YM4271 (MATa, ura3-52, his3-200, lys2-801, ade2-101, ade5, trp1-901, leu2-3, 112, tyr1-501, gal4-D512, gal80-D538, ade5::hisG) were used for Y2H assays and cultured on SD-uracil and YPAD plates, respectively. The Y2H interaction assay was performed essentially as described previously (Chang et al., 2013). Briefly, the appropriate bait and prey plasmids were cotransformed into the EGY48 strain by the lithium acetate method. Interaction analyses were done according to the yeast protocol handbook (Clontech). A total of 2.5 μL of liquid culture was dropped on the SD-uracil/His/Leu/Trp selection medium and incubated at 30°C for 72 to 96 h. For yeast protein accumulation analysis, overnight yeast cultures were grown in SD selection medium to the same OD and harvested by centrifugation at 3,000 rpm for 5 min. A total of 100 μL of 2× Laemmli buffer was added to the samples and boiled 5 min before SDS-PAGE. The bait and prey were detected with anti-LexA antibody (Santa Cruz Biotechnology; sc-7544) or anti-HA antibody (Roche; 11867423001).
Luciferase Complementation Imaging Assays
Luciferase complementation imaging assays were performed as described previously (Chen et al., 2008). The coding region of wild-type MIR1 and its variant H104A or P118A was ligated into the pCAMBIA-CLuc vector to generate the CLuc-MIR1/CLuc-H104A/CLuc-P118A constructs. The N-terminal region (amino acids 1–225) of MLA1 or MLA6 was fused to the NLuc domain by ligation of the respective coding region into the pCAMBIA-NLuc vector. The NLuc-/CLuc-derivative constructs were transformed into Agrobacterium tumefaciens strain GV3101. Overnight agrobacteria cultures were resuspended with infiltration buffer (2% Suc, 0.5% Murashige and Skoog medium, 100 µm acetosyringone, and 10 mm MES) to OD600 = 1. Equal volumes of agrobacteria resuspension carrying the nLUC and cLUC derivative constructs were mixed and coinfiltrated into N. benthamiana leaves. The infiltrated area was examined for luciferase activity 40 to 50 h post agroinfiltration with a cooled CCD imaging apparatus.
Pull-Down Assay
The N-terminal regions of MLA including MLA1(1-225), MLA6(1-225), and MLA10(1-225) was individually ligated into the pET32a vector (Novagen) for protein expression in Escherichia coli.
All the constructs were expressed in E. coli strain BL21DE3. The empty vector expressing MBP alone was used as a control. MBP, MBP-MIR1, and MBP-MIR1(H104A) fusion proteins on amylose resin beads (∼1 μg; New England Biolabs) were incubated with 500 μL of crude extracts containing MLA1(1-225), MLA6(1-225), and MLA10(1-225) fusion proteins, respectively, at 4°C for 1 h. After incubation, the beads were washed five times with 1 mL of column buffer. The beads with bound proteins were then resuspended with 30 μL of 2× Laemmli buffer and boiled for 5 min, and then the proteins were loaded onto the gel for immunoblotting using anti-His antibody. MBP-MIR1 and MBP-MIR1(H104A) proteins were detected by Ponceau staining.
Expression of the MBP-MIR1 Fusion Protein and in Vitro Self-Ubiquitination Assay
The coding region of wild-type MIR1 was ligated into the pMAL-c2x vector (New England Biolabs) to generate the MBP-MIR1 construct. The construct was transformed into E. coli strain BL21DE3. The fusion protein in E. coli was prepared according to the manufacturer’s instructions. Recombinant proteins of wheat (Triticum aestivum) E1 (GI:136632) and Arabidopsis (Arabidopsis thaliana) E2 (Ubc10) were prepared as described (Xie et al., 2002; Zhao et al., 2013). The monomer Ub of the Arabidopsis UBQ14 gene (At4g02890) was fused to the pET28a (Novagen) vector to generate His-Ub protein. Purified wheat E1 (∼100 ng), Arabidopsis Ubc10 (E2; ∼40 ng), UBQ14 (∼1 μg), and crude extracts containing recombinant MBP-MIR1 (E3; ∼500 ng) were prepared for the E3 Ub ligase activity assay as described (Xie et al., 2002; Zhao et al., 2013). The reaction was stopped by 2× Laemmli buffer and boiled before SDS-PAGE separation. The ubiquitinated protein was analyzed using anti-UBI antibody.
In Vitro Ubiquitination Assays
The coding regions of MIR1 and its mutants MIR1(H104A) and MIR1(P118A) were recombined into vector CTAPi-GW-3HA via Gateway technology to generate constructs expressing the HA fusion protein in N. benthamiana. The 6× His-tagged N-terminal regions of MLA, MLA1(1-225), and MLA10(1-225) were individually ligated into the pMALc2x vector (New England Biolabs) for protein expression in E. coli. The CTAPi and pMALc2x constructs were transformed into A. tumefaciens strain GV3101 and E. coli strain BL21DE3, respectively. The E. coli culture was treated with the 20 mm isopropylthio-β-galactoside (Takara; D9030B), and the recombinant protein was purified using Ni-NTA agarose (Qiagen; 30230). The agrobacterial culture was resuspended and infiltrated into N. benthamiana leaves. The crude protein was extracted from the infiltrated leaves using RB buffer (100 mm NaCl, 50 mm Tris-Cl, pH 7.8, 25 mm imidazole, 0.1% Tween 20, 10% glycerol, EDTA-free complete miniprotease inhibitor cocktail, and 20 mm β-mercaptoethanol).
The in vitro ubiquitination assay was as described previously with some modifications (Wang et al., 2011; Choi and Harhaj, 2014). Briefly, about 1 μg of purified MBP-His fusion protein bound on Ni-NTA agarose (Qiagen; 30230) was incubated with a protein extract from 5 g of the infiltrated area of N. benthamiana leaves for 2 h. After washing with RB buffer (for protein-protein interaction assay) or RB buffer supplemented with 8 m urea and 1% SDS (denaturing washing buffer for in vitro ubiquitination assay), the precipitates were resuspended in 2× Laemmli buffer and boiled for 5 min before SDS-PAGE. The precipitation of HA-tagged proteins with the amylose resin and ubiquitination of the MBP fusion protein were analyzed by immunoblotting with anti-HA antibody and anti-UBI antibody, respectively.
In Vivo Protein Degradation Assay
The in vivo protein degradation experiment was performed following the protocol described previously (Liu et al., 2010). Substrate MLA, internal control GFP, and the E3 ligase MIR1 or its mutant H104A plasmid was transformed into A. tumefaciens strain GV3101. The E3 ligase and substrate were mixed with a volume ratio of 3:1. After 60 h of infiltration, the protein samples were extracted by adding the SDS buffer and boiling 5 min before gel analysis.
Cell-Free Degradation Assay
For protein expression in N. benthamiana, the coding regions of MIR1 and its mutant H104A were cloned into CTAPi-GW-3HA vector via a Gateway reaction. The constructs were infiltrated into 4- to 5-week-old N. benthamiana leaves, and the proteins were isolated after 48 h using the degradation buffer (25 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm MgCl2, 4 mm PMSF, 5 mm DTT, and 10 mm ATP); 100 μm MG132 was added to the degradation buffer for the proteasome inhibition experiments. The amount of each indicated protein was detected with anti-HA antibody. For protein expression in E. coli, 6× His-fused MLA1(1-225) and MLA10(1-225) sequences were cloned into pMALc2x and purified with amylose resin beads (New England Biolabs). The protein degradation assay was performed essentially as described previously (Wang et al., 2009). Briefly, 100 ng of recombinant MBP-MLA1(1-225)-6His was incubated in 100 μL of N. benthamiana crude extracts containing the indicated constructs for individual detection (Fig. 3, A and B). The extracts were incubated at 22°C, and the samples were taken at the indicated times. Protein abundance was analyzed by immunoblotting using anti-His antibody.
Proteasome Inhibition Experiments
The inhibition experiments were conducted essentially as described (Matsushita et al., 2013). Briefly, detached barley leaves from the MLA1-HA transgenic line were incubated at 18°C for 24 h and then inoculated with corresponding B. graminis spores for 48 h. Samples were submerged in solution containing 0.01% Silwet L-77 and 100 μm MG132, vacuum infiltrated 15 min, and then incubated on a rotary shaker at 25°C. The leaves were harvested at the indicated times, and the sample was frozen in liquid nitrogen.
Single-Cell Transient Gene Expression Assay
For single-cell transient gene overexpression assay, the coding regions of MIR1 and its mutants H104A and P118A were cloned into a vector driven by the Ub promoter to create the constructs pUbi-MIR1/H104A/P118A. This transient assay was performed essentially as described (Shen et al., 2007). Briefly, the GUS reporter vector and the plasmid containing the candidate of interest gene were mixed and bombarded into the plant epidermal cells. Then, the leaves were stained using staining solution for GUS activity 48 h postinoculation, and the fungal HI in leaves was scored after challenge by B. graminis germlines.
BSMV-VIGS
The BSMV-VIGS assays were performed as described previously (Yuan et al., 2011). A 344-bp fragment of the barley MIR1 gene was obtained by RT-PCR and subcloned into pCa-γbLIC vector to generate the pCa-γbLIC:HvMIR1344 construct. For agroinfiltration, equal amounts of A. tumefaciens strain EHA105 harboring pCaBS-α, pCaBS-β, or pCa-gbLIC:HvMIR1344 were mixed and infiltrated into 3-week-old N. benthamiana leaves. After 8 to 12 d, the sap extracted from the infiltrated N. benthamiana leaves was inoculated onto barley leaves at the two-leaf stage. Three to 4 weeks after inoculation, when virus symptoms were visible, the infected leaves were harvested for real-time PCR analysis and immunoblot assay. These experiments were repeated three times.
A. tumefaciens-Mediated Transient Gene Expression and Electrolyte Leakage Assay
A. tumefaciens-mediated transient gene expression in N. benthamiana and electrolyte leakage assays were performed as described previously (van Ooijen et al., 2008; Bai et al., 2012). A. tumefaciens strain GV3101 was transformed with the indicated constructs, and the A. tumefaciens suspensions were infiltrated into 4- to 5-week-old N. benthamiana leaves. For MLA10-induced cell death experiments, A. tumefaciens suspensions expressing CTAPi-MIR1-3Myc and CTAPi-MLA10-3HA were mixed in a ratio of 3:1, and empty vector was used as a control. Samples were collected from the infiltrated leaves at 60 h post infiltration for immunoblotting. At 60 h post infiltration, four round patches from the infiltrated leaf area were punched and washed with distilled, deionized water for 30 min, and the leaf discs were subsequently placed into 10 mL of 0.001% Silwet L-77 solution in 60-mm petri dishes. Ion conductivity was determined using the B-173 ion conductivity meter (Horiba) at the indicated time points. These experiments were repeated at least twice.
Trypan Blue Staining
Trypan Blue staining was performed as described (Bai et al., 2012). N. benthamiana leaves were boiled for 5 min in staining solution mixed with an equal amount of ethanol. After staining, the leaves were destained in 2.5 g mL−1 chloral hydrate solution.
Gene Expression Analysis
For real-time PCR analysis, total RNA of 7-d-old barley primary leaves was isolated using Trizol solution (Invitrogen; 15596-026), and the cDNA was digested with RNase-free DNase I (Takara; 2270B). One microgram of total RNA was used to synthesize the first-strand cDNA. The cDNA was used as a template to analyze the expression of MIR1. Real-time PCR analysis was performed using the ABI Step-One Real Time PCR system with GoTaq qPCR Master Mix (Promega; A6001). The expression pattern of MIR1 was detected using the primers 5′-GGTCCATACGCTCTCAGT-3′ and 5′-CTGTCAACATCCGCAACTT-3′, and the expression of barley ACTIN was set as the internal control. Each real-time PCR assay was repeated with three independent biological replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers KY038422/AK359201.1 (Hordeum vulgare MIR1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic relationship of MIR1 and its homologs.
Supplemental Figure S2. Sequence alignment of MIR1 and its homologs.
Supplemental Figure S3. Analysis for MIR1 interaction with allelic MLAs.
Supplemental Figure S4. MIR1 can ubiquitinate the MLA10 N terminus.
Supplemental Figure S5. MIR1 promotes proteasomal degradation of MLAs.
Supplemental Figure S6. Transcript profiling of the MIR1 gene in barley.
Supplementary Material
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- NLR
Nod-like receptor
- ETI
effector-triggered immunity
- UPS
ubiquitin-26S proteasome system
- Ub
ubiquitin
- CC
coiled-coil
- Y2H
yeast two-hybrid
- VIGS
virus-induced gene silencing
- TPR
tetratricopeptide repeat
- LCI
luciferase complementation imaging
- BSMV
Barley stripe mosaic virus
- HI
haustorium index
Footnotes
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020400), the National Key Research and Development Program of China (2016YFD0100602), and the National Science Foundation of China (31530061) to Q.-H.S.
References
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, et al. (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, Shirasu K, et al. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16: 3480–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Caffier V, deVallavieille-Pope C, Brown JKM (1996) Segregation of avirulences and genetic basis of infection types in Erysiphe graminis f sp hordei. Phytopathology 86: 1112–1121 [Google Scholar]
- Caldo RA, Nettleton D, Peng J, Wise RP (2006) Stage-specific suppression of basal defense discriminates barley plants containing fast- and delayed-acting Mla powdery mildew resistance alleles. Mol Plant Microbe Interact 19: 939–947 [DOI] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen RAE, Rowan BA, Tenenboim H, et al. (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25: 1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Li X (2012) Ubiquitination in NB-LRR-mediated immunity. Curr Opin Plant Biol 15: 392–399 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Harhaj EW (2014) HTLV-1 tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog 10: e1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dou D, Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Duplan V, Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J (2012) The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Hackenberg T, Juul T, Auzina A, Gwizdz S, Malolepszy A, Van Der Kelen K, Dam S, Bressendorff S, Lorentzen A, Roepstorff P, et al. (2013) Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25: 4616–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Holt BF III, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309: 929–932 [DOI] [PubMed] [Google Scholar]
- Huang S, Chen X, Zhong X, Li M, Ao K, Huang J, Li X (2016) Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 19: 204–215 [DOI] [PubMed] [Google Scholar]
- Huang Y, Minaker S, Roth C, Huang S, Hieter P, Lipka V, Wiermer M, Li X (2014) An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 26: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan T, Seeholzer S, Schwizer S, Töller A, Somssich IE, Keller B (2011) The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J 65: 610–621 [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W (2010) The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lu D, Shan L (2014) Ubiquitination of pattern recognition receptors in plant innate immunity. Mol Plant Pathol 15: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu J, Wang G, Cha JY, Li G, Chen S, Li Z, Guo J, Zhang C, Yang Y, et al. (2015a) A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhong S, Li G, Li Q, Mao B, Deng Y, Zhang H, Zeng L, Song F, He Z (2011) Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res 21: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kapos P, Zhang Y (2015b) NLRs in plants. Curr Opin Immunol 32: 114–121 [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y (2010) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH (2014) The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet 10: e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park CH, He F, Nagano M, Wang M, Bellizzi M, Zhang K, Zeng X, Liu W, Ning Y, et al. (2015) The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog 11: e1004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011a) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P (2011b) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S (2012) Ubiquitination during plant immune signaling. Plant Physiol 160: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2013) Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73: 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24: 4748–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Shirsekar G, Bellizzi M, Chen S, Songkumarn P, Xie X, Shi X, Ning Y, Zhou B, Suttiviriya P, et al. (2016) The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog 12: e1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ (2004) Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695: 55–72 [DOI] [PubMed] [Google Scholar]
- Rodriguez E, El Ghoul H, Mundy J, Petersen M (2016) Making sense of plant autoimmunity and ‘negative regulators.’ FEBS J 283: 1385–1391 [DOI] [PubMed] [Google Scholar]
- Seeholzer S, Tsuchimatsu T, Jordan T, Bieri S, Pajonk S, Yang W, Jahoor A, Shimizu KK, Keller B, Schulze-Lefert P (2010) Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant Microbe Interact 23: 497–509 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K. (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K (2010) Ubiquitination in plant immunity. Curr Opin Plant Biol 13: 402–408 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MMA, Albrecht M, Cornelissen BJC, Takken FLW (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shao Q, Yu X, Kong W, Hildreth JE, Liu B (2011) N-terminal hemagglutinin tag renders lysine-deficient APOBEC3G resistant to HIV-1 Vif-induced degradation by reduced polyubiquitination. J Virol 85: 4510–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu F, Huang Y, Li L, Gannon P, Linster E, Huber M, Kapos P, Bienvenut W, Polevoda B, Meinnel T, et al. (2015) Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 27: 1547–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013) A plant-specific in vitro ubiquitination analysis system. Plant J 74: 524–533 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li Y, Fei F, Wang Z, Wang W, Cao A, Liu Y, Han S, Xing L, Wang H, et al. (2015) E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J 84: 154–168 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.