The ATP synthase is a molecular rotor that recycles ADP into ATP. Leone and Faraldo-Gómez use structural modeling to reinterpret and reconcile recent cryo-EM data for its membrane domain with other experimental evidence, gaining insights into its mechanism and the mode of inhibition by oligomycin.
Abstract
Two subunits within the transmembrane domain of the ATP synthase—the c-ring and subunit a—energize the production of 90% of cellular ATP by transducing an electrochemical gradient of H+ or Na+ into rotational motion. The nature of this turbine-like energy conversion mechanism has been elusive for decades, owing to the lack of definitive structural information on subunit a or its c-ring interface. In a recent breakthrough, several structures of this complex were resolved by cryo–electron microscopy (cryo-EM), but the modest resolution of the data has led to divergent interpretations. Moreover, the unexpected architecture of the complex has cast doubts on a wealth of earlier biochemical analyses conducted to probe this structure. Here, we use quantitative molecular-modeling methods to derive a structure of the a–c complex that is not only objectively consistent with the cryo-EM data, but also with correlated mutation analyses of both subunits and with prior cross-linking and cysteine accessibility measurements. This systematic, integrative approach reveals unambiguously the topology of subunit a and its relationship with the c-ring. Mapping of known Cd2+ block sites and conserved protonatable residues onto the structure delineates two noncontiguous pathways across the complex, connecting two adjacent proton-binding sites in the c-ring to the space on either side of the membrane. The location of these binding sites and of a strictly conserved arginine on subunit a, which serves to prevent protons from hopping between them, explains the directionality of the rotary mechanism and its strict coupling to the proton-motive force. Additionally, mapping of mutations conferring resistance to oligomycin unexpectedly reveals that this prototypical inhibitor may bind to two distinct sites at the a–c interface, explaining its ability to block the mechanism of the enzyme irrespective of the direction of rotation of the c-ring. In summary, this study is a stepping stone toward establishing the mechanism of the ATP synthase at the atomic level.
Introduction
As a result of cellular respiration or light harvesting, protons or sodium ions are actively exported across the inner membranes of mitochondria, chloroplasts, and prokaryotic cells, thus establishing an inward electrochemical potential gradient. How ATP synthases harness the reverse, downhill movement of these ions to power ATP production is a long-standing question in bioenergetics. From the general architecture of these enzymes (Fig. S1), which is largely conserved across species, it is apparent that ion translocation and ATP synthesis occur in two distinct, albeit mechanically coupled domains (Meier et al., 2011; Walker, 2013). It has also been established through structural, biochemical, and biophysical studies that two specific elements within the membrane domain, referred to as the c-ring and subunit a, are directly involved in facilitating ion permeation and that they do so through a rotary mechanism, which in turn powers the catalytic domain (Junge et al., 1997). However, insufficient information on the structure of the a–c complex has hindered a more detailed understanding of this unparalleled energy conversion mechanism. Multiple c-ring structures have been resolved at atomic resolution (Meier et al., 2005, 2009; Pogoryelov et al., 2009, 2010; Krah et al., 2010b; Preiss et al., 2010, 2015; Symersky et al., 2012b; Schulz et al., 2013; Matthies et al., 2014), consistently revealing a series of ion-binding sites along the outer circumference of the ring, each featuring a conserved acidic residue halfway across the transmembrane region. However, an atomic-resolution structure of subunit a, either isolated or in complex with the c-ring, has not been reported to date. This notwithstanding, the so-called two-half-channel model, proposed two decades ago (Vik and Antonio, 1994; Junge et al., 1997; Vik et al., 1998), still holds as the most plausible mechanistic hypothesis. According to this model, two aqueous half-channels or pathways would run through the a–c complex and reach to two ion-binding sites in the ring from either side of the membrane. Importantly, these two channels are noncontiguous and offset relative to each other; specifically, the half-channel open to the electrochemically negative (N) side of the membrane is clockwise with respect to that open to the positive (P) side (viewed from the catalytic domain). Similarly crucial is a conserved arginine residue on the surface of subunit a, believed to preclude ions from hopping directly between the two sites/channels (Mitome et al., 2010; Pogoryelov et al., 2010; Matthies et al., 2014). Therefore, an ion that binds the c-ring via the P channel can be released through the N channel only after the c-ring undergoes a near-complete revolution, counterclockwise, through a series of elementary rotational steps (Fig. S1). Individually, these steps would be stochastic and reversible, owing to the inherent symmetry of the c-ring structure; yet, a net directionality in the rotation of the c-ring would emerge in the presence of a proton-motive force, as forward and backward rates would become imbalanced (Matthies et al., 2014).
Although conceptually appealing, it has been challenging to verify or refute the two-half-channel hypothesis at the molecular level. In a remarkable breakthrough, the structure of a complete mitochondrial ATP synthase was recently determined at subnanometer resolution (7 Å), through single-particle cryo–electron microscopy (cryo-EM; Allegretti et al., 2015). This structure confirmed the expectation that subunit a lies adjacent to the c-ring and that it comprises at least four transmembrane helices, which at this resolution appear as rod-like density elements. Surprisingly, though, these helices differ noticeably in length and are highly tilted relative to the membrane perpendicular, by ∼70°, in stark contrast with earlier predictions based on a wealth of biochemical, biophysical, and bioinformatic analyses conducted to probe this structure (Rastogi and Girvin, 1999; Vik and Ishmukhametov, 2005; Moore and Fillingame, 2008; Hopf et al., 2014). Subsequent reports of two additional subnanometer cryo-EM structures (Zhou et al., 2015; Hahn et al., 2016) and of a partial crystal structure (Morales-Rios et al., 2015) have qualitatively verified this unprecedented architecture, but the proposed topologies for subunit a differ. Arguably, however, none of these structures is by itself of sufficient resolution to permit a conclusive assignment of the protein amino acid sequence. Given that none of these studies have provided an objective, quantitative assessment of alternative interpretations of the data, nor a comprehensive comparative evaluation of earlier biochemical studies, it seems both timely and pertinent to clarify these discrepancies, to establish a clear foundation for future mechanistic studies.
To this end, we sought to build and refine a structural model of the a–c complex that is optimally consistent not only with the abovementioned cryo-EM data but also with an evolutionary analysis of the primary sequences of both subunits and with existing biochemical and functional data. Specifically, we use a model-building protocol whereby knowledge-based methodologies are first used to create a large and diverse ensemble of putative models that are similarly compatible with the cryo-EM map of best resolution (Allegretti et al., 2015); these models are then ranked according to their consistency with inter-residue distances inferred from correlated mutation analyses, cysteine cross-linking experiments, and key functional experiments. This integrative procedure enabled us to conclusively establish the topology of subunit a and its relationship with the c-ring, despite the inherent uncertainty of each of the sources of data. The resulting model of the a–c complex provides clear insights into the mechanism by which proton permeation drives the rotation of the c-ring and into the mode of action of known bidirectional inhibitors. From a methodological standpoint, we posit that systematic, integrative molecular-modeling approaches such as that used here are ideally suited to leverage the impending growth in subnanometer-resolution structural data for membrane proteins, and particularly cryo-EM data.
Materials and methods
Modeling of the c-ring
An initial homology model of the c-ring from Polytomella sp. Pringsheim 198.80 was generated with MODELLER 9v8 (Fiser and Sali, 2003), based on the structure of the Saccharomyces cerevisiae c-ring (Symersky et al., 2012b) and a multiple-sequence alignment of c subunits generated with HHblits (Remmert et al., 2012). An ensemble of 2,000 models was initially produced and ranked in terms of the DOPE (Shen and Sali, 2006) and GA341 (Melo and Sali, 2007) scores. The top-ranking model was then refined and fitted into the relevant region of the Polytomella cryo-EM map (Allegretti et al., 2015), which had been previously carved out with CHIMERA (Pettersen et al., 2004). The refinement was performed with Rosetta, specifically with the so-called “relax” protocol (DiMaio et al., 2009), simultaneously using the high-resolution membrane and fit-to-density scoring functions (Yarov-Yarovoy et al., 2006; DiMaio et al., 2009). A total of 1,200 models were generated and scored. The transmembrane spans in the protein were translated from those predicted by OPM (Lomize et al., 2006) for the structure of the S. cerevisiae c-ring. During the refinement, the positions of the Cα atoms were restrained, and 10-fold symmetry was imposed along the ring axis. It was assumed that the resolution of the map was that originally estimated for the c-ring (Allegretti et al., 2015), namely 7.8 Å (the overall value is 7.0 Å). The voxel spacing was set to 3.0 Å. The fit-to-density score was based on the correlation between the experimental map and the map calculated for a model, for the complete c-ring structure. As has been prescribed for maps of similar resolution (DiMaio et al., 2009), the calculated density map was that of the Cα trace, assuming an 8-Å atomic radius that is meant to also encompass the side chain.
Modeling of subunit a
Initial models of the Cα trace of either the TM4-TM5 or TM2-TM3 hairpins were generated following a protocol previously described (Baker et al., 2010), using the versions of SSEHunter and SSEBuilder (Baker et al., 2007) implemented in Gorgon (https://gorgon.wustl.edu). The skeletonization used an algorithm described elsewhere (Ju et al., 2007) with a threshold of 0.1, assuming a resolution of 6.5 Å for the portion of the cryo-EM map corresponding to subunit a (Allegretti et al., 2015). Each hairpin was threaded into the cryo-EM map starting from the C terminus, guided by a consensus secondary structure prediction. Based on these initial models, a series of alternative threadings were generated by displacing the Cα trace in either direction in one-residue increments; in practice, these alternative threadings are “homology models” of the initial threading, in which the reference sequence alignment includes gaps artificially introduced to achieve the desired shift. Each of these Cα traces of the TM4-TM5 and TM2-TM3 hairpins was individually transformed into an all-atom model, using Rosetta, as described elsewhere (DiMaio et al., 2009). In brief, fragments of nine and three residues of known structure were considered for each of the helical regions of the Cα trace (note that the residues encompassed in these helical regions vary with the threading). After these fragments were built in, the resulting structures were perturbed in a Monte Carlo simulation, fostering displacements of 30° per 0.5 Å along the helix axis and 2° per 0.5 Å off axis. A constraint of 2 Å from the Cα initial model was applied with a penalty of 0.1 (arbitrary units) in the scoring function. The loops were then rebuilt for the lowest-energy model, and the complete hairpin model was fitted and refined into the cryo-EM density, using the same procedure used for the c-ring. The map resolution used for this refinement was 6.5 Å, with a voxel spacing of 3.0 Å. For each threading of each hairpin, an ensemble of 1,200 models was produced with this algorithm. Models with the best 20% total Rosetta score were extracted from each ensemble and evaluated according to their consistency with an evolutionary analysis of correlated mutations, as well as with preexisting cross-linking data (see below). After determining the most plausible threading for each of the two hairpins, we rebuilt and refined the complete four-helix bundle into the partial cryo-EM map in a manner identical to that used for each individual hairpin/threading.
Evolutionary couplings within subunit a and between subunits a and c
Correlations between naturally occurring mutations in subunit a were identified in an alignment of ∼9,500 sequences, which had been generated with HHblits (Remmert et al., 2012) using two iterations for the homologue search and an E-value of 10−5. The coupling score for each pair of mutations was computed using the EVcouplings server (Marks et al., 2011, 2012; Hopf et al., 2012). To score the degree of correlation in concurrent mutations of the c and a subunit, an analogous multiple sequence alignment was generated for the former, concatenated with that of the latter, and evaluated with the EVcomplex server (Hopf et al., 2014). Only residue pairs with an Evolutionary Coupling score of 0.15 or greater were considered.
Evaluation of model compliance with evolutionary couplings and cross-links
Two methods were used to evaluate the degree to which a given model is compatible with the set of Cα–Cα distances inferred from either the analysis of correlated mutations or the existing cross-linking data. The first method was to simply determine the number of distances that exceed the ideal values, i.e., the number of violations; e.g., in the case of evolutionary couplings, we defined a violation as an instance in which the distance between two residues in a correlated pair is >15 Å. In the second method, we used a scoring function that accounts for the fact some of the reference data are less reliable, either because the data are too sparse (e.g., correlated mutations between the c and a subunit), or not necessarily self-consistent (e.g., cross-linking data). This function takes the form
where the i and j indexes refer to the two Cα atoms in each residue pair considered and rij is the distance between them in the model being evaluated. The k index denotes whether the score pertains to the cross-linking data (k = 1) or to the evolutionary couplings (k = 2). Rk is the target distance value in each case. The values of R1 used were 8 Å for direct disulfide bonds and 12 Å for M2M-spaced cross-links, whereas that of R2 was 15 Å. Note these values imply that the maximum scores for a given model are F1 = 19 and F2 = 11. The more compliant a given model is with the data (i.e., the greater the number of pairs that fulfill rij ≤ Rk), the larger the values of Fk. However, unfeasible cross-links or couplings (rij >> Rk) in a given model do not contribute negatively to its Fk score. Hence, this ranking method is designed to reveal the models that are most globally consistent with the experimental data.
Online supplemental material
Fig. S1 shows the basic architecture of an ATP synthase. Fig. S2 shows a cryo-EM map of the subunit a–c-ring complex from the bovine mitochondrial ATP synthase. Fig. S3 shows alignment of the amino acid sequences of subunit c from Polytomella and Chlamydomonas reinhardtii against those of known atomic structure. Fig. S4 shows refinement of the c-ring homology model with Rosetta. Fig. S5 shows amino acid sequences of subunit a from Polytomella and Escherichia coli. Fig. S6 evaluates the consistency of the proposed structure of the a–c complex with previous cross-linking studies of the E. coli ATP synthase. Fig. S7 evaluates the consistency of the proposed structure of the a–c complex with cysteine accessibility data for the E. coli ATP synthase. Fig. S8 shows a comparison between different models of the subunit a structure. Table S1 evaluates the model of the Polytomella c10-ring structure before and after refinement with Rosetta. Table S2 shows cysteine cross-links engineered in the E. coli ATP synthase. Table S3 shows Cd2+ block sites in the E. coli ATP synthase.
Results and discussion
Structure of the c10-ring
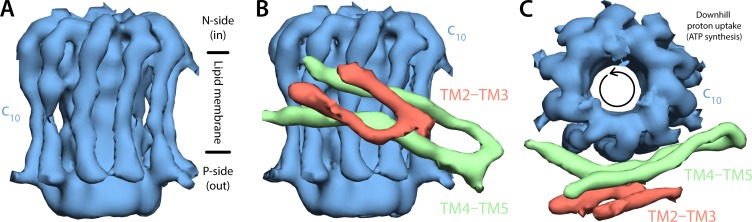
The cryo-EM map used in this study is that of the mitochondrial ATP synthase dimer from Polytomella sp. Pringsheim 198.80, which so far is that with the best resolution in the membrane domain (Allegretti et al., 2015). The portion of this map that is relevant to model the a–c complex is shown in Fig. 1 (for comparison, the cryo-EM map for the bovine ATP synthase [Zhou et al., 2015] is shown in Fig. S2). We began by constructing an atomic model of the c-ring, based not only on this cryo-EM map but also on its expected homology with other c-rings of known atomic structure. The cryo-EM map clearly shows that the c-ring of Polytomella comprises 10 copies of subunit c (Fig. 1 A), and hence the most suitable homology modeling template is the c10-ring of the mitochondrial ATP synthase of S. cerevisiae, whose structure is known at a resolution of 2.0 Å (Symersky et al., 2012b). Indeed, given the high similarity of the amino acid sequences of the Polytomella and S. cerevisiae c subunits (47% identical, without any gaps in their alignment [Fig. S3]), it is likely that this homology model differs from the true structure by <1 Å (root-mean-square deviation of the Cα trace in the transmembrane region; Forrest et al., 2006).
Figure 1.
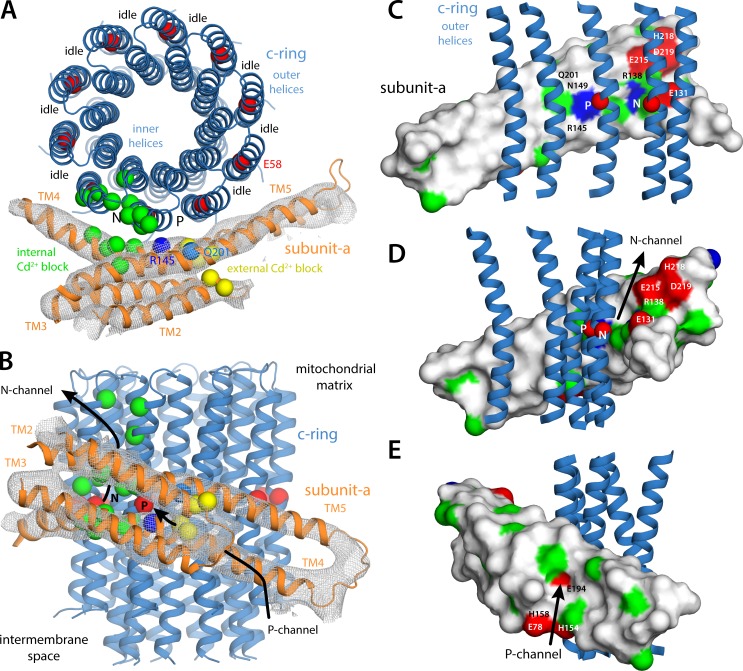
Cryo-EM map of the subunit a–c-ring complex from the Polytomella mitochondrial ATP synthase. The map shown is a selection of that previously reported for the complete ATP synthase dimer (Allegretti et al., 2015). (A) The c-ring, comprising 10 c subunits, viewed along the plane of the membrane. The proton-motive force drives protons inwards, from the electropositive or P side of the membrane to the electronegative or N side, i.e., inwards. (B) In the cryo-EM map, subunit a appears as two hairpins of two transmembrane helices each, adjacent to the c-ring, and highly tilted relative to the membrane perpendicular. (C) Same as B, viewed from the mitochondrial interior. To energize the synthesis of ATP, inward proton translocation downhill results in the counterclockwise rotation of the c-ring against subunit a, which is believed to remain static.
Because the Polytomella c subunit is encoded in the nuclear genome, a long portion of the N-terminal sequence is a mitochondrial transport signal, which is ultimately cleaved. To identify the starting point of the mature amino acid sequence, we compared the Polytomella c subunit sequence with the Edman degradation product of the c subunit from the closely related C. reinhardtii algae (Fig. S3; van Lis et al., 2003). We assigned the same starting residue to the Polytomella c subunit and constructed an initial homology model for the c10-ring using MODELLER (Fiser and Sali, 2003); we then improved and refined this initial prediction with Rosetta (Yarov-Yarovoy et al., 2006; DiMaio et al., 2009; see Materials and methods). During this refinement, the portion of the experimental cryo-EM map that corresponds to the c-ring (Fig. 1 A) was used as a three-dimensional density restraint, concurrently with the Rosetta energy function and sampling algorithm. As expected, the refined model does not differ significantly from the initial homology model in terms of the Cα trace, but a MolProbity (Davis et al., 2007) evaluation demonstrates that there are clear improvements in the overall quality of the structure otherwise (Table S1). The refined model also has low values of the Rosetta high-resolution membrane scoring function and, concurrently, the fit-to-density score, without any indication of over-fitting to the low-resolution map (Fig. S4).
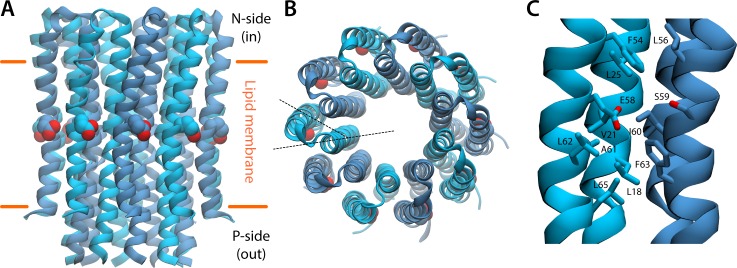
Fig. 2 A shows the overall structure of the Polytomella c10-ring. As in other c-rings of small stoichiometry, the outer helices of the c subunits are not arranged radially relative to the inner helices; instead, they are noticeably staggered (Fig. 2 B). This staggering seems to owe to the requirement that the outer helices remain at an approximately constant distance regardless of the curvature of the ring, as residues in neighboring outer helices must come together to form the ion-binding sites (Pogoryelov et al., 2012; Leone et al., 2015). In the Polytomella c10-ring, Glu58 marks the location of these binding sites (Fig. 2 C), halfway across the transmembrane span of the ring and on its outer face (Fig. 2, A–C). Aside from Glu58 and Ser59, on the adjacent outer helix, these binding sites are mostly hydrophobic, indicating a very high proton selectivity (Krah et al., 2010a; Leone et al., 2015). This high proton selectivity would ensure the Polytomella ATP synthase is coupled to the proton-motive force generated by the mitochondrial respiratory chain, despite the fact that the concentration of Na+ is multiple orders of magnitude greater than that of H+. By analogy with other c-rings (Leone et al., 2010), it is highly probable that proton binding to these sites entails the protonation of Glu58, further stabilized by a water-mediated bridge to Ser59.
Figure 2.
Refined cryo-EM structure of the Polytomella c-ring. (A) The c-ring, viewed along the membrane plane. The ring consists of 10 hairpin-like c subunits, with the N-terminal helix in the interior and the outer helix in the exterior. A conserved glutamate in each of the outer helices (red spheres) marks the location of the proton-binding sites. (B) The c-ring, viewed from the mitochondrial interior. Note that the outer helices are staggered relative to the inner helices, rather than projecting radially (dashed black lines). (C) Close-up of one of the proton-binding sites, at the interface between two adjacent c subunits, featuring the conserved glutamate side chain (Glu58).
Assignment and threading of TM4 and TM5 in subunit a
As mentioned, the cryo-EM map of the Polytomella ATP synthase reveals a bundle of four rod-like densities adjacent to the c-ring, oriented in the membrane at an unusually high tilt angle (Fig. 1, B and C). Although the map is not sufficiently resolved to assign any of these elements to a specific segment of protein amino acid sequence, a wealth of functional and biochemical data for the E. coli enzyme (Jiang and Fillingame, 1998; Vik and Ishmukhametov, 2005; Fillingame and Steed, 2014) demonstrate that these density elements correspond to subunit a and that the two elements closest to the c-ring are the two C-terminal helices, originally designated TM4 and TM5. We thus set out to build the structure of this region of the amino acid sequence into these rod-like densities.
To define the mature amino acid sequence of the Polytomella subunit a, we again compared it with that of the closely related C. reinhardtii, whose N terminus is known (van Lis et al., 2003). Alignment of the resulting sequence with that of E. coli and comparison of alternative secondary structure predictions (Fig. S5) indicate that TM4 and TM5 are encompassed by the 117-residue C-terminal fragment. There are, however, two possible ways to thread this sequence fragment into the cryo-EM map: either with TM5 closer to the mitochondrial matrix and TM4 closer to the intermembrane space or vice versa. Hereafter, these two alternatives will be referred as “TM4-out/TM5-in” and “TM4-in/TM5-out,” respectively. Owing to the limited resolution of the cryo-EM data, we decided to model both possibilities and assess which is the most probable by integrating bioinformatic and biochemical information into our analysis. Specifically, we devised a scoring scheme that considers (a) a newly conducted, family-wide analysis of evolutionary covariant amino acid pairs in subunits a and c (Materials and methods and Fig. 3) and (b) a comprehensive set of cysteine cross-linking data for both subunits a and c, which had been previously obtained for the E. coli ATP synthase (Table S2). The premise of this scoring scheme is that two residues that are in sequence positions that mutate in a correlated manner are likely to be in close proximity; a similar assumption is made if cysteine variants of those residues were found to form a cross-link, either directly or via a spacer.
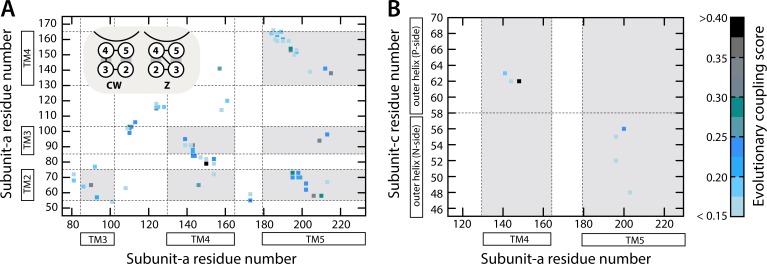
Figure 3.
Evolutionary analysis of correlated mutations within subunit a and between subunit a and the c-ring. (A) Matrix representation of the evolutionary couplings detected between pairs of residues within subunit a; the amino acid sequence and secondary structure elements of this subunit are indicated along the x and y axes. Each element in the matrix corresponds to a specific pair of residues, color-coded according to the degree to which these residues have evolved (i.e., mutated) in a correlated manner (see color scale on the right). The transmembrane regions are shaded in gray. The two possible topologies of subunit a, given a TM4-out/TM5-in assignment, i.e., clockwise and Z-like, are schematized in the top left inset (see Results and discussion). (B) Evolutionary couplings between the outer helix of the c subunit and helices TM4 and TM5 in subunit a. The horizontal dashed line divides the c subunit outer helix in two halves, before and after the proton-binding site, one closer to the P side of the membrane and the other to the N side.
In addition, for both the TM4-out/TM5-in and the TM4-in/TM5-out assignments, we considered a range of seemingly equally plausible threadings of the amino acid sequence, shifted relative to each other in 1-residue increments, up to an 11-residue span (i.e., approximately three turns of a helix). For each threading in each assignment of the TM4-TM5 hairpin, we generated an ensemble of 1,200 models by introducing smaller shifts in the protein backbone and a wide variety of side chain configurations. Collectively, we generated a total of 13,200 models of the TM4-TM5 hairpin for each assignment.
As mentioned, the resolution of the cryo-EM data is such that all of these models fit to the density comparably. However, we found that only a few of the threadings are truly compatible with the set of Cα–Cα distances that can be inferred from an analysis of evolutionary covariant residue pairs within the TM4-TM5 hairpin (Fig. 3 A and Materials and methods). Specifically, in only for four threadings in each of the two possible assignments did we detect a small percentage of distance violations in the majority of models in the corresponding ensemble (Fig. 4 A). Logically, though, the evolutionary couplings between TM4 and TM5 are not sufficient to discriminate between the TM4-out/TM5-in and TM4-in/TM5-out alternatives. However, if the evolutionary couplings between the TM4-TM5 hairpin and the outer helix of subunit c are considered (Fig. 3 B), a much greater consistency is apparent for three of the threadings in the TM4-out/TM5-in configuration (Fig. 5 A). Admittedly, the latter couplings are few in number, so the statistical significance of this result is less than ideal (Fig. 3 B). Reassuringly, though, if the experimental cross-linking data are used instead to evaluate the models (Materials and methods), the same three threadings in the TM4-out/TM5-in configuration are found to be also in better agreement with the set of Cα–Cα distances inferred from the measurements (Fig. 5 B).
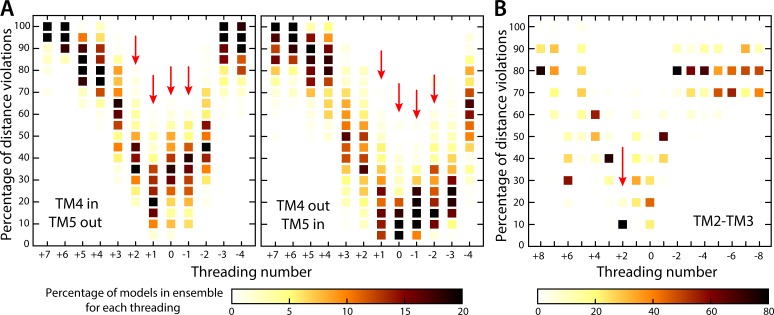
Figure 4.
Quantitative evaluation of alternative threadings of the transmembrane helices of subunit a into the cryo-EM map. (A) Analysis of the TM4-TM5 hairpin; the two possible assignments of TM4 (in/out) and TM5 (out/in) are evaluated separately in each panel (see Results and discussion). In each case, 11 alternative threadings were considered (as numbered), shifted relative to each other in one-residue increments. An ensemble of models was produced for each threading and analyzed individually, in terms of the consistency of the models with the Cα–Cα distances inferred from the evolutionary couplings between TM4 and TM5 (Fig. 3 A). Specifically, each column is a histogram of the percentage of distance violations calculated for each of the ensembles/threadings; the color scale indicates the population of each bin. The threadings marked with red arrows are the most compatible with the evolutionary analysis because the corresponding ensembles include the largest populations of models with the lowest percentage of violations. (B) Analysis of the TM2-TM3 hairpin. 17 different threadings of the TM2-TM3 hairpin (in a clockwise topology relative to TM4-TM5) were evaluated through an analysis identical to that summarized in A, except that the evolutionary couplings considered are those between TM2 and TM3 (Fig. 3 A). A single threading (marked with a red arrow) stands out as the most compatible with this evolutionary data.
Figure 5.
Quantitative evaluation of alternative threadings of TM4 and TM5 into the cryo-EM map. The eight most likely threadings of the TM4-TM5 hairpin, based on the analysis described in Fig. 4 A, are evaluated further. (A) The ensembles of models constructed for each threading are evaluated in terms of the evolutionary couplings between the TM4-TM5 hairpin and the outer helix of subunit c (Fig. 3 B). For clarity, a single, global score was calculated for each model from this evolutionary data (Materials and methods), and a histogram of that score across the ensemble is shown; by definition, the maximum value of this score (i.e., no distance violations) is 11. (B) Analogous analysis to A, instead based on Cα–Cα distances inferred from cross-linking experiments (Materials and methods and Table S2); by definition, the maximum value of this score is 19. (C) Analysis of the top three threadings according to A and B, in terms of the proximity between Glu58 in subunit c, Arg145 in TM4 of subunit a, and Gln201 in TM5 (see Results and discussion). The plots show the percentage of models in each ensemble for which the distance between these side chains is less than or equal to an increasing value between 0 and 20 Å.
In summary, this unbiased analysis makes clear that the resolution of the cryo-EM map of the Polytomella ATP synthase is not sufficient to assign TM4 and TM5 without any other inputs of information (even though this map is the best resolved of all available); however, this assignment becomes unequivocal when sequence analysis and biochemical data are integrated with the low-resolution structural information. Collectively, these data conclusively reveal that TM4 is closer to the electropositive (P) side of the membrane (i.e., the intermembrane space, or outside), whereas TM5 is closer to the electronegative side (N) side (i.e., the mitochondrial matrix, or inside).
This key result notwithstanding, neither the cryo-EM, bioinformatic, or cross-linking data are sufficiently resolved to discern alternative threadings of TM4 and TM5 differing by up to a three-residue shift. To narrow down this assignment further, we resorted to functional data, specifically pertaining to a conserved, functionally crucial arginine residue in TM4 of subunit a (Lightowlers et al., 1987; Cain and Simoni, 1989). Of particular relevance are functional assays performed for the E. coli enzyme (Ishmukhametov et al., 2008; Bae and Vik, 2009) that strongly indicated that (a) the conserved glutamate in the c subunit proton-binding site interacts with this arginine (here Arg145) and (b) that this direct interaction is preserved if that arginine is swapped with a conserved glutamine in TM5 (here Gln201). We reasoned that the concurrent proximity of Glu58 to Arg145 and Gln201 could be used as a metric to rank the three remaining threadings of the TM4-TM5 hairpin (Materials and methods). Indeed, this criterion led to a unique solution (Fig. 5 C), which therefore can be considered the most consistent with all available experimental and bioinformatic data. Having identified this most probable threading of TM4 and TM5, we next set out to complete the subunit a model by adding helices TM2 and TM3.
Threading the subunit a TM2-TM3 hairpin
Although helices TM2 and TM3 can be clearly discerned in the cryo-EM map of the Polytomella ATP synthase, the connectivity between TM4 and TM3 cannot (Fig. 1). Therefore, there are again two possible ways to thread TM2 and TM3 relative to TM4 and TM5. In a Z-like topology, TM2 would be closer to TM4, and TM3 to TM5 (Fig. 3 A, inset); alternatively, the four helices could be arranged clockwise. We reasoned that the analysis of evolutionary couplings within this four-helix bundle ought to reveal which of these topologies is the most probable. Indeed, as shown in Fig. 3 A, the number of covariant pairs for residues in TM2 and TM5 is much greater than that between TM2 and TM4; in addition, the diagonal pattern of the correlation matrix indicates that TM2 and TM5 are in a parallel arrangement. Similarly, many more pairs can be detected for TM3 and TM4 than for TM3 and TM5, and also in a diagonal pattern. It is therefore highly probable that TM2 is more proximal to TM5 than TM4, whereas TM3 is more proximal to TM5 than TM4.
Based on this result, we modeled the amino acid sequence of the TM2-TM3 hairpin (residues 52–115) into the cryo-EM map in a clockwise topology, following a procedure analogous to that used for TM4-TM5. That is, we considered 17 distinct threadings of the TM2-TM3 hairpin and produced a set of >20,000 models in total, which reflect a highly diverse ensemble of backbone and side chain conformations for each threading (Materials and methods). We then evaluated these models according to their consistency with the evolutionary couplings detected between TM2 and TM3. As shown in Fig. 4 B, this assessment clearly revealed one specific threading as the most probable, with ∼80% of the models in the corresponding ensemble in agreement with 90% of the expected Cα–Cα distances.
Structure of the a–c complex supports the two-half-channel hypothesis
Having identified the most probable threading of the subunit a TM2-TM3 and TM4-TM5 hairpins, among a wide range of alternatives, we again used Rosetta to refine a model of the TM2-TM5 bundle, through a protocol similar to that used to improve the homology model of the c-ring (Materials and methods). The two models were then fitted simultaneously into the cryo-EM map, resulting in the structure of the a–c complex shown in Fig. 6.
Figure 6.
Cryo-EM structure of the subunit a–c-ring complex. (A) The a–c complex is viewed from the mitochondrial matrix, i.e., the N side. The gray mesh is the cryo-EM map for subunit a; that for the c-ring is omitted for clarity. Red spheres indicate the Cα atoms of the conserved glutamate (Glu58) in the c-ring proton-binding sites. The dark and light blue spheres on subunit a indicate the conserved arginine (Arg145) in TM4 and the conserved glutamine (Gln201) in TM5. Green and yellow spheres indicate the positions in the E. coli subunit a for which a substituted cysteine is reactive with Cd2+, added from either the N or the P side, respectively, thereby blocking the proton transport mechanism (Gln201 is also a site of external Cd2+ block). (B) Same as A viewed along the membrane plane. (C–E) Smoothed surface of subunit a up to the Cγ atom of each side chain, colored as follows: blue, Lys or Arg; red, Asp, Glu, or His; green, polar; white, hydrophobic.
Despite its limited resolution, this structural model is squarely consistent with the two-half-channel hypothesis outlined above and also helps to rationalize a wide range of biochemical data pertaining to the mechanism and inhibition of the enzyme. By construction, the structure shows one of the proton-binding sites in the c-ring in proximity to both Arg145, the crucial arginine in TM4 of subunit a, and Gln201 in TM5 (Fig. 6 A). The Cα–Cα distance from Glu58 to Arg145 (8 Å) is indeed consistent with a salt bridge, and that to Gln201 (13 Å) implies this same interaction would be feasible if Arg145 and Gln201 were swapped, as has been suggested for the E. coli ATP synthase (Ishmukhametov et al., 2008; Bae and Vik, 2009). A nontrivial finding, however, is that this c-ring binding site is aligned with a series of residues on subunit a that have been inferred to be exposed to the aqueous half-channel on the P side of the membrane (Fig. 6 A). Specifically, these are positions whose equivalent in E. coli, upon cysteine substitution, have been shown to mediate binding of Cd2+, thereby blocking proton uptake from the periplasm (Table S3; Steed and Fillingame, 2008, 2009, 2014; Dong and Fillingame, 2010). In our structure, these residues group in the loop of the TM2-TM3 hairpin and near Arg145 and Gln201 in TM4 and TM5, respectively; together, these residues clearly delineate a pathway through the protein interior that leads to the abovementioned c-ring binding site, which we therefore refer to as the P site (Fig. 6, A and B). In close proximity to these Cd2+ block sites, four ionizable residues cluster together (Fig. 6 E), seemingly marking the entrance of this pathway (Fig. 6 B). Two of these residues, H154 and E194, are of special interest, as they are largely conserved. Indeed, analysis of correlated mutations across subunit a homologues shows that these two residues are highly covariant (Fig. 3 A); for example, in the native E. coli sequence, these residues are swapped for E219 and H245, respectively. Functional assays of the E. coli ATP synthase show these two residues contribute to determine the rate of proton uptake (Cain and Simoni, 1988), suggesting that they might serve as a buffering site for the incoming proton; such buffering sites exist in other proton transport systems of known structure (Eicher et al., 2014; Kaila et al., 2014). In the context of our model, it becomes straightforward to envisage how a proton transiently residing at E194/H154, deep inside the subunit a four-helix bundle, could transfer to the P site on the c-ring, for example via a water wire flanked by the polar side chains of N149 (TM4) and Q201 (TM5; Fig. 6 C). Indeed, the equivalent residues in E. coli (N214 and Q252) are among those accessible to Cd2+, and therefore water, from the exterior (Steed and Fillingame, 2009, 2014; Dong and Fillingame, 2010).
The model of the a–c complex reported here also indicates that the c-ring binding site adjacent to the P site, clockwise, is exposed to the solution on the negative side of the membrane, i.e., it is the N site. The N channel, however, appears to be a solvent-filled crevice at the a–c interface, as we proposed elsewhere (Pogoryelov et al., 2010), rather than a narrow pathway. Similarly to the P channel, though, positions known to be accessible by Cd2+ on the cytoplasmic side of the E. coli a–c complex (Table S3; Steed and Fillingame, 2009, 2014; Dong and Fillingame, 2010) cluster in this region, both on subunit a and on the c-ring (Fig. 6, A and B). Consistent with the notion that the N channel is an interfacial solvent-filled crevice, the surface of subunit a in this region features multiple polar and ionizable residues (Fig. 6, C and D). Two of these residues, E131 and R138, are worth noting as they are well conserved. The former corresponds to E196 in E. coli, whose replacement by, e.g., lysine slows down the proton transport activity of the enzyme but does not preclude proton binding to the c-ring (based on the reactivity of E58 to DCCD; Vik et al., 1988). Therefore, E131 might provide a transient binding site for the protons released via the N channel in a manner similar to E194 in the P channel. The proximity of the highly conserved R138 (K203 in E. coli), which is also one of the Cd2+ block sites in this pathway (Dong and Fillingame, 2010), suggests that this residue might also be involved in this process.
Our structural model reveals a striking difference in the nature of the surface of subunit a that is exposed to the c-ring at either side of the membrane center (Fig. 6, C–E). On the N side, this surface is highly polar and most probably well hydrated; on the P side, however, it is highly hydrophobic and most probably sealed. These features are consistent with the notion that the N channel is at the a–c interface, whereas the P channel runs across the interior of subunit a (Angevine et al., 2007; Steed and Fillingame, 2008, 2009; Dong and Fillingame, 2010). A second implication is that most of the 10 binding sites in the c-ring (all sites except 2) face a hydrophobic microenvironment; as we have discussed elsewhere, this environment restricts the conformational freedom of the binding sites and ensures that protons (or sodium ions) remain tightly bound (Pogoryelov et al., 2009, 2010; Symersky et al., 2012b; Matthies et al., 2014), as these sites travel within the membrane. Our model suggests that despite the unexpected length of the a–c interface, only two adjacent ion-binding sites in the c-ring, referred to here as P and N sites, are implicated in the mechanism that couples proton binding and release to the rotation of the c-ring (Fig. 7); hence, we do not concur with the three-site mechanism proposed elsewhere (Allegretti et al., 2015; Hahn et al., 2016).
Figure 7.
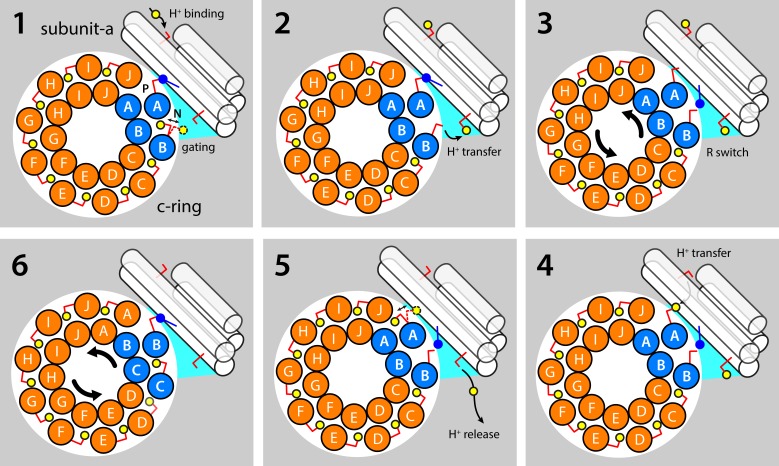
Hypothetical mechanism coupling the translocation of protons through the a–c complex to the rotation of the c-ring. (1) Most of the c-ring binding sites are exposed to the membrane interior, and the conserved glutamate residue (Glu58 in Polytomella) is protonated and retracted in a closed state. However, one binding site is exposed to the aqueous crevice at the a–c interface that is open to the interior/negative side of the membrane; as a result, this N site fluctuates between a closed and open state. The adjacent c-ring binding site, counterclockwise, or P site, forms a salt bridge with a conserved arginine in TM4 of subunit a (Arg145). A protonatable side chain exposed to the exterior/positive side of the membrane serves as a transient buffering site for H+. (2) The aqueous nature of a crevice at the a–c interface (on the N side) fosters the deprotonation of the N site; the released H+ might be transiently captured by a buffering residue analogous to that on the opposite side of the membrane. (3) The N and P sites compete for the same interaction with Arg145, as the c-ring rotates back and forth, stochastically. (4) When the N site is engaged by Arg145, the proton buffered in the P channel can transfer to the deprotonated Glu58 in the P site, through an aqueous pathway. (5) The P site, H+ loaded, can now exchange between open and closed states. The H+ buffered in the N channel is released into the internal solution. (6) The protonated, closed P site enters the hydrophobic membrane interior, as the c-ring rotates counterclockwise; the N site becomes the new P site and continues to engage Arg145, and a new binding site enters the a–c interface, resetting the transport cycle. Note the proposed mechanism is entirely reversible, consistent with the fact that ATP synthases can function as ATP-driven ion pumps.
Finally, the proposed structure seems to reaffirm the hypothesis that the role of the conserved arginine in TM4 of subunit a is not to induce the deprotonation of the c-ring binding sites, but rather to preclude the leakage of ions across the a–c interface, by acting as an electrostatic barrier between the P and N sites (Fig. 7; Pogoryelov et al., 2010; Matthies et al., 2014). Indeed, functional assays of the E. coli ATP synthase have shown that the a–c complex retains its ability to catalyze the permeation of H+ even when this arginine is neutralized by mutation; the acidic residue in the c-ring binding sites is, in contrast, essential (Valiyaveetil and Fillingame, 1997; Mitome et al., 2010). What these arginine mutations do, however, is to abolish the mechanical coupling between the catalytic and membrane domains; i.e., they short-circuit the mechanism that couples permeation and rotation. From our structure and previous experiments (Matthies et al., 2014), we envisage that the role of this arginine side chain is to sequester either the P or N site, already in their deprotonated state, freeing the other site to load or release H+, while simultaneously precluding those incoming or outgoing H+ from directly hopping from one half-channel to the other (Fig. 7). Similarly to secondary-active transporters, therefore, the a–c complex would drive the catalytic domain by virtue of an alternating access mechanism.
Lessons from failed predictions: The value of integrative modeling
The architecture and membrane orientation of subunit a observed in the cryo-EM structure of the Polytomella ATP synthase diverges dramatically from all earlier predictions (Rastogi and Girvin, 1999; Vik and Ishmukhametov, 2005; Moore and Fillingame, 2008; Hopf et al., 2014). These predictions had been based on biophysical and biochemical experimental data collected for over two decades for the E. coli enzyme, which is, admittedly, different from the mitochondrial enzyme in significant ways. For example, the membrane domain of the mitochondrial ATP synthase includes additional protein subunits that mediate its dimerization (Davies et al., 2012). In our view, however, the discrepancy between the actual and predicted structures of the a–c complex cannot be not explained by variations in the primary sequence across species. In fact, the sequence of the C-terminal four-helix bundle of subunit a is relatively well conserved, as is the c subunit, and so it is rather improbable that different species will feature entirely different folds (or mechanisms). Therefore, this discrepancy suggests that the structural inferences made from the biophysical and biochemical experimental data were often inaccurate. From a methodological standpoint, it is worth examining what lessons may be learned from these earlier efforts, if only to foster better predictions in the future.
In hindsight, it seems fair to conclude that one of the sources of error in previous predictions stems from an excessively stringent geometric interpretation of cysteine cross-linking data (Jiang and Fillingame, 1998; Schwem and Fillingame, 2006; Moore and Fillingame, 2008; Moore et al., 2008). As discussed above, this dataset is highly informative: it helped us to discern whether the TM2-TM5 bundle is arranged clockwise or in a Z-like pattern and also to determine the orientation of the TM4-TM5 hairpin relative to the c-ring. However, when interpreting such data, it is worth keeping in mind that that the cross-linking reaction is irreversible (in oxidizing conditions), and hence that a cross-link might reshape the inherent conformational distribution of a protein very significantly. In addition, cysteine pairs are typically engineered one at a time. Thus, we would argue that it is not self-evident that the structural constraints imposed by an individual cross-link, e.g., a Cα–Cα distance of up to ∼8 Å for a disulfide bond (Richardson, 1981), must also be feasible when other cross-links are considered simultaneously, e.g., when translating this data into a three-dimensional model. This assumption is particularly dubious when no functional data can be gathered for the cross-linked protein, to ascertain whether or not its fold has been significantly altered.
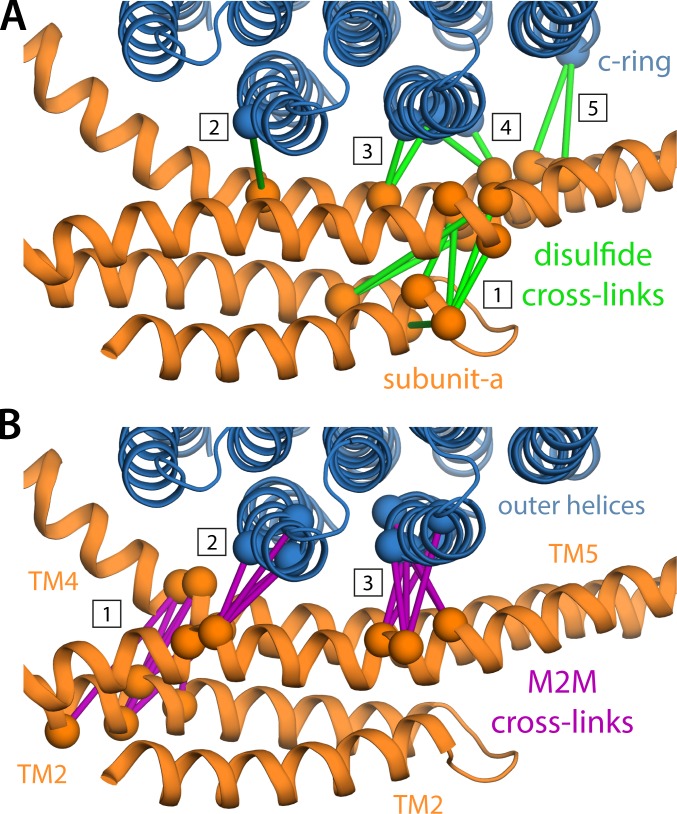
To illustrate this point, Fig. 8 highlights the residue pairs in our structure of the Polytomella a–c complex that are equivalent to those cross-linked in E. coli, via either a direct disulfide bond or an M2M spacer; a quantitative analysis of corresponding Cα–Cα distances is also provided in Fig. S6. It is apparent that it is not feasible for all of these distances to take simultaneously the values that would be expected for each disulfide bond individually, even if we were to assume that our model is inaccurate by one full turn of a helix. If instead we were to allow for an extra 5 Å in the expected Cα–Cα distances, as prescribed by a previous meta-analysis of cross-linking data for MFS transporters (Radestock and Forrest, 2011), all residue pairs would be within range, with the exception of G85-E196 (corresponding to N148-H245 in E. coli), for which the cross-linking reaction was only partial (Schwem and Fillingame, 2006). In summary, despite the unexpected features implied by the cryo-EM data, we would argue that the structure of the a–c complex reported here is in good agreement with the cross-linking results, providing a pragmatic interpretation of this type of experimental data.
Figure 8.
Evaluation of the consistency of the proposed structure of the a–c complex with previous cross-linking studies of the E. coli ATP synthase. See Fig. S6 for a quantitative representation. (A) Green bars connect residues for which disulfide cross-links could be engineered, partially or fully, in the E. coli ATP synthase (see Table S2 and references therein). These cross-links cluster in five groups: cluster #1 pertains to the internal arrangement of the TM2-TM5 bundle, whereas clusters #2 to #5 reflect its orientation relative to the c-ring; note the latter are entirely consistent with the high tilt of TM5, relative to the outer helices in the c-ring. (B) Similarly to A, purple bars indicate cross-links between subunit a and the c-ring, mediated by an M2M spacer, which also cluster in three groups.
A similar conclusion may be drawn in regard to the comprehensive set of cysteine accessibility measurements and other biochemical assays that were conducted to locate the transmembrane spans of subunit a along the protein sequence (Valiyaveetil and Fillingame, 1998; Vik et al., 2000; Vik and Ishmukhametov, 2005; Schwem and Fillingame, 2006; Moore and Fillingame, 2008). These data appear to have been originally interpreted under the premise that these transmembrane spans ought to be comparable in length and arranged perpendicularly to the membrane. In hindsight, though, it is not evident that these assumptions necessarily stem from the experimental data. On the contrary, as illustrated in Fig. S7, it could be argued that the biochemical data are more consistent with the cryo-EM structure, though admittedly in a nontrivial way. Note, for example, the TM3-TM4 region: what the biochemical data indicates is that the C terminus of TM3 and much of the TM3-TM4 loop are exposed to the solvent on the cytoplasmic (matrix) side. The structure indicates that this pattern owes to the fact that this portion of the protein lines the aqueous crevice at the a–c interface; however, TM4 does not traverse the membrane from side to side, as originally anticipated (Fig. S7). The biochemical data are also consistent with the N terminus of TM5 being hardly exposed and with transmembrane spans TM4 and TM5 being at least four to five turns of a helix longer than in the “canonical” definition.
Analogously to the cross-linking data, therefore, idealized geometric interpretations of accessibility data and transmembrane predictions are prone to error. Indeed, a recent model of the E. coli subunit a based on an evolutionary analysis analogous to that used here resulted in an incorrect structure (Hopf et al., 2014), precisely because the modeling algorithm was biased to reproduce the transmembrane spans defined originally. For similar reasons, it is now apparent that the structure of the E. coli a–c complex publicly available in the Protein Data Bank (entry 1C17; Rastogi and Girvin, 1999) is incorrect. That structural model, used in a recent simulation study (Mukherjee and Warshel, 2012), was based on some of the abovementioned biochemical data and on pioneering solution-NMR experiments on the isolated c subunit, using chloroform-methanol extractions. Although these NMR studies provided groundbreaking insights, a mechanistically realistic three-dimensional model of the a–c complex is arguably beyond the scope of that data. Thus, a salient feature of that subunit a model, aside from the matter of the transmembrane tilt, is that it features a continuous open pathway through the center of the bundle, which would render unfeasible any kind of conformational mechanism coupled to the proton-motive force, let alone a rotary mechanism. As we have discussed in detail elsewhere (Zhou et al., 2016), we also find the c-ring structure in that model to be implausible, not least because it features the wrong number of c subunits (12 instead of 10). A stark difference with the c-ring model proposed here, and with available crystal structures, is that the conserved carboxyl side chain in the binding sites (Asp51 in E. coli) is buried in between the inner and outer helices of the subunits c, rather than being exposed on the surface of the c-ring (Fig. 2). In that case, as the c-ring rotates against subunit a, a large structural change would be required in each of the c subunits to permit the interaction of Asp51 with the conserved arginine in TM4 (R210 in E. coli). The cryo-EM map is at odds with this notion, in that it reveals no sign of a structural asymmetry in the c-ring; instead, the cryo-EM data, and our structure based thereon, appears to be more consistent with the hypothesis that the sequential engagement of this key interaction primarily involves a reconfiguration of the side chains at the a–c interface and rotational displacements of the c-ring as a whole (Fig. 7; Pogoryelov et al., 2010; Matthies et al., 2014).
Paradoxically, the conclusion that emerges from this retrospective analysis is that the types of sparse biochemical and bioinformatic data underlying the earlier models of subunit a do actually have significant predictive value; to derive their full value, however, it seems critical that direct structural information be also available, even at nanometer resolution, and that systematic molecular-modeling approaches are used to integrate all available data in an unbiased, quantitative manner that also accounts for the inherent uncertainties of each source.
Lastly, we note that the subunit a topology resulting from this objective analysis of the cryo-EM data for the Polytomella ATP synthase corrects the initial assignment of this data (Allegretti et al., 2015), which had TM4 and TM5 reversed. Our assignment, however, coincides with that subsequently proposed for the bovine enzyme, also based on cryo-EM data (Zhou et al., 2015), and for the synthase from Paracoccus denitrificans, from low-resolution diffraction data (Morales-Rios et al., 2015). Nevertheless, the specific threading of the transmembrane helices differs with respect to ours, and therefore the relative positions of mechanistically key residues also vary (Fig. S8). For example, in the model of the bovine enzyme (PDB entry 5FIL), the equivalent of Arg145 seems to project away from the a–c interface, rather than toward the nearest proton-binding site in the c-ring, whereas Glu194 seems to face that interface, rather than project toward the interior of subunit a, and the P channel. Whether this variability owes to the fact that these enzymes are from different organisms or to inaccuracies in existing models will become clearer when high-resolution structural data are available.
Molecular basis for bi-directional inhibition of the membrane motor
Owing to its fundamental role in cellular metabolism, the ATP synthase is increasingly seen as a potential pharmacological target, particularly in the context of last-resort treatments against dormant or resistant bacterial strains (Ahmad et al., 2013; Cook et al., 2014). Indeed, the latest FDA-approved drug against multi-drug–resistant tuberculosis, which affects millions of people worldwide, targets the membrane domain of the ATP synthase (Andries et al., 2005; Palomino and Martin, 2013; Lu et al., 2014; Preiss et al., 2015). However, the mechanism of action of this and other membrane-permeable inhibitors of the ATP synthase is not well understood, hampering efforts to develop more potent and specific drugs. Crystal structures of isolated c-rings have shown that antibiotics such as oligomycin, which inhibits the membrane sector of the mitochondrial ATP synthase (Kagawa and Racker, 1966; Hong and Pedersen, 2008), can recognize the proton-binding sites in the c-ring (Symersky et al., 2012a). Together with the finding that genetic mutations conferring antibiotic resistance localize on both subunits c and a (John and Nagley, 1986; Nagley et al., 1986), this observation has lent support to the notion that oligomycin and others inhibitors bind to one of the c-ring sites at the a–c interface, thereby blocking proton translocation and further rotation (Symersky et al., 2012a). It is puzzling, however, that these inhibitors halt the rotation of the c-ring not only when the enzyme operates as an ATP synthase, but also when the enzyme functions as an ATPase and the c-ring rotates in the opposite direction (Kagawa and Racker, 1966; Förster et al., 2010). For example, if oligomycin was able to bind only to the N site (Fig. 7), it would block the rotation of the ring counterclockwise, but clockwise rotation would foster dissociation. Indeed, inhibitors that block the rotation of the central stalk (Fig. S1), such as IF1, do so only in one direction for this reason (Cabezón et al., 2003). Why then, are the inhibitors of the membrane motor bi-directional?
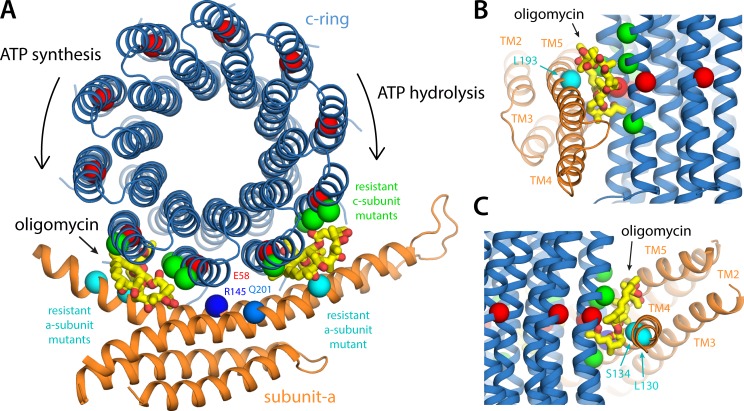
Our structure of the mitochondrial a–c complex offers a surprisingly simple but completely unexpected answer. If we model oligomycin molecules bound to the three c-ring binding sites closest to the a–c interface (i.e., those referred to above as N and P sites, plus that immediately adjacent) in the exact same conformation and pose observed in the abovementioned crystal structure of the S. cerevisiae c10-ring (Symersky et al., 2012a), we observe that the two sites at either side of the P site appear to be (at this resolution) sterically compatible with the inhibitor (Fig. 9). Furthermore, if we map the mutations on subunit a that are known to result in resistance to oligomycin inhibition, we find that these positions coincide exactly with those two c-ring binding sites. Collectively, these qualitative insights strongly indicate that oligomycin and possibly other inhibitors are able to block the rotary mechanism of the enzyme in a bi-directional manner because they can bind to two distinct binding sites at the a–c interface, namely at either side of the c-ring site engaged in the key glutamate-arginine salt bridge with subunit a.
Figure 9.
Insights into the mechanism of bi-directional inhibition of the ATP synthase by oligomycin. (A) Structure of the a–c complex, viewed as in Fig. 6 A, with two oligomycin molecules (yellow/red) bound to the c-ring. The binding pose of oligomycin is exactly that observed in a cocrystal structure of the c-ring from S. cerevisiae (in the absence of subunit a; see Results and discussion). This pose appears to be entirely feasible with two different sites at the a–c interface, to the left and to the right of the site where Glu58 interacts with Arg145 (i.e., the P site). An oligomycin molecule would not fit in this P site, implying that binding at the flanking sites shown would ultimately halt the rotation of the ring. The positions in the S. cerevisiae c and a subunits where mutations confer resistance to oligomycin inhibition (see Results and discussion) are marked with green and cyan spheres, respectively. These positions are Leu52, Ala55, Leu56, and Phe63 in the outer helix of the c subunit (Leu53, Ala56, Leu57, and Phe64 in S. cerevisiae), Leu130 and Ser134 in aTM4, and Leu193 in aTM5 (aILe171, aSer175, and aLeu232 in S. cerevisiae). (B) Side view of the oligomycin inhibition site when the c-ring rotates clockwise, transporting H+ uphill, driven by ATP hydrolysis. (C) Side view of the inhibition site when the c-ring rotates counterclockwise, driven by downhill H+ permeation, during ATP synthesis.
Conclusions
The ATP synthase is a highly paradigmatic system. Detailed investigations of its structure and mechanism have and will continue to reveal insights and principles that are broadly relevant to other enzymes, other molecular motors, and other membrane transport systems. Studies of this important system are also expanding our understanding of the seemingly crucial role that some proteins have in defining the morphology of the membranes where they reside (Davies et al., 2012), and they might also foster the development of novel last-resort antibiotics (Lu et al., 2014). Recent breakthroughs in the cryo-EM field have advanced our understanding of the structure of this fascinating protein enormously, particularly for the membrane motor, which has been the focus of this study. However, the cryo-EM structures have also seemingly put in question the conclusions derived from a myriad of biochemical and biophysical studies performed earlier. Here, we have used systematic molecular-modeling methods and bioinformatics analyses to arrive at what we believe is an objective interpretation of the available cryo-EM data and to show that it is also quantifiably consistent with the wealth of biochemical and functional information gathered over the past decades. Although at this point neither the experimental data or the computational tools permit us to define the structure of the membrane motor in atomic detail, the low-resolution structure of the a–c complex discussed here provides important insights into the mechanism that couples the translocation of protons to the rotation of the c-ring and into the mode of action of known bi-directional inhibitors.
Supplementary Material
Acknowledgments
We are thankful to Dr. Lucy Forrest for her comments on this manuscript and our progress throughout this study; to Dr. Karen Davies, Prof. Werner Kühlbrandt, and Prof. Thomas Meier for sharing and discussing their cryo-EM data before its publication; and above all, to Prof. Robert H. Fillingame for helping us to understand and integrate his pioneering biochemical work on the E. coli ATP synthase into this study.
This study was funded by the Division of Intramural Research of the National Heart, Lung, and Blood Institute, National Institutes of Health and previously by the Max Planck Society.
The authors declare no competing financial interests.
Toby Allen served as guest editor.
Footnotes
Abbreviation used:
- EM
- electron microscopy
References
- Ahmad Z., Okafor F., Azim S., and Laughlin T.F.. 2013. ATP synthase: A molecular therapeutic drug target for antimicrobial and antitumor peptides. Curr. Med. Chem. 20:1956–1973. 10.2174/0929867311320150003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti M., Klusch N., Mills D.J., Vonck J., Kühlbrandt W., and Davies K.M.. 2015. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 521:237–240. 10.1038/nature14185 [DOI] [PubMed] [Google Scholar]
- Andries K., Verhasselt P., Guillemont J., Göhlmann H.W., Neefs J.M., Winkler H., Van Gestel J., Timmerman P., Zhu M., Lee E., et al. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 307:223–227. 10.1126/science.1106753 [DOI] [PubMed] [Google Scholar]
- Angevine C.M., Herold K.A., Vincent O.D., and Fillingame R.H.. 2007. Aqueous access pathways in ATP synthase subunit a. Reactivity of cysteine substituted into transmembrane helices 1, 3, and 5. J. Biol. Chem. 282:9001–9007. 10.1074/jbc.M610848200 [DOI] [PubMed] [Google Scholar]
- Bae L., and Vik S.B.. 2009. A more robust version of the Arginine 210-switched mutant in subunit a of the Escherichia coli ATP synthase. Biochim. Biophys. Acta. 1787:1129–1134. 10.1016/j.bbabio.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Ju T., and Chiu W.. 2007. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 15:7–19. 10.1016/j.str.2006.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Zhang J., Ludtke S.J., and Chiu W.. 2010. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat. Protoc. 5:1697–1708. 10.1038/nprot.2010.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón E., Montgomery M.G., Leslie A.G., and Walker J.E.. 2003. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 10:744–750. 10.1038/nsb966 [DOI] [PubMed] [Google Scholar]
- Cain B.D., and Simoni R.D.. 1988. Interaction between Glu-219 and His-245 within the a subunit of F1FO-ATPase in Escherichia coli. J. Biol. Chem. 263:6606–6612. [PubMed] [Google Scholar]
- Cain B.D., and Simoni R.D.. 1989. Proton translocation by the F1FOATPase of Escherichia coli. Mutagenic analysis of the a subunit. J. Biol. Chem. 264:3292–3300. [PubMed] [Google Scholar]
- Cook G.M., Greening C., Hards K., and Berney M.. 2014. Energetics of pathogenic bacteria and opportunities for drug development. Adv. Microb. Physiol. 65:1–62. 10.1016/bs.ampbs.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Davies K.M., Anselmi C., Wittig I., Faraldo-Gómez J.D., and Kühlbrandt W.. 2012. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. USA. 109:13602–13607. 10.1073/pnas.1204593109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. III, Snoeyink J., Richardson J.S., and Richardson D.C.. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383. 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F., Tyka M.D., Baker M.L., Chiu W., and Baker D.. 2009. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392:181–190. 10.1016/j.jmb.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., and Fillingame R.H.. 2010. Chemical reactivities of cysteine substitutions in subunit a of ATP synthase define residues gating H+ transport from each side of the membrane. J. Biol. Chem. 285:39811–39818. 10.1074/jbc.M110.175844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher T., Seeger M.A., Anselmi C., Zhou W., Brandstätter L., Verrey F., Diederichs K., Faraldo-Gómez J.D., and Pos K.M.. 2014. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 3:e03145 10.7554/eLife.03145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame R.H., and Steed P.R.. 2014. Half channels mediating H+ transport and the mechanism of gating in the Fo sector of Escherichia coli F1Fo ATP synthase. Biochim. Biophys. Acta. 1837:1063–1068. 10.1016/j.bbabio.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Fiser A., and Sali A.. 2003. MODELLER: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461–491. 10.1016/S0076-6879(03)74020-8 [DOI] [PubMed] [Google Scholar]
- Forrest L.R., Tang C.L., and Honig B.. 2006. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys. J. 91:508–517. 10.1529/biophysj.106.082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster K., Turina P., Drepper F., Haehnel W., Fischer S., Gräber P., and Petersen J.. 2010. Proton transport coupled ATP synthesis by the purified yeast H+-ATP synthase in proteoliposomes. Biochim. Biophys. Acta. 1797:1828–1837. 10.1016/j.bbabio.2010.07.013 [DOI] [PubMed] [Google Scholar]
- Hahn A., Parey K., Bublitz M., Mills D.J., Zickermann V., Vonck J., Kühlbrandt W., and Meier T.. 2016. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell. 63:445–456. 10.1016/j.molcel.2016.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., and Pedersen P.L.. 2008. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 72:590–641. 10.1128/MMBR.00016-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A., Colwell L.J., Sheridan R., Rost B., Sander C., and Marks D.S.. 2012. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 149:1607–1621. 10.1016/j.cell.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A., Schärfe C.P.I., Rodrigues J.P.G.L.M., Green A.G., Kohlbacher O., Sander C., Bonvin A.M.J.J., and Marks D.S.. 2014. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife. 3:e03430 10.7554/eLife.03430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmukhametov R.R., Pond J.B., Al-Huqail A., Galkin M.A., and Vik S.B.. 2008. ATP synthesis without R210 of subunit a in the Escherichia coli ATP synthase. Biochim. Biophys. Acta. 1777:32–38. 10.1016/j.bbabio.2007.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., and Fillingame R.H.. 1998. Interacting helical faces of subunits a and c in the F1Fo ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc. Natl. Acad. Sci. USA. 95:6607–6612. 10.1073/pnas.95.12.6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U.P., and Nagley P.. 1986. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 207:79–83. 10.1016/0014-5793(86)80016-3 [DOI] [PubMed] [Google Scholar]
- Ju T., Baker M.L., and Chiu W.. 2007. Computing a family of skeletons of volumetric models for shape description. Comput. Aided Des. 39:352–360. 10.1016/j.cad.2007.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge W., Lill H., and Engelbrecht S.. 1997. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22:420–423. 10.1016/S0968-0004(97)01129-8 [DOI] [PubMed] [Google Scholar]
- Kagawa Y., and Racker E.. 1966. Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase. J. Biol. Chem. 241:2461–2466. [PubMed] [Google Scholar]
- Kaila V.R., Wikström M., and Hummer G.. 2014. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc. Natl. Acad. Sci. USA. 111:6988–6993. 10.1073/pnas.1319156111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah A., Pogoryelov D., Langer J.D., Bond P.J., Meier T., and Faraldo-Gómez J.D.. 2010a Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase Fo rotors. Biochim. Biophys. Acta. 1797:763–772. 10.1016/j.bbabio.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Krah A., Pogoryelov D., Meier T., and Faraldo-Gómez J.D.. 2010b On the structure of the proton-binding site in the Fo rotor of chloroplast ATP synthases. J. Mol. Biol. 395:20–27. 10.1016/j.jmb.2009.10.059 [DOI] [PubMed] [Google Scholar]
- Leone V., Krah A., and Faraldo-Gómez J.D.. 2010. On the question of hydronium binding to ATP-synthase membrane rotors. Biophys. J. 99:L53–L55. 10.1016/j.bpj.2010.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V., Pogoryelov D., Meier T., and Faraldo-Gómez J.D.. 2015. On the principle of ion selectivity in Na+/H+-coupled membrane proteins: experimental and theoretical studies of an ATP synthase rotor. Proc. Natl. Acad. Sci. USA. 112:E1057–E1066. 10.1073/pnas.1421202112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers R.N., Howitt S.M., Hatch L., Gibson F., and Cox G.B.. 1987. The proton pore in the Escherichia coli F0F1-ATPase: a requirement for arginine at position 210 of the a-subunit. Biochim. Biophys. Acta. 894:399–406. 10.1016/0005-2728(87)90118-6 [DOI] [PubMed] [Google Scholar]
- Lomize M.A., Lomize A.L., Pogozheva I.D., and Mosberg H.I.. 2006. OPM: orientations of proteins in membranes database. Bioinformatics. 22:623–625. 10.1093/bioinformatics/btk023 [DOI] [PubMed] [Google Scholar]
- Lu P., Lill H., and Bald D.. 2014. ATP synthase in mycobacteria: special features and implications for a function as drug target. Biochim. Biophys. Acta. 1837:1208–1218. 10.1016/j.bbabio.2014.01.022 [DOI] [PubMed] [Google Scholar]
- Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R., and Sander C.. 2011. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 6:e28766 10.1371/journal.pone.0028766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D.S., Hopf T.A., and Sander C.. 2012. Protein structure prediction from sequence variation. Nat. Biotechnol. 30:1072–1080. 10.1038/nbt.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies D., Zhou W., Klyszejko A.L., Anselmi C., Yildiz Ö., Brandt K., Müller V., Faraldo-Gómez J.D., and Meier T.. 2014. High-resolution structure and mechanism of an F/V-hybrid rotor ring in a Na+-coupled ATP synthase. Nat. Commun. 5:5286 10.1038/ncomms6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier T., Polzer P., Diederichs K., Welte W., and Dimroth P.. 2005. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 308:659–662. 10.1126/science.1111199 [DOI] [PubMed] [Google Scholar]
- Meier T., Krah A., Bond P.J., Pogoryelov D., Diederichs K., and Faraldo-Gómez J.D.. 2009. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 391:498–507. 10.1016/j.jmb.2009.05.082 [DOI] [PubMed] [Google Scholar]
- Meier T., Faraldo-Gómez J.D., and Börsch M.. 2011. ATP synthase - A paradigmatic molecular machine. In Molecular Machines in Biology. Frank J., editor. Cambridge University Press, Cambridge, UK: 208–238. 10.1017/CBO9781139003704.013 [DOI] [Google Scholar]
- Melo F., and Sali A.. 2007. Fold assessment for comparative protein structure modeling. Protein Sci. 16:2412–2426. 10.1110/ps.072895107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitome N., Ono S., Sato H., Suzuki T., Sone N., and Yoshida M.. 2010. Essential arginine residue of the Fo-a subunit in FoF1-ATP synthase has a role to prevent the proton shortcut without c-ring rotation in the Fo proton channel. Biochem. J. 430:171–177. 10.1042/BJ20100621 [DOI] [PubMed] [Google Scholar]
- Moore K.J., and Fillingame R.H.. 2008. Structural interactions between transmembrane helices 4 and 5 of subunit a and the subunit c ring of Escherichia coli ATP synthase. J. Biol. Chem. 283:31726–31735. 10.1074/jbc.M803848200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.J., Angevine C.M., Vincent O.D., Schwem B.E., and Fillingame R.H.. 2008. The cytoplasmic loops of subunit a of Escherichia coli ATP synthase may participate in the proton translocating mechanism. J. Biol. Chem. 283:13044–13052. 10.1074/jbc.M800900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Rios E., Montgomery M.G., Leslie A.G., and Walker J.E.. 2015. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc. Natl. Acad. Sci. USA. 112:13231–13236. 10.1073/pnas.1517542112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., and Warshel A.. 2012. Realistic simulations of the coupling between the protomotive force and the mechanical rotation of the F0-ATPase. Proc. Natl. Acad. Sci. USA. 109:14876–14881. 10.1073/pnas.1212841109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagley P., Hall R.M., and Ooi B.G.. 1986. Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance. FEBS Lett. 195:159–163. 10.1016/0014-5793(86)80152-1 [DOI] [PubMed] [Google Scholar]
- Palomino J.C., and Martin A.. 2013. TMC207 becomes bedaquiline, a new anti-TB drug. Future Microbiol. 8:1071–1080. 10.2217/fmb.13.85 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pogoryelov D., Yildiz O., Faraldo-Gómez J.D., and Meier T.. 2009. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 16:1068–1073. 10.1038/nsmb.1678 [DOI] [PubMed] [Google Scholar]
- Pogoryelov D., Krah A., Langer J.D., Yildiz Ö., Faraldo-Gómez J.D., and Meier T.. 2010. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat. Chem. Biol. 6:891–899. 10.1038/nchembio.457 [DOI] [PubMed] [Google Scholar]
- Pogoryelov D., Klyszejko A.L., Krasnoselska G.O., Heller E.M., Leone V., Langer J.D., Vonck J., Müller D.J., Faraldo-Gómez J.D., and Meier T.. 2012. Engineering rotor ring stoichiometries in the ATP synthase. Proc. Natl. Acad. Sci. USA. 109:E1599–E1608. 10.1073/pnas.1120027109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss L., Yildiz O., Hicks D.B., Krulwich T.A., and Meier T.. 2010. A new type of proton coordination in an F1Fo-ATP synthase rotor ring. PLoS Biol. 8:e1000443 (published errata appear in PLoS Biol. 2010.) 10.1371/journal.pbio.1000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss L., Langer J.D., Yildiz Ö., Eckhardt-Strelau L., Guillemont J.E., Koul A., and Meier T.. 2015. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci. Adv. 1:e1500106 10.1126/sciadv.1500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radestock S., and Forrest L.R.. 2011. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 407:698–715. 10.1016/j.jmb.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Rastogi V.K., and Girvin M.E.. 1999. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 402:263–268. 10.1038/46224 [DOI] [PubMed] [Google Scholar]
- Remmert M., Biegert A., Hauser A., and Söding J.. 2012. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 9:173–175. 10.1038/nmeth.1818 [DOI] [PubMed] [Google Scholar]
- Richardson J.S. 1981. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34:167–339. 10.1016/S0065-3233(08)60520-3 [DOI] [PubMed] [Google Scholar]
- Schulz S., Iglesias-Cans M., Krah A., Yildiz O., Leone V., Matthies D., Cook G.M., Faraldo-Gómez J.D., and Meier T.. 2013. A new type of Na+-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biol. 11:e1001596 10.1371/journal.pbio.1001596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwem B.E., and Fillingame R.H.. 2006. Cross-linking between helices within subunit a of Escherichia coli ATP synthase defines the transmembrane packing of a four-helix bundle. J. Biol. Chem. 281:37861–37867. 10.1074/jbc.M607453200 [DOI] [PubMed] [Google Scholar]
- Shen M.Y., and Sali A.. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15:2507–2524. 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed P.R., and Fillingame R.H.. 2008. Subunit a facilitates aqueous access to a membrane-embedded region of subunit c in Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 283:12365–12372. 10.1074/jbc.M800901200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed P.R., and Fillingame R.H.. 2009. Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 284:23243–23250. 10.1074/jbc.M109.002501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed P.R., and Fillingame R.H.. 2014. Residues in the polar loop of subunit c in Escherichia coli ATP synthase function in gating proton transport to the cytoplasm. J. Biol. Chem. 289:2127–2138. 10.1074/jbc.M113.527879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky J., Osowski D., Walters D.E., and Mueller D.M.. 2012a Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA. 109:13961–13965. 10.1073/pnas.1207912109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky J., Pagadala V., Osowski D., Krah A., Meier T., Faraldo-Gómez J.D., and Mueller D.M.. 2012b Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 19:485–491: S1. 10.1038/nsmb.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil F.I., and Fillingame R.H.. 1997. On the role of Arg-210 and Glu-219 of subunit a in proton translocation by the Escherichia coli F0F1-ATP synthase. J. Biol. Chem. 272:32635–32641. 10.1074/jbc.272.51.32635 [DOI] [PubMed] [Google Scholar]
- Valiyaveetil F.I., and Fillingame R.H.. 1998. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 273:16241–16247. 10.1074/jbc.273.26.16241 [DOI] [PubMed] [Google Scholar]
- van Lis R., Atteia A., Mendoza-Hernández G., and González-Halphen D.. 2003. Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii. A proteomic approach. Plant Physiol. 132:318–330. 10.1104/pp.102.018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik S.B., and Antonio B.J.. 1994. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 269:30364–30369. [PubMed] [Google Scholar]
- Vik S.B., and Ishmukhametov R.R.. 2005. Structure and function of subunit a of the ATP synthase of Escherichia coli. J. Bioenerg. Biomembr. 37:445–449. 10.1007/s10863-005-9488-6 [DOI] [PubMed] [Google Scholar]
- Vik S.B., Cain B.D., Chun K.T., and Simoni R.D.. 1988. Mutagenesis of the alpha subunit of the F1Fo-ATPase from Escherichia coli. Mutations at Glu-196, Pro-190, and Ser-199. J. Biol. Chem. 263:6599–6605. [PubMed] [Google Scholar]
- Vik S.B., Patterson A.R., and Antonio B.J.. 1998. Insertion scanning mutagenesis of subunit a of the F1F0 ATP synthase near His245 and implications on gating of the proton channel. J. Biol. Chem. 273:16229–16234. 10.1074/jbc.273.26.16229 [DOI] [PubMed] [Google Scholar]
- Vik S.B., Long J.C., Wada T., and Zhang D.. 2000. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim. Biophys. Acta. 1458:457–466. 10.1016/S0005-2728(00)00094-3 [DOI] [PubMed] [Google Scholar]
- Walker J.E. 2013. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41:1–16. 10.1042/BST20110773 [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V., Schonbrun J., and Baker D.. 2006. Multipass membrane protein structure prediction using Rosetta. Proteins. 62:1010–1025. 10.1002/prot.20817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Rohou A., Schep D.G., Bason J.V., Montgomery M.G., Walker J.E., Grigorieff N., and Rubinstein J.L.. 2015. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 4:e10180 10.7554/eLife.10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Leone V., and Faraldo-Gómez J.D.. 2016. Predicted structures of the proton-bound membrane-embedded rotor rings of the Saccharomyces cerevisiae and Escherichia coli ATP synthases. J. Phys. Chem. B. 10.1021/acs.jpcb.6b08051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.