Abstract
The eukaryotic multi-subunit RNA exosome complex plays crucial roles in 3′-to-5′ RNA processing and decay. Rrp6 and Ski7 are the major cofactors for the nuclear and cytoplasmic exosomes, respectively. In the cytoplasm, Ski7 helps the exosome to target mRNAs for degradation and turnover via a through-core pathway. However, the interaction between Ski7 and the exosome complex has remained unclear. The transaction of RNA substrates within the exosome is also elusive. In this work, we used single-particle cryo-electron microscopy to solve the structures of the Ski7-exosome complex in RNA-free and RNA-bound forms at resolutions of 4.2 Å and 5.8 Å, respectively. These structures reveal that the N-terminal domain of Ski7 adopts a structural arrangement and interacts with the exosome in a similar fashion to the C-terminal domain of nuclear Rrp6. Further structural analysis of exosomes with RNA substrates harboring 3′ overhangs of different length suggests a switch mechanism of RNA-induced exosome activation in the through-core pathway of RNA processing.
Keywords: exosome, RNA decay, Ski7, Rrp6, cryo-EM
Introduction
In eukaryotic cells, almost all RNA transcripts are under the surveillance of the RNA quality control system, which removes aberrant, misfolded, or damaged RNA molecules by inducing their degradation1,2. Whereas the 5′-to-3′ decay of RNA substrates is mainly mediated by the Xrn1 and Rat1 exonucleases, the 3′-to-5′ RNA degradation is executed by the exosome, a multi-subunit RNA degradation complex that trims RNA substrates continuously at their 3′ end3. Functional studies have revealed that the exosome is a critical player in the turnover of almost all RNA species, including mRNAs, rRNAs, tRNAs and other non-coding RNAs in both the nucleus and the cytoplasm4. The exosome is also responsible for removal of the 3′ tail of many RNA species during their maturation in the nucleus5.
The eukaryotic exosome complex shares a homologous 9-subunit core complex with the prokaryotic PNPase and the archaeal exosome complexes6,7,8,9. All have a barrel-shaped structure with a region at the top for RNA substrate recruitment and a central channel allowing single-stranded RNA substrate to access. Whereas the PNPase and the archaeal exosome both have phosphorolytic exoribonuclease activity on the inner surface of the channel, the eukaryotic exosome core has completely lost its exoribonuclease activity during evolution, though it has preserved the architecture. The sole RNase activity of the eukaryotic exosome comes from the tenth protein, Rrp44 (Dis3), which attaches to the bottom of the core complex. Rrp44 is a multi-domain protein with a processive hydrolytic exonuclease activity at its C-terminal region, which is homologous to bacterial RNase II, and a relatively weak endonuclease activity at its N-terminal region10,11,12. Previous structural studies have shown that Rrp44 specifically interacts with Rrp41, Rrp43 and Rrp45 of the core complex, aligning with the central channel's exit of the core complex13,14,15. The Rrp44-bound exosome complex is considered to be an important active core complex in eukaryotes and is termed Exo1016.
The architecture of Exo10 suggests multiple pathways for RNA substrates to be processed by the complex13. Similar to its prokaryotic and archaeal homologs, the eukaryotic Exo10 can recruit RNA substrates with single-stranded 3′ overhangs via the RNA-binding proteins at the top of its core and move the substrates entirely through the central channel and to the RNase active site of Rrp44 below the core. This through-core pathway, supported by the crystal structure of the holo-enzyme bound to an RNA substrate14, requires the RNA substrate to have a single-stranded 3′ overhang longer than 30 nt to reach the Rrp44 exonuclease active site from the entry point of the core. The solvent exposure of the Rrp44 active site in the apo structure of Exo10 also suggests another pathway where RNA substrates reach the Rrp44 active site without passing through the central channel of the core. This direct-access pathway does not require RNA substrates to have very long single-stranded 3′ overhangs and enables Exo10 to process RNAs with a tertiary structure17. Interestingly, our previous structural studies have indicated that Exo10 adopts distinct conformations in the through-core and direct-access pathways, of which the latter is very similar to the apo-state of the complex17. Our previous studies have also indicated that the through-core conformation of Exo10 is very likely to be induced by the RNA substrate within the central channel. How this conformational switch is triggered is not known yet.
Multiple protein cofactors have been found to regulate the cellular localization, RNA substrate specificity and activity of the exosome in vivo. In the best studied Saccharomyces cerevisiae system, Rrp6, an RNase D-type enzyme with distributive exonuclease activity, interacts physically and functionally with the exosome in the nucleus18. Rrp6 has been revealed to interact with the Exo10 complex at the top region, next to the entry site of the core14,15,19. In the cytoplasm, Ski7, a putative GTPase, bridges the exosome with the ribosome complex and the Ski2/3/8 complex, a cofactor complex, thus promoting recognition and degradation of aberrant mRNAs by the exosome20. Ski7 has been suggested to interact with Csl4 at the top of the Exo10 core21,22. To date, no structural information about the assembly of Exo10 with Ski7 is available.
In this work, we used single-particle cryo-electron microscopy (cryo-EM) to obtain the structures of the Exo10-Ski7 complexes in RNA-free and endogenous RNA-bound forms at resolutions of 4.2 Å and 5.8 Å, respectively. The high-resolution structures allowed us to depict the conformational difference between the two states at a near-atomic resolution level, providing insights into the mechanism of RNA-induced conformational change that activates the through-core RNA processing pathway. Furthermore, our structural analysis revealed that the N-terminal domain of Ski7 adopts a structural arrangement and interacts with the exosome core complex in a similar fashion to the C-terminal domain of Rrp6. This explains how distinct exosome complexes in vivo are exclusively distinguished in different RNA processing events.
Results
Purification of the endogenous Exo10-Ski7 complex
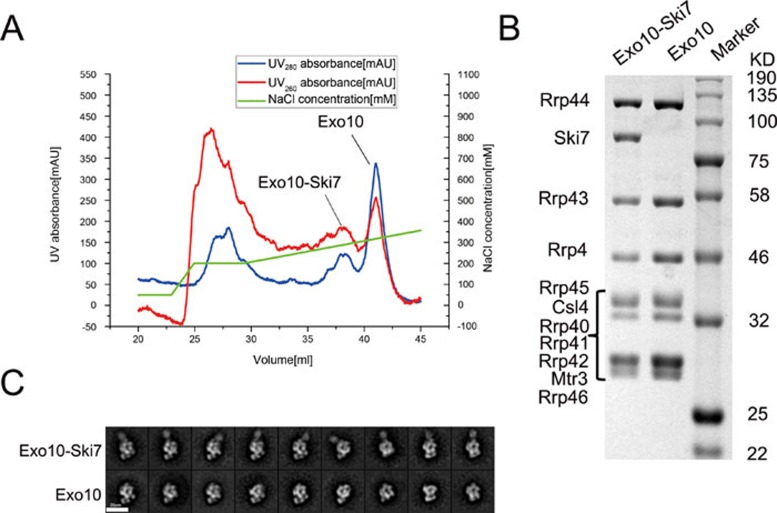
In our previous studies, we purified the endogenous Exo10 complex from an RRP6-knockout S. cerevisiae strain with TAP-tagged Rrp4613,17. Using the same protocol except for a minor modification at the washing step of the tandem affinity purification procedure, we recovered the exosome complex after elution from the IgG beads. Using Mono QTM ion-exchange chromatography, we were able to separate the Exo10 and Exo10-Ski7 complexes as indicated by the measurement of UV absorbances at 260 and 280 nm and the SDS-PAGE analysis (Figure 1A and 1B). Mass spectrometry verified that the Ski7-containing fraction was the Exo10-Ski7 complex, with Ski7 in a stoichiometric ratio with the other Exo10 components (Supplementary information, Tables S1 and S2).
Figure 1.
Exo10-Ski7 complex purification. (A) Chromatography of the Exo10-Ski7 and Exo10 complexes eluted from a Mono QTM ion-exchange column. Blue and red curves represent the UV absorbances at 280 nm and 260 nm, respectively. The green curve represents the corresponding concentration of NaCl in the elution. (B) SDS-PAGE of the Exo10-Ski7 and Exo10 fractions collected in ion-exchange chromatography. Protein components of the complexes are indicated on the left. The molecular weight of the standard marker is labeled on the right. (C) 2D class averages of the Exo10-Ski7 and Exo10 complexes by negative-stain EM. The additional density corresponding to Ski7 is clearly visible in the class averages in the upper row.
We used negative-stain EM to examine the Exo10-Ski7 and Exo10 complexes separated by ion-exchange chromatography. Both complexes were mono-disperse under EM and hence were suitable for single-particle two-dimensional (2D) classification analysis. As expected, the 2D class averages of the Exo10 complex showed classical Exo10 structures, as previously observed13,17. In contrast, the 2D class averages of the Exo10-Ski7 complex revealed a prominent additional density attached to the top of the Exo10 core (Figure 1C). Because mass spectrometry demonstrates that the Exo10-Ski7 fraction only contains Ski7 and components of the Exo10 complex, the additional density should correspond to the Ski7 protein and the RNA attached to it. The observation that the Ski7 protein sits on top of the Exo10 core complex corroborates the previous discovery that Ski7 interacts with Csl4, an RNA-binding protein located at the top region of the core complex21. In the 2D class averages, the additional density exhibited a globular shape, probably corresponding to the C-terminal GTPase domain of Ski723. The different orientations of the globular density relative to Exo10 in different 2D class averages indicate a flexible interaction of this part with the core complex.
Cryo-EM structure of the Exo10-Ski7 complex
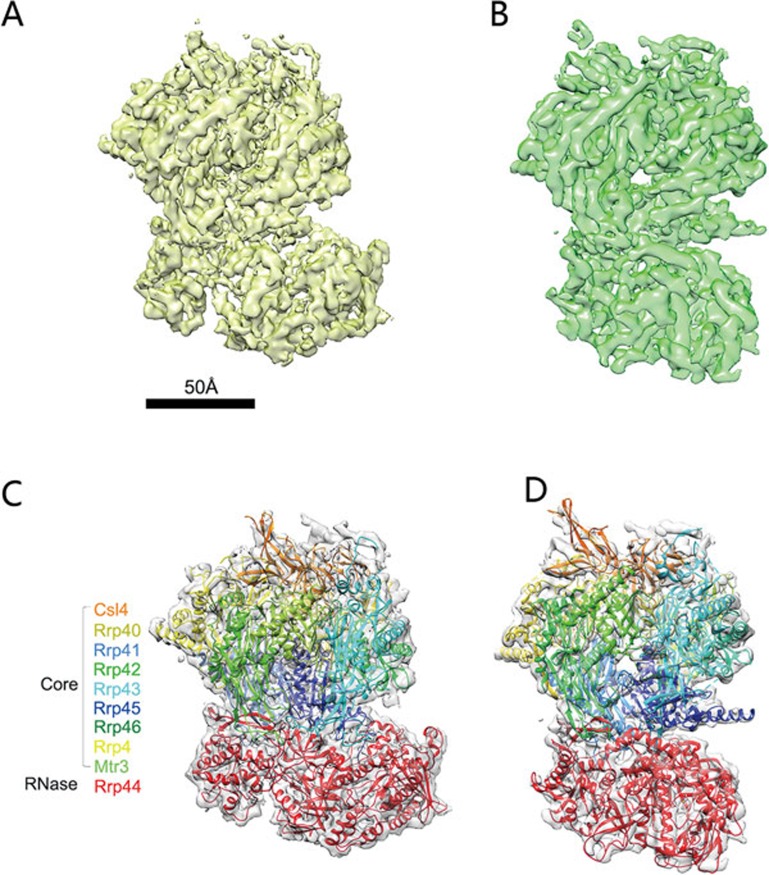
To analyze the high-resolution structure of the Exo10-Ski7 complex, we performed cryo-EM analysis of the Exo10-Ski7 complex by using a Titan Krios electron microscope equipped with a K2 Summit direct electron counting camera. This allowed us to detect fine details in the 2D class averages of the complex, calculated by the RELION software (Supplementary information, Figure S1). However, we did not observe any additional density on top of the core in the 2D class averages, which was observed in the negative-stain EM. Further classification of the particle images with the IMAGIC-4D image processing package revealed some additional weak densities attached to the top of the exosome (Supplementary information, Figure S1D), again reflecting the flexibility of Ski7's interaction with Exo10. We performed three-dimensional (3D) classification of the data set and found that there were primarily two types of conformations of the complex (Supplementary information, Figure S2A), resembling what we have previously observed as the apo and the RNA-bound states of Exo1017. Further refinement of the two populations of particles yielded a reconstruction of the complexes in apo-like and RNA-bound-like states at resolutions of 4.2 Å and 5.8 Å, respectively (Figure 2A, 2B and Supplementary information, Figure S2).
Figure 2.
3D cryo-EM reconstruction and atomic model of the Exo10-Ski7 complex in RNA-free and endogenous RNA-bound states. (A) 3D reconstruction of the RNA-free Exo10-Ski7 complex at 4.2 Å resolution. (B) 3D reconstruction of the endogenous RNA-bound Exo10-Ski7 complex at 5.8 Å resolution. (C) Atomic model of the Exo10 complex built in the 3D EM map of the RNA-free Exo10-Ski7 complex. (D) Docking of the atomic model of the Exo10 portion from the crystal structure of the RNA-bound Exo10-Rrp6 complex (PDB code: 4IFD) in the 3D EM map of the endogenous RNA-bound Exo10-Ski7 complex. The color scheme of the protein subunits in C and D is indicated in the left panel in C.
The much higher-resolution 3D cryo-EM reconstructions allowed us to visualize the details in the exosome complex in different states. We used the available atomic models of the reconstituted yeast Exo10-Rrp6-RNA complex (PDB code: 4IFD) and the reconstituted yeast sub-complex Rrp41-Rrp44-Rrp45 (PDB code: 2WP8) to build the atomic model of the apo-state Exo10 within the apo-like 3D map of the complex (Figure 2C). The model agreed with the EM map with a high cross-correlation coefficient of 0.9652, and some major side chains were depicted in the EM map (Supplementary information, Figure S2D and Movie S1). There are still some vacant portions of the map, in which the atomic models do not fit. We therefore calculated a difference map by subtracting the built-in atomic model of the apo-state Exo10 from the 4.2 Å apo-like 3D map (Supplementary information, Figure S3A). The difference map showed an extra density on top of the core complex (see Discussion), but did not show any additional densities along the central channel or close to the Ribo Nuclease Binding (RNB) domain of Rrp44, suggesting that the structure indeed represents an RNA-free Exo10-Ski7 complex. For the 5.8-Å RNA-bound-like Exo10-Ski7 EM map, we compared it with the atomic model of the reconstituted Exo10-Rrp6-RNA complex (PDB code: 4IFD) and found that the two structures highly resembled each other in the Exo10 portion. We extracted the Exo10 portion from the 4IFD coordinates and docked it into the 3D EM map. The docking was precise, with a cross-correlation coefficient of 0.9350, indicating the high fidelity between the EM and crystal structures (Figure 2D, Supplementary information, Figure S2C and Movie S2). We also calculated the difference map by subtracting the docked Exo10 model from the 3D EM map and found two stretches of prominent additional densities at the bottom of the central channel and in the activity pocket within the RNB domain of Rrp44 (Supplementary information, Figure S3B), in addition to a prominent density on top of the core complex (see Discussion). These results indicate that the 5.8 Å EM map represents an RNA-containing Exo10-Ski7 complex, and thus we designated this complex as endogenous RNA-bound Exo10-Ski7 complex.
Ski7 interaction with the Exo10 complex
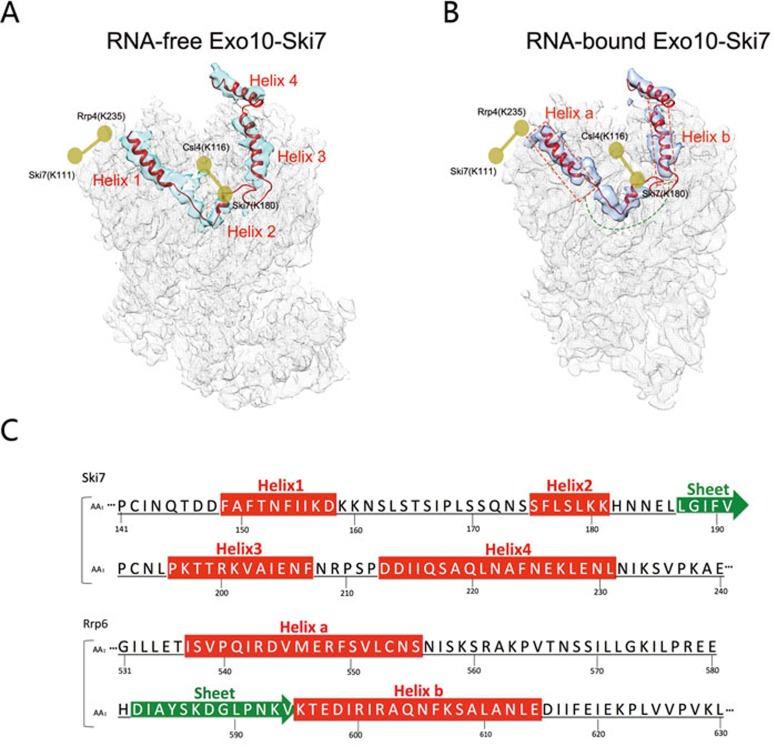
The difference maps calculated as described above from the RNA-free and RNA-bound Exo10-Ski7 EM maps had other prominent density features in addition to those within the central channel and the Rrp44 active site. A major additional density with a similar U-shaped structure in both maps is on the upper surface of the core complex (Supplementary information, Figure S3A and S3B). It is likely that this density, wrapping around the Csl4 and Rrp4 proteins of the core, corresponds to the N-terminal region of Ski7 (Figure 3A and 3B) as cross-linking mass spectrometry revealed cross-linked signals of the N-terminus of Ski7 with both Csl4 and Rrp4 (Supplementary information, Figure S4), in agreement with the density's location on the outer surface of the core. To further verify that this additional density corresponds to Ski7, we performed single-particle reconstruction of the Exo10 complex eluted in the second peak in the ion-exchange chromatography and obtained its 3D EM reconstruction at a resolution of 6.3 Å (Supplementary information, Figure S5). There was no additional density above the core complex of Exo10, which served well as the negative control.
Figure 3.
Architecture of the N-terminal portion of Ski7 binding with the exosome. (A) Difference map (solid cyan rendering) showing the Ski7 density on top of the RNA-free complex calculated by subtracting the atomic model of the RNA-free Exo10 from the RNA-free Exo10-Ski7 3D EM map (mesh rendering). (B) Difference map (solid blue rendering) showing the Ski7 density on top of the RNA-bound complex, calculated by subtracting the atomic model of Exo10 obtained from the RNA-Exo10-Rrp6 complex structure (PDB code: 4IFD) from the RNA-bound Exo10-Ski7 3D EM map (mesh rendering). In both A and B, the hypothetical atomic model of the N-terminal portion of Ski7 (residues 141-230; red) was docked into the difference map. The arrangement of the four helices is marked. The yellow ball and stick cartoons show the estimated positions of the two pairs of cross-linked residues between the Ski7 and Exo10 components. In B, the two red dashed boxes show the location of Rrp6 helices a and b sitting on Exo10, as determined in the structure of the RNA-Exo10-Rrp6 complex (PDB code: 4IFD). The green dashed line shows the location of residues 556-595 of Rrp6 on Exo10. (C) The secondary structure elements of Ski7 (residues 141-240) and Rrp6 (residues 531-630) that are responsible for the interaction with Exo10. The structural element arrangement of Ski7 was based on the secondary structure prediction by PSIPRED47, whereas that of Rrp6 was based on the RNA-Exo10-Rrp6 complex structure (PDB code: 4IFD).
Further examination of the density of Ski7's N-terminal region revealed that it shared similar features with Rrp6's C-terminal domain and even the similar interaction interface when bound to the core complex (Figure 3A and Supplementary information, Figure S3F). This unexpected structural similarity between the N-terminal portion of Ski7 and the C-terminal portion of Rrp6 motivated us to investigate the homology relationship between the Ski7 and Rrp6 proteins. Although no sequence homology was found between these two proteins, secondary structure prediction suggests the presence of several long helices within the Ski7 N-terminus (residues 141-240), with a similar distribution pattern as helices in the C-terminus of Rrp6 (residues 531-630) (Figure 3C). To determine whether Rrp6 and Ski7 competitively interact with Exo10, we purified the recombinant yeast Rrp6-Rrp47 complex from E. coli and performed binding assays of this complex with Exo10-Ski7- or Exo10-coated calmodulin beads. We found that excess Exo10 beads effectively pulled down all of the Rrp6-Rrp47 complexes from the supernatant, whereas the Exo10-Ski7 beads showed almost no binding of the Rrp6-Rrp47 complex (Supplementary information, Figure S3C). This result supports the hypothesis that Ski7 can effectively block the interaction of Rrp6 with Exo10.
Combining the secondary structural prediction, cross-linking results, and the feature of the difference maps, we were able to build the N-terminal region of Ski7 to fit into the U-shaped difference map as a hypothetical model (Figure 3A and 3B). This model indicates that Ski7 interacts with Csl4 and Rrp4 of the core complex mainly via residues 145-235 (Supplementary information, Figure S3D and S3E), consistent with a previous report21.
Conformational difference of Exo10-Ski7 complexes in the RNA-free and RNA-bound states
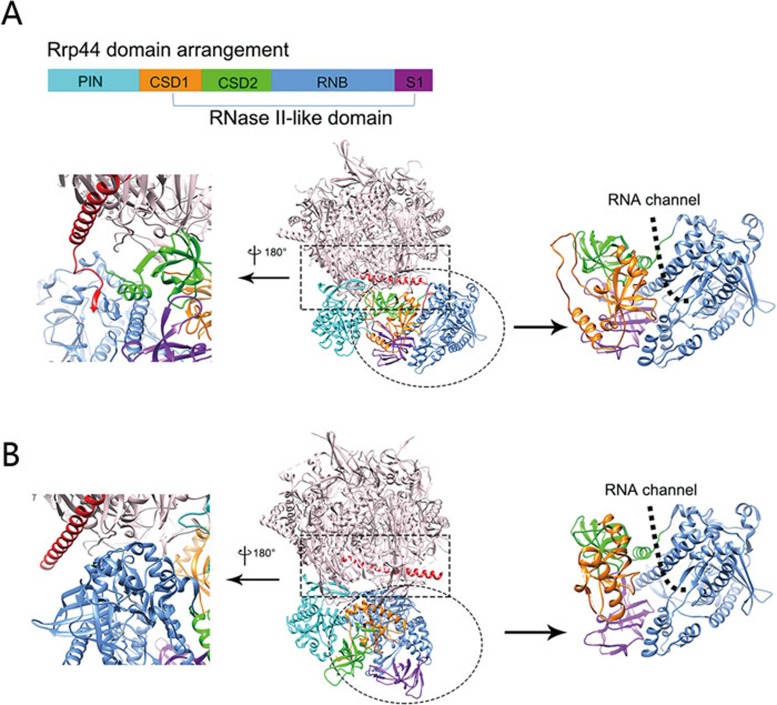
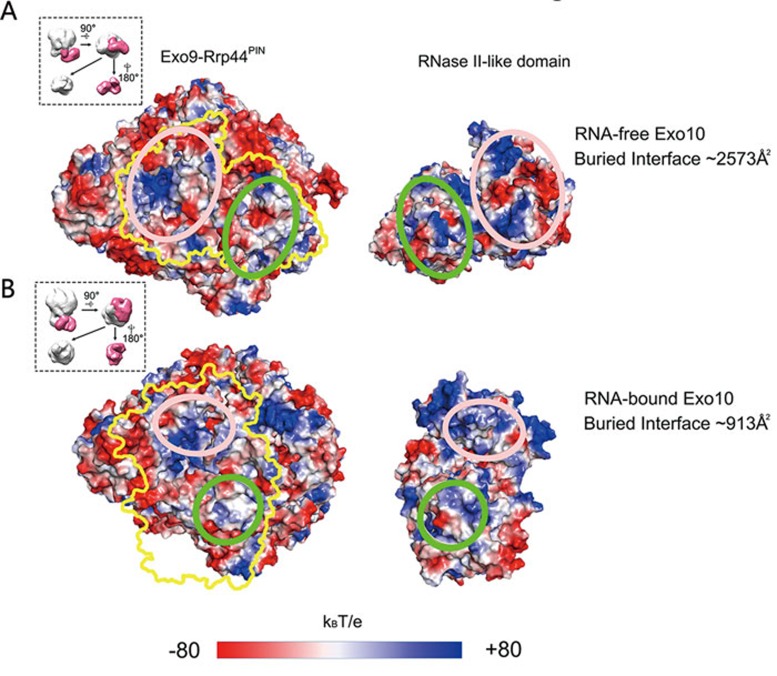
The atomic models of the RNA-free and RNA-bound Exo10-Ski7 complexes allowed us to elucidate the conformational differences between these two states in greater detail. The conformations of the core complex in these two states are very similar, except for Rrp44's RNase II-like domain, which comprises the CSD1, CSD2, RNB and S1 domains (Figure 4A). The major conformational difference between the two states occurred in the Rrp44 protein, in which the RNase II-like domain displays an inward and downward rotation relative to the core complex from the RNA-free state to the RNA-bound state (Figure 4A, 4B and Supplementary information, Movie S3). This rotation causes a change in the folding of the C-terminal tail of Rrp45. In the RNA-free complex, the C-terminal tail of Rrp45 (residues 297-303) inserts into a pocket between the CSD2 and RNB domains of Rrp44 and a newly formed β-strand element in Rrp45's C-terminal tail interacts with two strands within the RNB domain (Figure 4A). In the RNA-bound complex, this tail is released from the binding pocket and refolds to become the extension of Rrp45's C-terminal α-helix, which no longer interacts with Rrp44 (Figure 4B). A similar conformational change in the C-terminal tail of Rrp45 was observed through X-ray crystallography15. The dramatic conformational change of Rrp44's C-terminal portion also markedly decreased the interaction area between the RNase II-like domain and the rest of the Exo10 complex from ∼2 573 Å2 in the RNA-free state (Figure 5A) to ∼913 Å2 in the RNA-bound state (Figure 5B). Furthermore, the surface electrostatic potential distribution patterns of the interaction surfaces in the RNase II-like domain and the core complex were complementary with each other in the RNA-free complex (Figure 5A) but were rather repulsive in the RNA-bound complex (Figure 5B). All of the above results suggest that protein-protein interaction in the RNA-free complex is stronger than that in the RNA-bound complex. Therefore, in the RNA-bound state, the RNA substrate in the central channel spanning from the core to Rrp44 may play a major role in stabilizing the complex.
Figure 4.
Conformational change between the RNA-free and RNA-bound Exo10 complexes. The atomic models of the RNA-free Exo10 (A) and the RNA-bound Exo10 (B). In both models, the Exo9 core complex is in light pink, Rrp44 is labeled according to its domains: PIN in cyan, CSD1 in orange, CSD2 in green, RNB in blue, and S1 in purple, and the C-terminal helix of Rrp45 is in red. Rrp44's domain arrangement in the primary structure is shown in the upper panel of A. The overall atomic models of Exo10 in both states are shown in the center. A zoomed-in view of the interface between Rrp45 and Rrp44 boxed in the overall models is shown on the left with a 180-degree rotation. The zoomed-in view of the RNase II-like domains in both states with their RNB domains aligned is shown on the right. The RNA substrate recruitment path is shown as a black dashed line.
Figure 5.
The surface electrostatic potential distribution within the Exo9-Rrp44PIN and RNase II-like domain binding interface. The surface electrostatic potential distribution at the interface between the Exo9-Rrp44PIN and the RNase II-like domain of Rrp44 in RNA-free Exo10 (A) and RNA-bound Exo10 (B) complexes. For each complex structure, the RNase II-like domain of Rrp44 was dismantled from the Exo10 atomic model and rotated 180 degrees (as shown in the thumbnail panels) to reveal the interaction interface on the Exo9 core and Rrp44 surface. The interaction area of the RNase II-like domain on Exo9-Rrp44PIN is marked by a yellow contour. The oval circles with the same color in the pairing surfaces represent the same interface in the corresponding models.
We also observed another conformational change within the C-terminal portion of Rrp44. Compared with the RNA-free complex, the RNA-bound complex had its Rrp44-CSD1 domain moving away from the Rrp44-RNB domain, enlarging the RNA entry channel of Rrp44 so that its RNase active site is more accessible (right panels in Figure 4A, 4B and Supplementary information, Movie S4). There is also a loop (residues 705-720) within the RNB domain that moves away from the active site upon RNA binding (Supplementary information, Movie S4), as revealed earlier10. Therefore, Rrp44 undergoes an induced-fit enzymatic activation by its RNA substrates24.
Conversion between the two conformations induced by RNA
To understand the mechanism underlying the conformational conversion induced by RNA, we incubated the Exo10-Ski7 complex with RNA substrates in vitro and analyzed complex structures with cryo-EM. Our previous work has shown that the Exo10 complex can be purified in an RNA-free conformation but was converted into an RNA-bound conformation by single-stranded RNA substrates with 3′ overhangs longer than 14 nt in a buffer containing EDTA. In contrast, the Exo10-Ski7 complex purified in the present study adopted both conformations, suggesting a function of the Exo10-Ski7 complex in regulating mRNA degradation. We incubated the purified Exo10-Ski7 or Exo10 samples in a buffer containing 1 mM MgCl2 for 1.5 h on ice, and EM analysis revealed that this treatment converted all of the complex molecules into an RNA-free conformation, accompanied by the degradation of all endogenous RNAs (Supplementary information, Figure S6A and S6B). When we added RNA substrates with 3′ overhang of 48 nt (RNA48) to Mg2+-treated exosome sample and incubated the sample in the EDTA-containing buffer, all of the complex molecules adopted an RNA-bound conformation (Supplementary information, Figure S6H and S6I). These EM results verify that the conformational conversion is indeed solely induced by RNA substrates.
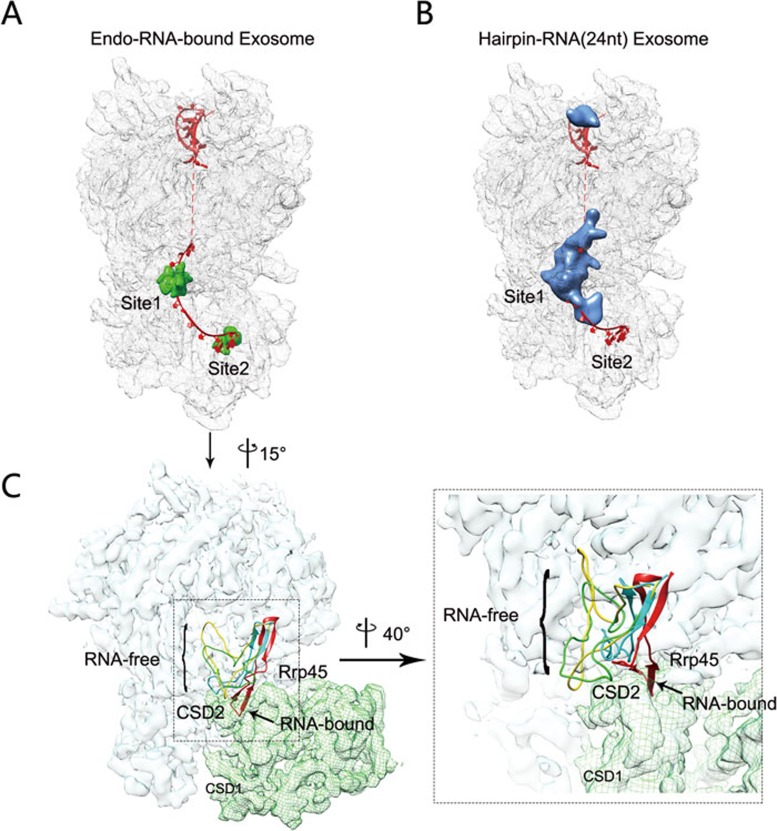
We further investigated the mechanism underlying RNA substrate-induced conformational change of the exosome complex. As shown above, the structure of the endogenous RNA-bound Exo10-Ski7 complex has two major densities corresponding to RNA molecules located at the bottom part of the central channel (Site 1) and the active site of Rrp44 (Site 2), respectively (Figure 6A and Supplementary information, Figure S3B). Because the reconstructed image was an average of many molecule images, the strong signal of RNA within the complex indicates that the presence of RNA in the complex is a common feature in most of the molecules contributing to the reconstruction, thus reflecting the stable interaction of the RNA substrates with the exosome proteins. We reasoned that these two sites might be responsible for RNA-induced conformational changes in the exosome. To determine which site is more critical for the conformational change, we designed two hairpin RNAs with single-stranded 3′ overhangs of 24 nt (RNA24) or 18 nt (RNA18), and examined their abilities to induce exosome conformational change. According to the crystal structure of the holo-Exo10-Rrp6-RNA complex14, RNA24 should be able to reach the bottom region of the central channel at Site 1 but should not reach Site 2, whereas the tail of RNA18 should be too short to even reach Site 1. We incubated these substrates with the Exo10 complex (in a preparation free of any endogenously bound RNAs, generated by the aforementioned MgCl2 treatment procedure) and performed single-particle EM analysis of the final complex. As predicted, RNA18 did not cause a conformational change in the Exo10 complex (Supplementary information, Figure S6C and S6D). In contrast, RNA24 incubation led to a major conformational alteration in the complex. Approximately 40% of the complexes adopted the RNA-bound conformation, whereas the rest appeared to have a very flexible arrangement in the RNase II-like domain of Rrp44 (Supplementary information, Figure S6E and S6F). Another round of 2D classification of images from the latter group by the IMAGIC-4D package revealed various orientations adopted by the RNase II-like domain relative to the core (Supplementary information, Figure S6G). We reconstructed the structure of the RNA24-Exo10 complex at a 6.8-Å resolution and calculated the difference map by subtracting the docked Exo10 atomic model from the 3D reconstruction (Figure 6B and Supplementary information, Figure S7). Compared with the endogenous RNA-bound Exo10-Ski7 complex, the RNA24-Exo10 complex had the additional density corresponding to RNA substrates only at Site 1 but not at Site 2 (Figure 6B). In summary, the above results from the analyses of RNA18- and RNA24-incubated exosome complexes indicate that Site 1 is both necessary and sufficient for the RNA substrate to induce a major conformational conversion of Exo10 from the RNA-free state to the RNA-bound state.
Figure 6.
Structural elements involved in the RNA-induced conformational change of Exo10 in the central channel pathway. (A) The additional density along the central channel, extracted from the difference map by subtracting the Exo10 atomic model (PDB code: 4IFD) from the 3D EM map of endogenous RNA-bound Exo10-Ski7 complex (mesh rendering), is shown in solid green rendering. The threshold is 5.7 sigma. (B) The additional density along the central channel, extracted from the difference map by subtracting the Exo10 atomic model (PDB code: 4IFD) from the 3D EM map of RNA24-bound Exo10-Ski7 complex (mesh rendering), is shown in solid blue rendering. The threshold is 9 sigma. The atomic model of the RNA substrate (red) located in the Exo10 channel shown in both A and B was extracted from the RNA-Exo10-Rrp6 complex structure (PDB code: 4IFD). (C) Multiple potential locations of the hairpin loop of Rrp43 in the Exo10 complex. Loops marked with yellow, cyan and green indicate the potential locations of residues 245-280 of Rrp43 in the RNA-free Exo10-Ski7 complex. The hairpin marked with red shows the location of this region in the endogenous RNA-bound Exo10-Ski7 complex. The interface between the CSD2 domain of Rrp44 and Rrp45 is disrupted by this red hairpin. The model in C has a 15-degree rotation relative to the models in A and B as indicated. A zoomed-in view of the interface between Rrp45 and Rrp44 is shown on the right with a 40-degree rotation.
We further examined the structural elements around Site 1 of Exo10 and found that a hairpin loop of Rrp43 might be involved in the RNA-induced conformational change. This loop, composed of residues 251-270 of Rrp43, folds as a β hairpin and is involved in RNA binding in the crystal structure of the Exo10-Rrp6-RNA complex (PDB code 4IFD). We also observed a clear density corresponding to this structural element in the 3D map of the endogenous RNA-bound Exo10-Ski7 complex near Site 1 (Figure 6C). In contrast, in the 4.2-Å 3D map of the RNA-free Exo10-Ski7 complex, after unambiguously modeling almost the entire Rrp43 protein, we were unable to find a significant density to model the hairpin loop structure, indicating that it is a highly flexible structural feature in the apo structure of the exosome complex. Using molecular dynamics simulations, we calculated a few possible conformations of this loop in the RNA-free complex, all sitting at Site 1 of the core channel's exit, in a location higher than the β hairpin in the RNA-bound conformation (Figure 6C). This analysis provides structural information to explain RNA-induced conformational changes in the Exo10 complex (Figure 4): the interaction between Rrp45 and Rrp44's CSD2 domain is well maintained in the apo state, but would clash with Rrp43's β hairpin in the RNA-bound state if the interaction is maintained. Furthermore, we constructed a mutant yeast strain in which residues 250-264 of Rrp43's hairpin loop were deleted and observed that the mutated yeast showed growth defect in comparison with the wild-type strain (Supplementary information, Figure S7C). Collectively, we hypothesize that the hairpin loop of Rrp43 might serve as a sensor for RNA substrates at Site 1 to trigger the conformational conversion of the exosome complex from apo to RNA-bound states in the through-core route of RNA degradation.
Discussion
The cytoplasmic exosome is responsible for both general mRNA turnover to degrade transcripts no longer needed and mRNA quality control to remove aberrant transcripts5,25. The degradation of these RNA substrates by the exosome requires Ski7 and its partner complex Ski2/3/8. Ski7 is thought to function in bridging the exosome with the ribosome complex in the nonstop decay pathway and bringing the Ski2/3/8 complex to the exosome to promote its RNA degradation activity26,27. Our current work isolated Ski7-bound exosomes from yeast cell lysates, showing that Ski7 directly and stably interacts with the cytoplasmic exosome. The Ski7 protein belongs to the eRF3/GSPT family based on its conserved GTPase domain. In this family, the function of two well-studied members, EF1A and eRF3, is to interact with the A site of the ribosome; thus, the GTPase domain of Ski7 is likely to be responsible for recognizing the ribosome's empty A-site during nonstop translation events, so that Ski7 can present the aberrant mRNA to Exo10 for degradation28. The observation that Ski7 sits on top of the exosome core indicates its role in guiding mRNA substrates to the Rrp44 RNase site via the through-core route. Of note, the orientation of the Ski7 GTPase domain relative to the Exo10 core is highly flexible (Figure 1C), indicating the dynamic nature of Ski7 in RNA or ribosome recognition with little steric hindrance.
The exosome executes RNA degradation and surveillance roles in both the cytoplasm and the nucleus, but through two distinct cofactors, Ski7 and Rrp6, respectively, with different activities. It is still unclear how the cytoplasmic and nuclear exosome complexes within a cell relate to each other in their assembly and functional and spatial separations20. Our observation that Ski7 interacts with the core exosome in a very similar fashion as Rrp6 provides new insight into the relationship between the two types of exosomes in a cell. Despite the lack of sequence homology, the N-terminal domain of Ski7 and the C-terminal domain of Rrp6 share similar secondary structural folding and bind almost exactly the same surface on the core exosome (Supplementary information, Figure S3D-S3F). Our in vitro binding experiments suggest that these two types of exosome complexes are exclusive to each other at the assembly stage (Supplementary information, Figure S3C). The assembly most probably occurs in the cytoplasm, accompanied by the synthesis or maturation of Rrp6 and Ski7. After the assembly, the exosome bound with nuclear localization sequence-containing Rrp629 enters the nucleus and is therefore separated spatially from the Ski7-exosome complex remaining in the cytoplasm. This also explains why Rrp6 and Ski7 have never been detected to bind to the exosome complex simultaneously. Thus, the two types of complexes carry out distinct functions in RNA processing, and the homeostasis of the nuclear and cytoplasmic exosome complexes within a cell may be regulated directly by the synthesis rate of the Ski7 and Rrp6 proteins. Furthermore, the competition between Ski7 and Rrp6 on the core exosome may reduce the leakage activity between the nuclear and cytoplasmic exosome species.
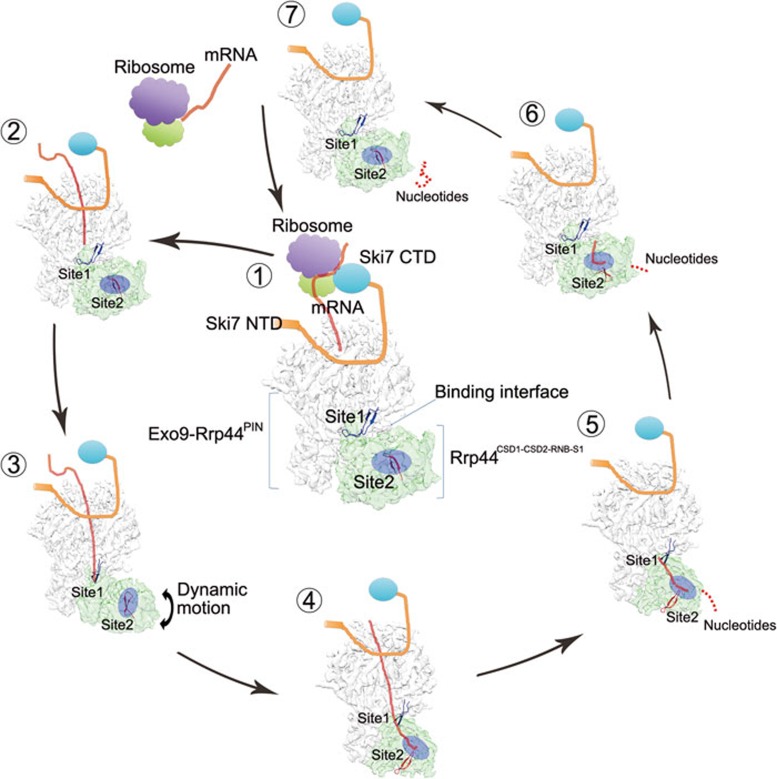
Our near-atomic resolution structure of the Exo10 complex in different RNA-binding states allowed us to depict the mechanism by which the enzymatic activity of the exosome is regulated by its RNA substrate in the through-core route (Figure 7). As we and others have previously discovered, RNA molecules with a long enough single-strand (ss) 3 overhang induce a major rearrangement of the Rrp44 RNase II-like domain relative to the core complex so that the ssRNA substrates coming out of the core directly thread into the exonuclease active site for degradation13,14,15,17. However, how this major conformational change in Rrp44 occurs upon RNA substrate binding was unknown. The comparison of the high-resolution structures of Exo10-Ski7 in its apo and RNA-bound states suggests that structural elements such as a hairpin loop of Rrp43 at Site 1 may serve as a switch sensor allowing the incoming RNA from the core chamber to interact and induce a cascade of structural rearrangements that weaken the interaction between Rrp45 and Rrp44. This weakened interaction may potentially liberate the Rrp44 RNase II-like domain so that it can rotate and rock more freely, sampling different orientations relative to the core complex and thus allowing RNA substrates to come out of it. Once there is a ssRNA 3′ tail of sufficient length emerging from the core's exit, the RNA tail may be captured by the RNA-binding tunnel between the CSD domains and the RNB domain of Rrp44, thus stabilizing Rrp44 into the RNA-bound conformational state. The positively charged tunnel of Rrp44 further recruits the RNA substrate to Site 2 for degradation. As revealed previously, the tension built up within the Rrp44 RNase II-like domain during RNA processing causes a burst that pulls the RNA substrate forward, thus leading to processivity and mild unwinding activity24. When the RNA is completely degraded, the electrostatic potential repulsion and small binding area of the two interfaces drive the reverse conformational change from the RNA-bound state to the apo state. The exosome is then ready to recognize new RNA substrates for the next cycle of processing (Figure 7).
Figure 7.
Model of RNA degradation by Exo10-Ski7 in the cytoplasm. During the mRNA decay process, the Ski7 protein recruits the exosome to the ribosome for effective targeting on aberrant RNAs (1). The RNA is loaded by Exo10-Ski7 from the top entry of the core (2), within which the RNA can be channeled through Site 1 and Site 2 along the central channel (3, 4, 5, and 6). When the 3′ end tail of the RNA reaches Site 1, it triggers a conformational change in Rrp43's hairpin loop that may weaken the interaction between the Rrp45 and the Rrp44 CSD2 domain (3). A conformational change in Rrp44 from the RNA-free to the RNA-bound state is stabilized when the RNA tail reaches Site2 (4). As the RNA substrate is shortened by the continuous degradation (5), the 5′ end of the RNA passes Site 1 and cannot hold Rrp44 in the RNA-bound state (6), thus resulting in the conformational recovery of the exosome for the next round of reaction (7). In the cartoon model, the exosome is divided into two parts, Exo9-Rrp44PIN (grey) and the RNase II-like domain (green). The N-terminal and C-terminal domains of Ski7 are marked with Ski7 NTD and Ski7 CTD, respectively. The loop of Rrp43 (aa 245-280) in Site 1 (the binding interface) is colored blue, and the loop of Rrp44 (aa 695-720) in Site 2 is colored red. The RNA degradation pocket (Site 2) is boxed by a blue semi-transparent circle.
We have previously shown that the Exo10 complex can process RNA in a direct-access route in addition to its canonical through-core route17. These results are in agreement with previous biochemical data suggesting that the exosome complex may use the direct-access route to process certain RNAs in the nucleus for their maturation. We now show that the Rrp44 subunit of the Exo10-Ski7 complex in the apo state adopts the same conformation as that of the Exo10 complex, indicating that the cytoplasmic complex may also be able to process RNA substrates through the direct-access pathway. The interaction of Ski7 with the core complex does not cause conformational changes in the Rrp44 subunit. The existence of a direct-access route for RNA processing in the cytoplasmic exosome complex requires further verification, and, if it exists, RNA processing through the direct-access pathway in the cytoplasm will be of interest in future investigations.
Materials and Methods
Affinity purification of exosome from IgG resin beads
2 ml Rabbit IgG resin (Sigma-Aldrich) was equilibrated with 200 ml washing buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol with 2 mM DTT, 2 mM EGTA, 1 mM PMSF and 1 tablet of complete EDTA-free protease inhibitor mixture from Roche Applied Science). Then, 100 ml supernatant of TAP-Rrp46-ΔRrp6 yeast lysate was loaded through the resin twice at 4 °C17. The resin was then gently washed with 150 ml high salt buffer (washing buffer plus 250 mM NaCl) and equilibrated with 150 ml TEV cleavage buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT). About 10 ml TEV buffer was retained in the chromatographic column with 1 ml TEV protease (homemade, 15.4 mg/ml) additive for overnight shaking at 4 C to release exosome from the resin.
Ion-exchange chromatography purification for exosome
The Mono Q column (5/50 GL; GE) is pre-washed by 20 ml 100% buffer B, and then equilibrated with 5% buffer B until the conductivity and UV curves are stable. The affinity purification eluent is loaded onto the Mono QTM column. After washing with 10 ml 20% buffer B, the column is eluted in gradient from 30 ml 45% buffer B to 20 ml 100% buffer B. The eluent is collected with 0.5 ml per fraction under the monitor of 260 nm and 280 nm wavelength UV detector. All the processes were carried on a GE AKTA chromatography system. Buffer A: 50 mM Tris-HCl (pH 8.0) and 2 mM EGTA. Buffer B: 50 mM Tris-HCl (pH 8.0), 2 mM EGTA and 1 M NaCl. Different percentage of Buffer B was made by diluting it with Buffer A.
Purification of the Rrp6-Rrp47 complex
The S. cerevisiae Rrp6 (His-tagged) and Rrp47 cDNAs were cloned into MCS1 and MCS2 of pRSFDuet-1(Novagen), respectively. Rrp6 and Rrp47 plasmids were co-transformed into chemically competent E.coli BL21 (DE3) pLysS cells. Then N-terminally His-tagged Rrp6 and untagged Rrp47 were co-expressed by overnight IPTG (Sigma) induction at 16 °C. Rrp6-Rrp47 was firstly enriched by Ni-affinity chromatography (GE Healthcare). Then, eluted fractions containing Rrp6/Rrp47 were pooled and further applied to a HiTrap Heparin HP column (GE Healthcare), and finally purified over a Superdex 200 column (GE Healthcare) with high-salt buffer (20mM Tris pH8.0, 500mM NaCl, 1mM β-Me).
Preparation of RNA-free exosome and RNA-bound exosome complexes
To remove the endogenously bound RNA substrates from the purified exosome complexes, the complex was incubated in reaction buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT, and 1 mM MgCl2) on ice for 1.5 h to ensure all the endogenous RNA was totally degraded. The reaction was then stopped by adding EDTA to a final concentration of 5 mM. RNA substrates were ordered from Takara company with following sequences: 5′-CCCCCGAGAGGGGGU18-3′ for RNA18, 5′-CCCCCGAGAGGGGGU24-3′ for RNA24, and 5′-CCCCGGGG(AUUU)12-3′ for RNA48. RNA-bound exosome samples were then prepared by incubating the MgCl2-treated exosome and RNA substrates together with a molar ratio of 1:10 in binding buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, and 1 mM DTT) on ice for 30 min.
Mutagenesis and growth assay of yeast cells
The yeast strain (BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0)30 was transformed with wild-type Rrp43 in a ura3-bearing p5472 vector and screened in medium lacking uracil. For each transformation, 2 colonies were selected for further analysis to get the strain overexpressing the p5472 construct. The DNA fragment containing wild-type Rrp43 from the upstream (500 bp) to the downstream (200 bp) of its open reading frame genomic sequences was amplified by PCR and cloned into the pJD8 vector carrying a hygromycin resistance gene. The construct was then used to generate the Rrp43 mutant with aa 250-264 deletion by site-directed mutagenesis. Then the DNA sequences encoding Rrp43 (wild-type or mutant) and hygromycin resistant gene were amplified by PCR from these plasmids, generating PCR products with 500 bases of homology to the region upstream and 60 bases of homology to the region downstream of the Rrp43 ORF by using corresponding primers (forward primer sequence: 5′-AAGCCTTGAAGGTAAAAGCTGGTGTTCG-3′ reverse Primer sequence: 5′-CGTTGAAAAAAGTTTTCCGTTTCCTTTCGACAGTCATCAGATAATTTTATCCGAGTCTTTCGACACTGGATGGCGGCGTTAG-3′). Yeast clones with the p5472 construct were transformed with either of these PCR products and screened on YPD plate plus 600 μg/ml hygromycin (Roche). For each batch of transformation, 6 colonies were selected for further analysis. The sequence of Rrp43 was amplified from the genome of individual clones and sequenced to confirm the mutagenesis. The identified strains grew in synthetic complete (SC) medium at 30 °C overnight before spotting serial dilutions onto the synthetic complete medium plates with and without 0.1% 5-fluoroorotic acid (5-FOA), which is used to remove ura3-bearing p5472 plasmid. The growth colonies on the plates were analyzed after 3 days of incubation at 30 °C.
In vitro protein binding assay
About 20 μl calmodulin beads (Sigma) were incubated with 10 μl 5 μM Exo10 or Exo10-Ski7 in 170 μl binding buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT, 2 mM CaCl2). The protein-coated beads were washed 3 times with 600 μl washing buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT). Then 10 μl Exosome-coated beads were mixed with 5 μl 2.5 μM Rrp6-Rrp47 complex solution and incubated in pull-down buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM DTT) on ice for 30 min. The incubation was stirred every 5 min manually to assure efficient interaction. After the incubation, the solution was centrifuged on a bucket-spinner for 5 min to separate the beads from the solution. After keeping the centrifuged sample sitting on ice for 10 min, we gently collected supernatant and washed the beads three times with 600 μl pull-down buffer. The supernatant and beads were then treated for subsequent SDS-PAGE analysis.
Negative-staining EM and image processing
Samples were diluted at a final concentration of ∼50 nM of the exosome and negatively stained in 2% (w/v) uranyl acetate (Electron Microscopy Sciences) solution following the standard deep-stain procedure on holey carbon-coated EM copper grids covered with a thin layer of continuous carbon. Then, negatively stained specimens were mounted on a transmission electron microscope holder and examined by a Tecnai Spirit electron microscope operated at 120-kV acceleration voltage. Magnified digital micrographs of the specimen were taken at a nominal magnification of 49 000 on a Gatan Ultrascan4000 CCD camera with a pixel size of 2.29-Angstroms at the specimen level. The defocus values used were about −1.0 to −1.5 μm, and the total accumulated dose at the specimen was about 70 electrons per Å2. We used EMAN231 to do the semi-automatic particle picking and IMAGIC-4D32 to perform CTF correction and reference-free 2D alignment and classification for all the particles33.
Cryo-EM and image processing
For each glow-discharged holey carbon grid (Quantifoil Cu R1.2/1.3), 3.5 μl sample with a particle concentration of ∼1 μM was applied. The grids were then blotted for 1.5 s by Whatman 55 mm filter paper and flashed frozen in liquid ethane slush cooled at liquid nitrogen temperature in FEI Vitrobot Mark IV. Grids were transferred to an FEI Titan Krios electron microscope operated at 300-kV acceleration voltage and equipped with a Gatan K2 Summit direct electron counting camera. Micrographs were recorded in super-resolution mode by semi-automated low-dose acquisition program UCSF-Image4 at a nominal magnification of 22 500×, corresponding to a pixel size of 1.31 Å on the specimen level. The total exposure time of each image was 8 seconds which was fractionated into 32 sub-frames. The total accumulated dose on specimen was about 50 electrons per Å2. The defocus values used for the image recording were about −1.2 to −3.2 μm. The 32 frames of each image stack were aligned, decimated and summed by frame-based motion correction algorithm34. Particle picking and coordinate exporting were performed in EMAN2. CTF values of the micrographs were determined by CTFFIND3 program35. 2D reference free classification and 3D classification procedures were performed in RELION 1.336. For certain 2D averages for some samples, we performed further reference-free 2D classification with the CTF-corrected particles exported from Relion1.3 by IMAGIC-4D. The auto-refinement, post-processing and auto-bfactor correction processes were all performed in RELION 1.3.
Cross-linking mass spectrometry analysis
The exosome samples were cross-linked with two cross-linkers – Bis (sulfosuccinimidyl) suberate (BS3, from Pierce) and an in house-developed cross-linker called Leiker (Tan et al., data to be published). Leiker targets lysine residues and eventually adds 316.1423 Da to a pair of cross-linked peptides. The cross-linking reactions containing 10 g of BS3 or Leiker and 10 μl (for BS3) or 40 μl (for Leiker) of 1 mg/ml protein sample in 20 mM HEPES, pH 8.0, 150 mM NaCl, were set at RT for 1 h and then quenched with 20 mM ammonium bicarbonate (JT Baker). Then, the cross-linked proteins were precipitated with acetone, resuspended in 8 M urea, 100 mM Tris, pH 8.5, and digested with trypsin (Promega) at 37 °C overnight. The peptides were analyzed by LC-MS/MS on an EASY-nLC 1000 system coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Peptides were separated on an analytical capillary column (75 μm × 10 cm) packed with 1.8 μm C18 resin, using a 100 min linear gradient at a flow rate of 200 nl/min. The mass spectrometer was operated in data-dependent mode with one MS1 event at 70 000 resolution followed by ten HCD MS2 events at 17 500 resolution. Precursors with a charge state of +1, +2, or unassigned charges were rejected. Dynamic exclusion time was set to 60 s. Cross-linked peptides were identified by searching the MS/MS spectra against a sequence database containing all the proteins in the sample (identified through a conventional LC-MS/MS experiment for protein identification) using the pLink software. The pLink search results were filtered by requiring FDR < 0.05 and E-value < 0.000137.
Molecular modeling and structure refinement of RNA-free exosome complex
We combined rigid body docking and molecular dynamic flexible fitting (MDFF)38 to model the structure in RNA-free Exo10-Ski7 complex. In order to generate the starting atomic model, we firstly superimposed the crystal structures of RNA-free Rrp41-Rrp44-Rrp45 tertiary complex (PDB code: 2WP8) and RNA-bound Exo10 complex (PDB code: 4IFD), aligning at Rrp41 and Rrp45 protein coordinates. We then built a merged model comprising of Rrp44 and Rrp45 from 2WP8 and the rest components from 4IFD. We deleted the loop region of 256-269 within Rrp43 due to the steric clash with Rrp44 in the model. This starting model was docked as a rigid body into the 4.2 Å resolution EM density map using UCSF Chimera39 and subsequently refined by the molecular dynamics flexible fitting which incorporated the EM density gradient as an external potential into MD simulation. All MDFF simulations were performed in NAMD 2.9 using CHARMM 22 all-atom force field with CMAP correction36,40,41,42. The simulations were carried at options of T = 300 K and 1 bar, employing the Langevin algorithm43 with an integration time-step of 1 fs and a scaling factor ζ = 0.3 kcal*mol−1 for 10 ns. Missing loops were modeled with MODELLER 9.1444. As for loop 256-269 in Rrp43, 10 typical generated configurations were further sampled and refined using Protein Local Optimization Program (PLOP)45. Top 1000 ranked configurations were clustered by NMRCLUST46 program according to the RMSD values of C-α atoms in the loop to select the representative models.
Ski7 model building
A difference map was calculated by subtracting the above MDFF-refined atomic model from the RNA-free Exo10-Ski7 EM map using UCSF Chimera. The secondary structure of the N terminal of Ski7 (residues 1-250) was predicted by PSIPRED47. Four regions predicted to be helixes were selected, which matched the length of the helix and loops in the difference map and were also consistent with the identified cross-linked pairs of Ski7 by cross-linking mass spectrometry analysis. The initial helical structures were built by MODELLER and docked into the EM density map. To maximally match the EM density map, these helical structures were further refined by truncating residues outside the density map or elongating the helix from neighboring residues based on unoccupied density. Then the loop configurations of the residues between helices were modeled and refined. A further round of MDFF simulation was carried out to obtain the structure of RNA-free Exo10-Ski7 complex.
Accession code
The 3D reconstruction maps obtained by Cryo-EM have been deposited into the Electron Microscopy Data Bank under accession codes, EMD-3366 (RNA-free Exo10-Ski7), EMD-3369 (RNA-bound Exo10-Ski7), EMD-3367 (RNA-free Exo10), EMD-3368 (RNA18-bound Exo10), EMD-3370 (RNA24-bound Exo10), EMD-3371 (RNA48-bound Exo10), and EMD-3372 (Untreated Exo10). The atomic coordinates of RNA-free Exo10-Ski7 have been deposited via PDB submission tool in the Protein Data Bank with the accession code 5G06.
Author Contributions
J-JL, C-YN and H-WW designed the experiments. J-JL and C-YN performed the biochemistry and EM analyses with assistance from YW and M-DY. J-JL and YW built the atomic models under supervision of NH. DT performed CXMS analysis under supervision of M-QD. C-YN, YL and WZ performed RNA sequencing analysis under supervision of XY. KZ and Q-SL provided experimental materials for in vitro biochemistry. JD supervised yeast genetic analysis. J-JL, C-YN and H-WW wrote the manuscript. All authors commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank JL Lei, Y Xu and XM Li for the EM support, NJ Zhou for atomic model validation, A Ke (Cornell University) for his helpful discussions and suggestions on the paper, and the Wang's group members and the Sui's group members (Tsinghua University) for their helpful discussions. We acknowledge the China National Center for Protein Sciences Beijing and “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology for providing the facility support. This work was supported by the National Natural Science Foundation of China (31270765 and 31530018 to HWW, 21375010 to MQD, 91540109 to XY, and 31170689 to QSL) and the National Basic Research Program of China (973 Program; 2014CB849802 to NH, 2014CB849801 to MQD, and 2012CB917201 to QSL).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Cryo-EM image of the Exo10-Ski7
3D reconstruction and atomic model building of the Exo10-Ski7
Interaction between the Ski7NTD and the Exo10
MS2 spectra of the two cross-linked peptides between the Ski7 and the Exo10
3D reconstruction of the RNA-free Exo10
RNA induced Rrp44 instability
Function of the Rrp43 loop in RNA processing.
Mass-spectrum ID results of the Exo10 sample.
Mass-spectrum ID results of the Exo10-Ski7 sample.
Summarized information of Cryo-EM data.
3D reconstruction and model of the RNA-free Exo10-Ski7
Atomic model of the RNA-free Exo10 built in this work is docked into the 3D EM map of the RNA-free Exo10-Ski7 complex (solid surface with 50% transparency). Different subunits in atomic model are colored as in Fig 2c. The models are firstly rotated 180 degrees along y-axis and 90 degrees along x-axis, then clipped to compare the model with the EM map.
3D reconstruction and model of the endogenous RNA-bound Exo10-Ski7
Atomic model of the Exo10 portion from the crystal structure of the RNA-bound Exo11 complex (PDB 4IFD) is docked in the 3D EM map of the endogenous RNA-bound Exo10-Ski7 (solid surface with 50% transparency). Different subunits in atomic model are colored as in Fig 2d. The models are firstly rotated 180 degrees along y-axis and 90 degrees along x-axis, then clipped to compare the model with the EM map.
Exo10 conformational change from the RNA-free to RNA bound states
Atomic models of the RNA-free Exo10 and the RNA-bound Exo10 are aligned based on the 9-subunit core. Different subunits in atomic model are colored as in Fig 4a. The simulated movement between the two states is generated by UCSF-Chimera with the Morph conformations tool.
Conformational change within the RNase II-like domain of Rrp44 from the RNA-free to RNA bound states
Atomic models of the RNase II-like domain of Rrp44 extracted from the RNA-free and RNA-bound exosome are aligned based on the RNB domain. Different domains in atomic model are colored as in Fig 4a. The loop from aa 705 to 720 is marked with red. The simulated movement between the two states is generated by UCSF-Chimera with the Morph conformations tool.
References
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→ 5′ exoribonucleases. Cell 1997; 91:457–466. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell 2007; 131:660–668. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113–126. [DOI] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide analysis of exosome targets. Mol Cell 2012; 48:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 2006; 7:529–539. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006; 127:1223–1237. [DOI] [PubMed] [Google Scholar]
- Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA 2008; 14:2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 2009; 139:547–559. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. Crystal structure of a 9-subunit archaeal exosome in pre-catalytic states of the phosphorolytic reaction. Archaea 2012; 2012: 721869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 2008; 29:717–728. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 2008; 456:993–996. [DOI] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 2009; 16:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-W, Wang J, Ding F, et al. Architecture of the yeast Rrp44–exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci USA 2007; 104:16844–16849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 2013; 495:70–75. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgärtner M, Basquin C, Conti E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 2015; 524:54–58. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. RNA exosome: Springer. 2010:9–28. [PMC free article] [PubMed]
- Liu JJ, Bratkowski MA, Liu X, Niu CY, Ke A, Wang HW. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat Struct Mol Biol 2014; 21:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Mitchell P. Rrp6, rrp47 and cofactors of the nuclear exosome. Adv Exp Med Biol 2010; 702:91–104. [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 2014; 511:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Clark A, Klauer AA, Tsanova B, van Hoof A. Functions of the cytoplasmic exosome. Adv Exp Med Biol 2010; 702:79–90. [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino Si, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 2001; 20:4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 2013; 154:814–826. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Schuller A, Green R, Conti E. Saccharomyces cerevisiae Ski7 Is a GTP-binding protein adopting the characteristic conformation of active translational GTPases. Structure 2015; 23:1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science 2012; 336:1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002; 295:2262–2264. [DOI] [PubMed] [Google Scholar]
- Klauer AA, van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA 2012; 3:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, et al. Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 2011; 18:715–720. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA 2011; 17:1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998; 14:115–132. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol 1996; 116:17–24. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang HW. Single particle electron microscopy reconstruction of the exosome complex using the random conical tilt method. J Vis Exp 2011; 49:e2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol 2015; 192:174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, et al. Identification of cross-linked peptides from complex samples. Nat Medthods 2012; 9:904–906. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 2008; 16:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 1998; 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 2004; 25:1400–1415. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 1995; 103:4613–4621. [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006; Chapter 5:Unit 5.6. [DOI] [PMC free article] [PubMed]
- Jacobson MP, Pincus DL, Rapp CS, et al. A hierarchical approach to all-atom protein loop prediction. Proteins 2004; 55:351–367. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng 1996; 9:1063. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000; 16:404–405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cryo-EM image of the Exo10-Ski7
3D reconstruction and atomic model building of the Exo10-Ski7
Interaction between the Ski7NTD and the Exo10
MS2 spectra of the two cross-linked peptides between the Ski7 and the Exo10
3D reconstruction of the RNA-free Exo10
RNA induced Rrp44 instability
Function of the Rrp43 loop in RNA processing.
Mass-spectrum ID results of the Exo10 sample.
Mass-spectrum ID results of the Exo10-Ski7 sample.
Summarized information of Cryo-EM data.
3D reconstruction and model of the RNA-free Exo10-Ski7
Atomic model of the RNA-free Exo10 built in this work is docked into the 3D EM map of the RNA-free Exo10-Ski7 complex (solid surface with 50% transparency). Different subunits in atomic model are colored as in Fig 2c. The models are firstly rotated 180 degrees along y-axis and 90 degrees along x-axis, then clipped to compare the model with the EM map.
3D reconstruction and model of the endogenous RNA-bound Exo10-Ski7
Atomic model of the Exo10 portion from the crystal structure of the RNA-bound Exo11 complex (PDB 4IFD) is docked in the 3D EM map of the endogenous RNA-bound Exo10-Ski7 (solid surface with 50% transparency). Different subunits in atomic model are colored as in Fig 2d. The models are firstly rotated 180 degrees along y-axis and 90 degrees along x-axis, then clipped to compare the model with the EM map.
Exo10 conformational change from the RNA-free to RNA bound states
Atomic models of the RNA-free Exo10 and the RNA-bound Exo10 are aligned based on the 9-subunit core. Different subunits in atomic model are colored as in Fig 4a. The simulated movement between the two states is generated by UCSF-Chimera with the Morph conformations tool.
Conformational change within the RNase II-like domain of Rrp44 from the RNA-free to RNA bound states
Atomic models of the RNase II-like domain of Rrp44 extracted from the RNA-free and RNA-bound exosome are aligned based on the RNB domain. Different domains in atomic model are colored as in Fig 4a. The loop from aa 705 to 720 is marked with red. The simulated movement between the two states is generated by UCSF-Chimera with the Morph conformations tool.