Summary
To achieve chromosome segregation during mitosis, sister chromatids must undergo a dramatic change in their behavior to switch from balanced oscillations at the metaphase plate to directed poleward motion during anaphase. However, the factors that alter chromosome behavior at the metaphase-to-anaphase transition remain incompletely understood. Here, we perform time-lapse imaging to analyze anaphase chromosome dynamics in human cells. Using multiple directed biochemical, genetic, and physical perturbations, our results demonstrate that differences in the global phosphorylation states between metaphase and anaphase are the major determinant of chromosome motion dynamics. Indeed, causing a mitotic phosphorylation state to persist into anaphase produces dramatic metaphase-like oscillations. These induced oscillations depend on both kinetochore-derived and polar ejection forces that oppose poleward motion. Thus, our analysis of anaphase chromosome motion reveals that dephosphorylation of multiple mitotic substrates is required to suppress metaphase chromosome oscillatory motions and achieve directed poleward motion for successful chromosome segregation.
Keywords: Mitosis, Kinetochore, Microtubule, Chromokinesin, Phosphorylation
Introduction
During mitosis in vertebrate cells, several sequential phases occur to distribute replicated sister chromatids to daughter cells. First, during prometaphase, chromosomes form attachments to spindle microtubules and are moved to the center of the cell in a process termed congression. At metaphase, chromosomes align at the metaphase plate where they undergo oscillations (Jaqaman et al., 2010; Skibbens et al., 1993). Finally, during anaphase, sister chromatids are separated and segregated towards opposite spindle poles (Maiato and Lince-Faria, 2010).
Chromosome congression and metaphase chromosome oscillations have been the subject of intense investigation (reviewed in Vladimirou et al., 2011). These studies have revealed that multiple factors acting on chromosomes are integrated to achieve observed chromosome motion. This includes the attachment of kinetochores to microtubules, changes in microtubule dynamics that act to push or pull on kinetochores (Dumont et al., 2012; Inoue and Salmon, 1995), chromosome cohesion between replicated sister chromatids that provides a spring-like connection between them, and chromokinesin-dependent polar ejection forces (Civelekoglu-Scholey et al., 2013; Joglekar and Hunt, 2002).
Despite extensive work on metaphase chromosome dynamics, the nature and molecular origin of anaphase chromosome motion are less well understood. Here, we established a procedure to image the metaphase-to-anaphase transition and anaphase chromosome motion with high temporal resolution in human cells to assess the dynamics of anaphase chromosome motion and the mechanisms that direct sister chromatid segregation. Our results indicate that changes in chromosome motion at anaphase onset do not result from the physical separation of sister chromatids. Instead, we find that poleward chromosome motion in anaphase requires critical changes to the global phosphorylation state of the cell. Our results suggest that a change in the phosphorylation state of factors required for kinetochore-derived forces and chromokinesin-dependent polar ejection forces provides a regulatory switch to alter chromosome motion between metaphase and anaphase. Thus, the regulatory changes that occur at anaphase onset and the precise timing of sister chromatid separation act together to ensure the proper segregation of sister chromatids to daughter cells.
Results
Tracking Analysis of anaphase chromosome motion
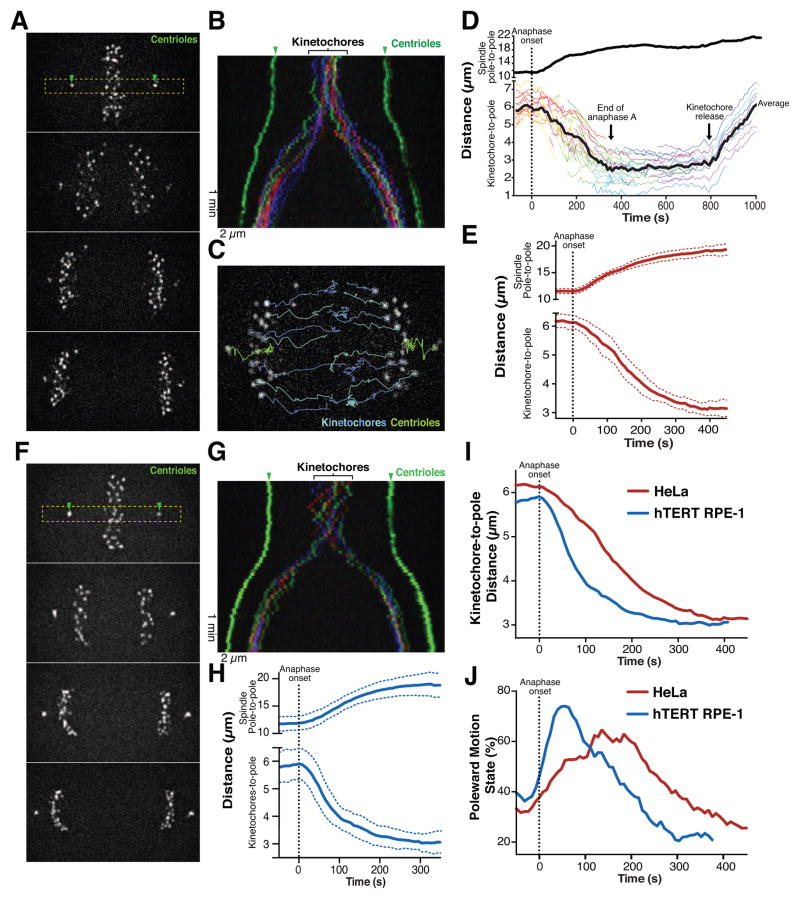
To analyze the behavior of anaphase chromosome motion in human cells, we generated human cancer (HeLa) and non-transformed (hTERT RPE-1) cell lines stably expressing the centromere protein CENP-A and the centriole-component Centrin (CETN1), each fused to 3x tandem repeats of GFP. We performed live-cell imaging of these cells progressing through mitosis (Figure 1) to visualize the trajectories of individual kinetochores and their motion relative to the spindle pole (Figure 1D; Figure S1A). Using single-particle tracking and trajectory analysis, we were able to assess anaphase chromosome segregation to distinguish the anaphase A-based motion of kinetochores towards the spindle poles (Figure 1D, E and H, lower graphs) and the anaphase B-based separation of the spindle poles (Figure 1D, E, H, upper graphs; Table S1) in a model-free manner (Monnier et al., 2015). We found that the overall dynamics of spindle pole separation and chromosome motion were similar in HeLa and hTERT-RPE1 cells (see Table S1).
Figure 1. Analysis of anaphase chromosome dynamics in human cells.
(A) Still images from a time-lapse movie of HeLa cells expressing 3xGFP-CENP-A, 3xGFP-centrin. Box indicates the section used to generate the kymograph. (B) Color-coded kymograph of the time-lapse movie from A. (C) Representative image of time-lapse series displayed in A overlaid with selected tracks of particles. (D) Graph showing the distances over time for the distance between spindle poles (top; to measure spindle elongation) and the kinetochore to pole distance (bottom; to visualize chromosome motion) using tracks of a HeLa cell. The average kinetochore to pole distance is indicated as a black line, with individual kinetochores indicated in color. The time of anaphase onset is indicated by the dashed line. (E) Average spindle pole-to-pole distance (upper graph) and kinetochore to pole distance (lower graph) for HeLa cells undergoing anaphase (n=10). Colored dotted lines indicate standard deviation between cells. (F–H) Still images from a time-lapse movie of an hTERT RPE-1 cell expressing 3xGFP-CENP-A, 3xGFP-centrin. (G) Color-coded kymograph of time-lapse displayed in F. (H) Average spindle pole to pole distance (upper graph) and kinetochore to pole distance (lower graph) of hTERT RPE-1 undergoing anaphase (n=10). Blue dotted lines indicate standard deviation between cells. (I) Direct comparison of average kinetochore to pole distances over time for HeLa (from E) and hTERT-RPE cells data (from H). (J) Percentage of poleward motion over time. Scale bars, 2 μm. See also Figure S1, Table S1, S2 and Movie1.
Prior work in other organisms has found that the forces acting on bi-oriented sister chromatids prevent spindle elongation until the sister chromatids are separated at anaphase onset (Desai et al., 2003). Consistent with this, we observed that spindle elongation initiated coincident with visually-discernible sister chromatid separation in both HeLa and hTERT RPE-1 cells (Figure 1D, E and H). We predicted that the loss of a physical connection between sisters at anaphase onset would also induce the rapid motion of chromatids towards their associated poles. However, although hTERT RPE-1 cells displayed a rapid increase in overall poleward motion shortly following anaphase onset, HeLa cells showed a delay of ~80 s in achieving a maximal rate of average poleward motion (Figure 1I). Once chromosomes reached a distance of ~3 μm from the spindle pole, they maintained this position as the spindle poles continued to elongate (Figure 1D, E and H). At the end of anaphase, the kinetochore-to-pole distance increased suddenly and synchronously (Figure 1D and S1B), indicating the release of the kinetochores from the spindle poles by eliminating kinetochore-microtubule interactions. This provides an assay to systematically analyze anaphase chromosome motion in human cells.
Disrupting the opposing forces acting on sister chromatids is insufficient to explain the suppression of chromosome oscillations at anaphase onset
The metaphase-to-anaphase transition is characterized by a switch between metaphase chromosome oscillations and directional anaphase poleward motion. To analyze this switch in behavior, we classified distinct periods of kinetochore motion as either poleward or anti-poleward (see Supplemental Experimental Procedures; Figure S1F). In cases where a kinetochore moved less than the experimentally determined localization error (see Figure S1C and D) between successive time points, it was classified as having “indeterminate” motion during this corresponding time-interval.
During metaphase, HeLa cells displayed an equivalent fraction of poleward and anti-poleward motion, with 33 ± 2% (mean ± s.d.; mean was measured as cell to cell variation after averaging kinetochore motion in individual cells; see Supplemental Methods for additional information) of events classified as poleward and 33 ± 3% anti-poleward events (Table S2). In contrast, during the early phase following anaphase onset (240 s in HeLa, 152 s in hTERT RPE1) we observed both poleward and anti-poleward motion, but the majority of kinetochore motion was poleward, as expected for anaphase A sister chromatid segregation (Figure 1J; Table S1). For example, kinetochores in HeLa cells moved poleward for 54 ± 2% of events detected during early anaphase, with only 20 ± 3% of their motion spent in the anti-poleward state. However, in comparison with hTERT RPE-1 cells, HeLa cells were delayed in achieving a maximal proportion of poleward motion (Figure 1J, Table S1).
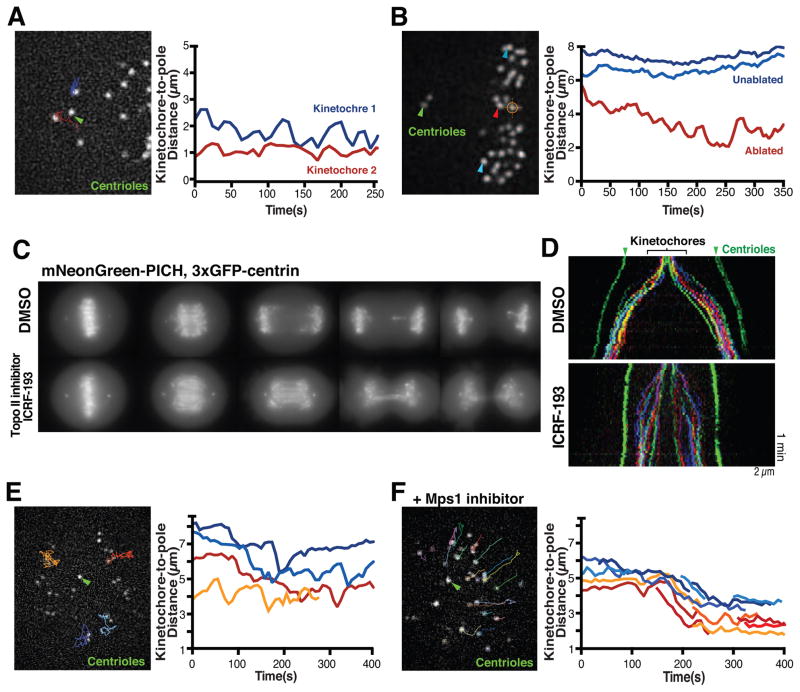
To determine the molecular origin of the transition from clearly distinct chromosome motions in metaphase versus anaphase, we first considered the physical connections that differ between these two phases. At anaphase onset, physical associations between sister chromatids are eliminated by the cleavage of cohesin molecules (Hauf et al., 2001). To disrupt sister chromatid cohesion prematurely in our human cell culture system, we depleted the cohesin complex subunit Rad21 by RNAi (Figure 2A). Rad21-depleted cells displayed separated sister chromatids that failed to congress to the metaphase plate, but these chromatids continued to exhibit both poleward (33 ± 5%) and anti-poleward (44 ± 8%) motions (Figure 2A and S2A), consistent with prior studies using TEV-induced cohesion cleavage in Drosophila embryos (Oliveira et al., 2010). We also used laser ablation to eliminate a single kinetochore from a pair of sister chromatids in a metaphase cell, thereby removing the pulling forces created by the connection to the opposing spindle pole (Figure 2B). In these laser ablation experiments, the released kinetochore initially moved away from the metaphase plate during the first 20 seconds after ablation, displaying net poleward motion (69 ± 25% poleward motion and 13 ± 18% anti-poleward motion). However, these released kinetochores then displayed a balance of poleward (40 ± 8%) and anti-poleward (35 ± 10%) motion (Figure S2A) that resembled the behavior of bi-oriented metaphase chromosomes (Figure 2B; S2B and Table S2, also see Rieder et al., 1986; Skibbens et al., 1995).
Figure 2. Physical connections between sister chromatids are not required for anti-poleward motion.
(A) Still image from a representative time-lapse movie of a HeLa cell (3xGFP-CENP-A, 3xGFP-centrin; n=20) following depletion of the cohesin subunit RAD21 (48 h) displaying tracks until current time point of selected kinetochores used to generate the kinetochore to spindle pole distance graph (right). (B) Image of a HeLa cell (3xGFP-CENP-A, 3xGFP-centrin) before laser ablation (orange hair cross) to inactivate one of 2 sister kinetochores (n=29 experiments). Arrowhead indicates the released kinetochore (red) or unaffected kinetochores (blue) which were tracked to generate spindle to pole distance graph (right). (C) Maximal intensity projections of still images from representative time-lapse sequences of HeLa cells expressing mNeonGreen-PICH, 3xGFP-centrin entering anaphase in presence of DMSO (n=10) or 1 μM of the topoisomerase inhibitor ICRF-133 (n=14). (D) Color-coded kymographs of HeLa cells (3xGFP-CENP-A, 3xGFP-centrin) from anaphase onwards treated with DMSO (n=5) or ICRF-193 (n=7). (E) Still image from a time-lapse movie of a HeLa cell (3xGFP-CENP-A, 3xGFP-centrin) treated with S-trityl-L-cysteine (STLC) to generate a monopolar spindle (n=11) showing tracks of the selected kinetochores used to generate kinetochore to spindle pole distance graph (right). (F) Image from time-lapse movie of a monopolar HeLa cell (3xGFP-CENP-A, 3xGFP-centrin) treated with STLC and the Mps1 inhibitor AZ3146 (n=8). Selected tracks were used to generate the kinetochore to spindle pole distance graph (right). Green arrowheads highlight spindle poles. t=0 is beginning of movie. Scale bars, 2 μm. See also Figure S2, Table S2 and Movie 2.
Reciprocally, to cause connections between sister chromatids to persist into anaphase we treated cells with the topoisomerase II inhibitor ICRF193, which prevents the resolution of ultra-fine DNA bridges (UFBs; Wang et al., 2008). UFBs are generated between sister chromatids during DNA replication, but are resolved in metaphase and early anaphase (Liu et al., 2014). Treatment with 1 μM ICRF193 significantly delayed UFB resolution as detected by the presence of the UFB marker mNeonGreen-PICH (Chan et al., 2007), resulting in decreased spindle elongation (Figure 2C and D). However, in cells treated with ICRF193, the majority of the kinetochores moved towards the spindle poles and we did not detect a noticeable increase in anti-poleward motion (Figure 2D). We note that UFBs do retard the rate of chromosome movement, possibly by acting to provide resistance similar to that created by sister chromatid cohesion. In summary, although a physical connection between sister chromatids controls the amplitude and period of metaphase chromosome oscillations (Burroughs et al., 2015; Wan et al., 2012), removing these connections is not sufficient to induce the change in the proportion of poleward and anti-poleward motion that occurs at anaphase onset.
Preventing protein dephosphorylation induces dramatic chromosome oscillations in anaphase
We next considered whether changes to the cell regulatory environment are responsible for the altered chromosome dynamics at anaphase onset. To test the relative contributions of the forces acting on the sister chromatid and the cell regulatory state, we generated monopolar spindles using S-trityl-L-cysteine (STLC) to inhibit the kinesin-5 motor, Eg5, which causes all poleward-pulling forces to emanate from a single origin. Despite the absence of bi-oriented attachments in STLC-treated cells, we observed both poleward and anti-poleward motion (Figure 2E). However, triggering anaphase onset by inactivating the spindle assembly checkpoint using an Mps1 inhibitor was sufficient to induce a synchronous directional motion towards the single pole (Figure 2F; also see Canman et al., 2003).
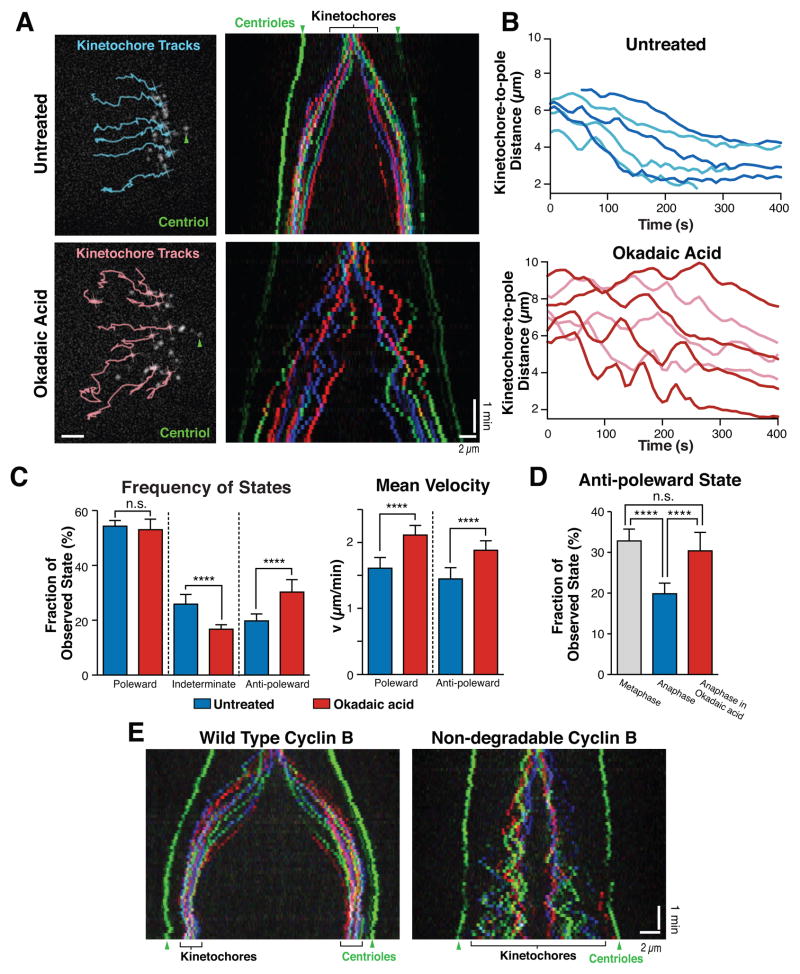
Protein dephosphorylation is a hallmark of mitotic exit (Wurzenberger and Gerlich, 2011) and alters microtubule dynamics required for chromosome segregation (Higuchi and Uhlmann, 2005). To inhibit protein dephosphorylation, we treated metaphase HeLa cells with 1 μ acid, a potent inhibitor of both PP1 and PP2A phosphatases. Okadaic acid treatment resulted in dramatic metaphase-like chromosome oscillations that persisted into anaphase (Figure 3A and B). These oscillations reflect a statistically significant increase in the proportion of anti-poleward motions (30 ± 5%: Figure 3C; Table S2), with a similar proportion of anti-poleward moving kinetochores to that observed in metaphase cells (33 ± 3%: Figure 3D and S3A). The change in chromosome movement was not due to the persistence of UFBs based on mNeonGreen-PICH fluorescence (Figure S3B). In addition to altering the proportion of poleward/anti-poleward motions, Okadaic acid treatment significantly increased the velocity of both poleward and anti-poleward moving kinetochores (Figure 3C; Figure S3C and D; Table S2). Interestingly, this rate was higher than that observed for metaphase kinetochores (Figure S3D; Table S2), likely due to opposing forces derived from the attached sister kinetochore in metaphase that act to retard chromosome motion. We also observed a similar effect following treatment with the phosphatase inhibitor cantharidic acid (data not shown). This effect is considerably more severe than that observed in prior work that inhibited a subset of PP1 function by the depletion of Sds22 or Repoman, which induced occasional pausing and infrequent anti-polar motion in anaphase (Wurzenberger et al., 2012). Previous work expressing high levels of a non-degradable version of cyclin B, to prevent the down regulation of CDK1 activity, found that this prevented normal anaphase progression after sister chromatid separation resulting in a metaphase-like arrest (Vazquez-Novelle et al., 2014). We found that expression of lower levels of non-degradable cyclin B permitted full progression into anaphase and cytokinesis, but resulted in dramatic anaphase chromatid oscillations (Figure 3E), similar to Okadaic acid treatment (Figure 3A). Therefore, dephosphorylation of target proteins downstream of CDK1 by PP1 and PP2A is essential for the changes in chromosome dynamics that occur at anaphase onset. Allowing a metaphase phosphorylation state to persist into anaphase results in dramatic metaphase-like chromosome oscillations despite the separation of sister chromatids.
Figure 3. Perturbing the cellular phosphorylation state induces anaphase anti-poleward chromosome motion.
(A) (left) Images of untreated HeLa cells (3xGFP-CENP-A, 3xGFP-centrin) or cells treated with Okadaic acid (n≥10 cells). Selected kinetochore tracks until current time point are displayed. (right) Color-coded kymographs from the corresponding movies starting at anaphase onset. (B) Selected representative curves of individual kinetochore to pole distances from the cells shown in A. Color shades are used to distinguish different tracks. t=0 was set to anaphase onset. (C) Comparison of distribution of motion stages and velocity for the 240 seconds post anaphase onset in untreated or Okadaic acid treated HeLa cells (3xGFP-CENP-A, 3xGFP-centrin; n≥10 each). (D) Comparison of the proportion of anti-poleward motion during metaphase, untreated anaphase (n=10), or Okadaic acid-treated (n=18) anaphase HeLa cells (3xGFP-CENP-A, 3xGFP-centrin). (E) Kymographs as in A for cells expressing either wild type Cyclin B, or a non-degradable Cyclin B mutant. Arrowheads highlight spindle poles (green). Unpaired t tests were applied for comparison (**** p<0.0001; ** p=0.0031; Not significant (n.s.) C: p=0.235, D: p=0.117). Standard deviations were measured across cells using the average behavior for kinetochores in each cell. Scale bars, 2 μm. See also Figure S3, Table S2 and Movie 3.
Both chromosome and kinetochore-derived forces contribute to anaphase anti-poleward motion in Okadaic acid-treated cells
We next sought to determine the origin of the induced chromosome oscillations that occur during anaphase when protein dephosphorylation is perturbed. To assess the sources of the force acting on the sister chromatids, we first tested whether Okadaic acid-induced anti-poleward motion require polar ejection forces. The chromokinesins KID and KIF4A act on the chromosome arms during metaphase to push chromosomes away from the spindle poles and thereby contribute to metaphase chromosome oscillations (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001; Wandke et al., 2012).
To test the role of polar ejection forces during anaphase, we generated CRISPR/Cas9-mediated knockout cell lines for the chromokinesins KID and KIF4A (Figure S4A). Individual elimination of KID or KIF4A resulted in a reduced distance between kinetochores and the spindle poles in cells with monopolar spindles, consistent with a role for these motors in generating polar ejection force. The double KID+KIF4A knockout cell line displayed an enhanced reduction in the kinetochore-spindle pole distance (Figure S4B), consistent with previous RNAi-based experiments (Barisic et al., 2014; Wandke et al., 2012). However, despite this strong effect on chromosome-pole distances during mitosis, the double KID+KI4A knockout cell line was viable (Figure S4A).
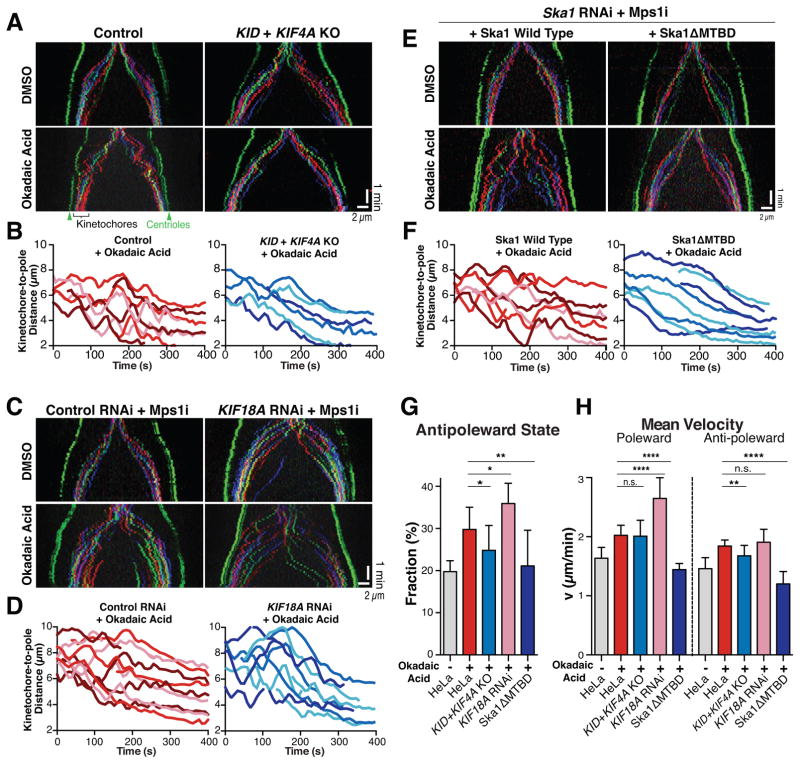
We next assessed whether polar ejection forces act during an unperturbed anaphase. As described above, control cells display a plateau in poleward motion during anaphase A such that they halt their next poleward motion when they reach a distance of ~3 μm away from the spindle pole. In contrast, we found that the KID+KIF4A double knockout cell line displayed a reduced kinetochore to pole distance at the end of anaphase A (Figure S4C). This suggests that the activity of these chromokinesins persists into anaphase where they contribute to the plateau in poleward motion (Figure 1D). However, KID+KIF4A double knockout cells did not otherwise display a striking difference in anaphase chromosome dynamics in untreated cells. Interestingly, we found that the proportion of anti-poleward motion in anaphase was modestly, but statistically significantly decreased in the KID+KIF4A double knockout in Okadaic acid-treated cells (Figure 4A, B and Table S2). Thus, chromokinesin-based polar ejection forces contribute to the Okadaic acid-induced, anti-poleward anaphase motions.
Figure 4. Chromosome and kinetochore-derived forces contribute to anaphase anti-poleward motion in Okadaic acid-treated cells.
(A) Representative color-coded kymographs of HeLa cells (3xGFP-CENP-A, 3xGFP-centrin) undergoing anaphase either for control cells (left) or KID and KIF4A double knockout cells (KID+KIF4A KO; right) treated with DMSO (upper panels; n=10 or 6) or Okadaic acid (lower panels; n≥10). (B) Graph showing selected representative kinetochore to pole distances from A. (C) Kymographs as A displaying cells treated with non-targeting control siRNAs (left) or KIF18A siRNA after 24h (right) incubated in DMSO (upper panels; n=6 or 18) or Okadaic acid (lower panels; n=6 or 5, respectively) and MPS1 inhibitor AZ3146. (D) Graph showing selected representative kinetochore to pole distances from Okadaic acid treated cells as displayed in C. (E) Kymographs as A displaying cells in which either wild type mCherry-Ska1 (left) or a Ska1ΔMTBD mutant (right) replaces endogenous Ska1(48h RNAi). Cells were treated with AZ3146 and DMSO (upper panels; n=3 or 4) or AZ3146 and Okadaic acid (lower panels; n=8 or 5, respectively). (F) Graph showing selected representative kinetochore to pole distances from E. (G) Diagrams display fraction of anti-poleward state of kinetochores 240 seconds post anaphase onset for conditions A–F. Unpaired t tests were performed to HeLa cells (n=18): KID+KIF4A KO (n=12) *p=0.0225; KIF18A RNAi (n=5) *p=0.0285; Ska1ΔMTBD (n=5) **p=0.0096. (H) Diagram display velocity of kinetochore motion 240 seconds post anaphase onset for conditions A–F. Unpaired t-tests were performed for poleward motion to HeLa cells (n=18): KID+KIF4A KO (n=12) n.s. p=0.854; KIF18A RNAi (n=5) ****p<0.0001; Ska1ΔMTBD (n=5) ****p<0.0001 and for anti-poleward motion: KID+KIF4A KO (n=12) **p=0.0033; KIF18A RNAi (n=5) n.s. p=0.3492; Ska1ΔMTBD (n=5) ****p<0.0001. Arrowheads highlight spindle poles (green) and examples of anti-poleward motion (white). Scale bars, 2 μm. See also Figure S4, Table S2 and Movie 4.
We next tested the contributions of the kinetochore-associated motor Kif18A, which acts to dampen the chromosome dynamics in metaphase (Du et al., 2010; Stumpff et al., 2008; Stumpff et al., 2012). HeLa cells depleted of Kif18A by RNAi displayed increased metaphase chromosome oscillations (Fig. S4D), defects in chromosome congression, and displayed a delay in the mitotic progression (Fig. S4E). Using the Mps1 inhibitor AZ3146 to control the timing of anaphase onset in Kif18A-depleted cells treated with 1 μM Okadaic acid, we observed a further increase of anaphase chromatid oscillations (Figure 4C, D and Table S2), similar to the enhanced metaphase oscillations that occur in Kif18A-depleted cells (Stumpff et al., 2008).
Finally, to test whether kinetochore-derived forces contribute to the observed anaphase motion, we used a mutant of the kinetochore protein Ska1 complex, which we have previously shown inhibits chromosome oscillations during metaphase (Schmidt et al., 2012). We generated stable cell lines expressing mCherry fused to RNAi resistant versions of wild type Ska1 or a mutant of Ska1 lacking the microtubule-binding domain (ΔMTBD). In cells in which Ska1 was replaced with the Ska1ΔMTBD mutant, we did not detect a significant change in chromosome dynamics during anaphase in untreated cells (Figure S4F). Strikingly, we observed a complete loss of the Okadaic-induced oscillations during anaphase in Ska1ΔMTBD mutant cells (Figure 4E–F). We observed a significant decrease in the fraction of anti-poleward motions and the rate of both polar and anti-poleward motions such that these were similar to anaphase cells in the absence of Okadaic acid (Figure 4E–H and Table S2). Together, these analyses indicate that both chromosome and kinetochore-derived forces are required for the Okadaic acid-induced chromosome oscillations during anaphase.
Discussion
A phospho-regulatory switch regulates anaphase chromosome dynamics
By analyzing the dynamics of chromosome movements under diverse conditions, including physical, pharmacological, and genetic perturbations, our work demonstrates that the movement behavior of mitotic chromosomes in human cells is determined primarily by the cellular regulatory environment (Figure 5). The physical connections between sister chromatids contribute to controlling the period and amplitude of sister chromatid oscillations during mitosis, but do not control the proportion of poleward and anti-poleward motion. Indeed, in prometaphase, the premature removal of cohesin or the loss of a connection to one of the spindle poles does not preclude anti-poleward and oscillatory motion (Figures 2A and B), similar to prior observations in Drosophila embryos (Oliveira et al., 2010; Parry et al., 2003). Reciprocally, causing a metaphase regulatory state to persist into anaphase using phosphatase inhibition or non-degradable cyclin B expression dramatically increases anti-poleward motions (Figure 3A and 3E; Table S2). We found that these anti-poleward motions require proteins that have been implicated in metaphase oscillations, including factors that contribute to kinetochore-derived forces and polar ejection forces. An overall change in the microtubule turnover takes place at the metaphase to anaphase transition (Zhai et al., 1995). Consistent with this, previous work found that Kif18A (Hafner et al., 2014), which acts to dampen microtubule dynamics, and the chromokinesin KID (Ohsugi et al., 2003) are regulated downstream of CDK. A change in their phosphorylation status at anaphase onset may act to dampen chromosome oscillations and reduce polar ejection forces. Similarly, kinetochore-derived forces that depend on the Ska1 complex must also be altered upon mitotic exit to suppress the persistence of oscillations into anaphase. Thus, a broad spectrum of targets is regulated directly and indirectly downstream of CDK and their combined action alters the dynamics of microtubules and chromosome motion. In summary, our work reveals that the switch of chromosome motion from metaphase to anaphase is not simply the result of a physical separation of sister chromatids, but additionally requires changes in the phosphorylation of multiple mitotic targets that collectively regulate chromosome poleward motion. This may ensure that chromosome segregation is precisely coordinated with other phosphorylation-regulated steps of mitotic exit, such as furrow ingression or nuclear membrane reformation, to ensure proper genome separation and integrity.
Figure 5. Model for the regulatory control of chromosome dynamics at the metaphase to anaphase transition.
In metaphase, chromosome oscillations are caused by chromokinesin-based polar ejection forces and kinetochore-derived forces. These activities are controlled by phosphorylation downstream of CDK1. At anaphase onset, CDK1 is inactivated and phosphatases reverse the phosphorylation of its substrates to downregulate polar ejection forces and kinetochore-derived forces that act through microtubule polymerization. This allows chromosomes to display net motion towards the spindle poles. In the contrast, in the presence of the phosphatase inhibitor Okadaic acid, dephosphorylation is delayed such that chromokinesins and kinetochore-derived forces remain active. This maintains a metaphase-like oscillatory chromosome behavior in anaphase even after sister chromatid separation. Thus, mitosis is characterized by two distinct phases of chromosome motion – metaphase oscillations to align the chromosomes and poleward anaphase motion to segregate the chromosomes – and the switch in movement behavior is controlled by a regulatory transition.
Experimental Procedures
Cell culture and cell line generation
HeLa and hTERT RPE-1 cells were maintained under standard tissue conditions (Schmidt et al., 2012). Cells expressing fluorescent tag fusions of Centrin (CETN1), CENP-A, PICH, Ska1 wild type or ΔMTBD (Schmidt et al., 2012) were generated using retroviral infection of cells with pBABE–based vectors as previously described (Cheeseman et al., 2004). CRISPR/cas9 mediated knock out cells were generated by co-transfection of px330 (Cong et al., 2013) targeting KID (GCAGAGGCGACGCGAGATGG) or KIF4A (GCTCTCCGGGCACGAAGGAA) with CS2+mCherry (1:10) using Fugene HD according to manufacturer’s instructions and sorting for single cells using mCherry signal after 2 days. Clones were verified via Western blotting using antibodies (Abcam) against KID (1:1000, ab69824) or KIF4A (1:2000, ab124903) and α-tubulin (1:2000, ab40742). For a list of cell lines used in this study, see Supplemental Experimental Procedures.
Drug treatment and cell transfection
Where indicated, cells were incubated in 1 μM ICRF193 (Santa Cruz), 1 μM Okadaic acid (Santa Cruz) or 2 μM AZ3146 (Tocris) for 5 min or 10 μM S-trityl-L-cysteine (Sigma) for 20 minutes (Figure 2E, F) or 2h (Figure S3C) before imaging. For RNAi, cells were transfected with 50 nM ON-TARGET plus siRNAs (Dharmacon) targeting RAD21 (AUACCUUCUUGCAGACUGUUU), KIF18A (GCCAAUUCUUCGUAGUUUU), Ska1 (pool targeting: GGACUUACUCGUUAUGUUA, UCAAUGGUGUUCCUUCGUA, UAUAGUGGAAGCUGACAUA and CCGCUUAACCUAUAAUCAA), or a nontargeting control using Lipofectamine RNAi MAX (Invitrogen) following instructions of the manufacturer. Plasmids containing wild type cyclin B1-mCherry or the non-degradable mutant (R42A and L45A) (Gavet and Pines, 2010; Vazquez-Novelle et al., 2014) were transfected into HeLa cells using Fugene HD (Promega) according to manufacturer’s instructions 24 hours prior to imaging.
Live cell imaging
Cells were imaged in CO2-independent media (Invitrogen) at 37°C. All images except laser microsurgery were acquired on a Nikon eclipse microscope equipped with a CCD camera (Clara, Andor) using a 40x Plan Fluor objective 1.3NA (Nikon) and appropriate fluorescence filters. Images of 3xGFP-CENP-A cell lines were acquired every 8 seconds using 3 (HeLa) or 5 (hTERT RPE-1 and Figure S3C) z sections at 0.7 μm intervals. Where indicated, cells were imaged at 4s interval using a single plane focus. mNeonGreen-PICH cells were imaged every 60s at 4 z-sections at 1 μm intervals. The laser microsurgery was conducted as described previously (Pereira et al., 2009) and is detailed in the Supplemental Experimental Procedures. An extended description of the analysis of the time-lapse movies and kinetochore motion is also included in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank António J. Pereira for assistance generating kymographs, and members of the Cheeseman laboratory for their support, input, and helpful discussions. This work was supported by a Scholar award to IMC from the Leukemia & Lymphoma Society, a grant from the NIH/National Institute of General Medical Sciences to IMC (GM088313), a Research Scholar Grant to IMC (121776) from the American Cancer Society, and a grant from the NSF (PoLS PHY 1305537) to MB. KS was funded by FP7 Marie Curie Actions/Erwin Schrödinger Fellowship of the Austrian Science Fund (J-3478). HM is supported by the 7th framework program grant PRECISE from the European Research Council, FLAD Life Science 2020 and The Louis-Jeantet Foundation Young Investigator Award 2015.
Footnotes
Author Contributions
Conceptualization, K.S. and I.M.C.; Software, Z.B.; Formal Analysis, K.S. and Z.B.; Investigation, K.S. and N.S.; Data Curation, Z.B.; Writing-Original Draft, K.S. Z.B. and I.M.C.; Writing- Review & Editing, N.S., H.M. and M.B.; Funding Acquisition and Supervision, H.M., M.B. and I.M.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Barisic M, Aguiar P, Geley S, Maiato H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat Cell Biol. 2014;16:1249–1256. doi: 10.1038/ncb3060. [DOI] [PubMed] [Google Scholar]
- Burroughs NJ, Harry EF, McAinsh AD. Super-resolution kinetochore tracking reveals the mechanisms of human sister kinetochore directional switching. eLife. 2015;4 doi: 10.7554/eLife.09500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. Embo J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, He B, Shen M, Wan X, Roscioli E, Bowden B, Cimini D. Dynamic bonds and polar ejection force distribution explain kinetochore oscillations in PtK1 cells. J Cell Biol. 2013;201:577–593. doi: 10.1083/jcb.201301022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner J, Mayr MI, Mockel MM, Mayer TU. Pre-anaphase chromosome oscillations are regulated by the antagonistic activities of Cdk1 and PP1 on Kif18A. Nat Commun. 2014;5:4397. doi: 10.1038/ncomms5397. [DOI] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, King EM, Amaro AC, Winter JR, Dorn JF, Elliott HL, McHedlishvili N, McClelland SE, Porter IM, Posch M, et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J Cell Biol. 2010;188:665–679. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Hunt AJ. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J. 2002;83:42–58. doi: 10.1016/S0006-3495(02)75148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev. 2014;26:1–5. doi: 10.1016/j.gde.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Maiato H, Lince-Faria M. The perpetual movements of anaphase. Cell Mol Life Sci. 2010;67:2251–2269. doi: 10.1007/s00018-010-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier N, Barry Z, Park HY, Su KC, Katz Z, English BP, Dey A, Pan K, Cheeseman IM, Singer RH, et al. Inferring transient particle transport dynamics in live cells. Nature methods. 2015;12:838–840. doi: 10.1038/nmeth.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Tokai-Nishizumi N, Shiroguchi K, Toyoshima YY, Inoue J, Yamamoto T. Cdc2-mediated phosphorylation of Kid controls its distribution to spindle and chromosomes. Embo J. 2003;22:2091–2103. doi: 10.1093/emboj/cdg208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Hickson GR, O’Farrell PH. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr Biol. 2003;13:647–653. doi: 10.1016/s0960-9822(03)00242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AJ, Matos I, Lince-Faria M, Maiato H. Dissecting mitosis with laser microsurgery and RNAi in Drosophila cells. Methods Mol Biol. 2009;545:145–164. doi: 10.1007/978-1-60327-993-2_9. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Rieder CL, Salmon ED. Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J Cell Sci. 1995;108(Pt 7):2537–2548. doi: 10.1242/jcs.108.7.2537. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Novelle MD, Sansregret L, Dick AE, Smith CA, McAinsh AD, Gerlich DW, Petronczki M. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr Biol. 2014;24:638–645. doi: 10.1016/j.cub.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirou E, Harry E, Burroughs N, McAinsh AD. Springs, clutches and motors: driving forward kinetochore mechanism by modelling. Chromosome Res. 2011;19:409–421. doi: 10.1007/s10577-011-9191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Cimini D, Cameron LA, Salmon ED. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol Biol Cell. 2012;23:1035–1046. doi: 10.1091/mbc.E11-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- Wurzenberger C, Held M, Lampson MA, Poser I, Hyman AA, Gerlich DW. Sds22 and Repo-Man stabilize chromosome segregation by counteracting Aurora B on anaphase kinetochores. J Cell Biol. 2012;198:173–183. doi: 10.1083/jcb.201112112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.