Abstract
The molecular mechanisms and genetic architecture that facilitate adaptive radiation of lineages remain elusive. Polymorphic chromosomal inversions, due to their recombination-reducing effect, are proposed instruments of ecotypic differentiation. Here we study an ecologically diversifying lineage of An. gambiae, known as the Bamako chromosomal form based on its unique complement of three chromosomal inversions, to explore the impact of these inversions on ecotypic differentiation. We used pooled and individual genome sequencing of Bamako, typical (non-Bamako) An. gambiae, and the sister species An. coluzzii to investigate evolutionary relationships and genome-wide patterns of nucleotide diversity and differentiation among lineages. Despite extensive shared polymorphism and limited differentiation from the other taxa, Bamako clusters apart from the other taxa, and forms a maximally supported clade in neighbor-joining trees based on whole genome data (including inversions) or solely on collinear regions. Nevertheless, FST outlier analysis reveals that the majority of differentiated regions between Bamako and typical An. gambiae are located inside chromosomal inversions, consistent with their role in the ecological isolation of Bamako. Exceptionally differentiated genomic regions were enriched for genes implicated in nervous system development and signaling. Candidate genes associated with a selective sweep unique to Bamako contain substitutions not observed in sympatric samples of the other taxa, and several insecticide resistance gene alleles shared between Bamako and other taxa segregate at sharply different frequencies in these samples. Bamako represents a useful window into the initial stages of ecological and genomic differentiation from sympatric populations in this important group of malaria vectors.
Keywords: adaptive radiation, Anopheles gambiae, ecological divergence, chromosomal inversions, genome scan, population genomics, selective sweeps
Introduction
Local adaptation to heterogeneous environments can produce genetically differentiated populations or ecotypes and potentially lead to ecological speciation (Nosil 2012, 2005; Schluter 2009). Although the genes and genetic mechanisms underlying local adaptation are elusive, empirical evidence and theoretical models suggest that initial genomic differentiation between ecotypes that are still exchanging genes should be confined to small chromosomal regions containing the loci directly responsible for phenotypes that are advantageous in one but not the other environment (Andrew & Rieseberg 2013; Flaxman et al. 2014; Nosil et al. 2009; Seehausen et al. 2014; Wu 2001; Yeaman & Whitlock 2011). Because migration, gene flow and recombination among diverging populations can break down locally adapted allelic combinations, genomic regions of reduced recombination—including chromosomal inversions—are likely to harbor loci that contribute to local adaptation and ecotypic differentiation (Butlin 2005; Hoffmann & Rieseberg 2008; Jackson et al. 2012; Joron et al. 2011; Kirkpatrick 2010; Kirkpatrick & Barton 2006; Lowry & Willis 2010; Nishikawa et al. 2015; Rieseberg 2001; Roesti et al. 2015; Yeaman 2013).
Structural rearrangements known as chromosomal inversions are now known to occur in most species, but they were first discovered in Drosophila melanogaster through their impact on recombination (Sturtevant 1917), which is suppressed between the breakpoints of alternative arrangements in inversion heterozygotes. Their recombination-reducing effect may be the basis for the role of chromosomal inversions in local adaptation. As modeled by Kirkpatrick and Barton (2006), if a set of locally adapted alleles is captured between the breakpoints of an inversion, the inversion can establish and spread in the population because the advantageous allelic combinations are maintained in the face of gene flow with migrant populations carrying other genetic backgrounds. Indeed, the adaptive significance of inversions is reflected in predictable shifts in polymorphic inversion frequencies in response to spatially or seasonally varying selection (Hoffmann & Rieseberg 2008; Krimbas & Powell 1992; Schaeffer 2008). Examples are clines in inversion frequencies related to climatic gradients that are replicated in different geographic locations, and in some cosmopolitan species, across continents (Etges & Levitan 2004; Knibb 1982; Krimbas & Powell 1992; Rako et al. 2006). Other examples are cyclical variations in inversion frequencies related to season (Dobzhansky 1948; Dubinin & Tiniakov 1946).
Twenty-five years before the formal mathematical modeling of Kirkpatrick and Barton (2006), Coluzzi (1982) proposed a conceptually related verbal model for ecotype formation in anopheline mosquitoes, via chromosomal inversions which are abundant in the genus. Anophelines possess only three pairs of chromosomes, and inversions frequently span more than 5% of the total chromosome complement, increasing the chance for an inversion to capture adaptively important variation. The high dispersal ability of anophelines, combined with the “boom-bust” nature of their population dynamics, can produce situations in which populations at the margins of suitable habitat become temporarily isolated—spatially and/or ecologically—and locally adapted. Such population isolates could be stabilized by the association of adaptive variation with inversions (Coluzzi 1982).
The Anopheles gambiae complex is an African group of at least eight species that radiated within the last 2 million years (Fontaine et al. 2015; White et al. 2011). Best known for its medical importance, this group contains three of the principal vectors of human malaria in Africa (An. arabiensis, An. coluzzii, and An. gambiae), which also are the most widely distributed. Although morphologically indistinguishable at all developmental stages, even the most closely related pair of species (An. coluzzii and An. gambiae) differ in aquatic larval ecology (Gimonneau et al. 2012; Kamdem et al. 2012), a factor that may have been instrumental in the radiation of the An. gambiae complex as a whole (Coluzzi et al. 2002). In addition, both interspecific fixed chromosomal inversion differences and intraspecific inversion polymorphisms are common and nonrandomly distributed in the genome, within and among chromosomes (Coluzzi et al. 2002; Pombi et al. 2008). Particularly striking is the disproportionate involvement of chromosome 2R in the autosomal inversions described in the An. gambiae complex. Although this arm comprises only one-third of the autosomal complement, it bears 21 of 34 main fixed or polymorphic autosomal rearrangements recorded in the species complex (Coluzzi et al. 2002).
Heterogeneities within what was initially considered to be a single panmictic species were uncovered in Mali through intensive longitudinal cytogenetic surveys of inversions on chromosome 2 (Toure et al. 1998). Whereas significant and stable heterozygote deficits were observed in sympatric samples considered as one randomly mating population, their partitioning into three reproductive units (“chromosomal forms”) restored Hardy-Weinberg equilibrium. Two of these forms, named Mopti and Savanna and later defined molecularly as M and S (della Torre et al. 2001), have been subsequently elevated to species and are now known as An. coluzzii and An. gambiae, respectively (Coetzee et al. 2013). The third, Bamako, is indistinguishable from An. gambiae based on rDNA sequence (Favia et al. 2001), a locus that contains fixed sequence differences between all recognized species in the An. gambiae complex (Besansky et al. 2006; Coetzee et al. 2013; Fettene & Temu 2003; Scott et al. 1993). However, Bamako carries a unique configuration of chromosomal inversions on chromosome 2R (Coluzzi et al. 1985). It is homokaryotypic (fixed) for inversions j, c, and u, with only inversion b segregating; thus, it is considerably less polymorphic chromosomally than sympatric typical (i.e. non-Bamako) An. gambiae populations and An. coluzzii from Mali, in which inversions b, c and u segregate at high frequencies (Della Torre et al. 2005; Toure et al. 1998). Inversion 2Rj is absent or very rare in An. coluzzii and generally rare in most An. gambiae populations, only reaching higher frequencies in a few localities in West Africa (Della Torre et al. 2005; Toure et al. 1998). Bamako also has a far more restricted distribution than typical An. gambiae populations or An. coluzzii, being endemic to the upper Niger River in southern Mali and northern Guinea. Although adults of all three taxa freely co-occur in the same Malian localities, often resting together inside the same buildings, their larvae are associated with different habitat types; for this reason, breeding is not always synchronous. Larvae of An. coluzzii are associated with rice fields and other permanent or semi-permanent irrigated sites in this part of Africa, and therefore this species can be abundant during the dry season (Gimonneau et al. 2012). Typical An. gambiae populations breed in ephemeral rain-dependent pools and puddles away from the river, but Bamako larvae are disproportionately found in more stable laterite rock pools beside the Niger River (Manoukis et al. 2008), which are only exposed as the river recedes at the end of the rains. With its peculiar karyotype, its association with a peripheral and ecologically marginal habitat, and cytogenetic evidence consistent with positive assortative mating (Coulibaly et al. 2007; Manoukis et al. 2008; Toure et al. 1998), Bamako appears to be in the earliest stages of ecological differentiation from sympatric typical An. gambiae populations.
The nature, distribution, and degree of genomic divergence between the Bamako ecotype and typical An. gambiae have been explored previously at increasing levels of genetic resolution, with puzzlingly inconsistent results. In 2006, analyses involving >10,000 AFLP bands uncovered no diagnostic differences between Bamako and typical An. gambiae, but revealed significant—albeit slight—genetic divergence in unknown genomic locations (Slotman et al. 2006). By 2010, a 400,000 SNP genotyping array (with median distance of ~300 bp between assayed SNPs) was available based on whole genome sequencing of An. gambiae and An. coluzzii (Lawniczak et al. 2010), and results indicated that the greatest genomic differentiation between Bamako and typical An. gambiae was found in inversions on 2R, as expected under the hypothesis that inversions are instruments of ecotypic divergence, while no significant differentiation was found on the X chromosome (Neafsey et al. 2010). However, in contradiction to these results, application of an even higher resolution whole genome tiling microarray suggested that the majority of genomic divergence between Bamako and typical An. gambiae was located on the X chromosome, leading to the inference that genes mediating isolation are likely on the X (Lee et al. 2013a), as may be the case for the sister species An. gambiae and An. coluzzii (Aboagye-Antwi et al. 2015). Both the genotyping array and the whole genome tiling microarray have technical and statistical limitations inherent in their design that may have biased or obscured measurements of genetic differentiation. To overcome these limitations and approach a more comprehensive understanding of the genetic architecture of ecotypic differentiation, we use whole genome re-sequencing of individuals and population pools and assess the pattern of genomic differentiation between Bamako, typical An. gambiae, and for comparison, An. coluzzii. As expected, our data reveal extensive differentiation between An. coluzzii and either Bamako or typical An. gambiae in the centromere-proximal region of the X chromosome. However, we observed virtually no genomic differentiation between the latter two taxa on the X chromosome. The majority of differentiation was associated with chromosome 2 inversions, implicating these rearrangements in the ecotypification process.
Materials and Methods
Sampling and sequencing strategy
We exploited prior indoor resting collections of adult An. gambiae and An. coluzzii from nearby villages in southern Mali, sampled in August–September of 2004 (Coulibaly et al. 2007). Cytogenetic karyotyping was performed using ovaries from females at the correct gonotrophic stage, and the corresponding carcass was molecularly identified to species (Coulibaly et al. 2007). Of the available population samples, a total of 87 adult female mosquitoes previously karyotyped cytogenetically and identified molecularly were used for this study: 39 An. gambiae Bamako, 16 typical An. gambiae, and 32 An. coluzzii (see Tables S1, S2, Supporting information, for sample provenances, karyotypes, and chromosomal inversion frequencies). For each of the three population samples, we used pooled sequencing (Pool-seq) to identify SNPs and estimate allele frequencies (Futschik & Schlotterer 2010). In addition to the pools, we analyzed individual whole genome sequences from each taxon, determined as part of a previous sequencing effort (Fontaine et al. 2015; Neafsey et al. 2015): 10 Bamako, 12 typical An. gambiae, and 12 An. coluzzii (Table S3).
Library preparation, sequencing, data processing and filtering of pools
A single DNA pool was prepared for each of the three population samples: Bamako (N=39), typical An. gambiae (N=16), and An. coluzzii (N=32). Each pool (200 ng total DNA) contained approximately equal contributions of genomic DNA from component mosquitoes. Pooled DNA was sheared into fragments with a Covaris S220 ultrasonicator (Covaris, Inc., Woburn, MA). After end-repair of the fragments, paired-end genomic libraries were constructed using the TruSeq DNA Nano Sample Preparation Kit (Illumina, Inc., San Diego, CA) and size-selected for an average fragment size of 550 bp using a BluePippin instrument (Sage Science™). Final average fragment library sizes were 539 bp for Bamako, 534 bp for typical An. gambiae, and 536 bp for An. coluzzii, corresponding to average insert sizes of 353 bp, 350 bp, and 350 bp, respectively. Each library was barcoded, and the three libraries were multiplexed such that the relative contribution of each pool to the combined total volume was proportional to the number of individuals in the pool: 45% Bamako, 18% typical An. gambiae, and 37% An. coluzzii. This was done so that sequencing coverage (per individual) would be comparable across pools, given the different pool sizes. Multiplexed barcoded libraries were sequenced with paired-end 100-bp runs on 2 lanes of an Illumina HiSeq 2000 at BGI (University of California, Davis). Raw sequencing data from the pools are available as SRA BioProject PRJNA311062.
Short reads from each pool were demultiplexed, and trimmed following the pipeline described in Fontaine et al. (2015). Reads were mapped to the An. gambiae PEST AgamP4 reference assembly [www.vectorbase.org; (Giraldo-Calderon et al. 2015)] using bwa-aln 0.7.12-r1044 (Li & Durbin 2009) with parameters (-I, -k 2, -o 1, -l 32). Further processing of the mapped paired reads followed Fontaine et al. (2015). This included soft clipping of any reads where part of the read extended beyond the end of the sequence to which it was aligned (using Picard Tools 1.102 CleanSam.jar; https://github.com/broadinstitute/picard), sorting of reads based on alignment location (using Picard Tools’ SortSam.jar), identification and marking of duplicate reads (using Picard Tools’ MarkDuplicates.jar), and localized realignment near insertion-deletions [with the Indel Realignment tool of the Genome Analysis Toolkit, GATK v3.3; (DePristo et al. 2011; McKenna et al. 2010)]. Quality of the resulting mapped reads was assessed with Qualimap v2.1.1 (Garcia-Alcalde et al. 2012). Reads were compiled by position into pileup files using SAMtools v.1.2 (Li et al. 2009), for each pool and collectively for all pools. We considered only bases with a minimum Phred-scaled mapping quality of 20 and a base quality of 30.
For each pool, we set a minimum coverage threshold of 10 reads. To avoid repetitive genomic regions with high coverage, a maximum coverage threshold per pool was imposed, equal to the top 1% coverage (corresponding to 106 for Bamako, 45 for typical An. gambiae, and 90 for An. coluzzii). For window-based analyses, any window with less than 60% of sites compliant with coverage requirements was excluded. For all analyses, we used a minimum minor allele count of 2. For all downstream analysis, only sites or windows that could be assigned to a chromosomal arm (2L, 2R, 3L, 3R, or X) were retained.
Data processing and filtering of individual whole genome sequences
Reads from the 34 individual whole genome sequences of Bamako, typical An. gambiae and An. coluzzii determined as part of a previous sequencing effort (Fontaine et al. 2015) were trimmed and aligned, following the workflow of Fontaine et al. (2015). Variants (SNPs) were called in a single cohort using the HaplotypeCaller walker of GATK v2.6–5. Variant calling was followed by variant quality score recalibration (VQSR) to differentiate true variants from false positives, as recommended for low-coverage samples (see www.broadinstitute.org/gatk/guide/article?id=3225). This recalibration produced a top tranche of SNPs (n=4,697,938 SNPs) defined as having variant quality score log odds greater than 2.007. Following VQSR, we retained biallelic SNPs with a minimum per-genotype depth of 3 and a minimum genotype quality of 15. We then constructed two data sets, one specific to Bamako individuals and used to estimate linkage disequilibrium (LD), and one for all 34 individuals from the three taxa. For the Bamako-only data set, sites with more than 3 missing genotypes (30%) were excluded. A total of 2,839,964 SNPs were retained after quality filtering. The second data set, after the exclusion of sites with more than 10 missing genotypes (29%) and the application of quality filters, consisted of 2,794,793 SNPs. For generating estimates of diversity, Tajima’s D, and differentiation, we retained only mosquito genomes sampled from the neighboring West African countries of Mali and Burkina Faso [i.e., we excluded the more geographically distant samples from Central Africa (Cameroon)]. For analysis of population clustering, we added the An. christyi reference genome (Neafsey et al. 2015), as An. christyi is the closest outgroup to the An. gambiae complex (Fontaine et al. 2015). Only sites that passed all previous filters and also had data for An. christyi (1,230,508 SNPs) were retained.
Population clustering
We used the R package adegenet v2.0.0 (Jombart 2008; Jombart & Ahmed 2011) to store SNP genotype data from individual genome sequences. We then used the R package ape v3.3 (Paradis et al. 2004) to compute a genotype-based Euclidian distance matrix between individuals and to construct neighbor-joining (NJ) trees. Node support was assessed using 100 bootstrap replicates. In addition, we used Admixture 1.23 (Alexander et al. 2009) to conduct an unsupervised estimate of ancestry using K=1–4 with cross-validation.
Estimation of population genetic parameters
Using a genome scan approach to compare patterns of polymorphism and divergence within and between taxa, we estimated standard population genetic parameters of within-population variation (average pairwise nucleotide diversity, π), deviation of the allele frequency spectrum from neutral expectation (Tajima’s D), and pairwise population differentiation (FST) and divergence (Dxy). From the Pool-seq data, parameter estimates were obtained using PoPoolation (Kofler et al. 2011a) and PoPoolation2 (Kofler et al. 2011b), as appropriate, based on non-overlapping windows of 1-kb. The window-based approach to parameter estimation was adopted in light of the unavoidably small sample sizes of diploid individuals included in our pools (<40 individuals), as recommended by Schlotterer (2014). Tajima’s D (Tajima 1989) within the pools was calculated using the correction of Achaz (2008), and because one of its components (the number of segregating sites) strongly depends on pool size, which differed between our taxa, we standardized the pooled data by randomly downsampling to a read coverage matching the smallest pool (typical An. gambiae) using Picard Tools’ DownsampleSam.jar, following Nolte et al. (2013). The average number of pairwise differences, Dxy (Nei 1987, equation 10.20), was calculated between reference sequences from each pool using PoPoolation 1.2.2. Reference sequences were generated with a custom Python script (accessible at github.com/rrlove/Bamako) that incorporates alleles with a probability proportional to their frequency in the population; progressiveMauve (Darling et al. 2010) was used to generate the input file required by Popoolation. From the individual sequences, parameters were estimated from larger (200-kb) non-overlapping windows using vcftools 0.1.11 and 0.1.12a (Danecek et al. 2011). All downstream analysis of population genetic parameter estimates was done in R (R Development Core Team 2016). Population genetic parameters were plotted in R using the packages ggplot2 (Wickham 2009), gridExtra (Auguie 2016), and scales (Wickham 2014).
Linkage disequilibrium
From individual sequences and within pools, we analyzed patterns of linkage disequilibrium (LD). From pools, LD was calculated using LDx (Feder et al. 2012) and custom R scripts. For each chromosome arm, we calculated the correlation coefficient r2 between pairs of SNPs spaced 200–300 bp apart in sliding windows of 1000 SNPs with step size of 100 SNPs. Because differences in pool size strongly affect LD estimates, we first down-sampled to a coverage matching the smallest (typical An. gambiae) pool, as described above. Variants used for LD calculations were called from the down-sampled data using the UnifiedGenotyper walker of GATK (due to the pooled nature of the data), and filtered to retain only variant sites that met the following criteria: minimum read depth of 10; maximum read depth fixed to the 99th percentile of coverage for the pool; PHRED-scaled base quality score of 20; minimum allele frequency of 0.1; insert sizes as reported above. LD calculated from pools was plotted using the R package zoo (Zeileis & Grothendieck 2005).
From individual Bamako sequences, LD (r2) was estimated on chromosome 2R with PLINK 1.90b (Purcell et al. 2007) and plotted with the R packages reshape2 (Wickham 2007) and LDheatmap (Shin et al. 2006). The heatmap was colored using RColorBrewer (Neuwirth 2011).
Functional enrichment analysis in outlier genomic regions
We used the Functional Annotation Clustering tool of DAVID 6.8 (Beta) (Huang et al. 2009) to determine whether certain pathways and functional annotation terms are statistically overrepresented in Bamako genomic regions with elevated differentiation from typical An. gambiae. The annotation databases used by this software (i.e., DAVID Knowledgebase) are current (updated May 2016). Input gene lists were compared to the background reference list (An. gambiae PEST AgamP4.3 gene set, 20 October 2015). Annotation clusters (groups of related terms that share gene members) whose overall enrichment score exceeds 1.3 (P<0.05) are considered significantly enriched. The overall enrichment score for a cluster is the geometric mean (in −log scale) of individual scores assigned to each annotation term in the cluster. Individual scores (known as EASE scores) are P-values based on a modified Fisher exact test (Huang et al. 2009).
Results
Sequencing of three population pools resulted in 682,456,384 paired-end reads; after adapter and quality trimming, we retained 579,119,542 paired-end reads (84.9%), with an average length of 95.5 bases. Of the retained reads, 434,130,146 (75.0%) mapped to the reference genome. These reads, after filtering for base and mapping quality, produced total mean coverage of 117X across all three pools, and mean coverage per pool (individual) of 51.1X (1.3X), 20.5X (1.3X), and 45.6X (1.4X), for Bamako, typical An. gambiae and An. coluzzii, respectively (Table 1 and Fig. S1, Supporting information). At least 80% of the chromosomally-assigned reference genome was accessible for analysis after implementation of quality filters in Bamako, typical An. gambiae and An. coluzzii: 88.1% (203,059 1-kb windows), 80.5% (185,544 1-kb windows), and 88.7% (204,484 1-kb windows), respectively.
Table 1.
Mean (median) [5–95 percentiles] for nucleotide diversity and Tajima’s Da from pools of An. gambiae s.l. from southern Mali.
| Bamako | An. gambiae | An. coluzzii | |
|---|---|---|---|
| Individuals per pool | 39 | 16 | 32 |
| Coverage | 51.1 (53.0) | 20.5 (20.0) | 45.6 (48.0) |
| π, Autosomes | 0.015 (0.014) [0.004 – 0.030] | 0.016 (0.015) [0.004 – 0.030] | 0.015 (0.014) [0.004 – 0.029] |
| π, X | 0.009 (0.008) [0.001–0.020] | 0.009 (0.008) [0.001–0.020] | 0.008 (0.007) [0.001 – 0.020] |
| D, Autosomes | −0.270 (−0.268) [−0.985 – 0.438] | −0.414 (−0.408) [−1.040 – 0.188] | −0.377 (−0.367) [−1.089 – 0.290] |
| D, X | −0.590 (−0.588) [−1.427 – 0.210] | −0.659 (−0.665) [−1.434 – 0.106] | −0.628 (−0.632) [−1.479 – 0.202] |
Tajima’s D calculations based on down-sampled data sets to achieve uniform coverage.
Previous resequencing of individual samples produced an average coverage depth of 8.9X across the three taxa, 9.5X for Bamako + typical An. gambiae, and 7.5X for An. coluzzii [see Table S5 of Fontaine et al. (2015) for further detail].
Population clustering
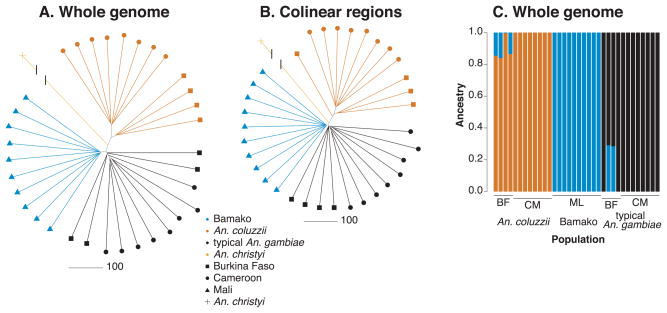
Using SNPs called from individual whole genome sequences representing our ingroup (Bamako, typical An. gambiae, and An. coluzzii) and the outgroup An. christyi, we constructed NJ distance trees (Fig. 1). When the NJ trees were constructed from the genome-wide SNP set (Fig. 1A), all Bamako individuals form a maximally supported cluster exclusive of the other two taxa. As expected, all internode distances—both within and between taxa—are very short, reflecting the low levels of differentiation in this group of taxa. To explore whether exclusive clustering of Bamako mosquitoes is driven solely by inverted regions on chromosome 2R that formally define this taxon, we next constructed trees based on SNPs from collinear parts of the genome. The Bamako mosquitoes again clustered together exclusive of the other taxa with maximal bootstrap support (Fig. 1B), suggesting an important degree of differentiation even outside of inverted genomic regions.
Fig. 1.
Population clustering of 34 individual samples of Bamako, typical An. gambiae, and An. coluzzii. Neighbor-joining trees based on whole genome data (A) or collinear (uninverted) genomic regions (B), rooted with the outgroup An. christyi. In both trees, Bamako clades are maximally supported by bootstrapping. Length of the branch leading to An. christyi is not to scale. (C) Unsupervised ancestry estimation from Admixture based on K=3 (BF, Burkina Faso; CM, Cameroon; ML, Mali).
As an independent assessment of population clustering, we used Admixture to conduct an unsupervised estimate of ancestry. Although cross-validation error was lowest for K=1 (Table S4, Supporting information), consistent with the very recent and modest differentiation of the taxa studied, K=3 correctly identified Bamako as a population group (ancestral component), as shown in Fig. 1C.
Genome-wide patterns of variation
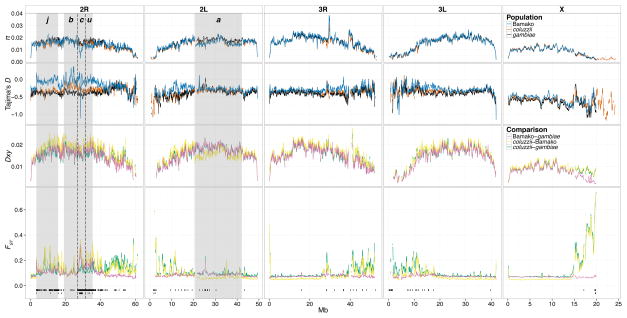
We conducted genome scans and generated summaries of genetic diversity in Bamako, typical An. gambiae, and An. coluzzii (Fig. 2, Table 1; Figs S2, S3, Table S5, Supporting information). Overall, nucleotide diversity (π) is lowest on the X chromosome in all three pools (Fig. S3b–e, Supporting information). In addition, all three taxa show sharp drops in nucleotide diversity near the centromeric regions where the recombination rate declines (Pombi et al. 2006), particularly on the X chromosome. This pattern is expected (Begun & Aquadro 1992) and was observed previously for species in the An. gambiae complex (Fontaine et al. 2015), although it was not observed in a recent population genomic study of Drosophila mauritiana (Nolte et al. 2013). In our data, this pattern may be exacerbated by lower coverage and missing data in centromere-proximal regions (Fig. S1, Supporting information). Overall, the patterns of diversity along chromosome arms are very similar among taxa, with the notable exception of inversions 2Rc and 2Ru, which show strongly decreased diversity in Bamako (Fig. 2; Fig. S2, Table S5, Supporting information).
Fig. 2.
Whole-genome scans of diversity and pairwise divergence for Bamako, typical An. gambiae and An. coluzzii population pools. Shaded boxes span genomic regions corresponding to known chromosome 2 inversions. All parameters are displayed in 200-kb windows slid 20-kb. (A) Mean nucleotide diversity, π; (B) Mean Tajima’s D; (C) Mean pairwise absolute divergence, Dxy; (D) Mean pairwise differentiation, FST. Below the line graph in (D), positions of 1-kb outlier FST windows (top row) and 1-kb outlier windows of putative positive selection (bottom row) are displayed as black vertical bars.
To identify deviations in the allele frequency spectrum from neutral expectation, we calculated Tajima’s D in windows across the genome (Fig. 2, Table 1). In general, all three taxa show negative Tajima’s D values genome-wide (Table 1), indicating an excess of rare variants, as observed in previous studies (e.g., Cohuet et al. 2008; Crawford & Lazzaro 2010; Donnelly et al. 2001; Donnelly et al. 2002; O’Loughlin et al. 2014). The lower level of variation observed on the X versus autosomes is paralleled by slightly more negative Tajima’s D values on the X chromosome in all taxa (Table 1). Across the rearranged regions of 2R, Bamako generally exhibits less strongly negative values for Tajima’s D relative to the other taxa. The exception to this trend is a strong negative spike in D inside the 2Rc inversion in the Bamako pool (Fig. 2), which was also captured in the genome scan from individually sequenced Bamako mosquitoes (Fig. S2, Supporting information).
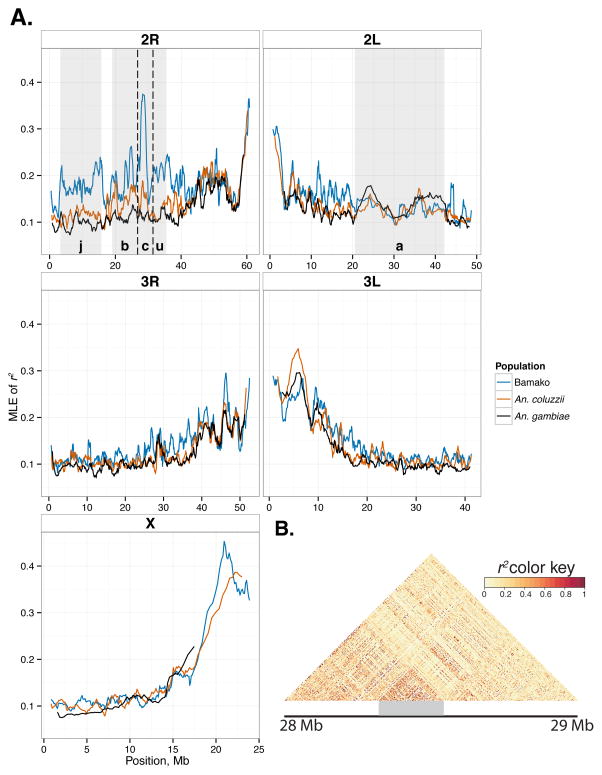
As expected owing to reduced recombination near centromeres, linkage disequilibrium (LD) is elevated in the centromere-proximal regions in all taxa. Patterns of LD across chromosome arms are comparable among taxa except on chromosome 2. On this chromosome, Bamako is distinguished by elevated LD inside inversions on 2R and strongly elevated LD inside inversion 2Rc; LD in Bamako is also moderately elevated on 2L, albeit outside of the 2La inversion in a ~6 Mb proximal region (Fig. 3). Inside the 2La inversion (fixed or at very high frequency in all three taxa in southern Mali; Table S2, Supporting information), LD does not appear to be elevated.
Fig. 3.
Analysis of linkage disequilibrium (r2) in Bamako, typical An. gambiae and An. coluzzii. (A) Sliding window analysis of LD between SNPs separated by 200–300 bp on the same set of reads in population pools (bin, 1000 SNPs; step 100 SNPs). Shaded boxes denote chromosomal inversions. (B) LD within 10 individual Bamako sequences, shown for the portion of chromosome 2R spanning 28 Mb to 29 Mb.
Genome-wide patterns of differentiation and divergence
We investigated how allelic frequencies in Bamako differ from the other two taxa (Fig. 2, Table 2; Figs S2, S4, Table S6, Supporting information). Mean absolute pairwise divergence (Dxy) is virtually identical between all pairs of taxa, ~0.015. Mean genome-wide differentiation (FST) is 0.084 between Bamako and typical An. gambiae, and slightly higher between An. coluzzii and either Bamako or typical An. gambiae (0.108 and 0.120, respectively). Overall, differentiation is slight among all three taxa, reflecting extensive shared ancestral polymorphism and likely some contemporary gene flow (Table 2). Of note, the strongly elevated levels of FST observed between An. coluzzii and either Bamako or typical An. gambiae near the centromeres [prompting the metaphor “speciation islands” (Turner et al. 2005)] are not echoed by Dxy (Fig. 2). Indeed, overall Dxy levels appear relatively constant across the chromosome arms, with one exception: in contrast to FST, Dxy is moderately depressed near all centromeres (and possibly also near telomeres), as expected (Cruickshank & Hahn 2014). The depression in Dxy near the centromeres is strongest between Bamako and typical An. gambiae—an effect most dramatic on the X chromosome. In the inverted regions on chromosome 2, especially on 2R, corresponding FST values between Bamako and typical An. gambiae are elevated.
Table 2.
Mean (median) [95 percentile] pairwise FST and Dxy values from pools of An. gambiae s.l. from southern Mali.
| Bamako-gambiae | Bamako-coluzzii | gambiae-coluzzii | ||
|---|---|---|---|---|
| FST: | Genome-wide | 0.084 (0.075) [0.146] | 0.108 (0.071) [0.298] | 0.120 (0.089) [0.287] |
| Autosomes, collinear | 0.077 (0.072) [0.121] | 0.095 (0.067) [0.244] | 0.115 (0.087) [0.264] | |
| Autosomes, rearranged | 0.112 (0.098) [0.215] | 0.125 (0.099) [0.300] | 0.111 (0.098) [0.203] | |
| X chromosome | 0.068 (0.066) [0.101] | 0.156 (0.055) [0.724] | 0.178 (0.080) [0.733] | |
| Dxy: | Genome-wide | 0.015 (0.013) [0.031] | 0.016 (0.014) [0.033] | 0.015 (0.013) [0.031] |
| Autosomes, collinear | 0.014 (0.013) [0.030] | 0.016 (0.014) [0.033] | 0.015 (0.013) [0.031] | |
| Autosomes, rearranged | 0.018 (0.016) [0.033] | 0.018 (0.016) [0.034] | 0.017 (0.016) [0.032] | |
| X chromosome | 0.008 (0.006) [0.020] | 0.010 (0.008) [0.022] | 0.009 (0.007) [0.021] |
Outlier loci between Bamako and typical An. gambiae
To gain more insight into the distribution and nature of genomic regions of differentiation between Bamako and typical An. gambiae, we focused on FST outliers based on non-overlapping 1-kb windows across the genome that passed all filters (N=182,174). Outliers were empirically defined as FST values in the top 1% of the genome-wide distribution (FST ≥ 0.233). Of the 1,822 outlier FST windows, the vast majority—92.5% (1,686)—are on chromosome 2R (Fig. 2, bottom panel), despite that 2R represents only 22.5% of the assembled genome (and only 27% of 1-kb windows in the analysis). Moreover, 87% of the outlier FST windows on 2R (1,474) are located inside of 2R chromosomal rearrangements (j, b, c, u), and of those, 81% (1,199) fell within the two smallest rearrangements, 2Rc (~4.67 Mb, 815 outlier windows) and 2Ru (~4.02 Mb, 384 outlier windows). The region corresponding to the 2Rb rearrangement (~7.73 Mb)—the only one in which both inverted and standard orientations are segregating in Bamako (the other three 2R rearrangements, j, c, and u, are fixed for the inverted orientation in this taxon)—contained only 20 outlier FST windows.
The remaining 7.5% (136) of outlier FST windows not located on 2R are distributed across the other chromosome arms: 65 on 2L, 37 on 3L, 28 on 3R, and 6 on the X chromosome. Even on these arms, the outlier windows are not dispersed uniformly. On 2L, 61.5% of the windows (40 of 65) are located in two small regions: a ~200-kb region inside of the ~22 Mb 2La inversion spanning chromosomal coordinates ~25.4–25.6 Mb that contains 30 windows, and a ~45-kb collinear region at position ~14.5 Mb that contains 10. Similarly, 60% of the windows on 3R are confined to three small regions totaling ~250-kb, and all but one of the six outlier windows on the X chromosome also are within a single ~250-kb region.
Genic differentiation between Bamako and typical An. gambiae
Candidate genes overlapping outlier FST windows were identified in two steps. An initial list, based on the gene models available in VectorBase (gene set AgamP4.3), was based on the 310 predicted protein-coding genes (and 8 noncoding RNAs) that overlap the 1,822 outlier FST windows identified in the genome-wide analysis reported above. This list was refined using a second conservative filtering step imposed in rearranged genomic regions, with the goal of reducing false positives in these regions that may be present owing to reduced recombination in inversion heterozygotes. Toward this end, we identified FST outlier windows separately inside each inversion, basing the empirical cutoff on the top percentile of values for windows spanning the focal inversion. The final list of 150 candidate genes (Table S7, Supporting information) is composed of the 59 genes overlapping outlier windows in collinear regions from the first list and the 91 genes overlapping windows in inverted regions that were identified as outliers in both the first and the second steps.
The functional annotation clustering tool of DAVID was able to cluster all but 19 of the 150 candidate genes, revealing two enriched pathways and four enriched annotation clusters with overlapping genic content (Table 3). The KEGG pathways ‘lysosome’ (ko04142) and ‘neuroactive ligand-receptor interaction’ (ko04080) were related to the annotation clusters, which were enriched with terms such as transmembrane helix (cluster 1), G-protein coupled receptor (GPCR; cluster 2), cytochrome P450 (CYP450; cluster 3), and immunoglobulin domain (cluster 4), a domain potentially involved in protein-protein and protein-ligand interactions. Many of the genes in these clusters are inferred to encode chemosensory receptors (including 12 GPCRs and three ionotropic receptors), ion channels and transporters putatively involved in neuroactive signaling, an activity that may be supported by additional candidate genes predicted to encode proton pumps, ion exchanger, and components of the endosomal pathway. It is noteworthy that among the ion channel genes are the GABA-gated chloride channel known as resistant to dieldrin (rdl), implicated in resistance to that insecticide, and the para sodium channel gene, implicated in resistance to DDT and permethrin. Oxidoreductase activity is represented by several desaturases as well as eight CYP450 genes, of which at least four have been previously implicated in insecticide resistance in An. gambiae and An. coluzzii (see below).
Table 3.
Enrichment analysis of genes in regions of exceptional Bamako-typical An. gambiae differentiation
| Pathway or Annotation Cluster | Score | Count |
|---|---|---|
| KEGG lysosome | P=1.7E-3 | 5 |
| KEGG neuroactive ligand-receptor interaction | P=3.8E-2 | 3 |
| Transmembrane helix | 4.2 (P=6.3E-5) | 53 |
| G protein coupled receptor | 4.1 (P=7.9E-5) | 12 |
| Cytochrome P450 | 2.8 (P-1.6E-3) | 17 |
| Immunoglobulin | 1.9 (P=1.3E-2) | 12 |
We used OrthoDB (Kriventseva et al. 2015) to identify the orthologous group—at the level of Diptera—to which each candidate An. gambiae gene belongs, and to retrieve the corresponding functional annotations (indicated in Table S7, Supporting information). The power of this approach is that OrthoDB provides not only integrated functional annotations from UniProt, InterPro and GO, but also (where available from FlyBase) the associated mutant phenotype(s) based on experimentation with the model organism D. melanogaster. We found that the D. melanogaster orthologs of many of our candidate genes have been implicated experimentally in neurogenesis, neuromuscular synapse formation, neuronal signaling and a variety of behaviors controlled by the nervous system (e.g., locomotion, sleep, learning/memory, courtship). The concentration of Bamako-An. gambiae differentiation in 2R inversions, and the enrichment of genes implicated in nervous system development and signaling in these regions, invite the hypothesis that changes in Bamako perception and response to external cues may underlie its adaptation to marginal rock pool larval habitat.
Localized footprints of positive selection
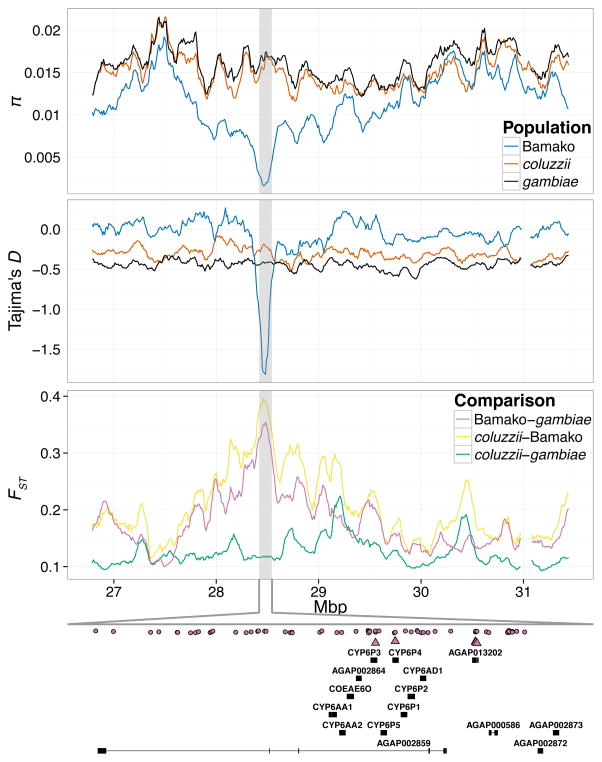
The subset of 1-kb outlier windows of FST that also are depauperate of nucleotide diversity within Bamako (i.e., they fall in the top and bottom 1% of genomic distributions of FST and π, respectively) may be associated with targets of positive selection in Bamako. Of 103 such windows, all except 5 are on chromosome 2R. The 98 windows on 2R are mainly (86%) distributed in two small clusters: a ~28-kb region near the distal breakpoint of the 2Rj inversion (N=20), and a ~1.9-Mb region in the distal half of 2Rc (N=64). As the 2Rj and 2Rc inversions are homokaryotypic in Bamako, the reduction in nucleotide diversity in these regions is unlikely to be due to suppressed recombination in the 2Rj and 2Rc regions of the Bamako genome. Nevertheless, LD within euchromatic regions of Bamako reaches its highest value in a 1000-SNP (~1.6 Mb) window centered at ~28.31 Mb inside 2Rc, where mean r2 is 0.375 in the Bamako pool (Fig. 3A) and individual Bamako mosquitoes show a noticeable haplotype block (Fig. 3B).
There are 24 candidate positively selected genes that overlap this set of 103 outlier windows and also are present in the refined list of genes from exceptionally differentiated genomic regions (Table S7). Several of the candidate CYP450s and para have been implicated repeatedly in insecticide resistance not only in An. gambiae and An. coluzzii, but also in other anopheline and insect species (Djegbe et al. 2014; Edi et al. 2014; Ranson et al. 2011; Silva et al. 2014; Tene Fossog et al. 2013; Toe et al. 2015), thus it is plausible that they may have been recent targets of positive selection in Bamako. CYP6P3 and CYP64 are two of eight consecutive CYP450 genes in the CYP6 subfamily, located within inversion 2Rc (Fig. 4). Both CYP6P3 and CYP64 genes carry nonsynonymous substitutions (CYP6P3, G151E: 2R 28,492,690; CYP6P4, P289L: 2R 28,497,809) that are apparently fixed in our pooled Bamako sample but are not detected in either of the other pools of typical An. gambiae or An. coluzzii; nor do these Bamako variants appear in other CYP6P3-4 gene sequences from An. gambiae s.l. deposited in VectorBase. Immediately downstream of the eight CYP6 genes inside 2Rc is another gene in this candidate set (AGAP13202), intriguing because it carries three nonsynonyomous substitutions that are apparently fixed or at high frequency in the Bamako pooled sample (2R: 28,518,614; 28,518,671; 28,519,004) but are not detected in the other pools. Unfortunately, AGAP13202 is one of several genes on this list devoid of any functional annotation, and this gene lacks known orthologs apart from those in other, similarly unannotated, anophelines.
Fig. 4.
Putative selective sweep on chromosome 2R in the Bamako population. Parameters were calculated in 100-kb windows slid 10-kb. (A) Mean nucleotide diversity π; (B) Mean Tajima’s D; (C) Mean pairwise differentiation, FST; (D) Genes wholly contained in the pictured region, with exons as black rectangles and introns as lines connecting them. Approximate location of SNPs occurring at high FST (≥ 0.9) between Bamako and typical An. gambiae is indicated by pink dots (synonymous or non-coding) and pink triangles (non-synonymous). SNPs were identified in the pooled samples at sites that met the coverage minimum and maximums described for the windowed analyses and had a minimum minor allele count of 2.
One of only two genes from this list on chromosome 2L—the voltage-gated sodium channel para—carries an insecticide resistance mutation well-known in West Africa (L1014F kdr) at low frequency in the Bamako pool (~3% of Bamako reads) and moderate frequency in typical An. gambiae (~35% of reads covering that site), congruent with previous studies from southern Mali (Fanello et al. 2003; Tripet et al. 2007). We did not detect the L1014F mutation in the An. coluzzii pool. An additional seven membrane channel/transporter genes (as well as other candidates on the list) carried no nonsysnonymous substitutions, but may have been associated with undetected regulatory mutations.
Although the GABA-gated chloride channel gene (rdl) was not included among the positive selection candidates, the known association of characteristic mutations in this gene with insecticide resistance in An. gambiae s.l. (Du et al. 2005) and other insects (Asih et al. 2012; Ffrench-Constant et al. 2000; Wondji et al. 2011), and the previous suggestion of recent independent selective sweeps centered on rdl in sympatric populations of An. gambiae and An. coluzzii from Mali (Lawniczak et al. 2010) prompted a closer examination. Two adjacent nonsynonymous substitutions affecting the same amino acid have been reported in An. gambiae s.l., one (Ala296Gly) observed in typical An. gambiae, the other (Ala296Ser) in An. coluzzii (Du et al. 2005; Lawniczak et al. 2010). We find that ~25% of reads in the typical An. gambiae pool match the Ala296Gly allele, which was not detected in the other pools of An. coluzzii and Bamako. The Ala296Ser allele was at high frequency in the An. coluzzii pool (~79%), and although it was sampled only once from typical An. gambiae, it was detected in ~40% of the pooled Bamako reads covering that site.
Discussion
Decades of cytogenetic evidence from Mali, based on both larval and adult samples, have supported the existence of an assortatively mating ecotype Bamako, despite the strict sympatry and partial syntopy of two additional reproductive units that are fully inter-fertile with Bamako: typical An. gambiae and An. coluzzii (Coluzzi et al. 1985; Toure et al. 1998). Our analyses based on whole genome sequencing of these taxa suggest that despite extensive shared variation, Bamako is a distinct evolutionary lineage. In neighbor-joining trees reconstructed from multiple individual genomic sequences of the three taxa, the Bamako individuals all cluster together exclusive of other taxa, even when chromosome 2R rearrangements are omitted from the analysis. Yet, the vast majority of differentiation between Bamako and typical An. gambiae occurs in the 2R inversions used to classify Bamako (2Rj, c and u), especially within the 2Rc region. This finding may have a larger evolutionary significance in the An. gambiae complex. A polytene chromosome analysis of this group of species revealed that the central part of chromosome 2R, corresponding to the 2Rc region, has been captured repeatedly by fixed and polymorphic chromosomal inversions in the species complex (Coluzzi et al. 2002). The species in this recent radiation are all morphologically indistinguishable at every developmental stage; their most obvious phenotypic differences are associated with preferred larval habitat (e.g., ephemeral versus stable; natural versus man-made; fresh water versus brackish). The centrality of the 2Rc region in chromosome changes within and between species prompted Coluzzi (2002) to speculate that genic content in this region may play an important role in the choice of oviposition site by gravid females and larval adaptation to breeding sites that differ in abiotic and biotic characteristics such as stability, biotic complexity, and salinity. The enrichment in this genomic region of functions related to nervous system development and signaling is broadly consistent with this hypothesis, which now calls for difficult follow-up studies aimed at connecting genotype to specific phenotypes and measuring fitness consequences (Barrett & Hoekstra 2011).
At a variety of loci associated with insecticide resistance (e.g., Cyp6P3, Cyp6P4, rdl, para), presumed resistance alleles in Bamako expected to confer a universal benefit upon exposure to particular classes of insecticides are nevertheless segregating at very different frequencies in sympatric samples of typical An. gambiae and An. coluzzii, underscoring the distinctiveness of the Bamako taxon (see also Main et al. 2015). Most striking is the Bamako-specific selective sweep in 2Rc centered on CYP6P3 and CYP6P4, genes fixed for (yet uncharacterized) nonsynonymous substitutions apparently absent from co-occurring population samples of An. coluzzii and typical An. gambiae. The rdl locus also supports the idea of Bamako as a reproductive unit distinct from typical An. gambiae. At this locus, a resistance allele common in typical An. gambiae is absent from Bamako, which instead carries an alternative resistance allele in common with An. coluzzii, albeit at a sharply lower frequency. Conceivably, frequency differences between Bamako and the other taxa with respect to insecticide resistance alleles may be the outcome of different degrees of xenobiotic exposure rather than reproductive isolation. This explanation seems unlikely for several reasons. Insecticides used in public health are applied indoors, on bed nets or walls, where the probability of contact by adults of all three taxa should be equivalent, given their equally high rates of anthropophily (Coluzzi et al. 1985). Although insecticides used in agriculture may differentially run off into spatially and ecologically distinct larval breeding sites utilized by these three taxa, habitat partitioning is not absolute (e.g., Manoukis et al. 2008). Actual larval habitat use overlaps considerably, implying at least some degree of common exposure to habitat-associated chemicals and pathogens. An alternative perspective, sometimes forgotten, is that resistance-associated mutations and the genes in which they occur may have other functions important to organismal fitness that are not necessarily related to insecticide resistance (Ffrench-Constant 2013). For example, although rdl is associated with resistance to dieldrin and fipronil insecticides, it is also involved in regulation of sleep, aggression and olfactory learning (Liu et al. 2014; Liu et al. 2009; Yuan et al. 2014). In the same vein, the TEP1 gene, whose complement C3-like product has been of great interest to vector biologists due to its role in anti-Plasmodium immunity, carries alternative alleles that render An. gambiae susceptible or refractory to Plasmodium infection (Blandin et al. 2009). However, it was recently shown that one of the same alleles associated with Plasmodium susceptibility (TEP1*S2) also functions during spermatogenesis to confer superior male fertility relative to the refractory (and other) TEP1 alleles, suggesting possible trade-offs between reproduction and immunity (Pompon & Levashina 2015). Interestingly, in our Pool-seq data the Bamako pool differs from typical An. gambiae and An. coluzzii at the TEP1 locus, in that the Bamako sample appears fixed for key nonsynonymous substitutions characteristic of the TEP1*S2 allele, in contrast to the other two taxa whose corresponding sequences are polymorphic but strongly skewed toward refractory alleles (data not shown). Taken together, our data suggest that differences in resistance allele frequencies among the co-occurring taxa reflect an ongoing process of ecological isolation.
Although the vast majority of genomic divergence between Bamako and typical An. gambiae coincides with inversions on 2R, all 2R inversions are shared among taxa. Indeed, with the exception of 2Rj (generally rare in typical An. gambiae and An. coluzzii populations), 2R inversions characteristic of (and fixed in) Bamako are polymorphic at moderate to high frequencies in the other taxa. For example, in a foundational synthesis of cytogenetic data from Mali (Toure et al. 1998), a 1983 collection of ~1400 mosquitoes sampled from the middle of the rainy season in the village of Banambani was karyotyped and analyzed in detail. The Bamako subsample carried two chromosome 2R karyotypes (jcu and jbcu) at frequencies of ~78% and ~22%. However, karyotypes that included the b, c, and u inversions also were common in typical An. gambiae (e.g., bcu, ~17%; cu, ~23%) and An. coluzzii (e.g., bc, ~56%). The most extreme frequency differences were found for the j inversion, which was fixed in Bamako while segregating at frequencies of only ~7% and ~3% in typical An. gambiae and An. coluzzii, respectively. Hence the role of inversions in helping to prevent suites of locally adapted alleles beneficial to the Bamako ecotype from recombining into other genetic backgrounds is most easily envisioned for 2Rj. In this light, it is striking that most differentiation observed between Bamako and typical An. gambiae was observed in 2Rc, but this observation does not negate an instrumental role for 2Rj, even if differentiation is difficult to detect. In addition, because inversion frequencies cycle seasonally (Toure et al. 1994; Toure et al. 1998), frequency differences may be more pronounced depending upon the time of year, and this factor may interact with other factors that may contribute to isolation, including spatial heterogeneity and asynchronous variation in relative abundance of the different populations (Toure et al. 1998).
The pattern of genomic differentiation that we find between Bamako and typical An. gambiae is concordant with previous results based on a 400,000 SNP genotyping array (Neafsey et al. 2010). That study, like ours, found that differentiation between this pair of taxa was almost exclusively located in rearranged regions of chromosome 2R (2Rj, ~3.2–3.6 Mb; 2Rb, ~19.6 Mb; 2Rc, ~27–29 Mb; 2Ru, 35.3 Mb) and 2L (2La, ~23.3–25.5 Mb); no differentiation was found on the X chromosome (Table S2 of Neafsey et al. 2010). On the other hand, a 2013 study employing a tiling microarray to examine the distribution of genomic differentiation between Bamako and typical An. gambiae reported strongly contradictory results (Lee et al. 2013a), in which the majority of divergence was on the X chromosome, particularly near the centromere—a pattern reminiscent of the X chromosome “speciation island” between An. gambiae and An. coluzzii (Turner et al. 2005). The tiling microarray employed a perfect-match probe design based on the An. gambiae (AgamP3) PEST reference genome, a design that could theoretically result in false negatives (if one or both taxa differ with respect to the probe sequence such that hybridization is prevented and true differentiation cannot be detected). However, it is unlikely that such a design could result in false positive detection of differentiation. To explain the discrepancy between the tiling array and the SNP array studies, Lee et al. (2013a) proposed that the SNP array study might have failed to detect true differentiation between Bamako and typical An. gambiae in the centromere-proximal X chromosome region due to bias in the array design. Indeed, in the design of the SNP genotyping array, SNPs near the X centromere were preferentially included that were fixed for different alleles between An. gambiae and An. coluzzii, instead of polymorphic in both taxa (Neafsey et al. 2010). Such SNPs, if they did not happen to be polymorphic between Bamako and typical An. gambiae, would have been uninformative. Our whole genome sequencing data do not suffer from that bias. In centromere-proximal regions, we are able to clearly show the expected centromere-proximal elevation of FST between An. coluzzii and either Bamako or typical An. gambiae. Nevertheless, comparable differentiation between Bamako and typical An. gambiae is entirely absent from the corresponding centromere-proximal region. Thus, rather than a technical issue, we favor the hypothesis that the difference in the genomic patterns of divergence between Lee et al. (2013a) on the one hand, and both Neafsey et al. (2010) and the present study on the other, has to do with differences between the mosquito samples. Although very speculative, we propose that at least a fraction of the “typical” An. gambiae samples hybridized to the tiling microarray by Lee et al. (2013a) may actually have represented An. gambiae-An. coluzzii hybrids (possibly advanced generation hybrids). These authors have recently shown that such hybrids occur in nature more frequently than had been appreciated, including in Mali (Lee et al. 2013b). Assuming that these hybrids carried some of the X chromosome alleles that are highly diverged between An. gambiae and An. coluzzii, their inadvertent inclusion in the microarray experiment as “pure” An. gambiae samples presumably would have produced the observed elevated divergence on the X chromosome that was interpreted as divergence between Bamako and An. gambiae.
In the present study, we compared patterns of genomic differentiation among ecotypes sampled at the same time from a common geographic area in Mali, a design that helps to minimize the impact of spatial or temporal variation on patterns of genomic variation. However, our study had a number of limitations that complicate interpretation of the genome scans. Perhaps the most obvious is that by their very nature, polymorphic chromosomal inversions reduce recombination, causing difficulties in distinguishing true targets of positive selection from linked false positives. Assuming that at least some important phenotypic differences among ecotypes are manifest at the level of gene expression (Fuller et al. 2016; Gilad et al. 2006; Mack et al. 2016), transcriptional profiling could constitute a complementary approach to identifying candidate genes that would not be subject to the same limitation. Another important limitation is the near-complete lack of prior phylogeographic or demographic information concerning the Bamako ecotype, and the absence of other geographic samples available for study. The Bamako ecotype has been understudied, at least in part due to its relative rarity (even in Mali) and its consequent minor contribution to the overall malaria vectorial system. However, in the absence of expanded sampling and attention to population demographic history, robust inferences about the nature and the genetic basis of ecotypic differentiation will not be possible.
Studying the genome architecture and genetic mechanisms that facilitate adaptive radiations is fundamental to understanding the origins of biological diversity. In the case of anopheline mosquito vectors of human malaria, this approach can also provide insights into the evolutionary forces that affect vectorial capacity (Cohuet et al. 2010) and malaria epidemiology. Although An. gambiae may be the most intensively studied, nearly all of the ~10% of anopheline species that transmit malaria in nature are the products of recent and rapid species radiations. A better understanding of their evolutionary ecology is important both for the development and the success of vector control strategies in the global fight against malaria.
Supplementary Material
Acknowledgments
We thank H. Uyhelji and the Notre Dame Genomics Core Facility for assistance with library construction; R. Waterhouse for assistance with OrthoDB; C. Witzig for programmatic advice and scripting assistance; M. Hahn for helpful discussion and feedback; and six anonymous reviewers. This work was supported by NIH grant R01AI076584 (NJB, RRL), the FNIH through the VCTR program of the Grand Challenges in Global Health Initiative (NJB, MCF), a Richard and Peggy Notebaert Premier Fellowship (RRL), and a Jack Kent Cooke Foundation Graduate Scholarship (RRL).
Footnotes
Data Accessibility
Pooled sequence data generated for this project has been deposited in the Sequence Read Archive under BioProject PRJNA311062. VCF files generated for this project are available on Dryad at doi:10.5061/dryad.m2821. Individual sequence data generated for this project was previously submitted to NCBI under BioProject ID PRJNA254046.
Author Contributions
Designed project: RRL, MCF, NJB; Contributed samples: MBC, ST; Designed alignment and variant calling pipelines: AS, MCF, SJE; Performed analysis: RRL, MCF; Wrote manuscript: RRL, MCF, NJB, with input from coauthors.
References
- Aboagye-Antwi F, Alhafez N, Weedall GD, et al. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X-chromosome divergence island in incipient sympatric speciation. PLoS genetics. 2015;11:e1005141. doi: 10.1371/journal.pgen.1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achaz G. Testing for neutrality in samples with sequencing errors. Genetics. 2008;179:1409–1424. doi: 10.1534/genetics.107.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew RL, Rieseberg LH. Divergence is focused on few genomic regions early in speciation: incipient speciation of sunflower ecotypes. Evolution. 2013;67:2468–2482. doi: 10.1111/evo.12106. [DOI] [PubMed] [Google Scholar]
- Asih PB, Syahrani L, Rozi IE, et al. Existence of the rdl mutant alleles among the anopheles malaria vector in Indonesia. Malar J. 2012;11:57. doi: 10.1186/1475-2875-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguie B. R package version 2.2.1. 2016. gridExtra: miscellaneous functions for “grid” graphics. [Google Scholar]
- Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Collins FH, Townson H. A species-specific PCR for the identification of the malaria vector Anopheles bwambae. Annals of Tropical Medicine and Parasitology. 2006;100:277–280. doi: 10.1179/136485906X105624. [DOI] [PubMed] [Google Scholar]
- Blandin SA, Wang-Sattler R, Lamacchia M, et al. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science. 2009;326:147–150. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK. Recombination and speciation. Molecular Ecology. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- Cohuet A, Harris C, Robert V, Fontenille D. Evolutionary forces on Anopheles: what makes a malaria vector? Trends Parasitol. 2010;26:130–136. doi: 10.1016/j.pt.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Cohuet A, Krishnakumar S, Simard F, et al. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics. 2008;9:227. doi: 10.1186/1471-2164-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M. Spatial distribution of chromosomal inversions and speciation in anopheline mosquitoes. In: Barigozzi C, editor. Mechanisms of Speciation. Alan R. Liss, Inc; New York: 1982. pp. 143–153. [PubMed] [Google Scholar]
- Coluzzi M, Petrarca V, Dideco MA. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Bollettino di Zoologia. 1985;52:45–63. [Google Scholar]
- Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Coulibaly MB, Pombi M, Caputo B, et al. PCR-based karyotyping of Anopheles gambiae inversion 2Rj identifies the BAMAKO chromosomal form. Malaria journal. 2007;6:133. doi: 10.1186/1475-2875-6-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, Lazzaro BP. The demographic histories of the M and S molecular forms of Anopheles gambiae s.s. Molecular Biology and Evolution. 2010;27:1739–1744. doi: 10.1093/molbev/msq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Torre A, Fanello C, Akogbeto M, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Molecular Biology. 2001;10:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochemistry and Molecular Biology. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Depristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djegbe I, Agossa FR, Jones CM, et al. Molecular characterization of DDT resistance in Anopheles gambiae from Benin. Parasit Vectors. 2014;7:409. doi: 10.1186/1756-3305-7-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of Natural Populations. Xvi. Altitudinal and Seasonal Changes Produced by Natural Selection in Certain Populations of Drosophila Pseudoobscura and Drosophila Persimilis. Genetics. 1948;33:158–176. doi: 10.1093/genetics/33.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Molecular Biology and Evolution. 2001;18:1353–1364. doi: 10.1093/oxfordjournals.molbev.a003919. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Simard F, Lehmann T. Evolutionary studies of malaria vectors. Trends in Parasitology. 2002;18:75–80. doi: 10.1016/s1471-4922(01)02198-5. [DOI] [PubMed] [Google Scholar]
- Du W, Awolola TS, Howell P, et al. Independent mutations in the Rdl locus confer dieldrin resistance to Anopheles gambiae and An. arabiensis. Insect Molecular Biology. 2005;14:179–183. doi: 10.1111/j.1365-2583.2005.00544.x. [DOI] [PubMed] [Google Scholar]
- Dubinin NP, Tiniakov GG. Seasonal cycle and inversion frequency in populations. Nature. 1946;157:23. doi: 10.1038/157023a0. [DOI] [PubMed] [Google Scholar]
- Edi CV, Djogbenou L, Jenkins AM, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etges WJ, Levitan M. Palaeoclimatic variation, adaptation and biogeography of inversion polymorphisms in natural populations of Drosophila robusta. Biological Journal of the Linnean Society. 2004;81:395–411. [Google Scholar]
- Fanello C, Petrarca V, Della Torre A, et al. The pyrethroid knock-down resistance gene in the Anopheles gambiae complex in Mali and further indication of incipient speciation within An. gambiae s.s. Insect Molecular Biology. 2003;12:241–245. doi: 10.1046/j.1365-2583.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Molecular Biology. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- Feder AF, Petrov DA, Bergland AO. LDx: estimation of linkage disequilibrium from high-throughput pooled resequencing data. PloS one. 2012;7:e48588. doi: 10.1371/journal.pone.0048588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettene M, Temu EA. Species-specific primer for identification of Anopheles quadriannulatus sp. B (Diptera: Culicidae) from Ethiopia using a multiplex polymerase chain reaction assay. Journal of Medical Entomology. 2003;40:112–115. doi: 10.1603/0022-2585-40.1.112. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant RH. The molecular genetics of insecticide resistance. Genetics. 2013;194:807–815. doi: 10.1534/genetics.112.141895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Anthony N, Aronstein K, Rocheleau T, Stilwell G. Cyclodiene insecticide resistance: from molecular to population genetics. Annual Review of Entomology. 2000;45:449–466. doi: 10.1146/annurev.ento.45.1.449. [DOI] [PubMed] [Google Scholar]
- Flaxman SM, Wacholder AC, Feder JL, Nosil P. Theoretical models of the influence of genomic architecture on the dynamics of speciation. Molecular Ecology. 2014;23:4074–4088. doi: 10.1111/mec.12750. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347:1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW. Genomics of Natural Populations: How Differentially Expressed Genes Shape the Evolution of Chromosomal Inversions in Drosophila pseudoobscura. Genetics. 2016;204:287–301. doi: 10.1534/genetics.116.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik A, Schlotterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Okonechnikov K, Carbonell J, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends in Genetics. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Gimonneau G, Pombi M, Choisy M, et al. Larval habitat segregation between the molecular forms of the mosquito, Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Medical and Veterinary Entomology. 2012;26:9–17. doi: 10.1111/j.1365-2915.2011.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderon GI, Emrich SJ, Maccallum RM, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Research. 2015;43:D707–713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annual Review of Ecology Evolution and Systematics. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jackson B, Kawakami T, Cooper S, Galindo J, Butlin R. A genome scan and linkage disequilibrium analysis among chromosomal races of the Australian grasshopper Vandiemenella viatica. PloS one. 2012;7:e47549. doi: 10.1371/journal.pone.0047549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C, Tene Fossog B, Simard F, et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PloS one. 2012;7:e39453. doi: 10.1371/journal.pone.0039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. How and why chromosome inversions evolve. PLoS biology. 2010;8:e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR. Chromosome Inversion Polymorphisms in Drosophila-Melanogaster .2. Geographic Clines and Climatic Associations in Australasia, North-America and Asia. Genetica. 1982;58:213–221. [Google Scholar]
- Kofler R, Orozco-Terwengel P, De Maio N, et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PloS one. 2011a;6:e15925. doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlotterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011b;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. Drosophila inversion polymorphism. CRC Press; London: 1992. [Google Scholar]
- Kriventseva EV, Tegenfeldt F, Petty TJ, et al. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Research. 2015;43:D250–256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, Emrich SJ, Holloway AK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Collier TC, Sanford MR, et al. Chromosome inversions, genomic differentiation and speciation in the African malaria mosquito Anopheles gambiae. PloS one. 2013a;8:e57887. doi: 10.1371/journal.pone.0057887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Norris LC, et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2013b;110:19854–19859. doi: 10.1073/pnas.1316851110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, et al. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron. 2014;82:151–166. doi: 10.1016/j.neuron.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Buchanan ME, Han KA, Davis RL. The GABAA receptor RDL suppresses the conditioned stimulus pathway for olfactory learning. Journal of Neuroscience. 2009;29:1573–1579. doi: 10.1523/JNEUROSCI.4763-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack KL, Campbell P, Nachman MW. Gene regulation and speciation in house mice. Genome Research. 2016;26:451–461. doi: 10.1101/gr.195743.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Lee Y, Collier TC, et al. Complex genome evolution in Anopheles coluzzii associated with increased insecticide usage in Mali. Molecular Ecology. 2015;24:5145–5157. doi: 10.1111/mec.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukis NC, Powell JR, Touré MB, et al. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proceedings of the National Academy of Science U S A. 2008;105:2940–2945. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Lawniczak MK, Park DJ, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330:514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, et al. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Neuwirth E. R package version 1.0–5. 2011. RColorBrewer: ColorBrewer palettes. [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nature Genetics. 2015;47:405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- Nolte V, Pandey RV, Kofler R, Schlotterer C. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Research. 2013;23:99–110. doi: 10.1101/gr.139873.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. Ecological Speciation. Oxford University Press; Oxford: 2012. [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- O’loughlin SM, Magesa S, Mbogo C, et al. Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Molecular Biology and Evolution. 2014;31:889–902. doi: 10.1093/molbev/msu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, et al. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC evolutionary biology. 2008;8:309. doi: 10.1186/1471-2148-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, Stump AD, Della Torre A, Besansky NJ. Variation in recombination rate across the X chromosome of Anopheles gambiae. American Journal of Tropical Medicine and Hygiene. 2006;75:901–903. [PubMed] [Google Scholar]
- Pompon J, Levashina EA. A New Role of the Mosquito Complement-like Cascade in Male Fertility in Anopheles gambiae. PLoS Biol. 2015;13:e1002255. doi: 10.1371/journal.pbio.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- Rako L, Anderson AR, Sgro CM, Stocker AJ, Hoffmann AA. The association between inversion In(3R)Payne and clinally varying traits in Drosophila melanogaster. Genetica. 2006;128:373–384. doi: 10.1007/s10709-006-7375-7. [DOI] [PubMed] [Google Scholar]
- Ranson H, N’guessan R, Lines J, et al. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends Parasitol. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Roesti M, Kueng B, Moser D, Berner D. The genomics of ecological vicariance in threespine stickleback fish. Nat Commun. 2015;6:8767. doi: 10.1038/ncomms9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8:336–352. [Google Scholar]
- Schaeffer SW. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution. 2008;62:3082–3099. doi: 10.1111/j.1558-5646.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- Schlotterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nature Reviews Genetics. 2014;15:749–763. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. American Journal of Tropical Medicine and Hygiene. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, et al. Genomics and the origin of species. Nature Reviews Genetics. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- Shin J-H, Blay S, Mcneney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. Journal of statistical software. 2006;16 Code Snippet 3. [Google Scholar]
- Silva AP, Santos JM, Martins AJ. Mutations in the voltage-gated sodium channel gene of anophelines and their association with resistance to pyrethroids - a review. Parasit Vectors. 2014;7:450. doi: 10.1186/1756-3305-7-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Mendez MM, Torre AD, et al. Genetic differentiation between the BAMAKO and SAVANNA chromosomal forms of Anopheles gambiae as indicated by amplified fragment length polymorphism analysis. American Journal of Tropical Medicine and Hygiene. 2006;74:641–648. [PubMed] [Google Scholar]
- Sturtevant AH. Genetic factors affecting the strength of linkage in Drosophila. Proceedings of the National Academy of Sciences U S A. 1917;3:555–558. doi: 10.1073/pnas.3.9.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Poupardin R, Costantini C, et al. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaounde Cameroon. PloS one. 2013;8:e61408. doi: 10.1371/journal.pone.0061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toe KH, N’fale S, Dabire RK, Ranson H, Jones CM. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genomics. 2015;16:146. doi: 10.1186/s12864-015-1342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure YT, Petrarca V, Traore SF, et al. Ecological genetic studies in the chromosomal form Mopti of Anopheles gambiae s.str. in Mali, West Africa. Genetica. 1994;94:213–223. doi: 10.1007/BF01443435. [DOI] [PubMed] [Google Scholar]
- Toure YT, Petrarca V, Traore SF, et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998;40:477–511. [PubMed] [Google Scholar]
- Tripet F, Wright J, Cornel A, et al. Longitudinal survey of knockdown resistance to pyrethroid (kdr) in Mali, West Africa, and evidence of its emergence in the Bamako form of Anopheles gambiae s.s. American Journal of Tropical Medicine and Hygiene. 2007;76:81–87. [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Collins FH, Besansky NJ. Evolution of Anopheles gambiae in relation to humans and malaria. Annual Review of Ecology Evolution and Systematics. 2011;42:111–132. [Google Scholar]
- Wickham H. Reshaping data with the reshape package. Journal of statistical software. 2007;21:1–20. [Google Scholar]
- Wickham H. ggplot2: Elegant graphics for data analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- Wickham H. scales: scale functions for graphics, R package version 0.2.4. 2014. [Google Scholar]
- Wondji CS, Dabire RK, Tukur Z, et al. Identification and distribution of a GABA receptor mutation conferring dieldrin resistance in the malaria vector Anopheles funestus in Africa. Insect Biochemistry and Molecular Biology. 2011;41:484–491. doi: 10.1016/j.ibmb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
- Yeaman S. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proceedings of the National Academy of Sciences U S A. 2013;110:E1743–1751. doi: 10.1073/pnas.1219381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Song Y, Yang CH, Jan LY, Jan YN. Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat Neurosci. 2014;17:81–88. doi: 10.1038/nn.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A, Grothendieck G. Zoo: S3 infrastructure for regular and irregular time series. Journal of statistical software. 2005;14:1–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.