Abstract
Targeting host cell membranes by M2 of influenza A virus is important for virus invasion and replication. We study the transmembrane domain of M2 (M2TM) interacting with mica supported planar bilayers and free-standing giant unilamellar vesicles (GUVs). Using solution atomic force microscopy (AFM), we show that the size of M2TM oligomers is dependent on lipid composition. Addition of M2TM to lipid bilayers containing liquid-ordered (Lo) and liquid-disordered (Ld) phases reveals that M2TM preferentially partitions into the Ld phase; phase dependent partitioning results in larger rigidity of the Ld phase. We next use fluorescence microscopy to study effects of M2TM on phase coexisting GUVs. In particular, M2TM is found to increase GUVs' miscibility transition temperature Tmix. The augmented thermodynamic stability can be accounted for by considering an enhanced energy barrier of lipid mixing between coexisting phases. Our GUV study also shows that M2TM can elicit an array of vesicle shapes mimicking virus budding. M2TM enhanced membrane curvature is consistent with our AFM data, which show altered membrane rigidity and consequently line tension at domain edges. Together, our results highlight that in addition to conducting proton, M2TM can actively regulate membrane heterogeneity and augment membrane curvature.
Keywords: lipid raft, curvature, phase coexistence, ion channel, miscibility transition temperature
Graphical Abstract
Introduction
Many enveloped viruses (e.g., influenza virus, HIV-1, and Ebola virus) hijack lipid rafts of host cell membranes to cluster fusion proteins and enzymes that are essential for virus assembly, budding, and scission 1–4. M2 of influenza A virus is a 97-residue protein; it is composed of an extracellular N-terminal domain (residues 1–23), a transmembrane (TM) helix (residues 24–46), and a cytoplasmic C-terminal domain (M2CT, residues 47–97). In its native form, the four TM helices of M2 (M2TM) line a low-pH activated proton channel 5–6. Acidification of virus interior through M2 channel is essential for dissociating M1 from ribonucleoprotein. In addition to its ion channel activity, M2 may also participate in virus packaging, assembly, and budding 7–8. In particular, it was found that proper budding and scission of influenza A virus requires M2 to reside near the edge of virus budding zones, from which virions egress 9. The newly formed viral membranes are often enriched with raft lipids and raft associating proteins 10. Based on these observations 9–10, it was proposed that M2 preferentially segregates at the edge of lipid rafts to facilitate virus budding and scission 11. Interestingly, a recent study found a similar role for the HIV fusion peptide, which might function by preferentially targeting domain interfaces 12.
Membrane proteins are susceptible to structural and functional modulations conferred by lipid matrix 13–14. For M2TM, many studies have shown that lipid membrane environment imparts a large effect on M2TM conformation 15, orientation 16–17, and oligomeric organization 18–19. Despite large interests, studies on how M2TM distributes in raft containing heterogeneous membranes are scarce. In addition to raft association/dissociation, another important question is what molecular feature of M2 is responsible for M2 induced membrane curvature that is essential during virus exit. Previous studies have focused on the amphipathic helix of M2CT 20–21, while assuming that M2TM only contributes as a passive ion channel. A recent study of the TM domain of parainfluenza virus 5 fusion protein showed that by adopting a flexible conformation, TM helix can actively promote curved membrane structures 22. This result prompts us to quest whether M2TM can play a similar role in augmenting membrane curvature.
Due to compositional complexity, lipid rafts in living cells are nanoscale and short-lived 23. These properties render lipid rafts not amenable to many conventional experimental approaches. The contemporary consensus is that lipid rafts are dictated by lipid–lipid and lipid–protein interactions 23. Therefore, studies of simplified model systems composed of a few lipid species and proteins with precise chemical compositions are instrumental in elucidating key properties associated with lipid rafts. In this study, we use a three-component lipid bilayer system composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), egg sphingomyelin (eSM), and cholesterol (Chol) to generate raft-mimicking liquid-ordered (Lo) and liquid-disordered (Ld) phases.
We first use high-resolution atomic force microscopy (AFM) in solution to investigate the structure and organization of M2TM in mica supported planar bilayers. Our AFM study shows that M2TM preferentially partitions into the Ld phase in Lo+Ld phase coexisting bilayers. Phase dependent partitioning of M2TM does not change the height difference between coexisting phases, but does modify domain line tension by increasing the rigidity of the Ld phase. Moreover, our solution AFM measurements reveal that the oligomeric state of M2TM is dependent on lipid composition, especially cholesterol. We also use fluorescence microscopy to study effects of M2TM on thermodynamic stability and curvature of giant unilamellar vesicles (GUVs). Specifically, M2TM is found to progressively increase the miscibility transition temperature Tmix of phase coexisting GUVs; incubation of M2TM with phase coexisting GUVs results in an array of vesicle shapes mimicking virus budding. Altogether, our experimental data highlight that M2TM can act as a raft sensor and a membrane curvature enhancer. Both properties are important in understanding the molecular basis of M2 facilitated virus budding and scission.
Materials and Methods
DOPC, eSM, cholesterol, and rhodamine-DPPE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine) are purchased as lyophilized powders from Avanti Polar Lipids (Alabaster, AL). M2TM (residues 22–46 of M2, SSDPLVVAASIMGILHLILWILDRL) is prepared by solid phase synthesis method, followed by HPLC purification and lyophilization. Unless noted otherwise, all lipid/peptide compositions are molar based.
AFM experiments
Stock solutions of lipids and M2TM peptide are prepared by dissolving them in organic solvents (chloroform or chloroform/methanol). Lipid/peptide mixtures are prepared by mixing appropriate ratios of stock solutions in glass test tubes. Organic solvents are removed by a gentle stream of argon gas using a 12-position N-EVAP evaprator, and then vacuum pumped for > 2 h. The resultant lipid/peptide films are hydrated in 10 mM HEPES pH 7.0. Small unilamellar vesicles (SUVs) are generated by ultrasonicating lipid/peptide suspension using a Sonic Dismembrator operated at 40 W with a total duration of 10 min. The obtained SUVs are centrifuged, and characterized by dynamic light scattering using a Dynapro Nanostar instrument (Wyatt Technology, Santa Barbra, CA).
All AFM measurements are performed at room temperature (23°C). Detailed experimental procedure has been described elsewhere 24–26. A fluid-compatible Multimode 8 AFM (Bruker, Santa Barbara, CA) operated at the PeakForce quantitative nanomechanic (QNM) mode is used for the study. SUVs are injected into a fluid cell formed by a freshly cleaved mica substrate, a silicone O-ring, and a fluid-compatible glass cantilever holder (model: MTFML-V2). Mica supported lipid bilayer is formed by the method of SUV rupture and fusion 27. Finally, ~200 µL buffer is injected to flush out excess vesicles. Bilayers are scanned using a silicon nitride probe designed to work with the PeakForce QNM mode (model: ScanAsyst-Fluid+). The maximum normal force applied on lipid bilayer is typically ~300 pN. In the PeakForce QNM mode, the probe oscillates at a frequency of 1 kHz while it scans across the bilayer. On fly analysis of the oscillating force curves at each pixel yields a map of mechanical property (i.e., the reduced Young’s modulus E*) that has the same resolution as the topographic (height) image. The reduced Young’s modulus is obtained by fitting the tip force Ftip during each oscillation to the DMT model 28: , where R is the tip radius, d is the tip-bilayer separation, and Fo is a constant background force. The algorithm is built into the commercially available Nanoscope software. Square images ranging from 100 nm to 2 µm are acquired at a scan rate of 0.5–1.0 Hz. Multiple maps including the height and the reduced Young’s modulus are collected in a single scan.
GUV experiments
Appropriate ratios of DOPC/eSM/Chol or DOPC/eSM/Chol/M2TM are prepared from stock solutions. For fluorescence imaging, 0.2mol% rhodamine-DPPE is added to each mixture. Lipid mixture in organic solvent is deposited on two one-side coated ITO glass slides. After solvent removal by vacuum pumping for > 2 h, a GUV production chamber is formed by inserting a Viton O-ring between the two slides. After filling the production chamber with ~600 µL buffer containing 100 mM sucrose and 10 mM HEPES pH 7.0, the assemblage is stationed in a homemade aluminum block, which is in thermal contact with a dry bath incubator. The temperature of the aluminum block and the GUV assemblage is controlled by a Digi-Sense T-type temperature controller. After heating the assemblage to 60°C, GUVs are produced by the electroformation method 29 with an AC field of ~2.0 V at 10 Hz (total duration of 2 h). Once the assemblage cools down to room temperature, GUVs are suspended in 3 mL GUV buffer containing 100 mM glucose and 10 mM HEPES pH 7.0. Two approaches are used for GUV study. In the first approach, an aliquot of lipid/M2TM GUVs (with variable peptide concentrations) is transferred into a silicone well, which is sandwiched by a cover slip and a microscope slide. In the second approach, M2TM is first dissolved in GUV buffer containing 1% (v/v) DMSO. An aliquot of lipid-only GUVs (without M2TM) is mixed with an equal volume of M2TM solution in a microcentrifuge tube. After equilibration for 20 min, the mixture is transferred to a silicone well for microscope examination. For control, the M2TM solution is replaced by the GUV buffer with 1% (v/v) DMSO. The final DMSO and peptide concentration used in the second approach is 0.5% (v/v) and 2.3 µM, respectively.
For temperature control, the silicone well assemblage with GUVs in the center is placed in an observation chamber made of two copper plates. The chamber is connected to a bath circulator. Thermal stability is enhanced by wrapping thermal insulation foam around the observation chamber and the microscope objective. GUV temperature is monitored by a T-type thermal probe, which is taped to the center of the silicone well assemblage using a thermal conductive aluminum tape. Temperature variation of our setup is < 0.1°C. Fluorescence images are acquired using an inverted Nikon Eclipse Ti-U microscope, a CFI Super Fluor ELWD 60× objective, and an Andor iXon Ultra 897 EM-CCD camera.
To explore the effect of DMSO on M2TM structure, we use attenuated total reflection Fourier transform infrared (FTIR) spectroscopy. M2TM in 1% (v/v) DMSO (0.2 mg/mL) is centrifuged at 266,000×g for 2 h (Sorvall WX80, Thermal Fisher Scientific Inc.). An aliquot of the supernatant (35 µL) is transferred into the fluid cell of a Vertex 70 Bio ATRII spectrometer (Bruker, Billerica, MA). FTIR spectrum is obtained by first averaging over 1500 scans, and then subtracting a background spectrum due to solvent absorption. The same supernatant is examined by dynamic light scattering measurement.
Results
Solution AFM experiments
Homogenous lipid bilayers
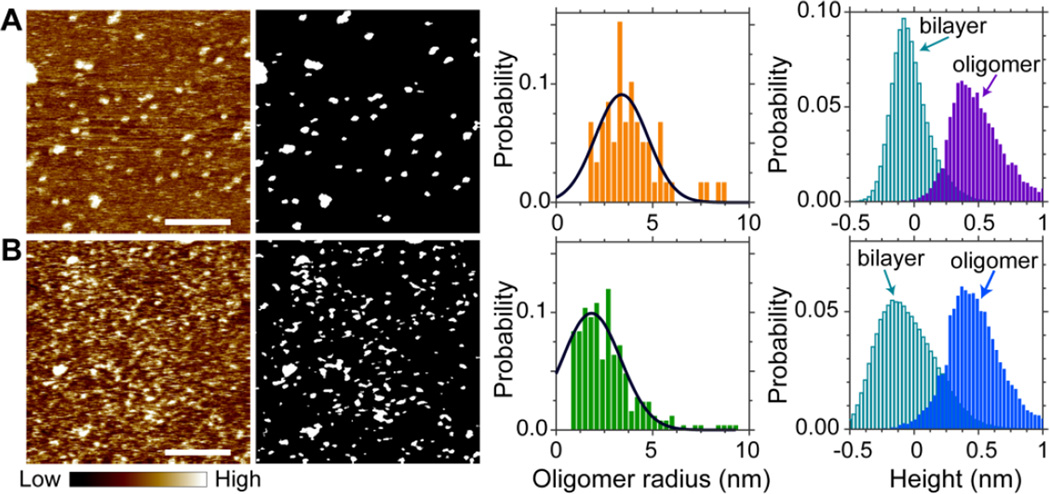
Using solution AFM, we first examine M2TM oligomeric structure in lipid bilayers that exhibit homogeneous organization: DOPC and DOPC + 20mol% Chol. In each bilayer the peptide concentration is 4mol% of the total lipid+peptide mixture. Circular dichroism spectroscopy measurement indicates that M2TM is properly folded into α-helix when incorporated into lipid bilayers (data not shown). Solution AFM height images are shown in Fig. 1. (Height images with different magnifications are also shown in Fig. S1, Supporting Information.) Isolated M2TM oligomers are clearly seen. To determine oligomer size, we extract oligomers by using Matlab Image Analysis toolbox. The results are shown as bright regions in gray-scale binary images. Oligomer sizes are determined by summing pixels corresponding to each oligomer; oligomer radius is then calculated by assuming that oligomers are round shaped. The normalized probabilities of oligomer radius distributions reveal that the most probable radius is 3.4 nm for the DOPC/M2TM bilayer, and 1.8 nm for the DOPC/Chol/M2TM bilayer. In addition to lateral sizes, we also determine oligomer heights. This is done by calculating height distributions of oligomers and the rest of the bilayer. The normalized probabilities of the two types of regions indicate that for both bilayers, the most probable height difference between oligomers and the intact bilayer is ~0.5 nm.
Figure 1.
Solution AFM height images of 4mol% M2TM in planar bilayers composed of (A) DOPC and (B) DOPC + 20mol% Chol. Height-scale indicated by the color bar is 1.5 nm. Scale bars = 80 nm. Gray-scale binary images show oligomers determined by Matlab image analysis. Gaussian fitting (solid lines) indicates that the most probable oligomer radius is 3.4 nm for the DOPC/M2TM bilayer, and 1.8 nm for the DOPC/Chol/M2TM bilayer. Height probabilities are calculated separately for oligomers and the rest of the bilayer.
Heterogeneous lipid bilayers
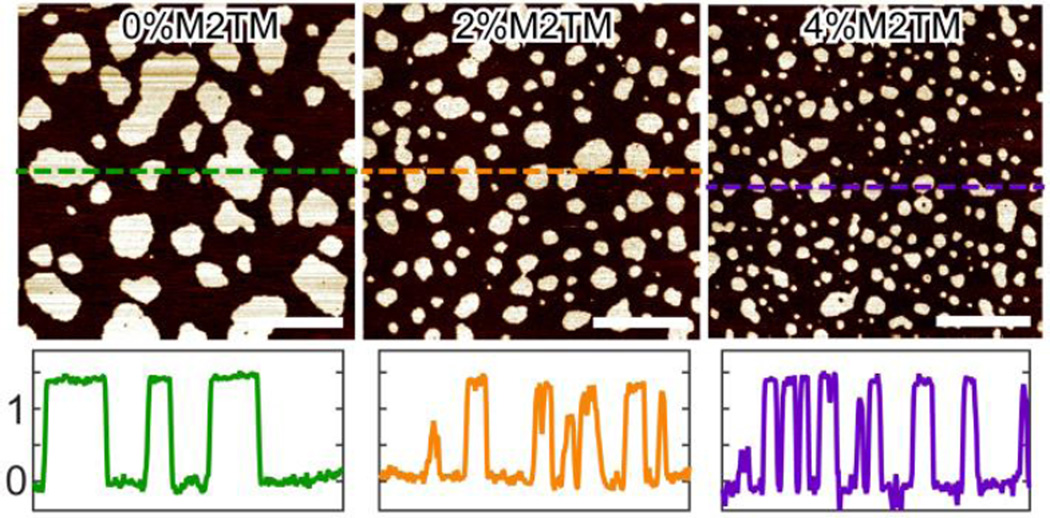
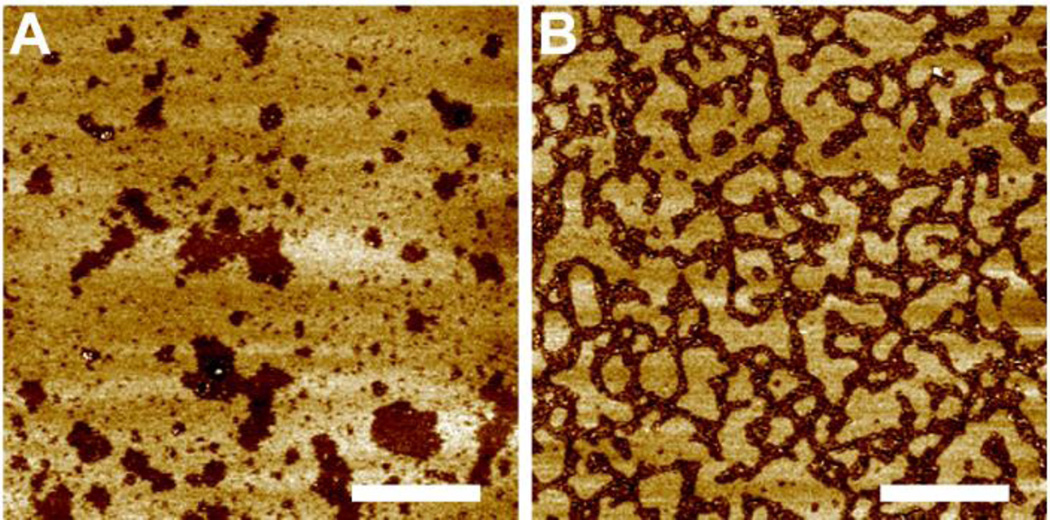
We next explore lipid bilayer structures of a three-component system exhibiting macroscopic Lo+Ld phase coexistence. Figure 2 shows an overall view of the topographic images of DOPC/eSM 3:2 + 20mol% Chol doped with different amount of M2TM. When no M2TM is present, the lipid bilayer exhibits Lo+Ld phase coexistence. Height profile indicates that the liquid coexisting phases have a height difference of ~1.4 nm. Addition of 2 and 4mol% M2TM successively decreases the size of Lo domains, while the height difference between the coexisting phases remains similar. To quantitatively correlate Lo domain size and M2TM concentration, we measure the average area of Lo domains at each M2TM concentration. The diameter of an equivalent circle having the same domain area is assigned as the average size of Lo domains. The resulting diameter of Lo domains is 190, 100, and 69 nm for bilayers containing 0, 2, and 4mol% M2TM, respectively. Moreover, the surface area fraction of Lo domains also decreases from 0.33 to 0.23 as M2TM concentration increases from 0 to 4mol% (area fraction is calculated from the number of pixels corresponding to Ld and Lo phases, respectively).
Figure 2.
Solution AFM height images of DOPC/eSM 3:2 + 20mol% Chol doped with different amount of M2TM. Scale bars = 500 nm. The bottom row shows height profiles (in nm) along dashed lines indicated in the height images.
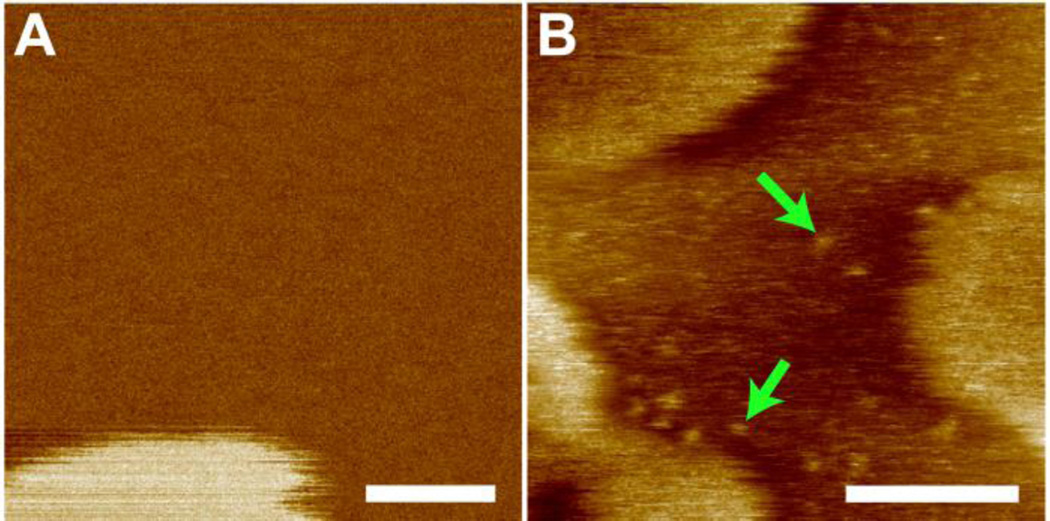
Isolated M2TM oligomers are also observed in the three-component bilayer system (Fig. 3). To determine oligomer size, a region of the Ld phase containing distinguishable oligomers is selected (Fig. S2, Supporting Information). Image analysis indicates that oligomers have an average radius of 1.9 nm; they are higher than the Ld phase by ~0.5 nm.
Figure 3.
Solution AFM height images of DOPC/eSM 3:2 + 20mol% Chol without (A) and with (B) 2mol% M2TM. Green arrows highlight M2TM oligomers in the Ld phase. Scale bars = 50 nm.
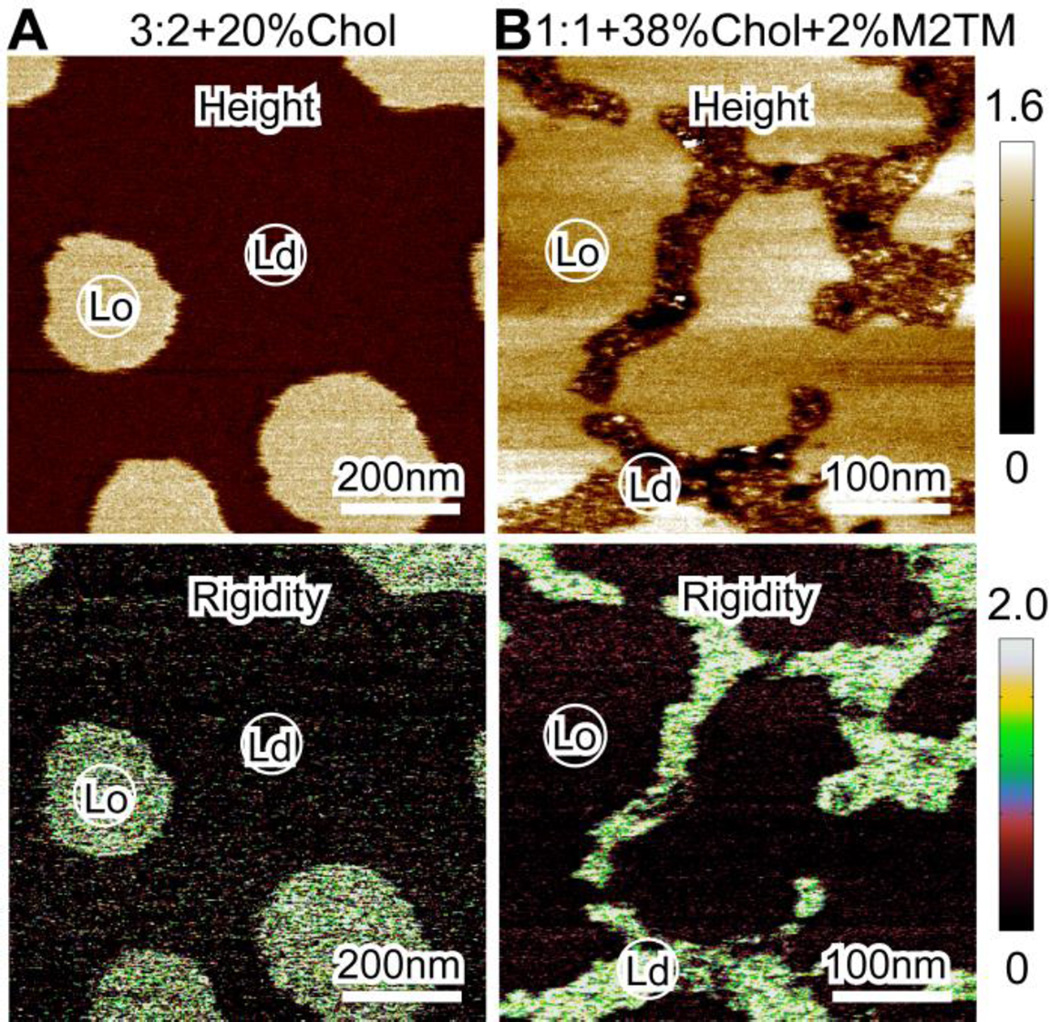
We also looked at a lipid bilayer with larger cholesterol content, DOPC/eSM 1:1 + 38mol% Chol (Fig. 4). In the absence of M2TM, Ld domains are surrounded by the bulk Lo phase. The height difference between coexisting phases is ~0.8 nm, which is smaller than that of the DOPC/eSM 3:2 + 20mol% Chol bilayer. This observation is consistent with the fact that DOPC/eSM 1:1 + 38mol% Chol is close to, while DOPC/eSM 1:1 + 20mol% Chol is far away from the upper phase boundary 24, 30.
Figure 4.
Solution AFM height images of DOPC/eSM 1:1 + 38mol% Chol without (A) and with (B) 2mol% M2TM. Scale bars = 500 nm.
Addition of 2mol% M2TM reorganizes the Lo phase, yielding isolated Lo domains with curvilinear shapes (Fig. 4B). The height difference between coexisting phases remains similar (i.e., ~0.8 nm). AFM images with larger magnifications (Fig. S3, Supporting Information) reveal additional details: (i) individual oligomers are observed in the Ld phase, and (ii) due to the small area fraction of the Ld phase, M2TM oligomers are densely packed. Since no individual oligomers are observed in the Lo phase, it seems that M2TM is primarily localized in the Ld phase. One caveat is that the height of M2TM oligomers might be very close to that of the Lo phase; therefore, oligomers would not be distinguishable in the Lo phase. This concern, however, is ameliorated by the observation that a few regions containing M2TM oligomers are also observed in the Lo phase (Fig. S3-A). Therefore, the (almost) absence of M2TM in the Lo phase cannot be accounted for by the argument that the height of M2TM is the same as or similar to that of the Lo phase. Our statement of phase dependent partitioning of M2TM is further supported by bilayer rigidity data shown below.
Our solution AFM measurements are performed using PeakForce QNM mode, which allows simultaneous mapping of bilayers’ topographical and material properties. Accurate rigidity of lipid bilayers can be obtained by careful system calibration (e.g., by using a reference sample with known mechanical properties). Here we are only interested in the relative rigidity of coexisting phases in heterogeneous bilayers. Figure 5 shows the height and reduced Young’s modulus maps of lipid bilayers with and without M2TM. In both cases, the Lo phase has a larger height than that of the Ld phase. In the absence of M2TM, the Lo phase has a larger reduced Young’s modulus than that of the surrounding (Fig. 5A). This observation is consistent with the general consensus that the Lo phase is stiffer than the Ld phase 31. A remarkable result is obtained when 2mol% M2TM is incorporated into the phase coexisting bilayer: the oligomer enriched Ld phase exhibits larger reduced Young’s modulus than that of the oligomer deficient Lo phase (Fig. 5B). The modified rigidity of coexisting phases can be reconciled by considering that protein helix (mainly through hydrogen bonding) has a larger compression spring constant than that of lipid bilayers (mainly through van der Waals interactions) 32; therefore, the rigidity of the Ld phase is significantly enhanced by sequestering M2TM oligomers. Since the rigidity of the Lo phase is not increased by the presence of M2TM, it is reasonable to argue that the Lo phase does not contain M2TM oligomers. Together, our AFM based height and rigidity images support that M2TM is mainly localized in the Ld phase.
Figure 5.
Solution AFM height images (top row) and reduced Young’s modulus (rigidity) maps (bottom row). (A) DOPC/eSM 3:2 + 20mol% Chol. (B) DOPC/eSM 1:1 + 38mol% Chol with 2mol% M2TM. The height is in unit of nm and the reduced Young’s modulus is in unit of MPa.
GUV experiments
Miscibility transition temperature
We use free-floating GUVs to investigate the effect of M2TM on lipid bilayer thermodynamics, particularly the miscibility transition temperature Tmix of phase coexisting bilayers composed of DOPC/eSM 3:2 + 32mol% Chol. (The lipid composition is chosen for easy access to Tmix.) Tmix is defined by the temperature above which phase coexisting bilayers become homogeneous 33–34. GUV lateral organization is revealed by the Ld phase marker rhodomine-DPPE. It is known that GUV lipid composition often exhibits vesicle-to-vesicle variation 35; therefore, different vesicles can have different Tmix. For this reason, we define two temperatures corresponding to (i) the majority of GUVs exhibit critical like fluctuations 36, and (ii) the majority of GUVs become homogeneous. Tmix is the average of the two temperatures.
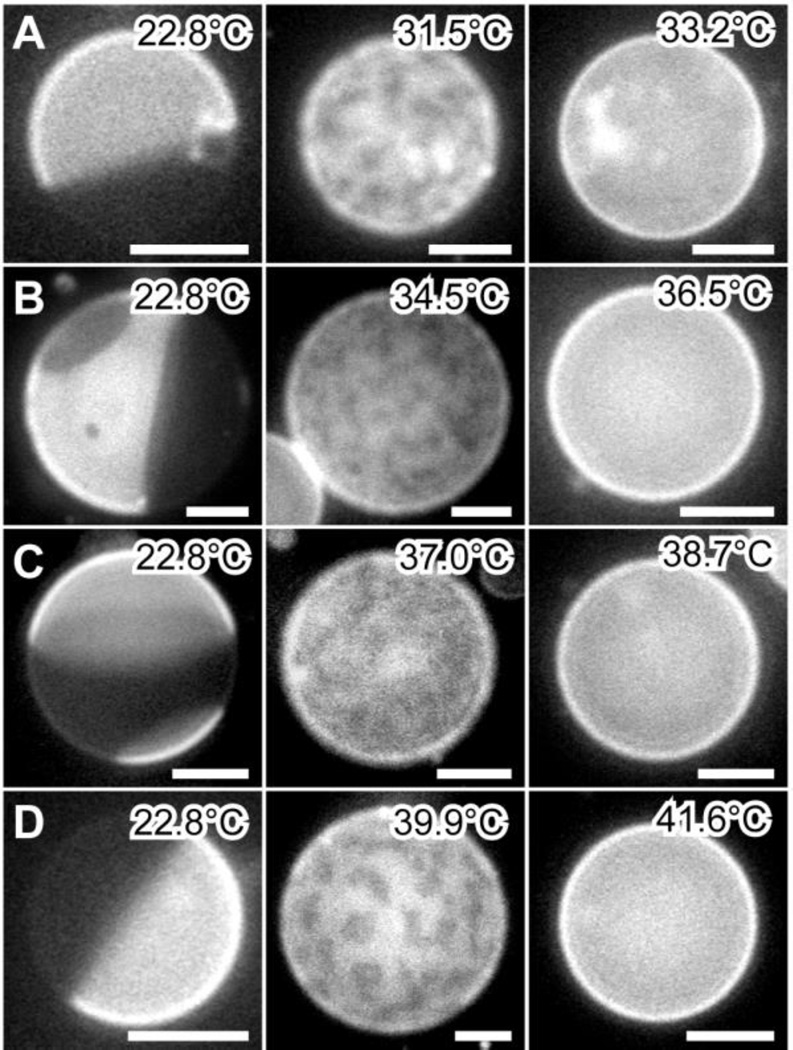
For the control sample without M2TM, macroscopic phase separation is observed at low temperature; critical like fluctuations emerge when temperature is raised; vesicles become homogeneous by an additional ~2°C increase (Fig. 6). The observed miscibility transition temperature Tmix is 32.4±1.2°C. This value is in good agreement with a previous study employing a different sphingomyelin species 30. Compared to the control sample, addition of M2TM ranging from 1 to 6mol% results in similar (i) macroscopic phase separation at low temperatures, and (ii) critical like fluctuations at elevated temperatures (Fig. 6). Importantly, Tmix is found to increase almost linearly with peptide concentration (Fig. 7). The largest Tmix is 40.8°C for GUVs with 6mol% M2TM.
Figure 6.
GUV miscibility transition as a function of M2TM concentration. GUVs are composed of DOPC/eSM 3:2 + 32mol% Chol with 0 (A), 2 (B), 4 (C), and 6mol% (D) M2TM. Scale bars = 10 µm.
Figure 7.
Miscibility transition temperature Tmix of GUVs composed of DOPC/eSM 3:2 + 32mol% Chol as a function M2TM concentration.
Vesicle shape
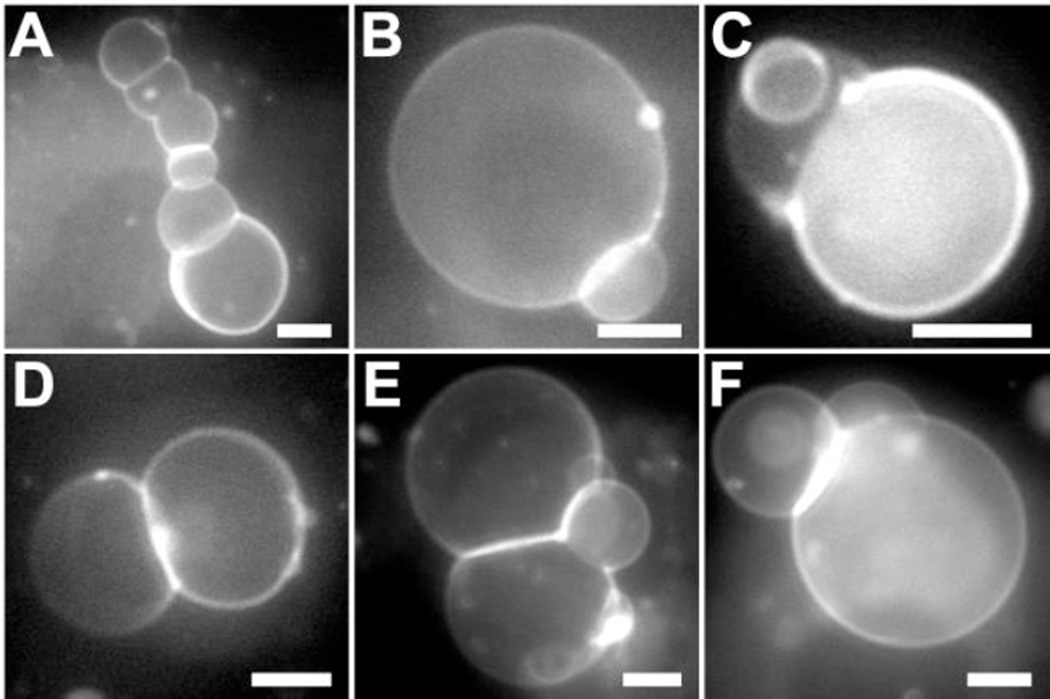
In the study of Tmix, we observe a small fraction of GUVs containing budding caps when M2TM is present. This prompts us to propose that M2TM might be able to enhance membrane curvature. To test this hypothesis, we incubate lipid-only GUVs with M2TM solubilized in 1% (v/v) DMSO. We first use attenuated total reflection FTIR to examine the secondary structure of M2TM dissolved in DMSO (Fig. S4, Supporting Information). The pronounced absorption band centered at 1651 cm-1 highlights that the secondary structure of M2TM is mainly α-helix. We also use dynamic light scattering to characterize the oligomeric state of M2TM dissolved in DMSO. The resulting hydrodynamic radius is ~3 nm. This value is consistent with the tetrameric organization of M2TM. We next explore the effect of M2TM on GUV curvature. For control, we find that DMSO buffer exerts no discernable effect on vesicle morphology. Introduction of M2TM to GUV exterior results in remarkable changes of vesicle shapes. Based on experiments replicated on different days (total number of GUVs > 100), we find more than 30% GUVs exhibiting budding morphology. When taking images, we mainly focused on GUVs with non-spherical shapes; therefore, the percentage could be biased. Nevertheless, our data unequivocally show that (i) M2TM can modify GUV morphology (at least for a significant portion of GUVs), and (ii) the change is not due to the solvent (control experiment with DMSO containing buffer shows no vesicle budding). Figure 8 shows a few examples of GUV budding induced by M2TM: a chain of vesicles is formed by juxtaposing neighboring GUVs (Fig. 8A); spherical caps with variable sizes bud from mother vesicles (Fig. 8B–D); three caps interconnect at the same site (Fig. 8E); and two caps bud from one mother vesicle (Fig. 8F). A special note is the chain morphology, which has some similarity to tubular structures observed during virus budding 9, 11. Compared to the Tmix study where GUVs are prepared by mixing lipids and M2TM before electroformation, the incubation study unveils that dynamic absorption and insertion of M2TM from one side of lipid bilayers are able to augment membrane curvature and generate a multitude of budding architectures.
Figure 8.
Fluorescence images of GUVs (DOPC/eSM 3:2 + 32mol% Chol) after being exposed to 2.3 µM M2TM. (A) Inter-connected vesicle chain, (B) small cap budding, (C) small and intermediate cap budding, (D) hemispherical budding, (E) three-cap interconnection, and (F) two-cap budding. Scale bars = 10 µm.
Discussion
Oligomer size
Our solution AFM study indicates that the average radius of M2TM oligomers is 3.4 nm in DOPC bilayer; the radius decreases to 1.8 nm when 20mol% cholesterol is added (Fig. 1); for bilayers with coexisting phases, the average radius is 1.9 nm (Fig. S2, Supporting Information). Compared to the size of M2TM tetramers (S5, Supporting Information), it is clear that M2TM forms oligomers larger than a tetramer in DOPC bilayers, while tetrameric structure dominates in DOPC/Chol and DOPC/eSM/Chol bilayers. Since helix-helix association is mainly mediated by weak van der Waals interactions, the same type of interactions taking place between helix and lipids, it is conceivable that modified lipid composition and consequently modified lipid-helix interactions can be translated into altered oligomeric state 18, 37. Indeed, helix-helix distance measurement using spin labeled M2TM indicates that M2TM can exist as monomer, dimer, or tetramer depending on peptide/lipid ratio 19. Similarly, dimer-tetramer conversion of full length M2 can be elicited by changing aqueous pH 38. The variable oligomer size suggests that membrane environment imparts a profound effect on the stochastic oligomeric state of M2TM. This result is consistent with the notion that lipid membrane can affect protein conformation, dynamics, oligomeric state, and function by altering lipid membrane structural and material properties 39.
Phase-dependent partitioning
Phase-based protein sorting is a hallmark of lipid rafts. A previous study using analytical immunogold electron microscopy showed that M2 is non-raft associated in infected cells 9. Similarly, M2 was found to localize in the Ld phase of plasma membrane spheres derived from living cells; mutation of M2CT does not affect Ld phase-association of M2 11. All these observations 9, 11 are consistent with our AFM data, which show that M2TM primarily partitions into the Ld phase of phase coexisting bilayers. Two mechanisms have been proposed to describe TM protein sorting in raft containing membranes. One is based on the thickness difference between raft and non-raft domains. It is energetically favorable to sort proteins with long TM segment into raft domains, and proteins with short TM segment into non-raft domains 40–42. The thickness-matching mechanism, however, was found to be inconsistent with many experimental observations, which showed that regardless of TM length, TM proteins/peptides were exclusively localized in the Ld phase 43–44. In addition, many canonical raft-associating proteins were found to be excluded from macroscopic Lo domains 45–46. To reconcile the discrepancy, a second mechanism was proposed that proper raft-based sorting requires a moderate stiffness difference between coexisting phases 47.
Both mechanisms could play a role during the initial stage of M2TM sorting. The thickness of M2TM might be better matched to that of the Ld phase; the Lo phase has a larger rigidity than that of the Ld phase (Fig. 5). However, at larger M2TM concentrations, the peptide enriched Ld phase exhibits a larger rigidity than that of the peptide deficient Lo phase. In this case, the second mechanism would predict Lo phase association of M2TM. To antagonize this tendency, counteracting mechanisms must take effect, such as favorable protein-protein interactions.
Our AFM data also reveal that both the size and area fraction of Lo domains become smaller by increasing M2TM content. The modified phase fraction is consistent with earlier findings that TM proteins/peptides can exert a large effect on lipid membrane phase behavior 48–50. Finally, we note that phase dependent sorting of membrane proteins could be potentially influenced by many factors, including the system under study (e.g., lipid bilayers vs. living cells) and experimental approaches. Our observation of Ld phase association of M2TM is based on AFM measurement of planar lipid bilayers. For future studies, it will be interesting to explore whether the partitioning behavior of M2TM remains the same in other systems that are more representative of biological membranes, such as giant plasma membrane vesicles 51.
Miscibility transition temperature
Our thermodynamic study of GUVs composed of DOPC/eSM 3:2 + 32mol% Chol shows that the miscibility transition temperature Tmix increases from 32.4 to 40.8°C when M2TM concentration increases from 0 to 6mol% (Fig. 7). By shifting Tmix to larger values, M2TM seems to stabilize Lo domains in phase coexisting bilayers. This result can be understood by considering unequal distribution of M2TM in heterogeneous bilayers. Specifically, by preferentially partitioning into the Ld phase, M2TM oligomers could increase the energy barrier of Lo phase enriched lipids (i.e., eSM and cholesterol) to cross domain edge and mix with Ld phase enriched lipids. Therefore, M2TM could impair the mixing propensity of lipids in phase coexisting bilayers, and consequently shift Tmix upward. Similar upward shifting of Tmix has been observed by connecting GUVs to actin network 52 or by crosslinking ganglioside lipid GM1 using exogenous toxin 45. Conversely, n-alcohols and liquid anesthetics were found to downshift Tmix of cell-derived giant plasma membrane vesicles 53. Although often considered as an intrinsic component of lipid membranes, cholesterol also decreases Tmix 30. The disparate effects of membrane additives on Tmix highlight their regulatory roles in mediating membrane heterogeneous organization.
Membrane curvature
Our GUV incubation study indicates that vesicle shape restructuring can be induced by introducing M2TM to GUV exterior (Fig. 8). Protein elicited membrane shape remodeling is important in a plethora of cellular processes, including endocytosis and exocytosis, vesicle trafficking, and cell division 54–55. For virus budding and scission, many viruses use the host endosomal sorting complex required for transport (ESCRT) machinery 56. Influenza A virus has developed an ESCRT-independent mechanism for virus egress, in which M2 protein was proposed to play a pivotal role 11, 57. Based on the notion of surface protein assisted membrane deformation 58–60, previous studies primarily focused on the amphipathic helix located at M2CT. The hydrophobic face of M2CT helix was proposed to partially insert into membranes at the cytoplasmic side. Asymmetric binding of M2CT stresses nearby lipid packing, creates local defects, and promotes membrane curvature. Indeed, incorporation of M2CT into 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) enriched lipid mixtures was found to favor cubic phase formation – a unique lipid organization containing negative Gaussian curvature 20.
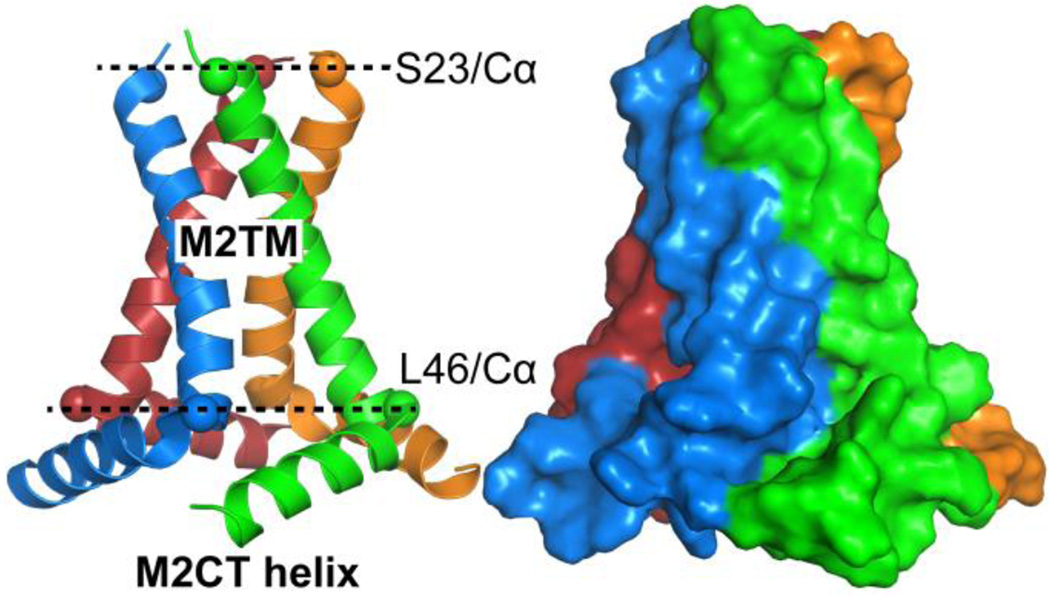
We find that M2TM is capable of inducing vesicle budding. This observation coincides with a recent finding that the TM domain of parainfluenza virus 5 fusion protein augments the tendency of forming curved bilayer structures 22. TM peptide induced nonlamellar phases have also been broadly observed 62–65. A similar GUV experiment to ours was performed using full-length M2 and M2CT helix 11. The authors found that daughter vesicles or small budding caps can be developed from mother vesicles. Interestingly, the authors also found that cholesterol content plays an important role in promoting or suppressing budding events. Figure 9 displays the structure of a tetrameric M2TM-M2CT complex resolved by solid state NMR in lipid environment 61. An overall thickness of 3.0 nm for the M2TM segment (Fig. 9) is consistent with its TM disposition. Importantly, the four TM helices tilt by 22–32° with respect to the bilayer normal 61, resulting in a conical tetramer with the N-terminal narrowed and the C-terminal widened. When the conical tetramer inserts into lipid membranes, its C-terminal will exert a larger stress on neighboring lipids than its N-terminal. The asymmetrically stressed lipid leaflets can then generate negative curvature. Addition of M2CT to the C-terminal of M2TM further increases of the dimension of the cone base (Fig. 9); therefore, the complex structure of M2TM-M2CT is expected to be more effective in augmenting membrane curvature than M2TM alone. We note that the orientation of M2TM (or M2) when inserting into host membranes is important for generating negative curvature. This is apparent that oppositely orientated conical tetramers will stress the opposite leaflets equally, resulting in a null effect on the overall membrane curvature. Asymmetrical insertion could explain our observation that many GUVs undergo shape change when M2TM is introduced to vesicle exterior (Fig. 8), while only a few GUVs have budding caps when GUVs are produced by mixing M2TM and lipids before electroformation.
Figure 9.
Solid state NMR structure of a tetrametic M2TM-M2CT complex 61 (PDB: 2L0J) shown in cartoon (left) and surface (right) representations. Cα atoms of S23 and L46 are shown as spheres. The distance between the two planes formed by the Cα atoms of S23 and L46 is 3.0 nm. The diameter of the circle outlined by the Cα atoms of S23 is 1.7 nm, and the diameter of the circle outlined by the Cα atoms of L46 is 3.0 nm.
In addition to the conical shape of M2TM oligomers, modulated membrane properties could also contribute to the enhanced curvature of phase coexisting lipid bilayers. Vesicle budding is mediated by two competing energy terms: one stems from the line tension energy located at budding edge and the other is related to the bending energy of the budding cap 66–69. Vesicle budding is favored when the increased bending energy in the budding cap is outweighed by the reduction of line tension energy at the budding edge. Our AFM data show that (i) M2TM preferentially partitions into the Ld phase, and (ii) phase-dependent localization increases the rigidity of the Ld phase. Using a continuum elasticity theory of lipid bilayers, Kuzmin et al. showed that line tension is governed by the thickness and bending modulus of coexisting phases 70. Our AFM measurements indicate that the height difference between coexisting phases is unchanged by M2TM. Therefore, the altered line tension by M2TM is primarily achieved by increasing the bending modulus of the Ld phase. Our observation of M2TM enhanced vesicle curvature is consistent with theoretical prediction that absorption and insertion of membrane inclusions can increase the effective line tension and induce vesicle budding 71.
Supplementary Material
Acknowledgments
J. Cai is supported by NSF 1351265, NIH 1R01GM112652-01A1 and R15GM117531. J. Pan is supported by NIH R15GM117531.
Footnotes
Supporting Information Available
Three AFM images, one FTIR spectrum, and two tetrameric structures of M2TM are shown in the supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol R. 2003;67(2):226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma K, Wang YJ, Wang JF. Influenza Virus Assembly and Budding in Lipid Rafts. Prog Biochem Biophys. 2015;42(6):495–500. [Google Scholar]
- 3.Veit M, Engel S, Thaa B, Scolari S, Herrmann A. Lipid domain association of influenza virus proteins detected by dynamic fluorescence microscopy techniques. Cell Microbiol. 2013;15(2):179–189. doi: 10.1111/cmi.12045. [DOI] [PubMed] [Google Scholar]
- 4.Barman S, Nayak DP. Lipid raft disruption by cholesterol depletion enhances influenza a virus budding from MDCK cells. J Virol. 2007;81(22):12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto LH, Holsinger LJ, Lamb RA. Influenza-Virus M2 Protein Has Ion Channel Activity. Cell. 1992;69(3):517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 6.Duff KC, Ashley RH. The Transmembrane Domain of Influenza-a M2 Protein Forms Amantadine-Sensitive Proton Channels in Planar Lipid Bilayers. Virology. 1992;190(1):485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 7.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411(2):229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Qiu JXY, Soto C, DeGrado WF. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr Opin Struc Biol. 2011;21(1):68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342(2):215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 10.Ohkura T, Momose F, Ichikawa R, Takeuchi K, Morikawa Y. Influenza A Virus Hemagglutinin and Neuraminidase Mutually Accelerate Their Apical Targeting through Clustering of Lipid Rafts. J Virol. 2014;88(17):10039–10055. doi: 10.1128/JVI.00586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossman JS, Jing XH, Leser GP, Lamb RA. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell. 2010;142(6):902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang ST, Kiessling V, Simmons JA, White JM, Tamm LK. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat Chem Biol. 2015;11(6):424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Bba-Biomembranes. 2012;1818(2):234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntosh TJ, Simon SA. Roles of bilayer material properties in function and distribution of membrane proteins. Annu Rev Bioph Biom. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 15.Hu FH, Luo WB, Cady SD, Hong M. Conformational plasticity of the influenza A M2 transmembrane helix in lipid bilayers under varying pH, drug binding, and membrane thickness. Bba-Biomembranes. 2011;1808(1):415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duong-Ly KC, Nanda V, Degrad WF, Howard KP. The conformation of the pore region of the M2 proton channel depends on lipid bilayer environment. Protein Sci. 2005;14(4):856–861. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Bba-Biomembranes. 2007;1768(12):3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schick S, Chen LR, Li E, Lin J, Koper I, Hristova K. Assembly of the M2 Tetramer Is Strongly Modulated by Lipid Chain Length. Biophys J. 2010;99(6):1810–1817. doi: 10.1016/j.bpj.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgieva ER, Borbat PP, Norman HD, Freed JH. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci Rep-Uk. 2015;5:11757. doi: 10.1038/srep11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GCL. Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission. J Am Chem Soc. 2013;135(37):13710–13719. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, Hong M. Investigation of the Curvature Induction and Membrane Localization of the Influenza Virus M2 Protein Using Static and Off-Magic-Angle Spinning Solid-State Nuclear Magnetic Resonance of Oriented Bicelles. Biochemistry-Us. 2015;54(13):2214–2226. doi: 10.1021/acs.biochem.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao H, Lee MW, Waring AJ, Wong GC, Hong M. Viral fusion protein transmembrane domain adopts beta-strand structure to facilitate membrane topological changes for virus-cell fusion. Proc Natl Acad Sci U S A. 2015;112(35):10926–10931. doi: 10.1073/pnas.1501430112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 24.Khadka NK, Ho CS, Pan JJ. Macroscopic and Nanoscopic Heterogeneous Structures in a Three-Component Lipid Bilayer Mixtures Determined by Atomic Force Microscopy. Langmuir. 2015;31(45):12417–12425. doi: 10.1021/acs.langmuir.5b02863. [DOI] [PubMed] [Google Scholar]
- 25.Ho CS, Khadka NK, Pan J. Sub-ten-nanometer heterogeneity of solid supported lipid membranes determined by solution atomic force microscopy. Bba-Biomembranes. 2015;1858:181–188. doi: 10.1016/j.bbamem.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Ho CS, Khadka NK, She F, Cai J, Pan J. Polyglutamine aggregates impair lipid membrane integrity and enhance lipid membrane rigidity. Bba-Biomembranes. 2016;1858:661–670. doi: 10.1016/j.bbamem.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reviakine I, Brisson A. Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir. 2000;16(4):1806–1815. [Google Scholar]
- 28.Derjaguin BV, Muller VM, Toporov YP. Effect of Contact Deformations on Adhesion of Particles. J Colloid Interf Sci. 1975;53(2):314–326. [Google Scholar]
- 29.Morales-Penningston NF, Wu J, Farkas ER, Goh SL, Konyakhina TM, Zheng JY, Webb WW, Feigenson GW. GUV preparation and imaging: Minimizing artifacts. Bba-Biomembranes. 2010;1798(7):1324–1332. doi: 10.1016/j.bbamem.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veatch SL, Keller SL. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys Rev Lett. 2005;94(14):148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 31.Kollmitzer B, Heftberger P, Podgornik R, Nagle JF, Pabst G. Bending Rigidities and Interdomain Forces in Membranes with Coexisting Lipid Domains. Biophys J. 2015;108(12):2833–2842. doi: 10.1016/j.bpj.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietz H, Berkemeier F, Bertz M, Rief M. Anisotropic deformation response of single protein molecules. P Natl Acad Sci USA. 2006;103(34):12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J. 2003;85(5):3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys J. 2004;86(5):2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezlyepkina N, Gracia RS, Shchelokovskyy P, Lipowsky R, Dimova R. Phase Diagram and Tie-Line Determination for the Ternary Mixture DOPC/eSM/Cholesterol. Biophys J. 2013;104(7):1456–1464. doi: 10.1016/j.bpj.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honerkamp-Smith AR, Veatch SL, Keller SL. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Bba-Biomembranes. 2009;1788(1):53–63. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cymer F, Veerappan A, Schneider D. Transmembrane helix-helix interactions are modulated by the sequence context and by lipid bilayer properties. Bba-Biomembranes. 2012;1818(4):963–973. doi: 10.1016/j.bbamem.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 38.Kawano K, Yano Y, Matsuzaki K. A Dimer Is the Minimal Proton-Conducting Unit of the Influenza A Virus M2 Channel. J Mol Biol. 2014;426(14):2679–2691. doi: 10.1016/j.jmb.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Bioph Biom. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 40.Lundbaek JA, Andersen OS, Werge T, Nielsen C. Cholesterol-induced protein sorting: An analysis of energetic feasibility. Biophys J. 2003;84(3):2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Planque MRR, Goormaghtigh E, Greathouse DV, Koeppe RE, Kruijtzer JAW, Liskamp RMJ, de Kruijff B, Killian JA. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry-Us. 2001;40(16):5000–5010. doi: 10.1021/bi000804r. [DOI] [PubMed] [Google Scholar]
- 42.van Duyl BY, Rijkers DTS, de Kruijff B, Killian JA. Influence of hydrophobic mismatch and palmitoylation on the association of transmembrane alpha-helical peptides with detergent-resistant membranes. Febs Lett. 2002;523(1–3):79–84. doi: 10.1016/s0014-5793(02)02939-3. [DOI] [PubMed] [Google Scholar]
- 43.Gandhavadi M, Allende D, Vidal A, Simon SA, McIntosh TJ. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent, resistant rafts. Biophys J. 2002;82(3):1469–1482. doi: 10.1016/S0006-3495(02)75501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fastenberg ME, Shogomori H, Xu XL, Brown DA, London E. Exclusion of a transmembrane-type peptide from ordered-lipid domains (Rafts) detected by fluorescence quenching: Extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry-Us. 2003;42(42):12376–12390. doi: 10.1021/bi034718d. [DOI] [PubMed] [Google Scholar]
- 45.Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. P Natl Acad Sci USA. 2005;102(18):6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sengupta P, Hammond A, Holowka D, Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Bba-Biomembranes. 2008;1778(1):20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, Simons K. Order of lipid phases in model and plasma membranes. P Natl Acad Sci USA. 2009;106(39):16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poveda JA, Fernandez AM, Encinar JA, Gonzalez-Ros JM. Protein-promoted membrane domains. Bba-Biomembranes. 2008;1778(7–8):1583–1590. doi: 10.1016/j.bbamem.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 49.Pathak P, London E. Measurement of Lipid Nanodomain (Raft) Formation and Size in Sphingomyelin/POPC/Cholesterol Vesicles Shows TX-100 and Transmembrane Helices Increase Domain Size by Coalescing Preexisting Nanodomains But Do Not Induce Domain Formation. Biophys J. 2011;101(10):2417–2425. doi: 10.1016/j.bpj.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domanski J, Marrink SJ, Schafer LV. Transmembrane helices can induce domain formation in crowded model membranes. Bba-Biomembranes. 2012;1818(4):984–994. doi: 10.1016/j.bbamem.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 51.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. P Natl Acad Sci USA. 2007;104(9):3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91(11):4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray E, Karslake J, Machta BB, Veatch SL. Liquid General Anesthetics Lower Critical Temperatures in Plasma Membrane Vesicles. Biophys J. 2013;105(12):2751–2759. doi: 10.1016/j.bpj.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sens P, Johannes L, Bassereau P. Biophysical approaches to protein-induced membrane deformations in trafficking. Curr Opin Cell Biol. 2008;20(4):476–482. doi: 10.1016/j.ceb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci. 2010;35(12):699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc T. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 57.Rossman JS, Lamb RA. Viral Membrane Scission. Annu Rev Cell Dev Bi. 2013;29:551–569. doi: 10.1146/annurev-cellbio-101011-155838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sc. 2006;7(6):539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 59.Antonny B. Membrane deformation by protein coats. Curr Opin Cell Biol. 2006;18(4):386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Shi Z, Baumgart T. Membrane tension and peripheral protein density mediate membrane shape transitions. Nat Commun. 2015;6:5974. doi: 10.1038/ncomms6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma M, Yi MG, Dong H, Qin HJ, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colotto A, Epand RM. Structural study of the relationship between the rate of membrane fusion and the ability of the fusion peptide of influenza virus to perturb bilayers. Biochemistry-Us. 1997;36(25):7644–7651. doi: 10.1021/bi970382u. [DOI] [PubMed] [Google Scholar]
- 63.Liu F, Lewis RNAH, Hodges RS, McElhaney RN. A differential scanning calorimetric and P-31 NMR spectroscopic study of the effect of transmembrane alpha-helical peptides on the lamellar-reversed hexagonal phase transition of phosphatidylethanolamine model membranes. Biochemistry-Us. 2001;40(3):760–768. doi: 10.1021/bi001942j. [DOI] [PubMed] [Google Scholar]
- 64.Siegel DP, Cherezov V, Greathouse DV, Koeppe RE, Killian JA, Caffrey M. Transmembrane peptides stabilize inverted cubic phases in a biphasic length-dependent manner: Implications for protein-induced membrane fusion. Biophys J. 2006;90(1):200–211. doi: 10.1529/biophysj.105.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tenchov BG, MacDonald RC, Lentz BR. Fusion Peptides Promote Formation of Bilayer Cubic Phases in Lipid Dispersions. An X-Ray Diffraction Study. Biophys J. 2013;104(5):1029–1037. doi: 10.1016/j.bpj.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lipowsky R. Budding of Membranes Induced by Intramembrane Domains. J Phys Ii. 1992;2(10):1825–1840. [Google Scholar]
- 67.Julicher F, Lipowsky R. Shape transformations of vesicles with intramembrane domains. Phys Rev E. 1996;53(3):2670–2683. doi: 10.1103/physreve.53.2670. [DOI] [PubMed] [Google Scholar]
- 68.Seifert U. Configurations of fluid membranes and vesicles. Adv Phys. 1997;46(1):13–137. [Google Scholar]
- 69.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425(6960):821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 70.Kuzmin PI, Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys J. 2005;88(2):1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allain JM, Ben Amar M. Budding and fission of a multiphase vesicle. Eur Phys J E. 2006;20(4):409–420. doi: 10.1140/epje/i2006-10030-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.