Here, Jin et al. determined the crystal structure of Gemin5 in complex with a 13-nt U4 snRNA oligo. Gemin5 is required for snRNP biogenesis and has been implicated in snRNA recognition. Their findings provide novel mechanistic insights into Gemin5's snRNA-binding specificity and the molecular mechanism of RNA binding by WD40 repeat proteins.
Keywords: Gemin5, WD40, snRNA, spliceosome, structure
Abstract
Assembly of the spliceosomal small nuclear ribonucleoparticle (snRNP) core requires the participation of the multisubunit SMN (survival of motor neuron) complex, which contains SMN and several Gemin proteins. The SMN and Gemin2 subunits directly bind Sm proteins, and Gemin5 is required for snRNP biogenesis and has been implicated in snRNA recognition. The RNA sequence required for snRNP assembly includes the Sm site and an adjacent 3′ stem–loop, but a precise understanding of Gemin5's RNA-binding specificity is lacking. Here we show that the N-terminal half of Gemin5, which is composed of two juxtaposed seven-bladed WD40 repeat domains, recognizes the Sm site. The tandem WD40 repeat domains are rigidly held together to form a contiguous RNA-binding surface. RNA-contacting residues are located mostly on loops between β strands on the apical surface of the WD40 domains. Structural and biochemical analyses show that base-stacking interactions involving four aromatic residues and hydrogen bonding by a pair of arginines are crucial for specific recognition of the Sm sequence. We also show that an adenine immediately 5′ to the Sm site is required for efficient binding and that Gemin5 can bind short RNA oligos in an alternative mode. Our results provide mechanistic understandings of Gemin5's snRNA-binding specificity as well as valuable insights into the molecular mechanism of RNA binding by WD40 repeat proteins in general.
Splicing of pre-mRNA is catalyzed by a large dynamic ribonucleoprotein complex known as the spliceosome, which not only faithfully churns out mature messengers but also is responsible for generating diverse proteomes in eukaryotes through alternative splicing. The spliceosome is assembled on pre-mRNA from U-type small nuclear ribonucleoparticles (snRNP) and additional splicing factors. The five major spliceosomal snRNPs—U1, U2, U4, U5, and U6—share some common features but differ in composition and structures, as required by their distinct roles in spliceosome assembly and splicing reactions (Will and Luhrmann 2011; Papasaikas and Valcarcel 2016). All spliceosomal snRNPs contain a characteristic noncoding RNA ranging in size between 100 and 200 nucleotides (nt) in humans, a heptameric Sm/Lsm protein complex, and varying numbers of snRNP-specific protein components. U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II (Pol II) and then travel to the cytoplasm for assembly of the snRNP core and snRNA maturation, including 2,2,7-trimethylation of the guanosine cap and 3′ end trimming (Battle et al. 2006a; Neuenkirchen et al. 2008; Li et al. 2014). The assembly of the snRNP core, composed of snRNA and the seven-member Sm protein complex, depends on a conserved short RNA sequence motif known as the Sm site. U6 snRNA differs from other major spliceosomal snRNAs in several aspects: It is transcribed by Pol III, contains a γ-monomethylated cap, and lacks a conserved Sm site, and the core snRNP is formed by the association of Lsm proteins, which are distinct members of the Sm family of proteins (Reddy et al. 1987; Singh and Reddy 1989; Achsel et al. 1999; Mayes et al. 1999).
The Sm-dependent assembly of snRNP in vivo requires coordinated actions of the PRMT5 and SMN (survival of motor neuron) complexes. In mammalian cells, the PRMT5 complex is composed of PRMT5 (the catalytic subunit of the arginine symmetric dimethylase complex), MEP50, and pICln. The PRMT5 complex is a 20S methylosome that catalyzes arginine symmetric dimethylation of the B, B′, D1, and D3 subunits of the Sm protein complex (Battle et al. 2006a; Neuenkirchen et al. 2008; Li et al. 2014). PRMT5 is an obligatory dimer, and, in the presence of MEP50, further dimerizes to form a hetero-octameric 4x(PRMT5–MEP50) complex (Sun et al. 2011; Antonysamy et al. 2012). MEP50 and pICln direct the enzymatic activity of PRMT5 toward Sm proteins. In particular, pICln alone can form a ring-shaped 6S complex composed of SmD1/D2 and SmF/E/G, while methylation of SmB/D3 is believed to take place with pICln in the methylosome complex (Friesen et al. 2001b; Meister et al. 2001b; Pesiridis et al. 2009; Grimm et al. 2013). A separate arginine symmetric dimethylase, PRMT7, can also methylate Sm proteins (Gonsalvez et al. 2007). Arginine methylation of Sm proteins is required for proper snRNP biogenesis. A conventional wisdom is that arginine symmetric dimethylation increases the binding affinity between Sm proteins and SMN, which contains a Tudor domain known to preferentially recognize symmetrically dimethylated arginines (Brahms et al. 2001; Friesen et al. 2001a; Liu et al. 2010; Tripsianes et al. 2011).
The SMN complex is composed of SMN, Gemin2–8, and UNRIP (Li et al. 2014). SMN is the product of the spinal muscular atrophy (SMA) disease gene. Loss or mutation of SMN results in motor neuron degeneration in the spinal cord, and the disease is the leading genetic cause of infant mortality. SMN interacts with Gemin2, which in turn binds the SmD1/D2/E/F/G pentamer to form an snRNP assembly intermediate complex (Chari et al. 2008; Zhang et al. 2011). Gemin3 is a DEAD-box protein believed to be responsible for the ATP-dependent snRNP assembly activity in early studies, but subsequent in vitro studies using purified systems have cast doubt on the ATP dependence during snRNP assembly (Meister et al. 2001a; Pellizzoni et al. 2002; Chari et al. 2008; Kroiss et al. 2008). Gemin3 interacts with both SMN and Gemin4, whose function in snRNP assembly remains unclear (Charroux et al. 1999, 2000). Similarly, little is known about the precise functions of Gemin6–8 and UNRIP apart from the observations that Gemin6 and Gemin7 both possess an Sm fold domain and form a heterodimer capable of binding Sm proteins in vitro (Ma et al. 2005). This structural feature of Gemin6 and Gemin7 suggests that they may serve as an Sm dimer surrogate during snRNP assembly.
Gemin5 is the snRNA-binding subunit of the SMN complex (Gubitz et al. 2002; Battle et al. 2006b). Based on sequence prediction, it contains 13 WD40 repeats, and the snRNA-binding region is mapped to the WD40 repeat domains (Lau et al. 2009). The snRNA region for binding the SMN complex has been mapped to the Sm site and an adjacent 3′ stem–loop (Yong et al. 2004; Golembe et al. 2005). An exception is found with U1 snRNA, which requires a 5′ SL1 stem–loop for binding the SMN complex (Yong et al. 2002). A recent study revealed a special U1 snRNP assembly pathway dependent on the U1-70K protein, which binds SL1 (So et al. 2016). This finding perhaps explains the observation that Gemin5 is not strictly needed for snRNP assembly in vitro, judged by an assay using purified systems (Neuenkirchen et al. 2015). The binding of the SMN complex and Gemin5 to the Sm site is sequence-specific, but the nucleotide sequence for the stem–loop appears to be unimportant. The Sm-dependent binding of Gemin5 has been attributed to its WD40 repeat domains (Lau et al. 2009). The WD40 repeat domain is a β-propeller structure traditionally considered to be a versatile protein–protein interaction module (Xu and Min 2011). Nevertheless, an increasing number of WD40 repeat domains have shown up as sequence-specific RNA-binding units (Tycowski et al. 2009; Chan et al. 2014; Schonemann et al. 2014). To date, only very limited information is available about the structural basis for sequence-specific RNA binding by WD40-like domains (Loedige et al. 2015), but more examples of RNA-interacting WD40 repeat domains are turning up in the rapidly developing high-resolution cryo-electron microscopy (cryo-EM) studies of spliceosomal complexes (Yan et al. 2015, 2016; Agafonov et al. 2016; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016). To facilitate mechanistic understanding of snRNP assembly and elucidate the structural basis for RNA recognition by the unorthodox RNA-binding module, we determined the structure of the WD40 repeat domains of Gemin5 in complex with RNA fragments encompassing the Sm site. Our result not only revealed the molecular basis for Sm site recognition by Gemin5 but also greatly advanced the understanding of sequence-specific RNA binding by the WD40 repeat protein in general.
Results
Structure of the WD40 repeat domains of Gemin5
Human Gemin5, consisting of 1508 amino acid residues, is the largest subunit of the SMN complex. The snRNA-binding region of Gemin5 is mapped to its N-terminal half, which is predicted to contain 13 WD40 repeats. We expressed a large Gemin5 fragment (amino acids 1–726)—termed G5N, encompassing the reported snRNA-binding region—using baculovirus-infected insect cells and crystallized the protein. The 2.0 Å structure shows that the G5N region encompassing residues 3–722 is folded into two seven-bladed WD40 repeat domains. The two WD40 repeat domains are juxtaposed next to each other, with the very N-terminal short strand joining the second WD40 repeat domain (amino acids 4–10 and 378–722) as the last strand, by anti-parallelly pairing with the very C-terminal strand (Fig. 1A). The first WD40 repeat domain is an all-β-strand structure, with the first blade formed by residues 11–61, constituting the blade “missing” from sequence prediction (Fig. 1A). Several short helices are found embedded in loops connecting neighboring blades and in those between adjacent strands within the same blade, more often in the second WD40 repeat domain. The two WD40 repeat domains are rigidly held together, with the apical surfaces of the two propeller domains, defined by the side of the N-terminal ends of their first β strands, placed at an angle of ∼70°. An extended surface region traversing the two apical surfaces forms a contiguous electropositive gorge where snRNA could potentially bind (Fig. 1B).
Figure 1.
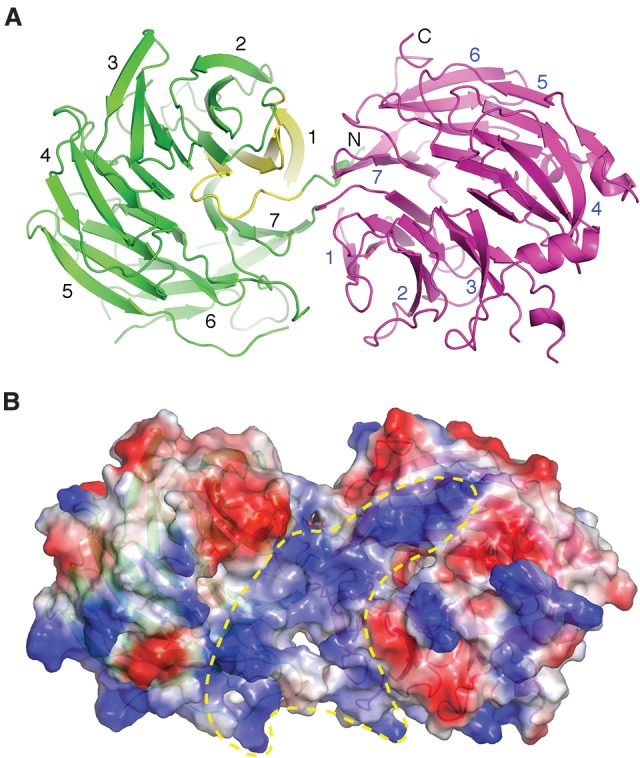
Overall structure of the WD40 repeat domains of Gemin5. (A) Ribbon diagram showing that an N-terminal short β strand completes the C-terminal β-propeller structure (magenta) and showing four following β strands (yellow) form the first blade of the N-terminal seven-bladed β-propeller structure (green). (B) The surface electrostatic potential of G5N shows a continuous positively charged region (enclosed in the yellow dashed line) potentially capable of binding RNA. The partially transparent surface is superimposed with the ribbon representation of G5N and is viewed from a direction similar to that in A.
RNA-binding property of G5N
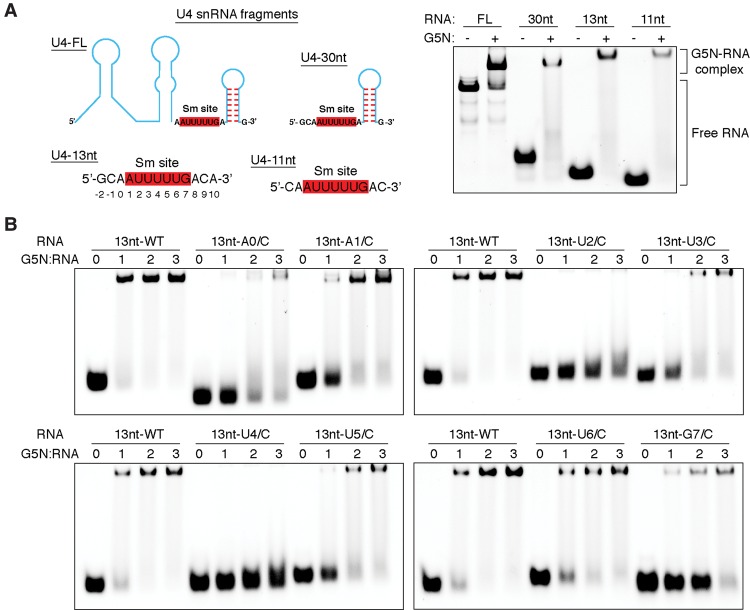
It was reported that Gemin5 binds the snRNA region encompassing the Sm site and an adjacent 3′ stem–loop, and its G5N domain binds snRNA in an Sm-dependent manner (Battle et al. 2006b; Lau et al. 2009). The C-terminal region of Gemin5 was also reported to harbor a noncanonical RNA-binding site (Fernandez-Chamorro et al. 2014). We first set out to isolate a minimal RNA segment for G5N binding and probe whether the G5N domain is necessary and sufficient for specific recognition of the Sm site using in vitro transcribed full-length U4 snRNA and chemically synthesized short oligos (Fig. 2A). As shown in Figure 2A, G5N binds well to the full-length U4 snRNA and a synthetic 30-nt U4 snRNA fragment containing the Sm site and an adjacent 3′ stem–loop, recapitulating the earlier result obtained using full-length Gemin5 (Battle et al. 2006b). Surprisingly, we found that G5N also binds well to shorter U4 snRNA fragments containing the Sm site but without the 3′ stem–loop (Fig. 2A). In particular, a 13-nt RNA fragment containing a centrally located Sm site (5′-G-2C-1A0A1U2U3U4U5U6G7A8C9A10-3′) binds G5N efficiently. Hence, we used this RNA fragment as the template to probe the binding requirement of individual bases (Fig. 2B). We found that replacement of the first or the third uracil (Ura2 or Ura4) within the heptameric Sm sequence with a cytosine most severely affected the binding of G5N, as also observed with full-length Gemin5 (Battle et al. 2006b), and cytosine replacement of other bases within the Sm sequence affected G5N binding to varying but less severe degrees. Interestingly, changing Ade0 outside of the Sm motif to a cytosine also compromised the binding by G5N (Fig. 2B).
Figure 2.
snRNA-binding properties of G5N. (A, left panel) Schematic diagram showing U4 snRNA fragments used for electrophoretic mobility shift assay (EMSA) experiments. The Sm sequence is highlighted with red-shaded boxes. The 13-nt RNA fragment used for subsequent studies is numbered following a scheme indicated below the nucleotide sequence. (Right panel) EMSA results of the binding of G5N to the indicated RNA molecules. A 3:1 protein to RNA molar ratio was used. (B) The binding of G5N to the 13-nt RNA fragment and its base substitution derivatives. Each nucleotide of the Sm site and a 5′ adenine were individually changed to a cytosine, and protein to RNA molar ratios of 1, 2, and 3 were used in EMSA.
Structure of the G5N–RNA complex
To determine the structural basis for Sm site recognition by Gemin5, we crystallized and solved a 1.9 Å structure of G5N in complex with the 13-nt U4 snRNA fragment. There are two G5N–RNA complexes in one crystallographic asymmetric unit (Supplemental Fig. S1). The two complexes superimpose well, with the proteins aligned with an RMSD (root mean squared deviaion) of 0.62 Å in Cα positions, while the two RNA molecules aligned with an RMSD of 0.81 Å for nucleotides from Ade0 to Ade10, encompassing the entire Sm site. In one complex, the very 5′ end guanine is disordered, while, in the other complex, both the first guanine and the second cytosine are ordered, and their bases stacked together. In the latter case, the first two bases also contact a neighboring symmetry-related protein molecule; hence, their exact conformation may be influenced by crystal packing. In both complexes, the first two nucleotides are not involved in tight interactions with their cognate protein molecules; hence, their precise conformation is immaterial for the remainder of our analyses.
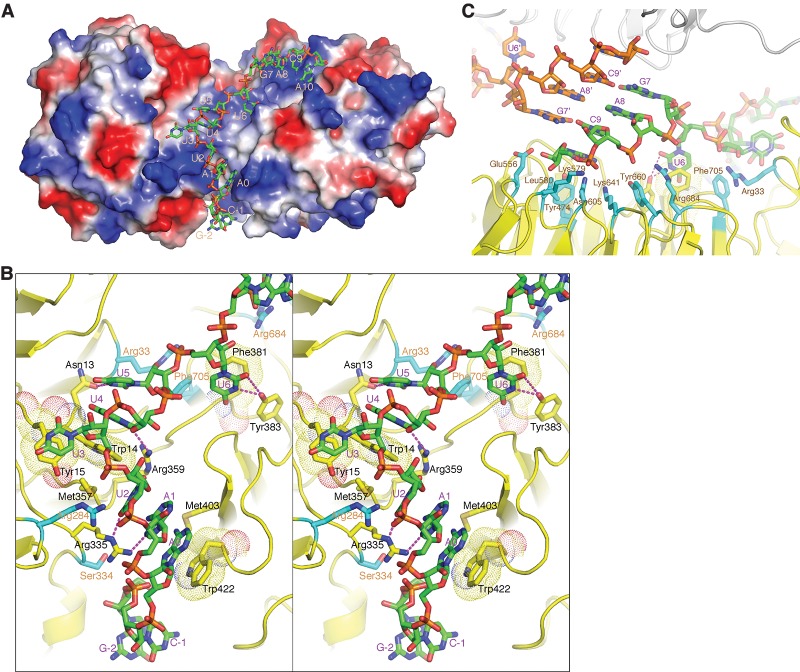
In the structure, the RNA adopts an extended conformation and is bound along the positively charged gorge formed by the tandem WD40 repeat domains (Fig. 3A). The 5′ end of RNA is inserted in the cleft between the two domains; the segment spanning Ade1–Ura5 primarily contacts the first WD40 repeat domain, and the 3′ end nucleotides starting from Ura6 interact mainly with the second WD40 repeat domain. Conspicuous base contacts are observed for Ade0 and Ade1–Ura6 of the Sm site (Fig. 3B). For Ade0, its adenine ring stacks with the indole ring of Trp422, and its extracyclic amino group contacts Met403. Both Ade1 and Ura2 make hydrogen bonds with the guanidino group of Arg335, and Ura2 also contacts Met357 via van der Waals interaction. Ura3 base stacks with Tyr15 in a rather open space, while Ura4 base-stacks with Trp14, and its extracyclic carbonyl groups form hydrogen bonds with the main chain amino group of Trp14 and the guanidino group of Arg359, respectively. The ring amino group of Ura5 and the carbonyl oxygen attached to the C2 atom of the pyrimidine ring form hydrogen bonds with the carboxylate group of Asn13; the same set of atoms of Ura6 interacts with the hydroxyl group of Tyr383 in addition to stacking its pyrimidine ring with Phe381 (Fig. 3B).
Figure 3.
G5N–RNA cocrystal structure. (A) The 13-mer U4 snRNA fragment is bound in the positively charged region at the interface of the tandem WD40 repeat domains and the C-terminal WD40 domain. The G5N structure is shown in a surface representation colored according to its surface electrostatic potential ([blue] positive; [red] negative; [white] neutral) and is viewed from the same direction as in Figure 1A. The RNA is shown in a stick model ([green] carbon; [blue] nitrogen; [red] oxygen; [orange] phosphorus) and is numbered as in Figure 2A. (B) A stereo diagram showing the interaction of the first 9 nt of RNA with G5N. The involved amino acid residues are shown in a stick model, with the carbon bonds involved in contacting RNA bases colored yellow, and the carbon bonds (labeled with orange letters) involved in non-base-contacting interactions colored cyan. Hydrogen bonds are indicated with magenta dashed lines, and aromatic residues involved in stacking with RNA bases are superimposed with a dot representation. (C) G5N–RNA interactions involving the last 4 nt. A neighboring G5N–RNA within the same asymmetric unit is shown with the protein in a gray ribbon representation, and the RNA is shown in a stick model with the bonds connected to phosphorus atoms colored orange.
The bases of Gua7, Ade8, and Cyt9 do not directly contact the protein (Fig. 3C). Gua7 and Cyt9 are stabilized by Watson-Crick base-pairing with Cyt9 and Gua7, respectively, from the other complex within the asymmetric unit (Fig. 3C). The base of Ade8 ladder-stacks in between Gua7 and Cyt9 and packs parallelly against the Ade8 base from the neighboring complex. At present, it is not clear whether the RNA–RNA contacts have any physiological meanings, but our finding indicates that the involved bases have flexible conformation in the absence of other binding partners. In addition to stacking with Cyt9, Ade10 makes protein contacts with several residues from the central lining of the second β propeller, including Tyr474, Leu580, Lys641, and Asn605, mostly via van der Waals interactions (Fig. 3C). Apart from the base interactions described above, a number of G5N interactions with the phosphate backbone and the ribose moieties are eminent. Of note is the involvement of several arginine residues, as their positively charged side chains favorably interact with the negatively charged phosphate groups of the RNA backbone (Fig. 3C). One arginine, Arg33, interacts with both the 2′-OH group of Ura5 and the phosphate group of Ura6 via its guanidine nitrogens. Two aromatic residues, Tyr660 and Phe705, interact with the RNA backbone via van der Waals interaction. Tyr660 interacts with the phosphate group of Cyt9, while Phe705 supports the binding of Ura5 by contacting its sugar moiety (Fig. 3C).
Biochemical analysis of determinants for RNA-binding specificity
To validate the structural findings and assess the degree of importance of the aforementioned Gemin5 residues in snRNA binding, we generated 11 point mutants of G5N and tested their bindings to the 13-mer U4 snRNA fragment and to in vitro transcribed full-length U4 snRNA by electrophoretic mobility shift assay (EMSA) and fluorescence polarization assay (FP) (Fig. 4). For the four aromatic residues stacking with RNA bases, we individually changed Trp422 and Phe381 to negatively charged glutamate and aspartate, respectively, and Trp14 and Tyr15 to alanines. Tyr383 was changed to a phenylalanine to probe the loss of the hydrogen bonds between Tyr383 and Ura6. The W422E and F381D mutants mimic the inactivating aromatic residue mutant of the canonical RNA recognition motif (RRM) protein SF2/ASF (Caceres and Krainer 1993); the W14A and Y15A mutants are intended to minimize perturbations to neighboring RNA-binding residues while abolishing stacking with Ura4 and Ura3, respectively. The rest of the involved residues are changed to either an alanine or a glutamate based on considerations of the nature of their interactions with RNA.
Figure 4.
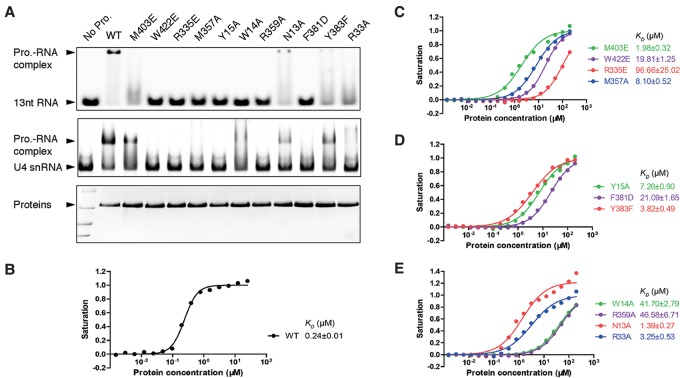
Determinants for RNA-binding specificity. (A, top panel) EMSA of the binding of the wild-type (WT) and indicated mutants of G5N to the 13-mer U4 fragment used for obtaining the cocrystal structure. A 2:1 protein to RNA molar ratio was used in the assay. (Middle panel) Binding of G5N and its mutants to in vitro transcribed full-length U4 snRNA. (Bottom panel) G5N and its mutants used for EMSA experiments. (B–E) FP measurements of binding affinities of the wild-type and mutant G5N proteins to the 13-mer RNA. The derived dissociation constant (KD) values are shown.
Figure 4A shows that all four aromatic residue mutants designed to disrupt base stacking essentially lost bindings to RNA. While it is easy to understand that disruption of π stacking results in diminished or loss of binding of RNA to protein, the contribution of stacking interaction to RNA-binding specificity is less intuitive. There are at least two factors that can contribute to the binding specificity: One is the stacking efficiency due to the differences between purines and pyrimidines and the substituent effect of distinct nucleotide bases, as exemplified in sequences-specific recognition of RNase T (Duh et al. 2015), and the other is the involvement of aromatic residues in shaping up the binding site (i.e., via shape complementarity and energetic coupling of adjacent amino acid residues), as exemplified in RNA recognition by U1A protein (Oubridge et al. 1994; Shiels et al. 2002). An examination of the binding sites involving Trp14, Tyr15, Phe381, and Trp422 (Fig. 3A,B) reveals that stacking between Ura3 and Tyr15 occurs in a relatively unrestrained open space, suggesting that this binding site may be more tolerant to base substitution—an observation relevant to our discussion of alternative modes of RNA binding later. In addition to the base-stacking aromatic residues, two base-contacting arginine mutants, R335E and R359A, also severely compromised RNA binding (Fig. 4A). Met357 (which, together with R335, coordinates the binding of Ura2) also showed its crucial role in RNA binding. The remaining four mutants—M403E, N13A, Y383F, and R33A—displayed reduced but detectable levels of RNA binding.
EMSA results using in vitro transcribed full-length U4 snRNA generally agree with that obtained using the 13-mer RNA (Fig. 4A). An interesting exception is that the W14A mutant appears to still be able to bind full-length U4 (Fig. 4A). The precise reason for this difference is as yet unexplained, but a possible scenario may be that when full-length U4 snRNA is bound, a conformation change of Arg359, which also directly contacts Ura4, remodels the binding site to enable a stronger interaction with Ura4, partly offsetting the loss of base stacking introduced by the W14A mutation (Fig. 3B).
To gain quantitative feelings about the contribution of the involved residues in RNA binding, we measured their RNA-binding affinities by FP using a 5′-fluorescien isothiocyanate (FITC)-labeled 13-mer RNA (Fig. 4B–E). The wild-type G5N binds the RNA oligo with a dissociation constant (KD) of 0.24 µM, while the four aromatic residue mutants W14A, Y15A, F381D, and W422E bind RNA with KD values of 41.7, 7.3, 21.1, and 19.8 µM, respectively, ranging from 30-fold to 170-fold reduction in binding affinities. The two base-contacting arginine mutants R335E and R359A, with KD values of 96.7 and 46.6 µM, also severely compromised RNA binding. The M357A mutant has a KD of 8.1 µM, while M403E, N13A, Y383F, and R33A bind RNA with KD values of 2.0, 1.4, 3.8, and 3.3 µM, respectively (Fig. 4B–E). The FP and EMSA results agree well and strongly corroborate our structural findings.
Alternative RNA-binding mode
The complex structure of G5N with the 13-mer RNA indicates that major protein contacts occur through the RNA region encompassing Ade0–Ura6. We therefore wondered whether G5N and its mutants could bind short RNA fragments in a similar fashion. Earlier, we demonstrated that G5N could bind an 11-mer RNA containing a centrally located Sm sequence (Fig. 2A). We chose the 11-mer RNA, a 9-mer containing a centrally located Sm site, and an 8-mer with one adenine added to the 5′ of the 7-mer Sm sequence for testing and used wild-type G5N and M403E, W422E R335E, and M357A mutants, which affect interactions with 5′ end nucleotides Ade0, Ade1, and Ura2, as the probes. Figure 5 shows that the 11-mer RNA binds G5N and its derivatives in a manner identical to that of the 13-mer RNA. Oddly, two shifted bands appeared in G5N bindings to the 9-mer and 8-mer RNA (Fig. 5, bottom panel). In the lanes with G5N mutants, the bottom band disappeared, but the top band largely remained. A close examination of EMSA results with 13-mer and 11-mer RNA also revealed a previously unnoticed faint top band (Fig. 5). We conclude that the bottom band represents RNA binding in a manner observed in the cocrystal structure, and the top band indicates an unexplained binding mode insensitive to G5N mutations known to affect RNA binding through its 5′ end nucleotides.
Figure 5.
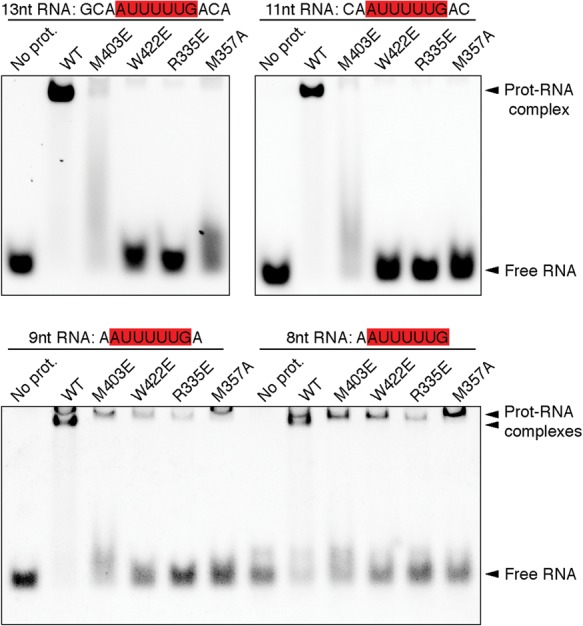
Binding of shorter RNA fragments to G5N. EMSA analysis of the binding of G5N and selected mutants to the indicated RNA fragments, all encompassing an intact Sm site. A 3:1 protein to RNA molar ratio was used. Please note the positions of the shifted bands when 9-mer and 8-mer oligos are used.
To reveal the alternative RNA-binding mode, we attempted to cocrystallize G5N with shorter RNA oligos but without success. We then tried to soak the native G5N crystal with shorter RNA fragments. The G5N–RNA complex structures obtained by soaking with 9-mer, 8-mer, and 7-mer (Sm sequence only) oligos are largely the same; hence, we refer to the structures collectively here using the 7-mer (bare Sm site) structure as a representative unless explicitly specified otherwise. The new structure shows that Ura5 is disordered, and Ura6 has a relatively weak density, while Ade1 base-stacks with Tyr15—a position occupied by Ura3 in the original 13-mer RNA cocrystal structure (Fig. 6A). Superposition of the two structures shows that Ura2, Ura3, and Ura4 are all bound in positions out of registry by 2 nt relative to that in the 13-mer structure; i.e., they are bound in the locations of Ura4, Ura5, and Ura6 of the 13-mer structure, respectively (Fig. 6A). Owing to the occupation by identical nucleotides in the new and old structures, the two sets of the uracil triplet interact with G5N identically. Ura6 has a weak density in the 7-mer structure, while it is disordered in several other soaked structures; hence, its conformation may be considered flexible. A main difference involves Gua7, which is situated in the central pore of the second WD40 domain, stacked by Tyr474 on one side and surrounded by Leu580 and Asn582 on the other side (Fig. 6A). In addition, the extracyclic amino group of Gua7 makes a hydrogen bond with the carboxylate group of Glu541, the amino group at ring 1 position hydrogen bonds with the main chain carbonyl of Thr540, and the N3 atom forms a hydrogen bond with the side chain carbonyl of Asn582 (Fig. 6C). A Y474A mutant does not affect the binding of G5N to the 13-mer RNA but does clear the minor top band in EMSA, indicating that the soaked-in 7-mer structure indeed represents an alternative, minor RNA-binding mode (Fig. 6B).
Figure 6.
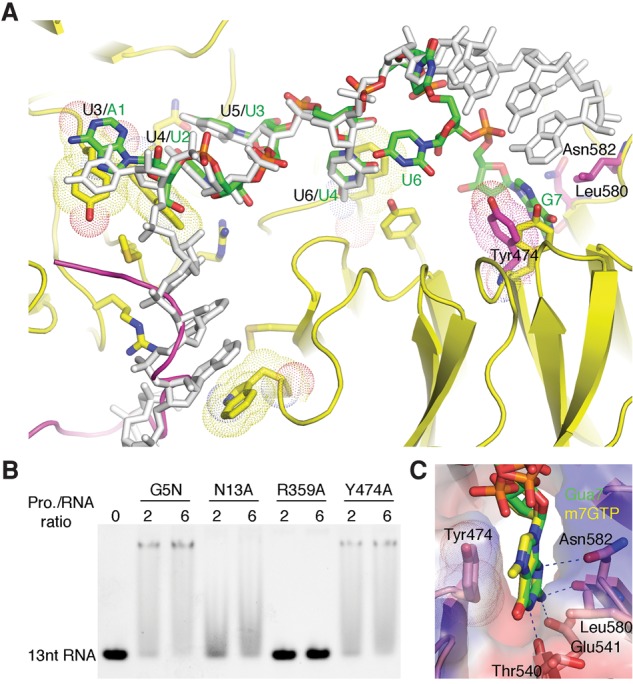
Alternative RNA-binding mode of G5N. (A) The soaked-in 7-mer Sm RNA is superimposed with the cocrystal structure with 13-mer RNA. The 7-mer RNA is shown in a stick model colored according to the scheme in Figure 3, while the cocrystallized 13-mer RNA is colored gray. The green labels indicate the identity of the 7-mer RNA nucleotides, while the preceding black labels separated by a slash indicate nucleotide positions in the cocrystallized 13-mer RNA. Tyr474, Asn582, and Leu580 from the soaked-in G5N structure are superimposed as a stick model, with carbon atoms colored magenta. Please note that the conformation of Tyr474 is changed from that in the cocrystallized structure (yellow) to accommodate the binding of Gua7. A loop connecting blades 5 and 6 of the first WD40 domain, which is ordered in native and soaked-in G5N structures (magenta) but disordered in the cocrystal structure, is superimposed. (B) EMSA analysis of the binding of the Y474A mutant to the 13-mer RNA. Wild-type G5N and N13A and R359A mutants were used as controls. (C) Binding of the m7GTP cap analog to G5N at the site that bound Gua7 in the soaked-in 7-mer RNA complex.
Binding of the 7-methylguanosine (m7G) cap to G5N
Gemin5 can exist in SMN-free forms alone or in complex with Gemin3 and Gemin4 (Battle et al. 2007), but little is known about the SMN-independent functions of Gemin5. Biochemical pull-down experiments identified Gemin5 as a m7G cap-binding protein (Bradrick and Gromeier 2009), an observation that could benefit the search for Gemin5's SMN-independent cellular functions. The m7G-binding site was also mapped to the WD40 repeat domains of Gemin5, thus enabling us to experimentally determine the m7G-binding site by soaking a m7G cap analog, m7GTP, into the native crystal of G5N. Interestingly, the base of m7G binds G5N in the same location and in the same manner as Gua7 in the 7-mer structure (Fig. 6C). This mode of cap binding differs from the canonical mode of cap recognition, in which two aromatic residues sandwich the cap (Fechter and Brownlee 2005). In the present structure, Tyr474 stacks m7G from one side, but the other side is contacted by Leu580 and Asn582. The methyl group of m7G points away from the WD40 domain and is not interacting with other residues. The specificity for m7G over unmethylated guanosine seen in pull-down experiments may be attributed to enhanced stacking interaction due to charge delocalization resulting from guanosine methylation (Hu et al. 1999). The rest of the interactions involving hydrogen bonds between Glu541, side chain carbonyl, and main chain carbonyl groups of Asn582 and the carbonyl of Thr540 conform to the general properties of a canonical m7G recognition site (Fig. 6C). Thus, the observed m7G location in the central pore of the second WD40 domain constitutes a novel type of cap-binding site. Clearly, it can also bind a nonmethylated guanine base in a proper context, such as Gua7 of the 7-mer RNA. It is also clear that m7G cap cannot bind Gemin5 simultaneously with snRNA in either the manner of 7-mer RNA binding (due to overlap of the Gua7-binding site) or the 13-mer binding mode (because of steric conflicts introduced by the phosphate moiety of m7G cap).
Discussion
Owing to the fundamental importance of snRNPs in eukaryotic lives, it is imperative to understand the processes governing their assembly and biogenesis in molecular details. Great progress has been made in understanding the structure and function of the central snRNP assembly machine, the SMN complex (Battle et al. 2006a; Neuenkirchen et al. 2008; Li et al. 2014). Considerable mechanistic insights about the formation of the Sm protein complex have been gained from the determination of structures of key assembly intermediates, including the complex of Gemin2, the Gemin2-binding domain of SMN, and the SmD1/D2/E/F/G pentamer as well as the 6S pICln-SmD1/D2/E/F/G complex and a stalled transfer intermediate, including the 6S complex together with SMN and Gemin2 (Zhang et al. 2011; Grimm et al. 2013). By comparison, the mechanism for snRNA recognition during snRNP assembly is less well understood. Although Sm proteins are capable of binding snRNA at the Sm site, they are trapped in a state unable to bind RNA by Gemin2 (Zhang et al. 2011; Grimm et al. 2013). This observation reinforces the notion that some components of the SMN complex need to bring the correct RNA to the vicinity of the SMN/Sm protein complex intermediate to further the assembly process. The most obvious and best-studied snRNA-binding component of the SMN complex is Gemin5, although some concerns about its precise function remain to be clarified. One concern arises from the absence of Gemin5 homologs in lower organisms, such as Caenorhabditis elegans and yeasts (Kroiss et al. 2008). However, it is not impossible that an as yet unidentified RNA-binding protein unrelated to Gemin5 may transiently associate with the SMN complex to fulfill the role of Gemin5. Another concern is that Gemin5 appears to be not needed for snRNP assembly in vitro (Neuenkirchen et al. 2015). There are several possible explanations for this observation. First, the efficiency of snRNP assembly may be different in in vitro and in vivo settings. Second, as has become clear recently, different snRNPs, such as U1, may have additional assembly pathways (So et al. 2016). Thus, Gemin5 remains the most viable candidate for snRNA recognition during snRNP core assembly, although additional functions of Gemin5 unrelated to its roles in snRNP assembly have begun to emerge (Pineiro et al. 2015).
The precise mechanism of snRNA recognition by Gemin5 was unknown until this study. The predicted WD40 repeat fold of Gemin5 did not help in understanding its RNA-binding mode, as WD40 repeat domains have not been well characterized as RNA-binding proteins. To date, the crystal structure of only one WD40-like protein bound to RNA has been determined (Loedige et al. 2015). Our results revealed for the first time the mechanism by which two WD40 repeat domains form an integral RNA-binding unit, as indicated by the importance of the interface between the two WD40 repeat domains in RNA binding. One common feature regarding RNA binding by WD40-like domains that resulted from this study is that RNA-contacting residues are located primarily on loops on the apical surface of the propeller-like structure—a characteristic perhaps useful for analyzing other WD40-like RNA-binding proteins in general. Recent cryo-EM structures of several spliceosomal complexes have revealed the involvement in RNA binding of several spliceosomal WD40 repeat proteins; e.g., Cwf17/U5-40K and Prp46 (Yan et al. 2015; Agafonov et al. 2016; Wan et al. 2016). However, both Cwf17/U5-40K and Prp46 contact dsRNA regions via surface areas on the perimeter of the β propellers, and, in both cases, the protein–RNA contacts do not appear to be base-specific.
Our analyses show that specific recognition of snRNA by the WD40 repeat domains of Gemin5 is mainly through seven contiguous nucleotides (a 5′ extra-Sm adenine followed by 6 nt of the Sm sequence), consistent with the updated U4 Sm recognition sequence of AAUUUUU from reanalysis of the spliceosomal U4 snRNP core domain structure (Li et al. 2016). In agreement with earlier results, our structural and biochemical analyses showed that the first and the third uracils of the Sm site are essential for snRNA recognition by Gemin5 (Battle et al. 2006b). Our study also revealed contributions of other nucleotides important for efficient recognition. One superficial discrepancy between our result and the reported snRNA-binding property of Gemin5 may be the role of a 3′ stem–loop for snRNA recognition (Battle et al. 2006b). However, it is worth pointing out that, in addition to the N-terminal WD40 repeat domains, the C-terminal domain of Gemin5 has been reported to possess RNA-binding activity (Fernandez-Chamorro et al. 2014). It is possible that the C-terminal domain can bind the 3′ stem–loop of snRNA. It is also possible that additional surface area of the WD40 repeat domains may bind the stem–loop to increase binding affinity, such as in the case of spliceosomal WD40 proteins, as it has been noted that the exact sequence of the stem–loop is unimportant. Regardless, it is fair to say that specific recognition of the Sm site by the WD40 repeat domains of Gemin5 is now backed by a solid mechanistic foundation.
The G5N–13-mer RNA complex was crystallized with two protein–RNA complexes in one asymmetric unit, and 3 nt from each complex base-paired with each other in the structure. Thus, a natural question is whether this structure is physiologically meaningful. There are strong reasons to believe that the mode of Sm site recognition revealed by the structure represents the manner by which the tandem WD40 repeat domains of Gemin5 bind snRNA in solution. First, the contacts between the two protein–RNA complexes are not due to crystal contacts, as they are contents within one crystallographic asymmetric unit. The base pairing indicates that the conformation of these three U4 bases, which are not the ones involved in recognition by the heptameric Sm ring in the U4 snRNP core structure (Li et al. 2016), is flexible when the 13-mer RNA is bound to G5N. In fact, we believe that the flexible conformation of Gua7 and Cyt9 may have important implications for snRNP core assembly, as we discuss below. Second, our mutagenesis results show that all of the base-interacting residues contacting Ade0 to Ura6 behave as expected from the structure. In particular, mutation of Gemin5 residues contacting Ade0, Ade1, and Ura2—namely, Trp422, Arg335, and Met357, which do not contact RNA in the alternative binding mode with 7-mer RNA—significantly affected the 13-mer and full-length U4 snRNA binding (Figs. 4, 5). Third, it may be more than coincidental that our identification of the AAUUUUU sequence (not the traditionally thought AUUUUUG Sm sequence) as the recognition site for the WD40 domains of Gemin5 agrees perfectly with the mode of U4 snRNA recognition by the Sm protein complex in the U4 snRNP core (Li et al. 2016). Altogether, we believe that there is compelling evidence supporting the notion that reading of the AAUUUUU sequence by G5N as revealed by the 13-mer RNA cocrystal structure represents a bona fide mode of snRNA recognition by Gemin5.
An important question is how our structural knowledge about the G5N–RNA complex may advance our understanding of the process of snRNP core assembly. An examination of the U4 snRNP core structure revealed that while the AAUUUUU sequence curls around to interact with Sm proteins with one base for every Sm subunit in the order of SmF–E–G–D3–B–D1–D2, the single-stranded region between the Sm site and the 3′ stem–loop of U4 snRNA is snugged into the central pore of the Sm ring (Fig. 7A). As pointed out earlier, binding of the Sm sequence to the assembly intermediate SMN–Gemin2–SmD1–D2–F–E–G (SMN–G2–Sm5) complex is blocked by the N-terminal region of Gemin2 (G2N), but G2N does not interfere with the binding of Sm proteins to the single-stranded region immediately 3′ to the Sm sequence. Therefore, it is an attractive hypothesis that the binding of Gemin5 to the Sm sequence, possibly in conjunction with the C-terminal domain binding to the 3′ stem–loop, delimits the single-stranded region of snRNA, which will then be presented to the Gemin2–Sm5 complex for initial binding through the single-stranded region (Fig. 7B). It is worth pointing out that the Sm sequence is in an extended conformation when it is bound to Gemin5; therefore, G2N may not need to come off the Sm5 unit when the Gemin5-bound snRNA approaches the Gemin2–Sm5 complex. It is likely that snugging the single-stranded region of snRNA onto the central channel of Sm5 via the opening reserved for SmB/D3 binding will bump Gemin5 off snRNA, leaving the Sm bases searching for the right Sm subunit to bind. At this stage, there is no hindrance to prevent the association of the SmB/D3 heterodimer with the Sm5 complex, as G2N is not in a position to interfere with the joining of SmB/D3 to the Sm protein complex. Closure of the Sm protein ring will drive the Sm nucleotides to bind their energetically favorable cognate sites in the Sm protein subunits, and this will conflict with the Gemin2 binding to the Sm proteins, hence resulting in the dissociation of Gemin2 and the completion of the snRNP core assembly. However, it should be cautioned that while our proposed mechanism is an attractive and testable scenario, alternative sequences of events may be equally possible, such as dissociation of Gemin2 prior to the binding of SmB/D3. A novel feature of our proposal is that Gemin5 binding delimits the free single-stranded region of snRNA, preparing the snRNA for efficient assembly of the snRNP core.
Figure 7.
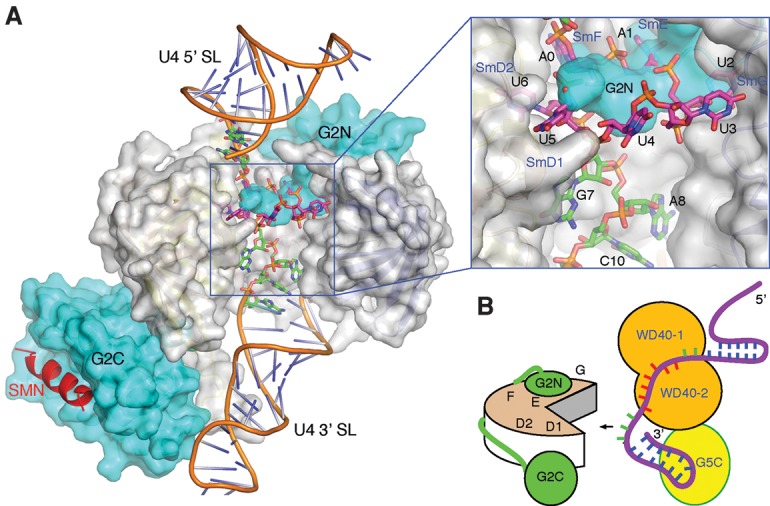
A stretch of nucleotides 3′ to the Sm site is important for snRNP assembly. (A) A model with Gemin2 and a short region of SMN (Protein Data Bank [PDB] ID 3s6n) bound to the U4 snRNA (PDB ID 4wzj). The SmB/D3 heterodimer was removed for viewing clarity and to mimick the pentameric Sm assembly intermediate. Gemin2 and SmD1–D2–F–E–G are shown in a surface representation, with Gemin2 colored in cyan, and the Sm proteins colored in gray. The SMN fragment is shown in a cartoon representation colored in red. The 13 nt used for cocrystallization is shown in a stick representation, with the carbon bonds colored magenta for nucleotides belonging to the heptameric A0A1U2U3U4U5U6 sequence involved in G5N binding and green for the remaining nucleotides. U4 snRNA outside of the 13-mer region is shown in a cartoon representation with the backbone colored brown. An inset at the top right displays an enlarged view of the boxed area of the main figure. (B) An illustration of how Gemin5 may delimit the free single-stranded region of U4 snRNA 3′ to the AAUUUUU G5N recognition sequence (colored red) for presentation to Gemin2 (labeled G2N and G2C) bound the Sm pentamer. A hypothetical C-terminal region of Gemin5 bound to the 3′ stem–loop is shown.
Our careful analyses also revealed that the WD40 repeat domains of Gemin5 could bind short RNA oligos containing the Sm site in an alternative mode, particularly with short oligos having a 3′ end guanine. The structural rationale for the alternative mode of RNA binding is that the central pore of the second WD40 repeat domain has a propensity for binding a guanine, and the binding site for the second Sm uracil (Ura3) in the 13-mer structure (i.e., stacking via Tyr15) has a relaxed nucleotide specificity due to their interaction by base stacking in a rather open environment. Furthermore, the five contiguous uracils in the Sm sequence can shift binding positions without introducing binding energy penalties. These features of Gemin5 enabled the binding of short Sm-containing RNA oligos in an alternative fashion. Stabilization of the loop connecting blades 5 and 6 (amino acids ∼270–283) of the first WD40 repeat domain in the preformed crystal lattice of the native Gemin5 crystal, which prevents the soaking in of RNA with long 5′ extensions, probably also made the RNA oligos more susceptible to the adaptation of the alternative binding mode (Fig. 6A). We think that this mode of RNA binding is reflective of Gemin5's general and versatile RNA-binding ability, which may be important for its broad roles in RNA-related biological processes, such as translational regulation (Bradrick and Gromeier 2009; Pineiro et al. 2015; Workman et al. 2015; Francisco-Velilla et al. 2016). Additionally, the guanine-preferred central pore of the second WD40 repeat domain of Gemin5 may be suitable for targeting small molecules to interfere with its functions in certain RNA-related biological processes.
Materials and methods
Protein expression and purification
The cDNA fragment encoding an N-terminal fragment of human Gemin5 encompassing the WD40 repeat domains (G5N, amino acids 1–726) was inserted between EcoRI and NotI sites of the pFastBac-HTC vector for generation of recombinant baculovirus. Wild-type and mutant G5N proteins were expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) in Sf21 cells. For protein purification, G5N or its derivatives were first purified by Ni-NTA (Novagen) affinity column and eluted in a buffer containing 20 mM HEPES (pH 7.5), 90 mM NaCl, 250 mM imidazole, and 5% glycerol. The 6× his tag was removed by TEV protease, and the proteins were further purified by heparin and gel filtration column chromatography (Superdex 200 16/60 XK column, GE Healthcare) in a buffer containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 5% glycerol, and 1 mM DTT. The peak fractions containing highly purified G5N or its mutants were concentrated to ∼10 mg/mL for later use in crystallization and RNA-binding assays.
Crystallization
Crystals of G5N were grown in 0.1 M sodium malonate (pH 7.0) and 12% polyethylene glycol (PEG) 3350 by hanging-drop vapor diffusion at 16°C. The complex of G5N and a 13-nt synthetic U4 snRNA oligo (5′-GCAAUUUUUGACA-3′) was formed by incubating the protein (∼7 mg/mL) with RNA in a molar ratio of 1:2 for 1 h on ice. Crystallization screening was carried out by sitting-drop vapor diffusion at 16°C. Diffracting crystals were grown in 4% tacsimate (pH 8.0) and 16% PEG 3350. Crystals of G5N in complex with 9-nt (5′-CAAUUUUUG-3′), 8-nt (5′-AAUUUUUG-3′), or 7-nt (5′-AUUUUUG-3′) RNA were obtained by soaking native G5N crystals with either 0.15 or 0.45 mM RNA for 20 h at 16°C in a solution containing 0.1 M HEPES (pH 7.5), 14% PEG 8000, and 25 mM MgCl2. G5N–m7GTP complex crystals were generated by soaking native G5N crystals with 0.5 mM m7GTP (Sigma, M6133) in 0.1 M sodium malonate (pH 7.0) and 12% PEG 3350 for 24 h at 16°C.
Data collection and structure determination
The native structure of G5N was solved by single-wavelength anomalous dispersion (SAD) using a Pt derivative prepared by soaking native G5N crystals with 1 mM K2Pt(NO2)4 in the crystallization solution for 24 h before data collection. A 2.0 Å native data set and a 3.0 Å platinum derivative SAD data set were collected at beamline BL18U1 of the Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 0.9786 Å using a Pilatus 6M detector, and the data were processed using HKL3000 (Otwinowski and Minor 1997). Four heavy atoms were located by SHELXD (Sheldrick 2010), and SAD phasing was performed using PHENIX (Adams et al. 2010). A polyalanine model of the structure was first built using COOT (Emsley and Cowtan 2004), and the final model was rebuilt with PHENIX using the high-resolution native data set and refined to 2.0 Å using PHENIX and COOT.
The 1.9 Å G5N–13-nt RNA cocrystal data set was collected at beamline BL17U of SSRF at a wavelength of 0.9792 Å using an Area Detector Systems Corporation (ADSC) Quantum 315r detector and was processed using HKL2000. The structure was solved by molecular replacement with PHASER (McCoy et al. 2007) using the protein-alone structure as the search model. There were two G5N–RNA complexes in one asymmetric unit. The density of the two RNA chains was clear, allowing unambiguous model building (Supplemental Fig. S2). The final model was built with COOT and refined using RefMac (Murshudov et al. 1997). The 2.1 Å G5N–7-nt RNA data set was collected at beamline BL18U1 of SSRF, and the structure was solved by molecular replacement with PHASER. The RNA model was built with COOT, and the structure was refined using PHENIX. Additional soaked-in structures with 8-nt and 9-nt RNA were solved by methods similar to that of the 7-nt RNA complex. The 2.5 Å G5N–m7GTP data set was collected at beamline BL17U of SSRF at a wavelength of 0.9788 Å using an ADSC Quantum 315r detector and processed using HKL2000. The structure of G5N–m7GTP was solved by molecular replacement using a native structure as the search model with PHASER. There was no density for the third phosphate group of m7GTP in this structure; thus, it was not modeled.
Details of X-ray data collection and structure refinement are in Supplemental Table S1. Atomic coordinates and X-ray diffraction data for the structure of native G5N and that of the 13-nt, 7-nt, and m7GTP complexes with G5N have been deposited in Protein Data Bank under the accession codes 5H1J, 5H1K, 5H1L, and 5H1M, respectively.
EMSA
Full-length human U4 snRNA was in vitro transcribed with the MEGAscript kit (Ambion) according to the manufacturer's instructions. Chemically synthesized short RNA oligos used for EMSA and crystallization were purchased from Takara. For EMSA experiments, 80 ng of each RNA was incubated with the indicated amounts of wild-type or mutant G5N proteins for 30 min on ice in a 10-µL reaction in 20 mM HEPES (pH 7.5), 100 mM NaCl, 5% glycerol, and 1 mM DTT. The RNA–protein complexes were separated by 10% native PAGE gel, stained with SYBR Green (Thermo Fisher Scientific) in 0.5× TBE for 10 min, and analyzed on a Gel Doc EZ imager system (Bio-Rad).
FP
Custom-synthesized 5′-FITC-labeled 13-mer RNA (Takara) was mixed at 100 nM with increasing amounts of G5N proteins in a buffer containing 20 mM HEPES (pH 7.5) and 100 mM NaCl. The mixtures were incubated for 20 min at room temperature. The measurements were performed on an Envision multimode plate reader (PerkinElmer). The binding affinity (KD value) was calculated by nonlinear regression fitting of a model of one site-specific binding with Hill slope using GraphPad Prism 5 software following a published protocol (Zhou et al. 2014).
Supplementary Material
Acknowledgments
We thank Dr. Jun Xiong for advice in FP experiments, and staff scientists at beamlines BL17U and BL18U1 of the Shanghai Synchrotron Radiation Facility for help with X-ray data collection. We also thank Dr. Chao Xu and Dr. Jinrong Min for communication of their data prior to publication. This work was supported by Natural Science Foundation of China grants 31570747, 31622020, 31430018, and 31521002; Beijing Natural Science Foundation grant 5164038; and Strategic Priority Research Program grants XDB08010100 and XDB08030201 of the Chinese Academy of Sciences (CAS).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.291377.116.
References
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18: 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Luhrmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. [DOI] [PubMed] [Google Scholar]
- Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, et al. 2012. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci 109: 17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. 2006a. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. 2006b. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23: 273–279. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Wang J, Dreyfuss G. 2007. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J Biol Chem 282: 27953–27959. [DOI] [PubMed] [Google Scholar]
- Bradrick SS, Gromeier M. 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4: e7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell 135: 497–509. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. 1999. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol 147: 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. 2000. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol 148: 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh Y, Hsiao YY, Li CL, Huang JC, Yuan HS. 2015. Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids. Protein Sci 24: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fechter P, Brownlee GG. 2005. Recognition of mRNA cap structures by viral and cellular proteins. J Gen Virol 86: 1239–1249. [DOI] [PubMed] [Google Scholar]
- Fernandez-Chamorro J, Pineiro D, Gordon JM, Ramajo J, Francisco-Velilla R, Macias MJ, Martinez-Salas E. 2014. Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res 42: 5742–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco-Velilla R, Fernandez-Chamorro J, Ramajo J, Martinez-Salas E. 2016. The RNA-binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Res 44: 8335–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. 2001a. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 7: 1111–1117. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. 2001b. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. 2005. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol 25: 10989–11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. 2007. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 178: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Chari A, Pelz JP, Kuper J, Kisker C, Diederichs K, Stark H, Schindelin H, Fischer U. 2013. Structural basis of assembly chaperone- mediated snRNP formation. Mol Cell 49: 692–703. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem 277: 5631–5636. [DOI] [PubMed] [Google Scholar]
- Hu G, Gershon PD, Hodel AE, Quiocho FA. 1999. mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc Natl Acad Sci 96: 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss M, Schultz J, Wiesner J, Chari A, Sickmann A, Fischer U. 2008. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc Natl Acad Sci 105: 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. 2009. Gemin5–snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16: 486–491. [DOI] [PubMed] [Google Scholar]
- Li DK, Tisdale S, Lotti F, Pellizzoni L. 2014. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol 32: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Leung AK, Kondo Y, Oubridge C, Nagai K. 2016. Re-refinement of the spliceosomal U4 snRNP core-domain structure. Acta Crystallogr D Struct Biol 72: 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. 2010. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, et al. 2015. The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila brain-tumor-mediated gene regulation. Cell Rep 13: 1206–1220. [DOI] [PubMed] [Google Scholar]
- Ma Y, Dostie J, Dreyfuss G, Van Duyne GD. 2005. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure 13: 883–892. [DOI] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. 2001a. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol 3: 945–949. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. 2001b. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11: 1990–1994. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. 2008. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 582: 1997–2003. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Englbrecht C, Ohmer J, Ziegenhals T, Chari A, Fischer U. 2015. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J 34: 1925–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438. [DOI] [PubMed] [Google Scholar]
- Papasaikas P, Valcarcel J. 2016. The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem Sci 41: 33–45. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775–1779. [DOI] [PubMed] [Google Scholar]
- Pesiridis GS, Diamond E, Van Duyne GD. 2009. Role of pICLn in methylation of Sm proteins by PRMT5. J Biol Chem 284: 21347–21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro D, Fernandez-Chamorro J, Francisco-Velilla R, Martinez-Salas E. 2015. Gemin5: a multitasking RNA-binding protein involved in translation control. Biomolecules 5: 528–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Reddy R, Henning D, Das G, Harless M, Wright D. 1987. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem 262: 75–81. [PubMed] [Google Scholar]
- Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels JC, Tuite JB, Nolan SJ, Baranger AM. 2002. Investigation of a conserved stacking interaction in target site recognition by the U1A protein. Nucleic Acids Res 30: 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Reddy R. 1989. γ-Monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci 86: 8280–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. 2016. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu RM. 2011. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci 108: 20538–20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. 2011. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 18: 1414–1420. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Kalda C, Patel A, Battle DJ. 2015. Gemin5 binds to the survival motor neuron mRNA to regulate SMN expression. J Biol Chem 290: 15662–15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell 2: 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yong J, Pellizzoni L, Dreyfuss G. 2002. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J 21: 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. 2004. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol 24: 2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, Dreyfuss G. 2011. Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell 146: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B, Xu RM. 2014. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol Cell 54: 879–886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.