Fantauzzo and Soriano show that ablation of Pdgfrb in the neural crest lineage results in increased nasal septum width, delayed palatal shelf development, and subepidermal blebbing. They reveal a physical interaction between PDGFRα and PDGFRβ in the craniofacial mesenchyme and demonstrate that the receptors form functional heterodimers with distinct signaling properties.
Keywords: PDGFRβ, craniofacial development, neural crest, PDGFRα, heterodimers
Abstract
Craniofacial development is a complex morphogenetic process, disruptions in which result in highly prevalent human birth defects. While platelet-derived growth factor (PDGF) receptor α (PDGFRα) has well-documented functions in this process, the role of PDGFRβ in murine craniofacial development is not well established. We demonstrate that PDGFRα and PDGFRβ are coexpressed in the craniofacial mesenchyme of mid-gestation mouse embryos and that ablation of Pdgfrb in the neural crest lineage results in increased nasal septum width, delayed palatal shelf development, and subepidermal blebbing. Furthermore, we show that the two receptors genetically interact in this lineage, as double-homozygous mutant embryos exhibit an overt facial clefting phenotype more severe than that observed in either single-mutant embryo. We reveal a physical interaction between PDGFRα and PDGFRβ in the craniofacial mesenchyme and demonstrate that the receptors form functional heterodimers with distinct signaling properties. Our studies thus uncover a novel mode of signaling for the PDGF family during vertebrate development.
The platelet-derived growth factor (PDGF) receptors (PDGFRs) are receptor tyrosine kinases (RTKs) that bind to a subset of growth factors on the surface of cells and elicit responses with broad roles in developmental cellular processes. The mammalian PDGF family consists of four ligands, PDGF-A–D, which variously signal through two RTKs: PDGFRα and PDGFRβ. Receptors in this family consist of an extracellular ligand-binding domain containing five immunoglobulin-like loops, a single transmembrane domain, and an intracellular domain harboring a split catalytic tyrosine kinase (Williams 1989). The two receptors share the highest amino acid homology in the N-terminal (85% identity) and C-terminal (75% identity) kinase domains, with considerably reduced homology in the extracellular (31% identity) and interkinase (27% identity) domains (Matsui et al. 1989). PDGFRs are activated by ligand binding that induces receptor dimerization and promotes tyrosine kinase activity, resulting in the autophosphorylation of intracellular tyrosine residues. Signaling molecules containing Src homology 2 (SH2) phosphotyrosine recognition motifs bind to specific phosphorylated residues in the cytoplasmic domains of the receptors and mediate downstream cellular responses through various intracellular signaling pathways (Heldin and Westermark 1999). These signaling molecules include a subset with intrinsic enzymatic activity, including Src family tyrosine kinases, phosphatidylinositol 3-kinase (PI3K), the tyrosine phosphatase SHP-2, phospholipase Cγ (PLCγ), and Ras GTPase-activating protein (GAP) as well as adaptor proteins such as Crk, Grb2, and Nck (Heldin and Westermark 1999). While both PDGFRα and PDGFRβ are capable of interacting with Src, PI3K, SHP-2, and PLCγ, PDGFRα is additionally able to bind Crk, and PDGFRβ is further capable of interacting with GAP, Grb2, and Nck (Heldin and Westermark 1999).
Although numerous ligand and receptor interactions have been demonstrated in vitro, the homodimers PDGF-AA and PDGF-CC have been shown to exclusively activate PDGFRα signaling in vivo during mammalian development (Boström et al. 1996; Soriano 1997; Ding et al. 2004), while PDGF-BB solely activates PDGFRβ signaling (Levéen et al. 1994; Soriano 1994). Targeted disruption of Pdgfra in mice results in embryonic lethality during mid-gestation, with homozygous null embryos exhibiting a wide range of phenotypes, including facial clefting, blebbing, edema, hemorrhaging, cardiac outflow tract defects, abnormalities in neural tube development, abnormally patterned somites, and extensive skeletal defects affecting cranial neural crest cell (NCC) derivatives in the frontonasal skeleton as well as non-NCC-derived skeletal elements such as the shoulder girdle, sternum, ribs, and vertebrae (Soriano 1997). These phenotypes are recapitulated in embryos lacking both Pdgfa and Pdgfc (Ding et al. 2004). Alternatively, both Pdgfrb- and Pdgfb-deficient mice die perinatally and exhibit edema, hemorrhaging, cardiac ventricular septal defects, thrombocytopenia, anemia, and kidney defects (Levéen et al. 1994; Soriano 1994). Recently, a Pdgfd-deficient mouse model has been reported with a mild vascular phenotype in adult animals, indicating that this ligand is not essential for embryonic development or postnatal life (Gladh et al. 2016).
Both PDGFRα signaling and PDGFRβ signaling have been shown to contribute to cranial and cardiac NCC development. Within the craniofacial region, Pdgfra is expressed in the cranial NCC-derived mesenchyme of the facial processes during mid-gestation, while its ligands, Pdgfa and Pdgfc, are reciprocally expressed in the overlying epithelium (Morrison-Graham et al. 1992; Orr-Urtreger and Lonai 1992; Ding et al. 2000; Hamilton et al. 2003; He and Soriano 2013). Conditional ablation of Pdgfra in the neural crest lineage using the Wnt1-Cre driver (Danielian et al. 1998) results in a subset of the null phenotypes, including facial clefting, midline hemorrhaging, aortic arch defects, and thymus hypoplasia (Tallquist and Soriano 2003; He and Soriano 2013). Pdgfrafl/fl;Wnt1-Cre+/Tg embryos exhibit a delay in the migration of NCCs into the frontonasal prominence and decreased proliferation in this structure (He and Soriano 2013). Furthermore, PDGFRα signaling has been shown to regulate survival and proliferation of the cranial NCC-derived mesenchyme contributing to the palatal shelves (Fantauzzo and Soriano 2014). Pdgfrb is also expressed in the embryonic mesenchyme, with high expression levels in the heart and diffuse expression in the cephalic mesenchyme, among other sites (Soriano 1994; McCarthy et al. 2016). Its ligand, Pdgfb, is expressed in the palatal shelves of mid-gestation embryos (Rahimov et al. 2008), with increased expression in the palatal epithelium as compared with the mesenchyme (P Mazot and P Soriano, unpubl.). Intriguingly, the expression patterns of PDGFRα and PDGFRβ are nonoverlapping in the mesenchyme of the developing murine frontonasal region. While PDGFRα is localized to the cartilage primordia of the nasal septum and the medial sides of the vomeronasal organs during mid-gestation, PDGFRβ is expressed laterally throughout the nasal septum (McCarthy et al. 2016). The PDGFRs have been demonstrated previously to genetically interact during craniofacial and heart development. Conditional ablation of both Pdgfra and Pdgfrb in the neural crest lineage leads to malformations in the basisphenoid, alisphenoid, and hyoid bones and defects in multiple cardiac NCC derivatives that are more severe than those observed in either single conditional homozygous mutant alone (Richarte et al. 2007; McCarthy et al. 2016).
Previous work by our laboratory identified PI3K as the main downstream effector of PDGFRα signaling during embryonic development in mice. Embryos homozygous for an autophosphorylation mutant knock-in allele (PdgfraPI3K) in which PDGFRα is unable to bind PI3K due to two tyrosine-to-phenylalanine mutations at residues Y731 and Y742 (Yu et al. 1991) die perinatally and display a cleft palate, among other defects (Klinghoffer et al. 2002; Fantauzzo and Soriano 2014). However, these embryos do not display a subset of skeletal defects found in Pdgfra-null embryos, such as incomplete fusion of the anterior facial bones that results in complete facial clefting (Soriano 1997). Intriguingly, embryos homozygous or hemizygous for an allele (PdgfraF7) containing five additional mutations at residues (Y572, Y574, Y720, Y988, and Y1018) required for association of PDGFRα with Src, SHP-2, and PLCγ (Rosenkranz et al. 1999) do not exhibit more severe abnormalities than those detected in PdgfraPI3K/PI3K embryos (Klinghoffer et al. 2002). One interpretation of these findings is that PDGFRβ may be able to compensate for the loss of PI3K signaling through PDGFRα if the two receptors form functional heterodimers in which PDGFRα is able to transphosphorylate PI3K-binding sites on PDGFRβ. In support of this hypothesis, whereas PdgfrbPI3K/PI3K mice do not display overt craniofacial defects (Heuchel et al. 1999), PdgfraPI3K/PI3K;PdgfrbPI3K/PI3K double-homozygous mutant embryos in which PI3K signaling cannot be engaged through PDGFRα/β heterodimers indeed exhibit complete facial clefting, thus phenocopying Pdgfra-null embryos (Klinghoffer et al. 2002).
Two main possibilities arise from these studies. The first is that PDGFRα and PDGFRβ independently regulate cell activity and are able to compensate for one another in the neural crest lineage. The second is that the two receptors form functional heterodimers. In addition to the in vivo findings described above, a handful of in vitro studies performed in porcine aortic endothelial cells using an exogenous expression model suggest that PDGFRα/β heterodimers have properties distinct from those of homodimeric receptor complexes in terms of signal molecule binding and mitogenic response (Rupp et al. 1994; Ekman et al. 1999). Currently, the prevalence of these heterodimers and their effect on cellular behavior during development are unknown.
In this work, we combined expression, genetic, and biochemical studies to demonstrate that PDGFRα and PDGFRβ genetically and physically interact to form functional heterodimers during murine embryogenesis, thus highlighting a novel mode of signaling for the PDGF family. Furthermore, we uncovered roles for PDGFRβ in nasal septum and palatal shelf development as well as basement membrane integrity and provide evidence that the subepidermal blebbing phenotype observed in Pdgfr mutants stems from mislocalization of various extracellular matrix proteins. Our findings thus reveal that previously observed requirements for PDGFRβ in human skeletal and skin development are evolutionarily conserved in mice and are cell-autonomous to the murine neural crest lineage.
Results
The PDGFRs are coexpressed in the craniofacial mesenchyme
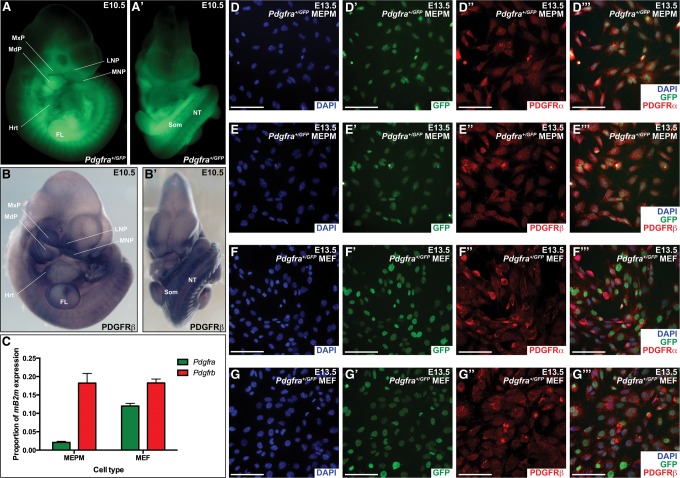
Consistent with its critical role in craniofacial development, Pdgfra is expressed in the mesenchyme of the facial processes of embryonic day 10.5 (E10.5) heterozygous reporter embryos in which an H2B-GFP construct had been knocked into the Pdgfra locus (Fig. 1A,A′; Hamilton et al. 2003; He and Soriano 2013). To determine the expression of PDGFRβ in these same structures, we performed whole-mount immunohistochemistry on wild-type E10.5 embryos using an anti-PDGFRβ antibody, revealing expression in the medial and lateral nasal processes, in the maxillary processes from which the palatal shelves will extend, and in the mandibular processes (Fig. 1B,B′). Outside of the craniofacial region, expression of both receptors was additionally detected in the heart, neural tube, somites, and limb buds at E10.5 (Fig. 1A–B′).
Figure 1.
The PDGFRs are coexpressed in the craniofacial mesenchyme. (A,A′) Expression of Pdgfra as assessed by GFP fluorescence in E10.5 Pdgfra+/GFP embryos viewed laterally (A) and frontally (A′). (B,B′) Expression of PDGFRβ as assessed by whole-mount immunohistochemistry with an anti-PDGFRβ antibody in E10.5 wild-type embryos viewed laterally (B) and frontally (B′). No signal was detected in a secondary antibody control embryo at E10.5 (data not shown). Expression of both receptors was detected in the facial process mesenchyme, heart, neural tube, somites, and limb bud mesenchyme. (MxP) Maxillary process; (MdP) mandibular process; (LNP) lateral nasal process; (MNP) medial nasal process; (Hrt) heart; (FL) forelimb; (NT) neural tube; (Som) somite. (C) Bar graph depicting quantitative RT–PCR (qRT–PCR) values revealing reduced expression of Pdgfra in mouse embryonic palatal mesenchyme cells (MEPMs) as compared with mouse embryonic fibroblasts (MEFs) and similar expression of Pdgfrb across the two cell types. Data are presented as mean ± SEM. (D–G′′′) Expression of PDGFRα (red; D′′,F′′) and PDGFRβ (red; E′′,G′′) as assessed by immunofluorescence analyses in E13.5 Pdgfra+/GFP-derived MEPMs (D–E′′′) and MEFs (F–G′′′). Expression of both receptors was detected in virtually every cell. (D′,E′,F′,G′) Pdgfra-expressing cells are additionally indicated by GFP fluorescence (green). (D,E,F,G) Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Bars, 100 µm.
Primary mouse embryonic palatal mesenchyme cells (MEPMs) and primary mouse embryonic fibroblasts (MEFs) have been shown previously to independently express both PDGFRα and PDGFRβ (Klinghoffer et al. 2001, 2002; He and Soriano 2013; Vasudevan et al. 2015), with the former cell type demonstrating responsiveness to stimulation with PDGF-AA (He and Soriano 2013; Fantauzzo and Soriano 2014; Vasudevan and Soriano 2014; Vasudevan et al. 2015), and the latter demonstrating responsiveness to stimulation with PDGF-AA and PDGF-BB (Klinghoffer et al. 2001, 2002). Quantitative RT–PCR (qRT–PCR) analyses comparing expression of the transcripts encoding the two receptors in primary MEPMs and primary MEFs revealed greater expression of Pdgfra in MEFs (2.087% ± 0.3069% and 12.00% ± 0.7276% of mB2m expression, respectively) and similar expression of Pdgfrb across the two cell types (18.22% ± 2.623% and 18.25% ± 1.078% of mB2m expression, respectively) (Fig. 1C). To assess coexpression of the two receptors in these cell types, low-passage primary MEPMs and primary MEFs were derived from E13.5 Pdgfra+/GFP embryos and subjected to immunofluorescence analyses with both anti-PDGFRα and anti-PDGFRβ antibodies. Our analyses revealed expression of both receptors in virtually every cell (Fig. 1D–G′′′).
Craniofacial skeletal defects in a subset of embryos lacking Pdgfrb in the neural crest lineage
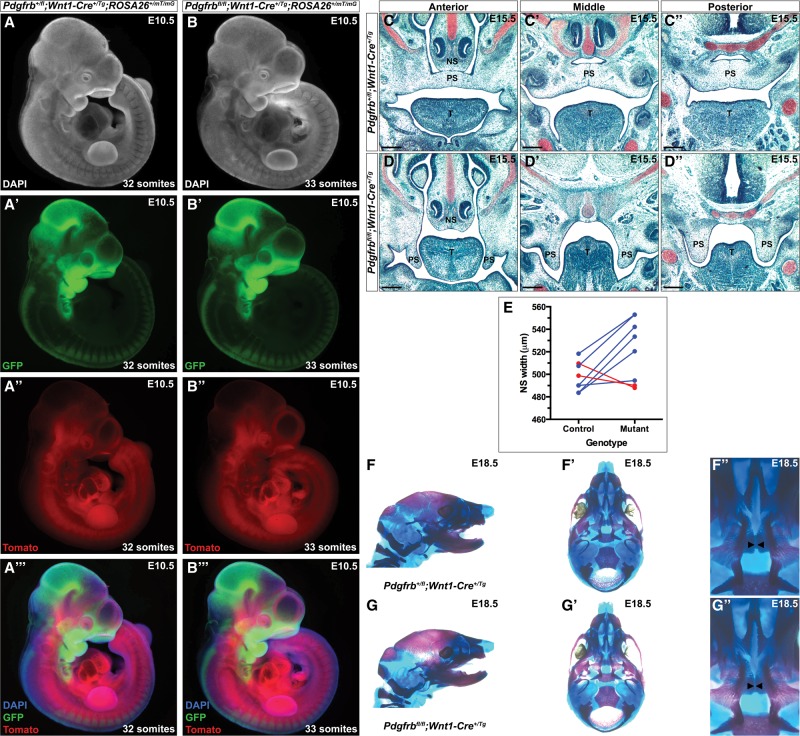
As PDGFRβ does not have an established role in murine craniofacial development beyond a genetic interaction with Pdgfra, we performed a series of experiments examining the effect of conditionally ablating Pdgfrb in the NCC lineage using the Wnt1-Cre driver. These analyses and all further experiments in this study were performed using mice maintained on a 129S4 coisogenic genetic background to prevent the effects of potential second site modifiers. We began by assessing the timing and extent of NCC migration in this context using the ROSA26mT/mG allele (Muzumdar et al. 2007), which expresses a double-fluorescent Cre reporter throughout the embryo, resulting in the expression of membrane-targeted tdTomato before Cre excision and membrane-targeted EGFP after Cre excision. This analysis revealed no obvious defects in the extent or pattern of NCC migration into the facial processes and pharyngeal arches of embryos lacking Pdgfrb in the NCC lineage at E8.5 (n = 3) (data not shown) or E10.5 (n = 4) (Fig. 2A–B′′′). Moreover, whole-mount 4′,6-diamidino-2-phenylindole (DAPI) staining of these same embryos revealed no morphological differences in the shape or size of neural crest-derived craniofacial structures in experimental versus control embryos at E10.5 (Fig. 2A,B).
Figure 2.
Conditional ablation of Pdgfrb in the neural crest lineage. (A–B′′′) Whole-mount fluorescence analysis of Pdgfrb+/fl;Wnt1-Cre+/Tg;ROSA26+/mT/mG (A–A′′′) and Pdgfrbfl/fl;Wnt1-Cre+/Tg;ROSA26+/mT/mG (B–B′′′) embryos at E10.5. DAPI staining revealed no morphological differences in the shape or size of neural crest-derived craniofacial structures in homozygous mutant (B) versus heterozygous control (A) embryos. (B′) GFP fluorescence analysis revealed no defects in the extent or pattern of NCC migration into the facial processes and pharyngeal arches of embryos lacking Pdgfrb in the NCC lineage. (C–D′′) Safranin O staining of coronal Pdgfrb+/fl;Wnt1-Cre+/Tg (C–C′′) and Pdgfrbfl/fl;Wnt1-Cre+/Tg (D–D′′) anterior (C,D), middle (C′,D′), and posterior (C′′,D′′) palatal shelf sections at E15.5. While the palatal shelves of heterozygous control embryos are in the process of fusing along the anterior–posterior axis, the palatal shelves of a subset of homozygous mutant embryos have not yet begun to elevate at this time point. (NS) Nasal septum; (PS) palatal shelves; (T) tongue. Bars, 100 µm. (E) Line graph depicting width of the nasal septum between E15.5 heterozygous control and homozygous mutant littermates. The majority of homozygous mutant embryos has an increased width of the nasal septum (blue) compared with heterozygous control littermates. (F–G′′) Lateral (F,G) and ventral (F′,F′′,G′,G′′) views of craniofacial skeletal preparations generated from Pdgfrb+/fl;Wnt1-Cre+/Tg (F–F′′) and Pdgfrbfl/fl;Wnt1-Cre+/Tg (G–G′′) littermate pups at 18.5. Pdgfrb conditional knockout mice have a secondary palate identical to that of their heterozygous littermates. Black arrowheads indicate medial edges of palatal shelves.
We next explored the effect of ablating Pdgfrb in the NCC lineage on development of the craniofacial skeleton. The majority of E15.5 homozygous mutant embryos has an increased width of the nasal septum compared with heterozygous control littermates (521.8 µm ± 9.818 µm vs. 497.7 µm ± 4.582 µm; P = 0.0781) (Fig. 2C–E). Examination of perinatal mice confirmed that this is a transient phenotype, as the nasal septum and associated structures of Pdgfrb conditional knockout mice (Fig. 2G–G′′) are indistinguishable from those of heterozygous littermates at E18.5 (Fig. 2F–F′′). Furthermore, while E15.5 heterozygous control embryos have palatal shelves that are in the process of fusing along the anterior–posterior axis (Fig. 2C–C′′), the palatal shelves of a subset (10%, n = 10) of homozygous mutant embryos had not yet begun to elevate at this time point (Fig. 2D–D′′). This likely reflects a delay in Pdgfrb conditional knockout palatal shelf development, as these mice are ultimately born at Mendelian ratios (44 pups vs. 49.5 expected pups out of 198 total; χ2 P = 0.3667) and are viable. Skeletal preparations of perinatal mice confirmed that palatal shelf development may be only temporarily delayed in Pdgfrbfl/fl;Wnt1-Cre+/Tg embryos, as all Pdgfrb conditional knockout mice examined to date (n = 17) (Fig. 2G–G′′) have a secondary palate identical to that of their heterozygous littermates (Fig. 2F–F′′).
Pdgfra and Pdgfrb genetically interact during craniofacial development
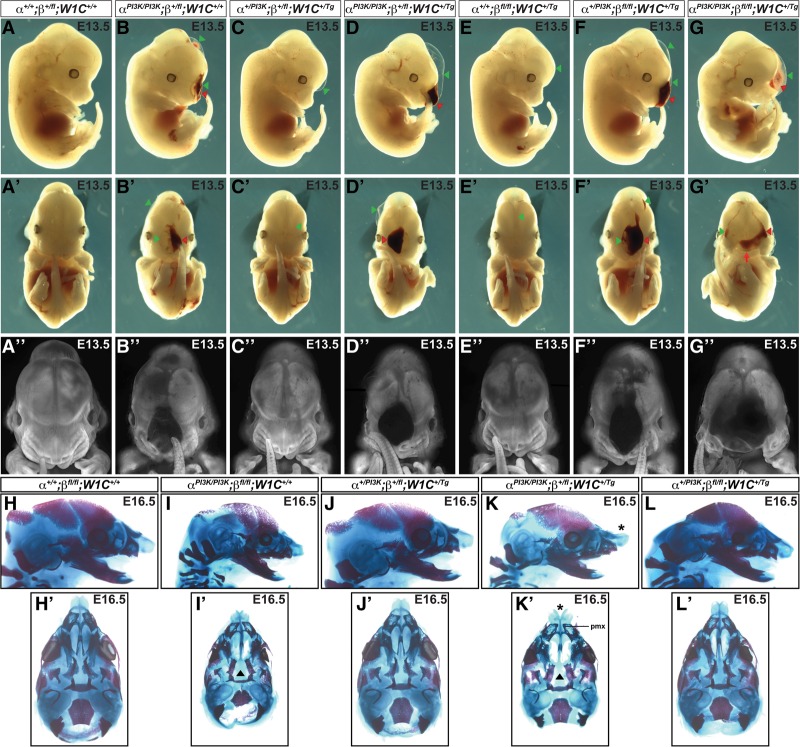
A previous skeletal analysis in which both Pdgfra and Pdgfrb were conditionally ablated in the NCC lineage did not detect additional frontonasal midline defects in double conditional mutant embryos beyond those observed in Pdgfrafl/fl;Wnt1-Cre+/Tg embryos (McCarthy et al. 2016). As PdgfraPI3K/PI3K embryos have a cleft palate phenotype (Klinghoffer et al. 2002; Fantauzzo and Soriano 2014) less severe than the complete facial clefting phenotype observed in Pdgfrafl/fl;Wnt1-Cre+/Tg embryos (Tallquist and Soriano 2003; He and Soriano 2013), we chose to use the PdgfraPI3K allele in our in vivo studies as a sensitized background in the hopes of uncovering a genetic interaction between the two receptors in frontonasal midline development. We thus used the constitutive PdgfraPI3K allele together with the conditional Pdgfrbfl allele and the Wnt1-Cre driver to ablate various allele combinations in the NCC lineage. We intercrossed Pdgfra+/PI3K;Pdgfrbfl/fl mice with Pdgfra+/PI3K;Pdgfrb+/fl;Wnt1-Cre+/Tg mice and harvested the resulting progeny at E13.5 for gross morphological examination (Fig. 3A–G′′). As observed previously (Fantauzzo and Soriano 2014), all embryos homozygous for the PdgfraPI3K allele were smaller than their littermates (Fig. 3B,B′,D,D′,G,G′). Double-homozygous mutant embryos were recovered at Mendelian frequencies at E13.5 (four embryos vs. six expected embryos out of 97 total; χ2 P = 0.3870) and exhibited an overt facial clefting phenotype (100%; n = 4) (Fig. 3G–G′′; Table 1) not observed among embryos possessing any of the other 11 allele combinations from the intercrosses (n = 93) (Table 1), indicating that the two receptors genetically interact in the NCC lineage.
Figure 3.
Pdgfra and Pdgfrb genetically interact during craniofacial development. (A–G′′) Gross morphology of E13.5 embryos resulting from intercrosses of Pdgfra+/PI3K;Pdgfrbfl/fl mice with Pdgfra+/PI3K;Pdgfrb+/fl;Wnt1-Cre+/Tg mice as viewed laterally (A–G) and frontally before (A′–G′) and after (A′′–G′′) whole-mount DAPI staining. (G–G′′) The two receptors genetically interact in the NCC lineage, as double-homozygous mutant embryos have an overt facial cleft (red arrow) not observed among embryos possessing any of the other allele combinations from the intercrosses. Facial blebbing (green arrowheads) and facial hemorrhaging (red arrowheads) were also detected among embryos possessing a variety of allele combinations. While hemorrhaging was always accompanied by blebbing, the latter phenotype also arose independently. (H–L′) Lateral (H–L) and ventral (H′–L′) views of craniofacial skeletal preparations generated from E16.5 embryos obtained from intercrosses of Pdgfra+/PI3K;Pdgfrbfl/fl mice with Pdgfra+/PI3K;Pdgfrb+/fl;Wnt1-Cre+/Tg mice. (I,I′) The vomer and palatal processes of the maxilla of PdgfraPI3K/PI3K skeletons were markedly reduced, and the palatal processes of the palatine had not elevated or grown toward the midline (black arrowhead). (K,K′) Midline defects were exacerbated in PdgfraPI3K/PI3K;Pdgfrb+/fl;Wnt1-Cre+/Tg skeletons, which additionally exhibited upturned and clefted nasal cartilage (black asterisks), a widening of the gap between the premaxilla bones, and generalized broadening of the skull. (pmx) Premaxilla.
Table 1.
Pdgfra and Pdgfrb genetically interact during craniofacial development
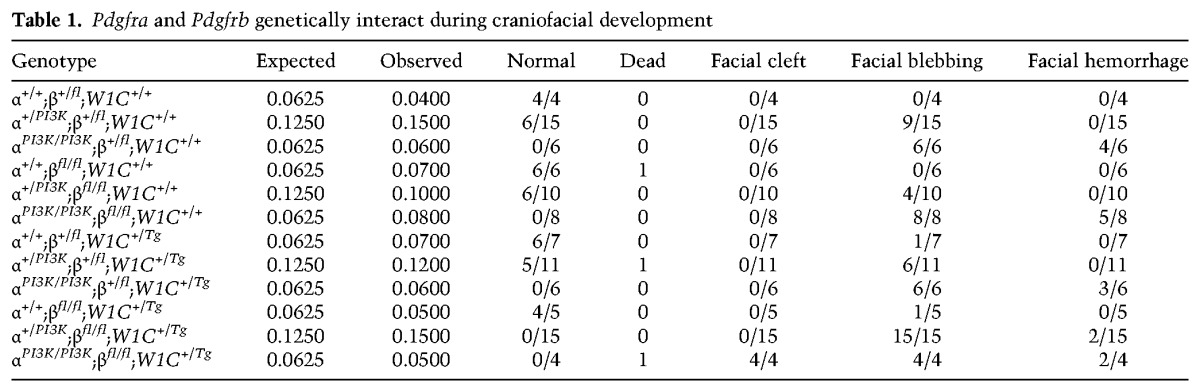
Moreover, blebbing of the surface ectoderm in the facial region was detected in approximately half of the embryos heterozygous for the Pdgfra+/PI3K allele (53%; n = 36) (Fig. 3C,C′; Table 1) and was fully penetrant among embryos homozygous for the PdgfraPI3K allele (100%; n = 24) (Fig. 3B,B′,D,D′,G,G′; Table 1) and among Pdgfra+/PI3K;Pdgfrbfl/fl;Wnt1-Cre+/Tg embryos (100%; n = 15) (Fig. 3F,F′; Table 1). Interestingly, this phenotype was also observed among a subset of embryos heterozygous (14%; n = 7) or homozygous (20%; n = 5) for the Pdgfrbfl allele in combination with the Wnt1-Cre transgene in the absence of Pdgfra mutation (Fig. 3E,E′; Table 1). Finally, facial hemorrhaging was frequently detected in embryos homozygous for the PdgfraPI3K allele (58%; n = 24) (Fig. 3B–B′′,D–D′′,G–G′′; Table 1) and occasionally in Pdgfra+/PI3K;Pdgfrbfl/fl;Wnt1-Cre+/Tg embryos (13%; n = 15) (Fig. 3F–F′′; Table 1), revealing that while hemorrhaging was always accompanied by blebbing, the latter phenotype also arose independently (Table 1).
To assess the genetic interaction of Pdgfra and Pdgfrb in craniofacial cartilage and bone development in vivo at later time points, we generated skeletal preparations of E16.5 embryos obtained from the same intercrosses. While double-heterozygous (Fig. 3J,J′) and Pdgfra+/PI3K;Pdgfrbfl/fl;Wnt1-Cre+/Tg (Fig. 3L,L′) skeletons were virtually indistinguishable from control skeletons (Fig. 3H,H′), the vomer and palatal processes of the maxilla of PdgfraPI3K/PI3K skeletons were markedly reduced, and the palatal processes of the palatine had not elevated or grown toward the midline (Fig. 3I,I′). Introduction of a single Pdgfrbfl allele exacerbated the midline defects observed in PdgfraPI3K/PI3K skeletons such that PdgfraPI3K/PI3K;Pdgfrb+/fl;Wnt1-Cre+/Tg skeletons additionally exhibited upturned and clefted nasal cartilage, a widening of the gap between the premaxilla bones, and generalized broadening of the skull (Fig. 3K,K′) reminiscent of the craniofacial skeletal defects observed upon conditional ablation of Pdgfra in the NCC lineage (Tallquist and Soriano 2003; He and Soriano 2013).
Mislocalization of extracellular matrix proteins in Pdgfra mutant facial blebs
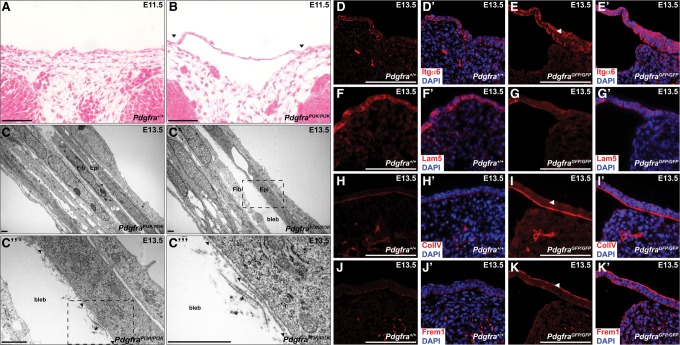
As discussed above, blebbing in the facial region was detected in a subset of embryos possessing at least one PdgfraPI3K or Pdgfrbfl allele, suggesting that each receptor contributes to this phenotype. While such fluid-filled blebs have been described as early as 1960 in patch (Ph) mutant mice (Grüneberg and Truslove 1960), which harbor a deletion of Pdgfra (Smith et al. 1991), this phenotype has not been described previously in Pdgfrb models or been examined in detail. To determine the time of onset of the blebbing phenotype, we performed histological analysis on earlier PdgfraPI3K/PI3K embryos, revealing blebs as early as E11.5 (Fig. 4A,B). We next used transmission electron microscopy to analyze this phenotype in more detail in E13.5 PdgfraPI3K/PI3K embryos. Outside of the bleb, a two-layered epithelium was observed overlying dermal fibroblasts (Fig. 4C). Moving toward the bleb, the epidermis was completely detached from the underlying mesenchyme (Fig. 4C′). Higher-magnification images revealed an intact basement membrane and what appeared to be loose collagen fibrils at the bleb interface (Fig. 4C′′,C′′′), consistent with previous findings in Pdgfc−/− (Ding et al. 2004) and Ph/Ph mutant (Erickson and Weston 1983) embryos, respectively. Immunofluorescence analyses of E13.5 Pdgfra-null craniofacial surface tissue using antibodies specific to the basal epidermis (keratin 14) (Supplemental Fig. S1A–B′) and the basement membrane (integrin α6) (Fig. 4D–E′) verified that these cell layers were completely detached from the underlying mesenchyme within the mutant blebs.
Figure 4.
Mislocalization of extracellular matrix proteins in Pdgfra mutant facial blebs. (A,B) Hematoxylin and eosin staining of coronal Pdgfra+/+ (A) versus PdgfraPI3K/PI3K (B) craniofacial sections at E11.5 revealing separation of the epidermis from the dermis (black arrowheads) in the mutant samples. Bars, 100 µm. (C–C′′′) Transmission electron micrographs of E13.5 PdgfraPI3K/PI3K facial sections outside (C) and within (C′–C′′′) the bleb. The epidermis was completely detached from the underlying mesenchyme within the bleb. An intact basement membrane (black arrowheads) and what appeared to be loose collagen fibrils (asterisk) were detected at the bleb interface. (Fib) Fibroblast; (Epi) epidermis. Dashed rectangles indicate the frame of the following panel. Bars, 1 µm. (D–K′) Expression of integrin α6 (red; D–E′), laminin 5 (red; F–G′), collagen type IV (red; H–I′), and Frem1 (Fras1/FRAS1-related extracellular matrix protein 1) (red; J–K′) as assessed by immunofluorescence analyses of E13.5 wild-type (D,D′,F,F′,H,H′,J,J′) versus PdgfraGFP/GFP (E,E′,G,G′,I,I′,K,K′) craniofacial surface tissue. Expression of integrin α6, collagen type IV, and Frem1 was increased throughout the mutant epidermis (white arrowheads), while expression of laminin 5 was slightly decreased. (D′,E′,F′,G′,H′,I′,J′,K′) Nuclei were stained with DAPI (blue). Bars, 100 µm.
Interestingly, we observed that integrin α6 expression was increased throughout the mutant epidermis (Fig. 4E,E′) as compared with that of wild-type littermates (Fig. 4D,D′). Similar immunofluorescence analyses revealed that expression of the extracellular matrix proteins collagen type IV (Fig. 4H–I′) and Frem1 (Fras1/FRAS1-related extracellular matrix protein 1) (Fig. 4J–K′) was also increased throughout the Pdgfra-null epidermis, suggesting that these proteins may not be properly localizing to the basement membrane in this setting. Expression of laminin 5 was slightly decreased throughout the Pdgfra-null epidermis (Fig. 4F–G′), while localization of collagen type III (Supplemental Fig. S1C–D′), collagen type VII (Supplemental Fig. S1E–F′), and Hspg2 (also known as perlecan) (Supplemental Fig. S1G–H′) was unchanged between wild-type and mutant tissues. qRT–PCR analyses comparing expression of the transcripts encoding these extracellular matrix proteins in RNA derived from E13.5 wild-type versus PdgfraGFP/GFP whole cranial skin revealed no significant changes in transcript expression in the mutant samples, with the exception of a modest decrease in Lamb3 expression (22.10% ± 4.211% decrease; P = 0.0344) (Supplemental Fig. S2A). Moreover, Western blotting demonstrated similar levels of integrin α6 (Supplemental Fig. S2B), collagen type IV (Supplemental Fig. S2C), and Frem1 (Supplemental Fig. S2D) protein in E13.5 wild-type and PdgfraGFP/GFP whole cranial skin lysates, further suggesting that the blebbing defect is associated primarily with mislocalization, and not misexpression, of these extracellular matrix proteins.
PDGFRα and PDGFRβ form functional heterodimers with properties distinct from those of homodimeric receptor complexes
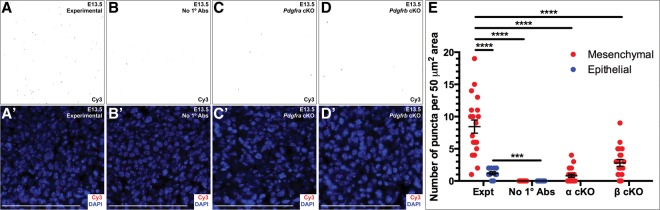
Having characterized a genetic interaction between Pdgfra and Pdgfrb during craniofacial development, we next explored whether PDGFRα and PDGFRβ heterodimerize in the craniofacial mesenchyme. We initially used the proximity ligation assay (PLA) (Söderberg et al. 2006) to examine the interaction of the proteins at single-molecule resolution. We chose antibodies for each receptor that generated minimal background staining in the craniofacial mesenchyme of embryos in which the receptor had been conditionally ablated in the NCC lineage (Supplemental Fig. S3B,B′,D,D′) as compared with control embryos (Supplemental Fig. S3A,A′,C,C′). Applying the PLA technique to E13.5 craniofacial tissue, PDGFRα/β interactions were detected in the craniofacial mesenchyme (Fig. 5A,A′). Quantification of these puncta revealed that these interactions were significantly increased in the mesenchyme (8.450 ± 1.027 per 50-µm2 area) as compared with the epithelia (1.200 ± 0.2494 per 50-µm2 area; P < 0.0001), consistent with the expression patterns of the PDGFRs, as well as compared with a technical negative control without the addition of primary antibodies (0.0 ± 0.0 per 50-µm2 area; P < 0.0001) (Fig. 5B,B′) and conditional knockout tissues in which Pdgfra (0.8000 ± 0.2575 per 50-µm2 area; P < 0.0001) (Fig. 5C,C′) or Pdgfrb (2.800 ± 0.5361 per 50-µm2 area; P < 0.0001) (Fig. 5D,D′) had been ablated in the NCC lineage (Fig. 5E).
Figure 5.
PDGFRα and PDGFRβ physically interact in the craniofacial mesenchyme. (A–D′) Interaction of PDGFRα and PDGFRβ in E13.5 craniofacial tissue as assessed by fluorescence analysis following application of the PLA. PDGFRα/β interactions were significantly increased in the mesenchyme (A,A′) as compared with the epithelia of experimental sections as well as compared with a technical negative control without the addition of primary antibodies (B,B′) and conditional knockout tissues in which Pdgfra (C,C′) or Pdgfrb (D,D′) had been ablated in the NCC lineage. Puncta are indicated in inverse grayscale (A,B,C,D) and by Cy3 fluorescence (red; A′,B′,C′,D′). Nuclei were stained with DAPI (blue; A′,B′,C′,D′). Bars, 100 µm. (E) Scatter dot plot depicting the number of puncta per 50-µm2 area detected in the mesenchyme (red) or epithelia (blue) for each condition. Data are presented as mean ± SEM. (***) P = 0.001; (****) P < 0.0001.
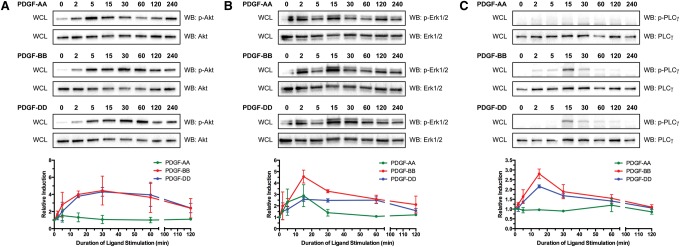
We next performed a series of in vitro biochemical experiments to assess whether PDGFRα and PDGFRβ are capable of forming functional heterodimers with the capacity to activate downstream signaling pathways. We chose to perform these experiments in primary MEFs, as the levels of Pdgfra and Pdgfrb expression are more similar in this cell type than in primary MEPMs (Fig. 1C). In view of the demonstrated roles of Akt, Erk1/2, and PLCγ signaling downstream from PDGFR activation during craniofacial development (Klinghoffer et al. 2002; Moenning et al. 2009; Fantauzzo and Soriano 2014; Vasudevan et al. 2015), we first assayed the ability of PDGF-AA, PDGF-BB, and PDGF-DD ligands to stimulate phosphorylation of these signaling proteins. To induce receptor signaling, primary MEFs were starved in medium containing 0.1% calf serum for 24 h followed by stimulation with 10 ng/mL PDGF-AA, PDGF-BB, or PDGF-DD ligand for up to 4 h. Western blot analysis of whole-cell lysates revealed that, while PDGF-AA treatment induced a modest increase in phospho-Akt levels by 5 min (1.521-fold ± 0.760-fold over baseline), PDGF-BB and PDGF-DD treatments generated sustained phospho-Akt responses that peaked at comparable levels by 30 min (4.451-fold ± 1.691-fold and 4.302-fold ± 0.513-fold over baseline, respectively) (Fig. 6A). A similar examination of the time course of Erk1/2 phosphorylation revealed peaks at 15 minutes for all three ligands, with PDGF-BB treatment generating a more robust response (4.568-fold ± 0.564-fold over baseline) than PDGF-AA (2.894-fold ± 0.964-fold over baseline) and PDGF-DD (2.571-fold ± 0.174-fold over baseline) (Fig. 6B). Finally, PDGF-AA treatment resulted in negligible increases in phospho-PLCγ levels, while PDGF-BB and PDGF-DD treatments induced peaks of PLCγ phosphorylation at 15 min, with PDGF-BB again generating a stronger response (2.805-fold ± 0.246-fold over baseline) than PDGF-DD (2.159-fold ± 0.078-fold over baseline) (Fig. 6C).
Figure 6.
PDGF-BB treatment induces robust phosphorylation of Akt, Erk1/2, and PLCγ. (A–C) Phosphorylation time courses of Akt (A), Erk1/2 (B), and PLCγ (C) in E13.5 primary MEFs following treatment with PDGF-AA (green), PDGF-BB (red), or PDGF-DD (blue) for up to 4 h. (A) Western blot analysis of whole-cell lysates revealed that, while PDGF-AA treatment induced a modest increase in phospho-Akt levels by 5 min, PDGF-BB and PDGF-DD treatments generated sustained phospho-Akt responses that peaked at comparable levels by 30 min. (B) An examination of the time course of Erk1/2 phosphorylation revealed peaks at 15 min for all three ligands, with PDGF-BB treatment generating a more robust response than PDGF-AA and PDGF-DD. (C) PDGF-AA treatment resulted in negligible increases in phospho-PLCγ levels, while PDGF-BB and PDGF-DD treatments induced peaks of PLCγ phosphorylation at 15 min, with PDGF-BB again generating a stronger response than PDGF-DD. (WCL) Whole-cell lysate; (WB) Western blot. Graph data are presented as mean ± SEM.
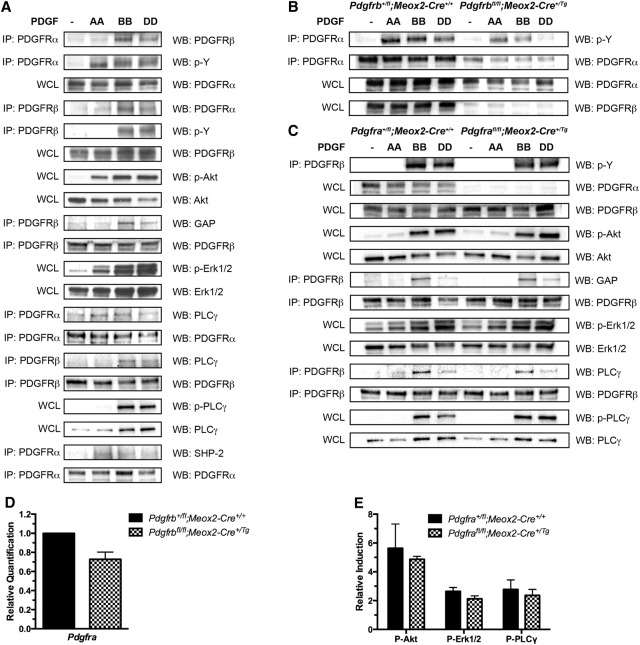
Having determined a general peak of intracellular signaling 15 min after PDGF ligand treatment, we used this time point to assay the ability of each PDGFR to bind the other as well as interact with known signaling molecules upon stimulation of primary MEFs with PDGF-AA, PDGF-BB, or PDGF-DD ligand. PDGFRα and PDGFRβ heterodimers were detected following treatment with PDGF-BB, a ligand that has been shown previously to bind both receptors in vitro (Andrae et al. 2008), and, to a lesser extent, upon stimulation with PDGF-DD (Fig. 7A). PDGFRα/β heterodimers were similarly observed in primary MEPMs predominantly in response to PDGF-BB treatment (data not shown). As expected, PDGF-AA stimulation of primary MEFs resulted in autophosphorylation of PDGFRα exclusively, while PDGF-BB and PDGF-DD treatments induced autophosphorylation of both receptors (Fig. 7A), consistent with the formation of functional heterodimers. GAP binding to PDGFRβ was noticeably increased upon PDGF-BB treatment and minimally increased upon PDGF-DD stimulation (Fig. 7A). PLCγ binding to PDGFRα, although increased over baseline levels, was unchanged between PDGF-AA and PDGF-BB treatments, while binding of this signaling molecule to PDGFRβ was increased upon PDGF-BB treatment over PDGF-DD treatment (Fig. 7A). Finally, stimulation with PDGF-BB led to decreased binding of SHP-2 to PDGFRα as compared with treatment with PDGF-AA (Fig. 7A). We were never able to detect SHP-2 binding to PDGFRβ under any conditions within our system (data not shown).
Figure 7.
PDGFRα/β heterodimers have properties distinct from those of homodimeric receptor complexes. (A) Biochemical analysis of signal molecule binding to PDGFRα and PDGFRβ in wild-type E13.5 primary MEFs following treatment with PDGF-AA, PDGF-BB, or PDGF-DD for 15 min. PDGFRα/β heterodimers were detected following treatment with PDGF-BB and, to a lesser extent, upon stimulation with PDGF-DD. PDGF-AA stimulation resulted in autophosphorylation of PDGFRα exclusively, while PDGF-BB and PDGF-DD treatments induced autophosphorylation of both receptors. GAP binding to PDGFRβ was noticeably increased upon PDGF-BB treatment and minimally increased upon PDGF-DD stimulation. PLCγ binding to PDGFRα, although increased over baseline levels, was unchanged between PDGF-AA and PDGF-BB treatments, while binding of this signaling molecule to PDGFRβ was increased upon PDGF-BB treatment over PDGF-DD treatment. Stimulation with PDGF-BB led to decreased binding of SHP-2 to PDGFRα as compared with treatment with PDGF-AA. (B) Biochemical analysis of control versus Pdgfrbfl/fl;Meox2-Cre+/Tg E13.5 primary MEFs following treatment with PDGF-AA, PDGF-BB, or PDGF-DD for 15 min. The extent of PDGFRα autophosphorylation upon PDGF-BB and PDGF-DD treatments was reduced in this setting as compared with levels in control cells. (C) Biochemical analysis of signal molecule binding to PDGFRβ in control versus Pdgfrafl/fl;Meox2-Cre+/Tg E13.5 primary MEFs following treatment with PDGF-AA, PDGF-BB, or PDGF-DD for 15 min. Ablation of PDGFRα had no effect on PDGFRβ protein levels or the autophosphorylation of PDGFRβ upon treatment with PDGF-BB and PDGF-DD ligands. The extent of GAP and PLCγ binding to PDGFRβ in response to the various PDGF ligands was consistent between control and Pdgfrafl/fl;Meox2-Cre+/Tg MEFs. Western blotting for a particular PDGFR served as a loading control for each immunoprecipitation of that receptor. (IP) Immunoprecipitation; (WB) Western blot; (WCL) whole-cell lysate. (D) Bar graph depicting qRT–PCR values revealing a modest reduction in Pdgfra transcript levels in the facial processes of E11.5 Pdgfrbfl/fl;Meox2-Cre+/Tg embryos compared with control littermates. Data are presented as mean ± SEM. (E) The amplitude of Akt, Erk1/2, and PLCγ phosphorylation following PDGF-BB stimulation was reduced in Pdgfrafl/fl;Meox2-Cre+/Tg MEFs as compared with levels in control MEFs. Data are presented as mean ± SEM, normalized to unstimulated samples for each genotype.
We next derived primary MEFs from embryos in which Pdgfrb had been ablated throughout the embryo using the Meox2-Cre driver (Tallquist and Soriano 2000) and thus could not engage in heterodimer formation. We confirmed near-complete knockdown of PDGFRβ in these cells (Fig. 7B) and further revealed that the extent of PDGFRα autophosphorylation upon PDGF-BB and PDGF-DD treatments was reduced in this setting as compared with levels in control cells (Fig. 7B). Consistent with the Western blotting results (Fig. 7B), qRT–PCR analysis revealed a modest reduction in Pdgfra transcript levels in the facial processes of E11.5 Pdgfrbfl/fl;Meox2-Cre+/Tg embryos compared with control littermates (27.26% ± 7.652% decrease; P = 0.0705) (Fig. 7D). To distinguish the binding properties of PDGFRα/β heterodimers from PDGFRβ homodimers upon PDGF-BB stimulation, we repeated the above experiments in primary MEFs derived from embryos in which Pdgfra had been ablated throughout the embryo. We confirmed knockdown of PDGFRα protein expression in these cells (Fig. 7C) and an inability of PDGFRβ to coimmunoprecipitate with PDGFRα (data not shown). Importantly, ablation of Pdgfra had no effect on PDGFRβ protein levels or the autophosphorylation of PDGFRβ upon treatment with PDGF-BB and PDGF-DD ligands (Fig. 7C). These findings confirm that PDGF-AA treatment results in the exclusive autophosphorylation of PDGFRα and that PDGF-BB and PDGF-DD treatments result in the preferential autophosphorylation of PDGFRβ. The extent of GAP and PLCγ binding to PDGFRβ in response to the various PDGF ligands was consistent between control and Pdgfrafl/fl;Meox2-Cre+/Tg MEFs (Fig. 7C), while the amplitude of Akt, Erk1/2, and PLCγ phosphorylation following PDGF-BB stimulation was reduced in Pdgfrafl/fl;Meox2-Cre+/Tg MEFs as compared with levels in control MEFs (Fig. 7E). These results reveal that the primary difference in signal molecule binding between PDGFR homodimers and heterodimers in primary MEFs is decreased binding of SHP-2 to PDGFRα upon heterodimer formation (Fig. 7A). As SHP-2 binding results in dephosphorylation of autophosphorylated PDGFRs (Klinghoffer and Kazlauskas 1995), decreased SHP-2 binding upon heterodimer formation may explain the robust, sustained intracellular signaling observed upon PDGF-BB treatment (Fig. 6) and the opposite trend detected in the absence of heterodimer formation (Fig. 7E).
Discussion
Our results indicate that the developmental roles of both PDGFRα and PDGFRβ in craniofacial development are conserved between humans and mice. Missense mutations in the PDGFRA-coding region that alter amino acids within the fourth immunoglobulin domain of the extracellular portion of the receptor, the transmembrane domain, or the cytoplasmic domain C-terminal to the tyrosine kinase domains as well as single base-pair substitutions in the 3′ untranslated region (UTR) are associated with nonsyndromic cleft palate (Rattanasopha et al. 2012). Moreover, single-nucleotide polymorphisms in the PDGFC regulatory region that repress transcriptional activity of the promoter are associated with cleft lip and palate (Choi et al. 2009). In the case of PDGFRB, heterozygous missense mutations that modify amino acids in the juxtamembrane and tyrosine kinase domains cause Kosaki overgrowth syndrome (OMIM 616592) and Penttinen syndrome (OMIM 601812), respectively, both of which are characterized by facial dysmorphism and fragile skin, among other defects (Johnston et al. 2015; Takenouchi et al. 2015). We now show that PDGFRβ also plays a role during craniofacial development in mice and that ablation of the gene encoding this RTK in the NCC lineage can lead to cartilage, bone, and skin phenotypes in mice that emulate those found in human patients with PDGFRB mutations.
Of note, no craniofacial abnormalities were observed in a previous study of late gestation embryos homozygous for a Pdgfrb-null allele or in adult mice homozygous for a Pdgfrb hypomorphic allele (McCarthy et al. 2016). The most likely explanation for this discrepancy is a difference in genetic background, as the mice in our study were maintained on a 129S4 coisogenic background, while those in the previous study were maintained on a mixed C57BL/6J × 129/Sv background (McCarthy et al. 2016). In support of this theory, mutant models of alternate RTKs such as Fgfr1 have been shown to have more severe developmental phenotypes on a 129S4 coisogenic background than on mixed genetic backgrounds (Brewer et al. 2015).
Several lines of evidence support a role for disrupted PI3K signaling in the manifestation of the blebbing phenotype in Pdgfra and Pdgfrb mutants. Ablation of all class Ia PI3K p85 regulatory subunit isoforms leads to subepidermal blebbing in the craniofacial and trunk regions (Brachmann et al. 2005). These p85a−/−;p55a−/−;p50a−/−;p85b−/− mutant embryos die during mid-gestation and additionally exhibit facial clefting, hemorrhaging, and a wavy neural tube (Brachmann et al. 2005), conspicuously similar to Pdgfra-null embryos (Soriano 1997). Subepidermal blebbing in the craniofacial region is commonly observed in PdgfraPI3K/PI3K embryos (Fantauzzo and Soriano 2014) as well as PdgfraPI3K/− and PdgfraPI3K/PI3K;PdgfrbPI3K/PI3K embryos (Klinghoffer et al. 2002). Interestingly, PdgfrbPI3K/PI3K mice develop normally but, when challenged with a mast cell degranulating agent, are unable to normalize interstitial fluid pressure or resolve the resulting edema. Moreover, PdgfrbPI3K/PI3K-derived MEFs exhibit defects in migration concomitant with an absence of circular actin ruffles and impaired collagen gel contraction (Heuchel et al. 1999). These deficiencies point to a role for PI3K-mediated PDGFRβ signaling in regulating the mechanical forces between fibroblasts and the extracellular matrix (Heuchel et al. 1999).
Among the extracellular matrix proteins with mislocalized expression in Pdgfra-null epidermis, Frem1 stands out as a particularly appealing potential mediator of PDGF signaling due to its expression pattern and null mouse model phenotypes. Frem1 is a member of the larger Fras1/Frem family of extracellular matrix proteins and is the only member of the family that is initially expressed in mesenchymal cells before stabilization in the epithelial basement membrane (Smyth et al. 2004), thus overlapping with PDGFRα and PDGFRβ expression in the mesenchyme. Fras1, Frem1, and Frem2 have been shown to form a physical complex that is essential for the basement membrane stabilization of each protein (Kiyozumi et al. 2006). As such, null mouse models of each of these family members share a subset of phenotypes that includes subepidermal blebbing, cryptophthalmos, syndactyly, and renal dysgenesis (Petrou et al. 2008). Individual mutants exhibit additional phenotypes that are also observed in Pdgfra-null embryos, such as craniofacial skeletal defects (Fras1, Frem1, and Frem2), hemorrhaging (Fras1 and Frem2), cardiac septal malformations (Frem2), abnormalities in neural tube development (Frem2), abnormal sternum and rib morphology (Fras1), and pigmentation defects (Frem2) (Vrontou et al. 2003; Jadeja et al. 2005; Timmer et al. 2005; Slavotinek et al. 2011; Vissers et al. 2011; Miller et al. 2013). Importantly, the timing of the blebbing in Fras1/Frem family mutant models coincides with that of Pdgfra mutants, as does the nature of the blebbing, which occurs between the lamina densa of the basement membrane and the underlying dermis (McGregor et al. 2003; Vrontou et al. 2003; Smyth et al. 2004; Jadeja et al. 2005; Kiyozumi et al. 2006). In humans, FREM1 is mutated in Manitoba oculotrichoanal syndrome (MOTA; OMIM 248450) (Slavotinek et al. 2011), which is characterized by a combination of eye malformations, an aberrant scalp hairline, a bifid or broad nasal tip, and gastrointestinal anomalies, as well as in bifid nose with or without anorectal and renal anomalies syndrome (BNAR; OMIM 608980) (Alazami et al. 2009).
Fras1/Frem family proteins possess chondroitin sulphate proteoglycan (CSPG) core repeats, among several other domains. In proteoglycan NG2, these same repeats have been shown to bind PDGF-AA (Goretzki et al. 1999). Intriguingly, Frem1 has been shown previously to physically interact with PDGF-C in mid-gestation mouse embryo lysates (Wiradjaja et al. 2013). Furthermore, PDGF-CC stimulation of MEFs derived from Frem1bat/bat embryos, which harbor a chemically induced mutation that introduces a premature stop codon in the C terminus of the CSPG repeat domain (Smyth et al. 2004), results in decreased phosphorylation of PDGFRα as compared with wild-type cells and reduced amplitude and duration of downstream Akt and Erk1/2 phosphorylation (Wiradjaja et al. 2013). These changes are accompanied by decreased secretion of the metalloproteinase inhibitor Timp1 following PDGF-CC stimulation of Frem1bat/bat MEFs and reduced collagen 1 localization in the Frem1bat/bat embryonic basement membrane (Wiradjaja et al. 2013). Whether these defects stem from structural alterations in the basement membrane and/or aberrant growth factor activity remains to be determined. Our current findings cannot distinguish whether the mislocalization of extracellular matrix proteins observed in Pdgfra mutant epidermis results from direct loss of the receptor or is a secondary effect of the blebbing phenotype. As PDGF ligands have been shown to interact with numerous basement membrane proteoglycans (Iozzo 2005) and as PDGFRs are known to associate with integrins (Schneller et al. 1997), which function as transmembrane receptors for extracellular matrix ligands, future studies will explore how disruption of the interactions between these proteins may underlie the blebbing phenotype in Pdgfr mutant mice.
While PDGFRα predominantly signals through PI3K during mouse embryonic development (Klinghoffer et al. 2002), PDGFRβ has been shown to additively signal through multiple intracellular pathways in at least one developmental context: formation of vascular smooth muscle cells and pericytes (Tallquist et al. 2003). Despite these apparent differences in signaling, knock-in mice in which the intracellular domain of one PDGFR has been replaced with that of the other exhibit remarkably normal development, with the exception of a particular requirement for normal PDGFRβ signaling in the vasculature (Klinghoffer et al. 2001). This result indicates that the divergent functions of the two PDGFRs during development likely stems from distinct patterns of receptor expression and/or ligand affinities and not from inherent differences in signal transduction capabilities (Klinghoffer et al. 2001).
Our results demonstrate that both PDGFRα and PDGFRβ are robustly expressed in the craniofacial mesenchyme and that each independently regulates development of the facial skeleton through cell-autonomous activity in the cranial neural crest. However, binding to a particular ligand, PDGF-BB, induces the formation of both PDGFRβ homodimers and PDGFRα/β heterodimers in this context, each with distinct signal molecule-binding properties. We reveal that binding of the tyrosine phosphatase SHP-2 to PDGFRα is decreased upon heterodimer formation, suggesting that PDGFRα/β heterodimers have the unique ability to generate a more robust intracellular signaling response (in terms of both amplitude and duration) than those generated by homodimeric receptor complexes. Interestingly, heterozygous missense mutations in the gene encoding SHP-2, PTPN11, underlie Noonan syndrome 1 (NS1; OMIM 163950) in humans (Tartaglia et al. 2001). NS1 is characterized by facial dysmorphism, cardiac defects, and short stature and is frequently associated with additional features such as bleeding diathesis and defects in non-NCC-derived skeletal elements, among others (Noonan 1968). The most prevalent of these PTPN11 mutations are found in the amino SH2 and phosphotyrosine phosphatase domains, and structural analysis has indicated that they result in a shift toward the active conformation of the protein and thus excessive SHP-2 activity (Tartaglia et al. 2001). Modeling of one such gain-of-function mutation in mice results in NS1-like phenotypes, including craniofacial abnormalities (Araki et al. 2004), further emphasizing the critical role of balanced SHP-2 signaling in this setting.
While the ErbB and VEGF receptors have been shown to form functional heterodimers that play roles in multiple lineages during embryogenesis (Klapper et al. 1999; Citri et al. 2003; Koch and Claesson-Welsh 2012), our results now demonstrate the presence of PDGFRα/β heterodimers during vertebrate craniofacial development and raise the possibility of context-specific heterodimer formation among further RTK families. Future studies will address whether PDGFRα/β heterodimers are present at other sites throughout the embryo (in particular the heart, neural tube, somites, and limb buds) and will explore the effect of heterodimer signaling on cell behavior during development as well as in disease settings such as cancer, vascular disorders, and fibrosis.
Materials and methods
Mouse strains
All animal experimentation was approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai. Pdgfratm11(EGFP)Sor mice (Hamilton et al. 2003), referred to here as PdgfraGFP mice; Pdgfrbtm11Sor mice (Schmahl et al. 2008), referred to here as Pdgfrbfl mice; Tg(Wnt1-Cre)11Rth mice (Danielian et al. 1998), referred to here as Wnt1-CreTg mice; Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo mice (Muzumdar et al. 2007), referred to here as ROSA26mT/mG mice; Pdgfratm5Sor mice (Klinghoffer et al. 2002), referred to here as PdgfraPI3K mice; Pdgfratm8Sor mice (Tallquist and Soriano 2003), referred to here as Pdgfrafl mice; and Meox2tm1(cre)Sor mice (Tallquist and Soriano 2000), referred to here as Meox2-CreTg mice were maintained on a 129S4 coisogenic genetic background.
Whole-mount immunohistochemistry
Whole-mount immunohistochemistry was performed according to a previously published protocol (Joyner and Wall 2008) using PDGFRβ primary antibody (1:200; 958; Santa Cruz Biotechnology, Inc.) and goat anti-rabbit IgG peroxidase-conjugated secondary antibody (1:500; Jackson ImmunoResearch Laboratories, Inc.). Stained embryos were photographed using a ProgRes C5 digital camera (Jenoptik Optical Systems GmbH) fitted onto a Nikon SMZ-U stereomicroscope (Nikon, Inc.).
Cell culture
Primary MEPMs were isolated from the palatal shelves of embryos dissected at E13.5 (day of plug considered 0.5 d) in 1× phosphate-buffered saline (PBS) and cultured in medium (Dulbecco's modified Eagle's medium [Gibco, Invitrogen] supplemented with 50 U/mL penicillin [Gibco], 50 µg/mL streptomycin [Gibco], 2 mM L-glutamine [Gibco]) containing 10% fetal bovine serum (FBS) (HyClone Laboratories, Inc.) as described previously (Bush and Soriano 2010). Primary MEFs were isolated from embryos dissected at E13.5 in PBS. Eviscerated trunks were transferred to 0.25% Trypsin/1 mM EDTA, manually homogenized, and incubated for 30 min at 37°C. Following manual dissociation, cells were cultured in medium containing 10% calf serum (HyClone Laboratories, Inc.).
qRT–PCR
For analysis of Pdgfra+/+ versus PdgfraGFP/GFP whole cranial skin samples and Pdgfrb+/fl;Meox2-Cre+/+ versus Pdgfrbfl/fl;Meox2-Cre+/Tg facial process samples, three independent sets of littermates were harvested. Total RNA was isolated from cultured cells and tissues using the RNeasy minikit (Qiagen, Inc.) according to the manufacturer's instructions. First strand cDNA was synthesized using a ratio of 2:1 random primers: oligo (dT) primer and SuperScript II RT (Invitrogen) according to the manufacturer's instructions. qRT–PCR was performed on a Bio-Rad iQ5 Multicolor real-time PCR detection system and analyzed with iQ5 optical system software (version 2.0; Bio-Rad Laboratories, Inc.). All reactions were performed with PerfeCTa SYBR Green FastMix for iQ (Quanta Biosciences, Inc.), 300 nM primers (Integrated DNA Technologies, Inc.), and 100 ng of cDNA in a 20-µL reaction volume. The following PCR protocol was used: step 1, 3 min at 95°C; step 2, 10 sec at 95°C; step 3, 30 sec at 60°C; repeat steps 2 and 3 for 40 cycles; step 4 (melting curve), 30 sec at 55°C; and repeat step 4 for 81 cycles. All samples were run in triplicate and normalized against an endogenous internal control, B2m. Statistical analyses were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed unpaired t-test. The qRT–PCR primers used are in Supplemental Table S1.
Immunofluorescence analysis
Cells were seeded onto glass coverslips. Alternatively, embryos were fixed in 4% paraformaldehyde (PFA) in PBS and infiltrated with 30% sucrose in PBS before being mounted in OCT compound (Sakura Finetek USA, Inc.). Sections (8 µm) were deposited on glass slides. Cells/sections were fixed in 4% PFA in PBS with 0.1% Triton X-100 for 10 min and washed in PBS with 0.1% Triton X-100. Cells/sections were blocked for 1 h in 5% normal serum (donkey or goat) in PBS and incubated overnight at 4°C in primary antibody diluted in 1% normal serum in PBS. After washing in PBS, cells/sections were incubated in Alexa fluor 546-conjugated secondary antibody (1:1000; Molecular Probes, Invitrogen) diluted in 1% normal serum in PBS with 2 µg/mL DAPI (Sigma-Aldrich Corp.) for 1 h. Cells/sections were mounted in Aqua Poly/Mount mounting medium (Polysciences, Inc.) and photographed using an Orca-Flash4.0 LT digital camera (Hamamatsu Photonics K.K.) fitted onto an AxioObserver.Z1 fluorescence microscope (Carl Zeiss, Inc.). The following antibodies were used for immunofluorescence analysis: PDGFRα (1:100; Thermo Fisher Scientific), PDGFRβ (1:25; 28; Becton Dickinson), integrin α6 (1:200; H-87; Santa Cruz Biotechnology, Inc.), laminin 5 (1:200; Abcam Plc), collagen IV (1:100; Bio-Rad Laboratories, Inc.), Frem1 (1:50; Thermo Fisher Scientific), keratin 14 (1:1000; Poly19053; BioLegend), collagen III (1:100; Abcam Plc), collagen VII (1:100; EMD Millipore Corporation), and Hspg2 (1:100; A7L6; Thermo Fisher Scientific).
Whole-mount DAPI staining
Whole-mount DAPI staining was performed according to a previously published protocol (Sandell et al. 2012), with the exception that staining was performed with 10 µg/mL DAPI (Sigma-Aldrich Corp.) for 1 h at room temperature. Embryos were photographed using an ORCA-Flash4.0 LT digital camera (Hamamatsu Photonics K.K.) fitted onto an AxioObserver.Z1 fluorescence microscope (Carl Zeiss, Inc.). Extended depth of focus was applied to z-stacks using Zen software (Carl Zeiss, Inc.) to generate images with the maximum depth of field.
Histology
Embryos were dissected at multiple time points in PBS, fixed in Bouin's fixative, dehydrated through a graded ethanol series, and embedded in paraffin. After deparaffinization and rehydration, 8-µm sections deposited on glass slides were stained with Weigert's iron hematoxylin, 0.05% Fast Green, and 0.1% Safranin O or Harris modified hematoxylin and eosin and permanently mounted with Permount (Thermo Fisher Scientific). Sections were photographed using a ProgRes CT3 digital camera (Jenoptik Optical Systems GmbH) fitted onto an Axioplan microscope (Carl Zeiss, Inc.). Statistical analyses of morphometric measurements were performed with Prism 6 (GraphPad Software, Inc.) using a Wilcoxon matched-pairs signed rank test.
Skeletal preparations
Perinatal pups were skinned and eviscerated. Perinatal pups and E16.5 embryos were fixed in 95% ethanol and stained in 0.015% Alcian blue, 0.005% Alizarin red, and 5% glacial acetic acid in 70% ethanol at 37°C. Pups and embryos were then cleared in 1% KOH and transferred to solutions of decreasing KOH concentration and increasing glycerol concentration. Skeletons were photographed using a ProgRes C5 digital camera (Jenoptik Optical Systems GmbH) fitted onto a Nikon SMZ-U stereomicroscope (Nikon, Inc.).
Transmission electron microscopy
E13.5 embryos were dissected in PBS, fixed in 3% glutaraldehyde in PBS overnight at 4°C, dehydrated in a graded ethanol series through propylene oxide, and embedded in Epon. Serial sections were cut at 1-µm thickness and stained with methylene blue and azure II for observation by light microscopy. Ultrathin sections were then cut from representative areas and stained with 4% uranyl acetate and Reynold's lead citrate. Electron micrographs were taken at 2500×–25,000× magnification using a Hitachi H-7650 transmission electron microscope (Hitachi High-Technologies).
Immunoprecipitation and Western blotting
To induce receptor signaling, low-passage (P1–P3) MEFs or MEPMs at ∼70% confluence were serum-starved for 24 h in medium containing 0.1% serum and stimulated with 10 ng/mL PDGF-AA, PDGF-BB, or PDGF-DD ligand (R&D Systems) for the indicated length of time at 37°C. This concentration results in a similar molarity within a twofold range across the three ligands. The above cells or whole cranial skin dissected from E13.5 embryos were harvested in ice-cold lysis buffer (20 mM Tris-HCl at pH 8, 150 mM NaCl, 10% glycerol, 1% nonidet P-40, 2 mM EDTA, 1× complete mini protease inhibitor cocktail [Roche Applied Science], 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate), and lysates were collected by centrifugation at 12,000 rpm for 20 min at 4°C. For immunoprecipitations, cell lysates were incubated with PDGFRα (1 µg per 500 µg of lysate; C-20; Santa Cruz Biotechnology, Inc.) or PDGFRβ (1 µg per 500 µg of lysate; 958; Santa Cruz Biotechnology, Inc.) primary antibody overnight at 4°C followed by incubation with 20 µL of Protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Inc.) for 2 h at 4°C the following day. Beads were washed with lysis buffer five times, and the precipitated proteins were eluted with Laemmli buffer containing 10% β-mercaptoethanol, heated for 5 min at 95°C, and separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was performed according to standard protocols using horseradish peroxidase-conjugated secondary antibodies (1:10,000; Jackson ImmunoResearch Laboratories, Inc.). Blots were photographed on a ChemiDoc MP system (Bio-Rad Laboratories, Inc.), and quantifications of signal intensity were performed using volume tools in Image Lab software (version 5.1 build 8, Bio-Rad Laboratories, Inc.). The following antibodies were used for Western blotting: integrin α6 (1:200; H-87; Santa Cruz Biotechnology, Inc.), collagen IV (1:2000; Bio-Rad Laboratories, Inc.), Frem1 (1:1,000; Thermo Fisher Scientific), β-tubulin (1:1,000; E7; Developmental Studies Hybridoma Bank), phospho-Akt (1:1,000; Ser473; Cell Signaling Technology, Inc.), Akt (1:1000; Cell Signaling Technology, Inc.), phospho-p44/42 MAPK (1:1000; Erk1/2; Thr202/Tyr204; Cell Signaling Technology, Inc.), p44/42 MAPK (1:1000; Erk1/2; Cell Signaling Technology, Inc.), phospho-PLCγ1 (1:1000; Tyr783; Cell Signaling Technology, Inc.), PLCγ1 (1:1,000; Cell Signaling Technology, Inc.), PDGFRβ (1:200; 958; Santa Cruz Biotechnology, Inc.), phosphotyrosine (1:1000; 4G10; EMD Millipore Corporation), PDGFRα (1:200; C-20; Santa Cruz Biotechnology, Inc.), GAP (1:5000; 70.3; gift of Andrius Kazlauskas), and SHP-2 (1:1000; 34.3; gift of Andrius Kazlauskas).
PLA
PLA was performed with the Duolink in situ orange starter kit mouse/rabbit (Sigma-Aldrich Corp.) according to the manufacturer's instructions for custom solutions. Briefly, E13.5 embryos were fixed in 4% PFA in PBS and infiltrated with 30% sucrose in PBS before being mounted in OCT compound (Sakura Finetek USA, Inc.). Sections (8 µm) deposited on glass slides were fixed in 4% PFA in PBS with 0.1% Triton X-100 for 10 min and washed in PBS with 0.1% Triton X-100. Sections were blocked for 1 h in 5% normal donkey serum in PBS and incubated overnight at 4°C in primary antibodies diluted in 1% normal donkey serum in PBS. The following antibodies were used for PLA: PDGFRα (1:100; Thermo Fisher Scientific) and PDGFRβ (1:25; 28; Becton Dickinson). Sections were washed in PBS prior to incubation in PLA probes diluted in 1% normal donkey serum in PBS. Sections were photographed using an ORCA-Flash4.0 LT digital camera (Hamamatsu Photonics K.K.) fitted onto an AxioObserver.Z1 fluorescence microscope (Carl Zeiss, Inc.). Extended depth of focus was applied to z-stacks using Zen software (Carl Zeiss, Inc.) to generate images with the maximum depth of field. The number of puncta per 10 (mesenchyme) and five (epithelium) 50-µm2 areas were counted in two independent experiments, and statistical analyses were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed unpaired t-test.
Supplementary Material
Acknowledgments
We thank Dr. Ronald Gordon of the Icahn School of Medicine at Mount Sinai for assistance with electron microscopy. We are grateful to Tony Chen and Matthew Hung for genotyping and technical assistance. We thank members of the Soriano laboratory and Dr. Avner Schlessinger for their helpful discussions and critical comments on the manuscript. This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research grants R01DE022363 (to P.S.) and R03DE025263 (to K.A.F.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.288746.116.
References
- Alazami AM, Shaheen R, Alzahrani F, Snape K, Saggar A, Brinkmann B, Bavi P, Al-Gazali LI, Alkuraya FS. 2009. FREM1 mutations cause bifid nose, renal agenesis, and anorectal malformations syndrome. Am J Hum Genet 85: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J, Gallini R, Betsholtz C. 2008. Role of platelet-derived growth factors in physiology and medicine. Genes Dev 22: 1276–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, et al. 2004. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med 10: 849–857. [DOI] [PubMed] [Google Scholar]
- Boström H, Willetts K, Pekny M, Levéen P, Lindahl P, Hedstrand H, Pekna M, Hellström M, Gebre-Medhin S, Schalling M, et al. 1996. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85: 863–873. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC. 2005. Role of phosphoinositide 3-kinase regulatory isoforms in development and actin rearrangement. Mol Cell Biol 25: 2593–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Molotkov A, Mazot P, Hoch RV, Soriano P. 2015. Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes Dev 29: 1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Soriano P. 2010. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph–ephrin boundaries. Genes Dev 24: 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Marazita ML, Hart PS, Sulima PP, Field LL, McHenry TG, Govil M, Cooper ME, Letra A, Menezes R, et al. 2009. The PDGF-C regulatory region SNP rs28999109 decreases promoter transcriptional activity and is associated with CL/P. Eur J Hum Genet 17: 774–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y. 2003. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res 284: 54–65. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Kim I, Tam PP, Koh GY, Nagy A. 2000. The mouse Pdgfc gene: dynamic expression in embryonic tissues during organogenesis. Mech Dev 96: 209–213. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, O'Rourke M, Koh GY, Soriano P, Betsholtz C, et al. 2004. A specific requirement for PDGF-C in palate formation and PDGFR-α signaling. Nat Genet 36: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Ekman S, Thuresson E, Heldin CH, Ronnstrand L. 1999. Increased mitogenicity of an αβ heterodimeric PDGF receptor complex correlates with lack of RasGAP binding. Oncogene 18: 2481–2488. [DOI] [PubMed] [Google Scholar]
- Erickson CA, Weston JA. 1983. An SEM analysis of neural crest migration in the mouse. J Embryol Exp Morphol 74: 97–118. [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P. 2014. PI3K-mediated PDGFRα signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev 28: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladh H, Folestad EB, Muhl L, Ehnman M, Tannenberg P, Lawrence A-L, Betsholtz C, Eriksson U. 2016. Mice lacking platelet-derived growth factor D display a mild vascular phenotype. PLoS One 11: e0152276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretzki L, Burg MA, Grako KA, Stallcup WB. 1999. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J Biol Chem 274: 16831–16837. [DOI] [PubMed] [Google Scholar]
- Grüneberg H, Truslove GM. 1960. Two closely linked genes in the mouse. Genet Res 1: 69–90. [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. 2003. Evolutionary divergence of platelet-derived growth factor α receptor signaling mechanisms. Mol Cell Biol 23: 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P. 2013. A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet 9: e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. 1999. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 79: 1283–1316. [DOI] [PubMed] [Google Scholar]
- Heuchel R, Berg A, Tallquist M, Ahlén K, Reed RK, Rubin K, Claesson-Welsh L, Heldin CH, Soriano P. 1999. Platelet-derived growth factor β receptor regulates interstitial fluid homeostasis through phosphatidylinositol-3′ kinase signaling. Proc Natl Acad Sci 96: 11410–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV. 2005. Basement membrane proteoglycans: from cellar to ceiling. Nat Rev Mol Cell Biol 6: 646–656. [DOI] [PubMed] [Google Scholar]
- Jadeja S, Smyth I, Pitera JE, Taylor MS, van Haelst M, Bentley E, McGregor L, Hopkins J, Chalepakis G, Philip N, et al. 2005. Identification of a new gene mutated in Fraser syndrome and mouse myelencephalic blebs. Nat Genet 37: 520–525. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Sanchez-Contreras MY, Keppler-Noreuil KM, Sapp J, Crenshaw M, Finch NCA, Cormier-Daire V, Rademakers R, Sybert VP, Biesecker LG. 2015. A point mutation in PDGFRB causes autosomal-dominant Penttinen syndrome. Am J Hum Genet 97: 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner A, Wall N. 2008. Immunohistochemistry of whole-mount mouse embryos. Cold Spring Harb Protoc 10.1101/pdb.prot4820. [DOI] [PubMed] [Google Scholar]
- Kiyozumi D, Sugimoto N, Sekiguchi K. 2006. Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects. Proc Natl Acad Sci 103: 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper LN, Glathe S, Vaisman N, Hynes NE, Andrews GC, Sela M, Yarden Y. 1999. The ErbB-2/HER2 oncoprotein of human carcinomas may function solely as a shared coreceptor for multiple stroma-derived growth factors. Proc Natl Acad Sci 96: 4995–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer RA, Kazlauskas A. 1995. Identification of a putative Syp substrate, the PDGFβ receptor. J Biol Chem 270: 22208–22217. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Mueting-Nelsen PF, Faerman A, Shani M, Soriano P. 2001. The two PDGF receptors maintain conserved signaling in vivo despite divergent embryological functions. Mol Cell 7: 343–354. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. 2002. An allelic series at the PDGFαR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell 2: 103–113. [DOI] [PubMed] [Google Scholar]
- Koch S, Claesson-Welsh L. 2012. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med 2: a006502–a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C. 1994. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev 8: 1875–1887. [DOI] [PubMed] [Google Scholar]
- Matsui T, Heidaran M, Miki T, Popescu N, La Rochelle W, Kraus M, Pierce J, Aaronson S. 1989. Isolation of a novel receptor cDNA establishes the existence of two PDGF receptor genes. Science (80-) 243: 800–804. [DOI] [PubMed] [Google Scholar]
- McCarthy N, Liu JS, Richarte AM, Eskiocak B, Lovely CB, Tallquist MD, Eberhart JK. 2016. Pdgfra and Pdgfrb genetically interact during craniofacial development. Dev Dyn 245: 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor L, Makela V, Darling SM, Vrontou S, Chalepakis G, Roberts C, Smart N, Rutland P, Prescott N, Hopkins J, et al. 2003. Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat Genet 34: 203–208. [DOI] [PubMed] [Google Scholar]
- Miller KA, Gordon CT, Welfare MF, Caruana G, Bertram JF, Bateman JF, Farlie PG. 2013. bfb, a novel ENU-induced blebs mutant resulting from a missense mutation in Fras1. PLoS One 8: e76342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenning A, Jäger R, Egert A, Kress W, Wardelmann E, Schorle H. 2009. Sustained platelet-derived growth factor receptor α signaling in osteoblasts results in craniosynostosis by overactivating the phospholipase C-γ pathway. Mol Cell Biol 29: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison-Graham K, Schatteman GC, Bork T, Bowen-Pope DF, Weston JA. 1992. A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development 115: 133–142. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li N, Luo L. 2007. A global double-fluorescent cre reporter mouse. Genesis 45: 593–605. [DOI] [PubMed] [Google Scholar]
- Noonan J. 1968. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am J Dis Child 116: 373–380. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Lonai P. 1992. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development 115: 1045–1058. [DOI] [PubMed] [Google Scholar]
- Petrou P, Makrygiannis AK, Chalepakis G. 2008. The Fras1/Frem family of extracellular matrix proteins: structure, function, and association with Fraser syndrome and the mouse bleb phenotype. Connect Tissue Res 49: 277–282. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, et al. 2008. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet 40: 1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanasopha S, Tongkobpetch S, Srichomthong C, Siriwan P, Suphapeetiporn K, Shotelersuk V. 2012. PDGFRa mutations in humans with isolated cleft palate. Eur J Hum Genet 20: 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richarte AM, Mead HB, Tallquist MD. 2007. Cooperation between the PDGF receptors in cardiac neural crest cell migration. Dev Biol 306: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz S, DeMali KA, Gelderloos JA, Bazenet C, Kazlauskas A. 1999. Identification of the receptor-associated signaling enzymes that are required for platelet-derived growth factor-AA-dependent chemotaxis and DNA synthesis. J Biol Chem 274: 28335–28343. [DOI] [PubMed] [Google Scholar]
- Rupp E, Siegbahn A, Ronnstrand L, Wernstedt C, Claesson-Welsh L, Heldin CH. 1994. A unique autophosphorylation site in the platelet-derived growth factor α receptor from a heterodimeric receptor complex. Eur J Biochem 225: 29–41. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Kurosaka H, Trainor PA. 2012. Whole mount nuclear fluorescent imaging: convenient documentation of embryo morphology. Genesis 50: 844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J, Rizzolo K, Soriano P. 2008. The PDGF signaling pathway controls multiple steroid-producing lineages. Genes Dev 22: 3255–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneller M, Vuori K, Ruoslahti E. 1997. αvβ3 integrin associates with activated insulin and PDGFβ receptors and potentiates the biological activity of PDGF. EMBO J 16: 5600–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM, Baranzini SE, Schanze D, Labelle-Dumais C, Short KM, Chao R, Yahyavi M, Bijlsma EK, Chu C, Musone S, et al. 2011. Manitoba-oculo-tricho-anal (MOTA) syndrome is caused by mutations in FREM1. J Med Genet 48: 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Seldin MF, Martinez L, Watson ML, Choudhury GG, Lalley PA, Pierce J, Aaronson S, Barker J, Naylor SL, et al. 1991. Mouse platelet-derived growth factor receptor α gene is deleted in W19H and patch mutations on chromosome 5. Proc Natl Acad Sci 88: 4811–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth I, Du X, Taylor MS, Justice MJ, Beutler B, Jackson IJ. 2004. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc Natl Acad Sci 101: 13560–13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. [DOI] [PubMed] [Google Scholar]
- Soriano P. 1994. Abnormal kidney development and hematological disorders in PDGF β-receptor mutant mice. Genes Dev 8: 1888–1896. [DOI] [PubMed] [Google Scholar]
- Soriano P. 1997. The PDGF α receptor is required for neural crest cell development and for normal patterning of the somites. Development 124: 2691–2700. [DOI] [PubMed] [Google Scholar]
- Takenouchi T, Yamaguchi Y, Tanikawa A, Kosaki R, Okano H, Kosaki K. 2015. Novel overgrowth syndrome phenotype due to recurrent de novo PDGFRB mutation. J Pediatr 166: 483–486. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26: 113–115. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. 2003. Cell autonomous requirement for PDGFRα in populations of cranial and cardiac neural crest cells. Development 130: 507–518. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, French WJ, Soriano P. 2003. Additive effects of PDGF receptor β signaling pathways in vascular smooth muscle cell development. PLoS Biol 1: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, et al. 2001. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29: 465–468. [DOI] [PubMed] [Google Scholar]
- Timmer JR, Mak TW, Manova K, Anderson KV, Niswander L. 2005. Tissue morphogenesis and vascular stability require the Frem2 protein, product of the mouse myelencephalic blebs gene. Proc Natl Acad Sci 102: 11746–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Soriano P. 2014. SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Dev Cell 31: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Mazot P, He F, Soriano P. 2015. Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. Elife 4: e07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, Cox TC, Maga AM, Short KM, Wiradjaja F, Janssen IM, Jehee F, Bertola D, Liu J, Yagnik G, et al. 2011. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet 7: e1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrontou S, Petrou P, Meyer BI, Galanopoulos VK, Imai K, Yanagi M, Chowdhury K, Scambler PJ, Chalepakis G. 2003. Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat Genet 34: 209–214. [DOI] [PubMed] [Google Scholar]
- Williams LT. 1989. Signal transduction by the platelet-derived growth factor receptor. Science 243: 1564–1570. [DOI] [PubMed] [Google Scholar]
- Wiradjaja F, Cottle DL, Jones L, Smyth I. 2013. Regulation of PDGFC signalling and extracellular matrix composition by FREM1 in mice. Dis Model Mech 6: 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JC, Heidaran MA, Pierce JH, Gutkind JS, Lombardi D, Ruggiero M, Aaronson SA. 1991. Tyrosine mutations within the α platelet-derived growth factor receptor kinase insert domain abrogate receptor-associated phosphatidylinositol-3 kinase activity without affecting mitogenic or chemotactic signal transduction. Mol Cell Biol 11: 3780–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.