SUMMARY
Despite the accepted health benefits of consuming dietary fiber, little is known about the mechanisms by which fiber deprivation impacts the gut microbiota and alters disease risk. Using a gnotobiotic mouse model, in which animals were colonized with a synthetic human gut microbiota composed of fully sequenced commensal bacteria, we elucidated the functional interactions between dietary fiber, the gut microbiota and the colonic mucus barrier, which serves as a primary defense against enteric pathogens. We show that during chronic or intermittent dietary fiber deficiency, the gut microbiota resorts to host-secreted mucus glycoproteins as a nutrient source, leading to erosion of the colonic mucus barrier. Dietary fiber deprivation, together with a fiber-deprived, mucus-eroding microbiota, promotes greater epithelial access and lethal colitis by the mucosal pathogen, Citrobacter rodentium. Our work reveals intricate pathways linking diet, the gut microbiome and intestinal barrier dysfunction, which could be exploited to improve health using dietary therapeutics.
Graphical abstract
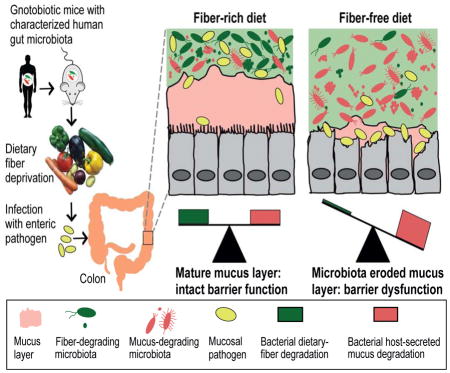
Regular consumption of dietary fiber helps prevent erosion of the intestinal mucus barrier by the gut microbiome, blunting pathogen infection and reducing the incidence of colitis.
INTRODUCTION
The diet of industrialized nations has experienced a decrease in fiber intake, which for many is now well below the recommended daily range of 28–35 g for adults and this deficit has been linked to several diseases (Burkitt et al., 1972; Sonnenburg and Sonnenburg, 2014). Fiber provides direct physical benefits, including increased fecal bulking and laxation (Burkitt et al., 1972). However, another feature of dietary fiber—a nutrient category that includes a broad array of polysaccharides that are not digestible by human enzymes—has also drawn it into the spotlight: it provides an important substrate to the community of microbes (microbiota) that inhabits the distal gut (Sonnenburg and Sonnenburg, 2014). Unlike humans, who produce ~17 gastrointestinal enzymes to digest mostly starch, our gut microbiota produce thousands of complementary enzymes with diverse specificities, enabling them to depolymerize and ferment dietary polysaccharides into host-absorbable short-chain fatty acids (SCFAs) (El Kaoutari et al., 2013). Thus, the physiology of the gut microbiota is geared towards dietary polysaccharide metabolism. At present, relatively little is known about how a fiber-deprived gut microbiota fulfils its energy demands and how low fiber-induced microbiota changes impact our health.
Apart from dietary fiber, an alternative energy source for the microbiota is the glycoprotein-rich mucus layer that overlies the gut epithelium as a first line of defense against both commensal microbes and invading pathogens (Johansson et al., 2013; McGuckin et al., 2011). The colonic mucus layer is a dynamic and chemically complex barrier composed largely of secreted mucin-2 glycoprotein (MUC2) (Johansson et al., 2008). Goblet cells secrete MUC2 as a disulfide cross-linked network that expands to form an inner layer, which is tightly adherent to the epithelium and is poorly colonized by commensal bacteria. As bacterial and host enzymes continuously hydrolyze the luminal edge of this layer, a looser outer layer is formed that supports a more dense and metabolically distinct (Li et al., 2015) community. A key nutritional aspect of the mucus layer for gut bacteria is its high polysaccharide content, with up to 80% of the mucin biomass being composed of mostly O-linked glycans (Johansson et al., 2013). However, only a distinct subset of gut microbiota species has evolved the capacity to utilize this nutrient source (Hoskins and Boulding, 1981; Png et al., 2010).
The direct impact of fiber polysaccharides on the microbiota, combined with the ability of at least one nutritional generalist (Bacteroides thetaiotaomicron) to shift from dietary polysaccharides to mucus glycan metabolism in the absence of fiber (Sonnenburg et al., 2005), suggests a connection between diet and the status of the colonic mucus barrier. Indeed, three previous reports have correlated reduced dietary fiber with thinner colonic mucus (Brownlee et al., 2003; Earle et al., 2015; Hedemann et al., 2009). Nevertheless, the underlying mechanisms with respect to involvement of the microbiota and, perhaps more importantly, consequences for the host remain largely unknown. Such knowledge is important as it could provide explanations for why deviations or imbalances in gut microbial community membership and physiology (“dysbiosis”) correlate with several negative health outcomes, including pathogen susceptibility, inflammatory bowel disease (IBD) and colon cancer (Cameron and Sperandio, 2015; McKenney and Pamer, 2015). Finally, such knowledge could inform therapeutic and preventative strategies to correct these conditions.
The integrity of the mucus layer is critical for health. Genetic ablation of Muc2 in mice brings bacteria into close contact with the epithelium, leading to inflammation and colon cancer (Van der Sluis et al., 2006). Additional studies have implicated reduced or abnormal mucus production or O-glycosylation in the development of intestinal inflammation (Fu et al., 2011; Larsson et al., 2011) and penetration of commensal bacteria in the inner mucus layer in murine models of colitis and ulcerative colitis patients (Johansson et al., 2014). Moreover, the mucus barrier—a reservoir of antimicrobial peptides and immunoglobulins—is the first structure that a mucosal pathogen must overcome to establish an infection (McGuckin et al., 2011). Given that the status of the mucus layer is precariously balanced between replenishment by goblet cells and degradation by gut bacteria, we hypothesized that a fiber-deprived microbiota would progressively forage on this barrier, leading to inflammation and/or increased pathogen susceptibility.
We aimed to investigate the mechanistic connections between chronic or intermittent dietary fiber deprivation on microbiota composition and physiology as well as the resulting effects on the mucus barrier. To create a model that facilitates functional interpretation, we assembled a synthetic gut microbiota from fully sequenced human gut bacteria in gnotobiotic mice. In the face of reduced dietary fiber, we examined changes in community physiology and susceptibility to Citrobacter rodentium, a murine pathogen that models human enteric E. coli infection (Collins et al., 2014). We demonstrate that a microbiota deprived of dietary fiber damages the colonic mucus barrier and promotes pathogen susceptibility. Our findings suggest a mechanism through which diet alters the activity of the gut microbiota and impacts health, which is important prerequisite knowledge for rationally designing future dietary interventions and therapeutics.
RESULTS
A synthetic human gut microbiota with versatile fiber polysaccharide degrading capacity
Diet changes are known to rapidly affect the composition of the microbiota in humans and rodents (David et al., 2014; Faith et al., 2011; McNulty et al., 2013; Rey et al., 2013). However, the full complexity of the gut microbiota is a barrier to deriving detailed conclusions because sequence-based approaches (16S rRNA gene and meta-genomics/-transcriptomics) suffer from substantial functional uncertainty. Thus, to test our hypothesis that specific members within a fiber-deprived gut microbiota cause damage by increasingly foraging for nutrients in the protective mucus layer, we designed a synthetic microbiota (SM) containing 14 species of fully sequenced commensal human gut bacteria (Fig. 1A). The selected species were chosen to represent the five dominant phyla and collectively possess important core metabolic capabilities (Fig. S1A).
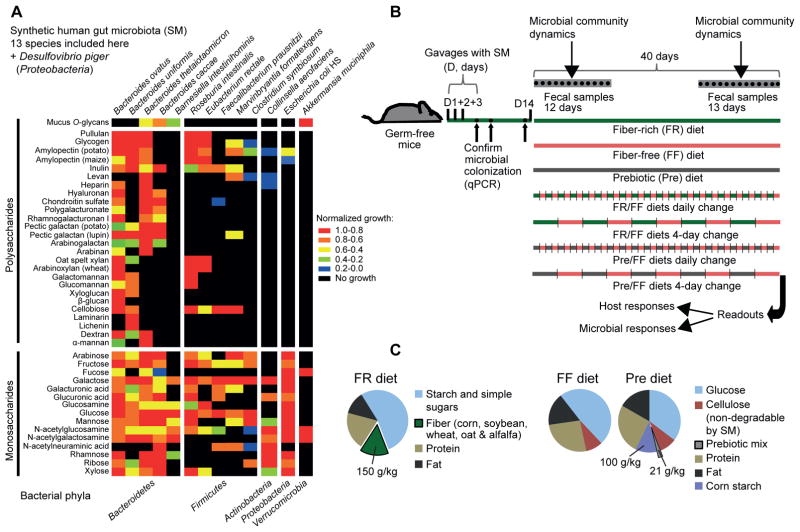
Figure 1. Carbohydrate utilization by the synthetic human gut microbiota (SM) members and gnotobiotic mouse treatments.
(A) Heat map showing normalized growth values of 13/14 SM members.
(B) Schematic of the gnotobiotic mouse model illustrating the timeline of colonization, feeding strategies and fecal sampling.
(C) Compositions of the three distinct diets employed in this study (common additives such as vitamins and minerals are not shown). The prebiotic mix contained equal proportions of 14 host indigestible polysaccharides (see Table S1). See also Fig. S1.
To provide an additional layer of functional knowledge about complex carbohydrate metabolism, we pre-evaluated our 14 species for growth in vitro on a panel of 42 plant- and animal-derived mono- and polysaccharides, including purified mucin O-glycans (MOGs) as sole carbon sources (Martens et al, 2011). These growth assays allowed us to determine that all major groups of dietary fiber and host mucosal polysaccharides could be used by one or more strains in our community as well as which bacteria target each glycan (Fig. 1A, Figs. S1A,B, Table S1). It is evident that the four mucin-degrading species fall into two categories: mucin specialists (A. muciniphila and B. intestinihominis), which only grow on MOGs as a sole polysaccharide source, and mucin generalists (B. thetaiotaomicron and B. caccae), which each grow on several other polysaccharides. Overall, our choice of species is physiologically and ecologically representative of the more complex native gut microbiota. Because our community is composed of bacteria with determined carbohydrate metabolic abilities, it allows us to address our central hypothesis in more precise, mechanistic detail.
To develop a gnotobiotic model, we assembled the SM in germfree mice, which were fed a standard fiber-rich (FR) laboratory diet that contains ~15% dietary fiber from minimally processed grains and plants (Figs. 1B,C). Colonized animals were maintained on the FR diet for 14 days to monitor reproducibility and stability of community assembly (Fig. 1B). All of the introduced species persisted in each mouse between 6 and 54 or 66 days of colonization depending on the length of the experiment (n = 37 total, 2 independent experiments; analyzed by both 16S rRNA sequencing (Table S2) and quantitative PCR approaches (Table S3)). Individual mice exhibited reproducible SM assembly irrespective of caging, mouse gender, experimental replicate or method of analysis (Fig. S2, Tables S2, S3). In addition to 29 germfree control animals, a total of 4 different gnotobiotic colonization experiments (51 SM-colonized mice in total; Experiments 1–4) were performed according to the timeline shown in Fig. 1B.
Both chronic and intermittent fiber deficiency promotes enrichment of mucus-degrading bacteria
Although dietary changes are known to perturb microbiota composition, the impact of diet variation, especially chronic or intermittent fiber deficiency, on the activities and abundance of mucin-degrading bacterial communities has not been studied in functional detail. After validating stable SM colonization, three groups of mice were maintained by constant feeding of one of three different diets: Fiber-rich (FR), Fiber-free (FF) or Prebiotic (Pre). In contrast to the FR diet that contained naturally milled food ingredients with intact fiber particles, the Pre diet was designed to study the effect of adding a mixture of purified, soluble glycans, similar to those used in prebiotic formulations (Fig. 2C). To imitate the fact that the human diet experiences fluctuating amounts of fiber from meal-to-meal, four other groups were alternated between the FR and FF or Pre and FF diets on a daily- or four-day basis (Fig. 1B).
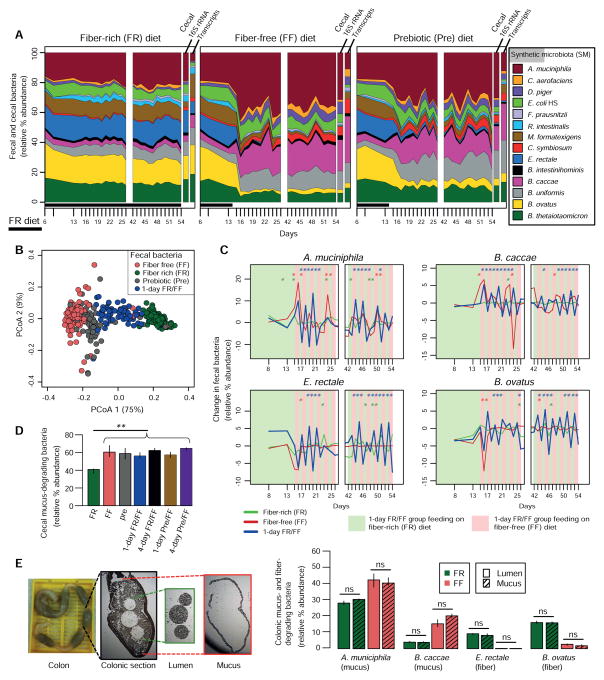
Figure 2. Complex dietary fiber deficiency leads to proliferation of mucus-degrading bacteria.
(A) Stream plots exhibiting fecal (over time, Fig. 1B) and cecal (end point) microbial community dynamics and average abundance of total species-specific transcripts from cecal RNA-Seq transcriptome mapping at the endpoint; for transcript abundance n = 3 mice/group.
(B) Principal coordinate analysis (PCoA) based on bacterial community similarity.
(C) Changes in relative bacterial abundance over time in mice oscillated for 1-day increments between FR and FF feeding. Changes in FR and FF control groups are shown for comparison. Asterisks (colored according to the dietary group) indicate a statistically significant difference in the change of relative abundance from the previous day within each group. Student’s t-test.
(D) Additive relative abundances of four mucus-degrading bacteria (Fig. 1A).
(E) Relative bacterial abundances in laser capture microdissected colonic lumen and mucus samples (images displayed on left). n = 3 mice/group.
Microbial community abundance data are based on Illumina sequencing of 16S rRNA genes (V4 region) and median values at each time point are shown. Unless specified, significance was determined using Kruskal–Wallis test and n = 4 for FR and FF groups, n = 3 for all other groups. All data in Fig. 2A–E is from Experiment 1.
Fecal microbial community dynamics showed that in mice switched to the FF diet, several species rapidly and reproducibly changed in abundance (Figs. 2A,B; Fig. S3). Four species—A. muciniphila, B. caccae, B. ovatus and E. rectale—were highly responsive to diet change. A. muciniphila and B. caccae are able to degrade MOGs in vitro. B. ovatus and E. rectale cannot metabolize MOGs, but together can use a broad range of polysaccharides found in dietary fiber (Fig. 1A). In the absence of fiber, the abundance of A. muciniphila and B. caccae increased rapidly with a corresponding decrease of the fiber-degrading species (Fig. 2A). The Pre diet, which contains purified polysaccharides and is otherwise isocaloric with the FF diet, had a similar effect on community composition as the FF diet but separated slightly by PCoA ordination from FF, likely due to increased Bacteroides abundance (Figs. 2A,B). Intriguingly, the abundances of the same four bacteria noted above fluctuated rapidly on a daily basis when the FR and FF diets were oscillated (Fig. 2C; Figs. S3), corroborating their ability to respond dynamically to variations in dietary fiber. The increase in mucin-degrading species observed in fecal samples matched with cecal abundances at the end of the experiment (Fig. 2D and panels to the right of plots in Fig. 2A). Moreover, similar levels of mucin-degrading bacteria were quantified in the colonic lumen and mucus layer using laser capture microdissection (Fig. 2E), indicating that proliferation of mucin-degrading bacteria in this model is a community-wide effect and not limited just to the mucus layer.
Many of the other bacteria (except R. intestinalis and B. intestinihominis) were sensitive to changes between the FR and FF diets on daily and four-day bases, albeit to lower degrees (Fig. S3B, Table S2). Two additional species especially sensitive to diet change were Desulfovibrio piger (increased on FF diet) and Marvinbryantia formatexigens (decreased on FF diet) (Fig. S3A). Population changes in the groups oscillated between Pre and FF diets were similar to the abundances observed in FF only diet regimen (Table S2). Thus, despite the pure polysaccharides contained in the Pre diet exerting a clear physiological impact on the microbiota (discussed below), the amount and composition of the purified polysaccharides in this diet exert little effect on species composition.
Community transcriptional and enzymatic readouts demonstrate enhanced degradation of mucus when fiber is absent
Since mucin-degrading bacteria were higher on both the FF and Pre diets that lack naturally complex plant fiber, we reasoned that this increased abundance is due to their ability to degrade mucus as an alternative nutrient. To test this, we measured changes in transcripts encoding carbohydrate active enzymes (CAZymes) that enable gut bacteria to utilize dietary fiber and mucosal polysaccharides (cecal samples from all but the 4-day oscillation groups were analyzed).
Based on new or existing genome annotations of the 14 species in our synthetic community, a total of 1,661 different degradative CAZymes belonging to 96 different families were detected (glycoside hydrolase, GH; polysaccharide lyase, PL; and carbohydrate esterase, CE, families were counted). This number is close to the total number of families (122) that was identified in a larger survey of 177 human gut bacterial reference genomes (El Kaoutari et al., 2013), indicating that our synthetic community retains much of the metabolic potential towards carbohydrates that is present in a more diverse microbiota. Of the 96 enzyme families in our community, members of 38 families, plus M60-like proteases (pfam13402), a group of enzymes previously shown to degrade mucin glycoproteins (Nakjang et al., 2012), showed variable expression in either FR/FF or Pre/FF community transcriptome comparisons (Fig. 3A). These differentially abundant degradative enzymes mapped to 770 different genes contributed from all species except D. piger (Table S4).
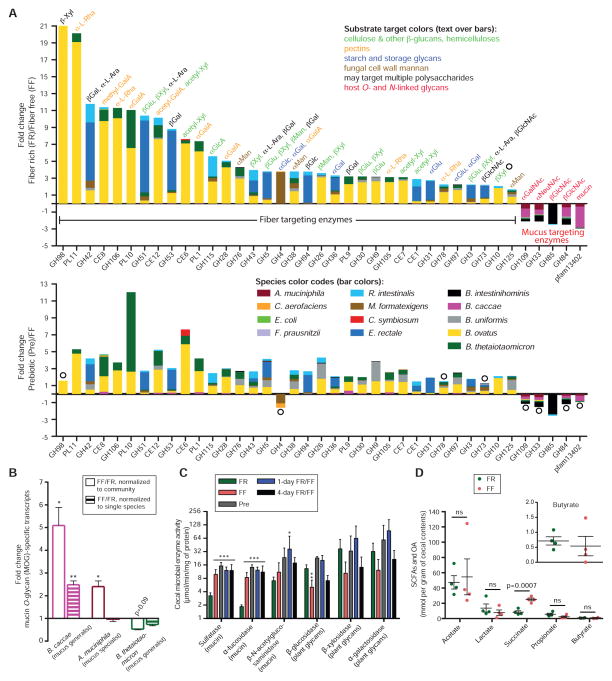
Figure 3. Diet-specific changes in carbohydrate active enzyme expression reveal a community shift from fiber to mucus degradation.
(A) Positive and negative fold-changes in transcripts encoding carbohydrate active enzymes (CAZymes) between either FR/FF (top) or Pre/FF (bottom) comparisons. Only CAZyme families (x-axis) in which >2-fold changes and p < 0.05 (Student’s t-test) were observed for all of the genes in that family in RPKM-normalized cecal community transcriptomes are shown as averages; open circles denote statistically insignificant differences. n = 3 mice/group, Experiment 1.
(B) Fold-change values of empirically validated (Table S5), MOG-specific transcripts of three mucus-degrading bacteria. n = 3 mice/group, Experiment 1. Data are shown as average ± SEM. Student’s t-test
(C) Activities of cecal enzymes determined by employing p-nitrophenyl-linked substrates. n = 4 for FR and FF groups and n = 3 for other groups, Experiment 1. Data are shown as average ± SD. One-way ANOVA, FR diet group vs. other groups.
(D) Concentrations of organic acid (OA, succinate) and short-chain fatty acids (SCFA) determined from cecal contents. n = 4 mice/group; 2 mice/dietary group in two independent experiments (#2A and 3). Student’s t-test.
Our transcriptomic data show that in the mice fed the FR diet, transcripts belonging to enzyme families that target dietary fiber polysaccharides were more abundant. In contrast, in mice fed the FF diet, transcripts encoding enzyme families known to release sugars from host substrates, including mucin O-glycans, were elevated (Fig. 3A). In line with our in vitro growth assays (Fig. 1A) and the microbial community abundance data (Fig. 2), we found that B. ovatus and E. rectale contributed a majority of CAZymes specific for plant polysaccharides (Fig. 3A). The four in vitro mucin degraders were the major contributors to the degradation of host glycans and mucus in vivo. Additionally, transcripts encoding M60-like proteases (pfam13402) were also more highly expressed in FF conditions (Fig. 3A). Expression of these putative mucin-targeting enzymes was primarily contributed by B. caccae, an organism that possesses 16 of these genes compared to just 4 in A. muciniphila and 5 in all other species combined. This observation suggests the tantalizing possibility that B. caccae is particularly equipped via its M60-like proteases to perform a key degradative step (cleavage of glycoprotein backbones) during mucin foraging and this ability may facilitate access to mucus carbohydrate structures by other bacteria.
In the group fed the Pre diet, similar transcripts as in the FR-fed group were elevated relative to FF, albeit to lower levels (Fig. 3A bottom histogram). Furthermore, transcripts for the same bacterial enzymes presumed to target mucus in the FR/FF diet comparison were observed in the Pre/FF comparison. Additional RNA-Seq analyses of cecal transcriptomes from mice oscillated between FR/FF and Pre/FF on a daily basis (collected after 1 day on FF diet) provided similar results to those obtained for the FF only diet mice (Table S4). Our transcriptomic readouts corroborate the increased abundance of mucin-degrading bacteria observed in these mice (Fig. 2D) and demonstrate that even intermittent fiber deficiency has the potential to alter the microbiota and favor mucin-degrading species.
To further connect the in vivo responses of B. caccae and A. muciniphila with degradation of mucin O-glycans, we performed additional transcriptional profiling of these two species on purified MOGs from porcine gastric mucus. We have previously shown that this mixture contains ~130 different structures (Hickey et al., 2015) that when metabolized by B. thetaiotaomicron stimulate a transcriptional response that overlaps substantially with genes expressed in vivo under fiber-restricted conditions (Martens et al., 2008). During growth on MOGs as the sole carbon source, B. caccae and A. muciniphila activated expression of 82 and 61 genes, respectively (Table S5). In previous microarray data from growth in the same substrate, B. thetaiotaomicron activated expression of 166 genes (Martens et al., 2008). Next, we examined expression of these validated O-glycan-responsive genes (for B. caccae, A. muciniphila and B. thetaiotaomicron) in the SM community from FF-fed mice compared to FR. In support of our hypothesis, validated B. caccae and A. muciniphila O-glycan-responsive genes were increased in the FF condition (Fig. 3B). B. caccae expression was increased irrespective of normalization by reads mapped to the whole community (i.e., including increased B. caccae abundance) or to just the B. caccae genome (discounts abundance change and examines changes in expression).
Consistent with its specialization for O-glycans, A. muciniphila mostly showed increased expression of O-glycan-responsive genes proportional to its increased population size (from ~20% to ~40%), indicating that it does not shift its substrate utilization in comparison to the FR diet (Fig. 3B, see also Table S6e). The idea that B. caccae is capable of broader transcriptional shifts, compared to A. muciniphila, is further supported by global analysis of its gene expression changes between the FF and FR diets and one-day alternations (Fig. S4). In response to the FF diet, B. caccae showed increased in vivo expression of 237 genes, including 29 degradative enzymes; whereas, A. muciniphila only showed increased expression of 43 genes, including 2 enzymes (Fig. S4). In contrast to previous monoassociation data (Sonnenburg et al., 2005), B. thetaiotaomicron mostly showed unchanged or slightly decreased expression of its known O-glycan utilization genes after a shift to FF (Fig. 3B, Table S6b).
Colonic mucin O-glycans contain glycosidic linkages distinct from plant fibers and also covalently linked sulfate. In further support of increased bacterial degradation of the host mucus on the FF diet, we detected significantly increased bacterial enzymes (sulfatase and α-fucosidase) in the mice subjected to the FF diet on either a chronic or intermittent basis (Fig. 3C). In contrast, enzymes targeting linkages in fiber polysaccharides (β-glucosidase) were significantly reduced in the mice fed the fiber-deficient diets, while others involved in xylan and α-galactan degradation trended similarly without significance (Fig. 3C).Despite the dramatic change in microbiota species abundance and transcriptional response, there was only one significant change (succinate) in SCFA and organic acids in the FF diet fed mice (Fig. 3D). Overall, the transcriptomic and enzyme analyses support the conclusion that a fiber-deprived gut microbiota synergistically and progressively expresses CAZymes, sulfatases and proteases to attack mucus polysaccharides when the diet lacks complex plant fiber.
Fiber deprivation leads the gut microbiota to degrade the colonic mucus barrier
The mucus layer is a dynamic barrier that is constantly replenished through the secretory activity of goblet cells (Johansson et al., 2013). We rationalized that if bacterial consumption of mucin-derived nutrients exceeds new production, the integrity of this critical barrier could be compromised. To explore this possibility, we performed blinded thickness measurements of the colonic mucus layer from proximal colon to rectum in each mouse using Alcian blue stained sections (Fig. 4A). We further validated thickness of the mucus layer by immunofluorescence staining of the Muc2 mucins using α-Muc2 antibody (Fig. 4B). To address the possibility that variations in thickness are directly influenced by the diets used, we measured mucus layer thickness in germfree mice fed the FR or FF diets.
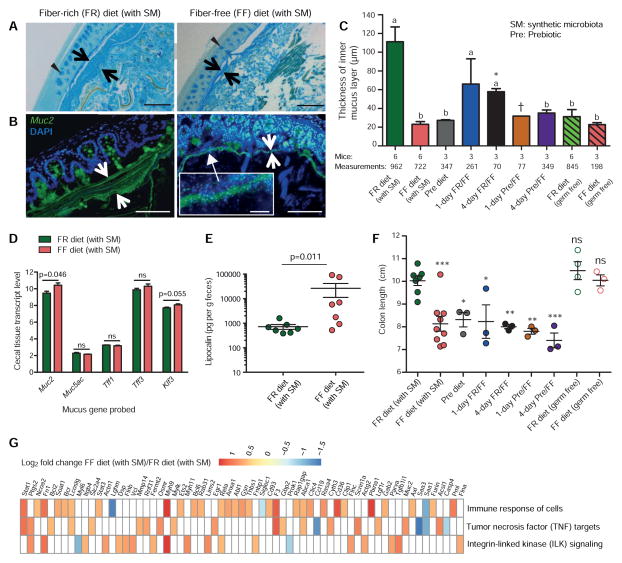
Figure 4. Microbiota-mediated erosion of the colonic mucus barrier and host responses.
(A) Alcian blue-stained colonic sections showing the mucus layer (arrows). Scale bars, 100 μm. Opposing black arrows with shafts delineate the mucus layer that was measured and triangular arrowheads point to pre-secretory goblet cells.
(B) Immunofluorescence images of colonic thin sections stained with α-Muc2 antibody and DAPI. Opposing white arrows with shafts delineate the mucus layer. Inset (FF diet group) shows a higher magnification of bacteria-sized, DAPI-stained particles in closer proximity to host epithelium and even crossing this barrier. Scale bars: 100 μm; inset, 10 μm.
(C) Blinded colonic mucus layer measurements from Alcian blue stained sections. Mice in the FR and FF fed colonized groups (Experiments 1 and 2A), and in the FR-diet fed germfree groups are from two independent experiments; all other colonized mice are from Experiment 1. Asterisk and dagger indicate that colons of only 2 and 1 mice contained fecal masses, respectively. Data are presented as average ± SEM. Statistically significant differences are annotated with different letters p < 0.01; One-way ANOVA with Tukey’s test.
(D) Microarray-derived transcript levels of genes involved in the production of colonic mucus (n = 4 for the FR diet group and n = 3 for the FF diet group). Data are from two independent experiments (#2A and 3). Values are shown as average ± SEM. Student’s t-test.
(E) Levels of fecal lipocalin (LCN2) measured by ELISA in the FR and FF diet fed groups (day 50, Figure S6A; Experiment 2A). n = 7 mice/group. Mann-Whitney test.
(F) Colon lengths of mice subjected to different dietary treatments. Data for the FR (with SM) and FF (with SM) are representative of 3 independent experiments (Experiments 1, 2A and 3). One-way ANOVA, FR diet group (with SM) vs. other groups.
(G) Changes in the host cecal transcriptome between FR and FF diet conditions. Heatmap shows statistically significant fold changes of genes identified from Ingenuity Pathway Analysis (FDR < 0.05 and absolute Log2 Fold-Change > 0.5). n = 4 for the FR diet group and n = 3 for the FF diet group; data are from two independent experiments (#2A and 3).
Colonic mucus measurements revealed that mucus thickness was highest in the colonized group fed the FR diet (Fig. 4C). In most other groups, including germfree (GF) controls, mucus thickness was significantly thinner than colonized FR mice. The observation that GF mice have thinner colonic mucus is consistent with previous studies in gnotobiotic mice and rats (Petersson et al., 2011; Wrzosek et al., 2013), which found that microbial colonization or exposure to cues such as peptidoglycan or lipopolysaccharides is required for mucus production. Notably, mucus in SM-colonized FF diet mice was 5–6X thinner than colonized mice fed the FR diet. From these data, we conclude that the mucus layer: i) is initially thinner in GF mice regardless of diet, ii) begins expanding upon microbial colonization, but iii) is disproportionately eroded back to a thinner layer due to the increased mucus foraging activity by the microbiota in the context of the FF diet. A similar reduction in mucus thickness was observed in the Pre diet and both 4-day oscillation groups, while an intermediate thickness was observed in the 1-day FR/FF oscillation group (Fig. 4C).
Next, we determined whether mucus production was altered in colonized mice fed the FF diet. We examined the abundance of transcripts encoding several key proteins involved in building and regulating the mucus barrier (Fig. 4D): Muc2 and Muc5ac, two building blocks of colonic mucus; Tff1 and Tff3, goblet cell proteins that promote mucosal repair and protection; and Klf3 a transcription factor involved in barrier function. Our results show that the transcription of the major colonic mucin gene (Muc2) was slightly elevated in the colonized FF diet group, suggesting a compensatory response of the host to offset the increased bacterial mucus degradation in this group; whereas other genes (Muc5ac, Tff1, Tff3 and Klf3) remained statistically unchanged (Fig. 4D). Qualitative visualization by Alcian blue (Fig. 4A) staining supports the conclusion that the colonic tissue of FF-fed colonized mice contained similar numbers of goblet cells that have yet to secrete their glycoproteins.
As expected, degradation of the mucus layer by the fiber-deprived gut microbiota brought luminal bacteria closer to the intestinal epithelium (Fig. 4B, inset), which could potentially trigger deleterious effects or other host compensatory responses. Histopathology (Fig. S5A) and body weight measurements over time (Fig. S5B) of mice from the groups with reduced mucus thickness did not reveal changes compared to the mice consuming the FR diet. However, measurements of three additional host parameters provided support for altered host responses in the face of mucus erosion: The first was fecal lipocalin – a neutrophil protein that binds bacterial siderophores and is associated with low-grade inflammation (Chassaing et al., 2015) – which was increased in the group of colonized mice fed the FF diet compared to those fed FR (Fig. 4E). A second readout, colon length, revealed shorter colons in colonized FF fed mice and other SM colonized groups when compared to colonized FR fed mice or GF mice on either diet (Fig. 4F). Additional analysis of host cecal tissue global transcriptional responses failed to reveal large-scale changes in the host; although, some compensatory responses were suggested by pathway analysis, which illuminated several immune responses as altered in the colonized FF fed mice (Fig. 4G, Fig. S5C, Table S7). Collectively, the data described above indicate that fiber-restricted, colonized mice experience erosion of the mucus barrier and some altered intestinal responses, albeit without overt signs of disease.
A fiber-deprived gut microbiota promotes heightened pathogen susceptibility
Since the mucus layer is a critical barrier against both commensal microbes and invading pathogens, we next hypothesized that the reduction in thickness associated with microbiota activity during low-fiber conditions would increase pathogen susceptibility. To test this idea, we chose the attaching/effacing pathogen Citrobacter rodentium (Cr) because it must traverse the mucus layer to access the epithelium and cause colitis (Collins et al., 2014). Towards this point, a previous study demonstrated that mice genetically lacking the dominant colonic mucin glycoprotein (Muc2−/−), but not wild-type mice, develop lethal colitis following infection with Cr, highlighting that the mucus layer is an important initial barrier to this pathogen (Bergstrom et al., 2010).
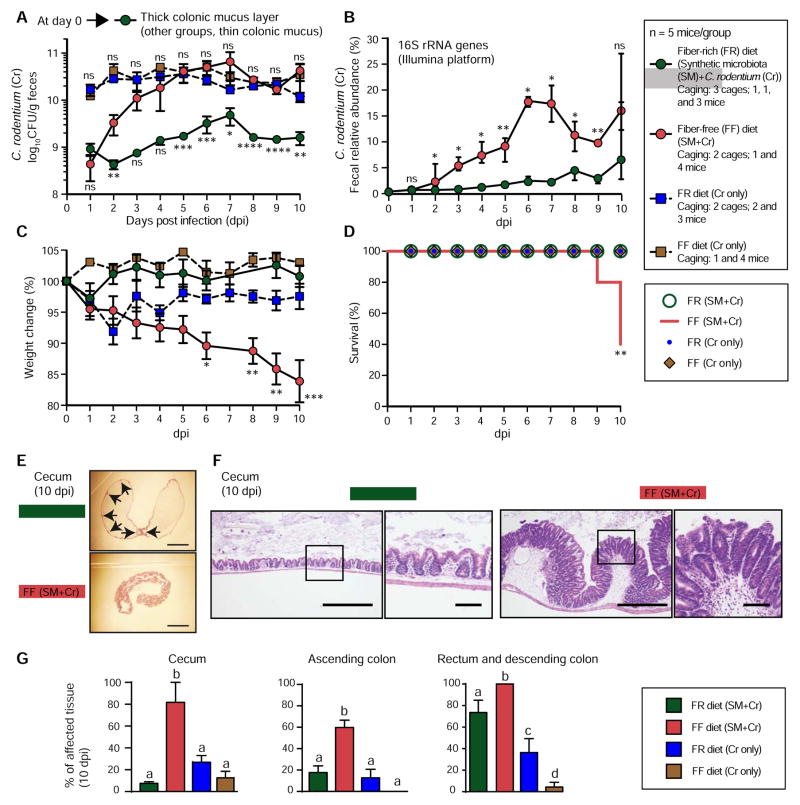
Therefore, we recreated the previously observed diet-modulated thick and thin mucus layer phenotypes in gnotobiotic mice, and infected both groups with Cr (Fig. S6A). To control for diet-specific effects on Cr pathogenesis in the absence of our SM, we infected two additional groups consisting of germfree (GF) mice fed a priori (~4 weeks before infection) the same FR or FF diets. We collected fecal samples each day post Cr infection to measure changes in pathogen colonization by both selective plating for Cr and 16S rRNA gene analysis (Figs. 5A,B, Fig. S6A). In the two groups colonized by the synthetic microbiota, Cr levels gradually increased but were significantly higher beginning at day 2 in mice fed the FF diet, and remained ~10-fold higher thereafter.
Figure 5. Fiber-deprived gut microbiota contributes to lethal colitis by Citrobacter rodentium.
(A) Fecal C. rodentium levels over time. Data are shown as average ± SEM. Student’s t-test; FR (SM+Cr) group vs. FF (SM+Cr) group (bottom statistics labels) and FR (Cr) vs. FF (Cr) (top statistics labels). Data in Figure 5A–5G are from Experiment 2B.
(B) Relative abundance of C. rodentium in fecal samples over time. Data are shown as median ± IQR. Wilcoxon test.
(C) Weight changes in the four groups of mice. Values are shown as average ± SEM. One-way ANOVA, FF diet group (with SM) vs. other groups.
(D) Survival curves for the four groups of mice. One-way ANOVA with Tukey’s test.
(E) Representative images of unflushed ceca after Hematoxylin and Eosin (H/E) staining highlighting major differences in hyperplasia (indicated with arrows in the FR group, where hyperplasia is patchy and infrequent). Scale bars, 5 mm.
(F) Images of representative H/E stained colonic thin sections depicting differences in hyperplasia between two groups. Scale bars: low power, 500 μm; high power, 50 μm.
(G) Measurements of inflamed tissue area in different intestinal segments. n = 5 mice/group except that n = 4 mice were used for FF (SM+Cr) group. Values are shown as average ± SEM. Statistically significant differences are shown with letters within each intestinal segment; p < 0.0002; One-way ANOVA with Tukey’s test.
The dramatic diet-specific increase of Cr levels in SM-colonized mice fed the FF diet was accompanied by weight loss that was specific to this group (Fig. 5C). Importantly, both GF+Cr groups that had high pathogen levels (Fig. 5A) failed to exhibit similar weight loss, illuminating that the pathogen alone is insufficient for this effect on either diet. The higher pathogen burdens in colonized mice fed the FF diet were associated with multiple signs of morbidity such as hunched posture and inactivity. Notably, by 10 days post infection, 60% of the mice from the SM colonized FF group had to be euthanized due to ≥20% loss of body weight (Fig. 5D). Mice in the other 3 groups did not show similar morbidity.
Histological scoring of the cecal and colonic tissue revealed that the SM-colonized FF diet group experienced inflammation that covered a significantly more expansive surface area (Figs. 5E–G). An exception was the descending colon/rectum, which showed larger areas of inflamed tissue in both FR and FF groups; although, the FF group was still significantly higher and 100% affected. When the tissue was inflamed, the level of hyperplasia was similar across all four groups (Fig. S6B). Importantly, there were overall lower levels of inflamed tissue in both of our GF+Cr control groups. To determine if increased mucus production post infection could explain the lower disease observed in the GF+Cr groups, we measured thickness of the colonic mucus and found that Cr triggered only a slight increase in GF mucus thickness; whereas the thick mucus layer associated with the FR diet in the context of microbiota colonization persisted (Fig. S7A,B).
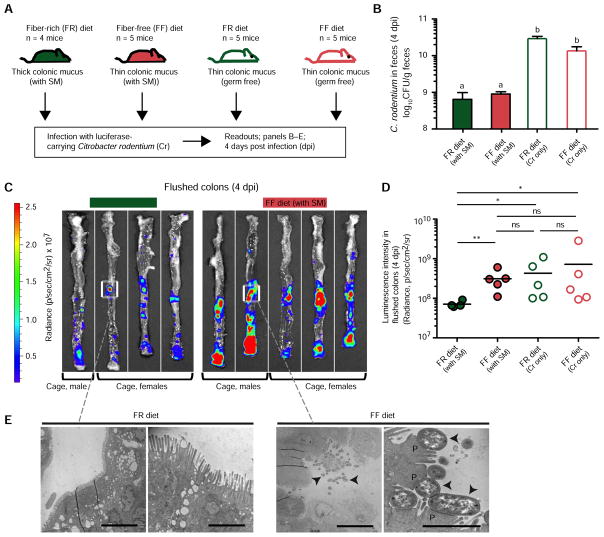
Based on the above results, we further hypothesized that the increased area of inflamed tissue in FF mice was due to earlier and increased pathogen access due to the microbiota-degraded mucus layer. To test this idea, we infected the same four treatment groups (SM-colonized or GF mice, fed either FR or FF diets) with a luciferase-expressing Cr strain (Fig. 6A). At 4 days post infection, we sacrificed all mice and conducted bioluminescent imaging of the colons after flushing out the luminal contents. In support of our hypothesis, and despite having similar levels of fecal Cr in FR- and FF-fed SM mice (Fig. 6B), we saw significantly higher pathogen signal adherent to the colonic tissue of SM colonized mice fed the FF diet as compared to those fed FR (Figs. 6C,D). The higher levels of attached Cr in FF fed SM mice were further validated by transmission electron microscopy, revealing increased appearance of the attaching and effacing lesions, pedestals and loss of microvilli that is typically associated with Cr infection (Fig. 6E). Notably, GF+Cr mice on either diet displayed similarly high adherent bacterial signal as the FF-fed SM mice (Fig. 6D, Fig. S7C). Taken together, these results suggest that the pathogen can more quickly traverse the thin colonic mucus layers in GF mice (irrespective of diet) and SM-colonized mice fed the FF diet. However, the commensal microbiota is also required in the context of increased pathogen access to elicit more severe disease, possibly by provoking co-inflammatory responses.
Figure 6. Fiber-deprived gut microbiota promotes faster C. rodentium access to the colonic epithelium.
(A) Experimental setup for luminescent C. rodentium experiment (Experiment 4).
(B) Fecal burdens of C. rodentium at 4 dpi. Data are shown as averages ± SEM; statistically significant differences are shown with different letters (p < 0.001). One-way ANOVA with Tukey’s test.
(C) Bioluminescence images of flushed colons showing the location and intensity of adherent C. rodentium colonization.
(D) Quantified bioluminescence intensities of C. rodentium from panel C and Fig. S7C. Kruskal-Wallis One-way ANOVA with Dunn’s test.
(E) Transmission electron microscopy images of the representative colonic regions from flushed colons; arrowheads denote individual C. rodentium cells and “P” denotes epithelial pedestals in high power/FF image. Scale bars, low power views 10 μm and high power views 2 μm.
See also Fig. S7.
DISCUSSION
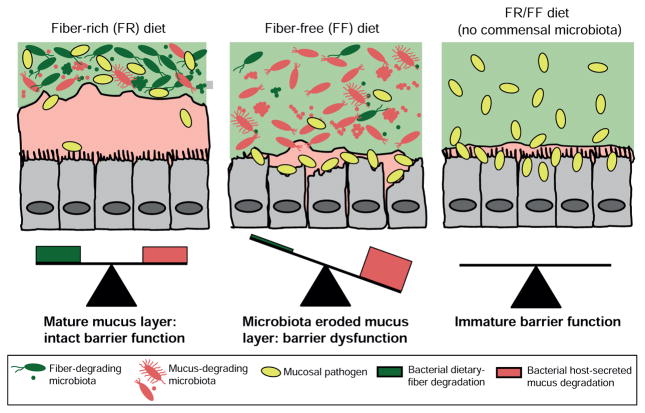
The health benefits of fiber consumption have been purported for decades, yet the influence of many different chemical and physical forms of fiber polysaccharides on the gut microbiota, and ways through which gut bacteria digest, sequester and share these chemically complex nutrients, are just now being unraveled in detail (Cuskin et al., 2015; Rakoff-Nahoum et al., 2016). Aside from loss of beneficial SCFA production, microbiota-mediated mechanisms that connect low fiber intake to poor gastrointestinal health have not been described. Using a gnotobiotic mouse model, our study provides a mechanism by which a diet deficient in complex plant fiber triggers a synthetic gut microbiota to feed on the colonic mucus layer that acts as a primary barrier against invading pathogens (Fig. 7).
Figure 7. Model of how a fiber-deprived gut microbiota mediates degradation of the colonic mucus barrier and heighted pathogen susceptibility.
Schemes derived from results shown in Figs. 1–6 illustrating the balance between fiber degradation and mucus degradation in FR diet-fed mice; whereas an FF diet leads to proliferation of mucus-degrading bacteria and microbiota-mediated degradation of the colonic mucus layer. The latter results in more severe colitis by C. rodentium.
Our findings reveal important implications regarding how our immediate diet history may modify susceptibility to some enteric diseases. Our approach highlights the power of using a tractable synthetic human gut microbiota, in which the individual members can be characterized or manipulated to support functional interpretations. We demonstrate that fiber deficiency allows the subset of mucin-degrading bacteria to increase their population and express mucin-degrading CAZymes to access mucin as a nutrient. While the ability to annotate CAZyme functions is well developed vis-a-vis many other metabolic functions that are important in the microbiome (El Kaoutari et al., 2013), there are still substantial ambiguities in connecting such predictions with precise catalytic roles. Here, we not only leverage knowledge of the substrate and enzyme specificities associated with some of the well-studied species in our SM (Table S4 and references therein), but we also employ new in vitro growth and transcriptional profiling experiments for key mucus-degrading bacteria (B. caccae and A. muciniphila). Our results point out a poignant example of how this evolving “bottom up” approach can increasingly resolve functional resolution in complex microbial systems: only very recently, the single B. ovatus GH98 enzyme that is increased in the FR diet (leftmost bar, Fig. 3A, top) was shown unequivocally to be an endo-β-xylosidase (Rogowski et al., 2015). Prior to this finding, the GH98 family was only known to contain blood group antigen-cleaving endo-α-galactosidases – a function that could have confusingly been associated with mucus O-glycan metabolism instead of its proper target, the plant fiber xylan.
Our results shed important light on the nature and amount of fiber that is required for the health of the colonic mucus layer. The prebiotic diet, which contains purified soluble fibers that are similar to common prebiotics (e.g., inulin, arabinoxylan, β-glucan), could not mitigate microbial erosion of the mucus barrier despite having a clear impact on the cecal community transcriptome. Since the FR diet contains complex plant fiber in its natural form (intact plant cell walls) and at higher concentration (~15% vs. 10% in the Pre diet), we cannot determine which variable (form or amount) is most important. However, the defined FF diet provides an ideal platform to which purified polysaccharides and even individual food items can be added to separately test both of these parameters for their ability to alleviate mucus degradation. Such an approach could help to design dietary therapeutics and next-generation prebiotics and will be particularly powerful given our existing knowledge of which polysaccharides the non-mucin-degrading species target (Fig. 1A) and our ability to implant new species with other defined functionalities in the SM.
The present work highlights that the gut microbiota plays significant positive and negative roles in the pathogenesis of C. rodentium. Whereas we previously showed that the presence of a microbiota blocks colonization by C. rodentium unless it possesses virulence traits (Kamada et al., 2012), here we demonstrate that diet-specific modulation of the gut microbiota can facilitate pathogen colonization and a fiber-deprived microbiota enhances disease susceptibility. Since the colonic mucus layer is an early barrier that a pathogen must transit (Collins et al., 2014), our data illustrate that a fiber-deprived microbiota has profound effects on the susceptibility to a gastrointestinal pathogen via reduction of this barrier. The contribution of mucus degradation to heightened pathogen susceptibility in our study is surprisingly parallel to a previous report that found a similar level of lethal colitis in mice with a genetically ablated (Muc2−/−) mucus layer (Bergstrom et al., 2010). From this perspective, it is striking that a dietary alteration in wild-type mice can imitate the phenotype of a mutation as severe as Muc2 loss. Given that Muc2 knockout mice experience inflammation and eventual colorectal cancer, it is reasonable to conclude that prolonged diet-driven mucus layer loss could result in similar outcomes. In this context, it is worth noting that higher levels of mucolytic bacteria have been found in IBD patients (Png et al., 2010). In light of our observations that mice subjected to intermittent (daily or 4-day) dietary fiber deprivation exhibit thinner mucus, it will be critical in future studies to investigate the impact of periodic fiber deprivation, which is more like real human dietary habits, on the status of the mucus layer and the many downstream health effects that may be connected to mucus barrier dysfunction.
Taken together, our findings support a model in which dynamic interactions between dietary fiber and metabolism of a synthetic microbiota composed of commensal bacteria influence the status of the colonic mucus layer and susceptibility to pathogens that traverse this barrier (Fig. 7). The current findings are likely applicable to gut microbial communities with higher numbers of species: three previous studies (see also Introduction) involving rats with native microbiota and mice with transplanted human gut microbiota found a correlation between fiber-deficient diets and thinner colonic mucus layer. However, it remains to be investigated whether a thin colonic mucus layer together with a complex microbiota would contribute to enhanced pathogen susceptibility or how this effect might vary between individual microbial communities. Moreover, to understand whether microbial degradation of the colonic mucus is required for its secretion by the host in order to achieve a thicker mucus layer, future experiments need to address the effects on the mucus thickness after exclusion of the 4 validated mucus degraders from our synthetic microbiota. Finally, since the strains used here are of human origin, and given the significant structural overlap between human and murine mucin (including glycosylation) (Johansson et al., 2013) and that C. rodentium uses similar pathogenesis mechanisms as human pathogenic E. coli strains, it is likely that such diet-induced disease susceptibility would extend to humans. Since E. coli infections are associated with high morbidity (Kaper et al., 2004) and health-care cost, our study emphasizes the need to consider a dietary perspective in fully understanding their transmission. With this in mind, efforts to find the optimal combinations of natural or prebiotic fiber polysaccharides and the minimum intake required to restore the integrity and resilience of the colonic mucus layer should be paramount.
METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information may be obtained from the Lead Contact Eric C. Martens (Email: emartens@umich.edu; address: University of Michigan Medical School, Ann Arbor, Michigan 48109, USA).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Gnotobiotic mouse model and diet treatments
All animal experiments followed protocols approved by the University of Michigan, University Committee for the Use and Care of Animals. Germfree male and female wild type Swiss Webster mice were colonized at 8–9 weeks of age and none of these mice were involved in any previous experiments/treatments. Mice were housed alone or in groups as appropriate for gender, litter and diet requirements and provided ad libitum with autoclaved distilled water and the diets described below.
Identities and culture purity of the bacterial species in the synthetic gut microbiota were confirmed by sequencing their 16S rRNA genes, followed by comparison to sequences in public databases. Bacteria were grown in their respective media (Table S1) for community assembly or in vitro growth evaluation on carbohydrates. Each individual bacterial member of the SM was grown anaerobically (atmosphere 85% N2, 10% H2, 5% CO2) in its respective medium (Table S1) at 37°C with final absorbance (600nm) readings ranging from about 0.5 to 1.0. Bacterial cultures were mixed in equal volumes and each individual inoculum sealed in its own tube with anaerobic headspace. Each mouse was gavaged with 0.2 ml of this mixture (freshly prepared each day) for three consecutive days at nearly the same time of the day.
Fiber-free (FF) and Prebiotic (Pre) diets were sterilized by gamma irradiation and the Fiber-rich (FR) diet (LabDiet 5010; autoclavable rodent diet) was sterilized by autoclaving. The FF diet was manufactured by Teklad/Envigo (WI, USA) and, as previously described (TD.140343) (Kamada et al., 2012), consisted of a modified version of Harlan TD.08810 in which starch and maldodextrin were replaced with glucose. The Pre diet was a new formulation based on the FF diet with 2.1% of a purified polysaccharide mixture (Table S1) added along with 10% cornstarch (each replacing an equivalent amount of glucose).
On day 14 after colonization (Figure 1B), mice were randomly assigned to groups by a technician, who was not aware of the details of the treatment groups. The mice were sometimes caged separately even within individual groups. For dietary oscillations, mice from their respective cage were transferred to a different cage with another diet. Bedding was replaced in each cage before the mice were transferred. To minimize the potential for circadian effects, the oscillation was carried out at nearly the same time of the day (±1.0 h between different days) and fecal samples were collected just prior to their transfer to another cage containing a different diet. The fecal samples were immediately stored at −20 °C until further use.
Citrobacter rodentium infection
A kanamycin (Km)-resistant wild-type C. rodentium strain (DBS120) and a luciferase-expressing strain of C. rodentium (DBS100; resistant to ampicillin (Amp)) were used (Kamada et al., 2012). Each mouse was gavaged with 0.2 ml of culture grown aerobically overnight at 37 °C (~109 CFU grown in Luria-Bertani broth without antibiotics). The exact same culture was used to gavage all mice in a single experiment to rule out effects of growth variation on pathogenesis. For experiments with luciferase expressing C. rodentium, mice were fed the FF diet for nearly the same duration (39 days instead of 42) as for the other experiment (Experiment 2B, Figure 5), prior to infecting them with C. rodentium. For the experiment with germfree (GF) mice, two groups of mice were separately pre-fed the FR and FF diets for ~4 weeks prior to infection with luciferase-expressing C. rodentium or the wild-type C. rodentium.
Formulation of the synthetic microbiota
We selected 12/14 bacterial species (Figure 1A) from the list of the most common/frequent 75–89 species in the human gut (Qin et al., 2010). Moreover, the selection of 14 species was based in part on carbohydrate utilization abilities of a larger pool of ~350 strains that were screened on the same platform as shown in Figure 1A (Urs, Pudlo and Martens, unpublished). Based on the in vitro assays, our synthetic microbiota (SM) is not biased towards mucin-degrading bacteria, as only 4/14 species possess this ability (Figure 1A).
METHOD DETAILS
Experimental design
A total of four gnotobiotic animal experiments (Experiments 1–4; also mentioned in figure legends) were performed – details of the experimental replication are provided in the corresponding figure legends. Both male and female mice were randomly used depending on the availability of animals. Gnotobiotic Experiment 1 contained 2 male mice in Fiber-rich (FR) group, 2 male mice in Fiber-free (FF) group and 1 male mouse in Prebiotic (Pre) group; all other animals in Gnotobiotic Experiment 1 were females. Gnotobiotic Experiment 2A and 2B had all male mice. All animals in Gnotobiotic Experiment 3 were females. Gender details of the animals in gnotobiotic Experiment 4 are shown in Figure 6 (both males and females were used). For infection with wild-type C. rodentium in germfree (GF) mice, all male mice were used. Gender details of GF mice used for infection with luciferase-expressing C. rodentium are included in Figure S7 (both males and females were used). Finally, all GF mice used for measurement of the colonic mucus layer (Figure 4C) were females. The researchers were not blinded to the identities of the treatment groups; however, the technician who assigned individual gnotobiotic animals to different treatment groups was not aware of the experimental details. Measurements of the colonic mucus layer were single blinded (see details below in the relevant section). The pathologist who devised the inflammation-scoring rubric was not blinded, and the pathologist who performed the histology scoring and the technician who performed electron microscopy were blinded for the identities of the treatment groups (see below for details of the methods). No data were excluded from the final analysis.
Sample size estimations were performed as follows in consultation with a statistician. Based on previous studies it was assumed an effect size (ratio of mean difference to within group standard deviation) of 3 would be reasonable for readouts such as mucus layer measurements, enzyme assays and measurement of transcript changes. With 3 animals in each group and a 5% significance level, two-sided, this would yield a power of 78% for the t-test. Therefore, for some of the feeding groups (those alternated between different diets), 3 animals were used. However, for other feeding groups that were more important for the central research question of the study (e.g., constant feeding on Fiber-rich (FR) and Fiber-free (FF) diets), at least 4 animals were used to obtain higher power. For Citrobacter rodentium infection experiments, in most cases 5 animals per group were used based on results of our previous study (Kamada et al., 2012).
Sample processing for animal experiments
All animals were killed using CO2 asphyxiation followed by cervical dislocation. The gastrointestinal tracts were quickly removed. The colons were gently separated, by cutting at the cecum-colon junction and the rectum, and immediately preserved in Carnoy’s fixative (dry methanol:chloroform:glacial acetic acid in the ratio 60:30:10) with slight modifications to a previous protocol (Johansson and Hansson, 2012). Note that the Carnoy’s fixative was made fresh with anhydrous methanol, chloroform and glacial acetic acid. The colons were fixed in Carnoy’s solution for 3 h followed by transfer to fresh Carnoy’s solution for 2–3 h. The colons were then washed in dry methanol for 2 h, placed in cassettes and stored in fresh dry methanol at 4 °C until further use. Cecal contents from each animal were divided into replicates; instantly flash-frozen in liquid nitrogen and were stored at −80 °C until further use. Immediately after squeezing out the cecal contents, the cecal tissues were transferred in separate screw-cap tubes and were rapidly flash-frozen in liquid nitrogen, followed by their storage at −80 °C until further use. Lengths of colons were measured immediately after fixation in Carnoy’s solution by photographing the colons in a reference cassette of identical size, followed by length measurement in Image J.
Purification of mucin O-glycans
Mucin O-glycans were purified from porcine gastric mucus as previously described in Martens et al., 2008, albeit with several modifications. Porcine gastric mucin (Type III, Sigma, USA) was suspended at 2.5% w/v in 100 mM Tris (pH 7.4): the mixture was immediately autoclaved for 5 min to increase solubility and reduce potential contaminating glycoside hydrolase and polysaccharide lyase activity, then cooled to 55°C. Proteinase K (Invitrogen, USA) was added to a final concentration of 0.1 mg/ml and the suspension was incubated at 55°C for 16–20 h with slow shaking. The proteolyzed solution was subsequently centrifuged at 21,000 × g for 30 min at 4°C to remove insoluble material, and NaOH and NaBH4 were added to final concentrations of 0.1 M and 1 M, respectively. This solution was incubated at 65°C for 18 h to promote selective release of O-linked glycans (mucin O-glycans and GAGs) from mucin glycopeptides by alkaline β-elimination. The pH was subsequently decreased to 7.0 with HCl and the neutralized mixture centrifuged at 21,000 × g for 30 min at 4°C, and then filtered through a 0.22 μm filter (Millipore) to remove remaining insoluble material. The filtrate was exhaustively dialyzed (1 kDa cutoff) against deionized distilled H2O to remove salts and contaminating small molecules. The collected mucosal glycans were further fractionated using anion exchange chromatography by passing them twice over a DEAE-Sepharose (Sigma) column (325 ml bed volume; equilibrated in 50 mM Tris 7.4; gravity flow). The flow through (neutral fraction) was collected and the column washed with 1L of 50 mM Tris, pH 7.4. This fraction was used in all growth experiments and was further prepared by dialyzing against ddH2O (1 kDa cutoff), lyophilized and resuspended in ddH2O at 20 mg/ml.
Bacterial growth assays in a custom carbohydrate array
All species except Desulfovibrio piger were evaluated in a custom carbohydrate array (n = 2 replicate cultures per glycan); D. piger failed to grow in any of the tested minimal media, but is not predicted to have extensive carbohydrate-degrading capacity based on its small genomic complement of only 30 carbohydrate active enzymes. The custom carbohydrate array was formulated according to Martens et al., 2011, but with a few modifications included in the following protocol: flat bottom 96-well plates (Costar) were used, to which 100 μl of a 2x concentrated solution (prepared in Milli-Q water) of each of the sterilized carbohydrate stocks (Table S1) were added. The plates were then transferred to the anaerobic chamber (10% H2, 5% CO2 and 85% N2) and were allowed to equilibrate with the anaerobic atmosphere for ~3–4 h. Growth assays for all carbohydrates were carried out in non-adjacent duplicates and all growth arrays contained two non-adjacent water only negative controls that were checked to ascertain that other medium components without added carbohydrates did not yield detectable growth. The cultures for the inoculation were grown overnight in their respective minimal media (MM)/regular growth media at 37°C under an anaerobic atmosphere (10% H2, 5% CO2 and 85% N2). MM for some bacterial species were used from previous studies, and for other members of the synthetic microbiota, MM with novel formulations were devised (see Table S1 for compositions of all growth media used in this study). MM were pre-reduced in the anaerobic chamber overnight by loosening the lids of the glass bottles containing the MM. 1 ml of the culture was centrifuged and the pellet was recovered – note that the centrifugation was performed inside the anaerobic chamber. The pellet was washed 2 times in the respective MM in order to remove carried over carbohydrates from the culture media and was then resuspended in 1 ml, 2x concentrated MM without any carbohydrates. This 1 ml culture was used to inoculate 50 ml of 2x concentrated MM without carbohydrates at a 1:50 ratio. 100 μl of the resulting cultures were then added to the individual wells of the carbohydrate solutions in the 96-well plates, resulting in 200 μl of final volumes. A gas permeable, optically clear polyurethane membrane (Diversified Biotech, USA) was then used to seal the well plates under the same anaerobic atmosphere. Next, the well plates were loaded in a Biostack automated plate-handling device (Biotek Instruments, USA) place inside the anaerobic chamber, which was coupled with a Powerwave HT absorbance reader (Biotek Instruments, USA).
Absorbance values were measured at 600nm (A600) at an interval of 10 min over 96 h for all species, except for Akkermansia muciniphila, for which the absorbance was measured over 144 h, owing to its relatively slow growth on mucin O-glycans (Figure S1B). To construct the heat map containing relative growth values (Figure 1A), absorbance data of all bacterial species were normalized as follows: only carbohydrate growth assays for which both replicate cultures produced an increase in absorbance of more than 0.1 were scored as positive (all other values were set to 0). Next, the maximum change in absorbance was normalized within each individual species by setting its best growth to 1.0 and normalizing all other positive growths to this maximum value (normalized values were thus between 0 and 1.0). Finally, growth on each substrate was normalized across species by setting the maximum (previously normalized) growth value on that substrate to 1.0 and then adjusting the growth values for other strains on that same substrate relative to the maximum value, yielding final normalized values between 0 and 1.0. Both raw and normalized values are provided in Table S1.
To perform RNA-Seq analysis on pure cultures, A. muciniphila and Bacteroides caccae were grown anaerobically in their respective minimal media (Table S1). A. muciniphila was grown separately on two different substrates N-acetylglucosamine (5 mg/ml final concentration) and purified mucin O-glycans (10 mg/ml final concentration). Cultures of A. muciniphila were grown to mid-log phase (A600 values between 0.45–0.6). Cultures of B. caccae were grown separately on glucose and mucin O-glycan as the carbon sources (10 mg/ml final concentration for both sugars) to mid-log phase (OD values between 0.7–0.8). Cultures of both species were treated with RNAprotect (Qiagen, USA) according to the manufacturer’s instructions. The RNAprotect treated bacterial pellets were stored at −80°C until extraction of RNA. Two replicate cultures per glycan (with closely matching ODs) were performed for each of the two bacterial species.
Citrobacter rodentium quantification
To determine the CFUs of C. rodentium, freshly collected fecal samples were weighed and homogenized in cold phosphate-buffered saline and were plated on LB agar plates with 50 μg/ml Km (for strain DBS120) or 200 μg/ml Amp (for strain DBS100) at serial dilutions up to 10−9. The plates were incubated aerobically overnight at 37 °C. Killing of E. coli HS, the only facultative anaerobe in our SM, to Km and Amp was confirmed by plating it on LB agar with Km or Amp.
Extraction of nucleic acids
DNA from fecal samples was isolated using the MoBio PowerSoil Isolation Kit (MoBio Laboratories, USA) adapted for use in the epMotion 5075 TMX or the DNA extraction protocol used for Collinsella aerofaciens (mentioned below). DNA was extracted from the bacterial pure cultures using DNeasy Blood & Tissue Kit (Qiagen, USA), except that the following bead beating and phenol–chloroform extraction protocol, was employed to better extract DNA from C. aerofaciens: 1–2 ml of the overnight grown culture was centrifuged and the resulting pellet was combined with acid-washed glass beads (212–300 μm; Sigma-Aldrich, USA), 500 μl Buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), 210 μl SDS (20% w/v, filter-sterilized) and 500 μl phenol:chloroform:isoamyl alcohol (25:24:1, pH 8.05; Thermo Fisher Scientific, USA). A Mini-BeadBeater-16 (Biospec Products, USA) was used to disrupt the bacterial cells for 5 min at room temperature, which was followed by cooling the samples for 1–2 min on wet ice. The samples were then centrifuged and the aqueous phase was recovered. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the aqueous phase and was mixed with the aqueous phase by gentle inversion. After centrifugation (12,000 rpm, 4°C, 3 min), the aqueous phase was removed. Next, 500 μl of pure chloroform was added to the aqueous phase, mixed by inversion and the tubes were centrifuged (12,000 rpm, 4°C, 3 min). The aqueous phase was removed in fresh tubes and 1 volume of −20°C chilled 100% isopropanol and 1/10 volume 3 M sodium acetate (pH 5.2) were added to the aqueous phase. The samples were mixed by gentle inversion and incubated at −20°C for 1 h, centrifuged for 20 minutes (12,000 rpm, 4°C) and the supernatants were discarded. The pellets were washed in 70% ethanol (v/v, prepared in nuclease-free water), air dried and then resuspended in nuclease-free water. The resulting DNA extracts were purified by using DNeasy Blood & Tissue Kit (Qiagen, USA).
RNA was extracted from cecal contents using a standard phenol–chloroform method with bead beating as mentioned earlier (Sonnenburg et al., 2006), but with a few modifications: 1 ml of RNAprotect (Qiagen, USA) stored at room temperature was added to 200–300 mg of cecal contents stored at −80 °C, followed by thawing the cecal samples on wet ice. After the cecal contents were thawed, 250 μl acid-washed glass beads (212–300 μm; Sigma-Aldrich, USA) were added to the samples. Next, 500 μl of a solution of Buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), 210 μl of 20% SDS (filter sterilized) and 500 μl of phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.3; Fischer Scientific, USA) were added to the samples. The mixture was then bead beaten (instrument same as above) for 5 min and centrifuged at 4 °C (3 min at 13000 rpm). The aqueous phase was recovered and mixed with 500 μl of the aforementioned phenol:chloroform:isoamyl alcohol solution. Afterwards, the mixture was centrifuged again at 4°C (3 min at 13,000 rpm) and the aqueous phase was recovered. 1/10 volume of a 3M sodium acetate (pH: 5.2) and 1 volume of −20°C chilled ethanol were added to the aqueous phase. The resulting solution was then mixed by gentle inversion and incubated for 20 min on ice. Afterwards, the mixture was centrifuged at 4°C (20 min at 13,000 rpm). The pellet was recovered and washed twice in 500 μl of cold 70% ethanol. The mixture was centrifuged at 4°C (5 min at 13,000 rpm) and the RNA pellet was recovered, air dried and then resuspended in nuclease-free water. The RNA extracts were then purified using an RNeasy Mini kit (Qiagen, USA) according to the manufacturer’s protocol. During extraction of RNA from the cecal contents, a portion (~100–200 μl) of homogenized material immediately after bead beating was removed and stored at −80°C for extraction of DNA. To extract DNA from these cecal-content derived samples, DNA extraction protocol described above (used for C. aerofaciens) was used, except that bead beating and inclusion of glass beads were skipped. RNA was extracted from the RNAprotect-treated cell pellets of bacterial pure cultures of B. caccae using RNeasy Protect Bacteria Mini Kit. For extraction of RNA from RNAprotect-treated cell pellets of A. muciniphila, the RNA extraction protocol used for cecal contents (see above) was used, except that the samples were not treated with RNAprotect after thawing. RNA was extracted from cecal tissue by thawing the samples in the presence of RNAprotect (as described above for cecal contents), followed by homogenization (OMNI International) involving metal beads. RNA was then extracted with Trizol (Invitrogen, USA) according to manufacturer’s instructions. All RNA extracts were subjected to digestion of DNA using TURBO DNase (Ambion, USA) according to the manufacturer’s instructions..
Bioluminescence imaging and transmission electron microscopy
For bioluminescence imaging of the luciferase-expressing Citrobacter rodentium, GI tracts were removed and luminal contents were gently flushed with a syringe by passing phosphate-buffered saline (PBS) through the colon. The GI tracts were then cut open flat and rinsed in PBS to remove loosely attached luminal contents. Bioluminescence was visualized (identical exposure across all samples) and photographed using the IVIS200, Xenogen system. The colonic tissue sections showing highest luciferase intensity (from both Fiber-rich (FR) and Fiber-free (FF) diet fed colonized mice) were then immediately fixed in 2.5% glutaraldehyde prepared in 0.1 M Sorensen’s buffer (pH 7.4). Thereafter, the samples were treated with 1% osmium tetroxide in 0.1 M Sorensen’s buffer and were sequentially dehydrated in graded alcohols and propylene oxide, followed by infiltration in Spurrs or Epon. Ultrathin sections of the tissue samples were made using a diamond knife, stained and were visualized with a transmission electron microscope (Philips CM-100).
Laser capture microdissection
To perform laser capture microdissection (LCM) on colonic thin sections, 4 and 3 fecal masses were analyzed for the Fiber-rich (FR) and Fiber-free (FF) groups, respectively. Colonic thin sections that were deposited on microscope slides were deparaffinized in xylene followed by dehydration by isopropanol (see details in the immunofluorescence staining protocol below). The sections were stored overnight in a container with Drierite dessicant (Drierite, USA). LCM was carried out using a Veritas Microdissection instrument (Arcturus, USA). DNA was extracted from the microdissected samples using the Arcturus Pico Pure DNA extraction kit and the accompanying protocol. In order to perform Ilumina sequencing, 16S rRNA genes were amplified from the LCM-derived samples using a low biomass-optimized touch down PCR protocol as follows: denaturation at 95°C for 2 min; a total of 20 cycles with a touch-down program: denaturation at 95°C for 20 s, extension at 72°C for 5 min, annealing starting at 60°C for 15 s which decreased 0.3°C per cycle; a total of 20 cycles: extension at 72°C for 5 min, annealing at 55°C 15 s and extension at 72°C for 5 min; final extension at 72°C for 5 min. Note that a 5 min extension was used in order to reduce chimera development. Library preparation and sequencing were carried out using similar protocols described for fecal and cecal samples (see below).
Illumina sequencing and data analysis
PCR and library preparation were performed by the University of Michigan Microbial Systems Molecular Biology Lab as described by Kozich et al., 2013. The V4 region of the 16S rRNA gene was amplified using the dual-index primers described by Kozich et al. with a few modifications to the PCR assay, which are included in the following protocol. Each of these dual-index primers contains an Illumina adapter, an 8-nt index sequence, a 10-nt pad sequence, a 2-nt linker, and the V4 primers F515 and R806. These primer sequences are listed in Table S2. For the PCR assays, 5 μl of each of the 4 μM primers, 0.15 μl AccuPrime High Fidelity Taq polymerase (Thermo Fisher Scientific, USA), 2 μl of 10x AccuPrime PCR II buffer (Thermo Fisher Scientific, USA), 11.85 μl of sterile PCR-grade water and 1 μl of the DNA template were mixed. The PCR cycles started with a 2 min of denaturation at 95°C, followed by 30 cycles each consisting of 95°C for 20 s, 55°C for 15 s and 72°C for 5 min, followed by a final step of 72°C for 10 min. The amplicons were normalized to the lowest concentration of the pooled plates using a SequalPrep normalization plate kit (Thermo Fisher Scientific, USA). A KAPA Library Quantification kit for Illumina platforms (Kapa Biosystems, USA) was used to determine the library’s concentration and an Agilent Bioanalyzer high-sensitivity DNA analysis kit (Agilent, USA) was employed to determine the amplicon size. The amplicons were sequenced using an Illumina MiSeq with a MiSeq Reagent 222 kit V2 (Illumina, USA). The libraries were prepared following the Illumina protocol for 2nM libraries: ‘Preparing Libraries for Sequencing on the MiSeq’ (part 15039740, Rev. D).
Raw sequences were analyzed using mothur (v1.33.3) (Schloss et al., 2009). The following control samples were included: 1) DNA extracted from the fecal samples collected from germfree mice, 2) a mixture of extracted DNA from pure cultures of the members of the synthetic microbiota (DNA samples from each strain were mixed in equal amounts) and 3) PBS negative controls during DNA extraction and PCR amplification. Following sequence barcode-trimming, sequences were aligned to a custom reference database, consisting of the V4 16S rRNA region from each of the 14 bacterial members and C. rodentium. UCHIME (Edgar et al. 2011) was used to remove sequence chimeras. The R package ‘vegan’ was used to calculate the principal coordinates analysis (PCoA) from the Bray-Curtis dissimilarity index based on phylotype classification of the 14 bacterial members. Standard R commands and the R package ‘plyr’ (Wickham, 2011) were used to generate median values of relative abundance or change in relative abundance over time, and the Wilcoxon signed-rank test (two-sample comparisons) or the Kruskal-Wallis test (multiple groups) was used to determine significance as indicated due to the nonparametric distribution of relative abundance data. R was used to visualize relative abundance of bacterial members in different groups, over time as streamplots, or in heatmaps. Change in relative abundance over time was determined by subtracting the relative abundance of the specified microbial member from the day prior within each animal, over time. Change in relative abundance followed a parametric distribution, and student t-tests were used to calculate significant differences in the change of relative abundance between diet groups. The R package ‘ggplots’ was used to generate heatmaps visualizing the Percent of Maximum Abundance (POMA) as previously described (McNulty et al., 2013). For this, the relative abundance of the different species was normalized by their maximum abundance observed for a given species across all time-points from the given animal. A detailed list of commands used to analyze the data, including the commands used in mothur, are included in https://github.com/aseekatz/mouse.fiber. Raw sequences have been deposited in the Sequence Read Archive under the study accession SRP065682 (BioProjectID PRJNA300261).
Microbial RNA-Seq and CAZyme annotation
Microbial RNA-Seq was performed on pure cultures of A. muciniphila and B. caccae that were grown separately on mucin O-glycans and on the respective simple sugars (see above). For Bacteroides thetaiotaomicron, gene expression data from previous studies was utilized (Martens et al., 2008; Sonnenburg et al., 2005). To perform RNA-Seq on cecal samples, 3 samples each (out of 4) were randomly selected from Fiber-rich (FR) and Fiber-free (FF) diet groups and all three samples each from the Prebiotic diet (Pre), FR–FF daily oscillation and Pre–FF daily oscillation groups were utilized. To remove ribosomal RNA, samples were subjected to Ribo-Zero rRNA Removal Kits (Bacteria) (Epicentre, Illumina, USA) according to the manufacturer’s instructions. The resulting residual mRNA concentrations were quantified using Qubit RNA Assay Kit (Life Technologies, USA).
Libray preparation and sequencing of RNA-Seq libraries was carried out using the Illumina HiSeq platform and TruSeq adaptors. Samples were multiplexed in groups of 24 per lane (see Tables S4, S5, S6 for quantification of reads mapped to each sample). The resulting data in fastq file format were demultiplexed and mapped to the respective species genomes or community metagenomes using RPKM normalization and default parameters, and were further analyzed for fold-change and statistics (moderated t-test with Benjamini-Hochberg correction) within the Arraystar software package (DNAStar, USA). Mapping reads to all genes in the 14 species community was intended to retain the contributions of community member abundance shifts while mapping only to individual genomes was intended to normalize abundance shifts of the same species between conditions and isolate gene expression changes between conditions. The diet-specific behaviour of known B. thetaiotaomicron and B. ovatus genes involved in fiber polysaccharide degradation (Table S6) was used as internal validation that biologically relevant changes in gene expression were indeed being detected. As stated above, three biological replicates were analyzed for each of the in vivo dietary conditions used.
p-Nitrophenyl glycoside-based enzyme assays
p-Nitrophenyl glycoside-based enzyme assays were carried out on cecal samples stored at −80°C. The cecal samples were thawed on wet ice and 500 μl buffer (50 mM Tris, 100 mM KCl, 10 mM MgCl2; pH 7.25) was added to 22–67 mg of cecal contents. The buffer additionally contained the following additives: lysozyme (tiny amount of powder/100 ml buffer), TritonX (100 μl, 12%/100 ml buffer), DNases (tiny amount of powder/100 ml buffer) and protease inhibitor (one tablet of EDTA-free, Protease Inhibitor Cocktail, Roche, USA/100 ml buffer). After adding the buffer to cecal samples, the samples were sonicated with an ultrasonic processor for 45 s (9 cycles of 5 s sonication followed by a break of 10 s; 35% amplitude; using a tapered microtip of 3 mm) on ice. Sonicated samples were subjected to centrifugation (10,000 g, 10 min, 4°C). Supernatants (~400 μl) were carefully pipetted and were stored at −20°C until further use. The following nitrophenyl-linked substrates (Sigma-Aldrich, USA) were employed: Potassium 4-nitrophenyl sulfate, 4-nitrophenyl α-D-galactopyranoside, 4-nitrophenyl N-acetyl-β-D-glucosaminide, 4-nitrophenyl β-D-glucopyranoside, p-nitrophenyl α-L-fucopyranoside and p-nitrophenyl β-D-xylopyranoside. Protein concentrations in the supernatants were determined using Pierce Microplate BCA Protein Assay Kit (Thermo Scientific, USA). Some samples were diluted with the buffer (same buffer as above) to obtain a homogenous range of protein concentrations across all samples. 5 μg of total protein was used in the 150 μl reactions inside flat-bottom, 96-well plates (Costar) with 10 mM nitrophenyl-based substrate in the buffer (same buffer as above). Absorbance measurements (405 nm) were started immediately in a plate reader (Biotek, USA) at 37°C and absorbance values were recorded every minute for 6–12 h duration depending on linearity of the kinetic curve. The enzyme activities were determined by plotting a standard curve of known concentrations of 4-nitrophenol and measuring the OD values at 37°C.
Thickness measurements of the colonic mucus layer
Post Carnoy’s fixation, the methanol-stored colon samples (see above) were embedded in paraffin and thin sections (~5 μm) were cut and deposited on glass slides. Alcian blue staining was performed by the following protocol: 1) deparaffinization and hydration to distilled water, 2) Alcian blue solution for 30 minutes, 3) washing in running tap water for 2 minutes, 4) rinsing in distilled water, 5) dehydration (2X changes) and treatment with absolute alcohol (2X changes), 3 minutes each, 6) clearance in xylene (3X changes), 3 minutes each 7) cover with coverslip. To measure the thickness of the colonic inner mucus layer, thousands of partially overlapping photographs were taken from nearly the entire length of each colon based from the Alcian blue stained slides after cross-validation using anti-Muc2 staining (Fig. 4A,B main text). The images captured all of the available fecal masses of all mice, although this number was variable and there were generally fewer colonic fecal masses in mice fed the FF diet alone or in any combination. Image sample names were blinded by M.S.D. and M.W., and the thickness of the colonic sections were then measured by E.C.M. using ImageJ. Only regions in which the mucus layer was sandwiched between epithelium on one side and luminal contents on the other were used; care was taken to measure regions that represented the average thickness in each blinded image; 2–3 measurements per image were taken and averaged over the entire usable colon surface. See Fig. 4A,B for representative images in which the region measured as the inner mucus layer is delineated in both Alcian blue and anti-Muc2 staining.
Measurements in Cr infected mice were conducted exactly as described for non-infected mice above, with the exception that only distal colon–rectal tissue was considered as this was uniformly a site at which inflammation was high and C. rodentium would be present. Since only sections in which luminal contents that could be visualized adjacent to the mucus layer were considered, only a few measurements were obtained for a single SM-colonized infected mouse fed the FF diet due to the fact that all mice in this group were extremely morbid and not eating.
Quantitative PCR
In addition to Illumina sequencing of the 16S rRNA genes (V4 region), as a second approach to quantifying relative bacterial abundance in fecal samples, phylotype-specific bacterial primers were designed. The primers were designed against randomly selected genes that were checked for homology against the other 13 species in each case. These primer sequences are listed in Table S3A. The primers were tested for specificities against the target strain by comparing the primer and target gene sequences against sequences in public databases. Moreover, specificity of each primer was validated by the following three approaches: 1) by quantitative PCR (qPCR) against target species genome and melting curve analysis (for a single peak), 2) by qPCR for each primer set against a non-target template comprising of genomic DNA from the 13 bacterial species in our synthetic microbiota, 3) by performing qPCR against DNA extracted from the fecal samples of germfree mice feeding on the Fiber-rich diet. qPCR was carried out in 384 wells (with each plate including known concentrations of template DNA included to plot a standard curve). The qPCR analyses were performed using KAPA SYBR FAST qPCR Kits (KAPA Biosystems, USA) on Applied Biosystems (ABI) Real Time PCR instrument (ABI, USA). The amount of DNA was quantified by plotting a standard curve of varying DNA concentrations of the target template.
Quantification of short chain fatty acids
Cecal samples stored at −80°C were used to quantify short chain fatty acids (SCFAs). Samples were first thawed on wet ice. Then, an equivalent amount of MilliQ water was added (100 ul per 100 mg of material) to cecal contents (≥ 0.05 g) and the samples were thoroughly homogenized by vortexing for 1 minute. The samples were then centrifuged at 13,000 g for at least 3 minutes (or for a longer time depending on time required to obtain a tight pellet). The supernatant was pipetted and filtered through a 0.22 μm filter (Millex-gv 4mm SLGV004SL). Samples were kept on ice or frozen until quantification of SCFAs by high-performance liquid chromatography (HPLC). Some samples were diluted to obtain enough liquid to inject onto the HPLC, or in certain cases they were diluted so that they could be filtered. A Shimadzu HPLC with an Agilent HP-87X column was utilized for separating compounds, with a mobile phase of 0.01 N H2SO4, a flow rate of 0.6 ml/min, and a column temperature of 50°C. A UV detector set to a wavelength of 214 nm was used to measure concentrations.
Immunofluorescence staining
The immunofluorescence staining for Muc2 mucin was performed on the colonic thin sections after several modifications to the protocols from Johansson and Hansson, 2012 and an immunohistochemistry/tissue section staining protocol from BD Biosciences, USA (http://www.bdbiosciences.com). The sections were deparaffinized by dipping in 50-ml Falcon conical tubes filled with xylene (Sigma-Aldrich, USA) for 5 min, followed by transfer to another tube with fresh xylene for 5 min – care was taken to completely immerse the tissue material in the liquid (also in the subsequent steps). This was followed by two dehydration steps of 5 min each using 100% isopropanol contained in conical tubes. The slides were then washed by dipping in conical tubes containing MilliQ-water. The antigens were retrieved by placing the slides in a glass beaker with enough BD Retrievagen A (pH 6.0; BD Biosciences, USA) to cover the slides. The sections were then heated by microwaving and holding at about 89°C for 10 min (microwaving was repeated during this time, as required). The slides were then cooled for 20 min at room temperature. Afterwards, the slides were washed 3 times with MilliQ-water. Excess liquid was gently blotted away and a PAP pen was used to draw a circle around the tissue area, in order to better hold liquid on the tissue area during subsequent steps. Blocking was performed by immersing the slides into blocking buffer (1:10 dilution of goat serum (Sigma, USA) in 1x Tris-buffered Saline (TBS; 500 mM NaCl, 50 mM Tris, pH 7.4)) and incubating them at room temperature for 1 h. For the primary antibody staining, the tissue sections were covered in a 1:200 dilution Mucin 2 antibody (H-300) (original concentration: 200 μg/ml; Santa Cruz Biotechnology, USA) in the aforementioned blocking buffer and incubated for 2 h at room temperature. After the incubation step, the excess liquid was blotted away and the slides were rinsed 3 times in 1x TBS (in conical tubes) for 5 min each. The secondary antibody staining was performed by covering the tissue sections with a 1:200 dilution of Alexa Fluor 488 conjugated goat anti-rabbit IgG antibody (original concentration: 2 mg/ml; Thermo Fisher Scientific, USA) in blocking buffer and the sections were incubated for 1 h at room temperature in dark. The excess liquid was blotted away and the sections were rinsed twice for 5 min each using TBS. Next, the sections were stained for 5 min at room temperature in dark using a 10 μg/ml of DAPI solution diluted in 1x TBS (Sigma-Aldrich, USA). The sections were then rinsed with Milli-Q water and blotted dry. Finally, the sections were covered with ProLong Gold Antifade Mountant (Invitrogen, USA), covered with coverslips and the edges of the coverslips sealed with nail polish. The slides were kept at room temperature in dark for at least 24 h and then visualized by Olympus BX60 upright fluorescence microscope (Olympus, USA).
ELISA for fecal Lipocalin
Frozen fecal samples (−20°C) were used to determine the levels of fecal Lipocalin (LCN-2). The assays were performed within 30 days of sample collection. The samples were prepared as mentioned previously (Chassaing et al., 2015), with a few modifications in the sample prepration protocol: fecal samples stored at −20°C were thawed on wet ice and 6.9–67.7 mg of samples were separated in fresh tubes, to which 0.5 ml of 1% (v/v) Tween 20 (Sigma-Aldrich, USA) prepared in PBS was added. To get a homogenous suspension, the samples were vortexed for 20 min. The suspension was then centrifuged at 4°C for 10 min at 12000 rpm. Next, the supernatant was carefully recovered and stored at −20°C until the analysis. To measure the LCN-2 levels, a mouse Lipocalin-2/NGAL DuoSet ELISA kit (R & D Biosystems, USA) was employed and the manufacturer’s protocol was followed.
Tissue histology
To perform histology analyses on GI tracts of C. rodentium infected mice: first the intestinal segments (cecum and colon together) were fixed in Carnoy’s fixative for 3 h, followed by transfer to fresh Carnoy’s fixative overnight. Next, the samples were washed in 100% methanol (2x) for 30 min each, which was followed by washing in 100% ethanol (2x) for 20 min each. The samples were then stored in 100% ethanol at 4°C until further use. After 100% ethanol washes, the intestinal tissue samples were divided into 3 sections for histology: cecum, ascending colon, and the descending colon/rectum. These sections were embedded, processed and cut by an experienced histology core (Washington University, USA), and then stained with hematoxylin and eosin (H and E) prior to analysis. An unblinded experienced pathologist (T.S.S.) examined the slides from each of the groups to determine a viable readout. The best readout was determined to be the extent of epithelial area showing crypt hyperplasia. After a scoring rubric was devised, an independent blinded evaluator (C.A.H.) then measured the total length of each intestinal segment with a ruler in millimeters. Areas of increased crypt hyperplasia were then determined by microscopy and the lengths of these areas were measured as a percentage of the total epithelial length on a single slide.
Mouse microarray analyses
Mouse microarrays were carried out with Affymetrix Mouse Gene ST 2.1 strips. Expression values for each gene were calculated using a robust multi-array average (RMA) approach (Irizarry et al., 2003). Linear models were fitted to the data by employing the limma bioconductor package in R version 3.1.1. Note that selected probesets had a fold change greater than 1.5 and an FDR adjusted p-value of 0.05 or less. Data were analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) using the default parameters. Input data correspond to significantly detected genes with FDR < 0.05 and absolute log Fold-Change > 0.5. ILK signalling was the number 1 canonical pathway detected (p-value = 8.33E-07). TNF targets corresponded to the number 1 (overlap p-value = 2.64E-06) upstream regulator detected by IPA. The category ‘Immune response of cells’ was predicted as the most significantly activated (p-value = 6.83E-06 and Z-score = 2.316) in the inflammatory response category.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
All experimental analyses were conducted in consultation with a statistician. Unless otherwise stated in individual method sections above, all statistical analyses were performed using Prism 5.04 (GraphPad Software, Inc.), except statistics for colony forming units (CFU) for C. rodentium were performed in Excel. Statistically significant differences are shown with asterisks as follows: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001; whereas, ns indicates comparisons that are not significant. Numbers of animals (n) used for individual experiments, details of the statistical tests used and pooled values for several biological replicates are indicated in the respective figure legends. A two-tailed t-test was employed in all cases. Since generally the microbiome data did not follow a normal distribution, for these data a nonparametric test such as the Wilcoxon test was used. An exception was the data in Figure 2C, which generally followed a normal distribution and hence a t-test was used (see details in the section: Illumina sequencing and data analysis). For the other data that appeared normally distributed, a t-test was used; otherwise, a non-parametric (Mann-Whitney) test was used (for example for Figure 4E). Finally, ANOVA (parametric) and Kruskal-Wallis (nonparametric) methods were used to describe differences between more than two groups. For data in Figure 6D, a non-parametric approach with Dunn’s test and involving pairwise comparisons were employed.
DATA AND SOFTWARE AVAILABILITY
Accession numbers
16S rRNA gene sequences and metadata have been deposited under the BioProjectID PRJNA300261 and in SRA under SRP065682. The commands used to analyze the 16S rRNA gene data can be found online at the following link: https://github.com/aseekatz/mouse.fiber. RNA-Seq data have been deposited at NCBI under BioProjectIDs PRJNA310052, PRJNA310058, PRJNA310062. Mouse microarray data are deposited to NCBI Geo under accession numbers GSM2084849–55.
Supplementary Material
(A) A schematic displaying abilities of the SM to degrade a wide variety of dietary and host-derived polysaccharides and possible metabolic interactions between members of the SM. GAGs, Glycosaminoglycans.
(B) Representative growth curves of selected members of the SM on several polysaccharides and glycans as sole carbon sources (n = 2 for each glycan; values are shown as averages). The absorbance was measured every 10 minutes. See Table S1 for raw and normalized growth values and growth media descriptions for the 13 members of the SM evaluated for carbohydrate growth ability (all except D. piger).
Summary of growth values of cultures included in synthetic microbiota, details of growth media and diets.
Microbial relative abundances based on Illumina sequencing of 16S rRNA genes are provided for all 1,114 samples (data are from Experiments 1 and 2).
Primer sequences and microbial relative abundances, based on qPCR measurements employing phylotype-specific primers, are provided (data are from Experiment 1).
RNA-seq-based microbial community transcriptomic data is presented for glycoside hydrolase (GH), polysaccharide lyase (PL) and carbohydrate esterase (CE) families that exhibited altered expression between the FR/FF or Pre/FF treatment comparisons (data are from Experiment 1).
The data are based on RNA-seq (data are from Experiment 1).
Principal coordinates analysis (PCoA) of microbial community dissimilarity (Bray-Curtis) in fecal samples (collected according to Figure 1B) as determined by 16S rRNA-based sequencing (V4 region). Samples from both Experiments 1 and 2 are shown, with samples coded by experiment (top panel) or sex (bottom panel) and by cage (legend) (Experiment 1: n = 4 mice/group; Experiment 2: n = 7 mice/group).
(A) Relative abundance of indicated bacteria in mice over time subjected to various dietary regimes as determined by Illumina-based 16S rRNA sequencing (Experiment 1). An explanation for the inverse relationship between the relative abundances of D. piger and M. formatexigens on FR and FF diets is their competition for the same electron donor (hydrogen). The increased proliferation of mucin-degrading bacteria in the FF diet indicates higher degradation of sulfated mucin O-glycans, which is supported by transcriptomic and enzyme assay data shown in Figure 3. The corresponding release of additional sulfate would preferentially feed the sulfate-reducer D. piger, leading to production of the toxic metabolite hydrogen sulfide (Fig. S1A). Increased D. piger has also been observed in IBD patients (Gibson et al., 1991; Loubinoux et al., 2002), which could result from enhanced sulfate release by mucinolytic bacteria. Values are shown as medians ± IQR. n = 4 for FR and FF groups; n = 3 for Pre and 1-day FR/FF groups.
(B) Heat map showing Percent of Maximum Abundance (POMA) values of all species for two of the feeding groups from Illumina-based sequencing (see Figure 1B); n = 3 mice/group (Experiment 1; according to timeline in Figure 1B). See also Table S2.
Figures are based on RNA-Seq measurements of B. caccae (A) and A. muciniphila (B) responses in vitro (minimal medium with simple sugars or MOG) and in vivo (constant feeding or daily alternation of FR and FF diets). In vivo samples are from the entire cecal community at the end of Experiment 1. Gene transcripts that were increased >5-fold relative to the corresponding simple sugar references are included for each bacterium. Venn diagrams show overlap and differences of the transcripts between various groups. Numbers indicate the total differentially regulated gene count for a given sector, while number in parentheses denote numbers of carbohydrate-degrading enzymes (glycoside hydrolase, polysaccharide lyase or carbohydrate esterase families counted toward this number; sulfatases and carbohydrate binding module, CBM, families were not counted). Note that A. muciniphila shows less regulatory versatility as manifest by most of its upregulated enzymes being confined to the core (dark pink) sector containing all of the in vivo samples. This suggests that MOG only triggers a small percentage of this species’ O-glycan degrading responses in vitro; although 8 enzymes were also triggered in vitro. The corresponding histograms display frequencies of related enzyme families (shown with matching colors to their respective Venn sectors). Possible mucin-related degradative functions are given above each family-specific histogram bar. For in vitro samples, n = 2 for each MOG and simple sugar grown condition; for in vivo samples, n = 3 mice/group (Experiment 1).
(C) Schematic mucin O-glycan structures, from among ~102 that can be found on human and murine Muc2, with the sites at which various enzymes noted in panels A. and B. would be expected, or are known, to cleave. See also Tables S4 and S5 for in vivo and in vitro transcript data.
(A) Depictive histology images (Hematoxylin and Eosin of colonic thin sections) showing no overt signs of inflammation between the two dietary regimens (Experiment 1), in the absence of C. rodentium. Scale bars, 500 μm.
(B) Weight change in mice over time. Values are shown as averages ± SEM. n = 4 for FR and FF groups; and n = 3 for Pre group (Experiment 1). ns, not significant; One-way ANOVA with Tukey’s test.
(C) Top 15 altered diseases and functions between two dietary regimens detected by Ingenuity Pathway Analysis of microarray data (cecal tissue mRNA). n = 4 for the FR diet group and n = 3 for the FF diet group (Experiments 2A and 3).
(A) Stream plots illustrating fecal microbial community dynamics over time. Stream plots are based on Illumina sequencing of the V4 region of 16S rRNA genes (Experiment 2A,B); see Figure 1B for timeline. See Table S2 for % relative abundance of each species in individual mice. Experimental setup for the gnotobiotic experiments 2A and 2B is also shown.
(B) Histological images illustrating the similar severity of C. rodentium-associated hyperplasia in SM-colonized mice from different feeding groups or germfree animals only exposed to pathogen. The images are Hematoxylin and Eosin (H and E) stained sections of unflushed cecal tissue all at 10 dpi (Experiment 2B). Scale bars, 500 μm; higher power inset bars, 50 μm.
(A) Periodic acid-Schiff (PAS)-Alcian Blue (AB) stained colonic thin sections showing the mucus layer (shown with opposing arrows with shafts) in recta of different groups of mice at 10 dpi (Experiment 2B). Scale bar, 50 μm.
(B) Mucus layer measurements in the recta of mice from PAS-AB stained thin sections (exemplified in panel A). Asterisk indicates that the FF-SM group had only one mouse, whose rectal mucus layer could be measured, because the other mice from this group were severely affected with colitis. Data are shown as average ± SEM; statistically significant differences are shown with different alphabets (p < 0.01); One-way ANOVA with Tukey’s test.
(C) Flushed colons showing intensity of adherent, luciferase-expressing C. rodentium in germ free (GF) mice pre-fed the FR and FF diets and infected with the pathogen (that is mice without the synthetic microbiota).
Diet treatment-specific RNA-seq-based transcriptional changes are provided for the 6 species that had a relative abundance of 3% or more in the in vivo conditions tested.
Changes in mouse gene expression (cecal samples) analyzed by mouse microarrays between colonized mice fed Fiber-rich and Fiber-free diets (data are from Experiments 2A and 3).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 488 goat anti-rabbit IgG | Life Technologies | Cat#A11008 |
| Mucin 2 antibody (H-300) | Santa Cruz Biotechnology | Cat#sc-15334 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-nitrophenyl N-acetyl-β-D-glucosaminide | Sigma-Aldrich | Cat#N9376 |
| 4-nitrophenyl α-D-galactopyranoside | Sigma-Aldrich | Cat#N0877 |
| 4-nitrophenyl β-D-glucopyranoside | Sigma-Aldrich | Cat#N7006 |
| 4-nitrophenol | Sigma-Aldrich | Cat#73560 |
| Acetic acid | Acros Organics | Cat#222140010 |
| Adenine | Sigma-Aldrich | Cat#A2786 |
| Alanine | Sigma-Aldrich | Cat#A7469 |
| Alcian blue | Sigma-Aldrich | Cat#A5268 |
| Alginate | Sigma-Aldrich | Cat#180947 |
| Ammonium chloride | Sigma-Aldrich | Cat#A0171 |
| Ammonium sulfate | Thermo Fisher Scientific | Cat#A702 |
| Arginine | Sigma-Aldrich | Cat#A8094 |
| Asparagine | Sigma-Aldrich | Cat#A4159 |
| Aspartic Acid | Sigma-Aldrich | Cat#A93100 |
| Barley β-glucan (Barliv Betafiber) | Cargill | Cat#BBF-100 |
| Beef extract | Sigma-Aldrich | Cat#B4888 |
| Biotin | Sigma-Aldrich | Cat#B4501 |
| Boric acid | Sigma-Aldrich | Cat#B6768 |
| Calcium chloride | Sigma-Aldrich | Cat#C1016 |
| Calcium pantothenate | Sigma-Aldrich | Cat#P2250 |
| Cellulose | International Fiber Corporation | Solka-Floc |
| Chloroform | Sigma-Aldrich | Cat#496189 |
| Chondroitin sulfate | Federal Laboratories | Cat#CSP1K |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#000000004693159001 |
| Copper(II) sulfate pentahydrate | Sigma-Aldrich | Cat#C7631 |
| Corn starch/amylopectin | Sigma-Aldrich | Cat#10120 |
| Cyanocobalamin | Sigma-Aldrich | Cat#V2876 |
| Cysteine | Sigma-Aldrich | Cat#C7352 |
| Cytosine | Sigma-Aldrich | Cat#C3506 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Dextran | Sigma-Aldrich | Cat#31389 |
| Dipotassium phosphate | Thermo Fisher Scientific | Cat#BP363-500 |
| EDTA | Sigma-Aldrich | Cat#ED4SS |
| Ethanol | Decon Labs | Cat#2701 |
| Fiber-Free diet | Teklad/Envigo | Cat#TD.130343 |
| Folic acid | Sigma-Aldrich | Cat#F7876 |
| Fructose | Sigma-Aldrich | Cat#F0127 |
| Galactose | Sigma-Aldrich | Cat#G0625 |
| Glucomannan | Konjac Foods | Konjac Glucomannan Powder |
| Glucose | Sigma-Aldrich | Cat#158968 |
| Glutamine | Sigma-Aldrich | Cat#G8540 |
| Glutaraldehyde | Electron Microscopy Sciences | Cat#16537 |
| Guanine | Sigma-Aldrich | Cat#G11950 |
| Guar gum galactomannan | Sigma-Aldrich | Cat#G4129 |
| Glutamic Acid | Sigma-Aldrich | Cat#G1501 |
| Hematin | Sigma-Aldrich | Cat#H3281 |
| Histidine | Sigma-Aldrich | Cat#H8000 |
| Hydrochloric Acid | Sigma-Aldrich | Cat#320331 |
| Inulin | Cargill | Oliggo-Fiber Instant Inulin |
| Iron(II) sulfate heptahydrate | Sigma-Aldrich | Cat#215422 |
| Isobutyric acid | Alfa Aesar | Cat#79-31-2 |
| Isoleucine | Sigma-Aldrich | Cat#I2752 |
| Isopropanol | Sigma-Aldrich | Cat#278475 |
| Isovaleric acid | Alfa Aesar | Cat#503-74-2 |
| Laboratory Autoclavable Rodent Diet - 5010 | LabDiet | Cat#0001326 |
| Larch arabinogalactan | Megazyme | Cat#P-ARGAL |
| Leucine | Sigma-Aldrich | Cat#L8000 |
| Lysine | Sigma-Aldrich | Cat#L5501 |
| Lysozyme | Thermo Fisher Scientific | Cat#BP535-1 |
| Magnesium chloride anhydrous | Sigma-Aldrich | Cat#M8266 |
| Magnesium sulfate anhydrous | Sigma-Aldrich | Cat#M7506 |
| Magnesium sulfate heptahydrate | Sigma-Aldrich | Cat#M5921 |
| Manganese(II) sulfate monohydrate | Mallinkrodt | Cat#6192 |
| Mannose | Acros Organics | Cat#150600 |
| Menadione | Sigma-Aldrich | Cat#M5625 |
| Methanol anhydrous | Thermo Fisher Scientific | Cat#A412-1 |
| Methionine | Sigma-Aldrich | Cat#M9625 |
| Mucin from porcine stomach | Sigma-Aldrich | Cat#M1778 |
| N-acetyl glucosamine | Sigma-Aldrich | Cat#A3286 |
| Nicotinic acid | Sigma-Aldrich | Cat#N4126 |
| Osmium tetroxide | Electron Microscopy Sciences | Cat#19100 |
| p-Aminobenzoic acid | Sigma-Aldrich | Cat#A9878 |
| Pancreatic digest of casein | BioWorld | Cat#30620060-1 |
| Phenol | Sigma-Aldrich | Cat#P4557 |
| Phenol:Chloroform:Isoamyl Alcohol (pH 8.05) | Thermo Fisher Scientific | Cat# 15593031 |
| Phenol:Chloroform:Isoamyl Alcohol (pH 4.3) | Fisher Scientific | Cat#BP1754I-400 |
| Phenylalanine | Sigma-Aldrich | Cat#P2126 |
| p-nitrophenyl α-L-fucopyranoside | Sigma-Aldrich | Cat#N3628 |
| p-nitrophenyl β-D-xylopyranoside | Sigma-Aldrich | Cat#N2132 |
| Polygalacturonic acid | Sigma-Aldrich | Cat#P3850 |
| Potassium 4-nitrophenyl sulfate | Sigma-Aldrich | Cat#N3877 |
| Potassium chloride | Sigma-Aldrich | Cat#P9333 |
| Potassium dihydrogen phosphate | Thermo Fisher Scientific | Cat#P284 |
| Potato pectic galactan | Megazyme | Cat#P-PGAPT |
| Proline | Sigma-Aldrich | Cat#P5607 |
| Propionic acid | Acros Organics | Cat#149300010 |
| Propylene oxide | Electron Microscopy Sciences | Cat#20412 |
| Pyridoxine HCl | Sigma-Aldrich | Cat#P9755 |
| Resazurin | Acros Organics | Cat#418900010 |
| Retrievagen A | BD Biosciences | Cat#550524 |
| Rhamnogalacturonic acid | Megazyme | Cat#P-RHAM1 |
| Riboflavin | Sigma-Aldrich | Cat#R7649 |
| RNAprotect | Qiagen | Cat#76506 |
| SDS | Sigma-Aldrich | Cat#L3771 |
| Serine | Sigma-Aldrich | Cat#84959 |
| Sodium acetate | Sigma-Aldrich | Cat#S2889 |
| Sodium bicarbonate | Sigma-Aldrich | Cat#S5761 |
| Sodium chloride | Sigma-Aldrich | Cat#S7653 |
| Sodium citrate | Sigma-Aldrich | Cat#S1804 |
| Sodium hydroxide | Sigma-Aldrich | Cat#S8045 |
| Sodium lactate | Thermo Fisher Scientific | Cat#S326-500 |
| Sodium molybdate dihydrate | JT Baker Chemical Company | Cat#3764 |
| Sugar beet arabinan | Megazyme | Cat#p-ARAB |
| Tamarind xyloglucan | Carbomer | Cat#4-00634 |
| Thiamine HCl | Sigma-Aldrich | Cat#T4625 |
| Thioctic acid | Sigma-Aldrich | Cat#T5625 |
| Threonine | Sigma-Aldrich | Cat#T8625 |
| Thymine | Sigma-Aldrich | Cat#T0895 |
| Tris | Thermo Fisher Scientific | Cat#BP152 |
| TritonX-100 | Sigma-Aldrich | Cat#T9284 |
| TRIzol | Invitrogen | Cat#15596026 |
| Tryptone | Thermo Fisher Scientific | Cat#BP1421 |
| Tyrosine | Sigma-Aldrich | Cat#T3754 |
| Uracil | Sigma-Aldrich | Cat#U1128 |
| Valeric acid | Alfa Aesar | Cat#109-52-4 |
| Valine | Sigma-Aldrich | Cat#V0500 |
| Wheat arabinoxylan | Megazyme | Cat#P-WAXYL |
| Xylene | Sigma-Aldrich | Cat#296333 |
| Xylene Substitute | Sigma-Aldrich | Cat#A5597 |
| Xylose | Sigma-Aldrich | Cat#X3877 |
| Yeast extract | Fluka Analytical | Cat#70161 |
| Zinc sulfate heptahydrate | JT Baker Chemical Company | Cat#4382 |
| Critical Commercial Assays | ||
| AccuPrimeTaq DNA Polymerase, high fidelity kit | Thermo Fisher Scientific | Cat#12346086 |
| Affymetrix Mouse Gene ST 2.1 strips | Affymetrix | Cat#902120 |
| Arcturus PicoPure DNA Extraction Kit | Arcturus | Cat#KIT0103 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69506 |
| epMotion 5075 TMX | Eppendorf | Cat#960020033 |
| High-sensitivity DNA analysis kit | Agilent | Cat#5067-4626 |
| KAPA SYBRFAST qPCR kit | KAPA Biosystems | Cat#KK4600 |
| KAPA Library Quantification Kit for Illumina platforms | KAPA Biosystems | Cat# KK4824 |
| Mouse Lipocalin-2/NGAL DuoSet ELISA kit | R & D Biosystems | Cat#DY1857 |
| Pierce Microplate BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#PI23252 |
| PowerSoil Isolation Kit | MoBio Laboratories | Cat#12888 |
| Qubit RNA Assay Kit | Thermo Fisher Scientific | Cat#Q32852 |
| Ribo-Zero rRNA Removal Kits (Bacteria) | Illumina | Cat#MRZB12424 |
| RNeasy Protect Bacteria Mini Kit | Qiagen | Cat #74524 |
| SequalPrep Normalization Plate Kit, 96-well | Thermo Fisher Scientific | Cat#A1051001 |
| TURBO DNase kit | Ambion | Cat#AM1907 |
| Deposited Data | ||
| 16S rRNA gene sequences and metadata | NCBI BioProjectID NCBI SRA; | PRJNA300261;SRP065682 |
| Mouse microarray data | NCBI Geo | GSM2084849–55 |
| RNA-Seq data | NCBI BioProjectID | PRJNA310052, PRJNA310058, PRJNA310062 |
| Experimental Models: Organisms/Strains | ||
| Akkermansia muciniphila: DMS 22959, type strain | DSMZ | Cat#DMS 22959 |
| Bacteroides caccae: DSM 19024, type strain | DSMZ | Cat#DSM 19024 |
| Bacteroides ovatus: DSM 1896, type strain | DSMZ | Cat#DSM 1896 |
| Bacteroides thetaiotaomicron: DSM 2079, type strain | DSMZ | Cat#DSM 2079 |
| Bacteroides uniformis: ATCC 8492, type strain | ATCC | Cat#ATCC 8492 |
| Barnesiella intestinihominis: YIT11860 | DSMZ | Cat#DSM 21032 |
| Citrobacter rodentium: DBS100 | David Schauer, Massachusetts Institute of Technology | N/A |
| Citrobacter rodentium: DBS120 | David Schauer, Massachusetts Institute of Technology | N/A |
| Clostridium symbiosum: DSM 934, type strain, 2 | DSMZ | Cat#DSM 934 |
| Collinsella aerofaciens: DSM 3979, type strain | DSMZ | Cat#DSM 3979 |
| Desulfovibrio piger: ATC 29098, type strain | ATCC | Cat#ATC 29098 |
| Escherichia coli HS | ATCC | N/A |
| Eubacterium rectale: DSM 17629, A1-86 | DSMZ | Cat#DSM 17629 |
| Faecalibacterium prausnitzii: DSM 17677, A2-165 | DSMZ | Cat#DSM 17677 |
| Marvinbryantia formatexigens: DSM 14469, type strain, I-52 | DSMZ | Cat#DSM 14469 |
| Roseburia intestinalis: DSM 14610 type strain, L1-82 | DSMZ | Cat#DSM 14610 |
| Sequence-Based Reagents | ||
| 16S rRNA gene Illumina sequencing primers | Kozich et al. 2013 | Table S2B |
| qPCR primers | This paper | Table S3A |
| Software and Algorithms | ||
| Arraystar | DNAStar | http://www.dnastar.com/t-sub-products-genomics-arraystar.aspx |
| Gen5 | Biotek | http://www.biotek.com/products/microplate_software/gen5_data_analysis_software.html |
| ImageJ | Schneider et al., 2012 | http://imagej.net/ |
| Microsoft Excel | Microsoft | https://products.office.com/en-us/excel |
| Mothur v1.33.3 | Schloss et al., 2009 | http://www.mothur.org/ |
| Multi-array average (RMA) method | Irizarry et al, 2003 | http://www.bioconductor.org/ |
| Prism v5.04 | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ |
| QIAGEN’s Ingenuity Pathway Analysis | Qiagen | http://www.ingenuity.com/products/ipa |
| R | The R Foundation | https://www.r-project.org/ |
| R - Limma package | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| R - Plyr package | Wickham, 2011 | https://CRAN.R-project.org/package=plyr |
| UCHIME | Edgar et al., 2011 | http://drive5.com/usearch/manual/uchime_algo.html |
| Other | ||
| Acid-washed glass beads (212–300 μm) | Sigma-Aldrich | Cat#G1277 |
| Anaerobic chamber | Coy manufacturing | Vinyl Type A + Type B |
| Biostack automated plate handling device | Biotek Instruments | BIOSTACK2WR |
| Bioluminescence reader | Xenogen | IVIS200 |
| Breathe-Easy polyurethane membrane | Biversified Biotech | Cat#BEM-1 |
| Flat bottom 96-well plates | Costar | |
| General Laboratory Homogenizer | Omni International | |
| Microdissection instrument | Arcturus | Veritas Microdissection Instrument |
| Millex-GV Syringe Filter Unit, 0.22 μm, PVDF, 4 mm, ethylene oxide sterilized | EMD Millipore | Cat#SLGV004SL |
| Mini-BeadBeater-816 | Biospec Products | Cat#607 |
| Powerwave HT absorbance reader | Biotek Instruments | PowerWaveHT |
| Synergy HT absorbance reader | Biotek Instruments | Synergy HT |
| Upright fluorescence microscope | Olympus | BX60 |
| Transmission electron microscope | Philips | Philips CM-100 |
HIGHLIGHTS.
Characterized synthetic bacterial communities enable functional insights in vivo
Low-fiber diet promotes expansion and activity of colonic mucus-degrading bacteria
Purified prebiotic fibers do not alleviate degradation of the mucus layer
Fiber-deprived gut microbiota promotes aggressive colitis by an enteric pathogen
Acknowledgments
We thank Lansing C. Hoskins for critical comments on this manuscript and the germfree animal facility of the University of Michigan for expert support. We also thank Markus Ollert and Rudi Balling for their encouragement and advice. This work was supported by: Luxembourg National Research Fund (FNR) INTER Mobility (13/5624108) and CORE (C15/BM/10318186) grants to M.S.D.; Luxembourg Ministry of Higher Education and Research support to M.S.D; FNR ATTRACT (A09/03), CORE (11/1186762) and European Union Joint Programming in Neurodegenerative Diseases (INTER/JPND/12/01) grants to P.W.; NIH R01 (GM099513) grant to E.C.M.; and financial support from the University of Michigan Host Microbiome Initiative and Center for Gastrointestinal Research (DK034933).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.S.D., E.C.M., P.W., T.S., and G.N. conceived the study; M.S.D. and E.C.M. designed the study, and M.S.D. performed the experiments; M.S.D, A.M.S., and E.C.M. analyzed data; M.S.D., A.M.S., C.H. and E.C.M. prepared figures; M.S.D. and E.C.M primarily wrote and edited the manuscript; M.S.D., N.M.K. and N.A.P. carried out bacterial in vitro growth assays; G.N., N.K. and S.K. assisted with C. rodentium infection and luminescence experiments; M. W. assisted with mucus measurements; C.H. and T.S.S. conducted blinded histology scoring; A.M. analyzed microarray data; N.T. and B.H. provided CAZy annotations; all authors discussed the results and provided comments on the manuscript.
References
- Bergstrom KSB, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee IA, Havler ME, Dettmar PW, Allen A, Pearson JP. Colonic mucus: secretion and turnover in relation to dietary fibre intake. Proc Nutr Soc. 2003;62:245–249. doi: 10.1079/pns2003206. [DOI] [PubMed] [Google Scholar]
- Burkitt DP, Walker ARP, Painter NS. Effect of dietary fibre on stools and transit-times, and its role in the causation of disease. Lancet. 1972;300:1408–1411. doi: 10.1016/s0140-6736(72)92974-1. [DOI] [PubMed] [Google Scholar]
- Cameron EA, Sperandio V. Frenemies: signaling and nutritional integration in pathogen-microbiota-host interactions. Cell Host Microbe. 2015;18:275–284. doi: 10.1016/j.chom.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, Finlay BB, Frankel G. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol. 2014;12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron Ea, Pudlo Na, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Earle KA, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative imaging of gut microbiota spatial resource quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015:1–11. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T, Johansson MEV, Liu X, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Lett. 1991;86:103–111. [Google Scholar]
- Hedemann MS, Theil PK, Bach Knudsen KE. The thickness of the intestinal mucous layer in the colon of rats fed various sources of non-digestible carbohydrates is positively correlated with the pool of SCFA but negatively correlated with the proportion of butyric acid in digesta. Br J Nutr. 2009;102:117–125. doi: 10.1017/S0007114508143549. [DOI] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins LC, Boulding ET. Mucin degradation in human colon ecosystems. J Clin Invest. 1981;67:163–172. doi: 10.1172/JCI110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjövall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubinoux J, Bronowicki J-P, Pereira IAC, Mougenel J-L, Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. 2002;40:107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell. 2015;163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS One. 2012;7:e30287. doi: 10.1371/journal.pone.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A. 2013;110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014:1–8. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Genomic and Metabolic Studies of the Impact of Probiotics on a Model Gut Symbiont and HostGlycan foraging in vivo by an intestine-adapted bacterial symbiont. PLoS Biol. 12, e413. Science. 2006;307:1955–1959. [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Büller Ha, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Wickham H. The Split-Apply-Combine Strategy for Data. J Stat Softw. 2011;40:1–29. [Google Scholar]
- Wrzosek L, Miquel S, Noordine M-L, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A schematic displaying abilities of the SM to degrade a wide variety of dietary and host-derived polysaccharides and possible metabolic interactions between members of the SM. GAGs, Glycosaminoglycans.
(B) Representative growth curves of selected members of the SM on several polysaccharides and glycans as sole carbon sources (n = 2 for each glycan; values are shown as averages). The absorbance was measured every 10 minutes. See Table S1 for raw and normalized growth values and growth media descriptions for the 13 members of the SM evaluated for carbohydrate growth ability (all except D. piger).
Summary of growth values of cultures included in synthetic microbiota, details of growth media and diets.
Microbial relative abundances based on Illumina sequencing of 16S rRNA genes are provided for all 1,114 samples (data are from Experiments 1 and 2).
Primer sequences and microbial relative abundances, based on qPCR measurements employing phylotype-specific primers, are provided (data are from Experiment 1).
RNA-seq-based microbial community transcriptomic data is presented for glycoside hydrolase (GH), polysaccharide lyase (PL) and carbohydrate esterase (CE) families that exhibited altered expression between the FR/FF or Pre/FF treatment comparisons (data are from Experiment 1).
The data are based on RNA-seq (data are from Experiment 1).
Principal coordinates analysis (PCoA) of microbial community dissimilarity (Bray-Curtis) in fecal samples (collected according to Figure 1B) as determined by 16S rRNA-based sequencing (V4 region). Samples from both Experiments 1 and 2 are shown, with samples coded by experiment (top panel) or sex (bottom panel) and by cage (legend) (Experiment 1: n = 4 mice/group; Experiment 2: n = 7 mice/group).
(A) Relative abundance of indicated bacteria in mice over time subjected to various dietary regimes as determined by Illumina-based 16S rRNA sequencing (Experiment 1). An explanation for the inverse relationship between the relative abundances of D. piger and M. formatexigens on FR and FF diets is their competition for the same electron donor (hydrogen). The increased proliferation of mucin-degrading bacteria in the FF diet indicates higher degradation of sulfated mucin O-glycans, which is supported by transcriptomic and enzyme assay data shown in Figure 3. The corresponding release of additional sulfate would preferentially feed the sulfate-reducer D. piger, leading to production of the toxic metabolite hydrogen sulfide (Fig. S1A). Increased D. piger has also been observed in IBD patients (Gibson et al., 1991; Loubinoux et al., 2002), which could result from enhanced sulfate release by mucinolytic bacteria. Values are shown as medians ± IQR. n = 4 for FR and FF groups; n = 3 for Pre and 1-day FR/FF groups.
(B) Heat map showing Percent of Maximum Abundance (POMA) values of all species for two of the feeding groups from Illumina-based sequencing (see Figure 1B); n = 3 mice/group (Experiment 1; according to timeline in Figure 1B). See also Table S2.
Figures are based on RNA-Seq measurements of B. caccae (A) and A. muciniphila (B) responses in vitro (minimal medium with simple sugars or MOG) and in vivo (constant feeding or daily alternation of FR and FF diets). In vivo samples are from the entire cecal community at the end of Experiment 1. Gene transcripts that were increased >5-fold relative to the corresponding simple sugar references are included for each bacterium. Venn diagrams show overlap and differences of the transcripts between various groups. Numbers indicate the total differentially regulated gene count for a given sector, while number in parentheses denote numbers of carbohydrate-degrading enzymes (glycoside hydrolase, polysaccharide lyase or carbohydrate esterase families counted toward this number; sulfatases and carbohydrate binding module, CBM, families were not counted). Note that A. muciniphila shows less regulatory versatility as manifest by most of its upregulated enzymes being confined to the core (dark pink) sector containing all of the in vivo samples. This suggests that MOG only triggers a small percentage of this species’ O-glycan degrading responses in vitro; although 8 enzymes were also triggered in vitro. The corresponding histograms display frequencies of related enzyme families (shown with matching colors to their respective Venn sectors). Possible mucin-related degradative functions are given above each family-specific histogram bar. For in vitro samples, n = 2 for each MOG and simple sugar grown condition; for in vivo samples, n = 3 mice/group (Experiment 1).
(C) Schematic mucin O-glycan structures, from among ~102 that can be found on human and murine Muc2, with the sites at which various enzymes noted in panels A. and B. would be expected, or are known, to cleave. See also Tables S4 and S5 for in vivo and in vitro transcript data.
(A) Depictive histology images (Hematoxylin and Eosin of colonic thin sections) showing no overt signs of inflammation between the two dietary regimens (Experiment 1), in the absence of C. rodentium. Scale bars, 500 μm.
(B) Weight change in mice over time. Values are shown as averages ± SEM. n = 4 for FR and FF groups; and n = 3 for Pre group (Experiment 1). ns, not significant; One-way ANOVA with Tukey’s test.
(C) Top 15 altered diseases and functions between two dietary regimens detected by Ingenuity Pathway Analysis of microarray data (cecal tissue mRNA). n = 4 for the FR diet group and n = 3 for the FF diet group (Experiments 2A and 3).
(A) Stream plots illustrating fecal microbial community dynamics over time. Stream plots are based on Illumina sequencing of the V4 region of 16S rRNA genes (Experiment 2A,B); see Figure 1B for timeline. See Table S2 for % relative abundance of each species in individual mice. Experimental setup for the gnotobiotic experiments 2A and 2B is also shown.
(B) Histological images illustrating the similar severity of C. rodentium-associated hyperplasia in SM-colonized mice from different feeding groups or germfree animals only exposed to pathogen. The images are Hematoxylin and Eosin (H and E) stained sections of unflushed cecal tissue all at 10 dpi (Experiment 2B). Scale bars, 500 μm; higher power inset bars, 50 μm.
(A) Periodic acid-Schiff (PAS)-Alcian Blue (AB) stained colonic thin sections showing the mucus layer (shown with opposing arrows with shafts) in recta of different groups of mice at 10 dpi (Experiment 2B). Scale bar, 50 μm.
(B) Mucus layer measurements in the recta of mice from PAS-AB stained thin sections (exemplified in panel A). Asterisk indicates that the FF-SM group had only one mouse, whose rectal mucus layer could be measured, because the other mice from this group were severely affected with colitis. Data are shown as average ± SEM; statistically significant differences are shown with different alphabets (p < 0.01); One-way ANOVA with Tukey’s test.
(C) Flushed colons showing intensity of adherent, luciferase-expressing C. rodentium in germ free (GF) mice pre-fed the FR and FF diets and infected with the pathogen (that is mice without the synthetic microbiota).
Diet treatment-specific RNA-seq-based transcriptional changes are provided for the 6 species that had a relative abundance of 3% or more in the in vivo conditions tested.
Changes in mouse gene expression (cecal samples) analyzed by mouse microarrays between colonized mice fed Fiber-rich and Fiber-free diets (data are from Experiments 2A and 3).