Abstract
Regulation of gene expression in response to nutrient availability is fundamental to the genotype-phenotype relationship. The metabolic-genetic make-up of the cell, as reflected in auxotrophy, is hence a likely determinant of gene expression. Here, we addressed the importance of the metabolic-genetic background by monitoring transcriptome, proteome, and metabolome in a repertoire of sixteen Saccharomyces cerevisiae laboratory backgrounds, combinatorially perturbed in histidine, leucine, methionine and uracil biosynthesis. The metabolic background affected up to 85% of the coding genome. Suggesting widespread confounding, these transcriptional changes showed, on average, 83% overlap between unrelated auxotrophs, and 35% with previously published transcriptomes generated for non-metabolic gene knock-outs. Background-dependent gene expression correlated with metabolic flux and acted, predominantly through masking or suppression, on 88% of transcriptional interactions epistatically. As consequence, the deletion of the same metabolic gene in a different background could provoke an entirely different transcriptional response. Propagating to the proteome and scaling up at the metabolome, metabolic background dependencies reveal the prevalence of metabolism-dependent epistasis at all regulatory levels. Urging for a fundamental change of the prevailing laboratory practice of using auxotrophs and nutrient supplemented media, these results reveal epistatic intertwining of metabolism with gene expression on the genomic scale.
Introduction
Metabolism represents the largest functional system within the cell, and as metabolic reactions are connected over the flux of metabolites, assembles in a highly connected network1–7. Metabolic activity needs constant adaptations to match cellular physiology, nutrition, growth rate and stress situations. This dual-dependency on both cell extrinsic and intrinsic properties renders metabolism a key mediator of gene-environment interactions, while its size represents a quantitative factor in physiology and gene expression8–10. Enumerating the total compendium metabolism-responsive genes is a difficult task, but transcriptional changes that follow S. cerevisiae metabolic oscillations suggest that it could be more than 50% of the genome11.
A difficulty in studying genetic- metabolic interactions is caused by a minimal redundancy within the metabolic network. Except secondary metabolic pathways, most metabolic systems cannot be perturbed without system-wide consequence. Exceptions are some metabolic pathways of amino acid and nucleobase biosynthesis, for which cells possess an uptake over self-synthesis preference for the product metabolite. These biosynthetic pathways can be perturbed as long as the product is provided extracellularly12. Single-gene auxotrophies in such pathways have established as effective selection markers for genetic experiments, so that they have been crossed into a large number of laboratory strains. In the present work we exploit such markers for studying the importance on the metabolic background on gene expression in Saccharomyces cerevisiae, and study combinatorial effects that result from a HIS313, LEU214, URA315 or MET1516 deletion on transcriptome, proteome and metabolome.
We report that metabolic background differences induce strong but adaptive molecular signatures even when growth is restored by external nutrient supplementation. Gene expression is affected in a metabolism dependent manner, and on 88% of transcriptional events that involve 77% of differentially expressed transcripts, we detect evidence for epistatic interactions that occur between the metabolic genes. These interactions are found to have a fundamental impact on the gene expression response that follows gene deletions. They reveal the metabolic genotype (or metabotype) to be, on a global scale, responsible for context-dependent biological responses on transcriptome, proteome and metabolome. The global dependency on metabotype substantiates an upstream and dynamic and key role of the cellular metabolic make-up in gene expression regulation. Metabolic-genetic background differences, hand-wavingly considered to be harmless in pre-genomic times, could hence have affected the outcome of a large number of experiments.
Results
Molecular signatures of the metabolic-genetic background
We have reported earlier that even when histidine, leucine, uracil and methionine auxotrophies are complemented, some physiological differences are retained. These manifest as small to moderate growth rate differences, some of which show evidence for epistatic interactions between the metabolic markers 17. Starting by an in-depth analysis of this growth data, we found that in supplemented media, LEU2 has a consistent effect on growth rate, while the other markers exert minor effects that reveal themselves only in a context (or background) dependent manner. The overall growth rates are therefore well explained by the leucine effect using both, by an additive model (Supplementary Fig. S1a), or by a multiple linear regression model that uses HIS3, LEU2, URA3 and MET15 as categorical predictors (adj. R2 = 0.86, P-value = 2.18e-05 (Supplementary Fig. S1a,b)).
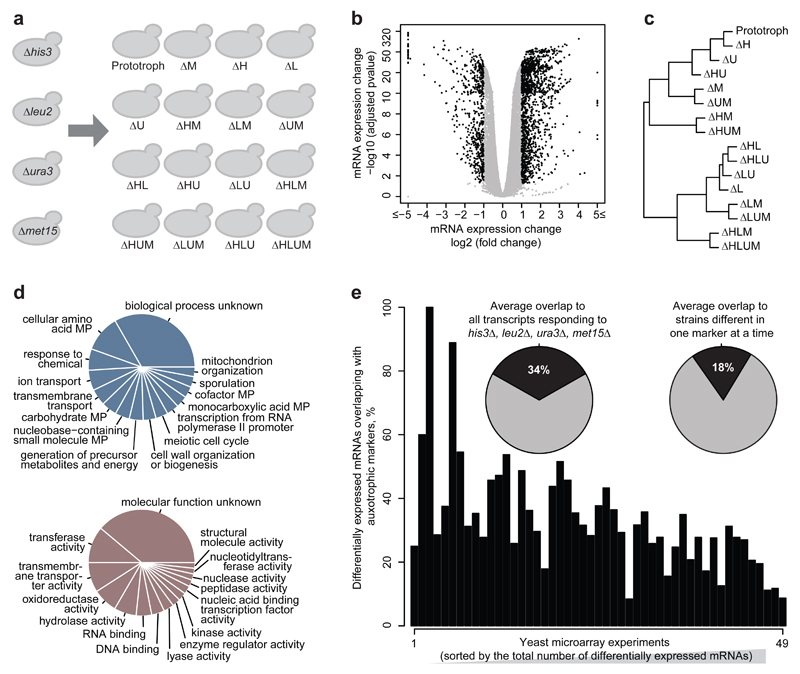
The molecular levels revealed a much more differentiated picture, however. mRNA expression profiles were obtained from the 16 strains in triplicate exponentially grown cultures each at identical cell density (OD600=0.8) by mRNA sequencing, resulting in highly reproducible expression profiles (Supplementary Fig. S2). 5011 transcripts out of total 5923 expressed mRNAs (85% of the transcriptome) were significantly differentially expressed (adj. P-values (BH method) <0.05) in auxotrophic strains compared to prototroph (Fig. 1b). A global transcriptional signature is corroborated by robust median normalization (Supplementary Fig. S3a). Hierarchical clustering revealed the strongest separation by the LEU2 followed by the MET15 genes, indicating that these two perturbations leave the most consistent signature in the transcriptome (Fig. 1c). 573 transcripts (9.7% of transcriptome) were differentially expressed not only significantly but more than 2-fold (Fig. 1b). These were enriched for metabolic activity (GO process terms) and enzymatic function (GO function terms) (Fig. 1d), and according to hypergeometric testing, for amino acid and carbohydrate metabolic pathways (Supplementary Fig. S3b). Thus, even-though histidine, leucine, uracil and methionine auxotrophy is complemented by external nutrition, the auxotrophic background is reflected by gene expression differences detected on three-fourth of the coding genome, with ~600 mostly metabolism-associated genes being strongly differentially expressed.
Fig. 1. The gene expression response to 16 combinatorial differences in the metabolic-genetic background.
(a) Schematic overview of otherwise isogenic 16 S. cerevisiae strains possessing nutrient-complemented auxotrophies in histidine (his3Δ), leucine (leu2Δ), uracil (ura3Δ) and methionine (met15Δ) metabolism
(b) Differential gene expression with reference to the prototrophic yeast (Volcano plot) illustrating gene expression (log2 scaling) and significance (-log10 scaling, BH adj. P-value) for the 16 metabotypes grown in synthetic complete medium, as determined by RNA sequencing (n = 3/strain).
(c) Hierarchical clustering of mRNA expression profiles by means of euclidean distance and complete linkage agglomeration, dividing the 16 strains first by leucine, followed by methionine auxotrophy
(d) Top 15 enriched Gene Ontology (GO):Process slim mapper (top) and GO:Function slim terms (bottom), across 573 differentially expressed transcripts changed > 2 fold change, adj. P-value <0.05 with reference to the prototrophic strain. MP = metabolic process.
(e) Confounding effects of metabotype on transcriptome. Overlap between genes highly and significantly (fold change >2, adj. P-value <0.05) differentially expressed in the auxotrophic strains, to those detected differentially expressed across previously published microarray experiments conducted upon single gene-deletion in the BY4741 auxotrophic background. Expression profiles have been sorted according to No of differentially expressed transcripts (ascending). Insets: Average overlaps between transcriptome obtained for all four markers, or those differing in just one marker at a time.
As HIS3, LEU2, MET15 and URA3 have been frequently exploited as genetic selection markers, these results implied that their transcriptional signatures could have confounded gene expression experiments. mRNA expression data conducted on a variety of single gene knock-outs (the vast majority being non-metabolic genes), in different context and laboratories, but all generated in auxotrophic BY4741 backgrounds (listed in Supplementary File 1) were obtained from ArrayExpress18 and re-processed to achieve identical cut-off criteria (fold change >2, BH adj. P-value < 0.05). In average 34% of differentially expressed mRNAs did overlap with those induced by HIS3, LEU2, MET15 or URA3 deletions > 2-fold (Fig. 1e). Even if we compare strains that differ only by a single marker, on average 18% of transcriptional changes do overlap (Fig. 1e). A similar picture was obtained for a further set of ~70 microarray experiments that studied different conditions (Supplementary Fig. S4a, Supplementary File 1). We noticed a significant correlation (r = –0.52, P-value = 0.0001186), that transcription profiles with a larger numbers of differentially expressed genes are better distinguished from the metabolic background signature (Fig. 1e).
Next we questioned whether this overlap would be the same, or larger, for metabolic genes. We compared each combination of a HIS3, LEU2, URA3, or MET15 deletion with differential gene expression induced by all other markers. Here a substantially larger overlap of 83% was detected (Supplementary Fig. S4b). Hence, a notable proportion of differential gene expression overlaps with the transcriptional signatures of metabolic markers. Comparing more than a hundred transcriptomes, we detect on average an overlap of ~1/3rd in case a non-metabolic gene is deleted, and, on the basis of our dataset, ~¾ in case a metabolic gene is deleted.
Metabolism-induced gene expression signatures are context dependent
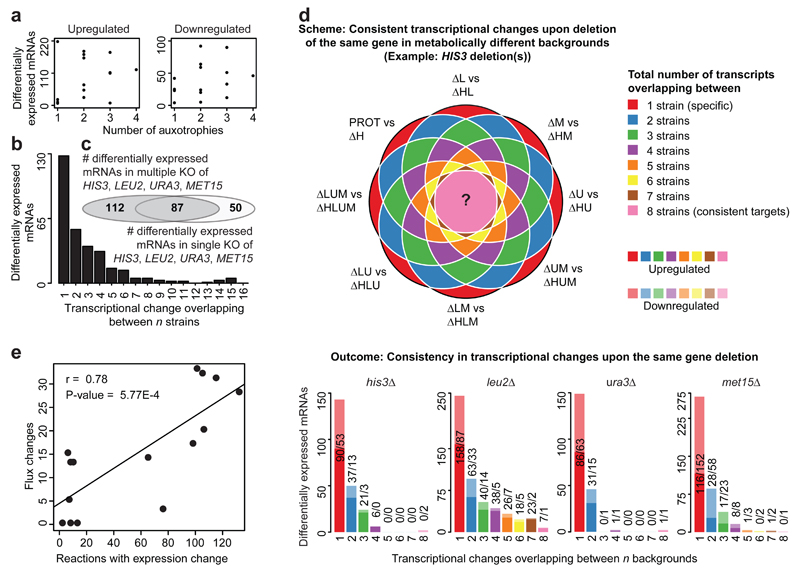
We noted that expression signatures were qualitatively and quantitatively dependent not only on the metabolic deficiencies, but also on their combination. First, strains possessing three or four auxotrophies did not have more transcripts induced compared to strains possessing one or two deficiencies (Fig. 2a, Supplementary Fig. S3c). This result was confirmed by normalization strategies referring to the median, ruling out a this result being a consequence of awild-type bias (Supplementary Fig. S3e,f). In addition, different transcripts responded to individual or to combinatorial perturbation. For instance, 112 transcripts were differentially expressed exclusively in the combinatorial knockouts, but not in the corresponding single knockouts (adj. P-value < 0.05, fold change 2, Fig. 2c), while 128 transcripts were differentially expressed solely in a unique strain (Fig. 2b). Qualitatively similar results were obtained without fold-change cut-offs (Supplementary Fig. S3g,h).
Fig. 2. The transcriptional response to a metabolic gene deletion is sensitive to the metabolic-genetic background.
(a) The number of auxotrophies is no predictor for total differential gene expression quantities. Numbers are given in relation to the prototrophic strain; the analysis relative to the expression median is given in Supplementary Fig. S3c.
(b) Specific transcriptional profiles dominate over commonly regulated genes; the majority of transcripts responds specifically in one or two of the 16 strains, despite these differ in just four metabolic genes to each other.
(c) Overlap of gene expression changes (adj. P-value <0.05, fold change 2) induced when HIS3, LEU2, URA3 or MET15 were deleted separately or in combination (adj. P-value <0.05, fold change >2 Please see Supplementary Fig. S3g without fold-change cut-off)
(d) Transcriptional response upon deleting the same gene (illustration example for HIS3) in a different metabotype reveals the dominance of context-specific transcriptional responses; Genes that consistently respond to the same deletion (in up to eight cases) are the exception. Lighter colors indicate down-regulated genes, whereas opaque colors represent up-regulated mRNAs. (adj. P-value <0.05, fold change > 2. Please see Supplementary Fig. S6c for the same analysis without fold-change cut-off)
(e) Flux variability analysis predicts context-dependent flux changes that correlate with strain-specific gene expression. Significant correlation (r = 0.78, P-value = 5.77E-4) between the number of reactions in which at least one gene is differentially expressed, and the number of reaction for which the flux-range is significantly different.
These results raised the question whether specific transcriptionally responsive genes could be assigned to HIS3, URA3, LEU2, and MET15 deletions, or whether the transcriptome responds differently dependent on metabolic background. We compared four times eight complementary strain pairs that differ from each other only in one of the auxotrophies (graphically exemplified for HIS3 (Fig. 2d)). Each gene deletion induced a strong transcriptional response (Supplementary Fig. S6a,b), but in each background, different gene-sets responded to the same gene deletion (Fig. 2d). Indeed, universal targets were the exception; the only consistent hits were the deleted genes themselves (Fig. 2d). Consistently, transcriptional changes induced by multiple deletions of the same gene did, except for a small subset of LEU2 responsive genes, not correlate with each other (Supplementary Fig. S6d). An analogous picture was obtained when considering all significantly differentially expressed transcripts, ruling out a thresholding bias (Supplementary Fig. S6b,c).
We speculated that strain specific transcriptional profiles might originate from different metabolic flux. Context-dependent gene expression changes did strongly correlate (r = 0.78, P-value = 5.77e-4) with flux as determined by flux variability analysis19,20 upon constraining the model with experimentally measured amino acid uptake and growth rates for the sixteen strains (Fig. 2e, Supplementary Note 2, Supplementary File 2). Metabolic reactions with highest correlation between gene expression and calculated fluxes were enriched for intermediate metabolic pathways (Supplementary Fig. S7).
Metabolic perturbations interact epistatically
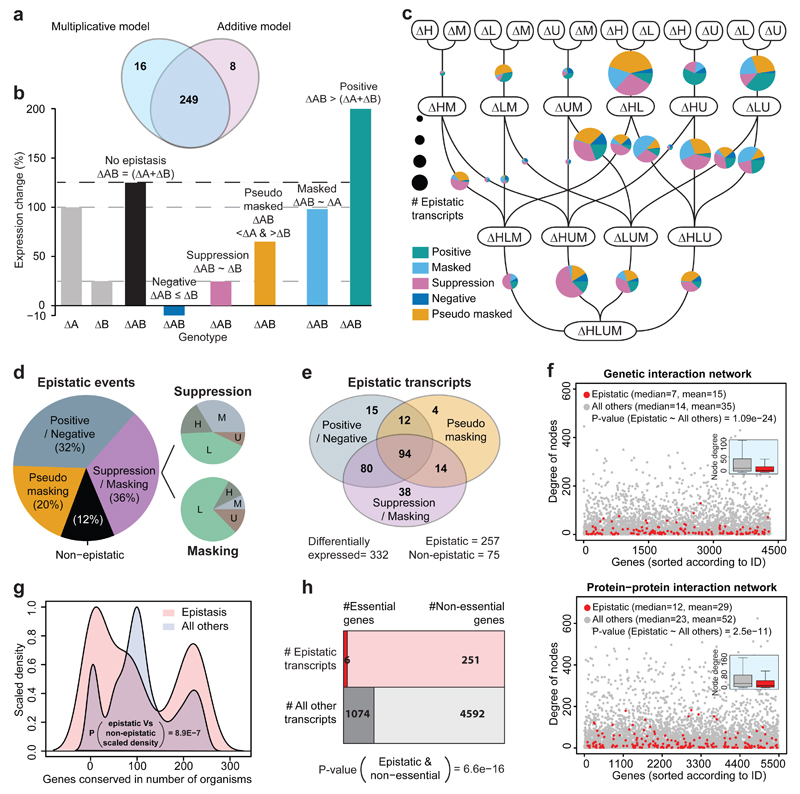
In principle, a target transcript could respond irrespectively of whether a metabolic pathway is perturbed alone or in combination, or the response could be sensitive to epistatic interactions between the pathways. 77% of differentially expressed transcripts did respond in more than one auxotroph and thus may fall into these categories (Fig. 2c). To identify epistatic interaction, be applied both additive models, as introduced by Fisher21, and multiplicative models. On our large and systematic dataset, both strategies yielded to a large extent (97%) the same transcripts, when applied on gene expression data scaled to fold-changes, and considering only genes both significantly (adj. P-value <0.05) and highly (>2 fold) differentially expressed (Fig. 3a). In accordance with the growth rates, we therefore sub-divided epistatic interactions using the additive model (Illustration Fig. 3b). 88% of gene expression interactions had epistatic impact on the target transcripts. Most frequent were suppressive or masking (= dominant) interactions (Fig. 3d). Suppressive interactions were distributed among all markers, while masking interactions where dominated by LEU2 (Fig. 3d, right panel). Second most frequent were positive and negative interactions, in which the effect of two alleles acting in concert- acting was stronger or weaker as expected from the individual alleles effect (Fig. 3d). Their case was complemented by a special case of suppression; we termed ‘pseudo-masking’, in which a second mutation weakened the effect of a masking allele. Illustrating the dynamic and context dependent nature of epistasis, 37% transcripts that were falling into a particular epistatic category in one case, could take any category depending on metabotype (Fig. 3e).
Fig. 3. Metabolic perturbations interact epistatically and dominate quantitative expression profiles.
(a) Multiplicative and additive models identify to 97% the same transcripts as being affected by metabolism-induced epistasis, when considering transcripts differentially expressed (>2 fold change; adj. P-value <0.05) compared to the median expression value of each gene.
(b) Schematic illustration: Classification of epistatic phenotypes on the basis of a linear model.
(c) The 16 strains aligned in a neighborhood network to illustrate the 38 neighborhood pairs that differ from each other in just one metabolic gene at a time. mRNA expression profiles were pairwisely compared to identify and classify epistatic gene expression changes (b). Size of the Pie charts represents the number of epistatically responding transcripts as identified in each neighborhood, and their distribution between the categories as defined in (b).
(d) Classification of epistatic events. Additive events (12%), considered non-epistatic according to21, contrast to epistatic interactions classified as masking and suppression, positive and negative epistasis, and pseudo-masking. Given is the sum of all events across the 38 neighborhood pairs illustrated in Fig. 3c. Suppressive interactions were found frequent for all four alleles, masking interactions are dominated by LEU2.
(e) Transcripts affected by epistatic interactions switch between subcategories dependent on metabotype. Compared are transcripts differentially expressed according to median (>2 fold change; adj. P-value < 0.05)
(f) Degree distribution of transcripts affected by metabolism-induced epistasis and all other transcripts in a genetic interaction network23 and protein-protein interaction network24. Genes encoding for transcripts epistatic transcripts are significantly less connected compared to the average gene.
(g) Density-distribution of PSI-BLAST identified orthologues of epistatically responding and all other genes. Those affected by metabolism-induced epistasis are significantly less conserved compared.
(h) Distribution of essential genes across epistatically responding and all other transcripts, according to viability in S. cerevisiae. Genes affected by metabolism-induced epistasis are less often essential.
Highly connected and conserved genes are buffered against metabolism-induced epistasis
It has been suggested that most connected nodes (‘hubs’) in genetic interaction network are more stable in terms of expression changes22, and would thus less likely to be affected by epistatic interactions. Indeed, epistatically responding transcripts were significantly less connected in genetic interaction23 and in protein-protein interaction networks (Fig. 3f), but more likely co-expressed24 (Supplementary Fig. S8g). In contrast, GO terms and functional classification (Supplementary Fig. S8a), co-citation frequency, co-occurrence of protein domains, nor 3-D protein structure networks of interacting orthologous proteins as obtained from YeastNet.v324 (Supplementary Fig. S8b-f) were indifferent between responding and non-responding transcripts.
It has further been observed that more important and conserved genes are more stable to expression changes, and that epistasis determines sequence conservation25,26. We compared evolutionary conservation and essentiality27 between affected or unaffected transcripts. Transcripts sensitive to metabolic epistasis had significantly less orthologues as identified by PSI-BLAST (P-value = 1.6E-07) (Fig. 3g) and were significantly less likely essential (P-value = 1.04E-17) (Fig. 3h). Epistatic metabolic interactions thus manifest prevalently on the least connected and conserved genes, while highly conserved genes and most connected genes appear to be buffered.
Metabolic epistasis translates into the proteome and amplifies at the metabolome
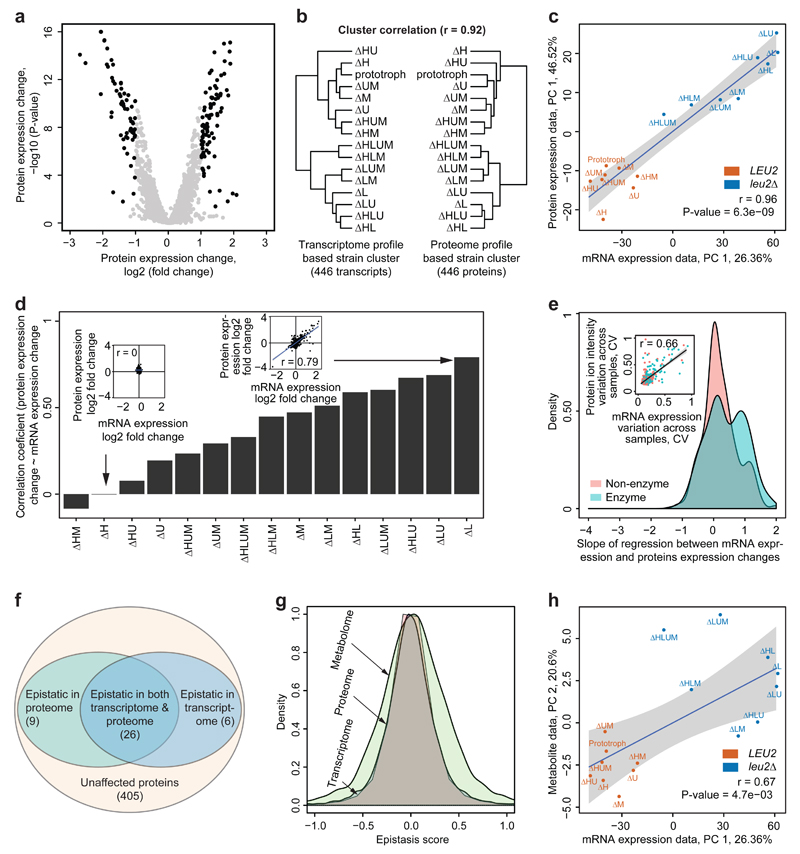
Next, we generated protein expression profiles by liquid chromatography tandem mass spectrometry (LC-MS/MS). We choose a data-independent acquisition (DIA) strategy (HDMSE)28,29 that identifies fewer proteins than shotgun methods, but is advantageous in large samples series as the same peptides are consistently quantified in each injection replicate. Further, facilitated by the large systematic dataset, we improved precision compared to conventional strategies by using covariance statistics to choose peptides best suited for protein quantification. Across the 48 proteomes, this strategy yielded precise and reproducible quantities for 442 proteins associated with 446 genes (Supplementary Fig. S9). Applying the same criteria as used for transcriptomics, 11% of proteins were found differentially expressed (Fig. 4a). Hierarchical clusters of transcriptome and proteome did fully correlate (cophylogenetic correlation = 0.92) and divided the 16 strains in a similar fashion (Fig. 4b). The correlation of principal components agreed with a coherent regulation at transcriptome and proteome; the first principal components, explaining 26.36% and 46.52% of the overall variation, were fully correlated (r = 0.96, P-value = 6.3e-09) (Fig. 4c). Both regulatory layers were correlated with growth rates (Supplementary Fig. S10), suggesting common regulatory response of growth and metabolism.
Fig. 4. Metabolism-induced epistasis propagates to the proteome and increases at the metabolome.
(a) Differential protein expression between 16 metabotypes grown in supplemented SC medium determined by HDMSE as illustrated in a volcano plot, expression values log2 in x axis, adj. P-values –log10 in y axis, n=3/strain.
(b) Hierarchical clusters based on euclidean distance and complete linkage agglomeration of gene and protein expression profiles correspond to each other (r = 0.92) Transcriptome and proteome cluster the strains in a similar fashion.
(c) Correlation of the major principal component of mRNA and protein expression. Strains are color coded according to presence or absence of the LEU2 gene, that has the most consistent (least epistatic) impact.
(d) Pearson's correlation coefficient of mRNA and protein expression values for 446 proteins and transcripts as identified in (a). Transcriptome and proteome correlate quantitatively in strains with differential protein expression (insets).
(e) Transcript and protein dynamic range correspond for metabolic enzymes. Slope of regression between gene and protein expression dynamic range, distinguishing enzymatic and non-enzymatic genes. Inset: fold-change induction across all differentially expressed genes, comparing mRNAs and protein level change.
(f) A majority of genes affected by epistasis on transcriptome are epistatic the proteome as well; P-value <0.05, fold change >2, Z score > 2s or < -2s.
(g) Metabolic epistasis affects transcriptome and proteome to a similar extent, but has broader impact on metabolite concentrations (represented by the absolute quantities in 50 essential metabolites (Supplementary Fig. S11)); the distribution is expressed as Epistatic score e, describing the relative deviation of an expression quantity from a linear relationship in pairwise interactions and illustrated as density plot of all quantified transcripts, proteins and metabolites. Transcriptome and proteome show the same distribution of e, which is increased on the metabolome.
(h) Correlation between first principal component of transcriptome data (26.36%) and the second principal component of metabolome data (20.6%). Strains are colored according to the presence and absence of LEU2 gene, which had strongest loading.
Furthermore, in strains with a high level of differential gene expression, proteome and transcriptome abundance values did correspond to each other (PCC > 0.7 (Fig. 4d). Weak correlation could be observed in the strains where few differentially expressed proteins were captured (Fig. 4d). Intriguingly, for differentially expressed genes also the fold-changes did significantly correlate. This quantitative correlation was better for metabolic enzymes as it was for other genes (Fig. 4e). On all assessed levels, the proteome was found similarly dependent on the metabolic background as the transcriptome. Also metabolic epistasis was observed at the proteome level to a similar extent as on the transcriptome. Out of 257 epistatic transcripts differentially expressed >2 fold, we were able to capture quantities for 32 proteins, of which 26 were significantly epistatic (P-value <0.05) (Fig. 4f).
To relate these results to the metabolome, we used liquid chromatography selected reaction monitoring (LC-SRM) to absolutely quantify 50 important metabolites (Supplementary Fig. S11a, Supplementary Note 1). 75% of metabolites differentially concentrated > two-fold it scored epistatic (Supplementary Fig. S11b,c). Due to the different nature of metabolite concentrations, a direct comparison to transcriptome/proteome data is limited. However, when calculating an epistasis score for all quantified transcripts, proteins or metabolites, metabolite concentrations revealed the broadest distribution (Fig. 4g). Metabolism-induced epistasis hence appears to manifest stronger on the metabolite concentrations compared to transcriptome and proteome. Of note, metabolite concentrations have with allostery and posttranslational regulation additional layers were epistasis can manifest. Consistently, transcriptome and proteome were only the secondary determinant of metabolite concentration changes. The first PCs of neither proteome nor transcriptome correlated with the first PC of metabolome (Supplementary Fig. S11d). Instead, the second principal component, explaining 20.6% of the variation on metabolite level, did show a significant agreement with transcriptome (r = 0.67, P-value = 4.7e-03) and proteome (r = 0.72, P-value = 1.6e-03 (Fig. 4h).
Discussion
Metabolic genes are often nicknamed ‘housekeeping’ genes, which incorrectly implies that metabolism would be static by nature. It is increasingly recognized that the chemical-physical environment of a cell is dynamic regarding metabolite load, redox potential, or pH, and that metabolic networks are flexible and dynamically regulated. The size and physiological importance of the metabolic network exclude that gene expression is inert to these adaptations 30–32. Here, we attempt a system wide analysis of the impact of the metabolic-genetic background on transcriptome, proteome and metabolome. We exploit sixteen combinatorial auxotrophs that would be tolerated as background in a typical Saccharomyces cerevisiae genetic and genomic experiment. Several studies have found evidence for physiological importance of metabolic background deficiencies17,33–36, but in the absence of a comparative, systems scale analysis, the magnitude and nature of these effects remained unclear. One objective of the present work was hence to provide systematic high-quality data, which allows assessing the impact of metabolic-genetic backgrounds by the scientific community. The obtained datasets are valuable for elaborating the relationship between transcriptome, proteome and metabolome in response to metabolic perturbation, and revealed a surprisingly high agreement between transcriptional and proteomics results. Transcriptome and proteome did agree on the level of mRNA and protein expression, in the dynamic range, in the correlation of the first principal component, as well concordant levels of epistasis (Fig. 4). At least upon inducing metabolic perturbations in exponentially growing cells, and when focusing on high precision technology, the correlation of transcriptome and proteome is thus better as concluded in some previous studies37,38.
Despite moderate growth rate differences, which were mostly driven by the LEU2 gene, all sixteen metabotypes and all four auxotrophic markers had strong molecular impact. In fact, studying just four out of many possible metabolic perturbations, we find 3/4 of the coding genome to be affected. It is thus worth speculating that in total, virtually no gene will be inert to the metabolic make-up of a cell. Moreover, metabolism-induced gene expression was characterized by a high degree of epistasis. Despite the deletion of HIS3, LEU2, MET15 or URA3 caused strong signatures with hundreds of differentially expressed genes, there were virtually no transcripts that would always respond to their deletion; transcriptional profiles were critically sensitive to the metabolic background. The analysis of 32 strain pairs that differ in one marker at a time, allowed to categorize the nature or these transcriptional interactions. 77% of induced transcripts and 88% of transcriptional events reflected epistatic interactions between the metabolic genes. The ‘epistatic transcriptome’ was dominated by the less conserved and less connected genes, which were most commonly affected by masking/suppressive interactions followed by positive or negative quantitative interactions. We noted that the nature of epistasis is dynamic as well; a transcript falling into one category in one strain pair, could obtain another type of epistasis in another situation. These context-dependent gene expression interactions did correlate with flux, implying that the molecular signatures are concordant with metabolic activity (Supplementary Note 2). The metabolic make-up of the cell, are hence systemic determinants of the outcome of a gene expression response.
The association with flux implied that metabolic background effects are strongest for metabolic genes. Indeed, the overlap of transcriptional changes between the metabolic gene deletion and the background (83%) was substantially larger compared to a random gene and the background (34%) (Supplementary Fig. 4, Fig. 1e). The metabolic background hence confounds gene expression profiles both for non-metabolic and metabolic genes, but for the later category, the effects are stronger. Potential consequences of this observation are illustrated by the following Gedankenexperiment: If a subset of our background data would be analyzed in isolation (i.e. one analysis would just concentrate o the data recorded in the his3Δ background, while the other on the data in the HIS3 backgrounds) one would identify different gene expression changes upon deletion of URA3, MET15 or LEU2. These differences could propagate, i.e. two studies could report alternate Gene Ontology, signaling- or gene regulatory networks, and claim eventually different functions for HIS3. Differences in the metabolic-genetic background could thus have negatively impacted cross-laboratory reproducibility. This finding is consistent with the importance of the genetic background to define gene essentiality and phenotype penetrance. 10% of gene deletions have a different phenotype in the closely related S. cerevisiae strains S288c and Sigma 1278b39, where 57 are essential in just one of the two strains40. We have shown previously that metabolism is implicated in such effects; 13 synthetic lethal phenotypes in the S288c background were rescued upon repairing just three auxotrophies17.
In summary, we here address the importance of the metabolic-genetic background on gene expression interactions found on transcriptome, proteome and metabolome. Sixteen combinatorial deficiencies in histidine, leucine, uracil and methionine biosynthesis, four in principle unrelated model metabolic pathways, retained a strong molecular signature upon complementation, and did cause a system wide response involving 85% of the coding genome. The metabolic genes interacted epistatically on 77% of differentially expressed genes, and these interactions determined to a large extent the transcriptional outcome of a (metabolic) gene deletion. Metabolism-induced epistasis manifested across all molecular levels, of which transcriptome and proteome were highly correlated and did in part influence an increasingly variable metabolome. It is hence essential to examine the role of background and metabolism-induced epistasis in genetic and genomic experiments, and to elucidate its role in gene regulatory networks. Overall, metabolic make-up of a cell is a key molecular factor in defining the consequence of gene loss across transcriptome, proteome and metabolome.
Methodology
Sampling for transcriptomics and proteomics
Main-cultures were inoculated in 50 ml synthetic complete media as described in17 at a starting OD600 of 0.15 (30°C, 180 rpm). Strains were then grown for 6-19 hour until an OD600 of 0.8, the cells collected by centrifugation (2 min, 3000 g) and the supernatant discarded. The pellet was re-suspended in 1 ml H2O and aliquoted. The cells were then collected (0.5 min, 5000 g), the supernatant removed with a pipette and the sample snap frozen and stored at –80°C until further processing.
Transcriptomics
(i). Sample preparation
RNA for transcriptomics was prepared using the yeast RNA mini kit (Zymo Research) followed by DNase treatment, and processed using the TruSeq RNA library preparation kit (Illumina) following the manufacturer’s instructions. Twelve samples each were pooled, loaded on a full lane and sequenced 2x50 bp paired end on a HiSeq 2000 (Illumina), yielding cluster densities between 730,000 and 806,000 (i.e. ~10,000,000 fragments/1 GB of data per sample). The paired end reads were aligned using tophat with default parameters against the yeast genome (ENSEMBL Version EF4). To generate gene-wise read counts for gene expression estimation the htseq tool was applied with the following parameters (htseq-count --mode=intersection-nonempty,--stranded=no) in average achieving median coverages between 294 and 942 per covered gene of iGenomes EF4 gene annotation.
Table SM1. Sequencing statistics of 4 pools of 12 samples each.
| Pool | Cluster density (*1000/mm2) | total fragments approx | |
|---|---|---|---|
| 1 | 794 | 142920000 | |
| 2 | 784 | 141120000 | |
| 3 | 806 | 145080000 | |
| 4 | 730 | 131400000 | |
(ii). RNAseq data processing
RNAs with very low read count values (<50 counts across all replicates, both raw and normalized read counts) were removed, so that in total 5923 genes were considered expressed in at least one strain. ‘DEseq’41 in R was used for normalization (leading to median coverages of 757 to 788 per RNA) and calculation of P-value for differential expression of RNAseq data. Gene expression fold change between strains was calculated by dividing average normalized readcount values from all replicates. Hierarchical clustering was based on Euclidean distance and complete linkage agglomeration.
(iii). Differential mRNA expression
Differential mRNA expression was calculated with reference a) to the prototrophic wild-type strain, b) to median of gene expression of all 16 strains, or c) complete auxotrophic strain, as indicated. P-values were adjusted for multiple test correction by using Benjamini and Hochberg (BH) method42, using the p.adjust function in R. Threshold value for differentially expressed mRNAs were adj. P-value < 0.05, fold change >2 for upregulated genes and fold change < ½ for downregulated genes, and readcount > 50 in both strains. A list of differentially expressed gene from any strain were annotated to GO terms (GO:Process and GO:Functions) by GO slim mapper tool (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl)43.
Proteomics
(i). Sample preparation
For data independent acquisition proteomics experiments, the 16 strains were grown and analysed in 3 biological replicates, adding to 48 samples. Proteins were extracted using SDS-containing extraction buffer described previously44, combined with protein precipitation by 10% TCA45. Protein pellets were re-suspended in 0.1% Rapigest and 50 mM triethylammonium bicarbonate buffer. 100 µg of protein was reduced by 5 mM DTT for 30 min at 37°C, alkylated with 15 mM iodoacetamide at room temperature for 1 hour and digested with trypsin at 1:20 protein to trypsin ratio overnight as described earlier46.
(ii). LC-MS/MS proteomics
Approximately 500 ng of peptides were separated chromatographically (NanoACQUITY, Waters). The LC aqueous mobile phase (buffer A) contained 0.1% formic acid in water and the organic mobile phase (buffer B) contained 0.1% formic acid in 100% acetonitrile. The samples were trapped on column (Symmetry C18 5 μm, 180 μm × 20 mm, Waters) and desalted for 5 min at 5 ul/min flow rate of aqueous mobile phase. The separation was performed on a T3 1.8 μM, 75 μm × 250 mm column (Waters) at 300 nL/min ow rates using a 90 min linear gradient elution (from 3% to 35% organic mobile phase). The column was then washed with 80% organic mobile phase for 5 min and re-equilibrated with 3% organic mobile phase for 15 min. The analytical column temperature was maintained at 40°C. Eluting peptides were analysed on SYNAPT G2 hybrid IMS-MS system (Waters). The data was acquired in IMS-MSE mode with low and high energy scans of 900 ms. Collision energy was linearly ramped from 21 to 44 V in the Transfer region of TriWave during high energy scans. The emitters employed were manufactured by etching fused silica line with hydrofluoric acid as described by47. [Glu-1]-brinopeptide B (500 fmol/μL) was infused via the lockspray ion probe at a flow rate of 500 nL/min using an auxiliary pump and was acquired once every 30 s for 1 s period.
(iii). Proteome data processing
The raw data was initially processed with the ProteinLynx Global Server (PLGS) 2.5.2 to generate a list of precursor and fragment EMRTs and associations between them based on similarity of retention time and drift time. The thresholds for low energy ions, high energy ions and low energy exact mass retention time (EMRT) pairs were set to 100, 15 and 750 counts respectively. The data was lock-mass corrected post-acquisition using [Glu-1]-fibrinopeptide as a lock mass compound with 785.84 m/z for z = 2 and 0.25 Da tolerance window. The EMRTs were then searched against UniProt S. cerevisiae database using ion accounting algorithm described previously48. A peptide required at least one fragment and a protein required at least three fragments and one peptide for identification. The database search was performed at 100% FDR to identify real and decoy peptides for subsequent filtering on q-value. Peptide identifications from strain 1 replicates 1 and 3 and strain 16 replicate 1 were combined to produce a master in silico run, that was used to transfer identifications to all other acquisitions using synapter algorithm described previously49. To account for potential sources of technical variation in proteomic experiments, we removed peptides, which were not detected in all 48 samples. Then, for each protein a Spearman's rank correlation coefficient was calculated between each pair of peptides across all samples. Peptides displaying overall low co-correlation (with shorth50,51 correlation < 0.3) were removed from subsequent analysis. Such procedure assumes that signals of peptides coming from the same protein has to be correlated. This selection thus identifies non-specific peptides, or peptides not linear for other reasons (i.e. posttranslational modifications present to a varying extend). Finally, for each protein the signals of all detected peptides were geometrically averaged and subsequently accounted for differential expression using modified eBayes t-test using the limma package52 implemented in R.
Metabolomics
Pre-cultures of the sixteen strains were prepared in synthetic media and cultured for 12-20 h in a 96-well plate (30°C, 250 rpm). Main-cultures were inoculated in fresh SC media and cultivation, sample collection, quenching and metabolite extraction were performed according to53. Specifically, the yeast strains were cultured in 96-well fritted plates (1 strain per two wells) (Nunc, USA) with a 4 mm glass bead in each well for mixing. For quenching a 48-well plate containing 3.6 ml -40°C quenching solution (60% methanol, 10 mM NH4-acetate) per well was prepared and put into a vacuum manifold. Mid-exponential cultures were put on top of the 48-well plate and sucked into the quenching solution. The quenched cultures were centrifuged for 5 min at 4000 rpm -9°C and the supernatant was discarded. Subsequently, the plate with the cell pellets was transferred into a -50°C ethanol bath with dry ice. For extraction the cell pellets were resuspended in 1 ml precooled extraction solution (75% ethanol, 10 mM NH4-acetate) and 50 µl 13C-yeast internal standard was added. To complete extraction, the plate was transferred into a 80°C water bath for 3 min with vortexing steps every 30-45 seconds. The extracts were stored at -80°C until they were dried overnight with a vacuum centrifuge (Christ-RVC 2-33 CD plus, Kuehner AG, Switzerland). LC-MS measurements were performed as described in54. Specifically, the dried extracts were dissolved in 50-100 µl ddH2O and 15 µl were injected for LC-MS analysis. The metabolites were separated with a Waters Acquity T3 end-capped reversed phase column (150mm x 2.1mm x 1.8 µm) on a Waters Acquity UPLC (Waters, Milford, MA, USA) system. For mass spectrometric detection of the metabolites we used a Thermo TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) with a heated electrospray ionization source operating in negative mode with selective reaction monitoring. Peak integration was done with in-house software (B. Begemann & N. Zamboni, unpublished). The metabolite peak areas were further normalized to the 13C internal standard and to biomass as determined by optical density (OD600).
Metabolic pathway enrichment
Metabolic pathway enrichment of differentially expressed or epistatic genes was analysed by hypergeometric testing by using phyper function in R. False discovery rate (FDR) was calculated according to55. Threshold for significant enrichment were: FDR < 0.05, P-value < 0.05, and the number of enriched genes 1/3rd of the pathway size.
Epistasis
To avoid an error propagation problem in the epistasis analysis, we normalize gene expression values given in Figure 3, and in several of the Supplementary Figures as indicated, to the median expression value of all strains. This strategy yields largely similar results, as normalization to the prototrophic strain, as its expression profile is close to that of the median (Supplementary Fig S3i). Normalization to the median is however better suited for the epistasis analysis in our experimental set-up as it, i) avoids unequal error propagation of the wild-type strain measurements, and ii) allows to compare all sixteen strains with each other).
To calculate epistasis from the median-normalized data, Fisher’s additive model was used as basis to elaborate an epistasis score (e) for mRNA expression, protein expression, metabolite concentration and growth data21. Epistasis score (e) = observed (WAB) – expected (WA + WB), where WAB, WA and WB are percentual changes in gene expression, protein expression, metabolite concentration or growth rate of knock-out strains56–58. The calculation was performed comparing strains representing closest neighbours in a genetic dependence network (Fig 3c), comparing all 38 possible strain pairs in total. P-value (BH adjusted) for differential expression of genes in all 38 strains pairs was calculated with respect to the respective product strains. Expression fold change was calculated by dividing average expression values in each strain with respect to the respective wild-type, median value, or any other reference strain as indicated. Epistatic score was transformed to a standard epistatic score (Z score). mRNAs, proteins or metabolites with significant P-value (<0.05) in at least one strain, plus a Z score > 2σ or < -2σ were considered epistatic. As illustrated in Fig 3b, the standard epistasis score and P-value (adjusted) for differential expression were used to subgroup epistatic genes into masking, suppression, pseudo-masking as well as positive and negative epistasis. Categorization was conducted for differential expression in all 38 strains pairs representing closest neighbourhoods (Fig 3c) and fold change with respect to median value were used for normalisation. (a) Masking: expression value is explained by the allele with quantitative stronger effect; (b) Suppression: expression value is explained by the allele with quantitative lower effect; (c) Pseudo-masking: expression value depends on both alleles, but the effect is lower as expected from the strong allele and larger as expected from the weak allele; (d) Positive epistasis: expression value is larger than expected from additivity of the single alleles (e) Negative epistasis: Expression values is lower than expected from additivity of the two single alleles and lower than each individual value.
Gene essentiality and conservation
A list of essential S. cerevisiae genes was taken from Database of Essential Genes (DEG) (http://www.essentialgene.org)59. Result of PSI-BLAST60 analysis of S. cerevisiae from SGD database43 was used to count the number of species in which a gene is conserved.
Confounding effects of metabolism-induced transcriptional changes in transcriptomics
Gene expression data for a total of 118 yeast knockout microarray experiments, conducted in 14 studies in different context and laboratories (ArrayExpress ID: E-GEOD-18644, E-GEOD-18994, E-GEOD-21571, E-GEOD-25582, E-GEOD-28794, E-GEOD-29530, E-GEOD-31176, E-GEOD-31326, E-GEOD-31774, E-GEOD-56702, E-MEXP-3150, E-MTAB-1059, E-MTAB-2539, E-TABM-63859,61–71) was downloaded from ArrayExpress. All of these studies are based on the (BY4741 background auxotroph in histidine, leucine, uracil and methionine, as used in our study. We categorize these datasets into knock-out experiments (49 arrays, used in Fig 1e) or all other conditions (69 arrays from 4 studies, Supplementary Fig. 4a). The raw data from Affymetrix GeneChip Yeast Genome 2.0 and S98 Arrays was re-analysed from applying the same stringent quality filtering criteria as used on our RNASeq data (FDR adjusted P-value < 0.05 and fold change >2). The lists of differentially expressed genes in 49 microarray experiments were compared with differentially expressed genes affected by the auxotrophic markers in the respective yeast strain background.
Flux variability analysis
Yeast genome-scale metabolic model (NAD corrected iMM90472,73) was used to perform a flux variability analysis (FVA)19,20. The model was constrained by setting the biomass flux according to the experimentally determined growth rates of the sixteen strains17. Metabolite uptake rates for amino acids and glucose are constrained according to experimentally observed uptake rates (Supplementary File 2). Upon including both growth rate and nutrient uptake rates, FVA predicts a feasible flux range for every reaction of the model and for every strain. Differences in metabolic flux (maximum flux – minimum flux) expressed as fold change and are calculated with reference to the prototrophic wild type strain. Flux-range is considered to be significantly different when it deviates 2 stdev from the mean.
Data Accessibility
-
(1)
Transcriptome data have been submitted to the ArrayExpress database18 (https://www.ebi.ac.uk/arrayexpress/) under accession number: E-MTAB-3991.
-
(2)
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository74 with the dataset identifier PXD001491.
-
(3)
Metabolome data have been submitted to the Metabolights database75 (http://www.ebi.ac.uk/metabolights/) with the accession number: MTBLS168.
-
(4)
Processed data: Supplementary File 2
Supplementary Material
Acknowledgments
We thank Uwe Sauer (ETH Zurich) for support in metabolite measurements and scientific discussion, and Martin Werber and Sven Klages (Max Planck Institute for Molecular Genetics) for support in RNA sequencing analysis. We thank the Wellcome Trust (RG 093735/Z/10/Z), the ERC (Starting grant 260809), the Isaac Newton Trust (RG 68998), and the Darwin Trust of Edinburgh for a studentship for P.V.S. A.Z. is EMBO fellow. M.R. is a Wellcome Trust Research Career Development and Wellcome-Beit Prize fellow.
Footnotes
Author’s contribution
MTA, AZ, RS, ER, SB performed data analysis, MM, PS, SC raw data processing, MM, PS, FC, JV, AK, EC, SM, SC experiments, KRP, BT, KSL and MR concepted the study, MR wrote the first draft, MTA, AZ and MR wrote the paper, all authors contributed in preparing the final version.
The authors declare no competitive interests.
References
- 1.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 2.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 3.Herrgård MJ, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási A-L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 5.Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark AG, Fucito CD. Stress tolerance and metabolic response to stress in Drosophila melanogaster. Heredity. 1998;81:514–527. doi: 10.1046/j.1365-2540.1998.00414.x. [DOI] [PubMed] [Google Scholar]
- 9.Ihmels J, Levy R, Barkai N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Li Y, Tollefsbol TO. Gene-Environment Interactions and Epigenetic Basis of Human Diseases. Curr Issues Mol Biol. 2008;10:25–36. [PMC free article] [PubMed] [Google Scholar]
- 11.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the Yeast Metabolic Cycle: Temporal Compartmentalization of Cellular Processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 12.Campbell K, et al. Self-establishing communities enable cooperative metabolite exchange in a eukaryote. eLife. 2015:e09943. doi: 10.7554/eLife.09943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fink GR. Gene-enzyme relations in Histidine biosynthesis in yeast. Science. 1964;146:525–527. doi: 10.1126/science.146.3643.525. [DOI] [PubMed] [Google Scholar]
- 14.Satyanarayana T, Umbarger HE, Lindegren G. Biosynthesis of branched-chain amino acids in yeast: regulation of leucine biosynthesis in prototrophic and leucine auxotrophic strains. J Bacteriol. 1968;96:2018–2024. doi: 10.1128/jb.96.6.2018-2024.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacroute F. Regulation of Pyrimidine Biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1968;95:824–832. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masselot M, De Robichon-Szulmajster H. Methionine biosynthesis in Saccharomyces cerevisiae. I. Genetical analysis of auxotrophic mutants. Mol Gen Genet MGG. 1975;139:121–132. doi: 10.1007/BF00264692. [DOI] [PubMed] [Google Scholar]
- 17.Mülleder M, et al. A prototrophic deletion mutant collection for yeast metabolomics and systems biology. Nat Biotechnol. 2012;30:1176–1178. doi: 10.1038/nbt.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brazma A, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher RA. The Correlation Between Relatives on the Supposition of Mendelian Inheritance. 1918 [Google Scholar]
- 22.Park S, Lehner B. Epigenetic epistatic interactions constrain the evolution of gene expression. Mol Syst Biol. 2013;9:645. doi: 10.1038/msb.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costanzo M, et al. The Genetic Landscape of a Cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, et al. YeastNet v3: a public database of data-specific and integrated functional gene networks for Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D731–D736. doi: 10.1093/nar/gkt981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 2012;490:535–538. doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- 26.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157:740–752. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 27.Alam MT, Medema MH, Takano E, Breitling R. Comparative genome-scale metabolic modeling of actinomycetes: the topology of essential core metabolism. FEBS Lett. 2011;585:2389–2394. doi: 10.1016/j.febslet.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res. 2013;12:2323–2339. doi: 10.1021/pr300775k. [DOI] [PubMed] [Google Scholar]
- 29.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 30.Grüning N-M, Lehrach H, Ralser M. Regulatory crosstalk of the metabolic network. Trends Biochem Sci. 2010;35:220–227. doi: 10.1016/j.tibs.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Patel A, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 32.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto S, et al. Isolation of Auxotrophic Mutants of Diploid Industrial Yeast Strains after UV Mutagenesis. Appl Environ Microbiol. 2005;71:312–319. doi: 10.1128/AEM.71.1.312-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kokina A, Kibilds J, Liepins J. Adenine auxotrophy--be aware: some effects of adenine auxotrophy in Saccharomyces cerevisiae strain W303-1A. FEMS Yeast Res. 2014;14:697–707. doi: 10.1111/1567-1364.12154. [DOI] [PubMed] [Google Scholar]
- 35.Low B. Rapid Mapping of Conditional and Auxotrophic Mutations in Escherichia coli K-12. J Bacteriol. 1973;113:798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol. 2002;68:2095–2100. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic. 2004;3:212–219. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]
- 38.Payne SH. The utility of protein and mRNA correlation. Trends Biochem Sci. 2015;40:1–3. doi: 10.1016/j.tibs.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan O, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 40.Dowell RD, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
(1)
Transcriptome data have been submitted to the ArrayExpress database18 (https://www.ebi.ac.uk/arrayexpress/) under accession number: E-MTAB-3991.
-
(2)
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository74 with the dataset identifier PXD001491.
-
(3)
Metabolome data have been submitted to the Metabolights database75 (http://www.ebi.ac.uk/metabolights/) with the accession number: MTBLS168.
-
(4)
Processed data: Supplementary File 2