Abstract
Recent progress in molecular understanding of the retinoid cycle in mammalian retina stems from painstaking biochemical reconstitution studies supported by natural or engineered animal models with known genetic lesions and studies of humans with specific genetic blinding diseases. Structural and membrane biology have been used to detect critical retinal enzymes and proteins and their substrates and ligands, placing them in a cellular context. These studies have been supplemented by analytical chemistry methods that have identified small molecules by their spectral characteristics, often in conjunction with the evaluation of models of animal retinal disease. It is from this background that rational therapeutic interventions to correct genetic defects or environmental insults are identified. Thus, most presently accepted modulators of the retinoid cycle already have demonstrated promising results in animal models of retinal degeneration. These encouraging signs indicate that some human blinding diseases can be alleviated by pharmacological interventions.
Keywords: retinoids, visual cycle, rhodopsin, cone pigments, RPE65, LRAT, retinol dehydrogenases, A2E, visual cycle modulator, age-related macular degeneration
1. DISCOVERY OF VISUAL PIGMENT REGENERATION PATHWAYS
The sole action of light in visual sensation is to photoisomerize the 11-cis-retinal (11-cis-RAL) chromophore of photoreceptor visual pigments in the retina to an all-trans configuration (Wald 1968). Conformational changes in the visual pigment elicited by this isomerization allow the receptor to couple to its cognate G protein and initiate the phototransduction cascade (Palczewski 2006). The absorption of light by visual pigments destroys their chromophore properties in a process referred to as photochemical bleaching (Wald 1968). Sustained vision thus relies on a mechanism for regenerating ground-state visual pigments (Kiser et al. 2014, McBee et al. 2001). This regeneration pathway was originally referred to as the visual cycle, although the more recent term retinoid cycle is probably a more accurate description of the process (the terms are used interchangeably in this article).
Research into this area of visual science was initiated in the late 1800s with the work of Böll and Kühne (Ripps 2008) who discovered the presence of a substance in the rod-dominant frog retina that they named visual purple, speculated to be the light-responsive substance of vision. Exposure of the purplish-red retina to light resulted in its progressive bleaching over the course of several seconds. Kühne made the critical discovery that the color of the retina could be restored if it was positioned back in contact with the retinal pigment epithelium (RPE) and placed in the dark, thus establishing the existence of a two-cell type system required for visual pigment regeneration.
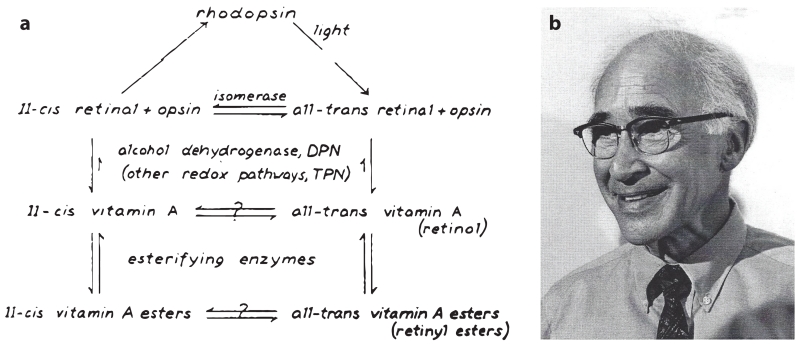
The molecular basis of these processes was revealed by the research of George Wald and his colleagues (Hubbard & Wald 1952, Wald 1968) in the mid-1900s (Figure 1). They found that the chromophore of the visual pigments is a vitamin A derivative known as retinaldehyde or retinene, as it was called then. This group also elucidated the multistep photochemistry of rhodopsin activation and laid out the first detailed scheme of chemical reactions involved in visual chromophore regeneration (Figure 1a) (see also the sidebar, Chemical Reactions of the Retinoid Cycle, and Figure 2). Following photoactivation, visual pigments release their chromophore as all-trans-RAL which is reduced to all-trans-retinol (all-trans-ROL) within photoreceptor cell outer segments (Dowling 1960). The latter compound is then transferred to the RPE where it undergoes esterification by a microsomal enzyme (Andrews & Futterman 1964), lecithin:retinol acyltransferase (LRAT) (Ruiz et al. 1999), to form all-trans-retinyl esters. During dark adaption, the retinyl esters are gradually converted back to 11-cis-RAL, which combines with apo-opsins to regenerate light-responsive, ground-state visual pigments (Dowling 1960). In the 1980s, a phospholipid membrane-dependent retinoid isomerase activity was discovered in RPE cellular fractions (Bernstein et al. 1987, Deigner et al. 1989), some 50 years after Wald’s first description of the visual cycle pathway. This retinoid isomerase activity was eventually linked to an RPE-specific protein called RPE65 (retinal pigment epithelium–specific 65 kDa protein) (Jin et al. 2005; Moiseyev et al. 2005; Redmond et al. 1998, 2005).
Figure 1.
The visual cycle pathway as proposed by George Wald. (a) Reactions involved in rhodopsin bleaching and regeneration of the visual chromophore. Panel a reproduced with permission from the George Wald Nobel lecture, copyright The Nobel Foundation. (b) A picture of George Wald following the announcement that he had won the 1967 Nobel Prize in Physiology or Medicine. Photograph courtesy of Dr. John Dowling (Harvard University).
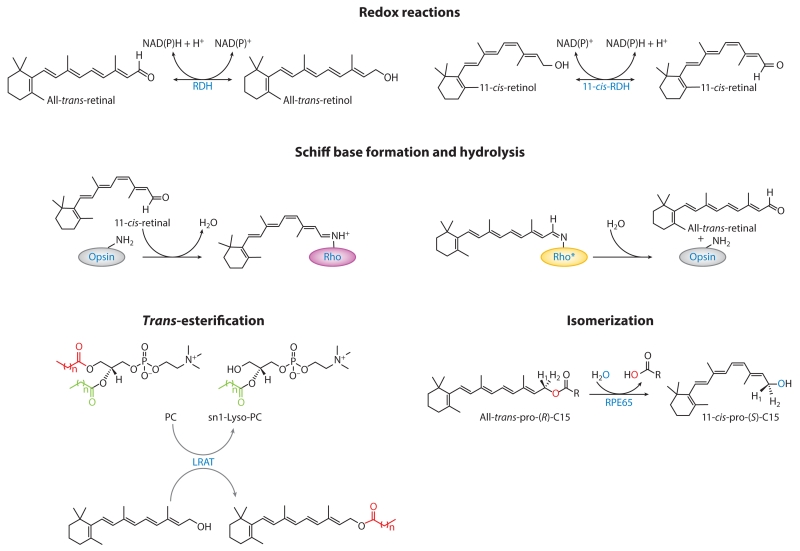
Figure 2.
Reactions that constitute the retinoid cycle. The asterisk denotes the active signaling state of rhodopsin (meta-II rhodopsin). Abbreviations: LRAT, lecithin:retinol acyltransferase; NAD, nicotinamide adenine dinucleotide; PC, phosphatidylcholine; RDH, retinol dehydrogenase; Rho, rhodopsin; RPE, retinal pigment epithelium.
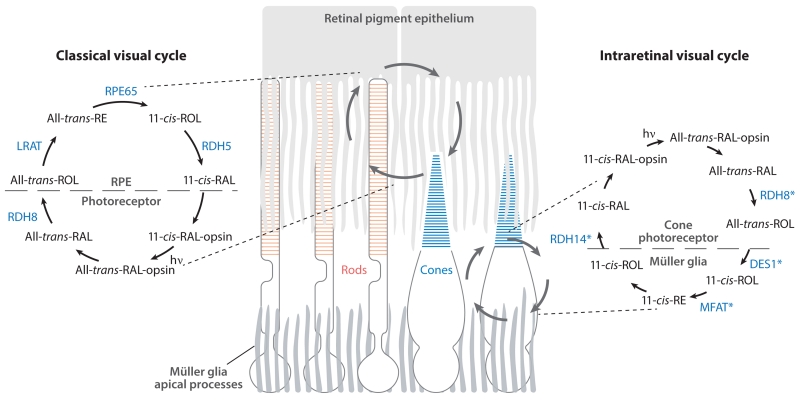
The reactions described above comprise the classical visual cycle essential for both rod and cone cell function in humans (Figure 3) (Jacobson et al. 2007, Saari 2012). Moreover, there is experimental support for the existence of a second pathway for 11-cis-RAL biosynthesis in the retina (Fleisch et al. 2008, Wang et al. 2009). This intraretinal visual cycle specifically supplies cone photoreceptors with quantities of visual chromophore sufficient for operation under photopic conditions (Wang & Kefalov 2011). The existence of such an alternative pathway was first suggested by experiments demonstrating that cone photoreceptors in isolated frog retinas regenerated in the dark, unlike rod photoreceptors which depend on the RPE for such regeneration (Goldstein 1970). Defined enzymatic activities thought important for cone-specific regeneration have been identified in the retinas of cone-dominant species (Figure 3) (Mata et al. 2002, Muniz et al. 2009). More recently, candidate enzymes of this pathway have been proposed, although their physiological relevance remains to be determined (Tables 1 and 2). In this article, we primarily focus on progress made in understanding the classical RPE-based retinoid cycle, as well as diseases that result from the disruption of this pathway.
Figure 3.
Retinoid transformations and enzymes of the classical and intraretinal visual cycles. (Left) The classical visual cycle involving enzymes located in the RPE and photoreceptor outer segments. This pathway supports both rod and cone cell function. (Right) Reactions comprising the putative intraretinal visual cycle that provides cones with a privileged source of 11-cis-RAL. This pathway is thought to involve enzymes located in Müller glia and cone photoreceptors. Candidate enzymes of the pathway are shown in blue and marked by asterisks to indicate that their physiological involvement in the pathway has not yet been established. Abbreviations: DES1, dihydroceramide desaturase 1; hv, light; LRAT, lecithin:retinol acyltransferase; MFAT, multifunctional O-acyltransferase; RAL, retinal; RDH, retinol dehydrogenase; RE, retinyl ester; ROL, retinol; RPE, retinal pigment epithelium.
Table 1. Genes of the retinoid cycle.
| Gene name | Gene symbol and structurea | Disabling mutations in humans: phenotypee,g |
Knockout mouse phenotype | Section in this review |
|---|---|---|---|---|
|
| ||||
| Rhodopsin |
RHO, 5 exons localized on chromosome 3q21-q24 |
Accounts for 20–30% of autosomal dominant RPb; more than 100 distinct mutations; the Pro23His causes ~10% of autosomal dominant RP in the United States; autosomal recessive RP mutations are rarec (≤1%) |
Rapid rod cell degeneration, lack of ROS, subsequent cone degeneration |
Section 2 |
|
| ||||
| Opsin 1f (cone pigments), long-wave sensitive; opsin 1 (cone pigments), medium-wave sensitive; opsin 1 (cone pigments), short-wave sensitive |
OPN1LW, 6 exons localized on chromosome Xq28; OPN1MW, 6 exons localized on chromosome Xq28; OPN1SW, 5 exons localized on chromosome 7q32.1 |
Protanopia (1 % of males) and protanomaly (1 % of males, 0.01% of females): lacking or blue-shifted long-wavelength-sensitive retinal cones; deuteranopia (1 % of males) and deuteranomaly (most common type: 6% of males, 0.4% of females): lacking or mutated medium-wavelength cones; tritanopia (< 1 % of males and females) and tritanomaly (equally rare for males and females, 0.01% for both): lacking or mutated short-wavelength cones |
Large differences noted between color vision of humans and rodents |
Section 2 |
|
| ||||
| Melanopsin |
OPN4, 11 exons localized on chromosome 10q22 |
Seasonal affective disorder (prevalence unknown) |
Affects the magnitude of circadian clock photic responses (but is not essential) |
Section 2 |
|
| ||||
| Retinal G protein–coupled receptor |
RGR, 7 exons localized on chromosome 10q23.1 |
Autosomal recessive RP (overall 1 in 3,000–7,000 people, but mutations in the RGR gene are rare at ≤1 %) |
Changes in retinoid content, specifically the isomeric composition of retinyl esters |
Section 2 |
|
| ||||
| ATP-binding cassette, subfamily A (ABC1), member 4 |
ABCA4, 50 exons localized on chromosome 1p22.1 |
Recessive Stargardt disease (prevalence of ~1 in 10,000); juvenile- and late-onset recessive macular dystrophy; autosomal recessive RP (2–5%); recessive fundus flavimaculatus; recessive cone–rod dystrophy; age-related macular degeneration |
A minor phenotype in mice; accumulation of all-trans-RAL adducts with age |
Section 3 |
|
| ||||
| Stimulated by retinoic acid 6 |
STRA6, 19 exons localized on chromosome 15q24.1 |
Microphthalmia, syndrome 9 (MCOPS9), a rare clinical entity that also causes pulmonary hypoplasia; only a few children survive |
Mice are viable when bred on diets replete with vitamin A, but display markedly reduced levels of ocular retinoids; malformations in the choroid and RPE; early cone photoreceptor cell death |
Section 3 |
|
| ||||
| Retinol dehydrogenase 5 |
RDH5, 5 exons localized on chromosome 12q13-q14 |
Recessive fundus albipunctatus; features late-onset recessive cone dystrophy (prevalence 1 in 2,000) |
Rdh5−/− mice have delayed dark recovery at high bleaching levels; a large increase in 11/13-cis-retinyl esters also has been noted |
Section 4 |
|
| ||||
| Retinol dehydrogenase 8 |
RDH8, 6 exons localized on chromosome 19p13.2 |
No human retinal disease has been associated with mutations of this gene |
Significant accumulation of all-trans-RAL, and delayed recovery of rod function has been noted following exposure to bright light |
Section 4 |
|
| ||||
| Retinol dehydrogenase 10 |
RDH10, 7 exons localized on chromosome 8q21.11 |
No human retinal disease has been associated with mutations of this gene |
Rdh10−/− leads to early embryonic lethality |
Section 4 |
|
| ||||
| Retinol dehydrogenase 11 |
RDH11, 7 exons localized on chromosome 14q24.1 |
One family identified with autosomal recessive RP displayed facial dysmorphology, developmental delay, and short stature |
Minor effects; single-flash ERGs of Rdh11−/− mice showed normal responses under dark- and light-adapted conditions, but exhibited delayed dark adaptation after bleaching levels of light |
Section 4 |
|
| ||||
| Retinol dehydrogenase 12 |
RDH12, 9 exons localized on chromosome 14q24.1 |
LCAd with severe childhood retinal dystrophy (4% of all cases); autosomal dominant RP (rare) |
Mice display slowed kinetics of all-trans-RAL reduction, delayed dark adaptation, increased susceptibility to light-induced photoreceptor apoptosis |
Section 4 |
|
| ||||
| Retinol dehydrogenase 13 |
RDH13, 12 exons localized on chromosome 19q13.42 |
No human retinal disease has been associated with mutations of this gene |
No obvious phenotype; in mice, intense light exposure caused swollen mitochondria with disrupted cristae |
Section 4 |
|
| ||||
| Retinol dehydrogenase 14 |
RDH14, 2 exons localized on chromosome 2p24.2 |
No human retinal disease has been associated with mutations of this gene |
Not generated | Section 4 |
|
| ||||
| Retinol dehydrogenase; dehydrogenase/ reductase (SDR family) member 3 (retSDR) |
DHRS3, 8 exons localized on chromosome 1p36.1 |
No human retinal disease has been associated with mutations of this gene |
Mice lacking this gene die before weaning and exhibit altered retinoid metabolism, and heart, craniofacial, and skeletal defects |
Section 4 |
|
| ||||
| Lecithin:retinol acyltransferase |
LRAT, 3 exons localized on chromosome 4q32.1 |
Early onset autosomal recessive RP; LCA(≤1% of all cases) |
Lrat−/− mice exhibit pupillary constriction and have trace levels of all-trans-retinyl esters in their liver, lung, eye, and blood; scotopic and photopic ERGs are severely attenuated at an early age |
Section 5 |
|
| ||||
| Retinoid isomerase |
RPE65, 14 exons localized on chromosome 1p31 |
LCA (or early onset autosomal recessive RP (2–5% of all cases) with an estimated prevalence of ~1 in 80,000; RPE65 LCA is thought to represent about 6% of all LCA cases |
Rpe65−/− mice exhibit poor rod function; they lack rhodopsin and overaccumulate all-trans-retinyl esters |
Section 6 |
|
| ||||
| Retinol-binding protein 4 |
RBP4, 6 exons localized on chromosome 10q23.33 |
Features RPE atrophy with night blindness and reduced visual acuity; scotopic and photopic ERGs are absent or reduced (extremely rare) |
Rbp4−/− mice have low serum retinol levels (12.5% of wild-type animals) with low retinoid levels in the eye, and impaired retinal function and visual acuity; they can recover normal vision only after several months on a vitamin A–sufficient diet |
Section 7 |
|
| ||||
| Retinaldehyde-binding protein 1 (CRALBP) |
RLBP1, 10 exons localized on chromosome 15q26 |
Autsomal recessive RP; recessive Newfoundland rod–cone dystrophy; recessive Bothnia dystrophy; recessive retinitis punctata albescens (≤1% of all cases) |
Rlbp1−/− mice display rhodopsin regeneration; 11-cis-RAL production and dark adaptation are delayed by > 10-fold; no photoreceptor degeneration has been observed |
Section 7 |
|
| ||||
| Cellular retinol-binding protein 1 (CRBP1) |
RBP1, 6 exons localized on chromosome 3q23 |
No human retinal disease has been associated with mutations of this gene |
Rbp1−/− mice are healthy and fertile, with about a 50% reduction of retinyl ester accumulation in their hepatic stellate cells; CRBP1 is required for efficient retinyl ester synthesis and storage |
Section 7 |
|
| ||||
| Interphotoreceptor (interstitial) retinol binding protein (IRBP) |
RBP3, 4 exons localized on chromosome 10q11.2 |
Autosomal recessive RP (≤1% of all cases) |
Irbp−/− mice reveal some loss of photoreceptor nuclei; the rates of recovery of 11-cis-RAL and of regeneration of rhodopsin in the dark in Irbp−/− mice are similar to those of wild-type mice; deletion of IRBP reduces the amplitude and slows the kinetics of mouse M- and L-cone photoresponses, but cone adaptation to bright, steady light and the kinetics of cone dark adaptation are unaffected; IRBP does not increase pigment regeneration and is not critical for mouse M- and L-cone function in bright light |
Section 7 |
Abbreviations: ATP, adenosine triphosphate; CRALBP, cellular retinaldehyde-binding protein; ERG, electroretinograph(y); His, histidine; LCA, Leber congenital amaurosis; Pro, proline; RAL, retinal; ROS, rod outer segments; RP, retinitis pigmentosa; RPE, retinal pigment epithelium; SDR, short-chain dehydrogenase/reductase.
Nomenclature according to HUGO Gene Nomenclature Committee.
Autosomal dominant retinitis pigmentosa (1 in 14,000).
Autosomal recessive retinitis pigmentosa (1 in 20,000).
Recessive Leber congenital amaurosis (2–3 cases per 100,000).
For updated information about mutations and the novelty of genes identified as being associated with retinal diseases, consult the curated RetNet (https://sph.uth.edu/retnet/).
For updated information about mutations causing changes in color vision, consult Causes of Color’s page on causes and incidence of color blindness (http://www.webexhibits.org/causesofcolor/2C.html).
Nonsyndromic RP by mode of inheritance: autosomal dominant RP, 15–25%; autosomal recessive RP, 5–20%; X-linked RP, 5–15%; unknown, 40–50%; digenic RP, very rare (from RetNet https://sph.uth.edu/retnet/).
Table 2. Proteins of the retinoid cycle.
| Protein | Localization | 3D structure | Function | Section in this review |
|---|---|---|---|---|
|
| ||||
| Rhodopsin | Rod cells; membrane-bound in rod outer segments |
X-ray structure available; a seven-transmembrane helical protein |
11-cis-retinylidene-binding protein; a rod photoreceptor light receptor that triggers phototransduction |
Section 2 |
|
| ||||
| Cone pigments | Cone cells; membrane-bound in cone outer segments |
Model based on bovine rhodopsin structure is available; seven- transmembrane helical proteins |
11-cis-retinylidene-binding proteins; cone photoreceptor light receptors that trigger phototransduction |
Section 2 |
|
| ||||
| Melanopsin | A subset of ganglion cells; cellular membrane-bound |
Model based on invertebrate rhodopsin structure is available; a seven- transmembrane helical protein |
A retinylidene-binding protein; a photoreceptor light receptor that triggers Gq phototransduction, controls sleep-awake pattern |
Section 2 |
|
| ||||
| RGR | RPE | Model based on invertebrate rhodopsin structure is available; a seven- transmembrane helical protein |
A retinylidene-binding protein; a photoreceptor light receptor that triggers Gq phototransduction; function still unclear |
Section 2 |
|
| ||||
| ABCA4 | Rod and cone outer segment rim regions |
An 18 Å resolution structure of ABCA4 isolated from bovine rod outer segments was determined by electron microscopy and single-particle reconstruction; a minimum of four domains is required for functional activity: two transmembrane domains, each with six predicted membrane-spanning helices and two nucleotide-binding domains, and two intradiscal domains, each linked to one of the transmembrane domains |
An ABC importer; transfers retinoid from the intradiscal space to the cytoplasm |
Section 3 |
|
| ||||
| STRA6 | In the RPE, STRA6 is localized to the basolateral membranes; also in brain, choroid plexus microvessels, testis, spleen, kidney, placenta, and the female reproductive tract |
9–11 transmembrane helical segments predicted; limited structural information available |
RBP4 receptor; transports all-trans-ROL driven by intracellular LRAT activity |
Section 3 |
|
| ||||
| RDH5a | Abundant expression in the RPE | From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the second binds substrate |
Redox reaction specific to 11-cis-RAL/11-cis-ROL; NAD-specific |
Section 4 |
|
| ||||
| RDH8a | Abundant in rod and cone outer segments |
From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL; NADP specific |
Section 4 |
|
| ||||
| RDH10a | Broadly expressed | From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP dinucleotide specificity |
Section 4 |
|
| ||||
| RDH11a | Broadly expressed; high levels in prostate epithelium |
From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP specific |
Section 4 |
|
| ||||
| RDH12a | Expressed in rod and cone inner segments, skin, cervical epithelium, liver, and platelets |
From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP specific |
Section 4 |
|
| ||||
| RDH13a | Suggested mitochondrial localization |
From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP specific |
Section 4 |
|
| ||||
| RDH14a | Not fully established | From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP specific |
Section 4 |
|
| ||||
| DHRS3a | Very broad expression, with highest in liver, smooth muscle, and cone outer segments |
From the SDR family; two domains: one binds NAD(P) (Rossmann fold) and the other binds substrate |
Redox reaction specific to all-trans-RAL/all-trans-ROL or cis-RAL/cis-ROL (dual specificity); NADP specific; low specific activity stimulated by RDH10 |
Section 4 |
|
| ||||
| LRATb | Very broad expression; enriched in hepatic stellate cells and the RPE |
Dimeric, with each monomer composed of a four-strand, antiparallel β-sheet and three α-helices, similar to NlpC/P60 thiol proteases, with a Cys-Elis-His catalytic triad |
Esterification of ROLs; specificity toward sn1 phospholipid transfer of fatty acid chains |
Section 5 |
|
| ||||
| RPE65 | Expression limited to the RPE | Seven-bladed β-propeller, monotopically bound to endoplasmic reticulum membranes |
A retinoid isomerase that catalyzes the conversion of all-trans-retinyl esters into 11-cis-ROL |
Section 6 |
|
| ||||
| RBP4 | Expressed in liver; lower expression in adipocytes |
The overall fold consists of a core β-barrel of eight up-and-down β-strands, an N-terminal coil, and a C-terminal α-helix followed by a coil region (lipocalin family) |
Carrier of all-trans-ROL in a complex with transthyretin |
Section 7 |
|
| ||||
| CRALBP | Expressed in RPE and Müller cells |
Overall protein folds similar to the prototypical SEC14-like domain structure exhibited by α-tocopherol transfer protein; the N-terminal α- domain comprises five helices; the C-terminal αβα domain contains a β-sheet with one antiparallel and four parallel strands (β1–β5) and six helices |
Highly selective carrier of 11-cis-RAL; also binds 11-cis-ROL and 9-cis-retinoids |
Section 7 |
|
| ||||
| CRBP1 | Expressed broadly at low levels | Compaction of two antiparallel β-sheets forms an orthogonal barrel |
Cellular RBP | Section 7 |
|
| ||||
| IRBPc | Expressed in photoreceptors, pineal gland |
Four homologous domains; the domain structure is similar to photosystem II D1 C-terminal processing protease and the enoyl-CoA isomerase/hydratase family (a three-helix bundle followed by a small β-strand connected to a large six-stranded, mixed β-sheet subdomain with four helices packed against one side and a fifth helix) |
An extracellular protein; transporter of retinoids between the RPE and photoreceptor cells; possibly involved in lipid binding |
Section 7 |
Abbreviations: ABCA4, adenosine triphosphate-binding cassette transporter 4; CRALBP, cellular retinaldehyde-binding protein; CRBP, cellular retinal-binding protein 1; Cys, cysteine; His, histidine; IRBP, interphotoreceptor retinoid-binding protein; LRAT, lecithin: retinol acyltransferase; NAD, nicotinamide adenine dinucleotide; RAL, retinal; RBP, retinol-binding protein; RDH, retinol dehydrogenase; RGR, retinal G protein–coupled receptor; ROL, retinol; RPE, retinal pigment epithelium; SDR, short-chain dehydrogenase/reductase; STRA6, stimulated by retinoic acid 6.
Structure determined only for several other members of the SDR family.
Structures of a hybrid or related proteins were determined.
Structure of the whole protein is unknown.
2. VISUAL PIGMENTS AND NONVISUAL OPSINS
Rhodopsin is the sole visual pigment of rod photoreceptors. Owing to its natural abundance in the rod-dominant retinas of many common laboratory animals and cattle (Palczewski 2006), this pigment is the most thoroughly characterized member of the mammalian opsin protein family. In rod outer segments, most of the rhodopsin resides in paracrystalline arrays within intracellular disks, of which in mice there are approximately 600 per outer segment. Rhodopsin constitutes an integral component of the rod outer segment structure as demonstrated by the absence of photoreceptor outer segments in Rho−/− mice (Humphries et al. 1997). The rod outer segment plasma membrane also contains a subpopulation of rhodopsin with distinct properties as compared with intradiscal rhodopsin (Kessler et al. 2014).
The lipophilic nature of rhodopsin was first noted by Kühne (1977), who observed that it could be solubilized in a spectrally intact form by treating rod outer segments with bile detergents. Determination of the bovine rhodopsin amino acid sequence (Hargrave et al. 1983, Nathans & Hogness 1983) has revealed seven distinct stretches of hydrophobic amino acids of appropriate length to be embedded as α-helices in a phospholipid membrane. This topology was confirmed by the projection structure of rhodopsin (Schertler et al. 1993), which provided the first detailed information concerning the configuration of the α-helical bundle of this protein. Shortly after elucidation of the rhodopsin primary structure, the gene encoding the β1-adrenergic receptor (β1-AR) was identified, which revealed that these two proteins belong to the G protein–coupled receptor superfamily (Dixon et al. 1986). Like the β1-AR, rhodopsin activation has been shown to stimulate downstream second messenger–generating enzymes (PDE6 and adenylyl cyclase for, respectively, rhodopsin and β1-AR) by catalyzing the exchange of guanosine triphosphate for guanosine diphosphate in an intermediary signaling, heterotrimeric G protein called transducin (Kwok-Keung Fung & Stryer 1980).
In addition to rhodopsin, there are a number of other light-activated opsin molecules expressed in the human retina. These include cone visual pigments, which mediate color vision, the retinal G protein–coupled receptor (RGR), which is involved in mobilizing retinyl esters within the RPE (Radu et al. 2008, Wenzel et al. 2005), melanopsin, expressed in photosensitive ganglion cells that trigger the pupillary light response (Lucas et al. 2003, Panda et al. 2003), and related proteins, which are involved in circadian rhythmicity (Van Gelder 2008). These light receptors belong to the broad type 2 opsin family and typically signal through G proteins (Ernst et al. 2014). Type 1 opsins are a second group of retinylidene proteins expressed in lower organisms that exhibit significant structural similarity to type 2 opsins, including a seven-transmembrane-spanning α-helical fold and a retinal-binding lysine (Lys) residue located at a position equivalent to Lys296 in rhodopsin (Spudich et al. 2000). Type 1 opsins function as light-driven ion channels or pumps, or mediate phototaxis through activation of transducer molecules. Type 1 and 2 opsins lack a discernible sequence identity to each other and differ in the configurations of their α-helical bundles. This finding seems to indicate that the two opsin families arose via convergent evolution, with the equivalent positioning of the lysine being a critical structural feature that had to be achieved twice during evolution from two ancestral sequences (Spudich et al. 2000). However, a recent study demonstrating that functional rhodopsin can be formed from mutants in which this lysine residue has been relocated within the helical bundle suggests that type 1 and 2 opsins descended from a common ancestral sequence (Devine et al. 2013).
Rhodopsin, like all known vertebrate visual pigments, senses light through a Schiff base–linked 11-cis-retinylidene chromophore (Hubbard & Wald 1952). The absorption of light by this chromophore promotes transition of electrons from ground-state to excited-state molecular orbitals. It is this change in electron density distribution that allows the cis–trans isomerization to proceed. Notably, photoisomerization of the 11-cis chromophore is extremely rapid, occurring within 200 fs after light absorption. Torsional vibrations of the bound 11-cis-retinylidene occur over a similar time scale, and vibrational coherence has been observed in the all-trans-retinylidene photoproduct (Wang et al. 1994). These findings imply that isomerization involves small movements, mainly within the center of the all-trans-RAL molecule, and that these movements are virtually unimpeded by the chromophore-binding pocket. Moreover, the vibronic coupling explains, in part, the high quantum yield (0.67) of rhodopsin photoisomerization.
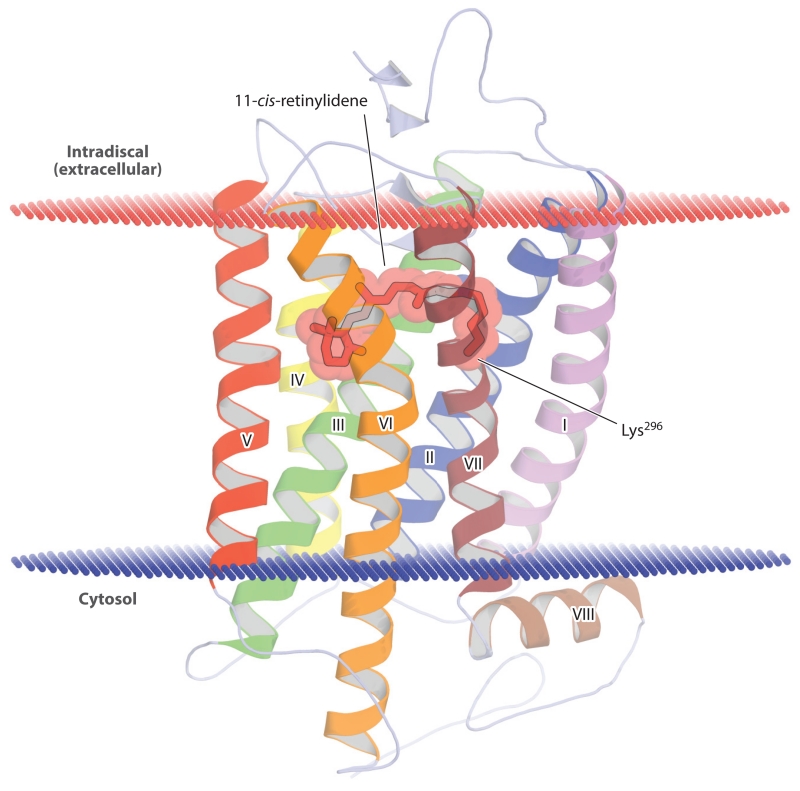
The ligand-binding pocket of rhodopsin, resolved in atomic detail by determination of its crystal structure (Figure 4) (Palczewski et al. 2000), exerts a profound influence on the optical absorbance properties of the bound retinylidene chromophore via distortions of the planar polyene structure, as well as by electrostatic influences, a phenomenon referred to as the opsin shift. The protonation state of the Schiff base is one well-characterized factor that determines the absorbance spectrum of rhodopsin. In rhodopsin’s ground state, the Schiff base is protonated, with the glutamate113 side chain carboxylate serving as the counterion (Nathans 1990, Sakmar et al. 1989). Upon photoisomerization, the Schiff base loses its proton to form the signaling-competent Meta II rhodopsin state, with a blue-shifted absorbance spectrum (Wald 1968).
Figure 4.
Crystal structure of ground-state bovine rhodopsin. The protein backbone is shown in a schematic representation with the 11-cis-retinylidene chromophore and covalently linked lysine (Lys296) side chain displayed as sticks and spheres. The α-helices are numbered based on their order in the polypeptide sequence. The red and blue lines demarcate the approximate width of the phospholipid membrane bilayer. The figure was generated with PyMol (Schrödinger, https://www.pymol.org/) using the rhodopsin atomic coordinates deposited in the Protein Data Bank under accession code 1U19.
Crystal structures of rhodopsin, including Meta II–like states, indicate that the isomerization of retinylidene is communicated to the G protein–interacting surface via alterations in a water-mediated hydrogen bonding network (Angel et al. 2009, Choe et al. 2011). The primary result of these changes is movement of the cytosolic ends of α-helices V and VI, making the binding site for transducin accessible. Crystal structures of rhodopsin in complex with transducin are needed to fully resolve the mechanism of information transfer from the ligand-binding pocket to the G protein.
Mutations in the opsin gene result in a spectrum of eyesight disorders, the most severe of which is referred to as retinitis pigmentosa (RP). A RHO mutation resulting in a Pro → His substitution at position 23 in the amino acid sequence (P23H) was the first reported pathological mutation in this gene, and it is a particularly common cause of inherited autosomal dominant RP (Dryja et al. 1990). This P23H rhodopsin is highly susceptible to degradation within the endoplasmic reticulum (ER), impeding rhodopsin trafficking to the outer segments, disrupting disc morphogenesis, and, ultimately, causing photoreceptor cell death (Sakami et al. 2011).
3. RETINOID TRANSPORTERS: STRA6 AND ABCA4
3.1. Stimulated by Retinoic Acid 6
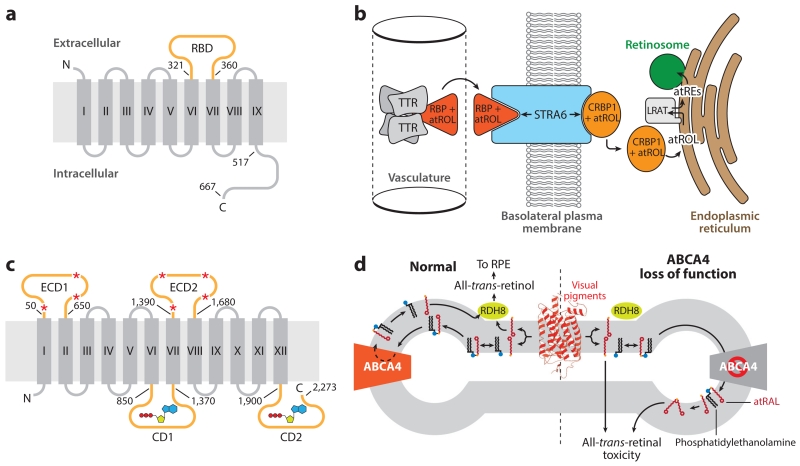
The passage of lipophilic retinoids, such as all-trans-ROL, between the two leaflets of phospholipid biological membranes can occur by simple diffusion. Nevertheless, there are two known locations within the retina where selective and efficient retinoid trafficking across phospholipid membranes relies on the activity of integral membrane transporter proteins, namely the apical plasma membrane of the RPE, where all-trans-ROL is obtained from the circulation, and the photoreceptor outer segment disks, where large quantities of all-trans-RAL are generated during light exposure (Figure 5).
Figure 5.
Structure and function of retinoid transporters essential for retinal health and function. (a) Topology diagram of the holo-RBP receptor, STRA6. The RBD (orange) resides between transmembrane helices VI and VII. Numbering indicates positions within the human amino acid sequence. (b) The role of STRA6 in the uptake of atROL from holo-RBP into a target tissue, such as the RPE. (c) Topological diagram of ABCA4. The large ECDs are glycosylated at multiple positions, as indicated by red asterisks. Two CDs contain Walker A and B motifs responsible for adenosine triphosphate binding and hydrolysis (shown in a schematic representation) that drive the transport function of the protein. Numbering indicates positions within the human amino acid sequence. (d) Role of ABCA4 in the metabolism of atRAL. atRAL (red sticks) released from photoactivated visual pigments (red schematic) into the exocytoplasmic leaflet of the disk membrane is inaccessible to RDH8, the enzyme that converts atRAL into atROL. atRAL readily forms a Schiff base conjugate with phosphatidylethanolamine (black sticks with blue headgroup). The Ret-PE adduct is translocated to the cytosolic leaflet through the action of ABCA4, where it dissociates to yield free atRAL that can be metabolized by RDH8 to nonelectrophilic atROL. Loss-of-function mutations in ABCA4, causative of Stargardt disease, result in the accumulation of atRAL and Ret-PE, which can undergo secondary reactions to form potentially toxic bis-retinoids and protein–retinoid conjugates that ultimately lead to retinal degeneration. Panels b and d adapted from Kiser et al. (2014). Abbreviations: ABCA4, adenosine triphosphate-binding cassette transporter 4; atRAL, all-trans-retinal; atRE, all-trans-retinol ester(s); atROL, all-trans-retinol; CD, cytosolic domain; CRBP, cellular retinal-binding protein; ECD, extracellular domain; LRAT, lecithin:retinol acyltransferase; RDH8, retinol dehydrogenase 8; RBP, retinol-binding protein; RBD, RBP-binding domain; Ret-PE, retinal-phosphatidylethanolamine Schiff base adduct; RPE, retinal pigment epithelium; STRA6, stimulated by retinoic acid 6; TTR, transthyretin.
After years of controversy regarding its existence, the receptor for holo-retinol-binding protein 4 (RBP4) was identified in 2007 as the protein known as stimulated by retinoic acid 6 (STRA6) (Kawaguchi et al. 2007), so named because of its retinoic acid (RA)-inducible nature (Bouillet et al. 1997). This multipass transmembrane protein is conserved in vertebrates, but bears no significant sequence homology to any known protein family (Figure 5a). All-trans-ROL (vitamin A) mainly circulates in the plasma in complex with RBP4, a 21 kDa protein, which in turn forms a complex with transthyretin (TTR) that prevents the renal excretion of RBP4 (Figure 5b). STRA6 contains an extracellular loop that mediates its interaction with holo-RBP4 to allow extraction of all-trans-ROL from the RBP4-binding pocket (Figure 5b) (Kawaguchi et al. 2008). Liberated all-trans-ROL then traverses a passageway formed by the STRA6 transmembrane α-helical segments, of which there are likely nine. Free all-trans-ROL then is picked up on the cytoplasmic side of the membrane by cellular retinal-binding protein 1 (CRBP1), which mediates the diffusion of all-trans-ROL to the ER, where it is metabolized (Figure 5b). Unlike most small molecule facilitative transporters, substrate flow is not governed by the concentration gradient of free substrate. Instead, the flow depends on the concentrations of the holo- and apo-forms of the retinoid-binding proteins RBP and CRBP1 (Kawaguchi et al. 2011). Besides its transport function, STRA6 also directly catalyzes the release of all-trans-ROL from holo-RBP into the transport passageway (Kawaguchi et al. 2011).
STRA6—which is expressed in several tissues, including the eye, brain, placenta, and testis, both during development as well as in adulthood—is particularly abundant in tight junction–linked epithelium and endothelial cell layers that form blood–organ barriers (Bouillet et al. 1997). In the RPE, STRA6 expression is restricted to the basolateral membrane, where it mediates vitamin A uptake from the choriocapillaris (Kawaguchi et al. 2007).
Mutations in STRA6 are associated with a severe developmental disorder known as the Matthew-Wood syndrome (Golzio et al. 2007, Pasutto et al. 2007). This disease, typically lethal in early childhood, is characterized by a range of developmental abnormalities, including anophthalmia or microphthalmia; heart, lung, and urogenital malformations; short stature; and facial dysmorphism. Similar malformations have been observed in a STRA6 knockdown zebrafish model of Matthew-Wood syndrome (Isken et al. 2008), whereas Stra6−/− mice exhibit a more mild and restricted phenotype, primarily affecting the eye (Amengual et al. 2014, Ruiz et al. 2012). Dominant negative mutations in RBP4 that enhance STRA6–apo-RBP4 binding affinity have also been linked to congenital eye malformations (Chou et al. 2015). Notably, maternal inheritance of this class of mutations is linked to greater disease penetrance, likely due to partial blockade of placental STRA6 adversely affecting maternal to fetal transfer of vitamin A.
3.2. Adenosine Triphosphate-Binding Cassette Transporter 4
After photobleaching, all-trans-RAL released from activated visual pigments can diffuse into the outer or inner leaflets of disk membranes. In the former case, all-trans-RAL is directly available to all-trans-retinol dehydrogenase (RDH) enzymes that quickly convert it to vitamin A (Rattner et al. 2000). RDH8 is such an enzyme, but all-trans-RAL released into the inner leaflet is not accessible and thus persists as a reactive electrophile in disk membranes. All-trans-RAL is prone to undergoing reactions with primary amine-containing compounds, such as proteins (which can directly cause cellular toxicity), small molecular drugs (Maeda et al. 2012), and phosphatidylethanolamine (PE). Although formation of the all-trans-RAL-PE Schiff base conjugate is reversible, a small portion of the conjugates undergo secondary reactions with an additional all-trans-RAL molecule that result in the irreversible formation of a number of bis-retinoid adducts, the most thoroughly characterized of which are A2E and all-trans-RAL-dimer (Kim et al. 2007). To prevent the accumulation of such potentially pathological side products, vertebrate photoreceptors express an approximately 250 kDa adenosine triphosphate (ATP)-driven transporter called ABCA4 (for ATP-binding cassette transporter 4, also known as ABCR) that belongs to the large ABC transporter family (Figure 5c) (Illing et al. 1997, Tsybovsky et al. 2010). This flippase is responsible for translocating all-trans-RAL-PE Schiff base conjugates from the intradiscal to the cytosolic side of the disk membrane, where they dissociate and the released all-trans-RAL can be reduced by all-trans-RDHs to promote retinoid cycle entry (Figure 5c,d) (Quazi et al. 2012). 11-cis-RAL-PE adducts also can accumulate during 11-cis-RAL excess and undergo condensation into toxic bis-retinoids (Quazi & Molday 2014). ABCA4 localizes specifically to the rim regions of rod photoreceptor disks, hence its prior designation as a Rim protein (Papermaster et al. 1978), possibly because of the large size of its extracellular domains, which could exclude the protein from the narrow interior of the disks’ central regions (Figure 5c) (Tsybovsky et al. 2013). ABCA4 is also expressed in cone photoreceptor outer segments and thus could participate in retinoid cycling within cones (Molday et al. 2000).
Loss-of-function mutations in ABCA4 result in a severe, juvenile-onset autosomal recessive retinopathy called Stargardt disease (SGD), characterized by the loss of central vision, disruption of retinal structure, and accumulation of retinoid-containing lipofuscin-like material (Figure 5d) (Allikmets et al. 1997b, Sun et al. 2000). Additionally, ABCA4 mutations have been associated with an increased risk of age-related macular degeneration (Allikmets et al. 1997a). Abca4−/− mice display some features associated with SGD, although the phenotypes are mild relative to the human disease. However, a combined knockout of Abca4 with Rdh8 greatly accentuates this retinal phenotype and the susceptibility to all-trans-RAL phototoxicity (Maeda et al. 2009a).
ABCA4 gene therapy for the treatment of SGD is an active and promising area of research (Cideciyan et al. 2009). The large size of the ABCA4 coding sequence precludes the use of traditional adeno-associated virus (AAV) vectors for gene delivery to the retina. This limitation has been circumvented by using dual AAV vectors containing portions of the ABCA4 coding sequence that reconstitute the complete coding sequence in situ via concatemerization (Trapani et al. 2014). Nonviral DNA nanoparticles, lacking the size limitations associated with AAV vectors, have also been successfully employed for delivering ABCA4 into the retinas of Abca4−/− mice (Han et al. 2012). The application of systems pharmacology has fostered the discovery of signaling pathway networks that can be pharmacologically modulated to treat SGD (Chen et al. 2013). This approach has been enabled by the advent of in vivo two-photon microscopy imaging of the retina, used in conjunction with traditional imaging and histological modalities, as a means to track disease pathogenesis and treatment efficacy (Maeda et al. 2014).
4. RETINOL DEHYDROGENASES: REVERSIBLE REDOX CHEMISTRY OF RETINOIDS
11-cis-RAL contains an electrophilic aldehyde needed for covalent binding to the lysine residue located in the orthosteric pocket of visual opsins. However, the chemical trans–cis reisomerization reaction of the visual cycle occurs at the alcohol level of oxidation (Bernstein & Rando 1986). Additionally, all-trans-RAL released from photoactivated visual pigments is cytotoxic at high concentrations and must be rapidly converted to nonelectrophilic all-trans-ROL to maintain the health of the retina (Chen et al. 2012, Maeda et al. 2009a). For these reasons, enzymes capable of catalyzing the interconversion of retinoids between their aldehyde and alcohol forms are essential components of the visual cycle (Table 3) (reviewed in Kiser et al. 2012b, Parker & Crouch 2010).
Table 3. Transcript reads across different RNA sequencing experiments with whole eye tissue from 1-month-old B6 and Rho−/− mice and the retina and cultured primary RPE cells from B6 micea.
| Genes | Whole eye from B6 mice |
Retina from B6 mice | Whole eye from Rho−/− mice |
RPE cells from B6 mice |
|---|---|---|---|---|
|
| ||||
| Retinol dehydrogenases | ||||
|
| ||||
| Rdh5 | 177.38 | 122.91 | 166.30 | 32.47 |
|
| ||||
| Rdh8 | 67.4 | 46.78 | 7.54 | 0.18 |
|
| ||||
| Rdh10 | 76.79 | 52.17 | 38.52 | 8.29 |
|
| ||||
| Rdh11 | 19.27 | 20.52 | 11.07 | 27.14 |
|
| ||||
| Rdh12 | 195.4 | 131.39 | 13.57 | 0.73 |
|
| ||||
| Rdh13 | 8.79 | 8.14 | 6.50 | 8.41 |
|
| ||||
| Rdh14 | 21.09 | 19.62 | 17.93 | 7.88 |
|
| ||||
| Dhrs3 | 104.7 | 41.73 | 34.11 | 8.28 |
|
| ||||
| Retinol-binding proteins | ||||
|
| ||||
| Rbp4 | 2.05 | 0.53 | 0.90 | 1.57 |
|
| ||||
| Rlbp1 | 354.00 | 226.98 | 137.88 | 36.86 |
|
| ||||
| Rbp1 | 90.74 | 12.36 | 103.43 | 26.73 |
|
| ||||
| Rbp2 | 6.20 | 0.08 | 122.79 | 0.00 |
|
| ||||
| Rbp3 | 654.90 | 1093.27 | 174.28 | 0.05 |
|
| ||||
| Control genes from the retina and RPE | ||||
|
| ||||
| Rpe65 | 114.07 | 57.02 | 25.44 | 1.94 |
|
| ||||
| Gcap1 | 1063.27 | 687.74 | 205.92 | 1.09 |
Abbreviations: Dhrs, dehydrogenase/reductase family member; Gcap, guanylate cyclase-activating protein; Rbp, retinol-binding protein; Rdh, retinol dehydrogenase; Rlbp, retinal binding protein; Rpe, retinal pigment epithelium.
All listed transcripts were detected at a level of 1 FPKM or higher (fragments per kilobase of exon per million fragments mapped) in RNA sequencing data (average of three biological replicates) from whole eye samples and indicated tissues.
All-trans-RAL and all-trans-ROL interconversion, using nicotinamide adenine dinucleotides (NADs) as cofactors, has a low energy barrier and is reversible under physiological conditions. Whether an enzyme acts as a RAL reductase or an RDH is thus governed primarily by mass action. In most cells, the ratio of [NAD+] to [NADH] is much greater than 1, such that in vivo enzymes preferentially using this cofactor tend to convert ROLs to RALs. The [NADP+]/[NADPH] ratio depends on the cell type, but is usually much lower than 1, meaning that enzymes with a preference for this cofactor, with some exceptions, act as reductases.
Although various oxidoreductase enzymes are capable of catalyzing these transformations, RDH members of the short-chain dehydrogenase/reductase (SDR) superfamily are the major in vivo contributors to these processes. SDRs constitute a large superfamily of oxidoreductases that metabolize a variety of endogenous and exogenous substrates—including retinoids, steroids, polyols, and xenobiotics—utilizing NAD(P)(H) as a source of redox equivalents. Based on sequence homology, all members of the RDH subfamily are predicted to adopt a classical SDR structure consisting of an N-terminal Rossmann fold that houses the nucleotide-binding site and a variable C-terminal domain where retinoids and other substrates bind. The topology of RDH membrane binding is an active area of research. These enzymes bind to membranes either through hydrophobic sequences at their N or C termini, or both, or by posttranslationally attached fatty acyl chains. Several members of this enzyme class are encoded in the human and mouse genomes and, in many cases, there is overlapping substrate specificity between orthologs. Such overlap has complicated the identification of the specific RDHs most responsible for catalyzing visual cycle redox chemistry in vivo. Nevertheless, knockout mouse studies and RDH mutations in humans with various retinal diseases have helped clarify the relative physiological importance of the various RDH members.
4.1. Reduction of All-trans-Retinal to All-trans-Retinol in Photoreceptor Cells
The first enzymatic step of the visual cycle is the conversion of all-trans-RAL into all-trans-ROL, which occurs mostly in photoreceptor outer segments (Zimmerman et al. 1975). RDH8 (also known as photoreceptor RDH) is an abundant enzyme in rod and cone photoreceptor outer segments. This protein displays selectivity for all-trans-retinoid substrates, and utilizes NADP(H) as a cofactor, signifying that it catalyzes reduction in cells (Rattner et al. 2000). In vitro experiments have indicated that RDH8 is the main contributor to RAL reductase activity in mouse photoreceptor membranes. Rdh8−/−mice accumulate all-trans-RAL and exhibit slightly delayed dark adaption following exposure to intense light. But when reared under nonextreme lighting conditions, these mice have structurally and functionally intact retinas, revealing that other enzymes can compensate for the loss of RDH8 enzymatic activity at this step of the retinoid cycle (Maeda et al. 2005). Indeed, in humans no RDH8 mutation has yet been associated with disease. In vivo, effective RDH8 function depends on the ABCA4 transporter, which flips N-retinylidene-phosphatidylethanolamine conjugates to the cytosolic leaflet of disks where they can dissociate, and the released all-trans-RAL becomes accessible to the RDH8 active site (Quazi et al. 2012). The partnership between these two proteins has been demonstrated in Rdh8−/−Abca4−/− mice, which exhibit a much greater susceptibility toward light-induced retinal degeneration and bis-retinoid accumulation (Maeda et al. 2009a). RDH12, a photoreceptor-specific RDH expressed in inner segments, accepts trans- and cis-retinoid isomers as well as some other lipophilic aldehydes and alcohols as substrates (Haeseleer et al. 2002). This RDH exhibits preferential utilization of NADP(H) as a cofactor and in vivo operates in the reductive direction (Haeseleer et al. 2002). The Rdh12−/− mouse exhibits delayed dark adaption and a paradoxical increase in the rate of 11-cis-RAL production. However, its most pronounced phenotype is an enhanced susceptibility to light-induced all-trans-RAL toxicity (Maeda et al. 2006b). Consistent with this strong murine phenotype, mutations in RDH12 cause RP in humans (Janecke et al. 2004). RDH12 probably protects photoreceptors from toxicity associated with electrophilic aldehydes. Dual knockout of Rdh8 and Rdh12 in mice eliminates more than 98% of all-trans-RAL reductase activity in mouse photoreceptors. These mice exhibit delayed dark adaptation and slowly progressive rod–cone dystrophy, but can still regenerate visual pigments, indicating the presence of additional RAL reductases in mouse retina (Maeda et al. 2007).
4.2. Oxidation of 11-cis-Retinol to 11-cis-Retinal in the Retinal Pigment Epithelium
Following trans–cis isomerization, the nascent 11-cis-ROL product must be oxidized to the aldehyde form to enable regeneration of photoreceptor visual pigments. In the classical retinoid cycle, this step takes place within the smooth ER of the RPE, catalyzed by cis-RDHs (Zimmerman et al. 1975), the most important of which is RDH5 (Simon et al. 1995). This integral membrane enzyme uses NAD(H) as a cofactor (Simon et al. 1995, Zimmerman 1976). Rdh5−/− mice exhibit a mild phenotype consisting of delayed dark adaptation at high bleaching light levels and the accumulation of ocular 11/13-cis-ROL/retinyl esters (Driessen et al. 2000). In humans, loss-of-function RDH5 mutations cause a mild retinal disease known as fundus albipunctatus, characterized by night blindness, delayed dark adaptation, and the presence of small, pale yellow deposits in the subretinal space (Yamamoto et al. 1999). That the retinoid cycle is slowed only in the setting of an RDH5 loss-of-function mutation indicates that additional enzymes are capable of carrying out oxidation of 11-cis-ROL in the retina (Jang et al. 2000). RDH11, a dual-substrate specificity RDH, is one such enzyme that contributes to 11-cis-RAL reduction in the RPE (Haeseleer et al. 2002, Kim et al. 2005). Recently, RDH11 mutations have been identified as a cause of syndromic RP, indicating that this enzyme could also have a role in controlling RA levels during development (Xie et al. 2014). RDH10 also is expressed in the RPE (Wu et al. 2002), where it contributes to 11-cis-ROL oxidation (Farjo et al. 2009). Additionally, RDH10 plays a decisive part in regulating RA levels and, hence, developmental processes, as evidenced by the embryonic lethality in Rdh10−/− mice (Farjo et al. 2011).
4.3. Retinol–Retinal Interconversion in Cone Photoreceptors and Müller Cells
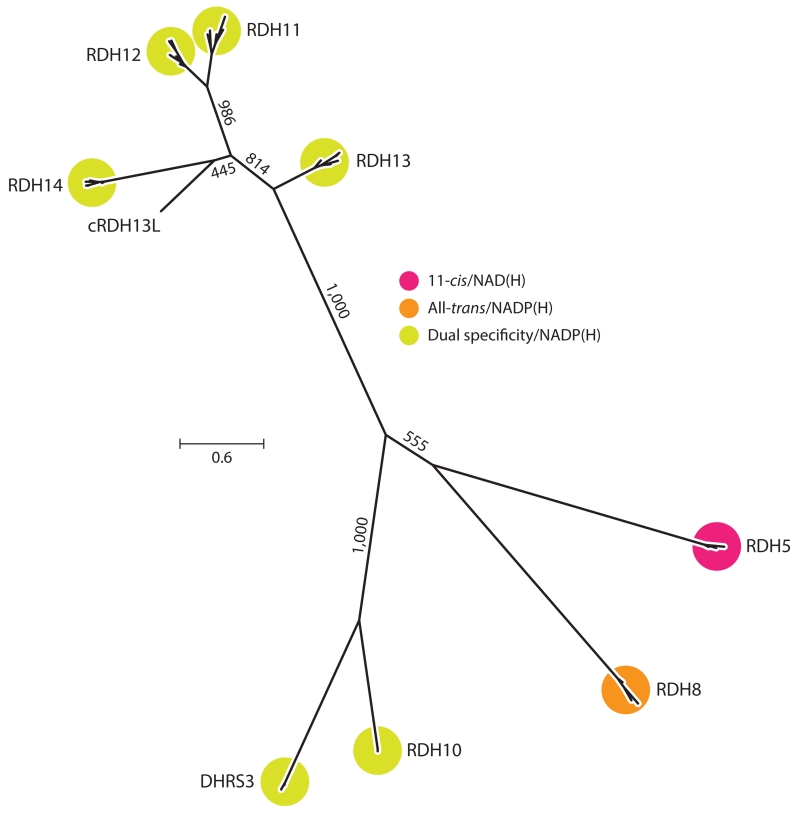
A current model of the Müller cell–based cone visual cycle posits key roles for RDHs in the selective delivery of visual chromophore to cone visual pigments (Kaylor et al. 2013). Retinal SDR1 (or RetSDR1), also known as DHRS3, is an all-trans-specific, NADP(H)-dependent RDH with retinal expression restricted to cone photoreceptor outer segments (Haeseleer et al. 1998). Its potential in vivo function in the retina is to reduce all-trans-RAL released from photoactivated cone visual pigments in conjunction with RDH8. RetSDR1 also metabolizes a number of nonretinoid compounds, such as steroids and various xenobiotics. This enzyme also is essential for controlling RA levels, as evidenced by the developmental defects and gestational lethality in Dhrs3−/− mice. Interestingly, retSDR1 and a related RDH also involved in RA biosynthesis, RDH10, modulate each other’s activity to precisely control RA levels (Figure 6) (Adams et al. 2014).
Figure 6.
An unrooted molecular phylogenetic tree illustrating relationships among groups of RDH homologs. Protein sequences (mammalian orthologs for each clade as well as carp RDH13L) were aligned with MUSCLE (Edgar 2004), and the tree was inferred using the phylogenetic maximum likelihood method as implemented in PhyML (ATGC Bioinformatics Platform, French National Institute of Bioinformatics, http://www.atgc-montpellier.fr/phyml/). Numbers on the internal branches indicate bootstrap support based on 1,000 pseudoreplicates. The scale bar indicates the average number of substitutions per site. The tree was generated using FigTree (Andrew Rambaut, http://tree.bio.ed.ac.uk/software/figtree/). Abbreviations: DHR, dehydrogenase/reductase family; NAD, nicotinamide adenine dinucleotide; RDH, retinol dehydrogenase.
RDH10 is expressed in Müller glial cells, although at much lower levels than in the RPE (Wu et al. 2004). It has been speculated that Müller cell RDH10 serves to oxidize the 11-cis-ROL generated through the alternative visual cycle for cones. Testing this hypothesis in vivo will require generating RAL-specific Rdh10−/− models because the global loss of RDH10 function is embryonically lethal in mice. However, such an activity may not be required because cones can directly take up 11-cis-ROL and oxidize it to 11-cis-RAL. Retinas from Nrl−/−mice, which have only S-cone photoreceptors, express RDH13 and RDH14 at high levels compared with rod-dominant, wild-type retinas, indicating that these candidate enzymes may be responsible for the cone RDH activity (Kanan et al. 2008). Carp RDH13L, which clusters with the mammalian RDH14 group (Figure 6), has been identified as a major 11-cis-RDH in cone photoreceptors. The in vivo relevance of RDH13 and RDH14 to mammalian cone photoreceptor function remains to be elucidated.
5. RETINYL ESTER SYNTHASE (LECITHIN: RETINOL ACYLTRANSFERASE)
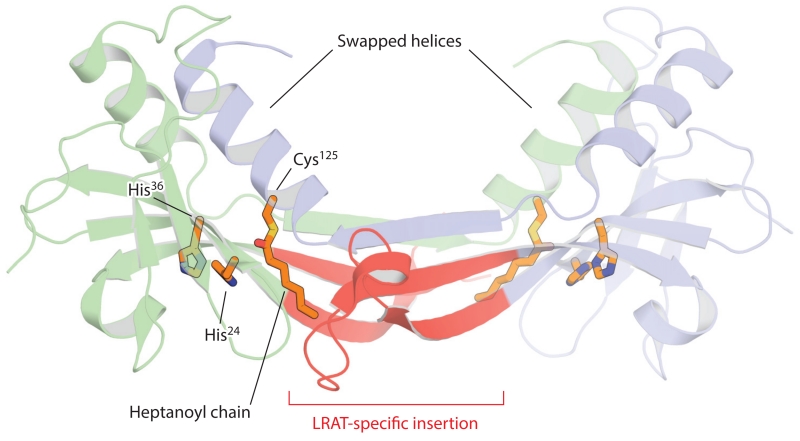
The RPE is the major site of vitamin A storage within the retina. Following photobleaching, vitamin A generated by the activity of photoreceptor RDHs is rapidly transferred from the retina to the RPE (Dowling 1960). Additionally, the RPE takes up vitamin A from holo-RBP4 circulating in the choriocapillaris through the STRA6 transporter expressed on its basolateral surface (Amengual et al. 2012, Isken et al. 2008, Kawaguchi et al. 2007). Both of these processes are driven by the esterification of vitamin A. The resulting all-trans-retinyl esters are efficiently stored in lipid droplet–like structures within the RPE referred to as retinosomes (Imanishi et al. 2004). These esters also serve as substrates for the retinoid isomerization step of the visual cycle. LRAT is the main enzyme responsible for catalyzing the formation of retinyl esters in the eye and many other tissues. LRAT activity, in which an acyl group is transferred from the sn1 position of phosphatidylcholine (lecithin) to ROLs, was first identified in microsomal membrane fractions from hepatocytes and the RPE (MacDonald & Ong 1988, Saari & Bredberg 1989). This reaction differs from that of a second retinyl ester synthase activity present in humans in which the retinyl ester is synthesized from a fatty-acyl-CoA instead of a phospholipid. Notably, LRAT is fairly promiscuous in its choice of acyl acceptors and can readily esterify or amidate a number of primary alcohol and amine compounds, including the retinoid cycle inhibitor retinylamine (Golczak et al. 2005a). LRAT-catalyzed acyl transfer occurs via a ping-pong mechanism featuring a covalent acyl-enzyme intermediate involving a conserved cysteine residue (Cys161 in the human sequence) (Mondal et al. 2000). Phylogenetic analysis has revealed that LRAT belongs to the NlpC/P60 protein superfamily, whose members feature a papain-like catalytic triad consisting of a nucleophilic cysteine residue, a histidine residue that acts as a general base, and a third polar residue (usually histidine or asparagine) (Anantharaman & Aravind 2003). Three-dimensional structures of LRAT family proteins have confirmed that they adopt a papain-like thiol peptidase fold, despite having no discernible sequence similarity to those enzymes (Figure 7). Compared with classical NlpC/P60 enzymes in which the nucleophilic cysteine residue resides on the N terminus, LRAT-like members are circularly permuted, with the catalytic cysteine relocated to the C-terminal end (Anantharaman & Aravind 2003). Besides LRAT, the human genome encodes seven other LRAT-like proteins: five H-Ras-like tumor suppressors (HRASLS) and two neurosensory proteins (NSE1 and 2). The former group of proteins exhibits phospholipase A2 and phospholipid acyltransferase activities, whereas the latter remains uncharacterized. LRAT and its homologs are bitopic integral membrane enzymes anchored to lipid bilayers via a C-terminal transmembrane α-helix. An amphipathic N-terminal region can also mediate monotopic membrane interactions (Moise et al. 2007). LRAT differs from the other LRAT-like family members by the presence of an approximately 30 residue insertion located near the middle of the protein sequence. Grafting this LRAT-specific region into the HRASLS3 sequence has resulted in a chimeric protein with sn1-specific LRAT activity akin to that of native LRAT (Golczak et al. 2015). The crystal structure of this chimeric protein has revealed that the LRAT-specific sequence mediates the formation of a homodimeric assembly in which the C-terminal α-helix containing the catalytic cysteine residue is domain swapped compared with native, monomeric HRASLS3 (Figure 7) (Golczak et al. 2015). The structure is consistent with biochemical work demonstrating that LRAT forms functional homodimers on microsomal membranes. Dimer formation results in structural changes around the catalytic cysteine residue in which the active site pocket with enhanced hydrophobicity is more deeply embedded in the protein. These characteristics have been postulated to impede water entry into the pocket such that acyltransferase activity is favored over hydrolase activity (Golczak et al. 2015).
Figure 7.
Crystal structure of the lecithin:retinol acyltransferase (LRAT)–H-Ras-like tumor suppressor 3 chimera. The two chains of the homodimer are shown in a schematic representation (blue and green). The LRAT-specific insertion (red) forms a β-sheet that mediates dimer formation. This region also provides a pathway for the exchange of the C-terminal α-helical regions within the dimer pair. The histidine (His) and cysteine (Cys) residues constituting the catalytic triad are shown as sticks. The catalytic Cys125 residue is acylated as a result of the crystals being grown in the presence of the acyl donor, diheptanoyl phosphatidylcholine. The figure was generated with PyMol (Schrödinger, https://www.pymol.org/).
Studies in Lrat−/− mice have revealed that this enzyme is critical for ocular retinoid homeostasis and photoreceptor health (Batten et al. 2004). Essentially, these mice lack all ocular retinoids, consistent with an indispensable role for LRAT in the uptake and retention of vitamin A by the eye. Only trace levels of retinyl esters in liver, the other major vitamin A storage site in the body, are present in these mice. Due to the lack of retinyl ester substrate needed for the retinoid isomerization step of the retinoid cycle, Lrat−/− mice have severely impaired electroretinographic (ERG) and pupillary light responses. These in vivo results are consistent with in vitro data showing that inhibition of LRAT activity blocks the biosynthesis of 11-cis-ROL by the retinoid isomerase RPE65. Lrat−/− mice have shortened rod outer segments at 8 weeks of age and develop a progressive loss of cone photoreceptors that can be rescued by administration of the artificial chromophore prodrug 9-cis-retinyl acetate (Maeda et al. 2009b). Despite the major disruption in vitamin A storage caused by the loss of LRAT function, these treated mice are otherwise viable and fertile. Consistent with the knockout mouse phenotype, loss-of-function mutations in LRAT cause RP in humans (Thompson et al. 2001). 9-cis-Retinyl acetate and LRAT gene replacement therapy are under investigation as treatments for RP associated with loss-of-function LRAT mutations (Batten et al. 2005, Koenekoop et al. 2014).
6. RETINOID ISOMERASE RPE65
Expressed largely in the RPE, RPE65 is an enzyme that catalyzes the formation of 11-cis-ROL from all-trans-retinyl ester substrates. This developmentally regulated microsomal protein was cloned in the early 1990s (Hamel et al. 1993), but its involvement in retinal health and function remained unclear until later in the decade when it was discovered that RPE65 mutations cause severe retinopathies in humans. The function of RPE65 as the visual cycle isomerase was first suggested by the phenotype observed in Rpe65−/− mice. This consisted of nearly flat ERG responses, a lack of ocular 11-cis-retinoids, and an overaccumulation of all-trans-retinyl esters in the RPE (Redmond et al. 1998). Owing to difficulties in observing enzymatic activity from purified RPE65 preparations, the identification of RPE65 as the visual cycle isomerase was not generally accepted until 2005, when three independent groups used reconstituted visual cycle systems in cultured cells to show that RPE65 is the key component conferring retinoid isomerase activity (Jin et al. 2005, Moiseyev et al. 2005, Redmond et al. 2005).
RPE65 belongs to a family of enzymes, termed carotenoid cleavage oxygenases, that typically catalyze the oxidation of carotenoid substrates to yield apocarotenoid products. Members of this family, which are found in all kingdoms of life, generally exhibit minimal amino acid sequence conservation, but almost all possess a set of four histidine residues and three glutamate residues that are involved in coordinating an iron cofactor essential for their catalytic function. Sequences belonging to the RPE65 clade are highly conserved and have a characteristic length of 533 ± 2 amino acid residues, with few exceptions. RPE65 activity appears to have been first acquired by a BCO II–like sequence in the last common ancestor of jawless and jawed vertebrates (Poliakov et al. 2012).
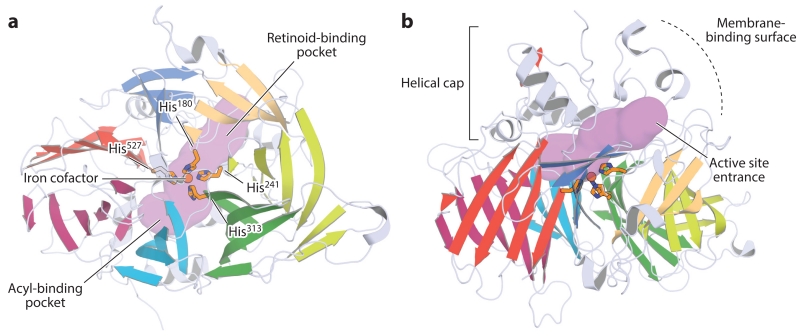
RPE65 adopts a seven-bladed β-propeller structure containing long insertions on its top face that form a mostly α-helical cap constituting the enzymatic active site (Figure 8) (Kiser et al. 2009, 2012a). The iron cofactor is axially bound on the top face of the β-propeller where it is covered by the helical cap. The non-heme iron center is accessible through a cavity formed at the junction between the β-propeller and dome structures. The retinoid-binding site is located within the membrane-proximal region of the cavity (Kiser et al. 2015), which exhibits a negative electrostatic potential likely needed to stabilize a carbocation intermediate of the isomerization reaction (Figure 8a) (McBee et al. 2000). The fatty acid–binding site begins at the iron-binding site and extends into the distal region of the cavity. In RPE65 it appears that iron facilitates catalysis by promoting ester cleavage to generate the transition state, which differs fundamentally from the role in activating dioxygen carried out by non-heme iron centers of other carotenoid cleavage oxygenases (Kiser et al. 2012a). The entrance to the active site cavity is surrounded by a patch of hydrophobic residues that mediate the binding of RPE65 to the ER (Figure 8b). Crystallographic data have indicated that RPE65 forms a homodimeric structure mediated by a sequence insertion found in vertebrate carotenoid cleavage oxygenases (Kiser et al. 2012a). This dimeric structure places the hydrophobic surfaces in a parallel orientation that reinforces membrane binding.
Figure 8.
Orthogonal views of the crystal structure of retinal pigment epithelium–specific 65 kDa protein. (a) View down the β-propeller axis, with the seven blades indicated by distinct colors. The iron cofactor (orange sphere) is coordinated on the propeller axis by four conserved histidine (His) residues. The purple surface delineates the active site pocket. (b) Rotating the structure by 90° along the horizontal axis shows the helical cap on the top face of the β-propeller. The active site entrance is surrounded by a patch of mostly hydrophobic residues that mediate the membrane binding critical for substrate uptake. The structural representations were generated with PyMol (Schrödinger, https://www.pymol.org/) using the coordinates deposited under Protein Data Bank accession code 4F2Z.
Loss-of-function mutations in RPE65 and the consequent retinoid cycle blockade result in the loss of visual function and retinal degeneration. In the most severe cases, RPE65 mutations cause the childhood retinopathy Leber congenital amaurosis (LCA), which results in childhood blindness (Gu et al. 1997, Marlhens et al. 1997, Morimura et al. 1998). In contrast to some other forms of LCA in which retinal degeneration occurs early in the course of the disease, patients with RPE65 LCA typically exhibit a preserved retinal structure for some time after visual function begins to decline (Cideciyan 2010). This structural preservation provides an opportunity for therapeutic intervention before vision loss becomes permanent. Two approaches are under investigation: RPE65 gene therapy to restore a functional RPE-based retinoid cycle (Acland et al. 2001, Bainbridge et al. 2008, Maguire et al. 2008) and treatment with an artificial chromophore (9-cis-RAL) to bypass the metabolic blockade caused by the lack of RPE65 activity (Koenekoop et al. 2014, Van Hooser et al. 2000). Both approaches have shown therapeutic promise in clinical trials but require further optimization.
7. THE RETINOID-BINDING PROTEINS
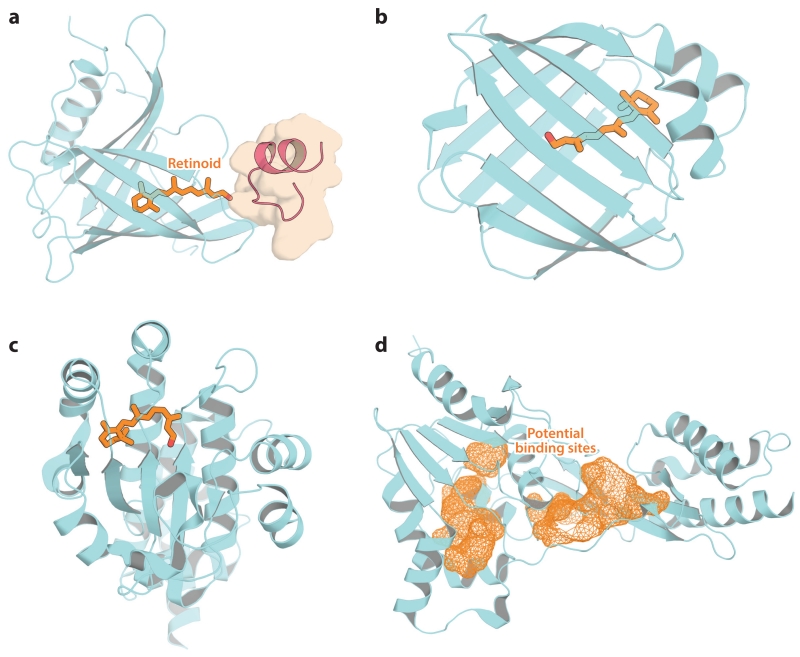
The efficient movement of retinoids between and within the photoreceptors, RPE, and Müller glia is required for proper visual cycle function (Table 3). Because retinoids are lipophilic compounds prone to destruction via off-pathway reactions, their diffusion within the retina depends critically on retinoid-binding proteins that mobilize these compounds from membrane stores and protect them from degradation. Four such binding proteins are known to be critical for visual function: serum RBP4, cellular RBP1 (CRBP1 or RBP1), interphotoreceptor retinoid-binding protein (IRBP or RBP3), and cellular retinaldehyde-binding protein (CRALBP) (Figure 9). Although these proteins are not homologous and are structurally diverse, they all are water soluble molecules that possess internal lipophilic pockets into which retinoids can bind and be protected from degradation.
Figure 9.
Crystal structures of retinoid-binding proteins important for visual function. (a) Retinol-binding protein 4 in complex with transthyretin (residues 76–87; red schematic with brown surface) and all-trans-retinol. (b) Cellular retinol-binding protein 1 in complex with all-trans-retinol. (c) Cellular retinaldehyde-binding protein in complex with 11-cis-retinal. (d) Module 2 from Xenopus laevis interphotoreceptor retinoid-binding protein. Potential retinoid-binding sites are shown as orange mesh. Retinoids are shown as orange sticks. The figure was generated with PyMol (Schrödinger, https://www.pymol.org/).
7.1. Retinol-Binding Protein 4
RBP4 is the major carrier responsible for distributing hepatic vitamin A obtained from dietary sources to peripheral tissues such as the eye. This approximately 21 kDa member of the lipocalin protein family circulates in the plasma in complex with the tetrameric protein TTR, which prevents its loss by glomerular filtration and renal excretion (Kanai et al. 1968). A single ROL-binding site is formed by the core, eight-stranded, β-barrel fold of RBP4 (Figure 9a) (Newcomer et al. 1984). All-trans-ROL is oriented with its hydroxyl moiety nearest to the binding cavity entrance, where residues involved in STRA6 binding have been localized. In the circulating RBP–TTR complex, the ROL hydroxyl group is covered by the tip of an extended loop region of TTR composed of residues 76–87 (Monaco et al. 1995). RBP4 facilitates efficient mobilization of ROL stored in the liver, but is not essential for vitamin A acquisition by peripheral tissues. Rbp4−/− mice are viable and fertile, but exhibit a non-developmental retinal dysfunction that is correctable by consumption of a vitamin A–enriched diet (Quadro et al. 1999). In humans, mutations in RBP4 are associated with iris coloboma, retinal dystrophy, and skin disorders. Microphthalmia has also been associated with dominant-negative RBP4 mutations in which the altered protein is inefficiently loaded with all-trans-ROL yet binds to STRA6 with a higher affinity than wild-type RBP4, as discussed in the section on STRA6 (Chou et al. 2015).
7.2. Cellular Retinol-Binding Protein 1
Following passage through the STRA6 transporter, or by other means, all-trans-ROL is transported inside the cell in complex with CRBP1. Like RBP4, CRBP1 is a small, water soluble protein that contains a single ROL-binding site formed by its β-barrel fold (Figure 9b) (Cowan et al. 1993). Of the retinoid isomers, it binds all-trans-ROL with the highest affinity, consistent with vitamin A being the physiological ligand (MacDonald & Ong 1987, Ong & Chytil 1975). In contrast to holo-RBP4, the hydroxyl moiety of all-trans-ROL is buried within the binding pocket of CRBP1, such that it is not solvent accessible. This structural finding indicates that all-trans-ROL must dissociate from the protein before it can be metabolized, although kinetic data have indicated that the holo–CRBP1 complex might be the substrate for all-trans-ROL esterification. CRBP1 is expressed in many sites throughout the body, whereas its paralog, CRBP2, is found mainly in the intestine. Within the retina, CRBP1 expression is highest in the RPE (Saari et al. 1977). Elevated levels of all-trans-ROL in the retina and a lack of retinyl ester production in the RPE of Rbp1−/− mice are consistent with a key role for this binding protein in retinoid metabolism within the eye (Saari et al. 2002).
7.3. Cellular Retinaldehyde-Binding Protein
CRALBP is the key protein responsible for binding 11-cis-retinoids generated via the RPE and Müller cell–based visual cycles and delivering them to the plasma membrane for transport to photoreceptors (Bunt-Milam & Saari 1983, Saari et al. 1977). As isolated from bovine neural retina, CRALBP contains both 11-cis-RAL and 11-cis-ROL as bound ligands, whereas 11-cis-RAL is the only retinoid associated with RPE-derived CRALBP (Saari et al. 1982). 9-cis-Retinoids are the only other isomers capable of binding CRALBP with high affinity. The strong binding selectivity for these two isomers has been explained by the presence of a curved binding pocket that has high complementarity to the 11-cis- and 9-cis-configurations (Figure 9c) (He et al. 2009). This structure also has revealed a cationic surface that likely interacts with negatively charged phospholipids to enable retinoid transfer to the plasma membrane. Rlbp1−/− mice exhibit delayed dark adaption consistent with a key role of CRALBP in efficient visual cycle function (Saari et al. 2001). Mutations in human RLBP1 are associated with several different retinopathies, including RP (Maw et al. 1997), Bothnia dystrophy, retinitis punctata albescens, and Newfoundland rod–cone dystrophy (Travis et al. 2007).
7.4. Interphotoreceptor Retinoid-Binding Protein
Photoreceptors secrete an approximately 136 kDa lipoglycoprotein called IRBP that mediates intercellular retinoid transfer (Bunt-Milam & Saari 1983, Lai et al. 1982). This protein has a complex structure consisting of four homologous modules that arose from an internal quadruplication (Fong & Bridges 1988). IRBP has an elongated appearance in negative-stained electron microscopy images and undergoes major conformational changes in the presence of retinoid ligands. Unlike the other retinoid-binding proteins, IRBP possesses three retinoid-binding sites and can bind both all-trans- and 11-cis-retinoids with high affinity. The structure of an IRBP module from Xenopus has revealed that the modules adopt a two-domain architecture (Loew & Gonzalez-Fernandez 2002). The retinoid-binding sites within the modules have not yet been revealed, but two candidate binding pockets are apparent from the crystal structure: one at the junction between the two domains and the other within the C-terminal domain of the module (Figure 9d). Consistent with an important role for IRBP in retinoid trafficking within the retina, mutations in the RBP3 gene are associated with autosomal recessive RP (den Hollander et al. 2009).
8. VISUAL CYCLE MODULATORS
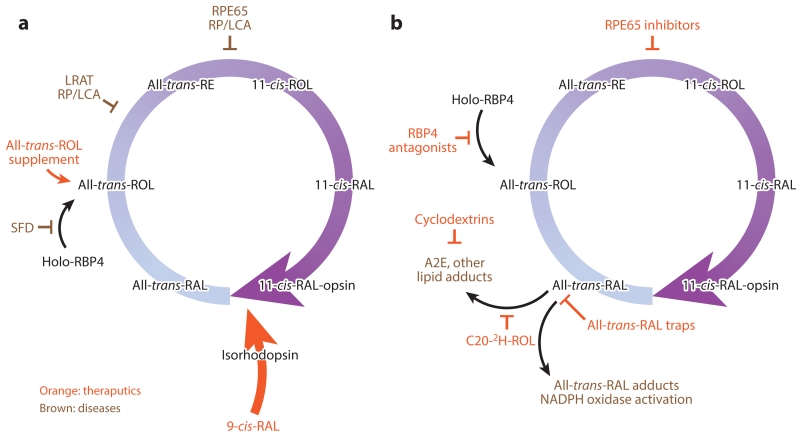
Defects in most genes involved in the retinoid cycle are associated with specific retinopathies (Tables 1 and 2). Here, we highlight research conducted on modulators of the visual cycle for the treatment of retinal diseases (Figure 10). Many of these agents are based on the retinoid structure; thus their specific delivery to the eye can employ the same retinoid mechanism, for example, transport by RBP4, delivery via STRA6, or enhancement of retention through esterification or amidation in the retina with the help of LRAT (reviewed in Palczewski 2010). These strategies are described below along with some key references (Figure 10). As this field rapidly evolves, further progress likely will be revealed in the literature.
Figure 10.
Therapeutic strategies for treating retinal diseases. (a) Retinoid supplementation for diseases such as SFD affecting ROL delivery to the RPE or generation of a visual chromophore (LRAT or RPE65 RP/LCA). (b) Strategies to reduce the formation of excess atRAL, its lipid condensation products, or both, to facilitate their clearance from the eye. Abbreviations: LCA, Leber congenital amaurosis; LRAT, lecithin:retinol acyltransferase; NAD, nicotinamide adenine dinucleotide; RAL, retinal; RBP, retinol-binding protein; ROL, retinol; RP, retinitis pigmentosa; RPE, retinal pigment epithelium; SFD, Sorsby’s fundus dystrophy.
8.1. Retinol Supplementation
Uncomplicated vitamin A deficiency can be overcome by providing therapeutic doses of the vitamin, either as free ROL or retinyl esters (reviewed in Kiser et al. 2014). Improvement in visual function upon vitamin A supplementation has also been observed in a mouse model with a specific rhodopsin mutation related to RP (Sibulesky et al. 1999). In these cases, all-trans-ROL or its active cis form acted as a chaperone for the mutated protein, although there was no follow-up to these initial studies (Figure 10a).
A mutation in the metalloproteinase inhibitor 3 gene (TIMP3) causes thickening of Bruch’s membrane, which is between the photoreceptor layer and its blood supply, leading to chronic deprivation of vitamin A in a disease known as Sorsby’s fundus dystrophy. A dose of 50,000 IU/day of vitamin A administered orally diminishes night blindness in patients with early stages of this disease (Jacobson et al. 1995). Similarly, treatment with pharmacological doses of vitamin A restored vision in Stra6−/− mice lacking adequate vitamin A supplementation to the eye (Amengual et al. 2014).
8.2. 9-cis-Retinal and 11-cis-Retinal Supplementation
When 11-cis-RAL cannot be generated because of retinoid cycle blockade, as with a nonfunctional or suppressed retinoid isomerase or LRAT, supplementation with pharmacological doses of 9-cis-RAL can dramatically improve visual function in animal models (Batten et al. 2004, Van Hooser et al. 2000, 2002). A successful clinical trial using this therapeutic strategy has recently been reported (Koenekoop et al. 2014). Treatment with cis-retinoid appears to be useful in preventing the deterioration of vision in aging mice (Maeda et al. 2006c), raising the possibility that a similar treatment could successfully preserve vision in older humans (Figure 10a).
8.3. Inhibitors of Retinal Pigment Epithelium–Specific 65 kDa Protein
Another idea is that slowing the regeneration of visual pigments by inhibiting the retinoid cycle will decrease the amount of free all-trans-RAL and its toxic condensation products (Radu et al. 2003). [Importantly, profound suppression of visual pigment regeneration is detrimental to retinal health because free opsin has constitutive signaling activity that eventually causes the death of photoreceptor cells, as mentioned in Section 5 (Woodruff et al. 2003).] 13-cis-RA was the initial compound found to decrease A2E formation (Radu et al. 2003). The first highly selective and potent inhibitor discovered was retinylamine (Golczak et al. 2005b). The systematic comparison of different inhibitors has been reported (Maeda et al. 2006a) and, based on those results, emixustat was developed by Acucela Inc. (Bavik et al. 2015), but human clinical trials have revealed significant side effects (Kubota et al. 2012). Finally, recent studies have questioned the mechanism of action of emixustat and suggested that sequestration of all-trans-RAL should be considered as discussed in Section 8.5 (Figure 10b) (Zhang et al. 2015).
8.4. Reducing the Supply of Vitamin A to the Eye
Dissociation of the RBP4–TTR complex allows filtration of RBP4 through renal glomeruli and elimination of vitamin A from the circulation. Thus, fenretinide, an amide of RA, had been hypothesized to have an antagonistic effect on the RBP4–TTR complex (Mata et al. 2013). However, it has been shown that the mechanism of action likely involves a slow hydrolysis of the amide along with upregulation of STRA6 and LRAT, thus decreasing circulating all-trans-ROL and increasing cellular vitamin A uptake by peripheral tissues. Nonretinoid antagonists of the RBP4 complex are being developed (Dobri et al. 2013).
8.5. Sequestration of All-trans-Retinal to Prevent Toxicity
A series of studies have demonstrated that one candidate toxin in the retinoid cycle is all-trans-RAL released from photoactivated visual pigments (reviewed in Kiser et al. 2014). All-trans-RAL toxicity can be lowered if a Schiff base is rapidly and reversibly formed with primary amines. In this case, the free aldehyde would be decreased, allowing RDHs to convert it into an alcohol. In mouse models of retinal degeneration, the peak concentration of free all-trans-RAL has been lowered with primary amine-containing, US Food and Drug Administration-approved drugs that did not inhibit chromophore regeneration because they were nonretinoid compounds. To validate this strategy, Schiff base adducts between all-trans-RAL and these amines have been identified by mass spectrometry (Maeda et al. 2012). This option is ready to be tested in humans (Figure 10b).
8.6. Elimination of A2E
Severe and rapid bleaching of visual pigments produces free all-trans-RAL that overwhelms the capacity of retinal cells to reduce this aldehyde to all-trans-ROL. During visual transduction, all-trans-RAL can react with itself and lipids. The resulting condensation products and their oxidized derivatives reportedly are toxic to the RPE (reviewed in Sparrow et al. 2012). Intravitreal treatment with β-cyclodextrins reduced bis-retinoids in the RPE of mice by 48% (Nociari et al. 2014). These novel prophylactic results are encouraging, but a beneficial effect on the health of the RPE has yet to be demonstrated (Figure 10b).
8.7. Use of Isotope Effects to Slow Production of A2E
What if A2E formation could be selectively blocked without affecting visual cycle kinetics? Consideration of the mechanistic details of A2E formation has revealed that one of the rate-limiting steps is the subtraction of a proton. Thus, the rate of this reaction could be changed if the hydrogen atom is replaced by a deuterium atom (a kinetic isotope effect). Long-term administration of deuterium-enriched vitamin A, namely C20-D3–vitamin A, slowed the formation of all-trans-RAL condensation products (Ma et al. 2011). However, this conceptually elegant idea has many caveats, including the difficulty in replacing all vitamin A in liver and eye, uncertainty about whether all condensation products will display the same isotopic effect as A2E formation, and whether slowing the reaction would make a significant impact on condensation product formation. Finally, if all-trans-RAL is the primary toxic agent that kills photoreceptor and RPE cells, then deuterium replacement would not be expected to have any therapeutic effect (Figure 10b).
9. CONCLUSIONS
The past three decades have led to a much greater molecular understanding of the retinoid cycle. Individual proteins have been well characterized by biochemical and biophysical methods. The physiological functions of these proteins have been clarified from analyses of human visual disease resulting from their inadequate expression, activation or altered properties and their corresponding transgenic animal models. Such animal models also are indispensable in testing new concepts for correcting genetic deficiencies by using pharmacological, genetic, or stem-cell-based approaches.
High-resolution structures provide detailed information on how the macromolecules of the retinoid cycle work. The utility of atomic information is exemplified by rhodopsin, the first G protein–coupled receptor whose structure was elucidated; by LRAT, through the use of hybrid approaches to expose its mechanism of catalysis; and with CRALBP, which in complex with RAL provided precise information as to its substrate specificity.
Historical knowledge about the chemistry of vision in the retina was obtained in three phases. First, the elegant work on the chemistry of vision by George Wald placed vision in a proper molecular perspective. Second, retinoid-binding proteins and enzymes were identified in the complex photoreceptor–RPE two-cell type system. Finally, molecular and structural characterizations of visual pathway components, supported by genetic approaches, have been, and continue to be, especially fruitful. This latest phase also has benefited greatly from mechanism-based pharmacology. As summarized in this review, visual cycle modulators will continue to advance to clinical trials, and confidence is building that these compounds will become efficacious therapies for some currently incurable retinal-blinding diseases.
CHEMICAL REACTIONS OF THE RETINOID CYCLE.
Reactions of the visual cycle take place at the membrane–cytoplasm interface. Vitamin A, a fat-soluble vitamin central to this cycle, is available to the enzymes because it partitions mostly into membranes. Yet access to other soluble substrates and products, for example, water, is also needed. Thus, these enzymes reside within membranes as transmembrane or membrane-associated proteins. The visual cycle depends on a few reactions that could occur partially in membranes at physiological temperatures. These include redox reactions, Schiff base formation and hydrolysis, esterification, and stereospecific isomerizations (Figure 2). The movement of retinoids between membranes is enabled by retinoid-binding protective proteins and active ATP-driven transporters (ABCA4), or it is driven by LRAT-mediated esterification. The transfer of all-trans-ROL from photoreceptors to the RPE occurs by passive diffusion because retinyl esters have a propensity to form insoluble lipid droplets, termed retinosomes. All-trans-ROL taken up from the circulation via the STRA6 transporter is also esterified and stored in retinosomes. 11-cis-RAL transfer from the RPE to photoreceptors is driven by a nearly irreversible reaction between this retinoid and opsins.
SUMMARY POINTS.
Structural and chemical bases for how the retinoid cycle components perform their physiological functions are presented.
Human retinal diseases that result from disruption of visual cycle components and animal models of these disorders are summarized.
Therapeutically aimed visual cycle modulation strategies include (a) supplementing the eye in animal models and humans that have defective retinoid cycles with all-trans- and -cis isomers, (b) reducing vitamin A delivery to the eye and inhibiting retinoid isomerase, (c) slowing A2E formation by involving isotope effects, (d) removing all-trans-RAL condensation products, and, finally, (e) sequestrating all-trans-RAL to eliminate its toxicity and reduce the formation of its condensation products.
FUTURE ISSUES.
High-resolution structures of IRBP, RDHs, ABCA4, and STRA6 are needed to understand how these proteins participate in the movement and metabolism of retinoids.
Additional visual cycle modulators should be identified that support retinal function without inhibiting RPE65 and prevent pathology resulting from genetic and environmental factors.
The precise sequence of events that leads to rhodopsin regeneration and chromophore hydrolysis after photoisomerization requires further characterization.
The molecular and genetic basis of cone chromophore regeneration must be characterized.
The factors governing the incorporation of retinyl esters into retinosomes must be elucidated as well as those governing their reentry into the retinoid cycle.
The molecular steps in recycling chromophores phagocytized by the RPE (Kevany & Palczewski 2010) must be defined as well as their utilization in regenerating visual pigments.
Cone visual pigment structures and the mechanisms of their spectral tuning should be elucidated.
ACKNOWLEDGMENTS
We thank Dr. Leslie T. Webster Jr., Dr. Grzegorz Palczewski, and members of the Palczewski laboratory for helpful comments on this manuscript. We thank Dr. Debarshi Mustafi and Dr. Brian Kevany for RNA sequencing data. This work was supported by funding from the US National Institutes of Health grants EY009339 and EY025451 (to K.P.), the Arnold and Mabel Beckman Foundation, the Foundation Fighting Blindness (to K.P.), and the US Department of Veterans Affairs grant BX002683 (to P.D.K.). K.P. is the John H. Hord Professor of Pharmacology.
Glossary
- RAL
retinal
- RPE
retinal pigment epithelium
- ROL
retinol
- LRAT
lecithin:retinol acyltransferase
- RPE65
retinal pigment epithelium–specific 65 kDa protein
- RP
retinitis pigmentosa
- RBP
retinol-binding protein
- STRA6
stimulated by retinoic acid 6
- RA
retinoic acid
- RDH
retinol dehydrogenase
- ABCA4
adenosine triphosphate (ATP)-binding cassette transporter 4
- SGD
Stargardt disease
- LCA
Leber congenital amaurosis
- IRBP
interphotoreceptor retinoid-binding protein
- CRALBP
cellular retinaldehyde-binding protein
Footnotes
DISCLOSURE STATEMENT
K.P. is an inventor of US Patent 8722669 (Compounds and methods of treating ocular disorders) and US Patent 20080275134 (Methods for treatment of retinal degenerative disease) issued to Case Western Reserve University (CWRU), whose values may be affected by this publication. CWRU may license this technology for commercial development. K.P. is a member of the scientific board of Vision Medicines, Inc., which develops visual cycle modulators. K.P. is Chief Scientific Officer at Polgenix, Inc., and an inventor of US Patent 7706863 (Methods for assessing a physiological state of a mammalian retina) and US Patent 8346345 B2 (Methods for assessing a physiological state of a mammalian retina), whose values may be affected by this publication.
LITERATURE CITED
- Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, et al. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 2001;28:92–95. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- Adams MK, Belyaeva OV, Wu L, Kedishvili NY. The retinaldehyde reductase activity of DHRS3 is reciprocally activated by retinol dehydrogenase 10 to control retinoid homeostasis. J. Biol. Chem. 2014;289:14868–80. doi: 10.1074/jbc.M114.552257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997a;277:1805–7. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 1997b;15:236–46. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- Amengual J, Golczak M, Palczewski K, von Lintig J. Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 2012;287:24216–27. doi: 10.1074/jbc.M112.353979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amengual J, Zhang N, Kemerer M, Maeda T, Palczewski K, Von Lintig J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014;23:5402–17. doi: 10.1093/hmg/ddu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JS, Futterman S. Metabolism of the retina. V. The role of microsomes in vitamin A esterification in the visual cycle. J. Biol. Chem. 1964;239:4073–76. [PubMed] [Google Scholar]
- Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. PNAS. 2009;106:14367–72. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–39. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, et al. Lecithin–retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 2004;279:10422–32. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten ML, Imanishi Y, Tu DC, Doan T, Zhu L, et al. Pharmacological and rAAV gene therapy rescue of visual functions in a blind mouse model of Leber congenital amaurosis. PLOS Med. 2005;2:1177–89. doi: 10.1371/journal.pmed.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavik C, Henry SH, Zhang Y, Mitts K, McGinn T, et al. Visual cycle modulation as an approach toward preservation of retinal integrity. PLOS ONE. 2015;10:e0124940. doi: 10.1371/journal.pone.0124940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PS, Law WC, Rando RR. Isomerization of all-trans-retinoids to 11-cis-retinoids in vitro. PNAS. 1987;84:1849–53. doi: 10.1073/pnas.84.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PS, Rando RR. In vivo isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state. Biochemistry. 1986;25:6473–78. doi: 10.1021/bi00369a020. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, et al. Developmental expression pattern of Stra6, a retinoic acid–responsive gene encoding a new type of membrane protein. Mech. Dev. 1997;63:173–86. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- Bunt-Milam AH, Saari JC. Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina. J. Cell Biol. 1983;97:703–12. doi: 10.1083/jcb.97.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, et al. Mechanism of all-trans-retinal toxicity with implications for Stargardt disease and age-related macular degeneration. J. Biol. Chem. 2012;287:5059–69. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Palczewska G, Mustafi D, Golczak M, Dong Z, et al. Systems pharmacology identifies drug targets for Stargardt disease–associated retinal degeneration. J. Clin. Investig. 2013;123:5119–34. doi: 10.1172/JCI69076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–55. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Chou CM, Nelson C, Tarle SA, Pribila JT, Bardakjian T, et al. Biochemical basis for dominant inheritance, variable penetrance, and maternal effects in RBP4 congenital eye disease. Cell. 2015;161:634–46. doi: 10.1016/j.cell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog. Retin. Eye Res. 2010;29:398–427. doi: 10.1016/j.preteyeres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Swider M, Aleman TS, Tsybovsky Y, Schwartz SB, et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum. Mol. Genet. 2009;18:931–41. doi: 10.1093/hmg/ddn421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan SW, Newcomer ME, Jones TA. Crystallographic studies on a family of cellular lipophilic transport proteins: refinement of P2 myelin protein and the structure determination and refinement of cellular retinol-binding protein in complex with all-trans-retinol. J. Mol. Biol. 1993;230:1225–46. doi: 10.1006/jmbi.1993.1238. [DOI] [PubMed] [Google Scholar]
- Deigner PS, Law WC, Canada FJ, Rando RR. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 1989;244:968–71. doi: 10.1126/science.2727688. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, McGee TL, Ziviello C, Banfi S, Dryja TP, et al. A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2009;50:1864–72. doi: 10.1167/iovs.08-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine EL, Oprian DD, Theobald DL. Relocating the active-site lysine in rhodopsin and implications for evolution of retinylidene proteins. PNAS. 2013;110:13351–55. doi: 10.1073/pnas.1306826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, et al. Cloning of the gene and cDNA for mammalian β-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Dobri N, Qin Q, Kong J, Yamamoto K, Liu Z, et al. A1120, a nonretinoid RBP4 antagonist, inhibits formation of cytotoxic bisretinoids in the animal model of enhanced retinal lipofuscinogenesis. Investig. Ophthalmol. Vis. Sci. 2013;54:85–95. doi: 10.1167/iovs.12-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JE. Chemistry of visual adaptation in the rat. Nature. 1960;188:114–18. doi: 10.1038/188114a0. [DOI] [PubMed] [Google Scholar]
- Driessen CA, Winkens HJ, Hoffmann K, Kuhlmann LD, Janssen BP, et al. Disruption of the 11-cis-retinol dehydrogenase gene leads to accumulation of cis-retinols and cis-retinyl esters. Mol. Cell. Biol. 2000;20:4275–87. doi: 10.1128/mcb.20.12.4275-4287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–66. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–97. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–63. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo KM, Moiseyev G, Nikolaeva O, Sandell LL, Trainor PA, Ma JX. RDH10 is the primary enzyme responsible for the first step of embryonic vitamin A metabolism and retinoic acid synthesis. Dev. Biol. 2011;357:347–55. doi: 10.1016/j.ydbio.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo KM, Moiseyev G, Takahashi Y, Crouch RK, Ma JX. The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Investig. Ophthalmol. Vis. Sci. 2009;50:5089–97. doi: 10.1167/iovs.09-3797. [DOI] [PubMed] [Google Scholar]
- Fleisch VC, Schonthaler HB, von Lintig J, Neuhauss SC. Subfunctionalization of a retinoid-binding protein provides evidence for two parallel visual cycles in the cone-dominant zebrafish retina. J. Neurosci. 2008;28:8208–16. doi: 10.1523/JNEUROSCI.2367-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SL, Bridges CD. Internal quadruplication in the structure of human interstitial retinol-binding protein deduced from its cloned cDNA. J. Biol. Chem. 1988;263:15330–34. [PubMed] [Google Scholar]
- Golczak M, Imanishi Y, Kuksa V, Maeda T, Kubota R, Palczewski K. Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle. J. Biol. Chem. 2005a;280:42263–73. doi: 10.1074/jbc.M509351200. [DOI] [PubMed] [Google Scholar]
- Golczak M, Kuksa V, Maeda T, Moise AR, Palczewski K. Positively charged retinoids are potent and selective inhibitors of the trans–cis isomerization in the retinoid (visual) cycle. PNAS. 2005b;102:8162–67. doi: 10.1073/pnas.0503318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczak M, Sears AE, Kiser PD, Palczewski K. LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3. Nat. Chem. Biol. 2015;11:26–32. doi: 10.1038/nchembio.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein EB. Cone pigment regeneration in the isolated frog retina. Vis. Res. 1970;10:1065–68. doi: 10.1016/0042-6989(70)90082-9. [DOI] [PubMed] [Google Scholar]
- Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, et al. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am. J. Hum. Genet. 2007;80:1179–87. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SM, Thompson DA, Srikumari CR, Lorenz B, Finckh U, et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet. 1997;17:194–97. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J. Biol. Chem. 1998;273:21790–99. doi: 10.1074/jbc.273.34.21790. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Jang GF, Imanishi Y, Driessen CA, Matsumura M, et al. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem. 2002;277:45537–46. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 1993;268:15751–57. [PubMed] [Google Scholar]
- Han Z, Conley SM, Makkia RS, Cooper MJ, Naash MI. DNA nanoparticle-mediated ABCA4 delivery rescues Stargardt dystrophy in mice. J. Clin. Investig. 2012;122:3221–26. doi: 10.1172/JCI64833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave PA, McDowell JH, Curtis DR, Wang JK, Juszczak E, et al. The structure of bovine rhodopsin. Biophys. Struct. Mech. 1983;9:235–44. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- He XQ, Lobsiger J, Stocker A. Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W. PNAS. 2009;106:18545–50. doi: 10.1073/pnas.0907454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard R, Wald G. Cis–trans isomers of vitamin A and retinene in the rhodopsin system. J. Gen. Physiol. 1952;36:269–315. doi: 10.1085/jgp.36.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997;15:216–19. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Illing M, Molday LL, Molday RS. The 220-kDa Rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J. Biol. Chem. 1997;272:10303–10. doi: 10.1074/jbc.272.15.10303. [DOI] [PubMed] [Google Scholar]
- Imanishi Y, Batten ML, Piston DW, Baehr W, Palczewski K. Noninvasive two-photon imaging reveals retinyl ester storage structures in the eye. J. Cell Biol. 2004;164:373–83. doi: 10.1083/jcb.200311079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, et al. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–68. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Aleman TS, Cideciyan AV, Heon E, Golczak M, et al. Human cone photoreceptor dependence on RPE65 isomerase. PNAS. 2007;104:15123–28. doi: 10.1073/pnas.0706367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Regunath G, Rodriguez FJ, Vandenburgh K, et al. Night blindness in Sorsby’s fundus dystrophy reversed by vitamin A. Nat. Genet. 1995;11:27–32. doi: 10.1038/ng0995-27. [DOI] [PubMed] [Google Scholar]
- Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, et al. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat. Genet. 2004;36:850–54. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- Jang GF, McBee JK, Alekseev AM, Haeseleer F, Palczewski K. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J. Biol. Chem. 2000;275:28128–38. doi: 10.1074/jbc.M004488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–59. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Raz A, Goodman DS. Retinol-binding protein: the transport protein for vitamin A in human plasma. J. Clin. Investig. 1968;47:2025–44. doi: 10.1172/JCI105889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanan Y, Kasus-Jacobi A, Moiseyev G, Sawyer K, Ma JX, Al-Ubaidi MR. Retinoid processing in cone and Müller cell lines. Exp. Eye Res. 2008;86:344–54. doi: 10.1016/j.exer.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–25. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Ter-Stepanian M, Zhong M, Cheng G, et al. Receptor-mediated cellular uptake mechanism that couples to intracellular storage. ACS Chem. Biol. 2011;6:1041–51. doi: 10.1021/cb200178w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J. Biol. Chem. 2008;283:15160–68. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor JJ, Yuan Q, Cook J, Sarfare S, Makshanoff J, et al. Identification of DES1 as a vitamin A isomerase in Müller glial cells of the retina. Nat. Chem. Biol. 2013;9:30–36. doi: 10.1038/nchembio.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C, Tillman M, Burns ME, Pugh EN., Jr. Rhodopsin in the rod surface membrane regenerates more rapidly than bulk rhodopsin in the disc membranes in vivo. J. Physiol. 2014;592:2785–97. doi: 10.1113/jphysiol.2014.272518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Jang YP, Jockusch S, Fishkin NE, Turro NJ, Sparrow JR. The all-trans-retinal dimer series of lipofuscin pigments in retinal pigment epithelial cells in a recessive Stargardt disease model. PNAS. 2007;104:19273–78. doi: 10.1073/pnas.0708714104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Maeda A, Maeda T, Heinlein C, Kedishvili N, et al. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: a role of RDH11 in visual processes in vivo. J. Biol. Chem. 2005;280:8694–704. doi: 10.1074/jbc.M413172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K. Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. PNAS. 2012a;109:E2747–56. doi: 10.1073/pnas.1212025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. PNAS. 2009;106:17325–30. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Maeda A, Palczewski K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta. 2012b;1821:137–51. doi: 10.1016/j.bbalip.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Palczewski K. Chemistry of the retinoid (visual) cycle. Chem. Rev. 2014;114:194–232. doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Zhang J, Badiee M, Li Q, Shi W, et al. Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat. Chem. Biol. 2015;11:409–15. doi: 10.1038/nchembio.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenekoop RK, Sui R, Sallum J, van den Born LI, Ajlan R, et al. Oral 9-cis retinoid for childhood blindness due to Leber congenital amaurosis caused by RPE65 or LRAT mutations: an open-label phase 1b trial. Lancet. 2014;384:1513–20. doi: 10.1016/S0140-6736(14)60153-7. [DOI] [PubMed] [Google Scholar]
- Kubota R, Boman NL, David R, Mallikaarjun S, Patil S, Birch D. Safety and effect on rod function of ACU-4429, a novel small-molecule visual cycle modulator. Retina. 2012;32:183–88. doi: 10.1097/IAE.0b013e318217369e. [DOI] [PubMed] [Google Scholar]
- Kühne W. Chemical processes in the retina. Vis. Res. 1977;17:1269–316. doi: 10.1016/0042-6989(77)90114-6. [DOI] [PubMed] [Google Scholar]
- Kwok-Keung Fung B, Stryer L. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. PNAS. 1980;77:2500–4. doi: 10.1073/pnas.77.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YL, Wiggert B, Liu YP, Chader GJ. Interphotoreceptor retinol-binding proteins: possible transport vehicles between compartments of the retina. Nature. 1982;298:848–49. doi: 10.1038/298848a0. [DOI] [PubMed] [Google Scholar]
- Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10:43–49. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- Lucas RJ, Hattar S, Takao M, Berson DM, Foster RG, Yau KW. Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science. 2003;299:245–47. doi: 10.1126/science.1077293. [DOI] [PubMed] [Google Scholar]
- Ma L, Kaufman Y, Zhang J, Washington I. C20-D3-vitamin A slows lipofuscin accumulation and electrophysiological retinal degeneration in a mouse model of Stargardt disease. J. Biol. Chem. 2011;286:7966–74. doi: 10.1074/jbc.M110.178657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. Binding specificities of cellular retinol-binding protein and cellular retinol-binding protein, type II. J. Biol. Chem. 1987;262:10550–56. [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem. Biophys. Res. Commun. 1988;156:157–63. doi: 10.1016/s0006-291x(88)80818-0. [DOI] [PubMed] [Google Scholar]
- Maeda A, Golczak M, Chen Y, Okano K, Kohno H, et al. Primary amines protect against retinal degeneration in mouse models of retinopathies. Nat. Chem. Biol. 2012;8:170–78. doi: 10.1038/nchembio.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Golczak M, Chou S, Desai A, et al. Involvement of all-trans-retinal in acute light-induced retinopathy of mice. J. Biol. Chem. 2009a;284:15173–83. doi: 10.1074/jbc.M900322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P, et al. Effects of potent inhibitors of the retinoid cycle on visual function and photoreceptor protection from light damage in mice. Mol. Pharmacol. 2006a;70:1220–29. doi: 10.1124/mol.106.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Imanishi Y, Kuksa V, Alekseev A, et al. Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J. Biol. Chem. 2005;280:18822–32. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Imanishi Y, Sun W, Jastrzebska B, et al. Retinol dehydrogenase (RDH12) protects photoreceptors from light-induced degeneration in mice. J. Biol. Chem. 2006b;281:37697–704. doi: 10.1074/jbc.M608375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Palczewski K. Improvement in rod and cone function in mouse model of Fundus albipunctatus after pharmacologic treatment with 9-cis-retinal. Investig. Ophthalmol. Vis. Sci. 2006c;47:4540–46. doi: 10.1167/iovs.06-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Sun W, Zhang H, Baehr W, Palczewski K. Redundant and unique roles of retinol dehydrogenases in the mouse retina. PNAS. 2007;104:19565–70. doi: 10.1073/pnas.0707477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Palczewska G, Golczak M, Kohno H, Dong Z, et al. Two-photon microscopy reveals early rod photoreceptor cell damage in light-exposed mutant mice. PNAS. 2014;111:E1428–37. doi: 10.1073/pnas.1317986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Cideciyan AV, Maeda A, Golczak M, Aleman TS, et al. Loss of cone photoreceptors caused by chromophore depletion is partially prevented by the artificial chromophore pro-drug, 9-cis-retinyl acetate. Hum. Mol. Genet. 2009b;18:2277–87. doi: 10.1093/hmg/ddp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr., Mingozzi F, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–48. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, et al. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat. Genet. 1997;17:139–41. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- Mata NL, Lichter JB, Vogel R, Han Y, Bui TV, Singerman LJ. Investigation of oral fenretinide for treatment of geographic atrophy in age-related macular degeneration. Retina. 2013;33:498–507. doi: 10.1097/IAE.0b013e318265801d. [DOI] [PubMed] [Google Scholar]
- Mata NL, Radu RA, Clemmons RC, Travis GH. Isomerization and oxidation of vitamin A in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 2002;36:69–80. doi: 10.1016/s0896-6273(02)00912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maw MA, Kennedy B, Knight A, Bridges R, Roth KE, et al. Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat. Genet. 1997;17:198–200. doi: 10.1038/ng1097-198. [DOI] [PubMed] [Google Scholar]
- McBee JK, Kuksa V, Alvarez R, de Lera AR, Prezhdo O, et al. Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry. 2000;39:11370–80. doi: 10.1021/bi001061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: the interlink of photo-transduction and retinoid metabolism in the vertebrate retina. Prog. Retin. Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- Moise AR, Golczak M, Imanishi Y, Palczewski K. Topology and membrane association of lecithin:retinol acyltransferase. J. Biol. Chem. 2007;282:2081–90. doi: 10.1074/jbc.M608315200. [DOI] [PubMed] [Google Scholar]
- Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. PNAS. 2005;102:12413–18. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday LL, Rabin AR, Molday RS. ABCR expression in foveal cone photoreceptors and its role in Stargardt macular dystrophy. Nat. Genet. 2000;25:257–58. doi: 10.1038/77004. [DOI] [PubMed] [Google Scholar]
- Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein. Science. 1995;268:1039–41. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- Mondal MS, Ruiz A, Bok D, Rando RR. Lecithin retinol acyltransferase contains cysteine residues essential for catalysis. Biochemistry. 2000;39:5215–20. doi: 10.1021/bi9929554. [DOI] [PubMed] [Google Scholar]
- Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or Leber congenital amaurosis. PNAS. 1998;95:3088–93. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz A, Betts BS, Trevino AR, Buddavarapu K, Roman R, et al. Evidence for two retinoid cycles in the cone-dominated chicken eye. Biochemistry. 2009;48:6854–63. doi: 10.1021/bi9002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff’s base counterion in bovine rhodopsin. Biochemistry. 1990;29:9746–52. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation, sequence analysis, and intron–exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–14. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Newcomer ME, Jones TA, Aqvist J, Sundelin J, Eriksson U, et al. The three-dimensional structure of retinol-binding protein. EMBO J. 1984;3:1451–54. doi: 10.1002/j.1460-2075.1984.tb01995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nociari MM, Lehmann GL, Perez Bay AE, Radu RA, Jiang Z, et al. Beta cyclodextrins bind, stabilize, and remove lipofuscin bisretinoids from retinal pigment epithelium. PNAS. 2014;111:E1402–8. doi: 10.1073/pnas.1400530111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DE, Chytil F. Specificity of cellular retinol-binding protein for compounds with vitamin A activity. Nature. 1975;255:74–75. doi: 10.1038/255074a0. [DOI] [PubMed] [Google Scholar]
- Palczewski K. G protein–coupled receptor rhodopsin. Annu. Rev. Biochem. 2006;75:743–67. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. Retinoids for treatment of retinal diseases. Trends Pharmacol. Sci. 2010;31:284–95. doi: 10.1016/j.tips.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, et al. Crystal structure of rhodopsin: a G protein–coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, et al. Melanopsin is required for non-image-forming photic responses in blind mice. Science. 2003;301:525–27. doi: 10.1126/science.1086179. [DOI] [PubMed] [Google Scholar]
- Papermaster DS, Schneider BG, Zorn MA, Kraehenbuhl JP. Immunocytochemical localization of a large intrinsic membrane protein to the incisures and margins of frog rod outer segment disks. J. Cell Biol. 1978;78:415–25. doi: 10.1083/jcb.78.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker RO, Crouch RK. Retinol dehydrogenases (RDHs) in the visual cycle. Exp. Eye Res. 2010;91:788–92. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet. 2007;80:550–60. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Gubin AN, Stearn O, Li Y, Campos MM, et al. Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: hint from jawless vertebrates. PLOS ONE. 2012;7:e49975. doi: 10.1371/journal.pone.0049975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadro L, Blaner WS, Salchow DJ, Vogel S, Piantedosi R, et al. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999;18:4633–44. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi F, Lenevich S, Molday RS. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 2012;3:925. doi: 10.1038/ncomms1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi F, Molday RS. ATP-binding cassette transporter ABCA4 and chemical isomerization protect photoreceptor cells from the toxic accumulation of excess 11-cis-retinal. PNAS. 2014;111:5024–29. doi: 10.1073/pnas.1400780111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu RA, Hu J, Peng J, Bok D, Mata NL, Travis GH. Retinal pigment epithelium–retinal G protein receptor-opsin mediates light-dependent translocation of all-trans-retinyl esters for synthesis of visual chromophore in retinal pigment epithelial cells. J. Biol. Chem. 2008;283:19730–38. doi: 10.1074/jbc.M801288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu RA, Mata NL, Nusinowitz S, Liu X, Sieving PA, Travis GH. Treatment with isotretinoin inhibits lipofuscin accumulation in a mouse model of recessive Stargardt’s macular degeneration. PNAS. 2003;100:4742–47. doi: 10.1073/pnas.0737855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner A, Smallwood PM, Nathans J. Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J. Biol. Chem. 2000;275:11034–43. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. PNAS. 2005;102:13658–63. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, et al. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 1998;20:344–51. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- Ripps H. The color purple: milestones in photochemistry. FASEB J. 2008;22:4038–43. doi: 10.1096/fj.08-1202ufm. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Mark M, Jacobs H, Klopfenstein M, Hu J, et al. Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Investig. Ophthalmol. Vis. Sci. 2012;53:3027–39. doi: 10.1167/iovs.11-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Winston A, Lim YH, Gilbert BA, Rando RR, Bok D. Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem. 1999;274:3834–41. doi: 10.1074/jbc.274.6.3834. [DOI] [PubMed] [Google Scholar]
- Saari JC. Vitamin A metabolism in rod and cone visual cycles. Annu. Rev. Nutr. 2012;32:125–45. doi: 10.1146/annurev-nutr-071811-150748. [DOI] [PubMed] [Google Scholar]
- Saari JC, Bredberg DL. Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J. Biol. Chem. 1989;264:8636–40. [PubMed] [Google Scholar]
- Saari JC, Bredberg L, Garwin GG. Identification of the endogenous retinoids associated with three cellular retinoid-binding proteins from bovine retina and retinal pigment epithelium. J. Biol. Chem. 1982;257:13329–33. [PubMed] [Google Scholar]
- Saari JC, Bunt AH, Futterman S, Berman ER. Localization of cellular retinal-binding protein in bovine retina and retinal pigment epithelium, with a consideration of the pigment epithelium isolation technique. Investig. Ophthalmol. Vis. Sci. 1977;16:797–806. [PubMed] [Google Scholar]
- Saari JC, Nawrot M, Garwin GG, Kennedy MJ, Hurley JB, et al. Analysis of the visual cycle in cellular retinol-binding protein type I (CRBPI) knockout mice. Investig. Ophthalmol. Vis. Sci. 2002;43:1730–35. [PubMed] [Google Scholar]
- Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, et al. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron. 2001;29:739–48. doi: 10.1016/s0896-6273(01)00248-3. [DOI] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, et al. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J. Biol. Chem. 2011;286:10551–67. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counter-ion in bovine rhodopsin. PNAS. 1989;86:8309–13. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R. Projection structure of rhodopsin. Nature. 1993;362:770–72. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Sibulesky L, Hayes KC, Pronczuk A, Weigel-DiFranco C, Rosner B, Berson EL. Safety of <7500 RE (<25000 IU) vitamin A daily in adults with retinitis pigmentosa. Am. J. Clin. Nutr. 1999;69:656–63. doi: 10.1093/ajcn/69.4.656. [DOI] [PubMed] [Google Scholar]
- Simon A, Hellman U, Wernstedt C, Eriksson U. The retinal pigment epithelial-specific 11-cis retinol dehydrogenase belongs to the family of short chain alcohol dehydrogenases. J. Biol. Chem. 1995;270:1107–12. [PubMed] [Google Scholar]
- Sparrow JR, Gregory-Roberts E, Yamamoto K, Blonska A, Ghosh SK, et al. The bisretinoids of retinal pigment epithelium. Prog. Retin. Eye Res. 2012;31:121–35. doi: 10.1016/j.preteyeres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 2000;16:365–92. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- Sun H, Smallwood PM, Nathans J. Biochemical defects in ABCR protein variants associated with human retinopathies. Nat. Genet. 2000;26:242–46. doi: 10.1038/79994. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Li Y, McHenry CL, Carlson TJ, Ding X, et al. Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nat. Genet. 2001;28:123–24. doi: 10.1038/88828. [DOI] [PubMed] [Google Scholar]
- Trapani I, Colella P, Sommella A, Iodice C, Cesi G, et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 2014;6:194–211. doi: 10.1002/emmm.201302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsybovsky Y, Molday RS, Palczewski K. The ATP-binding cassette transporter ABCA4: structural and functional properties and role in retinal disease. Adv. Exp. Med. Biol. 2010;703:105–25. doi: 10.1007/978-1-4419-5635-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsybovsky Y, Orban T, Molday RS, Taylor D, Palczewski K. Molecular organization and ATP-induced conformational changes of ABCA4, the photoreceptor-specific ABC transporter. Structure. 2013;21:854–60. doi: 10.1016/j.str.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN. Non-visual photoreception: sensing light without sight. Curr. Biol. 2008;18:R38–R39. doi: 10.1016/j.cub.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Van Hooser JP, Aleman TS, He YG, Cideciyan AV, Kuksa V, et al. Rapid restoration of visual pigment and function with oral retinoid in a mouse model of childhood blindness. PNAS. 2000;97:8623–28. doi: 10.1073/pnas.150236297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser JP, Liang Y, Maeda T, Kuksa V, Jang GF, et al. Recovery of visual functions in a mouse model of Leber congenital amaurosis. J. Biol. Chem. 2002;277:19173–82. doi: 10.1074/jbc.M112384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. The molecular basis of visual excitation. Nature. 1968;219:800–7. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- Wang JS, Estevez ME, Cornwall MC, Kefalov VJ. Intra-retinal visual cycle required for rapid and complete cone dark adaptation. Nat. Neurosci. 2009;12:295–302. doi: 10.1038/nn.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS, Kefalov VJ. The cone-specific visual cycle. Prog. Retin. Eye Res. 2011;30:115–28. doi: 10.1016/j.preteyeres.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. Vibrationally coherent photochemistry in the femtosecond primary event of vision. Science. 1994;266:422–24. doi: 10.1126/science.7939680. [DOI] [PubMed] [Google Scholar]
- Wenzel A, Oberhauser V, Pugh EN, Jr., Lamb TD, Grimm C, et al. The retinal G protein–coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J. Biol. Chem. 2005;280:29874–84. doi: 10.1074/jbc.M503603200. [DOI] [PubMed] [Google Scholar]
- Woodruff ML, Wang Z, Chung HY, Redmond TM, Fain GL, Lem J. Spontaneous activity of opsin apoprotein is a cause of Leber congenital amaurosis. Nat. Genet. 2003;35:158–64. doi: 10.1038/ng1246. [DOI] [PubMed] [Google Scholar]
- Wu BX, Chen Y, Fan J, Rohrer B, Crouch RK, Ma JX. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Investig. Ophthalmol. Vis. Sci. 2002;43:3365–72. [PubMed] [Google Scholar]
- Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX. Identification of RDH10, an all-trans retinol dehydrogenase, in retinal Müller cells. Investig. Ophthalmol. Vis. Sci. 2004;45:3857–62. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]
- Xie YA, Lee W, Cai C, Gambin T, Noupuu K, et al. New syndrome with retinitis pigmentosa is caused by nonsense mutations in retinol dehydrogenase RDH11. Hum. Mol. Genet. 2014;23:5774–80. doi: 10.1093/hmg/ddu291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat. Genet. 1999;22:188–91. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kiser PD, Badiee M, Palczewska G, Dong Z, et al. Molecular pharmacodynamics of emixustat in protection against retinal degeneration. J. Clin. Investig. 2015;125:2781–94. doi: 10.1172/JCI80950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman WF. Subcellular distribution of 11-cis retinol dehydrogenase activity in bovine pigment epithelium. Exp. Eye Res. 1976;23:159–64. doi: 10.1016/0014-4835(76)90199-8. [DOI] [PubMed] [Google Scholar]
- Zimmerman WF, Lion F, Daemen FJ, Bonting SL. Biochemical aspects of the visual process. XXX. Distribution of stereospecific retinol dehydrogenase activities in subcellular fractions of bovine retina and pigment epithelium. Exp. Eye Res. 1975;21:325–32. doi: 10.1016/0014-4835(75)90043-3. [DOI] [PubMed] [Google Scholar]
RELATED RESOURCES
- Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–63. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein–coupled receptor rhodopsin: a prospectus. Annu. Rev. Physiol. 2003;65:851–79. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu. Rev. Genom. Hum. Genet. 2014;15:151–71. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signaling. Nat. Rev. Mol. Cell Biol. 2014;15:411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Nathans J. The evolution of primate color vision. Sci. Am. 2009;300:56–63. doi: 10.1038/scientificamerican0409-56. [DOI] [PubMed] [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 2013;36:24–51. doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW. The First Steps in Seeing. Sinauer Assoc.; Sunderland, MA: 1998. [Google Scholar]
- Sundermeier TR, Palczewski K. The physiological impact of microRNA gene regulation in the retina. Cell. Mol. Life Sci. 2012;69:2739–50. doi: 10.1007/s00018-012-0976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]