Abstract
The host immune system offers a hostile environment with antimicrobials and reactive oxygen species (ROS) that are detrimental to bacterial pathogens, forcing them to adapt and evolve for survival. However, the contribution of oxidative stress to pathogen evolution remains elusive. Using an experimental evolution strategy, we show that exposure of the opportunistic pathogen Pseudomonas aeruginosa to sub-lethal hydrogen peroxide (H2O2) levels over 120 generations led to the emergence of pro-biofilm rough small colony variants (RSCVs), which could be abrogated by l-glutathione antioxidants. Comparative genomic analysis of the RSCVs revealed that mutations in the wspF gene, which encodes for a repressor of WspR diguanylate cyclase (DGC), were responsible for increased intracellular cyclic-di-GMP content and production of Psl exopolysaccharide. Psl provides the first line of defence against ROS and macrophages, ensuring the survival fitness of RSCVs over wild-type P. aeruginosa. Our study demonstrated that ROS is an essential driving force for the selection of pro-biofilm forming pathogenic variants. Understanding the fundamental mechanism of these genotypic and phenotypic adaptations will improve treatment strategies for combating chronic infections.
Keywords: biofilms, c-di-GMP, rough small colony variants, reactive oxygen species, Pseudomonas aeruginosa, adaptive evolution
1. Background
Bacterial pathogens can colonize human hosts for years and cause persistent/chronic infections [1,2]. These infections predominantly result from biofilm formation, whereby secreted extracellular-polymeric substances (EPS) such as adhesive proteins, biosurfactants, extracellular DNA and exopolysaccharides act as physical barriers to protect bacterial cells from the host immune clearance and antimicrobial treatments [3–5].
The transition between free-living planktonic and sessile biofilm lifestyles of most bacterial species is mediated by bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP), a global intracellular secondary messenger [6,7]. Increase in intracellular c-di-GMP content is caused by the activity of diguanylate cyclases (DGCs) [8,9], resulting in biofilm formation, whereas phosphodiesterases (PDEs) degrade c-di-GMP [10,11] and cause biofilm dispersal to the planktonic phase [12,13]. C-di-GMP signalling is involved not only in motility, surface attachment and production of EPS [14–16], but also in the sensing of chemicals and surfaces [8]. C-di-GMP signalling also regulates virulence and stress responses in pathogens [17,18].
Pseudomonas aeruginosa is the leading cause of chronic lung infection and morbidity of patients with cystic fibrosis (CF) [19,20]. It is able to survive and form biofilms despite the presence of a functional host immune system, antibiotic treatments and competing pathogens, such as Staphylococcus aureus in the CF lung environment [21–24]. Investigations of adaptive evolution of P. aeruginosa in CF lung infections have provided valuable information for our current understanding of chronic infections. P. aeruginosa can colonize the CF lungs for decades and usually gives rise to pro-biofilm sub-populations after adaptive evolution [25,26]. The occurrence of rough small colony variants (RSCVs) and mucoid strains is often reported from chronic CF infections, implying these variants have better fitness than their ancestors [27,28].
Although the characteristics of RSCVs have been studied [29,30], the stimuli and mechanisms leading to the evolution of such adapted sub-populations are unclear. Given the CF environment consists of oxidative stress, high antibiotic concentrations, high pro-inflammatory cytokine levels and poor nutrient conditions [31–35], it is highly likely that each of the CF-derived environmental factors can drive the adaptive evolution of pathogens differently.
Experimental evolution assays have been used in previous studies to investigate bacterial adaptation to various conditions such as antibiotic treatments and carbon sources [18,36]. Here, we employed the adaptive experimental evolution assay to evolve P. aeruginosa against an important host-derived antimicrobial, reactive oxygen species (ROS), resulting in the occurrence of RSCVs with a strong capability for biofilm formation and ROS stress resistance.
The RSCVs isolated from H2O2 treated cultures showed increased intracellular c-di-GMP content. Next-generation sequencing (NGS) analysis revealed that wspF mutation was associated with these isolated RSCVs. The wspF is part of the chemosensory-like system Wsp (wrinkly spreader phenotype) [8], whose product acts as a repressor against WspR (DGC), thus its mutagenesis leads to the de-repression of WspR. The increased production of exopolysaccharides (especially Psl) as the result of wspF mutation conferred resistance of RSCVs to H2O2 treatment. Hence, this work strongly suggests that exposure to ROS imposes a strong selective pressure on P. aeruginosa during chronic colonization and accounts for the occurrence of RSCVs in clinical isolates obtained from CF patients.
2. Material and methods
2.1. Bacterial strains, plasmids, media and growth conditions
Escherichia coli DH5α strain was used for standard DNA manipulations. Luria-Bertani (LB) medium was used to cultivate E. coli strains. Batch cultivation of P. aeruginosa strains was carried out at 37°C in ABTG (ABT minimal medium supplemented with 5 g l−1 glucose) or ABTGC (ABT minimal medium supplemented with 2 g l−1 glucose and 2 g l−1 casamino acids). For plasmid maintenance in E. coli, the medium was supplemented with 100 µg ml−1 ampicillin (Ap), 15 µg ml−1 gentamicin (Gm) and 15 µg ml−1 tetracycline (Tc). When appropriate, the marker selection in P. aeruginosa, 30 µg ml−1 Gm, 50 µg ml−1 Tc or 200 µg ml−1 carbenicillin (Cb), was used. The bacterial strains and plasmids are listed in the electronic supplementary material, table S1.
2.2. Imaging colony morphology of Pseudomonas aeruginosa
Cultures were streaked on LB agar together with 40 µg ml−1 Congo Red and 15 µg ml−1 Coomassie Brilliant Blue R at 37°C for 48 h as previously described [37]. Colony images were captured by the stereomicroscope (Zeiss Discovery V8, Germany) and processed using IMARIS software (Bitplane AG, Zurich, Switzerland).
2.3. Evolution assay of Pseudomonas aeruginosa PAO1 in H2O2 and GSH
Cultures were cultivated overnight from ancestral PAO1 at 37°C, 200 rpm in LB with three biological replicates. Each biological replicate with initially identical populations was then divided into three technical replicates, each grown in ABTGC only or ABTGC with 2 mM H2O2 + 5, 2.5, 1, 0.25, 0.125 or 0 mM GSH. One per cent of each replicate population was transferred to a new tube of fresh ABTGC with and without 2 mM H2O2 + GSH at 37°C, 200 rpm for 12 h, allowing each population to experience an estimated 6.67 generations every passage. This was then repeated for 15 days, so that there were an estimated 120 generations of cells grown in 2 mM H2O2.
The populations were then cryopreserved with 50% glycerol (cryoprotectant) for revival at a later time. To observe the emergence of unique phenotypes arising from treatment with 2 mM H2O2, the populations were grown on LB agar plates at 37°C overnight. Ten RSCV isolates were identified from the different replicates and frozen with 50% glycerol.
2.4. H2O2 resistance assay
The P. aeruginosa cultures were grown overnight and prepared by adjusting the optical density to OD600 = 0.3 in ABTGC with and without 4 mM H2O2. The cultures were grown at 37°C, 200 rpm for 4 h. The cell culture was serially diluted and plated on LB agar plates. The culture plates were incubated in 37°C for 16 h. The numbers of colonies were counted for tabulation of log10 CFU ml−1. The log10 CFU ml−1 is calculated by log10(average number of colonies × dilution factor × volume used to spread on LB agar plate). Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.5. Competitive mixed-treatment assay of rough small colony variant isolates and PAO1
The evolved RSCVs and PAO1 cultures were prepared by adjusting the optical density to OD600 = 0.5 in ABTGC. Each RSCV was mixed at equal volumes (1 : 1 ratio) with PAO1 in ABTGC with and without 4 mM H2O2 (1× MIC). The cultures were grown at 37°C, 200 rpm. Twenty microlitre samples were collected after a 4 h incubation. They were diluted serially in 0.9% NaCl and 100 µl of cell culture was cultivated on LB agar plates for 16 h at 37°C. The RSCVs and PAO1 were differentiated by the morphology of the colonies. The number of colonies was counted for tabulation of CFU ml−1. The competition ratio was then calculated by CFU ml−1 of RSCV/CFU ml−1 of PAO1. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.6. DNA sequencing of rough small colony variants isolates
Individual colonies of the ancestral PAO1, two random PAO1 isolates evolved in ABTGC only and 10 RSCV isolates were first streaked on the LB agar plates and incubated at 37°C for 16 h. Single colonies were picked by a 1 µl inoculation loop and grown in LB at 37°C, 200 rpm for 16 h. Genomic DNA of the P. aeruginosa strains was purified using QIAamp DNA Mini Kit (Qiagen, Venlo, The Netherlands). The quality control tests included NanoDrop (Thermo Fisher Scientific) and Qubit (Thermo Fisher Scientific) to ensure the concentration and quality of DNA, and DNA electrophoresis (1% agar) to determine DNA integrity.
The genomic DNA was then sequenced on an Illumina (San Diego, CA, USA) MiSeq V3 platform, generating 300 bp long paired-end reads using the method described in Chua et al. [38]. The average insert sizes were 490–544 bp, and average genomic coverage depths were 40–186 fold. Nucleotide differences were generated from the CLC Genomics Workbench 8.0 (CLC bio, Aarhus, Denmark). Briefly, adapters and low quality reads were trimmed. Paired-end reads in FASTQ format for RSCV and control genomes were mapped against the P. aeruginosa PAO1 genome (NC_002516). Both the mappings of RSCV and control strains were compared with their ancestor PAO1, and variants were detected using the quality based variant detection method with the required frequency of 35%.
2.7. Measurement of mutation rates
PAO1 was grown in ABTGC with or without 2 mM H2O2 until stationary phase, and plated on LB agar plates with and without 0.78 µg ml−1 ciprofloxacin, using the method described in Mandsberg et al. [39]. The mutated colonies cultured on LB agar with ciprofloxacin were enumerated to account for point mutations on the gyr gene that can easily develop in P. aeruginosa exposed to ciprofloxacin [40]. The mutation rate was then calculated by dividing the number of mutations by the final CFU of cells grown on LB agar plates without ciprofloxacin. Three independent experiments were performed in triplicate, the Student's t-test was used to determine statistical significance and the results were shown as the mean ± s.d.
2.8. The pcdrA-gfp reporter assay
The cultures of PAO1 with pcdrA-gfp reporter fusion were grown in ABTGC with or without H2O2. In total, 200 µl of cell culture was then transferred into each well of a 96-well plate (triplicates). The OD600 and GFP fluorescence (excitation 485 nm/emission 535 nm) were measured using the Tecan Infinite 200 microplate reader (Tecan, Austria). The relative fluorescence intensity count was calculated by dividing GFP values by OD600 values. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.9. Quantification of pyoverdine
The cultures were grown in ABTGC with or without H2O2. In total, 200 µl of cell culture was then transferred into each well of a 96-well plate (triplicates). As previously described [13], the OD600 and pyoverdine fluorescence (excitation 400 nm/emission 460 nm) were measured using the Tecan microplate reader, and the relative fluorescence intensity count calculated by dividing pyoverdine fluorescence values by OD600 values. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.10. C-di-GMP quantification by liquid chromatography–mass spectrometry
Fifteen millilitres of PAO1 cell culture treated with H2O2, RSCVs, Burkholderia cenocepacia strains and Klebsiella pneumoniae strains were harvested and washed twice with 1 mM ammonium acetate. An aliquot of cells was used for protein quantification. The remaining cells were lysed in 1 ml acetonitrile/methanol/ddH2O (v/v ratio 40 : 40 : 20) using a probe tip ultrasonicator (amplitude 30%; 5 s on, 5 s off) for 1 min on an ice slurry. The cell debris was removed by centrifuging at 13 000g, 4 °C for 3 min, rinsing twice. The supernatant containing the nucleotides was lyophilized with the vacuum concentrator. The lyophilized nucleotides were resuspended in 100 µl 1 mM ammonium acetate. The c-di-GMP standard was also used as a reference to identify the c-di-GMP peak and the concentration of c-di-GMP in the samples.
For the detection and quantification of c-di-GMP, a Thermo Accela 1250 series LC system fitted with EQuanMax autosampler and a Thermo Velos Pro Orbitrap mass spectrometer (Thermo Fisher Scientific) were used. Chromatographic separation was achieved using a Nucleodur C18 Pyramid (2 mm × 50 mm, 3 µm) column (Macherey-Nagel GmbH, Düren, Germany) at 40°C, with a solvent flow rate of 0.3 ml min−1 and an injection volume of 10 µl. Buffer A was 10 mM ammonium acetate buffer, containing 0.1% acetic acid, and buffer B was acetonitrile, containing 0.1% acetic acid. Solvent gradient conditions were as follows: 0% B from 0 to 3 min; 10% B at 3 min; 90% from 4th to 5th min; 0% B at 5.5th min and equilibrated for 4.5 min. Total run time was 10 min.
Detection was carried out in positive ion electrospray ionization (ESI+) mode. The heater and capillary temperatures were 300°C. Sheath, auxiliary and sweeper gas flows were 40, 15 and 1 arb. units, respectively. Source voltage was 3.5 kV. For quantitation, scan type in selected ion monitoring mode was used at high-resolution (60 000), with an AGC target of 1 × 106. Quantification was achieved via an MS/MS experiment using collision induced dissociation (CID) with normalized collision energy 20% (of maximum), with isolation width of 1 Da and activation time of 30 ms.
To quantify protein concentration, a small aliquot of cell culture was lysed in 100 µl 5 M sodium hydroxide at 95°C for 5 min. The protein concentration was then measured using a Qubit® 2.0 fluorometer (Invitrogen, Thermo Fisher Scientific, CA). The final concentration of c-di-GMP was then normalized with protein quantity. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used as to determine statistical significance and results were shown as the mean ± s.d.
2.11. Psl staining by fluorescent concanavalin-A
Planktonic MiniTn7-gfp-tagged cells were grown in ABTGC + 0, 0.5, 1 and 2 mM H2O2 at 37°C, 200 rpm for 4 h. Additionally, PAO1, mutants and evolved RSCVs with tagged MiniTn7-gfp were grown until stationary phase in ABTGC overnight at 37°C, 200 rpm. Ten microlitres of cell culture were then transferred to the glass slide and stained by 5 µM concanavalin-A Alexa Fluor® 647 conjugate (Thermo Fisher Scientific, Cat. No. C21421) [41].
To monitor fluorescence of GFP and Psl stain, the cells were imaged using an LSM780 confocal laser scanning microscope (CLSM; Carl Zeiss, Germany) with 40× objective or 63× oil objective (for planktonic cells) and the images were processed using IMARIS software (Bitplane AG, Zurich, Switzerland). Three independent experiments were performed in triplicate and representative images were shown.
2.12. Arabinose-inducible expression of Psl by PAO1/pBAD-psl
A starting culture of PAO1/pBAD-psl cells containing an l-arabinose-inducible promoter for psl operon expression was grown in ABTGC with increasing concentrations of l-arabinose at 37°C, 200 rpm for 3 h until OD600 = ∼0.3 was established, as described in Irie et al. [42]. The cultures were then treated with and without 4 mM H2O2 and continued to be grown at 37°C, 200 rpm for an additional 4 h. The cell culture was serially diluted and plated on LB agar plates. The culture plates were incubated at 37°C for 16 h. The colonies were counted for tabulation of CFU ml−1. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used as statistical tests and the results were shown as the mean ± s.d.
2.13. Biofilm quantification of PAO1/pBAD-psl by crystal violet assay
The PAO1/pBAD-psl biofilms were grown in 1 ml ABTGC with increasing concentrations of l-arabinose at 37°C in 24-well plates (Nunc, Denmark). The biofilms were washed three times with 0.9% NaCl and stained with 0.1% crystal violet for 15 min. Excess crystal violet was washed off with 0.9% NaCl. The stained biofilms were dissoluted by 100% ethanol and their OD595 values were quantified using a Tecan Infinite 200 microplate reader (Tecan, Austria). Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.14. Cellulase treatment of Pseudomonas aeruginosa cultures to degrade Psl
The P. aeruginosa cultures were grown overnight and prepared by adjusting the optical density to OD600 = 0.1 in ABTGC with 1 mg ml−1 cellulase (Sigma-Aldrich, Cat. No. C9748) [42]. The cultures were grown to OD600 = ∼0.3 at 37°C, 200 rpm for 3 h. The cultures were treated with 0 or 4 mM H2O2 and further grown at 37°C, 200 rpm for 4 h. The cell culture was serially diluted and plated on LB agar plates. The culture plates were incubated in 37°C for 16 h. Colonies were counted for tabulation of CFU ml−1. Three independent experiments were performed in triplicate. One-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.15. RAW264.7 macrophages culture
The murine RAW264.7 macrophage cell lines (ATCC No. TIB-71) were cultivated in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Thermo Fisher Scientific), supplemented with 10% fetal bovine serum (FBS) (Gibco™, Thermo Fisher Scientific). Cell cultures were incubated in 75 cm2 cell culture flasks at a density of 1.0 × 106 cells ml−1 at 37°C, 5% CO2 for 72 h.
2.16. Macrophage phagocytosis assay
As previously described [12], 5 × 105 macrophages were grown in each well of a 24-well culture plate (Nunc, Denmark). Macrophages were infected with bacterial suspension at a multiplicity of infection (MOI) of 100 : 1. The co-cultures were incubated at 37°C and 5% CO2 for 2 h. The macrophages were washed three times with phosphate-buffered saline (PBS) to remove extracellular bacteria, and then lysed with ddH2O containing 0.5% Triton X-100. The cell lysates were then serially diluted, and 100 µl of each dilution was plated on triplicate LB agar plates and incubated overnight at 37°C. The number of colonies was enumerated, and CFU ml−1 was tabulated. Experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to determine statistical significance and the results were shown as the mean ± s.d.
2.17. Macrophage cytotoxicity assay
RAW264.7 macrophages (5 × 105 ml−1 in each well) were grown in 24-well culture plates as previously described [12]. Triplicate cultures were infected with bacterial suspension at a MOI of 100 : 1. Macrophages were incubated for 4 h in 37°C, 5% CO2, then 20 µM propidium iodide (PI) was used to stain dead cells and assess macrophage cytotoxicity, leaving live cells unstained. Macrophages stained by PI (excitation 535 nm, emission 617 nm) under epifluorescent microscopy (20× objective) were counted as dead. Macrophages from five replicate images (on average 100 macrophages per image) of each sample were counted, enabling the ratio of dead cells to live cells to be calculated. Three independent experiments were performed in triplicate, one-way ANOVA and Student's t-tests were used to establish statistical significance and the results were shown as the mean ± s.d.
3. Results
3.1. Emergence of rough small colony variants during adaptive evolution of Pseudomonas aeruginosa against ROS
To determine if oxidative stress provokes the adaptive evolution of P. aeruginosa, P. aeruginosa was cultivated in the presence of a sub-lethal concentration of H2O2 (2 mM, 0.5 × minimal inhibitory concentration (MIC)) for 120 generations. This simulated the presence of ROS produced by polymorphonuclear leucocytes (PMNs) in the CF lung, which generate oxidative stress as a selective pressure on P. aeruginosa cells [43]. Although a millimolar range of H2O2 is required to kill most bacteria and bacteria internalized in the phagosome are usually exposed to micromolar concentrations, the rates of free radical generation within the phagosome are estimated to be 10 000 times faster, leading to higher concentrations of ROS than have been experimentally tested [44]. Hence, the sufficiently high levels of ROS present can be potentially damaging to bacterial cells.
After 120 generations, colonies with RSCV phenotype were observed on the agar plates inoculated with 2 mM H2O2 treatment (electronic supplementary material, figure S1), while no distinctive phenotypes were observed in the control populations without H2O2 treatment. The percentage of RSCVs in H2O2 exposed P. aeruginosa populations reached around 40% (figure 1a), implying that ROS could significantly contribute to the occurrence of RSCVs. We also showed that P. aeruginosa was able to evolve within 120 generations, when compared with a shorter 47.5 generation period in the evolution of RSCVs with antibiotics [18]. As P. aeruginosa can persist in CF lungs for decades, spanning 200 000 bacterial generations [33,45], there is more than sufficient time for the metabolically active [46] P. aeruginosa to adapt in CF lungs.
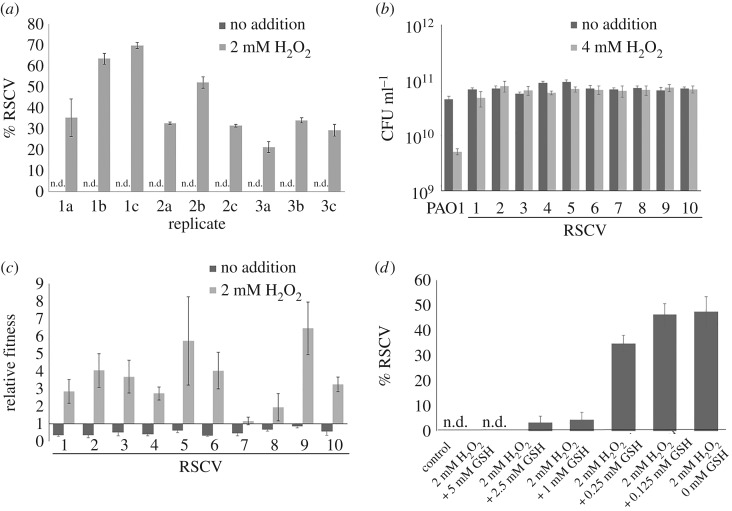
Figure 1.
Evolution assay of Pseudomonas aeruginosa in H2O2. (a) Percentage of rough small colony variants (RSCVs) after evolution in the presence of H2O2 compared with the control. (b) Resistance of RSCV isolates compared with PAO1 in H2O2. (c) Competitive fitness assay between RSCV isolates and PAO1 grown in H2O2. (d) Antioxidants (glutathione; GSH) reduced the proportion of RSCVs formed with ROS treatment, in a dose-dependent manner. Means ± s.d. from triplicate experiments are shown.
The resistance of randomly selected RSCV clones to H2O2 was then measured. The RSCVs showed up to 16-fold increase in MIC to H2O2 (table 1) compared with the ancestral PAO1 strain. The RSCV clones also survived better than PAO1 after treatment with 4 mM H2O2 (1× MIC) (figure 1b). Next, we measured the fitness of the selected RSCVs by co-culturing them with ancestral PAO1. In the absence of 2 mM H2O2, the ancestral PAO1 outcompeted the RSCVs (figure 1c), but RSCVs outcompeted PAO1 in the presence of 2 mM H2O2 (figure 1d), confirming that the RSCVs were favoured and selected for in the presence of oxidative stress. Interestingly, RSCV isolates ‘7’ and ‘8’ showed only slightly higher fitness against the PAO1 ancestral strain, compared with the rest of the RSCV isolates, implying different magnitudes of adaptation within the evolved populations.
Table 1.
MIC of evolved RSCV isolates to H2O2.
| strain/isolate | MIC (mM) |
|---|---|
| PAO1 | 4 |
| RSCV 1 | 32 |
| RSCV 2 | 64 |
| RSCV 3 | 64 |
| RSCV 4 | 64 |
| RSCV 5 | 64 |
| RSCV 6 | 32 |
| RSCV 7 | 64 |
| RSCV 8 | 64 |
| RSCV 9 | 8 |
| RSCV 10 | 64 |
We determined if the selection of RSCV by oxidative stress could be neutralized by the addition of l-glutathione, a commonly used antioxidant [47]. As expected, l-glutathione reduced the proportion of RSCVs formed in a dose-dependent manner, compared with that in the absence of antioxidants (figure 1d).
3.2. Increased c-di-GMP content confers resistance of rough small colony variants to H2O2
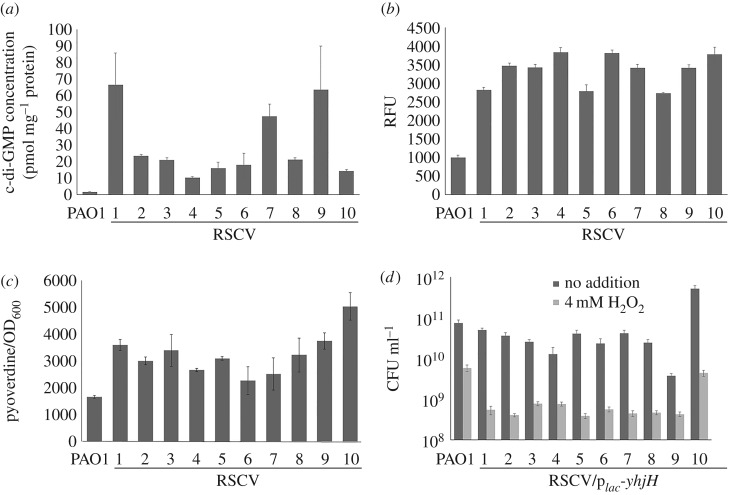
As c-di-GMP signalling is implicated in RSCV formation [27], we examined the intracellular c-di-GMP content in the selected RSCV isolates. Liquid chromatography–mass spectrometry (LC-MS) quantification of the whole cell extract showed that the RSCV isolates contained higher intracellular c-di-GMP than wild-type P. aeruginosa PAO1 (figure 2a). This was in accordance with their higher expression levels of the c-di-GMP reporter fusion pcdrA-gfp [48,49] (figure 2b) and higher pyoverdine production [50,51] (figure 2c), compared with that of ancestral PAO1. To confirm that the increased H2O2 resistance of RSCVs was attributed to enhanced intracellular c-di-GMP content, we transformed a plac-yhjH plasmid [52] into the RSCVs to deplete their intracellular c-di-GMP. As expected, constitutive expression of the YhjH PDE in the RSCVs attenuated their resistance to H2O2 (figure 2d). These results indicate that c-di-GMP signalling plays an important role in the evolutionary development of PAO1 to RSCVs under ROS stress and corroborated previous studies showing that CF RSCV isolates possessed high c-di-GMP levels [27].
Figure 2.
Adaptation of RSCVs to H2O2 is dependent on induction of c-di-GMP content of isolates. (a) C-di-GMP content of RSCVs quantified by HPLC. (b) The pcdrA-gfp expression in RSCVs. (c) Pyoverdine production in RSCVs. (d) Expression of plac-yhjH in RSCVs reduces their resistance to H2O2. Means ± s.d. from triplicate experiments are shown.
3.3. Comparative genomic analysis of rough small colony variants reveal parallel evolution traits
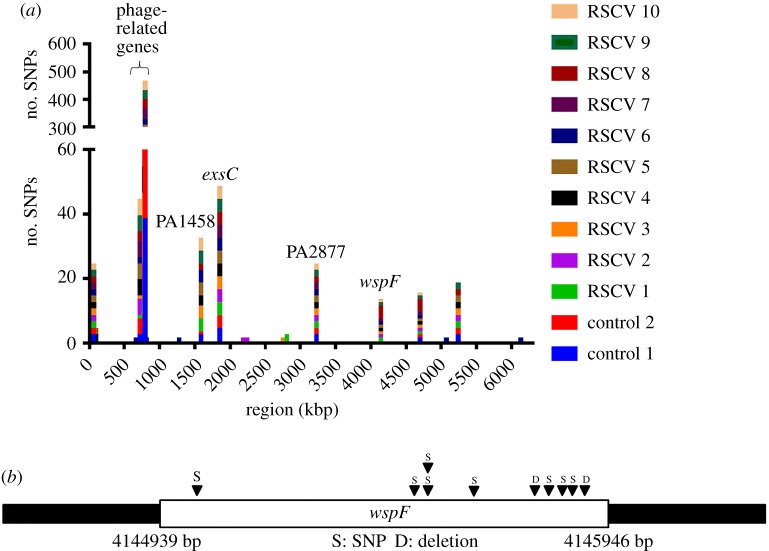
To gain insights into the underlying genetic changes contributing to c-di-GMP-linked RSCV phenotype development with prolonged oxidative stress via c-di-GMP signalling, we sequenced the genomes of 10 randomly selected RSCV isolates and 2 control PAO1 colonies using the Illumina MiSeq platform. Two random control PAO1 colonies were picked from cultures grown in a medium without H2O2 for 120 generations. A series of point mutations were evident in the sequences of 10 randomly selected RSCVs and control PAO1 colonies, compared with the ancestor PAO1 sequence (electronic supplementary material, tables S2 and S3). The distribution of the identified mutations across the PAO1 genome is illustrated in figure 3a. The functional annotation of these RSCV-associated mutations is listed in the electronic supplementary material, tables S2 and S3. Several parallel evolution traits were observed from both RSCVs and control PAO1 colonies, including mutations in PA0727 and PA0720 (encoding phage-related genes), PA1458 (encoding a two component sensor), exsC (encoding an anti-anti-activator of the type III secretion system), PA2877 (encoding a probable transcriptional regulator), intergenic region 4 699 910 bp and intergenic region 5 242 141 bp.
Figure 3.
Genetic sequencing for identification of mutation sites. (a) Number of mutations identified in evolved genotypes. (b) Location of mutations in the wspF gene of different RSCVs.
As P. aeruginosa commonly accumulate mutations even in planktonic cultures and the mutations are common between control and ROS-evolved strains, the above-mentioned mutations are not deemed to be generated by ROS pressure. We also found that the mutations were not attributed to highly mutagenic growth conditions, as there was no significant increase in mutation rates with ROS treatment (electronic supplementary material, figure S2) compared with control medium, with the mutation rates similar to previous reports [38]. Moreover, the cell populations did not develop mutations in the mutS gene, which is important in mismatch repair and its mutation can lead to the emergence of multiple phenotypic variants [53].
Interestingly, there was one parallel evolution trait, wspF gene mutation, which could only be detected in the genomes from the RSCVs and not in the control PAO1 colonies. All RSCV genomes had loss of function mutations in the wspF gene at different sites (figure 3b; electronic supplementary material, table S1). The ΔwspF mutation caused the de-repression of the WspR DGC, leading to increased c-di-GMP synthesis and high intracellular c-di-GMP content, resulting in bacterial cell clumping (electronic supplementary material, figure S1) [54]. This indicated that enhanced intracellular c-di-GMP content might play a positive role in P. aeruginosa's adaptation to oxidative stress.
3.4. Regulatory roles of wspF on ROS-mediated resistance of Pseudomonas aeruginosa
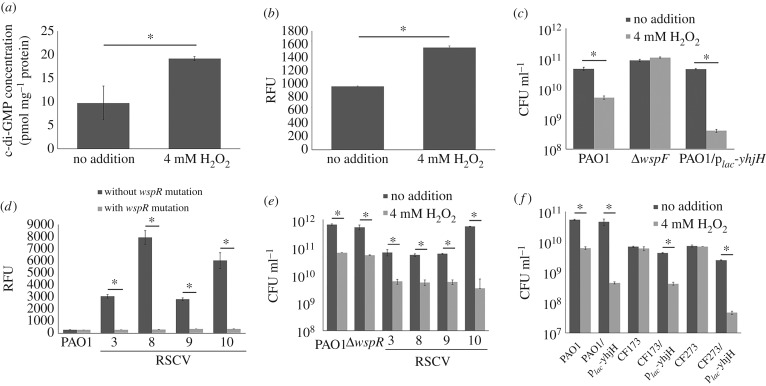
To investigate whether intracellular c-di-GMP content is increased under conditions of ROS stress, we measured the intracellular c-di-GMP content and expression of c-di-GMP reporter fusion pcdrA-gfp with H2O2 treatment. Both intracellular c-di-GMP content and pcdrA-gfp expression levels were increased in PAO1 strain after exposure to 4 mM H2O2 for 4 h (figure 4a,b). Given the increase in intracellular c-di-GMP content in adaptation to H2O2 stress and the presence of wspF mutations in our experimental evolution assay, we hypothesized that the genetic basis of adaptation to ROS was directly linked to the mutation in wspF, which resulted in the de-repression of wspR DGC [8] and a subsequent increased production of c-di-GMP. Hence, we used an isogenic ΔwspF mutant as previously described [51] to ascertain ROS resistance. We also used ‘locked’ high and low c-di-GMP content strains of PAO1 by using the plac-yedQ and plac-yhjH plasmids, respectively. The yedQ gene encodes an E. coli DGC that synthesizes c-di-GMP [50,55,56], thus the exogenous addition of the plac-yedQ would enable the constitutive production of c-di-GMP within the species, resulting in the highly aggregative nature of the colonies (electronic supplementary material, figure S1). The yhjH gene encodes an E. coli PDE that degrades c-di-GMP [56,57], so the presence of plac-yhjH plasmid would cause the reduction of intracellular c-di-GMP levels.
Figure 4.
ΔwspF mutation is important in the induction of c-di-GMP in the presence of ROS. (a) Induction of c-di-GMP in PAO1 by short term (4 h) H2O2 exposure (LC-MS quantification). (b) Induction of pcdrA-gfp expression level in PAO1 by short term (4 h) H2O2 exposure. (c) Resistance of PAO1, ΔwspF, PAO1/plac-yedQ and PAO1/plac-yhjH strains to 4 mM H2O2. (d) pcdrA-gfp expression of evolved RSCV isolates with epistatic wspR mutations to H2O2. (e) Resistance of evolved RSCV isolates with epistatic wspR mutations to H2O2. (f) Resistance of PAO1 and clinical RSCV isolates with known wspF mutations to H2O2 treatment. Means ± s.d. from triplicate experiments are shown; p < 0.05, one-way ANOVA.
The ΔwspF and PAO1/plac-yedQ mutants were more resistant to 4 mM H2O2 (1× MIC) than PAO1, while PAO1/plac-yhjH was more sensitive to 4 mM H2O2 than PAO1 (figures 4c and 5a). Epistatic mutations of wspR gene restored pcdrA-gfp expression (figure 4d) to that of the wild-type of a few selected RSCV isolates. This also caused the RSCV isolates to lose their resistance to H2O2 (figure 4e), confirming that PAO1 could adapt to ROS stress simply via the mutagenesis of wspF.
Figure 5.
C-di-GMP-mediated exopolysaccharides were required for resistance to ROS stress. (a) Resistance of PAO1, ΔwspFΔpelAΔpslBCD and ΔpelAΔpslBCD/plac-yedQ to H2O2. (b) Psl was more influential than Pel in conferring ROS resistance. (c) Psl staining showed formation of small aggregates and synthesis of larger amounts of Psl in PAO1 treated with 0.5, 1 and 2 mM H2O2 compared with control PAO1 cultures. (d) Psl staining revealed synthesis of larger amounts of Psl in representative RSCVs and strains with high intracellular c-di-GMP content than PAO1. (e) Addition of l-arabinose to PAO1/pBAD-psl increased biofilm formation in a dose-dependent manner. (f) Resistance of PAO1/pBAD-psl to H2O2 increased with increasing l-arabinose concentrations. (g) Cellulase treatment abolished ROS resistance in PAO1/pBAD-psl. Means ± s.d. from triplicate experiments are shown; p < 0.05, one-way ANOVA.
As clinical RSCV isolates with wspF mutations were isolated from CF patients [27], we also tested the ROS resistance of clinical RSCV isolates with known wspF mutations (CF173-2005 isolate of lineage H and a related isolate CF273-2002) [45]. Such RSCV isolates from the CF lungs possess higher c-di-GMP levels [8]. We showed that they were resistant to 4 mM H2O2 and the insertion of the plac-yhjH plasmid reduced their resistance to ROS (figure 4f). This confirmed that the clinical RSCV isolates were adapted to the presence of ROS in the human body and cemented the involvement of c-di-GMP regulation in biofilm ROS resistance.
3.5. C-di-GMP-dependent exopolysaccharides are required for Pseudomonas aeruginosa ROS resistance
To understand how the wsp operon mediates ROS resistance, we tested whether exopolysaccharides could interfere with the action of ROS on P. aeruginosa, as the wsp operon regulates the expression of Pel and Psl exopolysaccharide synthetic genes [8]. We showed that both ΔwspFΔpelAΔpslBCD and ΔpelAΔpslBCD/plac-yedQ mutants, which could not produce Pel and Psl exopolysaccharides, were highly sensitive to H2O2, even though they contain high intracellular c-di-GMP levels (figure 5a). To identify which of the two exopolysaccharides were more important in conferring ROS resistance, we further showed that the ΔpslBCD/plac-yedQ was more sensitive to ROS than ΔpelA/plac-yedQ, thus providing the first indication that Psl is more important than Pel in conferring ROS resistance (figure 5b). We then stained the Psl using a fluorescent lectin and observed the formation of small aggregates and enhanced production of Psl in PAO1 with 0.5, 1 and 2 mM H2O2 treatment (figure 5c). Moreover, the evolved RSCVs, ΔwspF and PAO1/plac-yedQ mutants produced higher amounts of Psl than PAO1, as indicated by the fluorescence intensity of the Psl stain (figure 5d).
We further validated the requirement of Psl for ROS resistance by using a PAO1/pBAD-psl mutant by replacing its native psl operon promoter with an l-arabinose-inducible promoter [58]. Upon addition of l-arabinose, there was an increase in biofilm formation via Psl production (figure 5e). Resistance of PAO1/pBAD-psl to ROS improved with increasing l-arabinose concentrations (figure 5f), indicating that Psl production is positively correlated with ROS resistance. Treatment with cellulase, which degrades Psl [58], rendered the PAO1/pBAD-psl sensitive to ROS treatment (figure 5g). Hence, Psl plays a major role in P. aeruginosa ROS resistance. Moreover, Psl acts as a signal for biofilm formation [43], thus could possibly provide positive feedback for biofilm formation and the emergence of RSCVs upon ROS exposure.
3.6. C-di-GMP signalling confers resistance to macrophage phagocytosis
ROS are mostly produced by host leucocytes, such as PMNs and macrophages, to kill the invading pathogens, which in turn induce c-di-GMP content to confer ROS resistance. Because the evolved RSCVs and the ΔwspF mutant showed increased resistance to H2O2 in vitro, we examined whether c-di-GMP signalling could protect P. aeruginosa from phagocytosis by RAW264.7 macrophages. Here, the ΔwspF mutant and evolved RSCVs were more efficient in evading phagocytosis by macrophages than PAO1 (figure 6a,b). Although the PAO1/plac-yhjH strain was subjected to lower phagocytosis levels by the macrophages than ΔwspF mutant (figure 6b), we found that it was highly cytotoxic to macrophages compared with PAO1 and the ΔwspF mutant (figure 6c). Hence, the lower macrophage phagocytosis level could be attributed to increased macrophage killing by PAO1/plac-yhjH. Moreover, the PAO1/plac-yhjH was exposed to higher ROS production from macrophages than PAO1 and the ΔwspF mutant (figure 6d).
Figure 6.
Adaptation to H2O2 provides further benefits by conferring protection against macrophages. (a) Quantification of phagocytosed PAO1 and evolved RSCV isolates by macrophages. (b) Quantification of phagocytosed PAO1, ΔwspF and PAO1/plac-yhjH strains by macrophages. (c) Cytotoxicity assay of macrophages by PAO1, ΔwspF and PAO1/plac-yhjH strains. (d) Quantification of ROS produced by macrophages in the presence of PAO1, ΔwspF, PAO1/plac-yedQ and PAO1/plac-yhjH strains using ROS detection assay. DCF (e) Quantification of phagocytosed PAO1 and ΔpslBCD strains by macrophages. (f) Cytotoxicity assay of macrophages by PAO1 and ΔpslBCD strains. Means ± s.d. from triplicate experiments are shown; p < 0.05, one-way ANOVA.
Further, Psl played a role in evading phagocytosis by macrophages, as ΔpslBCD/plac-yedQ was internalized at a higher rate than PAO1/plac-yedQ and ΔpelA/plac-yedQ (figure 6e). However, Psl was not involved in the cytotoxicity against macrophages, as Psl was not a virulence factor and there were no significant differences between PAO1/plac-yedQ and ΔpslBCD/plac-yedQ in killing the macrophage cells (figure 6f). Nonetheless, increases in c-di-GMP levels by plac-yedQ plasmid insertion reduced the cytotoxicity to macrophages, which corroborated the ΔwspF mutant results (figure 6f).
4. Discussion
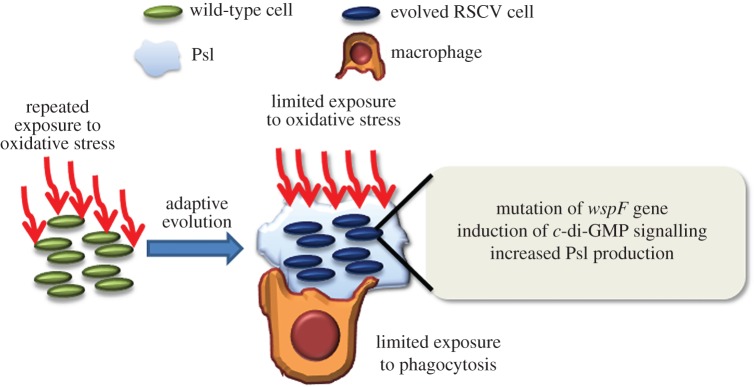
Resistance to oxidative stress is essential for pathogens to survive in the host environment. Here, we employed the adaptive experimental evolution assay to investigate P. aeruginosa's ROS resistance via the emergence of RSCVs with enhanced biofilm forming capacity (figure 7). The production of superoxide in the phagosome was estimated at 1–4 M [59,60], implying that the use of a millimolar range of H2O2 to induce pathogen evolution in our study was relevant to the host environment. We observed an adaptive evolution trait of mutagenesis in the wspF gene, resulting in c-di-GMP signal induction. Our findings are clinically significant as RSCVs with wspF mutations are frequently isolated from CF patients and these showed similar resistance to oxidative stress as our evolved RSCVs.
Figure 7.
Model of Pseudomonas aeruginosa adaptation to oxidative stress. Upon repeated exposure of oxidative stress, the cells evolved to form RSCVs via wspF mutation, resulting in the induction of c-di-GMP signalling and increased Psl production. This endowed RSCVs with the ability to resist oxidative stress and phagocytes.
The regulatory role of c-di-GMP signalling on oxidative stress response is extendable to other pathogenic species, including Burkholderia cenocepacia and Klebsiella pneumoniae (electronic supplementary material, figure S3).The ROS-induced evolution of RSCVs could potentially explain the emergence of RSCVs by B. cenocepacia and K. pneumoniae during infections [61,62]. The induction of c-di-GMP signalling has been linked to increased production of Psl exopolysaccharide in P. aeruginosa [50]. While Psl stiffens the biofilm structure, act as signal for biofilm formation and offers protection against antibiotics [43,63], the role of Psl as a barrier in the biofilm possibly reduces ROS penetration across bacterial membranes and causes cell damage. This confers bacterial cells with a biofilm-based protective mechanism from host immune cell generated ROS and/or antimicrobials.
As the oxidative stress resistance mechanism is commonly utilized by various pathogens, we propose the following strategies to modulate c-di-GMP signalling and treat biofilm-associated infections: (i) employing matrix-degrading enzymes, such as cellulase or the PslG hydrolase [64] to degrade Psl-like polysaccharides followed by effective killing of biofilm cells by antibiotics; and (ii) administering antioxidant drugs during early infection stages to prevent pathogen adaptation. To nullify P. aeruginosa's oxidative stress adaptation in our evolution assays, we used the antioxidant l-glutathione, which reduced the emergence of RSCVs. This also indirectly reduced the resistance of P. aeruginosa to host immune clearance strategies such as phagocytosis. Given the lower levels of glutathione and increased oxidative stress from neutrophils in CF patients' lungs, antioxidant therapy could potentially prove effective against inflammation [65,66].
In summary, the interactions between the immune system and pathogens are highly complex, driving the pathogen to adapt accordingly to different host immune system stimuli. Pseudomonas aeruginosa appears to draw a complex yet fine balance between its numerous DGCs and PDEs, thus leading to phenotypic variation, and ensuring the survival of its species. Hence, chemical manipulation of the c-di-GMP signalling pathway represents a promising strategy to manage chronic bacterial infections.
Supplementary Material
Acknowledgements
We thank Dr Gurjeet Singh Kohli for critical comments on our manuscript and fruitful discussions.
Data accessibility
The Whole Genome Shotgun bioproject for P. aeruginosa adaptation to ROS has been deposited at in the NCBI Short Read Archive (SRA) database with accession code SRP062804.
Authors' contributions
S.L.C., S.K., M.G. and L.Y. designed the research. S.L.C., Y.D. and J.Z. performed the experiments. Z.C. and S.S. performed and analysed LC-MS experiments. Y.L., D.I.D.-M. and S.C.S. performed and analysed DNA sequencing experiments. S.K. and M.G. analysed the data. S.L.C. and L.Y. wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing financial interests.
Funding
This research was supported by the National Research Foundation and the Ministry of Education of Singapore under its Research Centre of Excellence Program. Y.L. is supported by the Start-up Grant (M4330002.C70) from Nanyang Technological University and AcRF Tier 2 (MOE2014-T2-2-172) from Ministry of Education, Singapore. S.L.C. is supported by the Lee Kong Chian Medicine Postdoctoral Fellowship 2015.
References
- 1.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558. (doi:10.1002/ppul.21011) [DOI] [PubMed] [Google Scholar]
- 2.Young D, Hussell T, Dougan G. 2002. Chronic bacterial infections: living with unwanted guests. Nat. Immunol. 3, 1026–1032. (doi:10.1038/ni1102-1026) [DOI] [PubMed] [Google Scholar]
- 3.Domenech M, Ramos-Sevillano E, Garcia E, Moscoso M, Yuste J. 2013. Biofilm formation avoids complement immunity and phagocytosis of Streptococcus pneumoniae. Infect. Immun. 81, 2606–2615. (doi:10.1128/IAI.00491-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alhede M, Bjarnsholt T, Givskov M, Alhede M. 2014. Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. Adv. Appl. Microbiol. 86, 1–40. (doi:10.1016/B978-0-12-800262-9.00001-9) [DOI] [PubMed] [Google Scholar]
- 5.Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361. (doi:10.1128/AAC.00001-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. (doi:10.1038/nrmicro2109) [DOI] [PubMed] [Google Scholar]
- 7.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. (doi:10.1128/MMBR.00043-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl Acad. Sci. USA 102, 14 422–14 427. (doi:10.1073/pnas.0507170102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727. (doi:10.1101/gad.289504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781. (doi:10.1128/JB.187.14.4774-4781.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30 829–30 837. (doi:10.1074/jbc.M504429200) [DOI] [PubMed] [Google Scholar]
- 12.Chua SL, et al. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5, 4462 (doi:10.1038/ncomms5462) [DOI] [PubMed] [Google Scholar]
- 13.Chua SL, Hultqvist LD, Yuan M, Rybtke M, Nielsen TE, Givskov M, Tolker-Nielsen T, Yang L. 2015. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat. Protoc. 10, 1165–1180. (doi:10.1038/nprot.2015.067) [DOI] [PubMed] [Google Scholar]
- 14.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. (doi:10.1111/j.1365-2958.2007.05879.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim B, Beyhan S, Yildiz FH. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189, 717–729. (doi:10.1128/JB.00834-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchma SL, Delalez NJ, Filkins LM, Snavely EA, Armitage JP, O'Toole GA. 2015. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J. Bacteriol. 197, 420–430. (doi:10.1128/JB.02130-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua SL, et al. 2015. C-di-GMP regulates Pseudomonas aeruginosa stress response to tellurite during both planktonic and biofilm modes of growth. Sci. Rep. 5, 10052 (doi:10.1038/srep10052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong A, Rodrigue N, Kassen R. 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 8, e1002928 (doi:10.1371/journal.pgen.1002928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen SS, Koch C, Hoiby N, Rosendal K. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J. Antimicrob. Chemother. 17, 505–516. (doi:10.1093/jac/17.4.505) [DOI] [PubMed] [Google Scholar]
- 20.Valerius NH, Koch C, Hoiby N. 1991. Prevention of chronic Pseudomonas aeruginosa colonisation in cystic fibrosis by early treatment. Lancet 338, 725–726. (doi:10.1016/0140-6736(91)91446-2) [DOI] [PubMed] [Google Scholar]
- 21.Hartl D, et al. 2007. Cleavage of CXCR1 on neutrophils disables bacterial killing in cystic fibrosis lung disease. Nat. Med. 13, 1423–1430. (doi:10.1038/nm1690) [DOI] [PubMed] [Google Scholar]
- 22.Jensen PO, et al. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153, 1329–1338. (doi:10.1099/mic.0.2006/003863-0) [DOI] [PubMed] [Google Scholar]
- 23.Ren CL, Konstan MW, Yegin A, Rasouliyan L, Trzaskoma B, Morgan WJ, Regelmann W; Scientific advisory group, investigators, and coordinators of the epidemiologic study of cystic fibrosis. 2012. Multiple antibiotic-resistant Pseudomonas aeruginosa and lung function decline in patients with cystic fibrosis. J. Cyst. Fibros. 11, 293–299. (doi:10.1016/j.jcf.2012.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pernet E, et al. 2014. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat. Commun. 5, 5105 (doi:10.1038/ncomms6105) [DOI] [PubMed] [Google Scholar]
- 25.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64. (doi:10.1038/ng.3148) [DOI] [PubMed] [Google Scholar]
- 26.Huse HK, Kwon T, Zlosnik JE, Speert DP, Marcotte EM, Whiteley M. 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1, e00199-10. (doi:10.1128/mBio.00199-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starkey M, et al. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191, 3492–3503. (doi:10.1128/JB.00119-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65, 3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanka A, Duvel J, Dotsch A, Klinkert B, Abraham WR, Kaever V, Ritter C, Narberhaus F, Haussler S. 2015. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci. Signal. 8, ra36. (doi:10.1126/scisignal.2005943) [DOI] [PubMed] [Google Scholar]
- 30.Almblad H, Harrison JJ, Rybtke M, Groizeleau J, Givskov M, Parsek MR, Tolker-Nielsen T. 2015. The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic di-GMP. J. Bacteriol. 197, 2190–2200. (doi:10.1128/JB.00193-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams HD, Zlosnik JE, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71. (doi:10.1016/S0065-2911(06)52001-6) [DOI] [PubMed] [Google Scholar]
- 32.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087. (doi:10.1128/JB.01138-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Haagensen JA, Jelsbak L, Johansen HK, Sternberg C, Hoiby N, Molin S. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190, 2767–2776. (doi:10.1128/JB.01581-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriarty TF, McElnay JC, Elborn JS, Tunney MM. 2007. Sputum antibiotic concentrations: implications for treatment of cystic fibrosis lung infection. Pediatr. Pulmonol. 42, 1008–1017. (doi:10.1002/ppul.20671) [DOI] [PubMed] [Google Scholar]
- 35.Reid DW, Misso N, Aggarwal S, Thompson PJ, Walters EH. 2007. Oxidative stress and lipid-derived inflammatory mediators during acute exacerbations of cystic fibrosis. Respirology 12, 63–69. (doi:10.1111/j.1440-1843.2006.00962.x) [DOI] [PubMed] [Google Scholar]
- 36.Barrett RD, MacLean RC, Bell G. 2005. Experimental evolution of Pseudomonas fluorescens in simple and complex environments. Am. Nat. 166, 470–480. (doi:10.1086/444440) [DOI] [PubMed] [Google Scholar]
- 37.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. (doi:10.1111/j.1365-2958.2010.07320.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua SL, et al. 2016. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat. Commun. 7, 10750 (doi:10.1038/ncomms10750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandsberg LF, Ciofu O, Kirkby N, Christiansen LE, Poulsen HE, Hoiby N. 2009. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob. Agents Chemother. 53, 2483–2491. (doi:10.1128/AAC.00428-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouneimne H, Robert J, Jarlier V, Cambau E. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran CS, Rangel SM, Almblad H, Kierbel A, Givskov M, Tolker-Nielsen T, Hauser AR, Engel JN. 2014. The Pseudomonas aeruginosa type III translocon is required for biofilm formation at the epithelial barrier. PLoS Pathog. 10, e1004479 (doi:10.1371/journal.ppat.1004479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 109, 20 632–20 636. (doi:10.1073/pnas.1217993109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolpen M, et al. 2010. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62. (doi:10.1136/thx.2009.114512) [DOI] [PubMed] [Google Scholar]
- 44.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J. Biol. Chem. 281, 39 860–39 869. (doi:10.1074/jbc.M605898200) [DOI] [PubMed] [Google Scholar]
- 45.Yang L, et al. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl Acad. Sci. USA 108, 7481–7486. (doi:10.1073/pnas.1018249108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oberhardt MA, Goldberg JB, Hogardt M, Papin JA. 2010. Metabolic network analysis of Pseudomonas aeruginosa during chronic cystic fibrosis lung infection. J. Bacteriol. 192, 5534–5548. (doi:10.1128/JB.00900-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suttorp N, Toepfer W, Roka L. 1986. Antioxidant defense mechanisms of endothelial cells: glutathione redox cycle versus catalase. Am. J. Physiol. 251, C671–C680. [DOI] [PubMed] [Google Scholar]
- 48.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842. (doi:10.1111/j.1365-2958.2009.06991.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78, 5060–5069. (doi:10.1128/AEM.00414-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, et al. 2015. Multiple diguanylate cyclase-coordinated regulation of pyoverdine synthesis in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 7, 498–507. (doi:10.1111/1758-2229.12278) [DOI] [PubMed] [Google Scholar]
- 51.Frangipani E, Visaggio D, Heeb S, Kaever V, Camara M, Visca P, Imperi F. 2014. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16, 676–688. (doi:10.1111/1462-2920.12164) [DOI] [PubMed] [Google Scholar]
- 52.Chua SL, Tan SY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. 2013. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 2066–2075. (doi:10.1128/AAC.02499-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smania AM, Segura I, Pezza RJ, Becerra C, Albesa I, Argarana CE. 2004. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology 150, 1327–1338. (doi:10.1099/mic.0.26751-0) [DOI] [PubMed] [Google Scholar]
- 54.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184, 6481–6489. (doi:10.1128/JB.184.23.6481-6489.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez-Torres V, Hu H, Wood TK. 2011. GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl. Microbiol. Biotechnol. 90, 651–658. (doi:10.1007/s00253-010-3074-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hengge R, Galperin MY, Ghigo JM, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2016. Systematic nomenclature for GGDEF and EAL domain-containing cyclic Di-GMP turnover proteins of Escherichia coli. J. Bacteriol. 198, 7–11. (doi:10.1128/JB.00424-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22, 2434–2446. (doi:10.1101/gad.475808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5, e1000354 (doi:10.1371/journal.ppat.1000354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupre-Crochet S, Erard M, Nubetae O. 2013. ROS production in phagocytes: why, when, and where? J. Leukoc. Biol. 94, 657–670. (doi:10.1189/jlb.1012544) [DOI] [PubMed] [Google Scholar]
- 60.Segal AW. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23, 197–223. (doi:10.1146/annurev.immunol.23.021704.115653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernier SP, Nguyen DT, Sokol PA. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect. Immun. 76, 38–47. (doi:10.1128/IAI.00874-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi M, Yoshida K, San Clemente CL. 1977. Relation of colonial morphologies in soft agar to morphological and biological properties of the K-9 strain of Klebsiella pneumoniae and its variants. Can. J. Microbiol. 23, 448–451. (doi:10.1139/m77-066) [DOI] [PubMed] [Google Scholar]
- 63.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 9, e1003526 (doi:10.1371/journal.ppat.1003526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu S, et al. 2015. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 25, 1352–1367. (doi:10.1038/cr.2015.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galli F, et al. 2012. Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim. Biophys. Acta 1822, 690–713. (doi:10.1016/j.bbadis.2011.12.012) [DOI] [PubMed] [Google Scholar]
- 66.Ciofu O, Lykkesfeldt J. 2014. Antioxidant supplementation for lung disease in cystic fibrosis. Cochrane Database Syst. Rev. 8, CD007020 (doi:10.1002/14651858.CD007020.pub3) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Whole Genome Shotgun bioproject for P. aeruginosa adaptation to ROS has been deposited at in the NCBI Short Read Archive (SRA) database with accession code SRP062804.