Significance
The development of iPSCs provides unprecedented opportunities for life sciences, drug discovery, and regenerative medicine. The efficiency of iPSC generation is quite low: typically less than 1% of human primary somatic cells that have received reprogramming factors turn into iPSCs. Previous studies revealed that cellular senescence was a major barrier to iPSC generation. In this study using human FOP mutant cells, we provide evidence that the BMP-SMAD-ID signaling suppressed p16/INK4A-mediated cellular senescence during the early phase in iPSC generation. These results are unexpected because BMP-SMAD signaling has negative effects on the self-renewal of human iPSCs. Here, we show that a human natural mutation increases the efficiency of iPSC generation.
Keywords: reprogramming, pluripotency, BMP, senescence, FOP
Abstract
Fibrodysplasia ossificans progressiva (FOP) patients carry a missense mutation in ACVR1 [617G > A (R206H)] that leads to hyperactivation of BMP-SMAD signaling. Contrary to a previous study, here we show that FOP fibroblasts showed an increased efficiency of induced pluripotent stem cell (iPSC) generation. This positive effect was attenuated by inhibitors of BMP-SMAD signaling (Dorsomorphin or LDN1931890) or transducing inhibitory SMADs (SMAD6 or SMAD7). In normal fibroblasts, the efficiency of iPSC generation was enhanced by transducing mutant ACVR1 (617G > A) or SMAD1 or adding BMP4 protein at early times during the reprogramming. In contrast, adding BMP4 at later times decreased iPSC generation. ID genes, transcriptional targets of BMP-SMAD signaling, were critical for iPSC generation. The BMP-SMAD-ID signaling axis suppressed p16/INK4A-mediated cell senescence, a major barrier to reprogramming. These results using patient cells carrying the ACVR1 R206H mutation reveal how cellular signaling and gene expression change during the reprogramming processes.
Reprogramming somatic cells into pluripotent stem cells is an exciting paradigm in biology and has critical implications for transplantation medicine and disease modeling. We developed a method to generate induced pluripotent stem cells (iPSCs) by transducing defined factors, such as OCT4, SOX2, KLF4, and C-MYC (OSKM), into somatic cells (1, 2). These transcription factors regulate the expression of genes important for self-renewal and pluripotency. However, only a small proportion of cells become iPSCs after the introducing these defined factors (3), and this is a major roadblock toward applying this technology for biomedicine. Cytokine- and chemical-induced cell signaling affect the efficiency of iPSC generation (4, 5), but the precise effects and mechanisms in reprogramming are unclear.
The BMP-SMAD signal has important roles in the induction and maintenance of pluripotency. BMP promotes the self-renewal of mouse embryonic stem cells (mESCs) (6, 7). In addition, BMP-SMAD signaling facilitates mouse iPSC (miPSC) generation (8). Thus, BMP signaling has positive effects on both the induction and self-renewal of mouse pluripotent stem cells. In contrast, BMPs inhibit self-renewal of human PSCs (9–13). Recently, Hamasaki et al. (15) tried to generate human iPSCs (hiPSCs) from the human dermal fibroblasts (HDFs) of patients with fibrodysplasia ossificans progressiva (FOP; Online Mendelian Inheritance in Man no. 135100) who carried a missense mutation in ACVR1 (617G > A) that leads to hyperactivation of the BMP-SMAD signaling pathway (14), with little success; they obtained many differentiated colonies, but only a few undifferentiated ESC-like colonies. These results indicated that BMP-SMAD signaling negatively affects hiPSC generation as well as their self-renewal.
In this study, we independently generated hiPSCs from FOP patients. Although our primary motivation was to establish in vitro disease models of FOP (16, 17), we unexpectedly found that the efficiency of hiPSC generation from FOP HDFs was much higher than that of control HDFs without any BMP inhibitors. Thus, we explored the roles of the BMP-SMAD signaling during reprogramming to hiPSCs. Our findings show that patient-derived hiPSCs of human genetic diseases, such as FOP, are useful to understand how specific gene mutations affect reprogramming processes, in addition to their utilities to model human diseases.
Results
Increased Efficiency of HiPSC Generation from FOP HDFs Under Low Cell Density.
We used episomal vector-mediated iPSC generation with HDFs from FOP1–3, as well as four additional control HDFs (1323, WTa, WTb, and WTc). We determined the efficiency of hiPSCs by detecting colonies that were positive for a pluripotent stem cell marker, TRA-1-60 (18). After transfecting episomal plasmids containing OCT4, SOX2, KLF4, L-MYC, LIN28, and shRNA for p53 (epiY4) and replating at 10,000 cells per well of six-well plate, all three FOP HDFs produced significantly more TRA-1-60–positive colonies than the four normal HDFs (Fig. 1A). The ratio of TRA-1-60–positive cells in reprogrammed FOP cells was also higher than that in normal HDFs detected with flow cytometry (Fig. 1B). We also generated iPSCs from normal and FOP fibroblasts using retroviral vectors and found that the increased efficiency of iPSC generation from FOP HDFs was observed when plated at low density (SI Text and Fig. S1). These results indicated that hiPSC generation was more efficient from FOP HDFs than from control HDFs, regardless of reprogramming methods and factors. We then established hiPSC lines from FOP HDFs and characterized them as they maintained self-renewal and pluripotency. In brief, these lines had normal karyotypes, expressed pluripotency markers, including TRA1-60 and NANOG, and were able to differentiate into various cells of the three germ layers both in vitro and in teratomas (16).
Fig. 1.
Increased efficiency of hiPSC generation from FOP HDFs via the BMP-SMAD signaling pathway. (A and B) Number of TRA-1-60–positive colonies (A) or the ratio of TRA-1-60–positive cells (B) from reprogrammed FOP HDFs deposited in the Coriell Institute and collected at UCSF and four different normal HDF lines. TRA1-60–positive cells were analyzed 25 d after transfection from 10,000 cells per 100-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05 (t test). (C and D) Effects of BMP-SMAD signal inhibitors, Dorsomorphin (1 µM), LDN-193189 (1 µM), P38 MAPK inhibitor, SB203580 (10 µM), and vehicle (0.1% DMSO) on hiPSC generation from HDF-FOP1 (C) and HDF-FOP2 (D) with OSKM retroviral transduction. ESC-like colonies were counted 25 d after the transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with vehicle conditions). (E and F) Effects of the overexpression of inhibitory SMADs SMAD6 or SMAD7 on hiPSC generation from HDF-FOP1 (E) and HDF-FOP2 (F) with OSKM retroviral transduction. ESC-like colonies were counted 25 d after transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with GFP conditions). (G and H) Effects of ACVR1 knockdown on hiPSC generation from HDF-FOP1 (G) and HDF-FOP2 (H) with episomal plasmids. TRA-1-60–positive colonies were counted 25 d after transduction from 5,000 cells per 35-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with nontarget siRNA conditions).
Fig. S1.
FOP HDFs showed hyperactive BMP-SMAD signaling and increased efficiency of iPSC generation. (A) Direct DNA sequence analysis of 617 G (or A) in ACVR1 from genomic DNA of HDF-1323, HDF-FOP1, and HDF-FOP2. (B) Protein expression of phosphorylated SMAD1/5/8, total SMAD1/5/8, and α-tubulin in normal or FOP fibroblasts treated with 100 ng/mL BMP4 for 1 h and analyzed by Western blotting. (C) Protein expression levels of phosphorylated SMAD1/5/8 (normalized to the amount of α-tubulin expression) in normal or FOP fibroblasts. The results are mean and SE, n = 4. *P < 0.05 (Dunnett’s test with HDF-1323). (D) iPSC colonies have an ESC-like morphology with a flat, round shape and a distinct edge (Upper). Non–ESC-like colonies are epithelial with irregular edges (Lower). (Scale bars: 200 µm.) (E–H) The efficiency of ESC-like colonies from normal or FOP HDFs with retroviral transduction of OSKM in (E and G) or OSK in (F and H) at different seeding densities. Transduced cells were replated at 50,000 (E and F), 150,000, or 500,000 cells per 100-mm dish. Colony numbers were counted 25 or 30 d after transduction in OSKM or OSK conditions, respectively. Results are mean and SE, n = 4. *P < 0.05, **P < 0.01 (Dunnett’s test with HDF-1323). Hashtags (#) indicate that cells became confluent before ESC-like colonies appeared. (I) ALP staining of ESC-like colonies from FOP HDFs of three Japanese patients (FOP-J1, -J2, and -J3) and normal fibroblasts (TIG-120). The images were taken 25 d after OSKM retroviral transduction. (J and K) Numbers of ESC-like colonies from FOP HDFs of three Japanese patients with OSKM (J) and OSK (K) retroviral transduction. Colony numbers are automatically counted by ALP staining in every 5 d after transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 4.
Hyperactivated BMP-SMAD Signaling Contributes to Increased iPSC Generation.
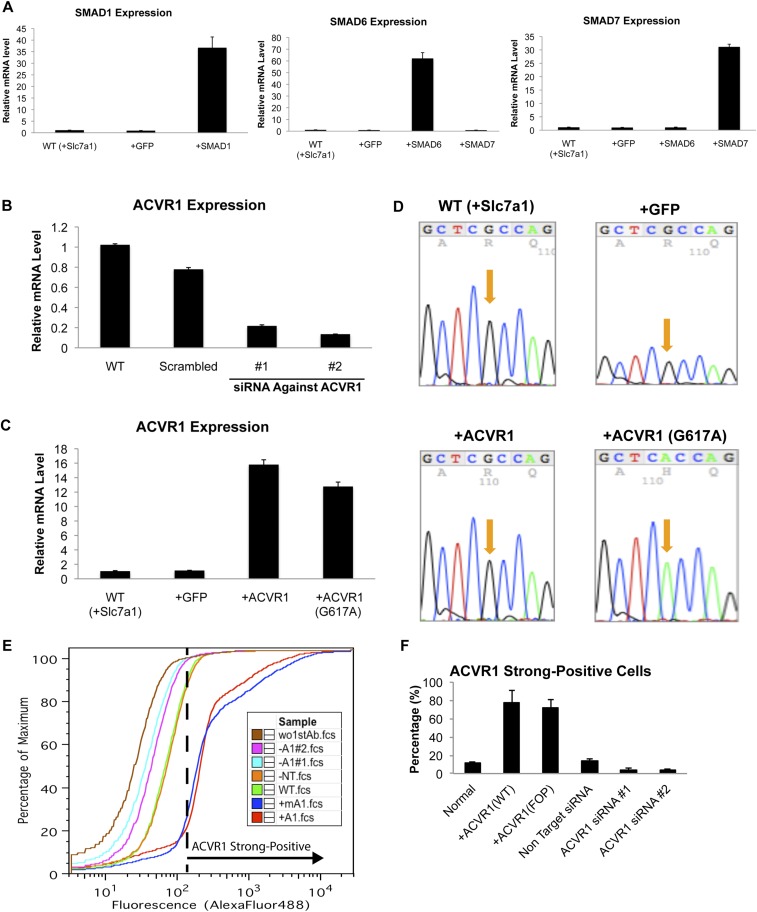
We next determined if the increased efficiency of hiPSC generation from FOP HDFs could be attributed to the FOP mutation itself and its signaling effect. We examined the effects of chemical inhibitors of BMP signaling on hiPSC generation from FOP HDFs under retroviral OSKM conditions. BMP-SMAD signaling inhibitors [i.e., Dorsomorphin (19) and LDN-193189 (20)] markedly decreased hiPSC generation from FOP HDFs, but a BMP-P38 MAPK signaling inhibitor [i.e., SB203580 (21)] had little effect (Fig. 1 C and D). However, Dorsomorphin and LDN-193189 are cytotoxic and/or inhibit protein kinases other than BMP-SMAD signaling (22). To confirm the direct effects of BMP-SMAD signaling on hiPSC generation from FOP HDFs, we overexpressed inhibitory SMADs (SMAD6 or SMAD7) by retroviral infection together with OSKM in hiPSC generation (Fig. S2A). Overexpressing SMAD6 or SMAD7 decreased the efficiency of hiPSC generation from FOP HDFs (Fig. 1 E and F). We also transfected siRNA against ACVR1 (targeting both the wild-type and FOP mutant) during iPSC generation with episomal plasmids (Fig. S2B). Knockdown of ACVR1 decreased the efficiency of hiPSC generation from FOP HDFs (Fig. 1 G and H). These results indicated that inhibition of BMP-SMAD signaling decreased the efficiency of iPSC generation from FOP HDFs.
Fig. S2.
Expression of SMAD6, SMAD7, ACVR1, and SMAD1 in HDFs. (A) Normal HDFs (HDF-1323 transduced with lentiviral Slc7a1) were transduced with retroviral GFP, SMAD1, SMAD6, or SMAD7 and cultured for 5 d. Results are mean and SE, n = 3. (B) Normal HDFs (HDF-1323) were transfected with scrambled, ACVR1 no. 1, or ACVR1 no. 2 siRNA and cultured for 3 d. Results are mean and SE, n = 3. (C) Expression levels of overexpressed ACVR1 (WT or G617A) by retroviral transduction in normal HDFs analyzed by RT-qPCR. Normal HDF (HDF-1323 transduced with lentiviral Slc7a1) were transduced with retroviral GFP, ACVR1, or mutant ACVR1 (G617A) and cultured for 5 d. Results are mean and SE, n = 3. (D) Direct DNA sequence analysis of position 617 in ACVR1 gene from RT-PCR products of ACVR1-overexpressed HDFs. (E) Flow cytometric analysis of ACVR1 protein expression in normal HDFs (HDF-1323) transfected with scrambled (−NT), ACVR1 no. 1 (−A1 no. 1), or no. 2 (−A1 no. 2) siRNA or transduced with retroviral wild-type ACVR1 (+A1), or mutant ACVR1 (G617A) (+mA1) and cultured for 3 d. Results are mean and SE, n = 3. (F) The ratio of transfected or transduced cells which strongly express ACVR1 protein analyzed by flow cytometry. Results are mean and SE, n = 3.
Activating BMP-SMAD Signaling Promotes iPSC Generation in the Early Reprogramming Phase.
We asked whether activating BMP-SMAD signaling increases the efficiency of iPSC generation from normal HDFs. To examine the effect of the FOP mutation on iPSC generation from normal HDFs, we overexpressed wild-type or FOP mutant ACVR1 (617G > A) by retroviral infection together with OCT4, SOX2, and KLF4 (OSK) or OSKM for generating iPSCs. Overexpressing FOP mutant, but not wild-type, ACVR1 has been reported to cause hyperactivation of BMP-SMAD signaling (23–25). The expression levels of wild-type and mutant ACVR1 in HDFs were similar as assessed by RT-quantitative PCR (qPCR) and flow cytometry (Fig. S2 C–F). Overexpressing FOP mutant, but not wild-type, ACVR1 generated more hiPSCs from normal HDFs under both OSKM and OSK retroviral conditions (Fig. 2 A and B).
Fig. 2.
BMP-SMAD signaling activation increases the efficiency of hiPSC generation from normal HDFs. (A and B) Effects of the overexpression of wild-type or mutant (G617A) ACVR1 on hiPSC generation from normal HDFs with OSKM (A) or OSK (B) retroviral transduction. ESC-like colonies were counted 25 d after transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 4. *P < 0.05 (Dunnett’s test with +GFP conditions). (C and D) Effects of the overexpression of SMAD1 on hiPSC generation from normal HDFs with OSKM (C) and OSK (D) retroviral transduction. ESC-like colonies were counted 25 d after transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05 (t test). (E and F) An ESC-like colony, epithelial with a round edge, made with BMP4 for first 1 wk (E) and a degenerated (or differentiated) colony with sac-like structure made with BMP4 for 3 wk (F). (Scale bars: 200 µm.) (G) Effects of recombinant BMP4 and ACTIVIN A proteins on hiPSC generation from normal HDFs with episomal plasmids. TRA-1-60–positive colonies were counted 25 d after electroporation of epiY4 from 10,000 cells per 100-mm dish at replating. Results are mean and SE, n = 4. *P < 0.05 (Dunnett’s test with no addition conditions).
Next, we determined the effects of elevated expression of SMAD1 on hiPSC generation from normal HDFs by overexpressing SMAD1 by retroviral infection with OSKM or OSK to generate iPSCs. SMAD1 overexpression caused hyperactive BMP-SMAD signaling (26). SMAD1 overexpression (Fig. S2A) increased the efficiency of hiPSC generation from normal HDFs under both OSKM and OSK retroviral conditions (Fig. 2 C and D).
To examine how the timing of BMP-SMAD signaling affects hiPSC generation, we added recombinant BMP4 protein at specific times during iPSC generation from normal HDFs under epiY4 conditions. Recombinant ACTIVIN A protein, which only activates TGFβ-SMAD signaling, but not BMP-SMAD signaling, served as a control. BMP4 proteins (10 ng/mL) or ACTIVIN A proteins (10 ng/mL) were added in weeks 1, 2, and 3, and 1–2, 2–3, or 1–3 wk after the cells were replated in hiPSC culture conditions. Adding BMP4 early during reprogramming (until week 1) increased the efficiency of iPSC generation (Fig. 2 E and F). However, late addition (week 3) of BMP4 decreased the efficiency of hiPSCs and produced degenerated or differentiated colonies with sac-like morphologies (Fig. 2G). In contrast, adding ACTIVIN A in any periods had no effect. These results indicated that BMP-SMAD signaling promotes reprogramming into iPS cells in the early phase of reprogramming, but promote differentiation in the late phase. These hiPSC lines from normal fibroblasts reprogrammed with transient BMP4-SMAD activation maintain their self-renewal and pluripotency (SI Text and Fig. S3 A–G).
Fig. S3.
Characterization of transgene-free hiPSC lines generated with transient BMP4 treatment. (A) An iPSC colony (HiPS-1323-epiY4 no. 1 at passage 20) from normal HDFs (HDF-1323) generated with episomal plasmids and BMP4 for 1 wk. (Scale bar: 100 µm.) (B) Karyotype analysis of HiPS-1323-epiY4 no. 1 at passage 5. (C) Protein expression of pluripotent stem cell markers (NANOG, SSEA3, and TRA1-60) detected by immunocytochemistry in HiPS-1323-epiY4 no. 1 at passage 11. Secondary antibodies were labeled with Alexa Fluor 488 (green). Nuclei were stained with DAPI (blue). (Scale bars: 100 µm.) (D) Protein expression of differentiation markers (α-fetoprotein as endoderm, cardiac troponin T as mesoderm, and MAP2 as ectoderm) detected by immunocytochemistry in HiPS-1323-epiY4 no. 2 at passage 17. Secondary antibodies were labeled with Alexa Fluor 488 (green). Nuclei were stained with DAPI (blue). (Scale bars: 100 µm.) (E) H&E staining sections of teratomas generated from HiPS-1323-epiY4 no. 1. Each image shows neural tissue (Left), cartilage (Center), and gut-like epithelial tissues (Right). (F) Endogenous and plasmid expression of OCT4, SOX2, KLF4, L-MYC, and LIN28 in hiPSC lines generated from HDF-1323 detected by RT-qPCR. (G) Genomic insertion of EBNA-1 gene in hiPSC lines generated from HDF-1323 detected by genomic qPCR. Copy number was estimated with the amount of pCXLE-EGFP as standards and 10 ng of genomic DNA (∼1.6 × 103 cells) of each sample.
Inhibitor of Differentiation Genes Regulated by BMP-SMAD Signaling Enhance iPSC Generation.
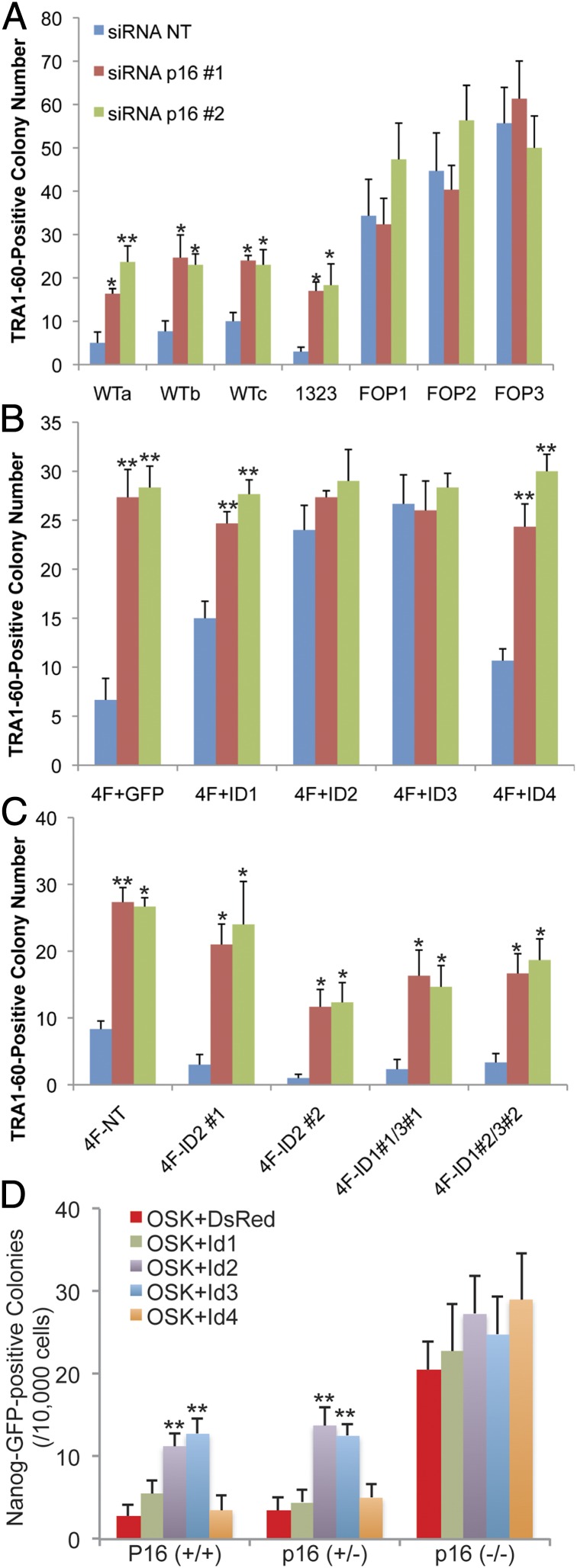
We asked whether inhibitor of differentiation (ID) genes regulated by BMP-SMAD signaling had a role during iPSC generation. ID genes are direct targets of BMP-SMAD signaling in various biological contexts (27–30), which maintain the pluripotency of mESCs (6, 31) and the stem cell identity of several somatic stem cells (32, 33). FOP-HDF alleviated the decrease of ID gene expression during iPSC generation (SI Text and Fig. S4B). To determine if exogenous ID genes promote iPSC generation, we overexpressed ID1, ID2, ID3, or ID4 with OSKM or OSK by retroviral vectors during iPSC generation from normal HDFs (Fig. S5A). Overexpressing any ID gene increased the efficiency of iPSC generation from normal HDFs (Fig. 3 A and B). Next, we examined the role of endogenous expression of ID genes by knockdown experiments. We designed siRNAs against ID1, ID2, and ID3 (ID4 was not expressed in HDFs as shown in Fig. S5A) (Fig. S5B). Because the expression patterns and functions of ID1 and ID3 are highly redundant (34), we used mixed siRNA oligos against ID1/ID3 during hiPSC generation from normal and FOP HDFs. Knockdown of ID1/3 markedly decreased the efficiency of hiPSC generation from normal and FOP HDFs (Fig. 3 C and D). Knockdown of ID2 also decreased the efficiency of iPSC generation from normal and FOP fibroblasts (Fig. 3 E and F). These results indicated that the expression of ID genes is critical for successful reprogramming into hiPSCs.
Fig. S4.
Expression of BMP-SMAD pathway genes during iPSC generation. (A) Expression levels of ID1-3, SMAD1, and ACVR1 in normal or FOP HDFs stimulated by BMP4 analyzed by RT-qPCR. Normal HDF (HDF-1323) were starved for serum overnight and then treated with 100 ng/mL BMP4 for 1 h. Results are mean and SE, n = 5. *P < 0.05, ***P < 0.001 (Dunnett’s test with HDF-1323). (B) Expression levels of ID1–3, SMAD1, EPCAM, OCLN, SNAI1, SNAI2, and ZEB1 in normal or FOP HDFs untransfected or after 5 d of transfection by epiY4 detected by RT-qPCR. Results are mean and SE, n = 3. *P < 0.05, ***P < 0.001 (t test). (C) Changes in expression levels of ID1, ID2, ID3, EPCAM, P16/INK4A, and NANOG by retroviral transduction. Normal HDFs (HDF-WTc) uninfected or after 7 d of retroviral transduction carrying O(OCT4), S(SOX2), K(KLF4), M(MYC), or their combinations were analyzed by RT-qPCR. Expression level of uninfected HDFs is defined as 1.0. Results are mean and SE, n = 3.
Fig. S5.
Expression of ID genes and p16/INK4A during iPSC generation. (A) Expression levels of ID genes in HDFs transduced by retroviral ID genes. Normal HDFs (HDF-1323 transduced with lentiviral Slc7a1) were transduced with retroviral GFP, ID1, ID2, ID3, or ID4 and cultured for 5 d. Results are mean and SE, n = 3. (B) Expression levels of ID genes in HDFs transfected with siRNAs against ID genes. Normal HDF (HDF-1323) were transfected with nontarget, ID1 no. 1, ID1 no. 2, ID2 no. 1, ID2 no. 2, ID3 no. 1, or ID3 no. 2 siRNA and cultured for 3 d. Results are mean and SE, n = 3. (C) Expression levels of Id genes in Nanog-GFP MEF transduced with retroviral Id genes. Nanog-GFP MEFs were transduced with retroviral DsRed, Id1, Id2, Id3, or Id4 and cultured for 5 d. For Id1–3 expression, results are mean and SE, n = 3. For Id4 expression, a DNA gel image of electrophoresis after 40 cycles of PCR is shown. (D) Expression levels of p16/INK4A, p15/INK4B, and p14/ARF in HDFs carrying siRNA against p16/INK4A analyzed by RT-qPCR. HDF-1323 were transfected with nontarget, p16 no. 1, p16 no. 2, p16 no. 3, or p16 no. 4 siRNA and cultured for 3 d. Results show mean and SE, n = 3. (E) Protein expression of p16/INK4A and GAPDH in HDFs carrying siRNA against P16/INK4A. OSKM-transduced HDF-1323 (at day 2) were treated with nontarget, P16 no. 1, P16 no. 2, p16 no. 3, or P16 no. 4 siRNA and cultured for 3 d and then analyzed with Western blotting. Secondary antibodies for p16/INK4A or GAPDH were labeled with IRdye680LT (red) or IRdye800CW (green), respectively. (F) Protein expression levels of p16/INK4A (normalized to the amount of GAPDH expression) in HDF-1323. Results are mean and SE, n = 4. (G and H) Cell proliferation rates of normal and FOP HDFs transduced with retroviral OSKM (G) and of normal HDFs transduced with retroviral OSKM and ID genes (H). At day 0, transduced cells (3 d after transduction) were replated at 10,000 cells per 35-mm dish and then counted daily. At day 1, the medium was changed to SNL-conditioned hESC medium. Results are mean and SE, n = 4. (I–L) p16/INK4A protein expression of FOP and normal HDFs transduced with retroviral OSKM or untransduced and HiPSCs (I and J) or of normal HDFs transduced with retroviral OSKM and ID genes or transfected with siRNA against ID genes or P16/INK4A (K and L). At day 0, transduced HDFs (3 d after transduction) were replated at 10,000 cells per 35-mm dish. At day 1, the medium was changed to SNL-conditioned hESC medium. The cells were collected for Western blot analysis at day 5. Secondary antibodies for p16/INK4A or GAPDH were labeled with IRdye680LT (red) or IRdye800CW (green), respectively. Results are mean and SE, n = 3. *P < 0.05 (t test in B), *P < 0.05, **P < 0.01 (Dunnett’s test with 4F + GFP conditions in D).
Fig. 3.
ID genes are crucial for iPSC generation. (A and B) Effects of overexpressed ID genes on hiPSC generation from normal HDFs with OSKM (A) and OSK (B) retroviral transduction. ESC-like colonies were counted 25 d after transduction from 50,000 cells per 100-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with +GFP conditions). (C and D) Effects of ID1/3 knockdown on hiPSC generation from HDF-1323 (C) and HDF-FOP1 (D) with episomal plasmids. TRA-1-60–positive colonies were counted 25 d after electroporation of epiY4 from 10,000 cells (of HDF-1323) or 5,000 cells (of HDF-FOP1)/35-mm dish at replating. Results are mean and SE, n = 3. **P < 0.01 [Dunnett’s test with nontarget (NT) conditions]. (E and F) Effects of ID2 knockdown on hiPSC generation from HDF-1323 (E) and HDF-FOP1 (F) with episomal plasmids. TRA-1-60–positive colonies were counted 25 d after electroporation of epiY4 from 10,000 cells (of HDF-1323) or 5,000 cells (of HDF-FOP1)/35-mm dish at replating. Results are mean and SE, n = 3. *P < 0.05 (Dunnett’s test with NT conditions). (G and H) Effects of overexpressed Id genes on generating miPSCs from Nanog-GFP MEF with OSKM (G) and OSK (H) retroviral transduction. Nanog-GFP–positive colonies were counted every 3 d after transduction from 10,000 cells (OSKM) or 50,000 cells (OSK)/35-mm dish at replating. Results are mean and SE, n = 4.
To confirm these findings in miPSC generation, we overexpressed mouse versions of Id1, Id2, Id3, or Id4 by retroviral infection with OSKM or OSK in Nanog-GFP MEF to generate miPSCs (35) (Fig. S5C). The efficiency and speed of generating Nanog-GFP positive colonies were much higher with Id genes, especially with the +Id2 and +Id3 conditions (Fig. 3 G and H). The efficiency of generating alkaline phosphatase (ALP)-positive colonies detected at 12 d after transduction was also higher with Id genes (Fig. S6 A–C). These results indicated that overexpressing Id genes promoted miPSC generation in the early phase of reprogramming. We confirmed that miPSC lines generated with Id genes had full developmental potential, including germline transmission (SI Text and Fig. S6 D–L).
Fig. S6.
Characterization of miPSC lines established with Id2/3. (A–C) ALP-positive colonies were counted at day 12 after transduction from Nanog-GFP MEF with OSKM and OSK retroviral transduction. Results are mean and SE, n = 3. *P < 0.05 (Dunnett’s test with +DsRed conditions). (D) A Nanog-GFP–positive miPSC colony of from Nanog-GFP MEFs generated with retroviral human OSK + mouse Id3. (Scale bar: 200 µm.) (E) Protein expression of pluripotent stem cell marker proteins (Nanog, SSEA1) detected by immunocytochemistry in miPS-Id3 no. 1 at passage 12. Secondary antibodies were labeled with Alexa Fluor 555 (red). Nuclei were stained with DAPI (blue). (Scale bar: 100 µm.) (F) Protein expression of differentiation marker proteins (ectoderm: Tuj; mesoderm: cTnT; endoderm: AFP; endoderm or neural crest: SMA) detected by immunocytochemistry in the embryoid bodies made from miPS-Id3 no. 1 at passage 12. Secondary antibodies were labeled with Alexa Fluor 555 (red). Nuclei were stained with DAPI (blue). (Scale bars: 100 µm.) (G) Genomic insertion of retroviral sequence used in generating miPSCs and detected by genomic PCR. PCR products were amplified from 10 ng of genomic DNA of Nanog-GFP MEF, mESC (E14 line), and Nanog-GFP miPSC lines generated with OSK, OSK + DsRed, OSK + Id2, or OSK + Id3. Primer sets are targeting for human OCT4, human SOX2, human KLF4, retroviral Id2, retroviral Id3, or mouse Oct4 promoter region (as a loading control). (H) Id2 and Id3 expression levels were determined by RT-qPCR with primers specific for endogenous transcripts (blue column) or those common to both endogenous and transgenic transcripts (red column). (I) Expression levels of Nanog, Oct4, Sox2, Klf4, human OCT4, human SOX2, human KLF4 in Nanog-GFP MEF (no treatment or transduced with OSK, OSK + DsRed, OSK + Id2, or OSK + Id3 for 5 d), mESC line (E14), Nanog-iPSC lines generated with OSK, OSK + DsRed, OSK + Id2, or OSK + Id3, detected by RT-qPCR. (J) Karyotype of a miPSC line made with OSK + Id3 (miPS − Id3 no. 1) at passage 10. (K) Chimeric mice (black and white mixed coat color) produced with miPS − Id3 no. 1. (L) F1 mice produced from a chimera made with miPS − Id3 no. 1. Black coat color indicates germline-transmitted mice.
BMP-SMAD-ID Signaling Axis Suppresses P16/INK4A-Mediated Cell Senescence During iPSC Generation.
We asked how the BMP-SMAD-ID signaling axis promotes iPSC generation. Activating the BMP-SMAD-ID signaling axis increases the cell proliferation rate and decreases cell senescence during iPSC generation (Fig. S5 G and H and Fig. 4 A–I). We determined if p16/INK4A (CDKN2A), which has a major role in cell senescence during iPSC generation (36, 37), was regulated by BMP-SMAD-ID signaling axis. ID genes suppress p16/INK4A-mediated cell senescence in other biological contexts (38–41). p16/INK4A protein expression was up-regulated in the samples transduced with retroviral OSKM for 5 d, but no or little expression could not be seen in established hiPSCs (Fig. S5 I and J). In contrast, FOP HDFs had lower protein levels of p16/INK4A than normal HDFs under these conditions. Overexpressing ID genes decreased the p16/INK4A protein levels and the knockdown of ID2 or ID1/3 increased the levels during iPSC generation (Fig. S5 K and L). These results indicated that BMP-SMAD-ID signaling axis negatively regulates p16/INK4A expression levels during iPSC generation.
Fig. 4.
BMP-SMAD-ID signaling axis regulates cell senescence during hiPSC generation. (A, D, and E) BrdU assay of normal HDFs transduced with OSKM and ID genes or mACVR1 or transfected with siRNA against ID genes or P16/INK4A. Three days after transduction, these cells were replated at 10,000 cells per 35-mm dish. The next day, the medium was changed to feeder cell-conditioned human ESC (hESC) medium. The cells were treated with siRNAs at day 2 and with BrdU at day 9. Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with 4F conditions in D and with 4F − NT conditions in E). (B, F, and G) SAβgal staining of normal HDFs transduced with OSKM and ID genes or mACVR1 or transfected siRNA against ID genes or P16/INK4A. The transduced cells were fixed to stain with SAβgal at day 12. (Scale bars: 100 µm.) Results are mean and SE, n = 3. *P < 0.05, **P < 0.01 (Dunnett’s test with 4F + GFP conditions in F with 4F − NT conditions in G). (C, H, and I) Immunocytostaining of reprogramming human fetal lung fibroblast line (MRC-5) transduced with OSKM and ID genes or mACVR1 or transfected siRNA against ID genes or P16/INK4A. The transduced cells were fixed to stain with antibodies targeting for HP1β and macroH2A at day 12. Secondary antibodies for HP1β or macroH2 were labeled with Alexa Fluor 488 (green) or Alexa Fluor 555 (red), respectively. Nuclei were stained with DAPI (blue). (Scale bars: 100 µm.) Results are mean and SE, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 (Dunnett’s test with 4F conditions in H with 4F − NT conditions in I).
We asked whether the BMP-SMAD-ID signaling axis was functionally epistatic to p16/INK4A-mediated cell senescence in iPSC generation. As reported (37), inhibiting p16/INK4A by siRNA (tested in Fig. S5 D–F) increased BrdU incorporation (Fig. 4 A and E) and decreased senescence associated β-galactosidase (SAβgal) activity (Fig. 4 A and G) and senescence associated heterochromatin foci (SAHF) formation (Fig. 4 A and I) during iPSC generation. Also as reported (36, 37), knockdown of p16/INK4A increased the efficiency of iPSC generation from normal HDFs; however, knockdown of p16/INK4A had no effect or only a slight increase in the efficiency of iPSC generation from FOP HDFs (Fig. 5A). Overexpressing ID genes in normal HDFs attenuated the increase of the efficiency of hiPSC generation by knocking down p16/INK4A (Fig. 5B). Furthermore, knockdown of p16/INK4A partially rescued the decrease of the efficiency of hiPSC generation from normal HDFs by the knockdown of ID2 or ID1/3 (Fig. 5C). These results indicated that BMP-SMAD-ID signaling was functionally epistatic to p16/INK4A-mediated cell senescence in iPSC generation.
Fig. 5.
BMP-SMAD-ID signaling axis is functionally epistatic to p16/INK4A in iPSC generation. (A–C) Effects of p16/INK4A knockdown during reprogramming on hiPSC generation from FOP and normal HDFs (A), normal HDFs (HDF-WTc) transduced with retroviral OSKM and ID1-4 (B), or normal fibroblasts (HDF-WTc) transfected with ID2 or ID1/3 siRNA (C). TRA-1-60–positive colonies were counted 25 d after electroporation of epiY4 from 10,000 cells per 100-mm dish at replating (A and C) or after transduction with retroviral OSKM and ID1-4 from 50,000 cells per 100-mm dish at replating (B). Results are mean and SE, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 (Dunnett’s test with each siRNA NT condition). (D) Effects of overexpressed Id genes on generating miPSCs from p16 (+/+), p16 (+/−) MEF, or p16 (−/−) MEFs (crossed with Nanog-GFP) with OSK retroviral transduction. Nanog-GFP–positive colonies were counted at 16 d after transduction from 10,000 cells per 35-mm dish at replating. Results are mean and SE, n = 4. **P < 0.01 (Dunnett’s test with each +DsRed condition).
We also used a miPSC generation system by crossing Nanog-GFP mice (35) and p16/ink4a null mice (42). We obtained Nanog-GFP MEF with p16/ink4a+/+, +/−, or −/− and reprogrammed them to generate Nanog-GFP–positive iPSC colonies by transducing retroviral OSK and Id genes. As reported, the efficiency of Nanog-GFP colonies was higher in p16/ink4a null MEF than that in p16/ink4a+/+ or +/− MEF, confirming that p16/ink4a-mediated cell senescence is important in mouse iPSC generation (36, 37). The efficiency of iPSC generation in p16/ink4a+/+ or +/− MEF was increased by transducing Id2 or Id3, but not in p16/ink4a−/− (Fig. 5D). These results suggested that ID genes are epistatic to p16/ink4a-mediated cell senescence in iPSC generation.
Discussion
In this study, we found a positive effect of BMP-SMAD signaling on human iPSC generation. However, the positive effect was easily masked by overgrowth of nonreprogrammed cells when initial cell densities were too high, or by alteration in the periods of BMP treatment because the positive effect requires BMP exposure in the early phases of iPSC generation. It is likely that these complex effects are responsible, at least in part, for the seemingly contradictory results from our study and those of Hamasaki et al. (15). Also, the differences of ingredients in the culture conditions might also modulate the results because recent studies showed that Activin A contributed the activation of BMP-SMAD signaling in FOP cells (43, 44).
We found that ID genes, induced by BMP-SMAD signaling or exogenous transduction, support successful reprogramming into iPSCs. In contrast, ID genes were down-regulated during iPSC generation, and further down-regulation by knockdown decreased the efficiency of iPSC generation. The expression of ID genes alleviates the p16/INK4A-mediated cell senescence barrier during iPSC generation. Conversely, the inhibition of ID genes during hiPSC generation induced p16/INK4A expression and increased cell senescence. Previous studies showed that, during iPSC generation, OSKM treatment up-regulates p16/INK4A expression, which causes cell senescence and inhibits reprogramming (37, 45); however, the mechanisms regulating p16/INK4A expression during iPSC generation remained unclear. Our results suggest that ID genes may be key factors that link transducing reprogramming factors and cell senescence.
We found that a genetic mutation found in FOP patients increased the efficiency of iPSC generation; this is contrary to the case of the FANCA or FANCD2 mutation in Fanconi anemia patients, which makes hiPSC generation more difficult (46, 47). Also, although our study is not primarily intended to clarify FOP pathology, the mechanism of cell senescence mediated by p16/INK4A, which is counterbalanced by BMP-SMAD signaling and ID gene expression, could be relevant to FOP pathology. In FOP patients, the pool of multipotent and proliferating cells plays a major role in the preossification stage (48, 49). The suppression of cell senescence mediated by P16/INK4A caused by the ACVR1 mutation and the hyperactive BMP-SMAD signaling could positively contribute to this process. Collectively, iPSC generation from patient cells carrying genetic mutation are useful for clarifying the reprogramming processes as well as disease modeling.
Materials and Methods
HiPSCs were generated with retrovirus or episomal plasmids as described (2) with some modifications, within six passages after receipt of fibroblasts (summarized in Table S1). Informed written consent was obtained from all donors. All procedures were approved by the Ethics Committee of the Department of Medicine and Graduate School of Medicine, Kyoto University, and the University of California, San Francisco, Institutional Review Board. All of the protocols of mouse experiments were approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco. See Table S2 for the siRNA sequences and Table S3 for DNA oligos and primers. Details of the materials, methods, and associated references are in SI Materials and Methods.
Table S1.
hiPSC lines generated and characterized in this study
| Name | Origin | Methods | Karyotype | Mutation | Silencing | Self-renewal markers | In vitro differentiation | Teratoma differentiation |
| FOP1-1 | HDF-FOP1 | Retro OSKM | 46 XX | ACVR1(617G > A) | KLF4 high | Yes | Yes | Yes |
| FOP1-4 | HDF-FOP1 | Retro OSKM | 46 XX | ACVR1(617G > A) | KLF4 high | Yes | Yes | Yes |
| FOP2-1 | HDF-FOP2 | Retro OSKM | 46 XY | ACVR1(617G > A) | KLF4 high | Yes | Yes | Yes |
| FOP2-2 | HDF-FOP2 | Retro OSKM | 46 XY | ACVR1(617G > A) | KLF4 high | Yes | Yes | Yes |
| FOP1-epiY4-2 | HDF-FOP1 | EpiY4 | 46 XX, t(18, 22) | |||||
| FOP1-epiY4-3 | HDF-FOP1 | EpiY4 | 46 XX | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP1-epiY4-4 | HDF-FOP1 | EpiY4 | 46 XX | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP1-epiY4-10 | HDF-FOP1 | EpiY4 | 46 XX | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP2-epiY4-2 | HDF-FOP2 | EpiY4 | 46 XY, 12 trisomy | |||||
| FOP2-epiY4-3 | HDF-FOP2 | EpiY4 | 46 XY | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP2-epiY4-4 | HDF-FOP2 | EpiY4 | 46 XY | ACVR1(617G > A) | OCT4 high | |||
| FOP2-epiY4-8 | HDF-FOP2 | EpiY4 | 46 XY | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP3-2 | HDF-FOP3 | EpiY4 | 46 XY | ACVR1(617G > A) | OK | Yes | Yes | Yes |
| FOP3-4 | HDF-FOP3 | EpiY4 | 46 XY | ACVR1(617G > A) | OK | |||
| FOP3-8 | HDF-FOP3 | EpiY4 | ACVR1(617G > A) | OK | Yes | Yes | ||
| 1323-epiY4-1 | HDF-1323 | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| 1323-epiY4-2 | HDF-1323 | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| 1323-epiY4-4 | HDF-1323 | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| WTa-1 | HDF-WTa | EpiY4 | NA | OK | Yes | |||
| WTa-2 | HDF-WTa | EpiY4 | NA | OK | Yes | |||
| WTa-5 | HDF-WTa | EpiY4 | NA | OK | Yes | |||
| WTa-7 | HDF-WTa | EpiY4 | NA | OK | Yes | |||
| WTb-3 | HDF-WTb | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| WTb-6 | HDF-WTb | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| WTb-8 | HDF-WTb | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| WTb-14 | HDF-WTb | EpiY4 | 46 XX | NA | OK | Yes | Yes | Yes |
| WTc-3 | HDF-WTb | EpiY4 | 46 XY | NA | OK | Yes | Yes | Yes |
| WTc-5 | HDF-WTb | EpiY4 | 46 XY | NA | OK | Yes | Yes | Yes |
| WTc-10 | HDF-WTb | EpiY4 | 46 XY | NA | OK | Yes | Yes | Yes |
| WTc-11 | HDF-WTb | EpiY4 | 46 XY | NA | OK | Yes | Yes | Yes |
Table S2.
siRNA sequences used in this study
| Product name | Product ID | Sequence of sense strand (5′ → 3′) | Sequence of antisense strand (5′ → 3′) |
| ACVR1 no. 1 | Custom | 5′-rGrGrA rUrUrA rCrArA rGrCrC rArCrC rGrUrU rCrUrA rCrGA T-3′ | 5′-rArUrC rGrUrA rGrArA rCrGrG rUrGrG rCrUrU rGrUrA rArUrC rCrUrC-3′ |
| ACVR1 no. 2 | Custom | 5′-rCrArG rUrUrA rGrCrU rGrCrC rUrUrC rGrArA rUrArG rUrGC T-3′ | 5′-rArGrC rArCrU rArUrU rCrGrA rArGrG rCrArG rCrUrA rArCrU rGrUrA-3′ |
| ID1 no. 1 | HSC.RNAI.N002165.12.10 | 5′-rGrCrA rArGrA rCrArG rCrGrA rGrCrG rGrUrG rCrGrG rGrCG A-3′ | 5′-rUrCrG rCrCrC rGrCrA rCrCrG rCrUrC rGrCrU rGrUrC rUrUrG rCrCrG-3′ |
| ID1 no. 2 | HSC.RNAI.N002165.12.8 | 5′-rCrGrC rCrArA rGrArA rUrCrA rUrGrA rArArG rUrCrG rCrCA G-3′ | 5′-rCrUrG rGrCrG rArCrU rUrUrC rArUrG rArUrU rCrUrU rGrGrC rGrArC-3′ |
| ID2 no. 1 | HSC.RNAI.N002166.12.1 | 5′-rGrCrU rArUrA rCrArA rCrArU rGrArA rCrGrA rCrUrG rCrUA C-3′ | 5′-rGrUrA rGrCrA rGrUrC rGrUrU rCrArU rGrUrU rGrUrA rUrArG rCrArG-3′ |
| ID2 no. 2 | HSC.RNAI.N002166.12.2 | 5′-rGrGrC rUrGrA rArUrA rArGrC rGrGrU rGrUrU rCrArU rGrAT T-3′ | 5′-rArArU rCrArU rGrArA rCrArC rCrGrC rUrUrA rUrUrC rArGrC rCrArC-3′ |
| ID3 no. 1 | Custom | 5′-rGrArG rGrCrA rCrUrC rArGrC rUrUrA rGrCrC rArGrG rUrGG A-3′ | 5′-rUrCrC rArCrC rUrGrG rCrUrA rArGrC rUrGrA rGrUrG rCrCrU rCrUrC-3′ |
| ID3 no. 2 | Custom | 5′-rGrCrU rGrCrC rUrGrU rCrGrG rArArC rGrCrA rGrUrC rUrGG C-3′ | 5′-rGrCrC rArGrA rCrUrG rCrGrU rUrCrC rGrArC rArGrG rCrArG rCrArC-3′ |
| p16 no. 1 | HSC.RNAI.N058195.12.1 | 5′-rCrUrA rCrCrG rUrArA rArUrG rUrCrC rArUrU rUrArU rArUC A-3′ | 5′-rUrGrA rUrArU rArArA rUrGrG rArCrA rUrUrU rArCrG rGrUrA rGrUrG-3′ |
| p16 no. 2 | HSC.RNAI.N058195.12.2 | 5′-rArCrC rUrCrG rGrGrA rArArC rUrUrA rGrArU rCrArU rCrAGT-3′ | 5′-rArCrU rGrArU rGrArU rCrUrA rArGrU rUrUrC rCrCrG rArGrG rUrUrU-3′ |
| Nontarget* | DS NC1 | Unknown | Unknown |
A universal negative control siRNA does not target any sequence in the human, rat, or mouse transcriptomes.
Table S3.
Primer sequences used in this study
| Species | Sequence name | TaqMan probe ID | Primer S | Primer AS |
| Human | ACVR1 | Hs00153836_m1 | ||
| SMAD1 | Hs00195432_m1 | |||
| SMAD6 | Hs00178579_m1 | |||
| SMAD7 | Hs00998193_m1 | |||
| ID1 | Hs03676575_s1 | |||
| ID2 | Hs00747379_m1 | |||
| ID3 | Hs00171409_m1 | |||
| ID4 | Hs00155465_m1 | |||
| INK4A/P16 | Hs02902543_mH | |||
| GAPDH | Hs99999905_m1 | |||
| Retro-Id2 in genomic DNA | GCAGGCTTGAAGGAATTCGGTACC (pMXs-s1923) | CTATCATTCGACATAAGCTCAGAAG | ||
| Retro-Id3 in genomic DNA | GCAGGCTTGAAGGAATTCGGTACC (pMXs-s1923) | GCGCGAGTAGCAGTGGTTCA | ||
| hOCT in genomic DNA | CTGAGGGCCAGGCAGGAGCACGAG | CTGTAGGGAGGGCTTCGGGCACTT | ||
| hSOX2 in genomic DNA | GGTTACCTCTTCCTCCCACTCCAG | TCACATGTGCGACAGGGGCAG | ||
| hKLF4 in genomic DNA | CACCATGGACCCGGGCGTGGCTGCCAGAAA | TTAGGCTGTTCTTTTCCGGGGCCACGA | ||
| OCT4 | Hs00999632_g1 | |||
| SOX2 | Hs01053049_s1 | |||
| KLF4 | Hs01034973_g1 | |||
| MYCL1 | Hs00420495_m1 | |||
| LIN28A | Hs00702808_s1 | |||
| Pla-OCT4 | CATTCAAACTGAGGTAAGGG | TAGCGTAAAAGGAGCAACATAG | ||
| Pla-SOX2 | TTCACATGTCCCAGCACTACCAGA | TTTGTTTGACAGGAGCGACAAT | ||
| Pla-KLF4 | CCACCTCGCCTTACACATGAAGA | TAGCGTAAAAGGAGCAACATAG | ||
| Pla-MYCL1 | GGCTGAGAAGAGGATGGCTAC | TTTGTTTGACAGGAGCGACAAT | ||
| Pla-LIN28 | AGCCATATGGTAGCCTCATGTCCGC | TAGCGTAAAAGGAGCAACATAG | ||
| Mouse | Id1 | GAACCGCAAAGTGAGCAAGG | GATCGTCGGCTGGAACACAT | |
| Endo Id2 expression | CACAAAGGTGGAGCGTGAATAC | CAGTAGGCTCGTGTCAAAAAGG | ||
| Total Id2 expression | ACTCGCATCCCACTATCGTCA | CTATCATTCGACATAAGCTCAGAAG | ||
| Endo Id3 expression | ACCTGGAGCCCGAGAGAAGG | AGGGTGGGGACAGAGTGACG | ||
| Total Id3 expression | CTGCCTGTCGGAACGTAGCC | GCGCGAGTAGCAGTGGTTCA | ||
| Id4 | GACTCACCCTGCTTTGCTGAGA | GAATGCTGTCACCCTGCTTGTT | ||
| Mouse Oct4 promoter | ATCCGAGCAACTGGTTTGTG | CAATCCCACCCTCTAGCCTT | ||
| GAPDH | Mm99999915_g1 | |||
| Oct4 | Mm00658129_gH | |||
| Sox2 | Mm00488369_s1 | |||
| Klf4 | Mm00516105_g1 | |||
| Nanog | Mm02019550_s1 | |||
| Others | EBNA-1 | ATCAGGGCCAAGACATAGAGATG | GCCAATGCAACTTGGACGTT |
SI Text
Increased Efficiency of HiPSC Generation from FOP HDFs with Retroviral Vectors Under Low Cell Density.
We obtained two FOP HDF lines (FOP1 and FOP2) from the Coriell Institute (GM00513 and GM00783, respectively) and confirmed that both carry a heterozygous mutation in ACVR1 (617G > A) that is typical in FOP patients (14) (Fig. S1A). As reported (23–25), BMP-SMAD signaling was hyperactivated in these FOP HDFs, as detected by phospho-SMAD1/5/8 expression after BMP4 treatment (Fig. S1 B and C).
We generated hiPSCs from the two FOP HDFs and a control HDF line (HDF-1323; from a 48-y-old Caucasian female) by transducing retroviral vectors containing OCT4, SOX2, KLF4, and C-MYC (OSKM) or vectors containing OCT4, SOX2, and KLF4 (OSK). After 25 d for OSKM conditions or 30 d for OSK conditions, we counted the ESC-like colonies, which were characterized by flat, round shapes with distinct edges (Fig. S1D, Upper), and the non–ESC-like colonies, which were characterized by granular or epithelial shapes with an irregular edge (Fig. S1D, Lower). The ratios of the ESC-like colonies to the total colony number were comparable among the three conditions, and no ESC-like colonies were obtained with any of the two-factor conditions (i.e., OS, OK, SK, OM, SM, or KM).
We found significantly more ESC-like colonies from FOP HDFs than from control HDFs under both OSK and OSKM conditions when cells were replated at 50,000 cells per 100-mm dish (Fig. S1 E and F). With higher cell densities at replating (i.e., 1.5 × 105 or 5 × 105 cells per 100-mm dish), FOP HDFs did not produce ESC-like colonies because they reached confluent before ESC-like colonies appeared. In contrast, the control HDF line produced more ESC-like colonies with increased seeding density (Fig. S1 G and H). These results indicated that the increased efficiency of iPSC generation from FOP fibroblasts was observed only when plated at low density.
In other experiments, we generated iPSCs from HDFs from three Japanese FOP patients at Kyoto University by retroviral OSKM transduction. After plating transduced HDFs at 50,000 cells per 100-mm dish, using an automatic image processing system, we detected ESC-like colonies by alkaline phosphatase (ALP) staining, a marker for undifferentiated pluripotent stem cells. We used the TIG-120 HDF line as a normal control (from a 6-y-old Japanese female). ALP-positive colonies were generated sooner and in greater numbers in FOP HDFs than in normal HDFs (Fig. S1 I–K).
FOP-HDF Alleviated the Decrease of ID Gene Expression During iPSC Generation.
We confirmed that BMP4 stimulated greater levels of expression of ID1, ID2, and ID3 in FOP fibroblasts than in normal HDFs after a 24-h serum starvation (Fig. S4A). Then, we checked the expression levels of ID1, ID2, and ID3 during iPSC generation. The expression levels of ID1, ID2, and ID3 decreased in the reprogramming cells 5 d after transfecting reprogramming factors (Fig. S4B). However, the rate of decrease was significantly smaller in FOP HDFs. These decreases might be due to the cell stress by transducing retrovirus or transfecting episomal plasmids, because transducing individual factors or GFP by retroviral vectors also decreased ID genes expression in HDF (Fig. S4C). Previous studies reported that BMP signaling promotes mesenchymal-to-epithelial transition (MET) occurred in iPSC generation from mouse embryonic fibroblasts (MEFs) (8, 50). We found no detectable changes in the expression of MET marker genes between FOP fibroblasts and normal HDFs in these samples (Fig. S4B). These results suggested that FOP-HDF alleviated the decrease of ID gene expression during iPSC generation specifically.
HiPSC Lines Generated with Transient BMP4 Treatment Maintained Pluripotency and Self-Renewal.
The hiPSC lines established from normal HDFs with episomal plasmids and recombinant BMP4 treated in week 1 had morphologies similar to ESCs (Fig. S3A) and normal karyotypes (Fig. S3B); they expressed undifferentiated ES–cell marker proteins (e.g., NANOG, SSEA3, and TRA1-60; Fig. S3C) and maintained pluripotency as shown by in vitro embryoid body differentiation (Fig. S3D) and in vivo teratomas injected in SCID mice testis (Fig. S3E); they also expressed endogenous pluripotent marker genes (e.g., OCT4, SOX2, KLF4, L-MYC, and LIN28) at levels similar to those of hESCs, but not detectable plasmid genes (Fig. S3F). There was no detectable insertion of EBNA-1 sequences in their genomic DNA (Fig. S3G). These results indicated that hiPSC lines from normal fibroblasts reprogrammed with transient BMP4-SMAD activation maintain their self-renewal and pluripotency.
miPSC Lines Generated with Id Genes Had Full Developmental Potential Including Germline Transmission.
We generated miPSC lines, including the ones transduced with OSK + Id2 and OSK + Id3 conditions, by selecting Nanog-GFP–positive colonies (Fig. S6D). miPSC lines, which we named OSK no. 1, OSK + DsRed no. 2, OSK + Id2 no. 2, OSK + Id2 no. 3, OSK + Id2 no. 3, OSK + Id3 no. 1, or OSK + Id3 no. 4, expressed undifferentiated ES–cell marker proteins, such as Nanog and SSEA1 (Fig. S6E), and they were pluripotent: they randomly differentiated in vitro in embryoid body formation assays (Fig. S6F). These miPSCs contained genomic insertions of retroviral sequences for human OCT4, human SOX2, human KLF4, and Id2 or Id3 detected by genomic PCR (Fig. S6G). Expression from these viral sequences was largely silenced in the established miPSC lines, as shown by RT-qPCR (Fig. S6 H and I). In contrast, the lines strongly expressed endogenous mouse Nanog, Oct4, Sox2, Klf4, Id2, and Id3 genes, as shown by RT-qPCR. OSK + Id3 no. 1 miPSC line maintained normal male karyotypes (Fig. S6J); to determine whether they contribute to in vivo development and tissue formation, we injected them into host blastocysts to produce chimeric mice. From the OSK + Id3 no. 1 miPSC line, we obtained nine viable chimeric mice from 14 pups (total 76 injected embryos) as shown by their fur coat colors (Fig. S6K). We crossed six of the chimeras from this clone. Some of the F1 mice had a black coat color (2 of 11 from no. 4 chimera, 11 of 11 from no. 5 chimera, 0 of 31 from no. 7 chimera, 0 of 28 from no. 8 chimera, and 0 from 19 from no. 11 chimera; Fig. S6L), indicating germline transmission. Thus, these miPSC lines generated with ID genes maintained their self-renewal and pluripotency with full developmental potential.
BMP-SMAD-ID Signaling Axis Increases the Cell Proliferation Rate and Decreases Cell Senescence During iPSC Generation.
We asked how the BMP-SMAD-ID signaling axis promotes iPSC generation. We found that activating BMP-SMAD-ID signaling axis increases the cell proliferation rate during iPSC generation and decreases cell senescence during iPSC generation (Figs. S5 G and H and 4 A–I). First, we found that the proliferation rates of FOP HDFs were much higher than those of normal HDFs during iPSC generation (Fig. S5G). Similarly, overexpressing ID genes enhanced cell proliferation rates during hiPSC generation (Fig. S5H). The BrdU incorporation ratio increased when BMP-SMAD-ID signaling was activated and decreased when the signal was inhibited (Fig. 4 A, D, and E). These results indicate that activating BMP-SMAD-ID signaling axis increases the cell proliferation rate during iPSC generation.
Next, we asked whether these proliferation rate changes resulted from cell senescence, a major barrier to iPSC generation (36, 37, 45, 51). We examined a cell senescence marker, senescent-associated β-galactosidase (SAβGAL) during hiPSC generation with retroviral OSKM. The ratio of SAβGAL-positive cells decreased when the BMP-SMAD-ID signaling axis was activated and increased when it was inhibited (Fig. 4 A, F, and G). We also examined the formation of SAHF during hiPSC generation. SAHFs were formed in a cell type-dependent manner and were hardly detected in any HDF lines during hiPSC generation (52). In contrast, SAHFs formed consistently in the human fetal lung fibroblast line, MRC-5, after transducing retroviral reprogramming factors as reported (37). SAHF-positive cells were clearly distinguished by the strong expression and colocalization of macroH2A.1 and by DAPI staining. The ratio of SAHF-positive cells decreased under conditions in which the BMP-SMAD-ID signaling axis was activated and increased when this axis was inhibited (Fig. 4 A, H, and I). These results indicated that activating the BMP-SMAD-ID signaling axis decreases cell senescence during iPSC generation.
SI Materials and Methods
Cell Culture.
The following human cells were used in this study: HDF-1323 from a 48-y-old Caucasian female (Cell Applications; 106-05a); TIG-120 from a 6-y-old Japanese female (Japanese Collection of Research Bioresources; JCRB0542); HDF-WTa, HDF-WTb, and HDF-WTc from a 48-y-old Latin female, 52-y-old Caucasian/Japanese male, and 30-y-old Japanese male, respectively, whose skin samples were collected at Gladstone; HDF-FOP1 and HDF-FOP2 from a 16-y-old Caucasian female and an unknown-year-old black male (Coriell Institute for Medical Research; GM00513 and GM00783, respectively); HDF-FOP3 from a 10-y-old male collected at the University of California, San Francisco. HDF-FOPJ1, J2, and J3 from 12-y-old, 50-y-old, and unknown-year-old Japanese males, respectively, collected at Kyoto University; and the human fetal lung fibroblast line, MRC-5 [American Type Culture Collection (ATCC); CCL-171]. We confirmed these FOP fibroblasts carry 617 G to A by PCR sequencing (16).
Human fibroblasts, Plat-E, Plat-GP (Cell Biolab), 293FT (Invitrogen), Nanog-GFP MEF, and SNL feeder cells [STO (ATCC, CRL-1503) cells expressing neomycin-resistant gene and leukemia inhibitory factor (LIF) gene] (Health Protection Agency Culture Collection) were cultured in DMEM containing 10% (vol/vol) FBS, 1% GlutaMAX, 0.1 mM MEM nonessential amino acids (Invitrogen), 1 mM sodium pyruvate, and penicillin-streptomycin (all from Invitrogen). A total of 10 µg/mL puromycin and 10 µg/mL Blasticidin, 10 µg/mL Blasticidin, and 500 µg/mL G418 for Plat-E, Plat-GP, 293FT, were added to each cell type, respectively. For passaging, cells were washed once with Dulbecco’s PBS (DPBS; calcium and magnesium-free) and incubated with TrypLE Express (Invitrogen) at 37 °C. When cells were dissociated, the TrypLE Express was removed, and the cells were washed with culture medium described above. The cells were collected into a 15-mL conical tube and spun down. The contents were transferred to a new dish. The split ratio was routinely 1:3–1:20.
Established hiPSC lines and the hESC line (H7 from WiCell Research Institute) were cultured in mTeSR1 medium (Stem Cell Technologies) on Matrigel (BD) or Synthemax (Corning)-coated dishes. For passaging, hiPSC and hESC cells were washed once DPBS and incubated with Accutase (Millipore) at 37 °C. When colonies at the edge of the dish started dissociating from the bottom, Accutase was removed, and the cells were washed with mTeSR1 medium. Cells were scraped and collected into a 15-mL conical tube. An appropriate volume of the medium with 10 µM ROCK inhibitor (Y-27632; Millipore) was added, and the contents were transferred to a new dish. The split ratio was routinely 1:3–1:10.
Established miPSC lines and mESCs (E14 line) were cultured in ESGRO Complete PLUS Clonal Grade Medium (Millipore). For passaging, miPSC and mESC cells were washed once with DPBS and incubated with Accutase at 37 °C. When cells were dissociated, the medium was added. The cells were collected into a 15-mL conical tube and spun down. The contents were transferred to a new dish. The split ratio was routinely 1:10–1:100.
Generation of HiPSCs with Retroviral Vectors.
Plat-E or Plat-GP cells were plated at 5 × 106 cells per 100-mm dish. The next day, pMXs-based retroviral vectors for each gene (and pCMV-VSVG for Plat-GP) were independently introduced into the Plat-E or Plat-GP cells with the FuGENE 6 transfection reagent (Promega). After 24 h, the medium was replaced. Human fibroblasts (expressing the mouse Slc7a1 (solute carrier family 7 member 1) for retroviral-transduction from Plat-E cells) were seeded at 1 × 105 cells per well of a six-well plate. The next day, virus-containing supernatants were recovered and mixed together. Fibroblasts were incubated in the virus containing supernatants added with Polybrene (Millipore) at a final concentration of 4 μg/mL overnight. At 3 d after the infection, fibroblasts were harvested by trypsinization and replated at 5 × 104 cells per 100-mm dish or 1 × 104 cells per six-well plate on SNL feeder cells. The next day, the medium was replaced with primate ES cell medium (ReproCELL), supplemented with 5 ng/mL recombinant human basic fibroblast growth factor (bFGF). Dorsomorphin (Stemgent), LDN-193189 (Stemgent), or SB203580 (Promega) solved in DMSO were treated in appropriate concentrations in the primate ES cell medium throughout hiPSC generation in specific experiments.
Generation of HiPSCs with Episomal Vectors.
Three micrograms of expression plasmid mixtures (epiY4) were electroporated into 3 × 105 HDFs with Neon Electroporation Device (Invitrogen) with a 100-μL kit, according to the manufacturer’s instructions. Conditions for electroporations were 1,650 V, 10 ms, and three pulses. The cells were trypsinized 4 d after electroporation and seeded at 5 × 103 or 1 × 104 cells per well of six-well plate covered with SNL feeder cells. The culture medium was replaced the next day (day 5) with the primate ESC medium containing bFGF. The colonies were counted 25 d after electroporation, and those colonies similar to hESCs were selected for further cultivation and evaluation. Human recombinant BMP4 or Activin A (both from R&D systems) dissolved in 4 mM HCl, 0.1% BSA were treated at specific periods and concentrations during hiPSC generation in specific experiments.
Generation of miPSCs with Retroviral Vectors.
Nanog-GFP MEFs were isolated from male mouse embryos 13.5 embryonic days from Nanog-GFP reporter mice (RBRC02290; RIKEN Bio Resource Center) crossed with the same strain (35). p16(Ink4a)+/+, +/−, or −/− MEFs with Nanog-GFP were isolated from mouse embryos 13.5 embryonic days from p16+/−, Nanog-GFP reporter mice crossed with the same strain mice. These p16+/−, Nanog-GFP reporter mice were obtained by crossing with Nanog-GFP reporter mice mentioned above and p16(Ink4a) null mice (01XE4, FVB.129-Cdkn2atm2.1Rdp/Nci; National Cancer Institute) (42). Genotyping assays were performed following the providers’ recommendations.
Plat-E cells were seeded at 5 × 106 cells per 100-mm dish. The next day, 9 µg of pMXs-based retroviral vectors for human OCT3/4, SOX2, KLF4, (MYC), or mouse Id genes were independently introduced into Plat-E cells with 27 μL of FuGENE 6 transfection reagent. After 24 h, the medium was replaced. MEFs were seeded at 1 × 105 cells per well of a six-well plate. The next day, virus-containing supernatants from these Plat-E cultures were recovered and filtered through a 0.45-µm cellulose acetate filter. Equal volumes of the supernatants were mixed and supplemented with polybrene at the final concentration of 4 µg/mL. MEFs were incubated in the virus/polybrene-containing supernatants for 24 h. Three days after infection, the cells were reseeded at 1,000 cells (OSKM conditions) or 5,000 cells (OSK conditions) per well of a six-well plate onto SNL feeder cells. The next day, medium was changed with mouse ESC medium supplemented with LIF. Medium was changed every 2 d. The GFP-positive colonies were counted every 3 d after 15 d of infection. ALP staining was performed 12 d after the transduction. GFP-positive, mESC-like colonies were picked up 24 d after infection and maintained in the mouse ESC medium supplemented with 2 µg/mL of puromycin.
Plasmid Construction.
The ORF of human ACVR1, SMAD1, SMAD6, SMAD7, ID1, ID2, ID3, ID4, mouse Id1, mouse Id2, mouse Id3, and mouse Id4 contained in pDONR vectors were purchased from GeneCopoeia. All of the genes were transferred to pMXs-Gateway (Addgene) using the Gateway cloning system (Invitrogen) according to the manufacturer’s instructions. The FOP mutant ACVR1 expression vector was generated as described (23). Briefly, the pMXs-ACVR1 (wild-type) plasmids were treated by site-directed mutagenesis of the FOP mutant ACVR1 sequence at cDNA position 617 (from G to A) with the GeneTailor Site-Directed Mutagenesis System (Invitrogen). The oligonucleotides used to generate the mutant construct were: forward 5′-GTACAAAGAACAGTGGCTCaCCAGATTACACTG-3′; reverse 5′-GTGAGCCACTGTTCTTTGTACCAGAAAAGGAAG-3′.
siRNA Transfection.
siRNA (Dicer-substrate RNAs, which are chemically synthesized 27-mer duplexes) for ACVR1, ID1, ID2, ID3, and p16/INK4A were purchased from Integrated DNA Technologies. At day 4 and day 10 after electroporation, 5 nM of siRNA were transfected using 5 µL of Lipofectamine RNAiMAX (Invitrogen) during iPSC generation in a six-well culture plate. siRNAs used in this study are listed in Table S2.
RNA and DNA Isolation, Reverse Transcription, and PCR.
Genomic DNA was extracted with a DNeasy Tissue and Blood Kit (Qiagen) according to the manufacturer’s recommendations. Total RNA was purified with RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s recommendations. One micron of total RNA was used for reverse transcription reaction with SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. qPCR was performed with Power SYBR Green PCR Master Mix or TaqMan Gene Expression Master Mix and analyzed with the 7900 real-time PCR system (all from Applied Biosystems). The expression levels of each gene are normalized with the amount of GAPDH expression in the same samples. Conventional PCR was performed with AccuPrime Taq DNA Polymerase System (Invitrogen) according to the manufacturer’s recommendations. We amplified PCR products by 30 cycles of denature (94 °C for 15 s), annealing (58 °C for 30 s), and extension (68 °C for 1 min). Ten nanograms of genomic DNA from each sample were used for PCR. Primer sequences and TaqMan probe numbers are shown in Table S3.
ALP and SAβGal Staining.
For ALP staining, cells were fixed with PBS containing 4% (vol/vol) paraformaldehyde for 2 min at room temperature. Staining was performed using the Leukocyte Alkaline Phosphatase Kit (Sigma) or Alkaline Phosphatase Detection Kit (Millipore) according to the manufacturer’s recommendations. For counting ALP-positive colony number in the generation of hiPSCs from Japanese FOP mutant HDFs, images of a whole 100-mm dish were taken with Sony DSLR-A350k with DT 18–70 mm, F/3, 5–5.6 zoom lens after the staining and were analyzed with Integrated Morphometry Analysis function in MetaMorph Software (Molecular Devices). SAβGal staining was performed using Cellular Senescence Assay Kit (Cell BioLabs) according to the manufacturer’s recommendations. Stained samples were analyzed in randomly selected images (10 randomized image fields for each sample).
BrdU Incorporation Assay.
A BrdU incorporation assay was performed using the FITC BrdU Flow Kit (BD Pharmingen) according to the manufacturer’s recommendation. Briefly, the cells were treated with 10 µM BrdU for 16 h. After dissociation, the cells were fixed and permeabilized. Then, the fixed cells were treated with 300 µg/mL DNase for 1 h at 37 °C. The cells were incubated with FITC-conjugated anti-BrdU antibodies for 20 min at room temperature. The stained cells were analyzed with FACSCalibur (BD) and FlowJo software (Tree Star).
Immunocytochemistry.
For counting the TRA-1-60–positive colonies in hiPSC generation experiments, cells were fixed with PBS containing 4% (vol/vol) paraformaldehyde for 10 min at room temperature and reacted with ImmunoPure Peroxidase Suppressor (Thermo Scientific). The cells were washed with PBS and treated with 1% BSA for blocking. Primary antibodies used were biotin-conjugated anti-TRA-1-60 antibody (1 µg/mL; eBiosciences). Immunostaining was performed using HRP-conjugated streptavidin (1 µg/mL; Thermo Scientific) and ImmunoHisto Peroxidase Detection Kit (Thermo Scientific) according to the manufacturer’s instructions.
For immunocytochemistry of pluripotency and differentiation markers, cells were fixed with PBS containing 4% (vol/vol) paraformaldehyde for 10 min at room temperature. The cells were permeabilized with PBS containing 0.1% Triton X-100 for 10 min at room temperature and then washed with PBS and treated with 1% BSA for blocking.
For hiPSCs or reprogramming HDFs, primary antibodies used were SSEA1 (0.5 µg/mL; eBiosciences), SSEA3 (0.5 µg/mL; eBiosciences), SSEA4 (0.5 µg/mL; eBiosciences), TRA-1-60 (0.5 µg/mL; eBiosciences), TRA-1-81 (0.5 µg/mL; eBiosciences), Nanog (2 µg/mL, AF1997; R&D Systems), βIII-tubulin (1:100, MAB1637; Millipore), glial fibrillary acidic protein (1:500, Z0334; DAKO), α-smooth muscle actin (prediluted, N1584; DAKO), α-fetoprotein (2 µg/mL, MAB1368; R&D Systems), tyrosine hydroxylase (1:100, AB152; Chemicon), anti-macroH2A.1 (1:1,000; Abcam), and anti-HP1β (1:1,000; Millipore).
For miPSCs, primary antibodies used were SSEA1 (0.5 µg/mL; eBiosciences), Nanog (2 µg/mL, AF2729; R&D Systems), α-fetoprotein (2 µg/mL, MAB1368; R&D Systems), cardiac troponin T (ready-to-use, clone 13-11; Thermo Scientific), βIII-tubulin (1:100, MAB1637; Millipore), and smooth muscle actin (ready-to-use, clone 1A4; DAKO).
Secondary antibodies used were Alexa Fluor 488 or 555-conjugated goat anti-mouse IgG (1:200; Invitrogen), Alexa 488 or 555-conjugated goat anti-rabbit IgG (1:200; Invitrogen), and Alexa 488 or 555-conjugated donkey anti-goat IgG (1:200; Invitrogen). Nuclei were stained with DAPI contained in the VectaShield set (Vector Laboratories). For SAHF detection, the samples were analyzed in randomly selected images (10 randomized image fields for each sample) with INcell Analyzer 2000 (GE Healthcare) and counted for the macroH2A.1- and SAHF (detected by DAPI staining)-positive cells.
Flow Cytometry.
Harvested cells were washed with PBS containing 1% FBS and stained with mouse monoclonal anti-ACVR1 antibody MAB637 (10 µg/mL; R&D Systems) for 1 h at 4 °C. Cells were then washed once with PBS containing 1% FBS, and goat anti-mouse IgG Alexa Fluor 448 (Thermo Fisher Scientific) and 7-AAD was used for detection. After two washes in PBS containing 2% (vol/vol) FBS, cells were filtered with 70-mm cell strainer (BD). 7-AAD-negative, live cells were sorted for surface expression of ACVR1. The stained cells were analyzed with FACSCalibur (BD) and FlowJo software (Tree Star).
In Vitro Differentiation.
For EB formation from hiPSCs, the cells were harvested by treating with Accutase. The clumps of the cells were transferred to ultra-low attachment dishes (Corning) in DMEM/F12 containing 20% (vol/vol) knockout serum replacement (Invitrogen), 2 mM L-glutamine, 100 mM nonessential amino acids, 100 mM 2-mercaptoethanol (Invitrogen), and penicillin-streptomycin. The medium was changed every other day. After 8 d as a floating culture, EBs were transferred to gelatin-coated plate and cultured in the same medium for another 8 d. For EB formation from miPSCs, the cells were harvested by treating with TrypLE Express (Invitrogen). The dissociated cells were transferred to ultra-low attachment dishes (Corning) in the MEF culture medium. The medium was changed every other day. After 8 d as a floating culture, EBs were transferred to gelatin-coated plate and cultured in the same medium for another 8 d.
Teratoma Formation.
hiPSCs cells were harvested by Accutase treatment, collected into tubes, and centrifuged, and the pellets were suspended in mTeSR1 complete medium with 10 µM Rock inhibitor, Y-27632. All of the cells from a confluent well of a six-well plate were injected into a testis or back area of a SCID mouse (Jackson Laboratory). miPSCs cells were harvested by trypsin/EDTA treatment, collected into tubes, and centrifuged, and the pellets were suspended in FP medium. All of the cells from a confluent well of a six-well plate were injected into a testis or back area of a SCID mouse. At 7–9 wk after injection, tumors were dissected, weighted, and fixed with PBS containing 4% (vol/vol) paraformaldehyde. Paraffin-embedded tissue was sliced and stained with H&E.
Western Blotting.
Cells at a semiconfluent state were lysed with RIPA buffer (50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS). Cell lysates were separated by electrophoresis on 4–20% (wt/vol) SDS/PAGE and transferred to a nitrocellulose membrane with the iBlot transfer system (Invitrogen). The membrane was blocked with Odyssey Blocking Buffer (Li-COR Biosciences) and incubated with primary antibody diluted in the blocking buffer in SNAP i.d. protein detection system (Millipore). After washing with TBST, the membrane was incubated with IRDye 680LT- or IRDye 800CW-conjugated secondary antibody (Li-COR Biosciences) diluted in the blocking buffer. Signals were detected with Odyssey Imaging Systems (Li-COR Biosciences). Primary antibodies used for Western blotting were rabbit anti-phospho Smad1/5/8 (1:1,000; Cell Signaling Technology), rabbit anti-Smad1/5/8 (1:1,000; Santa Cruz Biotechnology), mouse anti–β-actin (1:5,000, A5441; Sigma), chicken anti-GAPDH (1:5,000, AB2302; Millipore), and rabbit anti-p16 (1:200, C-20; Santa Cruz Biotechnology). Signal intensity was calculated with ImageJ software.
Karyotyping and Short Tandem Repeat (Genomic Fingerprinting) Analysis.
Chromosomal G-band analyses and short tandem repeat analysis were performed at Cell Line Genetics.
Chimeric Mice Production and Germline Transmission Experiments.
Chimeric mice production and germline transmission experiments were performed at Cornell Induced Pluripotent Stem Cell Core Laboratory.
Statistical Analyses.
All data are shown as the mean ± SE. The unpaired t test, one-factor ANOVA, and Dunnett’s test were used for statistical analysis of the data. Differences were considered to be statistically significant for P < 0.05 (*), P < 0.01 (**), or P < 0.001 (***). All statistical analyses were done with Excel 2008 for Mac (Microsoft) or R for Mac OS X GUI 1.40.
Acknowledgments
We thank Drs. Akiko Hata and Eileen Shore for scientific critical review; Gary Howard, Anna Lisa Lucido, and Mimi Zeiger for editorial review; Drs. Tim A. Rand and Kathleen A. Worringer for scientific comments and valuable discussion; Dr. Toshio Kitamura for the retroviral system components; Rie Kato, Yoko Miyake, Sayaka Takeshima, and Karena Essex for administrative support; and the Gladstone Stem Cell, Histology, and Microscopy Cores for technical support. This work was supported by NIH Grant R01 HL60664-07 (to B.R.C.); NIH Grant K08 AR056299-02; California Institute of Regenerative Medicine/Gladstone Institutes California Institute for Regenerative Medicine (CIRM) Fellowship T2-00003, the University of California, San Francisco (UCSF) Department of Medicine, and March of Dimes Basil O’Connor Starter Grant 5-FY12-167 (to E.C.H.); Duke-NUS Graduate Medical School Singapore Third-Year Research Program (C.R.S.); Grants-in-Aid for Scientific Research from the Japan Ministry of Education, Culture, Sports, Science, and Technology and the Leading Project for Realization of Regenerative Medicine (to M.I. and J.T.); the Uehara Memorial Foundation and USCF’s Program for Breakthrough Biomedical Research (Y.H.); and Kyoto University Grants, Grants-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS) and Ministry of Education, Culture, Sports, Science and Technology-Japan (MEXT), the Program for Promotion of Fundamental Studies in Health Sciences of National Institute of Biomedical Innovation (NIBIO) (Japan), the L. K. Whittier Foundation, and the Roddenberry Foundation (S.Y.). The Gladstone Institutes received support from National Center for Research Resources Grant RR18928. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest statement: S.Y. is a scientific advisor of iPS Academia Japan without salary. E.C.H. receives research funding from Clementia Pharmaceuticals to support clinical trials in FOP that are unrelated to this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603668113/-/DCSupplemental.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460(7251):49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 4.Ichida JK, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5(5):491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin T, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6(11):805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 7.Qi X, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci USA. 2004;101(16):6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton DA, Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2000;97(21):11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadwick K, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102(3):906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 11.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132(6):1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 12.Xu RH, et al. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2(3):185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109(7):2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shore EM, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38(5):525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 15.Hamasaki M, et al. Pathogenic mutation of Alk2 inhibits ips cell reprogramming and maintenance: mechanisms of reprogramming and strategy for drug identification. Stem Cells. 2012;30:2437–2449. doi: 10.1002/stem.1221. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto Y, et al. Induced pluripotent stem cells from patients with human fibrodysplasia ossificans progressiva show increased mineralization and cartilage formation. Orphanet J Rare Dis. 2013;8:190. doi: 10.1186/1750-1172-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto Y, et al. New protocol to optimize iPS cells for genome analysis of fibrodysplasia ossificans progressiva. Stem Cells. 2015;33(6):1730–1742. doi: 10.1002/stem.1981. [DOI] [PubMed] [Google Scholar]
- 18.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 19.Yu PB, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4(1):33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuny GD, et al. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18(15):4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuenda A, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364(2):229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 22.Vogt J, Traynor R, Sapkota GP. The specificities of small molecule inhibitors of the TGFß and BMP pathways. Cell Signal. 2011;23(11):1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Shen Q, et al. The fibrodysplasia ossificans progressiva R206H ACVR1 mutation activates BMP-independent chondrogenesis and zebrafish embryo ventralization. J Clin Invest. 2009;119(11):3462–3472. doi: 10.1172/JCI37412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song GA, et al. Molecular consequences of the ACVR1(R206H) mutation of fibrodysplasia ossificans progressiva. J Biol Chem. 2010;285(29):22542–22553. doi: 10.1074/jbc.M109.094557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dinther M, et al. ALK2 R206H mutation linked to fibrodysplasia ossificans progressiva confers constitutive activity to the BMP type I receptor and sensitizes mesenchymal cells to BMP-induced osteoblast differentiation and bone formation. J Bone Miner Res. 2010;25(6):1208–1215. doi: 10.1359/jbmr.091110. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381(6583):620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 27.Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274(28):19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 28.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277(7):4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 29.López-Rovira T, Chalaux E, Massagué J, Rosa JL, Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277(5):3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- 30.Miyazono K, Miyazawa K. Id: A target of BMP signaling. Sci STKE. 2002;2002(151):pe40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- 31.Romero-Lanman EE, Pavlovic S, Amlani B, Chin Y, Benezra R. Id1 maintains embryonic stem cell self-renewal by up-regulation of Nanog and repression of Brachyury expression. Stem Cells Dev. 2012;21(3):384–393. doi: 10.1089/scd.2011.0428. [DOI] [PubMed] [Google Scholar]
- 32.Benezra R, Rafii S, Lyden D. The Id proteins and angiogenesis. Oncogene. 2001;20(58):8334–8341. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
- 33.Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20(58):8308–8316. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- 34.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401(6754):670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 35.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 36.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23(18):2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohtani N, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409(6823):1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 39.Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci USA. 2001;98(14):7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickoloff BJ, et al. Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem. 2000;275(36):27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- 41.Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20(58):8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- 42.Sharpless NE, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413(6851):86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 43.Hatsell SJ, et al. ACVR1R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Sci Transl Med. 2015;7(303):303ra137. doi: 10.1126/scitranslmed.aac4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hino K, et al. Neofunction of ACVR1 in fibrodysplasia ossificans progressiva. Proc Natl Acad Sci USA. 2015;112(50):15438–15443. doi: 10.1073/pnas.1510540112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller LU, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119(23):5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medici D, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16(12):1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barruet E, et al. The ACVR1 R206H mutation found in fibrodysplasia ossificans progressiva increases human induced pluripotent stem cell-derived endothelial cell formation and collagen production through BMP-mediated SMAD1/5/8 signaling. Stem Cell Res Ther. 2016;7(1):115. doi: 10.1186/s13287-016-0372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosar M, et al. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell Cycle. 2011;10(3):457–468. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]