Significance
Because the SNP linking chromosome 17q21 to asthma is associated with increased gasdermin B (GSDMB) expression, we generated transgenic mice expressing increased levels of the human GSDMB transgene (hGSDMBZp3-Cre), which develop an asthma phenotype characterized by a spontaneous increase in airway responsiveness and airway remodeling (increased peribronchial smooth muscle) in the absence of the development of airway inflammation. These results challenge the current paradigm in asthma that airway inflammation induces smooth muscle remodeling and airway responsiveness, as these hGSDMBZp3-Cre mice develop increased airway-hyperresponsiveness and smooth muscle in the absence of airway inflammation. Furthermore, this study adds to our understanding of gene networks in asthma that we have identified can act in sequential pathways (i.e., GSDMB induces 5-lipoxygenase to induce TGF-β1).
Keywords: GSDMB, asthma, airway-hyperresponsiveness, remodeling, inflammation
Abstract
Gasdermin B (GSDMB) on chromosome 17q21 demonstrates a strong genetic linkage to asthma, but its function in asthma is unknown. Here we identified that GSDMB is highly expressed in lung bronchial epithelium in human asthma. Overexpression of GSDMB in primary human bronchial epithelium increased expression of genes important to both airway remodeling [TGF-β1, 5-lipoxygenase (5-LO)] and airway-hyperresponsiveness (AHR) (5-LO). Interestingly, hGSDMBZp3-Cre mice expressing increased levels of the human GSDMB transgene showed a significant spontaneous increase in AHR and a significant spontaneous increase in airway remodeling, with increased smooth muscle mass and increased fibrosis in the absence of airway inflammation. In addition, hGSDMBZp3-Cre mice showed increases in the same remodeling and AHR mediators (TGF-β1, 5-LO) observed in vitro in GSDMB-overexpressing epithelial cells. GSDMB induces TGF-β1 expression via induction of 5-LO, because knockdown of 5-LO in epithelial cells overexpressing GSDMB inhibited TGF-β1 expression. These studies demonstrate that GSDMB, a gene highly linked to asthma but whose function in asthma is previously unknown, regulates AHR and airway remodeling without airway inflammation through a previously unrecognized pathway in which GSDMB induces 5-LO to induce TGF-β1 in bronchial epithelium.
Chromosome 17q21 was initially linked to asthma in genome-wide association studies (GWAS) in 2007, with confirmation in multiple GWAS and non-GWAS studies in populations of diverse ethnic backgrounds (1, 2). Chromosome 17q21 contains a cluster of four genes [ORMDL3, gasdermin B (GSDMB), IKZF3, and ZPBP2] (2), which may, either individually or in combination, be responsible for its genetic association to asthma. A precedent for linkage of a cluster of genes in a single chromosomal region with asthma is evident from chromosome 5q31-33, where the genes IL-4, IL-5, IL-9, and IL-13 are located (3). Although there are many genetic epidemiologic studies linking chromosome 17q21 with asthma, there are limited functional studies of each of these four genes to understand how they contribute to the pathogenesis of asthma. We have previously demonstrated the importance of one of the 17q21 localized genes (i.e., ORMDL3) to asthma, airway responsiveness, and airway remodeling (4, 5). Mice overexpressing human ORMDL3 regulated downstream pathways (ATF6α, SERCA2b, metalloproteases) important in asthma. Interestingly, mice overexpressing human ORMDL3 developed spontaneous increased airway responsiveness and airway remodeling in the absence of airway inflammation (5).
In this study, we are focusing on a second gene on chromosome 17q21, namely GSDMB, because little is known about its expression and function in the normal lung or asthmatic lung, and published studies of GSDMB in asthma are limited to genetic association studies. The human GSDMB gene located on chromosome 17q21 belongs to the gasdermin (GSDM) protein family (6, 7). The human GSDM family consists of four members: GSDMA, GSDMB, GSDMC, and GSDMD. The GSDMA and GSDMB genes are located at 17q21, and the GSDMC and GSDMD genes are located at 8q24. Several genetic linkage studies have shown an association with GSDMB on chromosome 17q21 and asthma (2, 8), but none of the studies have linked asthma with chromosome 8q24 (where GSDMC and GSDMD are located). This finding suggests that GSDMB (and perhaps GSDMA) are the only GSDM family members linked to asthma. The function of GSDMB (a 416-amino acid protein) is largely unknown because GSDMB is a novel protein without characteristic domains related to known function (6, 7). The single-nucleotide polymorphism (SNP) linked to chromosome 17q21 and asthma is associated with increased expression of both GSDMB and ORMDL3 (genes located next to each other on chromosome 17q21) (2). Several studies indicate a strong linkage disequilibrium between the SNPs of ORMDL3 and GSDMB (1, 2), suggesting that these genes could be acting in concert toward asthma pathophysiology. However, at present the function of GSDMB in the lung and in asthma is largely unknown.
Because the SNP linking chromosome 17q21 to asthma is associated with increased GSDMB expression (1, 2), we generated transgenic mice expressing increased levels of the human GSDMB transgene (hGSDMBZp3-Cre), which develop a significant spontaneous increase in airway-hyperresponsiveness (AHR) and a significant spontaneous increase in airway remodeling (increased smooth muscle, increased fibrosis) in the absence of development of airway inflammation. This study, combined with our previous studies on ORMDL3 (4, 5), suggest that at least two genes localized to chromosome 17q21 (GSDMB and ORMDL3) contribute to airway remodeling in the absence of airway inflammation, and challenges the current paradigm that inflammation precedes lung remodeling and airway responsiveness in asthma.
Results
GSDMB Is Highly Expressed in Human Bronchial Epithelial Cells in Asthma.
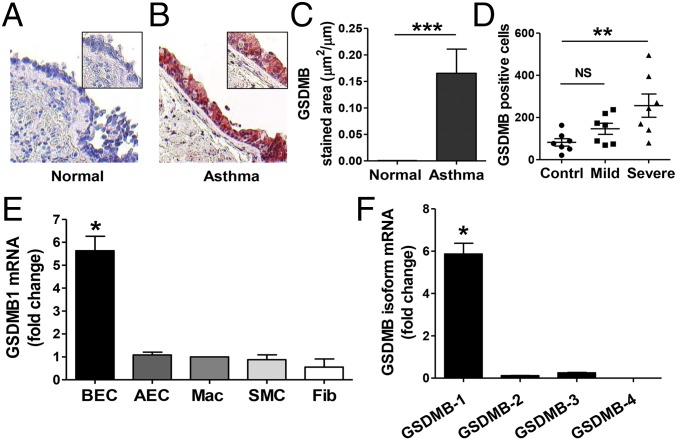
To determine which human lung cells express GSDMB, we examined human lung sections from asthmatics and nonasthmatic controls. Immunohistochemistry analyses showed that levels of GSDMB were significantly increased in the bronchial epithelial cells in the lungs of asthmatics compared with control lungs (Fig. 1 A–C). We also examined the effect of asthma severity on GSDMB expression and found GSDMB+ cells to be significantly elevated in bronchial biopsy specimens from severe asthmatics compared with nonasthmatic controls (Fig. 1D). We further investigated which human lung structural cells express GSDMB or its isoforms GSDMB-1, -2, -3, and -4 (Fig. S1A). We found that bronchial epithelial cells, but not alveolar epithelial cells, were a significant source of the GSDMB-1 isoform as quantitated by quantitative PCR (qPCR) (Fig. 1 E and F). In addition, we did not detect significant GSDMB-1 expression in other human lung structural cells (i.e., smooth muscle cells, fibroblasts), or in human lung macrophages. Moreover, GSDMB-1 was the most predominant isoform expressed in human bronchial epithelial cells (Fig. 1F).
Fig. 1.
GSDMB is highly expressed in human bronchial epithelial cells in asthma. Human lungs from (A) healthy controls and (B) asthmatic patients were examined by immunohistochemistry with an anti-GSDMB Ab (n = 7 subjects per group). (Magnification: A and B, 200×; Insets, 400×.) (C) The number of peribronchial GSDMB+ cells were quantitated in each group by light microscopy (LM) and image analysis. (D) GSDMB+ cells in bronchial biopsies from healthy controls, mild asthmatics, and severe asthmatics were quantitated by LM (n = 7 subjects per group). (E) Relative quantification of GSDMB-1 mRNA in primary human bronchial epithelial cells (BEC), alveolar epithelial cells (AEC), macrophages (Mac), smooth muscle cells (SMC), and fibroblasts (Fib). (F) RNA from bronchial epithelial cells was examined using primers specific to GSDMB-1, -2, -3, and -4 isoforms (Table S1). GAPDH mRNA was used as normalization control. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Fold-change is expressed as mean ± SEM and results are from three independent experiments.
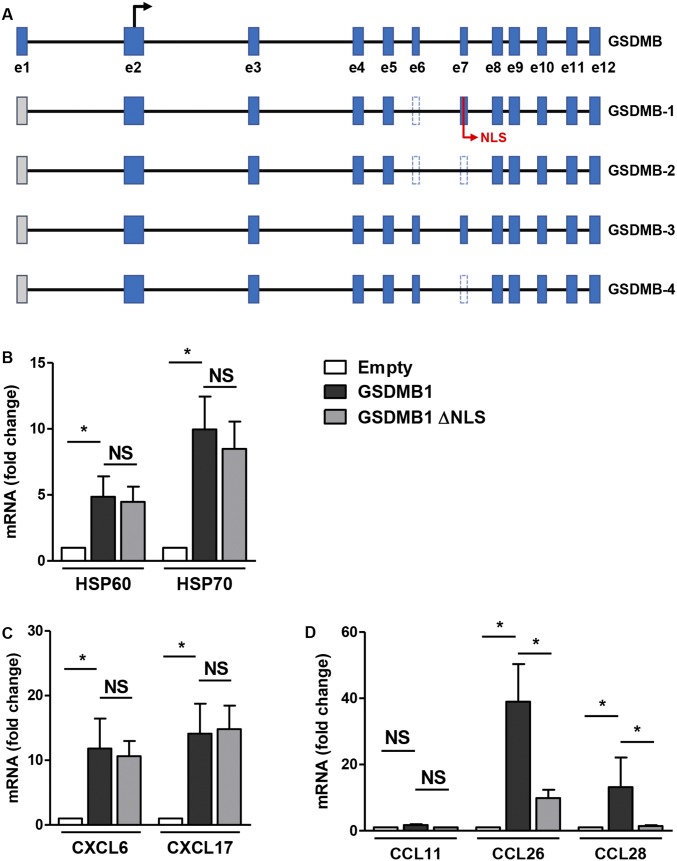
Fig. S1.
GSDMB-1 overexpression in primary human bronchial epithelium cells activates HSPs, CXC chemokines, and CC chemokines. (A) Schematic representation of the exon–intron structure and the alternative splicing isoforms of the human GSDMB gene, as predicted by the National Center for Biotechnology Information (NCBI) database. A black arrow marks the translational start site of GSDMB in exon 2 (e2). Blue boxes represent the 12 coding exons (e1–e12), gray boxes show untranslated regions, and the introns are indicated by solid lines. The alternative processing of exons 6 and 7 (e6 and e7) in the GSDMB isoforms 1, 2, and 4 are represented by dotted lines. The nuclear localization signal is depicted as NLS. (B–D) Primary bronchial epithelial cells were transfected with either empty, GSDMB-1, or GSDMB-1 vector lacking nuclear localization signal (GSDMB-1 ΔNLS) for 72 h. qPCR was performed to measure the mRNA levels of (B) HSPs, HSP60 and HSP70; (C) CXC chemokines, CXCL6 and CXCL17; and (D) CC chemokines, CCL11, CCL26, and CCL28. β-Actin mRNA was used as normalization control and fold-change is expressed as mean ± SEM from four independent experiments. *P < 0.05; NS, not significant.
GSDMB-1 Up-Regulates in Vitro Expression of Remodeling Genes (TGF-β1, 5-LO, MMP9), CC Chemokines (Eotaxin-3, CCL28), CXC Chemokines (CXCL6, CXCL17), and Heat-Shock Proteins (HSP60, HSP70) in Human Bronchial Epithelial Cells.
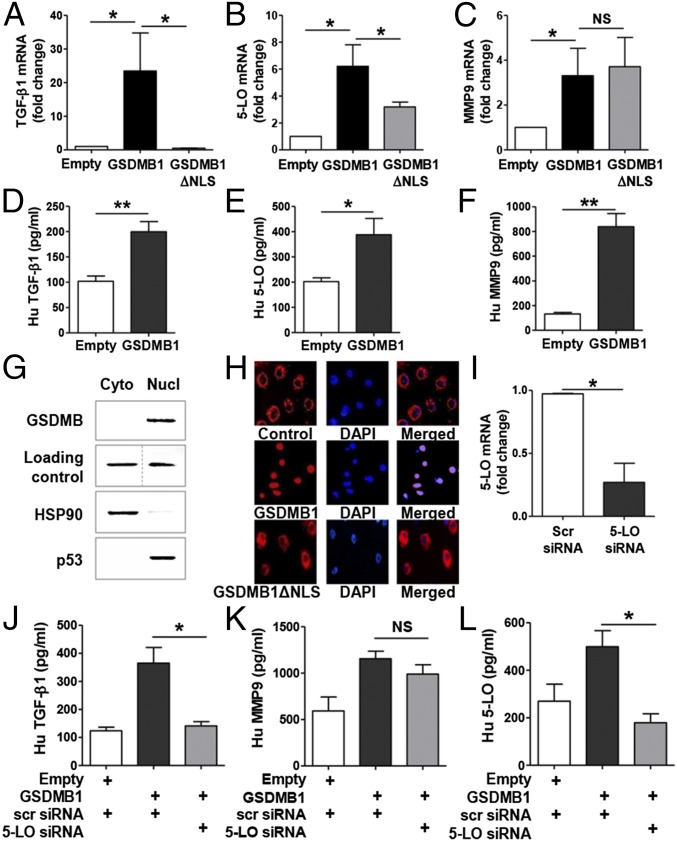
Because GSDMB-1 showed the highest level of expression among the four isoforms of GSDMB in bronchial epithelial cells, we focused our in vitro study on the GSDMB-1 isoform. We investigated the effect of GSDMB-1 overexpression in primary human bronchial epithelial cells on the levels of several genes important to airway remodeling and airway inflammation in asthma. There was a significant increase in levels of key remodeling genes, such as TGF-β1, 5-lipoxygenase (5-LO), and matrix metalloproteinase 9 (MMP9) mRNA as assessed by qPCR (Fig. 2 A–C), as well as a significant increase in TGF-β1, 5-LO, and MMP9 protein levels in GSDMB-1–overexpressing cells compared with empty vector-overexpressing control cells (Fig. 2 D–F).
Fig. 2.
Nuclear localization of GSDMB activates TGF-β1, 5-LO, and MMP9 in human bronchial epithelial cells transfected with GSDMB. Primary bronchial epithelial cells were transfected with either empty, GSDMB-1, or GSDMB-1 vector lacking nuclear localization signal (GSDMB-1 ΔNLS) for 72 h and qPCR was performed to measure (A) TGF-β1, (B) 5-LO, and (C) MMP9 mRNA. β-Actin mRNA was used as normalization control. Fold-change is expressed as mean ± SEM from four independent experiments. ELISA was performed to measure (D) active TGF-β1, (E) 5-LO, and (F) MMP9 levels in cells transfected with GSDMB-1 or empty vector. (G) Quantification of cellular localization of GSDMB in bronchial epithelial cells by Western blot. β-Tubulin and histone H3 were used as loading controls for cytoplasmic and nuclear extracts, respectively. The purity of these extracts was checked using HSP90, a cytoplasmic marker, and p53, a nuclear marker. Western blot image is a representative of three independent experiments. (H) Bronchial epithelial cells were transfected with RFP-tagged GSDMB-1, GSDMB-1 ΔNLS, or empty vector (red) and cells were mounted with DAPI (blue). (Magnification: 20×.) (I–L) Bronchial epithelial cells overexpressing GSDMB-1 or empty vector were transfected with scrambled (scr.) or 5-LO siRNA. (I) Efficiency of 5-LO mRNA knockdown was assessed by qPCR. β-Actin mRNA was used as normalization control. Protein levels of (J) active TGF-β1, (K) MMP9, and (L) 5-LO were measured by ELISA. Results are expressed as mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01; NS, not significant.
Because GSDMB has been previously demonstrated to interact with heat-shock protein 90 (HSP90) in cancer cells (7), we investigated whether GSDMB-1 could regulate the levels of HSPs in epithelial cells. Because HSP60 and HSP70 have been previously associated with asthma (9), we examined the effect of GSDMB-1 overexpression on these genes and found a significant increase in HSP60 and HSP70 mRNA levels (Fig. S1B). In addition, because ORMDL3 regulates expression of CC and CXC chemokines in epithelial cells (4), we examined whether GSDMB also regulated the expression of chemokines. GSDMB-1 overexpression in human bronchial epithelial cells induced a significant increase in expression of CXC chemokines, CXCL6 (granulocyte chemotactic protein 2 or GCP-2) and CXCL17 (VEGF coregulated chemokine 1 or VCC-1) (Fig. S1C). We also found an increase in expression of CC chemokines, CCL26 (eotaxin-3) and CCL28 (mucosa-associated epithelial chemokine or MEC) mRNA but no change in the levels of CCL11 (eotaxin-1) (Fig. S1D).
GSDMB-1 Localizes to the Nucleus in Human Bronchial Epithelium.
Our studies of the cellular localization of ORMDL3 in the endoplasmic reticulum (ER) provided important insights into ORMDL3’s function (4, 5), because we demonstrated that ORMDL3 activates only one of the three pathways of the ER unfolded response (i.e., the ATF-6α pathway) and regulates Serca2b (a calcium pump localized in the sarcoplasmic ER). We have used a similar approach to examine the cellular expression of GSDMB in human bronchial epithelial cells and made the observation that GSDMB-1 is localized in the nucleus by using two approaches: (i) fractionating epithelial cells into cytoplasmic and nuclear components (Fig. 2G), and (ii) transfection of red fluorescent protein (RFP)-labeled GSDMB-1 into bronchial epithelial cells to see where it localizes compared with the RFP-labeled empty vector (Fig. 2H).
Western blots of the fractionated epithelial cell compartments demonstrated that GSDMB-1 was highly expressed in the nucleus, but not expressed in the cytoplasm. We validated the purity of our nuclear and cytoplasmic fractions by demonstrating that the nuclear extracts contained high levels of the nuclear histone protein p53 (not detected in the cytoplasmic fraction), whereas the cytoplasmic extracts contained high levels of the cytoplasmic HSP (HSP90), which was not detected in the nuclear fraction (Fig. 2G). In addition, RFP-labeled GSDMB-1 localized to the nucleus, whereas the empty RFP-labeled vector remained in the cytoplasm (Fig. 2H).
GSDMB-1 Requires Nuclear Localization to Up-Regulate Key Remodeling Gene TGF-β1 in Human Bronchial Epithelial Cells.
Because previous studies have demonstrated the presence of a nuclear localization signal (NLS) in GSDMB-1 (10), we investigated whether the NLS is critical in regulating remodeling genes, chemokines, and HSPs. We observed that whereas overexpression of tagged GSDMB-1 in epithelial cells showed nuclear localization, the GSDMB-1 construct with the deleted NLS (GSDMB-1 ΔNLS) showed cytoplasmic localization similar to the control plasmid (Fig. 2H). Interestingly, overexpression of GSDMB-1 ΔNLS in bronchial epithelial cells failed to induce TGF-β1 mRNA compared with full-length GSDMB-1, indicating that nuclear localization of GSDMB-1 is critical for GSDMB-1–mediated induction of TGF-β1 (Fig. 2A). In contrast, nuclear localization of GSDMB-1 was only partially required for GSDMB-1 induction of 5-LO mRNA (Fig. 2B) and was not required for induction of MMP9 mRNA (Fig. 2C).
To understand the potential function of nuclear localized GSDMB-1, we performed GSDMB protein modeling and predictions using the GenBank GSDMB amino acid sequence, in conjugation with the Phyre2 bioinformatics tool (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The Phyre2 bioinformatics tools did not predict any DNA binding motifs in GSDMB, indicating that GSDMB may not function as a transcription factor. Protein sequence and secondary structure analysis using Phyre2 software predicted a plausible transcription initiation factor and chaperone domains in GSDMB with a confidence level of 42%, indicating that it might be involved in the regulation of expression of genes, such as TGF-β1, by acting as either a transcriptional coactivator or enhancer.
5-LO Is Critical for GSDMB-1 Mediated Up-Regulation of TGF-β1 in Human Bronchial Epithelial Cells.
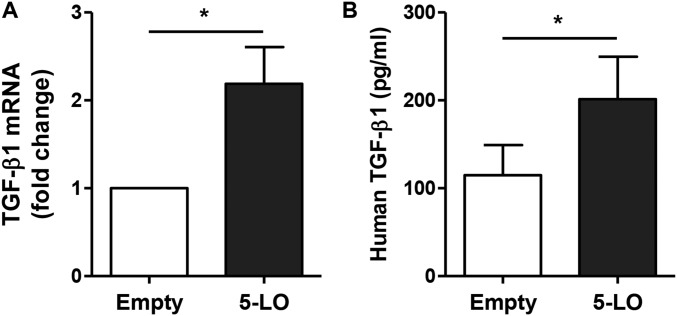
Because we demonstrated that GSDMB up-regulates remodeling genes, such as TGF-β, 5-LO, and MMP9, we performed small-interfering RNA (siRNA) knockdown of 5-LO to understand the relationship between TGF-β1, 5-LO, and MMP9 during GSDMB-1 up-regulation. We observed that 5-LO knockdown after GSDMB-1 overexpression in bronchial epithelial cells significantly down-regulates TGF-β1 protein levels (Fig. 2J). In contrast, 5-LO knockdown did not have a significant effect on MMP9 protein levels (Fig. 2K), indicating that 5-LO is critical for up-regulation of TGF-β1 but not required for MMP9. We also observed a significant down-regulation in 5-LO mRNA (Fig. 2I) and protein levels (Fig. 2L), thus demonstrating that 5-LO siRNA knockdown was efficient in primary bronchial epithelial cells. Additionally, 5-LO overexpression alone was sufficient to induce TGF-β1 mRNA and protein expression (Fig. S2).
Fig. S2.
Overexpression of 5-LO in primary human bronchial epithelium cells induces TGF-β1 expression. Primary human bronchial epithelial cells were transfected with either empty vector or 5-LO–overexpressing vector for 72 h. Transfected cells were examined to assess the levels of (A) TGF-β1 mRNA by qPCR and β-actin mRNA was used as the normalization control. (B) Active TGF-β1 protein levels were measured by ELISA. *P < 0.05 and results are expressed as mean ± SEM from two independent experiments.
Mice Expressing hGSDMB Transgene Exhibit Increased Airway Responsiveness Without Airway Inflammation.
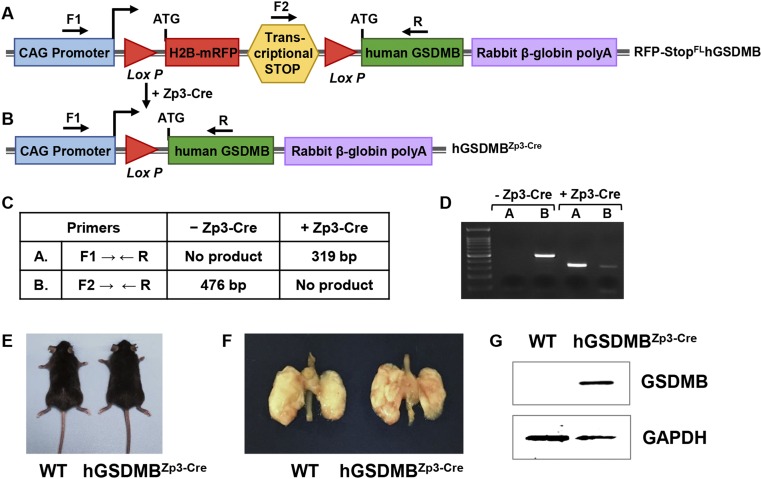
To understand the potential role of GSDMB in relation to asthma in vivo, we generated hGSDMBZp3-Cre mice, as detailed in SI Materials and Methods. Briefly, conditional hGSDMB transgenic (Tg) floxed mice (RFP-StopFLGSDMB-Tg) were crossed with zona pellucida 3 (Zp3) Cre mice, resulting in offspring expressing hGSDMB in all cells (hGSDMBZp3-Cre mice). The presence of loxP-flanked RFP and a transcriptional stop codon at the transcriptional initiation site of the hGSDMB transgene prevents transcription of hGSDMB until crossed with a Cre-mouse, which excises the transcriptional stop codon and histone H2B (H2B)-RFP (Fig. S3 A and B). DNA from hGSDMBZp3-Cre mice and littermate controls was used to confirm the presence or absence of the transgene by PCR (Fig. S3 C and D).
Fig. S3.
Generation of mice expressing human GSDMB transgene (hGSDMBZp3-Cre). (A–D) Generation of hGSDMBZp3-Cre mice. (A) The human GSDMB Tg construct contains a CAG promoter for universal overexpression (blue), H2B-RFP (red), followed by a transcriptional stop site (yellow), the human GSDMB ORF (green), and rabbit β-globin polyadenylation sequence (purple). The H2B-RFP and transcriptional stop sites are flanked by LoxP sites (red triangles). Cells containing this pCAGEN Lox RFP-H2B STOP Lox hGSDMB construct will express RFP and not hGSDMB, as the transcriptional stop codon (yellow) prevents transcription of hGSDMB. (B) Expression of the hGSDMB gene is Cre recombinase-dependent because the transcriptional stop codon preventing hGSDMB expression and H2B-RFP are excised by Cre recombinase via the LoxP sites. This results in overexpression of hGSDMB only in those cells that express Cre recombinase. (C) Primer sets (F1, R; F2, R) used to detect progeny of RFP-StopFLhGSDMB-Tg mice crossed with Zp3-Cre mouse and predicted sizes of transgene DNA as assessed by PCR. (D) PCR products run on 1.5% agarose gel, showing a PCR product band for hGSDMBZp3-Cre mice. (E) WT mice (Left) and hGSDMBZp3-Cre mice (Right) aged 12 wk appeared morphologically similar. (F) The gross appearance of lungs of WT mice (Left) and hGSDMBZp3-Cre mice (Right) aged 12 wk appeared morphologically similar. (G) Lung extracts from WT and hGSDMBZp3-Cre mice were examined by Western blot to quantitate the expression of human GSDMB protein. Mouse GAPDH was used as the loading control. Western blot image is a representative of two independent experiments.
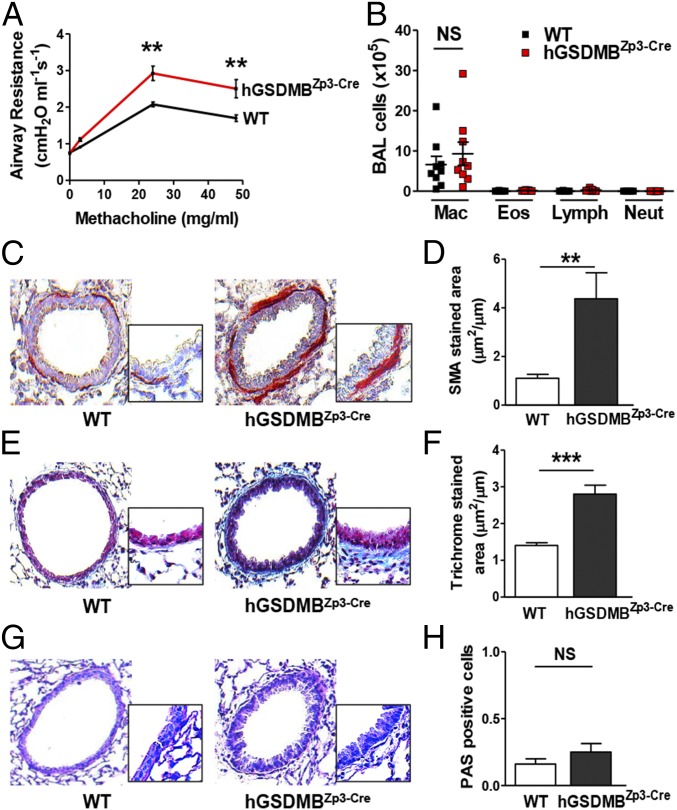
No developmental or morphologic defects were observed in hGSDMBZp3-Cre mice, and their lung size and weights were similar to those of WT mice (Fig. S3 E and F). A significant increase in hGSDMB protein levels was detected in hGSDMBZp3-Cre mouse lungs (Fig. S3G) but no changes in mouse Gsdma, -c, -d (mice do not have a GSDMB gene) was observed in hGSDMBZp3-Cre mice compared with WT mice (Fig. S4A). Interestingly, hGSDMBZp3-Cre mice exhibit a spontaneous increase in airway responsiveness to methacholine (MCh) compared with WT mice (Fig. 3A). However, we did not observe any significant changes in bronchoalveolar lavage (BAL) inflammatory cells associated with the increased AHR (Fig. 3B).
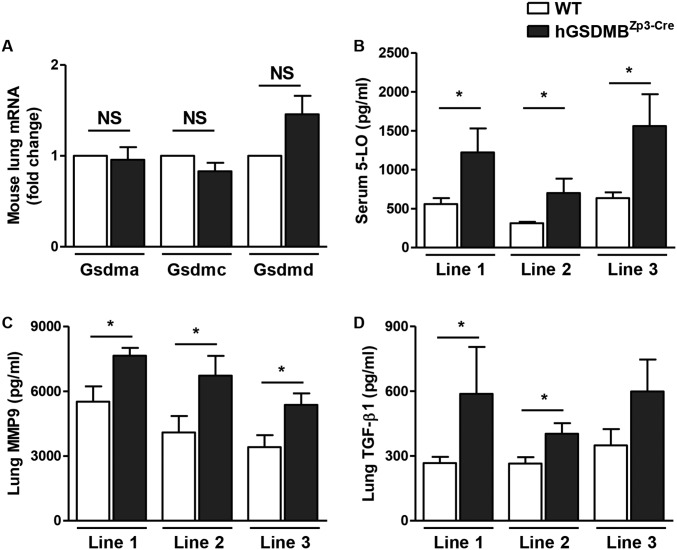
Fig. S4.
Multiple hGSDMBZp3-Cre mouse lines does not show any significant differences in pathways associated with airway remodeling in asthma. (A) Lungs of WT and hGSDMBZp3-Cre mice were examined to assess the levels of GSDM family genes, Gsdma, Gsdmc, and Gsdmd by qPCR. β-Actin mRNA was used as the normalization control. (B–D) To exclude the random genomic integration or copy number contributing to the observed phenotype in hGSDMBZp3-Cre mice, three lines of GSDMB transgenic animals (lines 1, 2, and 3) were examined to assess the protein levels of (B) serum 5-LO, (C) lung MMP9, and (D) lung TGF-β1 by ELISA. Results are expressed as mean ± SEM from four independent experiments. *P < 0.05; NS, not significant; n = 9–12 mice per group.
Fig. 3.
hGSDMBZp3-Cre mice exhibit increased AHR, with an increase in peribronchial smooth muscle and collagen deposition without airway inflammation. (A) AHR to MCh and (B) BAL inflammatory cells were assessed in intubated and ventilated WT and hGSDMBZp3-Cre mice. (C) Levels of peribronchial smooth muscle were quantitated by immunohistochemistry using an anti–α-smooth muscle actin Ab and (D) image analysis. Results are expressed as the α-smooth muscle actin-stained area (μm2) per circumference (μm) of basement membrane of bronchioles, 150- to 250-μm internal diameter in WT and hGSDMBZp3-Cre mice. (E) Levels of peribronchial trichrome staining were imaged using LM and (F) similarly quantitated by image analysis. (G) Levels of mucin were examined by PAS staining and (H) quantitated by image analysis. (Magnification: C, E, and G, 200×; Insets, 400×.) **P < 0.01; ***P < 0.001; NS, not significant. n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
Mice Expressing hGSDMB Transgene Exhibit Increased Peribronchial Smooth Muscle and Collagen Deposition.
Because we did not observe any changes in BAL inflammatory cells in hGSDMBZp3-Cre mice, we examined whether increased peribronchial smooth muscle or other pathways could contribute to AHR. Interestingly, we detected several features of airway remodeling in hGSDMBZp3-Cre mice, including an increase in the area of peribronchial smooth muscle as assessed by immunostaining with an anti–α-smooth muscle actin Ab (Fig. 3 C and D). In addition, there was an increase in peribronchial fibrosis as assessed by the area of peribronchial trichrome staining to detect lung collagen (Fig. 3 E and F). There was no significant increase in mucin expression as detected by periodic acid Schiff (PAS) staining (Fig. 3 G and H).
Mice Expressing hGSDMB Transgene Have Increased Expression of Pathways Associated with Airway Remodeling in Asthma: TGF-β1, MMP9, 5-LO, and Cysteinyl Leukotrienes.
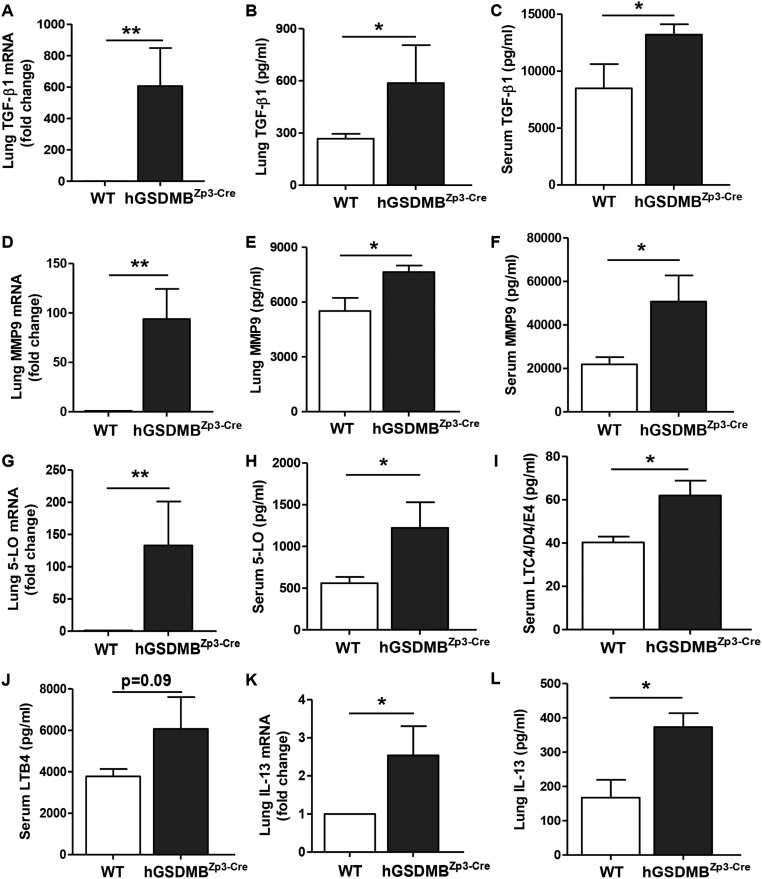
We quantitated levels of 5-LO, TGF-β1, and MMP9 mRNA in hGSDMBZp3-Cre mouse lungs (Figs. S4 B–D and S5 A–J) because these genes were up-regulated in human bronchial epithelial cells upon in vitro GSDMB-1 overexpression. We found a significant increase in TGF-β1 lung mRNA, lung protein, and serum levels in hGSDMBZp3-Cre mice lungs compared with WT mice (Fig. S5 A–C). hGSDMBZp3-Cre mice also exhibited a significant increase in MMP9 lung mRNA, lung protein, and serum levels (Fig. S5 D–F). We also observed a significant increase in 5-LO lung mRNA and 5-LO protein levels, in conjunction with a significant increase in downstream mediators of 5-LO pathway [i.e., cysteinyl leukotrienes (LT) LTC4, LTD4, and LTE4] (Fig. S5 G–I) and a trend for increase in LTB4 levels (P = 0.09) (Fig. S5J). Because previous studies demonstrated that the 5-LO pathway plays an important role in the pathogenesis of IL-13–induced remodeling (11), we examined the levels of IL-13 and found a significant increase in lung IL-13 mRNA and protein levels in hGSDMBZp3-Cre mice (Fig. S5 K and L). hGSDMBZp3-Cre mice also showed an increase in total serum IgE levels (Fig. S6A).
Fig. S5.
Lungs of hGSDMBZp3-Cre mice have increased expression of pathways associated with airway remodeling in asthma: TGF-β1, MMP9, 5-LO, and cysteinyl leukotrienes. WT and hGSDMBZp3-Cre mice were examined to assess the levels of (A) lung TGF-β1 mRNA by qPCR. ELISA was used to measure the levels of (B) lung TGF-β1 and (C) serum TGF-β1. Levels of (D) lung MMP9 mRNA were measured by qPCR, and the protein levels of (E) lung MMP9 and (F) serum MMP9 were measured by ELISA. (G) Levels of lung 5-LO mRNA in mouse lungs were measured by qPCR. (H) Levels of 5-LO and downstream pathway mediators, (I) cysteinyl leukotrienes (LTC4/D4/E4), and (J) LTB4 were measured in serum samples from WT and hGSDMBZp3-Cre mice by ELISA. Levels of (K) lung IL-13 mRNA were measured by qPCR and (L) lung IL-13 protein levels were measured by ELISA. β-Actin mRNA was used as normalization control for qPCR analyses and results are expressed as mean ± SEM from four independent experiments. *P < 0.05; **P < 0.01; NS, not significant; n = 9–12 mice per group.
Fig. S6.
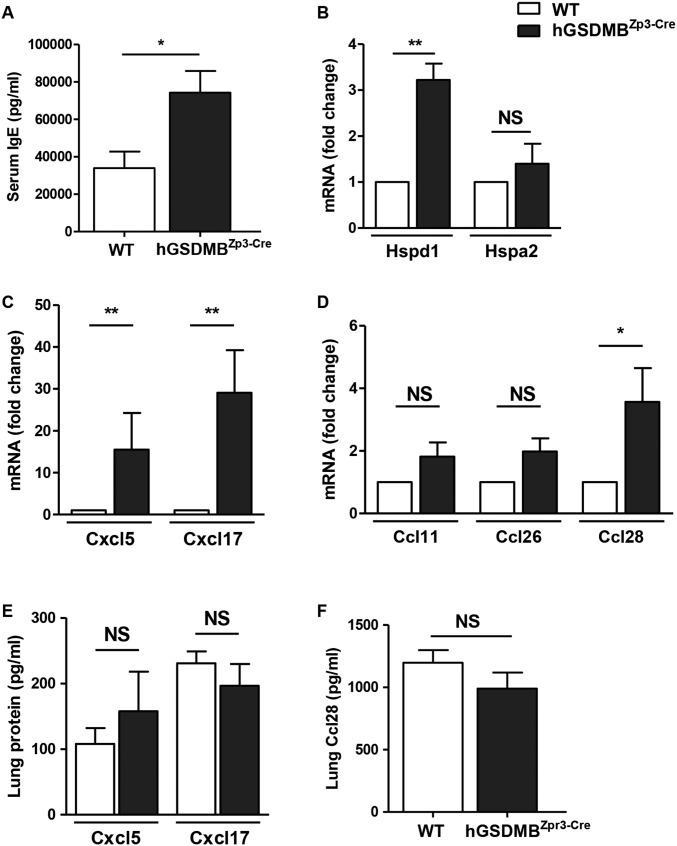
hGSDMBZp3-Cre mice have increased levels of serum IgE, lung HSP Hspd1, CXC chemokines Cxcl5 and Cxcl17, and CC chemokine Ccl28. (A) Levels of total IgE in WT and hGSDMBZp3-Cre mice were measured in serum samples by ELISA. (B–D) Lungs of WT and hGSDMBZp3-Cre mice were examined by qPCR to assess the levels of (B) mouse HSPs, Hspd1 (mouse ortholog of human HSP60) and Hspa2 (mouse ortholog of human HSP70), (C) mouse CXC chemokines Cxcl5 (mouse ortholog of human CXCL6) and Cxcl17, and (D) mouse CC chemokines Ccl11 (mouse ortholog of human eotaxin-1), Ccl26 (mouse ortholog of human eotaxin-3), and Ccl28. β-Actin mRNA was used as the normalization control for qPCR analyses. Lung protein extracts from WT and hGSDMBZp3-Cre mice were examined by ELISA to assess the protein levels of (E) mouse Cxcl5 and Cxcl17, and (F) mouse Ccl28. *P < 0.05; **P < 0.01; NS, not significant; n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
We also examined hGSDMBZp3-Cre mouse lungs for other genes (i.e., chemokines and HSPs) that were up-regulated in human bronchial epithelial cells upon in vitro GSDMB-1 overexpression. Levels of Hspd1 (mouse ortholog of HSP60) but not Hspa2 (mouse ortholog of HSP70) were found to be up-regulated in mouse lungs (Fig. S6B). We also found a significant increase in mRNA expression of CXC chemokines Cxcl5 (mouse ortholog of CXCL6) and Cxcl17 (Fig. S6C). There was a significant increase in the mRNA levels of CC chemokine Ccl28 (also known as MEC), but no change in eotaxin-1 (Ccl11) and eotaxin-3 (Ccl26) mRNA levels (Fig. S6D). Although we observed a significant increase in Cxcl5, Cxcl17, and Ccl28 mRNA, we did not observe any significant differences in Cxcl5, Cxcl17, and Ccl28 protein levels (Fig. S6 E and F).
Dust Mite Allergen-Challenged Mice Expressing hGSDMB Transgene Exhibit Increased AHR, Airway Inflammation, Th2 Cytokines, and IgE.
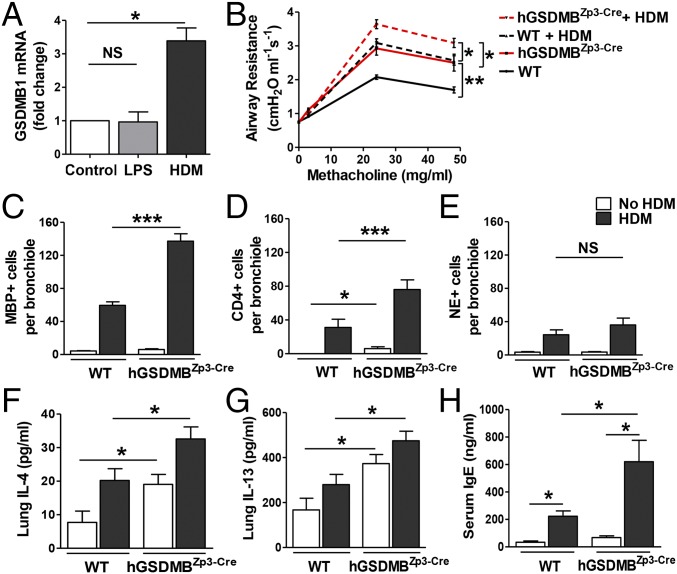
To examine the effect of house dust mite (HDM) allergen on GSDMB mRNA expression in epithelial cells, we stimulated primary human bronchial epithelial cells with HDM in vitro and found a significant increase in GSDMB-1 mRNA expression (Fig. 4A). Although LPS is a component in dust mites, LPS alone did not induce GSDMB-1 expression.
Fig. 4.
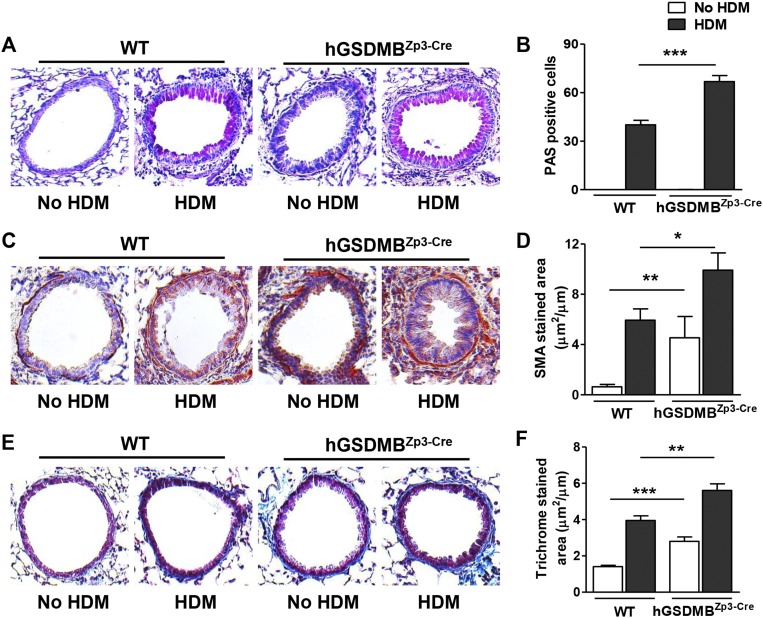
Mice expressing human GSDMB transgene exhibit increased AHR, airway inflammation, Th2 cytokines, and IgE upon dust mite allergen challenge. (A) Primary human bronchial epithelial cells were incubated with either HDM (25 μg/mL) or LPS (100 ng/mL) for 48 h and GSDMB-1 mRNA levels were measured by qPCR. Cells treated with media alone served as experimental control. β-Actin mRNA was used as normalization control and fold-change is expressed as mean ± SEM. (B) WT and hGSDMBZp3-Cre mice were administered either HDM extract (100 μg) or PBS via intranasal challenges on days 0, 7, 14, and 21. AHR to MCh was assessed on day 24 in intubated and ventilated mice. Lungs of WT and hGSDMBZp3-Cre mice challenged with or without HDM were examined for the number of lung (C) MBP+ peribronchial eosinophils, (D) CD4+ T cells, and (E) NE+ neutrophils per bronchiole of 150- to 250-μm internal diameter by immunohistochemistry and quantitated by image analysis. Levels of (F) lung IL-4, (G) lung IL-13, and (H) total serum IgE were quantitated by ELISA. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
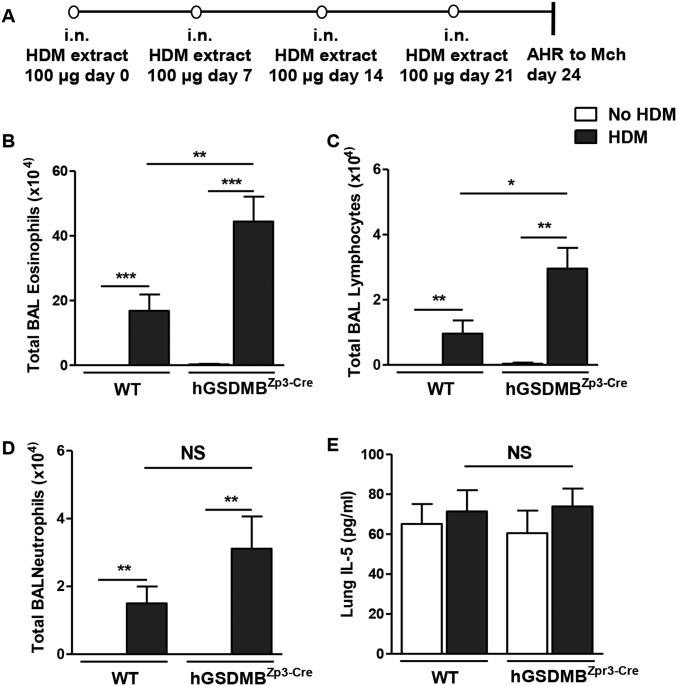
We used a mouse model to examine the effect of HDM allergen challenge on AHR, airway inflammation, and airway remodeling in hGSDMBZp3-Cre mice (Fig. S7A). There was a significant increase in AHR in hGSDMBZp3-Cre mice challenged with HDM compared with challenged WT mice (Fig. 4B). This increase in AHR was associated with a significant increase in peribronchial major basic protein (MBP)+ eosinophils (Fig. 4C) and CD4+ cells (Fig. 4D) in HDM-challenged hGSDMBZp3-Cre mice, indicating that increased GSDMB expression in vivo exacerbates HDM allergen-mediated airway inflammation. There was no significant difference in neutrophil elastase (NE+) lung neutrophils (Fig. 4E) in HDM-challenged hGSDMBZp3-Cre mice compared with WT mice. As observed in the lung tissue, there was a significant increase in BAL eosinophils and BAL lymphocytes, but not BAL neutrophils in HDM-challenged hGSDMBZp3-Cre compared with WT mice (Fig. S7 B–D).
Fig. S7.
hGSDMBZp3-Cre mice exhibit increased infiltration of inflammatory cells upon dust mite allergen challenge. (A) Schematic representation of mouse model of HDM-induced asthma. WT and hGSDMBZp3-Cre mice aged 8–9 wk were administered either HDM extract (100 μg) or PBS via intranasal (i.n.) challenges on days 0, 7, 14, and 21. AHR to MCh was assessed on day 24 in intubated and ventilated mice (flexiVent ventilator, Scireq). The number of (B) eosinophils, (C) lymphocytes, and (D) neutrophils in BAL was assessed by Giemsa staining. Differential cell counts were determined by light microscopy. (E) Levels of lung IL-5 in WT and hGSDMBZp3-Cre mice were quantitated by ELISA. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
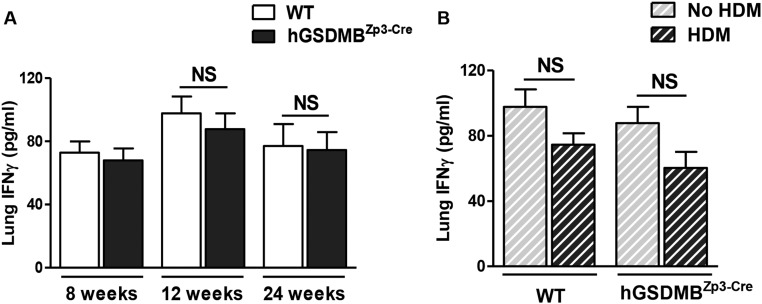
We measured lung Th2 cytokines, IL-4 and IL-13, and found a significant increase in HDM-challenged hGSDMBZp3-Cre mice compared with challenged WT mice (Fig. 4 F and G). However, we did not observe any significant difference in lung IL-5 levels (Fig. S7E). We also found a significant increase in total serum IgE levels in HDM-challenged hGSDMBZp3-Cre mice compared with challenged WT mice (Fig. 4H). There was no significant lung IFN-γ response in unchallenged or HDM-challenged hGSDMBZp3-Cre mice compared with WT mice (Fig. S8).
Fig. S8.
Lungs of hGSDMBZp3-Cre mice did not show a significant IFN-γ response with or without HDM allergen challenge. Lungs of WT and hGSDMBZp3-Cre mice were examined to assess the levels of IFN-γ. (A) Lung IFN-γ protein levels were measured by ELISA in mice aged 8, 12, and 24 wk of age without HDM-challenge. (B) Lung IFN-γ levels were measured in HDM-challenged mice at 12 wk of age by ELISA. NS, not significant; n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
HDM Challenge in hGSDMBZp3-Cre Mice Induced Mucin and Exacerbates Peribronchial Smooth Muscle and Collagen Deposition.
Interestingly, although unchallenged hGSDMBZp3-Cre mice did not exhibit increased mucin expression, HDM-challenged hGSDMBZp3-Cre mice showed an increase in mucin expression compared with WT mice, as detected by PAS staining (Fig. S9 A and B). HDM-challenged hGSDMBZp3-Cre mice had a significant increase in airway remodeling compared with hGSDMBZp3-Cre mice, as evident in the further increased thickness of the peribronchial smooth muscle layer (Fig. S9 C and D), and further increased level of peribronchial fibrosis (Fig. S9 E and F).
Fig. S9.
HDM challenge in hGSDMBZp3-Cre mice induced airway inflammation, and further increased peribronchial smooth muscle and collagen deposition. Lungs of WT and hGSDMBZp3-Cre mice challenged with or without HDM were examined for (A) the levels of mucin by PAS staining and (B) quantitated by image analysis. (C) Levels of peribronchial smooth muscle were quantitated by immunohistochemistry using an anti–α-smooth muscle actin Ab and (D) image analysis. Results are expressed as the α-smooth muscle actin-stained area (μm2) per circumference (μm) of basement membrane of bronchioles, 150- to 250-μm internal diameter in WT and hGSDMBZp3-Cre mice. (E) Levels of peribronchial trichrome staining were imaged using LM and (F) similarly quantitated by image analysis. (Magnification: A, C, and E, 200×.) *P < 0.05; **P < 0.01; ***P < 0.001; n = 9–12 mice per group. Results are expressed as mean ± SEM from four independent experiments.
Overall, our results indicate that allergen challenge in hGSDMBZp3-Cre mice exacerbates airway inflammation, IgE levels, the thickness of the peribronchial smooth muscle layer, collagen deposition, mucin, and AHR.
SI Materials and Methods
Expression of GSDMB in Human Asthma and Control Lungs.
Human lungs from asthmatics and normal individuals.
Postmortem human lungs from asthmatics and normal organ donors were procured by the Arkansas Regional Organ Recovery Agency and by the National Disease Research Interchange, and delivered to the Lung Cell Biology Laboratory at the Arkansas Children’s Hospital Research Institute under conditions identical to those used for transplant. The acquisition of deceased donor tissue was reviewed by the University of Arkansas for Medical Sciences Institutional Review Board and determined not to be human subject research. After processing, tissue was shipped to San Diego under a protocol approved by the University of California, San Diego Human Research Protections program.
Lung sections were stained by immunohistochemistry using an anti-GSDMB Ab (Abgent) or species-specific isotype control Ab, as previously described (30). Quantification of GSDMB+ staining in epithelial cells was performed for each subject using an image analysis system (Image-Pro plus, Media Cybernetics) as previously described (30), and results were expressed as GSDMB+ stained area per square micrometer.
Bronchial biopsy.
The protocols using bronchoscopy to obtain bronchial biopsies from patients with mild or severe asthma and control nonasthmatics at McGill University and Université de Montréal were performed with the approval of the respective Institutional Review Boards as previously described (30). The methods for processing the bronchial biopsies and immunostaining of lung sections using the ABC immunoperoxidase method have also been previously described (30). In this study, bronchial biopsy sections from severe asthmatics (n = 7), mild asthmatics (n = 7), and nonasthmatic controls (n = 7) were immunostained with an anti-GSDMB Ab (Abgent) or species-specific isotype control Ab. The number of bronchial biopsy GSDMB+ cells were quantitated with image analysis (Image-Pro) in each subject, and results expressed as GSDMB+ cells per square millimeter, as previously described (30).
GSDMB Expression in Human Lung Cells.
Primary human bronchial epithelial cells, primary human alveolar epithelial cells, primary human smooth muscle cells, and primary human lung fibroblasts were purchased from ScienCell and cultured in vitro according to the manufacturer’s instructions. Primary BAL macrophages from postmortem donor lungs were obtained as described previously (31).
Detection of GSDMB and GSDMB-Regulated Genes by qRT-PCR.
qRT-PCR was performed as previously described in our laboratory (30). In brief, total RNA was extracted with RNA-STAT-60 and reverse-transcribed with Oligo-dT and SuperScript II kit (Life Technologies). qPCR was performed with TaqMan PCR Master Mix and TaqMan primers (Thermo Fisher Scientific) for human GSDMB isoforms GSDMB-1, GSDMB-2, GSDMB-3, GSDMB-4 (Table S1), TGF-β1, 5-LO, MMP9, CXC chemokines (CXCL6/GCP-2, CXCL17/VCC-1), CC chemokines (CCL11/eotaxin-1, CCL26/eotaxin-3, CCL28/MEC), and HSPs (HSP60, HSP70). The relative amounts of transcripts were normalized to those of housekeeping gene (β-actin) mRNA and compared between the different genes by ΔΔCt method, as previously described (31).
Table S1.
Primers used for qPCR analyses of GSDMB isoforms
| GSDMB isoform | RefSeq sequence | Assay sequence | Exon boundary | Catalog # and source |
| GSDMB-1 | NM_001042471.1 | Hs00938445_m1 | 6–7 | 4351372 |
| Thermo Fisher Scientific | ||||
| GSDMB-2 | NM_018530.2 | Hs00939390_m1 | 4–5 | 4351372 |
| Thermo Fisher Scientific | ||||
| GSDMB-3 | NM_001165958.1 | AIQJC0K (custom primer) | 6–7 | 4331348 |
| Thermo Fisher Scientific | ||||
| GSDMB-4 | NM_001165959.1 | Hs00940509_m1 | 6–7 | 4351372 |
| Thermo Fisher Scientific |
In Vitro Transfection of GSDMB-1 in Airway Bronchial Epithelial Cells.
Primary human bronchial epithelial cells (ScienCell) were transduced with either empty control plasmid, adenoviral-GSDMB-1–RFP, or adenoviral-GSDMB-1ΔNLS–RFP plasmid (lacking a 60-bp nuclear localization domain in exon 7) according to the manufacturer’s instructions (Vector Biolabs). Briefly, primary bronchial epithelial cells were seeded in collagen-coated six-well plates at 70% confluency and overlaid with either empty control plasmid (570 × 106 pfu/mL), adenoviral-GSDMB-1–RFP (750 × 106 pfu/mL), or adenoviral-GSDMB-1ΔNLS–RFP plasmid (900 × 106 pfu/mL) at 500 multiplicity of infection. Media were replaced with complete media after 4 h and gene transduction were evaluated after 72 h. Transduction efficiency was calculated on the basis of RFP-tagged GSDMB-1 expression.
In Vitro Transfection of Human 5-LO in Airway Bronchial Epithelial Cells.
Primary human bronchial epithelial cells (ScienCell) were transfected with either empty control (pCMV6-AC-GFP) or 5-LO overexpressing plasmid (pCMV6-ALOX5–GFP) using TurboFectin 8.0 transfection reagent according to the manufacturer’s instructions (Origene). Briefly, primary bronchial epithelial cells were seeded in collagen-coated 12-well plates at 70% confluency and overlaid with TurboFectin-DNA complex (ratio 4:1). Media were replaced with complete media after 6 h and transfection efficiency was evaluated after 72 h on the basis of GFP-tagged 5-LO expression.
In Vitro Knockdown of Human 5-LO in Airway Bronchial Epithelial Cells.
Following transfection of GSDMB-1 as described above, primary bronchial epithelial cells were transfected with human 5-LO siRNA according to the manufacturer’s instructions (Dharmacon). Briefly, GSDMB-1–RFP or empty control plasmid was overexpressed in primary bronchial epithelial cells for 24 h followed by transfection with Accell 5-LO siRNA or scrambled control (1 μM) using Accell delivery media. The siRNA complexes were removed after 4 h and replaced with complete media. Cell culture supernatants were collected and total cell extracts were prepared after 48 h (72 h after GSDMB-1 transfection). Protein levels of MMP9, 5-LO, and active TGF-β1 were measured using ELISA, as described below.
Bioinformatics Tools for Modeling GSDMB Protein Domains.
Protein modeling and predictions was performed for human GSDMB protein using the GSDMB amino acid sequence from GenBank, in conjugation with the Phyre bioinformatics tool. (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). Protein sequence analysis was performed as previously described (32).
Detection of GSDMB by Western Blot.
Cytoplasmic and nuclear protein extraction from primary bronchial epithelial cells was performed using NE-PER Nuclear and cytoplasmic extraction reagents (Thermo Scientific) according to the manufacturer’s instructions.
Proteins were separated on SDS/PAGE gel and transferred to a PVDF membrane. Membranes were blocked in 5% (wt/vol) milk in 1× Tris-buffered saline with Tween for 1 h and then incubated with the primary antibody overnight at 4 °C. The primary antibodies used in this study were rabbit polyclonal anti-GSDMB (Abgent), rabbit monoclonal anti–β-tubulin (Cell Signaling) as cytoplasmic loading control, and rabbit monoclonal anti-histone H3 (Cell Signaling) as a nuclear loading control. The purity of cytoplasmic and nuclear extracts was checked using primary antibodies to rabbit monoclonal anti-HSP90 (Cell Signaling), a cytoplasmic HSP, and rabbit monoclonal anti-p53 (Cell Signaling), a nuclear histone protein.
Generation of hGSDMBZp3-Cre Mice.
Because mice express three of the four GSDM family members (GSDMA, GSDMC, GSDMD), but not GSDMB, it is not possible to use genetic approaches to generate GSDMB-deficient mice. However, because the SNP linking chromosome 17q21 to asthma is associated with increased expression of human GSDMB, we generated mice expressing increased levels of human GSDMB (hGSDMBZp3-Cre). All of the mouse experimental protocols were approved by the University of California, San Diego Institutional Animal Care and Use Committee.
Zp3-Cre mice.
Zp3-Cre mice (embryonic Cre expression) on a C57BL/6 background were acquired from The Jackson Laboratory.
Targeting plasmid construction.
The hGSDMB Tg construct with pCAGEN lox RFP-H2B STOP lox hGSDMB was generated by cloning the 1,248-bp hGSDMB ORF (which represents the entire length of the human GSDMB gene) from pCMV6-AC-GSDMB (Origene) with Agel/Notl into a construct previously developed and provided by A. J. Holland and D. W. Cleveland of the Ludwig Institute for Cancer Research at the University of California, San Diego, CA.
RFP-StopFL-hGSDMB-Tg mouse generation.
Spel/Pvul-linearized pCAGEN lox RFP-H2B STOP lox hGSDMB was microinjected into the pronuclei of fertilized mouse embryos and implanted into a pseudopregnant mouse (all on a C57BL/6 background) via an established protocol by the University of California, San Diego Transgenic mouse core. The conditional hGSDMB Tg floxed mice (RFP-StopFLGSDMB-Tg) with the pCAGEN lox RFP-H2B STOP lox hGSDMB transgene construct were crossed with Zp3 Cre mice. This process resulted in offspring expressing human GSDMB in all cells (hGSDMBZp3-Cre mice). The transgene construct has a loxP-flanked RFP and transcriptional stop codon positioned at the transcriptional initiation site of the hGSDMB transgene, which prevents transcription of hGSDMB. Thus, all cells in this RFP-StopFL hGSDMB-Tg mouse will not express hGSDMB until crossed with a Cre-expressing mouse, which excises the transcriptional stop codon and H2B-RFP (Fig. S3 A and B). The presence of the nonexpressed floxed transgene construct in RFP-StopFL hGSDMB-Tg mice tissue was assessed by red fluorescence before crossing to Zp3-Cre mice. Crossing RFP-StopFL hGSDMB-Tg mice with Zp3-Cre mice resulted in the loss of RFP expression and provided a rapid initial screen for successful expression of hGSDMB. Because the levels of RFP detected by immunofluorescence microscopy varied, we used DNA from hGSDMBZp3-Cre mice and littermate controls to confirm the presence of the transgene by PCR (Fig. S3 C and D). Subsequent mouse genotyping was performed using PCR with the following primers: F1 (flox−) GCA ACG TGC TGG TTA TTG TG; F2 (flox+) CCC CCT GAA CCT GAA ACA TA; R (common) TGA GGC CTG TTG TGT AGT GC. These RFP-StopFL hGSDMB-Tg mice (C57BL/6 background) were crossed with Zp3-Cre mice (C57BL/6 background) to generate hGSDMBZp3-Cre mice (C57BL/6 background).
Expression of GSDM-family genes in hGSDMBZp3-Cre mice.
Protein expression of GSDMB in WT and hGSDMBZp3-Cre mice was measured using Western blot. GAPDH was used as the loading control (Fig. S3G). To determine the effect of hGSDMB transgene overexpression on other Gasdermin family genes (i.e., Gsdma, Gsdmc, and Gsdmd) we examined WT and hGSDMBZp3-Cre mouse lungs by qPCR, as previously described. We did not observe any significant increase in the mRNA expression of mouse Gsdma, Gsdmc, or Gsdmd in GSDMB Tg mouse lungs (Fig. S4A).
Screening of multiple hGSDMBZp3-Cre mouse lines.
To exclude the random genomic integration or copy number contributing to the observed phenotype in hGSDMBZp3-Cre mice, we generated and examined three lines of GSDMB transgenic animals (lines 1, 2, and 3). Because our in vitro studies demonstrated that GSDMB transfection into epithelial cells induces the expression of 5-LO, TGF-β1, and MMP9, we screened the lungs of the three hGSDMBZp3-Cre lines we generated for levels of expression of these genes. All three hGSDMBZp3-Cre mouse lines showed a significant increase in the levels of these three GSDMB target genes (Fig. S4 B–D). These studies suggest that random integration or copy number variation of GSDMB is unlikely to explain our results. We decided to phenotype in detail hGSDMBZp3-Cre line 1 as this GSDMB transgenic mouse line expressed similar levels of the three target genes (5-LO, TGF-β1, and MMP9) as the other two hGSDMBZp3-Cre lines.
Processing of Lungs, BAL, and Blood.
hGSDMBZp3-Cre mice and littermate controls were killed at 12 wk of age to quantitate levels of airway inflammation, airway remodeling, as well as expression of lung cytokines, chemokines, and remodeling genes, as described below. Levels of IgE and selected cytokines were quantitated in peripheral blood as described below.
Lung processing.
Lungs were processed for RNA and protein extraction, as well as paraffin-embedded for immunohistology, as previously described in this laboratory (30). For protein and RNA extractions, lungs were snap-frozen in liquid nitrogen and stored at −80 °C. For paraffin-embedded sections, lungs were first equivalently inflated with an intratracheal injection using the same volume of 4% paraformaldehyde solution (Sigma-Aldrich) to preserve the lung architecture.
BAL fluid collection.
BAL fluid was collected by lavaging the lung with 1 mL PBS via a tracheal catheter, as previously described (30). BAL fluids were centrifuged and the supernatants were frozen at −80 °C for subsequent cytokine analysis.
Peripheral blood.
Peripheral blood was collected from mice in tubes without anticoagulant by mice cardiac puncture. Serum samples were separated as previously described (30) and frozen at −80 °C for quantitation of serum IgE levels, cytokines and leukotrienes.
Detection of Airway Remodeling.
Peribronchial smooth muscle layer.
The thickness of the airway smooth muscle layer was measured by α-smooth muscle actin immunohistochemistry, as previously described (30). Lung sections were immunostained with an anti–α-smooth muscle actin primary antibody (Sigma-Aldrich) to detect peribronchial smooth muscle. Species- and isotype-matched Abs were used as controls instead of primary Ab. The area of peribronchial α-smooth muscle actin staining in paraffin-embedded lungs was outlined and quantified under a LM (Leica DMLS) attached to an image analysis system (Image-Pro plus), as previously described (30). Results are expressed as the area of peribronchial α-smooth muscle actin staining per micrometer length of the basement membrane of bronchioles 150–250 μm of internal diameter.
Peribronchial trichrome staining.
Paraffin-embedded lungs were analyzed by trichrome staining and the positively stained area was outlined and quantified under a LM (Leica DMLS, Leica Microsystems). Quantitative analysis was performed using an image analysis system (Image-Pro plus, Media Cybernetics), as previously described (30). Results are expressed as the area of trichrome staining per micrometer length of the basement membrane of bronchioles 150–250 μm of internal diameter.
Airway Mucus Expression.
To quantitate the level of mucus expression in the airway, the number of PAS+ and PAS− epithelial cells in individual bronchioles was counted, as previously described in this laboratory (30). At least 10 bronchioles were counted in each slide. Results are expressed as the percentage of PAS+ cells per bronchiole, which is calculated from the number of PAS+ epithelial cells per bronchus divided by the total number of epithelial cells of each bronchiole.
Quantitation of Cytokines and Leukotrienes by ELISA.
In vitro experiments.
Human bronchial epithelial cells transfected with plasmids for overexpressing GSDMB-1 and 5-LO, with or without 5-LO siRNA, were lysed in PBS containing protein inhibitor mixture (Thermo Scientific) and cell extracts were used for human 5-LO and MMP9 ELISAs. Active TGF-β1 in cell culture supernatants was measured by ELISA after acid activation (HCl) followed by neutralization (NaOH), as per the manufacturer’s protocol.
In vivo experiments.
Mouse lung tissue (50 μg) was homogenized in 500 μL of RIPA buffer with protein inhibitor mixture and 100 mg of stainless-steel beads (Next Advance) for 5–20 min using the Bullet Blender homogenizer (Next Advance). The lysate was centrifuged at 16,200 × g for 20 min at 4 °C. The supernatant was collected and used for ELISA. Mouse serum samples were used for ELISA quantitation of peripheral blood TGF-β1, MMP9, total IgE, 5-LO, and cysteinyl leukotrienes.
The ELISA kits used in this study were human and mouse TGF-β1 (R&D Systems), human and mouse 5-LO (LifeSpan BioSciences), mouse leukotriene C4/D4/E4 (Amersham, GE healthcare), mouse LTB4 (R&D Systems), mouse total MMP9 (R&D Systems), mouse total IgE (BD Biosciences), mouse Cxcl5 (R&D Systems), mouse Cxcl17 (LifeSpan BioSciences), mouse Ccl28 (R&D Systems), and mouse IL-4, IL-5, and IL-13 (R&D Systems). Levels of lung 5-LO, MMP9, TGF-β1, Cxcl5, Cxcl17, Ccl28, IL-4, IL-5, and IL-13 are expressed as the amount of cytokines in picogram per milligram lung protein. Lung protein extracts were quantitated by the BCA method (Thermo Scientific).
Immunohistochemistry and Quantitation of Lung Inflammation.
Lung sections were processed for immunohistochemistry to detect peribronchial eosinophils expressing MBP (anti-mouse MBP Ab, kindly provided by James Lee, Mayo Clinic, Scottsdale, AZ), peribronchial neutrophils expressing NE (anti-mouse NE Ab; Santa Cruz Biotechnology), peribronchial macrophages expressing F4/80 (anti-mouse F4/80 Ab; Santa Cruz Biotechnology), and peribronchial T cells expressing CD4 (anti-mouse CD4 Ab; GeneTex). The number of individual cells staining positive for different cell types in the peribronchial space was counted using a LM. Results are expressed as the number of peribronchial cells staining positive per bronchiole with 150–250 μm of internal diameter. n = 9–12 mice per group. At least 10 bronchioles were counted per mouse in each slide.
Mouse Model of HDM-Induced Asthma.
hGSDMBZp3-Cre and littermate control mice aged 8–9 wk were anesthetized with isoflurane and challenged intranasally with HDM extract (Dermatophagoides pteronyssinus, Greer laboratories, Lenoir, NC) in 50 μL PBS. Non–HDM-challenged hGSDMBZp3-Cre and littermate groups of mice were sensitized and challenged with PBS only. Mice were exposed to 100 μg HDM on days 0, 7, 14, and 21. Seventy-two hours after the last challenge (day 24), BAL fluid, blood, and lungs were collected as described above.
AHR to MCh.
Airway responsiveness to MCh was assessed in intubated and ventilated mice aged 12 wk (9–12 mice per group) using flexiVent ventilator (Scireq). Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) intraperitoneally, as previously described (30). The dynamic airway resistance in mice exposed to either nebulized PBS or MCh (3, 24, 48 mg/mL) was determined using Scireq software. The following ventilator settings were used: tidal volume (10 mL/kg), frequency (150/min), and positive end-expiratory pressure (3 cm H2O). Increased airway resistance values signal an increased stiffness of the lungs.
Statistical Analysis.
All results are presented as mean ± SEM. A statistical software package (GraphPad Prism) was used for the analysis. P < 0.05 was considered statistically significant.
Discussion
Because the SNP linking chromosome 17q21 is linked to increased expression of GSDMB (2, 8), we generated mice expressing increased levels of GSDMB and made the observation that these hGSDMBZp3-Cre mice had spontaneous increased AHR and increased peribronchial smooth muscle in the absence of airway inflammation. Although the classic paradigm for airway remodeling in asthma suggests that repeated episodes of inflammation of the airways results in airway remodeling, our studies with GSDMB suggest that this concept may need to be revised, not solely based on the evidence that GSDMB can induce airway smooth muscle remodeling and AHR in the absence of inflammation, but also based on several clinical studies (12, 13). For example, in preschool children with established severe asthma, increased airway smooth muscle mass and reticular basement thickening are present and is dissociated from inflammation (12, 13). The presence of increased airway smooth muscle mass in large and small airways in asthma is well described and is considered to be a major contributor to bronchoconstriction of airways and persistent airflow obstruction (14).
To understand the mechanism by which GSDMB induces increased airway remodeling and AHR, we performed studies examining which pathways were activated by GSDMB overexpression in vitro and in vivo. Our studies of epithelial cells transfected with GSDMB in vitro, as well as our studies of hGSDMBZp3-Cre mice in vivo, identified that GSDMB induced high levels of expression of 5-LO and TGF-β1, mediators associated with AHR and smooth muscle remodeling. The 5-LO products can directly induce smooth muscle contraction with rapid onset kinetics, whereas TGF-β1 can directly increase smooth muscle contractility with slow onset kinetics (15). Previous studies have demonstrated that TGF-β1 and leukotrienes can produce synergistic effects to increase smooth muscle contraction, collagen synthesis, and epithelial cell proliferation (16). For example, LTC4 can stimulate airway epithelial cells to increase TGF-β1 expression, which in turn can induce the expression of MMPs and tissue inhibitors of metalloproteinases, thus affecting airway remodeling (16). In addition to leukotrienes, there is considerable evidence that other GSDMB-regulated genes, such as TGF-β1 and MMP9, can directly contribute to airway remodeling in asthma (17). Increased TGF-β1 mRNA levels have been observed in bronchial biopsies of asthmatics compared with normal subjects, and levels of TGF-β1 correlate with the depth of subepithelial fibrosis (17, 18). Levels of MMP9 are significantly increased in BAL fluid, blood, and sputum from allergic asthmatic patients (19).
To understand the relationship between GSDMB, 5-LO, and TGF-β1, we performed 5-LO knockdown studies in GSDMB-1–overexpressing epithelial cells and established that 5-LO knockdown led to a significant down-regulation in TGF-β1 protein levels. These studies demonstrate that GSDMB-induced TGF-β1 expression is critically dependent upon 5-LO, suggesting a remodeling pathway induced by GSDMB. Because nuclear localization of GSDMB is essential for induction of TGF-β1, we used bioinformatics approaches to determine whether GSDMB could function as a transcription factor but did not identify any such DNA binding motifs in GSDMB. However, secondary structure analysis predicted a plausible transcription initiation factor and chaperone domains in GSDMB, indicating that it might be involved in the regulation of genes, such as TGF-β1, by either acting as a transcriptional coactivator or enhancer. However, further studies are needed to define whether GSDMB’s nuclear localization regulates TGF-β1 gene expression by acting as either a transcription initiation factor, chaperone, coactivator, chromatin remodeler, histone acetylase, deacetylase, kinase, or methylase, which can all play crucial roles in gene regulation but lack DNA-binding domains, and therefore are not classified as transcription factors (20).
We demonstrate the importance of the GSDMB-1 isoform in asthma because it shows the highest expression among the four isoforms of GSDMB (GSDMB-1, -2, -3, -4). Interestingly, the GSDMB isoform expressed in epithelial cells from the lungs of asthmatics (GSDMB-1) differs from the isoform expressed in cancer-derived epithelial cells (GSDMB-2) (7). Alternative splicing can have diverse effects on protein folding and function (21) and can affect protein–ligand interactions. Recent studies have demonstrated an important role of alternative splicing in several human diseases (22), thus underscoring the need to further explore the role of GSDMB isoforms in asthma. The four GSDMB isoforms differ in the presence or absence of exons 6 and 7 of the GSDMB gene. In cancer, GSDMB-2 (lacking exons 6 and 7) has effects on tumor growth and development (7). In contrast we demonstrate that in asthma, GSDMB-1 (the only isoform that lacks only exon 6), is significantly expressed. We decided in our initial in vivo studies to overexpress the whole GSDMB gene rather than an individual isoform, as this would allow us to determine whether any GSDMB isoform influenced the development of an asthma phenotype. Further studies are needed to understand the roles of the individual GSDMB isoforms in asthma.
Although leukocytes, such as eosinophils, are a major source of 5-LO products in asthma, studies have demonstrated that human bronchial epithelial cells significantly express an active 5-LO and inducible biosynthetic pathway for LTC4, with levels similar to blood leukocytes (23). A higher number of bronchial epithelial cells relative to eosinophils in the lung may allow the bronchial epithelium to be the most significant source of leukotrienes in asthma. In our immunohistochemistry studies of GSDMB expression in human lungs from asthmatics, we observed that both epithelial cells and peribronchial inflammatory cells expressed GSDMB, indicating that epithelial cells might not be the sole source of GSDMB in human lungs. We also observed a mild increase in the number of CD4+ cells/bronchus in naïve hGSDMBZp3-Cre mice compared with WT mice, which is associated with an increase in IL-4 and IL-13 levels. GSDMB is known to be expressed in T cells (2) and may be expressed in other inflammatory cells in the lung. Because this study examines whether increased GSDMB expression results in an asthma phenotype, we elected to initially generate universal transgenic mice with increased expression of GSDMB in all cells, including epithelial cells and T cells known to express GSDMB. Future studies using cell specific-Cre mice (such as CC10-Cre, or CD4-Cre mice) might be helpful to delineate distinct functions for epithelial cell- and T-cell–expressed GSDMB.
GSDMB-1 overexpression led to a significant increase in HSP60 and HSP70 mRNA, which have been implicated in inflammation and asthma (9). Increased HSP60 is noted in alveolar macrophages of asthmatics, whereas HSP70 has demonstrated implications in inflammatory immune responses (9). Levels of the mouse orthologs of HSP60 (Hspd1) but not HSP70 (Hspa2) were up-regulated in the lungs of hGSDMBZp3-Cre mice. We also investigated the effect of GSDMB-1 overexpression on selected chemokines involved in asthma and found a significant increase in mRNA levels of CCL26 (eotaxin-3), CCL28 (MEC), CXCL6 (GCP-2), and CXCL17 (VCC-1). Eotaxin-3 and CCL28 are up-regulated during an asthmatic response and is expressed on immune cells, such as eosinophils and T-cell subsets (24). CXC chemokines (such as CXCL6) can activate and attract neutrophils to the asthmatic airways (25). Elevated CXCL17 levels have been reported in lung fibrosis (26). Lung mRNA levels of mouse orthologs of CCL28 (Ccl28), CXCL6 (Cxcl5), and CXCL17 (Cxcl17) were up-regulated in hGSDMBZp3-Cre mice. Epithelial cells are known cellular sources of chemokines (27), which are the likely source of Ccl28, Cxcl5, and Cxcl17 in hGSDMBZp3-Cre mouse lungs. However, no significant changes in the protein levels of these chemokines were observed in hGSDMBZp3-Cre mice.
Because the human transgene in hGSDMBZp3-Cre mice is not regulated by the allergen challenge, we hypothesized that HDM-induced inflammatory and immune response would synergize with GSDMB-regulated genes (such as chemokines) expressed spontaneously in hGSDMBZp3-Cre mice. This process would simulate an asthmatic with the chromosome 17q21 SNP (which is associated with increased GSDMB expression) when exposed to an allergen. These studies demonstrated that HDM allergen challenge induced a significant increase in airway responsiveness, Th2 response (Th2 cytokines, MBP+ eosinophils, CD4+ cells, IgE), airway smooth muscle, subepithelial fibrosis, and mucin in HDM-challenged hGSDMBZp3-Cre mice compared with HDM-challenged WT mice. Epidemiologic studies of asthma link IgE levels to chromosome 17q21 variants in some (28) but not all studies (29).
In summary, we have made the observation that GSDMB is expressed at increased levels in the lungs of human subjects with severe asthma, and that hGSDMBZp3-Cre mice exhibits significant spontaneous increases in AHR and airway remodeling, including an increase in peribronchial smooth muscle and increased peribronchial fibrosis in the absence of airway inflammation. We identified a pathway by which GSDMB regulates two genes previously implicated in asthma (i.e., TGF-β1 and 5-LO) and established that nuclear localization of GSDMB-1 was essential for the induction of TGF-β1. Overall, this study, in combination with previous data from our laboratory regarding ORMDL3 (4), suggests that at least two of the genes localized to chromosome 17q21 (GSDMB and ORMDL3) can contribute to increased AHR and increased airway remodeling in the absence of airway inflammation, which contrasts with the four chromosome 5q genes (IL-4, IL-5, IL-9, IL-13) that induce airway inflammation in asthma.
Materials and Methods
Methods and statistical analyses for qPCR, immunohistochemistry, immunofluorescence microscopy, Western blot, in vitro transfection, siRNA knockdown, generation of hGSDMBZp3-Cre mice, mouse model of HDM-induced asthma, AHR measurements, collection of mouse specimens including lungs, BAL and blood, histology, and ELISAs were performed as described in SI Materials and Methods. All of the mouse experimental protocols were approved by the University of California, San Diego Institutional Animal Care and Use Committee.
Postmortem human lungs from asthmatics and normal organ donors were procured by the Arkansas Regional Organ Recovery Agency and by the National Disease Research Interchange, and delivered to the Lung Cell Biology Laboratory at the Arkansas Children’s Hospital Research Institute under conditions identical to those used for transplant. The acquisition of deceased donor tissue was reviewed by the University of Arkansas for Medical Sciences Institutional Review Board and determined not to be human subject research. After processing, tissue was shipped to San Diego under a protocol approved by the University of California, San Diego Human Research Protections program. The protocols using bronchoscopy to obtain bronchial biopsies from patients with mild or severe asthma and control nonasthmatics at McGill University and Université de Montréal were performed with the approval of the respective Institutional Review Boards, as previously described (30).
Acknowledgments
Research reported in this publication was supported by NIH Grants AI 107779, AI 38425, AI 70535, AI 72115, and AI 242236 (to D.H.B.), and NIH Grant T32 AI 007469 (to M.R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610433113/-/DCSupplemental.
References
- 1.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Verlaan DJ, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85(3):377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh DG, et al. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264(5162):1152–1156. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 4.Miller M, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA. 2012;109(41):16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller M, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome. 2000;11(9):718–724. doi: 10.1007/s003350010138. [DOI] [PubMed] [Google Scholar]
- 7.Hergueta-Redondo M, et al. Gasdermin-B promotes invasion and metastasis in breast cancer cells. PLoS One. 2014;9(3):e90099. doi: 10.1371/journal.pone.0090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, et al. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64(4):629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madore A-M, et al. Alveolar macrophages in allergic asthma: An expression signature characterized by heat shock protein pathways. Hum Immunol. 2010;71(2):144–150. doi: 10.1016/j.humimm.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, et al. Expression of GSDML associates with tumor progression in uterine cervix cancer. Transl Oncol. 2008;1(2):73–83. doi: 10.1593/tlo.08112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim YM, et al. Role of 5-lipoxygenase in IL-13-induced pulmonary inflammation and remodeling. J Immunol. 2006;177(3):1918–1924. doi: 10.4049/jimmunol.177.3.1918. [DOI] [PubMed] [Google Scholar]
- 12.Bossley CJ, et al. Pediatric severe asthma is characterized by eosinophilia and remodeling without T(H)2 cytokines. J Allergy Clin Immunol. 2012;129(4):974–82.e13. doi: 10.1016/j.jaci.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Reilly R, et al. Increased airway smooth muscle in preschool wheezers who have asthma at school age. J Allergy Clin Immunol. 2013;131(4):1024–1032, 1032.e1–16. doi: 10.1016/j.jaci.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 14.Noble PB, et al. Airway smooth muscle in asthma: Linking contraction and mechanotransduction to disease pathogenesis and remodelling. Pulm Pharmacol Ther. 2014;29(2):96–107. doi: 10.1016/j.pupt.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Beppu LY, et al. TGF-β1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1100–1107.e4. doi: 10.1016/j.jaci.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perng DW, et al. Leukotriene C4 induces TGF-beta1 production in airway epithelium via p38 kinase pathway. Am J Respir Cell Mol Biol. 2006;34(1):101–107. doi: 10.1165/rcmb.2005-0068OC. [DOI] [PubMed] [Google Scholar]
- 17.Halwani R, Al-Muhsen S, Al-Jahdali H, Hamid Q. Role of transforming growth factor-β in airway remodeling in asthma. Am J Respir Cell Mol Biol. 2011;44(2):127–133. doi: 10.1165/rcmb.2010-0027TR. [DOI] [PubMed] [Google Scholar]
- 18.Das S, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50(5):882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mautino G, Oliver N, Chanez P, Bousquet J, Capony F. Increased release of matrix metalloproteinase-9 in bronchoalveolar lavage fluid and by alveolar macrophages of asthmatics. Am J Respir Cell Mol Biol. 1997;17(5):583–591. doi: 10.1165/ajrcmb.17.5.2562. [DOI] [PubMed] [Google Scholar]
- 20.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295(5556):813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 21.Xiong HY, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22(5):535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 23.Jame AJ, et al. Human bronchial epithelial cells express an active and inducible biosynthetic pathway for leukotrienes B4 and C4. Clin Exp Allergy. 2007;37(6):880–892. doi: 10.1111/j.1365-2222.2007.02733.x. [DOI] [PubMed] [Google Scholar]
- 24.Lukacs NW. Role of chemokines in the pathogenesis of asthma. Nat Rev Immunol. 2001;1(2):108–116. doi: 10.1038/35100503. [DOI] [PubMed] [Google Scholar]
- 25.Rohde G, et al. CXC chemokines and antimicrobial peptides in rhinovirus-induced experimental asthma exacerbations. Clin Exp Allergy. 2014;44(7):930–939. doi: 10.1111/cea.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burkhardt AM, et al. CXCL17 is a mucosal chemokine elevated in idiopathic pulmonary fibrosis that exhibits broad antimicrobial activity. J Immunol. 2012;188(12):6399–6406. doi: 10.4049/jimmunol.1102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loxham M, Davies DE, Blume C. Epithelial function and dysfunction in asthma. Clin Exp Allergy. 2014;44(11):1299–1313. doi: 10.1111/cea.12309. [DOI] [PubMed] [Google Scholar]
- 28.Galanter J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177(11):1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffatt MF, et al. GABRIEL Consortium A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller M, et al. Fstl1 promotes asthmatic airway remodeling by inducing oncostatin M. J Immunol. 2015;195(8):3546–3556. doi: 10.4049/jimmunol.1501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M, et al. Segmental allergen challenge increases levels of airway follistatin-like 1 in patients with asthma. J Allergy Clin Immunol. 2016;138(2):596–599.e4. doi: 10.1016/j.jaci.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]