Abstract
Alzheimer’s disease (AD), the most common form of dementia in the elderly, has no cure. Thus, the identification of key molecular mediators of cognitive decline in AD remains a top priority. As aging is the most significant risk factor for AD, the goal of this study was to identify altered proteins and pathways associated with the development of ‘normal’ aging and AD memory deficits, and identify unique proteins and pathways that may contribute to AD-specific symptoms. We used contextual fear conditioning to diagnose 8-month-old 5XFAD and non-transgenic (Ntg) mice as having either intact or impaired memory, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify hippocampal membrane proteins across groups. Subsequent analysis detected 113 proteins differentially expressed relative to memory status (intact vs impaired) in Ntg mice and 103 proteins in 5XFAD mice. Thirty-six proteins, including several involved in neuronal excitability and synaptic plasticity (e.g., GRIA1, GRM3, and SYN1), were altered in both ‘normal’ aging and AD. Pathway analysis highlighted HDAC4 as a regulator of observed protein changes in both genotypes and identified the REST epigenetic regulatory pathway and Gi intracellular signaling as AD-specific pathways involved in regulating the onset of memory deficits. Comparing the hippocampal membrane proteome of Ntg versus AD, regardless of cognitive status, identified 138 differentially expressed proteins, including confirmatory proteins APOE and CLU. Overall, we provide a novel list of putative targets and pathways with therapeutic potential, including a set of proteins associated with cognitive status in normal aging mice or gene mutations that cause AD.
Keywords: proteomics, aging, Alzheimer’s disease, ion channels, cognition
Graphical Abstract
1. Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease characterized, in part, by the pathogenic accumulation of neuritic beta-amyloid (Aβ) plaques accompanied by profound memory loss and neurodegeneration1. Although the cause of AD is unknown, studies comparing brain and/or peripheral tissues (e.g. blood) from individuals with AD to cognitively healthy individuals suggest that differences in mitochondrial function2, 3, calcium homeostasis4, 5, and immune function6, 7 may be critically involved in the development of AD. As the number of individuals with AD dementia in the United States is expected to reach 14 million by 20508, the identification of molecules and pathways whose dysfunction leads to AD dementia is critical for understanding the pathogenesis of the disease, as well as for the development of effective therapeutics.
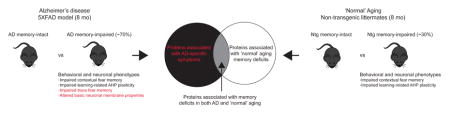
As aging is the most significant risk factor for AD, there is also a critical need to understand mechanisms that occur during ‘normal’ cognitive aging, as well as how they may contribute to the cognitive decline observed in AD. Our lab has previously shown that a subset of both middle-aged 5XFAD and non-transgenic (Ntg) mice exhibit aging-related hippocampal-dependent memory deficits relative to young ‘healthy’ mice9–11. In both AD and ‘normal’ aging, these memory deficits correspond to deficits in intrinsic plasticity of hippocampal neurons (e.g. impaired AHP plasticity after contextual fear conditioning9, 11). The extent to which these previously reported behavioral and neuronal changes observed in 5XFAD and Ntg mice correspond to dysfunction of the same molecular pathways is currently unknown. To identify altered proteins and pathways associated with the development of ‘normal’ aging and AD memory deficits, this study exploited individual differences in the memory abilities of 8-month-old B6SJL mice harboring human mutations in the genes for amyloid precursor protein (APP) and presenilin1 (PS1), which are known to cause early-onset familial AD (5XFAD model) and their Ntg littermates 12–16. Mice were diagnosed as having either ‘intact’ or ‘impaired’ memory based on their CFM performance relative to a previously tested group of 2-month-old controls9, 11, and LC-MS/MS was used to quantify hippocampal membrane proteins across groups. Based on the above evidence, we hypothesize some common molecular mediators contribute to the neuronal plasticity and memory deficits observed in AD and ‘normal’ aging.
While 5XFAD and Ntg mice do share some similarities (i.e. CFM deficits and impaired AHP plasticity), the neuronal and behavioral deficits observed in 5XFAD mice are exacerbated relative to those observed in ‘normal’ aging mice. Specifically, the incidence of CFM deficits in middle-aged 5XFAD mice is greatly increased, with ~70% of 5XFAD mice meeting ‘impaired’ criteria compared to only ~30% of Ntg mice. In addition, 5XFAD impaired mice exhibit both CFM deficits and trace-fear memory deficits, whereas age-matched Ntg impaired mice exhibit select CFM deficits9, 11 (see Figure 2C in Kaczorowski and Disterhoft, 2009 compared to Figure 6B in Kaczorowski et al., 2011). Furthermore, although both AD and ‘normal’ aging-related memory deficits in these mice correspond to deficits in intrinsic plasticity (e.g. impaired AHP plasticity after contextual fear conditioning), a reduction in hippocampal neuronal excitability at baseline is only observed in AD mice (see Supl Figure 2 in Kaczorowski et al., 2011). Therefore, we also hypothesize a number of proteins will be differentially expressed relative to memory status only in 5XFAD mice, and that these proteins and corresponding pathways may underlie the exacerbated disease-specific symptoms observed in 5XFAD mice.
As memory deficits in both ‘normal’ aging and AD correspond to deficits in hippocampal neuronal intrinsic plasticity9, 11, we hypothesize that changes in the expression of ion channels and/or receptors that play a key role in regulating neuronal excitability and plasticity17, 18 may underlie the onset of memory deficits. However, ion channels and receptors are under-represented in many proteomic studies due to the heterogeneous and low-abundant nature of these proteins19, especially in comparison to highly-abundant mitochondrial and cytosolic proteins. To reduce this under-sampling bias, we performed a targeted in-depth comparison of the hippocampal membrane proteome from 5XFAD and Ntg mice with either intact or impaired contextual fear memory (CFM). This approach allows for efficient detection of critical proteins that are hypothesized to directly mediate processes necessary for learning and memory that may have been overlooked in previous analyses of the entire hippocampal proteome20, 21. In addition, many plasma membrane proteins contain extracellular domains and can be easily manipulated by various chemical compounds or small molecules22, making them highly valuable drug targets. Overall, we hypothesize that the identification of hippocampal membrane proteins differentially expressed in AD mice that differ only in memory status may facilitate the prioritization of candidates for subsequent functional tests, as we recently reported in ‘normal’ aging models10.
2. Materials and Methods
2.1 Animals
The 5XFAD model of AD used in this study has been previously described16. Briefly, 5XFAD mice overexpress mutant human APP695 with the Swedish (K670N, M671L), Florida (I716V), and London (V717I) familial AD (FAD) mutations along with mutant human PS1 harboring two FAD mutations, M146L and L286V. Eight-month-old male 5XFAD mice and their Ntg littermates were group housed (2–5 per cage) and maintained in colony-housing (12-hour light/dark cycle) with ad libitum access to food and water in accordance with approval by the Medical College of Wisconsin Animal Care and Use Committee.
2.2 Contextual Fear Conditioning
Contextual fear conditioning was performed as previously described9, 11. Briefly, mice were placed in a training chamber, and after a 150s baseline period, received four foot shocks (0.7 mA, inter-trial interval 210 ± 10 s). Twenty-four hours later, mice were again placed into the training chamber and behavioral freezing during the 10 minute test session was measured as an index of conditional fear memory (CFM). Impaired mice were characterized as those whose CFM performance fell more than 3 standard deviations below that of a group of healthy 2-month-old mice using established criteria (cutoff = 61%, see Kaczorowski and Disterhoft, 2009, Figure 2)9, 11.
Figure 2.
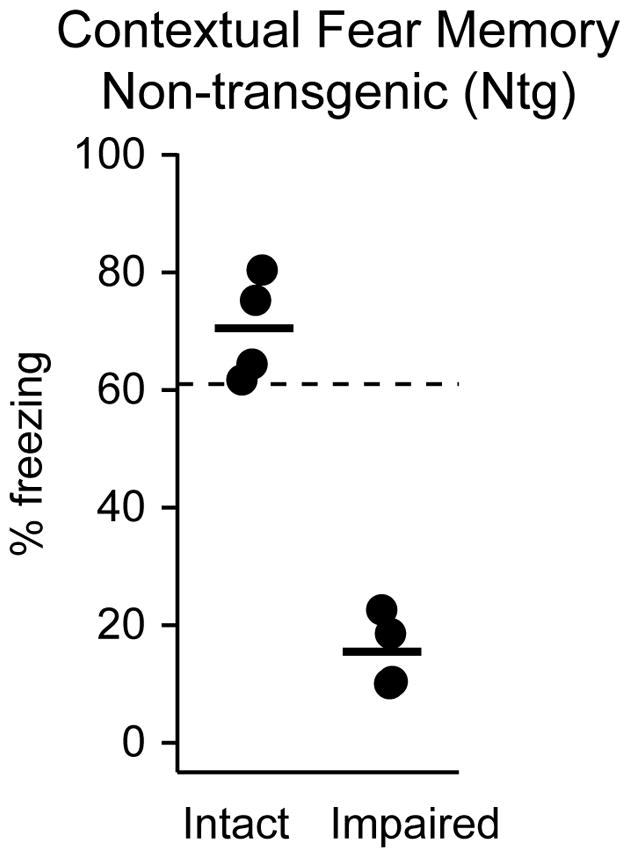
Identification of non-transgenic (Ntg) memory-intact vs memory-impaired individuals. Eight-month-old Ntg mice were trained on standard contextual fear conditioning. Memory-impaired mice performed below a predetermined criterion 3 standard deviations below the performance of healthy young mice (61%, dotted line)9, 11. Memory-intact mice performed above criterion. Four mice per group were used for proteomics analysis (3 technical replicates per mouse).
2.3 Proteomic Analysis
Immediately following CFM testing on Day 2, mice were anesthetized under isoflurane, decapitated, and the entire hippocampus was immediately dissected into ice cold lysis buffer containing protease inhibitors10. A membrane enrichment step was performed using freshly isolated hippocampus (n = 4 mice/grp)10 to reduce abundant mitochondrial and cytosolic proteins that impede detection of lesser-abundant proteins, such as ion channels and receptors20. Specifically, samples were homogenized in high salt buffer (2M NaCl, 10mM HEPES, 1mM EDTA, pH 7.4) and centrifuged at 4°C at 16,000g for 20 min. The remaining pellet was then homogenized in carbonate buffer (0.1M Na2CO3, 1mM EDTA, pH 11.3) and incubated at 4°C for 30 min to separate membrane fractions from free cytosolic proteins and organelles. After incubation, samples were centrifuged at 4°C at 16,000g for 20 min. Membrane proteins were then extracted from the lipid bilayer by re-dissolving pellet in ice-cold urea buffer (5M urea, 100mM NaCl, 10mM HEPES, 1mM EDTA, pH 7.4) and centrifuging at 4°C at 16,000g for 20 min. The remaining pellet was then disrupted and washed with ice-cold Tris/HCl buffer (pH 7.6), centrifuged 2x at 4°C at 16,000g for 20 min, and finally dissolved in 2% SDT-lysis buffer (4% SDS and 100mM DTT in 0.1M Tris/HCl buffer, pH 7.6). Samples were heated at 95°C for 1 min, sonicated for 2 min to shear DNA, and centrifuged at 14,000g for 1 min. The resulting supernatants were then processed using filter-assisted sample preparation (FASP) according to established methods23. Tryptic peptides generated from each individual mouse were then analyzed in triplicate using label-free spectral counting (ms/ms) on a ThermoFinnigan LTQ-Orbitrap Velos Mass Spectrometer coupled to a Waters nano-UPLC system10 (5 hr gradient per run) and ms/ms counts were added together to obtain final ms/ms counts per mouse. Using the Sequest and MASCOT Search algorithms, all sample spectral data was compared against the UniProtKB Mouse Database (generated January 26, 2015; 84,607 sequences, 36,971,481 residues) and a combined file was generated, keeping the best match for a given spectra from one algorithm to avoid redundancies. Search parameters included trypsin digestion, up to 3 missed cleavages, N-terminal Acetylation (+42-Da), oxidation of methione (+16-Da), and carbamidomethyl alkylation of cysteines (+57-Da). A total of 2,595 proteins were reliably detected in 3 of the 4 samples of at least one group using a protein probability False Discovery Rate (FDR) of 1%24. Of these, 780 had primary annotation as either localized to the plasma membrane or as an ion channel, receptor, or transporter according to Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, www.ingenuity.com), a 30% membrane enrichment.
2.4 Statistical Analysis
Proteomic sample comparisons were performed using Visualize proteomics analysis software with built-in statistical analysis for large proteomic dataset comparison24. The G (Goodness-of-fit) test, which is a log-likelihood ratio test widely accepted for the determination of protein abundance relationships among proteomic datasets, was used to determine statistical significance25. The test compares observed protein counts to expected, and calculates a test statistic based on deviation from expected counts. Specifically, the assumption that the expected proportion of scans for a given protein is related to the total scans in each group is used. Thus, each observed scan count is multiplied by this ratio or its inverse (depending on the group) to provide the expected frequency of detection. Scans from group zero (S0) are multiplied by the ratio of total scans in group zero (E0) over the total scans in group one (S1); conversely the inverse is performed for scans from group one (S1) and the total scans from group 1 (E1). Using these assumptions, the G-value calculation is 2 * ( S0 * ln(S0/E0) + S1 * ln(S1/E1)) and if observed frequencies perfectly fit expected frequencies, a G-value of zero is calculated. In contrast, a larger G-value indicates the observed frequency departs from the expected frequency. The G-value distribution can then be approximated by a chi-squared distribution with a single degree of freedom to determine significance25, 26. Excel comparison files were exported and protein abundances were determined by quantifying the number of spectra observed for each protein (protein identification FDR set to 1%). Differentially expressed proteins were defined as those proteins with > 20% abundance difference based on our prior work validating this cutoff as sufficient to nominate biologically relevant differences in protein expression10 when identified reliably in at least 3 of the 4 samples of one group (p < 0.05 level using the Bonferroni correction for multiple comparisons). Behavior data is presented at mean ± S.E.M. Data analysis in both behavioral and proteomic experiments were conducted blind to experimental groups.
2.5 Bioinformatics Analysis
In order to identify networks and pathways that were different between groups, proteins meeting statistical criteria for differential expression from each analysis, along with their log ratio abundance change, were imported into Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, www.ingenuity.com) for pathway analysis using the core analysis platform. Build version 366632M and content version 26127183 (release date: 11-30-2015) were used. The core analysis matched proteins in our data sets with those in the Ingenuity Knowledge Base (Genes Only), and only direct relationships were considered for network and upstream regulator analysis. The default settings of 35 molecules/network were retained. Only Ingenuity Expert Information, microRNA-mRNA interactions, and protein-protein interactions with experimentally observed or high confidence predictions were included (moderate confidence interactions were excluded). Data from both mammals (mouse, human, rat) and cell lines were considered. The p-value associated with a function or pathway is calculated using the Fisher Exact Test, which determines the likelihood that the association between experimental results and a given process or pathway is due to random chance. IPA corrects for multiple testing using the Benjamini-Hochberg method27 to control the rate of false discoveries. To independently validate IPA’s identification of HDAC4 as an upstream regulator of observed protein changes, we compared differentially expressed proteins in our analysis with a previous report of transcripts experimentally confirmed to be regulated by HDAC428. Specifically, the degree of overlap based on gene symbol was determined based on a hypergeometric test. The set of ~17,000 transcripts measured by the MoGene-1_0-st-v1 Affymetrix microarray was used as the background gene list.
3. Results
3.1 Identification of proteins differentially expressed relative to cognitive status
In order to identify specific proteins and pathways associated with the onset of memory deficits in middle-aged (8-month-old) 5XFAD and Ntg mice, as well as putative candidates that may underlie the exacerbated behavioral and neuronal deficits observed in AD, mice were first diagnosed as either memory-impaired or memory-intact using standard contextual fear conditioning. Specifically, mice were classified as ‘impaired’ if their CFM fell below our established criteria of three standard deviations away from the mean of 2-month old mice (cutoff = 61%, see Kaczorowski and Disterhoft, 2009, Figure 2). Four 5XFAD mice with intact CFM (AD intact) and 4 mice with impaired CFM (AD impaired, Fig. 1), as well as 4 Ntg mice with intact CFM (Ntg intact) and 4 Ntg mice with impaired CFM (Ntg impaired, Fig. 2), were selected for further hippocampal membrane proteome analysis. Mice were housed, phenotyped, sacrificed, processed, randomized, and analyzed in parallel to allow for well-controlled comparisons of differentially expressed proteins relative to memory status in Ntg ‘normal’ aging versus AD mice.
Figure 1.
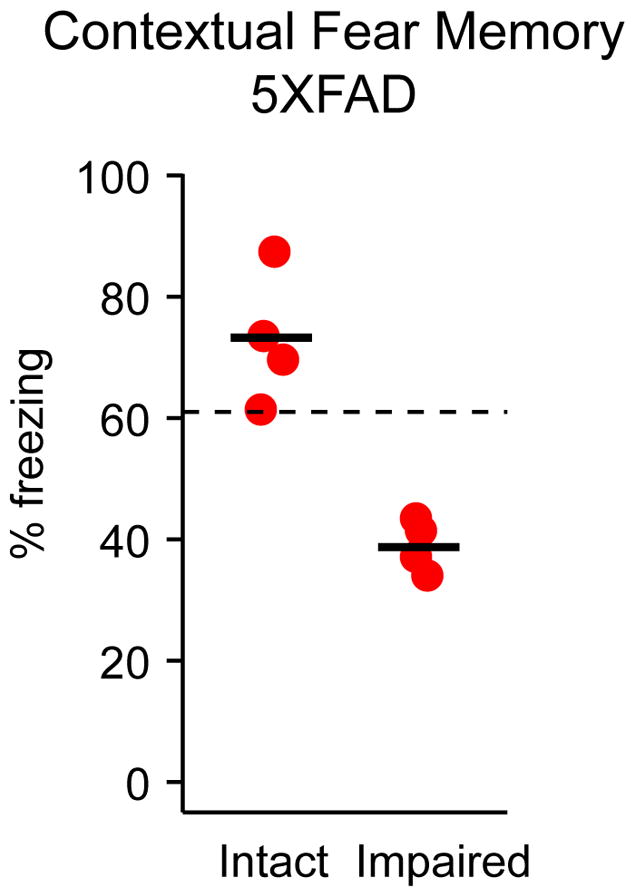
Identification of 5XFAD memory-intact vs memory-impaired individuals. Eight-month-old 5XFAD (AD) mice were trained on standard contextual fear conditioning. AD impaired mice performed below a pre-determined criterion 3 standard deviations below the performance of healthy young mice (61%, dotted line)9, 11. AD intact animals performed above criterion. Four mice per group were used for proteomics analysis (3 technical replicates per mouse).
A set of 113 proteins was identified as differentially expressed between Ntg intact vs Ntg impaired mice (Table 1, Supplementary Table 1) compared to 103 differentially expressed proteins between AD intact vs AD impaired mice (Table 2, Supplementary Table 2). Of these, 35/113 and 44/103 were annotated as either localized to the plasma membrane or as an ion channel, receptor, or transporter in Ntg and AD mice, respectively. This high proportion of ion channels and receptors that are differentially expressed relative to cognitive status support our initial hypothesis that these molecules located in the hippocampal plasma membrane likely modulate resilience or susceptibility to memory decline.
Table 1.
Top 20 proteins differentially expressed relative to cognitive status in Ntg mice
A comparison of Ntg mice with intact CFM (Ntg intact) versus those with impaired CFM (Ntg impaired) identified 113 proteins meeting statistical criteria for differential expression (n = 4/grp, 3 technical replicates per mouse). The top 20 with largest magnitude fold change are listed below. A positive log ratio indicates higher protein expression in Ntg intact animals. See Supplementary Table 2 for a full list of proteins.
| Protein | Description | Location | Type | Fold Change (log ratio) |
|---|---|---|---|---|
| CES2E | carboxylesterase 2E | Other | other | 4.64 |
| GLUD1 | glutamate dehydrogenase 1 | Cytoplasm | enzyme | 4.64 |
| CACNA1D | calcium channel, voltage-dependent, L type, alpha 1D subunit | Plasma Membrane | ion channel | −4.58 |
| MRPS7 | mitochondrial ribosomal protein S7 | Cytoplasm | other | −4.46 |
| DLGAP4 | discs, large (Drosophila) homolog-associated protein 4 | Plasma Membrane | other | 4.25 |
| TOMM70A | translocase of outer mitochondrial membrane70 homolog A (S. cerevisiae) | Cytoplasm | transporter | −4.17 |
| ILDR2 | immunoglobulin-like domain containing receptor 2 | Other | other | −3.91 |
| KRT1 | keratin 1, type II | Cytoplasm | other | 3.64 |
| RPS4X | ribosomal protein S4, Y-linked 1 | Cytoplasm | other | 3.52 |
| AURKB | aurora kinase B | Nucleus | kinase | −3.46 |
| IMPDH2 | IMP (inosine 5'-monophosphate) dehydrogenase 2 | Cytoplasm | enzyme | −3.46 |
| SLC19A3 | thiosulfate sulfurtransferase (rhodanese) | Cytoplasm | enzyme | 3.32 |
| APBB2 | amyloid beta (A4) precursor protein-binding, family B, member 2 | Cytoplasm | other | 2.91 |
| BCAN | brevican | Extracellular Space | other | −2.91 |
| TMPO | thymopoietin | Nucleus | other | 2.86 |
| KIF21A | kinesin family member 21A | Cytoplasm | other | −2.81 |
| GABRA5 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 | Plasma Membrane | ion channel | 2.72 |
| PSMD11 | proteasome 26S subunit, non-ATPase 11 | Cytoplasm | other | 2.58 |
| CCIN | calicin | Cytoplasm | other | -2.52 |
| AKAP14 | A kinase (PRKA) anchor protein 14 | Nucleus | other | 2.46 |
Table 2.
Top 20 proteins differentially expressed relative to cognitive status in 5FXAD mice
A comparison of 5XFAD mice with intact CFM (AD intact) versus those with impaired CFM (AD impaired) identified 103 proteins meeting statistical criteria for differential expression (n = 4/grp, 3 technical replicates per mouse). The top 20 with largest magnitude fold change are listed below. A positive log ratio indicates higher protein expression in AD intact animals. See Supplementary Table 1 for a full list of proteins.
| Protein | Description | Location | Type | Fold Change (log ratio) |
|---|---|---|---|---|
| ATG2B | Autophagy related 2B | Other | other | 4.32 |
| SMTNL1 | Smoothelin-like 1 | Cytoplasm | other | 3.87 |
| GSTM1 | Glutathione S-transferase mu 5 | Cytoplasm | enzyme | 2.74 |
| VPS35 | VPS35 retromer complex component | Cytoplasm | transporter | −2.70 |
| GLUD1 | Glutamate dehydrogenase 1 | Cytoplasm | enzyme | 2.39 |
| LTBP3 | Latent transforming growth factor beta binding protein 3 | Extracellular Space | other | 2.26 |
| MCM3 | Minichromosome maintenance complex component 3 | Nucleus | enzyme | −2.12 |
| NDUFA11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11 | Cytoplasm | enzyme | −2.00 |
| GNAI3 | Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | Cytoplasm | enzyme | 1.96 |
| TCP1 | T-complex protein 1 | Cytoplasm | other | −1.92 |
| IP3R1 | Inositol 1,4,5-triphosphate receptor, type 1 | Cytoplasm | ion channel | 1.88 |
| IMPDH2 | IMP dehydrogenase 2 | Cytoplasm | enzyme | −1.87 |
| RAB11B | RAB11B, member RAS oncogene family | Cytoplasm | enzyme | 1.75 |
| TPRN | Taperin | Extracellular Space | other | 1.73 |
| KCNQ2 | Potassium channel, voltage-gated KQT-like subfamily Q, member 2 | Plasma Membrane | ion channel | 1.69 |
| NRXN3 | Neurexin III | Plasma Membrane | other | 1.63 |
| NPTX1 | Neuronal pentraxin I | Extracellular Space | other | 1.60 |
| CMTM6 | CKLF-like MARVEL transmembrane domain containing 6 | Extracellular Space | cytokine | −1.53 |
| SCAMP1 | Secretory carrier membrane protein 1 | Cytoplasm | transporter | 1.52 |
| NDUFA7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 | Cytoplasm | enzyme | 1.49 |
3.2 Proteins and pathways that correspond to memory deficits in both 5XFAD and ‘normal’ aging mice
Of the total proteins identified, 36 proteins were associated with cognitive status in both 5XFAD and Ntg ‘normal’ aging mice (Table 3 and Supplementary Table 3), representing a significant degree of overlap between the sets (p = 1.35 x 10−26 based on a hypergeometric test). For example, glutamate receptors GRIA1 and GRM3, along with synaptic proteins SYN1 and SYNGAP1, were correlated with memory status regardless of genotype. Thus, altered expression of these four proteins, along with the additional 32 overlapping proteins, may contribute to CFM deficits and reductions in hippocampal neuronal excitability and intrinsic plasticity (e.g. impaired AHP plasticity after contextual fear conditioning) that are common to both AD and ‘normal’ aging mice.
Table 3.
Top 20 Proteins associated with memory status in both Ntg and 5XFAD mice.
Thirty-six proteins met statistical criteria for differential expression relative to memory status in both 5FXAD (AD intact vs AD impaired) and Ntg (Ntg intact vs Ntg impaired) analyses. The top 20 with the highest magnitude fold change in either group are listed below. A positive log ratio indicates higher protein expression in intact animals. See Supplementary Table 3 for a full list of proteins.
| Protein | Description | Location | Type | AD Fold Change (log ratio) | Ntg Fold Change (log ratio) |
|---|---|---|---|---|---|
| GLUD1 | glutamate dehydrogenase 1 | Cytoplasm | enzyme | 2.39 | 4.64 |
| MCM3 | minichromosome maintenance complex component 3 | Nucleus | enzyme | −2.12 | −2.43 |
| IMPDH2 | IMP (inosine 5'-monophosphate) dehydrogenase 2 | Cytoplasm | enzyme | −1.87 | −3.46 |
| CMTM6 | CKLF-like MARVEL transmembrane domain containing 6 | Extracellular Space | cytokine | −1.53 | −1.58 |
| NDUFA7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa | Cytoplasm | enzyme | 1.50 | −0.61 |
| GPR37 | G protein-coupled receptor 37 (endothelin receptor type B-like) | Plasma Membrane | G-protein coupled receptor | 1.27 | −0.95 |
| HSD1768 | hydroxysteroid (17-beta) dehydrogenase 8 | Cytoplasm | enzyme | 1.23 | 1.43 |
| KRT2 | keratin 2 | Cytoplasm | other | 1.22 | 1.12 |
| RPN1 | ribophorin I | Cytoplasm | enzyme | 1.20 | 0.84 |
| SLC1A7 | solute carrier family 1 (glutamate transporter), member 7 | Plasma Membrane | transporter | 1.10 | 0.57 |
| CHCHD6 | coiled-coil-helix-coiled-coil-helix domain containing 6 | Cytoplasm | other | 1.09 | 0.66 |
| MTOR | mechanistic target of rapamycin (serine/threonine kinase) | Nucleus | kinase | −1.06 | −1.23 |
| COX5B | cytochrome c oxidase subunit Vb | Cytoplasm | other | 0.97 | 0.60 |
| ANK2 | ankyrin 2, brain | Plasma Membrane | other | 0.88 | −0.34 |
| DLST | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | Cytoplasm | enzyme | 0.58 | 0.40 |
| GRIA1 | glutamate receptor, ionotropic, AMPA 1 | Plasma Membrane | ion channel | 0.55 | 0.34 |
| SYNGAP1 | synaptic Ras GTPase activating protein 1 | Plasma Membrane | other | 0.52 | 0.41 |
| GRM3 | glutamate receptor, metabotropic 3 | Plasma Membrane | G-protein coupled receptor | −0.34 | −0.51 |
| APOE | apolipoprotein E | Extracellular Space | transporter | 0.38 | 0.84 |
| NRCAM | neuronal cell adhesion molecule | Plasma Membrane | other | 0.84 | −0.73 |
To gain an unbiased perspective of the potential pathways and networks involved in mediating these common changes, this dataset of 36 common differentially expressed proteins was uploaded to IPA for pathway analysis. Based on observed protein abundance changes in our dataset, IPA used bioinformatics to identify glutamate synthesis and degradation, as well as glutamate receptor signaling, as top canonical pathways likely to be responsible for mediating observed differences (Table 4). This confirms previous results that highlight synaptic transmission as a core mediator of cognitive function29–31. In addition, IPA identified histone deactylase 4 (HDAC4) as a top predicted upstream regulator mediating observed protein abundance changes (p < 0.001). HDAC4 is known to cause deacetylation of histones and has been shown to regulate expression of genes critical to learning and memory. To independently validate IPA’s identification of HDAC4 as an upstream regulator, we compared differentially expressed proteins in our analysis with a previous report of transcripts experimentally determined to be regulated by HDAC428. Of the 36 proteins we as differentially expressed proteins relative to memory status, 3 correspond to transcripts experimentally determined to be regulated by HDAC4 in neurons by Sando and colleagues in 2012 (out of 210 total transcripts, p < 0.01). This converging evidence further supports a role for HDAC4 as an upstream regulator of proteins involved in the onset of memory deficits common to both AD and ‘normal’ aging.
Table 4.
Pathways associated with the onset of memory deficits in both ‘normal’ age-associated cognitive decline and AD.
Overlapping proteins (36) that met statistical criteria for differential expression in both AD (AD intact vs AD impaired) and Ntg (Ntg intact vs Ntg impaired) comparisons were uploaded in to Ingenuity Pathway Analysis (IPA) to identify enriched pathways and plausible upstream regulators.
| Top Canonical Pathways | |
|---|---|
| Name | p-value |
| Glutamate receptor signaling | 1.23e-04 |
| Glutamate Biosynthesis II | 3.39e-03 |
| Glutamate Degradation X | 3.39e-03 |
|
| |
| Top Upstream Regulators | |
| Regulator | p-value |
|
| |
| HTT | 7.44e-05 |
| HDAC4 | 3.41e-04 |
| MECP2 | 3.90e-04 |
| PARP | 1.57e-03 |
| PRKDC | 1.57e-03 |
3.3 Ion channels and receptors associated with memory status at the onset of decline in 5FXAD mice
Despite the significant degree of overlap between proteins associated with the onset of memory deficits in 5XFAD and Ntg mice, a number of unique differentially expressed proteins in each condition were identified. As both the incidence and severity of neuronal and memory deficits are increased in 5XFAD mice, this set of differentially expressed proteins may contribute to exacerbated disease-specific changes that are unique to AD impaired mice11. Proteins unique to the onset of AD memory deficits included ion channels and receptors with both excitatory and inhibitory roles in neuronal excitability were identified, confirming previous reports that disruption of the balance between excitation and inhibition within the hippocampus may be critical for AD-related memory and neurological deficits11, 32. Specifically, the alpha 1 subunit of the GABAA receptor (GABRA1) and hyperpolarization-activated cyclic nucleotide-gated channel 1 (HCN1) were all decreased in the hippocampus of impaired 5XFAD. Identified roles for these channels in the nervous system include plasticity of hippocampal neuronal excitability33, modulation of spatial orientation34–36, as well as oscillatory activity in neuronal networks18, 37–39, each of which have been hypothesized to contribute significantly to processes of learning and memory. Notably, levels of GABRA140, 41, and HCN142 have been reported as decreased in cognitively impaired humans with AD relative to healthy controls, suggesting our work in mouse models can nominate potential translationally relevant molecules.
In addition to changes in neuronal function and excitability, changes in intracellular calcium homeostasis have also been reported prior to the onset of overt clinical symptoms in AD, suggesting this pathway may play a causal role in cognitive decline4, 43. In support, ion channels and receptors with known functions in the regulation of calcium homeostasis were identified as misregulated between 5XFAD mice with intact versus impaired CFM. For example, expression of inositol 1,4,5-triphosphate receptor type 1 (IP3R1), which mediates calcium release from internal stores44, was increased in 5XFAD mice with intact CFM. IP3R1 has been shown to be co-localized to a specialized subcompartment of the endoplasmic reticulum (ER) called mitochondria-associated ER membranes, which would have also been enriched in our membrane preparation. APP, PS1, and presenilin2 (PS2) have also been shown to be localized to these mitochondria-associated ER membranes45, and along with IP3R1, have been shown to regulate calcium transport between ER and mitochondria45. As ER-mitochondrial communication has been proposed as an additional mechanism contributing to AD-related memory deficits46, the identification of IP3R1 as misregulated relative to memory status in our animal model provides an additional target that may aid in restoring calcium homeostasis and ER-mitochondrial calcium trafficking in AD.
3.4 Pathways associated with memory status at the onset of decline in 5XFAD mice
Similar to the analysis used to identify pathways associated with memory deficits in both ‘normal’ aging and AD, the list of 103 proteins differentially expressed between AD intact and AD impaired animals was uploaded for pathway analysis. IPA identified repressor element 1 silencing transcription factor (REST) as a top predicted upstream regulator that could explain the unique changes observed in 5XFAD mice (Table 5). REST, which interacts with chromatin remodeling factors to silence genes important to synaptic function47, was recently found to be positively associated with cognitive preservation in human patients who meet post-mortem pathological criteria for AD (i.e. patients with increased REST expression exhibited better cognition, even with AD pathology48). As REST was not identified in analyses using proteome changes associated with cognitive status across genotypes, our results highlight REST as a putative upstream regulator mediating disease-specific changes that contribute to the onset of memory deficits in AD.
Table 5.
Pathways associated with onset of memory deficits in 5XAD mice.
The list of 103 proteins differentially expressed relative to memory status in AD (AD intact mice vs AD impaired mice) was uploaded in to Ingenuity Pathway Analysis (IPA) to identify enriched pathways and plausible upstream regulators.
| Top Canonical Pathways | |
|---|---|
| Name | p-value |
| Huntington’s Disease Signaling | 1.83e-05 |
| Mitochondrial Dysfunction | 2.27e-05 |
| Gi signaling | 2.92e-05 |
|
| |
| Top Upstream Regulators | |
| Regulator | p-value |
|
| |
| HTT | 1.97e-12 |
| HDAC4 | 5.67e-12 |
| TP53 | 4.88e-08 |
| NRF1 | 1.35e-05 |
| REST | 9.74e-05 |
One of the top canonical pathways identified by IPA as unique to the onset of memory deficits in 5XFAD mice was Gi-mediated intracellular signaling (Table 5). The Gi protein is a heterotrimeric G protein subunit that inhibits the production of cAMP from ATP, and activation of this signaling cascade is known to reduce neuronal excitability in hippocampal neurons through activation of a G protein inward-rectifying potassium channel (GIRK)49, 50. In addition, this signaling pathway has been shown to inhibit presynaptic voltage-gated Ca2+ channels, inhibiting Ca2+ influx and subsequent neurotransmitter release51, 52. These results suggest that the exacerbated decrease in intrinsic neuronal excitability and learning-induced plasticity observed in AD impaired mice may be a result of Gi signaling and highlight Gi-protein coupled receptors (GPCRs) as potential therapeutic targets for early intervention. In particular, the GPCRs MGLUR3 and ADORA1, both identified as upregulated in AD impaired mice, may represent viable targets.
3.5 Proteins and pathways associated with memory status in ‘normal’ aging mice
In addition to proteins and pathways uniquely differentially expressed relative to memory status in 5XFAD mice, a number of proteins were differentially expressed relative to memory deficits only in Ntg mice. As the cognitive and neuronal deficits observed in the Ntg population are less severe than those identified in 5XFAD mice, these proteins may represent those associated with a maintenance of cognitive function in ‘normal’ aging. For example, a number of members of the NADH dehydrogenase complex (NDUFA13, NDUFA5, NDUFB7, and NDUFB8) were identified as upregulated in Ntg intact vs Ntg impaired animals (Supplementary Table 1). Further bioinformatics by IPA identified oxidative phosphorylation as one of the top canonical pathways that could explain observed protein abundance changes associated with onset of memory deficits in ‘normal’ aging (Table 6).
Table 6.
Pathways associated with the onset of memory deficits in Ntg mice.
The list of 113 proteins differentially expressed relative to memory status in ‘normal’ aging (Ntg intact mice vs Ntg impaired mice) was uploaded in to Ingenuity Pathway Analysis (IPA) to identify enriched pathways and plausible upstream regulators.
| Top Canonical Pathways | |
|---|---|
| Name | p-value |
| Oxidative Phosphorylation | 2.6e-05 |
| Mitochondrial Dysfunction | 3.70e-05 |
| Glutamate Receptor Signaling | 2.43e-04 |
|
| |
| Top Upstream Regulators | |
| Regulator | p-value |
|
| |
| HTT | 4.09e-04 |
| HDAC4 | 8.41e-04 |
| MECP2 | 9.96e-04 |
| TCF7L2 | 3.22e-03 |
| MED30 | 3.74e-03 |
3.6 Confirmatory analysis of proteins and pathways associated with 5XFAD genotype
Previous studies comparing differentially expressed proteins in the hippocampus of FAD mouse models to their corresponding Ntg controls have identified changes in the expression of proteins involved in processes such as autophagy, inflammation, and lipid metabolism, including APOE, PEA14, GFAP, and C1QB, among others53–55. In order to compare and contrast the results of our work with previous studies, we analyzed the hippocampal membrane proteome of AD and Ntg mice regardless of CFM status (n = 8/grp). Analysis of the results identified 139 proteins differentially expressed relative to genotype (Table 7 and Supplementary Table 4) and replicated numerous previously reported findings56–65. For example, the most highly overabundant protein in the hippocampus of 5XFAD mice relative to Ntg controls was clusterin (CLU). The CLU gene has been identified as a risk factor for AD in recent genome-wide association studies, associated particularly with the presence of plaque pathology59–62. Apolipoprotein E (APOE), mutations in which are the most well-characterized risk factor for sporadic AD66, was also highly enriched in the hippocampus of 5XFAD mice, demonstrating our ability to replicate known disease-specific changes. In addition to replicating differential expression of CLU and APOE in brain from FAD models compared to Ntg controls, we observed enrichment of adenylate cyclase-associated protein 2 (CAP2) in the hippocampus of 5XFAD relative to Ntg controls. CAP2 has been identified as differentially expressed in both animal models of AD56 as well as in human AD samples compared to healthy controls57, 58. CAP2 plays a role in regulating actin turnover67, and is posited to influence structural and molecular remodeling of synapses associated with disease63–65. Overall, both our approach and workflow confirmed previous results and provide new insight into additional proteins and pathways that may have been underrepresented and therefore overlooked in previous studies.
Table 7.
Top 20 proteins significantly differentially expressed relative to genotype.
A by-genotype comparison of AD vs Ntg mice, regardless of memory status (n = 8/grp) yielded 138 proteins meeting statistical criteria for differential expression. The 20 proteins with the largest magnitude fold change are listed below. A positive fold change indicates higher expression in AD mice. For a full list of proteins, see Supplementary Table 4.
| Protein | Description | Location | Type | Fold change (log ratio) |
|---|---|---|---|---|
| CLU | clusterin | Cytoplasm | other | 6.70 |
| CAP2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | Plasma Membrane | other | 4.93 |
| CES2E | carboxylesterase 2E | Other | other | −4.70 |
| APOE | apolipoprotein E | Extracellular Space | transporter | 4.32 |
| CRYM | crystallin, mu | Cytoplasm | enzyme | 4.30 |
| GPI | glucose-6-phosphate isomerase | Extracellular Space | enzyme | 3.97 |
| GFAP | glial fibrillary acidic protein | Cytoplasm | other | 3.86 |
| ACADL | acyl-CoA dehydrogenase, long chain | Cytoplasm | enzyme | −3.52 |
| APP | amyloid beta (A4) precursor protein | Plasma Membrane | other | 2.99 |
| PFKM | phosphofructokinase, muscle | Cytoplasm | kinase | 2.97 |
| PPIA | peptidylprolyl isomerase A (cyclophilin A) | Cytoplasm | enzyme | 2.87 |
| PFKL | phosphofructokinase, liver | Cytoplasm | kinase | 2.83 |
| PGAM1 | phosphoglycerate mutase 1 (brain) | Cytoplasm | phosphatase | 2.76 |
| PFN1 | profilin 1 | Cytoplasm | other | 2.75 |
| GABRA5 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 | Plasma Membrane | ion channel | −2.56 |
| TPI1 | triosephosphate isomerase 1 | Cytoplasm | enzyme | 2.48 |
| GDI1 | GDP dissociation inhibitor 1 | Cytoplasm | other | 2.37 |
| DNM1 | dynamin 1 | Cytoplasm | enzyme | 2.37 |
| ATP5D | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | Cytoplasm | transporter | −2.34 |
| EEF2 | eukaryotic translation elongation factor 2 | Cytoplasm | translation regulator | 2.31 |
4. Discussion
The goal of this study was to carefully characterize the hippocampal proteome of middle-aged mice relative to both genotype and memory status while controlling for age in order to identify a narrow set of proteins strongly associated with the development of memory deficits for subsequent functional validation. Here, we present an in depth report of the hippocampus membrane proteome and identify differentially expressed proteins, including plasma membrane ion channels, receptors, and transporters that may play a role in determining the onset and severity of memory deficits in either AD, ‘normal’ aging, or both.
4.1 HDAC4 as a regulator of cognitive deficits across genotypes
As similar behavioral and neuronal deficits are observed in 5XFAD and Ntg mice, we expected a certain degree of overlap in the proteins and pathways associated with the onset of memory deficits in both conditions. In support, bioinformatics analyses highlighted a role for HDAC4 as a key determinant of memory status in both AD and Ntg mice via epigenetic regulation of hippocampal protein expression patterns in response to a training event. It is well-known that epigenetic changes, such as histone modification and methylation, alter chromatin structure in order to facilitate transcription of genes necessary for memory formation28, 68, 69. Although it is possible changes in intrinsic neuronal excitability and synaptic transmission influence HDAC4 activity, changes in epigenetic regulation have been linked to specific memory disturbances in animal models of both aging70 and AD71. Although the exact mechanisms are still being elucidated, epigenetic changes appear to be relatively gene-specific and have been linked to intrinsic neuronal excitability, synapse formation, spine density, and dendritic complexity69, 72–76. Genes that have a well-characterized role in learning and memory that are also known to be epigenetically regulated include Bdnf77, Creb278, 79, Reelin80, and Pp181. Here, we provide an additional list of candidates that are differentially expressed relative to memory status and that may also respond to epigenetic regulation downstream of HDAC4 in order to maintain cognitive status in both aging and AD. As the mice used in this study are genetically similar, epigenetics provides a potential explanation for the individual differences in memory status in our models9, 11. In humans, a large portion of the heritability of sporadic AD remains unexplained by currently identified genetic variants as measured by GWAS studies2. Our data, along with that of others82, 83, lends support to the hypothesis that epigenetic changes, whether caused by environmental factors or polymorphisms that regulate methylation status, may explain some of the current ‘missing heritability’ of AD84, as well as underlying mechanisms mediating AD pathophysiology, including dementia symptoms.
4.2 Proteins associated with memory deficits in 5XFAD mice may contribute to disease-specific symptoms
It is important to note that although the behavioral and neuronal deficits observed in age-matched 5XFAD and Ntg are similar in some ways (i.e. CFM memory deficits and impaired AHP plasticity), memory deficits occur at a much greater frequency and to a greater extent in 5XFAD mice (see Figure 2C in Kaczorowski and Disterhoft, 2009 compared to Figure 6B in Kaczorowski et al., 2011; Supl Figure 2 in Kaczorowski et al., 2011, respectively). Thus, the identification of proteins and pathways that are specifically associated with the onset of memory deficits in AD models may help to explain the molecular mediators of exacerbated symptoms, ultimately contributing to the development of therapeutics designed to treat disease-specific symptoms. In addition to changes in intrinsic neuronal excitability and plasticity we have reported previously9, 11, evidence suggests that aberrant patterns of neuronal circuit and network activity may also be causally involved in the onset of dementia symptoms in AD85, 86. Thus, interventions that block or reverse these changes may prevent development of symptoms85. Our work highlights GABRA1 and HCN1, among others, as proteins that may represent viable targets for intervention to prevent or delay the dysfunction in network activity and cognitive deficits observed in AD. HCN1 has recently been linked to the cellular representation of space within the hippocampus via regulation of both place and grid cells34–36. Grid cells in particular have been found to be disrupted in human individuals at risk for Alzheimer’s87, suggesting early alterations in HCN1 may play a role in the pathophysiology of AD in humans as well as AD mouse models.
4.3 Pathways associated with memory deficits in Ntg mice
Our analyses also highlighted oxidative phosphorylation as a unique pathway associated with the onset of cognitive deficits in Ntg mice, which was not surprising based on previous literature linking mitochondrial dyfunction to symptoms of ‘normal’ aging 88, 89. Given that mitochondrial proteins are present in relatively high abundance 20, there has been concern that oversampling of mitochondrial proteins may have biased the results of pathway analysis toward mitochondrial dysfunction in prior studies20. However, since our approach incorporates a membrane enrichment step specifically designed to limit this oversampling bias, results here strengthen prior reports that changes in oxidative phosphorylation and overall mitochondrial dysfunction plays an important role in mediating cognitive deficits in ‘normal’ aging. Although these processes have been associated with memory deficits in AD as well90, we speculate that the addition of robust disease-specific factors such as beta-amyloid deposition and gliosis16 occluded the detection of this pathway in our AD samples, suggesting that other pathways are more strongly associated with the onset of cognitive deficits in AD.
4.4 Learning-related changes may be reflected in this reported proteome
Given that all mice in this study were trained on contextual fear conditioning prior to proteomics analyses, the membrane proteome reported here potentially reflects a modified proteome in response to a training event. As naïve untrained mice were not included in this analysis, it is difficult to say for sure whether observed proteome differences are a cause or effect of memory impairment. However, previous reports demonstrate that mice in both memory-impaired and memory-intact groups are capable of learning the task to the same extent on Day 1 of contextual fear conditioning9, 11, so differences likely are the result of either consolidation or retrieval processes. Successful consolidation and retrieval is well-known to depend on plasticity of intrinsic neuronal excitability and synaptic strength17, 91, 92, which are protein synthesis-dependent processes93–97. Given that protein synthesis and clearance are disrupted in aging and AD models94, 97, it is plausible that in AD and Ntg mice with impaired CFM there is either 1) a failure to appropriately remodel the hippocampal proteome, resulting in impaired protein synthesis dependent plasticity or 2) aberrant protein synthesis and remodeling of the proteome that impairs consolidation and/or recall. As ion channels are key mediators of neuronal excitability, the identification of numerous ion channels and receptors differentially expressed relative to memory status in both ‘normal’ aging and AD provides additional support to this hypothesis.
4.5 Considerations for proteomics analyses
Here we used strict protein identification criteria to identify observed proteins, including the identification of a “razor” peptide that uniquely matches the identified protein. Our FDR for protein identification was set at a strict 1% to minimize false positives. False negative results are also a known caveat inherent to proteomic studies, as current techniques do not yield complete coverage of the entire proteome due to a combination of biological and technical limitations98, 99. Therefore, caution should be used when interpreting the absence of a protein within a dataset. For example, a unique razor peptide produced by trypsin digestion may have inherent qualities that prevent ionization and make the peptide incompatible with detection by the mass spectrometer. In contrast, peptides that do ionize well may not be unique, and therefore cannot be reliably attributed to a specific protein98. In addition, peptides from a low-abundant protein may elute from the LC column at the same time as those from a much more highly abundant protein, and as such, will not be selected for sequencing by the mass spectrometer. For example, we observed a reliable overexpression of APP in samples from AD compared to Ntg mice (Table 7 and Supplementary Table 4), whereas peptides matched to PSEN1 were only detected in the hippocampus of one AD impaired and one AD intact animal (none were detected in hippocampi of Ntg mice). While expression of PSEN1 is expected in 5XFAD mice, as the gene encoding this protein is a component of the 5XFAD transgene, this result is consistent with prior observations of higher APP than PSEN1 in the 5XFAD model (Ref 16, Fig 1). However, for this reasons discussed above, the failure to reliably detect PSEN1 in 5XFAD mice may well be due to technical limitations, which suggests absolute quantitation using multiple reaction monitoring (MRM) proteomics may be necessary to reliably detect PSEN1 from hippocampal membrane samples. Overall, our analysis of proteins and pathways associated 5XFAD genotype identified a number of proteins previously associated with disease in animal models and humans, including APOE66 and CLU59–62, demonstrating that our approach and the statistical criteria used to mitigate the risk of both false positive and negatives is capable of capturing a high number of functionally relevant proteins.
5. Conclusion
Overall, our analyses provides a novel list of proteins that may play a functional role in cellular and molecular process underlying memory decline using methods that permit quantitation of low-abundance membrane proteins that are often underrepresented in proteomic analyses. Here we profile the hippocampal proteome of AD and normal Ntg aging mice following contextual fear training, and then stratify mice into well-defined groups based on their memory performance in order to identify novel proteins likely to be playing a causative role in memory deficits. Taken together, our results highlight novel putative mediators of cognitive decline, and examine the degree to which these molecular biomarkers and corresponding mechanisms overlap and/or diverge in both ‘normal’ aging and AD. Our results suggest a key role for epigenetic regulation, specifically via HDAC4, in the onset of memory deficits and impaired neuronal excitability that we reported in both AD and normal aging mouse models9–11. Broad HDAC inhibitors have been shown to be effective in treating AD-related memory deficits in mouse models100–102, but knowledge of which HDAC(s) are responsible for the majority of observed effects remains limited103. Our identification of HDAC4 as a key upstream regulator of changes in protein abundance relative to memory status in a mouse model of AD may aid in designing more effective and specific inhibitors while reducing off-target effects. In addition, the identification of REST as a disease-specific modulator of memory deficits provides an additional target that may explain exacerbated symptoms in AD compared to ‘normal’ aging. Ultimately, results here are poised to aid in the development of effective therapeutics for prevention and treatment of both AD and ‘normal’ cognitive decline and will be made publically available to the scientific community.
Supplementary Material
Highlights.
Proteomics detects 36 hippocampal proteins associated with AD and ‘normal’ aging memory deficits
Pathway analysis highlight HDAC4 as global regulator of memory deficits
103 proteins differ specifically in AD mice with intact vs impaired memory
Pathway analysis indicates disease-specific involvement of REST and Gi signaling
Publically available proteomics resource for hypothesis generation and testing
Acknowledgments
This work was supported by the National Institute on Aging (F31 AG050357 to SMN, R00 AG03511 to CCK) and the American Federation for Aging Research (PD15013 to LAW).
Abbreviations
- AD
Alzheimer’s disease
- CFM
contextual fear memory
- Ntg
Non-transgenic
- IPA
Ingenuity Pathway Analysis
- Aβ
beta-amyloid
Footnotes
Author Contributions: CCK conceived of the experiments. CCK, SMN, BH, KM, and LAW contributed to design, performance, and/or analyses of the experiments, as well as interpretation of results. SMN and CCK wrote the manuscript. All authors reviewed and contributed intellectually to the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Serrano-Pozo A, et al. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1(1):a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow RH, Burns JM, Khan SM. The Alzheimer's disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta. 2014;1842(8):1219–31. doi: 10.1016/j.bbadis.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, et al. Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim Biophys Acta. 2014;1842(8):1240–7. doi: 10.1016/j.bbadis.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Disterhoft JF, Moyer JR, Jr, Thompson LT. The calcium rationale in aging and Alzheimer's disease. Evidence from an animal model of normal aging. Ann N Y Acad Sci. 1994;747:382–406. doi: 10.1111/j.1749-6632.1994.tb44424.x. [DOI] [PubMed] [Google Scholar]
- 5.Jaworska A, et al. Analysis of calcium homeostasis in fresh lymphocytes from patients with sporadic Alzheimer's disease or mild cognitive impairment. Biochim Biophys Acta. 2013;1833(7):1692–9. doi: 10.1016/j.bbamcr.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16(6):358–72. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- 7.Zotova E, et al. Inflammatory components in human Alzheimer's disease and after active amyloid-beta42 immunization. Brain. 2013;136(Pt 9):2677–96. doi: 10.1093/brain/awt210. [DOI] [PubMed] [Google Scholar]
- 8.Hebert LE, et al. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology. 2013;80(19):1778–83. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaczorowski CC, Disterhoft JF. Memory deficits are associated with impaired ability to modulate neuronal excitability in middle-aged mice. Learn Mem. 2009;16(6):362–6. doi: 10.1101/lm.1365609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neuner SM, et al. TRPC3 channels critically regulate hippocampal excitability and contextual fear memory. Behav Brain Res. 2014 doi: 10.1016/j.bbr.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaczorowski CC, et al. Mechanisms underlying basal and learning-related intrinsic excitability in a mouse model of Alzheimer's disease. Neurobiol Aging. 2011;32(8):1452–65. doi: 10.1016/j.neurobiolaging.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–60. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 13.Levy-Lahad E, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269(5226):973–7. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 14.Goate A. Segregation of a missense mutation in the amyloid beta-protein precursor gene with familial Alzheimer's disease. J Alzheimers Dis. 2006;9(3 Suppl):341–7. doi: 10.3233/jad-2006-9s338. [DOI] [PubMed] [Google Scholar]
- 15.Sorbi S, et al. Missense mutation of S182 gene in Italian families with early-onset Alzheimer's disease. Lancet. 1995;346(8972):439–40. doi: 10.1016/s0140-6736(95)92809-x. [DOI] [PubMed] [Google Scholar]
- 16.Oakley H, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26(40):10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Linden DJ. The other side of the engram: experience-driven changes in neuronal intrinsic excitability. Nat Rev Neurosci. 2003;4(11):885–900. doi: 10.1038/nrn1248. [DOI] [PubMed] [Google Scholar]
- 18.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361(6407):31–9. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 19.Tan S, Tan HT, Chung MC. Membrane proteins and membrane proteomics. Proteomics. 2008;8(19):3924–32. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- 20.Vanguilder HD, Freeman WM. The hippocampal neuroproteome with aging and cognitive decline: past progress and future directions. Front Aging Neurosci. 2011;3:8. doi: 10.3389/fnagi.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CC, Yates JR., 3rd The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21(3):262–7. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 22.Grigoriadis DE, et al. Drugability of extracellular targets: discovery of small molecule drugs targeting allosteric, functional, and subunit-selective sites on GPCRs and ion channels. Neuropsychopharmacology. 2009;34(1):106–25. doi: 10.1038/npp.2008.149. [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski JR, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 24.Halligan BD, Greene AS. Visualize: a free and open source multifunction tool for proteomics data analysis. Proteomics. 2011;11(6):1058–63. doi: 10.1002/pmic.201000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5(11):2909–18. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 26.Sokal RRR, FJ . Biometry. New York: W.H. Freeman and Company; 1995. [Google Scholar]
- 27.Benjamini Y, HY Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J R Statist Soc. 1995;57(1):289–300. [Google Scholar]
- 28.Sando R, 3rd, et al. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151(4):821–34. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93(24):13445–52. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84(1):87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 31.Morris RG. Synaptic plasticity and learning: selective impairment of learning rats and blockade of long-term potentiation in vivo by the N-methyl-D-aspartate receptor antagonist AP5. J Neurosci. 1989;9(9):3040–57. doi: 10.1523/JNEUROSCI.09-09-03040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palop JJ, et al. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer's disease. Neuron. 2007;55(5):697–711. doi: 10.1016/j.neuron.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaczorowski CC. Bidirectional pattern-specific plasticity of the slow afterhyperpolarization in rats: role for high-voltage activated Ca2+ channels and I h. Eur J Neurosci. 2011;34(11):1756–65. doi: 10.1111/j.1460-9568.2011.07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giocomo LM, et al. Grid cells use HCN1 channels for spatial scaling. Cell. 2011;147(5):1159–70. doi: 10.1016/j.cell.2011.08.051. [DOI] [PubMed] [Google Scholar]
- 35.Hussaini SA, et al. Increased size and stability of CA1 and CA3 place fields in HCN1 knockout mice. Neuron. 2011;72(4):643–53. doi: 10.1016/j.neuron.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nolan MF, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119(5):719–32. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 37.Chan CS, et al. HCN2 and HCN1 channels govern the regularity of autonomous pacemaking and synaptic resetting in globus pallidus neurons. J Neurosci. 2004;24(44):9921–32. doi: 10.1523/JNEUROSCI.2162-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cobb SR, et al. Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature. 1995;378(6552):75–8. doi: 10.1038/378075a0. [DOI] [PubMed] [Google Scholar]
- 39.Whittington MA, Traub RD, Jefferys JG. Synchronized oscillations in interneuron networks driven by metabotropic glutamate receptor activation. Nature. 1995;373(6515):612–5. doi: 10.1038/373612a0. [DOI] [PubMed] [Google Scholar]
- 40.Hokama M, et al. Altered expression of diabetes-related genes in Alzheimer's disease brains: the Hisayama study. Cereb Cortex. 2014;24(9):2476–88. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan MG, et al. Genome wide profiling of altered gene expression in the neocortex of Alzheimer's disease. J Neurosci Res. 2010;88(6):1157–69. doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- 42.Saito Y, et al. Hyperpolarization-activated cyclic nucleotide gated channels: a potential molecular link between epileptic seizures and Abeta generation in Alzheimer's disease. Mol Neurodegener. 2012;7:50. doi: 10.1186/1750-1326-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stutzmann GE, et al. Enhanced ryanodine-mediated calcium release in mutant PS1-expressing Alzheimer's mouse models. Ann N Y Acad Sci. 2007;1097:265–77. doi: 10.1196/annals.1379.025. [DOI] [PubMed] [Google Scholar]
- 44.Carrasco MA, et al. Signal transduction and gene expression regulated by calcium release from internal stores in excitable cells. Biol Res. 2004;37(4):701–12. doi: 10.4067/s0716-97602004000400028. [DOI] [PubMed] [Google Scholar]
- 45.Cheung KH, et al. Mechanism of Ca2+ disruption in Alzheimer's disease by presenilin regulation of InsP3 receptor channel gating. Neuron. 2008;58(6):871–83. doi: 10.1016/j.neuron.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schon EA, Area-Gomez E. Mitochondria-associated ER membranes in Alzheimer disease. Mol Cell Neurosci. 2013;55:26–36. doi: 10.1016/j.mcn.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Rodenas-Ruano A, et al. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15(10):1382–90. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu T, et al. REST and stress resistance in ageing and Alzheimer's disease. Nature. 2014;507(7493):448–54. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armbruster BN, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104(12):5163–8. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62(3):544–52. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 51.Hille B. Modulation of ion-channel function by G-protein-coupled receptors. Trends Neurosci. 1994;17(12):531–6. doi: 10.1016/0166-2236(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 52.Reid CA, Bekkers JM, Clements JD. Presynaptic Ca2+ channels: a functional patchwork. Trends Neurosci. 2003;26(12):683–7. doi: 10.1016/j.tins.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Lv J, et al. Quantitative proteomics reveals that PEA15 regulates astroglial Abeta phagocytosis in an Alzheimer's disease mouse model. J Proteomics. 2014;110:45–58. doi: 10.1016/j.jprot.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 54.Hong I, et al. Quantitative proteomic analysis of the hippocampus in the 5XFAD mouse model at early stages of Alzheimer's disease pathology. J Alzheimers Dis. 2013;36(2):321–34. doi: 10.3233/JAD-130311. [DOI] [PubMed] [Google Scholar]
- 55.Sizova D, et al. Proteomic analysis of brain tissue from an Alzheimer's disease mouse model by two-dimensional difference gel electrophoresis. Neurobiol Aging. 2007;28(3):357–70. doi: 10.1016/j.neurobiolaging.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Vercauteren FG, et al. Early dysregulation of hippocampal proteins in transgenic rats with Alzheimer's disease-linked mutations in amyloid precursor protein and presenilin 1. Brain Res Mol Brain Res. 2004;132(2):241–59. doi: 10.1016/j.molbrainres.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 57.Andreev VP, et al. Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains. J Proteome Res. 2012;11(6):3053–67. doi: 10.1021/pr3001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emilsson L, Saetre P, Jazin E. Alzheimer's disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiol Dis. 2006;21(3):618–25. doi: 10.1016/j.nbd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Kok EH, et al. CLU, CR1 and PICALM genes associate with Alzheimer's-related senile plaques. Alzheimers Res Ther. 2011;3(2):12. doi: 10.1186/alzrt71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malkki H. Alzheimer disease: chaperone protein clusterin is involved in amyloid-beta-associated entorhinal atrophy in early AD. Nat Rev Neurol. 2014;10(2):60. doi: 10.1038/nrneurol.2014.1. [DOI] [PubMed] [Google Scholar]
- 61.Hardy J, Guerreiro R, Lovestone S. Clusterin as an Alzheimer biomarker. Arch Neurol. 2011;68(11):1459–60. doi: 10.1001/archneurol.2011.1000. [DOI] [PubMed] [Google Scholar]
- 62.Jones N. Alzheimer disease: plasma clusterin predicts degree of pathogenesis in AD. Nat Rev Neurol. 2010;6(9):469. doi: 10.1038/nrneurol.2010.122. [DOI] [PubMed] [Google Scholar]
- 63.Shankar GM, et al. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–42. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knobloch M, I, Mansuy M. Dendritic spine loss and synaptic alterations in Alzheimer's disease. Mol Neurobiol. 2008;37(1):73–82. doi: 10.1007/s12035-008-8018-z. [DOI] [PubMed] [Google Scholar]
- 65.Masliah E, Crews L, Hansen L. Synaptic remodeling during aging and in Alzheimer's disease. J Alzheimers Dis. 2006;9(3 Suppl):91–9. doi: 10.3233/jad-2006-9s311. [DOI] [PubMed] [Google Scholar]
- 66.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 67.Nakatani N, et al. Expression analysis of actin-related genes as an underlying mechanism for mood disorders. Biochem Biophys Res Commun. 2007;352(3):780–6. doi: 10.1016/j.bbrc.2006.11.101. [DOI] [PubMed] [Google Scholar]
- 68.Roth ED, et al. DNA methylation regulates neurophysiological spatial representation in memory formation. Neuroepigenetics. 2015;2:1–8. doi: 10.1016/j.nepig.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13(11):1319–23. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Penner MR, et al. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2011;32(12):2198–210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483(7388):222–6. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Azpurua J, Eaton BA. Neuronal epigenetics and the aging synapse. Front Cell Neurosci. 2015;9:208. doi: 10.3389/fncel.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meadows JP, et al. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci Signal. 2015;8(382):ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70(5):813–29. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guzman-Karlsson MC, et al. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim MS, et al. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32(32):10879–86. doi: 10.1523/JNEUROSCI.2089-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajasethupathy P, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149(3):693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartsch D, et al. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83(6):979–92. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 80.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89(4):599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Genoux D, et al. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418(6901):970–5. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- 82.Lord J, Cruchaga C. The epigenetic landscape of Alzheimer's disease. Nat Neurosci. 2014;17(9):1138–40. doi: 10.1038/nn.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Jager PL, et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17(9):1156–63. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ridge PG, et al. Alzheimer's disease: analyzing the missing heritability. PLoS One. 2013;8(11):e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer's disease: from synapses toward neural networks. Nat Neurosci. 2010;13(7):812–8. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D'Amelio M, Rossini PM. Brain excitability and connectivity of neuronal assemblies in Alzheimer's disease: from animal models to human findings. Prog Neurobiol. 2012;99(1):42–60. doi: 10.1016/j.pneurobio.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 87.Kunz L, et al. Reduced grid-cell-like representations in adults at genetic risk for Alzheimer's disease. Science. 2015;350(6259):430–3. doi: 10.1126/science.aac8128. [DOI] [PubMed] [Google Scholar]
- 88.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 89.Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6(3):361–70. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 91.Thompson LT, Moyer JR, Jr, Disterhoft JF. Transient changes in excitability of rabbit CA3 neurons with a time course appropriate to support memory consolidation. J Neurophysiol. 1996;76(3):1836–49. doi: 10.1152/jn.1996.76.3.1836. [DOI] [PubMed] [Google Scholar]
- 92.Whitlock JR, et al. Learning induces long-term potentiation in the hippocampus. Science. 2006;313(5790):1093–7. doi: 10.1126/science.1128134. [DOI] [PubMed] [Google Scholar]
- 93.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96(3):518–59. [PubMed] [Google Scholar]
- 94.Schimanski LA, Barnes CA. Neural Protein Synthesis during Aging: Effects on Plasticity and Memory. Front Aging Neurosci. 2010:2. doi: 10.3389/fnagi.2010.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McGaugh JL. Memory--a century of consolidation. Science. 2000;287(5451):248–51. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 96.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89(3):293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jarome TJ, Helmstetter FJ. Protein degradation and protein synthesis in long-term memory formation. Front Mol Neurosci. 2014;7:61. doi: 10.3389/fnmol.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Godoy LM, et al. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7(6):R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–99. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 100.Kilgore M, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35(4):870–80. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sung YM, et al. Mercaptoacetamide-based class II HDAC inhibitor lowers Abeta levels and improves learning and memory in a mouse model of Alzheimer's disease. Exp Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Govindarajan N, et al. Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26(1):187–97. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 103.Fischer A, et al. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31(12):605–17. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


