Abstract
Background
DNA methylation is a heritable covalent modification that is developmentally regulated and is critical in tissue-type definition. Although genotype-phenotype correlations have been described for different subtypes of renal cell carcinoma (RCC), it is unknown if DNA methylation profiles correlate with morphological or ontology based phenotypes. Here we test the hypothesis that DNA methylation signatures can discriminate between putative precursor cells in the nephron.
Experimental designs
We performed deep profiling of DNA methylation and transcriptome in diverse histopathological RCC subtypes and validated DNA methylation in an independent dataset as well as in The Cancer Genome Atlas Clear Cell and Chromophobe Renal Cell Carcinoma Datasets.
Results
Our data provide the first mapping of methylome epi-signature and indicates that RCC subtypes can be grouped into two major epi-clusters: C1 which encompasses clear-cell RCC, papillary RCC, mucinous and spindle cell carcinomas and translocation RCC; C2 which comprises oncocytoma and chromophobe RCC. Interestingly, C1 epi-cluster displayed three fold more hypermethylation as compared to C2 epi-cluster. Of note, differentially methylated regions between C1 and C2 epi-clusters occur in gene bodies and intergenic regions, instead of gene promoters. Transcriptome analysis of C1 epi-cluster suggests a functional convergence on Polycomb targets, whereas C2 epi-cluster displays DNA methylation defects. Furthermore, we find that our epigenetic ontogeny signature is associated with worse outcomes of patients with clear-cell RCC.
Conclusion
Our data defines the epi-clusters that can discriminate between distinct RCC subtypes and for the first time define the epigenetic basis for proximal versus distal tubule derived kidney tumors.
Keywords: renal cell carcinoma, subtypes, DNA methylation, polycomb, epigenetics, signature
Introduction
Renal cell carcinoma (RCC) is a heterogeneous disease with at least 12 subtypes of RCC based on the World Health Organization (WHO) classification, with distinct clinical outcomes ranging from malignant (ie clear cell RCC, papillary RCC) to benign (ie oncocytoma)(1). For both clear-cell RCC (ccRCC) and papillary RCC (pRCC), patients can present with either localized or metastatic disease and median overall survivals for the most common histology ccRCC is approximately 2 years (2, 3). Chromophobe RCC is usually indolent with a greater than 90% 10-year cancer specific overall survival, although some patients can develop metastases (3, 4).
Ontogenetically, each RCC subtype is classified on morphological and histological biomarkers; however some biomarkers can be expressed in more than one subtype making classification difficult (5-7). The precursor cell in the renal tubular system can impact the RCC subtype with studies suggesting that ccRCC and pRCC arise from the proximal tubules and oncocytoma and chromophobe arise from the distal tubules (8, 9). Given the common precursor cells that give rise to RCC, regulatory factors that maintain normal cell identity and cell proliferation may influence the RCC phenotype (5-7, 10). DNA methylation is a key DNA modification that allows proper regulation of gene expression and stable gene silencing allowing to define cell state (11). The importance of DNA methylation alterations in cancer has been extensively studied, with the discovery of both hypermethylation within promoter regions of certain tumor-suppressor genes and long-range hypomethylation which broadly contributes to cell transformation (12, 13). DNA methylation is considered to be stable, when compared to gene expression (14).
We hypothesized that we could identify a DNA methylation signature that discriminates between benign and malignant RCC. In this manuscript, we applied for the first time, to our knowledge, next-generation sequencing to study targeted DNA methylation in subtypes of RCC. Our analysis reveals two distinct RCC epi-clusters consistent each with their cell ontogeny.
Patients and Methods
Patient characteristics and tumor selection
Patient samples for the training set were related to 22 primary renal cell carcinomas with different histologies treated at the University of Texas, MD Anderson Cancer Center. Specimens were gathered in accordance with the institutional policies. All patients provided written informed consent. The study was approved by MD Anderson Cancer Center institutional review board. DNA was obtained exclusively from frozen nephrectomy, and none of the patient had been treated with chemotherapy or radiation prior to surgery. Tumors were selected solely on the basis of availability. Furthermore, DNA was extracted from 2 specimens of normal kidney tissue adjacent to renal cancers. Overall, our cohort included 3 ccRCC, 5 pRCC (3 type I and 2 type II), 5 tRCC confirmed cytogenetically, 2 oncocytomas, 3 chromophobes, 2 MTS and 2 RCC with sarcomatoid differentiation. All the hematoxylin and eosin-stained (HES) slides from surgical specimens have been reviewed by one uropathologist (F.T.). Out of the 22 cases, RNA was available for 11 cases including 2 oncocytoma, 3 chromophobe, 2 tRCC, 1 ccRCC, 2 papillary type 1 and 1 MTS. The clinical annotations of those samples are provided in Table 1.
Table 1.
Clinico-pathological annotations of renal cell carcinoma (RCC) samples selected in our study. All cases have been assessed for DNA methylation using DREAM technique. The details of sequencing tags and the read coverage are reported for DREAM. Selected RCC cases assessed by RNAseq are also specified.
| Histology | Sample-ID | sexe | TNM pathological stage | Total number of tags | RNAseq | Nb of CpG sites with at least 10 tags coverage per site | Nb of CpG sites with at least 100 tags coverage per site |
|---|---|---|---|---|---|---|---|
| MITF/TFE translocation RCC | T1 | F | T4N2M1 | 85 062 596 | Yes | 65 050 | 43 590 |
| T2 | M | T1N2M1 | 75 410 891 | no | 98 341 | 38456 | |
| T3 | M | T3bN0M0 | 71422259 | no | 139 443 | 44 191 | |
| T24 | F | T3aN0M0 | 99 019 782 | no | 162226 | 43 215 | |
| T34 | M | T3bN1M0 | 57537059 | yes | 158229 | 44375 | |
| Clear-cell RCC | T11 | F | T2N0M0 | 118 594 689 | yes | 156 156 | 42 552 |
| T12 | M | T2N0M0 | 42 845 148 | no | 53 989 | 33 982 | |
| T15 | F | T1bN0M0 | 86725973 | no | 65104 | 40630 | |
| Papillary RCC (Type I) | T14 | M | T1aN0M0 | 80 878 268 | no | 57 256 | 46 228 |
| T20 | M | T2aNxM0 | 125 615 959 | yes | 59059 | 42120 | |
| T21 | M | T4N1M0 | 91521140 | yes | 118971 | 45886 | |
| Papillary RCC (Type II) | T7 | M | T1bN0M0 | 78 384 802 | no | 151 757 | 37 930 |
| T8 | F | T3bN2M0 | 77404215 | no | 54928 | 45572 | |
| Muscinous and spindle cell RCC | T13 | M | T1N0M0 | 51 426 774 | no | 148 280 | 44 584 |
| T19 | F | T1bNxM0 | 27216549 | yes | 51127 | 38557 | |
| Sarcomatoid RCC | T22 | F | T4N1M1 | 59 402 641 | no | 121837 | 46 118 |
| T23 | M | T4N1M0 | 140885594 | no | 98104 | 44956 | |
| Oncocytoma | T45 | M | T1aNxM0 | 79 100 507 | yes | 82 991 | 36 167 |
| T46 | F | T1aNxM0 | 77335441 | yes | 87629 | 46205 | |
| chromophobe | T10 | F | T2N0M0 | 82 187 775 | yes | 61 940 | 61 940 |
| T16 | M | T2aNxM0 | 73 994 602 | yes | 78 618 | 45 828 | |
| T17 | F | T1bNxM0 | 38917059 | yes | 51653 | 34117 | |
| Normal | N4 | F | 87 459 889 | yes | 76 348 | 43 590 | |
| N8 | F | 84567697 | yes | 61256 | 41514 |
For the validation dataset, 41 new samples including 4 normal kidneys and 37 tumor samples were collected from Pitié-Salpêtrière Hospital. Specimens were gathered in accordance with the institutional policies. All patients provided written informed consent. The study was approved by Pitié-Salpêtrière ethical committee. Distribution of tumor samples in this independent dataset was as follows: clear-cell RCC (n=16), chromophobe RCC (n=6), oncocytoma (n=8), papillary type II RCC (n=6) and translocation RCC (n=1).
Digital Restriction Enzyme Analysis of Methylation
Analysis of DNA methylation was performed using Digital Restriction Enzyme Analysis of Methylation (DREAM), which is based on sequential DNA digestion with a pair of methylation-blocked and methylation-tolerant restriction enzymes SmaI/XmaI. These end sequences are repaired and then analyzed by ultra-deep next-generation sequencing; thereafter, the methylation status of each individual CCCGGG sites across the genome can be determined quantitatively (15). Sequencing was performed using Illumina Hiseq2000 at MD Anderson core facility. Mapping tags and algorithm of analysis have been previously reported (15, 16).
Genome annotation of DREAM data and statistical analysis
Genomic regions were defined according to NCBI coordinates downloaded from the UCSC web site (hg18 version). Promoters were defined as regions between −2000 bp from the transcription start site (TSS) to +2000 bp from TSS for each Refseq transcript. To calculate a gene promoter methylation, we averaged the methylation level of all CpG sites located between −2000bp and 2000bp from TSS.
To calculate CpG sites differentially methylated between C1 and C2 epi-clusters, a t-test was performed and CG sites were ranked according to their smallest p-value. A p-value less than 0.05 was considered as statistically significant.
RNA sequencing and analysis
RNA extraction for 11 RCC samples, for which materials were available, was done using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. RNA sequencing was performed using Illumina Hiseq2000 according to manufacturer instructions and after library preparation according to the NuGen Ovation RNA-Seq System V2 protocol. For the analysis, we first counted the overlaps between the mapped reads and genomic features, such as genes/exons using htseq-count script distributed with the HTSeq package. We then performed between-sample normalization when testing for differential expression. To do so, we used the scaling factor normalization method as it preserves the count nature of the data and has been shown to be an effective means of improving the detection of differential expression (17). To perform the normalization and the test for differential expression with a negative binomial model between conditions, we chose to use the Bioconductor package DESeq (version 1.11.0)(18). Specifically, for each gene, a generalized linear model (GLM) was fit to compare the expressions of the 2 epi-clusters C1 and C2. The Benjamini-Hochberg method was used to control the false discovery rate (FDR) (19).
Pathway analysis tools
GREAT prediction was performed using default settings (20). We first define the differentially methylated regions (DMR) as genomic regions located at + and −50 base pair from differentially methylated CpG sites. The background used was genomic regions covered by DREAM. A significantly enriched pathway was defined as the one with at least ten hyperforeground genes, a fold change of at least 2 and a qFDR less than 0.05.
For Ingenuity Pathway Analysis (IPA), default settings were used to analyze genes differentially expressed between C1 and C2 epi-clusters. Pathways differentially expressed were ranked according to their smallest p-values and were considered statistically significant if their p-value was less than 0.05.
Unsupervised clustering for promoter DNA methylation
Hierarchical clustering analysis was performed for CpG sites located in promoters CGI and outside CGI, with at least 10 tags of coverage across samples, using the Pearson correlation coefficient as the distance metric and Ward's linkage rule.
Analysis of gene expression between C1 and C2 epi-clusters
We performed unsupervised hierarchical clustering using all genes of the 11 RCC samples for which RNA seq were available. For Gene Set Enrichment Analysis (GSEA), gene sets were downloaded from the Broad Institute MSigDB website (21). Gene set permutations were used to determine statistical enrichment of the gene sets using the differentially expressed genes between C1 and C2 epi-clusters. Association of C1 and C2 epi-clusters expression patterns with those of specific regions of the nephron was done as previously reported (22) using data sets of Cheval et al. (23).
Validation using The Cancer Genome Atlas datasets
We extracted TCGA data from 271 samples of ccRCC and 66 chromophobes for which both DNA methylation data assessed by Infinium 450K and RNAseq were available (22, 24). Promoter DNA methylation was defined as regions spanning +/− 1000 base pair from TSS. Supervised clustering using the 56 genes epi-signature were performed on all the whole TCGA dataset (n=337) for both DNA methylation and gene expression. Accuracy of the method was then calculated for its ability to distinguish clear-cell RCC from chromophobe cases.
Analysis of associations between the epigenetic signature and overall survival of patients with clear-cell renal cell carcinomas
Supervised principal components (SPC) analysis was used to examine association between the gene expression of the 56 genes belonging to the epi-signature and overall survival of 463 ccRCC cases from TCGA (25, 26). This method has previously been used to examine associations between gene expression profiling data and survival in ccRCC (25, 26). We randomly divided TCGA ccRCC RNA-seq samples into two datasets, one of which was used as the training set (232 patients), and another as validation set (231 patients). Firstly, a modified univariate Cox score was calculated for the association between gene expression for each gene and overall survival, and genes whose Cox score exceeded a threshold that best predicted survival were used to carry out supervised principal components analysis. To determine the Cox threshold, the training set was split and principal components were derived from one half of the samples, and then used in a Cox model to predict survival in the other half. By varying the threshold of Cox scores and using twofold cross-validation, this process was repeated ten times, and a threshold of 0 (averaged over ten separate repeats of this procedure) was used to generate the principal components subsequently used to predict outcome, where all 56 genes were used to build the model.
For each case, we used the first principal component in a regression model to calculate a SPC risk score that represents the sum of the weighted gene expression levels for each of the 56 gene. To validate the SPC predictor, we computed risk scores for each of the 231 cases in validation set using the model developed in the 232 TCGA training set, and tested whether these scores were correlated with survival. To examine the role of individual genes in determining outcome, we computed importance scores for genes. The importance score is equivalent to the correlation between each gene and the first supervised principal component. Higher positive importance score means higher risk (worse survival, and lower negative importance score means lower risk (better survival).
Results
DNA methylation is conserved among normal kidney samples
To determine the variability of DNA methylation in normal kidney, we compared the differences in DNA methylation between normal kidneys from two different individuals using Digital Restriction Enzyme Analysis of Methylation (DREAM)(15). Using a minimum threshold of 10 tags/CG sites, we identified common 36,035 CG sites in the 2 normal kidneys. Of note, 91.6% (n=33,018/36,035) of those sites had at least a minimum coverage of 100 tags allowing us to analyze quantitatively subtle DNA methylation changes.
DNA methylation levels for each CG site were then plotted to identify correlations between the 2 normal kidneys. We found a high correlation (Spearman R=0.95, p<0.0001) suggesting there are few variations in DNA methylation in the normal kidneys. The estimated false discovery rate (FDR) of DREAM is 0.7% and 2.1%, using 15% and 20% as a difference methylation cut-off, respectively. This was consistent with our previous results using blood and breast samples (15, 16). To determine whether DNA methylation alters gene expression, we examined the association of DNA methylation with promoter activity. Consistent with our previous observations, we showed that promoters with no methylation (≤1%) were highly expressed (Figure S1A); conversely, those that had methylation levels >1% were repressed (Figure S1A). Interestingly, promoters with methylation ≥10% were completely repressed (Figure S1A). Although the frequency of methylated CG sites outside CGI was higher than in CGI, similar finding were observed for promoters located outside CGIs (Figure S1B), suggesting little difference between the correlation of DNA methylation and expression whether promoters were located in CGI or outside CGI.
Unsupervised clustering of DNA methylation profiles distinguishes renal cell carcinomas subtypes
To determine if RCC subtypes are associated with DNA methylation signatures, we analyzed DNA methylation of 22 RCC tumors with different subtypes using DREAM, with a median of 77 million tags obtained by sample (range: 27-141 millions) (Table 1). We assessed quantitative DNA methylation of a median 65,000 CpG sites (range: 51,000-156,000) per sample, with at least 10 tags coverage per CpG site; when we filtered for CpG sites with at least 100 tags coverage, we identified a median 43,000 CpG sites (range: 33,000-62,000) covered per sample (Table 1). After merging all the samples, we ended up with 36,035 CpG sites for which quantitative DNA methylation calculated on the basis of at least 10 tags coverage per CpG site (Supplementary Table 1); of note, the majority of those CpG sites had more than 100 tags coverage with a median sequencing depth of ~1,850 tags/CG site (range: 10-697,312). Out of those, 18,800 and 16,566 sites were located in CpG islands and outside CpG islands, respectively. We then filtered for CpG sites located in promoter CpG islands (n=13,593) and promoter outside CpG islands (n=3086).
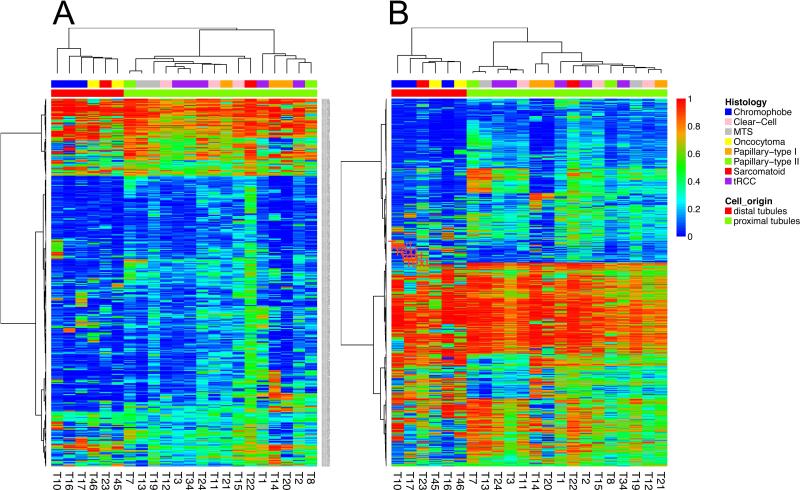
We then performed an unsupervised hierarchical clustering for promoter DNA methylation using CG sites. Our analysis revealed 2 similar epi-clusters independently of whether CG sites were located in CGI (CG islands) or outside (Figure 1A-B). Interestingly, the first epi-cluster “C1” was associated with ccRCC, pRCC, MITF/TFE translocation RCC (tRCC), mucinous and spindle cell carcinomas (MTS) and one sarcomatoid RCC case. On the hand, the second epi-cluster “C2” comprised oncocytoma, chromophobe, and one sarcomatoid sample. Sarcomatoid de-differentiation may occur in various RCC subtypes and after pathological re-review of the sarcomatoid cases, we identified the sarcomatoid case in the C1 epi-cluster arose from ccRCC. Similarly, the sarcomatoid case in the C2 epi-cluster arose from chromophobe RCC. Principal component analysis (PCA) confirmed the existence of the 2 epi-clusters (not shown).
Figure 1.
A) Unsupervised clustering of DNA methylation using CpG sites located in promoter CpG islands. B) Unsupervised clustering of DNA methylation using CpG sites located outside promotor CpG islands. Note that the analysis revealed 2 epi-clusters with C1 containing almost tumors with benign potential (except one sarcomatoid RCC case) and C2 containing tumors with potential malignant behavior
Unsupervised clustering of non-promoter CG sites located within and outside CGI yielded similar results (not shown), suggesting not only that tissue-specificity is related to CGI but that regulatory elements differentiating between proximal and distal tubules are dispersed across the genome. These finding support distinct DNA methylation signatures associated with histological RCC subtypes based on their cell ontogeny in the nephron.
C1 epi-cluster is characterized by coordinated methylation of gene promoters
To determine the gene pathways regulated by DNA methylation, we evaluated the differentially methylated promoters between the C1 and C2 epi-clusters. Using 15% difference of methylation as cut-off, we identified 40 CpG sites which harbored lower methylation in C2 epi-cluster as compared to C1 epi-cluster. Interestingly, there was no CG site unmethylated in C1 epi-cluster with concurrent gain of DNA methylation in the C2 epi-cluster, suggesting aberrant and coordinated DNA methylation in C1 epi-cluster. Importantly, the observed gain of DNA methylation in C1 epi-cluster was not associated with gene repression, except for the PLEK2 gene. This might be related to the fact that the majority of those CpG sites had measurable levels of methylation in C2 (≥1%) and were thus already repressed, consistent with previous reports (27).
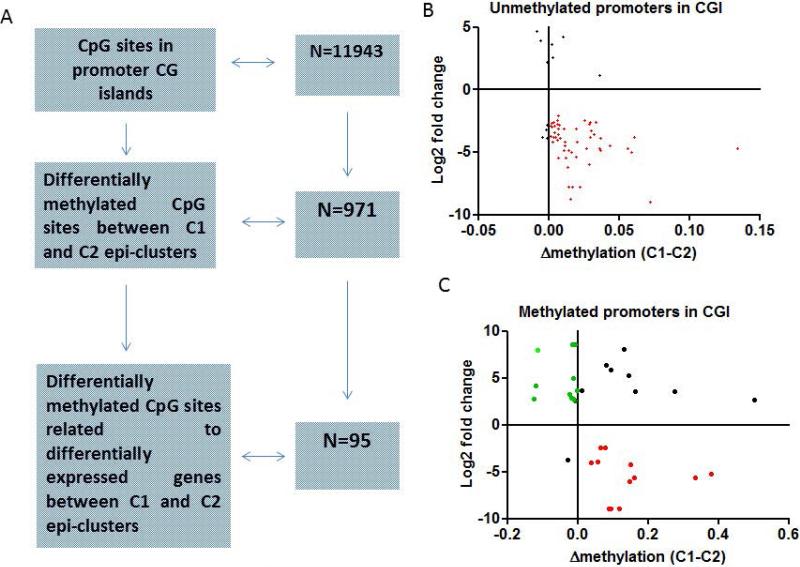
Our data indicates that promoters with some methylation (>1%) level were repressed when compared to those without any methylation level (Figure S1) suggesting that subtle changes in DNA methylation may impact gene expression. Using the following criteria, at least 2% methylation gain for unmethylated CG sites (≤1%) or demethylation below or equal to 1% for methylated CG sites (≥2%), we estimated the FDR of our method to be 0.01. In addition, as few CpG sites were identified using 15% difference methylation cut-off, we thus decided to analyze CpG sites in CGI which were differentially methylated between C1 and C2 epi-clusters (irrespective of their methylation difference levels) and with consistent changes in the expression of their related genes (Figure 2A). Overall, 971 out of 11,943 CG sites were differentially methylated (p-value <0.05), regardless of their methylation levels in C1 and C2 epi-clusters (Figure 2A). Out of those, 95 CpG sites related to 73 genes were also differentially expressed between C1 and C2 clusters (p<0.05) (Figure 2A) (Supplementary Table 2). To evaluate for an association between CpG site DNA methylation and gene expression, we divided the 95 CG sites in C2 epi-cluster into 2 subgroups: the first encompasses “unmethylated” (≤1% methylation) CpG sites (n=63) while the second one contained methylated (>1%) CpG sites (n=32). Interestingly, the majority of unmethylated CpG belonging to C2 epi-cluster showed gains of DNA methylation in C1 epi-cluster, consistent with gene expression repression (Spearman r=−0.46 (p<0.0001) (Figure 2B). Average methylation gain was estimated to be as low as 2.1% (p<0.0001), suggesting that small methylation gains might be correlated with gene repression. Likewise, we found an opposite correlation between genes that had “detectable level” of methylation in C2 epi-cluster and gene expression, although this was less strong then for our “unmethylated” subgroup (r=−0.37, p=0.03) (Figure 2C). Ingenuity Pathway Analysis (IPA) showed that those genes were enriched for interleukin-8 (p=4.6×10−3), CXCR4 (p=0.02) and mTOR signaling pathways (p=0.03) (Supplementary Table 3).
Figure 2.
A) Flow-chart showing the number of CG sites in promoter CG islands covered by DREAM and the number of differentially methylated CG between C1 and C2 epi-clusters. B) Correlation between differentially methylated CpG sites between C2 and C1 epi-clusters and gene expression changes. Herein, only unmethylated CpG sites (≤1%) of C2 epi-cluster located in promoter CG islands and both differentially methylated and expressed as compared to C1 epi-cluster are depicted. C) Correlation between differentially methylated CpG sites between C2 and C1 epi-clusters and gene expression changes. Herein, only methylated CpG sites (>1%) of C2 epi-cluster located in promoter CG islands and both differentially methylated and expressed as compared to C1 epi-cluster are depicted. Note that few of those methylated CG sites lost DNA methylation in C1 epi-cluster and become expressed (green), while the majority gains DNA methylation and get repressed.
RCC subtypes derived from the distal tubules display defects in DNA methylation
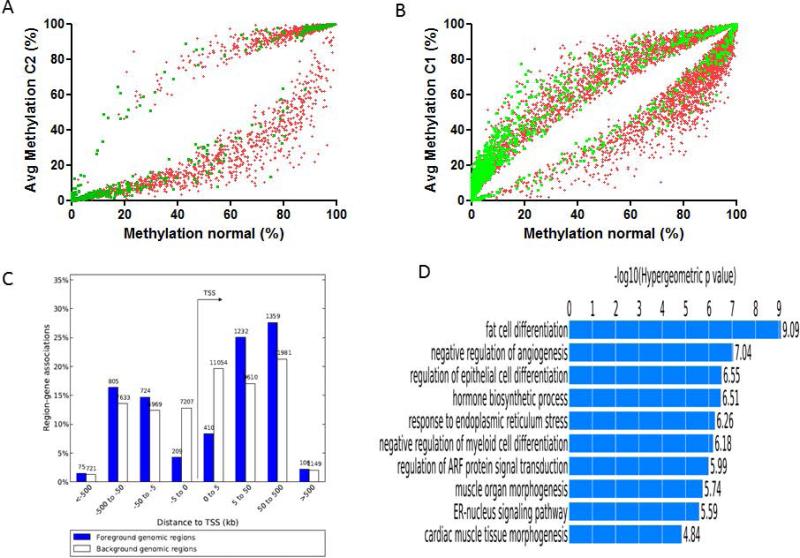
To identify differentially methylated CpG sites between the C2 epi-cluster relative to both normal kidney and C1 epi-cluster, we used a differential DNA methylation cut-off of 15% (FDR=0.03). We identified a trend towards lower DNA methylation in 2.4% (n=834/35366) of CG sites in C2 epi-cluster as compared to normal kidneys; in contrast, 0.6% (n=220/35366) of them gained DNA methylation as compared to normal kidneys (Figure 3A). This trend was not observed when we compared C1 epi-cluster to normal kidneys, as 3.8% (n=1363/35366) and 3.2% (n=1122/35366) of differentially methylated CG sites in C1 epi-cluster gained and lost DNA methylation, respectively, as compared to normal kidneys (Figure 3B).
Figure 3.
A) Square graph of CpG sites (green: CpG islands; red: outside CpG islands) with statistically significant difference of DNA methylation between C2 epi-cluster and normal kidneys. Note the tendency toward global hypomethylation of C2 epi-cluster with almost no gain of DNA methylation in CGI. B) Square graph of CpG sites with statistically significant difference of DNA methylation between C1 epi-cluster and normal kidneys. Note the tendency toward both hypomethylation of CpG sites located outside CGI and hypermethylation of CpG sites in CGI. C) Distribution of differentially methylated CpG sites between C1 and C2 epi-clusters according to the distance from transcription start site (TSS) as assessed by GREAT tool. D) Gene Ontology functional annotations for differentially methylated regions between C1 and C2 epi-clusters as identified by GREAT tool.
The same trend was maintained when differentially methylated CG sites were analyzed between C2 epi-cluster relative to C1 epi-cluster. Indeed, genome-wide, C1 epi-cluster displayed three fold more hypermethylation (5.6%; n=1872/33536) than hypomethylation (2.2%; n=773/35366) as compared to C2 epi-cluster. Of note, the majority of those CG sites were located far from the transcription start sites, mainly in gene bodies and intergenic regions (Figure 3C). GREAT Gene Ontology (GO) tools shows that differentially Methylated Regions (DMR) were enriched for genes involved in negative regulation of angiogenesis, regulation of epithelial cell differentiation and regulation of ARF protein signal transduction (Figure 3D) (Supplementary Table 4). This is interesting since genes involved in epithelial differentiation are key genes for the development of kidney, such as PAX2, PAX8, GATA3 and LHX1. Furthermore, genes involved in the regulation of angiogenesis are key therapeutic targets in ccRCC. Finally, GREAT identified enrichment for two motifs in those DMR; the first matches to the peroxisome proliferative activated receptor, alpha (PPARA) gene (p=1.46e-11) and the second to the CCAAT/enhancer binding protein (C/EBP), beta (CEBPB) gene (p=3.5e-4) (Supplementary Table 5). Importantly, CEBPB is one of the major transcription factors controlling the differentiation of a range of cell types (28). We thus conclude that DMRs between C1 and C2 epi-clusters contain key genes involved in epithelial differentiation, with several putative binding sites of enhancers.
C1 epi-cluster transcriptome is enriched for activation of genes related to Polycomb
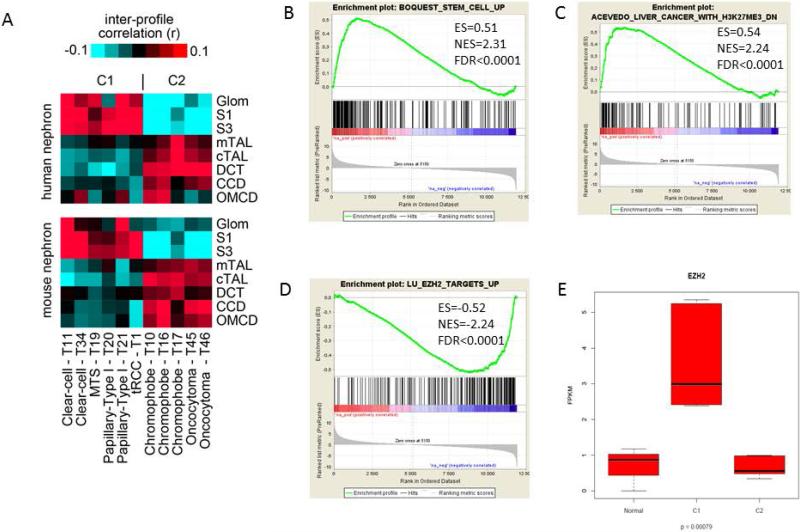
To define cell ontogeny of RCC subtypes, we examined our gene expression data using an external gene expression dataset of normal kidney cells microdissected from distinct regions of the nephron (23). Through supervised analysis, we globally compared the gene expression data of the C1 and C2 epi-clusters with that of each cell sample in the nephron structure, and found high mRNA expression correlations for C1 and C2 with proximal and distal tubules of the nephrons, respectively (Figure 4A).
Figure 4.
A) Heat maps showing inter-sample correlations (red, positive) between mRNA profiles of RCC (columns) belonging to C1 and C2 epi-clusters and mRNA profiles of nephron anatomical sites (rows). Association of C1 and C2 epi-clusters expression patterns with those of specific regions of the nephron are depicted. The 8 kidney nephron regions evaluated are : Glom, glomerulus; S1 and S3, the proximal tubule; mTAL, medullary thick ascending limb of Henle's loop; cTAL, cortical thick ascending limb of Henle's loop; DCT, distal convoluted tubule; CCD, cortical collecting duct; OMCD, outer medullary collecting duct. B-D) Gene Set Enrichment Analysis (GSEA) for differentially expressed genes in C1 as compared to C2 epi-clusters. E) Box-plot for EZH2 gene expression levels in normal kidneys and C1 and C2 epi-clusters.
To understand pathways which are specifically altered in our RCC epi-clusters, we thus compared differentially expressed genes between C1 and C2 epi-clusters using available RNAseq data. Overall, we identified 4.7% (n=606/12,877) of genes which were up-regulated in C1 epi-cluster as compared to 4.9% (n=627/12,877) of them which were down-regulated. Gene Set Enrichment Analysis (GSEA) revealed that up-regulated pathways in C1 epi-cluster were enriched for epithelial-mesenchymal transition (EMT) (FDR<0.0001), stem cell signature (Figure 4B) and Polycomb target genes (FDR<0.0001) (Figure 4C). Furthermore, EZH2 targets were found among the top altered pathways consistent with activation of EZH2 in the C1 epi-cluster (Figure 4D) (FDR<0.0001). We thus looked at EZH2 expression and confirmed its overexpression in C1 as compared to C2 epi-cluster (log2fold change=2.7, p=0.001) (Figure 4E). These data highly suggest that the difference between C1 and C2 epi-clusters might be related to the activation of the Polycomb repressive complex 2 (PRC2) within C1 epi-cluster.
Discovery of genes with promoter DNA hypermethylation in RCC
To discover genes repressed by DNA methylation and potentially novel candidate tumor suppressor genes in RCC, we analyzed genes which gain promoter DNA methylation in our dataset. Promoter DNA methylation was defined as genes with less than 2% methylation in normal kidney and which gain DNA methylation ≥10% methylation in cancer. Overall, 713 out of 4558 genes gain DNA methylation as compared to normal kidneys. Out of those, 126 genes gained DNA methylation in C2 epi-cluster while 688 genes gained DNA methylation in C1 epi-cluster. DAVID functional pathway analysis revealed that genes which gain DNA methylation are involved in development (p=1.8×10−9), cell differentiation (p=1.2×10−5). Furthermore, those were related to Homeobox genes (p=1.37×10−7) and displayed sequence-specific DNA binding (p=2.07×10−6) (Supplementary Table 6).
As genes that gain DNA methylation in cancer have been shown to be Polycomb targets in embryonic stem cells and adult stem/progenitor cells (29), we thus analyzed our results using the chromatin marks of the embryonic stem (ES) cells and found that 487 (68.3%) of gene promoters gaining DNA methylation in our RCC samples were marked by H3K27me3 in ES cells. These data are consistent with the existence of a “DNA hypermethylation module” in RCC and which has been previously shown to encompass a portion of the Polycomb target genes (29).
We then matched the list of our 126 genes methylated in C2 epi-cluster with the list of known tumor suppressor genes (30) and identified 11 candidate genes; Out of them, only IRX1 was methylated in 2 out of 3 chromophobe RCCs.
On the other hand, 47 genes out of the 688 genes which gained promoter DNA methylation in C1 epi-cluster C1 had been previously described as TSG. Out of the 205 genes methylated in at least 2 RCC samples, we identified 2 known TSGs, IRX1 and SOX11. IRX1 was methylated in 4 RCC cases and SOX11 was methylated in 3 RCC cases. The list of those 205 genes is reported in Supplementary Table 7. We then asked if any of the 205 genes which we identified as methylated at least 2 RCC samples are also methylated and repressed in a large independent cohort of ccRCC from The Cancer Genome Atlas (TCGA) project. Overall, 25 genes were identified as methylated and repressed in at least 2% of ccRCC samples, the majority of them never reported previously in ccRCC. Those genes were related to EMT (i.e. FOXC2 and OLMF1) and stimulation of endothelial cell proliferation and angiogenesis (i.e. SMOC2 and CLEC14A) pathways. The list of those genes including the prevalence of their methylation in ccRCC dataset is reported in Supplementary Table 8. Finally, we focused on the 5 tRCC and identified 85 genes which gained DNA methylation (Supplementary Table 7). Out of those, 15 genes were methylated in at least two samples (i.e. WNT3A, IRX4). Of note, the tumor suppressor gene IRX4 gained promoter DNA methylation in the 2 metastatic tRCC cases which was not the case for the three remaining cases.
Independent validation of the epi-signature in an independent dataset
We then asked if the 73 genes which we identified in our dataset as differentially methylated and expressed between C1 and C2 epi-clusters could distinguish chromophobe from ccRCC, using an independent data of renal cell carcinomas subtypes using Infinium 450K arrays. This dataset encompasses, 41 new samples including 4 normal kidneys and 37 tumor samples. Distribution of tumor samples in this independent dataset was as follows: clear-cell RCC (n=16), chromophobe RCC (n=6), oncocytoma (n=8), papillary type II RCC (n=6) and translocation RCC (n=1). To do so, we first filtered for gene promoters with data available for both DNA methylation β-values and gene expression; out of those 73 genes, data were available for 56 genes (Supplementary Table 9). Supervised clustering analysis using our 56 genes signature confirmed our previous findings about classification of kidney tumors in two major epi-clusters C1 and C2 (Figure S2). Of note, C2 contains the majority of oncocytoma and chromophobe RCC cases (n=11/13) as compared to C1 which encompasses the remaining RCC subtypes including clear-cell RCC, papillary RCC and translocation RCC (n=21/24) (p=0.0001).
Independent validation of the epi-signature in The Cancer Genome Atlas (TCGA) dataset
We furthermore performed a supervised clustering based on either DNA methylation (Figure 5A) or gene expression (5B) using this ontogeny epi-signature in an independent data sets from The Cancer Genome Atlas (TCGA) project encompassing 271 ccRCC and 66 chromophobe RCC. Importantly, hierarchical clustering revealed two distinct clusters with 94.4% and 97.6% accuracy for distinguishing chromophobe from ccRCC, respectively. Indeed, using gene expression signature, 97% (n=64/66) of chromophobe and 97.8% (n=265/271) ccRCC were classified correctly; in the other hand, using DNA methylation signature, 86.4% (n=57/66) chromophobe and 96.3% (n=261/271) ccRCC were classified correctly. Of note, several ccRCC which clustered with chRCC using our epi-genetic signature were misclassified and were chRCC or papillary clear-cell RCC features as we recently demonstrated in our long non-coding RNA subtype classification of ccRCC (6) (Supplementary Table 10).
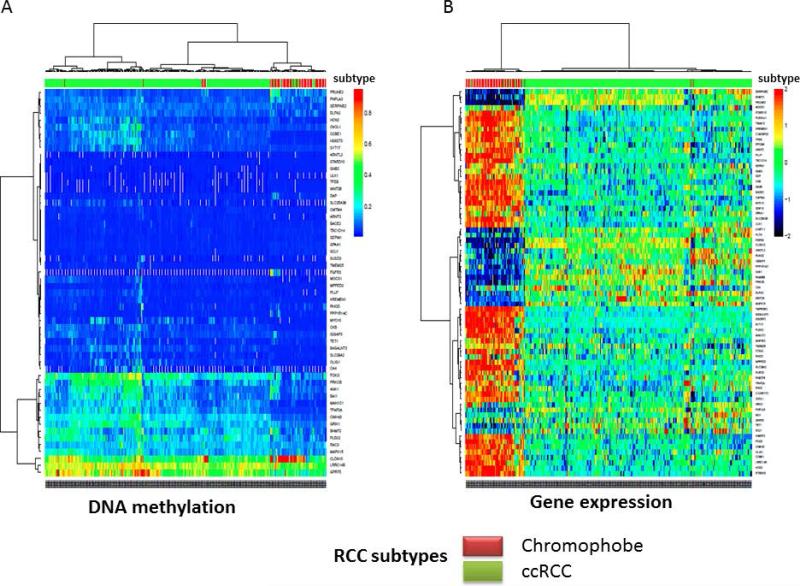
Figure 5.
A) Supervised clustering of promoter DNA methylation using the 56 genes epi-signature in The Cancer Genome Atlas (TCGA) dataset of clear-cell renal cell carcinomas and chromophobe samples B) Supervised clustering of gene expression using the 56 genes epi-signature in The Cancer Genome Atlas (TCGA) dataset of clear-cell cell carcinomas and chromophobe samples
Epigenetic signature is associated with outcome of patients with clear-cell renal cell carcinomas
When then asked whether epigenetic signature is capable of predicting outcome for patients with clear-cell RCC. To do so, we thus examined the association between the gene expression of the 56 genes defining the epi-signature and overall survival of 463 patients with ccRCC from TCGA. Firstly, we randomly divided TCGA ccRCC RNA-seq samples into two datasets, one of which was used as the training set (232 patients), and another as validation set (231 patients). For each case, we used the first principal component to calculate a supervised principal component (SPC) risk score (Supplementary Table S11). To validate the SPC predictor, we computed risk scores for each of the 231 cases in validation set using the model developed in the 232 TCGA training set (Supplementary Figure 3A). The risk score based on the supervised principal-components analysis was significantly associated with poor outcome in the validation cohort (Log–rank test p value = 0.0004) (Supplementary Figure 3B). Patients were then classified as being high or low risk according to the calculated SPC risk score. To examine the role of individual genes in determining outcome, we computed importance scores for genes. Finally, we asked whether there are associations between mutational load, ccRCC TCGA transcriptomic four-subgroup classification and somatic mutations; we discovered that our 56-gene signature was highly correlated with TCGA transcriptomic classification (supplementary Table 12) but not with mutational load. These data highly suggest that our epigenetic ontogeny signature is correlated with ccRCC transcriptomic classification and is useful in predicting outcome of patients with ccRCC.
Discussion
DNA methylation is a heritable covalent modification that is developmentally regulated, controls stemness, and is critical in tissue-type definition. Our data demonstrates the utility of analyzing the DNA methylation profiles of RCC subtypes that arise from either proximal or distal cells of the nephron to define an epigenetic basis of cell ontogeny in kidney cancers. Our key findings include 1) RCC subtypes can be grouped into two major epi-clusters; C1 which encompasses ccRCC, pRCC, MTS, tRCC; C2 which comprises oncocytoma, chromophobe RCC 2) differentially methylated regions between C1 and C2 occur in gene bodies and intergenic regions, instead of gene promoters with a functional convergence on Polycomb targets 3) methylation defects in C2 epi-clusters 4) the identification of a 56-gene epi-signature charting kidney cell ontogeny that was predictive of outcomes in ccRCC.
Historically, the cells of origin of different RCC subtypes have been controversial, and hypotheses are based on morphological similarities between RCC subtypes and proximal or distal cells of the nephron (7). Those observations have been corroborated by Heidelberg classification which added specific genetic aberrations to morphology (8). Recent studies that incorporate DNA methylation profiling have identified chromophobe RCC-specific methylation or differentially methylated regions between RCC tumors and adjacent uninvolved kidney (31, 32). Our data further defines the epi-clusters that can discriminate between additional RCC subtypes (ccRCC, pRCC, MTS, tRCC vs oncocytoma, chromophobe RCC) and for the first time, to our knowledge, define a epigenetic basis for proximal vs distal derived tumors using a training and an independent data sets.
The DREAM technique that incorporates next-generation sequencing to analyze DNA methylation allows accurate quantitation, thus allowing us to detect differences through the kidney methylome. First, we identified two main epi-clusters; C1 epi-cluster encompasses ccRCC, pRCC, MTS and tRCC. C2 epi-cluster comprises oncocytoma and chromophobe RCC. Interestingly, our data supports an epigenetic basis for the Heidelberg classification, suggesting that MTS and tRCC may also arise from proximal tubules. Second, we discovered that much of the differentially methylated regions distinguishing C1 and C2 epi-clusters occur outside of gene promoters; motif analysis uncovers binding sites related to transcription factors PPARA and CEBPB; of note, PPARA has been reported to be expressed predominantly in proximal tubules and medullary thick ascending limbs, in contrast to PPAR gamma which is exclusively expressed in medullary collecting duct and papillary urothelium (33). Furthermore, CEBPB is considered as one of the major transcription factors controlling the differentiation of a range of cell types (28). Altogether, these data corroborate the dynamic regulation of CpG DNA methylation during normal development which mainly occurs at distal regions from the transcription start sites (34). Third, we observed a methylation defect in C2 epi-cluster consistent with a recent report using Infinium arrays in a selected cohort of chromophobe (31), and most significantly we discovered that differentially expressed genes between C1 and C2 epi-clusters were targets of Polycomb; as expected, the methyltransferase EZH2 was overexpressed in C1 epi-cluster, which might be related to larger H3K27me3 domains in C1 epi-cluster as compared to C2. We speculate that activation of the PRC2 in the proximal tubules cell of origin might explain the aggressiveness of derived tumor subtypes in C1 epi-cluster, in contrast to distal tubules cell of origin giving rise to oncocytoma and chromophobe which have usually good outcome. We thus suggest that epigenetic therapies may have utility against a broad range of the most life-threatening kidney cancers, independent of genotype and morphological phenotype.
Finally, we have established an epi-signature ontogeny-based classifier composed of 56 genes. Using an independent cohort from TCGA comprising ccRCC and chromophobe samples, we validated the high accuracy of this ontogeny epi-signature for distinguishing tumors arising either from the proximal tubules or from the distal tubules of the nephron. Of note, this is consistent with the distinct pattern of genetic mutations according to kidney cancer ontogeny. Indeed, the mutation rates per exome for ccRCC (n=43) and pRCC (n=59) are by far higher than chromophobe (n=17) and oncocytoma (n=15) RCC (35). One explanation might be chromatin organization of kidney cell of origin as mutations rates in cancer genomes are associated with epigenetic architecture of human cells (36); indeed, heterochromatin-associated histone modification can account for more than 40% of mutation-rate variation (36). Our 56 genes epi-signature was also able to predict survival of patients with ccRCC. One explanation might be that ccRCC arising from distal part of the proximal nephron harbor a good outcome as recently showed by Buttner et al. using TCGA datasets (37).
In summary, we define herein an epigenetic basis of kidney tumors ontogeny and provide the first mapping of methylome epi-signature across different histological RCC subtypes. In addition, we showed that our epigenetic ontogeny signature is also capable of predicting patients outcome.
Supplementary Material
Translational relevance.
Genotype-phenotype correlations have been described for different subtypes of renal cell carcinoma (RCC), but it is unknown if epigenetics can define cell ontology across diverse histological kidney subtypes. Herein, we demonstrate that RCC can be divided epigenetically in 2 epi-clusters which correlate with kidney cell ontogeny. While C1 epi-cluster encompass clear-cell RCC and papillary RCC, C2 epi-cluster was composed of chromophobe RCC and oncocytomas. Of note, C1 epi-cluster displayed three fold more hypermethylation as compared to C2 epi-cluster consistent with functional convergence on Polycomb targets. Finally, our epigenetic ontogeny signature was associated with worse outcomes of patients with clear-cell RCC. These data provide clues for the epigenetic basis of proximal versus distal tubule derived kidney tumors and suggest that interest of using epigenetic therapy in the most threatening subtypes of kidney cancers.
Acknowledgments
Grant and Funding
The authors acknowledge AVEC Foundation for her support of research in kidney cancer. T.H.H. is supported by funding from the ASCO Young Investigator Award from the Kidney Cancer Association and NIH grant K12 CA90628. C.J.C is supported by NIH CA125123 grant.
Footnotes
Conflict of interest disclosure statement: The authors disclose no potential conflicts of interest
Disclosure declaration
The authors declare non conflict of interest
References
- 1.Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. 2004 WHO classification of the renal tumors of the adults. Eur Urol. 2006;49(5):798–805. doi: 10.1016/j.eururo.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 3.Amin MB, Paner GP, Alvarado-Cabrero I, Young AN, Stricker HJ, Lyles RH, et al. Chromophobe renal cell carcinoma: histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 145 cases. The American journal of surgical pathology. 2008;32(12):1822–34. doi: 10.1097/PAS.0b013e3181831e68. [DOI] [PubMed] [Google Scholar]
- 4.Przybycin CG, Cronin AM, Darvishian F, Gopalan A, Al-Ahmadie HA, Fine SW, et al. Chromophobe renal cell carcinoma: a clinicopathologic study of 203 tumors in 200 patients with primary resection at a single institution. The American journal of surgical pathology. 2011;35(7):962–70. doi: 10.1097/PAS.0b013e31821a455d. [DOI] [PubMed] [Google Scholar]
- 5.Malouf GG, Monzon FA, Couturier J, Molinie V, Escudier B, Camparo P, et al. Genomic heterogeneity of translocation renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(17):4673–84. doi: 10.1158/1078-0432.CCR-12-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malouf GG, Zhang J, Yuan Y, Comperat E, Roupret M, Cussenot O, et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol Oncol. 2014;9(1):32–43. doi: 10.1016/j.molonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuehn A, Paner GP, Skinnider BF, Cohen C, Datta MW, Young AN, et al. Expression analysis of kidney-specific cadherin in a wide spectrum of traditional and newly recognized renal epithelial neoplasms: diagnostic and histogenetic implications. The American journal of surgical pathology. 2007;31(10):1528–33. doi: 10.1097/PAS.0b013e318058818c. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, et al. The Heidelberg classification of renal cell tumours. The Journal of pathology. 1997;183(2):131–3. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 9.Oosterwijk E, Rathmell WK, Junker K, Brannon AR, Pouliot F, Finley DS, et al. Basic research in kidney cancer. Eur Urol. 2011;60(4):622–33. doi: 10.1016/j.eururo.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679–92. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 14.Issa JP. DNA methylation as a clinical marker in oncology. J Clin Oncol. 2012;30(20):2566–8. doi: 10.1200/JCO.2012.42.1016. [DOI] [PubMed] [Google Scholar]
- 15.Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, et al. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics : official journal of the DNA Methylation Society. 2012;7(12):1368–78. doi: 10.4161/epi.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, et al. Architecture of epigenetic reprogramming following Twist1 mediated epithelial-mesenchymal transition. Genome Biol. 2013;14(12):R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 20.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer cell. 2014;26(3):319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheval L, Pierrat F, Rajerison R, Piquemal D, Doucet A. Of mice and men: divergence of gene expression patterns in kidney. PLoS One. 2012;7(10):e46876. doi: 10.1371/journal.pone.0046876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2(4):E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bair EHH, Debashis P, Tibshirani R. Prediction by Supervised Principal Components. J Am Stat Assoc. 2006;101:119–37. [Google Scholar]
- 27.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 28.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends in cell biology. 2007;17(7):318–24. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome research. 2012;22(5):837–49. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41(Database issue):D970–6. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slater AA, Alokail M, Gentle D, Yao M, Kovacs G, Maher ER, et al. DNA methylation profiling distinguishes histological subtypes of renal cell carcinoma. Epigenetics : official journal of the DNA Methylation Society. 2013;8(3):252–67. doi: 10.4161/epi.23817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasseigne BN, Burwell TC, Patil MA, Absher DM, Brooks JD, Myers RM. DNA methylation profiling reveals novel diagnostic biomarkers in renal cell carcinoma. BMC Med. 2014;12(1):235. doi: 10.1186/s12916-014-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan Y, Zhang Y, Davis L, Breyer MD. Expression of peroxisome proliferator-activated receptors in urinary tract of rabbits and humans. Am J Physiol. 1997;273(6 Pt 2):F1013–22. doi: 10.1152/ajprenal.1997.273.6.F1013. [DOI] [PubMed] [Google Scholar]
- 34.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durinck S, Stawiski EW, Pavia-Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2014;47(1):13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488(7412):504–7. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 37.Buttner F, Winter S, Rausch S, Reustle A, Kruck S, Junker K, et al. Survival Prediction of Clear Cell Renal Cell Carcinoma Based on Gene Expression Similarity to the Proximal Tubule of the Nephron. Eur Urol. 2015;68(6):1016–20. doi: 10.1016/j.eururo.2015.05.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.