Abstract
We present drop-on-demand inkjet-based mammalian cell printing with a 30-μm nozzle diameter for cell-level accuracy. High-speed imaging techniques have been used to analyze the go-and-stop movement of cells inside the nozzle under a pulsed pressure generated by a piezo-actuator and the jet formation after ejection. Patterning of an array of 20 × 20 dots on a glass substrate reveals that each printed drop contains 1.30 cells on average at the cell concentration of 5.0 × 106 cells ml−1 for the very small nozzle, whereas larger nozzles with the diameter of 50 and 80 μm deliver 2.57 and 2.88 cells per drop, respectively. The effects of the size and concentration of printed cells on the number of cells have also been investigated. Furthermore, the effect of the nozzle diameter on printed cells has been evaluated through an examination of viability, proliferation, and morphology of cells by using a live/dead assay kit, CCK-8 assay, and cellular morphology imaging, respectively. We believe that the 30-μm inkjet nozzle can be used for precise cell deposition without any damages to the printed mammalian cells.
I. INTRODUCTION
The direct printing of living cells and biomaterials, also called “bioprinting,” has opened new doors for tissue engineering and regenerative medicine, representing a breakthrough in traditional pre-formed scaffold-based tissue engineering.1–4 Given that bioprinting is capable of placing cells and biomaterials in predetermined positions with appropriate cell compositions and density, there has been tremendous progress in the fabrication of two-dimensional (2D) cell patterns or three-dimensional (3D) structures, such as skin, ear, cartilage,5 and so on. Recently, beyond simple or thin structures, there emerged a demand for normally functioning but highly complex, hierarchical, and stratified architectures through accurate and precise cell printing in order to mimic both native complex and thick organs with heterogeneity in a length scale of natural tissues (10 μm–100 μm).6 A sophisticated vascular network also needs cell-level accuracy. These living tissues and organs are histologically multicellular constructs involving extracellular matrix proteins, and their respective functions could be expressed via a microenvironment, which is sensitive to the interactions among these components in 3D space. Therefore, stably and reliably controlling the spatial position and arrangement of cells and biomaterials in 3D space is crucial at high resolutions that are nearly equal to microscopic tissue structures of bio-structural and functional tissues.7
To ensure that bioprinting can lead to elaborately engineered architectures, various types of printers, such as drop-on-demand (DOD) inkjet,8–10 microextrusion,11,12 laser-assisted,13–15 and electrohydrodynamic (EHD)16,17 printers, have been introduced. Among these bioprinting techniques, DOD inkjet printing has been proven to be a promising approach in the accurate deposition of controlled volume of a cell-laden drop on demand.18,19 Given that spatial concentration gradients requiring accurate tiny droplet deposition play a pivotal role in biotechnology,20 such as in engineering microenvironments, cell behaviors on immobilized growth factors have been explored by printing spatial concentration gradients of growth factors using a 30-μm nozzle.21 In the investigation of multicellular interactions among synthetic bacterial cells, the concentration gradient of fluorescent proteins and Escherichia coli that is ∼2 μm-long has also been printed using a 21.5-μm nozzle.22 In addition, the stochastical23 or automatical24 ejection of a single cell could elicit zonal organization in 3D space with desired cell density.
However, in order to achieve high-resolution printing of mammalian cells with diameters ranging from 15–20 μm, the reputation of high resolution or tiny droplet volume has been derogated from the common adoption of a large nozzle diameter. As summarized in Table I, DOD inkjet nozzles having a diameter equal to or larger than 48 μm, which is approximately three times greater than that of cells (i.e., NIH/3T3 cells), have been typically utilized to avoid several practical problems, such as nozzle blockage and high shear stress. The Boland group used a thermal inkjet printer with a nozzle diameter of 48 μm to print Chinese Hamster Ovary cells and embryonic rat motoneurons25 as well as primary rat hippocampal neurons.26 Meanwhile, the Derby group used a piezoelectric inkjet printer in evaluating cell viability and proliferation with actuation parameters by printing with a 60-μm inkjet nozzle.9 Lorber et al. reported the absence of cell deformation during jet ejection through two nozzles with diameters of 50 and 80 μm.27 Nakamura and his colleagues electrostatically actuated a triangular-shaped nozzle with a 43-μm height, optically counted cell numbers in each drop, and observed the pre- and post-printed morphologies of cells.28 Moreover, Xu et al. studied the droplet formation processes of cell-laden bioink using a nozzle with a diameter of 120 μm.29 As reviewed, relatively large nozzle sizes have been used for inkjet-based cell printing in consideration of the level of stress that cells can experience during the printing process.
TABLE I.
Comparison of drop-on-demand inkjet-based mammalian cell printing. (n/a – Information not available.)
| Author | Inkjet type (model) | Nozzle size (μm) | Droplet volume (pl) | Cell | Cell density (cells/ml) | Post-printing cell viability (normalized)a (%) |
|---|---|---|---|---|---|---|
| Xu et al.25 | Thermal (HP 550C) | 48 | 85 | Chinese hamster ovary cell, rat embryonic motoneuron | 5 × 106 | 75 (90) |
| Xu et al.26 | Thermal (HP 550) | 48 | 85 | Primary rat hippocampal neuron, and so on | 2 × 106 | 74 |
| Saunders et al.9 | Piezoelectric (MicroFab) | 60 | 150–375 | HT1080 fibrosarcoma | 2 × 106 | >94 (>98) |
| Lorber et al.27 | Piezoelectric (MicroFab) | 50/80 | n/a | Glial cell/Retinal ganglion cell | 8 × 104/7 × 105 | 69 (88)/69 (93) |
| Xu et al.29 | Piezoelectric (MicroFab) | 120 | 598–1049 | NIH 3T3 mouse fibroblast | 1–10 × 106 | n/a |
| Nakamura et al.28 | Electrostatic (SEAJet) | 43 | n/a | Bovine vascular endothelial cell | 1–1.5 × 106 | n/a |
| Present work | Piezoelectric (MicroFab) | 30 | 34 | NIH 3T3 mouse fibroblast | 5 × 106 | 91 (94) |
Cell viability normalized by or compared to that of control.
In this work, we used a 30-μm nozzle to print NIH/3T3 and HEK293A cells with the aim of achieving cell-level accuracy and studied the printing process. After evaluating the fluid properties of the NIH/3T3 cell-laden bioink, such as shear viscosity and dynamic surface tension, we analyzed cell movements inside the nozzle and the jet formation with high-speed imaging techniques. With the optimized printing conditions for the formation of single drops without satellites, an array of cells was printed onto a glass substrate to count the number of cells delivered by a printed picoliter drop. Furthermore, the effects of cell concentration, cell type with different size, and nozzle diameter on cell number distribution were systematically studied. Finally, post-printing cell viability, proliferation, and morphologic cellular feature were examined with live/dead and CCK-8 assays as well as microscopic observation.
II. MATERIALS AND METHODS
A. Cell culture
NIH/3T3 mouse fibroblasts and HEK293A human embryonic kidney cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, USA) and 1% 100× antibiotic/antimycotic solution (Sigma Aldrich, USA). The fibroblasts were plated in 100-mm cell culture dishes (Falcon®, Corning Inc.), cultured at 37 °C in a humidified incubator with 5% CO2, and passaged every 2–3 days. Sterile 1× phosphate-buffered saline solution (PBS, Hyclone, USA) and 1× trypsin/EDTA solution (0.25%, 0.2 g l−1 EDTA) (Hyclone, USA) were used for rinsing and detaching, respectively. The cells were harvested at about 80% cell confluency stage.
B. Bioink preparation
Three types of bioinks were prepared to investigate printability: (1) serum-free DMEM (hereby designated as DMEM), (2) DMEM with 10% FBS and 1% antibiotic/antimycotic solution (hereby designated as 10% FBS/DMEM), and (3) cell suspension in the 10% FBS/DMEM solution (hereby designated as cell suspension). The preparation procedure of the cell suspension was as follows. NIH/3T3 cells were detached from 100-mm Petri dishes using trypsin/EDTA solution of 1 ml for 3 min; 4 ml 10% FBS/DMEM was added to stop the trypsin-EDTA activity. The cells were centrifuged at 800 rpm for 3 min. The supernatant was aspirated and discarded. The cell pellet was resuspended in 1 ml 10% FBS/DMEM and filtered through a 40-μm cell strainer to filter the cell aggregates. The filtered cells were quantified with a disposable hemocytometer (C-Chip DHC-N01, Digital Bio Technology Co.) to prepare the cell-laden bioink with a final concentration of 5.0 × 106 ± 0.5 × 106 cells ml−1. Sterile cartridges were used as ink reservoirs.
The dynamic shear viscosities of DMEM as control, 10% FBS/DMEM, and a cell-suspended bioink were measured with a rotational viscometer (DV2T, Brookfield Engineering). Each shear rate ranging from 187.5 s−1 to 1 500 s−1 was applied on the solutions for 90 s and the viscosities were derived automatically by averaging data during the next 10 s. The dynamic surface tensions of the bioinks were also measured by using a bubble pressure tensiometer (SITA pro line t15, SITA Messtechnik GmbH). Bubbles with lifetime ranging from 45 ms–20 s were applied to the solutions, and the surface tensions on all bubbles' lifetime were taken by averaging data five times within 3% tolerance. All measurements were performed thrice under a room temperature of 26 ± 1 °C.
In order to measure diameters of cells, the images of spherical cells in a hemocytometer were captured by an inverted microscope (ICX40, Ningbo Sunny Instruments Co.) at 200× magnification and a spatial resolution of 0.81 μm per pixel. Digital image processing and diameter analysis were conducted by using both MATLAB® and ImageJ. After excluding the small particles from the cell debris by size, the cell diameters were evaluated based on the average value of the maximal and minimal Feret diameters.
C. Inkjet printing apparatus with high-speed imaging
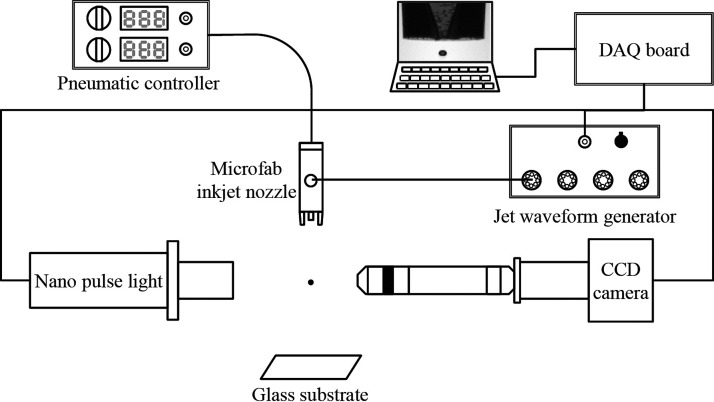
A home-made inkjet printing apparatus for the high-speed visualization of picoliter drops is illustrated in Fig. 1. The small drops were generated from a commercial piezoelectric printhead (MicroFab Inc., Plano, TX, USA) with a diamond-like carbon coated nozzle surface and nozzle apertures of 30, 50, and 80 μm. Two analogue waveforms were generated from the analog output (AO) ports of a data acquisition (DAQ) board (NI USB6351, National Instruments). One of the analogue waveforms (driving waveform) was generated by a voltage pulse controller (CT-M3-04, MicroFab Inc.). The driving waveform was controlled with the pulse duration and voltage amplitude ranging from 40 μs–61 μs and 30 V–68 V, respectively. After amplification, the pulse was applied to the piezo transducer in the printhead to eject drops. The other analogue pulse was sent to a high-resolution, 0.74 μm per pixel CCD camera (avA1000–100 gm, Basler) and a nano-flash light with a very short duration (150 ns, NP-1A, Sugawara Laboratories Inc.) via a digital delay for single-flash high-speed photography. Images were acquired at rates of 5–10 frames per second (fps). Continuous high-speed images of the inkjetting of cell-laden bioinks were also obtained by using a commercial high-speed camera imaging (Fastcam SA4 500 K C1, Photron) at the record rate of 100 000 fps with a fiber-coupled halogen lamp (LS-F150HS, Light Bank). The firing frequency was set at 50 Hz–200 Hz.
FIG. 1.
Schematic diagram of the cell printing and single-flash stroboscopic imaging apparatus. The bioink was ejected through a piezoelectric printhead with a nozzle diameter of 30 μm.
A commercial printing system (Jetlab II, MicroFab Inc.) with the same nozzle was used to count the number of cells in each printed drop. An array pattern of 20 × 20 dots was printed on a glass substrate using 30, 50, and 80-μm nozzles, respectively. An array pattern of 360 × 360 dots was also printed with the 30-μm nozzle to examine the long-term stability of the printing. All the experiments were carried out in a class 1 000 clean room to ensure a dust- and bacteria-free environment.
D. Cell viability and proliferation assays
A live/dead assay was performed to investigate the influence of shear stress caused by a very small nozzle on the viability of jetted cells. Approximately 50 000 cells were printed onto a flat-bottom 96-well microplate containing 100 μl DMEM in each well. Control groups were prepared by manual pipetting. The numbers of live and dead cells were automatically counted by using an automated cell counter (Countess II FL Automated Cell Counter, Invitrogen).
The proliferation rates of cells jetted at different nozzle diameters were assessed by using a cell counting kit-8 (CCK-8 kit, Dojindo Molecular Technologies). The jetted cells of three groups with three replicates were seeded into a 24-well plate at a low density of about 200 cells per well to avoid plateau phase at day 7. Cells of three groups were pre-incubated for 24 h to allow for complete adherence before conducting CCK-8 assay. At the scheduled time period (days 1, 3, 5, and 7) after seeding the cells, CCK-8 was added to each well. Briefly, the CCK-8 solution and DMEM (FBS 10%) were mixed at a ratio of 1:10 according to the manufacturer's instructions. After incubating for 2 h, the absorbance at 450 nm was measured as optical density (OD) values using a spectrophotometer (SPECTROstar Nano, BMG Labtech Inc.). We confirmed that the OD value or dye intensity was proportional to the number of viable cells. These OD values were gathered from three replicates of each well.
The cell morphologies of both the printed and control groups of NIH/3T3 cells were examined by staining with Bisbenzimide Hoechst 33342 labeling dye (Sigma Aldrich). Cells cultured at day 5 were pre-washed with PBS and incubated with Hoechst 33342 mixed 10% FBS/DMEM for 10 min. Both bright-field and blue fluorescent images were taken with 400× magnification using an inverted microscope (Eclipse Ti-S, Nikon) and then merged afterwards.
III. RESULTS AND DISCUSSION
A. Bioink characterization
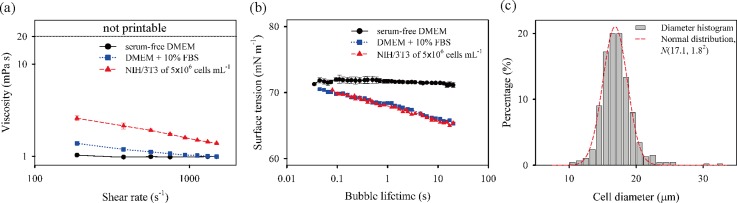
In order to investigate how fluid properties, such as shear viscosity and dynamic surface tension, affect the inkjet printing process, cell suspension as well as DMEM and 10% FBS/DMEM was used for inkjet printing in this study. Hence, shear viscosity and dynamic surface tension were characterized by using a rotational viscometer and a bubble tensiometer, respectively. The shear viscosities of the bioinks are shown in Fig. 2(a). As the base media, the DMEM ink showed Newtonian behavior with a constant viscosity of 0.99 ± 0.02 mPa s. The addition of 10% FBS into the DMEM resulted in weak shear-thinning behavior; its viscosity decreased with increasing shear rate, until it reached the infinite-shear viscosity value of about 1 mPa s, which corresponds to that of the DMEM. Moreover, the addition of cells into the combination strengthened the shear-thinning behavior. The measured viscosity of the bioink including cells did not reach the steady state in the measurement range of up to a shear rate of 1 500 s−1 due to the nominal limitation of the viscometer; nevertheless, the infinite-shear viscosity is expected to be close to the base-viscosity of the DMEM. The results showed that low and high viscosities of all the bioinks lay within the jettable range (η ≲ 20 mPa s) for DOD inkjet printing.
FIG. 2.
The characteristics of bioinks. (a) Viscosities and (b) dynamic surface tensions of three bioinks at different shear rates and bubble lifetimes, respectively, and (c) the histogram of the diameters of NIH/3T3 cells.
The measurement results on dynamic surface tension are shown in Fig. 2(b). The surface tension (γ0) of the DMEM was independent of surface age with equilibrium surface tension of (γ0) of 71.7 ± 0.3 mN m−1. The addition of 10% FBS in DMEM caused surface tension to continue decreasing from 70.5 mN m−1 down to 65.3 mN m−1 as the bubble lifetime increased from 45 ms to 20 s. The dynamic surface tension could reach the equilibrium state above 20 s, based on the approximate estimate of diffusion time (), which is about 20 s, at which a diffusion coefficient (D) of bovine serum albumin, a major component of FBS, is 1.84 × 10−11 m2 s−1 at 37 °C;30,31 here, a drop radius of about 20 μm is considered as a diffusion length (l). Despite the considerable variation of surface tension of the 10% FBS/DMEM ink according to surface age, there existed a little difference in jet formation between DMEM only and 10% FBS/DMEM ink. This finding can be attributed to the slow migration of the solute components of the FBS towards the new surfaces, while the jetting process finishes within 100 μs after the emergence of a jet at the nozzle.32
Meanwhile, the addition of cells into a 10% FBS/DMEM solution had a little influence on the dynamic surface tension of the solution, as seen in Fig. 2(b). Given that the cell suspension is regarded as a colloidal solution, we investigated the volume fraction (φ) of a monodisperse hard sphere system, which is determined as φ = 4/3πa3N/V, where a, N, and V are the cell mean radius, the cell number, and the total volume, respectively. As shown in Fig. 2(c), the diameters of NIH/3T3 cells were normally distributed with the mean diameter and standard deviation of 17.1 and 1.8 μm, respectively. In addition, the projected cell shape had an average eccentricity of 0.48. From the cell diameter and cell number (or concentration), the value of φ was calculated as 1.25%, similar to the value of 0.88% reported in a previous study.29 This value indicates that the cell-laden bioink belongs to a diluted suspension (φ ≤ 2%)33 and that the selected concentration of the cell is weakly associated with the behavior of dynamic surface tension and shear viscosity.
B. Cell trajectory inside a nozzle
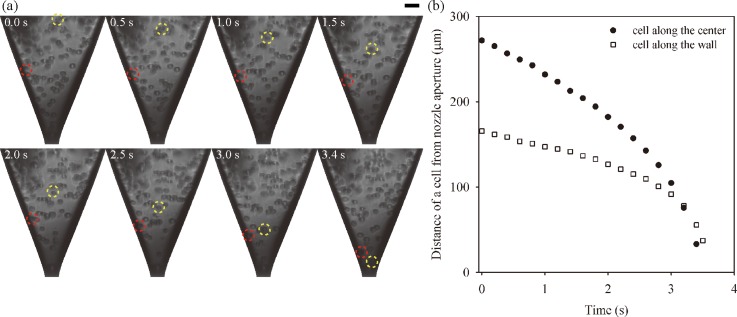
After transferring the cells to a glass capillary nozzle, their movements inside a nozzle were traced during the jetting process. Fig. 3 shows high-speed images and trajectory of cells moving down towards a nozzle aperture. The jetting frequency was set to 5 Hz to acquire the high-quality images by using a short duration flashlight and a high resolution CCD camera. The high-speed images, taken at different jet times, revealed that cells accelerated as they moved down toward the aperture of the nozzle, which narrowed down gradually by the venture effect or Bernoulli principle. We observed that a cell along the center (yellow dashed circle in Fig. 3(a)) moved to the aperture faster than a cell near the wall (red dashed circle in Fig. 3(b). The difference in the moving speed was caused by the velocity profile in the nozzle, which was conjectured as a parabolic distribution because of the no slip condition at the wall.
FIG. 3.
(a) Consecutive single-flash images of NIH/3T3 cells in a glass capillary nozzle during jetting. The yellow and red dotted circles indicate cells moving along the center and the wall, respectively. (Scale bar = 30 μm.) (b) Temporal variations on the distance of the cells on difference locations from the nozzle aperture.
In addition, cell migration was driven by pulsed pressure and gravitational force. Cell movements were not continuous, but the cells made go-and-stop movements as pulsed pressure was generated by the piezo-actuator for drop generation (see supplementary material for go-and-stop behavior of cells). When the printhead was actuated, and the droplet volume of ∼10 pl was dispensed, the bioink was refilled up to the orifice and the cells instantaneously travelled. After a drop ejection, along with the decay of meniscus oscillation, the cells slightly fluctuated. For one interval step of stroboscopy, the average and instantaneous velocities of the cell are expected as ∼ 10 μm/0.1 s = 100 μm s−1 and ∼ 10 μm/10 μs = 1 m s−1, respectively. On the contrary, although the cells under the influence of constant gravity in the reservoir slowly settled down, the gravitational effect on the cell movement in the nozzle proved to be negligible. Based on the assumption that the cells are regarded as a rigid sphere, the free-settling velocity (vg) of the cell is expected by Stokes' law34 to have a value of
where g, ρc, ρb, a, and μb are the gravitational acceleration, the densities of the cell and the bioink, the radius of the cell, and the dynamic viscosity of the bioink at a low shear rate, respectively. The vg is predicted as 2.5 μm s−1, implying that the gravity is not dominant and that the moving behavior of the cell is intimately related to the ejected volume.
C. Single-drop formation
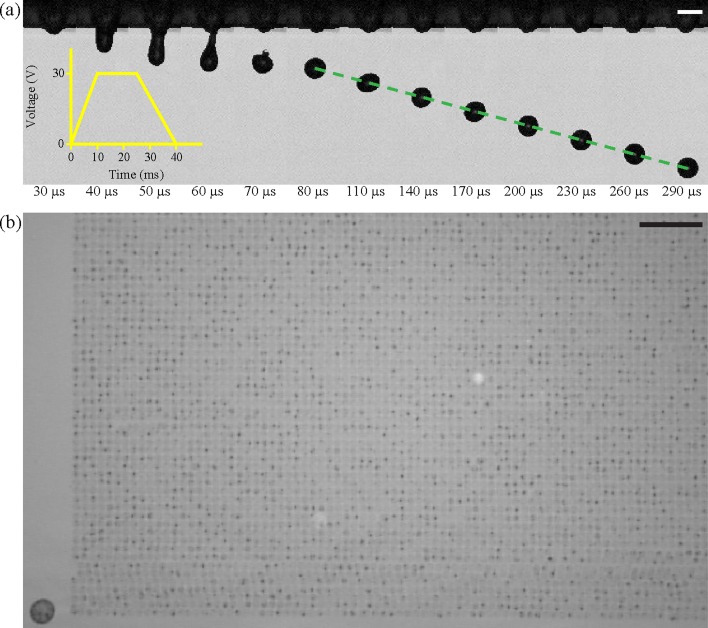
Fig. 4(a) shows single-flash images showing the evolution of single-drop formation of cell suspension under low voltage actuation. A jet head began to emerge from a nozzle during pulse duration, and after applying the waveform, the jet ligament became narrower until it eventually disintegrated. After breakup, the short ligament retracted into the head, and one stable spherical drop eventually formed and oscillated. Fig. 4(b) indicates that stable printing was maintained at about 1.3 × 105 drops. By using an X–Y moving stage, the single drops were patterned on to a glass substrate with a drop spacing of 120 μm.
FIG. 4.
(a) The consecutive high-speed images of the formation of a drop ejected from a 30-μm inkjet nozzle. The yellow inset illustrates a waveform to drive the actuator (Scale bar = 30 μm). (b) Partial pattern of a 360 × 360 array of printed cells on a bare glass substrate (Scale bar = 1 mm).
The single-drop formation is vital in ensuring stable and repeatable cell printing. In general, the cells affect the morphology of the jet with irregularity.17,29 Especially in the case of long ligaments, the morphology of the jet ligament, including cells, is anomalistic owing to the larger diameter of cells compared with that of a thinned ligament. However, if the cells are located at the main head, the morphology is stable and regular and is indiscernible from a jet morphology without a cell. The jet morphology and printing performance (e.g., droplet diameter and velocity) could be modulated by the shape of the input waveform. In the case of our bioinks, the single drop is formed under 68 V with a total pulse duration of 61 μs and a drop velocity of 2.5 m s−1.
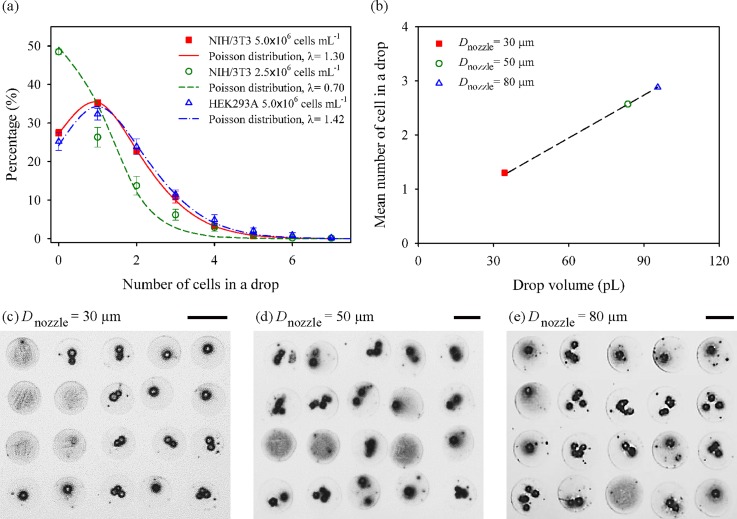
We investigated the influence of cell type, cell concentration, and nozzle diameter on the number of cells contained in a single drop by printing 20 × 20 dot array patterns on a glass substrate. Fig. 5(a) shows the influence of the cell type and cell concentration on the distribution of cell numbers in each dot. NIH/3T3 and HEK293A cells were printed by a 30-μm nozzle with the same cell concentration of 5.0 × 106 cells ml−1 to examine the influence of cell type with slightly different sizes of cells (17.1 ± 1.8 μm for NIH/3T3 and 15.6 ± 1.7 μm HEK293A). A negligible difference in the number of cells in a drop was observed between the printings of the two cells, and each distribution was in excellent agreement with the Poisson distribution with the mean and variance of = 1.30 and 1.42. The influence of the cell concentration was examined by varying the cell concentration of NIH/3T3 cells (5.0 × 106 and 2.5 × 106 cells ml−1). The mean number of cells was observed to be proportional to the cell concentration. The percentage of single-cell spots was 35.1% and 26.3% for 5.0 × 106 and 2.5 × 106 cells ml−1, respectively.
FIG. 5.
(a) Distributions of the number of cells in a printed drop with different NIH/3T3 cell concentrations (5.0 × 106 and 2.5 × 106 cells ml−1) and different cell types (NIH/3T3 and HEK293A). Each curve is fitted by Poisson distribution. (b) Effect of nozzle diameters on average cell number in one drop from Poisson distribution. (c)–(e) Dot array patterns formed by inkjet-printing with different nozzle diameters of 30, 50, and 80 μm, respectively. Scale bars are 100 μm.
We then studied the effect of the nozzle diameter by using different nozzles (30, 50, and 80-μm) at the concentration of 5.0 × 106 cells ml−1. The volume of ink ejected from the nozzles was approximately 34, 84, and 95 pl, respectively. The average number of cells in a drop was 1.30 for the 30-μm nozzle, while the number increased to 2.57 and 2.88 as the nozzle size changed to 50 and 80 μm (Fig. 5(b)). Figs. 5(c)–5(e) are the representative microscopic images of printed dots for the difference nozzle sizes. These results show that a higher percentage of single-cell printing could be achieved by the narrow nozzle diameter of 30 μm, compared to the larger one that has been commonly used.
D. Cell viability and proliferation
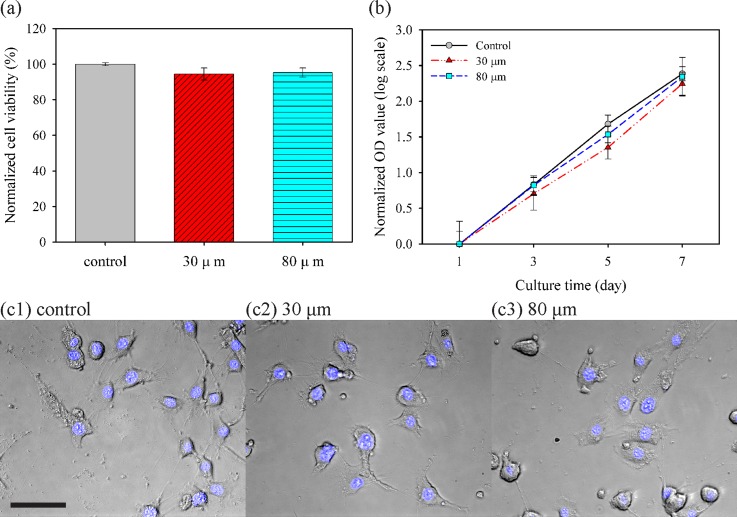
The effects of jetting through a narrow nozzle on cell viability and proliferation were investigated using live/dead assay for viability immediately post printing, CCK-8 assay for cell growth rate, and Hoechst 33342 staining for cellular morphology. First, Fig. 6(a) shows cell viability by automatically analyzing green- and red-stained cell populations based on the live/dead assay kit. As a control, the cells pipetted into a well revealed a survival rate of 96.2%, and survival ratios for the cells fired from 30- and 80-μm nozzles were quantitated as 94.4% and 96.3%, respectively, relative to that of the control group. This high survival rate is consistent with those reported in previous studies on inkjet-based cell printing with larger nozzle sizes. Next, CCK-8 assay was performed to investigate whether a small nozzle diameter affected the proliferation of cells in vitro for 7 days before reaching the plateau phase. Although cell populations differed among each group owing to the initially seeded cell number (data not shown), these cell culture groups showed the same tendency of cell growth as the slopes of curves in Fig. 6(b) representing cell growth rate. Based on a simple cell population model adopted from the law of natural growth expressed as
where t, P0, and k represented culture time (in days), initial population, and (per capita) growth rate, respectively; cells in each group exponentially grew with similar growth rates from each initial population at day 1. Finally, cellular morphological examination was carried out for qualitative analysis. As shown in Fig. 6(c), no obvious morphological abnormality and DNA fragmentation were observed; moreover, those printed cells still maintained good adhesion ability to the substrate. These results demonstrated that the 30-μm inkjet nozzle had negligible effects and damages to the printed mammalian cells.
FIG. 6.
Effects of nozzle diameter on viability, proliferation, and morphology of NIH/3T3 cells. (a) Normalized cell viability after pipetting for the control group and each printing through a live/dead assay kit. (b) Assessment of proliferation through the normalized optical density (OD) value at 450 nm by each OD value at day 1, corresponding to initial population with CCK-8 assay. Slopes of curves imply the growth rate. (c) Cell morphology and fluorescence of nuclei stained by Hoechst 33342 with cultured cells at day 5 (Scale bar = 50 μm).
IV. CONCLUSION
We present DOD inkjet-based cell printing with a 30-μm nozzle for cell-level accuracy. High-speed imaging techniques showed that cells inside the nozzle made go-and-stop movements inside the nozzle under pulsed pressure generated by a piezo-actuator. After evaluating the fluid properties of cell-laden bioinks, printing conditions were optimized to generate cell-laden single drops without satellites. Our results show that the number of cells in a drop was greatly influenced by the nozzle sizes (30, 50, and 80 μm) and cell concentrations (2.5 × 106 cells ml−1 and 5.0 × 106 cells ml−1), whereas printing different cells types made little variation in the number. The 30-μm inkjet nozzle was capable of printing 1.3 mammalian cells per drop on average at the cell concentration of 5.0 × 106 cells ml−1. The broader nozzle allowed more cells to pass through. Viability, proliferation, and morphology of cells were examined by using a live/dead assay kit, CCK-8 assay, and cellular morphology imaging, respectively. We found that cell survival rates after printing with the very small nozzle were consistent with those of unprinted controls. Inkjet printing is ideally suited to the deposition of living cells, because it is designed for the precise deposition of picoliter volumes of cell-laden liquid drops. Furthermore, the use of a 30-μm nozzle can help maximize inkjet technology for the precise deposition of bioinks with cell-level accuracy.
SUPPLEMENTARY MATERIAL
See supplementary material for multimedia on the go-and-stop movement of cells inside the nozzle under a pulsed pressure generated by a piezo-actuator.
ACKNOWLEDGMENTS
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Ministry of Health and Welfare, Republic of Korea (Grant No. HI15C0001), and was supported by research programs through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2015R1C1A1A01052904 and NRF-2016M1A5A1027599).
References
- 1. Mironov V., Boland T., Trusk T., Forgacs G., and Markwald R. R., Trends Biotechnol. , 157 (2003). 10.1016/S0167-7799(03)00033-7 [DOI] [PubMed] [Google Scholar]
- 2. Ringeisen B. R., Pirlo R. K., Wu P. K., Boland T., Huang Y., Sun W., Hamid Q., and Chrisey D. B., MRS Bull. , 834 (2013). 10.1557/mrs.2013.209 [DOI] [Google Scholar]
- 3. Murphy S. V. and Atala A., Nat. Biotechnol. , 773 (2014). 10.1038/nbt.2958 [DOI] [PubMed] [Google Scholar]
- 4. Ferris C. J., Gilmore K. G., Wallace G. G., and in het Panhuis M., Appl. Microbiol. Biotechnol. , 4243 (2013). 10.1007/s00253-013-4853-6 [DOI] [PubMed] [Google Scholar]
- 5. Cui X., Breitenkamp K., Finn M. G., Lotz M., and D'Lima D., Tissue Eng. , 1304 (2012). 10.1089/ten.tea.2011.0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ringeisen B. R., Othon C. M., Barron J. A., Young D., and Spargo B. J., Biotechnol. J. , 930 (2006). 10.1002/biot.200600058 [DOI] [PubMed] [Google Scholar]
- 7. Henmi C., Nakamura M., Nishiyama Y., Yamaguchi K., Mochizuki S., Takiura K., and Nakagawa H., Inflammation Regener. , 36 (2008). 10.2492/inflammregen.28.36 [DOI] [Google Scholar]
- 8. Calvert P., Science , 208 (2007). 10.1126/science.1144212 [DOI] [PubMed] [Google Scholar]
- 9. Saunders R. E., Gough J. E., and Derby B., Biomaterials , 193 (2008). 10.1016/j.biomaterials.2007.09.032 [DOI] [PubMed] [Google Scholar]
- 10. Xu C., Chai W., Huang Y., and Markwald R. R., Biotechnol. Bioeng. , 3152 (2012). 10.1002/bit.24591 [DOI] [PubMed] [Google Scholar]
- 11. Cohen D. L., Malone E., Lipson H., and Bonassar L. J., Tissue Eng. , 1325 (2006). 10.1089/ten.2006.12.1325 [DOI] [PubMed] [Google Scholar]
- 12. Norotte C., Marga F. S., Niklason L. E., and Forgacs G., Biomaterials , 5910 (2009). 10.1016/j.biomaterials.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Riggs B. C., Dias A. D., Schiele N. R., Cristescu R., Huang Y., Corr D. T., and Chrisey D. B., MRS Bull. , 1043 (2011). 10.1557/mrs.2011.276 [DOI] [Google Scholar]
- 14. Guillotin B., Souquet A., Catros S., Duocastella M., Pippenger B., Bellance S., Bareille R., Rémy M., Bordenave L., Amédée J., and Guillemot F., Biomaterials , 7250 (2010). 10.1016/j.biomaterials.2010.05.055 [DOI] [PubMed] [Google Scholar]
- 15. Guillemot F., Souquet A., Catros S., Guillotin B., Lopez J., Faucon M., Pippenger B., Bareille R., Rémy M., Bellance S., Chabassier P., Fricain J. C., and Amédée J., Acta Biomater. , 2494 (2010). 10.1016/j.actbio.2009.09.029 [DOI] [PubMed] [Google Scholar]
- 16. Zhang W. and He X., J. Biomech. Eng. , 74515 (2009). 10.1115/1.3153326 [DOI] [PubMed] [Google Scholar]
- 17. Jayasinghe S. N. and Townsend-Nicholson A., Lab Chip , 1086 (2006). 10.1039/b606508m [DOI] [PubMed] [Google Scholar]
- 18. Chen F., Zhang Y., Nakagawa Y., Zeng H., Luo C., Nakajima H., Uchiyama K., and Lin J. M., Talanta , 111 (2013). 10.1016/j.talanta.2012.12.058 [DOI] [PubMed] [Google Scholar]
- 19. Chen F., Lin L., Zhang J., He Z., Uchiyama K., and Lin J. M., Anal. Chem. , 4354 (2016). 10.1021/acs.analchem.5b04749 [DOI] [PubMed] [Google Scholar]
- 20. Kim M. and Kim T., Anal. Chem. , 9401 (2010). 10.1021/ac102022q [DOI] [PubMed] [Google Scholar]
- 21. Miller E. D., Fisher G. W., Weiss L. E., Walker L. M., and Campbell P. G., Biomaterials , 2213 (2006). 10.1016/j.biomaterials.2005.10.021 [DOI] [PubMed] [Google Scholar]
- 22. Choi W. S., Ha D., Park S., and Kim T., Biomaterials , 2500 (2011). 10.1016/j.biomaterials.2010.12.014 [DOI] [PubMed] [Google Scholar]
- 23. Liberski A. R., Delaney J. T., and Schubert U. S., ACS Comb. Sci. , 190 (2011). 10.1021/co100061c [DOI] [PubMed] [Google Scholar]
- 24. Yamaguchi S., Ueno A., Akiyama Y., and Morishima K., Biofabrication , 45005 (2012). 10.1088/1758-5082/4/4/045005 [DOI] [PubMed] [Google Scholar]
- 25. Xu T., Jin J., Gregory C., Hickman J. J., and Boland T., Biomaterials , 93 (2005). 10.1016/j.biomaterials.2004.04.011 [DOI] [PubMed] [Google Scholar]
- 26. Xu T., Gregory C. A., Molnar P., Cui X., Jalota S., Bhaduri S. B., and Boland T., Biomaterials , 3580 (2006). 10.1016/j.biomaterials.2006.01.048 [DOI] [PubMed] [Google Scholar]
- 27. Lorber B., Hsiao W.-K., Hutchings I. M., and Martin K. R., Biofabrication , 15001 (2014). 10.1088/1758-5082/6/1/015001 [DOI] [PubMed] [Google Scholar]
- 28. Nakamura M., Kobayashi A., Takagi F., Watanabe A., Hiruma Y., Ohuchi K., Iwasaki Y., Horie M., Morita I., and Takatani S., Tissue Eng. , 1658 (2005). 10.1089/ten.2005.11.1658 [DOI] [PubMed] [Google Scholar]
- 29. Xu C., Zhang M., Huang Y., Ogale A., Fu J., and Markwald R. R., Langmuir , 9130 (2014). 10.1021/la501430x [DOI] [PubMed] [Google Scholar]
- 30. Fardet A., Hoebler C., Djelveh G., and Barry J.-L., J. Agric. Food Chem. , 4635 (1998). 10.1021/jf980371n [DOI] [Google Scholar]
- 31. Ma L., Zhou C., Lin B., and Li W., Biomed. Microdevices , 753 (2010). 10.1007/s10544-010-9429-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang L., Kazmierski B. K., Hoath S. D., Jung S., Hsiao W.-K., Wang Y., Berson A., Harlen O., Kapur N., and Bain C. D., Phys. Fluids , 113103 (2014). 10.1063/1.4901823 [DOI] [Google Scholar]
- 33. Rutgers I. R., Rheol. Acta , 202 (1962). 10.1007/BF01983952 [DOI] [Google Scholar]
- 34. Guzman-Sepulveda J. R., Amin S., Lewis E. N., and Dogariu A., Langmuir , 10351 (2015). 10.1021/acs.langmuir.5b02665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for multimedia on the go-and-stop movement of cells inside the nozzle under a pulsed pressure generated by a piezo-actuator.