Abstract
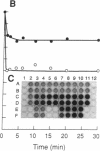
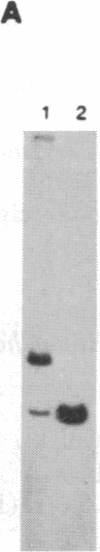
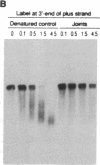
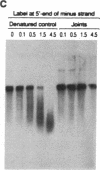
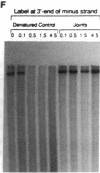
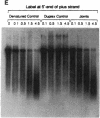
When RecA protein, in the form of a nucleoprotein filament containing circular single-stranded DNA (plus strand only), reacts with homologous linear duplex DNA, a directional transfer ensues of a strand from the duplex DNA to the nucleoprotein filament, resulting in the displacement of the linear plus strand in the 5' to 3' direction. The initial homologous synapsis, however, can occur at either end of the duplex DNA, or anywhere in between, and when homology is restricted to different regions of the duplex DNA, the joint molecules that form in each region show striking differences in stability upon deproteinization: distal joints greater than proximal joints much greater than medial joints. In the deproteinized distal joints, which are thermostable, 2000 nucleotide residues of the circular plus strand are resistant to P1 nuclease; both strands of the original duplex DNA remain resistant to P1 nuclease, and the potentially displaceable linear plus strand, which has a 3' homologous end, remains resistant to Escherichia coli exonuclease I. These observations suggest that RecA protein promotes homologous pairing and strand exchange via long three-stranded DNA intermediates and, moreover, that, once formed, such triplex structures in natural DNA are stable even when RecA protein has been removed.
Full text
PDF
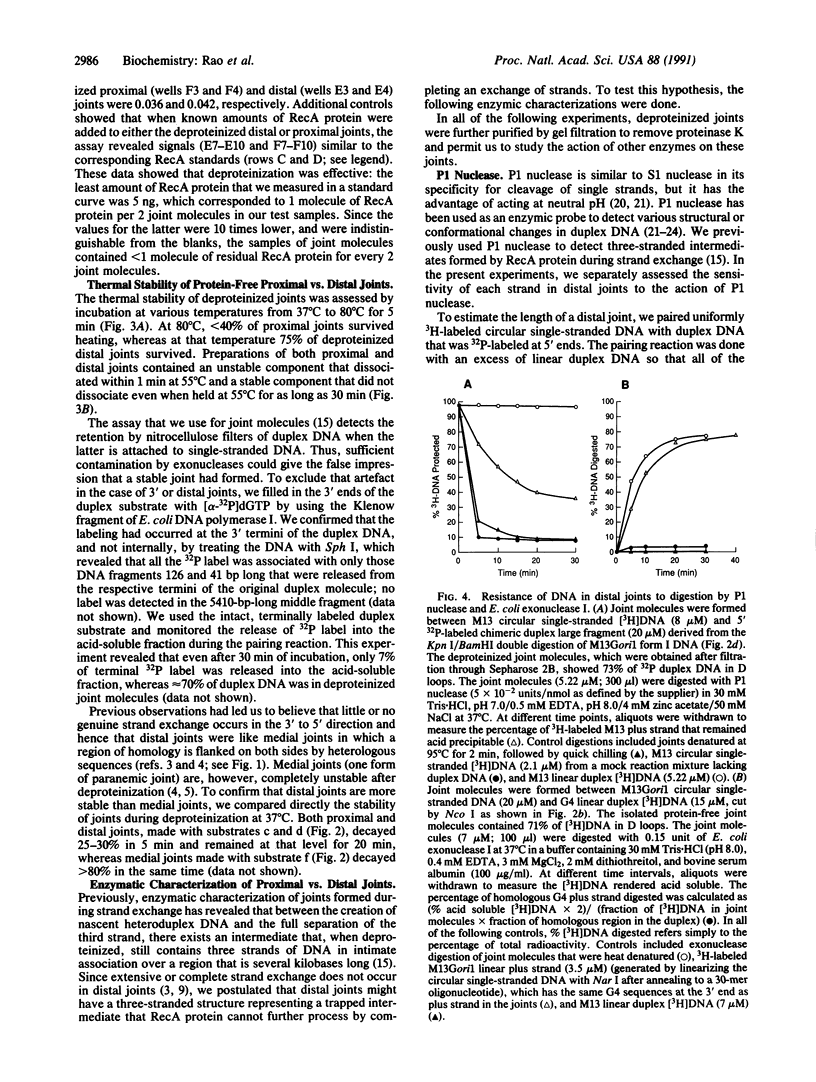
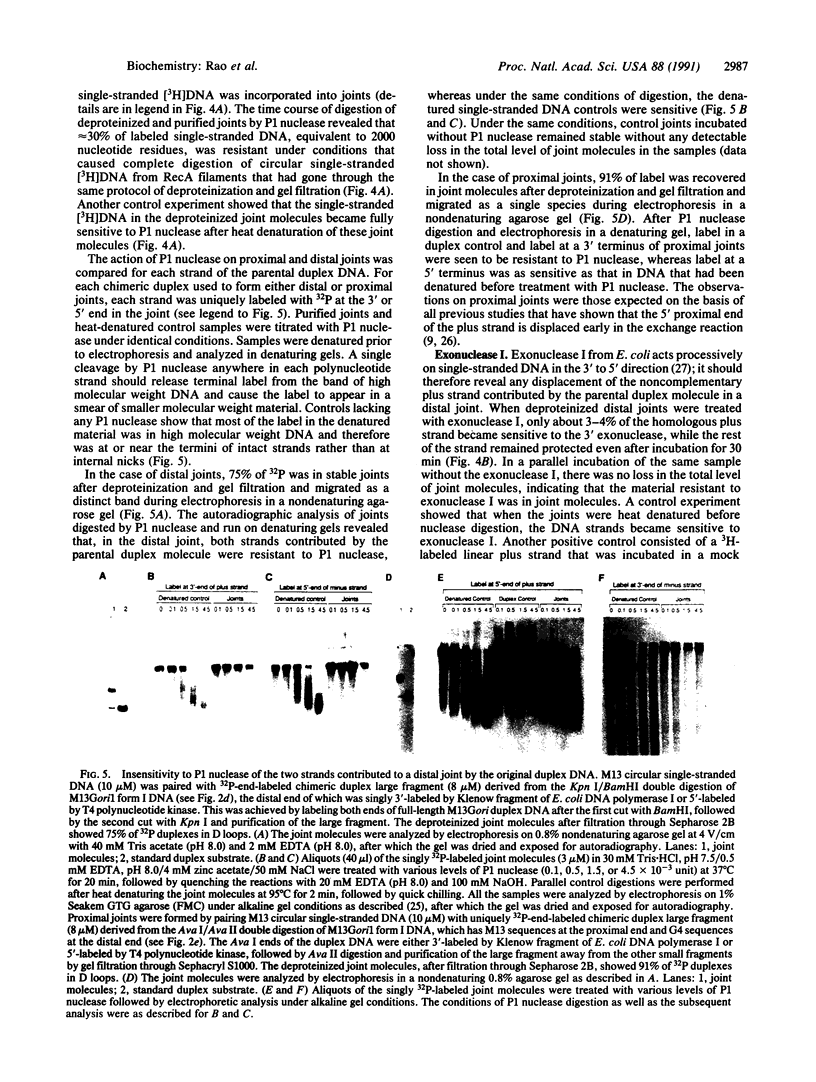

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bianchi M., DasGupta C., Radding C. M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983 Oct;34(3):931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Buchanan D., Kamarck M., Ruddle N. H. Development of a protein A enzyme immunoassay for use in screening hybridomas. J Immunol Methods. 1981;42(2):179–185. doi: 10.1016/0022-1759(81)90147-2. [DOI] [PubMed] [Google Scholar]
- Chow S. A., Rao B. J., Radding C. M. Reversibility of strand invasion promoted by recA protein and its inhibition by Escherichia coli single-stranded DNA-binding protein or phage T4 gene 32 protein. J Biol Chem. 1988 Jan 5;263(1):200–209. [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., DasGupta C., Shibata T., Radding C. M. Homologous pairing in genetic recombination: recA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell. 1980 May;20(1):223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., Wu A. M., Shibata T., DasGupta C., Radding C. M. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981 Apr;24(1):213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Shibata T., Cunningham R. P., Radding C. M. The topology of homologous pairing promoted by RecA protein. Cell. 1980 Nov;22(2 Pt 2):437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- Ey P. L., Prowse S. J., Jenkin C. R. Isolation of pure IgG1, IgG2a and IgG2b immunoglobulins from mouse serum using protein A-sepharose. Immunochemistry. 1978 Jul;15(7):429–436. doi: 10.1016/0161-5890(78)90070-6. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Kalinski A., Pritchard A. E., Kowalski D., Laskowski M., Sr Accessibility of some regions of DNA in chromatin (chicken erythrocytes) to single strand-specific nucleases. J Biol Chem. 1979 Aug 10;254(15):7405–7410. [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Hanley P. J., Carrigan C. M., Rowe D. B., Wake R. G. Breakdown and quantitation of the forked termination of replication intermediate of Bacillus subtilis. J Mol Biol. 1987 Aug 5;196(3):721–727. doi: 10.1016/0022-2836(87)90043-x. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero C. S., Camerini-Otero R. D. Pairing of homologous DNA sequences by proteins: evidence for three-stranded DNA. Genes Dev. 1990 Nov;4(11):1951–1963. doi: 10.1101/gad.4.11.1951. [DOI] [PubMed] [Google Scholar]
- Kaguni J., Ray D. S. Cloning of a functional replication origin of phage G4 into the genome of phage M13. J Mol Biol. 1979 Dec 25;135(4):863–878. doi: 10.1016/0022-2836(79)90516-3. [DOI] [PubMed] [Google Scholar]
- Kahn R., Cunningham R. P., DasGupta C., Radding C. M. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti B. B., Davis R. W. 3' homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Feb;84(3):690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti B. B., Davis R. W. The preference for a 3' homologous end is intrinsic to RecA-promoted strand exchange. J Biol Chem. 1990 Apr 25;265(12):6916–6920. [PubMed] [Google Scholar]
- Kowalski D. Changes in site specificity of single-strand-specific endonucleases on supercoiled PM2 DNA with temperature and ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7071–7086. doi: 10.1093/nar/12.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein U., Penhoet E. E., Linn S. An altered apurinic DNA endonuclease activity in group A and group D xeroderma pigmentosum fibroblasts. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1169–1173. doi: 10.1073/pnas.73.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEHMAN I. R., NUSSBAUM A. L. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE). J Biol Chem. 1964 Aug;239:2628–2636. [PubMed] [Google Scholar]
- Lohman T. M., Green J. M., Beyer R. S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986 Jan 14;25(1):21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Jwang B., Radding C. M. RecA protein reinitiates strand exchange on isolated protein-free DNA intermediates. An ADP-resistant process. J Mol Biol. 1990 Jun 20;213(4):789–809. doi: 10.1016/S0022-2836(05)80264-5. [DOI] [PubMed] [Google Scholar]
- Register J. C., 3rd, Christiansen G., Griffith J. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J Biol Chem. 1987 Sep 15;262(26):12812–12820. [PubMed] [Google Scholar]
- Riddles P. W., Lehman I. R. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985 Jan 10;260(1):165–169. [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., Radding C. M. Homologous pairing in genetic recombination. Purification and characterization of Escherichia coli recA protein. J Biol Chem. 1981 Jul 25;256(14):7557–7564. [PubMed] [Google Scholar]
- Stasiak A., Stasiak A. Z., Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf S. W., Cox M. M., Inman R. B. Triple-helical DNA pairing intermediates formed by recA protein. J Biol Chem. 1990 Oct 5;265(28):16898–16912. [PubMed] [Google Scholar]
- Wu A. M., Kahn R., DasGupta C., Radding C. M. Formation of nascent heteroduplex structures by RecA protein and DNA. Cell. 1982 Aug;30(1):37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]