Abstract
Both LARGE1 (formerly LARGE) and its paralog LARGE2 are bifunctional glycosyltransferases with xylosy- and glucuronyltransferase activities, and are capable of synthesizing polymers composed of a repeating disaccharide [-3Xylα1,3GlcAβ1-]. Post-translational modification of the O-mannosyl glycan of α-dystroglycan (α-DG) with the polysaccharide is essential for it to act as a receptor for ligands in the extracellular matrix (ECM), and both LARGE paralogs contribute to the modification in vivo. LARGE1 and LARGE2 have different tissue distribution profiles and enzymatic properties; however, the functional difference of the homologs remains to be determined, and α-DG is the only known substrate for the modification by LARGE1 or LARGE2. Here we show that LARGE2 can modify proteoglycans (PGs) with the laminin-binding glycan. We found that overexpression of LARGE2, but not LARGE1, mediates the functional modification on the surface of DG−/−, Pomt1−/− and Fktn−/− embryonic stem cells. We identified a heparan sulfate-PG glypican-4 as a substrate for the LARGE2-dependent modification by affinity purification and subsequent mass spectrometric analysis. Furthermore, we showed that LARGE2 could modify several additional PGs with the laminin-binding glycan, most likely within the glycosaminoglycan (GAG)-protein linkage region. Our results indicate that LARGE2 can modify PGs with the GAG-like polysaccharide composed of xylose and glucuronic acid to confer laminin binding. Thus, LARGE2 may play a differential role in stabilizing the basement membrane and modifying its functions by augmenting the interactions between laminin globular domain-containing ECM proteins and PGs.
Keywords: dystroglycan, glycosaminoglycan, laminin binding, LARGE2, proteoglycan
Introduction
Post-translational modification of α-dystroglycan (α-DG) by LARGE1 (formerly LARGE) is required for its binding to laminin globular (LG) domain-containing extracellular matrix (ECM) ligands such as laminin, agrin and neurexins (Michele et al. 2002). This cell-surface-ECM association is important for maintaining sarcolemmal integrity in skeletal muscle, central nervous system structure and function, and interactions between the basement membrane and epithelial cells (Barresi and Campbell 2006). Thus, this glycosylation is referred to as the “functional modification”. Mutations in the genes encoding LARGE1 (Longman et al. 2003) and a number of other proteins are involved, directly or indirectly, in O-mannosyl glycan synthesis on α-DG. These include protein O-mannosyltransferases 1 (POMT1) and 2 (POMT2) (Beltran-Valero de Bernabe et al. 2002; Manya et al. 2004; van Reeuwijk et al. 2005), protein O-mannose β2-N-acetylglucosaminyltransferase 1 (POMGnT1) (Yoshida et al. 2001), ribitol 5-phosphate (Rbo5P) transferases fukutin (FKTN) (Kobayashi et al. 1998; Kanagawa et al. 2016) and fukutin-related protein (FKRP) (Brockington et al. 2001; Kanagawa et al. 2016), the cytidyltransferase called isoprenoid synthase domain-containing (Roscioli et al. 2012; Willer et al. 2012; Riemersma et al. 2015; Kanagawa et al. 2016), protein O-linked mannose N-acetylglucosaminyltransferase 2 (POMGNT2) (Manzini et al. 2012; Yoshida-Moriguchi et al. 2013), β3-N-acetylglucosaminyltransferase/β4-glucuronyltransferase 1 (B3GNT1/B4GAT1) (Buysse, et al. 2013; Praissman et al. 2014; Willer et al. 2014), β3-N-acetylglucosaminyltransferase 2 (B3GALNT2) (Stevens et al, 2013), a UDP-xylose transferase called transmembrane protein 5 (TMEM5) (Vuillaumier-Barrot et al. 2012; Praissman et al. 2016) and protein O-mannose kinase (Jae et al. 2013, Yoshida-Moriguchi et al. 2013). Mutations in all of the encoding genes have been identified in the secondary dystroglycanopathies, a class of congenital muscular dystrophies characterized by perturbed glycosylation and reduced ligand binding of α-DG and are often associated with brain abnormalities (Godfrey et al. 2011).
Both LARGE1 and its paralog LARGE2 are bifunctional glycosyltransferases that have xylosyltransferase (Xyl-T) and glucuronyltransferase (GlcA-T) activities and are able to polymerize Xyl-GlcA repeats, thereby conferring the ability to bind laminin to α-DG (Inamori et al. 2012, 2013; Ashikov et al. 2013). Overexpression of LARGE2 has been shown to enhance the functional modification of α-DG, as is also the case for LARGE1 (Brockington et al. 2005; Fujimura et al. 2005; Grewal et al. 2005; Inamori et al. 2013). The LARGE2 gene was cloned from human and mouse kidney or testis RNA, and the two homologs have different tissue expression profiles (Fujimura et al. 2005; Grewal et al. 2005). While LARGE1 is widely expressed, with its highest levels in brain, heart and skeletal muscle, LARGE2 is most highly expressed in kidney and placenta, and is not detectable in brain (Fujimura et al. 2005; Grewal et al. 2005). Although Large2-deficient mice do not show overt kidney abnormalities, both LARGE1 and LARGE2 contribute to the efficient functional modification of α-DG in vivo (Inamori et al. 2014).
Prior to this study, α-DG was the only known substrate of LARGE1 or LARGE2. In this study, we identify an heparan sulfate proteoglycans (HS-PG) glypican-4 (GPC4) as a novel substrate for the functional modification by LARGE2. HS- and chondroitin/dermatan sulfate (CS/DS)-PGs are the major components of the basement membrane and ECM. Structurally, they consist of a core protein and one or more glycosaminoglycan (GAG) chains, each of which is a backbone polysaccharide with repeating disaccharide units. These GAGs are synthesized on specific serine residues of the core protein through a unique tetrasaccharide linkage region, GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser (Sugahara and Kitagawa 2000). We further show that both HS- and CS/DS-PGs serve as substrates for the LARGE2-mediated modification. Although the glycan structure has not yet been solved, we predict that the β1-3-linked GlcA in the tetrasaccharide linkage region is the most likely acceptor for LARGE2 in initiating synthesis of the laminin-binding glycan.
Results
Overexpression of LARGE2, but not LARGE1, mediates the functional modification on the surface of DG−/− cells
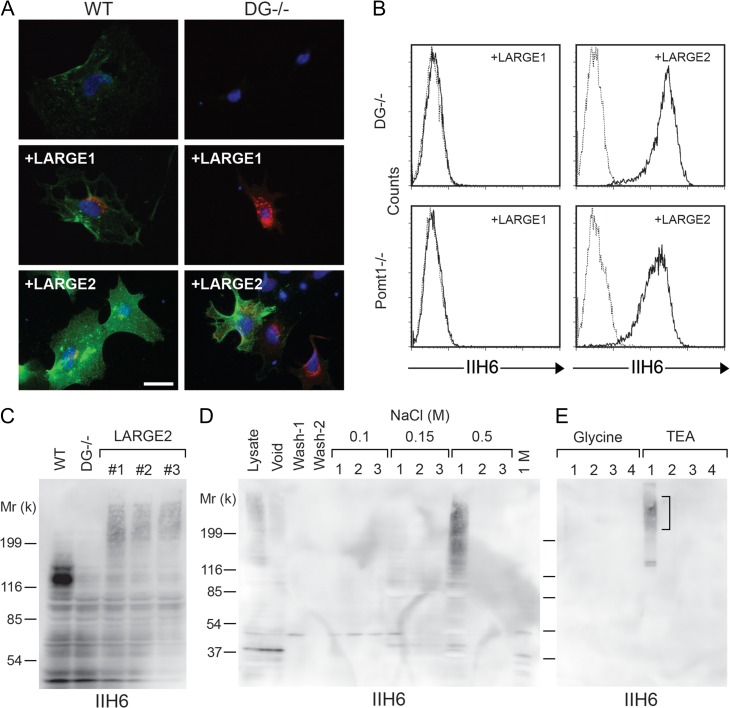
First we sought to investigate the functional difference between LARGE1 and LARGE2 by using adenovirus to overexpress LARGE1 or LARGE2 in wild-type (WT) and DG−/− mouse embryonic stem (ES) cells. When LARGE1 was overexpressed in the WT cells, there was a slight increase in the staining with IIH6, an antibody that recognizes the laminin-binding epitope of α-DG and stronger staining was observed when LARGE2 was overexpressed. Interestingly, we observed detectable IIH6 staining of varied intensity in the DG−/− cells infected with exogenous LARGE2, whereas no specific staining was observed with those infected with LARGE1 (Figure 1A). Since the efficiency of adenoviral infection in ES cells was low (flow cytometry revealed that <10% of cells were GFP+ following infection with CMV-LARGE2-internal ribosome entry site (IRES)-eGFP adenovirus, data not shown), we enriched the IIH6-immunoreactive (IIH6+) cells from the CMV-LARGE2-IRES-eGFP-infected DG−/− cells by magnetic cell separation and analyzed them by flow cytometry (Supplementary data, Figure S1). The bound fraction was double positive for GFP and IIH6, indicating that DG−/− cells transduced with the adenovirus construct expressed the functional glycan epitope at the cell surface judgment.
Fig. 1.
LARGE2 but not LARGE1 confers IIH6 immunoreactivity to DG−/− ES cells. (A) WT and DG−/− ES cells were infected with adenovirus to express LARGE1 or LARGE2. Cells were incubated with IIH6 (antibody that recognizes the functional glycan epitope of α-DG) and either anti-LARGE1 or anti-LARGE2 antibody, and then probed with Alexa-conjugated secondary antibodies (green pseudo-color: IIH6; red: LARGE1 or LARGE2; blue: DAPI). Scale bar, 50 μm. (B) Flow cytometry detecting surface IIH6 labeling of DG−/− cells or Pomt1−/− cells stably expressing either LARGE1 or LARGE2. Dashed line, secondary antibody alone. (C) Immunoblotting of whole cell lysate with IIH6. The WT sample shows a robust 120-kD band that corresponds to α-DG and is not present in DG−/− samples. Independent DG−/− cell clones stably expressing LARGE2 (#1–3) show a high-molecular weight smear that is absent in WT and the DG−/− cells. (D) IIH6 immunoblotting of eluted fractions from a diethylaminoethyl (DEAE)-cellulose column. Lysate of the DG−/− cells stably expressing LARGE2 was applied to the column. After two washes (Wash-1 and Wash-2), bound proteins were released by three sequential elutions with increasing concentrations of NaCl. (E) IIH6 immunoblotting of eluted fractions from a IIH6-Sepharose column. Bound proteins were eluted by four sequential glycine buffer washes, and then with triethanolamine (TEA) buffer. The high-molecular weight smear (square bracket) and two bands ~130-kD were excised from a SYPRO Ruby-stained gel, and were then subjected to in-gel trypsin digestion and mass spectrometric (MS) analysis (Table I). This figure is available in black and white in print and in color at Glycobiology online.
To investigate the LARGE2-mediated modification on DG−/− cells more clearly, we employed lentiviral gene transfer to generate cells stably expressing either LARGE1 or LARGE2, using an IRES-puromycin cassette for selection. Several drug-resistant cell clones were obtained and characterized by immunoblot analysis for the expression of LARGE1 or LARGE2 protein (Supplementary data, Figure S2), and by flow cytometry for surface IIH6 staining (Figure 1B). LARGE2-expressing cells showed a substantial level of the surface IIH6 staining, whereas LARGE1-expressing cells had no specific staining, consistent with the result from adenoviral expression. These results indicate that there are alternative substrate(s) that can be modified by LARGE2, but not by LARGE1, in mouse ES cells.
We also tested the effects of stable expression of LARGE1 or LARGE2 in Pomt1−/− and Fktn−/− ES cells. Since coexpression of POMT1 and POMT2 is required for protein O-mannosylation (Manya et al. 2004), Pomt1−/− cells lack O-mannosylation and the subsequent modification that is required for functional modification of α-DG. FKTN and FKRP are sequentially acting ribitol 5-phosphate transferases that form a tandem Rbo5P structure on the O-mannosyl glycan of α-DG, a scaffold required for formation of the ligand-binding moiety (Kanagawa et al. 2016). Neither the Pomt1−/− cells nor the Fktn−/− cells showed surface IIH6 immunoreactivity, but stable expression of LARGE2 could confer immunoreactivity in both null cell lines (Figure 1B, Supplementary data, Figure S3), as in the case of the DG−/− cells. No such staining was observed when LARGE1 was stably expressed. These results suggest that the LARGE2-dependent modification of the alternative substrate(s) is independent of both O-mannosylation and the FKTN-dependent modification.
Identification of GPC4 as a substrate of LARGE2
To identify the alternative substrate(s) that are modified by LARGE2, we attempted to purify IIH6+ protein(s) from DG−/− cells stably expressing LARGE2. Immunoblotting of whole cell lysates from WT cells using IIH6 showed a ~120-kD band corresponding to α-DG, and this was absent in the DG−/− cells (Figure 1C). In contrast, lysates from three independent clones expressing LARGE2 showed a high-molecular weight smear that was not seen in WT or DG−/− cells (Figure 1C). Next, pooled cell lysates (clone #1 in Figure 1C) were subjected to anion exchange diethylaminoethyl (DEAE)-cellulose column chromatography (Figure 1D). Following elution of the IIH6-immunoreactive fraction from this column with 0.5 M NaCl, the fraction was applied to a IIH6-immobilized affinity column (IIH6-Sepharose) and then eluted effectively from it using an alkaline triethanolamine (TEA) buffer (Figure 1E, fraction 1).
The eluted fraction was resolved by SDS-PAGE and SYPRO Ruby staining. Two bands of ~130-kD and the area corresponding to the high-molecular weight smear containing IIH6-immunoreactivity were excised from the gel for in-gel trypsin digestion and mass spectrometric (MS) analysis. From the two bands at 130-kD, we identified peptides derived from nuclear proteins facillitates chromatin transcription (FACT) complex subunit SPT16 and DEAH box polypeptide 9 (Table I). However, given that the candidate protein(s) should be glycosylated and expressed at the cell surface, we excluded these from further analysis. From the high-molecular weight smear, we identified 1 peptide of mouse GPC4 (Table I, Supplementary data, Figure S4). GPC4 is a member of the glypican family of HS-PGs, which are linked to the cell surface by a glycosylphosphatidylinositol (GPI) anchor (Watanabe et al. 1995). Six members of this family (GPC1–GPC6) have been identified in mammals. Each is composed of a 60–70 kD core protein that has 14 conserved cysteine residues, attachment sites for HS–GAG chains and a GPI-anchoring signal (Fransson et al. 2004; Filmus et al. 2008). GPC4 is highly expressed in kidney and developing brain (Watanabe et al. 1995), and has been shown to play a key role in regulating signaling and phenotypic responses to hepatocyte growth factor and endostatin through the HS chains (Karumanchi et al. 2001; Karihaloo et al. 2004). The banding pattern in the high-molecular weight smear observed by SDS-PAGE is consistent with previous reports of recombinant and native GPC4 proteins (Watanabe et al. 1995; Ford-Perriss et al. 2003).
Table I.
Proteins identified from IIH6-Sepharose fraction TEA-1 derived from the DG−/− cells stably expressing LARGE2 by MS analyses
| Protein name | Peptide sequences |
|---|---|
| High-molecular weight smear | |
| Glypican-4 | SISESAFSAR |
| ~130-kD bands | |
| FACT complex subunit SPT16 | LAESVEKAIEEK, GNENANGAPAITLLVR, QDSLVINLNR, LTEQKGEQQIQK, ELAAQLNEEAK, SYCSNLVR, LAESVEK, ELAAQLNEEAKR, DLYIRPNIAQK |
| DEAH box polypeptide 9 | ISAVAVAER, AAECNIVVTQPR, ILAKLPIEPR, LNQYFQK, DFVNYLVR |
LARGE2-dependent modification of GPC4
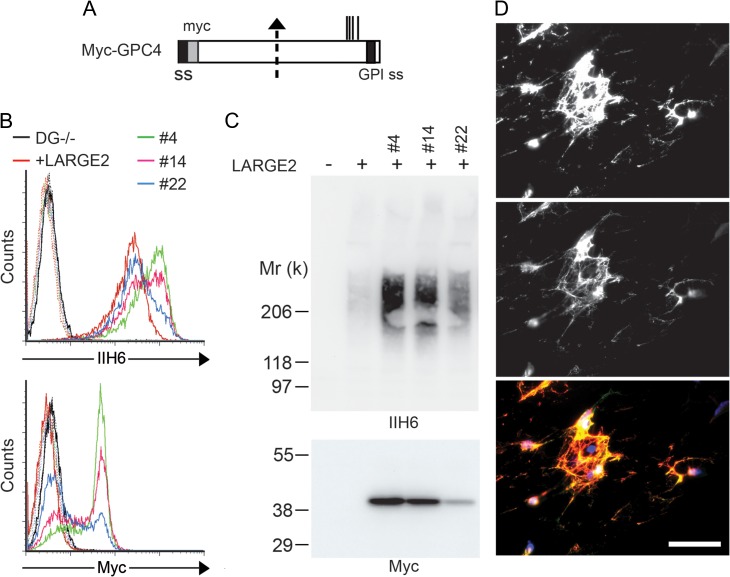
To confirm that GPC4 is indeed a substrate for functional modification by LARGE2, we examined whether overexpression of GPC4 could enhance the IIH6 immunoreactivity of DG−/− cells stably expressing LARGE2. We obtained several clones that stably express N-terminal myc-tagged GPC4 (Myc-GPC4, Figure 2A) and analyzed the surface expression by flow cytometry with anti-myc and IIH6 antibodies (Figure 2B). The Myc-GPC4 expressing clones showed increased IIH6 immunoreactivity, the intensity of which was positively correlated with the level of anti-myc detected. Immunoblotting of cell lysates enriched on a DEAE-cellulose column showed a high-molecular weight smear of IIH6 staining, with intensity greatly enhanced in the clones expressing Myc-GPC4, and a sharp 40-kD band representing myc (Figure 2C). The GPC4 core protein has been reported to undergo a proteolytic cleavage at its middle of the core protein (Watanabe et al. 1995; Ford-Perriss et al. 2003), generating two subunits that remain attached by one or more disulfide bonds. When Myc-GPC4 was expressed in human embryonic kidney (HEK293) cells, we observed a 40-kD band corresponding to the N-terminal fragment of GPC4 when using the anti-myc antibody under reducing conditions, and a high-molecular weight smear due to the HS chains when using nonreducing conditions (Supplementary data, Figure S5), consistent with the previous reports (Watanabe et al. 1995; Ford-Perriss et al. 2003). Therefore, under reducing conditions, while the N-terminal fragment was detected as a sharp band, the C-terminal portion of GPC4 was detected as a high-molecular weight smear containing LARGE2-mediated functional modification as well as HS chains. We also examined immunofluorescence staining in DG−/− cells stably expressing both LARGE2 and Myc-GPC4. Staining with both antibodies was strong and colocalization was robust, consistent with the notion that the IIH6 glycan modification occurs on cell surface-localized Myc-GPC4 (Figure 2D).
Fig. 2.
Overexpression of GPC4 enhances the IIH6 immunoreactivity of the LARGE2-expressing DG−/− cells. (A) Schematic representation of the myc-tagged GPC4 (Myc-GPC4) construct. The suggested proteolytic cleavage site and potential GAG attachment sites are indicated by an arrow and vertical lines, respectively. ss, signal sequence. GPI ss, GPI-anchoring signal sequence. The DG−/− cells stably expressing LARGE2 were transfected with Myc-GPC4. (B) Clones stably expressing myc (#4, #14 and #22) were analyzed by flow cytometry for cell-surface staining with IIH6 or anti-myc antibody. (C) Lysates of these cells enriched for Myc-GPC4 by DEAE-enrichment were immunoblotted with IIH6 or anti-myc antibody. Note that the enhanced intensity of the IIH6 staining is correlated with that of anti-myc staining. (D) Representative immunofluorescence micrograph of the DG−/− cells stably co-expressing LARGE2 and Myc-GPC4 (clone #4). The cell surface was stained with IIH6 and anti-myc antibody in the absence of permeabilization. Scale bar, 50 μm. The top image shows IIH6 immunofluorescence, the middle image shows myc immunofluorescence, and the bottom image shows IIH6 (red) merged with Myc (green) and DAPI (blue). All myc staining colocalizes with IIH6 as no ‘green only’ fluorescence is seen. This figure is available in black and white in print and in color at Glycobiology online.
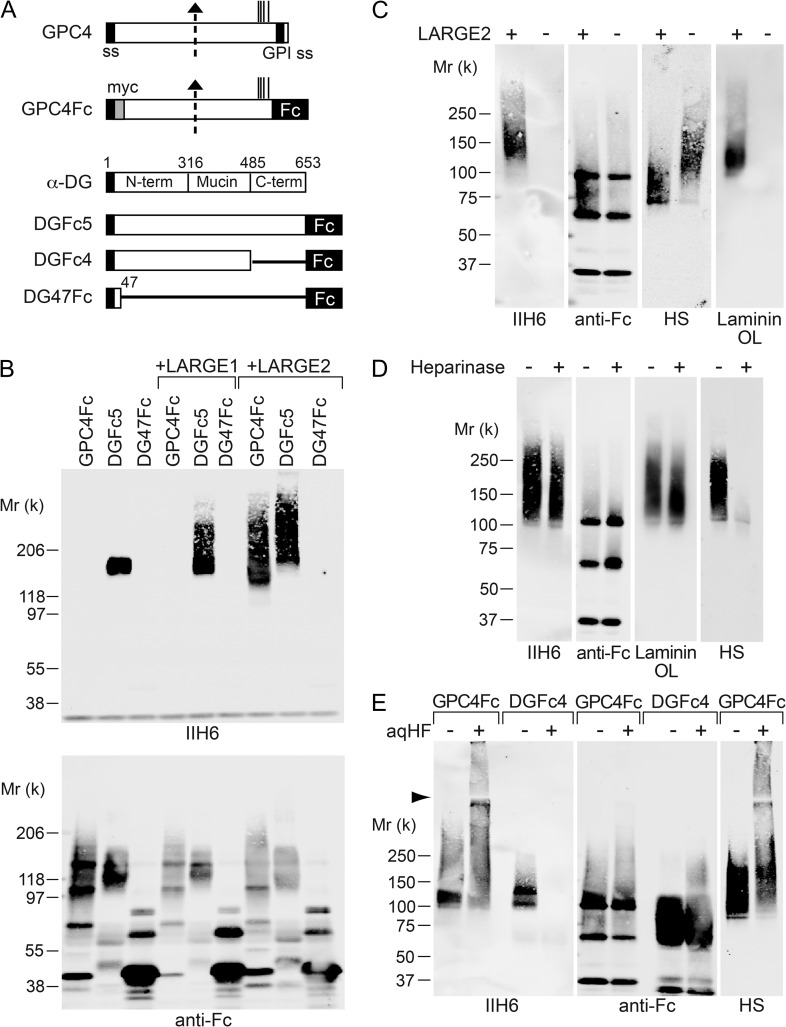
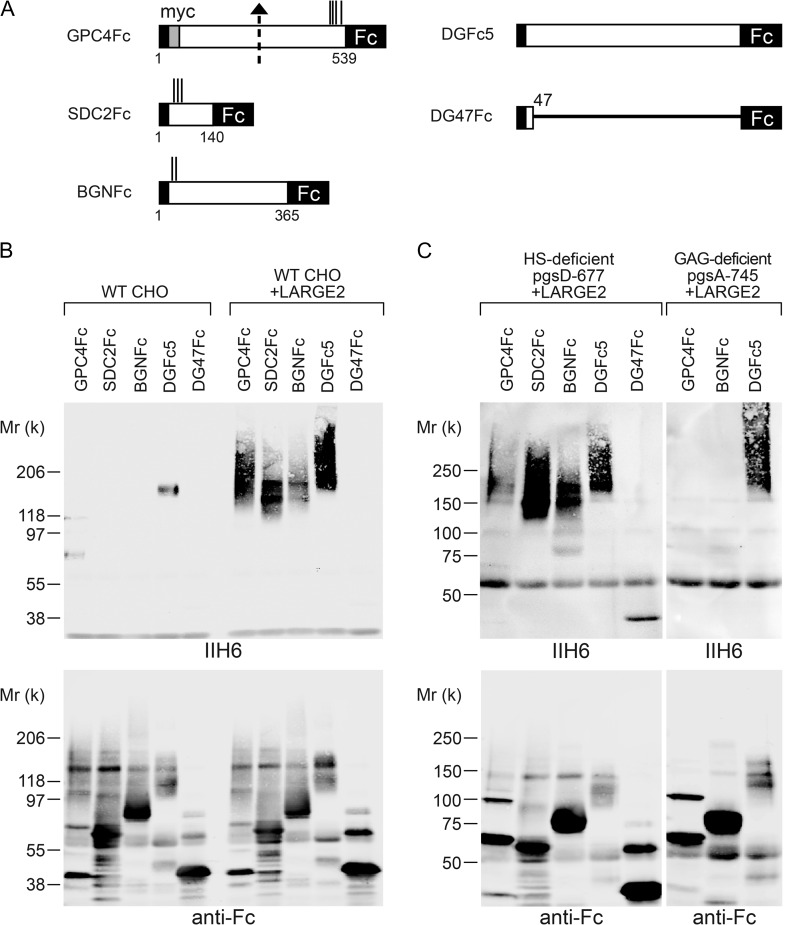
To investigate how GPC4 is modified by LARGE2, we generated an immunoglobulin G (IgG) Fc-fusion construct of GPC4 lacking its GPI-anchoring signal (GPC4Fc, Figure 3A) and expressed it in Chinese hamster ovary (CHO) cells stably expressing either LARGE1 or LARGE2. DGFc5 (α-DG-Fc fusion) and DG47Fc (DG amino acids 1–47, Fc fusion) were used as positive and negative controls for the functional modification, respectively (Figure 3A). The Fc protein secreted into the conditioned medium was purified using Protein-A agarose and analyzed by immunoblotting with IIH6 and anti-human IgG (anti-Fc). GPC4Fc in the medium was detected as a ~100–200-kD smear using anti-Fc (Figure 3B). DGFc5 expressed in CHO cells was functionally glycosylated and the IIH6 immunoreactivity was enhanced, as evident from the presence of high-molecular weight species in the cells stably expressing LARGE1 or LARGE2 (Figure 3B). In contrast, GPC4Fc was functionally modified only in the LARGE2-expressing cells, as is the case in the DG−/− cells expressing LARGE2. We also confirmed the requirement for LARGE2 in this process by stably expressing GPC4Fc in CHO cells in the presence or absence of stable expression of LARGE2. Cells were cultured in serum-free medium, and the purified Fc fusion protein was analyzed with IIH6 and anti-HS antibody as well as by laminin overlay assay (Figure 3C). GPC4Fc was functionally modified and bound laminin only when LARGE2 was expressed.
Fig. 3.
LARGE2 can modify GPC4 with the laminin-binding glycan. (A) Schematic representation of Fc-fusion constructs. Dotted arrow, suggested proteolytic cleavage site. Vertical lines, potential GAG attachment sites. ss, signal sequence. GPI ss, GPI-anchoring signal sequence. (B) LARGE2 can modify GPC4Fc in CHO cells. Immunoblotting of the Fc fusion proteins transiently expressed in, and purified from, the media of CHO cells with or without stable expression of LARGE1 or LARGE2. (C) Immunoblotting or laminin overlay (OL) of GPC4Fc purified from serum-free CHO culture with or without stable expression of LARGE2. Treatment with neither heparinase (D) nor aqHF (E) removed the functional modification from GPC4Fc.
Since the C-terminal part of GPC4 contains GAG attachment sites for HS–GAG chains and also undergoes functional modification by LARGE2, we examined the effects of heparinase treatment of GPC4Fc to test whether the HS chains could be modified by LARGE2. After the treatment of purified GPC4Fc, almost all the immunoreactivity to anti-HS antibody disappeared, whereas neither the IIH6 reactivity nor laminin-binding activity was diminished (Figure 3D).
It has been shown that the laminin-binding epitope of α-DG is attached to the O-mannosyl glycan via phosphodiester bonds and that the epitope can be removed by cold aqueous hydrofluoric acid (aqHF), which specifically cleaves phosphoester linkages (Yoshida-Moriguchi et al. 2010). We found that the IIH6 immunoreactivity of GPC4 was not diminished by aqHF treatment under conditions that completely abolished the epitope on DGFc4, even though the mobility of GPC4Fc as detected by SDS-PAGE was somewhat affected (Figure 3E). Thus, the functional modification on GPC4 is unlikely to be attached by a phosphodiester linkage.
Identification of GPC4 and other PGs as IIH6+ proteins from rabbit kidney extract
GPC4 was originally cloned from a mouse kidney cDNA library, and was reported to be highly expressed in the kidney and the developing brain (Watanabe et al. 1995). LARGE2 has also been shown to be abundant in the kidney (Fujimura et al. 2005; Grewal et al. 2005). We analyzed expression of the GPC4, LARGE1 and LARGE2 mRNAs in several mouse tissues by quantitative polymerase chain reaction (PCR), and confirmed that both GPC4 and LARGE2 are highly expressed in the kidney, whereas LARGE1 is not (Supplementary data, Figure S6), consistent with previous reports.
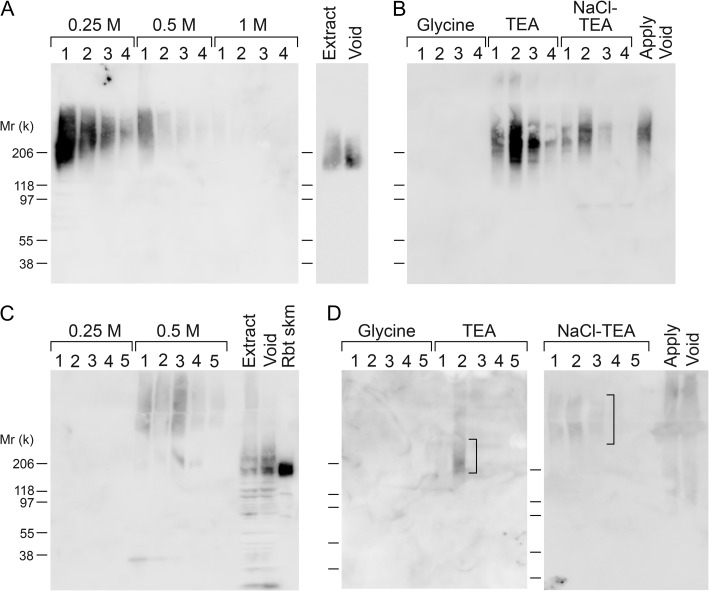
We attempted to determine whether native GPC4 is truly a substrate for LARGE2-mediated functional modification in tissues. For this purpose, we extracted proteins from rabbit kidney by sequential treatment with 1% Triton X-100 and 8 M urea, and purified IIH6+ proteins using a combination of DEAE-cellulose and IIH6 affinity columns. A considerable fraction of IIH6-immunoreactive protein, which is likely derived from α-DG, was detected in the void of the DEAE column and the higher molecular weight IIH6 immunoreactivity was retained on the column (Figure 4A for Triton extract, 4C for urea extract). The bound IIH6-immunoreactive protein was mainly eluted by 0.25 or 0.5 M NaCl, and the eluates were further separated on a IIH6-Sepharose column. Proteins bound to the IIH6 column were eluted using alkaline TEA buffer (Figure 4B for Triton extract, Figure 4D for urea extract). For samples derived from the Triton extract, each eluted fraction from the IIH6 column (TEA-1 to 3 and NaCl-TEA-1 to 3 in Figure 4B) was subjected to tryptic digestion followed by MS analysis and comparisons of the identified proteins among these fractions. The cutoff in the Scaffold software was set for 99% protein identification probability, and those proteins that were identified mainly in the fractions with weaker IIH6 immunoreactivity, as well as cytosolic proteins, nuclear proteins and unknown proteins, were excluded from the results. We were able to identify GPC4 and some other extracellular and cell-surface proteins (Table II) that were present only in the TEA-2 fraction or enriched in it (according to the numbers of assigned spectra for these proteins compared across fractions). Notably, no α-DG peptides were identified in any of these fractions. Intriguingly, agrin, collagen XVIII and perlecan are HS-PGs. As in the case of samples derived from the urea extract, we could detect IIH6 immunoreactivity in the TEA-2 and NaCl-TEA-1 to 3 fractions (Figure 4D). A 200-kD smear from the TEA-2 fraction and a high-molecular weight smear from fractions NaCl-TEA-1 to 3 were excised from the SYPRO Ruby-stained gel for in-gel trypsin digestion and MS analysis. Biglycan, syndecans 2 and 4 were identified in the 200-kD band (TEA-2), whereas agrin, perlecan and syndecan-2 were identified in the high molecular smear (NaCl-TEA-1 to 3) as specified in Table III. All of these proteins are PGs, with perlecan, agrin and syndecans representing the HS-PG class and biglycan being a CS/DS-PG.
Fig. 4.
Purification of IIH6+ proteins from rabbit kidney. IIH6 immunoreactivity of rabbit kidney extracts (A and B, Triton extract; C and D, urea extract) applied to columns (A and C, DEAE-cellulose; B and D, IIH6-Sepharose), and samples separated by the columns (the void and eluted fractions). The fractions TEA1-3 and NaCl-TEA1-3 derived from the Triton extract were subjected to trypsin digestion and analyzed by MS (Table II). In the cases of the TEA-2 and NaCl-TEA1-3 fractions derived from urea extract, the areas in the SDS-PAGE gels indicated by squared brackets were excised and analyzed by in-gel trypsin digestion and subsequent MS (Table III).
Table II.
Proteins identified from IIH6-Sepharose fraction TEA-2 derived from rabbit kidney Triton extract by MS analyses
| Protein name | Peptide sequences |
|---|---|
| Agrin | CEPGFWNFR, SELFGETAR |
| Beta-glucuronidase | WTQDTDMR, LVQSGPLTTCR, GFDWPLLVK, MFSEEYQK, TSAFILR |
| Collagen alpha-1(XII) | ITVDPTTDGPTK, VGVVQYSSDTR, VAIIITDGK, SQDEVEIPAR, SFEISPNR, VQISLVQYSR, VMILITDGK, VTWEPAPGEVK, VLVVVTDGR, TLSFFNK |
| Collagen alpha-1(XVIII) | ILNVAQGIR, EELYVR, ADDILANPPR, LQDLYSIVR |
| Galectin-3-binding protein | SLGWMVSR, VDAECMPVVR, IEVSMSSVK |
| Glypican-4 | DLFVELK, TFAQGLAVAR |
| Inter-alpha-trypsin inhibitor heavy chain H5 | TPLLSDIR |
| Perlecan | VTSYGGELR, SPAYTLVWTR, FDAGSGMATIR, YELGSGLAVLR |
| Short transient receptor potential channel 2 | (LGR)LLKIPVLK |
The table shows proteins identified in fraction TEA-2. The cutoff in the Scaffold software was set for 99% protein identification probability. Proteins identified mainly in fractions with weaker IIH6 immunoreactivity in Figure 4B, cytosolic proteins, nuclear proteins and unknown proteins were excluded from the results.
Table III.
Proteins identified from IIH6-Sepharose fraction TEA-2 and NaCl-TEA1-3 derived from rabbit kidney urea extract by MS analyses
| Protein name | Peptide sequences |
|---|---|
| Agrin | RPLQEHVR, ALQSNHFELSLR |
| Biglycan | IHEKAFSPLR(K), KLQKLYISK, DHLVEIPPNLPSSLVELR, KVPKGVFSGLR, ISEAKLTGIPK, IQAIELEDLLR, ELHLDNNKLSR, VPAGLPDLK, LAIQFGNYK |
| Perlecan | LHNGKLPSR, EGGSLPPQAR, ((R)GGSLPAR)HQTHGSLLR, (GGSLPSR)HQIVGSR, VVPYFTQTPYSFLPLPTIK |
| Syndecan-2 | (K)KDEGSYDLGER, APTKEFYA |
| Syndecan-4 | ETEVIDPQDLLEGR |
The table shows proteins identified in ~200-kD broad band of fraction TEA-2 and high molecular smear band of fraction NaCl-TEA-1-3 (Figure 4D). The former contained biglycan, syndecan-2 and 4, the latter contained agrin, perlecan and syndecan-2.
LARGE2-dependent modification of PGs
The data from the MS analysis for kidney IIH6+ proteins prompted us to examine whether LARGE2 could modify not only GPC4 but also other HS- and/or CS/DS-PGs. We expressed Fc-fusion constructs of HS-PG syndecan-2 (SDC2Fc) and CS/DS-PG biglycan (BGNFc) (Figure 5A) in CHO cells stably expressing LARGE2. Like GPC4Fc, both SDC2Fc and BGNFc could be functionally modified by LARGE2 (Figure 5B). Next we asked if the modification on those PGs depends on the synthesis of HS and/or GAG, using CHO mutant cell lines. In the CHO mutant pgsD-677, which is deficient for HS synthesis (Lidholt et al. 1992), the Fc fusion proteins were modified by LARGE2 (Figure 5C), consistent with the result of heparinase treatment of GPC4Fc (Figure 3D). Conversely, in the GAG-deficient mutant pgsA-745, which lacks the Xyl-T activity required for initiating GAG synthesis through the GAG-protein linkage region (Esko et al. 1985), neither GPC4Fc nor BGNFc was modified by LARGE2 (Figure 5C). Thus, the LARGE2-dependent modification of PGs requires the synthesis of GAG but not HS.
Fig. 5.
LARGE2 can modify not only GPC4 but also other PGs. (A) Schematic representation of Fc-fusion constructs. Dotted arrow, suggested proteolytic cleavage site. Vertical lines, potential GAG attachment sites. (B) LARGE2 can modify PGs. Immunoblotting of Fc fusion proteins transiently expressed in the WT CHO cells with or without stable expression of LARGE2. (C) Immunoblotting of Fc-fusion proteins transiently expressed in HS- or GAG-deficient CHO mutant cells that stably express LARGE2. In the GAG-deficient cells, LARGE2 could not modify GPC4Fc or BGNFc.
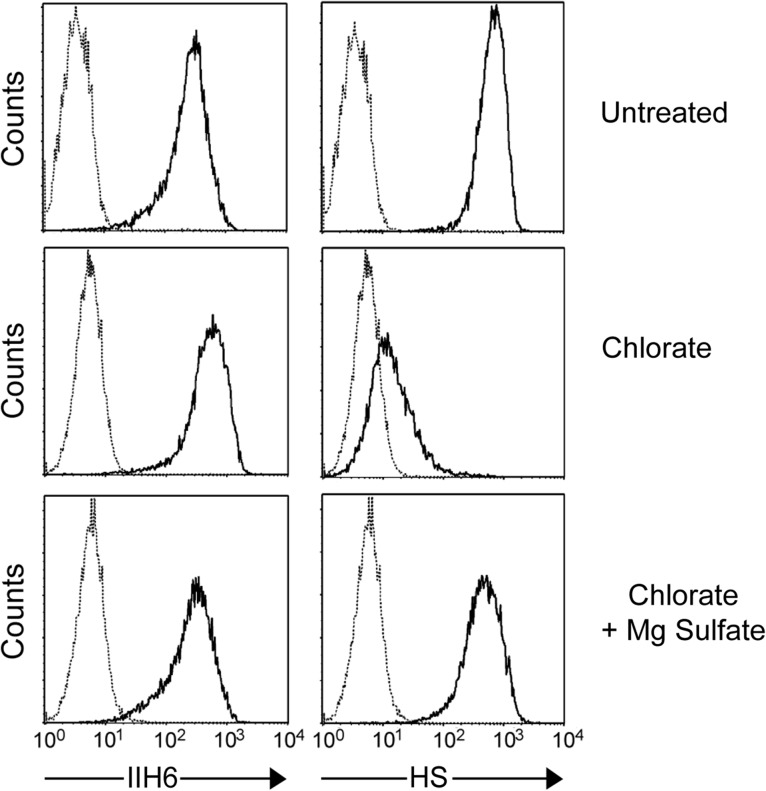
Given that the LARGE2-mediated modification occurs on both HS- and CS/DS-PGs, which share the common GAG-protein linkage tetrasaccharide GlcAβ1-3Galβ1-3Galβ1-4Xylβ1, it is likely that LARGE2 acts on the linkage region. Since it is known that galactose residues in the linkage region of CS/DS-GAGs undergo sulfation (Sugahara and Kitagawa 2000), we examined whether inhibiting sulfation could affect functional modification by LARGE2. Sodium chlorate inhibits the formation of 3′-phosphoadenosine 5′-phosphosulfate, the sulfate donor in cellular sulfation reactions (Baeuerle and Huttner 1986). Thus, DG−/− cells stably expressing LARGE2 were cultured in the presence of sodium chlorate and analyzed by flow cytometry with IIH6 or the anti-HS antibody 10E4. Since it has been reported that N-sulfation of glucosamine residues in HS is required for 10E4 binding (David et al. 1992), the inhibitory effect of chlorate was assessed by staining with this antibody. The 10E4 staining of chlorate-treated cells was greatly reduced, and this effect was sulfation-specific because it was reversed by the addition of magnesium sulfate (Figure 6). However, IIH6 reactivity was unaffected by the treatment, indicating that the LARGE2-dependent modification is independent of sulfation.
Fig. 6.
Sulfation is dispensable for the LARGE2-dependent modification. Flow cytometry of chlorate-treated DG−/− ES cells stably expressing LARGE2. HS staining was reduced by the chlorate treatment, and this effect was negated by the addition of magnesium sulfate. In contrast, IIH6 staining was not reduced by the treatment.
Discussion
LARGE1 and LARGE2 are highly conserved, both consisting of two catalytic domains that have Xyl-T and GlcA-T activities. LARGE1 is highly expressed in the tissues that are affected in dystroglycanopathies, such as brain and skeletal muscle. The distribution and pH optima for LARGE2 (both the Xyl-T and GlcA-T activities) have been shown to be distinct from those for LARGE1 (Inamori et al. 2013), suggesting that LARGE2 could play a different role in the functional modification of proteins.
Here we demonstrated that LARGE2, but not LARGE1, functionally modifies substrates other than α-DG. The LARGE2-mediated modification is independent of O-mannosylation or Fktn-dependent Rbo5P addition because both Pomt1−/− and Fktn−/− cells, regained the functional modification upon stable expression of LARGE2, but not in the presence of LARGE1, as is the case in DG−/− cells. This result conflicts with previous studies reporting that LARGE1 overexpression resulted in functional modification of proteins other than α-DG in the absence of O-mannosyl glycosylation (Zhang et al. 2011; Zhang and Hu 2012). In those studies, neural stem cells isolated from brain-specific DG knockout or DG and POMT2 double-knockout fetuses were analyzed after adenoviral overexpression of LARGE1. Although those studies did not examine LARGE2 activity, they indicated that transient adenovirus-mediated overexpression of LARGE1 can produce IIH6-immunoreactive epitopes with laminin-binding ability on non-α-DG proteins. In our study, we examined both transient, adenovirus-mediated expression and stable, lentivirus-mediated expression in DG−/− ES cells; however, we did not observe any detectable IIH6 immunoreactivity in the context of LARGE1 overexpression. Possible explanations for this discrepancy are that mouse ES cells might not express non-α-DG substrates of LARGE1 that are present in neural stem cells, and that the modification observed in the adenovirus-infected neural stem cells was caused by artificially high expression levels of LARGE1.
Other studies based on transient overexpression of LARGE1 in various CHO mutant cells have shown that LARGE1 can modify α-DG even in O-mannosylation-deficient mutant cells, and suggested that N- and mucin O-glycans could be modified by LARGE1 (Patnaik and Stanley 2005; Aguilan et al. 2009). However, another study showed that stable expression of LARGE1 in the O-mannosylation-deficient mutant CHO cells failed to alter the processing of α-DG, with only transient overexpression that causes extremely high levels of LARGE1 expression able to produce the functional modification (Hu et al. 2011). We speculate that when the expression level of LARGE1 is artificially high, this enzyme can modify unnatural acceptors such as N- or O-glycans, and possibly non-α-DG proteins. In fact, the enzymatic activities of LARGE1 were identified using labeled monosaccharide acceptors in in vitro assays, using a high concentration of the recombinant enzyme (Inamori et al. 2012). Since β-linked GlcA residues are present in the human natural killer-1 carbohydrate epitope (HSO3-3GlcAβ1-3Galβ1-4GlcNAc-), which is found in N-linked and O-mannosyl glycans of glycoproteins and also in some glycolipids, as well as in the GAG-protein linkage region tetrasaccharide of PGs, an extremely high level of LARGE1 may transfer Xyl to the GlcA terminus of even a nonnative substrate. Given that the disaccharide Xyl-GlcA is a much more potent acceptor of LARGE1 than are monosaccharide acceptors, as has been observed in in vitro enzymatic assays (Inamori et al. 2014), at high levels LARGE1 is likely to be able to elongate Xyl-GlcA repeats on the nonnative substrate.
In our study, only LARGE2 produced the functional modification on non-α-DG proteins. Since recognition of the N-terminal domain of α-DG by LARGE1 is required for the functional modification of the former (Kanagawa et al. 2004) and this occurs predominantly on specific Thr residues located at the extreme N-terminus of the mucin-like domain (Hara et al. 2011), LARGE1 recognizes the α-DG core protein and seems to have relatively high specificity for the acceptor. Although LARGE2 also recognizes the N-terminal domain of α-DG (Inamori et al. 2014), our observations may reflect a broader specificity of LARGE2 than LARGE1 for the acceptor substrate, which could enable LARGE2 to act on PGs as alternative substrates.
We demonstrated that LARGE2 can modify PGs with the laminin-binding glycan and that this modification requires, at least, initial O-xylosylation of core proteins but not HS synthesis. We predict that LARGE2 polymerizes from the β1,3-linked GlcA in the GAG-protein linkage region (Supplementary data, Figure S7). However, this proposed linkage has not yet been validated by structural analysis or in vitro synthesis. If LARGE2 does indeed extend from this GlcA moiety, then the synthesis of HS- and CS/DS-GAG would compete with the LARGE2-dependent modification, a notion that is supported by the immunoblot data of GPC4Fc expressed in the presence or absence of LARGE2 (Figure 3C). Stable expression of LARGE2 caused a reduction of the relative molecular weight of the band stained with anti-HS, suggesting that the LARGE2-mediated modification took over some of the GAG attachment sites on GPC4Fc in the cells. It will be of interest to determine the physiological function of the LARGE2-mediated modification of PGs. In the case of GPC4, it might have a function to regulate sublocalization at the cell surface through binding to LG-containing ECM proteins, and thereby could modulate GPC4 functions such as growth factor signaling. The modification of other PGs could also regulate basement membrane structure and function. Basement membranes are layered cell-adherent ECMs that form part of the tissue structure and contribute to tissue and organ morphogenesis and maintenance of adult functions, by serving as cell scaffolds and signaling platforms (Yurchenco 2011). ECMs are self-assembled on cell surfaces through binding interactions among laminins, type IV collagens, nidogens and PGs. Thus, LARGE2 might play a role in stabilizing ECM protein complexes and modifying their functions by augmenting the interactions between LG-containing ECM proteins and PGs.
Materials and methods
Antibodies to the α-DG glycan epitope (IIH6), core α-DG protein (5–2), LARGE1 and LARGE2 have been described previously (Michele et al. 2002; Kanagawa et al. 2004; Inamori et al. 2013; Fortunato et al. 2014). Anti-myc antibody 9E10 was purchased from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA). Anti-HS antibody 10E4 was purchased from US Biological (Salem, MA). SYPRO Ruby protein gel stain was purchased from Invitrogen (Carlsbad, CA). 4′,6-Diamidino-2-phenylindole (DAPI) was purchased from Sigma (St. Louis, MO). Mouse WT ES cells (R1) and DG−/− ES cells (354.B11) were described previously (Henry and Campbell 1998). WT CHO (CHO-K1), HS-deficient mutant pgsD-677 and GAG-deficient mutant pgsA-745 were obtained from American Type Culture Collection (Manassas, VA).
Generation and characterization of Pomt1−/− and Fktn−/− ES cells
For Pomt1−/− cells, Pomt1 genetrap mice (Lexicon Genetics, B6;129S5-Pomt1Gt(neo)205Lex, MMRRC (Chapel Hill, NC) strain 011649-UNC) were cryorecovered from the MMRRC, and heterozygotes were bred to derive Pomt1 null ES cell lines. Pregnant mice were euthanized and 3.5-d post-coital (d.p.c.) preimplantation embryos were flushed from the uterine horns with M2 solution (Specialty Media Inc. (Lavallette, NJ)). Isolated embryos were placed into wells of a 96-well plate with preplated irradiated fibroblasts, at 1 embryo per well. Embryos were cultured for ~7 d and fed every day with K-DMEM + 15% KSR (knockout serum replacement), and recombinant leukemia inhibitory factor (rLIF, chemicon 10 ng/mL). After 7 d, cell colonies were picked and expanded further on fibroblasts in DMEM with 15% fetal bovine serum (FBS) and 10 ng/mL rLIF. The expanded cells were used for PCR genotyping according to instructions from Lexicon Genetics (The Woodlands, TX).
For Fktn−/− cells, mice heterozygous for a Fktn exon 2 deletion were used. The null allele was generated by germline Cre recombination, as previously reported (Beedle et al. 2012). Initial attempts to obtain ES cell lines from embryos were unsuccessful, so a superovulation procedure (according to The Jackson Laboratory, Bar Harbor, ME) was implemented for the female Fktn+/− breeders. Briefly, 4.5-week old Fktn+/− females received a single intraperitoneal injection of pregnant mare serum. Forty-seven hours later, the females were injected with human chorionic gonadotropin and co-housed with Fktn+/− males for breeding. At 3.5 d.p.c., embryos were collected and Fktn+/− and Fktn−/− ES cells were obtained as described for the Pomt1 ES cell lines above. Genotyping for the Fktn WT and cre-recombined alleles was conducted as previously described (Beedle et al. 2012).
POMT1 and FKTN functions were assessed in the mouse Pomt1−/−, Fktn−/− and Fktn+/− ES cell clonal lines by western blotting of wheat germ agglutinin (WGA)-enriched cell lysates. Briefly, mouse ES cells were expanded on gelatin-coated plates in ES media for LIF-adapted cells (DMEM supplemented with 20% FBS, 1 mM nonessential amino acids (NEAA, Gibco, Carlsbad, CA), 2 mM L-glutamine, 1000 U/mL ESGRO (Millipore, Billerica, MA) and 0.001% v/v 2-mercaptoethanol). Cells were lysed in WGA solubilization buffer (50 mM tris pH 7.4, 150 mM NaCl, 1% nonident P-40, with protease inhibitors). Solubilized cell supernatants were incubated at 4°C overnight with wheat-germ agglutinin agarose (Vector Labs, Burlingame, CA), for the enrichment of N-glycosylated proteins (including α-DG). WGA-agarose was washed three times, after which protein was eluted with 0.3 M N-acetylglucosamine and analyzed by SDS-PAGE and western blotting. IIH6 was used to test for functionally glycosylated α-DG, and aDGct rabbit monoclonal 5–2 was used to detect α-DG core protein.
Adenovirus- and lentivirus-mediated gene transfer for expression of LARGE1 or LARGE2
Replication-deficient adenoviruses encoding the LARGE1 or LARGE2 protein tagged with an eGFP reporter gene (CMV-LARGE1/RSV-eGFP and CMV-LARGE2-IRES-eGFP, respectively) were generated by the Gene Transfer Vector Core at the University of Iowa (GTVC, Iowa City, IA) as described previously (de Bernabe et al. 2009). Mouse ES cells were infected with adenovirus at a multiplicity of infection of 100 in regular ES medium overnight. The next day the medium was replaced with fresh medium, and the cells were cultured for 1 d. For flow cytometry, the IIH6+ cells were separated by AutoMACS magnetic cell separation (Miltenyi Biotec, Auburn, CA) using IIH6 and anti-mouse IgM MicroBeads (Miltenyi Biotec, Auburn, CA) according to manufacturer's protocol. The generation of lentiviruses encoding LARGE1 and LARGE2 and the establishment of stable cell lines were described previously (Inamori et al. 2013).
Purification of IIH6+ proteins
For purification of IIH6+ proteins from the DG−/− ES cells stably expressing LARGE2, cells were lysed in lysis buffer: 1% Triton X-100, 5 mM EDTA in tris-buffered saline (TBS) containing protease inhibitors (0.6 μg/mL of pepstatin A, 0.5 μg/mL of aprotinin, 0.5 μg/mL of leupeptin, 0.75 mM of benzamidine, 0.1 mM of phenylmethanesulfonyl fluoride (PMSF), 0.4 μg/mL of calpain inhibitor and 0.4 μg/mL of calpeptin). The supernatant was obtained by centrifugation at 10,000 × g, and lysate diluted in 50 mM NaCl was applied to a DEAE-cellulose column equilibrated with 50 mM NaCl, 1% Triton X-100 in TBS. The column was washed with 10 volumes of the equilibration buffer (Wash-1 in Figure 1D) and then washed further with 0.1% Triton X-100 in the same buffer (Wash-2 in Figure 1D). Bound proteins were eluted stepwise with various concentrations of NaCl, and the eluted fractions were analyzed by immunoblotting with IIH6. The 0.5 M NaCl fraction was diluted with 2 volumes of 0.1% Triton X-100 in phosphate-buffered saline and applied to a IIH6-Sepharose column. The IIH6-Sepharose was prepared by coupling IIH6 (~3 mg in 2 mL) with 0.4 g dried powder of CNBr-activated Sepharose 4B (GE Healthcare) according to the manufacturer's protocol. After the column was washed, bound proteins were eluted with 0.5 M NaCl in 0.1 M glycine, pH 2.5 and then with 0.5 M NaCl in 0.1 M TEA, pH 11.5 in the presence of 0.1% Triton X-100. For purification of non-α-DG IIH6+ proteins from rabbit kidneys, the homogenate in the lysis buffer was centrifuged at 20,000 × g for 30 min, and the resultant supernatant was designated as Triton extract. The pellet was further extracted overnight with urea extraction buffer (8 M urea, 1% Triton X-100, 5 mM EDTA in TBS) and the supernatant was designated as urea extract.
GPC4 construct and generation of stable cell lines
The mouse GPC4 gene was cloned into pEF4/Myc-His (Invitrogen) with the native stop codon and a myc-tag sequence at the N-terminus, using the SacI site as described in the original study (Watanabe et al. 1995). First, a fragment containing the SacI site and a c-myc sequence with an internal BamHI site were amplified by PCR, using primers 5′-AGAGCTCGAACAAAAGCTGATTTCTGAAGAAGACCTCAAGTCGAAAAGTTGCTC-3′ and 5′-CACAGCCTTGGAAAACCTTC-3′ and the template mouse GPC4 cDNA (BC006622, OriGene, Rockville, MD). Two restriction fragments, a 0.3-kb SacI fragment containing the start codon excised from the template and a 0.9-kb SacI/BamHI fragment of the PCR product, were then cloned back into the same plasmid using SacI-BamHI sites, and this construct was designated as GPC4-Smyc-pCMV. A 0.5-kb EcoRI fragment and a 1.6-kb EcoRI/NheI fragment were subcloned into pEF4/Myc-His with EcoRI/XbaI sites, and this construct was designated as Myc-GPC4. The plasmid Myc-GPC4 was introduced into cells by transfection with LipofectAmine2000 (Invitrogen) or using an Amaxa Nucleofector device with Mouse ES Cell Nucleofector Solution according to the manufacturer's protocol. Transformants were selected in medium containing Zeocin (50 μg/mL, Invitrogen). Single colonies were isolated and surface expression of Myc-GPC4 was analyzed by flow cytometry with anti-myc antibody 9E10. Some of the positive clones were also analyzed by immunoblotting of proteins enriched on a DEAE-cellulose column. Briefly, cell lysate was incubated with DEAE-cellulose and bound proteins were eluted with lysis buffer containing 0.5 M NaCl.
Fc fusion proteins
The α-DG-Fc-fusion constructs DGFc4 and DGFc5 were described previously (Kunz et al. 2001). For construction of GPC4Fc, a fragment of mouse GPC4 lacking the GPI-anchoring signal was amplified by PCR, using primers 5′-GACTGCTGGAATGGCAAAGG-3′ and 5′-GAGGATCCGGCTTTGCCTCTGCATGG-3′, and the template GPC4-Smyc-pCMV. A 350-bp KpnI/BamHI fragment of the PCR product and a 1.5-kb KpnI fragment from the plasmid were cloned into the KpnI/BamHI arm of the DGFc5 plasmid after the α-DG sequence was excised. Similarly, fragments for SDC2, BGN and DG47 were amplified by PCR, using the following primers and templates: SDC2Fc: 5′-GCGAATCCCTGGGGAATATGCAG-3′ and 5′-CGAGATCTAGATTGTCTGAATGTTTCTC-3′, and mouse syndecan-2 cDNA (BC047144, Open Biosystems, Lafayette, CO); BGNFc: 5′-GAGAAT TCCCCAGGAACATTGACC-3′ and 5′-CA GGATCC CCAAATTGGATGGCCAGGC-3′, and mouse biglycan cDNA (BC019502, Open Biosystems); DG47Fc: 5′-CACTGCTTACTGGCTTATCG-3′ and 5′-CTGGATCCTCCAGCTGGTTCTCCCAGTC-3′ and DGFc5. Fragments KpnI/BglII (SDC2), EcoRI/BamHI (BGN) and BamHI (DG47) were cloned into the KpnI/BamHI, EcoRI/BamHI and BamHI arms, respectively, of the DGFc5 plasmid after the α-DG sequence was excised. Fc proteins were expressed transiently and purified from the conditioned media as described (Kanagawa et al. 2004). For large-scale expression of GPC4Fc, CHO cells or CHO cells stably expressing LARGE2 were transfected with the GPC4Fc plasmid and selected in medium containing Geneticin (500 μg/mL, Invitrogen). The GPC4Fc-expressing clone was adapted to serum-free suspension culture in ProCHO4 medium, and the recombinant protein was obtained in ProCHO5 medium (Lonza, Walkersville, MD).
Heparinase treatment and HF dephosphorylation
For heparinase treatment, samples were treated with a mixture of heparinases I and III (Sigma, 0.6 and 0.3 Sigma units, respectively) in 0.1 M sodium acetate, pH 7.0, 10 mM EDTA, 2 mM PMSF, 2 μg/mL pepstatin and 10 mM N-ethylmaleimide at 37°C for 4 h. Dephosphorylation by aqHF was performed as described previously (Yoshida-Moriguchi et al. 2010). Sample was desalted and incubated in precooled 48% aqHF. After 24 h on ice, the sample was dried using an N2 stream.
Chlorate treatment
For inhibition of sulfation, the DG−/− ES cells stably expressing LARGE2 were cultured in sulfate-free medium with 30 mM sodium chlorate, in the presence or absence of 10 mM magnesium sulfate, for 2 d. Joklik's modified MEM (Sigma) and dialyzed FBS (Invitrogen) were used for preparing the sulfate-free medium with supplementation of 2 mM calcium chloride.
Identification of IIH6+ proteins by MS
Identification of proteins from excised bands by LC–MS/MS analyses of enzymatic protein digests was conducted by the Tufts University Core Facility (Boston, MA). For samples purified from Triton extracts of rabbit kidney, the eluates from a IIH6-Sepharose column were subjected to LC–MS/MS on an Agilent 6520 Accurate-Mass Quadropole Time-of-Flight mass spectrometer interfaced with an HPLC Chip Cube. The samples were loaded onto the large-capacity C18 chip II, consisting of a 160 nL enrichment column and 150 mm analytical column (G4240-62010; Agilent Technologies, Santa Clara, CA). The samples were subjected to LC–MS/MS analysis using a 60-min gradient from 1.5 to 38% buffer B (100% acetonitrile, 0.8% acetic acid). The data-dependent settings (tandem MS) included a maximum of 20 ions per cycle and medium isolation width (~4 atomic mass units), and precursor masses were dynamically excluded for 30 s after 5 MS/MS in a 30-s time window. Mass spectrometry capillary voltage and capillary temperature settings were set to 1800 V and 330°C, respectively. The infused reference mass of 1221.99 was used to correct precursor m/z masses for each LC–MS/MS experiment. For protein identification, the raw.d files were searched against the UniProt mouse database using SpectrumMill Software version B.04.01.141 (Agilent Technologies) with the following settings: precursor mass tolerance of 50 parts per million (ppm), product mass tolerance of 300 ppm, and a maximum of two trypsin miss cleavages. Search modifications included static carbamidomethylation on cysteine residues (C = 57.02 AMU) and variable modifications for oxidized methionine (M = 15.99 AMU). The search results with a <1% false discovery rate were accepted and converted to a scaffold file for data interpretation.
Miscellaneous
Immunoblotting, laminin overlay, WGA enrichment of glycoproteins, immunofluorescence microscopy, quantitative PCR and flow cytometry were performed as described previously (Kanagawa et al. 2004; de Bernabe et al. 2009; Inamori et al. 2012).
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Acknowledgements
We thank all members of the Campbell laboratory and C. Blaumueller for fruitful discussions, and the University of Iowa Gene Transfer Vector Core, Gene Targeting Facility, and Flow Cytometry Facility, Tufts University Core Facility, and Iowa State University Hybridoma Facility for their services.
Conflict of interest statement
None declared.
Funding
Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (1U54NS053672 to K.P.C.); University of Georgia College of Pharmacy (to A.M.B.). K.P.C. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
BGN, biglycan; CS/DS, chondroitin/dermatan sulfate; DG, dystroglycan; ECM, extracellular matrix; ES, embryonic stem; FKRP, fukutin-related protein; FKTN, Fukutin; GAG, glycosaminoglycan; GPC, glypican; GPI, glycosylphosphatidylinositol; HS, heparan sulfate; MS, mass spectrometry; PG, proteoglycan; POMT, protein O-mannosyltransferase; SDC, syndecan; TBS, tris-buffered saline; TEA, triethanolamine; WT, wild-type.
References
- Aguilan JT, Sundaram S, Nieves E, Stanley P. 2009. Mutational and functional analysis of large in a novel CHO glycosylation mutant. Glycobiology. 19:971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikov A, Buettner FF, Tiemann B, Gerardy-Schahn R, Bakker H. 2013. LARGE2 generates the same xylose- and glucuronic acid-containing glycan structures as LARGE. Glycobiology. 23:303–309. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Huttner WB. 1986. Chlorate—A potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 141:870–877. [DOI] [PubMed] [Google Scholar]
- Barresi R, Campbell KP. 2006. Dystroglycan: From biosynthesis to pathogenesis of human disease. J. Cell Sci.. 119:199–207. [DOI] [PubMed] [Google Scholar]
- Beedle AM, Turner AJ, Saito Y, Lueck JD, Foltz SJ, Fortunato MJ, Nienaber PM, Campbell KP. 2012. Mouse fukutin deletion impairs dystroglycan processing and recapitulates muscular dystrophy. J Clin Invest. 122:3330–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, et al. . 2002. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 71:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, Ponting CP, Estournet B, Romero NB, Mercuri E, et al. . 2001. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 69:1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington M, Torelli S, Prandini P, Boito C, Dolatshad NF, Longman C, Brown SC, Muntoni F. 2005. Localization and functional analysis of the LARGE family of glycosyltransferases: Significance for muscular dystrophy. Hum Mol Genet. 14:657–665. [DOI] [PubMed] [Google Scholar]
- Buysse K, Riemersma M, Powell G, van Reeuwijk J, Chitayat D, Roscioli T, Kamsteeg EJ, van den Elzen C, van Beusekom E, Blaser S, et al. . 2013. Missense mutations in beta-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause Walker-Warburg syndrome. Hum Mol Genet. 22:1746–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. 1992. Developmental changes in heparan sulfate expression: In situ detection with mAbs. J Cell Biol. 119:961–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bernabe DB, Inamori K, Yoshida-Moriguchi T, Weydert CJ, Harper HA, Willer T, Henry MD, Campbell KP. 2009. Loss of alpha-dystroglycan laminin binding in epithelium-derived cancers is caused by silencing of LARGE. J Biol Chem. 284:11279–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Stewart TE, Taylor WH. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 82:3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus J, Capurro M, Rast J. 2008. Glypicans. Genome Biol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford-Perriss M, Turner K, Guimond S, Apedaile A, Haubeck HD, Turnbull J, Murphy M. 2003. Localisation of specific heparan sulfate proteoglycans during the proliferative phase of brain development. Dev Dyn. 227:170–184. [DOI] [PubMed] [Google Scholar]
- Fortunato MJ, Ball CE, Hollinger K, Patel NB, Modi JN, Rajasekaran V, Nonneman DJ, Ross JW, Kennedy EJ, Selsby JT, et al. . 2014. Development of rabbit monoclonal antibodies for detection of alpha-dystroglycan in normal and dystrophic tissue. PLoS One. 9:e97567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson LA, Belting M, Cheng F, Jonsson M, Mani K, Sandgren S. 2004. Novel aspects of glypican glycobiology. Cell Mol Life Sci. 61:1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Sawaki H, Sakai T, Hiruma T, Nakanishi N, Sato T, Ohkura T, Narimatsu H. 2005. LARGE2 facilitates the maturation of alpha-dystroglycan more effectively than LARGE. Biochem Biophys Res Commun. 329:1162–1171. [DOI] [PubMed] [Google Scholar]
- Godfrey C, Foley AR, Clement E, Muntoni F. 2011. Dystroglycanopathies: Coming into focus. Curr. Opin. Genet. Dev.. 21:278–285. [DOI] [PubMed] [Google Scholar]
- Grewal PK, McLaughlan JM, Moore CJ, Browning CA, Hewitt JE. 2005. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology. 15:912–923. [DOI] [PubMed] [Google Scholar]
- Hara Y, Kanagawa M, Kunz S, Yoshida-Moriguchi T, Satz JS, Kobayashi YM, Zhu Z, Burden SJ, Oldstone MB, Campbell KP. 2011. Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc Natl Acad Sci U S A. 108:17426–17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MD, Campbell KP. 1998. A role for dystroglycan in basement membrane assembly. Cell. 95:859–870. [DOI] [PubMed] [Google Scholar]
- Hu Y, Li ZF, Wu X, Lu Q. 2011. Large induces functional glycans in an O-mannosylation dependent manner and targets GlcNAc terminals on alpha-dystroglycan. PloS one. 6:e16866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamori K, Hara Y, Willer T, Anderson ME, Zhu Z, Yoshida-Moriguchi T, Campbell KP. 2013. Xylosyl- and glucuronyltransferase functions of LARGE in alpha-dystroglycan modification are conserved in LARGE2. Glycobiology. 23:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamori K, Willer T, Hara Y, Venzke D, Anderson ME, Clarke NF, Guicheney P, Bonnemann CG, Moore SA, Campbell KP. 2014. Endogenous glucuronyltransferase activity of LARGE or LARGE2 required for functional modification of alpha-dystroglycan in cells and tissues. J Biol Chem. 289:28138–28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamori K, Yoshida-Moriguchi T, Hara Y, Anderson ME, Yu L, Campbell KP. 2012. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 335:93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae LT, Raaben M, Riemersma M, van Beusekom E, Blomen VA, Velds A, Kerkhoven RM, Carette JE, Topaloglu H, Meinecke P, et al. . 2013. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 340:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa M, Kobayashi K, Tajiri M, Manya H, Kuga A, Yamaguchi Y, Akasaka-Manya K, Furukawa J, Mizuno M, Kawakami H, et al. . 2016. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14:2209–2223. [DOI] [PubMed] [Google Scholar]
- Kanagawa M, Saito F, Kunz S, Yoshida-Moriguchi T, Barresi R, Kobayashi YM, Muschler J, Dumanski JP, Michele DE, Oldstone MB, et al. . 2004. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell. 117:953–964. [DOI] [PubMed] [Google Scholar]
- Karihaloo A, Kale S, Rosenblum ND, Cantley LG. 2004. Hepatocyte growth factor-mediated renal epithelial branching morphogenesis is regulated by glypican-4 expression. Mol Cell Biol. 24:8745–8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumanchi SA, Jha V, Ramchandran R, Karihaloo A, Tsiokas L, Chan B, Dhanabal M, Hanai JI, Venkataraman G, Shriver Z, et al. . 2001. Cell surface glypicans are low-affinity endostatin receptors. Mol Cell. 7:811–822. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, et al. . 1998. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 394:388–392. [DOI] [PubMed] [Google Scholar]
- Kunz S, Sevilla N, McGavern DB, Campbell KP, Oldstone MB. 2001. Molecular analysis of the interaction of LCMV with its cellular receptor alpha-dystroglycan. J Cell Biol. 155:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholt K, Weinke JL, Kiser CS, Lugemwa FN, Bame KJ, Cheifetz S, Massague J, Lindahl U, Esko JD. 1992. A single mutation affects both N-acetylglucosaminyltransferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc Natl Acad Sci U S A. 89:2267–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman C, Brockington M, Torelli S, Jimenez-Mallebrera C, Kennedy C, Khalil N, Feng L, Saran RK, Voit T, Merlini L, et al. . 2003. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum Mol Genet. 12:2853–2861. [DOI] [PubMed] [Google Scholar]
- Manya H, Chiba A, Yoshida A, Wang X, Chiba Y, Jigami Y, Margolis RU, Endo T. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: Coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci U S A. 101:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, et al. . 2012. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 91:541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H, et al. . 2002. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 418:417–422. [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Stanley P. 2005. Mouse large can modify complex N- and mucin O-glycans on alpha-dystroglycan to induce laminin binding. J Biol Chem. 280:20851–20859. [DOI] [PubMed] [Google Scholar]
- Praissman JL, Live DH, Wang S, Ramiah A, Chinoy ZS, Boons GJ, Moremen KW, Wells L. 2014. B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of alpha-dystroglycan. Elife. 3:e03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praissman JL, Willer T, Sheikh MO, Toi A, Chitayat D, Lin YY, Lee H, Stalnaker SH, Wang S, Prabhakar PK, et al. . 2016. The functional O-mannose glycan on alpha-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 5:e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemersma M, Froese DS, van Tol W, Engelke UF, Kopec J, van Scherpenzeel M, Ashikov A, Krojer T, von Delft F, Tessari M, et al. . 2015. Human ISPD is a cytidyltransferase required for dystroglycan O-mannosylation. Chem Biol. 22:1643–1652. [DOI] [PubMed] [Google Scholar]
- Roscioli T, Kamsteeg EJ, Buysse K, Maystadt I, van Reeuwijk J, van den Elzen C, van Beusekom E, Riemersma M, Pfundt R, Vissers LE, et al. . 2012. Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of alpha-dystroglycan. Nat Genet. 44:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens E, Carss KJ, Cirak S, Foley AR, Torelli S, Willer T, Tambunan DE, Yau S, Brodd L, Sewry CA, et al. . 2013. Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of alpha-dystroglycan. Am J Hum Genet. 92:354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. 2000. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr Opin Struct Biol. 10:518–527. [DOI] [PubMed] [Google Scholar]
- van Reeuwijk J, Janssen M, van den Elzen C, Beltran-Valero de Bernabe D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Muntoni F, et al. . 2005. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet. 42:907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillaumier-Barrot S, Bouchet-Seraphin C, Chelbi M, Devisme L, Quentin S, Gazal S, Laquerriere A, Fallet-Bianco C, Loget P, Odent S, et al. . 2012. Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am J Hum Genet. 91:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Yamada H, Yamaguchi Y. 1995. K-glypican: A novel GPI-anchored heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J Cell Biol. 130:1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer T, Inamori K, Venzke D, Harvey C, Morgensen G, Hara Y, Beltran Valero de Bernabe D, Yu L, Wright KM, Campbell KP. 2014. The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated alpha-dystroglycan functional glycosylation. Elife. 3:e03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer T, Lee H, Lommel M, Yoshida-Moriguchi T, de Bernabe DB, Venzke D, Cirak S, Schachter H, Vajsar J, Voit T, et al. . 2012. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat Genet. 44:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Willer T, Anderson ME, Venzke D, Whyte T, Muntoni F, Lee H, Nelson SF, Yu L, Campbell KP. 2013. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 341:896–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. 2010. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 327:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Kobayashi K, Manya H, Taniguchi K, Kano H, Mizuno M, Inazu T, Mitsuhashi H, Takahashi S, Takeuchi M, et al. . 2001. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 1:717–724. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD. 2011. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 3:a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Hu H. 2012. Differential glycosylation of alpha-dystroglycan and proteins other than alpha-dystroglycan by like-glycosyltransferase. Glycobiology. 22:235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang P, Hu H. 2011. LARGE expression augments the glycosylation of glycoproteins in addition to alpha-dystroglycan conferring laminin binding. PloS one. 6:e19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.