Summary
The paper describes a targeted Aurora-A overexpressing transgenic mouse model of mammary adenocarcinoma, which displays genetic alterations shared with human breast tumors. The model should help elucidate Aurora-A pathways driving breast cancer and develop effective targeted therapeutics for the disease.
Abstract
Recent data from The Cancer Genome Atlas analysis have revealed that Aurora kinase A (AURKA) amplification and overexpression characterize a distinct subset of human tumors across multiple cancer types. Although elevated expression of AURKA has been shown to induce oncogenic phenotypes in cells in vitro, findings from transgenic mouse models of Aurora-A overexpression in mammary glands have been distinct depending on the models generated. In the present study, we report that prolonged overexpression of AURKA transgene in mammary epithelium driven by ovine β-lactoglobulin promoter, activated through multiple pregnancy and lactation cycles, results in the development of mammary adenocarcinomas with alterations in cancer-relevant genes and epithelial-to-mesenchymal transition. The tumor incidence was 38.9% (7/18) in Aurora-A transgenic mice at 16 months of age following 4–5 pregnancy cycles. Aurora-A overexpression in the tumor tissues accompanied activation of Akt, elevation of Cyclin D1, Tpx2 and Plk1 along with downregulation of ERα and p53 proteins, albeit at varying levels. Microarray comparative genomic hybridization (CGH) analyses of transgenic mouse mammary adenocarcinomas revealed copy gain of Glp1r and losses of Ercc5, Pten and Tcf7l2 loci. Review of human breast tumor transcriptomic data sets showed association of these genes at varying levels with Aurora-A gain of function alterations. Whole exome sequencing of the mouse tumors also identified gene mutations detected in Aurora-A overexpressing human breast cancers. Our findings demonstrate that prolonged overexpression of Aurora-A can be a driver somatic genetic event in mammary adenocarcinomas associated with deregulated tumor-relevant pathways in the Aurora-A subset of human breast cancer.
Introduction
Recent cancer genome profiling studies have revealed that recurrent amplification and overexpression of Aurora kinase A (AURKA; also known as STK15/BTAK) gene, encoding Aurora kinase A (hereafter referred to as Aurora-A), characterize a distinct subset of human tumors across multiple cancer types (1), with Aurora-A overexpression correlating with patient prognosis (2,3). Elevated expression of Aurora-A has been reported to induce oncogenic transformation along with centrosome amplification, chromosomal instability and chemoresistance in mammalian cell lines in vitro and xenografts in vivo (4–6). Genome-wide scans in mice and humans have also identified AURKA as a low-penetrance tumor-susceptibility locus (7). Aurora-A functional studies have demonstrated that Aurora-A phosphorylation of tumor-suppressor proteins p53 and p73 result in their loss of functions in high Aurora-A-expressing cells acquiring resistance to DNA- and spindle-damaging agents due to override of the respective checkpoint response pathways (8–10). Furthermore, Aurora-A phosphorylation of p53 was shown to be critical in the maintenance of self-renewal and pluripotency of embryonic stem cells as well as in the reprogramming of induced pluripotent stem cells (11). It has been proposed that cancer stem cells (CSCs) may be derived either from somatic stem cells or from differentiated cells through an epithelial-to-mesenchymal transition (EMT) reprogramming process (12,13), which may involve Aurora-A–p53 signaling axis because Aurora-A overexpression was correlated with resistance to anticancer drugs in tumors, a hallmark of CSCs (14). Aurora-A gain of function, therefore, appears important in the development of CSC phenotypes besides causing inactivation of tumor-suppressor pathways and activation of oncogenic pathways (15–17). Specific downstream pathway genes, which cooperatively drive Aurora-A tumorigenesis have not yet been well elucidated. Unbiased genome-wide screening for recurrently occurring genetic alterations in tumors developing in Aurora-A-overexpressing genetically engineered mouse models, not harboring other tumor-relevant genetic changes (e.g. p53 null or heterozygous genetic background), should help address this knowledge gap.
Aurora-A overexpression has frequently been observed at different stages of breast cancer progression from breast ductal carcinoma in situ to invasive ductal breast cancers correlating with chromosomal instability (18,19). Aurora-A transgenic mice overexpressing the kinase in mammary tissues have been reported to yield distinct phenotypes in various experimental models. Mammary glands targeted Aurora-A overexpression induced by the Wap-Cre system through a single pregnancy cycle revealed hyperplasia after a short latency in p53 wild-type background and atypical ductal hyperplasia like precancerous lesions following longer latency in p53-deficient background (20,21). On the other hand, Aurora-A overexpression through five cycles of pregnancy under the regulation of mouse mammary tumor virus (MMTV) promoter in p53 wild-type background was reported to induce mammary tumors in 40% mice at 20 months of age and in 70% mice with p53-heterozygous background at 18 months of age (22). In view of recent The Cancer Genome Atlas (TCGA) data identifying the Aurora-A subset of tumors across multiple human cancer types, including breast cancer, it is important that the role of Aurora-A overexpression in the context of deregulated genetic pathways underlying tumorigenesis be carefully investigated with appropriately designed genetically engineered Aurora-A overexpressing mouse models.
In this study, we investigated the role of Aurora-A overexpression and accompanying genomic changes contributing to mammary carcinogenesis by generating a novel Aurora-A transgenic mouse model in which Aurora-A is targeted to overexpress in mammary gland under the regulation of β-lactoglobulin (BLG) promoter (23,24). Following prolonged overexpression of Aurora-A through 4–5 pregnancy and lactation cycles, 7 of 18 mice (38.9%) developed mammary adenocarcinomas. Aurora-A-overexpressing tumors revealed activation of Akt and Cyclin D1 as well as upregulation of mitotic proteins Tpx2 and Plk1, the two well-characterized activator and substrate of Aurora-A, respectively. We also observed EMT phenotypes in tumors evident from downregulation of E-Cadherin and upregulation of Vimentin expression. Additionally, mammary tumors displayed copy number alterations in cancer-relevant genes, such as, gain of Glp1r and losses of Ercc5, Pten and Tcf7l2 loci, correlating with the findings from Aurora-A gain of function human breast cancers reported in TCGA data set. Interestingly, whole exome sequencing of Aurora-A transgenic mouse mammary tumors also revealed the presence of mutant genes detected in human breast cancers. The results of this study demonstrate that prolonged elevated expression of Aurora-A may be a driver genetic event underlying human mammary tumorigenesis. Additionally, the Aurora-A transgenic mouse model offers a unique opportunity toward developing novel prognostic markers and therapeutic targets for the Aurora-A overexpressing subset of human breast cancer.
Materials and methods
Genetically engineered AURKA transgenic mouse
To generate AURKA transgenic mouse model, a 2.3kb complementary DNA encoding full length of human Aurora-A complementary DNA sequence with intron 8 was cloned into pBJ41 vector at EcoRV site, placing it under the control of the ovine BLG promoter (23,24). Then, digestion with SalI and FspI released the 7.5kb transgene fragment from the vector. The transgene fragment was separated on a 0.8% low-melting agarose and purified with QIAEX II gel extraction kit (Qiagen). The BLG–Aurora-A transgenic founders were established by microinjecting the transgene fragment into C57BL/6 × SJL strain F2 mouse eggs and surgically transferred to pseudopregnant females. Offspring mice carrying the transgene (founders) were genotyped with tail genomic DNA (gDNA) by PCR using the following primers (forward: 5ʹ-CCTGTCCTTGTCTAAGAGGCTGAC-3ʹ in BLG promoter and reverse: 5ʹ-TTGAAGGACACAAGACCCGC-3ʹ in human Aurora-A). Then, the founders were mated to C57BL/6 mice.
Animal experiments
BLG–Aurora-A postpubertal female mice were bred continuously for four to five rounds and killed at day 15 of gestation, day 10 of lactation or day 10 of postweaning. The first abdominal (or #4) mammary glands on both sides of the animals were dissected for further analysis. Mice were maintained in accordance with guidelines set forth by the Association for Assessment and Accreditation of Laboratory Animal Care and the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. All animal experiments were approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center.
Semiquantitative RT–PCR analysis
Total RNA was extracted from frozen tissues using RNeasy kit (Qiagen) or Trizol reagent (Invitrogen), and RT–PCR was done as described previously (10) using the following primers, forward: 5ʹ-ACGTGTTCTCGTGACTCAGC-3ʹ and reverse: 5ʹ-GTGATCCAGGGGTGTTCAA-3ʹ for human Aurora-A, forward: 5ʹ-CACCATGGAGAAGGCCGGGGCCCAC-3ʹ and reverse: 5ʹ-ATCATACTT GGCAGGTTTCTCCAGG-3ʹ for glyceraldehyde-3-phosphate dehydrogenase (Gapdh), forward: 5ʹ-CAGTAGTGTGCTACAAGGCCA-3ʹ and reverse: 5ʹ-AGCCAACAAGGATGGCTGAA-3ʹ for glucagon-like peptide 1 receptor (Glp1r), forward: 5ʹ-GGTGCTGGCCGTGGATATTA-3ʹ and reverse: 5ʹ-AGCGG AGCATCACCATCAAA-3ʹ for excision repair cross-complementing rodent repair deficiency complementation group 5 (Ercc5), forward: 5ʹ-ACAA TTCCCAGTCAGAGGCG-3ʹ and reverse: 5ʹ-CCTTTAGCTGGCAGACCACA-3ʹ for phosphatase and tensin homolog (Pten), forward: 5ʹ-GCGG AAAGGGATTTAGCCGA-3ʹ and reverse: 5ʹ-TGAAAATGGAGGGTTCGGGC-3ʹ for T-cell-specific high mobility group box transcript variant 4 (Tcf7l2).
Droplet Digital PCR
Droplet Digital PCR Copy Number Assays, including primers and probes, were purchased from Bio-Rad. The amplified products of reference gene Rpp30 and other genes of interest were detected by fluorochromes HEX and FAM, respectively. For a 20 μL reaction, 0.1U HindIII and 40ng/μL gDNA were used. Otherwise, all procedures were performed according to the manufacturer’s instruction (Bio-Rad). Generating droplets, PCR and counting droplets were performed by QX200 Droplet Digital PCR System. Thresholds of ≤1.2 copies and ≥2.8 copies were cutoffs for copy loss and gain, respectively.
Western blot and antibodies
For western blots, frozen tissues were lysed in radioimmunoprecipitation assay buffer containing protease inhibitors (Roche Diagnostics), resolved in sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred and incubated with primary antibodies against human Aurora-A (4), Plk1, ERα, p53, Hsp90, β-actin (Santa Cruz Biotechnology), Tpx2 (Bethyl Laboratories), E-Cadherin (BD Biosciences), Her2/Neu, Cyclin D1, P-Akt (S473), P-Akt (T308), Akt and Vimentin (Cell Signaling).
Histology and immunohistochemical analysis
Ten mated non-transgenic female mice, 10 virgin BLG–Aurora-A transgenic mice and 18 mated BLG–Aurora-A transgenic mice were euthanized and necropsied at 16 months of age and were examined grossly and histologically for tumor development. Mammary glands from non-transgenic female mice and mammary glands with grossly detected tumors from BLG–Aurora-A transgenic female mice were fixed in 10% neutral-buffered formalin. Formalin-fixed tissues were processed and embedded in paraffin blocks from which 4 µm thick sections were mounted on glass slides and stained with hematoxylin and eosin.
Immunohistochemical analysis was done as described previously (2,4). Briefly, antigen retrieval was done in 0.1mol/L sodium citrate buffer at 100°C using a steamer. The sections were immersed in methanol containing 0.3% hydrogen peroxidase for 20min to block endogenous peroxidase activity, incubated in 2.5% blocking serum to reduce non-specific binding and incubated with antibodies described previously. Signals were then amplified with VECTASTAIN Elite ABC kit (Vector Laboratories) and visualized using Vector NovaRed substrate kit (Vector laboratories). Tissue histopathology and immunohistochemistry were examined independently by a veterinary pathologist (M.G.) and an expert human breast cancer pathologist (A.S.) at our institution.
Array CGH
One microgram of gDNA isolated by conventional phenol/chloroform method was digested with AluI and RsaI (Promega) and then labeled fluorescently using Agilent Genomic DNA labeling kit (Agilent Technology). Six sample DNAs from three individual tumors (duplicate each sample) were processed for the array CGH (aCGH). Cy5-labeled tumor gDNA and Cy3-labeled control reference gDNA were combined and hybridized to Agilent 4x180K aCGH slides (G4839A) using an Agilent CGH hybridization kit (Agilent Technologies). The probes were annotated based on NCBI Build 37. Washed slides were then scanned with Agilent Scanner G2505C using Agilent Feature Extraction Software V10.10. Data processing and analysis were performed using Bioconductor R packages (http://www.bioconductor. org) (25). The copy number alterations were detected using the Circular Binary Segmentation algorithm implemented in Bioconductor package DNA copy (26). The log2 ratio of signal intensity of each probe was visualized along each chromosome with the corresponding NCBI-annotated gene information. The degree of tumor heterogeneity was adjusted by rescaling the segment median using the estimated tumor percentage (27). Circle Manhattan Plot was generated using CIRCOS software. Regions of mouse genome showing significant gain or loss in aCGH analysis were visualized in a chromosome-wise manner using the cancer QTL viewer of Mouse Tumor Biology Database (28).
Whole exome sequencing
Freshly cut sections from formalin-fixed paraffin-embedded tissues were microdissected with the help of tissue-matched Hematoxylin and eosin-stained slide to locate the tumor area by an experienced pathologist (M.G.). Exome sequencing was performed by the Sequencing and Microarray Facility at MD Anderson Cancer Center. Briefly, indexed libraries were prepared from 1 µg of Biorupter ultrasonicator (Diagenode)-sheared gDNA using the KAPA Hyper Library Preparation Kit (Kapa Biosystems). Library quality was assessed using the Fragment Analyzer with High Sensitivity NGS Fragment Analysis Kit (Advanced Analyticals). The libraries were then prepared for capture with six cycles of preligation-mediated PCR amplification. Following preligation-mediated PCR, amplified libraries were assessed for (i) quality using the Fragment Analyzer with High Sensitivity NGS Fragment Analysis Kit and (ii) quantity using the Qubit dsDNA HS Assay Kit (ThermoFisher), then multiplexed four libraries per pool. Exome capture was performed using the NimbleGen SeqCap EZ Mouse Exome kit. The enriched libraries were PCR amplified 10 cycles post capture and assessed for the quality using the Fragment Analyzer. Enrichment of PCR products was assessed by quantitative PCR and quantified using the Qubit dsDNA HS Assay Kit. Sequencing was performed on the HiSeq2000 Sequencer (Illumina Inc), four samples per lane using the 76nt paired-end configuration. Data aligned to the reference were analyzed by C.C.
TCGA data analysis
Genomic alterations and protein expression changes detected in BLG–Aurora-A transgenic mouse mammary tumors were compared with the human breast cancer TCGA data available at cBioPortal (29) to see whether there was a similar trend in human breast cancer TCGA data. To this end, genetic/molecular alterations observed in BLG–Aurora-A transgenic mouse mammary tumors, such as upregulated Cyclin D1 (CCND1), were analyzed for their incidence in Aurora-A-overexpressing human breast cancer (for co-occurrence with Aurora-A overexpression) using TCGA data set. To input genetic/molecular events in a binary manner (up or down) into the online TCGA data query tool, we used the following criteria: for upregulated gene: or >2 copy gains, messenger RNA (mRNA) expression Z-score > mean + 2 SD; for downregulated gene: heterozygous or homozygous deletions, mRNA expression < mean − 2 SD. Then, these criteria were put in the TCGA data query tool (http://www.cbioportal.org/onco_query_lang_desc.jsp) and correlated with Aurora-A upregulation. The value of P < 0.05 was considered as statistically significant.
Correlation analysis of expressions of Aurora-A and putative Aurora-A pathway genes
Transcriptional data from TCGA and TRANSBIG human breast cancer data sets were used for this analysis (30,31). Pearson product–moment correlation was performed with R software. Correlation coefficient r values were shown with values from −1 to 1.
Results
BLG–Aurora-A transgene-induced myoepithelial-like adenocarcinoma in mouse mammary gland following prolonged activation through 4–5 cycles of pregnancy
Two independent transgenic founder lines harboring the BLG–Aurora-A transgene (Figure 1A) were developed for this study. The two lines of transgenic mice from these founders were maintained through at least four rounds of pregnancies. Seven of the 18 (38.9%) BLG–Aurora-A transgenic mice developed mammary tumors by the age of 16 months after four (two mice) or five cycles (five mice) of pregnancy, whereas no tumor was observed in mated wild-type mice (0/10) or virgin BLG–Aurora-A mice (0/10), maintained for the same period of time. Microscopic examination of the histological sections revealed mammary adenocarcinoma with adenomyoepithelial histologic phenotypes from mated BLG–Aurora-A female mice (Figure 1B). Focally, the neoplastic cells infiltrated the surrounding normal tissues (data not shown). The morphologic phenotypes of mammary adenocarcinoma were similar to those observed in previously published genetically engineered mouse mammary tumor models (32,33).
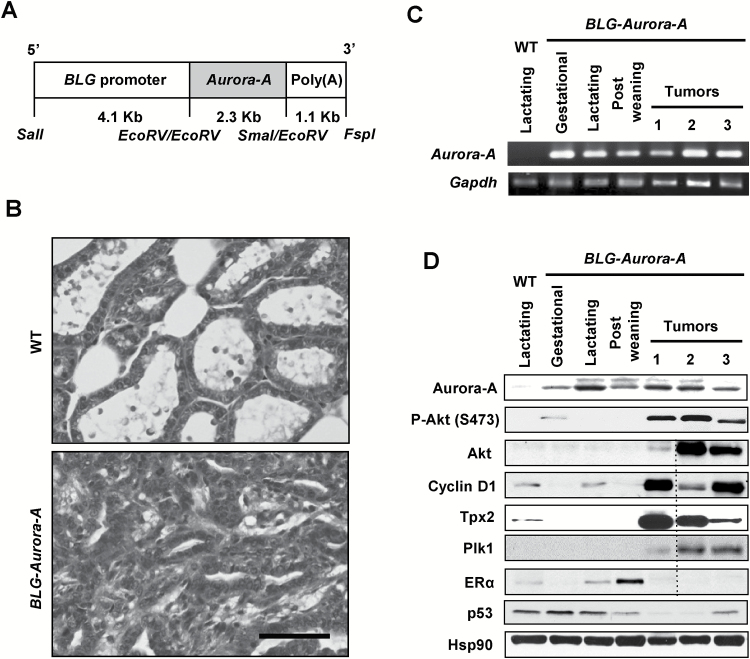
Figure 1.
BLG–Aurora-A transgene induced mouse mammary tumors. (A) Schema depicts the BLG–Aurora-A transgene containing BLG promoter, full-length Aurora-A coding sequence, followed by poly(A) tail. Restriction enzyme sites are mapped at the bottom. (B) Hematoxylin and eosin-stained sections depict lactating mammary glands from wild-type (WT) mouse and mammary adenocarcinoma from BLG–Aurora-A transgenic mouse. Scale bar, 50 µm. (C) Semiquantitative RT–PCR of Aurora-A mRNA of BLG–Aurora-A transgenic mouse mammary glands. Mammary gland tissues were collected from WT lactating mouse (day 10 of lactation), BLG–Aurora-A mice in gestation (day 15), in lactation (day 10), in postweaning (day 10). Three representative tumors were collected from BLG–Aurora-A mice bearing bulk mammary tumors. (D) Western blots for Aurora-A, P-Akt (S473), Akt, Cyclin D1, Tpx2, Plk1, ERα and p53 and Hsp90 (a loading control). Mammary gland tissues were collected from WT lactating mouse (day 10 of lactation), BLG–Aurora-A mice in gestation (day 15), in lactation (day 10), in postweaning (day10). Four representative tumors were collected from BLG–Aurora-A mice bearing bulk mammary tumors. One tumor sample run in the gel did not detect any proteins of interest except for the loading control (Hsp90). This lane is cropped in Fig. 1D (shown with a dash line) from the original blots shown in Supplementary Figure 5, available at Carcinogenesis Online.
To assess the expression of Aurora-A transgene and other genes of interest at various physiological stages of mouse mammary glands, total RNA was isolated at mid-gestation (day 15), mid-lactation (day 10), postweaning (day 10) and subjected to RT–PCR in parallel with non-transgenic mammary glands at day 10 lactation as a control. Consistent with previous studies of BLG-driven transgene expression system (23,24), Aurora-A transgene mRNA expression was detected during gestation through day 10 postweaning (Figure 1C). Mammary tumors developing in the parous BLG–Aurora-A mice also revealed elevated expression of the Aurora-A transgene protein (Figure 1D), albeit at varying levels, but not in the mammary gland of the virgin mice (data not shown). These data demonstrated that the Aurora-A transgene expression was faithfully regulated in vivo under BLG promoter in mammary tissues at specific time intervals from mid-pregnancy through lactation and postweaning. Furthermore, prolonged overexpression of Aurora-A transgene, together with physiologically regulated hormonal factors, through 4–5 cycles of pregnancy and lactation appeared to drive mammary carcinogenesis in about 39% mice.
BLG–Aurora-A transgene upregulated Akt, Cyclin D1 and Aurora-A-interacting proteins Tpx2 and Plk1 and downregulated ERα and p53
Activation of Akt due to phosphorylation at Ser473 by mammalian target of rapamycin and at Thr308 by PDK1 and accumulation of Cyclin D1 are associated with continued proliferation promoting hyperplasia and tumor formation (22,34). Therefore, we determined whether activation of Akt and accumulation of Cyclin D1 were detected in mammary tumors of BLG–Aurora-A mice by western blots of transgenic mouse tumor tissues compared with mammary tissue of wild-type mice. Data revealed the expression of activated Akt, evident from phosphorylated Akt at Ser473 in all tumors (Figure 1D) and at Thr308 in one of three tumors (data not shown), accompanying elevated expression of Cyclin D1 compared with lactating non-transgenic mammary tissues.
Aurora-A activation is regulated by the microtubule-binding protein TPX2, whereas its centrosomal localization is regulated by the centrosomal kinase PLK1, which is also involved in centrosome maturation and spindle assembly (35). Aurora-A, on the other hand, activates PLK1 controlling mitotic entry and checkpoint recovery (36,37). Aurora-A, TPX2 and PLK1 have been found frequently overexpressing in human cancers, and their deregulations have been implicated in the induction of genomic instability (38–40). We observed elevated expressions of Tpx2 and Plk1, at varying levels, in the transgenic mouse mammary tumors (Figure 1D). The functional inactivation of p53 and ERα proteins in Aurora-A overexpressing cells has been reported previously (8,41). Consistent with the earlier observation, western blot analyses revealed lower p53 and ERα levels in the BLG–Aurora-A transgenic mouse tumors (Figure 1D). The results, taken together, demonstrated activation of Akt and elevated expression of Cyclin D1 along with Aurora-A pathway proteins Tpx2 and Plk1 in the BLG–Aurora-A transgenic mouse mammary tumors.
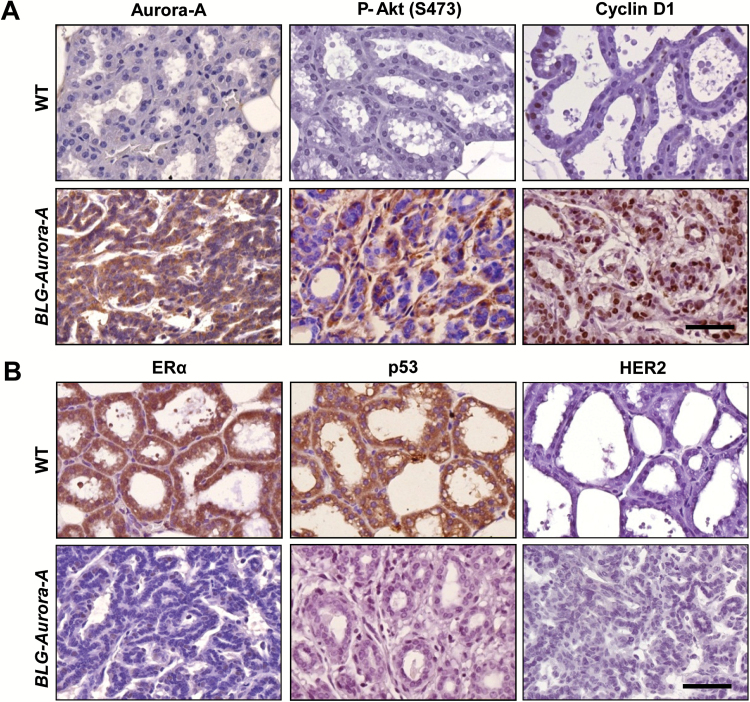
Similar to the results of western blotting analyses, immunohistochemical staining revealed higher levels of phosphorylated Akt and accumulation of Cyclin D1 besides reduced/undetectable levels of ERα and p53 in the mammary tumors of BLG–Aurora-A mice compared with the mammary glands of wild-type mice (Figure 2A, B). Of note, there was no p53 mutation observed in the tumors (data not shown). Her2/Neu, known to overexpress in ~30% of breast cancer, was undetectable in BLG–Aurora-A transgenic mouse tumors (Figure 2B). In summary, the molecular and immunohistochemical analyses of BLG–Aurora-A transgenic mouse mammary tumors revealed that Aurora-A overexpression-mediated tumorigenesis involved activation of Akt/Cyclin D1, upregulation of mitotic proteins, Tpx2 and Plk1 and downregulation of ERα and p53.
Figure 2.
Immunohistochemical analyses revealed distinct biological characteristics of BLG–Aurora-A mammary tumors from wild-type (WT) mouse mammary glands. (A) Representative stained sections with antibodies to P-Akt (S473) and Cyclin D1, markers for survival and proliferation respectively, and to Aurora-A depict overexpressions of P-Akt and Cyclin D1 as well as Aurora-A in BLG–Aurora-A mouse mammary tumor but not in WT mouse mammary glands (day 10 in lactation). Scale bar, 50 µm. (B) Representative stained sections with antibodies to ERα, p53 and HER2, all of which are clinical markers correlating with breast cancer prognosis, depict loss of ERα and p53 expressions in BLG–Aurora-A mouse mammary tumor compared with WT mouse mammary glands (day 10 in lactation). HER2 was also not stained in BLG–Aurora-A mouse mammary tumor as less as in WT mouse mammary glands. Scale bar, 50 µm.
BLG–Aurora-A transgene-induced EMT in mouse mammary tumor cells in vivo
Aurora-A has been reported to have an essential role in the development of tumorigenic potential and therapy resistance of CSCs (14). Aurora-A also has an essential role in the regulation of self-renewal of embryonic stem cells and in the development of induced pluripotent stem cells (11). These findings suggest that Aurora-A is critical in driving and/or maintenance of stem cell-like phenotype in pluripotent stem cells and CSCs. Because the induction of CSCs through cellular reprogramming process of EMT has been suggested (13), we analyzed for EMT-related biomarkers in BLG–Aurora-A transgenic mouse mammary tumors to see possible association of Aurora-A overexpression with induction of EMT.
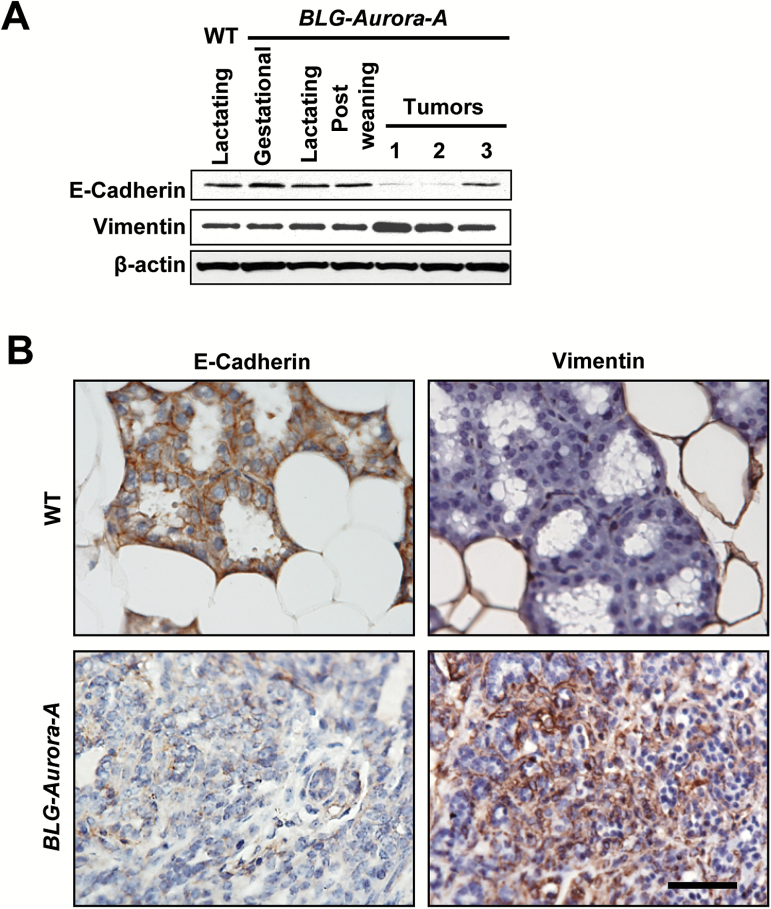
To this end, we analyzed the expression levels of E-Cadherin, an epithelial marker and Vimentin, a mesenchymal marker, in non-transformed mammary glands from wild-type and BLG–Aurora-A transgenic mice as well as in transgenic mice mammary tumors (Figure 3A, B). Results revealed downregulation of E-Cadherin and upregulation of Vimentin in tumors compared with wild-type mice and non-transformed mammary glands from BLG–Aurora-A transgenic mice although the pattern was variable among the tumors (Figure 3A). Immunohistochenical analysis of tumor sections also showed the same trend with downregulation of E-Cadherin and upregulation of Vimentin in tumors but not in wild-type mouse mammary glands (Figure 3B). These findings indicate that BLG–Aurora-A-driven tumorigenesis in vivo is accompanied with the induction of EMT.
Figure 3.
Transgene BLG–Aurora-A induced EMT along with tumorigenesis in mouse mammary glands. (A) Western blots for E-Cadherin, Vimentin and β-actin (a loading control). Mammary gland tissues were collected from wild-type (WT) lactating mouse (day 10 of lactation), BLG–Aurora-A mice in gestation (day 15), in lactation (day 10) and in postweaning (day 10). Three representative tumors were collected from BLG–Aurora-A mice bearing bulk mammary tumors. (B) Representative sections immunohistochemically stained with antibodies to E-Cadherin and Vimentin depict downregulated E-Cadherin and overexpressed Vimentin expressions in BLG–Aurora-A mouse mammary tumor compared with in WT mammary gland (day 10 in lactation). Scale bar, 50 µm.
BLG–Aurora-A transgene-driven mammary tumors revealed copy number alterations and mutations in genes associated with human breast cancer
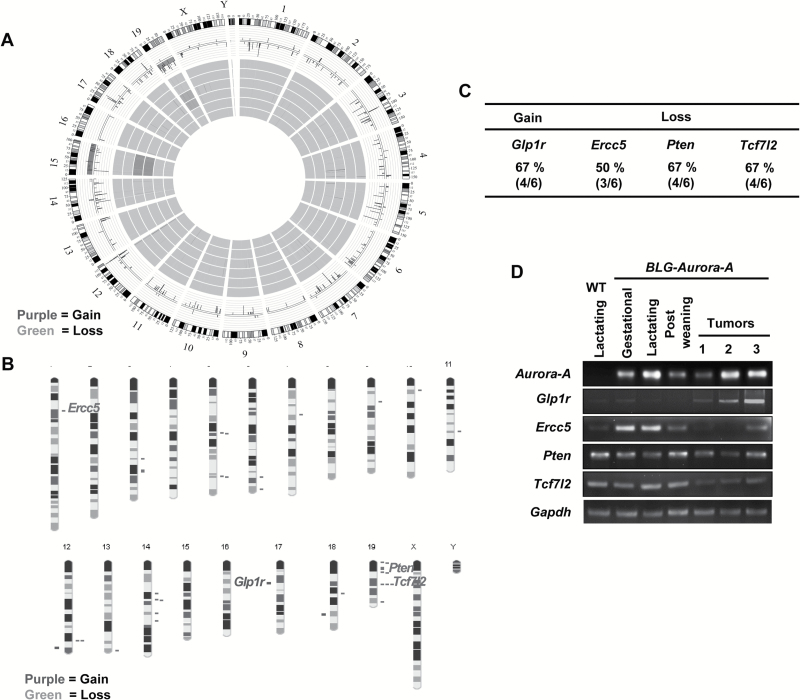
Aurora-A overexpression-mediated deregulation of oncogenic and tumor-suppressor pathways have been implicated in the induction of genomic instability in the Aurora-A transgenic mouse models (20,21). We thus assessed global genomic copy number alterations in BLG–Aurora-A transgenic mouse tumors by aCGH analysis to assess genomic rearrangements caused by BLG–Aurora-A transgene. The results revealed recurrent gDNA copy gains and losses in the tumors compared with the control mammary gland tissues (total of six samples representing duplicate samples of three tumors, Figure 4A). The genomic copy gains in the tumors were predominantly localized to chromosome 12 and 17 and deletions to chromosomes 5, 14 and 19, with one tumor showing gain and loss of the entire chromosome 15 and 19 (Figure 4A). The gDNA copy number alterations involved 141 genes; 10 genes showing gains and 131 genes showing losses of one or both alleles in the three tumors analyzed by aCGH (Supplementary Table 1, available at Carcinogenesis Online). To further comprehensively profile the genetic mutation events in Aurora-A-driven tumors, we performed whole exome sequencing of the three tumors analyzed by aCGH. Thirty six genes were found mutated in at least two of the three tumors, of which 31 have human orthologues (Supplementary Table 2, available at Carcinogenesis Online). To explore the biological significance of these mutations in the context of human breast cancer, we compared the mouse tumor mutant genes with those detected in human breast cancer, reported in TCGA, using the schema shown in Supplementary Figure 1, available at Carcinogenesis Online. Seven mutated genes detected in mouse mammary tumors (Supplementary Table 3, available at Carcinogenesis Online) were also reported in human breast cancers in the TCGA data set with their varying incidences, expressing elevated and basal levels of Aurora-A (Supplementary Figure 2, available at Carcinogenesis Online).
Figure 4.
BLG–Aurora-A transgene induced genomic instabilities. (A) Circle Manhattan plot depicts data summary derived from aCGH of six samples (duplicates of three individual BLG–Aurora-A tumors). The outer circle maps each mouse chromosome. The middle circle summarizes average genetic gains and losses of all tumor samples at subjected loci. The inner circles depict genetic gains and losses of individual samples at subjected loci. (B) Karyogram of a representative tumor depicts a genetic gain of Glp1r locus (chromosome 17, Ch17) and genetic losses of Ercc5 (Ch1), Pten (Ch19) and Tcf7l2 (Ch19) loci. (C) Table presents frequencies of genetic gains or losses of Glp1r, Ercc5, Pten and Tcf7l2 among BLG–Aurora-A mammary tumors. (D) RT–PCR analyses of Aurora-A, Glp1r, Ercc5, Pten and Tcf7l2. Total RNAs were extracted from mammary glands of wild-type (WT) lactating mouse (day 10 of lactation), BLG–Aurora-A mice in gestation (day 15), in lactation (day 10), in postweaning (day 10). Three representative tumors from BLG–Aurora-A mice were also subjected to total RNAs extraction followed by RT–PCR analyses.
Expression profiles of copy number-altered genes in BLG–Aurora-A transgenic mouse mammary tumors
We next investigated the expression profiles of the four copy number-altered cancer-relevant genes, Glp1r, Ercc5, Pten and Tcf7l2, identified in the Aurora-A-driven mouse mammary tumors (Figure 4A–C). Copy number alterations of these four genes were also analyzed in four additional tumors by Droplet Digital PCR. The results revealed gain of Glp1r gene in all additional four tumors and loss of Pten in one of the four tumors analyzed (Supplementary Figure 3, available at Carcinogenesis Online). Semiquantitative RT–PCR analysis was performed in the transgenic mice mammary tumors and non-transformed mammary glands using specific primer pairs for Glp1r, Ercc5, Pten and Tcf7l2 transcripts (Figure 4D). The results confirmed that Glp1r (with copy gain in BLG–Aurora-A mouse mammary tumor) was upregulated and the other three genes (with copy number loss) were relatively downregulated at transcriptional level in the tumors compared with the non-transformed BLG–Aurora-A and wild-type mammary glands. The data, thus, demonstrated that copy number alterations resulted in consequential changes in the expression of these genes in BLG–Aurora-A mouse mammary adenocarcinomas.
Co-occurrence of copy number-altered genes with Aurora-A copy gain/overexpression in human breast cancer
In view of the high prevalence of Aurora-A gain of function (copy gain and overexpression) in a subset of breast cancer (1), it is important that genetic alterations co-occurring with Aurora-A copy gain/overexpression in human breast cancer be identified and characterized for their role in tumorigenic transformation process. We, therefore, investigated whether copy number alterations and expression changes in the genes, Glp1r, Ercc5, Pten and Tcf7l2 (42–45), detected in the BLG–Aurora-A mouse mammary tumors also co-occur with Aurora-A copy gain/overexpression in human breast cancer. To address this question, we analyzed the publicly available TCGA breast cancer data set using online interface, cBioPortal query algorithm and also included the genes characterized in Figures 1 and 2 (CCND1, PLK1, TPX2, PTEN, ESR1 and TP53), which represent the biological hallmarks of human breast cancer. First, review of TCGA data set revealed that Aurora-A copy gain/elevated expression was present in about 53% of the 971 breast cancers analyzed (29). Second, altered copy number/expression profiles observed in BLG–Aurora-A mouse mammary tumors (GLP1R, CCND1, PLK1, TPX2, ERCC5, PTEN, TCF7L2, ESR1 and TP53) were found to significantly co-occur with Aurora-A copy gain/overexpression in human breast cancer TCGA data set (Table 1). We further correlated the mRNA expression levels of Aurora-A and the putative Aurora-A pathway genes, identified in the transgenic mouse mammary tumors, utilizing TCGA and TRANSBIG human breast cancer transcriptomic data sets, by Pearson product–moment correlation coefficient analysis (31). This analysis allowed measuring the strength and direction of association between the continuous variables of Aurora-A and its putative pathway gene transcript levels. Results revealed varying extent and direction of correlations between the mRNA expression levels in human breast cancer data sets (Supplementary Figure 4, available at Carcinogenesis Online). Varying association between the transcript levels of Aurora-A and the putative pathways genes possibly reflected stochastic nature of functional interactions of these genes with Aurora-A in the tumors analyzed. AURKA transcript showed strong positive correlation with the transcript levels of PLK1 and TPX2 reflecting their direct functional interaction in promoting cell proliferation. In contrast, relatively weaker negative association between the transcript levels of AURKA and the remaining putative pathway genes plausibly reflected their indirect inverse functional interactions with AURKA. In view of the transgenic mouse tumor data, it was surprising to see the low and discordant association between AURKA and GLP1R in the TCGA and the TRANSBIG data sets. This could be due to different tumor genomic backgrounds or varying breast cancer subtypes profiled in the human data sets. Negative association observed between AURKA and CCND1 transcripts in this analysis appeared inconsistent with previously published data on positive correlation observed between these two proteins in genetically engineered mouse models of breast cancer and also in the human breast cancer. There is, however, a possibility that posttranscriptional mechanisms may be responsible for positive correlation in the levels of the two proteins observed in mammary tumor tissues.
Table 1.
Co-occurrence of genetic alterations in human breast tumors reported in TCGA with Aurora-A gain of function (amplification and mRNA overexpression)
| Altered genes | % of Corresponding events of all samples | Odds ratio (log) | P value |
|---|---|---|---|
| AURKA (Up) | 53 | — | — |
| GLP1R (Up) | 26 | 1.163 | <0.001 |
| CCND1 (Up) | 40 | 1.045 | <0.001 |
| PLK1 (Up) | 55 | 0.870 | <0.001 |
| TPX2 (Up) | 46 | >3.000 | <0.001 |
| ERCC5 (Down) | 37 | 1.088 | <0.001 |
| PTEN (Down) | 31 | 1.181 | <0.001 |
| TCF7L2 (Down) | 31 | 1.326 | <0.001 |
| ESR1 (Down) | 31 | 0.501 | <0.001 |
| TP53 (Down) | 62 | 0.809 | <0.001 |
Discussion
Genetically engineered mouse models of cancer provide a unique opportunity for identifying true “driver” genetic events of tumorigenesis, which is a major challenge in the case of human tumors due to the presence of genomic complexity in late-stage tumors most often utilized for genomic profiling studies. A recent publication describing chromosomal copy number alterations in 45 mouse models of breast cancer has revealed distinct driver-dependent routes to development of breast tumorigenesis and also validated the role of a candidate gene promoting HER2-induced tumorigenesis in human breast cancer (42). The Aurora-A-driven mouse model of mammary adenocarcinoma described in this paper has also revealed a set of candidate genes possibly involved in Aurora-A overexpression-driven breast cancer. Furthermore, the mouse model makes it evident that elevated expression of Aurora-A for a prolonged period is necessary for the development of mammary tumors. The concept gains credence in view of the earlier published studies that Aurora-A overexpression for a relatively limited time interval in mammary glands with wild-type p53 and p53-deficient genetic backgrounds induced ductal hyperplasia and atypical ductal hyperplasia, respectively (20,21). The fact that another transgenic mouse model expressing Aurora-A driven by MMTV promoter through five pregnancy cycles was reported to develop mammary tumors in 40% of mice on a p53 wild-type background and in 70% of mice on a p53-heterozygous background further corroborates the idea (22). Our current findings on the incidence of mammary adenocarcinoma in about 39% of BLG–Aurora-A transgenic mouse following induction of Aurora-A through 4–5 cycles of pregnancy closely resembles the latter findings. It is relevant in this context that the MMTV–Aurora-A transgenic mouse mammary tumor model was generated in FVB/N strain, which is known to be more susceptible to develop tumors than the C57BL/6 strain utilized to generate the BLG–Aurora-A mammary tumor model described in this study. Development of mammary adenocarcinoma in the model derived with C57BL/6 strain, described in this study, indicates that Aurora-A gain of function induced tumorigenic transformation is primarily determined by the underlying genotypic changes rather than the strain of the mice.
It is noteworthy that high expression of P-Akt and Cyclin D1 detected in the BLG–Aurora-A mammary tumors were also observed in the MMTV–Aurora-A mouse mammary tumor model published earlier (22). Additionally, elevated expression of Tpx2 and Plk1 proteins, the activator and downstream effector of Aurora-A, in our BLG–Aurora-A mammary tumors suggest that aberrant high expressions of the major Aurora-A pathway proteins together play critical roles in driving the tumorigenic process through inactivation of tumor-suppressor pathways and activation of oncogenic pathways associated with genomic instability.
We and others have earlier reported that Aurora-A phosphorylation of p53 on Ser215 and Ser315 caused cytoplasmic sequestration and destabilization of p53, respectively (8,9). We also reported that Aurora-A sequestered the bound ERα resulting in the abrogation of its transactivation function (41). Low or undetectable expressions of p53 and ERα and induction of EMT in the BLG–Aurora-A mouse mammary tumors appear to corroborate the in vitro findings on Aurora-A–p53–ERα functional interactions. Although precise role of Aurora-A in the induction of EMT is not known, it is plausible that Aurora-A–p53 interaction plays an important role in the induction of EMT as well as CSCs. In fact, deregulation of p53 is involved in the development of aberrant self-renewal potential as well as cell polarity defects in CSCs (43,44). Stem cells with targeted mutation of p53 have been shown to possess enhanced self-renewal properties as do CSCs in a mouse mammary tumor model. Aurora-A also has been shown to have an essential role in the development of tumorigenic potential and therapy resistance of CSCs (14,44). Aurora-A gain of function mediated tumor formation has earlier been reported to be initiated with genomic instability (22,45). It is, therefore, logical to suggest that non-random gain and loss of cancer-relevant loci, Glp1r, Ercc5, Pten and Tcf7l2, detected in the BLG–Aurora-A mammary tumors resulted from genomic instability, which promoted and contributed to the tumorigenic process. Glp1r, a member of the G-protein-coupled receptor family involved in the activation of multiple signaling pathways, such as PI3K/AKT and MAPK, has been found amplified in human cancers including in breast cancer (46). Detection of both positive and negative association of GLP1R mRNA expression profile with the AURKA transcript in human breast cancer data sets, however, suggest that altered expression of this gene may have varying roles depending on the genomic context of Aurora-A overexpressing tumors. The loci showing copy number loss and mRNA downregulation in the BLG–Aurora-A mouse mammary tumors have been functionally implicated in cancer. Studies indicate that ERCC5 plays a key role in carcinogenesis and its deficiency causes DNA repair defects, genomic instability and deregulated gene transcription (47). In view of elevated Aurora-A-expressing cells being prone to DNA damage, possibly due to oxidative stress and inactivation of p53-mediated DNA damage response (8–10), it is likely that loss of ERCC5 promotes tumorigenic transformation of Aurora-A overexpressing cells through promoting DNA repair defects. PTEN functions as a tumor suppressor by negatively regulating AKT signaling pathway (48), expected to be activated in tumors deficient in PTEN, as observed in the BLG–Aurora-A tumors. Loss of PTEN is a frequent genetic anomaly associated with many human cancers. About 61% of mice heterozygous for a Pten-null mutation have been reported to develop mammary adenocarcinomas by 30–49 weeks of age (49). A recent publication also has shown that loss of Pten enhanced the stability of Aurora-A by attenuating Fbxw7-dependent degradation of Aurora-A-involving Akt/GSK3β pathway (50). The finding further indicates a cross talk between loss of PTEN and Aurora-A gain of function, as detected in the BLG–Aurora-A tumors. The aCGH analyses of the tumors also revealed loss of Tcf7l2, a member of the TCF/LEF family of transcription factors, which is a downstream effector of the Wnt/β-catenin signaling pathway (51). Tcf7l2 transcript was earlier reported to be highly downregulated in breast cancers relative to normal breast tissue, contributing to the activation of Wnt signaling pathway (51). Furthermore, Chd8 and Mcc, negative regulators of Wnt/β-catenin pathway (52,53), were also found deleted in these tumors (Supplementary Table 1, available at Carcinogenesis Online). The data taken together indicates that aberrant expressions of PI3K/AKT and Wnt/β-catenin signaling pathways involving the copy number-altered genes play roles in Aurora-A-driven mammary tumorigenesis. In addition to copy number-altered genes, the presence of cancer-relevant mutant genes, such as Ptch1 gene (in two of the three tumors), in the Aurora-A transgenic mouse mammary tumors implicate the cognate pathways, for example, Hedgehog pathway involving PTCH1, in Aurora-A gain-of-function-associated breast cancers. PTCH1 gene alterations have been reported in human breast cancer (30,54,55). Thus, although the findings from our BLG–Aurora-A transgenic mouse model of mammary adenocarcinoma reveal the identity of a set of genes contributing to Aurora-A-driven mammary tumorigenesis shared with humans, more in-depth functional genomic analyses are warranted to elucidate the significance of these genes in the Aurora-A overexpressing subset of human breast cancer. It would be interesting to investigate the functional cross talk of Aurora-A with the pathways identified in this mouse model of mammary adenocarcinoma in human breast cancer.
In conclusion, the BLG–Aurora-A mouse mammary tumor model, described in this study, demonstrates that prolonged Aurora-A overexpression may be a driver genetic event in the development of breast cancer. Furthermore, the presence of genetic alterations shared with human breast cancer helped identify a number of candidate “co-driver” genes and the cognate pathways playing roles in mammary carcinogenesis. The mouse model, thus, offers a unique opportunity for in-depth investigations of in vivo Aurora-A functional interactions in the initiation and progression of mammary tumors as well as for developing novel therapeutic strategies for Aurora-A overexpressing breast cancers.
Supplementary material
Supplementary Tables 1 to 3 and Supplementary Figures 1 to 4 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute grants (R01CA089716 and NCI/EDRN UO1CA111302 to S.S., NIH P30 CA125123 to C.C.); University Cancer Foundation, MD Anderson (to S.S.); Cancer Center Support grant (CA16672 to Sequencing and Microarray Facility).
Supplementary Material
Acknowledgements
Authors acknowledge Dr John Parant for his help in making the transgene construct, Dr Vibhuti Srivastava and Ms Yiqun Zhang for help with bioinformatics analysis and assistance of Ms Aimee LeBlanc and Ms Yvette Gonzales through the course of this project.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AURKA
Aurora kinase A
- BLG
β-lactoglobulin
- CSCs
cancer stem cells
- EMT
epithelial-to-mesenchymal transition
- gDNA
genomic DNA
- mRNA
messenger RNA
- TCGA
The Cancer Genome Atlas
References
- 1. Ciriello G., et al. (2013) Emerging landscape of oncogenic signatures across human cancers. Nat. Genet., 45, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sen S., et al. (2002) Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J. Natl. Cancer Inst., 94, 1320–1329. [DOI] [PubMed] [Google Scholar]
- 3. Aust D.E., et al. (2004) Prognostic relevance of 20q13 gains in sporadic colorectal cancers: a FISH analysis. Scand. J. Gastroenterol., 39, 766–772. [DOI] [PubMed] [Google Scholar]
- 4. Sen S., et al. (1997) A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene, 14, 2195–2200. [DOI] [PubMed] [Google Scholar]
- 5. Zhou H., et al. (1998) Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet., 20, 189–193. [DOI] [PubMed] [Google Scholar]
- 6. Bischoff J.R., et al. (1998) A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J., 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ewart-Toland A., et al. (2003) Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat. Genet., 34, 403–412. [DOI] [PubMed] [Google Scholar]
- 8. Katayama H., et al. (2004) Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet., 36, 55–62. [DOI] [PubMed] [Google Scholar]
- 9. Liu Q., et al. (2004) Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J. Biol. Chem., 279, 52175–52182. [DOI] [PubMed] [Google Scholar]
- 10. Katayama H., et al. (2012) Aurora kinase-A inactivates DNA damage-induced apoptosis and spindle assembly checkpoint response functions of p73. Cancer Cell, 21, 196–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee D.F., et al. (2012) Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell, 11, 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croker A.K., et al. (2008) Cancer stem cells: implications for the progression and treatment of metastatic disease. J. Cell. Mol. Med., 12, 374–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye X., et al. (2015) Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol., 25, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cammareri P., et al. (2010) Aurora-a is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer Res., 70, 4655–4665. [DOI] [PubMed] [Google Scholar]
- 15. Lim K.H., et al. (2010) Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol. Cell. Biol., 30, 508–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Otto T., et al. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell, 15, 67–78. [DOI] [PubMed] [Google Scholar]
- 17. Dar A.A., et al. (2009) The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene, 28, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka T., et al. (1999) Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res., 59, 2041–2044. [PubMed] [Google Scholar]
- 19. Miyoshi Y., et al. (2001) Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int. J. Cancer, 92, 370–373. [DOI] [PubMed] [Google Scholar]
- 20. Zhang D., et al. (2004) Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene, 23, 8720–8730. [DOI] [PubMed] [Google Scholar]
- 21. Zhang D., et al. (2008) Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice. Oncogene, 27, 4305–4314. [DOI] [PubMed] [Google Scholar]
- 22. Wang X., et al. (2006) Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene, 25, 7148–7158. [DOI] [PubMed] [Google Scholar]
- 23. Whitelaw C.B., et al. (1992) Position-independent expression of the ovine beta-lactoglobulin gene in transgenic mice. Biochem. J., 286 (Pt 1), 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Webster J., et al. (1995) Tissue-specific, temporally regulated expression mediated by the proximal ovine beta-lactoglobulin promoter in transgenic mice. Cell. Mol. Biol. Res., 41, 11–15. [PubMed] [Google Scholar]
- 25. Hofmann W.A., et al. (2009) Analysis of array-CGH data using the R and Bioconductor software suite. Comp Funct Genomics, 201325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venkatraman E.S., et al. (2007) A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics, 23, 657–663. [DOI] [PubMed] [Google Scholar]
- 27. Wang K., et al. (2009) Estimation of tumor heterogeneity using CGH array data. BMC Bioinformatics, 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krupke D.M., et al. (2008) The mouse tumor biology database. Nat. Rev. Cancer, 8, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciriello G., et al. (2015) Comprehensive molecular portraits of invasive lobular breast cancer. Cell, 163, 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Network. (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buyse M., et al. (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl Cancer Inst., 98, 1183–1192. [DOI] [PubMed] [Google Scholar]
- 32. Cardiff R.D., et al. (2000) The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene, 19, 968–988. [DOI] [PubMed] [Google Scholar]
- 33. Rosner A., et al. (2002) Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am. J. Pathol., 161, 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taga M., et al. (2009) Essential roles of mTOR/Akt pathway in Aurora-A cell transformation. Int. J. Biol. Sci., 5, 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Luca M., et al. (2006) A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle, 5, 296–303. [DOI] [PubMed] [Google Scholar]
- 36. Seki A., et al. (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science, 320, 1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macůrek L., et al. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature, 455, 119–123. [DOI] [PubMed] [Google Scholar]
- 38. Carter S.L., et al. (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet., 38, 1043–1048. [DOI] [PubMed] [Google Scholar]
- 39. Scotto L., et al. (2008) Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes. Chromosomes Cancer, 47, 755–765. [DOI] [PubMed] [Google Scholar]
- 40. Weichert W., et al. (2005) Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch., 446, 442–450. [DOI] [PubMed] [Google Scholar]
- 41. Katayama H., et al. (2011) Functional significance of Aurora kinase A regulatory interactions with p53-ERα complex in human breast cancer cells. Horm. Cancer, 2, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ben-David U., et al. (2016) The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat. Commun., 7, 12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prabhu V.V., et al. (2012) Therapeutic targeting of the p53 pathway in cancer stem cells. Expert Opin. Ther. Targets, 16, 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cicalese A., et al. (2009) The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell, 138, 1083–1095. [DOI] [PubMed] [Google Scholar]
- 45. Torchia E.C., et al. (2009) A genetic variant of Aurora kinase A promotes genomic instability leading to highly malignant skin tumors. Cancer Res., 69, 7207–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santarius T., et al. (2010) GLO1-A novel amplified gene in human cancer. Genes. Chromosomes Cancer, 49, 711–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walsh C.S., et al. (2008) ERCC5 is a novel biomarker of ovarian cancer prognosis. J. Clin. Oncol., 26, 2952–2958. [DOI] [PubMed] [Google Scholar]
- 48. Yin Y., et al. (2008) PTEN: a new guardian of the genome. Oncogene, 27, 5443–5453. [DOI] [PubMed] [Google Scholar]
- 49. Knobbe C.B., et al. (2008) The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene, 27, 5398–5415. [DOI] [PubMed] [Google Scholar]
- 50. Kwon Y.W., et al. (2012) Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol. Cancer Res., 10, 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shulewitz M., et al. (2006) Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer. Oncogene, 25, 4361–4369. [DOI] [PubMed] [Google Scholar]
- 52. Nishiyama M., et al. (2012) Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol. Cell. Biol., 32, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mukherjee N., et al. (2016) Frequent inactivation of MCC/CTNNBIP1 and overexpression of phospho-beta-catenin(Y654) are associated with breast carcinoma: clinical and prognostic significance. Biochim. Biophys. Acta, 1862, 1472–1484. [DOI] [PubMed] [Google Scholar]
- 54. Sinha S., et al. (2008) Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: pathological significance in early- and late-onset breast carcinoma. Mol. Cancer, 7, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bonilla X., et al. (2016) Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet., 48, 398–406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.