Abstract
Diseases and phenotypes caused by disrupted transcription factor (TF) binding are being identified, but progress is hampered by our limited capacity to predict such functional alterations. Improving predictions may be dependent on expanding the set of bona fide TF binding alterations. Allele-specific binding (ASB) events, where TFs preferentially bind to one of the two alleles at heterozygous sites, reveal the impact of sequence variations in altered TF binding. Here, we present the largest ASB compilation to our knowledge, 10 765 ASB events retrieved from 45 ENCODE ChIP-Seq data sets. Our analysis showed that ASB events were frequently associated with motif alterations of the ChIP'ed TF and potential partner TFs, allelic difference of DNase I hypersensitivity and allelic difference of histone modifications. For TF dimers bound symmetrically to DNA, ASB data revealed that central positions of the TF binding motifs were disproportionately important for binding. Lastly, the impact of variation on TF binding was predicted by a classification model incorporating all the investigated features of ASB events. Classification models using only DNase I hypersensitivity and sequence data exhibited predictive accuracy approaching the models with substantially more features. Taken together, the combination of ASB data and the classification model represents an important step toward elucidating regulatory variants across the human genome.
INTRODUCTION
With recent advances in DNA sequencing technology, comprehensive analysis of sequence variants in individual genomes is possible for the first time. The technology has enabled genetics researchers to systematically seek variations that contribute to disease phenotype. Up to now, clinical approaches using DNA sequencing have focused on about 2% of the human genome containing protein-coding exons. In contrast, most disease associated variants arising from genome-wide association studies are situated within non-coding regions (1). These regions are enriched with transcription factor (TF) binding sites (TFBSs) (2), critical sequences for the regulation of gene expression. Thus, there is a pressing need to predict the impact of genetic variations on TF binding.
The prediction of which DNA sequence alterations will alter TF binding is a long-standing challenge in bioinformatics. Progress is hampered by the limited number of reliable data sets for TF binding disruption. Although thousands of expression quantitative trait loci have been identified, they are not suitable for the study of TF binding alteration because TF binding information is not available. Only a few hundreds of naturally occurring variations have been experimentally validated to alter the binding of TFs, with low depth for any specific TF (3,4). Thus, current studies cannot directly train a model on true alteration data. Instead existing methods score the binding potential of the two alleles based on DNA sequence and then quantify the difference, with examples including is-rSNP (4), BayesPI-BAR (5) and deltaSVM (6). However, many TF binding alterations do not arise from genetic difference within the TFBSs, as other influences can contribute, such as epigenetic variation and disrupted binding of cooperative TFs (7). The lack of experimentally determined disruption data makes it difficult to capture multiple defining properties of disrupted TFBSs.
The availability of large-scale data obtained through the chromatin immunoprecipitation followed by sequencing (ChIP-Seq) technique has transformed the annotation of regulatory elements (8–10). Through the ENCODE project (11,12), there is a widespread access to millions of positions at which TFs are assumed to be present in at least one tissue or cell-type. The analysis of ChIP-Seq data for the purpose of regulatory variant discovery has been introduced. In short, by combining large-scale genotype data (such as whole genome sequencing, WGS) with ChIP-Seq, it is now feasible to identify the TF binding preference between the two alleles at heterozygous sites within bound regions (13–17). Heterozygous site binding events can be classified as allele specific binding (ASB) or non-ASB events specifying whether one allele is significantly preferred or not. The advantages of heterozygous site binding data are that: (i) it provides high-throughput compilation of altered TF binding data; and (ii) it compares TF binding at two similar sequences (1 nucleotide difference) in the same cell context, reducing technical and biological noise (18).
In this work, we focused on heterozygous site binding events to interpret the impact of variations on TF-DNA binding. Using genotype calls from WGS, we extracted heterozygous site binding events across 45 TF ChIP-Seq experiments from the ENCODE project (11). We identified a set of features correlated with TF heterozygous site binding events, including motif alterations of the ChIP'ed TFs and other potential partner TFs, allelic difference of DNase I hypersensitivity (DHS) and allelic difference of histone modifications. Finally, a classifier was trained to predict the variation impact on TF binding, revealing that combining DHS and WGS was an efficient approach to predict altered TF binding. Our results suggest that heterozygous site binding events provide a foundation to identify features that informed the detection of cis-regulatory variants.
MATERIALS AND METHODS
Genotype data of investigated cell lines
Genotype data for GM12878 and 6 other lymphoblastoid cell lines (Table 1) were obtained from the Complete Genomics website (as of June 2014, specific hyperlinks provided in Supplementary Table S1) (19). For HeLa-S3, NIH granted permission to access raw sequence data (accession number phs000640.v2.p1) (20). Encrypted SRA files of HeLa-S3 were converted to raw reads using fastq-dump command from sratoolkit (https://github.com/ncbi/sratoolkit, Version 2.3.2). Raw reads were mapped to the hg19 reference genome using bwa (version 0.7.10-r789) with the command bwa sampe with default parameters. The GATK tool (version 2.7-4-g6f46d11) IndelRealigner (21) was used to realign reads around indels. Finally, samtools (version 0.1.9-r783) mpileup (22) was used to call variations. Any variation with a quality of at least 30 was kept for subsequent analysis.
Table 1. Overview of heterozygous site binding data.
| Cell | TF | DHS and Histones | Peak Count | Heterozygous binding sites events | ASB |
|---|---|---|---|---|---|
| GM12878 | 16 | 12 | 405 427 | 17 222 | 2314 |
| HeLa-S3 | 23 | 12 | 518 558 | 18 481 | 5533 |
| GM12872 | 1 | 0 | 47 151 | 2496 | 488 |
| GM12873 | 1 | 0 | 51 005 | 2575 | 552 |
| GM19238 | 1 | 0 | 49 938 | 2909 | 500 |
| GM19239 | 1 | 0 | 41 085 | 2473 | 282 |
| GM19240 | 1 | 0 | 46 036 | 2972 | 573 |
| GM12864 | 1 | 0 | 46 798 | 2390 | 523 |
| 8 | 45 | 24 | 1 205 998 | 51 518 | 10 765 |
For each investigated cell line (first column), we reported the number of compiled TF ChIP-Seq experiments (second column) and DHS and histone modification data sets (third column). The corresponding total number of TF ChIP-Seq peaks was given in the fourth column. Finally, we provided the number of heterozygous sites supported by at least 10 reads within the ChIP-Seq peaks (fifth column) and the number of ASB events (sixth column). Note that the numbers were derived from the compilation of all the TF ChIP-Seq data for each cell line. Details for each TF can be found in Supplementary Table S2.
ChIP-Seq read alignment
We downloaded ChIP-Seq data for diverse TFs, DHS data and histone modification data, from the ENCODE project (11)(Supplementary Table S1). For each cell line, we built a personalized version of the hg19 reference genome in which the single nucleotide variation (SNV) sites were replaced with IUPAC degeneracy codes according to the genotype data. The downloaded ChIP-Seq reads were mapped to the personalized reference genome using Novoalign (version 3.01.00) with default parameters. We removed any reads with a mapping quality lower than 30.
Mapping bias simulation
Even though we used a personalized reference genome to improve the mapping sensitivity of alternative alleles, there remained a potential mapping bias toward certain alleles (13,17,23). To address this issue, we performed a read mapping simulation to estimate the mapping bias at each heterozygous site. For each heterozygous site within the TF ChIP-seq peak regions, we generated all the possible 36-bp reads overlapping with the heterozygous sites for each allele and each strand. Then, the generated reads were mapped to the personalized reference genome using the same settings as for the real ChIP-Seq data. Finally, we assessed the mapping bias and excluded the biased sites if the imbalance ratio of any allele was greater than 60%.
Retrieve heterozygous site binding events and call ASB events
Uniformly processed ChIP-Seq narrowPeaks were downloaded from ENCODE (24). In order to increase the confidence of TF-bound regions, we narrowed the peaks to the 100 base pairs (bp) core regions centered around the peak max positions (25,26). For each TF ChIP-Seq data set, we retrieved the read counts of the two alleles at heterozygous site binding events within the ChIP-Seq peak core regions. Replicates were pooled together to increase the overall read coverage (17,27). Peak core regions on sex chromosomes or overlapping with copy number variant regions (Supplementary Table S1) were filtered out. We excluded from the downstream analyses core peaks that harboured >1 heterozygous SNVs (8311 out of the 79 565 heterozygous core peaks) to ensure that the two alleles differed by a single nucleotide. For the sites supported by at least 10 reads, an ASB event was called if the read count on one allele was significantly different from the other allele based on a binomial test (false discovery rate (FDR) <0.05). As an aside, we explored the option of using replicate normalization and a beta-binomial method (see Supplementary Text for details). The hypothesized probability of the binomial test was set as the mapping imbalance detected in the above-mentioned read mapping simulation at each heterozygous site. For ASB events, we further required the favored allele to show at least 60% allele imbalance (proportion of reads mapped to one allele over the total) following (27), to remove extreme P-values caused by small changes at high read depth loci. We labeled the allele with higher number of mapped reads as the favored allele, and the lower one as the unfavored allele; in non-ASB events, if the numbers of mapped reads on the two alleles were equal, the reference allele was labelled as favored.
TFBS identification in ChIP-Seq peak regions
TF binding motifs were downloaded from the JASPAR database (version 2014) (28). The motif of each corresponding TF was scanned against the peak regions using the Biopython (version 1.65) motifs module (28,29). For each scanned site, the motifs module provided the motif score (position weight matrix, PWM, score) and the P-value of the score against a null uniform distribution of the four nucleotides (referred to as motif P-value). Sites with scores above the FDR threshold of 0.001 were predicted as TFBSs.
Define the ASB frequency within TFBS
We defined the frequency of ASB events at each TF motif position as the proportion of ASB events observed at this position over the total number of ASB events observed across all motif positions considering the predicted TFBSs; the same definition was applied to non-ASB events. Only the TFs with at least 10 ASB and 10 non-ASB events in the predicted TFBS are considered to calculate the ASB frequency within TFBS.
Identify comotifs within ChIP-Seq peak regions
We used the findMotifsGenome.pl script from the HOMER (30) package (version 4.6) with default settings to identify enriched known motifs in ChIP-Seq peak regions. The HOMER default analysis window of 200 bp was applied. Among the enriched motifs reported by HOMER, we identified the 5 most enriched motifs according to following criteria: (i) not similar to the motif of the ChIP'ed TF if available in JASPAR; and (ii) no similar motifs within the five identified motifs. Motif similarity was based on the compare-matrices command provided in the RSAT toolset (version 2011) (31) with an information content correlation threshold of 0.8. For the ASB SNVs not overlapping the predicted TFBSs of the ChIP'ed TF, we tested the correlation between motif alteration (log ratio of motif P-values between the two alleles) and allele imbalance of TF binding within the predicted TFBSs of each of the five enriched motifs (Spearman correlation, FDR < 0.05). The significantly correlated enriched motifs were identified as comotifs.
Identify cobound TFs associated with ASB events
To identify the distribution of ASB events within binding regions of other TFs, we used all the available TF ChIP-Seq peaks in the same cell line. Cobound TFs were identified if their peaks overlapped with the peaks of ASB TFs. For the heterozygous site binding events of each ASB TF, we investigated the association between the presence of ASB events and their overlap with the peaks of each cobound TF (two-sided Fisher's exact test, FDR < 0.005). The odds ratio of Fisher's exact test was used to interpret whether ASB events were enriched (odds ratio > 1) or depleted (odds ratio < 1) in cobound regions.
Classification of heterozygous site binding events
We used the randomForest package (32) and the recursive feature elimination function from the caret package (33) to train random forest classifiers (‘ntree’ parameter was set to 1000) and select key features. Since there were more non-ASB events than ASB events, non-ASB events were randomly downsampled to balance the training data set for each tree building process following the balanced random forest approach (34,35). We used a 5-fold cross-validation approach to assess the predictive power of the classifiers. Specifically, the predictive power corresponded to the average area under precision-recall curve (AUPRC) obtained through the 5-fold cross-validation. For determining the importance of each feature in a classifier, we took the ‘MeanDecreaseAccuracy’ (mean accuracy decrease over all trees) score reported by the random forest.
The input features, listed in Supplementary Table S5, spanned five categories: (i) motif-related features, for instance the motif scores of the two alleles, the best motif scores within the peak regions on two alleles; (ii) positional information, such as SNV distance to the ChIP-Seq peak max and SNV position within the predicted TFBS; (iii) enriched-motif related features (log ratio of motif P-values between the two alleles); (iv) cobound TFs, such as the overlapping of heterozygous site binding events with each available cobound TF peaks within the same cell line; and (iv) chromatin features, for instance the read counts on the two alleles from DHS and 11 histone modification data from the corresponding cell type.
We combined features across the five categories and trained three models: (i) a Seq model based on sequence features, including categories 1–3; (ii) a Seq+DHS model adding DHS data on top of the Seq model; and (iii) a Full model trained using all features.
We compared our classifiers to BayesPI-BAR (5) and deltaSVM (6). The deltaSVM score was calculated as the gkmSVM score difference between two alleles (6). For each TF, we trained a separate gkmSVM model (version 2.0) with default parameters using 5000 randomly selected ChIP-seq peaks following (36) and the associated tutorial (http://www.beerlab.org/gkmsvm/). One TF (PRDM1) had only 4577 peaks and we used all of them to train the gkmSVM model. The BayesPI-BAR package was downloaded from http://folk.uio.no/junbaiw/BayesPI-BAR/, and BayesPI-BAR scores were calculated with default parameters.
RESULTS
Compilation of heterozygous site binding events
We implemented a pipeline that combined ChIP-Seq and genotype data from the same cell types to extract heterozygous site binding events (Materials and Methods). Specifically, ChIP-Seq (and DHS) reads were mapped to personalized reference genomes in which the variants reported in the genotype data were incorporated. In total, we retrieved 51 518 heterozygous site binding events supported by at least 10 reads from 45 TF ChIP-Seq data sets from 8 cell lines. We also extracted read counts of 11 histone modifications and DHS on the two alleles of TF heterozygous site binding events in GM12878 and HeLa-S3 cell lines. We observed that 4.3% of the TF ChIP-Seq peak regions contained a single heterozygous site (Table 1 and Supplementary data). ASB events were defined if the number of mapped TF ChIP-Seq reads on one allele was significantly higher than the number of mapped reads on the other allele (Binomial test, FDR < 0.05) and with at least 60% allele imbalance for the favored allele as in (27). We found that 20.9% of heterozygous site binding events were classified as ASB events; others were classified as non-ASB events. Among the compiled data of 8 cell lines, GM12878 and HeLa-S3 (Tier 1 and Tier 2 cell lines from the ENCODE project) had data sets for all the investigated TFs, DHS and histone marks; the remaining 6 cell lines were restricted to ChIP-Seq data for CTCF. Therefore, we focused on GM12878 and HeLa-S3 for most of the study, using the additional cell lines for testing the classification models (see below).
TFBS alterations strongly correlate with ASB events
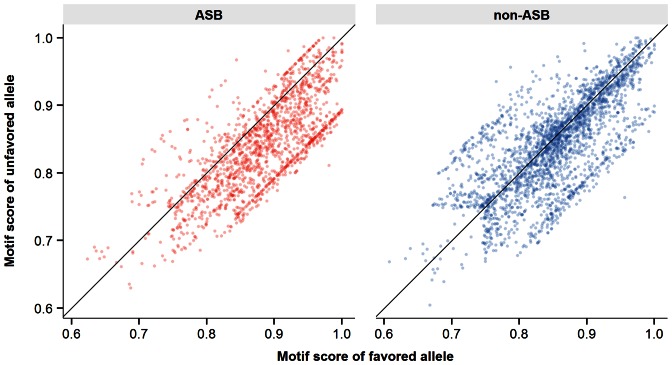
To understand the underlying genetic mechanisms of ASB, we considered the subset of SNVs overlapping with the predicted TFBSs (Materials and Methods). An initial analysis revealed that ASB SNVs were significantly enriched in predicted TFBSs compared with non-ASB events (Fisher's exact test, P-value = 1.8e-128). Next, we assessed the motif score alteration caused by the SNVs for ASB events. We found that motif scores of favored alleles (allele with higher read count) were significantly higher than those of unfavored alleles in predicted TFBSs (Figure 1, P-value = 8.9e-120, one-sided Wilcoxon signed-rank test), reflecting the contribution of motif score alteration to ASB events. In contrast, non-ASB events displayed a balanced score distribution between the two alleles. Our results agree with previous observations (18,37) but are based on data for a much larger number of TFs and TFBSs. However, only a portion of ASB SNVs (19.3%) overlapped with the predicted TFBSs, indicating that additional mechanisms beyond TFBS alteration contribute to the observed ASB events. A plot showing the total set of ASB and non-ASB events, including those outside the predicted TFBSs is provided in Supplementary Figure S1.
Figure 1.
Transcription factor binding sites (TFBSs) motif score analysis at heterozygous site binding events. In each panel, we plotted the motif score at heterozygous sites on the favored allele (harboring higher amount of mapped chromatin immunoprecipitation followed by sequencing (ChIP-Seq) reads, x-axis) and unfavored allele (y-axis) at predicted TFBSs. Allele-specific binding (ASB) (left panel) and non-ASB (right panel) events were plotted separately. The black diagonal lines indicated an identical motif score on the two alleles. Note that the figure was generated using all heterozygous site binding events for all compiled TFs in GM12878 and HeLa-S3.
ASB events show different positional distribution within TFBS compared with motif information content
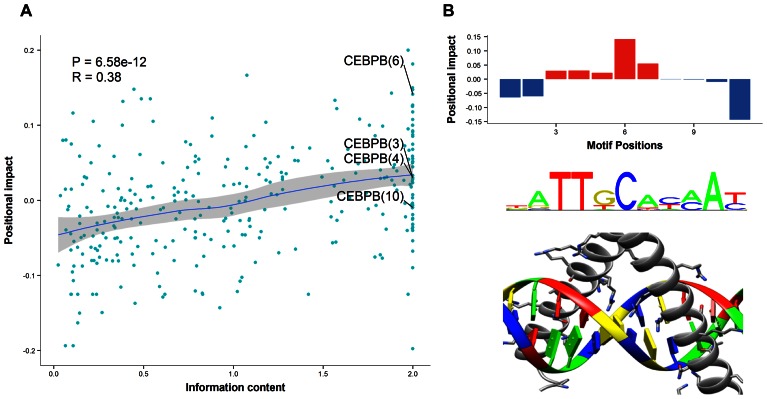
We next examined whether specific positions within TF binding motifs were more sensitive to ASB events and how such impactful positions related to their information content (IC) in the TFBS motif profiles. IC has been correlated with the strength of binding site preference for individual nucleotides in TF binding models (38). Given a TFBS motif, the positional impact was measured as the frequency difference between ASB and non-ASB events at each position (Materials and Methods). As expected, positional impact was significantly correlated with positional IC across motif positions of all investigated TFs (Spearman correlation coefficient = 0.38, P-value = 6.6e-12; Figure 2A). But most motif positions did not strictly follow this trend in Figure 2A, revealing a large variance of positional impact that cannot be attributed to IC.
Figure 2.
Information content and positional impact of each position within TFBS. (A) Correlation between positional impact and information content. Each point corresponded to a position (given in parenthesis) within TFBSs associated to ChIP'ed TFs. Positions were plotted with respect to their associated information content (x-axis) from the TF motif and positional impact (y-axis). The trend line was drawn by the locally weighted scatterplot smoothing method. (B) Exceptional example of CEBPB motif with its positional impact distribution (upper), TF binding motif logo (middle) and TF–DNA interface (lower; Protein Data Bank ID: 2e42).
The most extreme cases at the upper right corner of Figure 2A represented motif positions where TF binding was disproportionately impacted. We qualitatively observed that these positions tended to be centrally positioned within the TFBSs of the TFs which were dimers and bound symmetrically to DNA. When analyzing all four symmetric TF dimers in our data sets with known TF-DNA complex structures (CEBPB, MAX, TCF7L2 and USF1), we observed that central positions significantly showed high positional impact compared with other positions with similar IC (P-value = 0.02, one-sided Wilcoxon rank-sum test). As a specific example, CEBPB recognizes an 11 bp motif containing four positions with an IC of two bits (positions 3, 4, 6 and 10), which, according to the motif, would be expected to be equally important for binding (Figure 2B). However, the positional impact was particularly high at position 6, at the center of the motif, indicating that this position could be more critical for the disruption of TF binding (Figure 2B). Further structural analysis of a DNA-CEBPB dimer interaction revealed that position 6 was contacted by both monomers (Figure 2B). The critical role of central positions suggests that mutations at these positions might potentially affect the binding of the two monomers. Recently, the same position of the CEBPD motif was reported to display more somatic mutations within the predicted TFBSs than other positions in human cancer genomes (39), which is concordant with our findings. Other cases included the PAX5 motif at position 15 (Supplementary Figure S2), which was of low IC (0.4) but with high impact, suggesting that low IC positions could also be critical for TF binding (40). Taken together, IC derived from motifs partially explained the distribution of ASB events across the motif, while positional impact from ASB events provided deeper insights into the binding properties of TFs.
Disruption of enriched comotifs can lead to ASB events
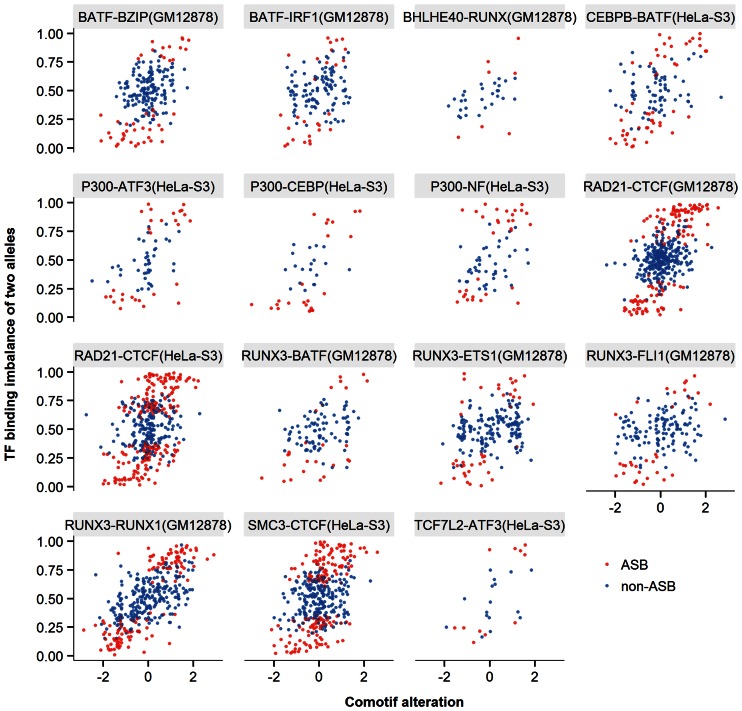
Since most variations at ASB events were outside of the predicted TFBSs (80.7%), we assessed whether disrupted TFBSs of potential partner TFs could be responsible for the observed events. We retrieved the five most enriched, non-redundant motifs within the peak regions of each TF ChIP-Seq experiment (Materials and Methods). Within the predicted TFBS of each enriched motif, we tested the correlation between the motif score change and the allelic binding imbalance of the ChIP'ed TF across all heterozygous site binding events (Materials and Methods). We found 15 significantly correlated enriched motifs for 9 TF ChIP-Seq experiments (based on the Spearman rank statistic, FDR < 0.05, Figure 3), hereafter referred to as comotifs. Decreased motif scores of comotifs were preferentially observed on unfavored alleles in ASB events, consistent with a cooperative binding model (41). The comotifs lay in three categories (Supplementary Table S3): (i) seven cases in which the TFs associated to the comotifs were known to interact with the ChIP'ed TF, for instance the comotif of P300 was CEBPB (P300-CEBPB); (ii) one case (RUNX3-RUNX1) in which the TF of comotif belonged to the same TF family as the ChIP'ed TF; and (iii) seven cases of novel relationships, from our knowledge, including CEBPB-BATF and P300-NF-E2.
Figure 3.
Alteration of comotif correlated with TF allelic imbalance. The name of each panel specified the ChIP'ed TF followed by the comotif name and the cell line in parentheses. Each dot represented one heterozygous site binding event (red for ASB and blue for non-ASB events) found within the predicted TFBSs of the comotif. The comotif alteration (x-axis) represented the log ratio of motif P-values between the reference and alternative alleles. The allelic binding imbalance (y-axis) indicated the fraction of reads mapped on the reference allele over the whole read coverage at that position. We tested the correlation between the two properties for each ChIP'ed TF and its enriched HOMER motifs, and only significantly correlated pairs were plotted (FDR < 0.05).
Moreover, 6 out of the 15 comotifs arose from the experiments in which the ChIP'ed TFs did not bind DNA directly, as for example P300. For these non-sequence specific TFs, 33.5% of ASB-SNVs overlapped the TFBSs of comotifs, significantly enriched compared with 17.4% for non-ASB events (Fisher's exact test, P-value = 7.1e-41, Supplementary Figure S3). Overall, ASB overlapping comotifs comprised 9.4% of ASB events.
ASB events associated with cobound TFs
Next we sought to understand how ASB events related to regions bound by additional TFs within the same cell using ChIP-Seq data. It has been observed that TF binding in cobound regions (cases where ChIP-Seq data for multiple distinct proteins have overlapping peaks) tends to be more conserved over evolution than isolated binding events (42). We tested the distribution difference between ASB and non-ASB events in the ChIP-seq peaks of each cobound TF (Materials and Methods), revealing 64 significant pairs (Supplementary Table S4, Fisher's exact test, FDR < 0.005). Of these, 27 were observed in GM12878 lymphoblastoid cells, and all of them displayed depletion of ASB (relative to non-ASB) in the cobound regions (odds ratio < 1). This pattern is concordant with the concept of variant buffering effects in motif-rich DHS regions (27). For instance, CTCF heterozygous site binding events were classified as ASB in 8.9% of cases where ZNF143 binding peaks were overlapping, while 18.3% of cases were classified as ASB if there were no overlapping ZNF143 peaks (Fisher's exact test, P-value = 3.6e-11). The ASB TF and cobound TF pairs included known TF–TF interactions, such as CTCF-ZNF143, and RUNX3-YY1 (43), suggesting functional interactions for the pairs observed. In HeLa-S3, a cancer cell line, we observed a reversed pattern where ASB events were enriched in cobound regions (odds ratio >1, not depleted as in GM12878) for 17 out of 35 cases (such as CEBPB-P300 and MAX-CMYC). The opposing pattern between normal and cancer cells suggests that binding site alterations in cobound regions of cancer cells may be functionally important for gene dysregulation. Further analyses would be required to test this hypothesis when more TF binding data become available.
Allelic chromatin properties coordinate with ASB events
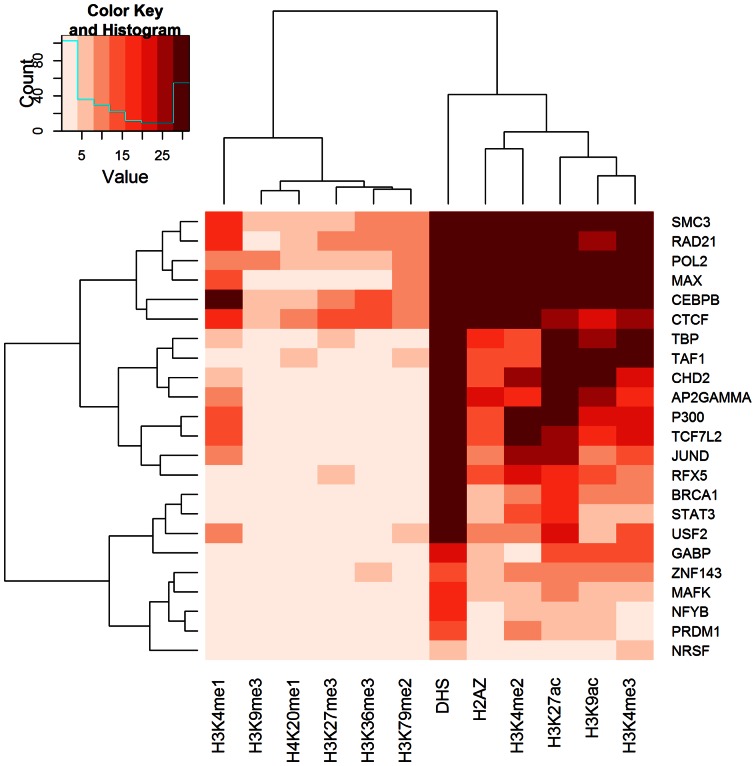
To further shed light to the mechanisms associated with ASB events, we investigated the non-genetic properties in proximity to ASB events. We extracted read counts from DHS and histone modification ChIP-Seq experiments on the two alleles at heterozygous site binding events. Next, we assessed the correlation between allelic imbalance of each chromatin property (DHS and 11 histone modifications) and TF binding. Overall, 196 significant correlations were observed (Pearson correlation, FDR < 0.05; Figure 4 and Supplementary Figure S4). DHS signal was significantly correlated with TF binding for 35 out of 39 TF ChIP-Seq experiments. DHS showed higher read counts on the TF favored allele for 73.4% of the ASB events compared with 52.5% for non-ASB events. Moreover, we found 161 TF-histone correlation pairs. Active histone modifications, such as H3K27ac, H3K4me2 and H3K3me3, exhibited positive correlation patterns with TF binding imbalance. Taken together, DHS and histone modifications widely correlated with ASB events, indicating their potential value for predictive modeling.
Figure 4.
Allelic coordination between heterozygous site binding events for multiple TFs and chromatin properties in HeLa-S3. The heatmap represented the -log(P-value) of Pearson correlation between allele imbalance of TF ChIP-Seq reads at heterozygous site binding events and chromatin properties (DHS and histone modifications).
DHS and sequence-derived properties are sufficient for cost-effective ASB event prediction
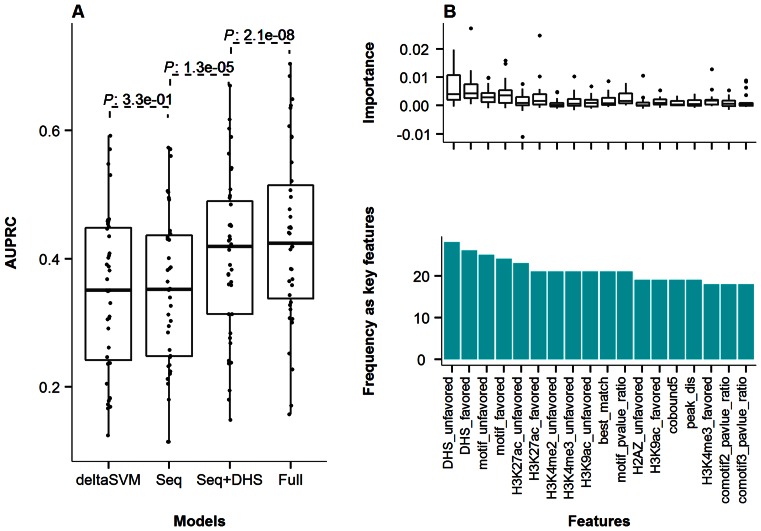
Building upon the observed associations between ASB events and properties of both sequence and experimental data, we constructed computational models to determine our capacity to predict SNVs disruptive of TF binding (that is to distinguish between ASB events and non-ASB events). We took ASB events as the positive training data, and non-ASB events as the negative set for model training. We constructed random forest classifiers using only DNA sequence information (that are the features derived from motif and comotifs, referred to as Seq model, see Materials and Methods) and assessed their predictive performances. Consistent with past literature (3–5), the Seq model had predictive value but the performance was quite limited across all the investigated TFs (average AUPRC of 0.35, Figure 5A). The Seq models allowed consistent performance across data from multiple individuals within the same cell type (Supplementary Text).
Figure 5.
Performance of ASB classification models and key features. (A) AUPRC of the deltaSVM, Seq, Seq+DHS and Full models across all the investigated TF ChIP-Seq experiments. Seq model was based only on sequence-related features; Seq+DHS model added DHS data on top of the Seq model; and Full model further added histone and cobound TFs. Details on each model and features can be found in Materials and Methods. (B) Top frequent key features in the Full models for all 27 TFs with known motifs. The suffix ‘favor’ and ‘unfavor’ referred to the favored and unfavored alleles at heterozygous sites. The ‘motif_pvalue_ratio’ was the log ratio between two alleles in terms of motif score P-value. The ‘peak_dis’ indicated the distance of the SNV to ChIP-Seq peak maximum position where the highest number of reads were mapped within the peak.
We compared our classifiers against two existing sequence-based models, deltaSVM (6) and BayesPI-BAR (5). Seq models outperformed BayesPI-BAR (Wilcoxon signed-rank test, P-value = 7.5e-09, Supplementary Figure S6) and showed similar performance with deltaSVM (Wilcoxon signed-rank test, P-value = 3.3e-01, Figure 5A) when predicting ASB events. The differences between deltaSVM and our Seq models are that deltaSVM uses k-mers to predict TF binding regions while our Seq models allow for combining positional feature on top of motif features (Materials and Methods). The ASB framework potentially can incorporate any features of two alleles into the discriminative model, e.g. adding deltaSVM scores to cover the k-mer changes.
Next, we took into account all the features analyzed in the previous sections into the model (Materials and Methods), which was hereafter referred to as the Full model. The Full model showed a mean AUPRC of 0.43 across all the tested TFs (Figure 5A). For those TFs with known binding motifs, the top ranked features highlighted two major categories contributing to the success of the model, DHS and motif sequence properties. Specifically, the top 5 features were DHS read count from the unfavored allele, DHS read count from the favored allele, motif score on the unfavored allele, motif score on the favored allele and H3K27ac read count from the favored allele (Figure 5B). For TFs lacking a motif model, the feature set could not include motif sequence properties of the ChIP'ed TF. Consequently DHS, H3K4me2 and H3K27ac were important for the success of the classifiers (Supplementary Figure S7).
Given that ChIP-Seq TF binding data were not available for most cell lines, while DHS was more likely to be available, we evaluated the performance of models limited to sequence-derived features and DHS (Seq+DHS model). Consistent with the number of features in the training sets for each model, results showed that the Full model outperformed the Seq+DHS model, which in turn outperformed the Seq model across all the tested TFs (P-values of 2.1e-8 and 1.1e-5, Wilcoxon signed-rank test) (Figure 5A). From a sequence-only baseline of 0.35 in terms of average AUPRC, the Seq+DHS model achieved 0.40 and the Full model achieved 0.43. Importantly, inclusion of DHS with sequence properties provided important value, representing 62.3% of the average improvement of the Full model over the sequence-only baseline. These results highlighted that ASB prediction could be pursued with few laboratory generated features cost-effectively by coupling sequence analysis with experimental genotyping (WGS) and DHS data.
DISCUSSION
Predicting variant impact on TF binding is amongst the biggest current challenges for genome interpretation. One of the main obstacles is the lack of sufficient and reliable TFBS alteration data, which are critical for the development of bioinformatics methods. We compiled 10 765 ASB events from 45 TF ChIP-Seq experiments from 8 cells lines. To the best of our knowledge, this is the largest experimentally defined ASB collection. While altered canonical TFBSs for the ChIP'd TFs were frequently observed (19.3%), most ASB SNVs did not overlap with the primary TF motif. When looking across positions within TFBS, we observed that central TFBS positions for symmetric TF dimers were more critical than other positions with similar information content. Alterations of comotifs, potentially bound by partner TFs, were observed for a portion of ASB events (9.4%). Taking the enlarged collection of data to train classification models, we demonstrated that baseline models using only genomic sequence data were improved by the incorporation of allelic DHS data that provided 62.3% of the performance improvement achieved by models using all available features (∼100 per cell type) from the ENCODE data.
There are multiple statistical approaches for the calling of ASB events, with most literature using the binomial test (18,27,37,44). Recent studies detected that experimental allelic imbalance was overdispersed compared to the binomial distribution (45,46). Beta-binomial tests have been proposed to correct the overdispersion in ASB calling under the assumption that most sites are balanced (17). We observed that non-ASB events with minor motif alterations exhibit higher overdispersion compared to other non-ASB events (Supplementary Text). Given this observation, we elected to use the binomial approach, as utilizing non-ASB events or all allelic events as the null distribution would over-estimate the over-dispersion parameter, which would increase false negatives. Finally, ASB events could also be called as differentially bound regions using general linear models (GLM), if replicates were available. However, GLM-based approaches tend to be conservative when calling differential binding regions (47), potentially missing a significant portion of true ASB events. Further comprehensive evaluation of the background null model will be needed.
Our results suggest that positions of SNVs within TFBSs should be considered when investigating SNV impact on symmetric TF dimers. The observed impact of SNVs within these central positions was not fully reflected by the information content of classic motif (position weight matrix) models (48). Classic PWM-based methods (3–5,49) did not capture such characteristics when predicting TF binding alteration. The importance of these central positions was supported by structures of DNA–TF dimer interactions showing them to be dual-contact points for both protein subunits, highlighting that structural information can be important for understanding the impact of SNVs on TF binding.
Our ASB classification model provides a novel supervised and integrative framework to model SNV impact on TF binding. To evaluate the impact of SNVs, most prior methods calculated binding score differences between altered alleles and reference alleles based on TF binding motifs (4,5,49) or enriched k-mers (6,50). Prediction of SNV impact was based on those cases where the difference exceeded a threshold. However, the selection of a threshold was difficult to justify. In contrast, our ASB model learned the optimal threshold (decision surface) from the data directly. Moreover, our method was not limited to sequence features (TF motifs and k-mers), with the capacity to incorporate diverse features (such as genetic features, DHS and histone modifications). We anticipate that such features will become increasingly available in the near future. In addition, the relative importance of each feature in the classification models provided insights into the mechanisms contributing to TF binding.
Only ∼30% of ASB events can be explained by motif or comotif alteration. Understanding how the altered binding arises in the remaining portion is likely to require advances in our knowledge and understanding of TF binding. First, the available TF binding models are insufficient. Most human TFs do not yet have binding models, although the coverage improves. Second, the existing binding models can be improved. For instance, CTCF has been shown to recognize flanking motifs that stabilize binding, but these are not yet well represented in the current PWM model (51). Moreover, there are properties outside the sequence-specific target that contribute to binding. Flanking sequences can influence binding strength (52–54), potentially involving the shape (topology) of DNA (55,56). As we advance our understanding, we can anticipate that the causally unexplained portion of ASB events will be decreased.
The predictive power (AUPRC) of the ASB classification models is limited, particularly when considered on the scale of analyzing a full genome. The inadequate performance might be attributable to multiple causes. For instance, the classification model may be under-fitted because the number of ASB events available for training was not sufficient. In the review stage of this manuscript, two studies compiled new ASB data sets in other cell lines to investigate GWAS loci or the variant impact on gene expression (17,57). In the future, we anticipate a rapidly growing body of ASB data will be critical in training more reliable models. Alternatively, the set of features available for modeling may have missing components, e.g. the limited set of TF binding models. Lastly, ASB events could be caused by multiple SNVs or distal SNVs. In our data compilation, we excluded the cases where multiple heterozygous SNVs situated within the same ChIP-Seq core peak regions to simplify the analysis. However, the accumulated effect of multiple SNVs proximal or distal to a TFBS could alter local TF binding according to the TF–TF interaction and chromatin interaction models (58,59). Further efforts needs to be devoted to these areas.
Identification of cis-regulatory variants is a critical need for understanding the genetic mechanisms contributing to diseases (60). Our compilation of heterozygous site binding data and ASB classification models provide unique data sets and a novel framework for modeling the impact of SNVs on TF–DNA interaction. Future advances in sequencing technology and enlarged ASB database will enable the reliable identification of cis-regulatory variants.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Wasserman lab members for helpful discussions, Miroslav Hatas for systems support and Dora Pak for management support. The research was enabled in part by the support provided by WestGrid (www.westgrid.ca) and Compute Canada Calcul Canada (www.computecanada.ca). The WGS data of HeLa-S3 cell line used in this research were derived from a HeLa cell line (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000640.v2.p1). Henrietta Lacks and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. The authors are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. This study was reviewed by the NIH HeLa Genome Data Access Working Group.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada/Genome BC [174DE]; Natural Sciences and Engineering Research Council of Canada [RGPIN355532-10]; National Institutes of Health (USA) [1R01GM084875]; PhD fellowship from China Scholar Council [201206110038 to W.S.]; Child and Family Research Institute, Vancouver; British Columbia Children's Hospital Foundation [to A.M. and W.W.W.]. Funding for open access charge: Genome Canada/Genome BC [174DE].
Conflict of interest statement. None declared.
REFERENCES
- 1. Li M.J., Wang L.Y., Xia Z., Sham P.C., Wang J.. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 2013; 41:W150–W158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khurana E., Fu Y., Colonna V., Mu X.J., Kang H.M., Lappalainen T., Sboner A., Lochovsky L., Chen J., Harmanci A. et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013; 342:1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersen M.C., Engstrom P.G., Lithwick S., Arenillas D., Eriksson P., Lenhard B., Wasserman W.W., Odeberg J.. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput. Biol. 2008; 4:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Macintyre G., Bailey J., Haviv I., Kowalczyk A.. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010; 26:i524–i530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J., Batmanov K.. BayesPI-BAR:A new biophysical model for characterization of regulatory sequence variations. Nucleic Acids Res. 2015; 43:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S., Beer M.A.. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015; 47:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kasowski M., Grubert F., Heffelfinger C., Hariharan M., Asabere A., Waszak S.M., Habegger L., Rozowsky J., Shi M., Urban A.E. et al. Variation in transcription factor binding among humans. Science. 2010; 328:232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herrmann C., Van de Sande B., Potier D., Aerts S.. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012; 40:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karczewski K.J., Dudley J.T., Kukurba K.R., Chen R., Butte A.J., Montgomery S.B., Snyder M.. Systematic functional regulatory assessment of disease-associated variants. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., Harmanci A., Leng J., Bjornson R., Kong Y., Kitabayashi N. et al. AlleleSeq: Analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 2011; 7:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Younesy H., Möller T., Heravi-Moussavi A., Cheng J.B., Costello J.F., Lorincz M.C., Karimi M.M., Jones S.J.M.. ALEA: a toolbox for allele-specific epigenomics analysis. Bioinformatics. 2014; 30:1172–1174. [DOI] [PubMed] [Google Scholar]
- 15. Bailey S.D., Virtanen C., Haibe-Kains B., Lupien M.. ABC: a tool to identify SNVs causing allele-specific transcription factor binding from ChIP-Seq experiments. Bioinformatics. 2015; 31:3507–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zuo C., Shin S., Keles S.. atSNP: transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics. 2015; 31:3355–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J., Rozowsky J., Galeev T.R., Harmanci A., Kitchen R., Bedford J., Abyzov A., Kong Y., Regan L., Gerstein M.. A uniform survey of allele-specific binding and expression over 1000-Genomes-Project individuals. Nat. Commun. 2016; 7:11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reddy T.E., Gertz J., Pauli F., Kucera K.S., Varley K.E., Newberry K.M., Marinov G.K., Mortazavi A., Williams B.A., Song L. et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012; 22:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010; 327:78–81. [DOI] [PubMed] [Google Scholar]
- 20. Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., Shendure J.. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013; 500:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Degner J.F., Marioni J.C., Pai A.A., Pickrell J.K., Nkadori E., Gilad Y., Pritchard J.K.. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009; 25:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilbanks E.G., Facciotti M.T.. Evaluation of algorithm performance in ChIP-seq peak detection. PloS One. 2010; 5:e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Worsley Hunt R., Mathelier A., Del Peso L., Wasserman W.W.. Improving analysis of transcription factor binding sites within ChIP-Seq data based on topological motif enrichment. BMC Genomics. 2014; 15:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurano M.T., Haugen E., Sandstrom R., Vierstra J., Shafer A., Kaul R., Stamatoyannopoulos J.A.. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015; 47:1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C. et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015; 43:W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breiman L. Random forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 33. Kuhn M. Building predictive models in R using the caret package. Journal of Statistical Software. 2008; 28:1–26.27774042 [Google Scholar]
- 34. Anaissi A., Kennedy P.J., Goyal M., Catchpoole D.R.. A balanced iterative random forest for gene selection from microarray data. BMC Bioinformatics. 2013; 14:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bekkar M., Alitouche T.A.. Imbalanced data learning approaches review. Int. J. Data Mining Knowl. Manag. Process. 2013; 3:15–33. [Google Scholar]
- 36. Ghandi M., Lee D., Mohammad-Noori M., Beer M.A.. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 2014; 10:e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kilpinen H., Waszak S.M., Gschwind A.R., Raghav S.K., Witwicki R.M., Orioli A., Migliavacca E., Wiederkehr M., Gutierrez-Arcelus M., Panousis N.I. et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013; 342:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wasserman W.W., Sandelin A.. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004; 5:276–287. [DOI] [PubMed] [Google Scholar]
- 39. Melton C., Reuter J.A., Spacek D.V., Snyder M.. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 2015; 47:710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathelier A., Lefebvre C., Zhang A.W., Arenillas D.J., Ding J., Wasserman W.W., Shah S.P.. Cis-regulatory somatic mutations and gene-expression alteration in B-cell lymphomas. Genome Biol. 2015; 16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karczewski K.J., Tatonetti N.P., Landt S.G., Yang X., Slifer T., Altman R.B., Snyder M.. Cooperative transcription factor associations discovered using regulatory variation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13353–13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefflova K., Thybert D., Wilson M.D., Streeter I., Aleksic J., Karagianni P., Brazma A., Adams D.J., Talianidis I., Marioni J.C. et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013; 154:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M.. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006; 34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leung D., Jung I., Rajagopal N., Schmitt A., Selvaraj S., Lee A.Y., Yen C.A., Lin S., Lin Y., Qiu Y. et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015; 518:350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harvey C.T., Moyerbrailean G.A., Davis G.O., Wen X., Luca F., Pique-Regi R.. QuASAR: quantitative allele-specific analysis of reads. Bioinformatics. 2014; 31:1242–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van de Geijn B., McVicker G., Gilad Y., Pritchard J.K.. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015; 12:1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steinhauser S., Kurzawa N., Eils R., Herrmann C.. A comprehensive comparison of tools for differential ChIP-seq analysis. Brief. Bioinform. 2016; bbv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000; 16:16–23. [DOI] [PubMed] [Google Scholar]
- 49. Manke T., Heinig M., Vingron M.. Quantifying the effect of sequence variation on regulatory interactions. Hum. Mutat. 2010; 31:477–483. [DOI] [PubMed] [Google Scholar]
- 50. Huang D., Ovcharenko I.. Identifying causal regulatory SNPs in ChIP-seq enhancers. Nucleic Acids Res. 2014; 43:236–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakahashi H., Kwon K.R., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A. et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013; 3:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maerkl S.J., Quake S.R.. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007; 315:233–237. [DOI] [PubMed] [Google Scholar]
- 53. Nutiu R., Friedman R.C., Luo S., Khrebtukova I., Silva D., Li R., Zhang L., Schroth G.P., Burge C.B.. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011; 29:659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Levo M., Zalckvar E., Sharon E., Dantas Machado A.C., Kalma Y., Lotam-Pompan M., Weinberger A., Yakhini Z., Rohs R., Segal E.. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015; 25:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R.. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013; 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou T., Shen N., Yang L., Abe N., Horton J., Mann R.S., Bussemaker H.J., Gordan R., Rohs R.. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4654–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cavalli M., Pan G., Nord H., Wallerman O., Wallen Arzt E., Berggren O., Elvers I., Eloranta M.L., Ronnblom L., Lindblad Toh K. et al. Allele-specific transcription factor binding to common and rare variants associated with disease and gene expression. Hum. Genet. 2016; 135:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ding Z., Ni Y., Timmer S.W., Lee B.K., Battenhouse A., Louzada S., Yang F., Dunham I., Crawford G.E., Lieb J.D. et al. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 2014; 10:e1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waszak S.M., Delaneau O., Gschwind A.R., Kilpinen H., Raghav S.K., Witwicki R.M., Orioli A., Wiederkehr M., Panousis N.I., Yurovsky A. et al. Population variation and genetic control of modular chromatin architecture in humans. Cell. 2015; 162:1039–1050. [DOI] [PubMed] [Google Scholar]
- 60. Mathelier A., Shi W., Wasserman W.W.. Identification of altered cis-regulatory elements in human disease. Trends Genet. 2015; 31:67–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.