Abstract
DNA of viral origin represents a ubiquitous element of bacterial genomes. Its integration into host regulatory circuits is a pivotal driver of microbial evolution but requires the stringent regulation of phage gene activity. In this study, we describe the nucleoid-associated protein CgpS, which represents an essential protein functioning as a xenogeneic silencer in the Gram-positive Corynebacterium glutamicum. CgpS is encoded by the cryptic prophage CGP3 of the C. glutamicum strain ATCC 13032 and was first identified by DNA affinity chromatography using an early phage promoter of CGP3. Genome-wide profiling of CgpS binding using chromatin affinity purification and sequencing (ChAP-Seq) revealed its association with AT-rich DNA elements, including the entire CGP3 prophage region (187 kbp), as well as several other elements acquired by horizontal gene transfer. Countersilencing of CgpS resulted in a significantly increased induction frequency of the CGP3 prophage. In contrast, a strain lacking the CGP3 prophage was not affected and displayed stable growth. In a bioinformatics approach, cgpS orthologs were identified primarily in actinobacterial genomes as well as several phage and prophage genomes. Sequence analysis of 618 orthologous proteins revealed a strong conservation of the secondary structure, supporting an ancient function of these xenogeneic silencers in phage-host interaction.
INTRODUCTION
Viral DNA, in the form of functional prophages or degenerated (cryptic) phage elements, is ubiquitously found in bacterial genomes and may constitute up to 20% of the host genome (1–3). The mosaic-like structure of bacterial genomes indicates that phage-mediated horizontal gene transfer is a pivotal driver of bacterial evolution (4). Recent studies demonstrated that these elements might contribute significantly to the fitness of their respective host by improving stress tolerance, antibiotic resistance, biofilm formation or virulence (5,6). Phage-mediated gene transfer may provide the cell with novel adaptive traits, improving the fitness of the receptor cell, but this does not occur without risks. The integration of selfish replicators, including transposable elements, integrative/conjugative elements (ICE) or phages, can lead to high transcriptional and translational costs or even cell death (7,8). Hence, bacteria possess a number of different systems that confer resistance to foreign genetic elements, e.g. CRISPR/Cas and restriction modification (R-M) systems (9,10).
However, to harness the adaptive potential of foreign DNA and enable its integration into the host regulatory circuitry, bacteria have evolved a rather mediative mechanism called xenogeneic silencing (XS) (11–13). This mechanism relies on the function of small nucleoid-associated proteins (NAPs) to target and inhibit the expression of foreign DNA, which is recognizable by its typically higher AT content in comparison to the host genome (1,14). The major role of XS proteins is the binding of foreign DNA elements and the inhibition of transcription by a complex formation of AT-rich DNA stretches causing either the occlusion or trapping of the RNA polymerase (15,16). Currently known XS proteins belong to one of four classes, consisting of H-NS-type proteins found in several proteobacteria (12,17), Lsr2-like proteins of the actinomycetes (18), MvaT of Pseudomonas species (16) and Rok of Bacillus subtilis (19).
To date, most studies have focused on host-encoded XS proteins acting as silencers of foreign DNA. However, it may also be of benefit for the foreign element to bring its own silencer protein to improve tolerance within the host cell. Here, we describe a novel prophage-encoded XS protein of the Lsr2-type in Corynebacterium glutamicum ATCC 13032. The genome of this important industrial amino acid producer contains three cryptic prophages (20,21). Whereas CGP1 and CGP2 are highly degenerated, CGP3 comprises almost 6% of the entire genome (187 kb) and is inducible in an SOS-dependent manner (22,23). Even under non-inducing conditions, spontaneous prophage induction (SPI) was observed, preceded by a spontaneous activation of the SOS response in >60% of cases (20,22,23). However, the precise regulatory control of CGP3 induction has not been studied thus far.
In this study, we demonstrate the essential role of a prophage-encoded NAP, which is a homolog to the mycobacterial Lsr2 protein and functions as a silencer of cryptic phage elements in C. glutamicum (CgpS, C. glutamicum prophage silencer). Genome-wide profiling of the CgpS–DNA interaction revealed its association with AT-rich DNA regions located primarily within prophage regions. Countersilencing of CgpS activity via the expression of its truncated oligomerization domain resulted in the induction of CGP3, causing cell death. A bioinformatics analysis revealed homologous proteins mainly in actinomycetes, but, interestingly, also in several phage and prophage genomes. These data demonstrate the importance of XS proteins for the tolerance of viral DNA and indicate that this mechanism is exploited by both the host and the virus.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Corynebacterium glutamicum ATCC 13032 was used as wild-type strain (24). E. coli DH5α was used as host for cloning procedures and cultivated in Lysogeny Broth (LB) medium or on agar plates at 37°C (25). For growth studies and fluorescence assays (e.g. preparation of cells for fluorescence microscopy), C.glutamicum cells were pre-cultivated in BHI (brain heart infusion, DifcoTM BHI, BD, Heidelberg, Germany) medium at 30°C for 6 h. This first preculture was used to inoculate an overnight culture in CGXII minimal medium (26) containing 2% (w/v) glucose and 30 mg·l−1 protocatechuat acid. The CGXII culture was finally used to inoculate the main culture in the same medium (CGXII with 2% (w/v) glucose) to a start OD600 of 1, unless specified otherwise. If necessary, 50 μg·ml−1 (E. coli) or 25 μg·ml−1 (C. glutamicum) kanamycin and/or 34 μg·ml−1 (E. coli) or 10 μg·ml−1 (C. glutamicum) chloramphenicol were added.
Recombinant DNA work
Plasmids and oligonucleotides used in this study are listed in Supplementary Table S2, respectively. Standard methods including PCR, DNA restriction and ligation, were performed according to established protocols (25). In some cases, Gibson assembly (27) was used for the constructions of plasmids. DNA sequencing and oligonucleotides synthesis were conducted by Eurofins MWG Operon (Ebersberg, Germany). The chromosomal integration of the Strep tagged cgpS gene variant was performed using the two-step homologous recombination method (28). The 500 bp up and downstream regions of cgpS were amplified using the oligonucleotides LF_cgpS_pK19_fw and LF_cgpS_rv and, accordingly, RF_cgpS_fw and RF_cgpS_pK19_rv. Amplification of the Strep-tagged cgpS gene was done by using the plasmid pAN6-cgpS-strep as template for the oligonucleotide pair cgpS_strep_fw and cgpS_strep_rv. The three resulting PCR products and the digested pK19mobsacB plasmid (with BamHI, EcoRI) were assembled using Gibson assembly (27). Correct integration into the cgpS locus was confirmed by sequencing of the colony PCR product with the oligonucleotides Cgps_indel-fw and CgpS_indel_rv.
Cultivation in the BioLector System
Growth experiments were performed predominantly in the BioLector® microcultivation system of m2p-labs (Aachen, Germany) as described by (29). Cultivation was performed in 48-well FlowerPlates (m2p labs, Germany) at 30°C and a shaking frequency of 1200 rpm. The cells were cultivated in 750 μl of CGXII minimal media with 2% (w/v) glucose containing different additives (e.g. Isopropyl β-D-1-thiogalactopyranoside (IPTG), MMC, kanamycin), as indicated. Measurements were taken at 15-min intervals.
DNA affinity chromatography with the promoter region of alpAC
The promoter region of alpAC was amplified by PCR with the oligonucleotides PalpAC-Biotin-Tag-fw and PalpAC rv (product size 516 bp). To flag the amplified product further PCRs were performed but with the Biotin-Primer (MWG Eurofins, Ebersberg, Germany) and the PalpAC rv. At least 220 pmol of the biotinylated products were purified by size exclusion chromatography with the usage of an 8 ml sepharose s400-HR column from GE Healthcare (Freiburg, Germany). A total of 5 mg of the M-280 Streptavidin Dynabeads® (Invitrogen, Carlsbad, CA, USA) were washed twice with the binding and wash (BW) buffer (10 mM Tris-HCl pH 7.5, 2 M NaCl), subsequently suspended in BW buffer containing biotinylated products and incubated for 1 h at room temperature. To eliminate unbound DNA fragments the beads were washed three times with the BW buffer and finally suspended in the binding and storage (BS) buffer (20 mM Tris-HCl pH 7.5, 1 mM EDTA, 10% (v/v) glycerin, 0.01% (v/v) Triton-X-100, 100 mM NaCl, 1 mM DTT). A total fo 500 ml of cells were grown in CGXII minimal media with glucose as carbon source (as described in bacterial strains and growth conditions) to an OD600 of ∼5. After the cells were harvested by centrifugation (20 min, 5300g and washed once with phosphate buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 20 mM Na2HPO4, 1.8 mM KH2PO4), cell pellets were suspended in BS buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell disruption was performed by five passages at 172 MPa through a French pressure cell (Heinemann, Schwaebisch Gmuend, Germany). The DNA binding reactions were set up with complete prepared crude extracts, the DNA-coupled beads and 500 μg of chromosomal DNA for 45 min at room temperature. After the binding reaction, beads were washed once with BS buffer, twice with BS buffer and 400 μg chromosomal DNA and, as a final washing step, again with BS buffer. The elution was fulfilled in two subsequent steps with BS buffer containing 2 M sodium chloride. After TCA precipitations (30) of the pooled elution fractions the samples were analyzed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (31). Identification of proteins was conducted by MALDI-ToF analysis as described in the section below.
Preparation of ChAP-Seq samples
Cells of the wild-type strain ATCC 13032 and the variant containing the Strep-tagged CgpS protein (WT::cgpS-strep) were first grown in BHI for 6 h and then 1 ml was used to inoculate minimal media cultures (CGXII with 2% (w/v) glucose). After cultivation overnight, these precultures were used to inoculate 500 ml of the same minimal medium, were grown to an OD600 5 to 6, and finally harvested by centrifugation (10 min, 11 325g at 4°C). After washing the cells with CGXII medium without (w/o) MOPS, the cells were resuspended in 10 ml MOPS-free CGXII containing 1% (v/v) formaldehyde. The fixation was conducted by incubation at room temperature for 20 min. Subsequently, glycine was added to a final concentration of 125 mM and the cells were incubated for further 5 min at room temperature. Then, the cells were washed twice with buffer A (100 mM Tris-HCl, pH 8.0, 1 mM EDTA) and resuspended in 10 ml buffer A supplemented with cOmplete Protease Inhibitor (Roche, Basel, Switzerland) and 5 mg RNase A. Cell disruption was performed as described in the DNA affinity chromatography section (five passages through a French Press cell). The chromosomal DNA of the lysates were sheared by sonication 3 × 30 s with a Branson sonifier 250 (Heinemann, Schwaebisch Gmuend, Germany) using a pulse length of 40% and an intensity of one to give an average fragment size of 200–1500 bp as confirmed by agarose gel electrophoresis. Cell debris was first removed by centrifugation at 5300g for 20 min and then centrifuged for 1 h at 150 000g both steps at 4°C. The supernatant was used for protein–DNA purification according to the standard Strep-tag® purification protocol (see below, protein purification). The pooled elution fractions were incubated overnight at 65°C, followed by a treatment with proteinase K (final concentration 400 mg·ml−1) for 3 h at 55°C. Finally, the DNA of the samples was purified by phenol–chloroform extraction (32), precipitated with ethanol, washed with 70% (v/v) ethanol, dried and resuspended in 50–100 μl ddH2O.
ChAP-Seq
The obtained DNA fragments of each sample (2 μg) were used for library preparation and indexing using the TruSeq DNA PCR-free sample preparation kit according to the manufacturer's instruction, yet omitting the DNA size selection steps (Illumina, Chesterford, UK). The resulting libraries were quantified using the KAPA library quant kit (Peqlab, Bonn, Germany) and normalized for pooling. Sequencing of pooled libraries was performed on a MiSeq (Illumina, San Diego, US) using paired-end sequencing with a read-length of 2 × 150 bases. Data analysis and base calling were accomplished with the Illumina instrument software and stored as fastq output files. The obtained sequencing data of each sample were imported into CLC Genomics Workbench (Version 7.5.1, Qiagen Aarhus A/S) for trimming and base quality filtering. The output was mapped to accession BX927147 as C. glutamicum reference genome (21). For peak detection the resulting mapping coverage of each sample was exported and imported into the in-house software Genome Data Viewer (unpublished). A peak was automatically annotated if the coverage of a region is above the 3-fold average of the averaged genome coverage. All peaks were inspected and confirmed manually.
qPCR
The relative amount of circular phage DNA was determined via quantitative PCR (qPCR). Therefore, C. glutamicum wild type cells containing empty pAN6 plasmid (control), pAN6-cgpS gene or pAN6-N-cgpS were grown in 48-well FlowerPlates containing CGXII minimal medium at 30°C and 900 rpm in a microtron (Infors-HT, Bottmingen, Switzerland). The overexpression of cgpS and the N-terminal part were induced with 150 μM IPTG (for control samples no IPTG was added). After 24 h, 750 μl of the cells were harvested and the DNA was extracted using the NucleoSpin microbial DNA Kit (Macherey Nagel, Dueren, Germany) and DNA concentration was quantified using a nanophotometer (Implen, München, Germany). Each sample contained 1 μg total DNA as a template. For the reaction an innuMIX qPCR MasterMix SyGreen (Analytic Jena, Jena, Germany) and a qTOWER 2.2 (Analytic Jena) was used. The reaction protocol was divided into two parts (i) polymerase chain reaction (PCR) ((a) 3 min preincubation at 95°C, (b) 5 s denaturation at 95°C, (c) 25 s elongation at 62°C, 40x repetition of step (b) to (c)) and a (ii) melting curve analysis (ΔT = 1°C/6 s). The PCR product size using oligonucleotides belonging to the circular phage product is 150 bp (listed in Supplementary Table S2). As reference gene ddh was used with the oligonucleotides listed in Supplementary Table S2 resulting in a 150 bp product. For data analysis the qPCR software qPCR 3.1 (Analytik Jena) and the Livak method were used (33) to determine the 2−ΔΔCt based on the measured CT-values.
DNA microarrays
For a comparative transcriptome analysis of C. glutamicum ATCC 13032/pAN6 with cells carrying the pAN6-N-cgpS- (used for countersilencing) were cultivated in CGXII with 2% (w/v) glucose and 100 μM IPTG as described in bacterial strains and growth conditions. The preparation of labeled cDNA and DNA microarray analysis was performed as described previously (34). Array data were deposited in the GEO database (ncbi.nlm.nih.gov/geo) under accession number GSE80674.
Cultivation and perfusion in microfluidic device
For single-cell analysis an in-house developed microfluidic platform was used (22,35–37). Phase-contrast and fluorescence time-lapse imaging was performed at 6 min intervals. Medium was supplied continuously to ensure stable and constant environmental conditions. CGXII minimal medium with 2% (w/v) glucose and 25 μg·ml−1 kanamycin was infused at a rate of 300 nl·min−1 using a high-precision syringe pump (neMESYS, Cetoni GmbH, Korbussen, Germany). For the expression of the N-terminal part of CgpS 150 μM IPTG were added to the medium. A constant cultivation temperature of 30°C was ensured (PeCon GmbH, Erbach, Germany). The cells were cultivated for 16 h.
Fluorescence microscopy
The cultivations were done as described in bacterial strains and growth conditions. After 6 h of cultivation, 1–3 μl were pipetted on a microscope slide coated with a thin 1% (w/v) agarose layer that was based on tris-acetate buffer. To stain the DNA with the Hoechst Dye, 33 342 1 ml cells were harvested (5300g, 5 min), subsequently resuspended in PBS buffer containing 100 ng·ml−1 Hoechst 33342 and incubated at room temperature for 20 min. Images were taken on an AxioImager M2 (Zeiss, Oberkochen, Germany) equipped with a Zeiss AxioCam MRm camera. Fluorescence was monitored with the filter set 46 HE YFP for eYFP, 63 HE filter was used for mCherry fluorescence and Hoechst fluorescence was examined with the filter set 49. An EC Plan-Neofluar 100x/1.3 Oil Ph3 objective was used. Images were acquired and analyzed with the AxioVision 4.8 software (Carl Zeiss).
Protein purification
CgpS tagged C-terminal with a Strep-tag® was heterologously produced in E. coli BL21 (DE3). Cells were grown to an OD600 of 0.4 at 37°C. Upon induction with 50 μM IPTG the cultivation was continued at 16°C overnight. Cells were harvested by centrifugation at 5300g and 4°C for 10 min and resuspended in buffer B (250 mM NaCl, 50 mM Tris-HCl, pH 7.5). Cell disruption was performed by two passages through a French pressure cell at 172 MPa. Cell debris was removed by centrifugation at 20 min, 5300g and 4°C, followed by an ultracentrifugation (60 min, 229 000g, 4°C). The supernatant was applied to an equilibrated 1 ml Strep-Tactin®-Sepharose® (IBA, Göttingen, Germany) column. It was subsequently washed with 10 ml buffer B and the protein was eluted with 10 ml buffer B containing 1 mM d-desthiobiotin (Sigma Aldrich).
Electrophoretic mobility shift assays (EMSA)
EMSA studies of CgpS and selected DNA regions identified by ChAP-Seq were performed with selected regions (500 bp fragments, for oligo sequences see Supplementary Table S3). The corresponding regions were amplified by PCR and purified by using the PCR clean-up Kit of Macherey Nagel (Dueren, Germany). The promoter region of gntK was used as control fragment (560 bp). A total of 90 ng DNA per lane were incubated with different concentrations (1 μM and 2 μM) of purified CgpS protein for 20 min in EMSA buffer (250 mM Tris-HCl pH 7.5, 25 mM MgCl2, 200 mM KCl, 25% (v/v) glycerol). Subsequently, samples were loaded onto a native 10% polyacrylamide gel (TBE-based, TBE (89 mM Tris base, 89 mM boric acid, 2 mM Na2EDTA, loading dye: 0.01% (w/v) xylene cyanol dye, 0.01% (w/v) bromophenol blue dye, 20% (v/v) glycerol, 1x TBE). The DNA was stained with SYBR Green I (Sigma Aldrich, St. Louis, MO, USA).
Protein pull down and MALDI-TOF analysis
C. glutamicum cells containing the plasmids pAN6, pAN6-cgpS-strep or pAN6-N-cgpS-strep were cultivated as described in bacterial strains and growth conditions. The cultures were grown in 500 ml CGXII with 2% (w/v) glucose to an OD600 of 5 and subsequently induced with 150 μM IPTG for further 4 h. The cells were harvested (5300g, 20 min, 4°C), washed in buffer B (see protein purification) and disrupted as descripted in the DNA affinity chromatography section. Purification was performed as described in the section above. The eluted fractions were analyzed by SDS-PAGE (31) using a 4–20% Mini-PROTEAN® gradient gel (Bio Rad, Munich, Germany). The gels were stained with a Coomassie dye based RAPIDstain solution (G-Biosciences, St. Louis, MO, USA). MALDI-TOF-MS measurements were performed with an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) for the identification of the proteins as described (38).
Homology search
BLAST ‘nr’ database (ver. February 2015) was downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). CgpS amino acid sequence was extracted from the GenBank file Corynebacterium glutamicum ATCC 13032, accession: NC_006958.1 and locus_tag: cg1966. A PSI-BLAST ((39)) search with CgpS sequence as the query was executed against the ncbi nr database. The e-value threshold was set to 0.005, the number of iteration was not limited and the search iteration was performed until it converged. A total of 5230 (1920 unique) homologous hits were achieved from which 618 could be allocated to a particular bacterial species or a phage. Sequence global identity was calculated by pairwise comparison between the CgpS sequence with all 618 PSI-BLAST hits using the Needleman–Wunsch algorithm (40) implemented in the EMBOSS package (41) needle.
Secondary structure prediction
The amino acid sequence of the CgpS protein and the sequences of the 618 homologous hits were used to predict the secondary structure by psipred (42). The visualization of the psipred output was done in R (43).
Statistics and visualization
All statistical analysis and data visualization from the bioinformatic section was performed in R (43).
RESULTS
A small nucleoid-associated protein encoded by a cryptic prophage element
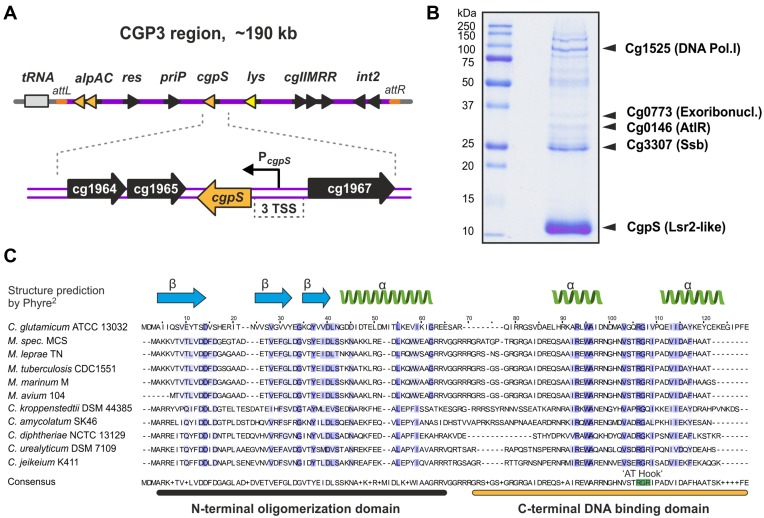
To decipher the control of prophage induction and activation of cryptic elements in C. glutamicum ATCC 13032, we performed DNA affinity chromatography with the promoter of the early phage operon alpAC using the crude extract of log-phase cells grown in glucose minimal medium ((34), Figure 1A). SDS-Page analysis of the proteins bound to the alpAC promoter revealed a prominent band corresponding to the 13.4 kDa protein Cg1966 encoded within the CGP3 prophage region (Figure 1B). In particular, the C-terminal domain of Cg1966 shares significant sequence similarity with the nucleoid-associated protein Lsr2 of Mycobacterium tuberculosis (Supplementary Figure S1). This domain corresponds to the DNA binding domain of Lsr2 (IPR024412), which was previously found to bind AT-rich DNA via an AT-hook motif and functions as a silencer of xenogeneic DNA (44,45). Based on the data described in the following sections, we renamed Cg1966 as CgpS (Corynebacterium glutamicum prophage silencer). Secondary structure predictions of CgpS as well as of CgpS homologs suggest a significant structural similarity with Lsr2 and reveal the presence of an AT-hook-like motif ‘RGI’ between the two predicted C-terminal alpha helices (Figure 1C) (18,45).
Figure 1.
CgpS is a prophage-encoded nucleoid-associated protein in C. glutamicum ATCC 13032. (A) Genomic organization of the CGP3 prophage region containing the cgpS gene in C. glutamicum strain ATCC 13032. (B) DNA affinity chromatography was used to identify putative regulatory proteins bound to the early phage promoter of alpAC (34). Protein samples were separated by SDS-PAGE, and enriched proteins were identified using MALD-ToF analysis. The nucleoid-associated protein CgpS (Cg1966) was detected as a prominent band with a molecular mass of 13.4 kDa. (C) Amino acid sequences of CgpS, five related proteins of Corynebacteria and five Lsr2 homologs of Mycobacteria were used for a multiple sequence alignment conducted using the Clustal Omega platform (73). The predicted domain organization is depicted above the sequence alignment. The Blosum62 identity score is highlighted in blue and matches between 21–26% across the species. The secondary structure of CgpS was predicted with 99.7% confidence of 54 residues (46% of CgpS sequence) by Phyre2 (74). The motif ‘RGI,’ which is similar to the AT hook motif ‘RGR’ of Lsr2 and H-NS (44,59), was identified between two predicted alpha helices.
CgpS functions as a silencer of CGP3 activity
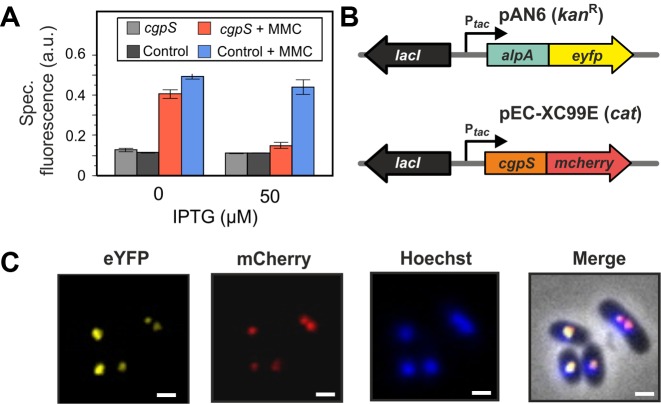
To study the impact of cgpS expression on the activity of the CGP3 prophage, we overexpressed cgpS in a strain carrying a reporter construct (WT-Plys-eyfp) indicative for the activation of CGP3 by the production of the yellow fluorescent protein eYFP under the control of a phage promoter (22). Upon induction with mitomycin C, the control strain carrying the empty plasmid displayed increased reporter activity. Consistent with our assumption, overexpression of cgpS reduced the reporter output to nearly the background level (Figure 2A).
Figure 2.
CgpS functions as a silencer of CGP3 prophage activity. (A) Silencing of CGP3 induction. The prophage reporter strain C. glutamicum ATCC 13032::Plys-eyfp containing the cgpS overexpression plasmid pAN6-cgpS-Strep was cultivated in CGXII minimal medium in the presence or absence of IPTG (50 μM) and MMC (600 μM). The prophage reporter strain carrying the empty plasmid pAN6 served as a control. EYFP fluorescence was measured after 20 h of microplate cultivation. (B and C) CgpS is located in the nucleoid and displays colocalization with the phage adaptor protein AlpA (34). For co-localization studies, a C-terminal CgpS-mCherry fusion and a C-terminal fusion of the prophage adaptor protein AlpA to eYFP (pEC-XC99E) were analyzed. Both gene fusions were under control of the inducible tac promoter. DNA was stained with Hoechst Dye 33342. Fluorescence images were taken after 2.5 h of cultivation in CGXII with 150 μM IPTG. Scale bar, 2 μm.
To study the intracellular localization of CgpS in C. glutamicum cells, we C-terminally fused this protein to mCherry and analyzed its distribution via fluorescence microscopy. As shown by Hoechst staining, this NAP appeared associated with the nucleoid but formed distinct foci in the cell (Figure 2B and C). Remarkably, CgpS-mCherry foci co-localized with foci of an AlpA-eYFP fusion that was previously described as a CGP3 DNA adaptor protein binding to the alpAC promoter region (34) (Figure 2C). The functionality of this CgpS-mCherry fusion was confirmed by the counteraction of CGP3 activation upon addition of MMC (Supplementary Figure S2).
Genome-wide binding profile of CgpS
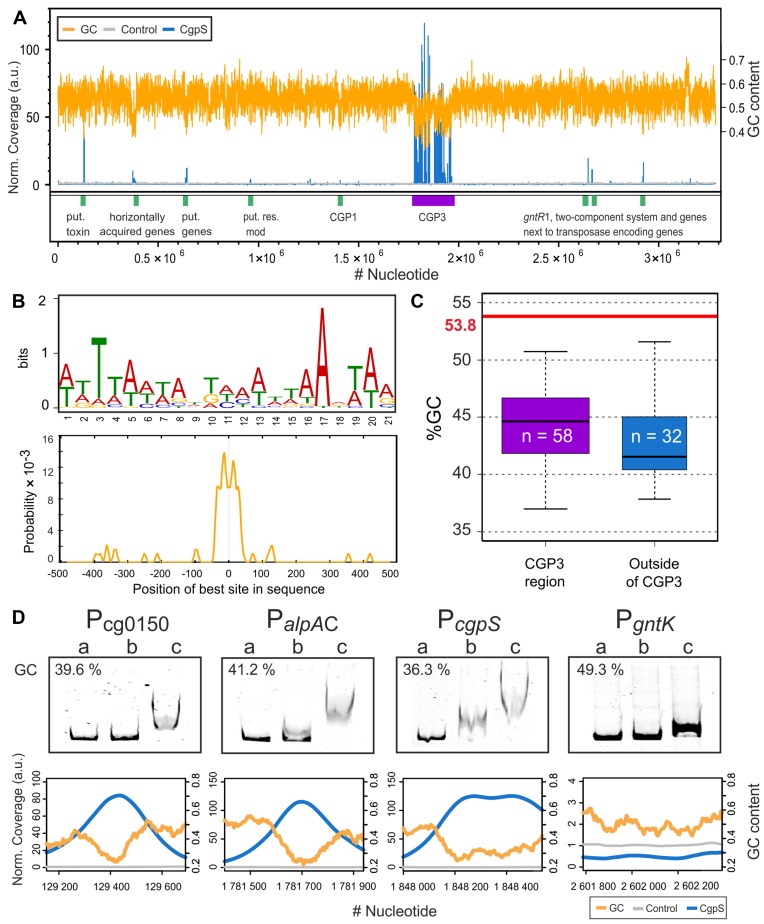
The data of the co-localization experiments suggest binding of CgpS to the CGP3 prophage region. In the following, the genome-wide binding profile was analyzed by combining affinity chromatography purification of crosslinked CgpS–DNA complexes followed by sequencing of associated DNA (ChAP-Seq). For this purpose, we replaced the native cgpS gene in the genome of ATCC 13032 with cgpS-Strep encoding a C-terminal Strep-tagged CgpS variant. This analysis revealed that CgpS associates with 1.5% of the ATCC 13032 genome and with ∼20.5% of the cryptic CGP3 prophage region (Supplementary Figure S3). In total, 90 peaks were detected, 58 of which were within and 32 were located outside the CGP3 prophage (Figure 3A, Supplementary Table S4). The majority of the peak maxima were located within promoter regions (60%), but CgpS binding was also observed within genes (31%) or intergenic regions (9%) (Supplementary Figure S4B and C). To deduce a binding motif of CgpS, sequences of the 90 peaks (Supplementary Table S4) were extracted and analyzed using the MEME-ChIP software platform (46). A 21-bp long AT-rich motif was predicted, which was present in 87 of 90 sequences (Figure 3B). The occurrences of the found DNA binding sites were validated using a FIMO search (Find Individual Motif Occurrences, (47)) in the ATCC 13032 genome, which revealed significant matches (>75%) of the predicted and experimentally identified CgpS binding sites (Supplementary Figure S5). Remarkably, the %GC content of the 90 peak sequences is considerably lower than the average GC content of the ATCC 13032 strain, indicating the preferred binding of CgpS to AT-rich DNA (Figure 3C). Moreover, the GC contents of the CgpS bound regions within the prophage revealed no significant differences from that of the regions bound outside the prophage (Figure 3C).
Figure 3.
Genome-wide profiling of CgpS-binding using ChAP-Seq. (A) Genome wide binding profile of CgpS obtained by ChAP-Seq experiments. Enriched DNA regions purified in complex with CgpS (blue) or with the control sample (gray, empty vector control) were normalized to their mean and plotted against the ATCC 13032 genome. For calculation of the GC content (orange) and the coverages, a step size of 50 bp and a window size of 500 bp were used. Regions with high coverage are indicated by green (outside of CGP3 region) and purple boxes (CGP3 region). A total of 90 peaks were detected by applying a threshold of 3-fold of the mean coverage (SD of the control sample = ± 0.28 (a.u.)). (B) Sequences of the 90 peaks were used to derive a DNA binding motif using MEME-ChIP (75). A 21-bp long AT-rich motif was identified (E-value = 1.1 × 10−58), and the highest probability was centered approximately ±38 bp around the maximum peak position. (C) A total of 58 detected signals were within and 32 were outside of the CGP3 region. GC contents of the peak sequences were calculated and compared to the average GC content of ATCC 13032 (21). (D) Electrophoretic mobility shift assays (EMSAs) were performed with promoter regions of the putative target genes. DNA fragments (around 500 bp) covering the promoter regions of cg0150, alpAC, cgpS and gntK (negative control) were incubated without (lane a) or with (lane b: 1 μM; c: 2 μM) purified CgpS protein. The corresponding ChAP-seq results of the particular regions are shown below the EMSA pictures (step size: 5 and windows size: 50).
Most of the identified CgpS targets were located within the CGP3 prophage and code for hypothetical proteins. The two strongest signals were found within transposase-encoding genes (cg1950-cg1951) and in the promoter region of cgpS itself, indicating a negative autoregulation similar to that of H-NS (48). Other potential target genes encode the actin-like protein and the corresponding adaptor protein (alpAC, cg1890 and cg1891 (34)), a resolvase (cg1929), a prophage primase (cg1959), a putative phage lysin (cg1974) and a phage integrase (cg2071), which are spread across the cryptic prophage element. In addition to regions within CGP3, CgpS target sites are located in the low GC island 1 (LCG1), in the cryptic phage element CGP1, or proximal to transposases encoding genes. Furthermore, promoter regions of genes coding for R-M systems (Pcg1028 and PcglIM, (Pcg1996)) are also bound by CgpS, which in several studies were shown to be transferred horizontally (49–52). A considerably high peak was observed for the promoter region of cg0150 that encodes a putative regulatory protein or toxin possessing a predicted fido domain (IPR003812).
The binding profile obtained by the ChAP-Seq analysis was validated by EMSAs (Supplementary Figure S6). For this purpose, CgpS was purified as a C-terminal Strep-tag fusion and incubated with DNA fragments covering selected putative CgpS binding sites as identified by ChAP-Seq (Figure 3D and Supplementary Figure S6). This in vitro approach confirmed the binding of CgpS for all selected target regions (including the promoters of cg0150, alpAC and cgpS itself) in comparison to the control fragment (gntK promoter) (Figure 3D). Overall, these data are consistent with CgpS acting as a xenogeneic silencer by targeting AT-rich DNA regions, several of which have likely been acquired by HGT.
Countersilencing of CgpS activity
Several independent efforts to inactivate the cgpS gene failed (data not shown), suggesting that cgpS represents an essential gene for C. glutamicum ATCC 13032. However, previous studies revealed that deletions of all three cryptic phage elements, including the cgpS gene, are possible and do not lead to a significant growth defect of the particular strain (53). In fact, trials to construct an in-frame deletion of cgpS resulted in the isolation of strains lacking large parts of the CGP3 prophage, indicating that the essentiality of cgpS is a consequence of the de-repression of toxic phage genes in the absence of CgpS.
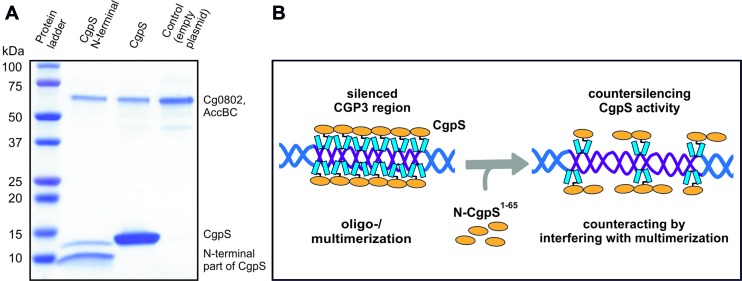
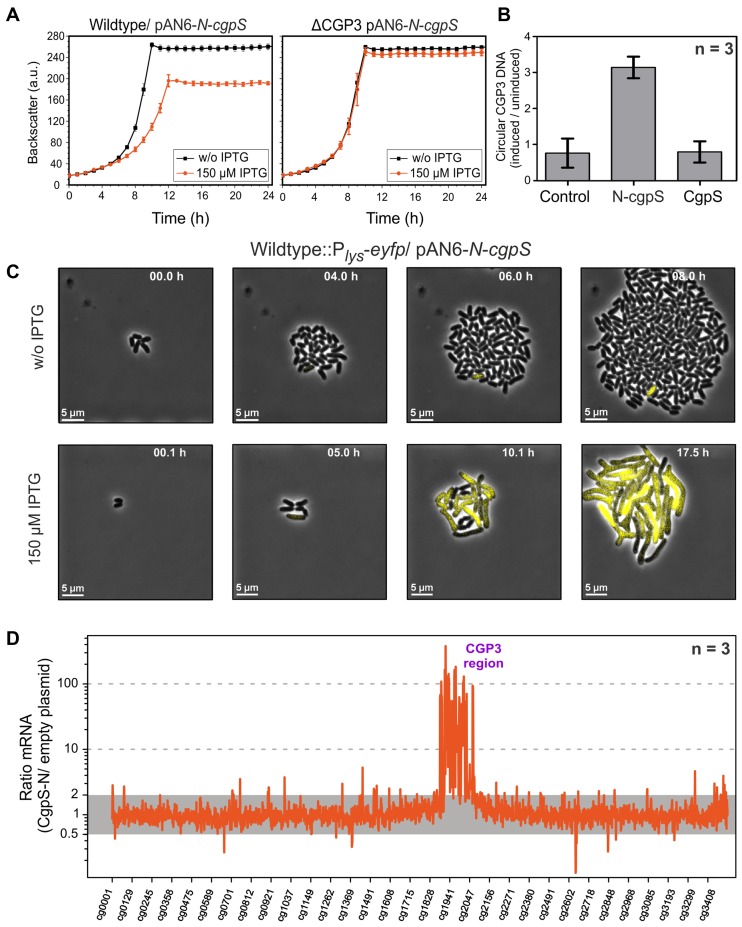
For the conditional inactivation of CgpS, we adapted a countersilencing approach similar to the H-NST system described by Williamson and Free (54). This protein was reported as a truncated H-NS derivative that antagonizes H-NS function by interfering with the multimerization of H-NS. Co-purification assays with the N-terminal domain of CgpS confirmed the interaction of this truncated variant with the full-length protein (Figure 4A). Based on previous data and the H-NST mechanism (Figure 4B), we constructed the pAN6-N-cgpS plasmid to overproduce a truncated variant of CgpS (amino acids 1–65) under the control of Ptac. Homology studies indicated that amino acids 1–65 cover the domain of CgpS required for the oligomerization of this NAP. Remarkably, production of the truncated CgpS-N domain in the wild-type strain resulted in a significant growth defect, whereas no impact on growth was observed in a strain lacking the CGP3 prophage (Figure 5A). This finding was supported by single-cell analysis of a strain containing a prophage reporter construct (Plys-eyfp) (22) and the countersilencing construct pAN6-N-cgpS. Production of the N-terminal domain of CgpS led to a strong increase in fluorescence accompanied by growth arrest and a branched cell morphology (Figure 5C, Video S1 and S2). Quantitative real-time PCR revealed a 3-fold increase in the level of circular CGP3 DNA in comparison to uninduced cells, which is consistent with the induction of this cryptic prophage (Figure 5B) (20).
Figure 4.
Principle of CgpS countersilencing. (A) Protein pulldown assays were conducted with C. glutamicum cells containing the plasmids pAN6-cgpS-Strep and pAN6-N-cgpS-Strep, which were used for the overexpression of Strep-tagged cgpS and its truncated variant. The pulldown of full-length CgpS by the truncated variant (aa 1–65) confirmed the N-terminal part of CgpS as oligo-/multimerization domain of CgpS proteins. (B) Model of CgpS silencing and countersilencing. The expression of genes depends on the accessibility of their particular promoter regions to the RNA polymerase. AT-rich regions such as CGP3 are bound by CgpS, likely resulting in an oligomerization of the CgpS protein (18,76,77), thereby interfering with the binding of RNA polymerase. The production of an N-terminal truncated CgpS variant interferes with the silencing ability of the native protein by binding to its N-terminal oligomerization domain while lacking the site for DNA binding.
Figure 5.
Countersilencing by overexpression of the N-terminal oligomerization domain of CgpS. (A) Growth studies of the wild-type ATCC 13032 and a strain lacking the CGP3 prophage (ΔCGP3), both carrying the pAN6-N-cgpS overexpression plasmid. Strains were grown in CGXII minimal medium with and without IPTG (150 μM). Data represent average values and standard deviations of three biological replicates. (B) Relative quantification of CGP3 excision using qPCR (20). The N-terminal domain of CgpS and the full-length protein were overproduced as described in (A). Samples for qPCR analysis were taken after 24 h. The relative amounts of circular phage DNA of induced and uninduced samples were compared. As a control wild-type cells with the empty plasmid were used. Data represent average values and standard deviations of three biological replicates. (C) Time-lapse fluorescence microscopy of the C. glutamicum prophage reporter strain (ATCC 13032::Plys-eyfp) carrying the pAN6-N-cgpS. Cells were grown in PDMS-based microfluidic chip devices under continuous supply of CGXII with 25 μg·ml−1 kanamycin and with or without 150 μM IPTG to induce the expression of the truncated CpgS variant (36) (300 nl·min−1) (Video S1 and S2). (D) Comparative transcriptome analysis of C. glutamicum ATCC 13032 containing the overexpression plasmid pAN6-N-cgpS and a strain containing the empty vector control was performed as described in the Materials and Methods section.
To monitor the impact of countersilencing CgpS activity on gene expression, we performed a comparative transcriptome analysis (Figure 5D, Supplementary Table S5). More than 194 genes were affected, 12 of which exhibited a reduced mRNA level (mRNA ratio ≤ 0.5, P-value < 0.05), and 182 genes were upregulated (mRNA ratio ≥ 2, P-value < 0.05). The majority of upregulated genes (148) were genes of the prophage CGP3. Additional genes that displayed an increased mRNA level were the ferritin gene (ftn, cg2782) and cg1517 of the CGP1 prophage (Supplementary Table S5), both of which were also identified as putative CgpS targets by ChAP-Seq. Together, these data demonstrate that CgpS is an essential NAP due to its function as a silencer of cryptic phage elements inC. glutamicum.
CgpS homologs are found in actinomycetes and their phages
Our data support a function for CgpS as a xenogeneic silencer that binds to AT-rich DNA similar to the Lsr2 of M. tuberculosis as well as the H-NS of E. coli. This is underlined by the fact that both proteins, Lsr2 and CgpS, are able to complement the phenotype of an hns mutant strain ((18), Supplementary Figure S7). These findings highlight the conserved mechanism of a highly diverse set of proteins.
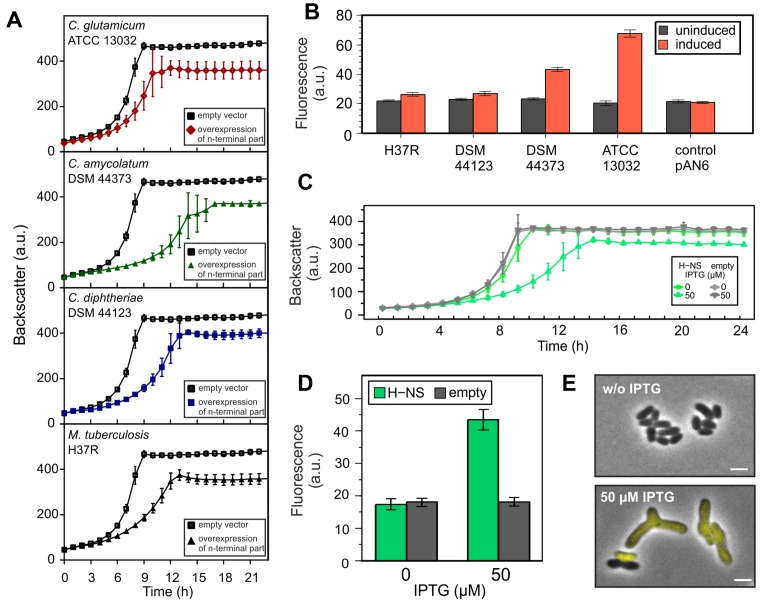
In the following, we overexpressed the N-terminal oligomerization domains of CgpS orthologs from Corynebacterium amycolatum DSM 44737 (CORAM0001_2081) and Corynebacterium diphtheria DSM 44123 (CDC7B_2240) and the Lsr2 from M. tuberculosis H37R (Rv3597c; Lsr2) (Figure 6A and B). Whereas the production of the oligomerization domain strongly affected cellular growth in all cases (Figure 6A), only the N-terminal domain of the ortholog of C. amycolatum (DSM 44737) led to a significant induction of CGP3 (Figure 6B). No significant reporter output was observed with production of the truncated orthologs of C. diphtheria or M. tuberculosis, suggesting a high level of plasticity within this family of xenogeneic silencers (Figure 6B).
Figure 6.
Impact of H-NS and Lsr2/Lsr2-like truncated variants on CGP3 prophage induction. (A and B) Impact of truncated CgpS orthologs on the growth and CGP3 prophage activity using the reporter strain ATCC 13032::Plys-eyfp. Shown is the cultivation in microtiter plates in CGXII minimal media with 25 μg·ml−1 kanamycin and 150 μM IPTG. As a control the reporter strain containing the empty plasmid was used. In (B), the fluorescence output after 20 h is shown in comparison to the uninduced samples. (C and D) Growth experiments were performed with WT::Plys-eyfp cells carrying hns on the overexpression plasmid pAN6 under the conditions described in (A). Expression of hns was induced with 50 μM IPTG. Fluorescence output of the prophage reporter after 20 h is shown in (D). (E) After 24 h, fluorescence images were taken of cells (of C and D) placed on agar pads. Scale bar is 2 μm. Data represent average values and standard deviations of three biological replicates.
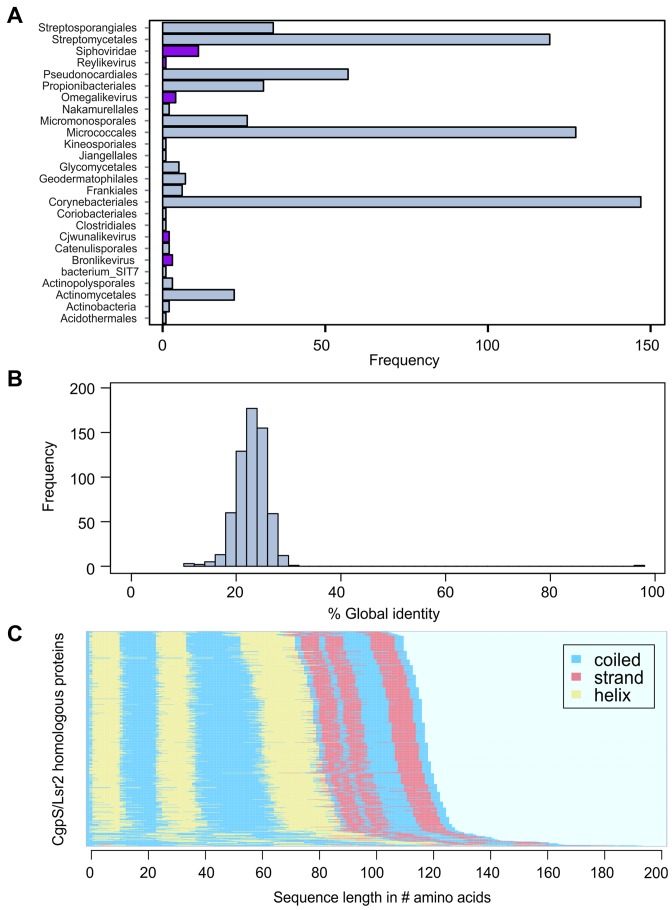
Furthermore, we used a bioinformatics approach to obtain a more general overview of the distribution of CgpS orthologous proteins. For this purpose, a PSI-BLAST (Position-Specific Iterated BLAST) search was performed on CgpS and resulted in 5230 hits, of which 1920 protein sequences were unique (threshold e-value ≤ 0.005). Of these, 98.3% were found in the domain of bacteria and 1.7% in phages, mostly belonging to the Siphoviridae (Figure 7A, Supplementary Table S6). Of 302 bacterial genomes containing prophage regions predicted by PhiSpy (55), 22 contain cgpS orthologs (Supplementary Table S6). The remaining 280 hits were found outside of any predicted prophage region. Moreover, secondary structure predictions were performed for 618 unique sequences, which were clearly assigned to bacterial or phage species, exhibiting high resemblances. The structural similarity suggests a common function, although the identity of the amino acid sequences is low (∼23%) (Figure 7B, C and Supplementary Figure S10).
Figure 7.
Orthologous sequences of the CgpS protein in actinomycetes and their phages. (A) Bar chart depicting the frequency of orthologous hits (n = 618) as predicted by PSI-BLAST (39) (e-value ≤ 0.005) across several orders of the phylum Actinobacteria and phages as annotated in the NCBI database (http://www.ncbi.nlm.nih.gov/). Orthologous found in phages are highlighted in purple. (B) Histogram of pairwise global identities between the CgpS amino acid sequence and its orthologous counterparts. The distribution reveals an overall low similarity (mean x = 23.07 and standard deviation σ = 4.05) to the orthologous sequences. Global identity was calculated using the Needleman–Wunsch (40) algorithm from the EMBOSS package (41). (C) Secondary structure prediction calculated by psipred (42) shows conserved protein structure for CgpS and the orthologous amino acid sequences. The corresponding secondary structure of each sequence was ordered in the direction from C-Terminus to N-terminus. Predicted coiled structures are shown in blue, strand regions in red and helices are colored in yellow.
XS exclusion hypothesis
A recent bioinformatics study on the distribution of XS genes revealed that members of the same family can appear within a particular species but that members of different families are never found together (56). To test the proposed exclusion mechanism, we expressed the hns gene from E. coli MG1655 in a C. glutamicum ATCC 13032 strain containing the prophage reporter (::Plys-eyfp). As expected, the overexpression of hns caused a severe growth defect, coinciding with a highly increased output of the prophage reporter (Figure 6C and D). The effect of hns overexpression was comparable to the countersilencing of CgpS activity with the production of a truncated CgpS variant (Figure 6E). When hns was expressed in a ΔCGP3 background the effect on growth was only moderate (Supplementary Figure S8). However, hns expression still negatively affected the growth of the CGP3 mutant strain which can likely be explained by unspecific binding and interference of H-NS at other genomic regions. These findings are in agreement with the hypothesis that different XS proteins interfere at AT-rich DNA regions, leading to a disruption of silencing complexes and thereby to an activation of foreign DNA elements. Nevertheless, in some cases the scenario is clearly more complex, as illustrated by the finding that the expression of cgpS in the E. coli wild-type strain was not able to counteract H-NS expression at the bgl operon (Supplementary Figure S9).
DISCUSSION
CgpS functions as a silencer of cryptic phage elements
In this study, we identified the prophage-encoded XS protein CgpS that inherits an essential role as a silencer of cryptic prophages in C. glutamicum. Genome-wide profiling of CgpS binding sites reveals an association of this protein to AT-rich DNA stretches primarily located within horizontally acquired genomic islands and shows a remarkable accumulation of binding sites within the large and cryptic CGP3 prophage. Countersilencing of CgpS activity by overproduction of its N-terminal oligomerization domain resulted in a strong increase in CGP3 activity leading to cell death. Furthermore, several CgpS binding sites were identified outside the CGP3 region, and the essentiality of the cgpS gene was attributed to the presence of the CGP3 prophage. This is consistent with the finding that the cgpS gene is located on the CGP3 island, suggesting that evolution favored a physical association between this XS and its main target.
Sequence analysis of CgpS revealed a low sequence identity (27%, Supplementary Figure S1) with the mycobacterial Lsr2 protein that was described in previous studies as an H-NS-like protein targeting AT-rich sequences in M. tuberculosis (18). Both XS proteins, Lsr2 and CgpS, complemented the bgl-based phenotype (57) of an Escherichia coli Δhns strain, supporting the overall analogous functions of these XS proteins (Supplementary Figure S7) (18). Whereas both lsr2 and cgpS are essential for viability in their native hosts, E. coli hns mutant strains are viable although exhibiting severe growth defects (58). Salmonella Typhimurium null mutants of hns are not viable unless mutations in rpoS (general stress response) or phoP (virulence gene regulator) counteract this deletion (12). Because the presence and diversity of phage elements contributes to major strain-specific differences within a bacterial species, our study illustrates that the essentiality of XS genes is highly dependent on the particular strain background. The C. glutamicum strain MB001, cured of all prophage regions as well as the cgpS gene located on prophage CGP3, displays wild-type-like growth behavior (53).
CgpS binds AT-rich xenogeneic DNA regions
Secondary structure predictions of CgpS-related proteins evince two α-helices flanking an ‘RGI’ motif (Figures 1C and 7C). This motif resembles the prokaryotic AT-hook motif ‘Q/RGR’ found in H-NS and Lsr2 and may also be responsible for the binding of AT-rich DNA as a general rule for XS functioning (44,59). A certain plasticity of the AT-hook motif is supported by experiments with AT-hook muteins of H-NS and Lsr2, showing that the exchange of a single arginine residue to an alanine reduces DNA binding but does not completely abolish it (59). Moreover, another member of the H-NS family, the Ler protein, has a hydrophobic amino acid (‘VGR’ motif) instead of an arginine at this position (60).
However, significant differences were observed for the number of target genes affected by the binding of the particular XS proteins. ChIP-on-Chip analysis revealed a direct influence of S. Typhimurium H-NS on the expression of more than 740 ORFs (12,61), and the binding of Lsr2 affected more than 800 regions within the M. tuberculosis genome and >900 in Mycobacterium smegmatis (45). ChAP-Seq profiling of CgpS binding, however, yielded only 90 potential target regions. Typical for XS function, an AT-rich DNA motif was derived from the ChAP-Seq results, which clusters at a high density within the CGP3 prophage region (Supplementary Figure S5). In general, promoter regions are more often bound by CgpS than genes or intergenic regions (Supplementary Figure S4), which is not surprising because promoter regions usually possess a higher AT content (62,63). CgpS targets outside the CGP3 region show a similar or lower GC content (Figure 3C) but less altered expression levels, and this may suggest the importance of motif density for XS function. Here, a variation of the AT-hook motif likely represents a mechanism to adjust the binding behavior of the XS protein to meet the needs of a particular host species.
In addition to CGP3 as a main CgpS target, further targets were identified which were also likely acquired by horizontal gene transfer, such as the LCG1 island, the cryptic prophage CGP1 (21), R-M systems, transposases and also regulatory proteins such as putative transcriptional regulators (Cg0725, Cg1340, Cg2426), the gluconate-responsive repressor GntR1 (Cg2783) (64) and an operon encoding the two-component system CgtSR6 (Cg3060) (Supplementary Table S4). Several previous studies reported similar target genes or regions for H-NS, Lsr2 and MvaT, demonstrating the convergent evolution of XS in bacterial species (12,47,61,65).
Overall, more that 80% of CgpS-bound regions also exhibited a more than 2-fold altered expression level under countersilencing conditions (Figure 5D) confirming the postulated silencing effect of CgpS. Several potential targets outside of the CGP3 region, however, showed only a moderate impact on the expression level suggesting a more complex regulatory scheme at the corresponding promoter regions. Therefore, the role of CgpS for the control of these potential targets, including, e.g. the gntR1 gene or the cgtSR6 operon, remains to be elucidated in further studies.
How to overcome CgpS silencing?
Several different mechanisms were described to counteract H-NS-mediated silencing, including structural interference with H-NS-bound nucleoids by transcription factors, temperature or osmolarity effects, and the binding of alternative sigma factors or other NAPs preventing multimerization of the XS protein (11,66,67). To interfere with CgpS XS activity, we produced a truncated part of the native protein covering the N-terminal oligomerization domain of CgpS (Figure 5). This overcomes the problem of cgpS being essential in the presence of CGP3 and was inspired by the study of Williamson and Free, who described the antagonistic function of a truncated H-NS variant found in an enteropathogenic E. coli strain (54). As expected, production of the N-terminal CgpS domain resulted in strong activation of CGP3, leading to cell death.
In recent studies we described the spontaneous induction of the CGP3 prophage occuring in the absence of an external trigger (20,22,23). Single-cell analysis demonstrated that a considerable fraction of this SPI is preceded by an activation of the SOS response, which is likely the result of spontaneous DNA damage during replication (68,69). However, these studies also highlighted a certain (>30%) fraction of SOS-independent SPI, suggesting that other factors influence this common phenomenon of bacterial populations (5). The present study shows the sensitive reaction of C. glutamicum cells to the downregulation of CgpS activity (Video S2). It is therefore interesting to determine whether cells can adjust the level of XS proteins to manipulate the frequency of SPI according to their particular requirements.
CgpS homologs in phage genomes
Sequence analysis revealed the presence of CgpS/Lsr2 homologs in phage and prophage genomes displaying a low sequence identity but highly conserved secondary structure prediction (Figure 7). This finding is not surprising because bacterial evolution has been shaped by a tight interaction with bacteriophages. For the integration of viral DNA into the host genome, both the bacterium and phage benefit from tolerance and a smooth integration into the host genetic circuitry. Because the activation of silent prophages or mobile elements often causes serious detrimental effects to host cells (11,70,71), the stringent control of xenogeneic elements is required.
Several examples of XS proteins involved in the control of mobile elements or phages have been described in the recent literature, including H-NS of S. Typhimurium (12), Rok from B. subtilis (19) and MvaT from P. aeruginosa (72). Their corresponding genes, however, are all located on the host chromosome and are characterized as a type of immunity system protecting hosts against foreign DNA (11,66). A PSI-BLAST search of CgpS-related proteins revealed that the majority (>98% of all hits, >92% of prophage containing strains) are found in bacterial genomes (Supplementary Table S6). However, several examples located in phages or prophage regions were identified. The functions of these phage-encoded XS-like proteins remain to be studied, but their presence suggests the following: (i) like CgpS, they may be required to secure tolerance of their carrier DNA within the respective host; (ii) they may, however, also function as antagonistic proteins, interfering with the host XS protein similar to the situation described for H-NST (54); or (iii) they may interfere with the function of another class of XS proteins. This hypothesis is based on the exclusion theory suggested by Perez-Rueda and Ibarra, who postulated that XS from different families do not appear in the same bacterial organism (56). Consistent with this bioinformatics study, our data show that the expression of E. coli hns results in strong activation of the cryptic prophage CGP3 and consequently cell death. The finding that expression of the C. glutamicum cgpS gene in E. coli MG1655 does not counteract H-NS-mediated silencing at the bgl operon shows, however, that the scenario is more complex and strongly depends on the particular strain and its regulatory equipment. However, our data on prophage activation in C. glutamicum provide evidence for an interference of analogous XS proteins at AT-rich DNA regions. Here, likely the incompatibility of the oligomerization domains inhibits the formation of XS multimeric structures required for silencing. Considering the presence of XS encoding genes in phage and prophage genomes, this principle is likely to be harnessed by any phage predator by encoding an interfering XS.
Supplementary Material
Acknowledgments
The authors thank Karin Schnetz (University of Cologne) for helpful advice and for providing us with the E. coli Δhns mutant strain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft priority program SPP1617 [FR 2759/2-2 and KO 4537/1-2]; Helmholtz Association [VH-NG-716]. Funding for open access charge: Helmholtz Association [VH-NG-716].
Conflict of interest statement. None declared.
REFERENCES
- 1.Juhas M., van der Meer J.R., Gaillard M., Harding R.M., Hood D.W., Crook D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canchaya C., Proux C., Fournous G., Bruttin A., Brussow H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casjens S. Prophages and bacterial genomics: what have we learned so far. Mol. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 4.Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 5.Nanda A.M., Thormann K., Frunzke J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 2015;197:410–419. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d'Hérelle F. Sur un microbe invisible antagoniste des bacilles dysentérique. Acad. Sci. Paris. 1917;165:373–375. [Google Scholar]
- 8.Diaz Ricci J.C., Hernandez M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000;20:79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- 9.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 10.Samson J.E., Magadan A.H., Sabri M., Moineau S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 11.Navarre W.W., McClelland M., Libby S.J., Fang F.C. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 12.Navarre W.W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S.J., Fang F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 13.Will W.R., Navarre W.W., Fang F.C. Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr. Opin. Microbiol. 2015;23:8–13. doi: 10.1016/j.mib.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langille M.G., Hsiao W.W., Brinkman F.S. Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 2010;8:373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 15.Dame R.T., Luijsterburg M.S., Krin E., Bertin P.N., Wagner R., Wuite G.J. DNA bridging: A property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding P., McFarland K.A., Jin S., Tong G., Duan B., Yang A., Hughes T.R., Liu J., Dove S.L., Navarre W.W., et al. A novel AT-rich DNA recognition mechanism for bacterial xenogeneic silencer MvaT. PLoS Pathog. 2015;11:e1004967. doi: 10.1371/journal.ppat.1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 18.Gordon B.R., Imperial R., Wang L., Navarre W.W., Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits W.K., Grossman A.D. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frunzke J., Bramkamp M., Schweitzer J.E., Bott M. Population Heterogeneity in Corynebacterium glutamicum ATCC 13032 caused by prophage CGP3. J. Bacteriol. 2008;190:5111–5119. doi: 10.1128/JB.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B.J., Gaigalat L., et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 22.Helfrich S., Pfeifer E., Krämer C., Sachs C.C., Wiechert W., Kohlheyer D., Nöh K., Frunzke J. Live cell imaging of SOS and prophage dynamics in isogenic bacterial populations. Mol. Microbiol. 2015;98:636–650. doi: 10.1111/mmi.13147. [DOI] [PubMed] [Google Scholar]
- 23.Nanda A.M., Heyer A., Krämer C., Grünberger A., Kohlheyer D., Frunzke J. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J. Bacteriol. 2014;196:180–188. doi: 10.1128/JB.01018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita S., Udaka S., Shimono M. Studies on the amino acid fermentation - Part I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 1957;50:331–343. [PubMed] [Google Scholar]
- 25.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 26.Keilhauer C., Eggeling L., Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 28.Niebisch A., Bott M. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 2001;175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- 29.Heyer A., Gätgens C., Hentschel E., Kalinowski J., Bott M., Frunzke J. The two-component system ChrSA is crucial for haem tolerance and interferes with HrrSA in haem-dependent gene regulation in Corynebacterium glutamicum. Microbiology. 2012;158:3020–3031. doi: 10.1099/mic.0.062638-0. [DOI] [PubMed] [Google Scholar]
- 30.Kim S.C., Chen Y., Mirza S., Xu Y., Lee J., Liu P., Zhao Y. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 2006;5:3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Evans G.A. Molecular cloning: A laboratory manual. Second edition. Volumes 1, 2, and 3. Current protocols in molecular biology. Volumes 1 and 2. Cell. 61:17–18. [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Donovan C., Heyer A., Pfeifer E., Polen T., Wittmann A., Krämer R., Frunzke J., Bramkamp M. A prophage-encoded actin-like protein required for efficient viral DNA replication in bacteria. Nucleic Acids Res. 2015;43:5002–5016. doi: 10.1093/nar/gkv374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grünberger A., Paczia N., Probst C., Schendzielorz G., Eggeling L., Noack S., Wiechert W., Kohlheyer D. A disposable picolitre bioreactor for cultivation and investigation of industrially relevant bacteria on the single cell level. Lab Chip. 2012;12:2060–2068. doi: 10.1039/c2lc40156h. [DOI] [PubMed] [Google Scholar]
- 36.Grünberger A., Probst C., Helfrich S., Nanda A., Stute B., Wiechert W., von Lieres E., Nöh K., Frunzke J., Kohlheyer D. Spatiotemporal microbial single-cell analysis using a high-throughput microfluidics cultivation platform. Cytometry. 2015;87A:1101–1115. doi: 10.1002/cyto.a.22779. [DOI] [PubMed] [Google Scholar]
- 37.Grünberger A., van Ooyen J., Paczia N., Rohe P., Schiendzielorz G., Eggeling L., Wiechert W., Kohlheyer D., Noack S. Beyond growth rate 0.6: Corynebacterium glutamicum cultivated in highly diluted environments. Biotechnol. Bioeng. 2013;110:220–228. doi: 10.1002/bit.24616. [DOI] [PubMed] [Google Scholar]
- 38.Bussmann M., Baumgart M., Bott M. RosR (Cg1324), a hydrogen peroxide-sensitive MarR-type transcriptional regulator of Corynebacterium glutamicum. J. Biol. Chem. 2010;285:29305–29318. doi: 10.1074/jbc.M110.156372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 41.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 42.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 43.R Development Core Team. R Foundation for Statistical Computing. Vienna: 2016. R: A language and environment for statistical computing.http://www.R-project.org [Google Scholar]
- 44.Ali S.S., Xia B., Liu J., Navarre W.W. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 2012;15:175–181. doi: 10.1016/j.mib.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Gordon B.R., Li Y., Wang L., Sintsova A., van Bakel H., Tian S., Navarre W.W., Xia B., Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant C.E., Bailey T.L., Noble W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dersch P., Schmidt K., Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 49.Khan F., Furuta Y., Kawai M., Kaminska K.H., Ishikawa K., Bujnicki J.M., Kobayashi I. A putative mobile genetic element carrying a novel type IIF restriction-modification system (PluTI) Nucleic Acids Res. 2010;38:3019–3030. doi: 10.1093/nar/gkp1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nobusato A., Uchiyama I., Kobayashi I. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene. 2000;259:89–98. doi: 10.1016/s0378-1119(00)00455-8. [DOI] [PubMed] [Google Scholar]
- 51.Jeltsch A., Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 52.Furuta Y., Kobayashi I. Bacterial Integrative Mobile Genetic Elements. Austin: 2014. [Google Scholar]
- 53.Baumgart M., Unthan S., Rückert C., Sivalingam J., Grünberger A., Kalinowski J., Bott M., Noack S., Frunzke J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 2013;79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williamson H.S., Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol. Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 55.Akhter S., Aziz R.K., Edwards R.A. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez-Rueda E., Ibarra J.A. Distribution of putative xenogeneic silencers in prokaryote genomes. Comput. Biol. Chem. 2015;58:167–172. doi: 10.1016/j.compbiolchem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Dole S., Kühn S., Schnetz K. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 2002;43:217–226. doi: 10.1046/j.1365-2958.2002.02734.x. [DOI] [PubMed] [Google Scholar]
- 58.Yamada H., Yoshida T., Tanaka K., Sasakawa C., Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 59.Gordon B.R., Li Y., Cote A., Weirauch M.T., Ding P., Hughes T.R., Navarre W.W., Xia B., Liu J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cordeiro T.N., Schmidt H., Madrid C., Juarez A., Bernado P., Griesinger C., Garcia J., Pons M. Indirect DNA readout by an H-NS related protein: structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 2011;7:e1002380. doi: 10.1371/journal.ppat.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucchini S., Rowley G., Goldberg M.D., Hurd D., Harrison M., Hinton J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedersen A.G., Jensen L.J., Brunak S., Staerfeldt H.H., Ussery D.W. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- 63.Ussery D.W., Tindbaek N., Hallin P.F. Genome update: promoter profiles. Microbiology. 2004;150:2791–2793. doi: 10.1099/mic.0.27504-0. [DOI] [PubMed] [Google Scholar]
- 64.Frunzke J., Engels V., Hasenbein S., Gätgens C., Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 2008;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castang S., McManus H.R., Turner K.H., Dove S.L. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarre W.W. In: Bacterial Chromatin. Dame RT, Dorman CJ, editors. Netherlands: Springer; 2009. [Google Scholar]
- 67.Stoebel D.M., Free A., Dorman C.J. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 68.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 69.Pennington J.M., Rosenberg S.M. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckling A., Rainey P.B. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S.W., Edlin G. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene. 1985;39:173–180. doi: 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- 72.Li C., Wally H., Miller S.J., Lu C.D. The multifaceted proteins MvaT and MvaU, members of the H-NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2009;191:6211–6218. doi: 10.1128/JB.00888-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma W., Noble W.S., Bailey T.L. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 2014;9:1428–1450. doi: 10.1038/nprot.2014.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cam E.L., Culard F., Larquet E., Delain E., Cognet J.A. DNA bending induced by the archaebacterial histone-like protein MC1. J. Mol. Biol. 1999;285:1011–1021. doi: 10.1006/jmbi.1998.2321. [DOI] [PubMed] [Google Scholar]
- 77.Chen J.M., Ren H., Shaw J.E., Wang Y.J., Li M., Leung A.S., Tran V., Berbenetz N.M., Kocincova D., Yip C.M., et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 2008;36:2123–2135. doi: 10.1093/nar/gkm1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.