Abstract
Mutations in human zinc-finger transcription factor WT1 result in abnormal development of the kidneys and genitalia and an array of pediatric problems including nephropathy, blastoma, gonadal dysgenesis and genital discordance. Several overlapping phenotypes are associated with WT1 mutations, including Wilms tumors, Denys-Drash syndrome (DDS), Frasier syndrome (FS) and WAGR syndrome (Wilms tumor, aniridia, genitourinary malformations, and mental retardation). These conditions vary in severity from individual to individual; they can be fatal in early childhood, or relatively benign into adulthood. DDS mutations cluster predominantly in zinc fingers (ZF) 2 and 3 at the C-terminus of WT1, which together with ZF4 determine the sequence-specificity of DNA binding. We examined three DDS associated mutations in ZF2 of human WT1 where the normal glutamine at position 369 is replaced by arginine (Q369R), lysine (Q369K) or histidine (Q369H). These mutations alter the sequence-specificity of ZF2, we find, changing its affinity for certain bases and certain epigenetic forms of cytosine. X-ray crystallography of the DNA binding domains of normal WT1, Q369R and Q369H in complex with preferred sequences revealed the molecular interactions responsible for these affinity changes. DDS is inherited in an autosomal dominant fashion, implying a gain of function by mutant WT1 proteins. This gain, we speculate, might derive from the ability of the mutant proteins to sequester WT1 into unproductive oligomers, or to erroneously bind to variant target sequences.
INTRODUCTION
Wilms tumor suppressor protein (WT1; UniProt: P19544) is a zinc-finger transcription factor central to embryonic development of the genitourinary system (reviewed in (1–3)). The gene for human WT1, located in band 13 on the short arm of chromosome 11 (11p13), comprises 10 exons. Exons 1–6 encode N-terminal domains responsible for protein dimerization and for transcriptional regulation (4,5); exons 7–10 encode four C-terminal zinc fingers (ZF), the last three of which are primarily responsible for DNA sequence-discrimination and target binding (Supplementary Figure S1a). Individuals with aberrant WT1 are invariably heterozygous, with copies of both normal and mutated Wt1 genes, and they exhibit a spectrum of unusual features typically early in life. Truncations of WT1 due to frame-shift or chain-termination mutations leads to pediatric renal malignancies termed Wilms tumors, the fifth most common childhood malignancy (6,7). Although the action of WT1 is not well understood, these mutations, whether inherited or de novo, give rise to Wilms tumors following somatic loss of the normal copy of the Wt1 gene resulting in hemizygous cells with no normal WT1 at all (8–10).
Another class of mutations in Wt1 causes Denys-Drash Syndrome (DDS). These are predominantly missense mutations in ZF2 and ZF3 that alter either the Cys2-His2 structural amino acids that coordinate the zinc ions, or the sequence-recognition amino acids at the protein-DNA interface (Supplementary Figure S1b). DDS mutations result in an array of severe problems, including a high probability of Wilms tumor, mesangial sclerosis and early renal failure, gonadal dysgenesis, and ambiguous or female genitalia in 46XY males. The most common mutation found in individuals with DDS is arginine 394 to tryptophan (R394W) (11), but many additional singletons are known as summarized in Supplementary Figure S1b.
Alternative splicing between exons 9 and 10 of Wt1 leads to the presence or absence of three additional amino acids—lysine (K), threonine (T) and serine (S)—between ZF3 and ZF4 of WT1 (Supplementary Figure S1a and b). The usual 2:1 balance between the two isoforms, ‘+KTS’ and ‘-KTS’, is critical for normal genitourinary development, and splice junction mutations that invert this ratio result in Frasier Syndrome (FS), a condition similar to but distinct from DDS, characterized by nephropathy, genital anomalies and gonadoblastoma, but only rarely Wilms tumor (8,12,13). Wilms tumors can arise from mutations in a number of different genes (14,15), but WT1-derived Wilms tumors, DDS, and FS all stem from mutations in the same, Wt1, gene. The resulting phenotypes correlate well with the position and type of mutation present, although there are individual differences in the extents to which these are manifest, and overlaps in diagnoses (8,9,16).
The varied consequences of Wt1 mutations appear to depend upon the functionalities of the mutant WT1 proteins. Truncations starting near the beginning of the gene are expected to lead to inactive proteins. Those starting further down the polypeptide chain could retain the ability to dimerize and, when paired with normal WT1 protein, to bind to DNA and affect regulation. C-terminal mutations in the zinc fingers should behave in a similar way when the mutations abolish DNA-binding, but if their effect is to change sequence-specificity instead, the proteins might bind to new DNA target sites as monomers or homodimers, or as heterodimers in combination with normal WT1. Mis-association with other transcription factors is also feasible (1,17). Numerous regulatory perturbations could result from such interactions. To better understand these, we have investigated the DNA-binding behavior of the zinc-finger domain of normal WT1, and of three missense mutations reported to cause DDS in which glutamine 369 (Q369) is replaced by arginine (18) (see Supplementary text), histidine (19) or lysine (20). Unlike R394W, Q369 mutations are rare—each has been reported only once—and if generalizations based on single examples have validity, their consequences are somewhat different. The Q369H patient displayed genitourinary anomalies, and the early onset renal failure characteristic of DDS (19). The Q369R patient displayed only mild mesangial proliferation that progressed slowly, and did not develop early renal failure (18). And the third, Q369K, patient developed gonadoblastoma, which is more typical of Frasier Syndrome than DDS (20). These patients exemplify the phenotypic variation and overlap that can result from mutations in WT1, and the challenges these pose to current diagnostic classification.
Eukaryotic proteins that interact with DNA often do so by means of ZFs (21–23) (reviewed in (24)). These comprise a tandem series of approximately 30-amino acid zinc finger modules, each of which interacts in the major DNA groove with three or four adjacent bases (the recognition ‘triplet’) of mainly one DNA strand. The C-terminal ZF domain of WT1 comprises four zinc fingers, but only three of these—ZF2, 3 and 4—are thought to determine specificity; ZF1 is reported to contribute to binding affinity, but in a relatively non-specific manner (25–27). The consensus DNA binding sequence of WT1 (Figure 1A) is 5′-GCG-(T/G)(G/A)G-G(C/A)G-3′ (28). ZF4 interacts with the 5′ triplet in this sequence (GCG); ZF3 interacts with the central triplet; and ZF2 interacts with the 3′ triplet (G-C/A-G) (27,29). Arginine 394 of ZF3 recognizes the conserved (3′) Gua of the central triplet (Supplemental Figure S1f). The R394W DDS mutation is reported to abolish DNA binding (30–32), precluding structural analysis of its interactions with DNA. Glutamine 369 recognizes the central base (C or A) of the 3′ triplet, and as we report below, the three Q369 mutants continue to bind to DNA, providing us an avenue to investigate the interactions of WT1 DDS associated mutants with DNA. We report our findings here, together with X-ray crystallographic structural analyses that provide molecular explanations for these results.
Figure 1.
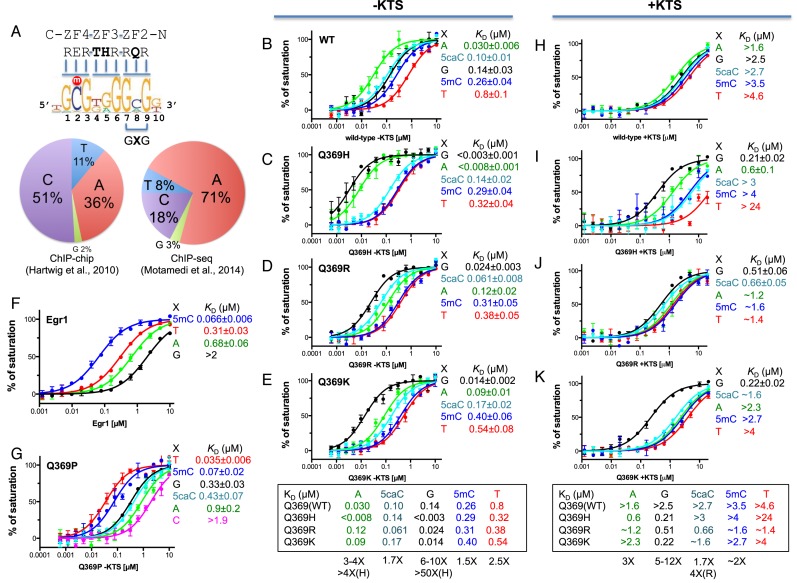
Normal WT1 prefers adenine and the three Q369 mutants prefer guanine in the 3′ triplet. (A) The consensus DNA binding sequence of WT1, adopted from (28). The pie-diagrams show the distribution of the base in the middle position of the 3′ triplet (GXG) from the ChIP-chip (28) and ChIP-seq (40). (B–E) Binding affinities of normal WT1 and three Q369 mutants (in the –KTS isoform) with oligos containing various base pairs in the middle position of the 3′ recognition triplet (position ‘X’ in panel a). (H–K) Binding affinities of normal WT1 and three Q369 mutants in the +KTS isoform with various oligos. (F) Binding affinities of the equivalent three-finger Egr1, which contacts a Glu (E) in the corresponding position of WT1 Q369 (29). (G) The Q369P of WT1 prefers T, followed by 5mC, to all other bases by a factor of 10 or more.
The WT1 consensus sequence overlaps the target sequence of another transcription factor, early growth response protein 1 (Egr1; also known as Zif268 or Krox-24): 5′-GCG-(T/G/C)GG-GCG-3′ (25,26,33,34). The 5′ and 3′ triplets of this consensus sequence include CpG dinucleotides, which are the primary sites of epigenetic DNA modification. In a previous study of binding to modified-cytosine forms of the Egr1 sequence, WT1 displayed markedly different affinities according to the type of modification present, suggesting that this protein might be responsive to epigenetic signals during development (29). In particular, WT1 bound with highest affinity when the modified base in the 3′ triplet was 5-carboxylcytosine (5caC—the terminal oxidation product of 5-methylcytosine), due to electrostatic attraction between the side chain of Q369 and the negatively charged carboxylate group (29). This observation prompted us to include in the present study of the Q369 mutations not only target nucleotide sequence variants, but also cytosine modification variants.
MATERIALS AND METHODS
Site-directed mutagenesis
GST-tagged human WT1 residues 350–437 (UniProt: P19544.2) for the -KTS isoform (pXC1295) and residues 350–440 for the +KTS isoform (pXC1329) were mutated and generated WT1-KTS Q369H (pXC1305), WT1+KTS Q369H (pXC1361), WT1-KTS Q369K (pXC1387), WT1+KTS Q369K (pXC1388), WT1-KTS Q369R (pXC1302), WT1+KTS Q369R (pXC1360), WT1-KTS Q369P (pXC1303) by QuikChange site-directed mutagenesis kit (Stratagene). All mutants were verified by sequencing.
Protein expression and purification
All proteins were expressed in the Escherichia coli strain of BL21-CodonPlus(DE3)-RIL (Stratagene) and purified as described (29). Typically, 2 l of cultures were grown at 37°C to log phase (OD600 = 0.5–0.8) and then shifted to 16°C. ZnCl2 was added to a final concentration of 25 μM, expression was induced by the addition of ß-d-1-thiogalactopyranoside to 0.2 mM, and the cultures were incubated overnight at 16°C. Cells were harvested by centrifugation, resuspended in lysis buffer containing 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 25 μM ZnCl2, and then lysed by sonication. Lysates were mixed with polyethylenimine (Sigma) to a final concentration of 0.4% (w/v) and clarified by centrifugation at 18 000 rpm (35). Cleared extracts were loaded onto a glutathione-Sepharose 4B column (GE Healthcare) pre-equilibrated with the lysis buffer. The GST fusion proteins were eluted with 20 mM glutathione in the elution buffer containing 100 mM Tris–HCl (pH 8.0), 5% (v/v) glycerol, 25 μM ZnCl2 and 500 mM NaCl. The GST tag was removed using PreScission protease (purified in-house), leaving five additional N-terminal residues (GPLGS). The proteins were diluted two-fold with 20 mM Tris–HCl (pH 7.5), 5% (v/v) glycerol, 25 μM ZnCl2, and 0.5 mM TCEP and loaded onto tandem HiTrap-Q/HiTrap-SP columns (GE Healthcare) (35). Most proteins flowed through the Q column onto the SP column from which they were eluted using a linear gradient of NaCl from 250 mM to 1 M. Finally, the pooled protein was concentrated and loaded onto a size exclusion column and eluted as a single peak in 500 mM NaCl, 20 mM Tris–HCl (pH 7.5), 5% (v/v) glycerol, and 25 μM ZnCl2. Final protein concentrations were estimated by absorbance at 280 nm (absorbance coefficient of 9.66 for 1 mM WT1). The protein yields (∼10 mg from 2 l culture) were approximately the same for normal WT1 protein and for the mutants (Supplementary Figure S1g).
Fluorescence-based DNA binding assay
Fluorescence polarization measurements were carried out at 25°C on a Synergy 4 microplate reader (BioTek). The 6-carboxy-fluorescein (FAM)-labeled dsDNA probe (5 nM) was incubated for 10 min with increasing amounts of protein in 20 mM Tris–HCl (pH 7.5), 5% (v/v) glycerol, 0.5 mM TCEP and NaCl (300 or 340 mM). No change in fluorescence intensity was observed with the addition of protein. The sequences of the oligonucleotides used in Figure 1 were 3′-TGXGGGTGMGA-5′ and FAM-5′-TACZCCCAMGC-3′ (where M = 5mC, X = G, A, T, 5mC or 5caC and Z is the corresponding base paired with X). The sequences of the oligonucleotides used in Figure 3 were 3′-TGXGGGTGXGA-5′ and FAM-5′-TAZGCCCAZGC-3′ (where X and Z = C, 5mC, 5hmC, 5fC or 5caC as defined in Figure 3). Curves were fit individually using GraphPad Prism 5.0 software (GraphPad Software, Inc.). Binding constants (KD) were calculated as [mP] = [maximum mP] × [C]/(KD + [C]) + [baseline mP] and saturated [mP] was calculated as saturation = ([mP] − [baseline mP])/([maximum mP] [baseline mP]), where mP is millipolarization and [C] is protein concentration. Averaged KD and its standard error are reported.
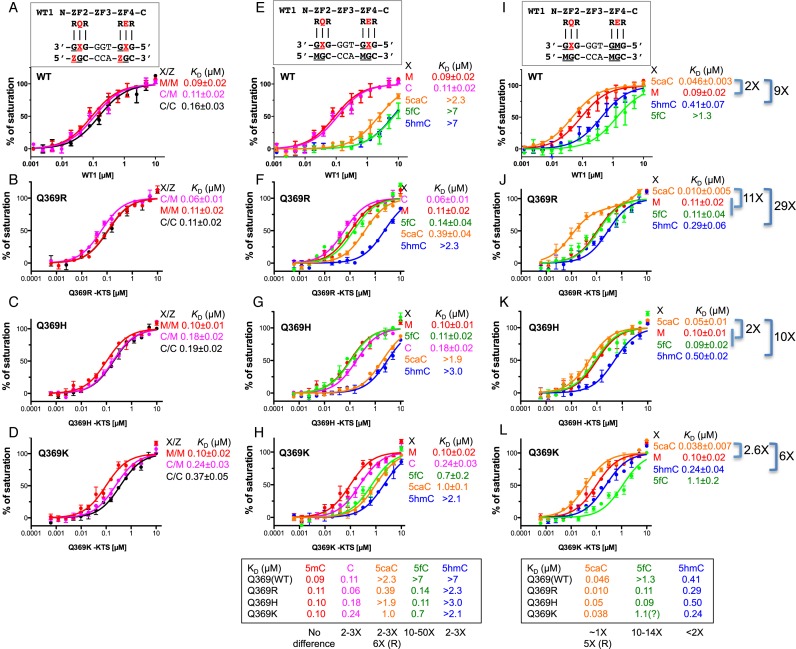
Figure 3.
Binding affinities of normal WT1 the three Q369 mutants with oligos containing varied forms of cytosine. (A–D) Oligos fully methylated at both CpG sites (M/M), unmethylated at both sites (C/C), or methylated in only the bottom strand at both sites (C/M). (E–H) Oligos modified in only the top strand at both sites with 5mC (M), C, 5hmC, 5fC or 5caC. 5mC was present in the bottom strand at both sites in all cases. (I–L) Oligos modified in only the top strand at the 3′ triplet. All other sites contained 5mC (M).
Crystallography
We crystallized WT1 in the presence of DNA by the sitting-drop vapor diffusion method at 16°C using equal amounts of protein–DNA mixtures (1 mM) and well solution (20% (w/v) polyethylene glycol (PEG) 3350, 40 mM citric acid, 60 mM Bis–Tris propane, pH 6.4 for Q369R–5caC complex; 25% (w/v) PEG 3350, 0.2 M ammonium acetate, 0.1 M Bis–Tris HCl, pH 6.5 for Q369H–5caC complex). The well solutions were 20 mM Citric Acid, 80 mM Bis-Tris propane, pH 8.8 for normal WT1 Q369-A:T complex; 12% (w/v) PEG 3350, 4% (v/v) Tacsimate, pH 8.0 for Q369H-A:T complex; 20% PEG 3350, 0.2 M Na-malonate, pH 7.0 for Q369R-G:C complex. Protein–DNA mixtures in equimolar ratios were incubated for 30 min at 16°C before crystallization. Crystals were cryo-protected by soaking in mother liquor supplemented with 20% (v/v) ethylene glycol before plunging into liquid nitrogen. X-ray diffraction data were collected at 100 K at the SER-CAT beamlines (22BM-D and 22ID-D) at the Advanced Photon Source, Argonne National Laboratory, and processed using HKL2000 (36). Initial crystallographic phases were determined by molecular replacement using the coordinates of the WT1-5mC (PDB 4R2E) as search models. Phasing, molecular replacement, map production, and model refinement were performed using PHENIX (37,38). The two mutant structures were solved, built, and refined independently. The statistics were calculated for the entire resolution range (Supplementary Table S1). The Rfree and Rwork values were calculated for 5% (randomly selected) and 95%, respectively, of the observed reflections. Molecular graphics were generated using PyMol (DeLano Scientific, LLC).
RESULTS
All experiments were performed using purified C-terminal binding domains corresponding to ZF2-4 of human WT1 (amino acids 350–437 for the –KTS isoform) and its corresponding +KTS isoform rather than full-length proteins. These were affinity-purified from Escherichia coli recombinants using a GST-tag, which was subsequently removed as described in Materials and Methods. Fluorescence polarization was used to measure the dissociation constant (KD) between these binding domains and double-stranded oligonucleotides (oligos) bearing fluorescent 5′-FAM labels.
Normal WT1 has highest affinity for adenine in the 3′ triplet
We compared the binding affinities of the -KTS and +KTS isoforms of WT1 to duplex oligos containing various base pairs in the middle position of the 3′ recognition triplet (position ‘X’ in Figure 1A). For all oligos examined, affinity of the +KTS isoform was at least one order of magnitude lower than for the −KTS isoform, confirming previous observations (29,39) that WT1 (+KTS) binds the consensus DNA sequence far less well than WT1 (−KTS) (compare Figure 1B and H).
WT1 (−KTS) showed the strongest binding to adenine at position X (KD = 0.03 μM); approximately 5-fold weaker binding to guanine (0.14 μM); and ∼25-fold weaker binding to thymine (0.8 μM) (Figure 1b). Previous analysis of cytosine modifications in the 3′ GCG triplet, showed that the affinity of WT1 (−KTS) for this site is 5caC > 5mC∼C > 5hmC > 5fC (29). The most strongly bound form, 5caC (KD = 0.1 μM), is bound with ∼3-fold lower affinity than adenine. This finding is in agreement with independent reports that WT1 binds with highest affinity when the 3′ triplet is GAG than when it is GCG (26,31). By comparison, anti-WT1 chromatin immunoprecipitation (ChIP) studies of embryonic mouse kidney tissue found that the base in the middle position of the 3′ triplet was 51% C, 36% A (ChIP-chip (28)), and 71% A, 18% C (ChIP-seq (40)). In both studies, G occurred least frequently (Figure 1A, left and right pie-charts).
The amino acid in ZF2 that juxtaposes the central base of the 3′ triplet is glutamine 369 (Q369) (27,29). Earlier crystallography studies revealed that the side chain of Q369 adopts different conformations with different forms of modified cytosine. This adaptability is the likely reason ZF2 can tolerate different bases at this position. 5-carboxylcytosine (5caC) is bound especially well among the five forms of cytosine, we found, due to attraction between the side chain of Q369 and the exocyclic carboxyl group (29).
The three Q369 mutants have highest affinity for guanine in the 3′ triplet
In further binding-affinity experiments, all three Q369 mutants were found to bind most strongly to guanine (G). For Q369H, in which histidine (His) replaced Gln, the -KTS isoform bound both guanine and adenine with very high affinity (KD < 0.003 and 0.008 μM; Figure 1C), resulting in the discrete specificity, G(G/A)G. Schumacher et al speculated that the Q369H mutation might change the secondary structure of ZF2 resulting in a loss of DNA-binding capacity (19), but this is evidently not the case. Histidine also occurs naturally at this position in ZF3 of WT1 (H397), which also recognizes G or A at the center of triplet 2. X-ray crystallography revealed that H397 donates a hydrogen bond (H-bond) to the N7 ring atom of the purine base (29). This N7 acceptor atom is present in both adenine and guanine, but absent in cytosine and thymine, accounting in part for why G or A are bound at this position, but not C or T.
Q369K and Q369R (−KTS) also exhibited high-affinity binding with Guanine (KD = 0.014 and 0.024 μM), but binding to adenine was somewhat reduced, resulting in the predominant specificity, GGG (Figure 1D and E). Affinity for 5caC was substantial in all three mutants, particularly for Q369R (KD = 0.06 μM), as well as for normal WT1 (0.10 μM). In all instances thymine was bound with lowest affinity (Figure 1B–E). Each mutant displayed unique behavior. Q369H discriminates purines (G and A; high-affinity substrates) from pyrimidines (C and T; low-affinity substrates) (Figure 1c). Q369R exhibited progressively weaker binding for the five bases tested: G > 5caC > A > 5mC > T (Figure 1d). And Q369K discriminates G from all of the rest (Figure 1E).
We also measured the effects of two other amino acid substitutions of Q369 (−KTS), namely glutamate (Glu; E) and proline (Pro; P) (Figure 1F and G; see also Supplementary text). The DNA-binding domain of early growth response protein 1 (Egr1) comprises three ZFs that are similar to ZF2–4 of WT1 (29), and bind the consensus sequence: 5′-GCG-(T/G/C)GG-GCG-3′. WT1 uses glutamate (E427) to contact the central base in the 5′ triplet, and glutamine (Q369) to contact the central base in the 3′ triplet. Egr1, in contrast, uses glutamate for both contacts (E410 and E354). The Egr1 binding domain exhibits a markedly different response to changes in the 3′ triplet than WT1 (compare Figure 1F and B). The presence of Glu rather than Gln in the first ZF results in 5mC and T being bound with highest affinity (both have a methyl group at the same, carbon-5, position) and A and G being bound with least affinity—the opposite behavior to WT1 (Figure 1B and F). In an earlier study, we generated a Q369 to proline mutant (Q369P) of WT1 (−KTS) with a strong preference for 5mC relative to both unmodified C and to the three oxidized forms of cytosine (29). We find Q369P binds T with even higher affinity than 5mC, again the opposite of normal WT1 (Figure 1G). In their discussion of WT1 mutations, Stoll et al suggested that Q369P might destabilize the ZF (27) but our finding, here, that Q369P binds as tightly to DNA as normal WT1 (compare Figure 1B and G)—albeit to a different triplet sequence, G-(T/5mC)-G rather than GAG—indicates, perhaps surprisingly, that this is not the case.
The Q369 (+KTS) mutants have substantial affinity for guanine in the 3′ triplet
We expressed and purified the ZF2–4 (+KTS) isoforms of WT1 and the three Q369 mutants and compared their binding affinities to various sequences. WT1 (+KTS) displayed much reduced affinity for all sequences tested compared with WT1 (–KTS), and made less of a distinction between them (compare Figure 1H and B). The three extra amino acids present in the +KTS isoform increase the length and flexibility of the linker between ZF3 and 4, and this structural perturbation is thought to disrupt binding of ZF4 to the 5′ triplet (41). All three mutants retained considerable affinity (KD = 0.2–0.5 μM) for the substrate with G at the center of the 3′ triplet (Figure 1I–K), roughly the same affinity as the –KTS isoform of normal WT1 (KD = 0.14 μM; Figure 1b).
Structural analysis of purine binding
To understand why Q369 and the mutants preferentially bind adenine and guanine, we determined the co-crystal structures of the –KTS isoform of ZF2–4 of normal WT1, Q369R and Q369H in complex with 10-bp oligos containing either A:T or G:C base pairs in the middle of the 3′ triplet (Supplementary Table S1). We were unable to grow sufficiently large crystals of the Q369K-DNA complex to analyze the lysine mutant. The structures were solved at the high-resolution range of 1.45–1.7 Å (Supplementary Table S1). The protein and DNA components of all three complexes were structurally similar, with a root-mean-squared (rms) deviation of 0.3 Å. We focus our discussion on ZF2 and its interaction with the 3′ G-(A/G)-G triplet (Figure 2).
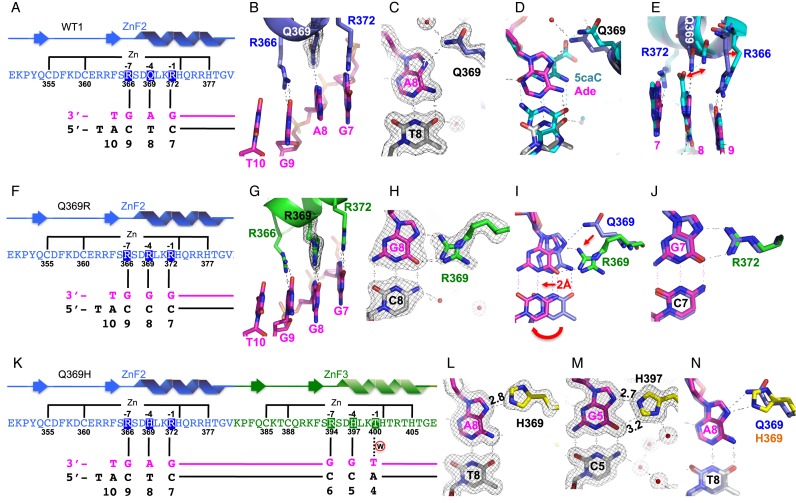
Figure 2.
Structural analysis of purine (adenine or guanine) binding in the 3′ triplet. (A–E) Structure of normal WT1 in complex with the A:T base pair. (A) Schematic representation of the ZF2 (from N-to-C termini) and the recognition sequence (from 3′-to-5′ of the top strand). The complementary strand (bottom) has a 5′ overhanging thymine used for the crystallization study. (B) R372, Q369, and R366 of ZF2 (in blue) interact, respectively, with Gua7, Ade8, and Gua9 of DNA (in magenta). (C) Q369-Ade8 interaction. The 2Fo-Fc electron density, contoured at 1σ above the mean, is shown in gray. (D) Structural superimposition of Q369-Ade (in blue for Q369, magenta for Ade, and gray for Thy) and Q369-5caC (in cyan; PDB: 4R2R). (E) The side chains of Q369 and R366 displaced shifts from binding of 5caC (in cyan) to Ade (in magenta). (F–J) Structure of Q369R in complex with the G:C base pair. (F) Schematic representation of the ZF2 with Q369R mutant. (G) The three arginines (R372, R369, and R366) align with three adjacent guanines along the DNA major groove. (H) R369-Gua8 interaction. (I) Structural superimposition of Q369-Ade (in blue) and R369-Gua (in green for R369 and magenta for Gua). (J) R372-Gua7 interaction. (K–N) Structure of Q369H in complex with the A:T base pair. (K) Schematic representation of the ZF2 (Q369H mutant) and ZF3 in recognition of the corresponding triplets. (L) H369-Ade8 interaction. (M) H397-Gua5 interaction. (N) Structural superimposition of Q369-Ade (in cyan) and H369-Ade (yellow for H369, magenta for Ade8).
The ZF domain of normal WT1 was crystallized with the A:T-containing oligo. R372, Q369, and R366 of ZF2 interacted specifically with Gua7, Ade8, and Gua9, respectively (Figure 2A and B) with the side-chain amide of Q369 forming an H-bond with the ring N7 atom of Ade8 (Figure 2C). Comparing this Q369-Ade structure with that of Q369-5caC (pdb: 4R2R) revealed that (i) the N7 atom superimposes on the C5 atom of 5caC (Figure 2D); (ii) the Q369 side chain is displaced slightly to accommodate the large 5caC carboxylate group (Figure 2E) and (iii) as a consequence, the side chain of R366 also shifts slightly, while the position of R372 remains unchanged (Figure 2E).
The side-chain conformation that Q369 adopts in our structure is somewhat unexpected because it results in only a single H-bond to adenine, with the Ade N7 atom (Figure 2C), instead of bidentate contacts involving one with the N7 atom and the other with the 6-amino group. In principal, this H-bond can form equally well with Gua N7, implying that WT1 should display the same affinity for G in the middle of the 3′-triplet as it displays for A. This is not the case, however, as A is preferred by a factor of 4 or 5 (Figure 1B). This might indicate that Q369 can rearrange in solution and form the second H-bond, one that can only arise with Ade, and not with Gua. Q369 also adopts a different conformation when juxtaposed with cytosine (see PDB: 2PRT (27).
The Q369R ZF domain was crystallized with the G:C-containing oligo. The three arginine residues, R372, R369 and R366 aligned along the DNA major groove and formed two H-bonds each with the N7 and O6 atoms the three adjacent guanines (Figure 2f, g). We could also model an alternative side-chain conformation for R369, in which the guanidinium group H-bonds with only the O6 atom of Gua8 (Figure 2h). Comparing this R369-Gua structure with Q369-Ade revealed that both the side chains and the base pairs had undergone significant changes of position (Figure 2i). Movement of R369 from the conformation that Q369 adopts with A:T requires rotation of all three side chain torsion angles. To accommodate the larger size of R369, the G:C base pair swings approximately 2 Å from the position of the A:T. Despite this structural perturbation, the adjacent G:C base pairs and arginine residues are barely disturbed (Figure 2J). Only one H-bond can form between arginine and adenine (donated to N7), whereas two can form with guanine (donated to N7 and O6), explaining perhaps why Q369R exhibits 5-fold lower affinity for A than for G (Figure 1D).
Q369H binds both G and A with exceptionally high affinities (Figure 1C). Q369H was crystallized with the A:T-containing oligo, because the natural H397 of ZF3 juxtaposes a G:C base pair within the oligo (Figure 2K), and so this structure revealed the interactions of histidine with both adenine and guanine. The imidazole ring of H369 forms one H-bond with Ade N7 via the Nϵ2 ring atom (Figure 2l). H397 forms a similar H-bond with Gua N7, and in addition by ring rotation forms a second, weaker (3.2 Å), C–H…O type H-bond (42) to Gua O6 via the ring Cϵ1 atom (Figure 2M). This latter bond cannot form with adenine because an amino group is present at the 6-position, and this already bears covalently attached hydrogen atoms. The Nδ1 ring atom of H397 also participates in water-mediated H-bonds involving the exocyclic 4-amino group of the partner cytosine (Figure 2m). Histidine engages in more electrostatic contacts with Gua than with Ade, then, which might explain why Q369H has a 2–3-fold higher binding affinity to G than to A (Figure 1C). Because of the similar sizes of their side chains, H369 and Q369 interact with A in a similar manner (Figure 2N).
Binding to epigenetically modified cytosine
We compared binding of the –KTS isoforms of WT1 and the mutants to double-stranded oligonucleotides (oligos) containing epigenetic modifications within 5′ and 3′ GCG triplets. In the first set of experiments (Figure 3A–D), the CpG sequences were unmethylated in both strands (C/C), methylated in both strands (M/M), or methylated in the bottom strand but not in the top strand (C/M). The presence of methyl groups enhanced binding affinity modestly in each case. In contrast to some proteins such as Myc (43,44), CTCF (45,46), STAT1 (47) and CREB (48) whose DNA-binding is inhibited by methylation, CpG-methylation does not reduce binding by normal WT1 or the three mutants, but instead either increases it somewhat or has no effect (Figure 3B–D).
In turn, we replaced the cytosines in the top (‘recognition’) strand of the oligo with all three oxidized forms of 5mC—5-hydroxymethylC (5hmC), 5-formylC (5fC) and 5-carboxylC (5caC)—while retaining 5mC in the bottom strand. For normal WT1, all three oxidized forms reduced binding greatly, by 25-fold or more (Figure 3E), confirming our previous observation that WT1 distinguishes un-oxidized cytosine from oxidized cytosine rather than un-methylated cytosine from methylated cytosine (29). For the mutants, the effects of the oxidized forms differed. For Q369R and Q369H, 5fC had little or no effect; for Q369K it reduced binding moderately. 5caC reduced binding substantially for Q369H and Q369K, but less so for Q369R. And 5hmC proved uniformly detrimental (Figure 3f–h). Overall, cytosine oxidation was found to reduce binding affinity, with 5fC being least detrimental, 5hmC being most detrimental and 5caC being in between.
These results represented the combined effects of modifications at both CpG sites, and of their interactions with two amino acids, the one at position 369, and the glutamate at position 427. To focus more specifically on position 369, we examined the consequences of oxidized cytosine in the top strand of just the 3′ GCG triplet, keeping the other cytosines in the preferred, 5mC, state (Figure 3I). The effects varied, as before, but normal WT1 and all three mutant binding domains now displayed very high affinity for 5caC—several-fold higher than for 5mC—and up to ten-fold increases in their affinities for 5hmC (Figure 3I–L).
Structural analysis of 5caC and 5fC binding
To understand the molecular basis for these results, we co-crystallized ZF2-4 (–KTS) of the mutants with 10-bp oligos containing 5caC or 5fC in the top strand of the 3′ GCG triplet (Supplementary Table S1). As before, the crystals of the Q369K-DNA complex proved too small to analyze, and so we solved only the structures of the Q369R and Q369H mutants. We compare these with the previously determined structure of normal WT1 bound to the same oligo (pdb: 4R2R), focusing on ZF2 and the 3′ G-5caC-G triplet (Figure 4a). The protein and DNA components of all three complexes were similar, with rms deviations of 0.3 Å.
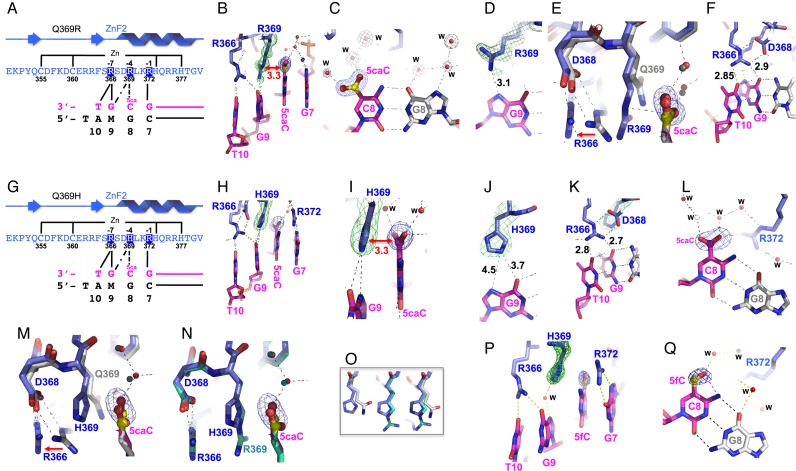
Figure 4.
Structural analysis of oxidized cytosine binding in the 3′ triplet. (A–F) Structure of Q369R in complex with 5caC DNA. (A) Schematic representation of the ZF2 with Q369R mutant. (B) The R369 positively charged guanidinium group stacks on the negatively charged carboxylate of 5caCyt 8. (C) The 5caC carboxylate goup H-bonds with water molecules. (D) R369 H-bond with Gua9. (E) The movement of the side chain of R366 from normal WT1 (Q369) to the mutant R369. (F) R366 H-bonds with Gua9 and the adjacent Thy10. (G–L) Structure of Q369H in complex with 5caC DNA. (G) Schematic representation of the ZF2 with Q369H mutant. (H) The three positively changed residues (R366, H369, and R372) span four bases. (I) The H369 imidazole ring stacks on the 5caC carboxylate group. (J) H369 is too far removed to form H-bonds with Gua9. (K) R366 forms H-bonds with both Gua9 and Thy10. (L) The 5caC carboxylate goup H-bonds with water molecules. (M) Superimposing the Q369 (in gray) and Q369H (in blue) structures in complex with 5caC DNA. (N) Superimposing the Q369H (in blue) and Q369R (in cyan) structures in complex with 5caC DNA. (O) The main-chain conformations at position 369 are virtually identical, and the side chain conformations are very similar with only small rotational differences in torsion angles among Q369, H369, and R369. (P-Q) Structure of Q369H in complex with 5fC DNA. A water molecule mediates the interaction between H369 and Gua9 (P). Both 5fC (panel Q) and 5caC (panel L) exhibit an intrabase hydrogen bond between their formyl or carboxyl oxygen atoms, respectively, and the adjacent cytosine N4 exocyclic amine nitrogen atom.
As shown in Figure 2, the side chains of the amino acids at positions 372, 369 and 366 of ZF2 normally interact with the bases 7, 8 and 9 (G-A/C-G), of the top strand of the 3′ triplet. In the Q369R structure with high-affinity 5caC (Figure 4A–F), the interaction between R372 and Gua7 (Figure 2B) remains unchanged, but the two other arginines undergo rearrangements (compare Figure 2F and G with Figure 4A and B). The side chain of R369 adopts an extended conformation and its positively charged guanidinium group stacks on the negatively charged carboxylate of 5caCyt 8 (Figure 4B), and rather than H-bonding with R369, the carboxylate H-bonds with water molecules, instead (Figure 4c). R369 forms a de novo H-bond with the N7 atom of the adjacent Gua 9 (Figure 4D), and in doing so pushes the side chain of R366 partly out of the register (Figure 4E) so that it now H-bonds not only with the Gua 9 O6 atom, but also with the O4 atom of the adjacent base, Thy 10 (Figure 4F). R366 continues to be stabilized by electrostatic interactions with the side chain of D368, the position of which changes little (Figure 4D).
Similar rearrangements are present in the structures of Q369H with 5caC (Figure 4G–L) and with 5fC (Figure 4P and Q). The R372–Gua7 interaction again remains unchanged. The imidazole ring of the H369 side-chain stacks on the 5caC carboxylate group (Figure 4H, I) and aligns with the purine ring of Gua 9, although it is perhaps too far removed to form H-bonds (Figure 4J). The stacking interaction likewise displaces the side chain of R366, which again forms H-bonds with both Gua 9 and Thy 10 (Figure 4K). In the structure with 5fC, a water molecule mediates the interaction between H369 and Gua9 N7 (Figure 4P). Superimposing the Q369, Q369R and Q369H structures revealed that the main-chain conformations at position 369 are virtually identical, and the side chain conformations are very similar with only small rotational differences in torsion angles (Figure 4E, M, N and O).
DISCUSSION
Mutations in human zinc-finger transcription factor WT1 cause an array of pediatric health problems that stem from aberrant development and functioning of the genitourinary system. The problems include nephroblastoma and renal failure; gonadal dysgenesis, sterility and gonadoblastoma; anomalous or ambiguous genitalia and male to female sex reversal; and possibly more. The N-terminus of WT1 comprises domains for dimerization, and for transcription activation and repression (1); the C-terminus comprises four zinc fingers (ZF) in tandem, which to varying degrees determine the DNA sequences to which WT1 binds. For its small size of around 450 amino acids, WT1 is a surprisingly complicated protein with multiple physiological effects (2).
Several overlapping conditions have been linked to mutations in Wt1, the gene that codes for WT1, including Wilms tumors, WAGR syndrome (Wilms tumor, aniridia, genitourinary malformations and mental retardation), Denys-Drash syndrome (DDS), and Frasier syndrome (FS). The phenotypes manifested depend largely on the type of mutation present, and its position within the gene. WAGR results from gross deletion of Wt1 and the adjacent Pax6 gene (49–52). FS is caused by mutations at the end of exon 9 of Wt1 that lead to an overabundance of the stronger DNA-binding –KTS isoform of WT1 versus the weaker binding +KTS isoform (8,12,13). And DDS is caused predominantly by missense mutations in ZFs 2 and 3 that change amino acids involved in zinc-coordination, or in sequence-recognition in the major DNA groove (Supplementary Figure S1). DDS mutations are generally considered to abolish DNA binding by WT1 (19,27,31,32,53), but what we find here is that those at position 369 in ZF2 continue to bind DNA very well indeed, only now they bind to different sequences instead of, or in addition to, the original sequence. ZF2 of normal WT1 has the predominant specificity G-(A/C)-G. The predominant specificity of ZF2 mutants Q369R and Q369K is G-G-G, and of Q369H it is G-(G/A)-G. Likewise, the predominant specificity of ZF2 Q369P is G-T-G.
In addition to changing nucleotide-specificity, we find these mutations also change affinity for epigenetically modified forms of cytosine within the WT1 target sequence. The effects vary according to the type of modification, and whether both CpG sites are modified, or only one. Normal WT1, for example, binds the target sequence well when either cytosine or 5-methylcytosine (5mC) is present at both sites, but binds poorly when any of the three oxidized forms of cytosine are present instead. Q369H and Q369R, by comparison, are very much more tolerant of the presence of 5-formylcytosine (5fC) at both sites, and Q369R is also more tolerant of 5-carboxylcytosine (5caC), probably due to the increased positive charge. Affinity changes such as these, we speculate, might allow DDS mutants of WT1 to bind, with unpredictable consequences, to an array of new genomic sites in addition to, or instead of, those normally bound. The common effect of such ‘off-target’ binding could be to reduce the amount of normal WT1 available to bind to correct sites, an effect that would be exacerbated if WT1 bound as a dimer rather than a monomer (5,54) due to sequestration of normal WT1 into dysfunctional heterodimeric complexes.
It is interesting to note that WT1 physically interacts with Tet2 (55,56), one of the three mammalian 5mC-dioxygenases that generates oxidized cytosine (57–59). It is possible that WT1 either recruits Tet2 to its target sites and/or binds the sequences modified by Tet2 enzymatic activity. Accumulating evidence supports the view that 5hmC exists as a relatively stable modification, constituting a distinct epigenetic signal (60,61) that marks lowly expressed genes, gene bodies, and intragenic regions (reviewed in (62,63)). 5fC and 5caC are found at much lower levels and at fewer sites, but are enriched at mono-methylated histone H3 lysine 4 (H3K4me1)-marked enhancers (64–69), implying that these higher oxidized forms might play an important role in enhancer activity.
Despite an enormous amount of work on the biochemistry, genetics, and physiology of WT1 spanning several decades, much remains unclear. From our perspective as specialists in protein-DNA structural interactions, the role of ZF1, for example, continues to puzzle us. The C-terminal ZF domain of WT1 is essentially identical in mammals, birds, amphibians and fish, whereas numerous differences occur in the N-terminal regulatory regions. This high degree of conservation implies that all four ZFs are essential for normal vertebrate development and are immutable. ZF1 has all the features of a regular zinc finger unit, and the identification of DDS missense mutations located in this ZF (70) suggest that it participates in sequence-discrimination and binding. Using in vitro selection approaches, ZF1 was found to have affinity for Thy as the first base of its putative triplet (26), and in a separate study it was found to preferentially bind the ambiguous triplet (T/G)-(G/A/T)-(T/G) (25) in which cytosine was excluded as the central base. Nevertheless, NMR and X-ray crystallographic analyses of ZFs 1–4 in complex with an oligonucleotide failed to reveal sequence-specific contacts by ZF1, leading to the conclusion that ZF1 ‘…does not contribute significantly to binding specificity’ (27). We are investigating whether this discrepancy could be due to the oligo used in this analysis, which was based on the Egr1 consensus rather than the WT1 consensus. We speculate that WT1 ZF1-4 might have higher affinity for the sequence of triplets 5′-GCG-TGG-GAG-TGT, than for 5′-GCG-GGG-GCG-TCT used by Stoll et al. (27), and that complexes with this oligo instead might reveal specific interactions with ZF1.
Another question concerns the behavior of the predominant, +KTS, isoform of WT1 (71), depletion of which leads to Frasier Syndrome. This protein has three extra amino acids—lysine, threonine, and serine—in the linker between ZF3 and ZF4, which greatly reduce its affinity for the consensus DNA sequence (72). It has been suggested that the +KTS isoform might play a role in RNA splicing (73), but from a structural point of view a change in overall affinity from DNA to RNA is difficult to rationalize. Lysine, threonine and serine are benign in the context of DNA-binding, and can interact favorably with both the bases and the backbone phosphate groups of DNA. It seems plausible to us, then, that the +KTS isoform has affinity for a variant of the WT1 consensus, one in which triplets 1 and 2 are separated by one or more base pairs to compensate for the increased length of the linker, for example, GCG-N1-3-TGG-GAG-TGT. Although analysis of natural WT1 (+KTS) binding-sites cast some doubt on this possibility (74), there is precedent for such changes among the sequence-specificity subunits of certain restriction enzymes, where insertion or deletion of four amino acids between pairs of DNA-binding domains increases or reduces the separation between the sequences recognized by one base pair (75).
Many apparently opposite activities have been ascribed to WT1 (reviewed in (76)). These include transcriptional activation and repression (77); roles in RNA processing and translation (73,78,79); localization in nucleus or cytoplasm (80–82); a tumor suppressor in the formation of Wilms tumor, and an oncogene in adult tumors (83–86); a role in controlling active (H3K4me3) and repressive (H3K27me3) histone modification marks (87); and a capacity to differentially bind to epigenetically modified DNA (29). WT1 is expressed in cells that are undergoing epithelial-to-mesenchymal transition (88–90) or the reverse, mesenchymal-to-epithelial transition (87,91). It is possible that the opposing activities of WT1 maintain the balance between epithelial and mesenchymal states (92) and the potential to transition in either direction. Mutations in the Wt1 gene, or altered expression could perturb this balance and lead to disease.
Supplementary Material
Acknowledgments
We thank B. Baker for synthesizing the oligonucleotides and Yusuf Olatunde Olanrewaju for performing protein purification.
Author contributions: H.H. performed protein purification, crystallographic and DNA-binding experiments; X.Z. corrected the error related to the Q369P mutation in the Human Gene Mutation Database; Y.Z. analyzed published WT1 ChIP profiles; X.Z. and X.C. organized and designed the scope of the study; G.G.W. performed data analysis and assisted in preparing the manuscript; and all were involved in analyzing data and preparing the manuscript.
ACCESSION NUMBERS
The X-ray structures (coordinates and structure factor files) of WT1 (Q369) (PDB: 5KL2), H369 (PDB: 5Kl3, 5KL4 and 5KL5) and R369 (PDB: 5KL6 and 5KL7) have been submitted to PDB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (NIH) [GM049245-23 to X.C.]; Department of Biochemistry of Emory University School of Medicine supported the use of the Southeast Regional Collaborative Access Team (SERCAT) synchrotron beamlines at the Advanced Photon Source of Argonne National Laboratory; Georgia Research Alliance Eminent Scholar (to X.C.). Funding for open access charge: NIH and New England Biolabs, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee S.B., Haber D.A. Wilms tumor and the WT1 gene. Exp. Cell Res. 2001;264:74–99. doi: 10.1006/excr.2000.5131. [DOI] [PubMed] [Google Scholar]
- 2.Wagner K.D., Wagner N., Schedl A. The complex life of WT1. J. Cell Sci. 2003;116:1653–1658. doi: 10.1242/jcs.00405. [DOI] [PubMed] [Google Scholar]
- 3.Niaudet P., Gubler M.C. WT1 and glomerular diseases. Pediatric Nephrol. 2006;21:1653–1660. doi: 10.1007/s00467-006-0208-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z.Y., Qiu Q.Q., Deuel T.F. The Wilms' tumor gene product WT1 activates or suppresses transcription through separate functional domains. J. Biol. Chem. 1993;268:9172–9175. [PubMed] [Google Scholar]
- 5.Moffett P., Bruening W., Nakagama H., Bardeesy N., Housman D., Housman D.E., Pelletier J. Antagonism of WT1 activity by protein self-association. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11105–11109. doi: 10.1073/pnas.92.24.11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidoff A.M. Wilms tumor. Adv. Pediatr. 2012;59:247–267. doi: 10.1016/j.yapd.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dome J.S., Fernandez C.V., Mullen E.A., Kalapurakal J.A., Geller J.I., Huff V., Gratias E.J., Dix D.B., Ehrlich P.F., Khanna G., et al. Children's Oncology Group's 2013 blueprint for research: renal tumors. Pediatr. Blood Cancer. 2013;60:994–1000. doi: 10.1002/pbc.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koziell A., Grundy R. Frasier and Denys-Drash syndromes: different disorders or part of a spectrum? Arch. Dis. Childhood. 1999;81:365–369. doi: 10.1136/adc.81.4.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heathcott R.W., Morison I.M., Gubler M.C., Corbett R., Reeve A.E. A review of the phenotypic variation due to the Denys-Drash syndrome-associated germline WT1 mutation R362X. Hum. Mutat. 2002;19:462. doi: 10.1002/humu.9031. [DOI] [PubMed] [Google Scholar]
- 10.Kucinskas L., Rudaitis S., Pundziene B., Just W. Denys-Drash syndrome. Medicina. 2005;41:132–134. [PubMed] [Google Scholar]
- 11.Royer-Pokora B., Beier M., Henzler M., Alam R., Schumacher V., Weirich A., Huff V. Twenty-four new cases of WT1 germline mutations and review of the literature: genotype/phenotype correlations for Wilms tumor development. Am. J. Med. Genet. A. 2004;127:249–257. doi: 10.1002/ajmg.a.30015. [DOI] [PubMed] [Google Scholar]
- 12.Barbaux S., Niaudet P., Gubler M.C., Grunfeld J.P., Jaubert F., Kuttenn F., Fekete C.N., Souleyreau-Therville N., Thibaud E., Fellous M., et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 13.Klamt B., Koziell A., Poulat F., Wieacker P., Scambler P., Berta P., Gessler M. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/-KTS splice isoforms. Hum. Mol. Genet. 1998;7:709–714. doi: 10.1093/hmg/7.4.709. [DOI] [PubMed] [Google Scholar]
- 14.Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat. Rev. Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinkes B.G., Mucha B., Vlangos C.N., Gbadegesin R., Liu J., Hasselbacher K., Hangan D., Ozaltin F., Zenker M., Hildebrandt F., et al. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2) Pediatrics. 2007;119:e907–e919. doi: 10.1542/peds.2006-2164. [DOI] [PubMed] [Google Scholar]
- 16.Fencl F., Malina M., Stara V., Zieg J., Mixova D., Seeman T., Blahova K. Discordant expression of a new WT1 gene mutation in a family with monozygotic twins presenting with congenital nephrotic syndrome. Eur. J. Pediatr. 2012;171:121–124. doi: 10.1007/s00431-011-1497-3. [DOI] [PubMed] [Google Scholar]
- 17.Dong L., Pietsch S., Englert C. Towards an understanding of kidney diseases associated with WT1 mutations. Kidney Int. 2015;88:684–690. doi: 10.1038/ki.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta S., Ozawa T., Izumino K., Sakuragawa N., Fuse H. A novel missense mutation of the Wt1 gene causing Denys-Drash syndrome with exceptionally mild renal manifestations. J. Urol. 2000;163:1857–1858. [PubMed] [Google Scholar]
- 19.Schumacher V., Thumfart J., Drechsler M., Essayie M., Royer-Pokora B., Querfeld U., Muller D. A novel WT1 missense mutation presenting with Denys-Drash syndrome and cortical atrophy. Nephrol. Dial. Transplant. 2006;21:518–521. doi: 10.1093/ndt/gfi285. [DOI] [PubMed] [Google Scholar]
- 20.Patel P.R., Pappas J., Arva N.C., Franklin B., Brar P.C. Early presentation of bilateral gonadoblastomas in a Denys-Drash syndrome patient: a cautionary tale for prophylactic gonadectomy. J. Pediatr. Endocrinol. Metab.: JPEM. 2013;26:971–974. doi: 10.1515/jpem-2012-0409. [DOI] [PubMed] [Google Scholar]
- 21.Pei J., Grishin N.V. C2H2 zinc finger proteins of the SP/KLF, Wilms tumor, EGR, Huckebein, and Klumpfuss families in metazoans and beyond. Gene. 2015;573:91–99. doi: 10.1016/j.gene.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadimpalli S., Persikov A.V., Singh M. Pervasive variation of transcription factor orthologs contributes to regulatory network evolution. PLoS Genet. 2015;11:e1005011. doi: 10.1371/journal.pgen.1005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 24.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton T.B., Barilla K.C., Romaniuk P.J. High affinity binding sites for the Wilms' tumour suppressor protein WT1. Nucleic Acids Res. 1995;23:277–284. doi: 10.1093/nar/23.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagama H., Heinrich G., Pelletier J., Housman D.E. Sequence and structural requirements for high-affinity DNA binding by the WT1 gene product. Mol. Cell. Biol. 1995;15:1489–1498. doi: 10.1128/mcb.15.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoll R., Lee B.M., Debler E.W., Laity J.H., Wilson I.A., Dyson H.J., Wright P.E. Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J. Mol. Biol. 2007;372:1227–1245. doi: 10.1016/j.jmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Hartwig S., Ho J., Pandey P., Macisaac K., Taglienti M., Xiang M., Alterovitz G., Ramoni M., Fraenkel E., Kreidberg J.A. Genomic characterization of Wilms' tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development. 2010;137:1189–1203. doi: 10.1242/dev.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto H., Olanrewaju Y.O., Zheng Y., Wilson G.G., Zhang X., Cheng X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014;28:2304–2313. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier J., Bruening W., Kashtan C.E., Mauer S.M., Manivel J.C., Striegel J.E., Houghton D.C., Junien C., Habib R., Fouser L., et al. Germline mutations in the Wilms' tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell. 1991;67:437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 31.Borel F., Barilla K.C., Hamilton T.B., Iskandar M., Romaniuk P.J. Effects of Denys-Drash syndrome point mutations on the DNA binding activity of the Wilms' tumor suppressor protein WT1. Biochemistry. 1996;35:12070–12076. doi: 10.1021/bi960758o. [DOI] [PubMed] [Google Scholar]
- 32.Little M., Holmes G., Bickmore W., van Heyningen V., Hastie N., Wainwright B. DNA binding capacity of the WT1 protein is abolished by Denys-Drash syndrome WT1 point mutations. Hum. Mol. Genet. 1995;4:351–358. doi: 10.1093/hmg/4.3.351. [DOI] [PubMed] [Google Scholar]
- 33.Lemaire P., Vesque C., Schmitt J., Stunnenberg H., Frank R., Charnay P. The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol. Cell. Biol. 1990;10:3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christy B., Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel A., Hashimoto H., Zhang X., Cheng X. Characterization of how DNA modifications affect DNA binding by C2H2 zinc finger proteins. Methods Enzymol. 2016;573:387–401. doi: 10.1016/bs.mie.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z., Borek D., Majewski W., Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 37.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Laity J.H., Chung J., Dyson H.J., Wright P.E. Alternative splicing of Wilms' tumor suppressor protein modulates DNA binding activity through isoform-specific DNA-induced conformational changes. Biochemistry. 2000;39:5341–5348. doi: 10.1021/bi9926678. [DOI] [PubMed] [Google Scholar]
- 40.Motamedi F.J., Badro D.A., Clarkson M., Lecca M.R., Bradford S.T., Buske F.A., Saar K., Hubner N., Brandli A.W., Schedl A. WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat. Commun. 2014;5:4444. doi: 10.1038/ncomms5444. [DOI] [PubMed] [Google Scholar]
- 41.Laity J.H., Dyson H.J., Wright P.E. Molecular basis for modulation of biological function by alternate splicing of the Wilms' tumor suppressor protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11932–11935. doi: 10.1073/pnas.97.22.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horowitz S., Trievel R.C. Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem. 2012;287:41576–41582. doi: 10.1074/jbc.R112.418574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prendergast G.C., Lawe D., Ziff E.B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 44.Perini G., Diolaiti D., Porro A., Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 46.Renda M., Baglivo I., Burgess-Beusse B., Esposito S., Fattorusso R., Felsenfeld G., Pedone P.V. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 47.Chen B., He L., Savell V.H., Jenkins J.J., Parham D.M. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 2000;60:3290–3298. [PubMed] [Google Scholar]
- 48.Iguchi-Ariga S.M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 49.Miller-Hodges E. Clinical Aspects of WT1 and the Kidney. Methods Mol. Biol. 2016;1467:15–21. doi: 10.1007/978-1-4939-4023-3_2. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto T., Togawa M., Shimada S., Sangu N., Shimojima K., Okamoto N. Narrowing of the responsible region for severe developmental delay and autistic behaviors in WAGR syndrome down to 1.6 Mb including PAX6, WT1, and PRRG4. Am. J. Med. Genet. A. 2014;164:634–638. doi: 10.1002/ajmg.a.36325. [DOI] [PubMed] [Google Scholar]
- 51.Miles C., Elgar G., Coles E., Kleinjan D.J., van Heyningen V., Hastie N. Complete sequencing of the Fugu WAGR region from WT1 to PAX6: dramatic compaction and conservation of synteny with human chromosome 11p13. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13068–13072. doi: 10.1073/pnas.95.22.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crolla J.A., Cawdery J.E., Oley C.A., Young I.D., Gray J., Fantes J., van Heyningen V. A FISH approach to defining the extent and possible clinical significance of deletions at the WAGR locus. J. Med. Genet. 1997;34:207–212. doi: 10.1136/jmg.34.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmes G., Boterashvili S., English M., Wainwright B., Licht J., Little M. Two N-terminal self-association domains are required for the dominant negative transcriptional activity of WT1 Denys-Drash mutant proteins. Biochem. Biophys. Res. Commun. 1997;233:723–728. doi: 10.1006/bbrc.1997.6545. [DOI] [PubMed] [Google Scholar]
- 54.Englert C., Vidal M., Maheswaran S., Ge Y., Ezzell R.M., Isselbacher K.J., Haber D.A. Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11960–11964. doi: 10.1073/pnas.92.26.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rampal R., Alkalin A., Madzo J., Vasanthakumar A., Pronier E., Patel J., Li Y., Ahn J., Abdel-Wahab O., Shih A., et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Xiao M., Chen X., Chen L., Xu Y., Lv L., Wang P., Yang H., Ma S., Lin H., et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell. 2015;57:662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bachman M., Uribe-Lewis S., Yang X., Burgess H.E., Iurlaro M., Reik W., Murrell A., Balasubramanian S. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 2015;11:555–557. doi: 10.1038/nchembio.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashimoto H., Zhang X., Vertino P.M., Cheng X. The mechanisms of generation, recognition, and erasure of DNA 5-methylcytosine and thymine oxidations. J. Biol. Chem. 2015;290:20723–20733. doi: 10.1074/jbc.R115.656884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasmussen K.D., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A.C., Fung H.L., Zhang K., Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H., et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H., Wu X., Shen L., Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu X., Han D., Zhao B.S., Song C.X., Zhang L.S., Dore L.C., He C. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Z., Dai N., Borgaro J.G., Quimby A., Sun D., Correa I.R., Jr, Zheng Y., Zhu Z., Guan S. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell. 2015;57:750–761. doi: 10.1016/j.molcel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 69.Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., Medana C., Dal Bello F., Oliviero S. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015;10:674–683. doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 70.Bruening W., Bardeesy N., Silverman B.L., Cohn R.A., Machin G.A., Aronson A.J., Housman D., Pelletier J. Germline intronic and exonic mutations in the Wilms' tumour gene (WT1) affecting urogenital development. Nat. Genet. 1992;1:144–148. doi: 10.1038/ng0592-144. [DOI] [PubMed] [Google Scholar]
- 71.Hastie N.D. Life, sex, and WT1 isoforms–three amino acids can make all the difference. Cell. 2001;106:391–394. doi: 10.1016/s0092-8674(01)00469-x. [DOI] [PubMed] [Google Scholar]
- 72.Rauscher F.J., 3rd, Morris J.F., Tournay O.E., Cook D.M., Curran T. Binding of the Wilms' tumor locus zinc finger protein to the EGR-1 consensus sequence. Science. 1990;250:1259–1262. doi: 10.1126/science.2244209. [DOI] [PubMed] [Google Scholar]
- 73.Larsson S.H., Charlieu J.P., Miyagawa K., Engelkamp D., Rassoulzadegan M., Ross A., Cuzin F., van Heyningen V., Hastie N.D. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- 74.Drummond I.A., Rupprecht H.D., Rohwer-Nutter P., Lopez-Guisa J.M., Madden S.L., Rauscher F.J., 3rd, Sukhatme V.P. DNA recognition by splicing variants of the Wilms' tumor suppressor, WT1. Mol. Cell. Biol. 1994;14:3800–3809. doi: 10.1128/mcb.14.6.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price C., Lingner J., Bickle T.A., Firman K., Glover S.W. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J. Mol. Biol. 1989;205:115–125. doi: 10.1016/0022-2836(89)90369-0. [DOI] [PubMed] [Google Scholar]
- 76.Hohenstein P., Hastie N.D. The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 2006;15:R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 77.Roberts S.G. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 78.Caricasole A., Duarte A., Larsson S.H., Hastie N.D., Little M., Holmes G., Todorov I., Ward A. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davies R.C., Calvio C., Bratt E., Larsson S.H., Lamond A.I., Hastie N.D. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes Dev. 1998;12:3217–3225. doi: 10.1101/gad.12.20.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vajjhala P.R., Macmillan E., Gonda T., Little M. The Wilms' tumour suppressor protein, WT1, undergoes CRM1-independent nucleocytoplasmic shuttling. FEBS Lett. 2003;554:143–148. doi: 10.1016/s0014-5793(03)01144-x. [DOI] [PubMed] [Google Scholar]
- 81.Niksic M., Slight J., Sanford J.R., Caceres J.F., Hastie N.D. The Wilms' tumour protein (WT1) shuttles between nucleus and cytoplasm and is present in functional polysomes. Hum. Mol. Genet. 2004;13:463–471. doi: 10.1093/hmg/ddh040. [DOI] [PubMed] [Google Scholar]
- 82.Dudnakova T., Spraggon L., Slight J., Hastie N. Actin: a novel interaction partner of WT1 influencing its cell dynamic properties. Oncogene. 2010;29:1085–1092. doi: 10.1038/onc.2009.444. [DOI] [PubMed] [Google Scholar]
- 83.Loeb D.M., Evron E., Patel C.B., Sharma P.M., Niranjan B., Buluwela L., Weitzman S.A., Korz D., Sukumar S. Wilms' tumor suppressor gene (WT1) is expressed in primary breast tumors despite tumor-specific promoter methylation. Cancer Res. 2001;61:921–925. [PubMed] [Google Scholar]
- 84.Koesters R., Linnebacher M., Coy J.F., Germann A., Schwitalle Y., Findeisen P., von Knebel Doeberitz M. WT1 is a tumor-associated antigen in colon cancer that can be recognized by in vitro stimulated cytotoxic T cells. Int. J. Cancer. 2004;109:385–392. doi: 10.1002/ijc.11721. [DOI] [PubMed] [Google Scholar]
- 85.Oji Y., Yano M., Nakano Y., Abeno S., Nakatsuka S., Ikeba A., Yasuda T., Fujiwara Y., Takiguchi S., Yamamoto H., et al. Overexpression of the Wilms' tumor gene WT1 in esophageal cancer. Anticancer Res. 2004;24:3103–3108. [PubMed] [Google Scholar]
- 86.Amini Nik S., Hohenstein P., Jadidizadeh A., Van Dam K., Bastidas A., Berry R.L., Patek C.E., Van der Schueren B., Cassiman J.J., Tejpar S. Upregulation of Wilms' tumor gene 1 (WT1) in desmoid tumors. Int. J. Cancer. 2005;114:202–208. doi: 10.1002/ijc.20717. [DOI] [PubMed] [Google Scholar]
- 87.Essafi A., Webb A., Berry R.L., Slight J., Burn S.F., Spraggon L., Velecela V., Martinez-Estrada O.M., Wiltshire J.H., Roberts S.G., et al. A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev. Cell. 2011;21:559–574. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pritchard-Jones K., Fleming S., Davidson D., Bickmore W., Porteous D., Gosden C., Bard J., Buckler A., Pelletier J., Housman D., et al. The candidate Wilms' tumour gene is involved in genitourinary development. Nature. 1990;346:194–197. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- 89.Armstrong J.F., Pritchard-Jones K., Bickmore W.A., Hastie N.D., Bard J.B. The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 1993;40:85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- 90.Martinez-Estrada O.M., Lettice L.A., Essafi A., Guadix J.A., Slight J., Velecela V., Hall E., Reichmann J., Devenney P.S., Hohenstein P., et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 2010;42:89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore A.W., Schedl A., McInnes L., Doyle M., Hecksher-Sorensen J., Hastie N.D. YAC transgenic analysis reveals Wilms' tumour 1 gene activity in the proliferating coelomic epithelium, developing diaphragm and limb. Mech. Dev. 1998;79:169–184. doi: 10.1016/s0925-4773(98)00188-9. [DOI] [PubMed] [Google Scholar]
- 92.Miller-Hodges E., Hohenstein P. WT1 in disease: shifting the epithelial-mesenchymal balance. J. Pathol. 2012;226:229–240. doi: 10.1002/path.2977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.