Abstract
PhIP is an abundant heterocyclic aromatic amine (HCA) and important dietary carcinogen. Following metabolic activation, PhIP causes bulky DNA lesions at the C8-position of guanine. Although C8-PhIP-dG adducts are mutagenic, their interference with the DNA replication machinery and the elicited DNA damage response (DDR) have not yet been studied. Here, we analyzed PhIP-triggered replicative stress and elucidated the role of the apical DDR kinases ATR, ATM and DNA-PKcs in the cellular defense response. First, we demonstrate that PhIP induced C8-PhIP-dG adducts and DNA strand breaks. This stimulated ATR-CHK1 signaling, phosphorylation of histone 2AX and the formation of RPA foci. In proliferating cells, PhIP treatment increased the frequency of stalled replication forks and reduced fork speed. Inhibition of ATR in the presence of PhIP-induced DNA damage strongly promoted the formation of DNA double-strand breaks, activation of the ATM-CHK2 pathway and hyperphosphorylation of RPA. The abrogation of ATR signaling potentiated the cell death response and enhanced chromosomal aberrations after PhIP treatment, while ATM and DNA-PK inhibition had only marginal effects. These results strongly support the notion that ATR plays a key role in the defense against cancer formation induced by PhIP and related HCAs.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide, with dietary factors, such as the intake of processed and red meat, accounting for over 30% of total CRC cases (1,2). Very recently, the consumption of processed and red meat has been classified as carcinogenic to humans (3), which was attributed to the presence and/or generation of food-borne carcinogens, including N-nitroso compounds and heterocyclic aromatic amines (HCAs) (4,5). HCAs are powerful DNA-damaging compounds formed in meat and fish cooked at high temperature (6). 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most abundant HCA in food (4). It undergoes metabolic activation by cytochrome P450 1A2 (CYP1A2) to yield the toxic intermediate 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH-PhIP), which is further acetylated by N-acetyl-transferases (NATs) or sulfonated by sulfotransferases (SULTs) (7). It has been postulated that these compounds reach the distal colon via the bloodstream and are actively secreted from the distal colon mucosa into the lumen, thereby passing through the stem cell compartment in the colon crypts (8). Owing to the instability of these esters, a DNA-reactive arylnitrenium ion is released, which forms primarily C8-PhIP-dG adducts (see Supplementary Figure S1A) (9). C8-dG-PhIP adducts are bulky DNA lesions that undergo nucleotide excision repair (NER) (10,11). Unrepaired C8-PhIP-dG adducts are pre-mutagenic, predominantly causing G:T transversion mutations and single nucleotide deletions (12,13). It was reported that cells progressively undergo cell death with increasing PhIP doses, whereas surviving cells exhibit increased mutation frequencies as measured at the hprt locus (14). In addition, an increase in thymidine kinase forward mutations was observed in MCL5 cells at PhIP doses ≥10 μM (15). Several in vivo studies further demonstrated that PhIP induces mammary, prostate and gastrointestinal tumors in rodents (16–18), which is consistent with results from epidemiological studies (19,20).
Molecular modeling and in vitro studies suggest that C8-PhIP-dG adducts block replicative polymerases, enhance the infidelity of replication and may engage error-prone translesion synthesis (21–23). Interference with the DNA replication machinery can trigger a cellular stress response, referred to as replication stress. A known trigger of replication stress is DNA damage by halting the replicative polymerase, while the MCM helicase continues unwinding the DNA duplex (24). This results in the generation of single-stranded DNA (ssDNA), which is rapidly coated by replication protein A (RPA). The ssDNA–RPA complex is then sensed by ATR-interacting protein (ATRIP), which recruits the protein kinase ATR (ATM- and Rad3-related), thereby leading to its activation as key event in the DNA damage response (DDR) (25,26). ATR phosphorylates several downstream effector molecules such as the histone 2AX (H2AX), the checkpoint kinase CHK1 and the cell cycle checkpoint protein RAD17 (27). ATR together with RPA thereby stabilizes stalled or damaged replication forks, activates repair pathways and facilitates the restart of stalled forks (28). Persistent replication stress can result in fork collapse and generation of DNA double-strand breaks (DSBs) (27). DSBs are recognized by the tripartite MRN complex, which consists of MRE11, RAD50 and NBS1 (29). The MRN complex then recruits the apical kinase ATM, which is activated by autophosphorylation at Ser1981 (30). DNA-PKcs is another apical DDR kinase attracted to DSBs by its interaction with DSB-bound Ku70/80 heterodimer, thereby forming the DNA-PK holoenzyme (31). Both ATM and DNA-PKcs are integral components of the DDR (31).
In the present study, we set out to analyze the PhIP-dependent activation of the DDR and the role of the apical DDR kinases ATM, ATR and DNA-PKcs in cell survival and genomic stability. Using different cell models, we show that PhIP, upon metabolic activation, generates C8-PhIP-dG DNA adducts and DNA strand breaks. Western blot analyses, confocal microscopy and DNA fiber assays revealed that PhIP and its metabolite N-OH-PhIP provoke replication stress and trigger the ATR-driven DDR. Subsequently, the role of ATR and the other apical DDR kinases ATM and DNA-PKcs were characterized, demonstrating that ATR inhibition together with N-OH-PhIP treatment strongly promotes DSB formation and concomitant ATM-CHK2 activation. Finally, we provide evidence that ATR, but not ATM or DNA-PKcs, confers protection against detrimental replication stress, cell death and chromosomal instability in response to (N-OH)-PhIP.
MATERIALS AND METHODS
Materials
Calf intestine alkaline phosphatase, micrococcal nuclease, calf spleen phosphodiesterase and ribonuclease A (RNase A) were purchased from Sigma (Steinheim, Germany). Proteinase K, HPLC-grade methanol, formic acid and acetic acid were from Carl Roth GmbH (Karlsruhe, Germany). The synthesis of the isotope-labeled reference standard [15N5,13C10]C8-PhIP-dG was previously described (9). The CHK1 inhibitor UCN-01 was obtained from Sigma. The ATR inhibitor VE821, the ATM inhibitor KU-55933 and the DNA-PKcs inhibitor NU7026 were from Selleck Chemicals (USA).
Cell lines and culture conditions
V79 Chinese hamster cells and V79-derived cells stably expressing both human cytochrome P450 1A2 (CYP1A2) and human sulfotransferase 1A1 (SULT1A1) (32), designated V79 CS, were kindly provided by Hans-Ruedi Glatt (German Institute of Human Nutrition, Potsdam-Rehbrücke, Germany). Cells were obtained in 2014 and authenticated by their fibroblast-like morphology and differential response to PhIP. Cells were maintained in DMEM-Ham's F12 medium supplemented with 5% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Caco-2 human colorectal cancer cells were obtained from CLS Cell Lines Service (Eppelheim, Germany) in 2012. Caco-2 cells were re-authenticated by their p53 status and typical enterocyte-like morphology, which was regularly assessed by light microscopy. The colorectal carcinoma cell line HCT116 (p53+/+, DNA-PKcs+/+; designated as wildtype (WT)) as well as their isogenic counterparts HCT116-p53−/− and HCT116-DNA-PKcs−/− were provided by Dr Bert Vogelstein (John Hopkins University, Baltimore, USA) and Dr Hans Peter Rodemann (Department of Radiation Oncology, University of Tübingen, Germany) in 2012 and 2015, respectively. Cells were re-authenticated by p53 and DNA-PKcs immunoblotting and by their characteristic differential response to 5-FU (WT versus p53−/−) and radiomimetics (WT versus DNA-PKcs−/−). HCT116 cells were maintained in DMEM supplemented with 10% FCS and antibiotics (100 U/mL penicillin and 100 μg/ml streptomycin). Non-transformed human colonic epithelial cells (HCEC) were kindly provided by Dr Jerry W. Shay (Department of Cell Biology, UT Southwestern Medical Center, Dallas, USA) in 2015 and maintained as previously described (33). All cell lines were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air, except for HCEC that were grown in a nitrogen incubator with reduced oxygen levels. Cell culture medium and supplements were obtained from Gibco Life Technologies (Darmstadt, Germany) unless stated otherwise.
Drugs and drug treatments
PhIP and its metabolite N-OH-PhIP were kindly provided by Dr Albrecht Seidel (Biochemical Institute of Environmental Carcinogens, Großhansdorf, Germany). Both compounds were dissolved in DMSO at a final stock concentration of 30 and 10 mM, respectively. Etoposide (ETO) was obtained from Sigma-Aldrich and dissolved in DMSO to give a stock solution of 10 mM. DMSO was used as solvent control according to its concentration in the respective (N-OH-)PhIP or ETO dose. All inhibitors (ATRi, ATMi, DNA-PKcsi, CHK1i) were dissolved in DMSO at a stock concentration of 10 mM and added to the cells 2 h before and 24 h after treatment with N-OH-PhIP as indicated.
For UV irradiation, medium was removed and cells were irradiated with UV-C (20 J/m2) with a radium NSE 11–270 low pressure UV-C lamp (Philips, Netherlands) (34). Thereafter, the removed medium was added again and cells were incubated for up to 24 h as indicated.
Detection of C8-PhIP-dG adducts by slot blot analysis
DNA adducts were determined in genomic DNA isolated from V79 CS cells using an immunological slot blot assay (35), which is based on the application of a polyclonal antibody directed against C8-PhIP-dG DNA adducts (Squarix, Marl, Germany). Briefly, cells were treated with (N-OH-)PhIP as described, lysed and subjected to RNase A digestion followed by proteinase K incubation overnight. Genomic DNA was isolated by phenol–chloroform extraction, precipitated with 70% ice-cold ethanol and dissolved in TE buffer pH 7.4. Purity and concentration were determined by UV absorbance using a NanoDrop 2000 (Thermo Scientific, Dreieich, Germany). To determine C8-PhIP-dG DNA adducts, a slot blot assay was performed. Sample DNA was denatured by heating for 10 min at 99°C followed by the addition of 2 M ammonium acetate. 500 ng of DNA were immediately vacuum-aspirated as triplicates onto a positively charged nylon membrane (GE Healthcare, Munich, Germany) using a slot-blot manifold. The membrane was fixed for 90 min at 90°C and blocked with 5% (w/v) non-fat dry milk in TBS-T. C8-PhIP-dG DNA adducts were visualized with a polyclonal antibody followed by a secondary peroxidase-coupled antibody and enhanced chemoluminescence detection.
Detection of C8-PhIP-dG adducts by mass spectrometry
Samples containing 10 to 30 μg isolated genomic DNA were dried together with 5413 fmol C8-PhIP-dG. The residues were taken up in 28 μl water and 8 μl of a 100 mM sodium succinate (pH 6.0)/50 mM CaCl2 solution. Aliquots of 6 μl calf spleen phosphodiesterase (4.0 mU/μl) and 6 μl micrococcal nuclease (200 mU/μl) were added and the samples were incubated for 6 h at 37°C. A volume of 19 μl 0.5 M Tris (pH 10.9) and 3 of μl shrimp alkaline phosphatase (1 U/μl) were added and the incubation was resumed at 37°C for 14 h. The DNA digests were diluted with 200 μl methanol. The mixture was centrifuged and the pellet was extracted with another 200 μl methanol. After evaporation of the solvents the residuals were taken up in 50 μl methanol. Samples were centrifuged at 15 000 g for 15 min and the supernatant was subjected to isotope dilution UPLC–MS/MS for quantification of C8-PhIP-dG. To this end, an Acquity ultra performance liquid chromatography (UPLC) System (Waters, Eschborn, Germany) with an HSS T3 column (1.8 μm, 2.1 × 100 mm, Waters) was used for sample separation. Aliquots of 8 μl were injected and eluted with a gradient of water (solvent A) and acetonitrile (solvent B). Both eluents were acidified with 0.25% acetic acid and 0.25% formic acid. Following a washing interval of 1 min at 90% solvent A, the eluent mixture was changed in a linear gradient to 20% solvent A (1–7 min) at 0.35 ml/min flow rate. The UPLC was connected to a Quattro Premier XE tandem quadrupole mass spectrometer (Waters) with an electrospray interface operating in the positive ion mode. The adduct quantification was based on a characteristic molecule fragmentation, the neutral loss of the furanose ring (C8-PhIP-dG, m/z = 490.1 → 374.1; [15N5,13C10]C8-PhIP-dG, m/z = 505.1 → 384.1). The tune parameters were as follows: temperature of the electrospray source: 110°C; desolvation temperature: 485°C; desolvation gas: nitrogen (950 l/h); cone gas: nitrogen (50 l/h); collision gas: argon (indicated cell pressure ∼ 5 × 10–3 mbar). For the fragmentation of C8-PhIP-dG the collision energy was set to 35 eV. The dwell time was 100 ms and the capillary voltage was 1.0 kV. The cone and RF1 lens voltages were 25 V and 0.1 V, respectively. Data acquisition and handling were performed with MassLynx 4.1 software (Waters). Representative UPLC–MS/MS chromatograms of a digested DNA sample from V79 CS cells incubated with PhIP are shown in the supplementary information (Supplementary Figure S2A).
Alkaline and neutral comet assay
Both alkaline and neutral Comet assays were conducted as reported (36). Comets were analyzed by fluorescence microscopy using a Nikon Microphot-FXA microscope (Perceptive Instruments Ltd., Bury St Edmunds, UK). Fifty cells were scored in each experiment using Comet IV software (Perceptive Instruments Ltd., Bury St Edmunds, UK).
Annexin V/PI staining and analysis of subG1 induction by flow cytometry
Cell cycle distribution and subG1 population were assessed as previously described (37). Briefly, Caco-2 and HCT116 cells were treated with N-OH-PhIP (5 and 2.5 μM, respectively) in the absence or presence of the ATR inhibitor (5 μM) for 48 h. Detached cells and trypsinized adherent cells were pooled, harvested by centrifugation and washed twice in PBS. After ethanol precipitation at −20°C for 1 h, the pellet was resuspended in PBS supplemented with RNase A (20 μg/ml) and incubated for 1 h at RT. PI was then added to a final concentration of 10 μg/ml and cells were analyzed for DNA content by flow cytometry using FACS Canto II (Becton Dickinson, Heidelberg, Germany). Cell cycle distribution and subG1 population was evaluated with FACS Diva software (BD, Heidelberg, Germany). In order to perform Annexin V/PI staining (38), cells were treated as described above. After 48 h, detached and adherent cells were harvested, combined and pelleted by centrifugation. Cells were then resuspended in binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2, 0.1% BSA) containing Annexin V labeled with Alexa Fluor 488 (1:20). After incubation on ice for 15 min, PI (0.5 μg/ml) was added and cells were analyzed by flow cytometry using FACS Canto II (Becton Dickinson, Heidelberg, Germany). Gating of healthy living cells (Annexin V/PI-double negative), early apoptotic cells (Annexin V-positive, PI-negative) and late apoptotic/necrotic cells (Annexin V/PI-double positive) was performed with FACS Diva software (see Supplementary Figure S5A and B).
Immunofluorescence
V79 and V79 CS cells grown on cover slips were treated with 5 and 50 μM PhIP for 24 h, while Caco-2 cells were incubated with 1 and 10 μM N-OH PhIP for up to 44 h. Ionizing radiation (IR) with a Cs137 source was used as positive control. The analysis of γ-H2AX foci was performed as previously described (38). Briefly, cells were fixed in 4% paraformaldehyde/PBS followed by ice-cold methanol. The samples were blocked in PBS containing 5% BSA and 0.3% Triton X-100. After incubation with a primary antibody targeting histone H2AX phosphorylated at Ser139 (#ab81299; Abcam, Cambridge, UK), the cells were incubated with a secondary antibody coupled to Alexa Fluor 488 (Life Technologies, Darmstadt, Germany). Finally, nuclei were counterstained with TO-PRO-3 (Life Technologies, Darmstadt, Germany) and cells were mounted using Vectashield® (Vector Labs, Burlingame, CA, USA). Confocal microscopy was performed using a Zeiss Axio Observer.Z1 microscope equipped with a LSM710 laser-scanning unit (Zeiss, Oberkochen, Germany). Images were acquired in optical sections of 1 μm and processed with ImageJ (NIH, USA). The number of γ-H2AX-foci per nucleus was quantified with ImageJ (50–100 cells/treatment) and data was evaluated by using the GraphPad Prism software. To detect RPA or phospho-Thr21-RPA foci, cells were fixed in ice-cold methanol/acetone (70:30) at −20°C followed by another fixation with 4% PFA for 10 min. After three washing steps, cells were blocked with 10% normal goat serum and incubated with a monoclonal RPA antibody (#NA19L; Merck Millipore, Billerica, USA) or a phospho-specific RPA antibody (#ab109394; Abcam, Cambridge, UK). Subsequently, the samples were incubated with an appropriate secondary antibody conjugated with Alexa Fluor 488 (Life Technologies, Darmstadt, Germany), processed and analyzed as described above.
Western blot analysis
SDS-PAGE and subsequent western blot analysis of cells treated with (N-OH-)PhIP were performed as previously described (39). The following primary antibodies, which were previously validated (38), were used: anti-heat shock protein (Hsp90) α/β (F8, mouse monoclonal; #sc-13119), anti-p21 (rabbit polyclonal; #sc-397) and anti-p53 (DO-1, mouse monoclonal; #sc-126) (all from Santa Cruz, Heidelberg, Germany), anti-γ-H2AX (Ser139, rabbit monoclonal; #ab81299) and anti-phospho-RPA (Thr21, rabbit monoclonal; #ab109394) (both from Abcam, Cambridge, UK), anti-phospho-CHK1 (Ser345, rabbit monoclonal; #2348), anti-CHK1 (mouse monoclonal; #2360), anti-phospho-CHK2 (Thr68, rabbit monoclonal; #2197), anti-CHK2 (rabbit polyclonal; #2662) and anti-ATM (rabbit monoclonal; #2873) (all from Cell Signaling Technology, Denvers, UK) and anti-phospho-ATM (Ser1981, mouse monoclonal; #05-740; Millipore, Billerica, USA).
DNA fiber assay
The DNA fiber assay was performed as previously described (40). Briefly, exponentially growing cells were treated with N-OH-PhIP or ETO in the absence or presence of ATR inhibitor for 14 h, and then pulse-labeled with 25 μM 5-chloro-2′-deoxycytidine (CldU; Sigma, Steinheim Germany) followed by labeling with 250 μM 5-Iodo-2′-deoxycytidine (IdU; TCI Deutschland, Eschborn, Germany) for 30 min each. Labeled cells were harvested and DNA fiber spreads prepared. Fiber spreads were fixed in methanol:acetic acid mixture (3:1, v/v) and air-dried. For DNA denaturation, slides were rehydrated in H2O followed by incubation with 2.5 M HCl for 75 min. After neutralization fiber spreads were incubated for 60 min in blocking buffer (5% goat serum + 1% bovine serum albumin (BSA) in PBS with 0.1% Tween-20). CldU was detected with a monoclonal rat anti-BrdU antibody (#OBT0030G; AbD Serotec, Puchheim, Germany). After post-fixation in 4% paraformaldehyde, the primary antibody was detected by a goat anti-rat fragment [ab′]2 specific Cy3-coupled secondary antibody (Jackson ImmunoResearch, Europe). After a washing step, a monoclonal mouse anti-BrdU ((#347580; Becton–Dickinson, UK) was used to detect IdU, followed by a goat anti-mouse fragment [ab′]2 specific Alexa488-coupled secondary antibody (Life Technologies, Darmstadt, Germany). Fibers were examined and images captured using a LSM 710 confocal microscopy unit supplied with ZEN 2009 software (Zeiss). CldU (red) and IdU (green) tracks were measured using LSM Image Browser (Zeiss) and micrometer values were converted into kilo base pairs. Different classes of labeled tracks were assessed: green-red-green (first pulse origin), red-green (ongoing replication), red (stalled forks), red-green-red (termination) and green (second pulse origin). At least 150 forks from three independent experiments were analyzed.
Real time cell analysis (RTCA) for monitoring cell growth
Cell growth was continuously monitored in an E-Plate 8 for at least 96 h using the iCELLigence RTCA system (ACEA Biosciences, USA). The background impedance signal was measured with 150 μl of cell culture medium followed by the addition of 400 μl cell suspension (V79 and V79 CS cells, respectively). 24 h after seeding, 400 μl of the cell culture supernatant were replaced by 400 μl fresh medium containing different PhIP doses (1, 10 and 50 μM) or the DMSO solvent control. After compound administration, impedance was measured every minute for 2 h and then every 30 min until the end of the experiment. The growth curves were evaluated by RTCA Data analysis software 1.0 and were depicted by GraphPad Prism 5.0.
Determination of cell viability by MTS assay
Cell viability was determined with the Cell Titer 96® AQueous One Solution Cell Proliferation Assay (Promega, Mannheim, Germay) according to the manufacturer's instructions. To this end, cells grown on 96-well plates were incubated with 10 μM N-OH-PhIP for 72 h or the solvent control in the absence or presence of the DDR inhibitors (ATRi, ATMi, DNA-PKi, CHK1i) as indicated. Viability was measured using a 96-well plate reader (Tecan, Crailsheim, Germany).
Analysis of chromosomal aberrations
HCEC were treated with N-OH-PhIP in the absence or presence of ATR inhibitor for 24 h. Subsequently, demecolcine (75 ng/ml) was added and cells were harvested after additional 14 h of incubation. Chromosome preparations were done according to standard protocols. Briefly, cells were harvested and resuspended in pre-warmed (37°C) 75 mM KCl solution and incubated for 10 min. Cells were pelleted and fixed in ice-cold methanol:acetic acid mixture (3:1, v/v). The pelleted fixed cells were resuspended in a small volume of freshly prepared ice-cold fixative and dropped onto pre-cleaned, wet ice-cold slides. After being air-dried, the slides were stained in 5% phosphate buffered Giemsa solution. Fifty metaphases per treatment group were evaluated for chromosomal aberrations (n = 3). The following aberrations were scored: chromatid breaks, chromatid translocations (triradials, quadriradials) and intercalary deletions. Gaps were not included in the final evaluation. Aberration frequencies were expressed as aberrations per chromosome and aberrations per metaphase (normalized to the mean number of chromosomes in 50 metaphases of each treatment variant).
Statistical analysis
All experiments were performed independently three times, except otherwise stated. Results from representative experiments are shown. Values are displayed as means + standard errors of the means (SEM) using GraphPad Prism 5.0 Software. Statistical analysis was performed using two-sided Student's t-test and statistical significance was defined as P < 0.05.
RESULTS
PhIP generates C8-dG DNA adducts and subsequent DNA strand breaks in metabolically competent fibroblasts and Caco-2 cells
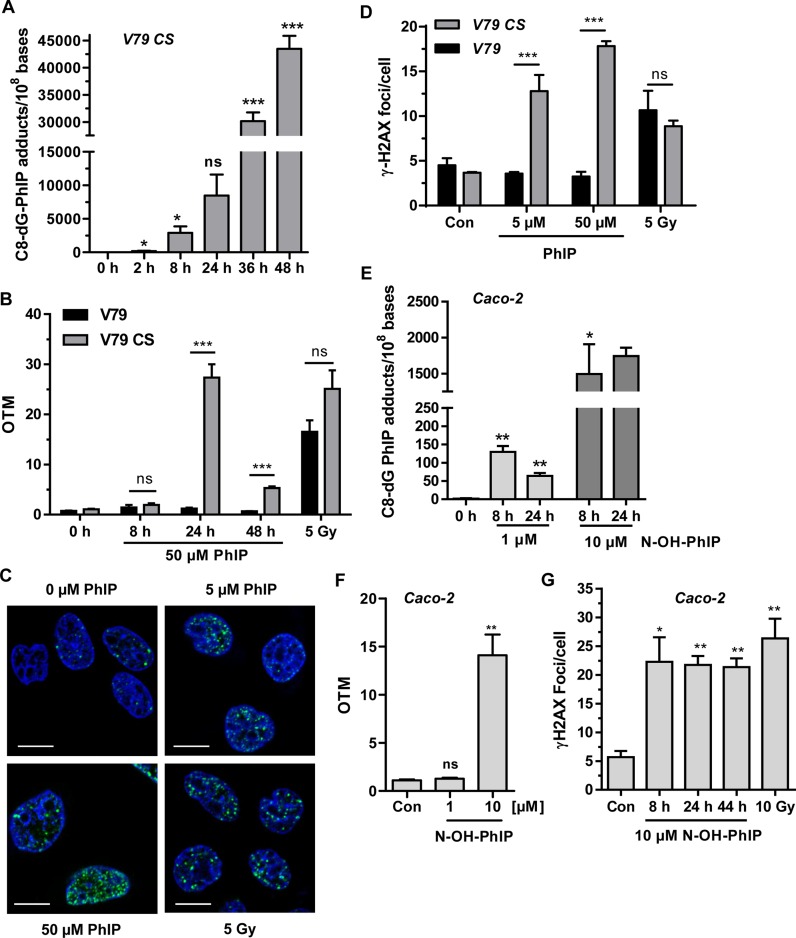
Two V79 hamster fibroblast cell lines, which differ in their ability to catalyze the metabolic activation of PhIP, were used. Parental V79 cells are metabolically deficient, while the derived cell line V79 CS displays both CYP1A2 and SULT1A1 activity required for PhIP activation (Supplementary Figure S1) (32). Initially, V79 CS cells were exposed to 50 μM PhIP for up to 48 h and DNA adduct formation was assessed by quantitative LC-MS/MS analysis with an internal isotope-labeled standard. A time- and concentration-dependent increase of C8-PhIP-dG lesions was observed (Figure 1A and Supplementary Figure S2B), which was confirmed by slot blot analysis (Supplementary Figure S2C and S2D). Next, we analyzed whether the induction of C8-PhIP-dG adducts is associated with the generation of DNA strand breaks. Using the alkaline Comet assay, no DNA strand breaks were detectable in both V79 and V79 CS cells following incubation for 8 h (Figure 1B). However, after treatment for 24 h, a strong (∼25-fold) increase in DNA strand breaks was observed in V79 CS cells treated with 50 μM PhIP, whereas no effect was seen in parental V79 cells (Figure 1B). After 48 h, DNA strand break formation markedly dropped in PhIP-treated V79 CS cells, but was still significantly higher than PhIP-treated V79 cells (Figure 1B). A dose of 5 μM PhIP also caused a higher level of DNA strand breaks in V79 CS cells, which persisted over 48 h (Supplementary Figure S2E). Subsequently, the formation of γ-H2AX, which is catalyzed by both ATM and ATR, was monitored by confocal immunofluorescence microscopy. PhIP (up to 50 μM, 24 h treatment) caused a dose-dependent formation of γ-H2AX foci in V79 CS cells (Figure 1C and D), whereas parental V79 cells displayed no increase in the number of γ-H2AX foci (Supplementary Figure S3B). To further assess the contribution of DSBs to the observed generation of γ-H2AX foci, a neutral Comet assay was conducted. As compared to the control, a 7-fold increase in DSB level was found in V79 CS cells exposed to 50 μM PhIP (Supplementary Figure S2F). This indicates that PhIP is able to generate DSBs and, to an even larger extent, SSBs (see Figure 1B).
Figure 1.
DNA damage induction by PhIP in metabolically competent V79 cells and Caco-2 cells. (A) Time-dependent induction of C8-PhIP-dG adducts in V79 CS cells treated with 50 μM PhIP. Genomic DNA was analyzed for C8-PhIP-dG adducts by mass spectrometry (n = 3). ***P < 0.001, **P < 0.01, *P < 0.05, ns: not significant. (B) Detection of DNA strand breaks by alkaline Comet assay. Parental V79 and metabolically competent V79 CS cells were challenged with 50 μM PhIP as indicated. OTM: olive tail moment. n ≥ 4; ns: not significant; ***P < 0.001. (C) Analysis of γ-H2AX foci in V79 CS cells treated with PhIP for 24 h. Cells were stained with a γ-H2AX antibody followed by an Alexa 488-coupled secondary antibody (green). Nuclei were counterstained with TO-PRO-3 (blue) and images were obtained by confocal microscopy. The scale bar represents 10 μm. (D) Quantitative evaluation of γ-H2AX in parental V79 and V79 CS cells. The number of γ-H2AX foci per nucleus were determined by ImageJ software and evaluated with GraphPad Prism 5.0 (50–100 cells per experiment; n = 3); ns: not significant. ***P < 0.001. (E) Dose- and time-dependent generation of C8-PhIP-dG adducts in Caco-2 cells upon treatment with N-OH-PhIP as revealed by mass spectrometry (n = 3; except for 24 h 10 μM N-OH-PhIP, n = 2); ns: not significant, ***P < 0.001, **P < 0.01, *P < 0.05, as compared to control cells (0 h). (F) Induction of DNA strand breaks by N-OH-PhIP. Caco-2 cells exposed to N-OH PhIP for 24 h were analyzed by alkaline Comet assay (n = 3); ns: not significant, ***P < 0.001, **P < 0.01, *P < 0.05, versus control cells. (G) Analysis of γ-H2AX foci in Caco-2 cells treated with 10 μM N-OH-PhIP. Cells were stained for γ-H2AX and nuclei were counterstained with TO-PRO-3. Images were recorded by confocal microscopy. The number of γ-H2AX foci per nucleus were determined as described above (50–100 cells per experiment; n = 3); **P < 0.01, *P < 0.05 versus control.
In order to translate the data to human colon cells, the DNA damage induction of PhIP was analyzed in Caco-2 cells, an intestinal cell line frequently applied for monitoring luminal gut uptake. These cells are proficient for SULT1A1 activity, but display a low basal level of CYP1A2 (41,42). We therefore used the PhIP metabolite N-OH-PhIP, which does not require CYP activity for its activation. Caco-2 cells were incubated with N-OH-PhIP and C8-PhIP-dG adducts were determined by mass spectrometry. A dose of 1 μM N-OH-PhIP already induced PhIP adducts in Caco-2 cells after 8 h, followed by a 50% decrease after 24 h, which likely reflects ongoing DNA repair (Figure 1E). A higher dose of 10 μM triggered about 10-fold higher adducts levels, which persisted over 24 h. DNA adduct formation was clearly associated with the occurrence of DNA strand breaks at a dose of 10 μM PhIP (Figure 1F). In agreement with the latter finding, a dose of 10 μM N-OH-PhIP caused a pronounced increase in the number of γ-H2AX foci, which did not change over 44 h (Figure 1G and Supplementary Figure S3A). Caco-2 cells treated with 1 μM N-OH-PhIP showed an initial increase in γ-H2AX foci formation after 8 h and a subsequent gradual decrease over the next 36 h (Supplementary Figure S3B). Taken together, these results demonstrate that, once metabolically activated, PhIP generates C8-PhIP-dG adducts and subsequently DNA strand breaks with DDR activation.
PhIP provokes replication stress and activates the ATR-CHK1 pathway
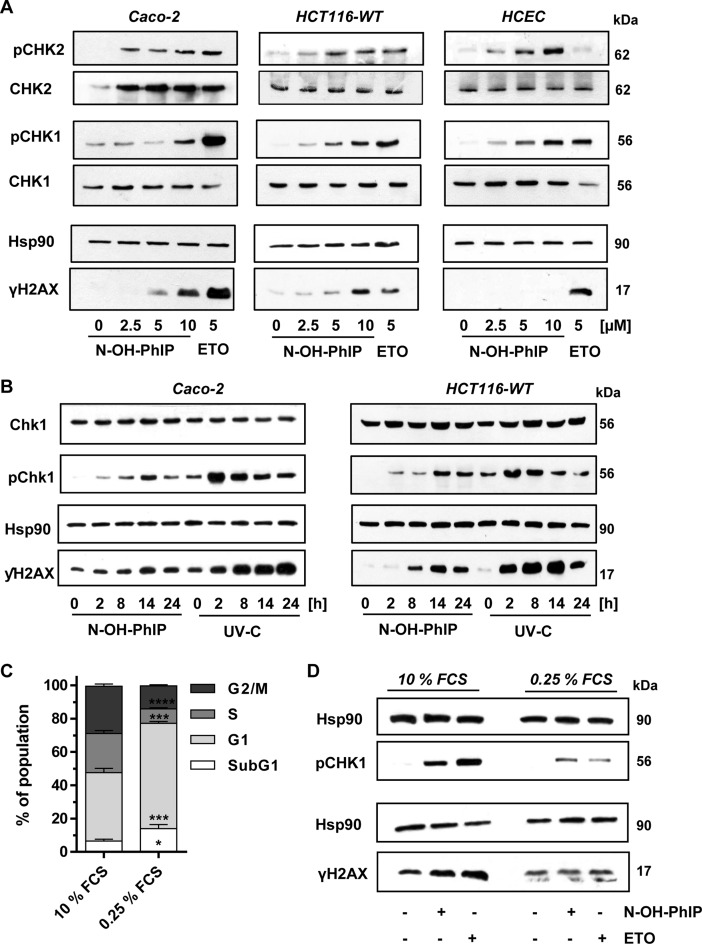
In view of the observed PhIP–DNA adduct and subsequent strand break formation, we hypothesized that PhIP may cause replication stress, thereby triggering ATR-dependent signaling. Parental V79 cells and V79 CS cells were incubated with 50 μM PhIP over 24 h. Phosphorylation of the primary ATR substrate CHK1 and induction of γ-H2AX were not detectable in parental V79 cells (Supplementary Figure S3C, left panel). Conversely, PhIP clearly triggered CHK1 phosphorylation in V79 CS cells, which was observed as early as 4 h after treatment. This was followed by γ-H2AX formation that peaked after 24 h and was accompanied by the induction of the cell cycle regulator p21 (Supplementary Figure S3C, right panel). As in the case of V79 CS cells, N-OH-PhIP caused a dose-dependent phosphorylation of CHK1 in Caco-2 cells (Figure 2A, left panel). In addition, we observed phosphorylation of CHK2, a known substrate of ATM, and a dose-dependent increase in γ-H2AX levels (Figure 2A). As a positive control, we included the topoisomerase II-inhibitor etoposide (ETO) at a dose of 5 μM. ETO is an anticancer agent known to activate both ATM and ATR signaling due to the generation of DNA strand breaks and interference with the replication machinery (43). Furthermore, a comparable pattern of CHK1 and CHK2 activation following N-OH-PhIP treatment was observed in HCT116 colorectal cancer cells (Figure 2A, middle panel) and in non-transformed human colonic epithelial cells (HCEC; Figure 2A, right panel). Notably, γ-H2AX levels were very low in HCEC exposed to N-OH-PhIP. The total level of CHK1 was not changed by N-OH-PhIP treatment in all cell lines tested, whereas ETO reduced the total CHK1 level in HCEC despite a strong increase in pCHK1. The level of total CHK2 was not affected in HCT116 cells and HCEC by incubation with N-OH-PhIP or ETO, while a lower basal CHK2 level was detected in untreated Caco-2 cells.
Figure 2.
N-OH-PhIP triggers the DNA damage response in a proliferation-dependent manner. (A) Dose-dependent activation of the DDR by N-OH-PhIP in Caco-2 cells, HCT116 cells and human colonic epithelial cells (HCEC) after 14 h. Samples were analyzed by SDS-PAGE and western blotting as indicated. Hsp90 was visualized as loading control. Etoposide (ETO) was used as positive control (B) Time-dependent activation of ATR-CHK1 signaling in Caco-2 and HCT116 cells exposed to N-OH-PhIP or UV-C light. Cells were treated with 5 μM N-OH-PhIP or irradiated with UV-C (20 J/m2) and incubated for up to 24 h. Samples were subject to SDS-PGE followed by Western blot analysis as indicated. (C) Cell cycle distribution of HCT116 cells upon serum deprivation. Cells were maintained in normal growth medium (10% FCS) or in serum deprived medium (0.25% FCS) for 24 h. Cell cyle distribution was determined by flow cytometry. (n = 4); ****P < 0.0001, ***P < 0.001, *P < 0.05 versus control (10% FCS). (D) Effect of cell proliferation on ATR-mediated DDR following N-OH-PhIP. Cells were pre-conditioned in medium with 10 or 0.25% FCS for 10 h, treated with N-OH-PhIP (10 μM) or ETO (5 μM) in the conditioned medium and harvested after 14 h (total 24 h). Cells were analyzed as described above.
In order to assess the kinetics of the ATR-CHK1 signaling, Caco-2 and HCT116 cells were exposed to N-OH-PhIP or irradiated with UV-C (20 J/m2) and incubated for up to 24 h. In both cell lines, phosphorylation of CHK1 was detected as early as 2 h following N-OH-PhIP treatment and peaked after 14 h (Figure 2B). yH2AX levels increased in a time-dependent manner with a maximum 14 h after the addition of N-OH-PhIP. This kinetic correlated well with N-OH-PhIP-induced DNA damage (see Figure 1). In contrast, UV irradiation caused an immediate phosphorylation of CHK1, which reached very high levels already after 2 h and then declined (Figure 2B). γ-H2AX increased in a time-dependent manner, reaching a plateau after 24 h in Caco-2 cells, while levels dropped in HCT116 cells (Figure 2B). Collectively, the data show that ATR-mediated DDR following UV exposure occurred much earlier than that induced by N-OH-PhIP, which is attributable to the immediate generation of DNA damage by UV light, such as cyclobutane pyrimidine dimers and (6–4) photoproducts.
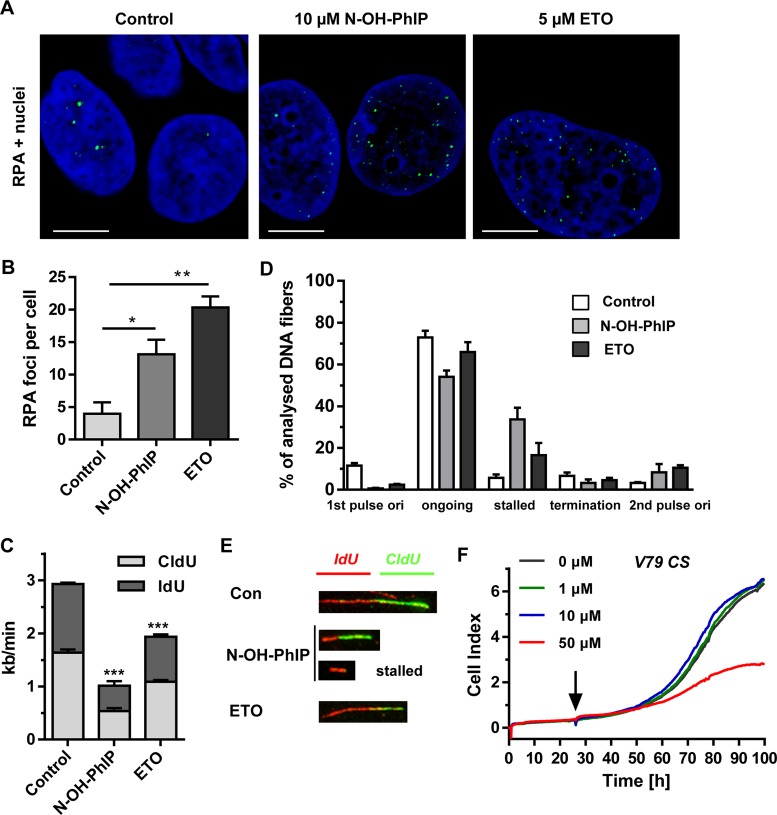
To address the role of replication in ATR activation and yH2AX formation, cells were maintained in serum-deprived medium with 0.25% FCS for 24 h, causing an accumulation of cells in G1 phase with a concomitant decrease in S-phase (24% to 9%) (Figure 2C and Supplementary Figure S4A). The serum-starved cells showed reduced pCHK1 and γH2AX levels following both N-OH-PhIP and ETO treatment (Figure 2D) as compared to cells grown in normal growth medium, indicating that ATR activation occurs in a replication-dependent manner. To detail the role of N-OH-PhIP-induced DNA damage during replication, the formation of RPA foci was studied. RPA rapidly binds to single stranded DNA (ssDNA) generated at stalled replication forks and is necessary for ATR activation (25). While untreated cells displayed only a small number of RPA foci, N-OH-PhIP caused a considerable increase in the number of RPA foci (Figure 3A and B) supporting the notion that PhIP induces replication stress. To address this aspect in more detail, a DNA fiber assay was used. Caco-2 cells were incubated with 10 μM N-OH-PhIP for 14 h and then pulse-labeled with the thymidine analogues CldU and IdU for 30 min each. The prepared DNA fibers were labeled with antibodies directed against CldU and IdU followed by confocal microscopy, which allows for the determination of replication fork speed and the assessment of different replication structures. Incubation with N-OH-PhIP caused a strong reduction in the average fork speed, the effect being even higher than in ETO-treated cells (Figure 3C and Supplementary Figure S3D). Further analysis revealed that N-OH-PhIP increased the number of stalled replication forks, while it reduced the number of ongoing forks (Figure 3E and F). In line with these findings, a dose of 50 μM PhIP potently decreased proliferation in metabolically proficient V79 CS cells as shown by real-time analysis of cell growth (Figure 3F). In contrast, parental V79 cells incubated with PhIP exhibited normal growth and were only marginally affected at the highest PhIP dose (50 μM) (Supplementary Figure S3E). Collectively, the results provide evidence that PhIP-induced DNA damage blocks DNA replication, which elicits ATR-CHK1 signaling with a concomitant formation of RPA foci and an activation of the ATM-CHK2 signaling.
Figure 3.
N-OH-PhIP induces replication stress. (A) Detection of RPA foci. Caco-2 cells were treated as indicated for 14 h, stained with a RPA antibody and subjected to confocal microscopy. Representative images are shown. RPA is depicted in green and nuclei are blue. The scale bar represents 10 μm. (B) Quantitative evaluation of RPA foci in Caco-2 cells shown in (B). The number of RPA foci per nucleus were determined by ImageJ software and evaluated with GraphPad Prism 5.0 (> 100 cells per experiment; n = 3); **P<0.01, *P<0.05 versus control. (C) Impact of N-OH-PhIP on average replication fork speed. Cells were treated for 14 h as described and replication speed was determined by DNA fiber assay followed by confocal microscopy (n = 3); ***P < 0.001 versus control. (D) Distribution of replication structures assessed by the DNA fiber assay in Caco-2 cells exposed to N-OH-PhIP or ETO for 14 h. (E) Representative DNA fiber tracks showing ongoing replication (red-green) and stalled forks (red). (F) Impact of PhIP on the proliferation of V79 CS cells. After 24 h, cells were treated with increasing doses of PhIP (black arrow) and growth was monitored by an impedance-based analysis over the following 72 h.
Inhibition of ATR potentiates PhIP-induced DSBs and promotes ATM activation and RPA phosphorylation
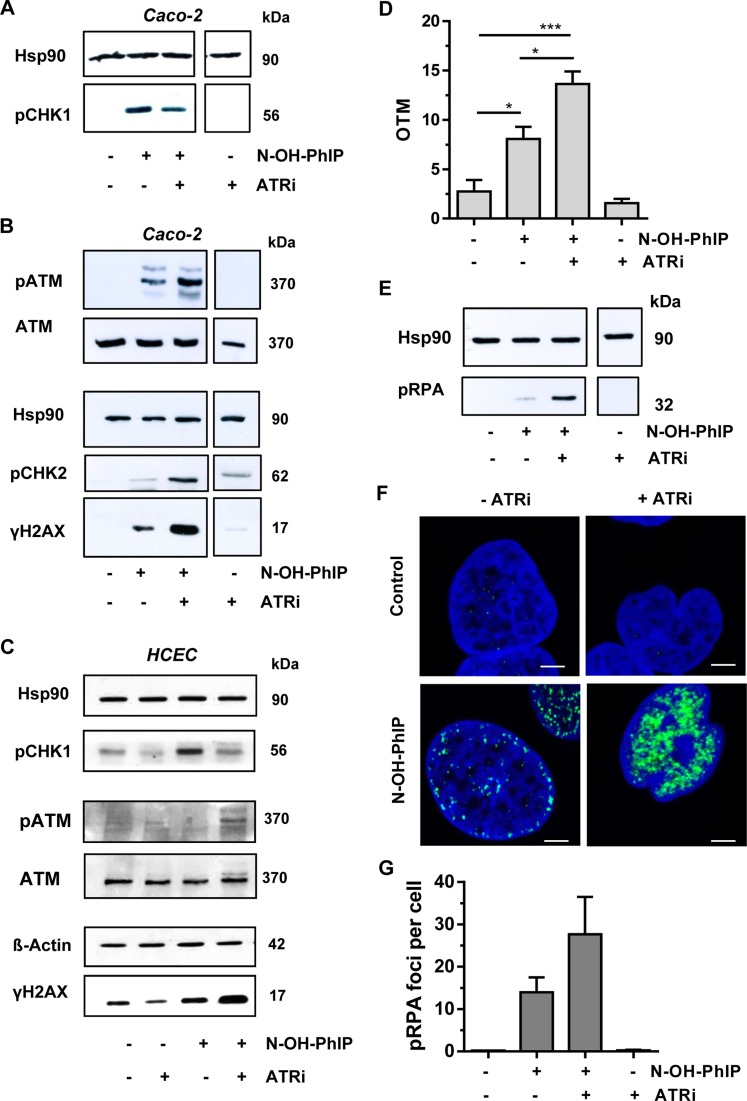
Since ATR plays an important role in the cellular response to replication stress, we set out to determine the effects of ATR inhibition. Caco-2 cells were incubated with N-OH-PhIP in the presence or absence of the selective ATR inhibitor VE821. Western blot analysis revealed that ATR inhibition strongly reduced the phosphorylation of CHK1 upon N-OH-PhIP treatment, demonstrating the effectiveness of the inhibitor (Figure 4A and C). Next, we determined the influence of ATR inhibition on DDR signaling and found a strong activation of the ATM pathway, as evidenced by increased phosphorylation of both ATM and CHK2 (Figure 4B). In agreement with this observation, the levels of γ-H2AX also greatly increased in drug and inhibitor co-treated cells (Figure 4B). The same set of experiments was conducted in non-transformed HCEC, also showing an elevated ATM and H2AX phosphorylation following ATR inhibition (Figure 4C).
Figure 4.
ATR inhibition potentiates DSB induction and activates ATM signaling in response to N-OH-PhIP. (A and B) Impact of ATR inhibition on N-OH-PhIP-induced DDR. Caco-2 cells were incubated with N-OH-PhIP (10 μM) in the absence or presence of the ATR inhibitor (ATRi) VE821 for 14 h. Samples were then subjected to SDS-PAGE followed by western blot analysis as indicated. Hsp90 served as loading control. (C) ATR inhibition in human colonic epithelial cells (HCEC) exposed to N-OH-PhIP. Cells were treated and analyzed as described above. (D) DSB induction upon ATR inhibition. Caco-2 cells were treated as stated above and DSB formation was measured using the neutral Comet assay (n = 4); ns: not significant. *P < 0.05, ***P < 0.001. (E) Influence of ATR inhibition on threonine-21 RPA phosphorylation. Caco-2 cells were treated as described in (A) and analyzed for pRPA. Hsp90 was used as loading control. (F) Confocal microscopy of RPA Thr21 phosphorylation. Caco-2 cells were incubated for 14 h as described above, fixed and stained with a p-Thr21-RPA antibody followed by an Alexa 488-coupled secondary antibody (green). Nuclei were counterstained with TO-PRO-3 (blue). Samples were then analyzed by confocal microscopy and processed by ImageJ software. The scale bar represents 5 μm. (G) Quantitative evaluation of pRPA staining. The number of pRPA foci per nucleus were assessed by ImageJ software and evaluated with GraphPad Prism 5.0 (>100 cells per experiment; n = 2).
Given that ATM is activated by DSBs, we hypothesized that ATR inhibition leads to enhanced DSB formation due to the collapse of stalled replication forks. The DSB level was already elevated by exposure of Caco-2 cells to N-OH-PhIP (Figure 4D). However, upon ATR inhibition, DSB induction by N-OH-PhIP was almost doubled. We also analyzed the phosphorylation of the RPA32 subunit at Thr21, which is mainly catalyzed by DNA-PK (43). DNA-PK is known to be activated following recruitment to the Ku heterodimer at the free ends of a DSB (44). Cells exposed to N-OH-PhIP showed a slight increase of phosphorylated RPA (pRPA), while concomitant inhibition of ATR by VE821 highly stimulated RPA phosphorylation at Thr21 (Figure 4E). The same response was observed in HCEC (Supplementary Figure S4B). The findings were further substantiated by confocal immunofluorescence microscopy. An enhanced formation of pRPA foci was observed after the combined treatment of Caco-2 cells with N-OH-PhIP and VE821 (Figure 4F and G). Taken together, the inhibition of ATR signaling resulted in significantly enhanced DSB levels and upregulation of the ATM pathway following N-OH-PhIP treatment.
Involvement of the DDR kinases ATM and DNA-PKcs in PhIP-triggered replication stress
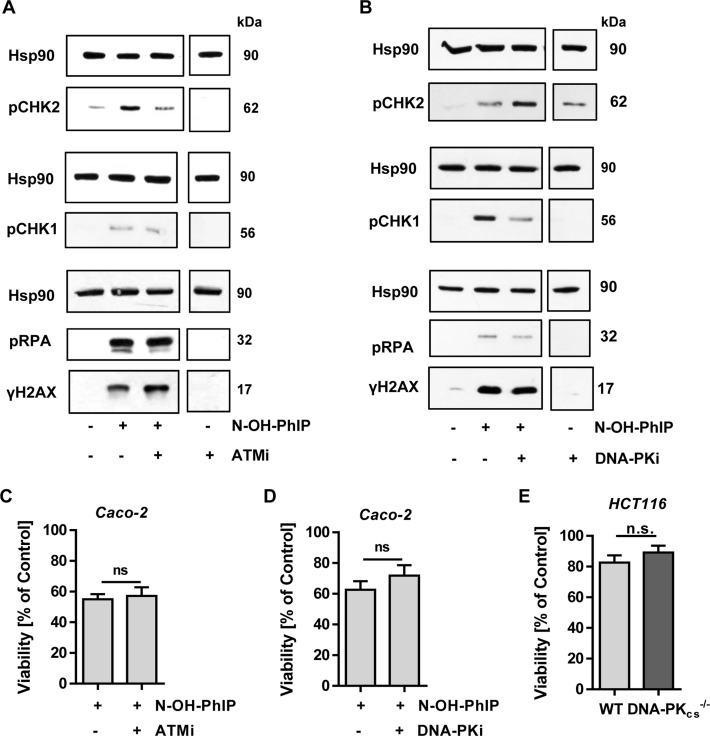
Next, we tested whether inhibition of the other PIKKs, ATM and DNA-PKcs, may affect the DDR triggered by N-OH-PhIP in Caco-2 cells. Abrogation of ATM signaling by the specific inhibitor KU55933 did not alter CHK1 and RPA phosphorylation in response to N-OH-PhIP, but suppressed CHK2 phosphorylation as expected (Figure 5A). N-OH-PhIP-induced phosphorylation of H2AX rather increased after treatment of cells with the ATM inhibitor (Figure 5A, bottom panel), which may point to the involvement of the DDR kinases ATR and DNA-PK. Furthermore, blocking of DNA-PK with the inhibitor NU7026 revealed an attenuated CHK1 phosphorylation following N-OH-PhIP treatment, while CHK2 phosphorylation was increased (Figure 5B). Phospho-modification of RPA and H2AX was not altered by the combined treatment of N-OH-PhIP and the DNA-PK inhibitor (Figure 5B, bottom panel). We also determined whether the inhibition of ATM and DNA-PK would have an impact on cell viability following N-OH-PhIP treatment. The ATM inhibitor KU55933 did not sensitize cells to N-OH-PhIP, thereby indicating that ATM is dispensable for cell survival after N-OH-PhIP-triggered replication stress (Figure 5C). Next, the role of DNA-PKcs was addressed in Caco-2 cells using the DNA-PK inhibitor NU7026 (Figure 5D), and in isogenic HCT116 cells proficient or deficient for DNA-PKcs (Figure 5E), revealing no significant differences in viability (Figure 5D and E). Taken together, these experiments revealed a minor role of both ATM and DNA-PKcs in the PhIP-induced replication stress response.
Figure 5.
Impact of ATM and DNA-PK inhibition on N-OH-PhIP triggered DDR and cytotoxicity. (A) Influence of ATM inhibition on N-OH-PhIP-induced DDR. Caco-2 cells were treated with N-OH-PhIP (10 μM) in the absence or presence of the ATM inhibitor (ATMi) KU-55933 for 14 h. Samples were then separated by SDS-PAGE followed by Western Blotting as indicated. Hsp90 served as loading control. (B) Effect of DNA-PK inhibition on N-OH-PhIP-induced DDR. Cells were treated with N-OH-PhIP (10 μM) with or without DNA-PK inhibitor (DNA-PKi) NU7026 for 14 h. Samples were then subjected to SDS-PAGE and western blot analysis was performed as indicated. Hsp90 was used as loading control. (C) Impact of ATM inhibition on cell viability. Caco-2 cells were incubated with 10 μM N-OH-PhIP with or without ATM inhibitor KU-55933 for 72 h and viability was determined (n = 5); ns: not significant. (D) Impact of DNA-PK inhibition on cell viability. Cells were incubated as described above with or without DNA-PK inhibitor (DNA-PKi) NU7026 for 72 h and viability was determined (n = 6); ns: not significant. (E) Impact of DNA-PKcs deficiency on cell viability. HCT116 cells proficient in DNA-PKcs (WT) and deficient in DNA-PKcs (DNA-PKcs−/−) were treated with or without 10 μM N-OH-PhIP and viability was measured after 72 h (n = 3); ns: not significant.
ATR promotes cell survival upon PhIP-triggered replication stress
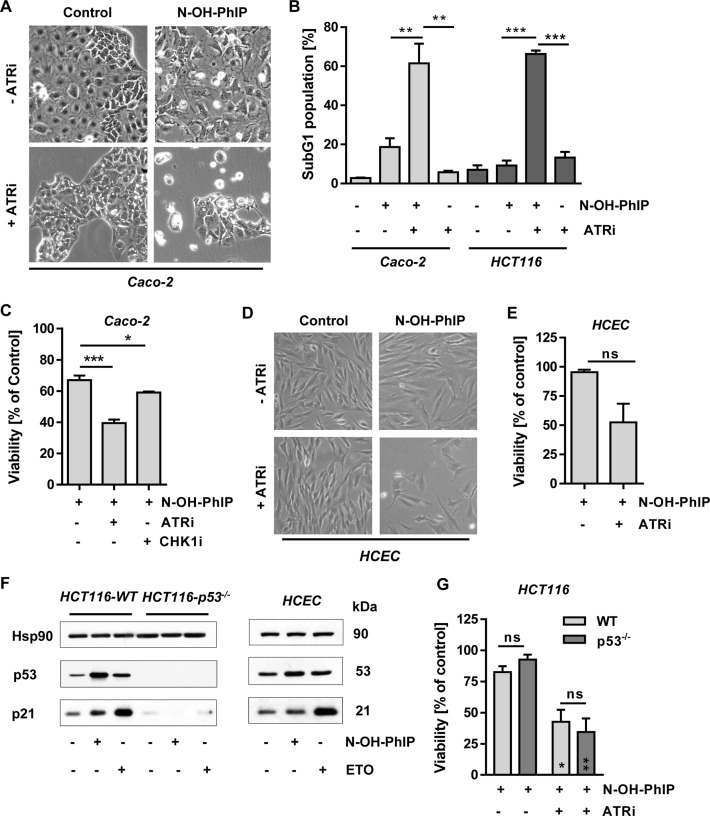
We then wished to address the cellular consequences of ATR inhibition. Caco-2 cells were treated with 5 μM N-OH-PhIP for 48 h with or without the ATR inhibitor VE821. N-OH-PhIP caused some cell rounding and detachment, which was greatly enhanced in the presence of the ATR inhibitor, thus pointing to the activation of cell death pathways (Figure 6A). These dramatic morphological changes were also observed in HCT116 cells treated with N-OH-PhIP and ATRi (Supplementary Figure S5A). Moreover, N-OH-PhIP led to an increase of the subG1 population, which is indicative of apoptotic cell death and was more pronounced in Caco-2 cells. This effect was strongly increased in both cell lines by the concomitant ATR inhibition, 60% of cells being in the subG1 phase (Figure 6B and Supplementary Figure S5A and SB). Annexin V/PI staining revealed that N-OH-PhIP increased both early apoptotic and late apoptotic/necrotic cell populations. Concomitant ATR inhibition potentiated cell death and particularly enhanced the population of Annexin V/PI-double positive cells, representing late apoptotic and necrotic cells (Supplementary Figure S5C and D), In line with these findings, the viability of Caco-2 cells strongly decreased after incubation with N-OH-PhIP and the ATR inhibitor for 72 h (Figure 6C). Consistent with these results, inhibition of the ATR substrate CHK1 by the compound UCN-01 also moderately enhanced the cytotoxicity of N-OH-PhIP (Figure 6C). As observed in the CRC cell lines, pharmacological abrogation of ATR signaling greatly sensitized non-transformed HCEC to N-OH-PhIP, as reflected by morphological changes and a strong decrease in viability (Figure 6D and E). Finally, the role of the tumor suppressor p53 was addressed using isogenic HCT116 cells proficient or deficient in p53. Both N-OH-PhIP and ETO clearly led to a p53 accumulation and concomitant induction of its downstream target p21, which was completely absent in p53-lacking cells (Figure 6F, left panel). Induction of p53 and p21 by N-OH-PhIP was also confirmed in HCEC (Figure 6F, right panel). p53-deficient HCT116 cells displayed a slightly higher viability upon N-OH-PhIP exposure as compared to their wildtype counterparts (93% vs. 83%), which was not statistically significant (Figure 6G). This little difference disappeared in the presence of ATRi, which strongly reduced viability in both cell lines to the same extent. In summary, the data revealed that ATR is crucial for the protection against PhIP-induced replication stress and cytotoxicity.
Figure 6.
ATR promotes cell survival following N-OH-PhIP-dependent replication stress. (A) Effect of ATR inhibition on cell morphology. Caco-2 cells were exposed to N-OH-PhIP (5 μM) with or without ATR inhibitor for 48 h and morphology was monitored by phase-contrast microscopy. (B) Impact of ATR inhibition on cell death induction. Caco-2 and HCT116 cells were exposed to N-OH-PhIP (5 μM and 2.5 μM, respectively) with or without ATR inhibitor for 48 h and the percentage of cells in the subG1 fraction was determined (n = 4); **P < 0.01, ***P < 0.001. (C) Determination of cell viability in response to ATR inhibition. Caco-2 cells were treated with 10 μM N-OH-PhIP in the absence or presence of ATR and CHK1 inhibitors for 72 h and viability was assessed (n = 4); *P < 0.01, ***P < 0.001. (D) Effect of ATR inhibition on cell morphology in non-transformed human colonic epithelial cells (HCEC). HCEC were treated with N-OH-PhIP (2.5 μM) with or without ATR inhibitor for 48 h and morphology was monitored by phase-contrast microscopy. (E) Determination of cell viability in response to ATR inhibition. HCEC were incubated with 10 μM N-OH-PhIP in the absence or presence of ATR inhibitor for 72 h and viability was assessed (n = 3); ns: not significant. (F) Activation of p53 and p21 by N-OH-PhIP, HCT116 cells and HCEC were exposed to N-OH-PhIP (10 μM) for 14 h. Etoposide (ETO) was used as positive control. Samples were analyzed by SDS-PAGE and Western Blotting as indicated. Hsp90 was visualized as loading control. (G) Impact of p53 status on cell survival. HCT116 cells proficient in p53 (WT) and deficient in p53 (p53−/−) were treated or not with 10 μM N-OH-PhIP in the absence or presence of ATR inhibitor. Viability was monitored after 72 h (n = 3); **P < 0.005, *P < 0.05 versus the respective N-OH-PhIP single treatment.
ATR maintains chromosomal stability following PhIP-induced replication stress
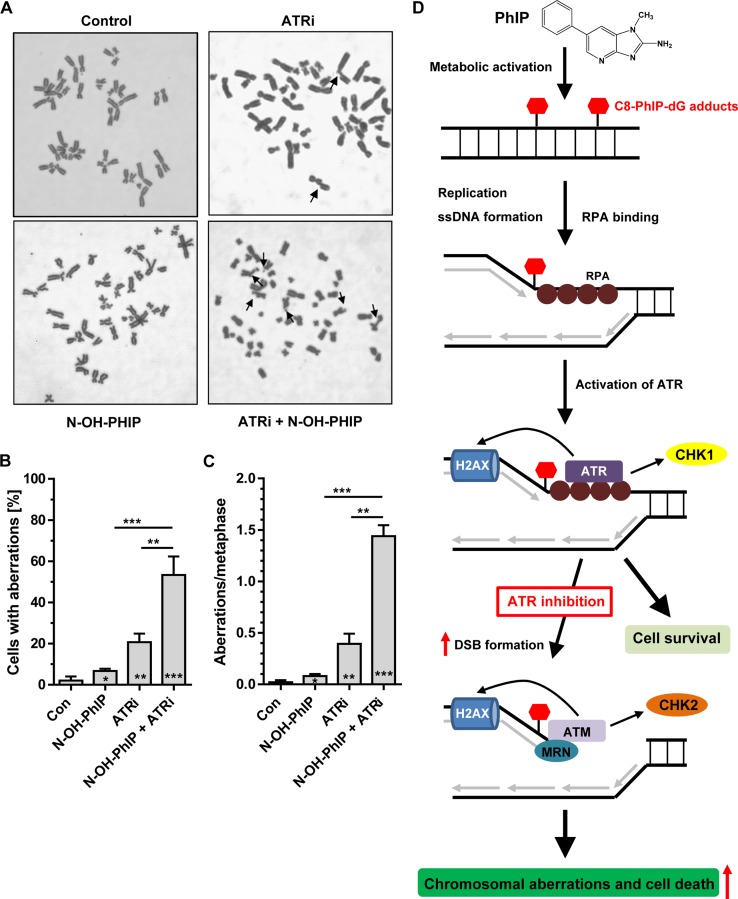
In order to assess the significance of ATR in the maintenance of chromosomal stability, non-transformed HCEC displaying a normal karyotype were incubated with 2.5 μM N-OH-PhIP in the presence or absence of ATRi. After 24 h, cells were treated with demecolcine for 14 h and harvested. Prepared metaphase spreads were stained with Giemsa and analyzed for chromosomal aberrations by light microscopy. Representative metaphase spreads are depicted in Figure 7A. Control cells displayed a very low frequency of aberrations, while N-OH-PhIP alone slightly increased the cell population with aberrations and the number of aberrations per metaphase (Figure 7B and C). The pharmacological inhibition of ATR markedly increased the frequency of chromosomal aberrations (Figure 7B and C). This highlights the important role of ATR in maintaining genomic stability even in the absence of induced replication stress. Intriguingly, concomitant ATR inhibition and N-OH-PhIP treatment strongly potentiated both the frequency of cells with aberrations and the number of aberrations per metaphase spread (Figure 7B and C). This finding together with the observed elevated DSB levels upon ATR inhibition (Figure 4) demonstrates that ATR is essential to limit replication fork collapse, DSB formation and subsequent chromosomal instability following PhIP exposure. A model of PhIP-triggered replication stress and DNA damage response is outlined in Figure 7D, which highlights the importance of ATR in preserving chromosomal stability and promoting cell survival in response to PhIP.
Figure 7.
Chromosomal aberrations induced by N-OH-PhIP in human colonic epithelial cells (HCEC) and the proposed model of PhIP-induced replication stress. (A) HCEC were incubated with 2.5 μM N-OH-PhIP for 24 h in the absence or presence of 2.5 μM ATR inhibitor prior to the addition of demecolcine. After 14 h, the cells were fixed and processed as described. Metaphase spreads (50 per treatment group, n = 3) were evaluated by phase-contrast microscopy and representative pictures thereof are shown. (B and C) Aberration frequencies expressed as aberrations per cell and aberrations per metaphase (normalized to the mean number of chromosomes in 50 metaphases of each treatment variant); *P < 0.05, **P < 0.01, ***P < 0.001 versus control cells. (D) Proposed model of PhIP-induced replication stress. Upon metabolic activation, PhIP induces the formation of C8-PhIP-dG adducts in DNA. These bulky lesions stall replicative polymerases, leading to ssDNA formation and subsequent accumulation of RPA. In turn, ATR is activated and phosphorylates its downstream targets CHK1 and H2AX, thereby conferring protection against PhIP-induced replication stress and promoting cell survival. Inhibition of ATR potentiates the formation of DSBs, which are recognized by the DNA damage sensor MRN, leading to the recruitment and activation of ATM. The enhanced DSB level results in both increased chromosomal aberrations and cell death.
DISCUSSION
This study focused on the food-borne carcinogen PhIP, which was previously shown to be genotoxic, giving rise to bulky C8-dG DNA adducts upon metabolic activation (9,45,46). Although PhIP is involved in dietary cancer formation (16,20,47), little is known about the DDR elicited by this carcinogen. We hypothesized that PhIP DNA adducts may induce replication stress by stalling replicative polymerases during S-phase. First, we demonstrate that the PhIP-induced DNA adducts and DNA strand break formation strictly rely on the metabolic activation of PhIP by CYP1A2 and SULT1A1, supporting previous studies (45,48). At high doses of this dietary carcinogen (>1 μM), C8-PhIP-dG adducts accumulated over time. In contrast, a low dose of 1 μM N-OH-PhIP caused a maximum adduct level after 8 h followed by a 50% reduction after 24 h, thus indicating ongoing NER. The generation of C8-PhIP-dG adducts, which are the main PhIP-DNA adducts (49), was accompanied by the induction of DNA strand breaks in V79 CS and Caco-2 cells. The level of DNA strand breaks strongly declined in PhIP-treated V79 CS cells after 48 h, despite a further increase of the adduct levels after 36 and 48 h. This may be a result of the decreased cell proliferation due to PhIP-dependent growth arrest and might imply that DNA replication is necessary for PhIP-dependent DNA strand break formation.
PhIP as well as its metabolite N-OH-PhIP triggered an ATR-dependent DDR, as evidenced by the phosphorylation of CHK1 and H2AX, and was shown to correlate closely with the kinetics of DNA damage induction. The ATR-mediated DDR showed a later onset than after UV irradiation, which immediately activated ATR-CHK1 signaling. This is in agreement with a previous study in HeLa cells exposed to UV-C light (50). These N-OH-PhIP-dependent effects were observed in metabolically proficient V79 cells, Caco-2 and HCT116 colorectal cancer cells as well as in non-transformed human colonic epithelial cells (HCEC). The reduced γH2AX formation observed in HCEC might be attributable to a lower SULT1A1 expression, resulting in weaker metabolic activation of N-OH-PhIP and DNA adduct formation. Another explanation might be a smaller population of S-phase cells in HCEC, which would affect the levels of N-OH-PhIP induced replication stress and strand breaks.
It was previously shown that, apart from replication-dependent activation, bulky adducts are capable of directly activating the apical DDR kinases. Using human purified proteins, plasmid DNA with bulky adducts induced by N-acetoxy-2-acetylaminofluorene (N-acetoxy-AAF) and UV light was revealed to activate ATR in a TopBP1 (topoisomerase IIb binding protein 1)-dependent manner and stimulated CHK1 phosphorylation (51,52). Further studies performed with cell free extracts provided evidence that plasmid DNA harboring bulky adducts introduced by N-acetoxy-AAF or benzo[a]pyrene diol epoxide can also directly engage ATM and DNA-PKcs, contributing to CHK1 phosphorylation (53). To address this issue, we conducted experiments with serum-starved cells treated with either N-OH-PhIP or ETO and compared those with cells grown under normal conditions. Our findings revealed a reduction of both CHK1 and H2AX phosphorylation, indicating that ATR activation occurs in a rather replication-dependent manner. This is also supported by our DNA fiber assay, which unveiled the N-OH-PhIP-dependent induction of stalled replication forks, a well-known trigger of ATR activation (27). Whether C8-PhIP-dG adducts also directly activate ATR or other PIKKs remains to be shown in future studies using cell extracts and plasmid DNA harboring PhIP adducts.
In line with the activation of ATR-CHK1 signaling, N-OH-PhIP led to the formation of RPA foci and strongly reduced the average replication fork speed with a concomitant increased number of stalled forks as shown by a DNA fiber assay. At the same time, CHK2 phosphorylation was detected. This effect could be due to a direct ATM activation or could be generated by the endonuclease-mediated cleavage of stalled replication forks, leading to DSBs and ATM activation. In support of this notion, N-OH-PhIP moderately caused ATM phosphorylation and induced DSBs. Our findings show that (N-OH-)PhIP triggers both the ATR-CHK1 and, to a lesser extent, the ATM-CHK2 pathway, both of which are known to converge on the tumor suppressor protein p53 (54). However, Caco-2 cells harbor one deleted and one mutated p53 allele, resulting in the absence of p53 protein expression (55). Therefore, isogenic HCT116 cells proficient and deficient for p53 were used, revealing a p53 induction upon N-OH-PhIP exposure, which was associated with the upregulation of the cell cycle regulator p21 in a p53-dependent manner. Notably, the stabilization of p53 and the increase of p21 levels were also detected in HCEC. This observation is in agreement with previous studies performed in TK6 lymphoblastoid cells and MCF10A human mammary epithelial cells co-cultured with metabolically competent cells (14,56). Interestingly, HCT116-p53−/− cells displayed a slightly higher viability than their wildtype counterpart following PhIP exposure, which might be attributed to decreased p53-mediated apoptosis or to the loss of p21 induction.
We further demonstrate that the pharmacological abrogation of ATR signaling largely blocked the N-OH-PhIP-dependent CHK1 phosphorylation, this effect being accompanied by a robust activation of the ATM-CHK2 pathway and elevated DSB levels. This is in agreement with another report showing an accelerated formation of DSBs upon replication stress in ATR-deficient cells, resulting in the engagement of ATM and DNA-PK (57). Furthermore, an increased RPA phosphorylation at Thr-21 was observed following an incubation of Caco-2 cells with N-OH-PhIP and the ATR inhibitor. This points to an activation of DNA-PK, since phospho-modification of RPA at Thr-21 is almost exclusively catalyzed by DNA-PK, while ATM plays, if at all, a minor role (43). To determine the role of the other PIKKs ATM and DNA-PKcs in PhIP-dependent DDR, specific pharmacological inhibitors were used. As expected, ATM inhibition hardly affected RPA-Thr21 and CHK1 phosphorylation in response to N-OH-PhIP. However, ATM inhibition slightly increased γ-H2AX formation following N-OH-PhIP treatment, thereby suggesting an upregulation of DNA-PK activity. In turn, we found that DNA-PKcs inhibition resulted in a marked reduction of CHK1 phosphorylation induced by N-OH-PhIP. This indicates a contribution of DNA-PK to N-OH-PhIP-induced CHK1 phosphorylation. Interestingly, a recent study demonstrated that DNA-PK is required for full activation of ATR-CHK1 signaling upon replication stress and a compromised CHK1 phosphorylation in DNA-PKcs defective cells (58). While ATM inhibition does not impact on the viability in N-OH-PhIP treated cells, pharmacological or genetic ablation of DNA-PKcs slightly elevated viability, which might point to an involvement of DNA-PK in N-OH-PhIP induced cytotoxicity. In a next step, we analyzed whether ATR protects against the detrimental consequences of PhIP-induced replication stress. PhIP and its metabolite N-OH-PhIP reduced the viability of V79 CS, Caco-2 and HCT116 cells, which is consistent with previous studies using co-cultures of a metabolically active cell line and a target cell line (14,56). Intriguingly, inhibition of ATR greatly sensitized cells towards the cytotoxic effects of N-OH-PhIP as confirmed by viability assays and cell death measurements in both CRC cell lines and non-transformed HCEC. This is in agreement with the notion that ATR is indispensable for fork stability and fork restart upon sustained replication stress. ATR signaling abrogation or failure causes RPA depletion due to the continuous production of ssDNA and finally results in the formation of DSBs and replicative catastrophe (59). The increased sensitivity upon ATR inhibition was independent of the p53 status, which was demonstrated in isogenic p53-wt and p53-null HCT116 cells. Cell death measurements provided evidence that N-OH-PhIP triggers mainly apoptotic cell death, which is potentiated in the presence of ATR inhibition. We therefore conclude that ATR-CHK1 signaling is a survival pathway that limits detrimental replication stress induced by N-OH-PhIP, which is line with the notion that both ATR and ATM balance pro-survival and pro-death pathways upon DNA damage (60). Further studies are required to reveal the precise mechanism of PhIP-induced cell death and to analyze the role of alternative death pathways, such as necrosis.
To elucidate the contribution of PhIP-induced replication stress to chromosomal instability, chromosomal aberrations were assessed in HCEC carrying a normal karyotype. These experiments showed that N-OH-PhIP moderately increased both the frequency of cells with aberrations and the number of aberrations per metaphase. The concomitant pharmacological abrogation of ATR dramatically increased the chromosomal instability induced by N-OH-PhIP, thus highlighting the pivotal role of ATR in the PhIP-triggered replication stress response and the maintenance of genome integrity. Interestingly, replication stress has recently emerged as an important factor leading to chromosomal instability (CIN) during malignant transformation, as shown in CIN+ versus CIN– colorectal carcinoma cells (61). By triggering replication stress, PhIP is able to induce DSBs and thus promotes chromosomal instability, in particular if ATR signaling is compromised as demonstrated here. Heterozygous loss of ATR renders cells hypersensitive to genotoxic stress, leading to fragile site instability and chromosomal aberrations (62). Furthermore, somatic mutations in ATR and CHK1 were identified in human tumors with microsatellite instability (63,64). Therefore, it is reasonable to posit that ATR might play a role in the protection against PhIP-induced colorectal cancer.
In conclusion, we demonstrate for the first time that the dietary carcinogen PhIP triggers DNA damage-dependent replication stress, with ATR, but not ATM and DNA-PK, playing a central role in the protection against the detrimental effects induced by this food-borne carcinogen. Screenings of colorectal tumor specimens as to a deregulation in the ATR-CHK1 signaling axis are warranted, which may provide further insight into the mechanism underlying dietary-induced CRC formation.
Supplementary Material
Acknowledgments
We are grateful to Anna Frumkina for performing the alkaline Comet assay, to Dr Wynand Roos for critically reading the manuscript and Dr Marcus Eich for fruitful discussions (all from the Department of Toxicology, Mainz). We would like to thank Dr Hans-Peter Rodemann (Department of Radiation Oncology, University of Tübingen, Germany) and Dr Bert Vogelstein (John Hopkins University, Baltimore, USA) for providing cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University Medical Center Mainz (MAIFOR) [972 8716]; German Research Foundation (DFG-FA1034). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parkin D.M., Boyd L., Walker L.C. 16. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br. J. Cancer. 2011;105(Suppl. 2):S77–S81. doi: 10.1038/bjc.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Bouvard V., Loomis D., Guyton K.Z., Grosse Y., Ghissassi F.E., Benbrahim-Tallaa L., Guha N., Mattock H., Straif K. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16:1599–1600. doi: 10.1016/S1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 4.Sugimura T., Wakabayashi K., Nakagama H., Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahrer J., Kaina B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis. 2013;34:2435–2442. doi: 10.1093/carcin/bgt275. [DOI] [PubMed] [Google Scholar]
- 6.Turesky R.J., Le Marchand L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem. Res. Toxicol. 2011;24:1169–1214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turesky R.J. Formation and biochemistry of carcinogenic heterocyclic aromatic amines in cooked meats. Toxicol. Lett. 2007;168:219–227. doi: 10.1016/j.toxlet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Nicken P., Schroder B., von Keutz A., Breves G., Steinberg P. The colon carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is actively secreted in the distal colon of the rat: an integrated view on the role of PhIP transport and metabolism in PhIP-induced colon carcinogenesis. Arch. Toxicol. 2013;87:895–904. doi: 10.1007/s00204-012-1006-0. [DOI] [PubMed] [Google Scholar]
- 9.Lin D., Kaderlik K.R., Turesky R.J., Miller D.W., Lay J.O., Jr, Kadlubar F.F. Identification of N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem. Res. Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 10.Stevnsner T., Frandsen H., Autrup H. Repair of DNA lesions induced by ultraviolet irradiation and aromatic amines in normal and repair-deficient human lymphoblastoid cell lines. Carcinogenesis. 1995;16:2855–2858. doi: 10.1093/carcin/16.11.2855. [DOI] [PubMed] [Google Scholar]
- 11.Klein J.C., Beems R.B., Zwart P.E., Hamzink M., Zomer G., van Steeg H., van Kreijl C.F. Intestinal toxicity and carcinogenic potential of the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in DNA repair deficient XPA-/- mice. Carcinogenesis. 2001;22:619–626. doi: 10.1093/carcin/22.4.619. [DOI] [PubMed] [Google Scholar]
- 12.Morgenthaler P.M., Holzhauser D. Analysis of mutations induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in human lymphoblastoid cells. Carcinogenesis. 1995;16:713–718. doi: 10.1093/carcin/16.4.713. [DOI] [PubMed] [Google Scholar]
- 13.Yadollahi-Farsani M., Gooderham N.J., Davies D.S., Boobis A.R. Mutational spectra of the dietary carcinogen 2-amino-1-methyl-6- phenylimidazo[4,5-b]pyridine(PhIP) at the Chinese hamsters hprt locus. Carcinogenesis. 1996;17:617–624. doi: 10.1093/carcin/17.4.617. [DOI] [PubMed] [Google Scholar]
- 14.Gooderham N.J., Zhu H., Lauber S., Boyce A., Creton S. Molecular and genetic toxicology of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Mutat. Res. 2002;506-507:91–99. doi: 10.1016/s0027-5107(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 15.David R., Ebbels T., Gooderham N. Synergistic and antagonistic mutation responses of human MCL-5 cells to mixtures of benzo[a]pyrene and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine: dose-related variation in the joint effects of common dietary carcinogens. Environ. Health Perspect. 2015;124:88–96. doi: 10.1289/ehp.1409557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung C., Loy S., Li G.X., Liu A.B., Yang C.S. Rapid induction of colon carcinogenesis in CYP1A-humanized mice by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and dextran sodium sulfate. Carcinogenesis. 2011;32:233–239. doi: 10.1093/carcin/bgq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhary S., Sood S., Donnell R.L., Wang H.C. Intervention of human breast cell carcinogenesis chronically induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Carcinogenesis. 2012;33:876–885. doi: 10.1093/carcin/bgs097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Wang H., Liu A.B., Cheung C., Reuhl K.R., Bosland M.C., Yang C.S. Dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced prostate carcinogenesis in CYP1A-humanized mice. Cancer Prev. Res. (Phila.) 2012;5:963–972. doi: 10.1158/1940-6207.CAPR-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross A.J., Peters U., Kirsh V.A., Andriole G.L., Reding D., Hayes R.B., Sinha R. A prospective study of meat and meat mutagens and prostate cancer risk. Cancer Res. 2005;65:11779–11784. doi: 10.1158/0008-5472.CAN-05-2191. [DOI] [PubMed] [Google Scholar]
- 20.Voutsinas J., Wilkens L.R., Franke A., Vogt T.M., Yokochi L.A., Decker R., Le Marchand L. Heterocyclic amine intake, smoking, cytochrome P450 1A2 and N-acetylation phenotypes, and risk of colorectal adenoma in a multiethnic population. Gut. 2013;62:416–422. doi: 10.1136/gutjnl-2011-300665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Shapiro R., Broyde S. Molecular dynamics of a food carcinogen-DNA adduct in a replicative DNA polymerase suggest hindered nucleotide incorporation and extension. Chem. Res. Toxicol. 2005;18:1347–1363. doi: 10.1021/tx050132b. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Rechkoblit O., Wang L., Patel D.J., Shapiro R., Broyde S. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): modeling and dynamics studies. Nucleic Acids Res. 2006;34:3326–3337. doi: 10.1093/nar/gkl425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda H., Takamura-Enya T., Masuda Y., Nohmi T., Seki C., Kamiya K., Sugimura T., Masutani C., Hanaoka F., Nakagama H. Translesional DNA synthesis through a C8-guanyl adduct of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in Vitro: REV1 inserts dC opposite the lesion, and DNA polymerase kappa potentially catalyzes extension reaction from the 3′-dC terminus. J. Biol. Chem. 2009;284:25585–25592. doi: 10.1074/jbc.M109.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 26.Ball H.L., Myers J.S., Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamarche B.J., Orazio N.I., Weitzman M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 31.Woods D., Turchi J.J. Chemotherapy induced DNA damage response: convergence of drugs and pathways. Cancer Biol. Ther. 2013;14:379–389. doi: 10.4161/cbt.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glatt H., Pabel U., Meinl W., Frederiksen H., Frandsen H., Muckel E. Bioactivation of the heterocyclic aromatic amine 2-amino-3-methyl-9H-pyrido [2,3-b]indole (MeAalphaC) in recombinant test systems expressing human xenobiotic-metabolizing enzymes. Carcinogenesis. 2004;25:801–807. doi: 10.1093/carcin/bgh077. [DOI] [PubMed] [Google Scholar]
- 33.Roig A.I., Eskiocak U., Hight S.K., Kim S.B., Delgado O., Souza R.F., Spechler S.J., Wright W.E., Shay J.W. Immortalized Epithelial Cells Derived From Human Colon Biopsies Express Stem Cell Markers and Differentiate In Vitro. Gastroenterology. 2010;138:U1012–U1272. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 34.Christmann M., Tomicic M.T., Origer J., Kaina B. Fen1 is induced p53 dependently and involved in the recovery from UV-light-induced replication inhibition. Oncogene. 2005;24:8304–8313. doi: 10.1038/sj.onc.1208994. [DOI] [PubMed] [Google Scholar]
- 35.Fahrer J., Frisch J., Nagel G., Kraus A., Dörsam B., Thomas A.D., Reissig S., Waisman A., Kaina B. DNA repair by MGMT, but not AAG, causes a threshold in alkylation-induced colorectal carcinogenesis. Carcinogenesis. 2015;36:1235–1244. doi: 10.1093/carcin/bgv114. [DOI] [PubMed] [Google Scholar]
- 36.Dörsam B., Wu C.F., Efferth T., Kaina B., Fahrer J. The eucalyptus oil ingredient 1,8-cineol induces oxidative DNA damage. Arch. Toxicol. 2015;89:797–805. doi: 10.1007/s00204-014-1281-z. [DOI] [PubMed] [Google Scholar]
- 37.Dörsam B., Göder A., Seiwert N., Kaina B., Fahrer J. Lipoic acid induces p53-independent cell death in colorectal cancer cells and potentiates the cytotoxicity of 5-fluorouracil. Arch. Toxicol. 2015;89:1829–1846. doi: 10.1007/s00204-014-1434-0. [DOI] [PubMed] [Google Scholar]
- 38.Fahrer J., Huelsenbeck J., Jaurich H., Dörsam B., Frisan T., Eich M., Roos W.P., Kaina B., Fritz G. Cytolethal distending toxin (CDT) is a radiomimetic agent and induces persistent levels of DNA double-strand breaks in human fibroblasts. DNA Repair (Amst.) 2014;18:31–43. doi: 10.1016/j.dnarep.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Fahrer J., Schweitzer B., Fiedler K., Langer T., Gierschik P., Barth H. C2-streptavidin mediates the delivery of biotin-conjugated tumor suppressor protein p53 into tumor cells. Bioconjug. Chem. 2013;24:595–603. doi: 10.1021/bc300563c. [DOI] [PubMed] [Google Scholar]
- 40.Parplys A.C., Petermann E., Petersen C., Dikomey E., Borgmann K. DNA damage by X-rays and their impact on replication processes. Radiother. Oncol. 2012;102:466–471. doi: 10.1016/j.radonc.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Ciolino H.P., Bass S.E., MacDonald C.J., Cheng R.Y., Yeh G.C. Sulindac and its metabolites induce carcinogen metabolizing enzymes in human colon cancer cells. Int. J. Cancer. 2008;122:990–998. doi: 10.1002/ijc.23218. [DOI] [PubMed] [Google Scholar]
- 42.Meinl W., Ebert B., Glatt H., Lampen A. Sulfotransferase forms expressed in human intestinal Caco-2 and TC7 cells at varying stages of differentiation and role in benzo[a]pyrene metabolism. Drug Metab. Dispos. 2008;36:276–283. doi: 10.1124/dmd.107.018036. [DOI] [PubMed] [Google Scholar]
- 43.Liu S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., Oakley G.G. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawelczak K.S., Bennett S.M., Turchi J.J. Coordination of DNA-PK activation and nuclease processing of DNA termini in NHEJ. Antioxid. Redox Signal. 2011;14:2531–2543. doi: 10.1089/ars.2010.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buonarati M.H., Turteltaub K.W., Shen N.H., Felton J.S. Role of sulfation and acetylation in the activation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine to intermediates which bind DNA. Mutat. Res. 1990;245:185–190. doi: 10.1016/0165-7992(90)90048-o. [DOI] [PubMed] [Google Scholar]
- 46.Brown K., Hingerty B.E., Guenther E.A., Krishnan V.V., Broyde S., Turteltaub K.W., Cosman M. Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka T., Suzuki R., Kohno H., Sugie S., Takahashi M., Wakabayashi K. Colonic adenocarcinomas rapidly induced by the combined treatment with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and dextran sodium sulfate in male ICR mice possess beta-catenin gene mutations and increases immunoreactivity for beta-catenin, cyclooxygenase-2 and inducible nitric oxide synthase. Carcinogenesis. 2005;26:229–238. doi: 10.1093/carcin/bgh292. [DOI] [PubMed] [Google Scholar]
- 48.Boobis A.R., Lynch A.M., Murray S., de la Torre R., Solans A., Farre M., Segura J., Gooderham N.J., Davies D.S. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 49.Nauwelaers G., Bessette E.E., Gu D., Tang Y., Rageul J., Fessard V., Yuan J.M., Yu M.C., Langouet S., Turesky R.J. DNA adduct formation of 4-aminobiphenyl and heterocyclic aromatic amines in human hepatocytes. Chem. Res. Toxicol. 2011;24:913–925. doi: 10.1021/tx200091y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi J.H., Kim S.Y., Kim S.K., Kemp M.G., Sancar A. An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J. Biol. Chem. 2015;290:28812–28821. doi: 10.1074/jbc.M115.690354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J.H., Lindsey-Boltz L.A., Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J.H., Lindsey-Boltz L.A., Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 2009;37:1501–1509. doi: 10.1093/nar/gkn1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kemp M.G., Lindsey-Boltz L.A., Sancar A. The DNA damage response kinases DNA-dependent protein kinase (DNA-PK) and ataxia telangiectasia mutated (ATM) Are stimulated by bulky adduct-containing DNA. J. Biol. Chem. 2011;286:19237–19246. doi: 10.1074/jbc.M111.235036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kralj M., Husnjak K., Korbler T., Pavelic J. Endogenous p21WAF1/CIP1 status predicts the response of human tumor cells to wild-type p53 and p21WAF1/CIP1 overexpression. Cancer Gene Ther. 2003;10:457–467. doi: 10.1038/sj.cgt.7700588. [DOI] [PubMed] [Google Scholar]
- 56.Creton S., Zhu H., Gooderham N.J. A mechanistic basis for the role of cycle arrest in the genetic toxicology of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Toxicol. Sci. 2005;84:335–343. doi: 10.1093/toxsci/kfi075. [DOI] [PubMed] [Google Scholar]
- 57.Chanoux R.A., Yin B., Urtishak K.A., Asare A., Bassing C.H., Brown E.J. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y.F., Shih H.Y., Shang Z., Matsunaga S., Chen B.P. DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 2014;42:4463–4473. doi: 10.1093/nar/gku116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 60.Roos W.P., Thomas A.D., Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 61.Burrell R.A., McClelland S.E., Endesfelder D., Groth P., Weller M.C., Shaikh N., Domingo E., Kanu N., Dewhurst S.M., Gronroos E., et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang Y., Tsao C.C., Goodman B.K., Furumai R., Tirado C.A., Abraham R.T., Wang X.F. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004;23:3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menoyo A., Alazzouzi H., Espin E., Armengol M., Yamamoto H., Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 64.Vassileva V., Millar A., Briollais L., Chapman W., Bapat B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002;62:4095–4099. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.