Abstract
Cytokines utilize the transcription factor STAT5 to control cell-specific genes at a larger scale than universal genes, with a mechanistic explanation yet to be supplied. Genome-wide studies have identified putative STAT5-based mammary-specific and universal enhancers, an opportunity to investigate mechanisms underlying their differential response to cytokines. We have now interrogated the integrity and function of both categories of regulatory elements using biological and genetic approaches. During lactation, STAT5 occupies mammary-specific and universal cytokine-responsive elements. Following lactation, prolactin levels decline and mammary-specific STAT5-dependent enhancers are decommissioned within 24 h, while universal regulatory complexes remain intact. These differential sensitivities are linked to STAT5 concentrations and the mammary-specific Stat5 autoregulatory enhancer. In its absence, mammary-specific enhancers, but not universal elements, fail to be fully established. Upon termination of lactation STAT5 binding to a subset of mammary enhancers is substituted by STAT3. No STAT3 binding was observed at the most sensitive STAT5 enhancers suggesting that upon hormone withdrawal their chromatin becomes inaccessible. Lastly, we demonstrate that the mammary-enriched transcription factors GR, ELF5 and NFIB associate with STAT5 at sites lacking bona fide binding motifs. This study provides, for the first time, molecular insight into the differential sensitivities of mammary-specific and universal cytokine-sensing enhancers.
INTRODUCTION
Mammary alveoli consist of highly specialized milk-secreting epithelial cells, expressing ∼1400 genes that show a minimum 2-fold change between pregnancy day six and day one of lactation (1). Genetic studies have demonstrated that expression of at least ∼400 of these genes during pregnancy is dependent on the transcription factors STAT5A and 5B (referred to as STAT5) (1). Notably, several mammary-specific genes are activated up to 1000-fold by prolactin through STAT5. Mouse genetics has demonstrated that STAT5 is critical not only for the expression of these genes but also for the formation of the alveolar compartment during pregnancy (2–4). In other cell types, STAT5 is activated by cytokines, such as erythropoietin and interleukins, and thereby controls the biology of hematopoietic cells (5–7).
Although ChIP-seq experiments have revealed the entity of STAT5 binding coordinates (1,8,9), the mechanisms behind the establishment of cell-specific and common regulatory units remain to be understood. It can be hypothesized that the exceptionally high concentration of STAT5 in differentiated milk secreting epithelium is key to the extraordinary activation of mammary-specific genes during pregnancy. Notably, a mammary-specific STAT5 autoregulatory enhancer located in the intergenic region separating the two Stat5 genes is responsible for the exceptional STAT5 levels in mammary tissue (10).
Based on genetic and genomic data, STAT5 activates mammary-specific genes as pregnancy progresses, opening the possibility that increasing STAT5 levels are critical not only for the establishment but also the maintenance of a mammary-specific genetic program throughout lactation. Sustaining a robust differentiation program during lactation, despite fluctuating prolactin levels, is essential to avoid premature and aberrant tissue remodeling prior to weaning and thereby ensures survival of offspring. Although the autoregulatory Stat5 enhancer is critical to attain maximum STAT5 levels in mammary tissue (10), its role in sustaining epithelial differentiation during lactation and in decommissioning enhancers upon loss of differentiation in post lactation tissue remains to be established. Upon weaning circulating levels of prolactin decline rapidly, which is accompanied by loss of active (phosphorylated) STAT5 and gain of active STAT3, a trigger of epithelial cell death and remodeling of the tissue compartment (11–14). It has been suggested that a shift in the balance from active STAT5 to active STAT3 distinguishes lactating and post-lactating mammary tissue (15). Maintaining high STAT5 levels during lactation would therefore ensure unabated epithelial differentiation by minimizing recruitment of STAT3, which otherwise would result in loss of differentiated epithelium.
We have now addressed the question whether high STAT5 levels imposed by the autoregulatory Stat5 enhancer contribute to the maintenance of mammary-specific enhancers and ensure the coordinated loss of differentiation upon weaning. With this in mind we have focused on enhancers of STAT5-dependent genes that are highly induced during pregnancy and rapidly inactivated upon loss of differentiation. The integrity of mammary-specific enhancers and their dependence on STAT5 concentrations was evaluated in mammary tissue from wild-type mice and mice lacking the Stat5 autoregulatory enhancer (10). Lastly, we investigated the consequences of the shifting balance from active STAT5 to STAT3 (16,17) at the intersection of lactation to involution on mammary-specific and common cytokine controlled regulatory units. The consequences of such a balance were interrogated in mice lacking the Stat5 enhancer.
MATERIALS AND METHODS
Mice
Eight-week-old female mice of wild-type (Stat5+/+), heterozygous for the Stat5 locus (Stat5+/−), homozygous for the Stat5 enhancer mutation (Stat5ΔE/ΔE) and carrying one Stat5-null allele and one mutant allele (Stat5ΔE/−) (10) were used. All animal procedures were in accordance with NIH, NIDDK guidelines for the care and use of laboratory animals.
ChIP-seq
Frozen-stored mammary tissues harvested at day 1 of lactation (L1), 12 h (I12) and 24 h (I24) upon termination of lactation in wild-type and mutants were ground into powder then fixed with 1% formaldehyde at room temperature for 10 min. Samples were processed as previously described (10). The following antibodies were used for immunoprecipitation: anti-STAT5A (Santa Cruz, sc-1081x), anti-GR (Thermo Scientific, PA1-511A), anti-H3K27ac (Abcam, ab4729) and anti-STAT3 (Santa Cruz, sc-482). Libraries for next generation sequencing were prepared and sequenced with HiSeq 2000 (Illumina) (10).
ChIP-seq data analysis
ChIP-seq signals were trimmed using trimmomatic (18) (version 0.33) to filter low-quality reads and subsequently aligned to the mouse reference genome (mm10) using Bowtie (19) aligner (version 1.1.2) with the parameter -m 1 to obtain only uniquely mapped reads. The correlation of all replicates was calculated using deepTools (20) with default parameters. Spearman correlation was chosen as it is more reliable if outliers occur, with the caveat that the correlation value is less sensitive. ChIP-seq data used in this study were highly reproducible (Supplementary Table S8). The correlation of ELF5 and NFIB (GSE74826) is shown in Shin et al. (21). HOMER software (22) (default settings) and Integrative Genomics Viewer (23) were used for visualization. To identify regions of ChIP-seq enrichment over the background, MACS2 (24) peak finding algorithm (version 2.1.0) was used. As the data were from different resources, the q-value/P-value parameter was adjusted individually for each file to optimize STAT5A, STAT3, GR and H3K27ac peak calling. Chip-seq peaks of two or more samples were overlapped applying R (https://www.R-project.org/; R version 3.2.3) and the package DiffBind (25). For detecting underlying Chip-seq peaks based on a known set of Chip-seq peaks bedOpts (26) was used. In order to obtain high-confident peaks, the peak calling result of the replicates for WT L1 (STAT5A, GR, H3K27ac), WT I12 (STAT3), WT I24 (STAT5A, STAT3) and Stat5ΔE/ΔE L1 STAT5A were overlapped and the consensus peaks were chosen for analysis. Only for WT I12 STAT5A the peaks of the replicates were merged, to capture all peaks within the time window. Coverage plots and heat maps were generated using Homer software (22) as well as motif analysis with default background. Graph plotting was performed using R with the packages dplyr (https://CRAN.R-project.org/package=dplyr) and ggplot2 (27).
RNA-seq data analysis and gene annotation
RNA-seq data (GSE37646) were trimmed in the same manner as the ChIP-seq data. Mapping was carried out using the STAR RNA-seq aligner (28) with default settings and mus musculus GRCm38.84 as a GTF file. To assign the enhancers only to high-confident genes, the GTF file was filtered to retain only protein-coding genes, predicted genes (LOC, Rik and BC) were excluded. R (https://www.R-project.org/; R version 3.2.3), Bioconductor (29) and the packages Rsubread ((30); default settings) and DESeq2 ((31); default settings) were used for RNA-seq analysis. Enhancers were annotated to the closest transcription start site using R and ChIPpeakAnno (32). Additional analyses of annotated ChIP-seq data and RNA-seq data were done using R and dyplr (https://CRAN.R-project.org/package=dplyr).
Histological analysis and immunofluorescence
Mammary tissues from wild-type and mutant mice were harvested at L1 and fixed in 10% formalin and dehydrated in ethanol. Paraffin sections were stained with hematoxylin and eosin by standard methods (Histoserve). For immunofluorescence, unstained sections were incubated with the following antibodies overnight at 4°C: anti-phosphorylated STAT5 (Cell Signaling, 9314) and anti-phosphorylated STAT3 (Cell Signaling, 9145).
Western blotting
Total protein was extracted in RIPA buffer (50 mM Tris-HCl (PH7.5), 150 mM NaCl, 10% Glycerol, 50 mM NaF, 0.5 mM EDTA and 1% NP-40) and 20 μg of proteins was loaded on 7.5% Mini-PROTEAN® TGX™ Precast Gel (Bio-Rad, #4561023EDU) for STAT5 and in NuPAGE® Novex 4–12% Bis-Tris Gel (Invitrogen, NP0321BOX) for GAPDH. Primary antibodies were incubated overnight at 4°C (anti-STAT5A (Santa Cruz Biotechnology, sc-1081X, 1:1000) and GAPDH (Novus Biologicals, NB300-221, 1:10000)).
RNA isolation and quantitative RT-PCR
RNA was isolated using the PureLink RNA Mini kit (Ambion) according to the manufacturer's protocol. Complementary DNA was synthesized from total RNA using SuperScript II (Invitrogen) and quantitative PCR was performed using the Taqman probe-based system (Stat5a, Mm00839861_m1; mouse Gapdh endogenous control, 4352339E, Applied Biosystem) on the CFX384 Real-Time PCR Detection System (Bio-Rad). Stat5a mRNA levels were measured by qRT-PCR and normalized to Gapdh.
Statistical analyses
All samples used for qRT-PCR and ChIP-seq were randomly selected. Statistical power was calculated using R and the package pwr (https://CRAN.R-project.org/package=pwr). The effect size was calculated for each comparison based on the estimated means and standard deviations of each group. The sample size for each group was therefore determined via power analysis of Stat5a expression change with significance level 0.05 and power level 0.9. Gene expression data were presented with the mean of independent biological replicates. To evaluate gene expression data that are statistically different between wild-type and each mutant group, a two-tailed unpaired t-test was used. Significance for box plots was determined using a Welch's t-test with a two-sided alternative hypothesis.
Bioinformatics
The workflow is illustrated in Supplementary Figure S3.
RESULTS
Rapid decommission of mammary enhancers upon termination of lactation
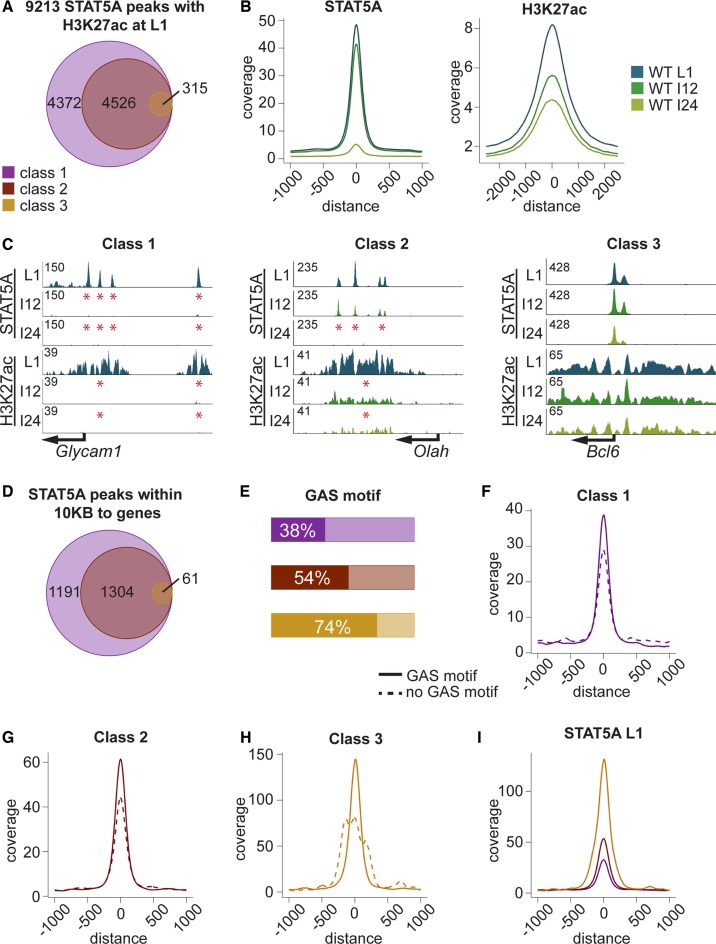
Precipitously declining prolactin levels at the end of lactation result in a rapid loss of epithelial cell differentiation and diminishing expression of milk protein genes, processes governed, at least in part, by STAT5-dependent super-enhancers (21). This is followed by a complete loss of milk secreting cells during involution. To determine to what extent these events are linked to decommission of enhancers, we analyzed genome-wide STAT5 occupancy at day one of lactation (L1) and at the interface between lactation and involution upon forced termination of lactation. For the subsequent analyses ChIP-seq ‘peaks’ were identified and as proven in Shin et al. they are putative enhancers (21). Therefore, the term enhancer is used throughout the manuscript. ChIP-seq for STAT5 and H3K27ac demonstrated the presence of ∼9200 structural enhancers within the mammary genome at L1 (Figure 1A). Out of these, ∼4370 were lost within 12 h (I12) after termination of lactation and they are referred to as ‘class 1’ enhancers. Approximately 4520 ‘class 2’ enhancers were lost after 24 h (I24), and 315 ‘class 3’ enhancers were retained after 24 h (Figure 1A). These data demonstrate that acute disruption of prolactin signaling results in the sequential eradication of putative cytokine-controlled enhancers. The three classes of enhancers likely reflect their differential sensitivities toward STAT5 signaling, with class 3 being the most resilient one that is retained outside lactation. Peak calling results were further validated by peak profiles. The peak coverage of the 9200 STAT5 enhancers decreased by ∼15% at I12 and 90% at I24 (Figure 1B). Loss of H3K27ac at I12 paralleled the STAT5 pattern but a significant degree of H3K27ac was retained at I24, suggesting that complete disassembly of enhancers was lagging. Glycam1 with its four mammary-specific STAT5 sites represents ‘very sensitive’ class 1 enhancers and all sites had been decommissioned within 12 h (Figure 1C). Loss of H3K27ac paralleled the elimination of STAT5 binding, further supporting the concept that the enhancers had been disassembled. STAT5 binding in Olah, a representative gene characterized by the ‘sensitive’ class 2 enhancers, was retained after 12 h but lost after 24 h (Figure 1C). STAT5 binding to Bcl6, a representative of the class 3 ‘resilient’ enhancers, was retained at 24 h (Figure 1C). As a note of caution, transcription factor peaks at promoter sequences, especially those lacking respective binding motifs and not changing upon loss of prolactin signaling, could coincide with phantom peaks (33).
Figure 1.
(A) Using peak calling on two replicates, 9213 STAT5A enhancers were found to coincide with H3K27ac (±500 bp) at day one of lactation (L1). A total of 4372 of them decommissioned within 12 h upon termination of lactation (class 1), 4526 within 24 h (class 2) and 315 were resilient (class 3). Since termination of lactation coincides with the onset of involution, these two time points were labeled I12 and I24. (B) Peak profile of STAT5A and H3K27ac based on the 9213 STAT5A peaks and STAT5A ChIP-seq data from wild-type tissue at L1 (blue), I12 (dark green) and I24 (light green). The coverage at I12 was reduced by 15% and by 90% at I24. H3K27ac ChIP-seq data from wild-type tissue at L1 (blue), I12 (dark green) and I24 (light green) are shown. The reduction in acetylation was larger at I12 than at I24. (C) A representative gene is shown for each of the enhancers groups. The left panel illustrates Glycam1, a class 1 gene, with ‘very sensitive’ enhancers. The four enhancers completely decommissioned within 12 h, as did H3K27ac. The middle panel shows Olah, a ‘less sensitive’ class 2 enhancer. Enhancer height decreased at I12, but all of them were still present. Also acetylation had decreased at I12. However, all STAT5A enhancers as well the acetylation were non-existent after 24 h. The right panel shows Bcl6, a class 3 gene with ‘resilient enhancers’, which were not affected by involution. The STAT5A enhancers as well as the acetylation remained stable throughout involution. (D) STAT5 enhancers within 10 kb of the nearest TSS of an annotated gene. By selecting enhancers within 10 kb of TSS, a region associated with enhancers (1), a total of 1191 class 1 enhancers, 1304 class 2 enhancers and 61 class 3 enhancers were identified. (E) The bar plot illustrates that the most stable enhancers were more likely associated with GAS motifs, as only 38% of the class 1 have a GAS motif within 200 bp. This percentage increased to 54% for the 1304 class 2 enhancers and 74% for class 3 enhancers, which were still present at I24. (F) Coverage of class 1 enhancers was higher in the presence of GAS motifs than without. (G) Enhancer height increased in class 2 enhancers, but the coverage of enhancers with an underlying GAS motif was still higher. (H) Class 3 enhancers displayed the highest coverage and the ones with GAS motif were again higher than those without. (I) Peak coverage at L1 of the three classes, independent of an underlying GAS motif. The resilient class 3 enhancers had the highest coverage, followed by class 2 and class 1 enhancers. Color code in panels A, D-I, class 1 (purple), class 2 (brown) and class 3 (yellow).
Since gene activation is likely controlled by STAT5 binding sites within 10 kb of promoter sequences (1) the analysis was further restricted to 2556 STAT5 enhancers identified during lactation within these boundaries. Out of these, ∼1190 were decommissioned within 12 h (class 1) and an additional 1300 after 24 h (class 2). Sixty-one enhancers were retained after 24 h (class 3) (Figure 1D, Supplementary Table S1). This suggests that the majority of putative cytokine-sensing STAT5 enhancers are abandoned swiftly upon hormone decline. This global analysis provides evidence that subsets of STAT5 binding sites display differential sensitivities toward hormone changes occurring within a 24-h window at the interphase between lactation and involution.
The mechanism controlling differential STAT5 occupancy at the transition of lactation to involution might involve the presence of specific GAS motifs (TTCnnnGAA). To address this possibility we carried out a motif analysis using the three different enhancer classes and determined the percentage of enhancers containing GAS motifs. While 38% of enhancers within class 1 enhancers coincided with a GAS motif, so did 54% of the class 2 enhancers and 74% of class 3 enhancers (Figure 1E). Importantly, enhancers with GAS motif show a higher coverage in all three categories (Figure 1F–H). Next we analyzed enhancers from classes 1, 2 and 3 in wild-type tissue at lactation day one. In general, enhancer height was larger at the resilient class 3 sites. The size of the most stable enhancers, those associated more frequently with GAS motifs, exceeded those that were decommissioned after 12 and 24 h (Figure 1I). These findings support the notion that the most resilient STAT5 binding sites are preferentially associated with GAS motifs and withstand rapid enhancer decommission upon declining hormone levels.
Interplay between STAT5 and GR
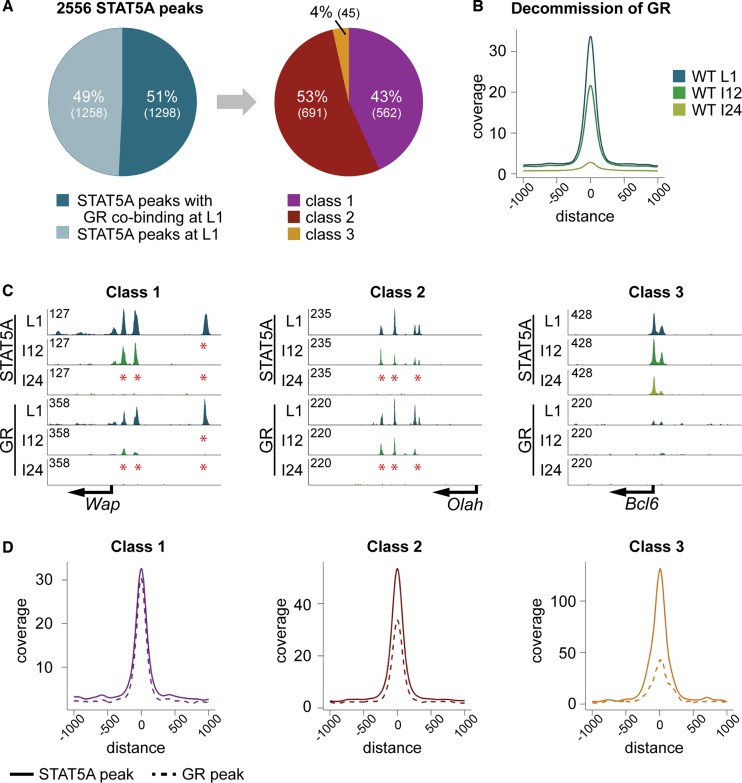
Glucocorticoids, in conjunction with prolactin, control the expression of mammary-specific genes (34) and the glucocorticoid receptor (GR) has been shown to act in concert with STAT5 in tissue culture cells (35). To gain additional insight into the in vivo interplay between GR and STAT5 we performed GR ChIP-seq during lactation and determined their global co-binding (Figure 2A). Out of 2556 STAT5 enhancers 51% coincided with GR binding and the majority of them were associated with class 1 (43%) or class 2 enhancers (53%). Only 4% were associated with class 3 enhancers (Figure 2A). Peak coverage analysis demonstrated coinciding decommission of GR and STAT5 upon disrupting lactation (Figure 2B). The enhancer coverage decreased by 35% within 12 h and 91% within 24 h. Decommission was exemplified at three different enhancers, Wap, Olah and Bcl6. GR binding to Wap was largely lost within 12 h, as was the most distal STAT5 enhancer (Figure 2C). The GR enhancers at the sensitive Olah enhancers were lost in parallel with STAT5 enhancers within 24 h (Figure 2C). Compared to Wap and Olah, GR binding at the Bcl6 enhancer was very weak (Figure 2C).
Figure 2.
(A) Co-binding of STAT5A and GR. Fifty-one percent of the STAT5A enhancers coincided with GR and out of those 43% (562) were classified as class 1 enhancers (30% contain GAS motif), 53% (691) as class 2 (51% contain GAS motif) and 4% (45) as class 3 enhancers (67% contain GAS motif). (B) Temporal decommission of GR. The peak coverage was reduced by 35% within 12 h, and by 91% within 24 h, equivalent to that observed for STAT5. (C) Genes representing the three differential STAT5 enhancers. Wap (class 1), with its three enhancers, showed the same decommission pattern for STAT5A and GR. The most distal enhancers (E3) was lost within 12 h (I12) and no enhancers remained at 24 h (I24) of involution. All enhancers in Olah (class 2) were intact at I12 but completely decommissioned at I24. However, the enhancer height for STAT5 and GR was already reduced at I12. Bcl6 (class 3) showed no GR co-binding compared to the classes 1 and 2. (D) Coverage plots for STAT5A and GR enhancers in each of the enhancer categories. STAT5A and GR enhancer coverage was equivalent at class 1 sites. At class 2 sites GR coverage was smaller than that of STAT5A and class 3 sites had relatively the smallest GR enhancers.
Since GR binding does not necessarily parallel that of STAT5 we compared the coverage of these two transcription factors. Notably, while the height of the GR enhancers in all three enhancer categories was similar, STAT5 enhancer height increased 4-fold between class 1 and class 3 (Figure 2D). GR and STAT5 coverage was virtually identical at class 1 enhancers but the relative STAT5 coverage was 1.6-fold higher at class 2 sites and 3-fold higher at class 3 sites. The GR peak coverage at class 3 sites was 43, 33 at class 2 sites and 30 at class 1 sites, those most sensitive to decommission upon cessation of lactation. These results suggest that enhancers bound by STAT5 and GR were particularly sensitive to prolactin signaling and those preferentially bound by STAT5 were resilient to hormonal changes.
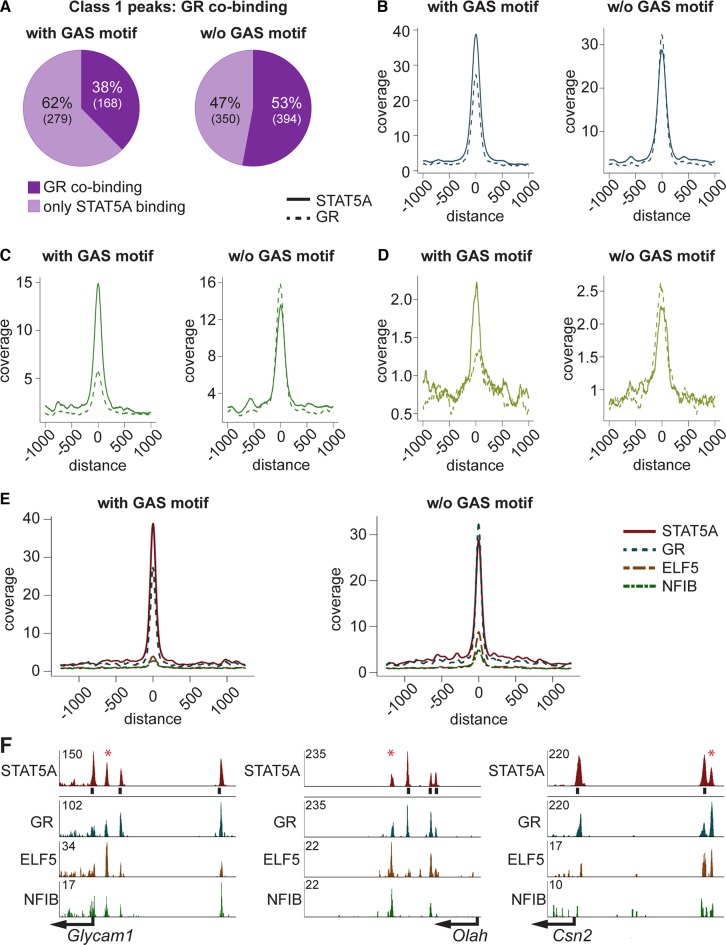
To address the possibility that GR selectively supports STAT5 binding to sites without GAS motif, we separately analyzed class 1 enhancers with and without GAS motifs (Figure 3A). While in the presence of GAS motifs, 38% of STAT5 sites coincide with GR, 53% coincide at enhancers without GAS motifs. Notably, while STAT5 coverage was lower at enhancers without GAS motifs, GR coverage was equivalent in both groups (Figure 3B). Preferential loss of STAT5 and GR binding was observed at sites containing GAS motifs (Figure 3C). A de novo motif search at class 1 sites lacking bona fide GAS motifs revealed the presence of GAS and NFIB half sites and GR binding motifs (Supplementary Figure S2), supporting the notion that additional transcription factors aid the binding of STAT5. To further examine this we integrated ChIP-seq data from STAT5, GR, ELF5 and NFIB (Figure 3E). Elevated binding of GR, ELF5 and NFIB was detected at STAT5 enhancers lacking GAS motifs (Figure 3E). The GR enhancer coverage was 15% reduced in STAT5 enhancers with GAS motif. Additionally, ELF5 was 56% and NFIB 40% lower than in enhancers without underlying GAS motifs. The concept that NFIB and ELF5 preferentially bind to STAT5 enhancers lacking a GAS motif was investigated at individual genes containing enhancers composed of class 1 sites with and without GAS motifs (Figure 3F). Both ELF5 and NFIB bind at STAT5 enhancers lacking GAS motifs.
Figure 3.
(A) Class 1 STAT5A enhancers were separated into enhancers with and without GAS motif. Thirty-eight percent of the enhancers with, and 53% without, a GAS motif co-bound GR. (B) STAT5 enhancers were higher in the presence of GAS motifs. Coverage of GR was independent of the GAS motif status. (C) Preferential reduction of class 1 enhancers containing a GAS motif occurred at 12 h of involution (I12). GR coverage at enhancers with GAS motifs is less than 45% of that obtained at enhancers lacking a GAS motif. At sites without GAS motif GR enhancers continue to exceed STAT5A enhancers. (D) Peak coverage at 24 h of involution (I24). At this time point most enhancers have been decommissioned but the pattern is reminiscent to that seen at I12. (E) Peak coverage of STAT5A class 1 enhancers with and without GAS motifs and GR, ELF5 and NFIB co-binding. Enhancers without GAS motif showed stronger co-binding of GR, ELF5 and NFIB as compared to those with a GAS motif. (F) Three representative genes and their STAT5A enhancers illustrate co-binding. Glycam1, GR co-bound at all STAT5A enhancers. Strongest ELF5 binding was at the enhancer lacking a GAS motif (asterix). The STAT5A enhancer without GAS motif at Olah showed the strongest ELF5 and NFIB co-binding. All STAT5A enhancers co-bound GR. Csn2 had also one STAT5A enhancer without GAS motif, which had the strongest co-binding by GR and ELF5.
Linking STAT5-sensitive enhancers to highly-induced genes
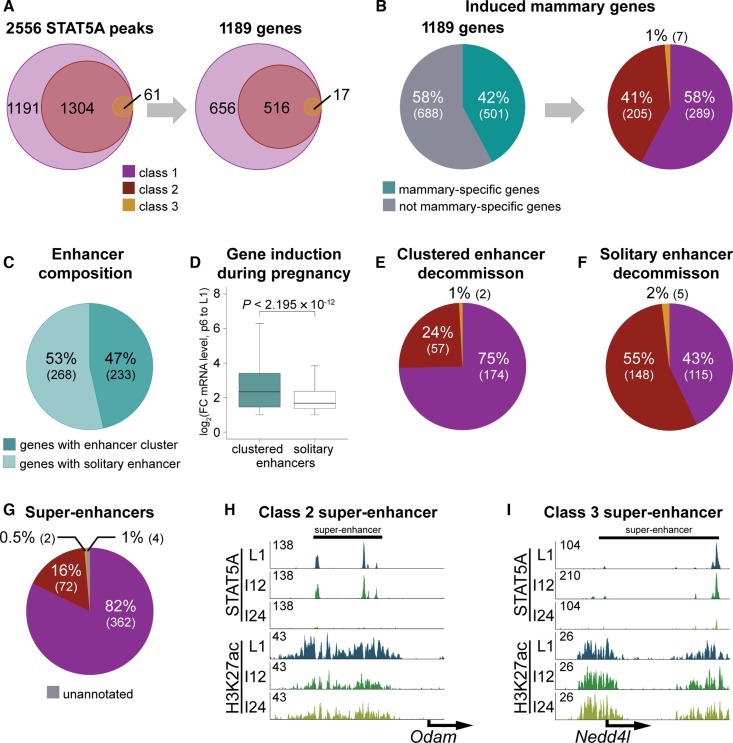
Next we investigated to what extent different classes of STAT5 binding sites were associated with highly activated genes. The 1191 class 1 sites were annotated to 656 unique genes, the 1304 class 2 sites to 516 unique genes and the 61 class 3 sites to 17 unique genes (Figure 4A). Genes were categorized based on the decommission time of the first enhancer. Class 1 genes have at least one enhancer decommissioned within 12 h, class 2 genes at least one within 24 h and in class 3 genes all enhancers were intact after 24 h. Representative illustrations for each enhancer category are shown in Figure 1C and Supplementary Figure S1. To further understand to what extent these 1189 genes are controlled by hormone-sensitive STAT5 enhancers, we integrated them with RNA-seq data (Supplementary Table S2, Figure 4B). Approximately 40% of these genes are mammary-specific (2-fold induction from pregnancy day 6 to lactation day 1) and 58% of them were associated to class 1, 41% with class 2 and 1% with class 3 genes.
Figure 4.
(A) Assignment of annotated enhancers to genes. The 1191 class 1 enhancers were assigned to 656 unique genes, the 1304 class 2 enhancers to 516 genes and the 61 class 3 enhancers to 17 genes. (B) Forty-two percent of the genes are associated with mammary-specific genes, which have a minimum of 2-fold induction between day six of pregnancy and day one of lactation. Out of those 58% (289) were categorized as class 1, 41% (205) as class 2 and 1% (7) as class 3. (C) Forty-seven percent (233) of the mammary-specific genes were associated with clustered enhancers and 53% (268) with solitary enhancers. (D) Genes assigned to clustered enhancers showed a significantly higher induction during pregnancy compared to those with solitary enhancers. Median, middle bar inside each box; IQR (interquartile range), the box containing 50% of the data; whiskers, 1.5 times the IQR. (E) Seventy-five percent of clustered enhancers belonged to class 1 enhancers with at least one enhancer decommissioned within 12 h; 24% (57) belonged to class 2 where at least one enhancer was decommissioned within 24 h, and two enhancer clusters belonged to class 3. (F) Forty-three percent (115) of solitary enhancers were decommissioned within 12 h (class 1), 55% (148) within 24 h (class 2) and 2% (5) were resilient during involution (class 3). (G) Mammary-specific super-enhancers are composed of differential enhancer classes. Eighty-two percent of the 440 identified mammary-specific super-enhancers had at least one enhancer decommissioned within 12 h; 16% (72) belonged to class 2 and two to class 3. (H) A class 2 super-enhancer, where all enhancers were intact after 12 h of involution, but decommissioned within 24 h of involution. (I) Representative of a class 3 super-enhancer with resilient enhancers after 24 h of involution.
ChIP-seq experiments performed during lactation have demonstrated that mammary-specific genes are associated with either clusters of STAT5 binding sites, named super-enhancers, or solitary enhancers (21). Forty-seven percent of mammary-specific genes (233) under STAT5 control were associated with clustered enhancers and 53% (268) with solitary enhancers (Figure 4C). Genes associated with enhancer clusters are in induced to a greater degree during pregnancy (Figure 4D). Having observed enhancer clusters at highly induced genes, it is possible that individual enhancer units within these clusters display differential sensitivities to cytokines and STAT5. ChIP-seq data conducted 12 and 24 h after termination of lactation provide compelling evidence that rapidly decommissioned STAT5 enhancers are highly enriched for genes associated with clustered enhancers (Supplementary Tables S3, S5, S7) as compared to solitary enhancers (Supplementary Tables S4, S6, S7). In clustered enhancers 75% of the genes (174 genes) are of class 1 (Supplementary Table S3) with at least one enhancer decommissioned within 12 h, 24% are of class 2 (Supplementary Table S5), with at least one enhancer decommissioned within 24 h and two genes belonged to class 3 (Supplementary Table S7) with resilient enhancers (Figure 4E). In genes with solitary enhancers 43% decommissioned within 12 h (class 1, Supplementary Table S4) and further 55% within 24 h of involution (class 2, Supplementary Table S6) and five enhancers were class 3 (Supplementary Table S7) and stable after 24 h (Figure 4F).
Recently we conducted H3K27ac, GR, MED1 and STAT5 ChIP-seq experiments and identified 440 super-enhancers specific to the mammary genome, half of which were associated with genes induced during pregnancy (21). Since 95% of these super-enhancers are established during pregnancy we hypothesized that they are composed of class 1 enhancers, i.e. those most sensitive to cytokine stimulation. Indeed, 82% of the mammary-specific super-enhancers contain at least one class 1 enhancer (Figure 4G). The super-enhancer of Wap is a class 1 representative, where one enhancer decommissioned within 12 h and the others within 24 h of involution (Supplementary Figure S1B). A representative of class 2 is shown in Figure 4H, where all enhancers were present after 12 h involution, but decommissioned within 24 h. A resilient super-enhancer is shown in Figure 4I, as the enhancers are still present after 24 h of involution even if they are reduced in height.
Mammary enhancers sense STAT5 concentrations
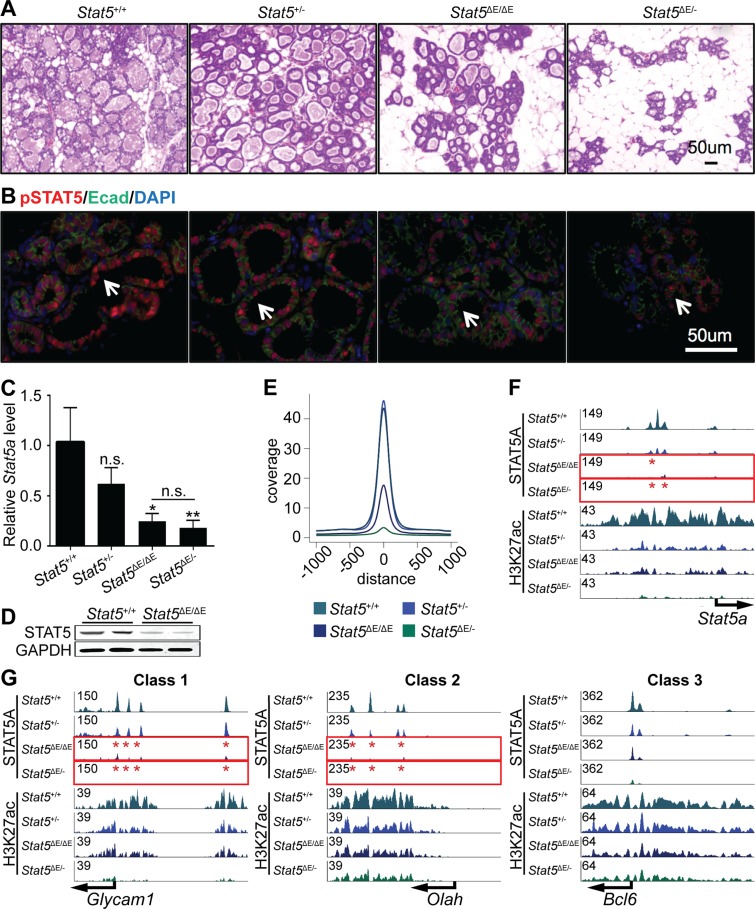
The instantaneous loss of STAT5 binding to class 1 enhancers at the interface of lactation and involution suggested that their integrity might be exquisitely dependent on STAT5 levels or activity. Having identified a mammary-specific autoregulatory enhancer within the Stat5 locus (10) we surmised that STAT5 concentrations might define class 1 enhancers. To address this possibility we investigated the integrity of STAT5-dependent enhancers in mice lacking the Stat5 autoregulatory loop. We analyzed mammary tissue and its defining enhancers in four distinct genotypes. Mice carrying two intact Stat5 loci (Stat5+/+) expressing 100% STAT5, mice heterozygous for Stat5 (Stat5+/−), mice homozygous for the Stat5 enhancer mutation (Stat5ΔE/ΔE) and mice carrying one Stat5-null allele and one mutant allele (Stat5ΔE/−). Histologically mammary tissue appeared normal in the presence of one intact Stat5 allele (Stat5+/−) (Figure 5A). The intensity of pSTAT5 staining was equivalent in wild-type and Stat5+/− tissue (Figure 5B). Stat5 expression in mammary tissue lacking the autoregulatory enhancer (Stat5ΔE/ΔE) was reduced by approximately 75% (10) and histological analyses revealed reduced alveolar expansion, although alveoli displayed hallmarks of differentiation (Figure 5A and B). In contrast, mammary tissue from Stat5ΔE/− mice was underdeveloped (Figure 5A and B). QRT-PCR data demonstrated an ∼75% reduction of Stat5a RNA levels in Stat5ΔE/ΔE tissue (Figure 5C), which is en par with our recently published data (10). Western blot analyses also showed a reduction of STAT5 (Figure 5D). These data emphasize the importance of the Stat5 autoregulatory enhancer for the expression of STAT5 and the physiology of mammary tissue.
Figure 5.
(A) Alveolar development is dependent on the Stat5 autoregulatory enhancer. Histologically mammary tissue appeared normal in the presence of only one intact Stat5 allele (Stat5+/−). Mammary tissue with homozygous deletion of the autoregulatory enhancer (Stat5ΔE/ΔE) appeared less differentiated and Stat5ΔE/− was severely underdeveloped. (B) Qualitatively, pSTAT5 staining of wild-type (Stat5+/+) tissue appeared similar to Stat5+/−. In contrast, pSTAT5 staining was greatly reduced in Stat5ΔE/ΔE, Stat5ΔE/− tissues. (C) Stat5a mRNA levels in wild-type and mutant mammary tissue (Stat5+/+ n = 6; Stat5+/- n = 2; Stat5ΔE/ΔE n = 4; Stat5ΔE/− n = 3). (D) The Western blot showed reduced STAT5 levels in Stat5ΔE/ΔE tissue. GAPDH served as control, and showed no reduction. (E) Peak coverage in wild-type and mutant tissue. (F) STAT5A enhancers were reduced in Stat5+/− tissue and completely absent in Stat5ΔE/ΔE and Stat5ΔE/− tissue. Reduction of H3K27ac was delayed in all genotypes except Stat5ΔE/− where it was absent. (G) Representative genes with class 1, class 2 and class 3 enhancers. STAT5 binding in mutant tissue was most severely affected in class 1 enhancers (Glycam1) and the least in class 3 enhancers (Bcl6).
The biological impact of ablating the Stat5 autoregulatory enhancer was also demonstrated in a coverage plot. While the peak coverage of wild-type and Stat5+/− tissue was equivalent, it decreased 60% in Stat5ΔE/ΔE tissue and more than 90% in Stat5ΔE/− tissue (Figure 5E). Importantly, STAT5 binding at the autoregulatory enhancer was still observed in the presence of one wild-type Stat5 allele but absent in Stat5ΔE/ΔE and Stat5ΔE/− tissues (Figure 5F). An equivalent behavior was observed at class 1 enhancers and STAT5 enhancers were smaller at the Glycam1 and Olah loci in Stat5ΔE/ΔE and Stat5ΔE/− tissues (Figure 5G). However, residual STAT5 binding in the absence of the autoregulatory enhancer was sufficient to maintain limited H3K27ac. The class 3 enhancer of Bcl6 was marginally affected by the mutations, even though the enhancer heights were reduced in Stat5ΔE/- tissue.
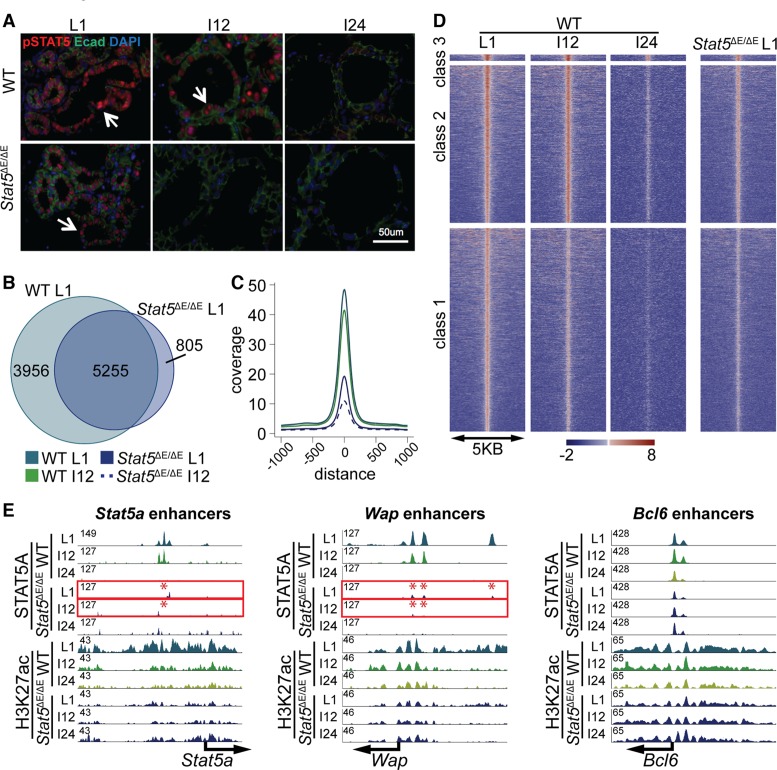
Based on these experiments we predicted an impaired stability of class 1 enhancers in the absence of the Stat5 autoregulatory loop, which was investigated in Stat5ΔE/ΔE tissue at the interface of lactation and involution (Figure 6). While active STAT5 was homogeneously detected in wild-type epithelium during lactation it was lost in most cells 12 h after forced ending lactation and completely absent after 24 h (Figure 6A). Phospho-STAT5 staining in Stat5ΔE/ΔE epithelium at L1 was greatly reduced and completely absent after 12 h (Figure 6A), likely a reflection of the greatly reduced STAT5 levels in mutant cells. Out of the 9213 STAT5 enhancers observed in wild-type tissue at L1 only 5225 coincided with enhancers in mutant tissue (Figure 6B) indicating a failure to establish the full complement of structurally intact enhancers.
Figure 6.
(A) Decline of active STAT5 in wild-type and mutant mammary tissue. Phospho-STAT5-positive cells in wild-type tissue decreased within 12 h (I12) after terminating lactation and were absent at 24 h (I24). In contrast no pSTAT5-positive cells were observed at I12 in tissue lacking the autoregulatory enhancer Stat5ΔE/ΔE. (B) A total of 5255 out of 9213 STAT5A binding sites (enhancers) in wild-type tissue were shared with Stat5ΔE/ΔE tissue at L1, suggesting that the full establishment of enhancer was not accomplished at lower STAT5 levels. (C) The coverage plot illustrates that the wild-type L1 sample had the highest coverage. Even wild-type tissue at I12 showed a higher coverage than the Stat5ΔE/ΔE samples at L1. Stat5ΔE/ΔE at involution 12 h showed the lowest coverage. (D) Heat map comparing STAT5A coverage in wild-type and Stat5ΔE/ΔE tissue in the three different enhancer categories. (E) Representative examples from the heat map. The STAT5A enhancer in the Stat5a locus was disrupted in Stat5ΔE/ΔE tissue. H3K27ac coverage was reduced but still present. STAT5 binding to the Wap enhancers was greatly reduced in Stat5ΔE/ΔE tissue. Class 3 enhancers were the least affected in mutant tissue. The height of the STAT5A enhancers was lower in the Stat5ΔE/ΔE sample, but H3K27ac remained unaltered.
The coverage plot demonstrated highest coverage in wild-type L1 samples, followed by wild-type samples 12 h upon disrupting lactation (I12), and Stat5ΔE/ΔE samples had the lowest coverage (Figure 6C). Heat map analyses further demonstrated the progressive loss of class 1 and class 2 enhancers within a 12 to 24 h window upon terminating lactation (Figure 6D). Notably, the intensity of the ‘very sensitive’ class 1 enhancers was greatly reduced in Stat5ΔE/ΔE tissue at day 1 of lactation compared to wild-type tissue (Figure 6D). It was more reminiscent to wild-type tissue at involution. These findings are exemplified at the Stat5a locus where STAT5 enhancers in mutant tissue are already significantly reduced compared to wild-type tissue (Figure 6E). In contrast, the peak coverage at the universal STAT5 target locus Bcl6 was equivalent between wild-type and mutant tissue and remained stable upon cessation of lactation (Figure 6E). For Wap, the enhancer height was also reduced about 3-fold but the kinetics of decommission is similar in wild-type and mutant tissue (Figure 6E).
Homeostasis between STAT5 and STAT3 binding
Since in vitro STAT3 and STAT5 recognize an identical GAS motif we hypothesized that their relative abundance and activity would determine the extent of in vivo binding at any given STAT5 enhancer. Based on this assumption, a shift from STAT5 to STAT3 binding should occur at the transition of lactation to involution, which is characterized by a decline of STAT5 activity and an increase of STAT3 activity (15). Immunohistochemical analyses demonstrated a loss of pSTAT5 in mammary epithelium within 12 h upon termination of lactation (I12) and a concomitant increase of pSTAT3 staining (Figure 7A). It can also be hypothesized that loss of the autoregulatory Stat5 enhancer could result in an imbalance of STAT5/STAT3 binding with physiological consequences.
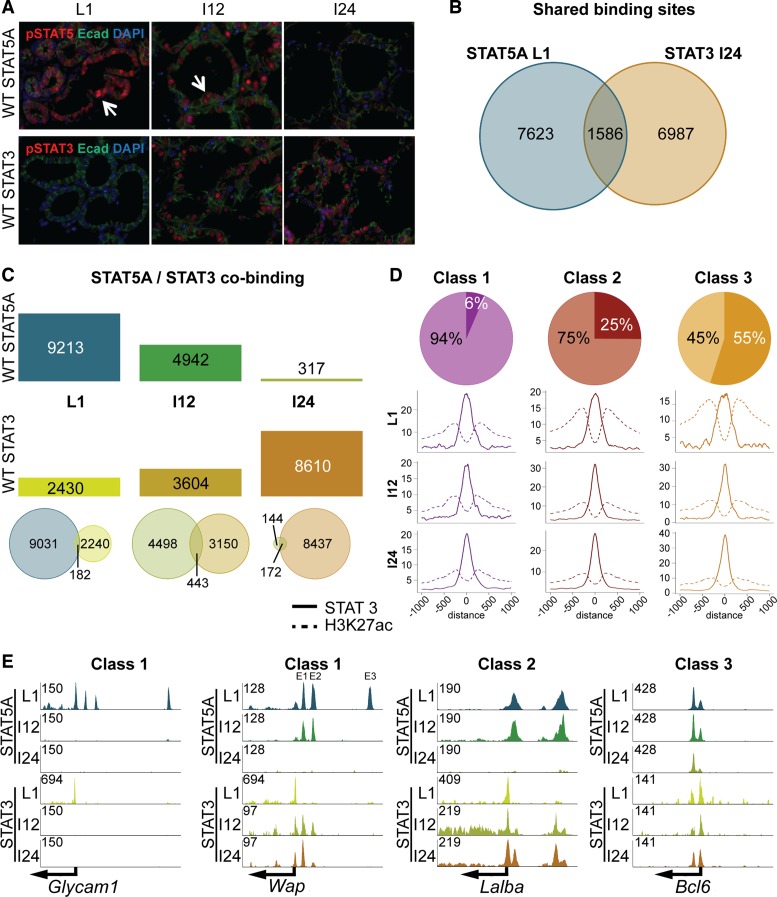
Figure 7.
(A) Homeostasis between STAT5 and STAT3. While pSTAT5 levels declined upon termination of lactation, pSTAT3 levels increased. (B) Binding sites shared by STAT5 and STAT3 were identified by integrating STAT5A data at L1 and STAT3 data at I24. (C) Bar plot showing the reduction of STAT5A binding compared to the increase of STAT3 binding during involution. This indicates that STAT3 replaced STAT5, but not necessarily at the same binding sites. The venn diagrams illustrate the overlap of STAT5A and STAT3 enhancers for L1, I12 and I24. Due to the replacement of those two factors most overlaps could be detected at involution 12 h. (D) STAT3 binding to the three enhancer categories. Six percent (280) of class 1 STAT5A sites overlapped with STAT3. Thereby, STAT3 and H3K27ac peak height was reduced from L1 to I12, but not from I12 to I24. Twenty-five percent (1137) of class 2 enhancers co-bound STAT3, but by looking at the peak coverage during involution the H3K27ac decreased and STAT3 increased. The last group of common enhancers had 173 enhancers (55%) overlapping STAT5A and STAT3 enhancers and the coverage plot shows that the peak coverage of STAT3 increased and H3K27ac decreased slightly during involution. (E) Representative genes to demonstrate the STAT5/3 homeostasis. Glycam1 (class 1) displayed STAT3 co-binding at the promoter at L1, but no increase of STAT3 at the individual enhancers was observed. Wap (class 1) had no STAT3 co-binding at the third enhancer (E3). However, E1 and E2, which decommissioned at I24, co-bound STAT3 at I12 and even stronger at I24. The promoter showed continuous co-binding. The class 2 representative, Lalba, displayed constant co-binding at the promoter, and the enhancers showed a co-binding starting at involution 12 h, getting stronger at I24, when the STAT5A enhancers were absent. The resilient Bcl6 gene (class 3) showed continuous co-binding during involution.
To investigate to which extent STAT3 and STAT5 bind to identical genomic coordinates, we conducted STAT3 ChIP-seq experiments at L1 and at 12 and 24 h after forced involution. While the strongest pSTAT5 signal was detected at lactation, STAT3 activity was highest after 24 h (Figure 7A). Thus, we compared genome-wide STAT5 coverage at L1 with STAT3 coverage at I24 (Figure 7B). Out of the ∼9200 STAT5 enhancers, 1586 coincided with STAT3 enhancers at I24 (Figure 7B). While the number of STAT5 enhancers precipitously declined during involution with only ∼300 remaining 24 h after terminating lactation, the number of STAT3 enhancers increased from ∼2400 at lactation to more than 8600 at I24 (Figure 7C). The largest overlap between STAT5 and STAT3 enhancers was at I12 when both members were in an active state (Figure 7C, bottom panel).
As shown, the three classes of enhancers were characterized by different affinities to STAT5, with class 1 representing the most sensitive ones and class 3 the resilient ones. Based on the unique association of class 1 and class 2 enhancers with mammary-specific enhancers that greatly depend on STAT5 and class 3 with universal regulatory elements we propose that the two former enhancer classes would be less frequently associated with STAT3. STAT5 and STAT3 ChIP-seq were analyzed and only 6% of class 1 enhancers coincided with STAT3 and STAT5 (Figure 7D). On the other side of the spectrum 55% of class 3 enhancers were bound by STAT3 and STAT5. Notably these were the ones most highly enriched for GAS motifs. Figure 7E illustrates STAT5A and STAT3 binding for all three enhancer classes. Glycam1 (class 1) lost all three enhancers within 12 h involution, but none of them was replaced by STAT3 binding. In contrast some of the enhancers of Wap, also a class 1 gene, showed a stepwise replacement with STAT3. The most distal enhancer disappeared at I12 and did not co-bind STAT3, whereas the two other enhancers showed STAT3 co-binding at I24. At I24 those two enhancers were fully decommissioned, but STAT3 binding remained and even increased at E1. A representative of class 2 is Lalba. Its enhancers co-bound STAT3 and STAT5 at I12 and STAT3 binding increased at I24 upon loss of STAT5 binding. Bcl6, a class 3 gene bound both STAT3 and STAT5 at all stages.
DISCUSSION
Molecular mechanisms responsible for different degrees of gene activation induced by cytokines through STAT transcription factors remain elusive. In mammary epithelium, a biological system with exceptional gene regulatory dynamics, prolactin induces lineage-specific genes more than 1000-fold during pregnancy. In contrast, activation of universal cytokine-induced STAT5 targets, such as Cish and Socs2 is in the single digits. Our study now demonstrates the presence of three distinct classes of regulatory elements that differentially sense STAT5 concentrations. While genes highly induced in mammary epithelium are preferentially associated with enhancers requiring high levels of STAT5, genes subject to a modest activation are linked to regulatory elements that strongly bind STAT5 at low levels.
Genes linked to lactation, such as those encoding milk proteins, are highly induced during pregnancy. Maintaining their maximum expression throughout lactation is essential to uphold epithelial differentiation and mammary function. In turn, genetic wiring in mammary epithelium also needs to respond speedily to declining prolactin levels at the termination of lactation to ensure a rapid inactivation of milk protein genes and the induction of tissue remodeling. Failure to execute this program would result in massive milk accumulation in a dying organ with severe consequences, such as mastitis. Prolactin, and its downstream transcription factor STAT5, likely are key driver of these processes, and the extraordinary high level of STAT5 found only in mammary tissue (10) controls a subset of genes critical for the establishment and maintenance of epithelial differentiation. Enhancers in differentiation-associated genes are highly sensitive to STAT5 levels and they rapidly decommission upon weaning when prolactin levels, and thereby active STAT5, decline. In contrast regulatory elements of cytokine-induced genes, such as Bcl6 and members of the Socs family, required for common cellular functions, are rather resilient and bound by STAT5 in the mammary gland at all biological stages. This ensures that the extreme fluctuations of active STAT5 preferentially affects genes associated with epithelial differentiation and milk production. While mammary tissue is characterized by its extraordinarily high levels of STAT5 other cytokine-responsive cell types, such as hepatocytes and hematopoietic cells, express lower levels of STAT5 and gene induction is at least one order of magnitude lower (36–40). Moreover, in most cell types cytokines and STAT5 have a more modulating function (7,41), which is distinct from mammary epithelium (2,42) and T regulatory cells (6), where STAT5 is essential for the establishment and maintenance of the respective lineages.
We have recently identified mammary-specific super-enhancers and mouse genetics has revealed that the seed enhancer in the Wap super-enhancer was reliant on the combined function of STAT5, NFIB and ELF5 (21). The concept that these super-enhancers rely on a specific set of mammary-enriched transcription factors was further supported by our findings that STAT5, GR, ELF5 and NFIB bind to the majority of these enhancers. Approximately two thirds of the highly sensitive class 1 enhancers do not contain bona fide GAS motifs suggesting that STAT5 binding is aided by above mentioned mammary enriched transcription factors. In addition to GR, NFIB is a prime candidate to facilitate the binding of transcription factors as it has been shown to facilitate chromatin accessibility (43). On a transcriptional level, NFIB cooperates with STAT5 in mammary tissue (44,45) and also associates with the androgen receptor and FOXA1 in prostate (46). All together these findings support the concept that mammary-specific enhancers are platforms for mammary enriched transcription factors, and possibly other components, which promote the induction of genes up to several thousand fold during pregnancy. Notably, individual enhancers within super-enhancers are not necessarily equivalent (21) with distinct transcription factor composition. Similarly to the Wap super-enhancer, the constituent enhancers in the α-globin super-enhancer are bound by different combinations of cell lineage enriched transcription factors and also distinct levels of MED1 (47). However, constituent enhancers in the α-globin super-enhancer appear to act independently and additively, suggesting that there is likely no unifying concept of super-enhancers.
STAT3 and STAT5 are largely activated by distinct cytokines and frequently play opposing roles, both in mammary epithelium (11,13) and hematopoietic cells (17). Notably, in T cells IL-2 activates STAT5 and inhibits differentiation of Th17 cells through directly competing with STAT3 binding to the IL-17 locus (16). Similarly, the ratio of IL-21 induced STAT3 and GM-CSF-induced STAT5 is critical for dendritic cell expansion (17). Thus, at least in the immune system, the balance between STAT3 and STAT5 is critical for lineage decision-making. In mammary epithelium STAT5 binding to a set of highly sensitive enhancers is rapidly and specifically replaced by STAT3 within hours following termination of lactation, coinciding with the swift inactivation of these genes. It can be hypothesized that cytokine activation of STAT3 (12,14) at the end of lactation and preceding involution aids to rapidly replace STAT5 from mammary-specific enhancers and is required to effectively inactivate these enhancers and shut down milk production. However, some rapidly decommissioned enhancers, such as one in the Wap super-enhancer, become refractory to STAT3 binding suggesting that their chromatin immediately acquires a closed status upon loss of STAT5 binding.
In summary, the mammary gland uses at least three distinct mechanisms to uniquely regulate cytokine-responsive genes, an autoregulatory enhancer to control STAT5 levels, regulatory elements that differentially respond to STAT5 levels and possibly the ability to inactivate enhancers through the replacement of STAT5 by STAT3. These mechanisms permit the induction of milk-producing genes up to 1000-fold and their rapid inactivation without compromising expression of universal cytokine-regulated genes required for maintaining cell metabolism.
Supplementary Material
Acknowledgments
The authors thank H. Smith from the NIDDK genomics core for help with next-generation sequencing. M.W. is a graduate student of the Individual Graduate Partnership Program (GPP) between NIH/NIDDK and the Medical University of Innsbruck. This work was performed in partial fulfillment of the graduation requirements for M.W.
Contributions: M.W. designed experiments, analyzed ChIP-seq and RNA-seq data, performed computational and statistical analyses. K.H.Y. designed experiments, conducted all mouse work, performed ChIP-seq and all molecular experiments and analyzed data. C.W. performed ChIP-seq experiments. Z.T. is the PhD advisor of M.W. L.H. designed the project and analyzed data. M.W. and L.H. wrote and finalized the manuscript, and all authors reviewed and approved the submitted version.
ACCESSION NUMBERS
The RNA-seq data from wild-type mice at day 6 of pregnancy and day 1 of lactation are available under GSE37646. ChIP-seq samples for ELF5 and NFIB at L1 are available under GSE74826. All ChIP-seq datasets are available under GSE84115.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IPR of the NIDDK/NIH. Funding for open access charge: Intramural research program of the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yamaji D., Kang K., Robinson G.W., Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013;41:1622–1636. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X., Robinson G.W., Wagner K.U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi K., Shillingford J.M., Smith G.H., Grimm S.L., Wagner K.U., Oka T., Rosen J.M., Robinson G.W., Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell. Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu B.M., McLaughlin S.K., Na R., Liu J., Cui Y., Martin C., Kimura A., Robinson G.W., Andrews N.C., Hennighausen L. Hematopoietic-specific Stat5-null mice display microcytic hypochromic anemia associated with reduced transferrin receptor gene expression. Blood. 2008;112:2071–2080. doi: 10.1182/blood-2007-12-127480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang K., Yamaji D., Yoo K.H., Robinson G.W., Hennighausen L. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol. Cell. Biol. 2014;34:464–473. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang K., Robinson G.W., Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metser G., Shin H.Y., Wang C., Yoo K.H., Oh S., Villarino A.V., O'Shea J.J., Kang K., Hennighausen L. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 2016;44:1052–1063. doi: 10.1093/nar/gkv999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman R.S., Lourenco P.C., Tonner E., Flint D.J., Selbert S., Takeda K., Akira S., Clarke A.R., Watson C.J. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L., Hart S., Cheng J., Melenhorst J.J., Bierie B., Ernst M., Stewart C., Schaper F., Heinrich P.C., Ullrich A., et al. Mammary gland remodeling depends on gp130 signaling through Stat3 and MAPK. J. Biol. Chem. 2004;279:44093–44100. doi: 10.1074/jbc.M313131200. [DOI] [PubMed] [Google Scholar]
- 13.Humphreys R.C., Bierie B., Zhao L., Raz R., Levy D., Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology. 2002;143:3641–3650. doi: 10.1210/en.2002-220224. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L., Melenhorst J.J., Hennighausen L. Loss of interleukin 6 results in delayed mammary gland involution: a possible role for mitogen-activated protein kinase and not signal transducer and activator of transcription 3. Mol. Endocrinol. 2002;16:2902–2912. doi: 10.1210/me.2001-0330. [DOI] [PubMed] [Google Scholar]
- 15.Li M., Liu X., Robinson G., Bar-Peled U., Wagner K.U., Young W.S., Hennighausen L., Furth P.A. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3425–3430. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X.P., Ghoreschi K., Steward-Tharp S.M., Rodriguez-Canales J., Zhu J., Grainger J.R., Hirahara K., Sun H.W., Wei L., Vahedi G., et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan C.K., Oh J., Li P., West E.E., Wong E.A., Andraski A.B., Spolski R., Yu Z.X., He J., Kelsall B.L., et al. The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity. 2013;38:514–527. doi: 10.1016/j.immuni.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L. Hierarchy within the mammary STAT-5-driven Wap super-enhancer. Nat. Genet. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S., et al. BEDOPS: High-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham H. ggplot2: Elegant Graphics for Data Analysis. NY: Springer-Verlag; 2009. [Google Scholar]
- 28.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain D., Baldi S., Zabel A., Straub T., Becker P.B. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015;43:6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burdon T., Sankaran L., Wall R.J., Spencer M., Hennighausen L. Expression of a whey acidic protein transgene during mammary development. Evidence for different mechanisms of regulation during pregnancy and lactation. J. Biol. Chem. 1991;266:6909–6914. [PubMed] [Google Scholar]
- 35.Stocklin E., Wissler M., Gouilleux F., Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 36.Park S.H., Liu X., Hennighausen L., Davey H.W., Waxman D.J. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J. Biol. Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- 37.Holloway M.G., Cui Y., Laz E.V., Hosui A., Hennighausen L., Waxman D.J. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148:1977–1986. doi: 10.1210/en.2006-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J.X., Leonard W.J. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 39.Liao W., Lin J.X., Wang L., Li P., Leonard W.J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villarino A.V., Tato C.M., Stumhofer J.S., Yao Z., Cui Y.K., Hennighausen L., O'Shea J.J., Hunter C.A. Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals. J. Exp. Med. 2007;204:65–71. doi: 10.1084/jem.20061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandiyan P., Yang X.P., Saravanamuthu S.S., Zheng L., Ishihara S., O'Shea J.J., Lenardo M.J. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J. Immunol. 2012;189:4237–4246. doi: 10.4049/jimmunol.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaji D., Na R., Feuermann Y., Pechhold S., Chen W., Robinson G.W., Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denny S.K., Yang D., Chuang C.H., Brady J.J., Lim J.S., Gruner B.M., Chiou S.H., Schep A.N., Baral J., Hamard C., et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson G.W., Kang K., Yoo K.H., Tang Y., Zhu B.M., Yamaji D., Colditz V., Jang S.J., Gronostajski R.M., Hennighausen L. Coregulation of genetic programs by the transcription factors NFIB and STAT5. Mol. Endocrinol. 2014;28:758–767. doi: 10.1210/me.2012-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S., Rosen J.M. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol. Cell. Biol. 1995;15:2063–2070. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabowska M.M., Kelly S.M., Reese A.L., Cates J.M., Case T.C., Zhang J., DeGraff D.J., Strand D.W., Miller N.L., Clark P.E., et al. Nfib regulates transcriptional networks that control the development of prostatic hyperplasia. Endocrinology. 2016;157:1094–1109. doi: 10.1210/en.2015-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hay D., Hughes J.R., Babbs C., Davies J.O., Graham B.J., Hanssen L.L., Kassouf M.T., Oudelaar A.M., Sharpe J.A., Suciu M.C., et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016;48:895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.