Abstract
Muscleblind-like (MBNL) proteins are critical RNA processing factors in development. MBNL activity is disrupted in the neuromuscular disease myotonic dystrophy type 1 (DM1), due to the instability of a non-coding microsatellite in the DMPK gene and the expression of CUG expansion (CUGexp) RNAs. Pathogenic interactions between MBNL and CUGexp RNA lead to the formation of nuclear complexes termed foci and prevent MBNL function in pre-mRNA processing. The existence of multiple MBNL genes, as well as multiple protein isoforms, raises the question of whether different MBNL proteins possess unique or redundant functions. To address this question, we coexpressed three MBNL paralogs in cells at equivalent levels and characterized both specific and redundant roles of these proteins in alternative splicing and RNA foci dynamics. When coexpressed in the same cells, MBNL1, MBNL2 and MBNL3 bind the same RNA motifs with different affinities. While MBNL1 demonstrated the highest splicing activity, MBNL3 showed the lowest. When forming RNA foci, MBNL1 is the most mobile paralog, while MBNL3 is rather static and the most densely packed on CUGexp RNA. Therefore, our results demonstrate that MBNL paralogs and gene-specific isoforms possess inherent functional differences, an outcome that could be enlisted to improve therapeutic strategies for DM1.
INTRODUCTION
Cell development and fate is guided by a multitude of RNA binding proteins (RBPs) that affect the processing, localization, translation and turnover of RNAs. Tissue-specific proteome complexity arises from a relatively low number of about 20 000 protein coding genes due to the production of multiple mRNA isoforms generated by alternative splicing (AS) from >90% of protein coding genes (1). In many cases, the profile of alternative isoforms is modified during the course of tissue development and cell-specific transcriptome maturation is adjusted by RBPs functioning as trans-acting splicing factors. Proper AS depends on normal RBP function and abnormalities in the activity of some splicing factors lead to a number of human diseases (1).
Muscleblind-like (MBNL) proteins are conserved multifunctional RBPs which influence AS and alternative polyadenylation (APA), mRNA stability and trafficking as well as microRNA biogenesis (2–9). In mammals, there are three MBNL paralogs, MBNL1, MBNL2 and MBNL3 (10,11). All MBNL paralogs contain two N-terminal tandem zinc finger (ZnF) domains which bind preferentially to specific RNA sequences and/or structures containing two or more clustered GC steps flanked by pyrimidines (YGCY) (12–15). Each MBNL paralog may contain variable amino acid sequences encoded by alternative exons that modulate its cellular localization, the number of ZnF domains and the distance between them, multimerization capacity, affinity to RNA sequence motifs and AS activity (16–19). A comparison of MBNL activities might be reflected by splicing activities. However, AS events are affected by the total expression level of MBNL paralogs and the distribution of multiple splicing isoforms. Both depends on tissue type and developmental stage (4,6,10,20,21). Mutual downregulation of MBNL1 and MBNL2 causes more prominent changes in the pattern of specific AS events and the selection of APA sites compared to the depletion of a single paralog (4,6,20). The analysis of MBNL3 is more difficult due to its low expression level in the majority of adult tissues (10). Mbnl3 isoform knockout mice (Mbnl3ΔE2) show abnormalities in muscle regeneration and functionality but not in the AS pattern (18). In contrast, both Mbnl1 and Mbnl2 knockout mice (Mbnl1ΔE3/ΔE3, Mbnl2ΔE2/ΔE2) exhibit global AS changes mainly in muscles and brain, respectively (3,6,22,23). The activity of recombinant MBNL paralogs has been analyzed in cellular models but few studies have compared their splicing activity in the context of selected exogenous AS events (24–27). Nevertheless, the activities of the MBNL1, MBNL2, MBNL3 proteins and their isoforms have never been directly compared and understanding the differences between MBNL activities might shed light on their impact on RNA metabolism in normal and pathological stages.
In myotonic dystrophy type 1 (DM1) and type 2 (DM2), the three MBNL paralogs are specifically bound to, and sequestered by, expanded CUG (CUGexp) and CCUG repeats (CCUGexp), respectively (10,28,29). These toxic RNAs are composed of multiple UGCU that in vitro form stable hairpin structures and concentrate in nuclear foci in multiple cell types of DM patients (29–37). In living cells, these dynamic structures undergo formation and dispersion events that are affected by MBNL proteins (38,39) and other factors (40). Depletion of MBNL1 and MBNL2 decreases CUGexp foci size and MBNL proteins bind to these structures (24,38,41). The reduction of free MBNL in the nucleoplasm leads to global AS and APA changes (3,4,6,20,22,23,42). The mouse model that overexpresses ∼250 CUG repeats (HSALR) shares more than 80% of the AS and APA abnormalities with the Mbnl1ΔE3/ΔE3 model (3,4,22,43), but the nature of the interaction between CUGexp RNA and MBNL proteins in nuclear foci remains unclear.
In this study, we compared the activity of MBNL paralogs and their splicing isoforms in different cellular models and discovered novel features of MBNL proteins responsible for their function. MBNL paralogs and splicing isoforms differed significantly in subcellular localization and they bound the same regulatory sequences in pre-mRNAs in vitro and in vivo evoking splicing changes of the same AS events with different strengths. MBNL paralogs also bound to toxic CUGexp RNA with high affinity forming densely packed complexes and associated/dissociated from CUGexp foci freely but to different extents. We also identified several factors that have an impact on both AS activity and CUGexp foci formation, including MBNL sequences encoded by alternative exons.
MATERIALS AND METHODS
MBNL1, MBNL2 and MBNL3 constructs were generated using pEGFP-C1 and MB1-41, MB1-42, MB1-43 and MB1-N used in this study will be described elsewhere (Konieczny et al., unpublished data). MB1-40 was produced by partial digestion and cloning of an EcoRI-Alw44I fragment from MB1-42 into MB1-41 while MB1-C was generated by the deletion of a region between MB1-41 HindII restriction sites. MBNL2 (MB2-38, MB2-39, MB2-40, MB2-41) and MBNL3 (MB3-37, MB3-39) isoforms were amplified from a human adult kidney and liver cDNA library, respectively (Human MTC Panel I, Clontech cat no. 636742) and amplified fragments were inserted into BamHI and EcoRI sites. The sequences of these constructs were verified by sequencing. For more information, see Supplementary Table S1 and Supplementary Material & Methods. The coding sequences of mCherry or Dendra2 were amplified and cloned between NheI-XhoI (MBNL1) and NheI-HindIII (MBNL2 and MBNL3) restriction sites instead of enhanced green fluorescent protein (EGFP or GFP). For generation of an EGFP-mCherry construct, an amplified mCherry sequence was inserted downstream of EGFP into HindIII-EcoRI sites. The final constructs were sequenced. All primers and polymerase chain reaction (PCR) conditions for Pfx50 high fidelity DNA polymerase amplifications (Invitrogen) are specified below. To generate recombinant MBNL1, MBNL2 and MBNL3 proteins, the construct for MBNL1 expression was described previously (44) while MBNL2 and MBNL3 were amplified (using primers listed in Supplementary Material & Methods) digested with BamHI and EcoRI, inserted into pGEX-6P-GST-His12 and the final constructs were sequenced. Purification of recombinant GST and His12-tagged MBNL1, MBNL2 and MBNL3 was performed as described (45) and protein concentration was measured using both the Bradford Assay and Sypro Ruby staining on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gels (Supplementary Figure S6A).
Minigenes
Human TNNT2 ex.4 minigene and a DT960 construct encoding CUGexp were a gift (Thomas Cooper, Baylor College of Medicine) and have been described previously (39,46). The mouse Atp2a1 ex.22 minigene was prepared on the pEGFP-C1 background as previously described (47). The generation of mutated Atp2a1, Nfix also Ldb3 minigenes will be described elsewhere (Cywoniuk et al., unpublished data).
Cell cultures and transfection
HeLa cells were grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 1× MEM non-essential amino acid solution (Sigma), 1× antibiotic and antimycotic (Sigma) at 37°C with 5% CO2. Before transfection cells were seeded on 12-well plates filled with 1 ml of medium and allowed to reach up to 60-70% confluence. For endogenous splicing and protein expression analysis, HeLa cells were transfected with 2 μg of MBNL constructs using X-tremeGENE HP DNA Transfection Reagent (1:2 ratio, Roche) and harvested after 42 h. For exogenous splicing analysis, HeLa cells were co-transfected with 100 ng of splicing minigene, and 75, 150 and 300 ng of MBNL constructs, which were supplemented by empty pEGFP to the highest total DNA amount and cells were harvested after 24 h. In order to study pre-mRNA-MBNL interactions, HeLa cells were cotransfected with 500 ng of MBNL constructs and 200 ng of the Atp2a1 minigene. After 4 h, cells were transfected with 125 nM AONs (Supplementary Material & Methods). For both transfections, Lipofectamine 2000 (Invitrogen) was used. In fluorescentin situ hybridization (FISH) and fluorescence recovery after photobleaching (FRAP) experiments, HeLa cells were transfected with 200 ng of DT960 and 500 ng of MBNL constructs. For the Dendra2 analysis, the amount of MBNL1 reached up to 1 μg while for fluorescence-lifetime imaging microscopy (FLIM), 200 ng of DT960 and 375 ng of each GFP and mCherry fused to MBNL were used.
RNA isolation and RT-PCR
Total RNA from HeLa cells was isolated using TRI Reagent (Sigma) and total RNA (2 μg) was reverse transcribed using GoScript Reverse Transcription System (Promega) and random primers (Promega) according to the manufacturer's protocol. Samples transfected with splicing minigenes were treated with RQ1 DNase (Promega). For human tissues, Human MTC Panel I and Human Fetal MTC Panel (Clontech cat no. 636742 and 636747) were while muscle samples were a gift (Charles Thornton, University of Rochester). All PCRs were conducted using GoTaq Flexi DNA Polymerase (Promega) and detailed PCR conditions are described in Supplementary Table S2. PCR products were resolved on agarose gels with USB dye (Syngene) and gels were visualized on G:BOX and analyzed using GeneTools software (Syngene).
Western blotting
HeLa cells were lysed with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% NP-40, 0.5% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS)) supplemented with SIGMAFAST Protease Inhibitor Cocktail (Sigma). Lysates were sonicated at 4°C and centrifuged at 18 000 × g at 4°C for 10 min. Samples were heated to 95°C for 5 min, separated on 10% SDS polyacrylamide gels and transferred to nitrocellulose (Protran BA 85, Whatman) using a wet transfer apparatus (1 h, 100 V, 4°C). Membranes were blocked for 1 h with 5% skim milk in PBST buffer (phosphate buffered saline (PBS), 0.1% Tween-20) and incubated with a primary antibody against GFP (1:1000, Santa Cruz cat. no. sc-8334) or GAPDH (1:1000, Santa Cruz cat. no. sc-47724). Anti-rabbit (1:20 000, Sigma cat. no. A9169) and anti-mouse (1:2000, Millipore cat. no. 12-349) secondary antibodies were conjugated with horseradish peroxidase and detected using the Pierce ECL Plus Western Blotting Substrate (Thermo Scientific) detection kit.
CLIP-seq and RIP-seq
Crosslinking and immunoprecipitation (CLIP) experiments were performed on mouse skeletal muscles and hearts as well as transiently transfected C2C12 cells. RIP-seq experiments were performed on skeletal muscles. Step-by-step protocols and the MBNL Interactome Browser (MIB.amu.edu.pl) is described in Supplementary Material & Methods.
Microscopy
Confocal microscopy experiments were performed within 48 h after HeLa cell transfection. For live cell imaging, time-lapse sequences, FRAP, photoconversion of Dendra2 fluorescent protein and FLIM cells were seated on 96-well glass-bottom plates (Greiner Bio-One, cat no. 82051-531). Images were processed with Imaris software (intensity thresholds, gamma correction, scale bars) and presented as maximum intensity projection from all z slices.
Live-cell imaging was performed on Nikon A1Rsi confocal microscope with objectives Nikon Plan Apo VC 60x/1.4 Oil DIC N2 (FRAP, dual-channel imaging, FLIM) and Nikon Plan Apo 40x/0.95 DIC N2 (time-lapse acquisition). GFP and mCherry fluorescent proteins were excited with 488 nm Argon-Ion and 561 nm pumped-diode laser, respectively. For detection dichroic mirror 405/488/561 nm with spectral filters 525/50 nm (green channel) and 595/50 nm (red channel) were used. To avoid spectral bleed-through channels were scanned sequentially. For each cell several optical slices covering majority of the cell volume were acquired. Time-lapse sequences were acquired through 12-16 h with 2-5 min step between scans. Z-stacks were covering cells and space above and below to avoid movement of the cells to out of focus planes.
FRAP was performed on single focal plane using a sequence of 5 pre-bleach, 2 bleach and 294 post-bleach image acquisition (overall experiment time set to 5 min). A few GFP foci were bleached in each nucleus using 488 nm laser at full power and fluorescence intensities were measured by confocal PMT detector. Results were analyzed with usage easyFRAP software (48). For analysis, the intensities of bleached region of interests (ROIs) from single nucleus were averaged and the entire nucleus was set as reference ROI and the extracellular medium was used as background. The raw data were normalized by full scale method. The curve was fitting to single exponential equation to calculate the mobile fraction and t-half. Statistical significance was determined by Mann-Whitney U test; ∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001.
Photoconversion of the Dendra2 fluorescent protein was done using a 405 nm diode laser set to 12% power, and each ROI was bleached for 66 ms. Image acquisition was performed on single focal plane through 5 min with 10 s interval and 2 pre-bleach/conversion images. Results were normalized by full scale method in Excel software.
FLIM measurements were performed using the PicoQuant LSM Upgrade Kit. Fluorescence resonance energy transfer (FRET) donor molecule (GFP) was excited by the 485 nm pulsed-diode laser at a 40 MHz repetition rate. Photons were counted by single-photon avalanche diode (SPAD) detector with 520/35 nm spectral filter and 256 × 256 pixel FLIM images were acquired as long as average number of photon counts per pixel reached at least 100. FLIM analysis was performed with SymPhoTime64 software from PicoQuant. Statistical significance was determined by Student's t-test; ∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001.
Fluorescence in situ hybridization
FISH was conducted as described (49) with some modifications. Cells were fixed in 2% paraformaldehyde (PFA)/PBS at 4°C for 5 min and washed three times in cold PBS. Pre-hybridization was performed in 30% formamide and 2× saline-sodium citrate (SSC) buffer at 4°C for 10 min, followed by hybridization in buffer containing 30% formamide, 2× SSC, 0.02% BSA, 66 μg/ml yeast tRNA, 10% dextran sulfate, 10 U Rnasin (Promega) and 2 ng/μl DNA/locked nucleic acid (LNA) probes (CAG)6-CA, labeled at the 5′-end with Cy3 and modified at positions 2, 5, 8, 13, 16 and 19 with LNA. Post-hybridization washing was done in 30% formamide and 2× SSC at 37°C for 30 min followed by 1× SSC at 37°C for the next 30 min. Microscopic slides were mounted using medium containing 2% propyl gallate (Sigma), 10% glycerol and 4′,6-diamidino-2-phenylindole (DAPI) and sealed with fingernail polish. All FISH images were acquired on Nikon A1Rsi microscope using the following excitation conditions: diode lasers 405, 488 and 561 nm, dichroic mirror 405/488/561, emission filters 450/50 for DAPI, 525/50 for GFP and 595/50 for Cy-3. All images were acquired with sequential scanning to avoid spectral bleed-through. A total of 18-25 optical sections were acquired. Foci volume was estimated by Imaris. Statistical significance was determined in GraphPad Prism software by Mann-Whitney U test; ∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001. The Spearman's rank correlation coefficient was calculated in Python (scipy.ststs).
Transcription in vitro and radiolabeling
DNA templates for Atp2a1, Mbnl1, Mbnl2, Tnnt3, Calm3, Clcn1 and Mfn RNAs were obtained in two PCR reactions. First, longer products were amplified using genomic DNA and primer sets (see Supplementary Material & Methods) at the indicated annealing temperature (Ta) which were subsequently used as template for a second PCR reaction carried out at 55°C and utilizing primer sets (see Supplementary Material & Methods) with a 5′ promoter sequence for T7 RNA polymerase (Ambion). The only exception was Insr, which was amplified from a synthesized DNA template using specific primers. Double-stranded (CCAG)14 DNA was prepared as described (35) using a GGCCC(CAGG)14GGGCCTATAGTGAGTCGTATTA ssDNA and a T7 oligomer TAATACGACTCACTATAGG. The transcription reaction was performed in 50 μl composed of 10 μl of DNA template, 0.15 mM nucleoside triphosphates (NTPs) (Invitrogen) 0.45 mM guanosine (Sigma-Aldrich), 50 U T7 RNA Polymerase (Ambion), 1× T7 transcription buffer (Ambion), 40 U Rnasin Plus RNase Inhibitor (Promega). Purification of the transcript was conducted on a denaturing 6% polyacrylamide gel (19:1 acrylamide:bisacrylamide) followed by ethanol precipitation. For radiolabeling, 2-4 pmol of transcript was incubated with 2-4 pmol of [γ-32P]ATP, 1 U Rnasin Plus, 10 U OptiKinase (Affymetrix), 1× reaction buffer (Affymetrix) and ddH20 up to 10 μl, at 37°C for 1 h. Labeled RNA was subsequently run on a denaturing 8% polyacrylamide gel (19:1) in 0.5× Tris-borate-EDTA (TBE) buffer, at 100 V for 1 h, the RNA was visualized using FLA-5100 (FujiFilm) and the RNA was cut out followed by ethanol precipitation and resuspended in 20-40 μl ddH2O. (CUG)20 and (CAG)20 were a gift (Włodzimierz Krzyżosiak, Polish Academy of Sciences).
Quantification of RNA-MBNL interaction and its inhibition by AONs in vitro
Filter binding assay was performed in 30 μl volume. To assess the MBNL1, MBNL2 and MBNL3 affinity to RNAs, 5′-labeled transcripts (0.05 nM) were incubated with the indicated concentrations of the proteins (ranging from 0 to 250 nM) in buffer B containing 250 mM NaCl, 15 mM KCl, 50 mM, Tris-HCl pH 8.0, 0.05% Tween-20, 1 mM MgCl2 at 37°C for 25 min. DNA template preparation and in vitro transcription were described in Supplementary Material & Methods. To estimate the inhibitory property of AONs, 0.05 nM of labeled transcripts, incubation underwent a three step with 20 μM of AONs, first at 90°C for 1 min, then on ice for 10 min and at 37°C for 25 min. Subsequently, the indicated concentrations of MBNLs were added to each sample and incubated at 37°C for 25 min. A total of 25 μl of samples were loaded onto filter binding apparatus with nitrocellulose (Protran BA 85, Whatman) and nylon (Hybond N+, Amersham) membranes previously wetted in buffer B. The signal from membranes was visualized on IP through FLA-5100 and quantified using Multi Gauge software (FujiFilm). Kd values were calculated in GraphPad Prism based on two experimental replicas, using the equation for one site-specific binding and standard error of the mean. Other statistical analyses were performed with GraphPad Prism, Python (scipy.stats) and Microsoft Excel software and significance was determined by the appropriate parametric or nonparametric statistical test.
RESULTS
Platform to evaluate the activities of MBNL paralogs and their splicing isoforms
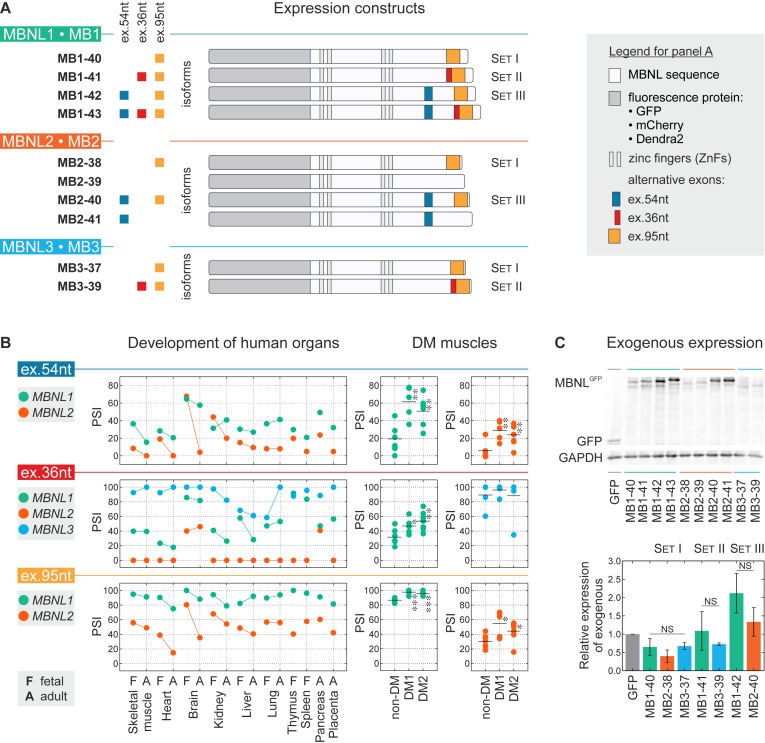
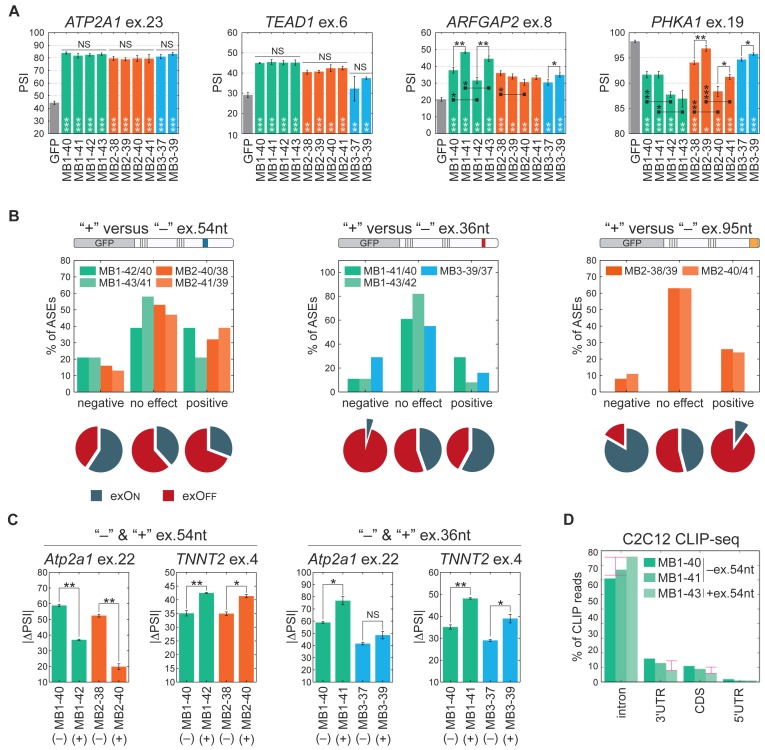
The main aim of our project was to explain whether three protein paralogs with very similar primary structure have similar or distinguishable activity in the same conditions? The MBNL1, MBNL2 and MBNL3 genes have various expression profiles in different tissue types and developmental stages (11,50) and there are several isoforms of individual MBNL paralogs containing different combinations of a few alternatively spliced exons (Supplementary Figure S1A). Thus, to directly compare the activity of the three MBNL paralogs and their spliced isoforms, we generated and transiently overexpressed 10 different proteins in HeLa cells in which the expression of endogenous MBNL isoforms is relatively low (Supplementary Figure S1B). All experiments were conducted within 48 h post-transfection using four isoforms of MBNL1 (MB1-40, MB1-41, MB1-42 and MB1-43 kDa), four isoforms of MBNL2 (MB2-38, MB2- 39, MB2-40 and MB2-41 kDa) as well as two isoforms of MBNL3 (MB3-37 and MB3-39 kDa) (Figure 1A and Supplementary Table S1). All of them represent the most common protein variants detected in different fetal and adult tissues (50). The similarity of amino acid sequences in the core region of MBNL paralogs is 65-73% and the highest between MBNL1 and MBNL2 (Supplementary Figure S1C). The variance in amino acid sequences of different isoforms of MBNL paralogs depends on the presence or absence of three highly conserved alternative exons which consist of 54, 36 and 95 nucleotides, herein called ex.54nt, ex.36nt and ex.95nt (previously numbered ex.5, ex.7 and ex.8, respectively) (2) (Figure 1A and Supplementary Figure S1A). The protein sequences encoded by MBNL1 and MBNL2 ex.54nt, which contains a nuclear localization signals, are 72% identical whereas no such sequence exists in MBNL3 (Supplementary Figure S1A) (16,17,19). The amino acid sequences encoded by ex.36nt and ex.95nt are 60-82% and 81-87% identical between the three paralogs, respectively, and form a C-terminus of the protein. Ex.36nt is believed to be responsible for homotypic MBNL1-MBNL1 dimerization (16,51).
Figure 1.
Distinct splicing and expression patterns for MBNL paralogs. (A) Scheme of 10 expression constructs containing sequences of fluorescence proteins (GFP, mCherry or Dendra2) and MBNL1, MBNL2 and MBNL3 paralogs possessing or lacking three alternative exons (ex.54nt, ex.36nt, ex.95nt). For details on MBNL exonic composition see Supplementary Table S1. Constructs for comparable isoforms having the same alternative exons are divided into Set I, Set II and Set III. (B) Splicing profiles for MBNL ex.54nt, ex.36nt and ex.95nt in human tissues analyzed by RT-PCR. Dots represent PSI values. The adult and fetal samples from particular tissue types are indicated by a bar. Splicing results obtained for non-DM (N = 7), DM1 (N = 5) and DM2 (N = 7) muscle samples. Statistical significance was determined by the Student's t-test (∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). (C) Relative expression level of exogenous MBNL paralogs. Exogenous proteins were detected by anti-GFP antibody and normalized to GAPDH. Bars represent average expression level and standard deviations are from two independent biological experiments. Statistical significance was determined by the Student's t-test (NS for P ≥ 0.05).
RT-PCR analysis revealed that the splicing of ex.54nt, ex.36nt and ex.95nt undergoes significant changes during organ development (Figure 1B). As a measure of AS changes, we used the percent-spliced-in (PSI) parameter which indicates the mRNA fraction that includes the specific alternative sequence. Interestingly, all these exons are also significantly misspliced in both DM1 and DM2 in which the pool of functional MBNL protein is reduced due to sequestration on toxic RNAs containing CUGexp or CCUGexp, respectively (Figure 1B). Cumulatively, these data indicate that during tissue development, and in some pathological stages, the quantity and/or quality of MBNL1, MBNL2 and MBNL3 expression undergo significant regulation.
All tested MBNL isoforms were C-terminal fusions to fluorescent proteins (Figure 1A). The effectiveness of exogenous protein production in HeLa cells was assessed using western blotting with an anti-GFP antibody and normalization to GAPDH. We noticed that the presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (∼2.7 and ∼4.0-times, respectively). We observed marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (∼1.3-times) and ex.95nt (∼0.8-times) (Figure 1C and Supplementary Figure S1D). We also compared the expression level of MBNL paralogs in three sibling splicing isoform sets differing in the presence or absence of one of three alternative exons. Set I consisted of MB1-40, MB2-38 and MB3-37 having only ex.95nt, Set II consisted of MB1-41 and MB3-39 having both ex.36nt and ex.95nt while Set III contained MB1-42 and MB2-40 having ex.54nt and ex.95nt (Figure 1A). Differences in the expression level of GFP fused MBNLs within these groups were very low (Figure 1C).
Cumulatively, these data indicate that this cellular model provides a reliable platform to evaluate the activities of MBNL paralogs and their alternatively spliced isoforms. Next, we focused on MBNL splicing activity in normal and pathological states.
MBNL1 possesses the strongest, while MBNL3 the weakest, splicing activity
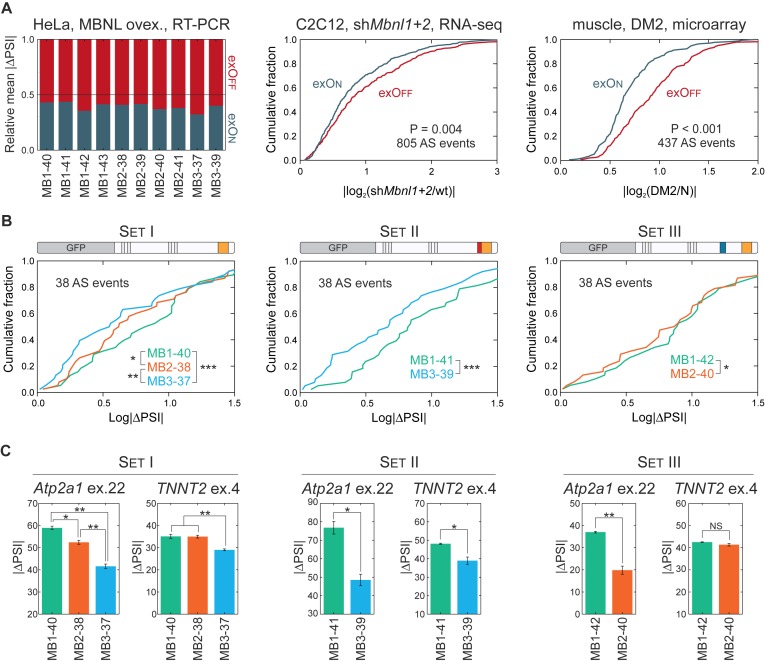
To study the impact of MBNL paralogs on AS regulation for individual AS events and globally for the transcriptome, we first selected 38 AS events reported previously to be MBNL-sensitive and analyzed them in HeLa cells using RT-PCR (Supplementary Table S2). To standardize alternative exon enumeration, we applied the numbering system from FasterDB (52). We observed that 10 MBNL isoforms regulate selected individual AS events always in the same direction, either toward exon inclusion (herein termed exOn) or exclusion (exOff) (Supplementary Figure S3A). However, we noticed that taking all 38 AS events into consideration, the average strength of exOff was significantly higher than exOn for all MBNL isoforms (Figure 2A). We decided to confirm this observation based on the splicing changes of hundreds of AS events selected from other available data sets (6,22,53). MBNL silencing in C2C12 myoblasts and mouse Mbnl knockout muscles induced a stronger effect on exOffs (Figure 2A; Supplementary Figure S3C and D). Even though C(C)UGexp RNAs may perturb the activity of several splicing factors, the same phenomenon was also observed in skeletal muscles of the HSALR mouse DM1 model as well as DM2 patients (Figure 2A and Supplementary Figure S3E).
Figure 2.
MBNL1 is the strongest, while MBNL3 is the weakest, AS regulator. (A) Comparison of the relative mean for exOn/exOff splicing changes (|ΔPSI|) of 38 endogenous AS events in HeLa cells transfected with 10 different MBNL isoforms (left chart). Global analysis of exOn and exOff changes in C2C12 with silencing of MBNL determined by RNA-seq (central chart) (6) and DM2 skeletal muscles analyzed by microarrays (right chart) (53). For all analyses, the fold change was used as a measure of splicing strength for the indicated number of AS events. The statistical significance was determined by the Mann-Whitney U test. All comparisons revealed higher average changes for exOff than exOn. For more examples see Supplementary Figure S3. (B) Combined analysis of splicing changes (expressed as |ΔPSI|) of 38 tested AS events induced by MBNL paralogs from Set I, Set II and Set III. Statistical significance was assessed by Wilcoxon signed rank test (∗ for P <0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). (C) Differences in the regulation of exogenous mouse Atp2a1 ex.22 and human TNNT2 ex.4 by MBNL paralogs. The represented mean and standard deviation come from two biological replicas. Statistical significance was determined by the Student's t-test (∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). For MBNL dose dependent changes, see Supplementary Figure S4.
Subsequently, we looked at differences between the activities of the three MBNL paralogs within comparable groups of Set I, Set II and Set III having the same alternative exons (Figure 1A). Surprisingly, we discerned that for combined splicing changes of all 38 AS events, MBNL1 had the strongest, whereas MBNL3 had the weakest, splicing activity (Figure 2B). MBNL1 showed ∼28% more splicing changes compared to MBNL3 and 30 of 38 tested AS events fit this rule while eight (21%) AS events did not, including ATP2A1 ex.23 and NASP ex.7 that showed similar PSI values for each MBNL group (Supplementary Figure S2). To validate this observation, we coexpressed three Sets of paralogs with two splicing minigenes, mouse Atp2a1 and human TNNT2, containing MBNL regulated exons. For the Atp2a1 minigene, the level of ex.22 (orthologue of human ATP2A1 ex.23) inclusion was always the highest for MBNL1, and the weakest for MBNL3, isoforms (Figure 2C). Similar results were obtained for the exclusion of TNNT2 ex.4 although contrary to Atp2a1, MBNL1 and MBNL2 isoforms induced similar splicing changes. These results demonstrated that MBNL paralogs influence pre-mRNA splicing to varying degrees and MBNL1 has the strongest alternative processing whereas MBNL3 has the weakest. These results suggest that even relatively small differences in primary structures of MBNL paralogs influence their activities. MBNL proteins regulate hundreds of AS events (4,6,23) thus even moderate differences in their activity might have a cumulative and significant impact.
MBNL paralogs regulate same AS events differentially binding the same motifs
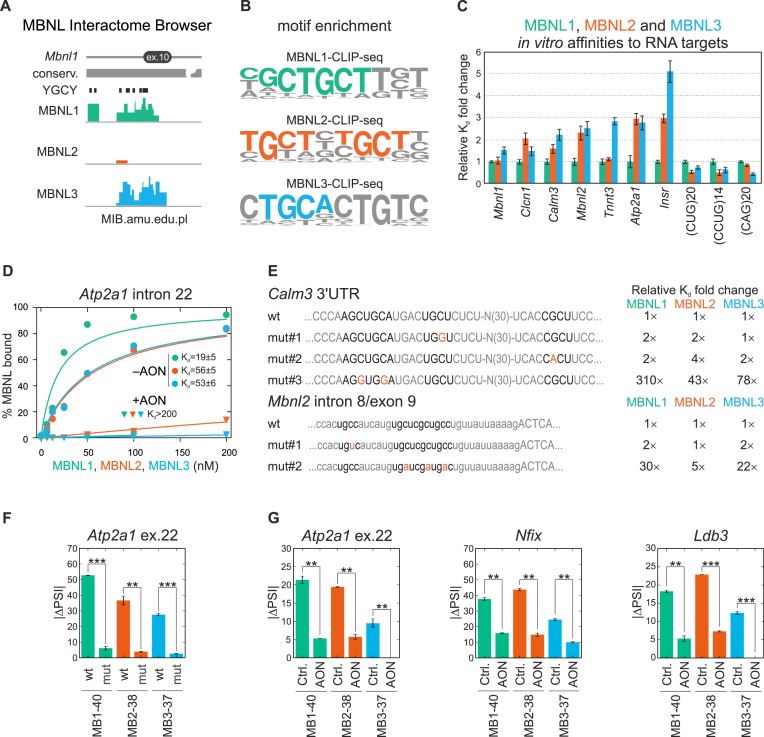
Potential explanations for the observed differences in AS regulation between the tested MBNL paralogs are target RNA binding properties-binding sites and affinity. To test this hypothesis, we first ascertained whether all three MBNL paralogs bind to the same or diverse RNA binding sites. To answer this question, global potential binding sites for the three MBNL paralogs were identified by combining all CLIP experiments followed by deep sequencing (CLIP-seq) results coming from our own (Supplementary Figure S5) as well as previously published datasets for MBNL1 (4,6,8), MBNL2 (4,20,23) and MBNL3 (4,18). The total number of mapped CLIP-seq unique reads amounted to about 1.0, 0.1 and 0.8 million for MBNL1, MBNL2 and MBNL3, respectively, and we developed the MBNL Interactome Browser (MIB) (available at MIB.amu.edu.pl) to analyze the localization of CLIP-seq reads specific for MBNL1 and MBNL3 within alternative exons or in neighboring introns (±250 nt) of 38 AS events (results for MBNL2 were underrepresented). Approximately half of the reads overlapped between the MBNL1 and MBNL3 CLIP-seq data (Figure 3A and Supplementary Table S2), which suggests that splicing regulation requires binding to the same RNA target sites. We also noticed that the consensus sequences for all MBNL proteins are very similar and include 1-2 YGCY/A motifs (Figure 3B).
Figure 3.
MBNL binding to RNA targets. (A) An example of overlapping MBNL1, MBNL2 and MBNL3 specific CLIP-seq clusters located in the region of intron 9 and alternative ex.10 (known also as ex.5; Supplementary Figure S1A) of mouse Mbnl1 based on the MBNL Interactome Browser (MIB.amu.edu.pl). For additional AS examples, see Supplementary Table S2. The presence of overlaid reads was detected for 81, 21 and 71% of AS events for MBNL1, MBNL2 and MBNL3, respectively. All YGCY sequence motifs located in this region, as well as evolutionary conservation are also shown. Note that Multi MIB also contains crosslinking induced mutations (CIMs), MBNL1-RIP-seq and CLIP-seq results for CELF1 (8) and NOVA (58) proteins. (B) A 10-mer sequence motif enrichment analysis from MBNL1, MBNL2 and MBNL3 CLIP-seq data determined by HOMER software. (C) The comparison of binding affinity of MBNL paralogs for 10 RNA fragments, indicating the strongest binding properties of MBNL1 and about 3-fold weaker of the other paralogs. However, in three examined instances, the binding affinity of all paralogs was alike. The dissociation constants (Kd) for MBNL1 constituted the normalization values. (D) MBNL paralogs have slightly different affinities for the 145 nt RNA fragment of Atp2a1 intron 22 in vitro. The Kd values significantly increase after blocking the MBNL1-specific binding site by 21nt-long antisense oligonucleotide (+AON) compared to control experiments (−AON). Dots represent the average percent of MBNL bound ± SD from two technical repetitions. For Mbnl1 exon 3, see Supplementary Figure S6C. For more details, see Supplementary Material & Methods. (E) Point mutations in crucial YGCY motifs compromise MBNL1, MBNL2 and MBNL3 binding efficiency in vitro. (Left panel) Nucleotide sequences of wild-type (wt) and mutated (mut#1-3) Calm3 3′UTR fragment and Mbnl2 intron 8/exon9 RNA fragment containing several YGCY motifs. (Right panel) Fold change of Kd values normalized to wt samples. (F) Removing the 111 nt sequence having the MBNL1-binding site in the Atp2a1 minigene (mut) also vastly affects MBNL2 and MBNL3 activity. (G) Blocking the MBNL-specific binding site in intron 22 of Atp2a1 minigene using AON reduces ex.22 inclusion for three MBNL paralogs in HeLa cells compared to the control AON (Ctrl.). Analogs results are represented for AONs designed against two other MBNL-binding sequences in Nfix and Ldb3. (F and G) Bars represent average PSI ± SD from two independent experiments and statistical significance was determined by the Student's t-test (∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001).
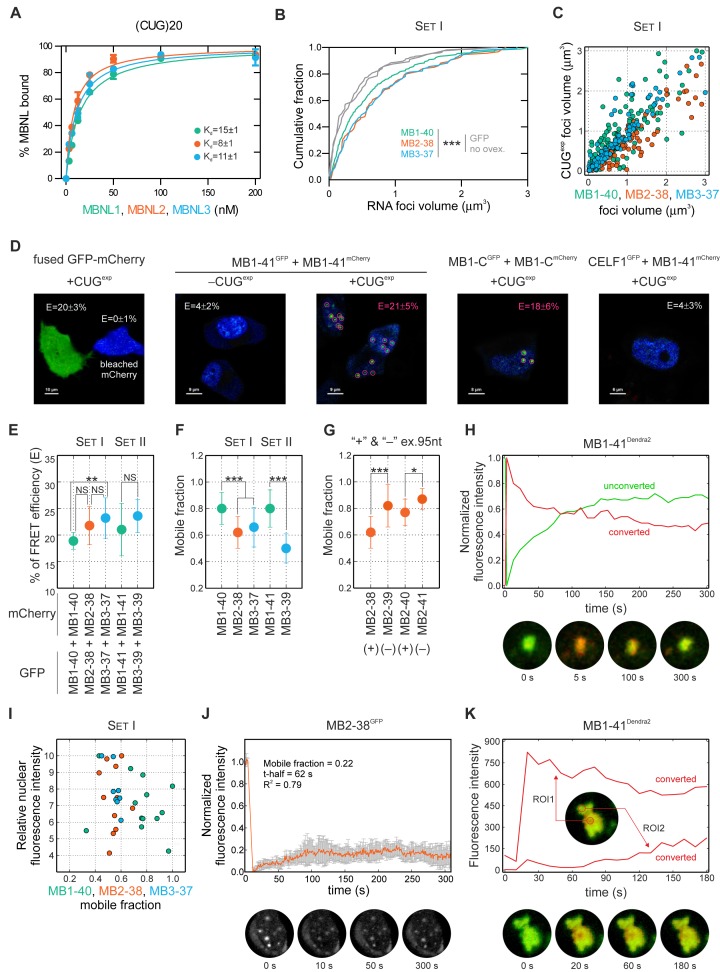
To check whether all MBNL paralogs efficiently bind to the same RNA molecules with the similar or different affinity we compared the binding affinity of MBNL proteins to short synthetic RNA fragments, which were known from previous studies to be MBNL1 targets (50). Using the in vitro assay we determined the dissociation constant (Kd) values for interaction between MBNL paralogs and 10 RNAs and found 3-fold differences in binding affinity. However, for some RNAs, including Mbnl1 exon 3 and (CUG)20 repeats, all MBNL paralogs showed similar, <2-fold Kd fold change values (Figure 3C and Supplementary Figure S6C-E).
To confirm that all MBNL paralogs interact with exactly the same binding sites, we utilized Atp2a1 intron 22 and Mbnl1 exon 3 RNA fragments and identified the YGCY sequences, which are predicted to be MBNL recognition motifs. Having designed antisense oligonucleotides (AONs) complementary to selected YGCY motifs, we noticed that for RNA fragments bound to these specific AONs the Kd was at least 100-times greater for all MBNL paralogs (Figure 3D; Supplementary Figure S6B and C). Next, we prepared two sets of Calm3 3′UTR and Mbnl2 intron 8/exon 9 RNA fragments with point substitutions into 1-3 YGCY motifs. Based on in vitro experiments we concluded that the binding affinity of each MBNL paralog is compromised by mutation in the same crucial binding motifs mut#3 and mut#2 for Calm3 and Mbnl2, respectively (Figure 3E). On the other hand affinity was not affected by mutations in other YGCY motifs.
Next, we asked whether MBNL paralogs regulate AS events interacting with the same YGCY motifs in living cells. To answer this question, we deleted 111 base pair sequence encoding a previously defined MBNL1 binding site in the intron 22 of Atp2a1 minigene and we cotransfected it with MBNL paralogs. As we expected, neither MBNL protein was able to efficiently regulate ex.22 splicing compared to the wild-type (wt) minigene (Figure 3F). In the next experiment, we cotransfected the wt Atp2a1 minigene also with the same AONs as used in vitro. For three tested paralogs, identical and strong inhibitory effects of these AONs on ex.22 inclusion was observed (Figure 3G). Similar results were obtained for AONs designed against two other MBNL1-binding sequences from Nfix and Ldb3 (Figure 3G).
These results indicate that a few determinants influence MBNL AS activity. MBNL paralogs recognize the same sequence motifs but the differences in splicing activity observed between paralogs are influenced by different affinity to RNA target, probably due to distinct sequence or structure sensitivity.
MBNL paralogs splicing activities correlate well with their subcellular localization
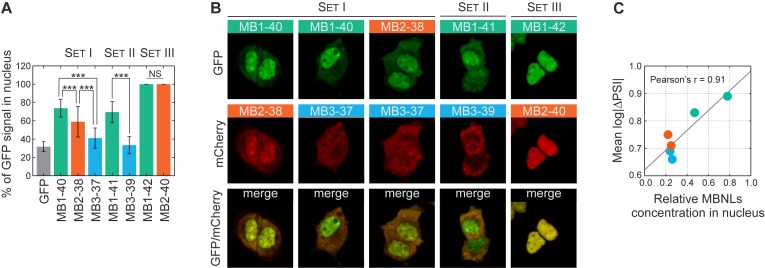
Subsequently, we addressed the question what is the the effective concentration of MBNL proteins in the nucleus. Firstly, we checked if there were any differences in subcellular localization of the studied proteins using quantitative confocal microscopy on HeLa cells transfected with three MBNL paralogs and their splicing isoforms. Surprisingly, we observed a significantly different distribution of the fluorescence signal between cytoplasm and nucleoplasm for cells transfected with different GFP or mCherry tagged MBNL constructs. The fluorescence intensity was measured from total volumes of ∼100 cells for each construct and the nuclear signal was normalized to the total cellular signal (Figure 4A and Supplementary Figure S7A). The comparison of paralogs from Set I and Set II revealed that MBNL1 predominates in nucleoplasm (∼72%), MBNL3 was primarily cytoplasmic (only 36% in nucleoplasm) and MBNL2 showed an intermediate localization pattern (∼55% in nucleoplasm). As we expected, for Set III, the presence of the amino acid sequence encoded by ex.54nt influenced the localization of MBNL proteins in the nucleus (16–17,19). We confirmed these observations with coexpression of MBNL isoform pairs fused with mCherry and GFP (Figure 4B and Supplementary Figure S7B). Interestingly, we also observed reduced nucleoplasmic distribution of both C-truncated (MB1-C) and N-truncated (MB1-N) MBNL1 proteins (Supplementary Figure S8).
Figure 4.
Distinct localization patterns for MBNL paralogs. (A) The percentage of nuclear signal for GFP-MBNL fusions belonging to Set I, Set II and Set III determined by quantitative confocal microscopy analysis of the GFP fluorescence signal. Bars represent the mean from the whole volume of about 50 cells. Statistical significance was assessed by the Student's t-test (NS for P ≥ 0.05 and ∗∗∗ for P < 0.001). (B) Distinct nucleoplasmic and cytoplasmic distribution patterns for isoform pairs visualized by coexpression of MBNL proteins fused with GFP and mCherry (Set I and Set II). Isoforms having ex.54nt localize exclusively to the nucleus (Set III). (C) The Pearson correlation coefficient of relative nuclear concentration (normalized MBNLs expression level × relative nucleoplasmic distribution) with mean strength of 38 AS event changes (log|ΔPSI|) for six MBNL paralogs without alternative ex.54nt.
Then, we investigated whether the different strength of AS changes reflected the expression level of MBNL paralogs. The Pearson correlation coefficient between the mean log|ΔPSI|, which reflected the strength of splicing changes, and the total MBNL level was moderate (r = 0.59). However, when the differences in localization pattern of the MBNL paralogs was assessed, a much stronger correlation was achieved for comparing log|ΔPSI| versus the MBNL nuclear concentration (r = 0.91) (Figure 4C).
These results indicate that MBNL nuclear concentration influences the MBNL-mediated regulation of AS. It is worthy to mention that instead of this phenomenon some AS events were equally regulated by MBNL paralogs (Supplementary Figure S2).
Alternative MBNL exons modulate splicing activity
Based on AS results, we also noted significant differences in the activity of splicing isoforms of individual MBNL isoforms (Figure 5A and Supplementary Figure S2). In the next step, we asked the question whether the presence of amino acid sequences encoded by ex.54nt, ex.36nt and ex.95nt influence MBNL splicing regulatory properties. We compared splicing changes of 38 AS events for the comparable pairs of MBNL paralogs differing in one alternative exon (Figure 5B). In spite of the significantly higher expression of MBNL1 and MBNL2 isoforms possessing ex.54nt and their exclusive nuclear localization, we observed a similar or even weaker effect on splicing regulation for the majority of AS events (∼67%) compared to isoforms lacking ex.54nt (Figure 5B). For all AS events with significant differences in splicing patterns affected by the presence or absence of ex.54nt, there was a relationship between the MBNL mechanism of action (exOn or exOff) and the strength of splicing changes. For AS events with a negative effect of ex.54nt, the exOn events predominated (59%, P = 0.058) (Figure 5B). To confirm our observation, we coexpressed mouse Atp2a1 and human TNNT2 minigenes undergoing exOn and exOff, respectively, with comparable MBNL isoforms. In agreement with our previous observation, the presence of ex.54nt sequence induced a negative effect on exOn of Atp2a1 but a positive effect on exOff of TNNT2 (Figure 5C).
Figure 5.
Specific MBNL alternative exons modulate splicing activity. (A) Compared splicing activity (expressed as PSI values) of all tested MBNL proteins for four selected AS events analyzed by RT-PCR. Bars represent average PSI ± SD from three independent experiments and statistical significance was determined by the Student's t-test (∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). White stars refer to statistical differences between a particular MBNL isoform and GFP control. Black stars refer to statistical differences between comparable MBNL isoforms having or lacking a sequence encoded by alternative ex.54nt, ex.36nt and ex.95nt. Supplementary Table S2 contains results for all 38 AS events. (B) Combined analysis of the 38 tested AS events for the comparable MBNL isoforms differing in the presence or absence of sequences encoded by ex.54nt, ex.36nt or ex.95nt. The comparison was made for nine pairs of isoforms, four for ex.54nt, three for ex.36nt and two for ex.95nt. AS events were divided into categories showing stronger, weaker or no splicing changes for individual isoform pairs taking statistical significance into consideration (‘no change’, P ≥ 0.05; weaker and stronger, P < 0.05; Student's t-test). Note that the mean strength of splicing changes for isoforms having ex.95nt was 2.6-time higher for exons under positive control compared to ex.36nt. The pie charts represent percentage of exOns and exOffs for each category represent (more information in text) (C) The influence of ex.54nt and ex.36nt on the splicing regulation of exogenous mouse Atp2a1 ex.22 and human TNNT2 ex.4. Bars represent average PSI ± SD from three independent experiments and statistical significance was determined by the Student's t-test (∗ for P < 0.05, ∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). (D) Differences in MBNL1 isoform binding site distribution determined by CLIP-seq analysis. Percentage of unique CLIP-seq reads mapped to different regions of transcripts, namely introns located only in pre-mRNA (nucleus) and coding sequences (CDS), 5′UTRs and 3′UTRs located in both pre-mRNA and mRNA (nucleus and cytoplasm) for three MBNL1 isoforms (MB1-40 and MB1-41 without ex.54nt and MB1-43 with ex.54nt showing exclusively nuclear localization). Pink arrows indicate differences between percentage of reads for MBNL1 isoforms with and without ex.54nt.
A similar analysis was performed for MBNL isoforms containing or lacking ex.36nt or ex.95nt. There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, we observed that exOff events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010) (Figure 5B). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA. The majority of tested AS events was altered by at least one isoform (Figure 5A and Supplementary Figure S2) and thus various isoforms can either positively or negatively regulate different AS events dependent on the MBNL mode of action (exOn and exOff mechanism).
To further investigate the functions of MBNL1 isoforms, we conducted CLIP-seq experiments for overexpressed MB1-40, MB1-41 and MB1-43 (Figure 1A) that differed in cellular localization in C2C12 myoblasts (Supplementary Figure S5). Since MBNL proteins function in both the nuclear and cytoplasmic compartments (6), we expected that the frequency of CLIP-reads for MBNL1 isoforms without ex.54nt (MB1-40 and MB1-41), which localize in both cytoplasm and the nucleus, should be higher for exonic sequences (UTRs and protein coding regions) compared to the nuclear MB1-43 isoform containing ex.54nt. Indeed, looking globally we found more reads within mature mRNA elements for both MBNL1 isoforms with nuclear and cytoplasmic localization (Figure 5D).
MBNL paralogs are crowded in CUGexp RNA foci
In DM1 and DM2, MBNL proteins are sequestered by transcripts containing expanded CUGexp and CCUGexp repeats, which aggregate in discrete nuclear foci in vivo (50,54). Therefore, we tested, whether different MBNL paralogs and their isoforms are capable of interacting with CUGexp and CCUGexp RNA and forming nuclear foci in a similar manner. Using the in vitro binding assay, we observed a very high affinity for all MBNL paralogs to non-pathogenic (CUG)20 and (CCUG)14 repeat transcripts (Kd in 4-15 nM range; the lowest among 10 tested RNA molecules) with just slight differences between paralogs (Figure 6A and Supplementary Figure S6E). Next, we coexpressed different GFP-MBNL fusions together with CUGexp in HeLa cells. The combined analysis of ribonucleoprotein foci, visualized by FISH (to detect CUGexp), and the signals from GFP-MBNL proteins revealed that all tested MBNL isoforms colocalized with repeat transcripts in nuclear foci (Supplementary Figure S9A). MBNL overexpression caused the average CUGexp foci volume to increase by up to 2-fold, which we termed foci pumping, without increasing expression of this RNA and this strongly correlated with the volume of the fluorescence protein signal within foci (Figure 6B and C; Supplementary Figure S9B-D). These observations led us to elucidate the nature of protein-protein and RNA-protein interactions in nuclear foci.
Figure 6.
MBNL on CUGexp RNA foci. (A) High affinity of recombinant MBNL1, MBNL2 and MBNL3 for (CUG)20 RNA in vitro. The Kd values are indicated. For (CCUG)14 and (CAG)20 transcripts, see Supplementary Figure S6E. (B) Overexpression of MBNL paralogs significantly increase CUGexp foci volume (‘foci pumping’) measured by FISH for ∼130 foci. There is no difference in CUGexp foci volume distribution between cells transfected with GFP and without protein overexpression (no ovex.). Statistical significance was assessed by Mann-Whitney U test (∗∗ for P < 0.01 and ∗∗∗ for P < 0.001). (C) Correlation between RNA foci volume with MBNL volume (measured by the GFP signal). The Spearman's ρ is between 0.87 and 0.95. FISH and GFP fluorescence measurements were performed using quantitative confocal microscopy, see Supplementary Figure S9. (D) Analysis of FRET efficiency (E) between GFP and mCherry either fused together (left picture) or with two MBNL proteins or with CELF1 in a control experiment (right picture) in the absence or presence of a CUGexp transcript. The E-values which are average from 10-20 analyses for pairs of full length MB1-41 and truncated MB1-C in CUGexp foci are as high as for a positive GFP-mCherry control in an entire cell. In the absence of CUGexp, the E-value is slightly above the background and similar to the value observed for a CELF1 and MB1-41 pair. For more results, see Supplementary Figure S10. (E) MBNL paralogs differ in E. Statistical significance was determined by Student's t-test (NS for P ≥ 0.05, ∗∗ for P < 0.01). (F) MBNL paralogs differ in their mobile fraction in FRAP experiments performed in cells saturated with MBNL protein. The results are mean from about 20 nuclei ± SD and statistical significance was determined by Mann-Whitney U test (∗ for P < 0.05 and ∗∗∗ for P < 0.001). See Supplementary Video S3. (G) The presence of a sequence encoded by alternative ex.95nt reduces MBNL2 mobility in FRAP experiments. For more examples, see Supplementary Figure S11A. (H) Photoswitching of Dendra2 fused with MB1-41 in cells with saturated levels of MBNL protein. Photoconverted (red, emission 573 nm) protein is shifting away whereas unconverted (green, emission 507 nm) protein is associating with CUGexp foci (see Supplementary Video S4). The same analysis for MB1-40 was shown in Supplementary Figure S11B. (I) There is no correlation between the relative nuclear fluorescence intensity of GFP fused MBNL proteins and the mobile MBNL fraction. (J) FRAP experiment for MB2-38 in cells with the very low fluorescence signal of the fusion protein in nuclei. Note that the mobile fraction is three-times lower in the unsaturated compared to saturated state of MBNL (see Supplementary Figure S11A for comparison). (K) The diffusion of photoconverted Dendra2 fused with MBNL1 within individual CUGexp focus in a cell with the low fluorescence signal in nucleoplasm (unsaturated MBNL state). Quantification was performed from two regions of interest (ROI1 and ROI2). The ROI1 is an area that was photoconverted by a laser and the red signal decreases during the course of time. On the other hand, in the distant ROI2 of the same CUGexp focus in which only green signal was detected in time 0, the red signal from photoconverted Dendra2 increases for several seconds. In this situation, MBNL proteins are moving to other binding sites on CUGexp RNA and are not dissociating from the foci.
We coexpressed different combinations of two fluorescence protein-MBNL fusions in the presence or absence of CUGexp transcripts. To measure the proximity of fusion proteins in the cytoplasm, nucleoplasm or in CUGexp foci, we applied FLIM and measured the efficiency of FRET between GFP (donor) and mCherry (acceptor). FRET occurs if the range between donor and acceptor is <10 nm and fused GFP-mCherry protein constituted a positive control for which 20% of FRET was detected (Figure 6D and Supplementary Figure S10A). For further experiments we selected cells only with an equal expression level (Supplementary Figure S10I). In HeLa cells, with or without the expression of CUGexp, neither combination of MBNL isoforms exhibited efficient FRET in cytoplasmic or nucleoplasmic compartments (Figure 6D; Supplementary Figure S10B and H). Thus, this population of MBNL proteins does not undergo FRET detectable homotypic or heterotypic interactions. On the other hand, in nuclear CUGexp foci FRET efficiency for each combination of MBNL protein, but not for a control MBNL1 and CELF1 pair, was very high and surpassed the positive GFP-mCherry control (average FRET efficiency 16-26%) (Figure 6; Supplementary Figure S10B-F and H). The most prominent difference between FRET efficiencies were observed for homotypic MBNL1 and homotypic MBNL3 interactions (Figure 6E). These results suggest that transcripts containing expanded CUG repeats induce an increase of local MBNL concentration leading to multimerization. To test this hypothesis, we utilized the MBNL protein truncated for the C-terminal domain (MB1-C) (Supplementary Figure S8A). Although, previous studies have shown that the C-terminal region is required for MBNL multimerization (16,26,51), we did not observe significant differences between FRET efficiency for the full-length and C-truncated MBNL1 isoforms (Figure 6D; Supplementary Figure S10G and H). This would imply that the proximity of MBNL proteins in CUGexp foci is very high for all tested proteins, including MB1-C. Our data indicate that MBNL proteins bind to CUG repeats with high affinity and CUGexp constitutes a nucleation center for MBNL accumulation, which leads to increased RNA foci size. Although the nuclear localization of MBNL3 was the lowest among MBNL paralogs, MBNL3 formed even more compacted foci due to more effective CUGexp-protein and/or protein-protein interactions.
Rapid exchange of MBNL proteins between the nucleoplasm and CUGexp foci
To investigate whether MBNL isoforms differ in their mobility and accumulation in CUGexp foci, we visualized the dynamic nature of MBNL-CUGexp complexes by performing 3D time-lapse sequences over 16 h. RNA foci containing exogenous fluorescently labeled MBNL proteins were formed de novo and then observed for dispersion as well as structural and volumetric changes (Supplementary Videos S1 and S2). While the results were consistent with previous observations (38), longer CUG repeat expansions formed more stable foci and the half-life of foci formation was measured in either minutes or hours, depending on the CUG repeat tract length, which was (CUG)145 in (38) versus (CUG)960 in our study.
In both DM1 and mouse models of this disease, there is a threshold of CUG repeat length and/or expression level which influences the degree of MBNL sequestration (43). This suggests the importance of stoichiometry between the number of MBNL binding sites on CUGexp RNA and the number of MBNL proteins in a single nucleus. Thus, we examined MBNL protein mobility in both saturated and unsaturated states in which there are more, or less, MBNL binding sites than MBNL proteins and also characterized differences in the mobility of MBNL proteins between nucleoplasm and foci. GFP-MBNL fusion proteins were coexpressed with CUGexp RNA in HeLa cells and protein mobility assessed using FRAP. We first examined cells with a saturated MBNL protein level in which proteins were present in both nucleoplasm and foci (Supplementary Video S3). For FRAP, we calculated two parameters, the mobile fraction and the half-time of fluorescence recovery. The mobile fraction defines the percent of the GFP fluorescence signal which diffuses from the nucleoplasm to RNA foci after photobleaching (2 μm diameter of selected foci) and the half-time (t ) is defined as the half maximal fluorescence recovery time. For each model we observed high heterogeneity of the mobile fraction and t
) is defined as the half maximal fluorescence recovery time. For each model we observed high heterogeneity of the mobile fraction and t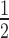 (Supplementary Figure S11A). For proteins representing Set I and Set II groups, the average mobile fractions for MBNL1 proteins were ∼39% higher compared to MBNL3 (Figure 6F) and this phenomenon was not affected by the nuclear concentration of GFP fusion proteins (Figure 6I). As anticipated, MBNL1 lacking the first ZnF tandems (MB1-N) (Supplementary Figure S8A) exhibited the lowest affinity for CUG repeats (55), had the highest mobility and the shortest t
(Supplementary Figure S11A). For proteins representing Set I and Set II groups, the average mobile fractions for MBNL1 proteins were ∼39% higher compared to MBNL3 (Figure 6F) and this phenomenon was not affected by the nuclear concentration of GFP fusion proteins (Figure 6I). As anticipated, MBNL1 lacking the first ZnF tandems (MB1-N) (Supplementary Figure S8A) exhibited the lowest affinity for CUG repeats (55), had the highest mobility and the shortest t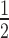 (Supplementary Figure S11A) suggesting a very high rate of diffusion between CUGexp foci and nucleoplasm. In contrast, MBNL3 proteins had the lowest mobile fraction. Moreover, a significant difference in the mobile fraction between MBNL2 proteins containing or lacking ex.95nt was observed and the inclusion of this exon reduced the efficiency of fluorescence recovery in foci (Figure 6G). These results show that MBNL3 proteins have the lowest, whereas MBNL1 (especially truncated MB1-Ns) proteins have the highest, ability to dissociate from CUGexp RNA and exit RNA foci and some alternative MBNL exons modulate this mobility rate.
(Supplementary Figure S11A) suggesting a very high rate of diffusion between CUGexp foci and nucleoplasm. In contrast, MBNL3 proteins had the lowest mobile fraction. Moreover, a significant difference in the mobile fraction between MBNL2 proteins containing or lacking ex.95nt was observed and the inclusion of this exon reduced the efficiency of fluorescence recovery in foci (Figure 6G). These results show that MBNL3 proteins have the lowest, whereas MBNL1 (especially truncated MB1-Ns) proteins have the highest, ability to dissociate from CUGexp RNA and exit RNA foci and some alternative MBNL exons modulate this mobility rate.
To confirm that MBNL proteins diffuse freely between the nucleoplasm and CUGexp foci, we used a photoconvertible Dendra2 fluorescence protein fused to MBNL1 in cells expressing a high level of this protein. After laser-induced photoswitching of Dendra2 from unconverted green (507 nm) to converted red (573 nm), we observed a rapid decrease of the red signal intensity in favor of an elevation of the green signal (Figure 6H, Supplementary Figure S11B and Supplementary Video S4). This indicates that the photoconverted MBNL1 fusion protein migrating within foci was shifted away whereas the unconverted green version of the same protein was simultaneously approaching foci. The quantity of proteins exiting and entering the foci was similar. Moreover, within a few minutes after laser induction, we monitored the red fluorescence signal enhancement at other foci in the same nucleus. Subsequently, we addressed the question of the rate of MBNL protein mobility in cells with a very low nucleoplasmic MBNL concentration. For this purpose, we performed FRAP in HeLa cells with weak nucleoplasmic fluorescence and relatively intense CUGexp foci signals. As expected, there was almost no FRAP in these cells (Figure 6J). We hypothesized that MBNL proteins, present in the unsaturated state, are not able to escape from CUGexp foci but circulate between available RNA binding sites within these structures. To test this hypothesis, we performed the experiments with a photoconvertible Dendra2 protein fused to MBNL1 in cells with a low nucleoplasmic signal and after photoswitching of Dendra2 we observed diffusion of the red fluorescence signal of the fusion protein within individual CUGexp foci (Figure 6K). Cumulatively, these data indicated that when CUGexp foci are saturated with MBNL, the proteins dissociate from the foci and are rapidly exchanged by nucleoplasmic MBNL proteins. In contrast, MBNL proteins circulate within RNA foci when unoccupied binding sites are available. Additionally, nuclear foci are mobile and dynamic structures, capable of slowly increasing and decreasing their volume and fusing with each other.
DISCUSSION
MBNL protein family consists of three paralogs that regulate RNA processing and localization (5,6,9–11) and undergo significant quantitative and qualitative changes during human tissue development. In spite of the strong sequence similarity between these paralogs, Mbnl1, Mbnl2 and Mbnl3 mouse knockouts develop strikingly different phenotypes. Mbnl1ΔE3/ΔE3, Mbnl2ΔE2/ΔE2 and Mbnl3ΔE2 exhibit muscle pathology and ocular cataracts, central nervous system disorders and impaired muscle regeneration, respectively (3,18,23). Moreover, depletion of only MBNL1 and MBNL2 cause the global metabolic RNA changes observed in DM1 adults (4,6,20). An explanation for these differences is the diverse MBNL expression pattern in specific tissues with MBNL1 predominately expressed in skeletal muscles whereas MBNL2 is expressed at a higher level in many neuronal cells (10). However, the differences in the activity of MBNL paralogs have never been directly tested. Thus, we addressed the basis of these MBNL activity differences, including the roles of alternatively spliced isoforms, by studying selected endogenous and exogenous MBNL transcripts. We focused on a comparison of MBNL1, MBNL2 and MBNL3 paralogs, as well as their splicing isoform activities, to determine the structural features which influence AS regulation (Figure 7A). All MBNL paralog activities were regulated by either promotion (exOn) or repression (exOff) of alternative exon inclusion. Notably, MBNL1 was the most active splicing factor and MBNL2 has intermediate activity whereas MBNL3 possessed the weakest influence on AS changes and individual AS events differed in their sensitivity to MBNL overexpression. We conclude that all three MBNL paralogs preferentially recognize the same RNA binding motifs on their RNA targets but often with different affinity. This difference is likely caused by the presence of linker sequences between the ZnF tandem motifs, which are quite different between the three paralogs, that have been shown to influence MBNL activity (16,26).
Figure 7.
Intracellular determinants of MBNL activity and localization. (A) The three main determinants of MBNL activity determined in current study. (B) Deterministic nature of MBNL-CUGexp foci formation is disrupted by the chaos within individual focus. Two distinct stages, differing in the number of MBNL-binding sites on CUGexp were shown. MBNLs would be in saturated (left panel) or unsaturated state (right panel) in CUGexp foci. The latter enables efficient MBNL sequestration. See the text for more details.
We also showed that the different splicing activity of MBNLs may depend, at least partially, on subcellular localization. MBNL paralogs differed significantly in nucleoplasmic and cytoplasmic distribution (Figure 7A). MBNL1 predominated in nucleoplasm whereas MBNL3 predominated in cytoplasm. It may have a significant impact on nuclear and cytoplasmic functions of MBNLs such as AS/polyadenylation and mRNA stability/trafficking. Previous studies have demonstrated that MBNLs truncated for the N-terminal sequence of protein containing the first tandem of ZnFs (MB1-N) and C-terminal (MB1-C) sequence containing KRPALE motif had a negative effect on nuclear localization (16,18,19). Both regions vary in the amino acid sequence between MBNL paralogs leading to relevant consequences. For instance, the difference in the substitution of a third amino acid in the KRPALE motif between all MBNLs was previously shown to effect MBNL1 nuclear localization (19). Moreover, each MBNL paralog expresses multiple protein isoforms (2,16,56) and our results indicate that that the inclusion of amino acid sequences encoded by the three tested alternative exons (ex.54nt, ex.36nt, ex.95nt) are either neutral or can increase or decrease splicing activity. However, the final effects on splicing depend primarily on the specific RNA targets and the mechanism of MBNL action (exOn/exOff). AS of these exons changes during normal human tissue development and in myotonic dystrophy (Figure 1B) and their aberrant regulation modulates spliceopathy evoked by MBNL sequestration in DM.
Currently, there is no consensus about the effect of the sequence encoded by MBNL1 and MBNL2 alternative ex.54nt on RNA splicing regulation (16,17,19). Our results suggest that this sequence reduces MBNL interactions with other factors participating in the exOn mechanism since the inclusion of ex.54nt resulted in mainly negative effects on exOn (e.g. Atp2a1 ex.22). Previously ex.36nt was described to encode the sequence prominent for MBNL1-MBNL1 homotypic interactions (16,51). Here, we show that the sequence encoded by this exon, but also ex.95nt, also modulates MBNL splicing activity. The negative effect of ex.36nt and the positive effect of ex.95nt were almost exclusively specific for exOffs. This observation suggests that the sequences encoded by both alternative exons are important for the interaction with other trans-acting factors and/or to enhance protein affinity for RNAs during the regulation of specific exon exclusion.
All MBNL paralogs have a very high affinity for expanded CUG and CCUG repeat in vitro and in cell models. We observed that overexpression of these RNAs significantly increased the volume of ribonuclear foci consistent with previously described results showing a reduction in RNA foci size due to the silencing of endogenous MBNL1 and MBNL2 (24,38,41). All tested isoforms form CUGexp RNA foci that are heterogeneous in size although the average size of CUGexp foci remained the same. RNA foci volume depends primarily on the amount of accumulated CUG repeat transcripts; however, they slightly differ in the density of MBNL proteins loaded on RNAs. MBNL1 is less densely packed with a lower FRET efficiency but it is also significantly more mobile and efficiently dissociates from, and associates with, CUGexp RNA and the former feature is under control of the sequence encoded by alternative ex.95nt. In good agreement with previous data (38), these results suggest that MBNL-CUGexp foci formation resembles a stochastic process (Figure 7B) although are some determinants, including the sequences encoded by alternatively spliced MBNL exons, that modulate the affinities of the three MBNL paralogs for CUGexp RNA. Therefore, we conclude that foci formation is also deterministic and chaotic nature (deterministic chaos). Generally, MBNL paralogs bind to CUG repeats and dissociate from them when MBNL is in a saturated state (Figure 7B, left panel). In DM1 and DM2, the sequestration of proteins on C(C)UGexp leads to a significant reduction in the free MBNL pool available for normal RNA splicing targets and this situation impacts global AS and APA regulation (57). Effective sequestration of MBNL activity depends on the length of the repeat tract which is modulated by somatic expansion during the lifespan of DM1 patients. When we examined cells designed to reproduce the impaired status of MBNL proteins in DM1, we noted that these proteins are unable to dissociate from RNA foci and instead continuously change intra-foci binding sites on CUGexp transcripts. Since somatic expansion in tissues of DM1 patients alters the stoichiometry between MBNL proteins and their RNA binding sites leading to the unsaturated state, we conclude that phenomenon underlies MBNL sequestration and the progression of the DM1 disease phenotype (Figure 7B right panel).
How do these results impact our understanding of the roles of MBNL proteins in DM1 disease and therapeutic development? Current disease models suggest that age-related expansion of the DMPK CTGexp leads to eventual titration and loss of MBNL splicing activity resulting in the appearance of more severe pathological manifestations, including muscle wasting. Ongoing clinical trials are evaluating the use of AON gapmers to target and degrade DMPK mutant allele transcripts (50). Our results shed light on the mechanism of action of these therapeutic agents as well as small molecule compounds that target MBNL-CUGexp RNA interactions. MBNL proteins are mobile in RNA foci and freely dissociate from these structures so both gapmers and small molecule compounds may actively compete for RNA binding sites and modulate CUGexp RNA structure to make these sites less accessible to circulating MBNL proteins. Since MBNL3 is more prone to be tightly packed on CUGexp RNA, but is primarily expressed only during embryogenesis and tissue regeneration, overexpression of this MBNL paralog should be considered for the future gene therapy development for myotonic dystrophy.
Supplementary Material
Acknowledgments
We thank M.S. Swanson (University of Florida) for critical comments and manuscript corrections, J.A. Berglund (University of Florida) for critical comments, T.A. Cooper (Baylor College of Medicine) for TNNT2 and DT960 minigenes, W.J. Krzyzosiak (Polish Academy of Sciences) for (CUG)20, (CAG)20 molecules and (CAGG)14 template, I. Makalowska (Adam Mickiewicz University) for MIB hosting and cooperation agreement, C.A. Thornton (University of Rochester Medical Center) for A2764 antibody and RNA samples from human muscles, M. Figiel (Polish Academy of Sciences) for mouse muscles, P. Wojtaszek (Adam Mickiewicz University) for cooperation agreement, M. Taylor for manuscript proofreading.
Author Contributions: Conception and design: Ł.J.S. and K.S. Acquisition of data: Ł.J.S. (all without in vitro assays, cellular experiments with AONs and CLIP-seq in C2C12), M. Michalak (confocal microscopy without FISH), K.T. (in vitro assays), P.C. (cellular experiments with AONs, AS events in human tissues), A.W-S. (FISH), M. Matłoka (in vitro assays) and P.K. (CLIP-seq in C2C12). Analysis and interpretation of data: Ł.J.S. (all), M. Michalak (confocal microscopy without FISH), M.K. (RIP-seq, CLIP-seq), K.T. (in vitro assays), P.C. (cellular experiments with AONs) and K.S. (all-supervision). Drafting or revising the article: Ł.J.S and K.S.
Footnotes
Present addresses:
Łukasz J. Sznajder, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, 2033 Mowry Road, Gainesville, FL 32610, USA.
Magdalena Matłoka, Sorbonne Universités, UPMC Univ Paris 06, INSERM UMRS974, CNRS FRE3617, Center for Research in Myology, Institut de Myologie, GH Pitié-Salpêtrière, F-75013 Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Foundation for Polish Science TEAM program cofinanced by the European Union within the European Regional Development Fund [TEAM/2011-7/10 to K.S.]; Polish National Science Centre [2011/01/B/NZ1/01603, 2014/15/B/NZ2/02453 to K.S., 2014/12/T/NZ2/00516 to Ł.J.S.]; National Multidisciplinary Laboratory of Functional Nanomaterials NanoFun [POIG.02.02.00-00-025/09]. Funding for open access charge: Ministry of Science and Higher Education of the Republic of Poland, from the quality promoting subsidy, under the Leading National Research Centre (KNOW) programme for the years 2014-2019.
Conflict of interest statement. None declared.
REFERENCES
- 1.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pascual M., Vicente M., Monferrer L., Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 3.Kanadia R., Johnstone K., Mankodi A., Lungu C., Thornton C., Esson D., Timmers A., Hauswirth W., Swanson M. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 4.Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A., et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang E.T., Ward A.J., Cherone J.M., Giudice J., Wang T.T., Treacy D.J., Lambert N.J., Freese P., Saxena T., Cooper T.A., et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E., Cody N., Jog S., Biancolella M., Wang T., Treacy D., Luo S., Schroth G., Housman D., Reddy S., et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rau F., Freyermuth F., Fugier C., Villemin J.-P., Fischer M.-C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D., et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 8.Masuda A., Andersen H., Doktor T., Okamoto T., Ito M., Andresen B., Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci. Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taliaferro J.M., Vidaki M., Oliveira R., Olson S., Zhan L., Saxena T., Wang E.T., Graveley B.R., Gertler F.B., Swanson M.S., et al. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell. 2016;61:821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fardaei M., Rogers M., Thorpe H., Larkin K., Hamshere M., Harper P., Brook J. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 11.Kanadia R., Urbinati C., Crusselle V., Luo D., Lee Y.-J., Harrison J., Oh S., Swanson M. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns. 2003;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- 12.Teplova M., Patel D. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He F., Dang W., Abe C., Tsuda K., Inoue M., Watanabe S., Kobayashi N., Kigawa T., Matsuda T., Yabuki T., et al. Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci. 2009;18:80–91. doi: 10.1002/pro.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goers E., Purcell J., Voelker R., Gates D., Berglund J. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B. RNA bind-n-seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran H., Gourrier N., Lemercier-Neuillet C., Dhaenens C.-M., Vautrin A., Fernandez-Gomez F., Arandel L., Carpentier C., Obriot H., Eddarkaoui S., et al. Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J. Biol. Chem. 2011;286:16435–16446. doi: 10.1074/jbc.M110.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terenzi F., Ladd A. Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity. RNA Biol. 2010;7:43–55. doi: 10.4161/rna.7.1.10401. [DOI] [PubMed] [Google Scholar]
- 18.Poulos M., Batra R., Li M., Yuan Y., Zhang C., Darnell R., Swanson M. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum. Mol. Genet. 2013;22:3547–3558. doi: 10.1093/hmg/ddt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kino Y., Washizu C., Kurosawa M., Oma Y., Hattori N., Ishiura S., Nukina N. Nuclear localization of MBNL1: splicing-mediated autoregulation and repression of repeat-derived aberrant proteins. Hum. Mol. Genet. 2015;24:740–756. doi: 10.1093/hmg/ddu492. [DOI] [PubMed] [Google Scholar]
- 20.Lee K.-Y., Li M., Manchanda M., Batra R., Charizanis K., Mohan A., Warren S., Chamberlain C., Finn D., Hong H., et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013;5:1887–1900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jog S., Paul S., Dansithong W., Tring S., Comai L., Reddy S. RNA splicing is responsive to MBNL1 dose. PLoS One. 2012;7:e48825. doi: 10.1371/journal.pone.0048825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du H., Cline M., Osborne R., Tuttle D., Clark T., Donohue J., Hall M., Shiue L., Swanson M., Thornton C., et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charizanis K., Lee K.-Y., Batra R., Goodwin M., Zhang C., Yuan Y., Shiue L., Cline M., Scotti M., Xia G., et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 25.Sen S., Talukdar I., Liu Y., Tam J., Reddy S., Webster N. Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J. Biol. Chem. 2010;285:25426–25437. doi: 10.1074/jbc.M109.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grammatikakis I., Goo Y.-H., Echeverria G., Cooper T. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011;39:2769–2780. doi: 10.1093/nar/gkq1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpentier C., Ghanem D., Fernandez-Gomez F.J., Jumeau F., Philippe J.V., Freyermuth F., Labudeck A., Eddarkaoui S., Dhaenens C.M., Holt I., et al. Tau exon 2 responsive elements deregulated in myotonic dystrophy type I are proximal to exon 2 and synergistically regulated by MBNL1 and MBNL2. Biochim. Biophys. Acta. 2014;1842:654–664. doi: 10.1016/j.bbadis.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Fardaei M., Larkin K., Brook J., Hamshere M. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mankodi A., Urbinati C., Yuan Q., Moxley R., Sansone V., Krym M., Henderson D., Schalling M., Swanson M., Thornton C. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 30.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 31.Fu Y.H., Pizzuti A., Fenwick R.G., Jr, King J., Rajnarayan S., Dunne P.W., Dubel J., Nasser G.A., Ashizawa T., de Jong P., et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 32.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O'Hoy K., et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 33.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 34.Napierała M., Krzyzosiak W. CUG repeats present in myotonin kinase RNA form metastable "slippery" hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 35.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H., Mankodi A., Swanson M., Moxley R., Thornton C. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 38.Querido E., Gallardo F., Beaudoin M., Ménard C., Chartrand P. Stochastic and reversible aggregation of mRNA with expanded CUG-triplet repeats. J. Cell Sci. 2011;124:1703–1714. doi: 10.1242/jcs.073270. [DOI] [PubMed] [Google Scholar]
- 39.Ho T., Savkur R., Poulos M., Mancini M., Swanson M., Cooper T. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 40.Pettersson O.J., Aagaard L., Andrejeva D., Thomsen R., Jensen T.G., Damgaard C.K. DDX6 regulates sequestered nuclear CUG-expanded DMPK-mRNA in dystrophia myotonica type 1. Nucleic Acids Res. 2014;42:7186–7200. doi: 10.1093/nar/gku352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith K., Byron M., Johnson C., Xing Y., Lawrence J. Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J. Cell Biol. 2007;178:951–964. doi: 10.1083/jcb.200706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X., Miller J., Mankodi A., Kanadia R., Yuan Y., Moxley R., Swanson M., Thornton C. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 43.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 44.Chen C., Sobczak K., Hoskins J., Southall N., Marugan J., Zheng W., Thornton C., Austin C. Two high-throughput screening assays for aberrant RNA-protein interactions in myotonic dystrophy type 1. Anal. Bioanal. Chem. 2012;402:1889–1898. doi: 10.1007/s00216-011-5604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojciechowska M., Taylor K., Sobczak K., Napierala M., Krzyzosiak W.J. Small molecule kinase inhibitors alleviate different molecular features of myotonic dystrophy type 1. RNA Biol. 2014;11:742–754. doi: 10.4161/rna.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philips A.V. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 47.Hino S.-I., Kondo S., Sekiya H., Saito A., Kanemoto S., Murakami T., Chihara K., Aoki Y., Nakamori M., Takahashi M., et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 48.Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., Giakoumakis N.N., Taraviras S., Lygerou Z. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 2012;28:1800–1801. doi: 10.1093/bioinformatics/bts241. [DOI] [PubMed] [Google Scholar]
- 49.Wojtkowiak-Szlachcic A., Taylor K., Stepniak-Konieczna E., Sznajder L.J., Mykowska A., Sroka J., Thornton C.A., Sobczak K. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 2015;43:3318–3331. doi: 10.1093/nar/gkv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan Y., Compton S., Sobczak K., Stenberg M., Thornton C., Griffith J., Swanson M. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallinjoud P., Villemin J.P., Mortada H., Polay Espinoza M., Desmet F.O., Samaan S., Chautard E., Tranchevent L.C., Auboeuf D. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014;24:511–521. doi: 10.1101/gr.162933.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamori M., Sobczak K., Puwanant A., Welle S., Eichinger K., Pandya S., Dekdebrun J., Heatwole C., McDermott M., Chen T., et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 2013;74:862–872. doi: 10.1002/ana.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersson O.J., Aagaard L., Jensen T.G., Damgaard C.K. Molecular mechanisms in DM1— a focus on foci. Nucleic Acids Res. 2015;43:2433–2441. doi: 10.1093/nar/gkv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell J., Oddo J., Wang E., Berglund J. Combinatorial mutagenesis of MBNL1 zinc fingers elucidates distinct classes of regulatory events. Mol. Cell. Biol. 2012;32:4155–4167. doi: 10.1128/MCB.00274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanadia R., Shin J., Yuan Y., Beattie S., Wheeler T., Thornton C., Swanson M. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chau A., Kalsotra A. Developmental insights into the pathology of and therapeutic strategies for DM1: back to the basics. Dev. Dyn. 2015;244:377–390. doi: 10.1002/dvdy.24240. [DOI] [PubMed] [Google Scholar]
- 58.Licatalosi D., Mele A., Fak J., Ule J., Kayikci M., Chi S., Clark T., Schweitzer A., Blume J., Wang X., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.